Background: α1(V) is an extensively modified collagen chain important in disease.
Results: Comprehensive mapping of α1(V) post-translational modifications reveals unexpectedly large numbers of X-position hydroxyprolines in Gly-X-Y amino acid triplets.
Conclusion: The unexpected abundance of X-position hydroxyprolines suggests a mechanism for differential modification of collagen properties.
Significance: Positions, numbers, and occupancy of modified sites can provide insights into α1(V) biological properties.
Keywords: Collagen, Extracellular Matrix, Extracellular Matrix Proteins, Glycosylation, Hydroxylase, Hydroxylysine, Hydroxyproline, Mass Spectrometry (MS), Proteomics, Post-translational Modification
Abstract
Aberrant expression of the type V collagen α1(V) chain can underlie the connective tissue disorder classic Ehlers-Danlos syndrome, and autoimmune responses against the α1(V) chain are linked to lung transplant rejection and atherosclerosis. The α1(V) collagenous COL1 domain is thought to contain greater numbers of post-translational modifications (PTMs) than do similar domains of other fibrillar collagen chains, PTMs consisting of hydroxylated prolines and lysines, the latter of which can be glycosylated. These types of PTMs can contribute to epitopes that underlie immune responses against collagens, and the high level of PTMs may contribute to the unique biological properties of the α1(V) chain. Here we use high resolution mass spectrometry to map such PTMs in bovine placental α1(V) and human recombinant pro-α1(V) procollagen chains. Findings include the locations of those PTMs that vary and those PTMs that are invariant between these α1(V) chains from widely divergent sources. Notably, an unexpectedly large number of hydroxyproline residues were mapped to the X-positions of Gly-X-Y triplets, contrary to expectations based on previous amino acid analyses of hydrolyzed α1(V) chains from various tissues. We attribute this difference to the ability of tandem mass spectrometry coupled to nanoflow chromatographic separations to detect lower-level PTM combinations with superior sensitivity and specificity. The data are consistent with the presence of a relatively large number of 3-hydroxyproline sites with less than 100% occupancy, suggesting a previously unknown mechanism for the differential modification of α1(V) chain and type V collagen properties.
Introduction
Collagen type V (col(V))2 is a low abundance fibrillar collagen that is widely distributed in vertebrate tissues as an α1(V)2α2(V) heterotrimer (1). It is also found with a more limited tissue distribution as an α1(V)α2(V)α3(V) heterotrimer and as rare α1(V)3 homotrimers (1, 2). Thus, all forms of col(V) contain the α1(V) chain. Additionally, the closely related collagen type XI, first described as an α1(XI)α2(XI)α3(XI) heterotrimer in fetal cartilage (3), incorporates α1(V) chains into heterotypic α1(XI)α1(V)α3(XI) trimers as cartilage matures (4), implying additional roles for the α1(V) chain.
It is generally accepted that α1(V)2α2(V) heterotrimers are incorporated into growing fibrils of the more abundant collagen type I and are involved in regulating the geometry and properties of the resulting type I/V heterotypic fibrils (5, 6). Thus, mutations in the genes encoding either α1(V) or α2(V) chains result in the human connective tissue disorder classic Ehlers-Danlos syndrome, characterized by collagen I fibrils of abnormal shapes and diameters and deficient tensile strength (7, 8). More recently, it has been demonstrated that anti-col(V) autoimmune responses can underlie chronic lung transplant rejection in both humans (9) and animal models (10) and that pre-transplant col(V)-specific autoimmunity is also a significant risk factor for primary graft dysfunction, the leading cause of early morbidity and mortality after lung transplantation (11, 12). Col(V) autoimmunity has also been identified as a consistent feature in both late stage human coronary artery disease and a mouse model of atherosclerosis (13). In both human lung transplant rejection and coronary artery disease, immune responses have been shown to be specific to the α1(V) chain, with an absence of such responses to the α2(V) chain (9, 13).
The α1(V) chain contains extensive post-translational modifications (PTMs) relative to other fibrillar collagen chains consisting of 4-hydroxyproline (4-Hyp) and 3-hydroxyproline (3-Hyp) residues and of hydroxylysine (Hyl) residues, most of which are decorated with di- and monosaccharides (14, 15). Functionally, 4-Hyp residues have well characterized effects in stabilizing triple helices (16), whereas Hyl residues are important to the formation of stable covalent cross-links between collagen chains (17), and Hyl glycosylation may be important in assembly and secretion of at least some collagens (18). The function of 3-Hyp residues is unclear, with conflicting reports on small stabilizing (19) or destabilizing (20) effects on triple helix stability. However, loss of 3-Hyp residues in collagens can have catastrophic phenotypic consequences (21), implying an important biological function(s). In addition to functional roles, previous studies show various PTMs are capable of affecting immune responses against collagenous antigens (22–29). For example, Pro hydroxylation can contribute to the antigenicity of collagen IV (22), whereas lysine hydroxylation and glycosylation are important to epitopes that affect the antigenic roles of collagen II in rheumatoid and experimentally induced arthritis (23–29).
Recent studies investigating PTMs on collagenous proteins have utilized tandem mass spectrometry (MSn) on low resolution, low mass accuracy ion trap mass spectrometers or mass measurements of large peptides using matrix-assisted laser desorption ionization time-of-flight (MALDI-TOF) MS (14, 30–32). However, performing MSn on instruments with low mass accuracy or relying on MS measurements of peptides that have not been subjected to MSn makes confident peptide identification and unambiguous PTM site localization challenging. The analysis of collagenous proteins by MS is complicated by the high degree of modification and the large number of proline residues. The former leads to generation of isobaric, but differentially modified, peptides that often co-elute, producing chimeric spectra difficult to interpret without the aid of high mass accuracy and abundant product ions from MSn. The latter hampers MSn sequencing efforts due the propensity of Pro/Hyp to preferentially cleave during collisional activation (the “proline effect”) (33, 34). This can limit the generation of a ladder of b- and y-type product ions most commonly used by database search algorithms for peptide identification and PTM localization of spectra generated by collision-activated dissociation (33, 35–37).
In the current study we used state-of-the-art proteomics workflow centered on the use of high resolution MS and nanoflow chromatographic separations to precisely map hydroxylation and glycosylation modifications in bovine placental α1(V) and human recombinant pro-α1(V) procollagen chains. Instead of relying on a single enzyme or chemical treatment to produce peptides amenable to MS, we used five different proteases (individually or sequentially) to maximize protein sequence coverage and facilitate PTM discovery. Peptides were subsequently separated by nanoflow liquid chromatography and introduced into an electron transfer dissociation (ETD-enabled) hybrid dual cell linear ion trap-orbitrap mass spectrometer by electrospray ionization. The orbitrap recorded the masses of eluting peptides with low ppm mass accuracy (<5 ppm) affording confident peptide identification and localization of PTMs. The hybrid orbitrap mass spectrometer permitted the use of several dissociation techniques; resonant excitation collision-activated dissociation, beam-type collision-activated dissociation (higher energy collisional dissociation (HCD)), and ETD (38, 39). The availability of multiple dissociation techniques and the use of MS2 and MS3 for peptide interrogation often allowed us to pinpoint the exact residue(s) carrying the individual PTMs. Such comprehensive, experiment-based PTM localization has not previously been achieved for collagenous proteins. Recent analyses of collagenous proteins by MS relied heavily on a priori biological knowledge to assess prolyl hydroxylation (i.e. PTM assignment based upon hydroxylation motifs) (30, 32) rather than PTM assignment based upon localizing fragments from MSn spectra.
We report comprehensive mapping of all PTMs involving hydroxylated residues on bovine placenta α1(V) and human recombinant pro-α1(V) collagen chains and provide manually verified mass spectral evidence for each modified site. Our analyses reveal PTMs that vary or are invariant between the bovine tissue α1(V) and human recombinant pro-α1(V) chains and also reveal all hydroxylated residue PTMs in pro-α1(V) sequences NH2-terminal to the COL1 major collagenous domain. We also identify an unexpectedly large number of Hyp residues in the X-position of Gly-X-Y triplets, attributed to our ability to identify modified peptides present in low stoichiometric amounts that go undetected by classic amino acid analyses or Edman sequencing. Another unexpected, and striking finding was X-position Hyp residues discovered in the unusual contexts of Gly-Hyp-Val and Gly-Hyp-Ala triplets in the bovine placental α1(V) COL1 domain. Findings presented herein may aid in characterizing and locating α1(V) autoimmune epitopes and may provide further insights into col(V) function.
EXPERIMENTAL PROCEDURES
Preparation of Human Pro-α1(V) Procollagen and Bovine α1(V) Collagen Chains
Human recombinant pro-α1(V) homotrimers were produced via expression from a modified pCEP-Pu vector in 293-EBNA human embryonic kidney cells (Invitrogen) followed by dialysis of conditioned media against 50 mm Tris-HCl, pH 8.6, buffer containing 0.1 mm phenylmethylsulfonyl fluoride, 1 mm N-ethylmaleimide, 0.1 mm p-aminobenzoic acid, and 5 mm EDTA for low salt precipitation of collagen chains as previously described (40). Bovine α1(V) chains were prepared essentially as previously described (41). Briefly, minced and washed amnion stripped off placenta from the area close to attachment of the umbilicus was suspended in 0.5 m acetic acid, 0.2 m NaCl and digested with pepsin at 4 °C. Col(V), which is soluble in 0.7 m NaCl and precipitates in 1.2 m NaCl, was purified from supernatants via differential NaCl precipitation (42). α1(V) chains were separated from α2(V) chains via chromatography on diethylaminoethyl cellulose.
Prolyl 3-Hydroxylase Expression
Breast carcinoma cell line MB 436 (ATCC catalogue no. HTB-130) and HEK293 T-REx cells were maintained in Dulbecco's modified Eagle's media (Cellgro, Manassas, VA) supplemented with 10% FBS and penicillin/streptomycin in 5% CO2. Total RNA was extracted from cell lines using TRIzol (Invitrogen). cDNA was synthesized from 1 μg of RNA using the SuperScript II Reverse Transcriptase kit (Invitrogen) with random hexamers. Subsequently, PCR was performed for 35 cycles with primer sets for human prolyl 3-hydroxylase 1 (P3H1), P3H2, P3H3, cartilage-associated protein (CRTAP), and PPIB previously described by Fernandes et al. (31) and for GAPDH previously described by Shah et al. (43).
Amino Acid Analysis
Bovine α1(V) collagen chains were hydrolyzed in 6 n HCl, 1% phenol at 110 °C for 48 h and analyzed by means of a Hitachi L 8800 A amino acid analyzer at the University of California-Davis Molecular Structure Facility.
Pro-α1(V) Sequences for Comparison to MS Data
For human pro-α1(V) sequences, MS data were compared with sequences available in the databases. For bovine α1(V) sequences, a BLAST search of bovine genome databases using human pro-α1(V) sequences (44) identified mRNA sequence XM_002691720.1, predicted by automated analysis of genomic sequences, which encodes the pro-α1(V) C-propeptide and ∼38% of the α1(V) major triple helical (COL1) domain interspersed with gaps and insertions of non-α1(V) sequences probably due to annotative misidentification of intron/exon junctions. Toward obtaining complete bovine α1(V) COL1 sequences, first strand cDNA was synthesized from bovine placenta total RNA (Zyagen, San Diego, CA) using oligo(dT)20 primers. Primers 5′-AGGCCCCCCAGGCGAGGTC-3′ (forward) and 5′-ATTCTGGCCCCTTCAGACTT-3′ (reverse), based on XM_002691720.1 COL1 sequences, were then used to amplify an ∼1.8-kb fragment of COL1 sequences that was directly sequenced using the same forward primer. Another predicted mRNA sequence (XM_617549.3), identified via subsequent BLAST search, encodes partial pro-α1(V) NH2-terminal globular sequences and some inserted intronic sequences. To obtain the remaining bovine α1(V) COL1 sequences, forward primer 5′-GTGGCACAGAATTGCTCTCA-3′, based on XM_617549.3 sequences, was used with primer 5′-GCCTTCACTGCCTTTCAGTC-3′ (reverse primer A) based on XM_002691720.1 sequences to amplify a ∼2.7-kb fragment. The ∼2.7-kb fragment could not be amplified from first strand cDNA synthesized using oligo(dT)20 primers as above and was instead amplified from first strand cDNA obtained using reverse primer A. The ∼2.7-kb PCR fragment was directly sequenced sequentially using reverse primers 5′-GTGGCACAGAATTGCTCTCA-3′, 5′-CTCCAAGTTTGCCCTTCTCC-3′, and 5′-GCCCCTTTCTCCGTCTTC-3′. The full-length bovine α1(V) COL1 sequences have been submitted to the GenBankTM/EMBL Data Bank with accession number JQ611730.
Digestion of Pro-α1(V) and α1(V) Samples for Mass Spectrometry
Pro-α1(V) and α1(V) samples were desalted using 50 mg tC18 SepPak cartridges (Waters Corp., Milford, MA). Eluates were dried down and resuspended in digestion buffer (see below) optimized for each protease. Cysteine residues were reduced and alkylated by incubation in 5 mm dithiothreitol for 45 min at 37 °C followed by a 30-min incubation at room temperature in 15 mm iodoacetamide in the dark. Alkylation was quenched by a 15-min incubation in 5 mm dithiothreitol at room temperature. This was followed by a multiple protease approach, utilized to maximize protein sequence coverage (45), and involved addition of five different proteases (individually or sequentially) (see the details in the supplemental Experimental Procedures).
Liquid Chromatography Electrospray Tandem Mass Spectrometry (LC-MS/MS)
A Waters nanoACQUITY UPLC and autosampler (Waters Corp.) were used to load samples onto a fused-silica capillary precolumn (75-μm inner diameter × 360-μm outer diameter). Precolumns with cast chemical frits were slurry-packed to 5 cm in length with a stationary phase consisting of a 5-μm diameter, 100 Å pore size, C18 particles (Magic C18AQ, Michrom Bioresources, Inc., Auburn CA) (46). Reversed-phase LC separation was achieved across a 13-cm-long, 50-μm inner diameter × 360-μm outer diameter fused-silica analytical column packed with the same C18 stationary phase. An electrospray ionization emitter was integrated into the analytical column by use of a laser puller (Sutter Instrument Co., P-2000, Novato, CA). An estimated 1–4 μg of protein digest was loaded onto the precolumn and chromatographed over a 60-min linear gradient at 300 nl/min (2 to 30% B, buffer A (0.2% formic acid in water); buffer B (0.2% formic acid in acetonitrile)). Eluate was introduced into the mass spectrometer via electrospray ionization (+2.0 kV), and peptide cations were subjected to tandem mass spectrometry using an LTQ Orbitrap Velos mass spectrometer (Thermo Fisher Scientific, Bremen, Germany) enabled for ETD (39, 47, 48). Typical experiments consisted of MS1 analysis in the Orbitrap mass analyzer using a resolving power of 60,000 followed by 10 data-dependent HCD MS2 events. Product ion mass analysis was also conducted in the Orbitrap using resolving powers of 7,500–15,000. Precursor and product ion mass error was typically <10 ppm. Similar experiments were conducted using ETD, where charge state-dependent ETD activation times were utilized. To obtain additional sequence information of glycosylated peptides, an additional set of LC analyses were conducted using an MS3 mass spectrometry method. For MS3 analyses, three data-dependent collision-induced dissociation or ETD MS2 events were executed followed by data-dependent HCD MS3 events, where the most intense peak in each MS2 spectrum was dissociated. For all experiments, an automatic gain control target value of 1,000,000 charges was used for MS1. An automatic gain control target of 50,000 charges was used for both MS2 and MS3. Precursors were dynamically excluded from data-dependent MS2 for 30–60 s. A precursor isolation width of 2–3 m/z was typically used.
Proteomic Data Analysis
Spectral reduction from raw data was performed by DTA Generator, a program available in the COMPASS software suite (freely available online) (49). The ETD preprocessing option was used to remove known neutral loss peaks from ETD spectra (50, 51). The OMSSA (Open Mass Spectrometry Search Algorithm) search algorithm was used for database correlation (52). Spectra were searched against a concatenated target-decoy version of human Pro-α1 (V) or bovine α1 (V) sequence databases (53). Several variable PTMs were considered: hydroxylation of proline and lysine residues (+15.9949 Da, monoisotopic mass), glycosylation of lysine (monosaccharide, +178.0473 Da, disaccharide, +340.0995), and glycosylation-associated neutral losses upon activation. Finally, oxidation of methionine (+15.9949 Da) residues was specified as a variable modification, and carbamidomethylation of cysteine (+57.0215 Da) residues was searched as a fixed modification. A mass tolerance of ±5 Da from the average mass was used for precursors, whereas a mass tolerance of ± 0.01 Da from the monoisotopic mass was used for product ions (38). Spectra for all the modified sites were manually validated (see details in the supplemental Experimental Procedures) and are provided in supplemental Spectra S1 for bovine and supplemental Spectra S2 for human.
RESULTS
PTMs of Bovine Placental α1(V) Chains
Chromatographically purified α1(V) chains extracted with acetic acid and pepsin from bovine placenta were subjected to tandem mass spectrometry (MSn) to map the positions of hydroxylated residues and of galactosylhydroxylysyl (Gal-Hyl) and glucosylgalactosylhydroxylysyl (Glc-Gal-Hyl) residues. Treatment of fibrillar collagens with pepsin removes NH2- and COOH-terminal non-triple helical sequences, leaving only the pepsin-resistant major triple helical COL1 domain. At the time this study began, bovine α1(V) cDNA sequences were unavailable, and only a small portion (C-propeptide sequences and about the COOH-terminal third of COL1 sequences) of bovine α1(V) coding sequences were available from annotated bovine genome databases. Thus, full-length bovine α1(V) cDNA sequences were generated as described under “Experimental Procedures,” for comparison to MS data. At the time of submission of this study, database annotated genomic sequences were mostly complete but still had 29 amino acid differences compared with the cDNA sequences reported here (GenBankTM accession number JQ611730). The cDNA sequences reported here have been validated via comparison of MS analyses of bovine placental α1(V) protein.
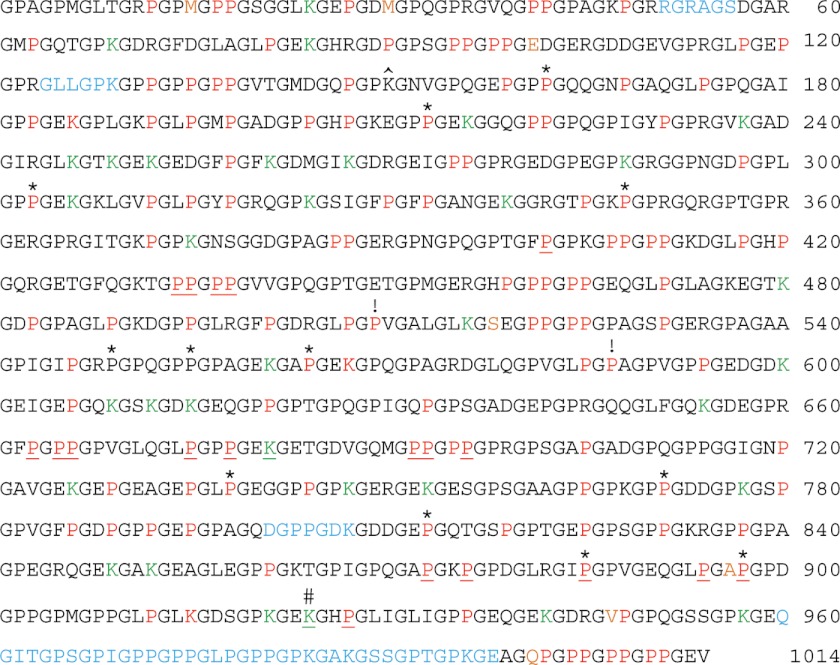
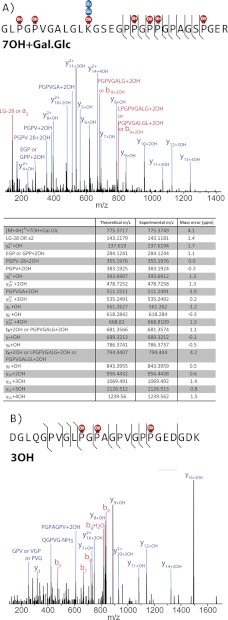
MSn of AspN, GluC, chymotrypsin, ArgC, and trypsin-generated peptides of bovine placental α1(V) chains produced 94% sequence coverage and identified the positions of 106 Hyp residues in the Y-position of Gly-X-Y triplet repeats and 22 Hyp residues in the X-position of Gly-Hyp-Hyp triplet repeats (Fig. 1). Y-position Pro residues in Gly-X-Y triplets are hydroxylated to 4-Hyp residues by the enzyme prolyl 4-hydroxylase, for which the minimum substrate appears to be the tripeptide X-Pro-Gly (54). Thus, the 106 Y-position Hyp residues reported here are likely 4-Hyp. Gly-Pro-Hyp triplets are thought to constitute the substrate sequence for P3Hs, of which there are three (55, 56). We conclude the 22 Hyp residues in the X-position of Gly-Hyp-Hyp triplet repeats in the COL1 domain of bovine placental α1(V) chain are likely to be 3-Hyp residues. Unexpectedly, Hyp residues were detected in the X-positions of a Gly-X-Val triplet and a Gly-X-Ala triplet (residues 509 and 587, respectively) (Figs. 1 and 2). As prolyl 4-hydroxylase is thought to hydroxylate only Y-position Pro residues and as P3H is thought to hydroxylate only Pro residues within Gly-Pro-Hyp triplets, it is not clear which enzyme has hydroxylated these sites or whether these hydroxylated prolines are 3-Hyp or 4-Hyp residues. Our analysis also identified the positions of 3 Hyl residues and 34 Glc-Gal-Hyl residues (Fig. 1), Hyl residues linked to the disaccharide glucosylgalactose.
FIGURE 1.
Hydroxylated amino acid residues and saccharide attachments of the bovine placenta α1(V) collagen chain. Sequence coverage was 94%, with identification and mapping of 106 Y-position Hyp, 22 Gly-Hyp-Hyp X-position Hyp, 1 Gly-Hyp-Ala, 1 Gly-Hyp-Val, 3 Hyl, and 34 Glc-Gal-Hyl residues. Red, hydroxylated, non-glycosylated residues; green, Glc-Gal-Hyl residues; underlined, hydroxylated/glycosylated residues mapped in previous studies (4, 32). Sequences not identified in the course of MS analysis are in light blue. The seven COL1 amino acid residues that differ between human and bovine are orange. *, 12 Y-position Pro residues hydroxylated in human or bovine α1(V) COL1 domain but not the other. ^, Lys residue hydroxylated in human recombinant pro-α1(V) but not in bovine placenta α1(V). #, Hyl residues identified by Wu et al. (4) as being involved in interchain covalent cross-links. !, Hyp residues detected in the X-position of Gly-X-Y triplets lacking a Y-position Hyp. Amino acid residues are numbered from 1 to 1014, with residue number 1 being the first amino acid and residue number 1014 being the final amino acid residue of the COL1 triple helical domain.
FIGURE 2.
X-position Hyp residues in Gly-X-Val and Gly-X-Ala triplets. Peptides were prepared by trypsin digestion of bovine placenta α1 (V) collagen chains. Mass error from the expected product ion monoisotopic mass is typically less than 5 ppm. y14+4OH fragment ion and internal fragment ions (PGPV-28 + 2OH, PGPV+2OH, and PGPVGA+2OH) confirm that one Hyp is in the Gly-X-Val triplet in the upper spectrum (A). y11+OH, y12+OH, and y14+2OH fragment ions confirm that one Hyp is in the Gly-X-Ala triplet in the lower spectrum (B). OH, one hydroxylation.
Hydroxylated Residues and Glycosylated Hyl Residues of Human Recombinant Pro-α1(V) Chains
A number of previous studies on the biology of pro-α1(V) collagen chains have been performed on human recombinant pro-α1(V) collagen chains produced in 293-HEK (human embryonic kidney) cells (40, 57–62). As previous partial characterizations of fibrillar collagen chain PTMs have shown tissue- and cell type-specific variations in distributions of 3-Hyp residues (30–32), we mapped the hydroxylated residues and saccharide-bound Hyl residues of the experimentally important human recombinant full-length pro-α1(V) collagen chains produced in 293-HEK cells. This study of procollagen chains also allowed characterization of hydroxylated amino acid residues in sequences NH2-terminal to the COL1 domain and in COOH-propeptide sequences, both of which are lost in pepsin extraction of α1(V) chains from tissues.
MSn of AspN, GluC, chymotrypsin, ArgC, and trypsin-generated peptides of human recombinant pro-α1(V) collagen chains produced 90% sequence coverage of the entire protein minus signal peptide sequences and 96% of the COL1 domain and identified the positions of 98 Hyp residues in the Y-position of Gly-X-Y triplet repeats and 9 Hyp residues in the X-position of Gly-Hyp-Hyp triplet repeats in the COL1 domain (Fig. 3). As described above, Hyp residues in the Y-position of Gly-X-Y repeats or in the X-position of Gly-Hyp-Hyp triplet repeats are likely 4- and 3-Hyp residues, respectively. Interestingly, within pro-α1(V) sequences NH2-terminal to the COL1 domain, MS analysis mapped six Hyp residues in the Y-position of Gly-X-Y triplets, 1 Hyp residue in the X-position of a Gly-Hyp-Hyp triplet, one Hyl, and two Glc-Gal-Hyl residues. Five of the Y-position Hyp residues exist within a hypothetical (44) short, interrupted collagenous (COL2) subdomain (Fig. 3), consistent with the likelihood that this subdomain actually forms a triple helix, as the latter would be stabilized by these five 4-Hyp residues. Possible functional significance of the sixth 4-Hyp, which lies upstream of this hypothetical collagenous domain, is unclear. All NH2-terminal domain Glc-Gal-Hyl, Hyl, and X-position Hyp residues lie within the COL2 subdomain, suggestive of functional roles for these residues within this subdomain as well. Supportive of our findings, the single COL2 domain Hyl mapped here was previously identified by MS analysis as an “NH2 telopeptide” Hyl residue involved in covalent cross-linking of α1(V) chains to α1(XI) chains in bovine cartilage (4). MSn analysis here of the human recombinant pro-α1(V) chain also localized the positions of 6 Hyl, 1 Gal-Hyl, and 22 Glc-Gal-Hyl residues and 1 residue found as both Gal-Hyl and Glc-Gal-Hyl within the COL1 domain. It seems unlikely that species-specific differences in α1(V) sequences contributed much to differences in numbers and placement of Y-position Hyp residues, as bovine and human α1(V) COL1 domain sequences are 99.3% identical, differing by only 7 primary amino acids, none of which is a Pro (Figs. 1 and 3, orange residues). Modifications could not be localized for Y-position Pro residues 642, 714, 903, or 909 in either human recombinant pro-α1(V) or bovine placenta α1(V) chains.
FIGURE 3.
Hydroxylated amino acid residues and glycosylated Hyl residues of recombinant human pro-α1(V) collagen chains produced in 293-EBNA human embryonic kidney cells. Sequence coverage was 90% of the entire pro-α1(V) chain and 96% of the COL1 domain. Within the COL1 domain 98 Y-position Hyp, 9 Gly-Hyp-Hyp X-position Hyp, 1 Gly-Hyp-Val, 1 Gly-Hyp-Ala, 1 Gly-Hyp-Thr, 7-Hyl, and 23 Glc-Gal-Hyl residues were identified. Red, hydroxylated, non-glycosylated residues; green, Glc-Gal-Hyl residues; dark blue, Gal-Hyl residue; purple, residue found as both Glc-Gal-Hyl and Gal-Hyl. Underlined, hydroxylated/glycosylated residues mapped in previous studies (4, 32). Sequences not identified in the course of MS analysis are in light blue. The seven COL1 amino acid residues that differ between human and bovine are orange. *, 12 Y-position Pro residues hydroxylated in human or bovine α1(V) COL1 domain but not the other. #, Hyl residues identified by Wu et al. (4) as being involved in interchain covalent cross-links. ^, Lys residues hydroxylated in bovine placenta α1(V) but not in human recombinant pro-α1(V). A vertical arrow marks the site of proteolytic removal of the signal peptide, as previously determined by Edman degradation NH2-terminal amino acid sequencing (40) and as confirmed by MS analysis in the present study. The brackets indicates limits of the small COL2 hypothetical triple helical domain, in which interruptions of Gly-X-Y repeats are underlined. Amino acid residues NH2-terminal to the COL1 domain are numbered 1 to 558, starting with the initial Met residue of the signal peptide. Amino acid residues of the COL1 domain are numbered 1 to 1014, for easy comparison to the similarly numbered residues of the bovine α1(V) COL1 domain in Fig. 1. Amino acids of the COOH-propeptide are shown for the sake of completeness, but are not numbered, due to lack of identified hydroxylated residues.
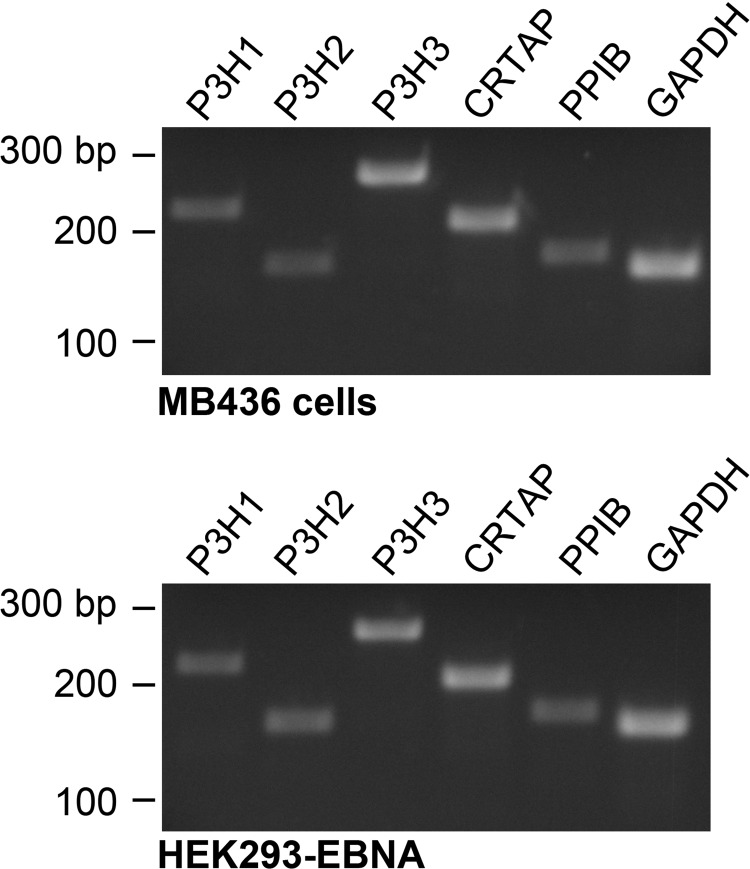
Expression of Prolyl 3-Hydroxylases in 293-HEK Cells
Differential 3 hydroxylation of prolines at some, but not other sites in clade A fibrillar collagen chain COL1 domains can be due to differential expression of the enzyme prolyl 3-hydroxylase 2 (P3H2) in different cell types and tissues (31). To test whether the absence of 3-Hyp residues at some sites in human recombinant pro-α1(V) chains that had been 3-hydroxylated in α1(V) chains from bovine placenta might be due to deficiency in levels of P3H2, we tested for P3H2 expression in 293-HEK cells. We also tested for expression of the other two prolyl 3-hydroxylases, P3H1 and P3H3, and for CRTAP and PPIB (peptidyl prolyl cis-trans isomerase B), which are components of the prolyl 3-hydroxylation complex (63). As can be seen (Fig. 4), all P3Hs are clearly expressed in 293-HEK cells at levels similar to those of a cell line previously shown positive for expression of all three enzymes (43). CRTAP and peptidyl prolyl cis-trans isomerase B expression levels were readily detectable as well. These findings are thus consistent with the likelihood that the decreased numbers of 3-Hyp residues in human recombinant pro-α1(V) chains, as compared with the tissue form of α1(V) chain obtained from bovine placenta, is unlikely due to decreased levels of P3Hs or other prolyl 3-hydroxylation complex components in 293-HEK cells. Rather, the reduction in 3-Hyp residues in the recombinant pro-α1(V) chains is likely due to some other variable(s). It is possible that even normal levels of endogenous enzymes are insufficient to fully modify the overexpressed pro-α1(V)chains produced in 293-HEK cells, perhaps contributing to some of the differences in PTM levels noted between the human recombinant pro-α1(V) and bovine placenta α1(V) chains. However, the distribution of X-position Hyp residues in the recombinant pro-α1(V)chains suggests that reduced numbers of such residues in these chains may involve the changed kinetics with which pro-α1(V) chains are incorporated into pro-α1(V)3 homotrimers, the form in which they were produced in HEK-293 cells for this study, compared with the kinetics of incorporation of pro-α1(V) chains into pro-α1(V)2pro-α2(V) heterotrimers, the predominant form in which they are found in tissues (see “Discussion”).
FIGURE 4.
Expression of P3H1, P3H2, P3H3, and prolyl 3-hydroxylation complex components CRTAP and peptidyl prolyl cis-trans isomerase B (PPIB) in 293-HEK cells. Expression of P3H1, P3H2, P2H3, CRTAP, and GAPDH expression was ascertained by RT-PCR analysis of RNA from MB436 cells (top panel), a cell line previously shown to be positive for expression of all three P3Hs (43), and from 293-HEK cells (bottom panel). Positions of size markers, given in base pairs (bp), are shown to the left of each panel.
DISCUSSION
A High Mass Accuracy, High Resolution Tandem Mass Spectrometric Approach for Analysis of Collagenous Proteins
Study of the PTMs of collagens has primarily involved traditional amino acid analysis, a methodology that determines amino acid compositions by hydrolysis of purified proteins/peptides followed by measurement of their individual amino acid constituents. Ordinarily, such analyses yield quantitative measurements of amino acid abundance, but not sequence information or the location of PTMs. More recently, MS has been employed to investigate PTMs in collagens (14, 30–32). Henkel and Dreisewerd (14) utilized ultraviolet MALDI MS to analyze fetal calfskin collagens I, III, and V, chemically cleaved by cyanogen bromide (CNBr) to generate a limited number of peptides with distinct masses. PTMs were deduced for each CNBr peptide by comparing the difference between experimental and theoretical masses, the mass difference representing the mass of the modification(s). However, MSn was not performed to confirm sequence identity or to locate the positions of PTMs. This approach provided valuable information on the total PTM state, but without the use of MSn, specific residues carrying the PTMs remained unknown. Eyre and co-workers have characterized collagen peptides by performing MS2 using a low resolution, low mass accuracy ion trap mass spectrometer (30–32). They identified several novel sites of prolyl 3-hydroxylation and proposed a biological function for these PTMs in collagen. However, many of the modifications were not experimentally localized by MS2. Because the acquired tandem mass spectra lacked the requisite information for localization, assignment of Pro hydroxylation was often inferred from known collagen motifs. Because of the highly modified nature of collagenous proteins, we contend that PTM mapping via MS should require spectral evidence in the form of localizing fragments to have the highest confidence in the validity of sequence-specific modifications.
Our strategy, founded on a high resolution MS platform, allowed characterization of the α1(V) chain with unparalleled sensitivity and PTM localization precision. The enabling technology was an ETD-enabled LTQ Orbitrap Velos hybrid mass spectrometer exhibiting low ppm mass accuracy, high resolution MS, and MSn capabilities combined with multiple options for performing peptide dissociation (47). We began by using multiple proteases to generate a diverse pool of peptides amenable to MSn (45). The combination of mass accuracy and a capability to perform multistage activation using three different dissociation techniques often allowed us to pinpoint the exact residue(s) carrying individual PTMs. However, we were not always able to unambiguously define the site of localization. Thus, we classified PTM assignments into 1 of 3 categories, 1) localized (MSn product ions support PTM assignment to a specific residue), 2) unlocalized (insufficient product ions to unambiguously assign a PTM to a specific residue), or 3) pseudolocalized (insufficient product ions to unambiguously assign a PTM to a specific residue), but localization can be inferred from known collagen modification motifs (supplemental Tables S1 and S2). Note that hydroxylation modification was only considered for Pro and Lys residues and that glycosylation (mono- and disaccharide) modifications were only considered for Hyl residues (15). The numbers of PTMs reported under the “Results” and “Discussion” are exclusively for localized sites.
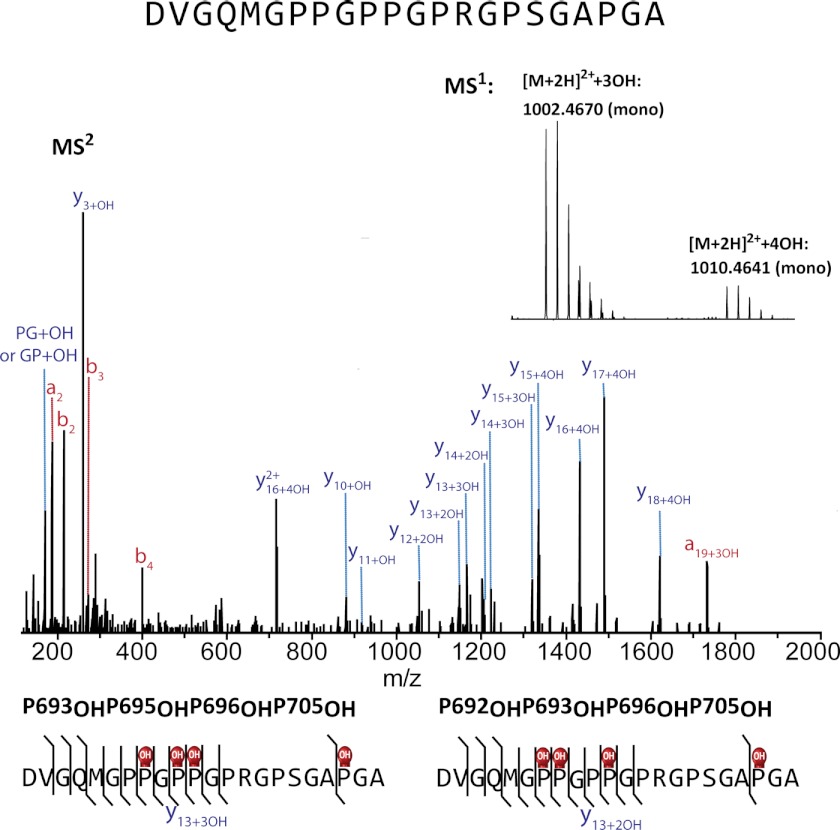
α1(V) and pro-α1(V) chains were cleaved using five different individual proteases or by applying two proteases sequentially. Trypsin, the most common protease used for MS experiments, has substantially reduced cleavage rates for Lys and Arg residues COOH-terminal to Pro, thus posing a problem for collagens, which have high Pro/Hyp content (64, 65). Such is also the case with chymotrypsin and Glu-C. However, Arg-C will cleave at Arg residues (also at Lys, but at lower rates) adjacent to Pro residues. Peptides were separated by on-line nanoflow reversed-phase liquid chromatography, where peptides with identical primary sequences differing only in degree of hydroxylation eluted across a wide elution window, whereas peptide isomers differing only in the position of hydroxylation typically coeluted and were cofragmented to produce chimeric MSn spectra (Fig. 5). Glycopeptides and short (<∼7 residues), highly modified peptides often eluted early during the chromatographic gradient, demonstrating poor and inconsistent retention. A direct injection style of sample loading was employed to enable analysis of highly polar and poorly retained peptides rather than the vented-style trapping normally used for rapid sample loading/concentrating (66).
FIGURE 5.
A chimeric MS2 spectrum revealing a pair of isomeric differently modified α1(V) chain peptides that co-eluted and co-fragmented. Peptides were generated by AspN cleavage of bovine placenta α1(V) chains. MS1 in the insert was obtained by averaging MS1 across the elution window. MS2 revealed that the two separate cluster peaks in MS1 belong to peptides with identical primary sequence but to differing degrees of hydroxylation. Peptides with a total of three hydroxylations contain Hyp693, Hyp696, and Hyp705 (MS2 for peptides with a total of 3 hydroxylation in the MS1 of the inset is not shown here). MS2 for peptides with a total of four hydroxylations (shown here) reveals that the extra Hyp can be either Hyp692 or Hyp695.
Because of high α1(V) Pro content and decreased efficacy of ETD-induced cleavage of Pro N-Cα bonds, ETD MS2 analyses occasionally did not provide sufficient numbers of sequence-informative product ions to permit PTM localization (67). To complement ETD data, data were also acquired utilizing HCD collected at different collision energies. Low HCD energies favor the generation of moderate mass fragment ions, whereas high HCD energies favor the production of low m/z fragments. Low mass fragments enabled screening of y1-y3, b1-b3, and immonium ions, improving the ability to localize PTMs residing on peptide termini. The high α1(V) Pro content also resulted in an unusually large number of internal fragments (multiple peptide backbone cleavages of a single ion) in spectra collected at moderate and high HCD energies (35). Many of these internal fragments were useful for PTM localization in the absence of localizing product ions produced by a single backbone cleavage event (supplemental Spectra 1 and 2). Despite our best efforts using multiple dissociation techniques and MS3, neutral loss of glycosylation was still common. In these cases neutral loss product ions were used to aid in localization, as reported for phosphorylation localization (supplemental Spectra S1 and S2) (68).
Comparison of the Col1 PTMs of Bovine Placental α1(V) and Human Recombinant Pro-α1(V) Chains to Each Other and to Previous Findings
Previous amino acid analyses of α1(V) chains from human skin (15) or placenta (69) or produced in 293-HEK cells (61) produced estimates of 4-Hyp content of from ∼106 to ∼111 per 1000 amino acids. The 106 and 98 Y-position Hyp residues directly localized here by MSn for the COL1 domains of bovine placenta α1(V) and human recombinant pro-α1(V) chains, respectively, are in the same range as these numbers, consistent with identification of the Y-position Hyp residues mapped here as 4-Hyp residues. Additional Y-position Hyp residues were either not directly localized or are likely located on COL1 proteolytic fragments not recovered here by MSn, as such fragments contain five additional Y-position Pro residues for both the bovine and human. Eighty-six Y-position Hyp residues mapped to identical positions in the bovine and recombinant human COL1 domains (Figs. 1 and 2), suggesting that these Hyp residues may be relatively invariant in α1(V) chains from various sources. Of the 12 Y-position Hyp residues that differ between the bovine α1(V) and human recombinant pro-α1(V) samples, only 2 are found in the latter but not the bovine. Thus, Y-position Hyp residues found in the human recombinant pro-α1(V) COL1 domain are for the most part a subset of those found in the bovine α1(V) chain. Differences in Y-position Hyp residues between the two COL1 domains may be due to tissue-/cell type-specific differences in modifying enzymes, although the pro-α1(V) chains studied here were synthesized as overexpressed pro-α1(V)3 homotrimers rather than as the endogenous pro-α1(V)2pro-α2(V) heterotrimers commonly found in tissues, which might also affect levels and placement of Hyp residues.
Previous estimates of the amino acid composition of α1(V) chains from human skin (15) suggested α1(V) to be more similar to nonfibrillar basement membrane collagen IV chains than to other fibrillar collagen chains in possessing a high content of Hyl residues, the majority of which are glycosylated. Specifically, human skin α1(V) chains (15) were estimated to comprise ∼39 hydroxylysines, 29 in the form of Glc-Gal-Hyl residues and 5 in the form of Gal-Hyl residues, suggesting Glc-Gal and Gal to be frequently and infrequently occurring PTMs, respectively. Henkel and Dreisewerd (14), employing MALDI MS analysis of fetal calf skin α1(V) chains, estimated that most, if not all, Hyl residues are likely glycosylated, with ∼87% predicted to be Glc-Gal-Hyl. However, they lacked MSn sequence data to support their assignments of PTMs, making it difficult to confidently determine the nature of saccharide species (e.g. two residues with monosaccharide modifications and one residue with a disaccharide modification share the same mass such that these PTMs cannot be distinguished based solely on analysis of intact peptide masses). Rhodes and Miller (69) employed amino acid analysis to estimate human placenta α1(V) chains to contain 35 Hyls but did not analyze glycosylation. Here, we directly localized 37 hydroxylysines in bovine placenta α1(V) chains, similar in number to the 34–39 hydroxylysines previously detected by amino acid analysis in α1(V) chains from bovine placenta and uterus (70). Furthermore, we identified 34 of the hydroxylysines mapped here as Glc-Gal-Hyl residues. Thus, overall numbers of Hyl and Glc-Gal-Hyl residues mapped here in bovine α1(V) chains accord well with previous estimates of amino acid content. Our estimates of Gal-Hyl modification of bovine placenta α1(V) may differ from those of previous reports because 1) extremely low levels of Gal-Hyl modifications were beyond our limit of detection, 2) we exclusively reported localized modifications, disregarding peptides with multiple potential glycosylation sites that could not be localized to a specific residue, and/or 3) previous reports did not attempt to localize modifications and may have overestimated Gal-Hyl numbers.
We found the COL1 domain of human recombinant pro-α1(V) chains produced in 293-HEK cells to contain 30 hydroxylysines: 22 as Glc-Gal-Hyl residues, 1 as Gal-Hyl, and 1 as both Glc-Gal-Hyl and Gal-Hyl. These hydroxylated Lys residues are a subset of those found in bovine α1(V), except for Hyl150, which is not hydroxylated in the bovine chain (Figs. 1 and 3). Thus, 29 hydroxylated Lys residues found in α1(V) COL1 domains from two very different sources may be relatively invariant in α1(V) chains. The reduced numbers of Hyl and Glc-Gal-Hyl residues in the human recombinant protein suggests that 293-HEK cells, which produce little or no endogenous extracellular matrix proteins (61), may possess reduced levels of the multifunctional enzyme lysyl hydroxylase 3, which has lysyl hydroxylase activity, and collagen galactosyltransferase and glycosyltransferase activities that allow it to glycosylate Hyl residues hydroxylated by itself and by other lysl hydroxylase isoforms (18), although even normal levels of enzymes might be insufficient to fully modify the overexpressed recombinant pro-α1(V)chains. Interestingly, COL1 residue 84 was detected here as both a Glc-Gal-Hyl and a Gal-Hyl residue, showing glycosylation at this site to be dynamic, with partial occupancy by both Glc-Gal and Gal saccharides. Our direct localization of 30 COL1 Hyl residues within the human recombinant pro-α1(V) supersedes a previous amino acid analysis estimate of 6 COL1 Hyl residues for human recombinant pro-α1(V) chains produced in 293-HEK cells (61) and is more congruent with the observed efficient secretion of recombinant pro-α1(V)3 homotrimers from 293-HEK cells (40, 61), as threshold levels of glycosylated Hyl residues are necessary for efficient secretion of at least some collagenous molecules (18).
Previous amino acid analysis of α1(V) chains from human placenta (69) or human skin (15) estimated 3-Hyp content at four or ten 3-Hyp residues, respectively, per 1000 amino acids, levels higher than those detected by similar analyses of the major fibrillar collagen chains (71–74). The nine Hyp residues mapped here to the X-positions of Gly-Hyp-Hyp repeats in the COL1 domain of human recombinant pro-α1(V) chains falls within this range (15, 69). However, the 22 X-position Hyp residues mapped here within the bovine placenta α1(V) chain lies beyond this range and is instead reminiscent of the range of 10 to 20 3-Hyp residues previously estimated by amino acid analysis to lie within the collagen IV chains of some tissues (75, 76).
Using low resolution MS2 and assignments based on known collagen motifs (see above), Eyre and co-workers (32) recently mapped three X-position Hyp residues in α1(V) chains from human bone to positions 434 and 665 and to a site variously identified by them as 692 or 695 of the COL1 domain. They also predicted the tissue-specific occurrence of 3-Hyp residues within Gly-Pro-Pro repeats at the COOH termini of the COL1 domains of α1(V) and other fibrillar collagen chains, based on mapping of such residues to Gly-Pro-Pro repeats at the COL1 COOH termini of α1(I) and α2(I) chains from tendon but not from skin or bone (30). Here, we confirm the presence of X-position Hyp residues at sites 434, 665, and 695 and in all three COL1 COOH-terminal Gly-Pro-Pro repeats (sites 1004, 1007, and 1010) in bovine placenta α1(V) chains (supplemental Spectra 1), whereas human recombinant pro-α1(V) chains contained an X-position Hyp residue only at site 434. Thus, we have confirmed the positions previously identified or predicted for six α1(V) COL1 X-position Hyp residues and found one of these (residue 434) to be, thus far, invariable. In addition, we identified the positions of 15 additional X-position Hyp residues in bovine placental α1(V) chains. The nine human recombinant pro-α1(V) COL1 X-position Hyp sites are also found in the bovine α1(V) chain and provide confirmatory evidence for Hyp occupancy of these nine X-position sites. Interestingly, COL1 X-position Hyp sites in human recombinant pro-α1(V) chains are limited to the NH2-terminal half of the domain. Although the reason for this is unknown, it may relate to the COOH to NH2 terminus direction of triple helix formation and to the ability of prolyl 3-hydroxylases to modify unfolded but not triple helical procollagen chains. Thus, because the recombinant pro-α1(V) chains are produced as pro-α1(V)3 homotrimers, which form triple helices with apparently increased kinetics than do pro-α1(V)2pro-α2(V) heterotrimers (77), prolyl 3-hydroxylases may be particularly limited in the ability to hydroxylate residues in the COOH-terminal portions of these recombinant chains. In fact, the NH2-terminal COL1 distribution of X-position Hyp residues in recombinant pro-α1(V)3 homotrimers is consistent with the suggestion that 3-Hyp content may be related to the speed of triple helix formation (55) but may be inconsistent with the suggestion (31) that 3-hydroxylation of fibrillar collagens begins at the COOH terminus and diminishes in the more NH2-terminal portion of the COL1 domain. Other PTMs were not limited to the NH2-terminal portion of recombinant pro-α1(V) COL1 domains, perhaps reflecting differences in the kinetics of prolyl 3-, prolyl 4-, and lysyl hydroxylases.
We report here the unexpected finding of X-position Hyp residues in the context of a Gly-X-Val and a Gly-X-Ala triplet in the bovine placenta α1(V) COL1 domain. It is unclear at this time whether these Hyp residues might be 3-Hyp or 4-Hyp or whether their appearance represents a lapse in fidelity of at least one of these enzymes or whether such sites represent previously unknown substrates at which hydroxylation serves specific functions. Supportive of our findings, Pro hydroxylation in an Gly-Pro-Ala motif has previously been reported for bovine-derived collagen I (78). It should be noted that although X-position 4-Hyp residues are thought to destabilize triple helices when in the context of Gly-Hyp-Pro triplets (79), X-position 4-Hyp residues do not necessarily destabilize the triple helix when in the context of triplets that lack Pro or Hyp in the Y-position (80). Thus, the significance of the unexpected finding of X-position Hyp in these non-canonical sites remains to be determined.
Numbers of Hyl, Glc-Gal-Hyl, and Y-position Hyp residues mapped here by MSn in the bovine placenta α1(V) chain are consistent with numbers of 4-Hyp, Hyl, and Glc-Gal-Hyl residues in estimates of the amino acid compositions of hydrolyzed α1(V) chains from various tissues. In contrast, the number of X-position Hyp and likely 3-Hyp residues detected here by tandem MS in Gly-Hyp-Hyp triplets in bovine placenta α1(V) chains is markedly higher than numbers of 3-Hyp residues previously estimated by amino acid analyses. Importantly, the numbers of PTMs predicted by early amino acid analyses were based on a presumption of 100% occupancy at a fixed number of sites. Thus, although congruence in numbers of PTMs predicted by previous amino acid analyses and the MSn localization results presented here suggests that this presumption holds true for most α1(V) PTMs, this does not appear to be the case for 3-Hyp residues. Rather, the large difference between the numbers of 3-Hyp predicted by previous amino acid analyses and the number of X-position Hyp residues in Gly-Hyp-Hyp triplets mapped here by high resolution MS is best explained by the concept of a relatively large number of 3-Hyp sites that have less than 100% occupancy in a population of α1(V) chains from a given tissue. This conclusion is supported by amino acid analysis performed on bovine α1(V) chain samples used in this study (Table 1), which estimated overall Hyp levels of these samples to be similar to, and not higher than, Hyp levels estimated for α1(V) chains in earlier amino acid analysis studies (70). It is not surprising that with the improved sensitivity and specificity afforded by our MS-driven proteomics workflow that we have identified numerous PTMs previously undiscovered, including those present at relatively low stoichiometric abundances. Interestingly, 3-Hyp residues point away from the triple helix (81), implying roles in protein-protein interactions as the bases of biological function for these residues. The presence of numbers of sites that can be differentially modified by P3Hs in a given tissue suggests a previously unknown mechanism for dynamic modification of the functions and intermolecular interactions of α1(V) chains and col(V) and perhaps other collagen types as well.
TABLE 1.
Amino acid composition of bovine placenta α1(V) chains
Data are estimated by amino acid analysis of acid-hydrolyzed protein and are consistent within the error rate for amino acid analysis, with previous results of Abedin et al. (70).
| Amino acid | No. detected/1000 residues |
|---|---|
| Hyp | 106 |
| Asx | 47 |
| Thr | 22 |
| Ser | 22 |
| Glx | 113 |
| Pro | 113 |
| Gly | 337 |
| Ala | 44 |
| Val | 22 |
| Ile | 22 |
| Leu | 49 |
| Tyr | |
| Phe | 14 |
| Hyl | 32 |
| Lys | 17 |
| His | 10 |
| Arg | 41 |
Supplementary Material
Acknowledgments
Y. G. and M. J. L. acknowledge the Wisconsin Partnership Fund for the establishment of the University of Wisconsin Human Proteomics Program.
This work was supported, in whole or in part, by National Institutes of Health Grants GM080148 (to J. J. C.) and AR047746 and AI084853 (to D. S. G.).

This article contains supplemental Experimental Procedures, Tables S1 and S2, and Spectra S1 and S2.
The nucleotide sequence(s) reported in this paper has been submitted to the GenBankTM/EBI Data Bank with accession number(s) JQ611730.
- col(V)
- collagen type V
- PTM
- post-translational modification
- Hyp
- hydroxyproline
- Hyl
- hydroxylysine
- COL1
- the major collagenous domain of fibrillar collagen chains
- MS2
- (MS/MS) a precursor measurement followed by precursor fragmentation to produce product ions
- MS3
- (MS/MS/MS) a product ion from MS2 re-isolated and fragmented
- MSn
- tandem mass spectrometry (n > 1)
- ETD
- electron-transfer dissociation
- HCD
- higher energy collisional dissociation
- CRTAP
- (cartilage-associated protein)
- P3H
- prolyl 3-hydroxylase.
REFERENCES
- 1. Fichard A., Kleman J. P., Ruggiero F. (1995) Another look at collagen V and XI molecules. Matrix Biol 14, 515–531 [DOI] [PubMed] [Google Scholar]
- 2. Imamura Y., Scott I. C., Greenspan D. S. (2000) The pro-α3(V) collagen chain. Complete primary structure, expression domains in adult and developing tissues, and comparison to the structures and expression domains of the other types V and XI procollagen chains. J. Biol. Chem. 275, 8749–8759 [DOI] [PubMed] [Google Scholar]
- 3. Morris N. P., Bächinger H. P. (1987) Type XI collagen is a heterotrimer with the composition (1α, 2α, 3α) retaining non-triple-helical domains. J. Biol. Chem. 262, 11345–11350 [PubMed] [Google Scholar]
- 4. Wu J. J., Weis M. A., Kim L. S., Carter B. G., Eyre D. R. (2009) Differences in chain usage and cross-linking specificities of cartilage type V/XI collagen isoforms with age and tissue. J. Biol. Chem. 284, 5539–5545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Birk D. E., Fitch J. M., Babiarz J. P., Linsenmayer T. F. (1988) Collagen type I and type V are present in the same fibril in the avian corneal stroma. J. Cell Biol. 106, 999–1008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Birk D. E., Fitch J. M., Babiarz J. P., Doane K. J., Linsenmayer T. F. (1990) Collagen fibrillogenesis in vitro. Interaction of types I and V collagen regulates fibril diameter. J. Cell Sci. 95, 649–657 [DOI] [PubMed] [Google Scholar]
- 7. Toriello H. V., Glover T. W., Takahara K., Byers P. H., Miller D. E., Higgins J. V., Greenspan D. S. (1996) A translocation interrupts the COL5A1 gene in a patient with Ehlers-Danlos syndrome and hypomelanosis of Ito. Nat. Genet. 13, 361–365 [DOI] [PubMed] [Google Scholar]
- 8. Richards A. J., Martin S., Nicholls A. C., Harrison J. B., Pope F. M., Burrows N. P. (1998) A single base mutation in COL5A2 causes Ehlers-Danlos syndrome type II. J. Med. Genet. 35, 846–848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Burlingham W. J., Love R. B., Jankowska-Gan E., Haynes L. D., Xu Q., Bobadilla J. L., Meyer K. C., Hayney M. S., Braun R. K., Greenspan D. S., Gopalakrishnan B., Cai J., Brand D. D., Yoshida S., Cummings O. W., Wilkes D. S. (2007) IL-17-dependent cellular immunity to collagen type V predisposes to obliterative bronchiolitis in human lung transplants. J. Clin. Invest. 117, 3498–3506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Sumpter T. L., Wilkes D. S. (2004) Role of autoimmunity in organ allograft rejection. A focus on immunity to type V collagen in the pathogenesis of lung transplant rejection. Am. J. Physiol. Lung Cell. Mol. Physiol. 286, L1129–L1139 [DOI] [PubMed] [Google Scholar]
- 11. Iwata T., Philipovskiy A., Fisher A. J., Presson R. G., Jr., Chiyo M., Lee J., Mickler E., Smith G. N., Petrache I., Brand D. B., Burlingham W. J., Gopalakrishnan B., Greenspan D. S., Christie J. D., Wilkes D. S. (2008) Anti-type V collagen humoral immunity in lung transplant primary graft dysfunction. J. Immunol. 181, 5738–5747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Bobadilla J. L., Love R. B., Jankowska-Gan E., Xu Q., Haynes L. D., Braun R. K., Hayney M. S., Munoz del Rio A., Meyer K., Greenspan D. S., Torrealba J., Heidler K. M., Cummings O. W., Iwata T., Brand D., Presson R., Burlingham W. J., Wilkes D. S. (2008) Th-17, monokines, collagen type V, and primary graft dysfunction in lung transplantation. Am. J. Respir. Crit. Care Med. 177, 660–668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Dart M. L., Jankowska-Gan E., Huang G., Roenneburg D. A., Keller M. R., Torrealba J. R., Rhoads A., Kim B., Bobadilla J. L., Haynes L. D., Wilkes D. S., Burlingham W. J., Greenspan D. S. (2010) Interleukin-17-dependent autoimmunity to collagen type V in atherosclerosis. Circ. Res. 107, 1106–1116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Henkel W., Dreisewerd K. (2007) Cyanogen bromide peptides of the fibrillar collagens I, III, and V and their mass spectrometric characterization. Detection of linear peptides, peptide glycosylation, and cross-linking peptides involved in formation of homo- and heterotypic fibrils. J. Proteome Res. 6, 4269–4289 [DOI] [PubMed] [Google Scholar]
- 15. Chung E., Rhodes K., Miller E. J. (1976) Isolation of three collagenous components of probable basement membrane origin from several tissues. Biochem. Biophys. Res. Commun. 71, 1167–1174 [DOI] [PubMed] [Google Scholar]
- 16. Shoulders M. D., Raines R. T. (2009) Collagen structure and stability. Annu. Rev. Biochem. 78, 929–958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kivirikko K. I. (1993) Collagens and their abnormalities in a wide spectrum of diseases. Ann. Med. 25, 113–126 [DOI] [PubMed] [Google Scholar]
- 18. Sipilä L., Ruotsalainen H., Sormunen R., Baker N. L., Lamandé S. R., Vapola M., Wang C., Sado Y., Aszodi A., Myllylä R. (2007) Secretion and assembly of type IV and VI collagens depend on glycosylation of hydroxylysines. J. Biol. Chem. 282, 33381–33388 [DOI] [PubMed] [Google Scholar]
- 19. Mizuno K., Peyton D. H., Hayashi T., Engel J., Bächinger H. P. (2008) Effect of the -Gly-3(S)-hydroxyprolyl-4(R)-hydroxyprolyl-tripeptide unit on the stability of collagen model peptides. FEBS J. 275, 5830–5840 [DOI] [PubMed] [Google Scholar]
- 20. Jenkins C. L., Bretscher L. E., Guzei I. A., Raines R. T. (2003) Effect of 3-hydroxyproline residues on collagen stability. J Am Chem Soc 125, 6422–6427 [DOI] [PubMed] [Google Scholar]
- 21. Morello R., Bertin T. K., Chen Y., Hicks J., Tonachini L., Monticone M., Castagnola P., Rauch F., Glorieux F. H., Vranka J., Bächinger H. P., Pace J. M., Schwarze U., Byers P. H., Weis M., Fernandes R. J., Eyre D. R., Yao Z., Boyce B. F., Lee B. (2006) CRTAP is required for prolyl 3-hydroxylation and mutations cause recessive osteogenesis imperfecta. Cell 127, 291–304 [DOI] [PubMed] [Google Scholar]
- 22. Arbogast B. W., Gunson D. E., Kefalides N. A. (1976) The role of hydroxylation of proline in the antigenicity of basement membrane collagen. J. Immunol. 117, 2181–2184 [PubMed] [Google Scholar]
- 23. Brand D. D., Myers L. K., Terato K., Whittington K. B., Stuart J. M., Kang A. H., Rosloniec E. F. (1994) Characterization of the T cell determinants in the induction of autoimmune arthritis by bovine α 1(II)-CB11 in H-2q mice. J. Immunol. 152, 3088–3097 [PubMed] [Google Scholar]
- 24. Corthay A., Bäcklund J., Broddefalk J., Michaëlsson E., Goldschmidt T. J., Kihlberg J., Holmdahl R. (1998) Epitope glycosylation plays a critical role for T cell recognition of type II collagen in collagen-induced arthritis. Eur. J. Immunol. 28, 2580–2590 [DOI] [PubMed] [Google Scholar]
- 25. Myers L. K., Miyahara H., Terato K., Seyer J. M., Stuart J. M., Kang A. H. (1995) Collagen-induced arthritis in B10.RIII mice (H-2r). Identification of an arthritogenic T-cell determinant. Immunology 84, 509–513 [PMC free article] [PubMed] [Google Scholar]
- 26. Myers L. K., Myllyharju J., Nokelainen M., Brand D. D., Cremer M. A., Stuart J. M., Bodo M., Kivirikko K. I., Kang A. H. (2004) Relevance of posttranslational modifications for the arthritogenicity of type II collagen. J. Immunol. 172, 2970–2975 [DOI] [PubMed] [Google Scholar]
- 27. Van den Steen P. E., Proost P., Brand D. D., Kang A. H., Van Damme J., Opdenakker G. (2004) Generation of glycosylated remnant epitopes from human collagen type II by gelatinase B. Biochemistry 43, 10809–10816 [DOI] [PubMed] [Google Scholar]
- 28. Van den Steen P. E., Proost P., Grillet B., Brand D. D., Kang A. H., Van Damme J., Opdenakker G. (2002) Cleavage of denatured natural collagen type II by neutrophil gelatinase B reveals enzyme specificity, post-translational modifications in the substrate, and the formation of remnant epitopes in rheumatoid arthritis. FASEB J. 16, 379–389 [DOI] [PubMed] [Google Scholar]
- 29. Michaëlsson E., Malmström V., Reis S., Engström A., Burkhardt H., Holmdahl R. (1994) T cell recognition of carbohydrates on type II collagen. J. Exp. Med. 180, 745–749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Eyre D. R., Weis M., Hudson D. M., Wu J. J., Kim L. (2011) A novel 3-hydroxyproline (3Hyp)-rich motif marks the triple-helical C terminus of tendon type I collagen. J. Biol. Chem. 286, 7732–7736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Fernandes R. J., Farnand A. W., Traeger G. R., Weis M. A., Eyre D. R. (2011) A role for prolyl 3-hydroxylase 2 in post-translational modification of fibril-forming collagens. J. Biol. Chem. 286, 30662–30669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Weis M. A., Hudson D. M., Kim L., Scott M., Wu J. J., Eyre D. R. (2010) Location of 3-hydroxyproline residues in collagen types I, II, III, and V/XI implies a role in fibril supramolecular assembly. J. Biol. Chem. 285, 2580–2590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Vaisar T., Urban J. (1996) Probing the proline effect in CID of protonated peptides. J Mass Spectrom 31, 1185–1187 [DOI] [PubMed] [Google Scholar]
- 34. Huang Y., Triscari J. M., Pasa-Tolic L., Anderson G. A., Lipton M. S., Smith R. D., Wysocki V. H. (2004) Dissociation behavior of doubly-charged tryptic peptides: correlation of gas-phase cleavage abundance with ramachandran plots. J. Am. Chem. Soc. 126, 3034–3035 [DOI] [PubMed] [Google Scholar]
- 35. Loo J. A., Edmonds C. G., Smith R. D. (1993) Tandem mass spectrometry of very large molecules. 2. Dissociation of multiply charged proline-containing proteins from electrospray ionization. Anal. Chem. 65, 425–438 [DOI] [PubMed] [Google Scholar]
- 36. Tang X. J., Thibault P., Boyd R. K. (1993) Fragmentation reactions of multiply protonated peptides and implications for sequencing by tandem mass spectrometry with low-energy collision-induced dissociation. Anal. Chem. 65, 2824–2834 [DOI] [PubMed] [Google Scholar]
- 37. Breci L. A., Tabb D. L., Yates J. R., 3rd, Wysocki V. H. (2003) Cleavage N-terminal to proline. Analysis of a database of peptide tandem mass spectra. Anal. Chem. 75, 1963–1971 [DOI] [PubMed] [Google Scholar]
- 38. McAlister G. C., Phanstiel D., Wenger C. D., Lee M. V., Coon J. J. (2010) Analysis of tandem mass spectra by FTMS for improved large-scale proteomics with superior protein quantification. Anal. Chem. 82, 316–322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Syka J. E., Coon J. J., Schroeder M. J., Shabanowitz J., Hunt D. F. (2004) Peptide and protein sequence analysis by electron transfer dissociation mass spectrometry. Proc. Natl. Acad. Sci. U.S.A. 101, 9528–9533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Imamura Y., Steiglitz B. M., Greenspan D. S. (1998) Bone morphogenetic protein-1 processes the NH2-terminal propeptide, and a furin-like proprotein convertase processes the COOH-terminal propeptide of pro-α1(V) collagen. J. Biol. Chem. 273, 27511–27517 [DOI] [PubMed] [Google Scholar]
- 41. Mares D. C., Heidler K. M., Smith G. N., Cummings O. W., Harris E. R., Foresman B., Wilkes D. S. (2000) Type V collagen modulates alloantigen-induced pathology and immunology in the lung. Am. J. Respir. Cell Mol. Biol. 23, 62–70 [DOI] [PubMed] [Google Scholar]
- 42. Seyer J. M., Kang A. H. (1989) Covalent structure of collagen, Amino acid sequence of three cyanogen bromide-derived peptides from human α 1(V) collagen chain. Arch. Biochem. Biophys. 271, 120–129 [DOI] [PubMed] [Google Scholar]
- 43. Shah R., Smith P., Purdie C., Quinlan P., Baker L., Aman P., Thompson A. M., Crook T. (2009) The prolyl 3-hydroxylases P3H2 and P3H3 are novel targets for epigenetic silencing in breast cancer. Br. J. Cancer 100, 1687–1696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Greenspan D. S., Cheng W., Hoffman G. G. (1991) The pro-α 1(V) collagen chain. Complete primary structure, distribution of expression, and comparison with the pro-α 1(XI) collagen chain. J. Biol. Chem. 266, 24727–24733 [PubMed] [Google Scholar]
- 45. Swaney D. L., Wenger C. D., Coon J. J. (2010) Value of using multiple proteases for large-scale mass spectrometry-based proteomics. J. Proteome Res. 9, 1323–1329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Ficarro S. B., Zhang Y., Lu Y., Moghimi A. R., Askenazi M., Hyatt E., Smith E. D., Boyer L., Schlaeger T. M., Luckey C. J., Marto J. A. (2009) Improved electrospray ionization efficiency compensates for diminished chromatographic resolution and enables proteomics analysis of tyrosine signaling in embryonic stem cells. Anal. Chem. 81, 3440–3447 [DOI] [PubMed] [Google Scholar]
- 47. Olsen J. V., Schwartz J. C., Griep-Raming J., Nielsen M. L., Damoc E., Denisov E., Lange O., Remes P., Taylor D., Splendore M., Wouters E. R., Senko M., Makarov A., Mann M., Horning S. (2009) A dual pressure linear ion trap Orbitrap instrument with very high sequencing speed. Mol. Cell. Proteomics 8, 2759–2769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Good D. M., Wirtala M., McAlister G. C., Coon J. J. (2007) Performance characteristics of electron transfer dissociation mass spectrometry. Mol. Cell. Proteomics 6, 1942–1951 [DOI] [PubMed] [Google Scholar]
- 49. Wenger C. D., Phanstiel D. H., Lee M. V., Bailey D. J., Coon J. J. (2011) COMPASS. A suite of pre- and post-search proteomics software tools for OMSSA. Proteomics 11, 1064–1074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Good D. M., Wenger C. D., McAlister G. C., Bai D. L., Hunt D. F., Coon J. J. (2009) Post-acquisition ETD spectral processing for increased peptide identifications. J. Am. Soc. Mass Spectrom. 20, 1435–1440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Good D. M., Wenger C. D., Coon J. J. (2010) The effect of interfering ions on search algorithm performance for electron-transfer dissociation data. Proteomics 10, 164–167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Geer L. Y., Markey S. P., Kowalak J. A., Wagner L., Xu M., Maynard D. M., Yang X., Shi W., Bryant S. H. (2004) Open mass spectrometry search algorithm. J. Proteome Res. 3, 958–964 [DOI] [PubMed] [Google Scholar]
- 53. Elias J. E., Gygi S. P. (2007) Target-decoy search strategy for increased confidence in large-scale protein identifications by mass spectrometry. Nat. Methods 4, 207–214 [DOI] [PubMed] [Google Scholar]
- 54. Gorres K. L., Raines R. T. (2010) Prolyl 4-hydroxylase. Crit. Rev. Biochem. Mol. Biol. 45, 106–124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Risteli J., Tryggvason K., Kivirikko K. I. (1977) Prolyl 3-hydroxylase. Partial characterization of the enzyme from rat kidney cortex. Eur. J. Biochem. 73, 485–492 [DOI] [PubMed] [Google Scholar]
- 56. Tiainen P., Pasanen A., Sormunen R., Myllyharju J. (2008) Characterization of recombinant human prolyl 3-hydroxylase isoenzyme 2, an enzyme modifying the basement membrane collagen IV. J. Biol. Chem. 283, 19432–19439 [DOI] [PubMed] [Google Scholar]
- 57. Unsöld C., Pappano W. N., Imamura Y., Steiglitz B. M., Greenspan D. S. (2002) Biosynthetic processing of the pro-α 1(V)2pro-α2(V) collagen heterotrimer by bone morphogenetic protein-1 and furin-like proprotein convertases. J. Biol. Chem. 277, 5596–5602 [DOI] [PubMed] [Google Scholar]
- 58. Gopalakrishnan B., Wang W. M., Greenspan D. S. (2004) Biosynthetic processing of the Pro-α1(V)Pro-α2(V)Pro-α3(V) procollagen heterotrimer. J. Biol. Chem. 279, 30904–30912 [DOI] [PubMed] [Google Scholar]
- 59. Delacoux F., Fichard A., Geourjon C., Garrone R., Ruggiero F. (1998) Molecular features of the collagen V heparin binding site. J. Biol. Chem. 273, 15069–15076 [PubMed] [Google Scholar]
- 60. Chanut-Delalande H., Fichard A., Bernocco S., Garrone R., Hulmes D. J., Ruggiero F. (2001) Control of heterotypic fibril formation by collagen V is determined by chain stoichiometry. J. Biol. Chem. 276, 24352–24359 [DOI] [PubMed] [Google Scholar]
- 61. Fichard A., Tillet E., Delacoux F., Garrone R., Ruggiero F. (1997) Human recombinant α1(V) collagen chain. Homotrimeric assembly and subsequent processing. J. Biol. Chem. 272, 30083–30087 [DOI] [PubMed] [Google Scholar]
- 62. Kessler E., Fichard A., Chanut-Delalande H., Brusel M., Ruggiero F. (2001) Bone morphogenetic protein-1 (BMP-1) mediates C-terminal processing of procollagen V homotrimer. J. Biol. Chem. 276, 27051–27057 [DOI] [PubMed] [Google Scholar]
- 63. Pyott S. M., Schwarze U., Christiansen H. E., Pepin M. G., Leistritz D. F., Dineen R., Harris C., Burton B. K., Angle B., Kim K., Sussman M. D., Weis M., Eyre D. R., Russell D. W., McCarthy K. J., Steiner R. D., Byers P. H. (2011) Mutations in PPIB (cyclophilin B) delay type I procollagen chain association and result in perinatal lethal to moderate osteogenesis imperfecta phenotypes. Hum. Mol. Genet. 20, 1595–1609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Olsen J. V., Ong S. E., Mann M. (2004) Trypsin cleaves exclusively C-terminal to arginine and lysine residues. Mol. Cell. Proteomics 3, 608–614 [DOI] [PubMed] [Google Scholar]
- 65. Rodriguez J., Gupta N., Smith R. D., Pevzner P. A. (2008) Does trypsin cut before proline? J. Proteome Res. 7, 300–305 [DOI] [PubMed] [Google Scholar]
- 66. Licklider L. J., Thoreen C. C., Peng J., Gygi S. P. (2002) Automation of nanoscale microcapillary liquid chromatography-tandem mass spectrometry with a vented column. Anal. Chem. 74, 3076–3083 [DOI] [PubMed] [Google Scholar]
- 67. Wiesner J., Premsler T., Sickmann A. (2008) Application of electron transfer dissociation (ETD) for the analysis of posttranslational modifications. Proteomics 8, 4466–4483 [DOI] [PubMed] [Google Scholar]
- 68. Savitski M. M., Lemeer S., Boesche M., Lang M., Mathieson T., Bantscheff M., Kuster B. (2011) Confident phosphorylation site localization using the Mascot Delta Score. Mol. Cell. Proteomics M110.003830-1–M110.003830-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Rhodes R. K., Miller E. J. (1979) The isolation and characterization of the cyanogen bromide peptides from the B chain of human collagen. J. Biol. Chem. 254, 12084–12087 [PubMed] [Google Scholar]
- 70. Abedin M. Z., Ayad S., Weiss J. B. (1982) Isolation and native characterization of cysteine-rich collagens from bovine placental tissues and uterus and their relationship to types IV and V collagens. Biosci. Rep. 2, 493–502 [DOI] [PubMed] [Google Scholar]
- 71. Butler W. T., Piez K. A., Bornstein P. (1967) Isolation and characterization of the cyanogen bromide peptides from the α-1 chain of rat skin collagen. Biochemistry 6, 3771–3780 [DOI] [PubMed] [Google Scholar]
- 72. Click E. M., Bornstein P. (1970) Isolation and characterization of the cyanogen bromide peptides from the α1 and α2 chains of human skin collagen. Biochemistry 9, 4699–4706 [DOI] [PubMed] [Google Scholar]
- 73. Miller E. J., Lunde L. G. (1973) Isolation and characterization of the cyanogen bromide peptides from the α1(II) chain of bovine and human cartilage collagen. Biochemistry 12, 3153–3159 [DOI] [PubMed] [Google Scholar]
- 74. Seyer J. M., Kang A. H. (1981) Covalent structure of collagen. Amino acid sequence of α 1(III)-CB9 from type III collagen of human liver. Biochemistry 20, 2621–2627 [DOI] [PubMed] [Google Scholar]
- 75. Kefalides N. A. (1975) Basement membranes. Structural and biosynthetic considerations. J. Invest. Dermatol. 65, 85–92 [DOI] [PubMed] [Google Scholar]
- 76. Gryder R. M., Lamon M., Adams E. (1975) Sequence position of 3-hydroxyproline in basement membrane collagen. Isolation of glycyl-3-hydroxyprolyl-4-hydroxyproline from swine kidney. J. Biol. Chem. 250, 2470–2474 [PubMed] [Google Scholar]
- 77. Roulet M., Välkkilä M., Chanut-Delalande H., Hämäläinen E. R., Kessler E., Ala-Kokko L., Männikkö M., Bonod-Bidaud C., Ruggiero F. (2010) The collagen V homotrimer [α1(V)](3) production is unexpectedly favored over the heterotrimer [α1(V)](2)α2(V) in recombinant expression systems. J. Biomed. Biotechnol. 2010, 376927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Zhang Y., Liu T., Wang Q., Chen L., Lei J., Luo J., Ma G., Su Z. (2009) Mass spectrometric detection of marker peptides in tryptic digests of gelatin. A new method to differentiate between bovine and porcine gelatin. Food Hydrocoll. 23, 2001–2007 [Google Scholar]
- 79. Inouye K., Kobayashi Y., Kyogoku Y., Kishida Y., Sakakibara S., Prockop D. J. (1982) Synthesis and physical properties of (hydroxyproline-proline-glycine)10. Hydroxyproline in the X-position decreases the melting temperature of the collagen triple helix. Arch. Biochem. Biophys. 219, 198–203 [DOI] [PubMed] [Google Scholar]
- 80. Bann J. G., Bächinger H. P. (2000) Glycosylation/hydroxylation-induced stabilization of the collagen triple helix. 4-Trans-hydroxyproline in the Xaa position can stabilize the triple helix. J. Biol. Chem. 275, 24466–24469 [DOI] [PubMed] [Google Scholar]
- 81. Schumacher M. A., Mizuno K., Bächinger H. P. (2006) The crystal structure of a collagen-like polypeptide with 3(S)-hydroxyproline residues in the Xaa position forms a standard 7/2 collagen triple helix. J. Biol. Chem. 281, 27566–27574 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.