Spliceosome assembly and/or splicing of a nascent transcript may be crucial for proper isoform expression and gene regulation in higher eukaryotes. It has been shown that cotranscriptional splicing occurs efficiently in Drosophila, but there are not comparable genome-wide nascent splicing data from mammals. To provide this comparison, the authors analyzed a recently generated, high-throughput sequencing data set of mouse liver nascent RNA. Cotranscriptional splicing is approximately twofold less efficient in mouse liver than in Drosophila. Absolute gene length correlates positively with cotranscriptional splicing efficiency independently of intron location and position, in flies as well as in mice. The gene length and distance effects indicate that more “nascent time” gives rise to greater cotranscriptional splicing efficiency in both systems.
Keywords: nascent sequencing, cotranscriptional, pre-mRNA splicing, mammalian splicing
Abstract
Spliceosome assembly and/or splicing of a nascent transcript may be crucial for proper isoform expression and gene regulation in higher eukaryotes. We recently showed that cotranscriptional splicing occurs efficiently in Drosophila, but there are not comparable genome-wide nascent splicing data from mammals. To provide this comparison, we analyze a recently generated, high-throughput sequencing data set of mouse liver nascent RNA, originally studied for circadian transcriptional regulation. Cotranscriptional splicing is approximately twofold less efficient in mouse liver than in Drosophila, i.e., nascent intron levels relative to exon levels are ∼0.55 in mouse versus 0.25 in the fly. An additional difference between species is that only mouse cotranscriptional splicing is optimal when 5′-exon length is between 50 and 500 bp, and intron length does not correlate with splicing efficiency, consistent with exon definition. A similar analysis of intron and exon length dependence in the fly is more consistent with intron definition. Contrasted with these differences are many similarities between the two systems: Alternatively annotated introns are less efficiently spliced cotranscriptionally than constitutive introns, and introns of single-intron genes are less efficiently spliced than introns from multi-intron genes. The most striking common feature is intron position: Cotranscriptional splicing is much more efficient when introns are far from the 3′ ends of their genes. Additionally, absolute gene length correlates positively with cotranscriptional splicing efficiency independently of intron location and position, in flies as well as in mice. The gene length and distance effects indicate that more “nascent time” gives rise to greater cotranscriptional splicing efficiency in both systems.
INTRODUCTION
Eukaryotic messenger RNA (mRNA) undergoes processing steps, including the removal of introns and ligation of exons via splicing, prior to nuclear export and translation. Work over several decades has determined that pre-mRNA splicing can occur cotranscriptionally, while the nascent RNA molecule is still covalently attached to RNA polymerase II (Pol II) and therefore to the DNA template (Perales and Bentley 2009). Previous work also shows kinetic coupling between Pol II transcription and spliceosome assembly and splicing (Beyer and Osheim 1988; LeMaire and Thummel 1990; Bauren and Wieslander 1994; Kiseleva et al. 1994; Zhang et al. 1994; Wetterberg et al. 1996; Das et al. 2006; Carrillo Oesterreich et al. 2010; de la Mata et al. 2010; Ip et al. 2011; Khodor et al. 2011).
Substantial evidence indicates that transcription and splicing are coupled and not merely two independent processes that occur at the same time. For example, the C-terminal domain (CTD) of Pol II may be required for splicing in higher eukaryotes (McCracken et al. 1997). In vitro assays also show that Pol II transcription aids in efficient assembly of an active spliceosome (e.g., Das et al. 2006). The rate of Pol II transcription can help determine splice-site choice (Kadener et al. 2001; de la Mata et al. 2003) and change the extent of cotranscriptional splicing (Khodor et al. 2011).
Splicing and splicing factors can also affect transcription. For example, U1 snRNP has been shown to recruit TFIIB, D, and H factors to the transcriptional initiation site even in the absence of active splicing (Kwek et al. 2002; Damgaard et al. 2008). Additionally, previous work has shown the direct recruitment of spliceosome components to nascent RNA in yeast and human cell lines (Gornemann et al. 2005; Lacadie and Rosbash 2005; Lacadie et al. 2006; Listerman et al. 2006; Tardiff et al. 2006). Moreover, recruitment occurs in a stepwise manner: U1 snRNP binds first, then U2, then the U4/U5/U6 tri-snRNP (Gornemann et al. 2005; Lacadie and Rosbash 2005; Tardiff et al. 2006). Further work in yeast and Drosophila confirmed widespread cotranscriptional splicing in these organisms (Alexander et al. 2010; Carrillo Oesterreich et al. 2010; Khodor et al. 2011). Our work in Drosophila also established that intron length negatively correlates with cotranscriptional splicing efficiency, which supports a model of the intron as the unit across which splice sites are defined in this organism (Mount et al. 1992; Fox-Walsh et al. 2005; Khodor et al. 2011). In addition, cotranscriptional splicing efficiency was highest for constitutive, internal introns (Khodor et al. 2011).
However, there is no comparable genome-wide study in mammals, and the few studies performed on individual genes show variability in cotranscriptional splicing efficiency between specific introns (Kessler et al. 1993; Listerman et al. 2006; Pandya-Jones and Black 2009). Moreover, mammalian genomic architecture is completely different from that of flies, making it unwise to extrapolate from the Drosophila picture (Khodor et al. 2011). For example, the median mammalian gene is sixfold longer than the median Drosophila gene and features short exons (∼150 bp) flanked by long introns (∼1000 bp) (Berget 1995; Waterston et al. 2002; Flicek et al. 2012). In addition, splice-site sequences are more degenerate in mammals, which allows for more alternative splicing and exon creation and loss (Ast 2004). These complexities of mammalian gene architecture compromise the bioinformatic recognition of splice sites and splicing in mammalian systems. In contrast to the apparent intron-definition pattern applicable to fly splicing, previous work in mammalian cell lines supports an exon definition model: The U1 snRNP present at an internal 5′ splice site (5′SS) interacts with spliceosome components on the 3′ splice site (3′SS) region of the previous intron. This cross-exon interaction is often buttressed by the presence of other splicing factors within the bridged exon (Robberson et al. 1990; Kuo et al. 1991; Talerico and Berget 1994; Berget 1995). Exon length is restricted to ∼50–500 bp by exon definition, and artificially extending an exon past the 500-bp mark often results in exon skipping (Robberson et al. 1990; Berget 1995; Sterner et al. 1996; Fox-Walsh et al. 2005).
Functional coupling of splicing to transcription is thought to occur principally via two mechanisms. One is through the interaction of splicing factors with the nascent template and the transcriptional machinery (Bourquin et al. 1997; Kim et al. 1997; Tanner et al. 1997; Hirose et al. 1999; Robert et al. 2002; de la Mata and Kornblihtt 2006). The other is kinetic coupling, i.e., a “race” between the time it takes a transcript to be transcribed and cleaved from Pol II by the 3′-end formation machinery versus the assembly of the spliceosome and the splicing reactions (de la Mata et al. 2003, 2010; Lacadie et al. 2006; Tardiff et al. 2006; Carrillo Oesterreich et al. 2010; Khodor et al. 2011). The kinetic coupling model is reinforced by data that decreasing the Pol II elongation rate enhances cotranscriptional splicing efficiency (de la Mata et al. 2003, 2010; Khodor et al. 2011). Similarly, a Pol II pause near the 3′SS of a yeast intron is thought to allow time for splicing to complete cotranscriptionally on high-efficiency genes (Alexander et al. 2010; Carrillo Oesterreich et al. 2010). Studies on the relationship between histone placement, modification, and splicing (Kolasinska-Zwierz et al. 2009; Spies et al. 2009; Kim et al. 2011) may provide insight into how modulating the elongation rate of Pol II within a gene affects the cotranscriptional splicing efficiency of specific introns (Kessler et al. 1993; Pandya-Jones and Black 2009; Khodor et al. 2011). Although gene-specific studies support the kinetic coupling model (de la Mata et al. 2003; Pandya-Jones and Black 2009), there are no genome-wide mammalian data relevant to this issue.
Recently, high-throughput sequencing (HTS) has emerged as an invaluable tool for the studies of global transcription and splicing. HTS of nascent RNA in particular (Nascent-Seq) has been used to examine the circadian regulation of transcription in the fly head as well as in mouse liver (Menet et al. 2012; J Rodriguez, CHA Tang, YL Khodor, S Vodala, JS Menet, and M Rosbash, in prep.). We previously characterized Drosophila S2 cell and fly head RNA by Nascent-Seq, which allowed an assessment of genome-wide cotranscriptional splicing (Khodor et al. 2011). The key metric was intron retention, which assessed the amount of nascent intron sequence compared with exon sequence within individual genes. Low retention was interpreted to indicate a high fraction of spliced nascent RNA.
Because no comparable genome-wide study has been done in mammals, we used our previously published data sets (Khodor et al. 2011; Menet et al. 2012) to assess cotranscriptional splicing in mouse liver as well as to compare mammalian and fly cotranscriptional splicing. Mouse introns are twofold less likely to be cotranscriptionally spliced than fly introns. Similar to fly introns, however, mouse introns annotated as “alternative” are more likely than generic (constitutive) introns to be retained in the nascent RNA fraction, i.e., less likely to be cotranscriptionally spliced. Also like fly introns, mouse introns within single-intron genes are also less efficiently cotranscriptionally spliced. Importantly, intron length in mouse does not negatively correlate with cotranscriptional splicing efficiency like in flies (Khodor et al. 2011). Rather, mouse cotranscriptional splicing is optimal for introns with 5′ exons of 50–500 bp, consistent with the exon definition model. There were striking effects of intron position and gene length on mouse cotranscriptional splicing efficiency, strongly indicating a gene length/transcription time effect on cotranscriptional splicing, i.e., kinetic coupling (Oesterreich et al. 2011). Because similar gene length effects were seen in the fly data, kinetic coupling appears to be general, independent of other species-specific differences in splicing.
RESULTS
Global cotranscriptional splicing efficiency is lower in mouse liver than in fly tissues
Recent work from our laboratory generated HTS data sets using NUN fractionation to sequence nascent pre-mRNA still attached to elongating Pol II (Khodor et al. 2011; Menet et al. 2012). One of these studies was from Drosophila tissue culture cells and heads, and it focused on cotranscriptional splicing (Khodor et al. 2011). The other assayed time points from mouse liver and focused on the circadian regulation of transcription (Menet et al. 2012). We therefore decided to analyze these liver data for cotranscriptional splicing efficiency, because there is little literature on global cotranscriptional splicing efficiency in mammals, and to compare the results with our recent analysis of Drosophila global cotranscriptional splicing.
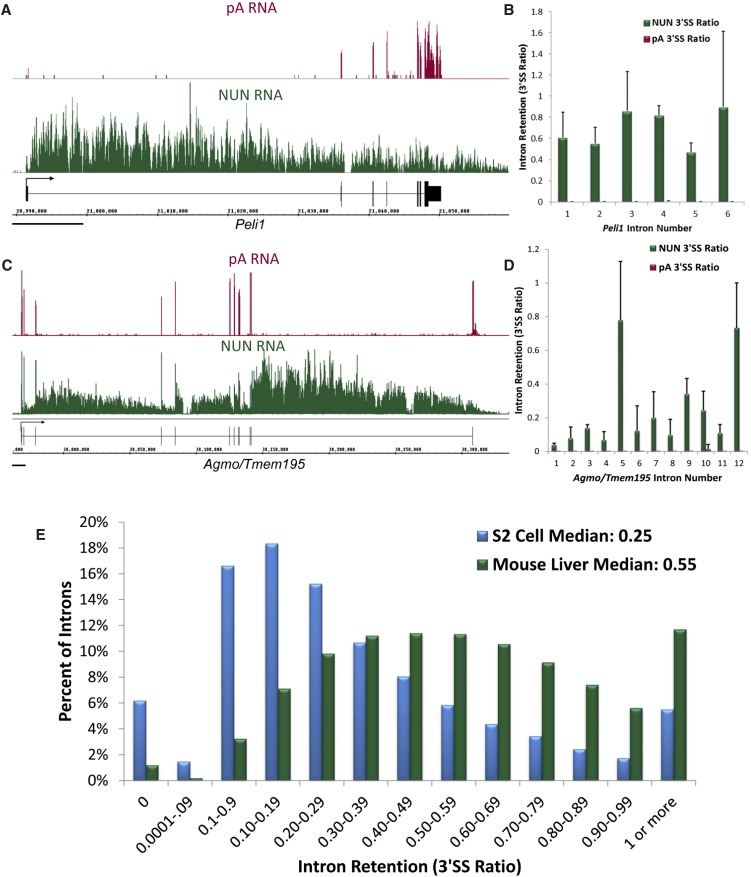
Visual inspection of the mouse liver Nascent-Seq with the Integrated Genome Browser (IGB) indicated that the liver nascent RNA data has features in common with the fly nascent RNA data (Fig. 1A,C; Supplemental Fig. S1A; Khodor et al. 2011). For example, there is often a 5′-to-3′ decrease in read abundance across the length of the gene, consistent with the cotranscriptional nature of the RNA: All nascent transcripts should share a 5′ end, but only a small fraction of Pol II molecules reach the 3′ end of the gene, resulting in a lower abundance of reads at this end of the transcript. Also evident by visual inspection in IGB is a notable difference in the two data sets, namely, the extent of cotranscriptional splicing. A typical fly gene manifests a high degree of cotranscriptional splicing, as noted by the dearth of sequence reads in the intron regions of the typical gene shown, gp210 (Supplemental Fig. S1A; Khodor et al. 2011). In contrast, there are many more intron reads in typical mouse genes, e.g., Peli1 (Fig. 1A), suggesting a considerably lower cotranscriptional splicing efficiency in mouse than in fly. Some mouse genes, like Agmo/Tmem195, show greater cotranscriptional splicing efficiency in 5′ introns and virtually no splicing in the last intron (Fig. 1C). For all of these genes (Fig. 1A,C; Supplemental Fig. S1A) and in the vast majority of cases, the control poly(A) (pA) tracks show little-to-no signal in the intron regions.
FIGURE 1.
Cotranscriptional splicing is twofold less efficient in mouse liver than in Drosophila S2 cells. (A) An image of a typical mouse gene, Peli1, in the Integrated Genome Browser (IGB). (Magenta) pA RNA; (green) NUN RNA; (black) gene structure. (Black bar) 10,000 bp. Note 5′-to-3′ abundance gradient in NUN fraction and abundant read signal within introns. NUN average reads/bp in the exons ∼5.5. (B) Quantitation of intron retention for the introns of Peli1. Intron Retention as 3′SS Ratio = Reads in last 25 bp of Intron/Reads in the first 25 bp of the 3′ exon. (C) Another gene, Agmo/Tmem195, which illustrates greater splicing in the introns closer to the 5′ end of the gene and a mostly unspliced last intron. NUN average reads per base pair in the exons ∼17.4. (D) Quantitation of intron retention for the introns of Agmo/Tmem195. (E) A histogram of intron retention as measured by the 3′SS ratio of all introns in abundantly transcribed genes in mouse and fly. (Blue) Fly NUN RNA; (green) mouse NUN RNA. Total sample size: mouse NUN = 58,493; fly NUN = 20,335 introns.
To quantify cotranscriptional splicing efficiency, we calculated the number of reads in the last 25 bp of the intron and divided it by the number of reads in the first 25 bp of the adjacent exon, a ratio of reads across the 3′ splice site (3′SS ratio) of introns from the mouse data as we had done from fly data (Khodor et al. 2011). We also calculated a 5′ splice site (5′SS) ratio, which was very similar to the 3′SS ratio (Supplemental Fig. S2A) (see Discussion). A ratio of 1 denotes a completely unspliced intron, suggesting poor cotranscriptional splicing efficiency, and a ratio of 0 denotes an intron that is efficiently and perhaps rapidly cotranscriptionally spliced. Quantitation of the 3′SS ratio for the introns in gp210 (Supplemental Fig. S1B), Peli1 (Fig. 1B), and Agmo/Tmem195 (Fig. 1D) shows that there is indeed a big difference in splicing efficiency between the representative cases of the two species.
We then calculated the 3′SS ratio for all mouse introns that have an average of at least three reads per base pair (reads/bp) in their exons and compared the numbers with those previously calculated for fly introns (Fig. 1E; Khodor et al. 2011). The median mouse ratio (0.55) is approximately twofold higher than the median fly ratio (0.25). One simple interpretation is that the median fly intron is cotranscriptionally spliced twice as efficiently as the median mouse intron. There were no mouse genes in which all introns had a 3′SS ratio of 0.1 or less, unlike in the fly (Khodor et al. 2011). This observation suggests that all mouse genes undergo some percentage of post-transcriptional splicing. Global pA sequencing indicates minimal intron presence for all introns and genes, which is consistent with efficient splicing upstream of nuclear export, independent of cotranscriptional splicing (Supplemental Fig. S2B,C).
Alternatively annotated introns are less efficiently spliced in both organisms
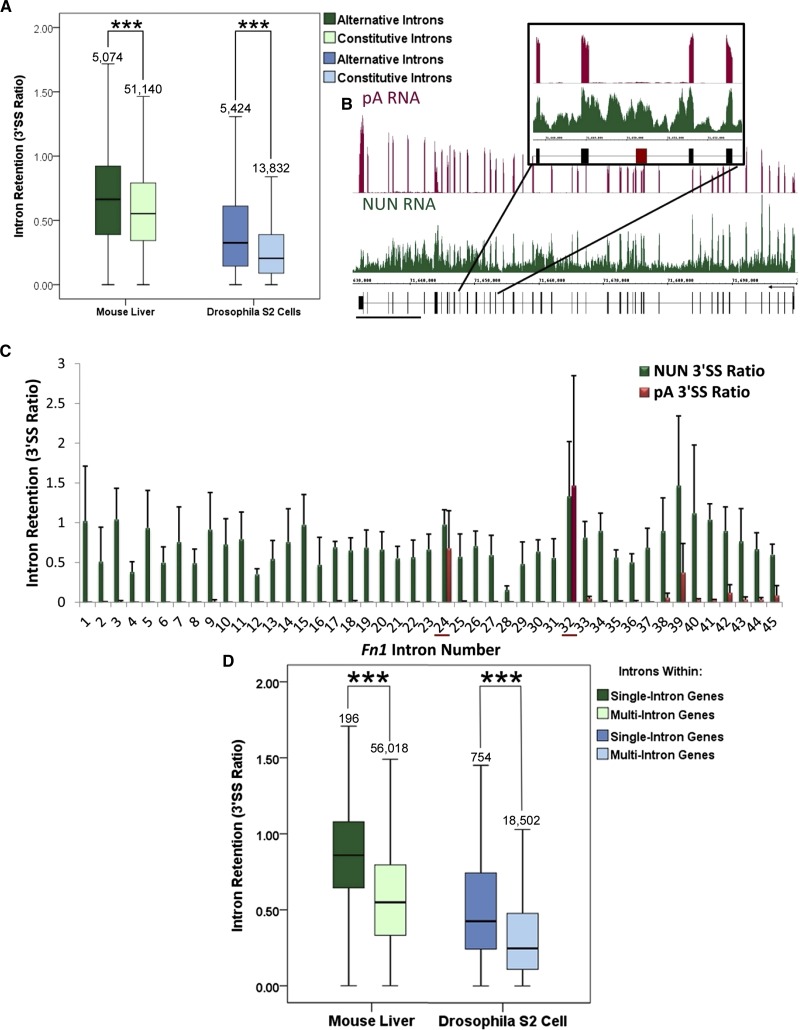
Previous work on select human genes showed that alternative exons are less efficiently cotranscriptionally spliced than their constitutive neighbors (Pandya-Jones and Black 2009). Consistent with this observation, our global sequencing of Drosophila nascent RNA showed that alternatively annotated introns are more retained in the nascent fraction when compared with constitutively annotated introns (Khodor et al. 2011). The same analysis done here on liver nascent RNA comes to an identical conclusion (P < 0.001, Mann-Whitney U-Test) (Fig. 2A).
FIGURE 2.
Special cases: Alternatively annotated introns and introns within single-intron genes correlate with poorer cotranscriptional splicing efficiency in both Drosophila and mouse. (A) Alternatively annotated introns (dark green, dark blue) show significantly greater 3′SS ratios than constitutively annotated introns (light green, light blue) in mouse (green) and fly (blue) (***, P < 0.001, Mann-Whitney U-Test). (B) An image of the mouse Fibronectin (Fn1) gene in IGB. (Magenta) pA RNA; (green) NUN RNA; (black) gene structure. (Black bar) 10,000 bp. The structure does not note alternative isoforms that exclude alternative exons, such as the one shown in the inset. NUN average reads/bp in the exons ∼29. (C) A quantitation of intron retention of all introns in Fn1. Note that introns 24 and 32 have artificially high 3′SS ratios in the pA fraction due to lack of signal in the 3′ exon. (D) Introns located within single-intron genes (dark green, dark blue) show significantly higher 3′SS ratios than introns within multi-intron genes (light green, light blue) in both mouse (green) and fly (blue) (***, P < 0.001, Mann-Whitney U-Test). Box plots span the 95th–fifth percentiles.
However, only 5385 introns of the almost 60,000 analyzed are annotated as alternative in the mouse reference sequence (Pruitt et al. 2009). This number severely underestimates the number of alternative splicing events believed to occur in this organism (Sharov et al. 2005; Kim et al. 2007; Harr and Turner 2010). For example, the 33rd exon of Fibronectin1 (Fn1) is the alternatively spliced exon homolog of the human exon, also known as EDI, which is frequently used to study alternative splicing (de la Mata et al. 2003, 2010). Recent work indicated that the intron downstream from this human EDI exon is spliced faster than the upstream intron (de la Mata et al. 2010). Indeed, the EDI exon is virtually missing from the pA RNA sequencing, and the downstream intron has a lower 3′SS ratio than the upstream intron in the nascent sequencing (Fig. 2B,C). The 3′SS ratio from pA RNA is artificially high due to the dearth of reads across the EDI exon in this sample (Fig. 2C); this result suggests that isoforms including this exon are not stably expressed in mouse liver. Because the reference sequence only has one annotated isoform of Fn1, these introns and exons are perhaps improperly annotated as constitutive. Given this limitation in defining alternative splicing, it is possible that the difference between the cotranscriptional splicing of alternative and constitutive mouse introns is even more dramatic.
Introns within single-intron genes are less efficiently cotranscriptionally spliced in both species
We also examined the special case of introns present in single-intron mouse genes. These introns are cotranscriptionally spliced less efficiently than those present in multi-intron genes, and this is also the case for single-intron fly genes (Fig. 2D). In fact, the cotranscriptional splicing of introns within single-intron mouse genes does not correlate with any other effects (discussed below), such as intron, 3′-exon, and 5′-exon lengths (Supplemental Fig. S3A–C). This result may indicate that introns within single-intron mouse genes, like select yeast introns, serve a regulatory function, such as limiting the amount of mature transcript (Dabeva et al. 1986; Barta and Iggo 1995; Preker et al. 2002; Parenteau et al. 2008). In contrast, both 3′- and 5′-exon length but not intron length of single-intron genes in the fly correlate with 3′SS ratio (Supplemental Fig. S3D–F). This result indicates that longer exons promote cotranscriptional splicing efficiency.
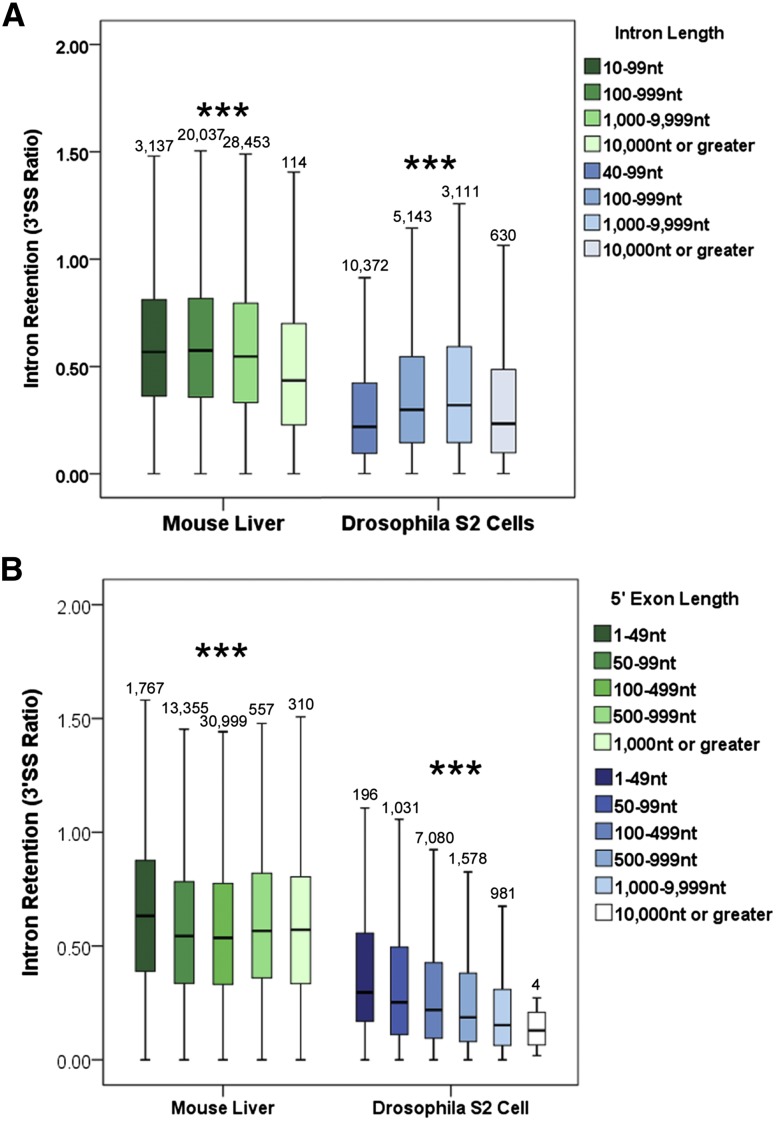
Intron length negatively correlates with cotranscriptional splicing efficiency in the fly but not in the mouse
Intron size is thought to play a role in determining splicing efficiency (Fox-Walsh et al. 2005) as well as in determining how an intron is defined (Mount et al. 1992; Sterner et al. 1996; Burnette et al. 2005; Fox-Walsh et al. 2005; Farlow et al. 2012; Gelfman et al. 2012). Indeed, our previous work determined that longer fly introns manifest poorer cotranscriptional splicing efficiencies (Khodor et al. 2011). Because the median mouse intron is 16-fold larger than the median fly intron (1369 bp vs. 82 bp), we asked whether intron length accounts for the twofold difference in cotranscriptional splicing efficiency between fly and mouse.
Intron length does not correlate with poorer cotranscriptional splicing in mouse liver, nor does it account for the decrease in efficiency between the two species (Fig. 3A). The median 3′SS mouse ratio is much higher than that of the fly for introns of comparable size (Fig. 3A). Moreover, the median 3′SS ratio slightly but significantly decreases with increasing intron size in the mouse (P < 0.001, Krustal-Wallis test). These results indicate that intron length plays no more than a minor role in determining cotranscriptional splicing efficiency in the mouse or in determining differences between species.
FIGURE 3.
Intron definition and exon definition in mouse and fly: correlations of 3′SS ratios with intron and 5′-exon lengths. (A) Intron length does not correlate with 3′SS ratio in mouse (green) but does in fly (blue) (***, P < 0.001, Kurstal-Wallis Test, all pairwise). (B) The 3′SS ratio in mouse (green) is lowest when the length of the 5′ exon is 50–500 bp. In fly (blue), the length of the 5′ exon is negatively correlated with the 3′SS ratio of a given intron (***, P < 0.001, Krustal-Wallis Test, all pairwise). Box plots span the 95th–fifth percentiles.
Cotranscriptional splicing efficiency and exon size: Investigating exon definition
Work over many decades has established two models for splice site definition, intron definition, and exon definition (Robberson et al. 1990; Kuo et al. 1991; Talerico and Berget 1994; Berget 1995). Intron definition is thought to occur when introns are relatively short: U1 snRNP binds the 5′ splice site (5′SS) and associates across the short intron with factors that bind the 3′SS such as U2AF (Mount et al. 1992; Berget 1995; Farlow et al. 2012; Gelfman et al. 2012). Exon definition is thought to occur with larger introns and smaller exons: U1 binds to the 5′SS and associates across the short exon with spliceosome components on the 3′ end of the previous intron and with splicing-relevant proteins bound to the exon (Robberson et al. 1990; Kuo et al. 1991; Talerico and Berget 1994; Berget 1995). If mouse cotranscriptional splicing efficiency does not correlate with intron size, might it be constrained by the size of neighboring exons? Because there are indications that first and last introns require interactions with capping and cleavage/polyadenylation machinery to be efficiently spliced, respectively (Niwa and Berget 1991; Lewis et al. 1996; Cooke et al. 1999; Dye and Proudfoot 1999; Rigo et al. 2005; Khodor et al. 2011), we excluded them from our analyses.
Strikingly, there are marked interspecific differences in the correlation of cotranscriptional splicing efficiency with upstream exon size in mouse versus fly. There is a slight but statistically significant decrease in the 3′SS ratio for mouse introns with 5′ exons 50–500 bp long (P < 0.001, Krustal-Wallis Test) (Fig. 3B). This size range includes the optimal exon size (∼150 nt) found in previous experimental studies with artificial constructs (Robberson et al. 1990; Berget 1995; Sterner et al. 1996) and supports an exon-definition model.
Surprisingly, upstream exon length in Drosophila correlates with a decrease in the 3′SS ratio for the intron (P < 0.001, Krustal-Wallis Test) (Fig. 3B). These results indicate that the longer the exon the better is the cotranscriptional splicing of the following intron. This surprising relationship is independent of other parameters such as 3′ exon length, gene length, or the position of the intron relative to the end of the gene (discussed below) (Supplemental Fig. S4). However, it does not apply to larger introns, those >1000 bp in length and >10% of the total gene size (Supplemental Fig. S5). The relationship may indicate that short introns, which are predominant in Drosophila, are optimally spliced cotranscriptionally when flanked by larger exons.
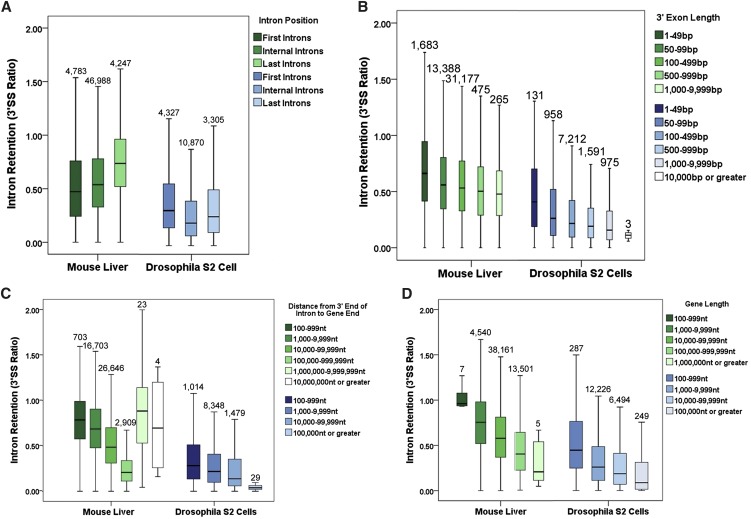
Intron position plays a major role in cotranscriptional splicing efficiency in both organisms
We previously reported that first and last Drosophila introns manifest greater intron retention than internal introns, likely indicating poorer cotranscriptional splicing efficiency (P < 0.001, Krustal-Wallis Test) (Fig. 4A; Khodor et al. 2011). These results are in contrast to a study of select human genes, which showed a general 5′-to-3′ gradient in cotranscriptional intron excision (Pandya-Jones and Black 2009). We therefore examined the global mouse liver data by intron position. As predicted by the gene-specific study (Pandya-Jones and Black 2009) and unlike what was observed in the fly, mouse introns indeed show a 5′-to-3′ gradient in 3′SS ratio (P < 0.001, Krustal-Wallis Test). The gradient includes first introns, indicating more efficient splicing of more 5′-proximal introns (Fig. 4A).
FIGURE 4.
Cotranscriptional splicing efficiency differs dramatically with intron position and the size of the gene. (A) Intron retention, as measured by the 3′SS ratio, differs significantly between first, internal, and final introns in both mouse (green) and fly (blue) (P < 0.001, Krustal-Wallis Test, all pairwise). (B) For internal introns in both mouse (green) and fly (blue), the length of the 3′ exon is negatively correlated with the 3′SS ratio (P < 0.001, Krustal-Wallis Test, all pairwise). (C) For internal introns in both mouse (green) and fly (blue), the distance of the intron from the end of the gene is negatively correlated with the 3′SS ratio. The exception is about 30 introns in mouse greater than a megabase away from the end of the gene (P < 0.001, Krustal-Wallis Test, all pairwise). (D) For all introns in both mouse (green) and fly (blue), the size of a gene containing a given intron is negatively correlated with the 3′SS ratio (P < 0.001, Krustal-Wallis Test, all pairwise). Box plots span the 95th–fifth percentiles.
These intron position effects were reminiscent of previously reported 3′-exon length effects in yeast splicing (Tardiff et al. 2006; Carrillo Oesterreich et al. 2010). We therefore examined whether 3′-exon length influences cotranscriptional splicing efficiency. Indeed, the 3′SS ratio is strongly correlated with the length of the exon 3′ to that intron, and that is true for Drosophila as well as mouse (P < 0.001, Krustal-Wallis Test) (Fig. 4B). The interpretation is that a longer 3′ exon allows more time for cotranscriptional splicing of the upstream intron.
To further explore this phenomenon, we measured mouse intron location, the absolute distance from its 3′SS to the 3′ end of the gene. Remarkably, introns show a very strong gradient: The farther away the 3′SS of a given intron is from the gene 3′ end, the more likely that intron is to be cotranscriptionally spliced (lower 3′SS ratio). The results are consistent with a prior report in Chironomus (Wetterberg et al. 1996). Although the 3′SS ratio of Drosophila internal introns at all distances from the 3′ end is dramatically lower than that of mouse internal introns (P < 0.001, Krustal-Wallis Test), the Drosophila introns show a similarly strong gradient (Fig. 4C). The exceptions to this “distance to the end of the gene rule” are the handful of mouse introns (27 out of ∼47,000) that are greater than a megabase away from the gene 3′ end. These introns are spliced even more poorly than those nearest their gene 3′ end (P < 0.001, Krustal-Wallis Test) (Fig. 4C). Surprisingly, these introns are all annotated as alternative (Supplemental Fig. S6). Consistent with this correlation, the farther away an alternatively annotated intron is from the 3′ end of its gene, the more likely the intron is to be retained than a constitutively annotated intron at the same distance (Supplemental Fig. S6).
We also examined the special case of single-intron fly genes for this “distance to the end of the gene” rule and found another difference from mouse: The fly introns continue to obey this rule (P = 0.004, Krustal-Wallis Test), whereas mouse single introns do not or much less well (P = 0.052, Krustal-Wallis Test) (Supplemental Fig. S7).
Gene size is positively correlated with cotranscriptional splicing efficiency in both mouse and fly
In addition to the difference in median intron size, there is a sixfold difference in median gene size between the mouse and fly genomes. The recent genome-wide studies of cotranscriptional splicing (Carrillo Oesterreich et al. 2010; Khodor et al. 2011) have not examined the effect of gene size.
Increasing gene size strikingly correlates with lower 3′SS ratios in both mouse and fly (P < 0.001, Krustal-Wallis Test) (Fig. 4D), an effect that is not dependent on the absolute position of the intron relative to the end of the gene (Supplemental Fig. S8). Single-intron genes are the only exception to this rule (Supplemental Fig. S9). Taken together with the 3′ exon length and distance to the 3′ end effects, these data reinforce a kinetic view of cotranscriptional splicing: The more time Pol II spends in active transcription prior to 3′-end formation, the more likely cotranscriptional splicing is to occur.
DISCUSSION
To understand the differences in cotranscriptional splicing efficiency between metazoans, we compared previously published nascent sequencing analyses of mouse liver and Drosophila S2 cells (Khodor et al. 2011; Menet et al. 2012). Cotranscriptional splicing in mouse liver is approximately twofold less efficient than in Drosophila S2 cells (3′SS ratio of 0.55 vs. 0.25). Another difference between the species is that intron length does not correlate with poorer cotranscriptional splicing efficiency in mouse. Moreover, 5′-exon length is optimal between 50 and 500 bp only in the mouse, a finding that recalls exon definition. Similarities between the two species include the fact that alternatively annotated introns are more likely to be cotranscriptionally retained than constitutively annotated introns and that introns within single intron genes are more retained than introns found in multi-intron genes. The most striking effect in both species is intron position: The further an intron is from the 3′ end of a gene, the lower is the 3′ss ratio, indicating better cotranscriptional splicing. Additionally, absolute gene length correlates with cotranscriptional splicing efficiency independent of intron position.
The twofold increase in intron retention between fly and mouse is surprising, especially given the low intron signal in the mouse pA sequence data. This may indicate that total splicing in mouse liver, post-transcriptional as well as cotranscriptional, is very efficient (Supplemental Fig. S2). However, unspliced transcripts may be efficiently degraded by the nuclear exosome or by nonsense-mediated decay (NMD) in the cytoplasm. An argument against a major effect by NMD is that sequencing of the fly nuclear pA fraction, which is not chromatin-associated (“nucleoplasm”), shows splicing patterns very similar to those seen in the total pA (data not shown). We have not performed a similar assay in the mouse system, so it is possible that considerable splicing occurs post-transcriptionally in this species, i.e., after 3′-end formation. This view is consistent with a revised first-come, first-served model (de la Mata et al. 2010), namely, commitment complex formation always occurs cotranscriptionally, but substantial splicing only occurs after 3′ cleavage and polyadenylation of the transcript.
Our results are compatible with the abundant data suggesting that transcripts are retained at their transcription site until splicing completes. Fluorescence in situ hybridization of human β-globin genes showed transcripts in intranuclear foci colocalizing with the template gene locus; splicing was shown to be limiting for release from the transcription site (Custodio et al. 1999). More recent experiments using real-time imaging of human cells demonstrated that incompletely spliced pre-mRNAs are retained at the transcription site in “dots” (Brody et al. 2011; Vargas et al. 2011). Splicing events of this nature would not be detectable in our assay, because they would be removed by the dT wash we include in our procedure prior to library preparation and sequencing. Although our fly paper interpreted this material as “contamination,” it is possible that it contains RNA destined to be post-transcriptionally spliced.
We previously determined that alternatively annotated fly introns are cotransciptionally spliced relatively poorly (Khodor et al. 2011). Despite an overall lower cotranscriptional splicing efficiency, mouse introns show a similar phenomenon. Notably, the few introns that are far from the 3′ end of their genes and show poor cotranscriptional splicing are all annotated as alternative (Supplemental Fig. S6). However, the mouse results appear complicated by the fewer introns that are correctly annotated as alternative in the reference sequence used for analysis. The median difference between alternative and constitutive intron retention could therefore be even greater. Nonetheless, the results indicate that removal of alternative introns is slower, perhaps due to the increased difficulty in determining appropriate splice site partners; inefficient splicing is especially true for alternatively annotated introns far from the 3′ end of the gene. Alternatively, the delay may be “by design” and upstream of splice site partner assignment, to uncouple this regulation from the temporal issues inherent in cotranscriptional events. In support of this notion, we could not find any mouse genes whose introns were all better than 90% spliced—something observed for fly genes. Therefore, some splicing of all mouse genes appears to complete post-transcriptionally.
Single-intron genes are another special class of retained introns. Although present in both species, this class is much rarer in mouse than in fly (0.3% and 4% of introns analyzed, respectively). These mouse single-intron genes are not affected by intron length, exon length, or intron position. Single-intron mouse genes may therefore be special with the splicing delay serving or reflecting post-transcriptional regulation. A precedent is autoregulation, which has been shown for select yeast introns (Dabeva et al. 1986; Barta and Iggo 1995; Preker et al. 2002; Parenteau et al. 2008). Signaling could also play a role, because the GO annotation for mouse single intron genes indicates significant enrichment for G-proteins and receptors as well as transmembrane and zinc-finger proteins (DAVID GO 3.2 × 10−8 for G-proteins; 3.8 × 10−5 for zinc-finger proteins). In contrast, the Drosophila S2 cell single-intron GO enrichment is much less striking with only a slight enrichment for structural proteins and ribosome components (DAVID GO 0.01 for ribosome and structural proteins). Given the much larger average number of mouse introns/gene, perhaps only the single-intron mouse genes constitute a bona fide cohort.
Our previous fly analysis determined that intron length correlates with poorer cotranscriptional splicing efficiency (Khodor et al. 2011). The exceptions were introns 10 kb or longer, which are not only rare in the fly genome but are also recursively spliced (Burnette et al. 2005; Khodor et al. 2011). In contrast, mouse cotranscriptional splicing efficiency did not decrease with intron length. Rather, cotranscriptional splicing was optimal with 5′ exons of 50–500 bp in length, supporting exon definition (Robberson et al. 1990; Kuo et al. 1991; Talerico and Berget 1994; Berget 1995; Sterner et al. 1996). This splicing mode has been shown to be less efficient than intron definition (Fox-Walsh et al. 2005), suggesting that a switch in mechanism may partly underlie the twofold difference in global cotranscriptional splicing efficiency.
In addition to overall better cotranscriptional splicing efficiency, longer 5′ as well as 3′ exons positively correlate with better cotranscriptional splicing efficiency of short fly introns. These data are in line with previous work indicating that short introns are difficult to recognize by the splicing machinery when flanked by short exons; i.e., short introns are better spliced when surrounded by longer exons under intron definition conditions (Sterner et al. 1996; Fox-Walsh et al. 2005).
3′-Exon length also correlates with better cotranscriptional splicing efficiency in mouse, which may reflect a simple timing mechanism: The greater the distance that Pol II transcribes from the end of an intron to the end of the gene, the more time there is for splicing to cotranscriptionally complete (de la Mata et al. 2003, 2010; Lacadie et al. 2006; Tardiff et al. 2006; Khodor et al. 2011). We previously found that this kinetic competition model could not explain all Drosophila splicing events, because first introns were more likely to be cotranscriptionally retained than internal or last introns in the fly (Khodor et al. 2011). Preferential retention of the first intron could have a number of explanations, including interactions of the U1 snRNP bound to the first 5′SS with transcription initiation factors (Kwek et al. 2002; Damgaard et al. 2008). Another possibility is that first intron retention facilitates a desired delay in nuclear export, dependent on cap- and splicing-mediated recruitment of the TREX complex to the 5′ end of transcripts (Cheng et al. 2006). Despite the fly first intron exception, however, the analysis shown here indicates that the 3′-exon length rule applies to internal fly introns and to almost all mouse introns. The only exceptions are alternative mouse introns that are very far (more than a megabase) from the 3′ end of a gene. These introns could be misannotated or they could be special, with their own splicing rules.
In contrast to the fly situation, mouse first introns are slightly less retained than internal introns, which are less retained than terminal introns. This simple 5′-to-3′ gradient model fits with previously published data on select human genes (Pandya-Jones and Black 2009). The lack of exceptional first introns in the mouse system may reflect the longer time required to transcribe the average gene in mammals, which might counteract a general delay in the splicing of first introns. Alternatively, the inferred delay in flies may not exist in mammals.
In addition to the effect of intron position, there is a gene-length effect in both organisms: Introns of longer genes are more likely to be cotranscriptionally spliced, i.e., show less intron retention, independent of intron location. This surprising effect is statistically significant even for last introns, the splicing of which has been linked in some cases to cleavage and polyadenylation of the nascent transcript (Cooke et al. 1999; Dye and Proudfoot 1999; Rigo et al. 2005). The recruitment or function of some splicing factor(s) may be more likely as a function of time spent by elongating Pol II. This time includes pausing, which should occur more frequently on longer genes and further increase elongation time, splicing factor accumulation, and function. Recent work has shown that histones are enriched in exons over introns (Spies et al. 2009), and an increase in nucleosome density at intron–exon boundaries could promote nucleosome-induced Pol II pausing, which has been shown to occur in yeast (Churchman and Weissman 2011).
To examine the relative kinetics of splicing and transcriptional elongation, we calculated the 5′SS ratio as well as the 3′SS ratio. In mouse, the two were highly and significantly correlated (Spearman's ρ = 0.566, P > 0.01), and there was no significant difference in their distributions (P = 1.0, Wilcoxon signed-rank test). These results led us to speculate that the actual catalytic events of splicing take place quickly relative to the times required for transcription and splice site recognition.
This analysis provides a link between the high-efficiency cotranscriptional splicing observed in yeast and flies (Carrillo Oesterreich et al. 2010; Khodor et al. 2011) and previous gene-specific work in mammals, which hinted at less efficient cotranscriptional splicing (de la Mata et al. 2010). Despite the increase in transcription time afforded by longer mammalian genes, the slower exon definition process (Fox-Walsh et al. 2005) may decrease the extent of cotranscriptional splicing. This may even be “by design,” because annotated alternative splicing is more frequent in mammals (Sharov et al. 2005; Kim et al. 2007; Wang et al. 2008; Harr and Turner 2010; Nilsen and Graveley 2010) and appears to be preferentially spliced post-transcriptionally in both organisms (Fig. 2A). Additionally, nuclear export of mRNA in mammals has been shown to be dependent on splicing (Kohler and Hurt 2007), and a shift toward post-transcriptional splicing may reflect this relationship. Despite these quantitative differences, several other gene features affect cotranscriptional splicing similarly in flies and mice, including 3′-exon length and distance to the end of the gene. This suggests that major features of the functional coupling between transcription and splicing existed 550 million years ago in the common ancestor of flies and mice.
MATERIALS AND METHODS
RNA isolation from Drosophila S2 cells
Nascent and pA RNA fractions were isolated as described previously (Khodor et al. 2011). Briefly, for nascent RNA, S2 cells were collected and nuclei isolated by douncing cells and spinning the resulting lysate through a sucrose cushion. Collected nuclei were resuspended and washed with 2× NUN buffer. After an incubation on ice, the chromatin-associated fraction was collected via centrifugation, and RNA was isolated using TRIzol Reagent (Invitrogen). For the pA, total RNA was isolated from S2 cells with TRIzol Reagent following the manufacturer's protocol, and pA RNA was collected with a 2× wash with oligo(dT) beads (Invitrogen).
RNA isolation from mouse liver
Nascent RNA fractions were isolated as described previously (Wuarin and Schibler 1994; Menet et al. 2012). mRNA fractions were collected from mouse liver samples using TRIzol Reagent (Invitrogen) and processed using Truseq RNA sample kits (Illumina).
RNA sequencing and alignment
Sequencing was performed on the Illumina Genome Analyzer II and HiSeq and samples were analyzed using TopHat with Bowtie (http://www.tophat.cbcb.umd.edu) (Langmead et al. 2009; Trapnell et al. 2009) as described previously (Khodor et al. 2011; Menet et al. 2012). Briefly, for the nascent and fly mRNA libraries were prepared using the standard Solexa protocol. Libraries were size-selected on a gel to be 200–300 bp long. Fly nascent RNA was subject to rRNA removal as described previously (Khodor et al. 2011). Both fly and mouse nascent RNA was washed across an oligo(dT) column to remove any polyadenylated RNA prior to library generation. All fly samples and mouse nascent RNA libraries were sequenced on the GAII. Mouse mRNA libraries were prepared using Truseq RNA sample kits (Illumina) and sequenced on a HiSeq2000.
Sequencing generated 72-bp reads, except for one fly replicate that was trimmed to 64 bp, and these were mapped to the Drosophila dm3 and mouse mm9 genome alignments downloaded from the UCSC Genome Browser database (http://genome.ucsc.edu/index.html). Non-unique reads were discarded and unique reads analyzed using custom scripts.
To avoid any bias resulting from circadian collection of the original mouse liver samples, only ZT08 and ZT20 replicates were averaged and analyzed for splicing efficiencies. The control pA samples averaged and analyzed were ZT06, ZT10, ZT18, and ZT22.
3′SS ratio determination
Custom scripts were used to determine splicing efficiency by the ratio of reads about the 3′SS. We determined the number of reads at each base pair for the last 25 bp of a given intron and the first 25 bp of the 3′ exon. The numbers were then divided. Alternative introns with overlapping exons in this region were excluded from this analysis. Constitutive introns were defined as introns appearing in all isoforms of the gene, and their retention was calculated as an average retention for all isoforms of the gene (Khodor et al. 2011).
Statistical analysis
Selected introns with an average of greater than or equal to three reads per base pair in all exons of the transcript were analyzed by length of intron and flanking exons, position within the gene, and prior annotation as alternative or constitutive in RefSeq using the PASW Statistics 18 software (IBM). Box plots generated show the 95th–fifth percentile distributions between the whiskers, and the box spans the 75th–25th percentiles. Nonparametric analysis was used, because the distributions were not normal. Significance marks above distributions analyzed using the Kurstal-Wallis Test reflect significant differences between all distributions in those groupings.
DATA DEPOSITION
Raw and processed sequencing data used in this work are available for download from Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo), accession number GSE32950 for Drosophila and GSE36916 for mouse liver.
SUPPLEMENTAL MATERIAL
Supplemental material is available for this article.
ACKNOWLEDGMENTS
We thank our colleagues in the Rosbash laboratory for helpful suggestions, particularly Joseph Rodriguez and Katharine Abruzzi. We also thank Douglas Black for his feedback during the preparation of this manuscript. This research was supported by the Howard Hughes Medical Institute and NIH grants GM023549 and NS044232 (to M.R.).
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.034090.112.
REFERENCES
- Alexander RD, Innocente SA, Barrass JD, Beggs JD 2010. Splicing-dependent RNA polymerase pausing in yeast. Mol Cell 40: 582–593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ast G 2004. How did alternative splicing evolve? Nat Rev Genet 5: 773–782 [DOI] [PubMed] [Google Scholar]
- Barta I, Iggo R 1995. Autoregulation of expression of the yeast Dbp2p ‘DEAD-box’ protein is mediated by sequences in the conserved DBP2 intron. EMBO J 14: 3800–3808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauren G, Wieslander L 1994. Splicing of Balbiani ring 1 gene pre-mRNA occurs simultaneously with transcription. Cell 76: 183–192 [DOI] [PubMed] [Google Scholar]
- Berget SM 1995. Exon recognition in vertebrate splicing. J Biol Chem 270: 2411–2414 [DOI] [PubMed] [Google Scholar]
- Beyer AL, Osheim YN 1988. Splice site selection, rate of splicing, and alternative splicing on nascent transcripts. Genes Dev 2: 754–765 [DOI] [PubMed] [Google Scholar]
- Bourquin JP, Stagljar I, Meier P, Moosmann P, Silke J, Baechi T, Georgiev O, Schaffner W 1997. A serine/arginine-rich nuclear matrix cyclophilin interacts with the C-terminal domain of RNA polymerase II. Nucleic Acids Res 25: 2055–2061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brody Y, Neufeld N, Bieberstein N, Causse SZ, Bohnlein EM, Neugebauer KM, Darzacq X, Shav-Tal Y 2011. The in vivo kinetics of RNA polymerase II elongation during co-transcriptional splicing. PLoS Biol 9: e1000573 doi: 10.1371/journal.pbio.1000573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burnette JM, Miyamoto-Sato E, Schaub MA, Conklin J, Lopez AJ 2005. Subdivision of large introns in Drosophila by recursive splicing at nonexonic elements. Genetics 170: 661–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrillo Oesterreich F, Preibisch S, Neugebauer KM 2010. Global analysis of nascent RNA reveals transcriptional pausing in terminal exons. Mol Cell 40: 571–581 [DOI] [PubMed] [Google Scholar]
- Cheng H, Dufu K, Lee CS, Hsu JL, Dias A, Reed R 2006. Human mRNA export machinery recruited to the 5′ end of mRNA. Cell 127: 1389–1400 [DOI] [PubMed] [Google Scholar]
- Churchman LS, Weissman JS 2011. Nascent transcript sequencing visualizes transcription at nucleotide resolution. Nature 469: 368–373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooke C, Hans H, Alwine JC 1999. Utilization of splicing elements and polyadenylation signal elements in the coupling of polyadenylation and last-intron removal. Mol Cell Biol 19: 4971–4979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Custodio N, Carmo-Fonseca M, Geraghty F, Pereira HS, Grosveld F, Antoniou M 1999. Inefficient processing impairs release of RNA from the site of transcription. EMBO J 18: 2855–2866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dabeva MD, Post-Beittenmiller MA, Warner JR 1986. Autogenous regulation of splicing of the transcript of a yeast ribosomal protein gene. Proc Natl Acad Sci 83: 5854–5857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damgaard CK, Kahns S, Lykke-Andersen S, Nielsen AL, Jensen TH, Kjems J 2008. A 5′ splice site enhances the recruitment of basal transcription initiation factors in vivo. Mol Cell 29: 271–278 [DOI] [PubMed] [Google Scholar]
- Das R, Dufu K, Romney B, Feldt M, Elenko M, Reed R 2006. Functional coupling of RNAP II transcription to spliceosome assembly. Genes Dev 20: 1100–1109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de la Mata M, Kornblihtt AR 2006. RNA polymerase II C-terminal domain mediates regulation of alternative splicing by SRp20. Nat Struct Mol Biol 13: 973–980 [DOI] [PubMed] [Google Scholar]
- de la Mata M, Alonso CR, Kadener S, Fededa JP, Blaustein M, Pelisch F, Cramer P, Bentley D, Kornblihtt AR 2003. A slow RNA polymerase II affects alternative splicing in vivo. Mol Cell 12: 525–532 [DOI] [PubMed] [Google Scholar]
- de la Mata M, Lafaille C, Kornblihtt AR 2010. First come, first served revisited: Factors affecting the same alternative splicing event have different effects on the relative rates of intron removal. RNA 16: 904–912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dye MJ, Proudfoot NJ 1999. Terminal exon definition occurs cotranscriptionally and promotes termination of RNA polymerase II. Mol Cell 3: 371–378 [DOI] [PubMed] [Google Scholar]
- Farlow A, Dolezal M, Hua L, Schlotterer C 2012. The genomic signature of splicing-coupled selection differs between long and short introns. Mol Biol Evol 29: 21–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flicek P, Amode MR, Barrell D, Beal K, Brent S, Carvalho-Silva D, Clapham P, Coates G, Fairley S, Fitzgerald S, et al. 2012. Ensembl 2012. Nucleic Acids Res 40: D84–D90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox-Walsh KL, Dou Y, Lam BJ, Hung SP, Baldi PF, Hertel KJ 2005. The architecture of pre-mRNAs affects mechanisms of splice-site pairing. Proc Natl Acad Sci 102: 16176–16181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gelfman S, Burstein D, Penn O, Savchenko A, Amit M, Schwartz S, Pupko T, Ast G 2012. Changes in exon–intron structure during vertebrate evolution affect the splicing pattern of exons. Genome Res 22: 35–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gornemann J, Kotovic KM, Hujer K, Neugebauer KM 2005. Cotranscriptional spliceosome assembly occurs in a stepwise fashion and requires the cap binding complex. Mol Cell 19: 53–63 [DOI] [PubMed] [Google Scholar]
- Harr B, Turner LM 2010. Genome-wide analysis of alternative splicing evolution among Mus subspecies. Mol Ecol (Suppl 1) 19: 228–239 [DOI] [PubMed] [Google Scholar]
- Hirose Y, Tacke R, Manley JL 1999. Phosphorylated RNA polymerase II stimulates pre-mRNA splicing. Genes Dev 13: 1234–1239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ip JY, Schmidt D, Pan Q, Ramani AK, Fraser AG, Odom DT, Blencowe BJ 2011. Global impact of RNA polymerase II elongation inhibition on alternative splicing regulation. Genome Res 21: 390–401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadener S, Cramer P, Nogues G, Cazalla D, de la Mata M, Fededa JP, Werbajh SE, Srebrow A, Kornblihtt AR 2001. Antagonistic effects of T-Ag and VP16 reveal a role for RNA pol II elongation on alternative splicing. EMBO J 20: 5759–5768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kessler O, Jiang Y, Chasin LA 1993. Order of intron removal during splicing of endogenous adenine phosphoribosyltransferase and dihydrofolate reductase pre-mRNA. Mol Cell Biol 13: 6211–6222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khodor YL, Rodriguez J, Abruzzi KC, Tang CH, Marr MT II, Rosbash M 2011. Nascent-seq indicates widespread cotranscriptional pre-mRNA splicing in Drosophila. Genes Dev 25: 2502–2512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim E, Du L, Bregman DB, Warren SL 1997. Splicing factors associate with hyperphosphorylated RNA polymerase II in the absence of pre-mRNA. J Cell Biol 136: 19–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim E, Magen A, Ast G 2007. Different levels of alternative splicing among eukaryotes. Nucleic Acids Res 35: 125–131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S, Kim H, Fong N, Erickson B, Bentley DL 2011. Pre-mRNA splicing is a determinant of histone H3K36 methylation. Proc Natl Acad Sci 108: 13564–13569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiseleva E, Wurtz T, Visa N, Daneholt B 1994. Assembly and disassembly of spliceosomes along a specific pre-messenger RNP fiber. EMBO J 13: 6052–6061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohler A, Hurt E 2007. Exporting RNA from the nucleus to the cytoplasm. Nat Rev Mol Cell Biol 8: 761–773 [DOI] [PubMed] [Google Scholar]
- Kolasinska-Zwierz P, Down T, Latorre I, Liu T, Liu XS, Ahringer J 2009. Differential chromatin marking of introns and expressed exons by H3K36me3. Nat Genet 41: 376–381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo H, Nasim FH, Grabowski PJ 1991. Control of alternative splicing by the differential binding of U1 small nuclear ribonucleoprotein particle. Science 251: 1045–1050 [DOI] [PubMed] [Google Scholar]
- Kwek KY, Murphy S, Furger A, Thomas B, O'Gorman W, Kimura H, Proudfoot NJ, Akoulitchev A 2002. U1 snRNA associates with TFIIH and regulates transcriptional initiation. Nat Struct Biol 9: 800–805 [DOI] [PubMed] [Google Scholar]
- Lacadie SA, Rosbash M 2005. Cotranscriptional spliceosome assembly dynamics and the role of U1 snRNA:5′ss base pairing in yeast. Mol Cell 19: 65–75 [DOI] [PubMed] [Google Scholar]
- Lacadie SA, Tardiff DF, Kadener S, Rosbash M 2006. In vivo commitment to yeast cotranscriptional splicing is sensitive to transcription elongation mutants. Genes Dev 20: 2055–2066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg SL 2009. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol 10: R25 doi: 10.1186/gb-2009-10-3-r25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- LeMaire MF, Thummel CS 1990. Splicing precedes polyadenylation during Drosophila E74A transcription. Mol Cell Biol 10: 6059–6063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis JD, Izaurralde E, Jarmolowski A, McGuigan C, Mattaj IW 1996. A nuclear cap-binding complex facilitates association of U1 snRNP with the cap-proximal 5′ splice site. Genes Dev 10: 1683–1698 [DOI] [PubMed] [Google Scholar]
- Listerman I, Sapra AK, Neugebauer KM 2006. Cotranscriptional coupling of splicing factor recruitment and precursor messenger RNA splicing in mammalian cells. Nat Struct Mol Biol 13: 815–822 [DOI] [PubMed] [Google Scholar]
- McCracken S, Fong N, Yankulov K, Ballantyne S, Pan G, Greenblatt J, Patterson SD, Wickens M, Bentley DL 1997. The C-terminal domain of RNA polymerase II couples mRNA processing to transcription. Nature 385: 357–361 [DOI] [PubMed] [Google Scholar]
- Menet JS, Rodriguez J, Abruzzi KC, Rosbash M 2012. Nascent-seq reveals novel features of mouse circadian transcriptional regulation. eLife (in press). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mount SM, Burks C, Hertz G, Stormo GD, White O, Fields C 1992. Splicing signals in Drosophila: Intron size, information content, and consensus sequences. Nucleic Acids Res 20: 4255–4262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsen TW, Graveley BR 2010. Expansion of the eukaryotic proteome by alternative splicing. Nature 463: 457–463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niwa M, Berget SM 1991. Mutation of the AAUAAA polyadenylation signal depresses in vitro splicing of proximal but not distal introns. Genes Dev 5: 2086–2095 [DOI] [PubMed] [Google Scholar]
- Oesterreich FC, Bieberstein N, Neugebauer KM 2011. Pause locally, splice globally. Trends Cell Biol 21: 328–335 [DOI] [PubMed] [Google Scholar]
- Pandya-Jones A, Black DL 2009. Co-transcriptional splicing of constitutive and alternative exons. RNA 15: 1896–1908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parenteau J, Durand M, Veronneau S, Lacombe AA, Morin G, Guerin V, Cecez B, Gervais-Bird J, Koh CS, Brunelle D, et al. 2008. Deletion of many yeast introns reveals a minority of genes that require splicing for function. Mol Biol Cell 19: 1932–1941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perales R, Bentley D 2009. “Cotranscriptionality” - the transcription elongation complex as a nexus for nuclear transactions. Mol Cell 36: 178–191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preker PJ, Kim KS, Guthrie C 2002. Expression of the essential mRNA export factor Yra1p is autoregulated by a splicing-dependent mechanism. RNA 8: 969–980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruitt KD, Tatusova T, Klimke W, Maglott DR 2009. NCBI Reference Sequences: Current status, policy and new initiatives. Nucleic Acids Res 37: D32–D36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rigo F, Kazerouninia A, Nag A, Martinson HG 2005. The RNA tether from the poly(A) signal to the polymerase mediates coupling of transcription to cleavage and polyadenylation. Mol Cell 20: 733–745 [DOI] [PubMed] [Google Scholar]
- Robberson BL, Coté GJ, Berget SM 1990. Exon definition may facilitate splice site selection in RNAs with multiple exons. Mol Cell Biol 10: 84–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robert F, Blanchette M, Maes O, Chabot B, Coulombe B 2002. A human RNA polymerase II–containing complex associated with factors necessary for spliceosome assembly. J Biol Chem 277: 9302–9306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharov AA, Dudekula DB, Ko MS 2005. Genome-wide assembly and analysis of alternative transcripts in mouse. Genome Res 15: 748–754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spies N, Nielsen CB, Padgett RA, Burge CB 2009. Biased chromatin signatures around polyadenylation sites and exons. Mol Cell 36: 245–254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sterner DA, Carlo T, Berget SM 1996. Architectural limits on split genes. Proc Natl Acad Sci 93: 15081–15085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talerico M, Berget SM 1994. Intron definition in splicing of small Drosophila introns. Mol Cell Biol 14: 3434–3445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanner S, Stagljar I, Georgiev O, Schaffner W, Bourquin JP 1997. A novel SR-related protein specifically interacts with the carboxy-terminal domain (CTD) of RNA polymerase II through a conserved interaction domain. Biol Chem 378: 565–571 [DOI] [PubMed] [Google Scholar]
- Tardiff DF, Lacadie SA, Rosbash M 2006. A genome-wide analysis indicates that yeast pre-mRNA splicing is predominantly posttranscriptional. Mol Cell 24: 917–929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Pachter L, Salzberg SL 2009. TopHat: Discovering splice junctions with RNA-Seq. Bioinformatics 25: 1105–1111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vargas DY, Shah K, Batish M, Levandoski M, Sinha S, Marras SA, Schedl P, Tyagi S 2011. Single-molecule imaging of transcriptionally coupled and uncoupled splicing. Cell 147: 1054–1065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang ET, Sandberg R, Luo S, Khrebtukova I, Zhang L, Mayr C, Kingsmore SF, Schroth GP, Burge CB 2008. Alternative isoform regulation in human tissue transcriptomes. Nature 456: 470–476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterston RH, Lindblad-Toh K, Birney E, Rogers J, Abril JF, Agarwal P, Agarwala R, Ainscough R, Alexandersson M, An P, et al. 2002. Initial sequencing and comparative analysis of the mouse genome. Nature 420: 520–562 [DOI] [PubMed] [Google Scholar]
- Wetterberg I, Bauren G, Wieslander L 1996. The intranuclear site of excision of each intron in Balbiani ring 3 pre-mRNA is influenced by the time remaining to transcription termination and different excision efficiencies for the various introns. RNA 2: 641–651 [PMC free article] [PubMed] [Google Scholar]
- Wuarin J, Schibler U 1994. Physical isolation of nascent RNA chains transcribed by RNA polymerase II: Evidence for cotranscriptional splicing. Mol Cell Biol 14: 7219–7225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang G, Taneja KL, Singer RH, Green MR 1994. Localization of pre-mRNA splicing in mammalian nuclei. Nature 372: 809–812 [DOI] [PubMed] [Google Scholar]