Background: Lens degeneration in Fpr1−/− mice prompted us to search for functional FPR1 expression directly on lens epithelial cells.
Results: FPR1 is functionally expressed on human lens epithelial cells but has atypical properties compared with hematopoietic cell FPR1.
Conclusion: Lens epithelial cell FPR1 may be involved in development and maintenance of the lens.
Significance: This is the first link between non-hematopoietic expression of FPR1 and an ophthalmologic phenotype.
Keywords: Cataract, Epithelial Cell, Eye, Lens, Leukocyte, Receptor Regulation, Formyl Peptide, G Protein-coupled Receptor (GPCR), Ligand Binding Assays, Ophthalmology
Abstract
Formyl peptide receptor 1 (FPR1) is a G protein-coupled chemoattractant receptor expressed mainly on leukocytes. Surprisingly, aging Fpr1−/− mice develop spontaneous lens degeneration without inflammation or infection (J.-L. Gao et al., manuscript in preparation). Therefore, we hypothesized that FPR1 is functionally expressed directly on lens epithelial cells, the only cell type in the lens. Consistent with this, the human fetal lens epithelial cell line FHL 124 expressed FPR1 mRNA and was strongly FPR1 protein-positive by Western blot and FACS. Competition binding using FPR1 ligands N-formyl-Nle-Leu-Phe-Nle-Tyr-Lys (Nle = Norleucine), formylmethionylleucylphenylalanine, and peptide W revealed the same profile for FHL 124 cells, neutrophils, and FPR1-transfected HEK 293 cells. Saturation binding with fluorescein-labeled N-formyl-Nle-Leu-Phe-Nle-Tyr-Lys revealed ∼2500 specific binding sites on FHL-124 cells (KD ∼ 0.5 nm) versus ∼40,000 sites on neutrophils (KD = 3.2 nm). Moreover, formylmethionylleucylphenylalanine induced pertussis toxin-sensitive Ca2+ flux in FHL 124 cells, consistent with classic Gi-mediated FPR1 signaling. FHL 124 cell FPR1 was atypical in that it resisted agonist-induced internalization. Expression of FPR1 was additionally supported by detection of the intact full-length open reading frame in sequenced cDNA from FHL 124 cells. Thus, FHL-124 cells express functional FPR1, which is consistent with a direct functional role for FPR1 in the lens, as suggested by the phenotype of Fpr1 knock-out mice.
Introduction
The formyl peptide receptor (FPR)3 family consists of Gi-coupled receptors that are activated by diverse agonists, including eponymous N-formylated peptides originating from bacteria and mitochondria. In humans the FPR family consists of three receptors, namely the “high affinity FPR,” FPR1, the “low affinity FPR,” FPR2/ALX (previously known as FPRL1 or ALX), and FPR3. All are expressed by phagocytic cells of the immune system (1). Human FPR1 is important for phagocyte chemotaxis, superoxide production, and degranulation and helps direct phagocytes to sites of infection. In mice, there are at least eight members of the FPR gene family. Mouse Fpr1 is important for antibacterial host defense but binds the prototype formyl peptide fMLF with much lower affinity than human FPR1 (1, 2).
Although FPR1 is expressed predominantly in phagocytic leukocytes, it has also been reported in glioblastoma (3), where it may promote a more aggressive phenotype (4). The evidence for functional FPR1 expression in non-cancerous non-hematopoietic cells in healthy humans is limited to a few scattered reports, most of them of uncertain biological significance. Very early reports showed a role of formyl peptide receptors in the modulation of the tone of coronary and pulmonary arteries (5, 6). The first systematic investigation of FPR1 expression in a broad range of human tissues (neuromuscular, vascular, endocrine, and immune) was limited to a description of immunoreactivity using an FPR1-specific antibody (7). Functional FPR1 expression has been demonstrated on fibroblasts (8), human bone marrow mesenchymal stem cells (9–11), A549 lung cells (12), HEP-G2 (13) hepatoma cells, and for several kinds of epithelial cells including Beas2B lung epithelial cells (14), SK-CO15 intestinal epithelial cells (15, 16), MKN-28, and AGS gastric epithelial cells (17) and most recently human retinal pigment epithelial cells (18). In several of these reports, it was proposed that FPR1 may play a role in wound healing and tissue repair, and for some of the epithelial cells this was supported with data from in vitro models of wound healing. However, except for a study on the role of FPR1 in zebrafish and rabbit bone formation (11), evidence for a function of FPR1 in non-cancerous tissues under in vivo conditions has been lacking. A role for FPR1 on non-hematopoietic cells is supported indirectly by the discovery of FPR2 on non-hematopoietic cells and tissues, such as astrocytes and lung (1) and expression of more distantly related members of the mouse FPR family in the vomeronasal organ of mice (19, 20).
Recently we observed that as Fpr1−/− mice age, the lens of the eye degenerates and forms severe cataracts without any inflammation or infection.4 The mechanism may involve Fpr1 function directly on lens epithelial cells, as they express Fpr1 mRNA and are the only cell type in the lens. However, mouse lens epithelial cell lines are not available, and it is not feasible to obtain sufficient primary mouse lens cells for biochemical analysis. For these reasons and to begin to test whether human lens function may also be regulated directly by FPR1, we have investigated FPR1 expression and function using human lens epithelial cell lines.
EXPERIMENTAL PROCEDURES
Reagents and Buffers
DMEM, lymphocyte separation medium, G418, HEPES (1 m), PBS, and HBSS were from Cellgro (Manassas, VA). Medium 199 and trypsin/EGTA were from Invitrogen. FBS was from Gemini Bio-Products (West Sacramento, CA). fNLFNYK was from Genscript (Piscataway, NJ), fMLF was from Sigma, peptide-W (WKYMVm) was from Macromolecular Resources (Ft. Collins, CO), and fNLFNYK-Fl was from Invitrogen. PE-labeled mouse anti-FPR1 mAb (25 μg/ml stock solution, catalog no. FAB3744P, clone 350418) and PE-labeled IgG2A control (10 μg/ml, catalog no. IC003P, clone 20102) were from R&D Systems (Minneapolis, MN).
Cells
The infant human lens epithelial cell line SRA 01/04 (21) and the fetal human lens epithelial cell line FHL 124 (22) were maintained in Medium 199 supplemented with 10% FBS and 20% fresh keratinocyte growth medium (Lonza, Walkersville, MD). HEK 293 cells (ATCC, Manassas, VA) were cultured in DMEM plus 10% FBS. HEK 293 cells stably expressing FPR1 (23) were selected with 2 mg/ml G418 (Cellgro). Cells were cultured at 37 °C in 5% CO2 and 100% humidity. Human neutrophils were prepared as previously described (24) from whole blood obtained from the Department of Transfusion Medicine, Clinical Center, NIH. If not indicated otherwise, adherent cells were detached with trypsin (0.05%, 2 min).
Flow Cytometry
Cells (0.25–0.5 × 106/sample) were suspended in 25 μl of FACS buffer (HBSS plus 0.5% BSA, 0.1% NaN3, and 20 mm HEPES) plus 10 μl of anti-FPR1 or isotype control mAb. Samples were incubated for 45–60 min on ice, then centrifuged (5 min, 4 °C, 500 × g) and washed 1–2× with FACS buffer. Cells were resuspended in 300 μl of FACS buffer, and PE fluorescence was determined in the Fl-2 PMT of a FACSCaliburTM flow cytometer. Data were analyzed with WinMDI 2.9.
Western Blot
For membrane preparations, cell monolayers were detached with PBS plus 1 mm EDTA at 37 °C, centrifuged, and resuspended in 2–4 ml of lysis buffer (10 mm Tris-HCl, pH 7.5, 1 mm EDTA, and Complete- or Complete/Mini EDTA-free protease inhibitor mixture (Roche Diagnostics)). Cells were then homogenized on ice by 50–100 strokes in a Dounce homogenizer and centrifuged (5 min, 4 °C, 500 × g). The supernatant was then centrifuged (60 min, 4 °C, 16,000 × g), and the membrane pellet was resuspended in PBS and frozen at −80 °C. Protein concentration was determined using the Quick StartTM Protein Assay (Bio-Rad).
For alkali-stripping, membranes were incubated in 200–500 μl of 10 mm NaOH for 10 min on ice. After centrifugation (1–2 h, 4 °C, 16,000 × g), the pellet was resuspended in PBS, and protein concentration was determined. For PNGase F digestion, membrane samples were incubated for 10–20 min at RT in 1× SDS-containing glycoprotein denaturation buffer (New England Biolabs, Ipswich, MA). G7 reaction buffer, Nonidet P-40, PNGase F (New England Biolabs), and water were added to yield 1× G7 reaction buffer, 1% Nonidet P-40, and 75,000 units/ml PNGase F. After 1–2 h of incubation at 37 °C, 4× NuPAGE® LDS sample buffer (Invitrogen) was added, and the samples were stored at −80 °C. SDS PAGE was performed for 60–90 min at ∼100 V on Mini-Protean TGX 4–15% gels (Bio-Rad). The samples were mixed with 2× sample buffer prepared as previously described (25) or with 4× NuPAGE® LDS sample buffer, both containing 2-mercaptoethanol (35 μl/ml). Immunoblotting was performed with Immobilon-P PVDF membranes (Millipore, Billerica, MA), blocked overnight with 5% milk in TBST. Blocking time can be reduced to 1–2 h with essentially the same results. After a brief wash with TBST, the membrane was incubated with primary antibody NFPR2 (1:750 in 5% dry milk/TBST, 45 min). Incubation was also performed overnight (at 4 °C) without a considerable increase in background. The membranes were washed with TBST (3 × 10 min) and incubated for 1–2 h at RT with a 1:3000–1:5000 diluted (5% dry milk/TBST) polyclonal HRP-labeled rabbit anti-mouse IgG antibody (Abcam, Cambridge, MA). Then the blots were washed again with TBST (3 × 10 min) and developed with chemiluminescence reagent. Luminescence was detected with a HyBlot CL autoradiography film (Denville Scientific Inc., Metuchen, NJ).
For β-actin, the chemiluminescence reagent was removed with TBST, and the membrane was incubated twice (30 min at RT) with stripping buffer preheated to 55 °C. After another wash with TBST, the blot was blocked with 5% BSA/TBST (up to 8 h) and then incubated for 1 h with 1:1000 diluted (5% BSA/TBST) β-actin antibody (Abcam). Ponceau S (Sigma) staining (30 min at RT) was performed between stripping and re-blocking.
For the MAP kinase assays, the membranes were blocked for 1 h with 5% dry milk in TBST, washed 3 times 5 min with TBST, and then incubated overnight with the corresponding antibody in 5% BSA/TBST at 4 °C. For ERK 1/2 activation we used the anti-phospho-ERK 1/2 antibody (D13.14.4E, catalog no. #4370, 1:2000, Cell Signaling Technologies, Beverly, MA) and the anti-total ERK 1/2 antibody (137F5, catalog no. #4695, 1:1000, Cell Signaling). For p38 activation, we incubated with anti-phospho p38 (D3F9, catalog no. 4511, 1:1000, Cell Signaling) and anti-total p38 (catalog no. 9212, Cell Signaling). On the next day the blots were washed with TBST and incubated with secondary antibody (1:2000–1:3000, HRP conjugated goat anti rabbit, catalog no. 7074, Cell Signaling). After washing with TBST, the blots were incubated with chemiluminescence reagent as described above.
Confocal Microscopy
Cells were grown to log phase in DMEM + 10% FBS, washed with HBSS, and resuspended in FACS buffer at 107 cells/ml. 25 μl of this suspension were mixed with either 10 μl of undiluted anti-FPR1 mAb or undiluted IgG2A isotype control. After incubation for 30 min on ice, cells were washed with 400 μl of FACS buffer, centrifuged, and resuspended in 1000 μl of FACS buffer. For fluorescent FPR1 ligand labeling, cells were suspended in binding buffer (HBSS with Ca2+ and Mg2+, HEPES 20 mm, BSA 0.1%) containing 10 nm fNLFNYK-Fl (total binding) ± a 100-fold excess unlabeled fNLFNYK (nonspecific binding) and incubated for 15 min on ice in the dark. Fluorescently labeled cells were placed on the stage of a Leica SP5 inverted fluorescent microscope and imaged after 10 min at RT.
Determination of Receptor Density
Receptor density was determined with fluorescein-coated calibration beads (QuantumTM FITC-5 Premix, lot #9974, Bangs Laboratories, Fishers, IN). Beads and cells (binding samples with 10 nm fNLFNYK-Fl) were measured with a FACSCaliburTM flow cytometer (Fl-1 = 550 V). The MESF number (molecular equivalents of soluble fluorescein) per cell was converted to the number of fNLFNYK-Fl molecules by multiplying by 1.22 fNLFNYK-Fl eq/MESF as previously reported (26). Because 10 nm fNLFNYK-Fl occupy only 75% of FPR1 on HEK 293-FPR1+ cells and PMNs, the result for these cells (but not for FHL 124 cells) was multiplied by 1.33.
Internalization Experiments
Human neutrophils and FHL 124 and HEK 293-FPR1+ cells were suspended in binding buffer (0.5–1 × 106 cells/ml) prepared as described for confocal microscopy. After incubation ± fMLF (30 min at 37 °C or, as an additional control, on ice), the cells were centrifuged (4 °C, 5 min, 500 × g), washed with binding buffer, resuspended in 25 μl of FACS buffer, and stained with antibodies as described under “Flow Cytometry” above.
Flow Cytometric Binding Assays
In general, 150 μl of a cell suspension (∼106 cells/ml) in binding buffer (cf. confocal microscopy section) were mixed with 150 μl of ice-cold binding buffer containing 2× concentrated ligands. After incubation on ice (30–60 min), the samples were immediately measured with a FACSCaliburTM flow cytometer (Fl-1) under equilibrium conditions as previously described (27).
For saturation binding cells were incubated with increasing concentrations (0–100 nm) of fNLFNYK-Fl in the absence of other ligands (total binding) or in the presence of a 1000-fold excess of peptide W (nonspecific binding). For competition binding, cells were incubated with fNLFNYK-Fl (3 nm or 10 nm) and increasing concentrations (0.1 nm–10 μm) of fMLF, fNLFNYK, or peptide W. Total binding was determined in the absence of competing ligands. KD and Ki values were calculated with Graph Pad Prism 5.03 (GraphPad Software, La Jolla, CA).
Calcium Flux Assay
Cells were suspended in BSA-free binding buffer (2.5 × 106 cells/ml) and mixed with an equal volume of component A (Calcium 3 assay kit, Molecular Devices, Sunnyvale, CA) in a 96-well plate. After 30–45 min of incubation (37 °C, 5% CO2), the plate was centrifuged (5 min, 1000 rpm), and the Ca2+ assay was performed with a FlexStation 3 plate reader (Molecular Devices). Emission was detected at 525 nm after excitation at 485 nm. Data were analyzed with SoftMax Pro 5.4 (Molecular Devices).
Homologous desensitization was determined by FlexStation as follows; after 30 s of base-line recording, an appropriate volume of 10× agonist solution was added. A second agonist addition occurred at t = 210 s. The measurement was stopped at t = 450 s.
Alternatively, Ca2+ assays were performed by flow cytometry. Cell suspensions were mixed with an equal volume of indicator dye solution. After 30–45 min of incubation (37 °C, 5% CO2) the cells were washed twice with BSA-free binding buffer and resuspended in BSA-free binding buffer (0.5 × 106 cells/ml). The Ca2+ responses were analyzed for 90–120 s. The data were exported to a text file (with WinMDI). The graphs were generated with GraphPad Prism 5.03 (averaging and smoothing with a 2nd order polynomial with 20 neighboring points).
The effect of pertussis toxin (PTX, List, Campbell, CA) on signaling was tested using a final concentration of 750 ng/ml PTX (PBS buffer for negative control). After incubation for 4–5 h (37 °C, 5% CO2), cells were trypsinized and tested for calcium flux responses.
MAP Kinase Activation Assay
FHL 124 cells were seeded in 100 × 20-mm Petri dishes (400,000 cells/dish) or in 6-well plates (300,000 cells/well). 24 h before the experiment, supplemented medium 199 was replaced by serum- and growth factor-free medium 199. In ERK 1/2 activation assays, the MEK inhibitor U0126 in DMSO was added to one sample at a concentration of 10 μm. The corresponding volume of DMSO was added to all other samples that did not contain MEK inhibitor. Stimulation of the cells was performed by removing the medium and replacing it with 1.5 ml (6-well plates) or 5 ml (Petri dishes) medium 199 containing 10 μm fMLF. The stimulation was stopped after 2, 5, 10, and 30 min by removing the medium, washing the monolayer once with HBSS, and lysing the cells with 150 μl (6-well plate) or 350 μl (Petri dishes) of M-PER mammalian protein extraction reagent (Thermo Scientific, Rockford, IL). Protease inhibitor (1× Roche Complete protease inhibitor mixture) and phosphatase inhibitor (Roche PhosStop, 1 Tbl/10 ml) were added to the M-PER reagent immediately before use. The 0 value was stopped after 30 min (6-well plate experiment) or after 7–8 min (Petri dish experiment). The lysates were transferred to Eppendorf tubes and kept on ice until all samples were stopped. Then the lysates were centrifuged for 10 min at 14,000 × g and 4 °C. The supernatant was removed and frozen at −80 °C. For the Western blots, the protein concentration was determined by the Bradford method. Samples containing loading buffer with SDS and β-mercaptoethanol were heated for 10–15 min at 100 °C, and 10 or 25 μg of protein were applied per lane of the gel.
mRNA Extraction and qPCR
mRNA was isolated with the RNeasy Mini kit (Qiagen, Valencia, CA) and reverse-transcribed using a Biometra T-Gradient thermocycler (Biometra, Göttingen, Germany). PCR reagents including Moloney murine leukemia virus reverse transcriptase were from Promega (Madison, WI). Real time PCR was performed with a FAM-MGB-labeled probe (20×, Hs00181830_m1, Applied Biosystems, Carlsbad, CA) and TaqMan Gene Expression Master Mix (Applied Biosystems) using a 7900HT Fast Real-time PCR system (Applied Biosystems). Data were analyzed with SDS 2.2.2 (Applied Biosystems).
Determination of mRNA Half-life with Actinomycin D
One day before the experiment, cells were seeded at a density of 300,000/well in a 6-well plate. On the next day, actinomycin D stock solution (actinomycin D-mannitol, 0.4 mg/ml, catalog no. A5156, Sigma) was added to yield a final actinomycin D concentration of 2 μg/ml. The cells were incubated at 37 °C, and mRNA was isolated from the negative control (without actinomycin D). The other 5 wells were stopped at 5 appropriate time points between 0 and 24 h. The isolation of mRNA was either performed by direct lysis of the cells in the plate or by lysis of a pellet of trypsinized cells. The mRNA was reverse-transcribed to cDNA, and qPCR was performed as described under “mRNA Extraction and qPCR” above.
Small Interfering RNA Analysis
Cells were nucleofected in buffer from AMAXA nucleofector kit V (Lonza) at a density of 107 cells/ml. 100 pmol of FPR1 siRNA (three 20–25-nucleotide siRNAs, catalog no. sc-40121, Santa Cruz Biotechnology, Santa Cruz, CA), 100 pmol of scrambled fluorescein-labeled siRNA (Santa Cruz Biotechnology, catalog no. sc-36869), or water was added to 100 μl of cell suspension and nucleofected with program X-005 (FHL 124) or Q-001 (HEK 293-FPR1+) of the Nucleofector II Device (Lonza). After culturing of the cells in 6- or 12-well plates for 24 h, mRNA isolation and flow cytometric experiments were performed.
For the shRNA experiments, the plasmids were dissolved in 10 mm Tris, 1 mm EDTA at a concentration of 0.1 μg/μl. FHL 124- and HEK 293 cells were suspended in supplemented Nucleofection Kit V buffer to yield a final density of 1.1 × 106 cells/100 μl. Then 22 μl of FPR1 shRNA (Santa Cruz, catalog no. sc-40121-SH), scrambled shRNA (Santa Cruz, catalog no. sc-108060), or H2O (DNA-free water control) were added to 200 μl of the cell suspension. The final transfection samples contained 1 μg of plasmid per 100 μl and 1 × 106 cells/ml. The transfections were performed using program X-005 (FHL 124) or Q-001 (HEK 293-FPR1+) of the Lonza nucleofection machine. The cells were seeded in 25- or 75-cm2 culture flasks. After culturing the cells for 48 h, puromycin (2.5 μg/ml for FHL 124- and 10 μg/ml for HEK 293 cells) was added as selection antibiotic. Four weeks after the transfection, both cell lines were maintained in the presence of 10 μg/ml of puromycin.
Sequencing of FPR1 mRNA from FHL 124 cells
Preparation of mRNA and reverse transcription were performed as described in the mRNA extraction and qPCR paragraph. The target sequence was amplified using the forward primer hFPR1f (5′-AGA CCT AGA ACT ACC CAG AGC AA-3′) and the reverse primer hFPR1-S6-r (5′-GGA GCT CGA AAG TGT CCC-3′), both of which hybridize with cDNA sequences that correspond to parts outside the open reading frame. The following PCR program was used: (1) 2 min at 95 °C, (2) 0.5 min at 95 °C, (3) 0.5 min at 55 °C, (4) 2.0 min at 72 °C, (5) 10 min at 72 °C. Steps 2–4 were repeated 34 times. The PCR product was isolated using the PCR purification kit (Qiagen) and according to the manufacturer's instructions. Dye terminator sequencing was performed by ACGT (Wheeling, IL) using the forward primers hFPR1-S1 (5′-GAG CAA GAC CAC AGC TGG-3′), hFPR1-S2 (5′-GGC TGG TTC CTG TGC AAA-3′), hFPR1-S4 (5′-GAG AGG CAT CAT CCG GTT-3′), hFPR1-S5 (5′-AGG CTG ATC CAC GCC CTT-3′), and the reverse primer hFPR1-S6r (5′-GGA GCT CGA AAG TGT CCC-3′). All PCR reactions were performed using PfuUltra DNA Polymerase (Agilent, Santa Clara, CA). Sequence analysis was performed with GeneRunner V3.05 (Hastings Software, Inc., Hastings, NY) or Sequencher (Gene Codes Corp., Ann Arbor, MI).
RESULTS
FPR1 Is Expressed by Human Lens Epithelial Cells
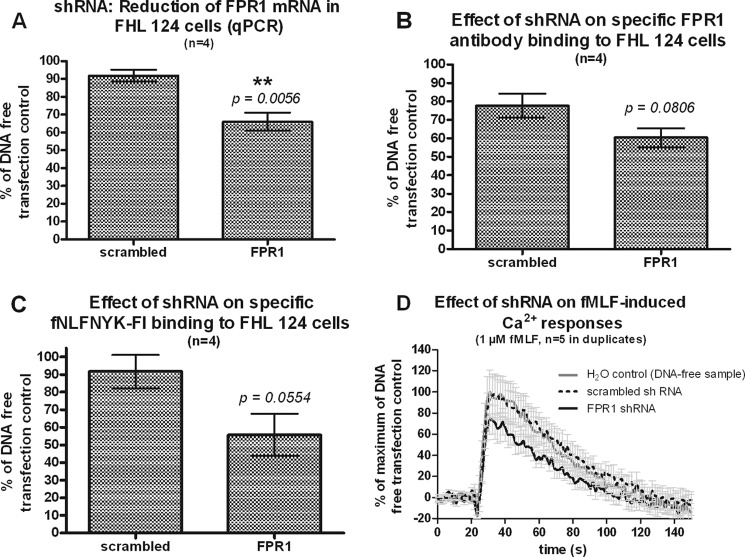
FPR1 mRNA was detected by PCR in both FHL 124 (Fig. 1A) and SRA 01/04 (data not shown) human lens epithelial cell lines. Target specificity of the qPCR reactions was assured by predigestion of RNA samples with DNase and by using primers that span an FPR1 intron. Moreover, stable transfection of FHL 124 cells with FPR1-specific shRNA inhibited the PCR signal, causing a moderate but significant reduction by ∼30%, compared with cells transfected with scrambled shRNA (Fig. 1A).
FIGURE 1.
Lens epithelial cells constitutively express FPR1; RNA and immunophenotypic evidence. A, RNA is shown. FHL 124 cells nucleofected with the shRNA indicated on the x axis were analyzed for FPR1 mRNA expression by PCR. B, immunophenotype is shown. FHL 124 cells nucleofected with the shRNAs indicated on the x axis were analyzed by FACS with a PE-labeled anti-FPR1 mAb. C, ligand binding; FHL 124 cells nucleofected with the shRNAs indicated on the x axis were analyzed by FACS with the fluorescent agonist ligand fNLFNYK-Fl (10 nm). D, Ca2+ assay; FHL 124 cells nucleofected with the shRNAs indicated in the legend were stimulated with 1 μm fMLF, and the Ca2+ response was followed for 150 s. The base line (first 20 s) of each individual curve and a background curve recorded with only buffer (no fMLF) were subtracted from each signal. Then all experiments were pooled and transformed to % of the peak signal intensity of the siRNA-free (only H2O) control. All data are the means ± S.E.
Consistent with this, we found that FHL 124 cells stained specifically with anti-FPR1 mAb. Similar results were obtained for SRA 01/04 cells (supplemental Figs. S1 and S2). Transfection with FPR1-specific shRNA reduced FPR1 antibody staining of FHL 124 cells to a similar magnitude (∼20%) as FPR1 mRNA (Fig. 1B). Binding of the fluorescent ligand fNLFNYK-Fl (10 nm) to FHL 124 cells was also affected by FPR1 shRNA to a similar magnitude (∼40% reduction) as FPR1 mRNA (p = 0.0554, Fig. 1C). Consistent with this, FPR1 shRNA reduced fMLF-induced calcium flux in FHL124 cells by ∼20% as compared with cells transfected with scrambled shRNA (Fig. 1D) or DNA-free H2O control.
We also performed experiments with short-acting FPR1-specific siRNA. In preliminary experiments with cells cultured in the presence of actinomycin D (data not shown), we found that the FPR1 transcript in FHL 124 cells has a half-life of ∼6 h, which should result in a >90% reduction of FPR1 mRNA within 24 h. In fact, 24 h after transfection of FHL 124 cells with FPR1-specific siRNA, the FPR1 mRNA was reduced by 75%, which is a 3-fold stronger reduction than achieved by shRNA (data not shown). The siRNA apparently induces higher effective concentrations in our system than the continuously expressed shRNA. However, we did not observe any effect on FACS antibody staining and only a weak non-significant effect on fluorescent ligand binding (data not shown). Thus, in contrast to shRNA, FPR1 siRNA was only effective at the mRNA level, not the protein level. This may be due to the fact that siRNA acts only transiently and is quickly degraded, whereas shRNA is constantly produced and replenished by the cell. If the target protein is very stable, the time window of the siRNA effect may be too short to produce a significant decrease in protein levels. The shRNA, however, is continuously present, and even if the concentration is low, it acts for a sufficiently long time to produce a new equilibrium between mRNA and protein where even small reductions in mRNA are reflected by corresponding reductions in protein levels.
We addressed the structural state of FPR1 protein from FHL 124 cell membranes, as defined by Western blot detection using a previously described second FPR1-specific antibody, the monoclonal reagent NFPR2 (25). This antibody strongly detected a broad band between 35 and 60 kDa in FPR1-transfected HEK 293 cells (Fig. 2A, left lane), in good agreement with published data for FPR1 in neutrophils and FPR1-transfected CHO cells using the same antibody (25) and in human HL-60 neutrophils detected by radioligand affinity labeling (28). No corresponding band was found in wild type HEK 293 cells (Fig. 2A, right lane). FPR1 is known to be glycosylated at the N terminus (28, 29), and this has been shown to account for the broad distribution of the immunoreactive protein on Western blot analysis. We also found an additional immunoreactive band at 120 kDa in FPR1 transfectants and wild type HEK 293 cells (Fig. 2A).
FIGURE 2.
Lens epithelial cells constitutively express FPR1. Western blots were probed in panels A and B and the top portion of C with NFPR2, a mAb to FPR1. PMN stands for polymorphonuclear leukocytes (neutrophils). The bottom portion of panel C depicts the blot from the top of panel C reprobed with anti-β-actin after stripping. Panel D depicts lanes 5–8 from the blot in panel C stained with Ponceau S as an additional loading control. The amount of membrane protein loaded is indicated below each lane. The cell types tested and cell treatments are indicated at the top. HEK hFPR1+, FPR1-transfected HEK 293 cells. Western blots in A, B, and C are from separate experiments. For A and B, the Super Signal West Pico Chemiluminescent Substrate (Thermo Scientific) was used, and for C the chemiluminescent Amersham Biosciences ECL Plus detection reagent (GE Healthcare) was used.
Because anti-FPR1 surface staining by FACS was similar in magnitude for FHL 124 cells and FPR1-transfected HEK 293 cells, we expected to find similar results for FPR1 protein for the two cell types by Western blot. We did in fact observe the expected immunoreactive band for FHL 124 cell membranes at ∼60 kDa; however, the relative intensities were the converse of the FPR1-transfected HEK 293 cells (Fig. 2B, lanes 1–3). The 120-kDa band was consistently found with high intensity in FHL 124 cell membranes and with variable intensity in several batches of wild type and hFPR1-transfected HEK 293 membranes.
We hypothesized that the 120-kDa band may represent a second FPR1 species that exists either as a dimer or as a complex with one or several other proteins. Thus, we treated the cell membranes with NaOH (10 mm), which has previously been used to remove peripheral membrane proteins (30, 31) and should also be able to break protein complexes that are held together by electrostatic interactions. This treatment completely removed the 120-kDa species from the Western blot of FHL 124- and wild type HEK 293 cells. When the same amount of protein from NaOH-treated and -untreated FHL 124 cell membranes was applied to the Western blot, the immunoreactive protein at 60 kDa was concentrated, resulting in a stronger band (Fig. 2B, fourth lane). The distribution of this signal in FHL 124 membranes was much narrower than for FPR1-transfected HEK 293 cell membranes, possibly because of conformational differences or differences in glycosylation pattern. Interestingly, neutrophil membranes probed with the same anti-FPR1 antibody reagent contained only the 60-kDa species and not the 120-kDa band (Fig. 2B, fifth lane). Treatment of the membranes with NaOH did not change the results (Fig. 2B, sixth lane).
To provide further evidence that the 60-kDa band is in fact FPR1, we treated the membranes with PNGase F. This enzyme selectively removes N-linked glycosylation moieties from proteins, the type that has previously been demonstrated to post-translationally modify the N-terminal extracellular domain of FPR1 (28, 29). Efficiency of the digestion procedure was shown with membranes from FPR1-transfected HEK 293 cells (positive control) and wild type HEK 293 cells (negative control). As shown in Fig. 2C, first and second lanes, PNGase F digestion of FPR1-transfected HEK 293 cell membranes shifted the FPR1 band from ∼60 to ∼35 kDa. This corresponds to the size of the deglycosylated core protein of human neutrophil FPR1 previously reported in the literature (28, 32, 33). Wild type untransfected HEK 293 cell membranes showed no signal (Fig. 2C, third and fourth lanes). Digestion of FHL 124 cell membranes also shifted the 60-kDa band to ∼35 kDa (Fig. 2C, fifth and sixth lanes). However, the 120-kDa band was resistant to PNGase F treatment. We also applied membranes from wild type untransfected HEK 293 cells to the gel at the same protein amount as the FHL 124 cell membranes. Surprisingly, we saw faint staining at ∼60 kDa (Fig. 2C, seventh lane), which was partially shifted to 35 kDa after PNGase F treatment (Fig. 2C, eighth lane). Thus, even wild type HEK 293 cells may express a very low background level of FPR1. Ponceau S staining after stripping of the membranes was used to verify equal protein loading (34) (Fig. 2D) as the cells had intrinsic differences in the amount of β-actin attached to the membranes.
The strong band stained by Ponceau S in the PNGase F-treated samples is most likely the enzyme protein added to the assay (36 kDa, according to the manufacturer's information). This poses the potential problem that the new species observed in PNGase F-treated membranes may be non-specifically stained PNGase F and not deglycosylated FPR1. Therefore, we also analyzed a sample containing only the enzyme. In fact, the enzyme was stained, most likely by the secondary HRP-conjugated antibody (data not shown). However, in all of our experiments this staining was consistently weaker than the staining in the PNGase F-treated membrane samples from both FPR1-transfected HEK 293 cells and FHL 124 cells. We stripped the membranes and re-stained them with FPR1 antibody using a longer incubation time. After this procedure, the enzyme staining was completely removed, whereas the FPR1 signal was retained at its original intensity (data not shown). Therefore, the 35-kDa species observed in PNGase F-treated membranes from FPR1-transfected HEK 293 cells and FHL 124 cells represents deglycosylated receptor protein and not just the added enzyme.
FPR1 Is Functional on Human Fetal Lens Epithelial Cells
To test whether FPR1 on FHL 124 cells is functional, we first investigated agonist-induced receptor down-regulation. fMLF induced at least 80% reduction of FPR1 on the surface of neutrophils (Fig. 3, B and C) and FPR1-transfected HEK 293 cells (Fig. 3C) but only ∼20% reduction on FHL 124 cells (Fig. 3, A and C). There was no significant difference between the fMLF-free 37 °C control and the controls at 4 °C (data not shown). Moreover, the controls confirm that there was no competition between fMLF and the fluorescent antibody, which could lead to a reduction of staining after incubation with fMLF. Similar results were obtained using SRA 01/04 cells (supplemental Fig. S1).
FIGURE 3.
FPR1 is resistant to ligand-induced internalization in lens epithelial cells. Cells were incubated for 30 min at 37 °C in the presence (dashed line) or absence (solid line) of 1 μm fMLF and then stained with anti-FPR1. Unstimulated control cells were also stained with isotype control mAb (dotted line). A and B represent individual Fl-2 histograms showing the results of one representative experiment for the cell types indicated at the top. C, summary data from 2–5 independent experiments (1–2 samples for each individual condition). The number of experiments for each cell type is given above the bars.
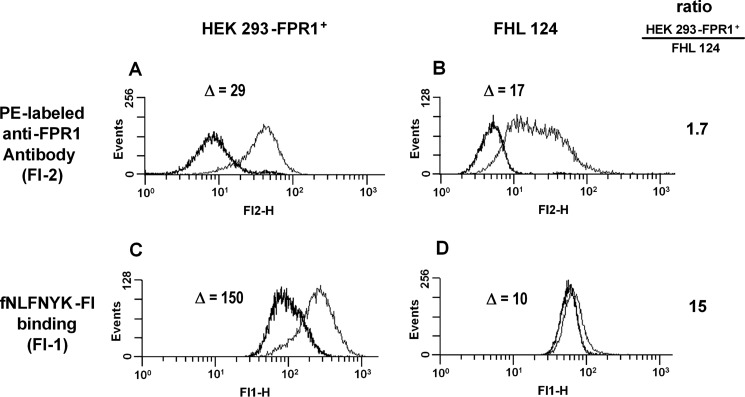
We next tested whether the lens cells could bind the fluorescent agonistic FPR1 ligand fNLFNYK-Fl. Surprisingly, although antibody staining of FPR1-transfected HEK 293 cells and FHL 124 cells was comparable (Fig. 4, A and B), FHL 124 cells showed much less fluorescent ligand binding than FPR1-transfected HEK 293 cells (Fig. 4, C and D). A similar discrepancy between antibody staining and fNLFNYK-Fl binding was observed in SRA 01/04 lens epithelial cells (supplemental Fig. S2).
FIGURE 4.
FPR1 adopts distinct states in lens epithelial cells and FPR1-transfected HEK 293 cells; evidence from flow cytometry. A and B, staining of HEK 293-FPR1+ cells (A) and FHL 124 cells (B) with anti-FPR1 is shown. Gray, anti-FPR1; black, isotype control. C and D, specific binding of fNLFNYK-Fl (10 nm) to HEK 293-FPR1+ cells (C) and FHL 124 lens epithelial cells (D) is shown. Gray, total binding; black, nonspecific binding. Specific binding was calculated by subtraction of nonspecific binding (in the presence of 10 μm fMLF) from total binding (10 nm fNLFNYK-Fl without fMLF). Δ represents the difference between anti-FPR1 staining and isotype control background (A and B) or between total and nonspecific binding (C and D). The ratio of Δ(HEK 293-FPR1+)/Δ(FHL 124) is shown on the right. The data are from one representative experiment that was reproduced twice.
These results were confirmed by confocal microscopy (Fig. 5). We were unable to detect specific fNLFNYK-Fl binding to FHL 124 cells, whereas anti-FPR1 antibody clearly bound to the surface of FHL 124 cells. Both reagents bound specifically to the surface of FPR1-transfected HEK 293 cells.
FIGURE 5.
FPR1 adopts distinct states in lens epithelial cells and FPR1-transfected HEK 293 cells; evidence from confocal microscopy. Cells were stained with either antibody (anti-FPR1 or isotype control) or with 10 nm fNLFNYK-Fl (with or without excess unlabeled ligand) as indicated at the top of each column. The cell types are indicated on the left side of each row. Data are from representative experiments that were reproduced once.
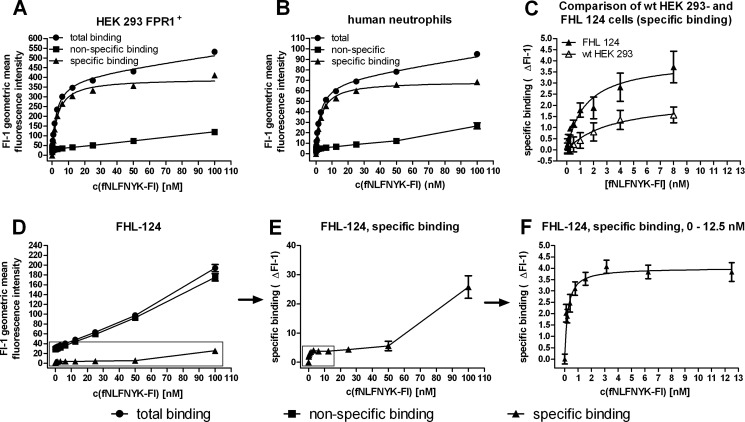
Despite the extremely low specific binding of fNLFNYK-Fl detected for FHL 124 cells by flow cytometry, we attempted to determine the KD. To validate the assay, we first performed saturation binding experiments with human neutrophils and HEK 293-FPR1+ cells. Specific binding was saturable in both cases, and nonspecific binding never exceeded 30% of the total binding (Fig. 6, A and B). Untransfected HEK 293 cells (Fig. 6C) showed very low specific binding, confirming the Western blot results that also had shown the presence of very few FPR1 molecules. We calculated an fNLFNYK-Fl KD value of 3.2 ± 0.1 nm (n = 2, mean ± S.E.) for human neutrophils and 2.7 ± 0.3 nm (n = 2, mean ± S.E.) for HEK 293-FPR1+ cells. In contrast, saturation binding assays with FHL 124 cells revealed extremely low specific binding of fNLFNYK-Fl with a nonspecific background of 80–90% (Fig. 6D). Nevertheless, subtraction of nonspecific binding resulted in a curve with the typical shape of a monophasic saturation binding curve in the initial part. However, there was a sudden increase in specifically bound fluorescence at higher concentrations of fNLFNYK-Fl (Fig. 6E). In Fig. 6F, only the first part of the curve between 0 and 13 nm fNLFNYK-Fl is shown, demonstrating a high affinity binding site that is saturated in this concentration range. The KD value calculated from this initial portion of the curve is 0.5 ± 0.2 nm (n = 5, mean ± S.E.). A direct comparison with wild type HEK 293 cells shows a 2–3-fold higher specific binding of fNLFNYK-Fl to FHL 124 cells (Fig. 6C).
FIGURE 6.
Characterization of high affinity binding sites for fluorescent FPR1 ligand fNLFNYK-Fl on lens epithelial cells. Specific binding (▴) was calculated by subtraction of nonspecific binding (■, presence of peptide W at a 1000-fold excess compared with fNLFNYK-Fl) from total binding (only fNLFNYK-Fl, ●). A and B, total, nonspecific, and specific binding of fNLFNYK-Fl to HEK 293-FPR1+ cells (A) and human neutrophils (B) is shown. C, comparison of specific binding of fNLFNYK-Fl to FHL 124 cells and untransfected HEK 293 cells is shown. D–F, saturation binding of fNLFNYK-Fl to FHL 124 lens epithelial cells shows the complete curves for total, nonspecific, and specific binding (D) as well as the complete specific binding curve alone (E) and the first part of the specific binding curve between 0 and 12 nm (F). All fluorescence data were recorded in the FL-1 PMT of a FACSCaliburTM flow cytometer at the same Fl-1 PMT settings (510 V). A–C show representative experiments with 2–3 replicates; the curves shown in D–F are the average of three independent experiments with 2–3 replicates.
We quantitated receptor expression for each cell type with 10 nm fNLFNYK-Fl by calibration using fluorescein-coated beads (Table 1). FHL 124 cells expressed ∼2500 receptors per cell, less than 10% that of expression on HEK 293-FPR1+ cells (∼70,000 receptors/cell) and human neutrophils (∼40,000 receptors/cell).
TABLE 1.
Determination of fNLFNYK-FL binding site number on the surface of human neutrophils, HEK 293-FPR1+ cells and FHL 124 lens epithelial cells using FITC-coated standard beads
Data are the means ± S.E.
| Cell type (no. of experiments) | ΔMESFa | No. of receptors occupied (×1.22)b | Total receptor number per cell |
|---|---|---|---|
| PMN (n = 2) | 25,514 ± 2,033 | 31,127 ± 2480 | ∼41,400 ± 3300c |
| HEK 293-FPR1+ (n = 3) | 43,094 ± 201 | 52,574 ± 245 | ∼69,900 ± 330c |
| FHL 124 (n = 4) | 2,077 ± 309 | 2,533 ± 377 | ∼2,500 ± 380 |
a MESF, molecular equivalents of soluble fluorescein.
b Multiplication with 1.22 fNLFNYK-FL equivalents per MESF.
c Multiplication with 1.33 to obtain Bmax (only 75% saturation with 10 nm of fNLFNYK-Fl).
To investigate the pharmacology of the FHL 124 cell FPR1 in more detail, we determined IC50 values for three standard FPR1 agonists (fMLF, fNLFNYK, and peptide W) in competition binding assays in the presence of 3 nm fNLFNYK-Fl. All three FPR1 agonists displaced fNLFNYK-Fl with the same rank order of potency on HEK 293-FPR1+ cells, FHL 124 cells, and human neutrophils (Fig. 7 and Table 2). As expected, the non-FPR1 binding chemokine MIP-1α (CCL3; MIP, macrophage inflammatory protein) caused no displacement. Table 2 shows similar pIC50 values for all three cell systems. The pIC50 values were converted to pKi values by means of the Cheng-Prusoff equation using the KD values mentioned above and a ligand concentration of 3 nm. We also performed competition binding with 10 nm fNLFNYK-Fl and increasing concentrations of fMLF with SRA 01/04 cells, demonstrating a concentration-dependent displacement of fNLFNYK-Fl by fMLF (supplemental Fig. S3).
FIGURE 7.
fNLFNYK-Fl binding sites are pharmacologically similar on lens epithelial cells, neutrophils, and FPR1-transfected HEK 293 cells. A–C, competition for binding of fNLFNYK-Fl (3 nm) by increasing concentrations of the FPR1 ligands fMLF (■), fNLFNYK (□), or peptide W (●). MIP-1α (CCL3; MIP, macrophage inflammatory protein) (×) served as a non-binding negative control. Competition curves were recorded for HEK 293-FPR1+ cells (A), FHL 124 lens epithelial cells (B), and human neutrophils (C). The curves contain the data of two independent experiments with duplicates. In each experiment all four ligands were tested at all three cell types on the same day under the same conditions.
TABLE 2.
Comparison of the pIC50/pKi values of fMLF, fNLFNYK-Fl, and peptide W at the FPR1 in FHL 124 cells, HEK 293-FPR1+ cells, and human neutrophils (PMN)
The data are the mean values ± S.E. from two independent experiments with duplicates.
| Ligand | FHL 124 |
HEK 293-FPR1+ |
PMN |
|||
|---|---|---|---|---|---|---|
| pIC50 | pKia | pIC50 | pKib | pIC50 | pKic | |
| fMLF | 6.80 ± 0.17 | 7.65 ± 0.17 | 6.81 ± 0.06 | 7.14 ± 0.06 | 6.67 ± 0.06 | 6.96 ± 0.06 |
| fNLFNYK | 7.91 ± 0.23 | 8.76 ± 0.23 | 7.82 ± 0.08 | 8.15 ± 0.08 | 7.74 ± 0.05 | 8.03 ± 0.05 |
| Peptide W | 7.33 ± 0.20 | 8.18 ± 0.20 | 7.55 ± 0.05 | 7.88 ± 0.05 | 7.17 ± 0.06 | 7.46 ± 0.06 |
a Calculated with a KD of 0.5 ± 0.2 nm.
b Calculated with a KD of 2.7 ± 0.3 nm.
c Calculated with a KD of 3.2 ± 0.1 nm.
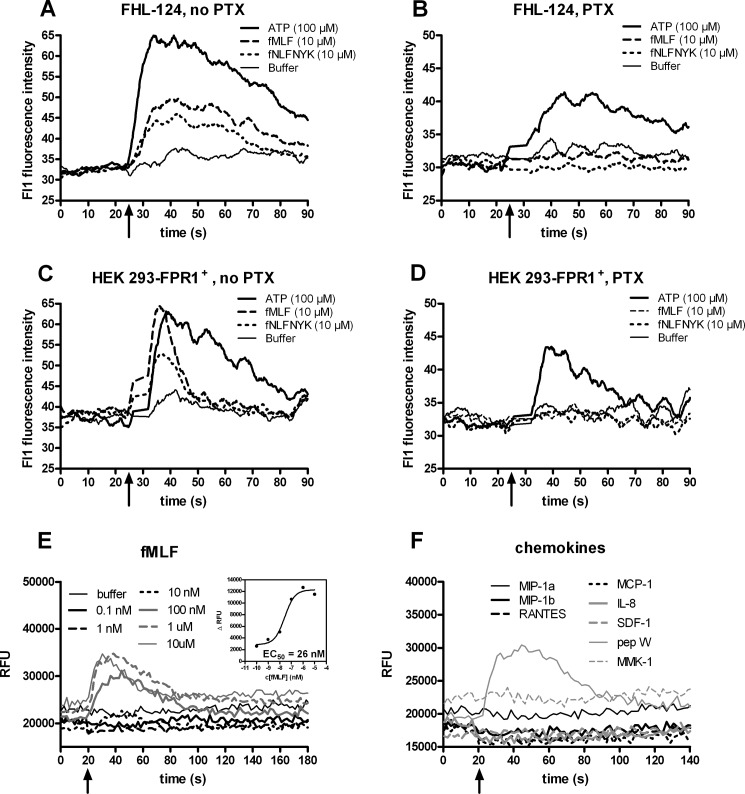
We next investigated the functionality of the FHL 124 cell formyl peptide receptor in Ca2+ flux assays. As shown in Fig. 8, A and C, 10 μm fMLF and fNLFNYK induced a transient rise of intracellular Ca2+concentration in both FHL 124 and HEK 293-FPR1+ cells. Repeated addition of the agonist fMLF to the samples revealed that FPR1 and its signal transduction pathway undergo homologous desensitization. FPR1 signaling was completely desensitized by 1 μm fMLF. Subsequent addition of a concentration of 100 nm fMLF was able to induce two Ca2+ signals. However, the second one is considerably reduced compared with the first one (supplemental Fig. S4). When the cells were treated with pertussis toxin, they failed to respond to FPR1 agonists, whereas the ATP-induced Ca2+ signal was retained, albeit with lower intensity (Fig. 8, B and D). The fMLF-induced Ca2+ flux response was concentration-dependent and saturable. The EC50 value for fMLF was ∼26 nm (Fig. 8E), which aligns well with the pKi value of 7.65 (Fig. 7 and Table 2). We found a similar concentration-dependent effect of fMLF in Ca2+ assays with SRA 01/04 cells (supplemental Fig. S5). None of the chemokines tested induced Ca2+ flux in FHL 124 lens epithelial cells (Fig. 8F).
FIGURE 8.
FPR1 agonists induce Ca2+ flux responses in lens epithelial cells in a pertussis toxin-sensitive manner. A–D, the cell types indicated at the top of each panel were incubated without (A and C) or with PTX (B and D), stimulated with substances listed in the insets, and analyzed by flow cytometry. ATP (100 μm) was used as a positive control. Arrows identify the time when ligand was added. Each trace represents the average of 2–5 calcium flux responses. The effect of PTX on FPR1 agonist-mediated calcium signaling was confirmed by three additional independent plate reader experiments. E, fMLF potency and efficacy in FHL 124 cells (plate reader based experiment) is shown. The boxed inset in E shows the corresponding concentration-peak effect relationship generated from the kinetic curves with the resulting EC50 value. F, chemokines do not induce calcium flux in FHL 124 cells. RFU, relative fluorescence units.
In the literature it has been repeatedly described that FPR1 leads to phosphorylation of MAP kinases like ERK 1/2 or p38 (35–37). Therefore, we stimulated serum-starved FHL 124 cells with 10 μm fMLF and determined MAPK phosphorylation at different time points between 0 and 30 min. In fact, fMLF stimulated ERK 1/2 and p38 phosphorylation with a signal maximum after ∼10 min (Fig. 9). Formyl peptide-induced ERK 1/2 phosphorylation could be completely blocked by a 2 h preincubation with the MEK inhibitor U0126 (Fig. 9A, sixth and seventh lane). Despite serum starvation for 24 h before fMLF stimulation, in some of our experiments FHL 124 cells showed a high basal MAPK activity, which made it impossible to see the fMLF-induced signal. The reasons for this are not clear to us, but MAPK activity may have been influenced by the passage of the cell line, the density of the culture, or the age of the medium.
FIGURE 9.
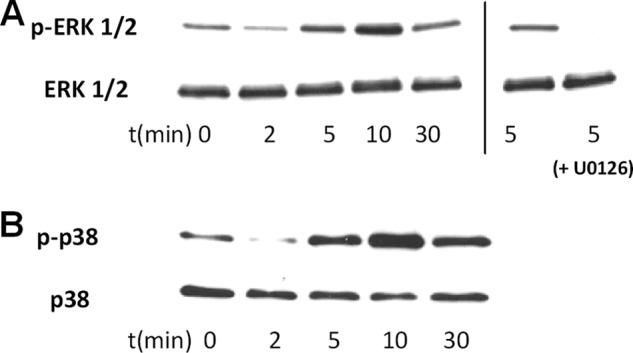
Phosphorylation of MAP kinases ERK 1/2 and p38 after stimulation of FHL 124 cells with 10 μm of fMLF. A, phosphorylated ERK 1/2 (upper row) and total ERK 1/2 (lower row) at different times between 0 and 30 min (lanes 1–5) and after 5 min without (lane 6) or with (lane 7) the MEK inhibitor U0126 is shown. B, phosphorylated p38 (upper row) and total p38 (lower row) at different times between 0 and 30 min (lanes 1–5) is shown.
FHL-124 Cells Express Full-length FPR1 mRNA
Because our results from binding and antibody staining experiments indicate an uncommon behavior of FPR1 in FHL 124 cells, we sequenced FPR1 from cDNA to identify potential mutations and splice variants of FPR1 mRNA. Our results (Table 3) show that the full-length FPR1 open reading frame is expressed in FHL 124 cells represented by the previously described haplotypes H-11 and H-19, both of which seem to be relatively rare (38). According to Ref. 38, H-11 was not detected in samples from Caucasian individuals and in only 11% of the investigated black population, whereas H-19 was completely absent in the samples that were completely sequenced in Ref. 38.
TABLE 3.
Determination of the FPR1 sequence expressed in FHL 124 cells and comparison with the FPR1 haplotypes previously described in the literature (38)
| Codon numbera | 11 | 101 | 116 | 182 | 190 | 192 | 346 |
|---|---|---|---|---|---|---|---|
| Codons with corresponding amino acidsb | ACC (T) | GTC (V) | ATC (I) | CCC (P) | AGG (R) | AAG (K) | GCG (A) |
| ATC (I) | CTC (L) | ATT (I) | CCA (P) | TGG (W) | AAC (N) | GAG (E) | |
| AAT (N) | |||||||
| Genotype in FHL 124 cells | C, C | C, G | C, C | C, A | A, A | T, T | A, C |
| H-11 haplotype | C | G | C | A | A | T | C |
| H-19 haplotype | C | C | n.d.c | n.d.c | A | C/T | A |
a Underlined codon numbers indicate positions with amino acid differences between the haplotypes.
b Bases that vary between the haplotypes are indicated by double underlines.
c n.d., not determined.
DISCUSSION
We have demonstrated at the RNA-, protein-, and functional level that the classic leukocyte chemoattractant receptor FPR1 is expressed endogenously by human lens epithelial cells. This expands the list of non-hematopoietic cell types reported to express FPR1 and provides the first detailed biochemical and functional characterization of such a receptor. Importantly, our work is consistent with a direct role in the lens for FPR1 in lens homeostasis, as evidenced by aging Fpr1−/− mice, which develop severe cataracts.4
At first glance the evidence for functional expression of the receptor in this cell line appears straightforward and consistent. 1) Full-length FPR1 mRNA was amplified and sequenced by PCR using specific primers and knocked down using specific si- and shRNAs. 2) FPR1 protein could be detected as strong cell surface staining by FACS using an FPR1-specific monoclonal antibody and as immunoreactive bands of the expected size and sensitivity to the deglycosylating enzyme PNGase F by Western blot using a second independent FPR1-specific antibody. 3) The cells bind known FPR1 ligands. 4) The same ligands tested as agonists induce classic pertussis toxin-sensitive calcium flux responses, and fMLF stimulates the MAP kinases ERK 1/2 as well as p38. Despite these similarities, however, several properties attributed to the receptor were strikingly different in lens epithelial cells compared with hematopoietic cells or FPR1-transfected HEK 293 cells.
First, FHL 124 cell FPR1 appears to be atypical in Western blots stained with the previously characterized FPR1-specific mAb NFPR2. We found four related but distinct immunoreactive banding patterns of cell membrane proteins in the four cell types studied: 1) a single strong 60-kDa band in human neutrophils, consistent with previous reports; 2) a much weaker 60-kDa band and a second very strong 120-kDa band in FHL-124 cells; 3) a strong group of bands from 35–60 kDa and a 120-kDa band with varying intensity in HEK293-FPR1+ cells; 4) in wt HEK 293 cells, a 120-kDa band of varying intensity and a very faint 60-kDa staining that, however, was much weaker than the signals from FHL 124- or HEK293-FPR1+ samples. For two reasons the FHL 124 cell 60-kDa band likely represents FPR1. First, this is the size of both the only immunoreactive band identified in neutrophils and a major band found in HEK 293-FPR+ cells. Western blots of wild type HEK 293 membranes show only a very weak staining at 60 kDa compared to the transfected cells and to FHL 124 cells. Second, PNGase F treatment, which removes N-linked glycosylation from proteins, eliminates the 60-kDa band and produces a 35-kDa immunoreactive band in both HEK 293-FPR1+ and FHL 124 cells (and to a lesser extent in wt HEK 293 cells), consistent with the predicted size of the core unglycosylated FPR1 protein. The 60-kDa band probably represents the receptor species binding fNLFNYK-Fl, as it is the only form found in neutrophil membranes, and its relative intensity in neutrophils, FHL 124 cells, and FPR1-transfected HEK293 cells corresponds well to the relative number of receptors/cell determined by ligand binding analysis.
The identity of the 120-kDa band is less clear. Two observations suggest that it could represent an FPR1 dimer or an FPR1 complex with a peripheral membrane protein. First, the band is double the size of the main monomer detected in all three cell types and also recognized by the alternative anti-FPR1 antibody, NFPR1, which like NFPR2 also recognizes a C-terminal epitope (25) (data not shown). Second, alkali treatment of cell membranes, which is known to remove proteins loosely associated with membrane and to disrupt protein-protein interactions, removed this band completely from FHL 124 cells while increasing the intensity of the 60 kDa band. Because the analysis was performed using a reducing gel, receptor dimerization may be the most likely possibility. Alternatively, the 120-kDa band may simply represent a cross-reacting protein with structural similarities to the FPR1 C terminus. This is supported by the fact that this species occurs at very high intensity in some membrane batches from non-transfected HEK 293 cells, showing the same properties as in FHL-124 cells (alkali sensitivity and staining with both NFPR1 and NFPR2; data not shown). Moreover, the 120-kDa band was not present in neutrophils, which are known to express high levels of FPR1, and it was not affected by PNGase F treatment either in FHL 124 cells or in wt HEK 293 cells. Unfortunately, the PE-labeled FACS antibody did not produce any staining in Western blots, not even at a dilution of only 1:250 (data not shown).
The second atypical property of the FHL 124 cell FPR1 is the extremely low level of specific binding relative to total binding attributable to the FPR1 ligand fNLFNYK-Fl. This is inconsistent with FACS antibody staining, which produced a similarly strong signal in FHL 124 cells, neutrophils, and HEK 293-FPR1+ cells. In fact, from fNLFNYK-Fl binding alone, we would have concluded that the receptor was not expressed by these cells. The low level of specific binding is consistent with the low level of immunoreactive 60-kDa FPR1 monomer detectable by Western blot. At this time we have not identified the factor responsible for this greatly limited specific binding of this ligand and for the divergent results from ligand binding and FACS antibody staining. We speculate that two FPR1 receptor populations may exist, a large one stained by the FACS antibody but inaccessible to ligand binding, and a small one, binding both the fluorescent ligand and the PE-labeled FACS antibody. The observation that fNLFNYK-Fl binding seemed to be more sensitive to shRNA (∼40% reduction) than antibody staining (∼20% reduction) may also point to the existence of two receptor populations on FHL 124 cells.
On the other hand, our observations could at least partially be explained by an FPR1 population on HEK 293-FPR1+ cells that is stained by the fluorescent ligand but not by the FACS antibody. Maybe the conformation of some of the FPR1 receptors is altered in HEK 293-FPR1+ cells, where the receptor protein is produced at an unphysiologically high level.
The third atypical property of FPR1 on FHL 124 cells is its high resistance to fMLF-induced internalization (detected with the PE-labeled FPR1 antibody). By contrast, FPR1 on neutrophils and in transfected HEK 293 cells was almost completely internalized. A very stable integration of the receptor in the cell membrane and a slow trafficking behavior may also explain why the short acting siRNA could not reduce FPR1 protein in FHL 124 cells despite a strong effect on mRNA.
An interesting question is how the properties of FPR1 in FHL 124 cells may be caused by the haplotypes (H-11/H-19) detected in our sequencing experiment (Table 3). It has been reported for FPR1 mutants expressed in Sf9 cell membranes that the V101L and the E346A mutation, which also occur in the H-19 and H-11 haplotype, respectively, reduce G protein coupling and decrease constitutive activity (39). The high affinity of fNLFNYK-Fl to the FHL 124 cell FPR1 and the ability to induce Ca2+ signaling, however, do not point to a G protein coupling defect of FPR1 in FHL 124 cells. Moreover, E346A was shown to increase the tendency of FPR1 to form oligomers (39). However, apart from the still unidentified 120-kDa band, which may be an FPR1 dimer, the main FPR1 band in Western blots with FHL 124 cell membranes was the 60-kDa monomer (Fig. 2B).
Despite the low Bmax value, high affinity direct binding (subnanomolar KD) by fNLFNYK-Fl and potent competition for fNLFNYK-Fl binding by a series of FPR1 ligands, but not by the irrelevant leukocyte chemoattractant MIP 1α (CCL3), confirms the expected pharmacologic profile for FPR1. In all three cell types unlabeled fNLFNYK had the highest affinity followed by peptide W and fMLF. The fMLF Ki value for FHL 124 cells is about 22 nm (pKi = 7.65, Table 2), which agrees well with 69 nm reported for fMLF displacing fNLFNYK-Fl from FPR1-transfected CHO cells (40). The high affinity of unlabeled fNLFNYK to FHL 124 cells (Ki ∼ 2 nm, pKi = 8.76) corresponds well to a Ki of 3 nm previously reported for human neutrophils (41).
Surprisingly, the fNLFNYK-Fl saturation binding to FHL 124 cells is monophasic only at low concentrations (0–20 nm) but suddenly increases at concentrations >20 nm. We can exclude the presence of the “classic” low affinity FPR, FPR2/ALX because it was not detectable on FHL 124 cells by a specific antibody (data not shown), and the FPR2/ALX agonist MMK-1 did not induce a calcium response. Maybe it is a second FPR1 receptor population with low affinity or simply a consequence of partial ligand degradation, which also can cause non-saturable specific binding (27). Due to the low signal intensity, this effect may be more strongly visible with FHL 124 cells compared with neutrophils or HEK 293-FPR1+ cells, where the signal-to-noise ratio is higher.
The initial part of the FHL 124 cell saturation curve revealed a KD value of 0.5 ± 0.2 nm, which agrees well with the fNLFNYK-Fl KD value of 0.6 ± 0.2 nm reported for human neutrophils (42) and with KD values reported for two non-hematopoietic FPR1-expressing (fibroblast) cell lines (0.22 and 0.67 nm) (8). However, we cannot explain why the fNLFNYK-FL KD value at the neutrophil FPR1 (3.2 ± 0.1 nm) is 5–10-fold higher in our hands than reported in the literature. Because we used a different isolation procedure, which may activate the neutrophils to a higher degree, neutrophil FPR1 may be uncoupled from G protein, resulting in a reduced agonist affinity. The KD value we determined for FPR1 on HEK 293-FPR1+ cells (2.7 ± 0.3 nm) is in good agreement with the reported KD value of 6 nm for CHO-FPR1 cells, which also was attributed to the presence of uncoupled FPR1 (40).
Interestingly, despite the low number of formyl peptide binding sites on FHL 124 cells, FPR1 agonists induced strong PTX-sensitive Ca2+ mobilization responses. This suggests that FPR1 on human lens epithelial cells efficiently stimulates Gαi/o. It is known from neutrophils that activation of as few as 50 receptors per cell can induce actin polymerization. Such a low receptor expression level has also been reported for fNLFNYK-Fl saturation binding with two non-hematopoietic (fibroblast) cell lines (600–1600/cell) (8). Maybe, a low receptor density with highly efficient G protein coupling is a common feature of “extra-hematopoietic” FPR1. The extremely efficient G protein coupling of the FHL 124 cell FPR1 was also demonstrated by the fact that in our shRNA experiments a 20–40% reduction in protein expression was reflected by a similar reduction of Ca2+ signaling. This suggests that virtually all receptor proteins contribute to fMLF-induced signaling and that the uncoupled “receptor reserve” is negligible.
In the past several years some evidence for non-hematopoietic FPR1 expression has accumulated, including ileum, hepatocytes, and thyroid follicular cells (7). Moreover, FPR1 is functionally expressed by fibroblasts and may support migration of these cells to sites of tissue injury (8). Functional expression of FPR1 has also been shown for human bone marrow mesenchymal stem cells, suggesting a potential role for migration and engraftment of such cells into injured tissues (9, 10) or for osteoblast differentiation (11). Moreover, FPR1 has been described to be involved in the regulation of acute-phase protein production by A549 lung cells (12) and human HepG2 hepatoma cells (13). Furthermore, FPR1 has been detected in several kinds of epithelial cells, e.g. Beas2B lung epithelial cells (14), SK-CO15 intestinal epithelial cells (15, 16), MKN-28 and AGS gastric epithelial cells (17), and human retinal pigment epithelial cells (18). Several papers (14, 16, 18) report that FPR1 agonists induced an FPR1 antagonist-sensitive acceleration of wound healing in an in vitro model using a scratch-injured cell monolayer. Mostly, FPR1 was investigated either in cancer cells or in non-cancerous tissues under in vitro conditions. Only in Ref. 11, were in vivo experiments performed with zebrafish and rabbits, suggesting a role of FPR1 in bone formation. The significance of our study is that to our knowledge it provides the first example of a non-hematopoietic setting for FPR1 expression linked directly to a biological phenotype in vivo, namely severe degeneration of lens structure in aging Fpr1−/− mice.4 Our characterization of the human FPR1 in human lens epithelial cells suggests that this property of Fpr1 may also be relevant to humans. How FPR1 functions to maintain the lens is not yet known. The lens continuously grows throughout life (43). Lens epithelial cells proliferate in the equatorial zones and elongate toward the anterior and posterior side of the lens, differentiating into lens fiber cells that form an onion-shaped structure. FPR1 could serve as a chemoattractant receptor guiding the elongation of lens epithelial cells in response to endogenous FPR1 ligands, such as mitochondrial peptides that have previously been reported to stimulate FPR1 (44). These peptides could be released from lens epithelial cells as they lose their organelles during differentiation to lens fiber cells. This function of FHL 124 cell FPR1 is supported by the observed fMLF-mediated activation of ERK 1/2 and p38 in FHL 124 cells. These MAP kinases were recently reported to be important for the regulation of neutrophil chemotaxis (36). It is intriguing to hypothesize that they may also regulate the directed growth of lens epithelial cells, albeit on a much longer time scale.
In conclusion, we have demonstrated that FPR1 is functionally expressed on human lens epithelial cells. The biological significance of FPR1 expression on these cells is suggested by the phenotype of severe cataracts in aging Fpr1−/− mice.4 Thus FPR1 appears to have an important biological role in innate immunity through its expression on phagocytic leukocytes and in development through expression directly in the lens.
Supplementary Material
This work was supported, in whole or in part, by the Division of Intramural Research of the NIAID, National Institutes of Health. This work was also supported by German Research Foundation Grant SCHN 1192/1-1 (to E. H. S.).

This article contains supplemental Figs. S1–S5.
J.-L. Gao et al., manuscript in preparation.
- FPR
- formyl peptide receptor
- fMLF
- formylmethionylleucylphenylalanine
- fNLFNYK
- N-formyl-Nle-Leu-Phe-Nle-Tyr-Lys
- fNLFNYK-Fl
- fluorescein-labeled N-formyl-Nle-Leu-Phe-Nle-Tyr-Lys
- RT
- room temperature
- PTX
- pertussis toxin
- HBSS
- Hanks' balanced salt solution
- PE
- phycoerythrin
- PMN
- polymorphonuclear leukocytes
- PNGase F
- peptide N-glycosidase F
- TBST
- Tris-buffered saline-Tween
- MAP
- mitogen-activated protein
- qPCR
- quantitative PCR.
REFERENCES
- 1. Ye R. D., Boulay F., Wang J. M., Dahlgren C., Gerard C., Parmentier M., Serhan C. N., Murphy P. M. (2009) International Union of Basic and Clinical Pharmacology. LXXIII. Nomenclature for the formyl peptide receptor (FPR) family. Pharmacol. Rev. 61, 119–161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Hartt J. K., Barish G., Murphy P. M., Gao J. L. (1999) N-Formyl peptides induce two distinct concentration optima for mouse neutrophil chemotaxis by differential interaction with two N-formyl peptide receptor (FPR) subtypes. Molecular characterization of FPR2, a second mouse neutrophil FPR. J. Exp. Med. 190, 741–747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Huang J., Chen K., Gong W., Zhou Y., Le Y., Bian X., Wang J. M. (2008) Receptor “hijacking” by malignant glioma cells. A tactic for tumor progression. Cancer Lett. 267, 254–261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Huang J., Chen K., Chen J., Gong W., Dunlop N. M., Howard O. M., Gao Y., Bian X. W., Wang J. M. (2010) The G-protein-coupled formyl peptide receptor FPR confers a more invasive phenotype on human glioblastoma cells. Br. J. Cancer 102, 1052–1060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Crowell R. E., Van Epps D. E., Reed W. P. (1989) Responses of isolated pulmonary arteries to synthetic peptide F-Met-Leu-Phe. Am. J. Physiol. 257, H107–H112 [DOI] [PubMed] [Google Scholar]
- 6. Keitoku M., Kohzuki M., Katoh H., Funakoshi M., Suzuki S., Takeuchi M., Karibe A., Horiguchi S., Watanabe J., Satoh S., Nose M., Abe K., Okayama H., Shirato K. (1997) FMLP actions and its binding sites in isolated human coronary arteries. J. Mol. Cell. Cardiol. 29, 881–894 [DOI] [PubMed] [Google Scholar]
- 7. Becker E. L., Forouhar F. A., Grunnet M. L., Boulay F., Tardif M., Bormann B. J., Sodja D., Ye R. D., Woska J. R., Jr., Murphy P. M. (1998) Broad immunocytochemical localization of the formyl peptide receptor in human organs, tissues, and cells. Cell Tissue Res. 292, 129–135 [DOI] [PubMed] [Google Scholar]
- 8. VanCompernolle S. E., Clark K. L., Rummel K. A., Todd S. C. (2003) Expression and function of formyl peptide receptors on human fibroblast cells. J. Immunol. 171, 2050–2056 [DOI] [PubMed] [Google Scholar]
- 9. Viswanathan A., Painter R. G., Lanson N. A., Jr., Wang G. (2007) Functional expression of N-formyl peptide receptors in human bone marrow-derived mesenchymal stem cells. Stem Cells 25, 1263–1269 [DOI] [PubMed] [Google Scholar]
- 10. Kim M. K., Min do S., Park Y. J., Kim J. H., Ryu S. H., Bae Y. S. (2007) Expression and functional role of formyl peptide receptor in human bone marrow-derived mesenchymal stem cells. FEBS Lett. 581, 1917–1922 [DOI] [PubMed] [Google Scholar]
- 11. Shin M. K., Jang Y. H., Yoo H. J., Kang D. W., Park M. H., Kim M. K., Song J. H., Kim S. D., Min G., You H. K., Choi K. Y., Bae Y. S., Min do S. (2011) N-Formyl-methionyl-leucyl-phenylalanine (fMLP) promotes osteoblast differentiation via the N-formyl peptide receptor 1-mediated signaling pathway in human mesenchymal stem cells from bone marrow. J. Biol. Chem. 286, 17133–17143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Rescher U., Danielczyk A., Markoff A., Gerke V. (2002) Functional activation of the formyl peptide receptor by a new endogenous ligand in human lung A549 cells. J. Immunol. 169, 1500–1504 [DOI] [PubMed] [Google Scholar]
- 13. McCoy R., Haviland D. L., Molmenti E. P., Ziambaras T., Wetsel R. A., Perlmutter D. H. (1995) N-Formyl peptide and complement C5a receptors are expressed in liver cells and mediate hepatic acute phase gene regulation. J. Exp. Med. 182, 207–217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Shao G., Julian M. W., Bao S., McCullers M. K., Lai J. P., Knoell D. L., Crouser E. D. (2011) Formyl peptide receptor ligands promote wound closure in lung epithelial cells. Am. J. Respir. Cell Mol. Biol. 44, 264–269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Babbin B. A., Lee W. Y., Parkos C. A., Winfree L. M., Akyildiz A., Perretti M., Nusrat A. (2006) Annexin I regulates SKCO-15 cell invasion by signaling through formyl peptide receptors. J. Biol. Chem. 281, 19588–19599 [DOI] [PubMed] [Google Scholar]
- 16. Babbin B. A., Jesaitis A. J., Ivanov A. I., Kelly D., Laukoetter M., Nava P., Parkos C. A., Nusrat A. (2007) Formyl peptide receptor-1 activation enhances intestinal epithelial cell restitution through phosphatidylinositol 3-kinase-dependent activation of Rac1 and Cdc42. J. Immunol. 179, 8112–8121 [DOI] [PubMed] [Google Scholar]
- 17. de Paulis A., Prevete N., Rossi F. W., Rivellese F., Salerno F., Delfino G., Liccardo B., Avilla E., Montuori N., Mascolo M., Staibano S., Melillo R. M., D'Argenio G., Ricci V., Romano M., Marone G. (2009) Helicobacter pylori Hp(2–20) promotes migration and proliferation of gastric epithelial cells by interacting with formyl peptide receptors in vitro and accelerates gastric mucosal healing in vivo. J. Immunol. 183, 3761–3769 [DOI] [PubMed] [Google Scholar]
- 18. Zhang X. G., Hui Y. N., Huang X. F., Du H. J., Zhou J., Ma J. X. (2011) Activation of formyl peptide receptor-1 enhances restitution of human retinal pigment epithelial cell monolayer under electric fields. Invest. Ophthalmol. Vis. Sci. 52, 3160–3165 [DOI] [PubMed] [Google Scholar]
- 19. Rivière S., Challet L., Fluegge D., Spehr M., Rodriguez I. (2009) Formyl peptide receptor-like proteins are a novel family of vomeronasal chemosensors. Nature 459, 574–577 [DOI] [PubMed] [Google Scholar]
- 20. Yang H., Shi P. (2010) Molecular and evolutionary analyses of formyl peptide receptors suggest the absence of VNO-specific FPRs in primates. J. Genet. Genomics 37, 771–778 [DOI] [PubMed] [Google Scholar]
- 21. Ibaraki N., Chen S. C., Lin L. R., Okamoto H., Pipas J. M., Reddy V. N. (1998) Human lens epithelial cell line. Exp. Eye Res. 67, 577–585 [DOI] [PubMed] [Google Scholar]
- 22. Reddan J. R., Lindemann C. B., Hitt A. L., Bagchi M., Raphtis E. M., Pena J. T., Dziedzic D. C. (1999) Invest. Ophthalmol. Vis. Sci. 40, S970 (Abstr. 5110) [Google Scholar]
- 23. Hartt J. K., Liang T., Sahagun-Ruiz A., Wang J. M., Gao J. L., Murphy P. M. (2000) The HIV-1 cell entry inhibitor T-20 potently chemoattracts neutrophils by specifically activating the N-formyl peptide receptor. Biochem. Biophys. Res. Commun. 272, 699–704 [DOI] [PubMed] [Google Scholar]
- 24. McDermott D. H., De Ravin S. S., Jun H. S., Liu Q., Priel D. A., Noel P., Takemoto C. M., Ojode T., Paul S. M., Dunsmore K. P., Hilligoss D., Marquesen M., Ulrick J., Kuhns D. B., Chou J. Y., Malech H. L., Murphy P. M. (2010) Severe congenital neutropenia resulting from G6PC3 deficiency with increased neutrophil CXCR4 expression and myelokathexis. Blood 116, 2793–2802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Riesselman M., Miettinen H. M., Gripentrog J. M., Lord C. I., Mumey B., Dratz E. A., Stie J., Taylor R. M., Jesaitis A. J. (2007) C-terminal tail phosphorylation of N-formyl peptide receptor. Differential recognition of two neutrophil chemoattractant receptors by monoclonal antibodies NFPR1 and NFPR2. J. Immunol. 179, 2520–2531 [DOI] [PubMed] [Google Scholar]
- 26. Fay S. P., Posner R. G., Swann W. N., Sklar L. A. (1991) Real-time analysis of the assembly of ligand, receptor, and G protein by quantitative fluorescence flow cytometry. Biochemistry 30, 5066–5075 [DOI] [PubMed] [Google Scholar]
- 27. Harvath L., Aksamit R. R., Cunningham R. E. (1999) Assay for chemoattractant binding. Methods Mol. Biol. 115, 299–308 [DOI] [PubMed] [Google Scholar]
- 28. Malech H. L., Gardner J. P., Heiman D. F., Rosenzweig S. A. (1985) Asparagine-linked oligosaccharides on formyl peptide chemotactic receptors of human phagocytic cells. J. Biol. Chem. 260, 2509–2514 [PubMed] [Google Scholar]
- 29. Wenzel-Seifert K., Seifert R. (2003) Critical role of N-terminal N-glycosylation for proper folding of the human formyl peptide receptor. Biochem. Biophys. Res. Commun. 301, 693–698 [DOI] [PubMed] [Google Scholar]
- 30. Jesaitis A. J., Erickson R. W., Klotz K. N., Bommakanti R. K., Siemsen D. W. (1993) Functional molecular complexes of human N-formyl chemoattractant receptors and actin. J. Immunol. 151, 5653–5665 [PubMed] [Google Scholar]
- 31. Neubig R. R., Krodel E. K., Boyd N. D., Cohen J. B. (1979) Acetylcholine and local anesthetic binding to Torpedo nicotinic postsynaptic membranes after removal of nonreceptor peptides. Proc. Natl. Acad. Sci. U.S.A. 76, 690–694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Perez H. D., Kelly E., Elfman F., Armitage G., Winkler J. (1991) Defective polymorphonuclear leukocyte formyl peptide receptor(s) in juvenile periodontitis. J. Clin. Invest. 87, 971–976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Remes J. J., Petäjä-Repo U. E., Rajaniemi H. J. (1991) Rat and human neutrophil N-formyl-peptide chemotactic receptors. Species difference in the glycosylation of similar 35–38 kDa polypeptide cores. Biochem. J. 277, 67–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Romero-Calvo I., Ocón B., Martínez-Moya P., Suárez M. D., Zarzuelo A., Martínez-Augustin O., de Medina F. S. (2010) Reversible Ponceau staining as a loading control alternative to actin in Western blots. Anal. Biochem. 401, 318–320 [DOI] [PubMed] [Google Scholar]
- 35. Kim S. D., Kim J. M., Jo S. H., Lee H. Y., Lee S. Y., Shim J. W., Seo S. K., Yun J., Bae Y. S. (2009) Functional expression of formyl peptide receptor family in human NK cells. J. Immunol. 183, 5511–5517 [DOI] [PubMed] [Google Scholar]
- 36. Liu X., Ma B., Malik A. B., Tang H., Yang T., Sun B., Wang G., Minshall R. D., Li Y., Zhao Y., Ye R. D., Xu J. (2012) Bidirectional regulation of neutrophil migration by mitogen-activated protein kinases. Nat. Immunol. 13, 457–464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Rane M. J., Carrithers S. L., Arthur J. M., Klein J. B., McLeish K. R. (1997) Formyl peptide receptors are coupled to multiple mitogen-activated protein kinase cascades by distinct signal transduction pathways: role in activation of reduced nicotinamide adenine dinucleotide oxidase. J. Immunol. 159, 5070–5078 [PubMed] [Google Scholar]
- 38. Sahagun-Ruiz A., Colla J. S., Juhn J., Gao J. L., Murphy P. M., McDermott D. H. (2001) Contrasting evolution of the human leukocyte N-formyl peptide receptor subtypes FPR and FPRL1R. Genes Immun. 2, 335–342 [DOI] [PubMed] [Google Scholar]
- 39. Wenzel-Seifert K., Seifert R. (2003) Functional differences between human formyl peptide receptor isoforms 26, 98, and G6. Naunyn Schmiedebergs Arch. Pharmacol. 367, 509–515 [DOI] [PubMed] [Google Scholar]
- 40. Miettinen H. M., Mills J. S., Gripentrog J. M., Dratz E. A., Granger B. L., Jesaitis A. J. (1997) The ligand binding site of the formyl peptide receptor maps in the transmembrane region. J. Immunol. 159, 4045–4054 [PubMed] [Google Scholar]
- 41. Sklar L. A., Sayre J., McNeil V. M., Finney D. A. (1985) Competitive binding kinetics in ligand receptor competitor systems. Rate parameters for unlabeled ligands for the formyl peptide receptor. Mol. Pharmacol. 28, 323–330 [PubMed] [Google Scholar]
- 42. Sklar L. A., Finney D. A., Oades Z. G., Jesaitis A. J., Painter R. G., Cochrane C. G. (1984) The dynamics of ligand-receptor interactions. Real-time analyses of association, dissociation, and internalization of an N-formyl peptide and its receptors on the human neutrophil. J. Biol. Chem. 259, 5661–5669 [PubMed] [Google Scholar]
- 43. Augusteyn R. C. (2007) Growth of the human eye lens. Mol. Vis. 13, 252–257 [PMC free article] [PubMed] [Google Scholar]
- 44. Rabiet M. J., Huet E., Boulay F. (2005) Human mitochondria-derived N-formylated peptides are novel agonists equally active on FPR and FPRL1, while Listeria monocytogenes-derived peptides preferentially activate FPR. Eur. J. Immunol. 35, 2486–2495 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.