Abstract
Genetically engineered mouse models (GEMM) have made major contributions to a molecular understanding of several adult cancers and these results are increasingly being translated into the pre-clinical setting where GEMM will very likely make a major impact on the development of targeted therapeutics in the near future. The relationship of pediatric cancers to altered developmental programs, and their genetic simplicity relative to adult cancers provides unique opportunities for the application of new advances in GEMM technology. In neuroblastoma the well-characterized TH-MYCN GEMM is increasingly used for a variety of molecular-genetic, developmental and pre-clinical therapeutics applications. We discuss: the present and historical application of GEMM to neuroblastoma research, future opportunities, and relevant targets suitable for new GEMM strategies in neuroblastoma. We review the potential of these models to contribute both to an understanding of the developmental nature of neuroblastoma and to improved therapy for this disease.
Keywords: MYCN, Neuroblastoma, Mouse models
1. Introduction
Recent advances in genetically engineered mouse modeling (GEMM) technology have provided unique opportunities to address fundamental questions about the developmental origin, molecular pathology and therapeutic susceptibility of pediatric cancers. As expertise grows in understanding of neural crest development and specification, and in the function of oncogenic drivers of the childhood cancer neuroblastoma, GEMM approaches will provide the means to validate these drivers and to generate important future models. This article reviews current models and GEMM strategies aimed at elucidating genes that alter neural crest fate specification and neuroblast function, and that thereby contribute to the formation of NB tumors.
In general, GEMM are most commonly developed to provide mechanistic data elucidating causal roles for particular genes in developmental fate specification or oncogenesis. Developmentally focused approaches examine whether an observed phenotype is associated with tissue- and/or time-specific mis-expression of a gene that regulates differentiation program of the target tissues. For this reason, targeted knock-in strategies incorporating some form of imaging, and the ability to regulate expression of the gene of interest, are most commonly used. The ability to control expression of the target gene in a time-dependent manner and to visualize expression are equally critical to the elucidation of molecular oncogenic mechanisms and to efforts to develop novel therapeutics, respectively.
Cancer focused GEMM strategies seek to determine more specifically: (1) whether altered expression of a cancer-associated gene or mutation can initiate and sustain tumorigenesis, (2) what clinical phenotype if any, is recapitulated by the model, (3) whether the oncoprotein target itself or changes in molecular signaling pathways induced by its mis-expression represent therapeutic targets, and (4) whether secondary genomic alterations evolve and are required for oncogenesis, and 5, whether these validated genes also be therapeutic targets. In neuroblastoma in particular, the sequence of genetic events underlying normal maturation of neural crest precursors is now understood in some detail. Manipulated expression of such genes (PHOX2B, Sox10, Hand2, and TH) promises to provide great insights into where these initiating defects occur, and how they impact the ultimate biology of neural crest-derived malignancy.
Given that many, if not most, pediatric (embryonal) cancers represent aberrations of normal developmental programs, GEMM approaches are ideal in that they combine elements of both controlled and visualized expression. In pediatric cancer the question of whether a particular gene is oncogenic often relates to its role in development, and this by nature demands the ability to manipulate gene expression, ideally in a fully reversible manner, and requires the capacity to confirm gene expression using a surrogate marker. Additional imaging techniques that serve as rapid and practical screens for presence of tumor, and are capable of quantitating volume or bulk responses in the setting of a pre-clinical drug trial are equally critical. Appropriate credentialing of a murine model for therapeutic use is vital and challenging. Below, we review some of the technologies and approaches available in these areas.
2. How murine genetic models could contribute to a complete understanding of NB
Major differences exist between pediatric and adult cancers in the genesis of the most common non-germline cancers. Spontaneous adult malignancies largely result from stochastic accumulation of somatic genetic mutations and epigenetic changes over a relatively long timescale [1–3]. In contrast, transformation in many pediatric cancers is initiated by comparably few “driver” mutations that occur during a short window of programmed differentiation [4]. Clones that acquire a survival advantage in this manner can in theory retain significant dependence on expression of the driver mutation and this will continue to impact the ultimate biologic phenotype and clinical behavior of derivative tumors. The clinical consequence is that relatively few mutations might determine the distinct clinical behavior of clinically definable NB subgroups. This is an ideal scenario for murine modeling strategies, and highlights three areas in which GEMM can significantly contribute to our understanding and treatment of NB: (1) manipulated expression of disease-associated mutations in mice should eventually result in GEMM that model all distinct subtypes of NB enabling development of tailored therapy, (2) time-dependent, reversible expression of neural crest associated genes in GEMM could localize the origin of distinct NB subtypes to fate-change alterations within specific phases of neural crest development program, and (3) collectively, results from 1 and 2 should identify and validate candidate stem, progenitor and tumor-initiating cell populations, leading to new therapeutic targets.
3. Existing GEMM models of NB–TH-MYCN
The crucial role of MYC genes in development of the central and peripheral nervous systems and their derived cancers has been recognized since the identification of MYC and MYCN as cellular homologues of v-MYC (avian myelocytomatosis inducing virus derived oncogene) [5–10]. Initial attempts using mice to model the role of MYC genes in specification of brain and neural crest derived structures used targeted knock-in approaches to ablate endogenous expression of MYC or MYCN, resulting in hypoplastic development of the brain and other tissues [11–13]. Subsequent attempts utilizing Cre-recombinase driven systems recapitulated the microcephaly associated with Feingold syndrome (in which various deletions of MYCN lead to haploinsufficiency) [14,15]. In these mouse-based studies, a critical role for MYC genes in embryonal fate-specification and development of neuronal precursor cells was first established. Significantly, MYCN gain could reciprocally compensate for defects arising from targeted deletion of MYC, implying that the high homology of these two genes relates to a critical but overlapping role in driving embryonal development [16]. Finally, excellent GEMM approaches now are available to study the role of micro-RNAs (regulable RNAi mice [17]) or epigenetic modifications that either target MYCN or are MYCN-targets can be modeled and functionally established. For example, the oncogenic miR-17-92 cluster is frequently overexpressed or amplified and is a transcriptional target of both MYC and MYCN [18–20].
The recognition that amplification of the MYCN gene was a common event in NB, medulloblastoma, rhabdomyosarcoma, retinoblastoma and other pediatric cancers, and data implying a significant role for MYCN in the development of the neural crest, prompted the initial modeling approach leading to construction of the TH-MYCN model of high-risk, MYCN-amplified NB in 1997 [21]. This GEMM is currently the only well-characterized native model of NB available and is extensively used within the NB research community. Complementary GEMM approaches have been undertaken and are in progress (personal communications) [22,23], but as yet no additional high-penetrance NB models have emerged. TH-MYCN was constructed using a first-generation, “transgenic” approach involving introduction of exogenous DNA into the nucleus of fertilized murine oocytes, resulting in random integration of the transgenic construct into genomic DNA. In this case, expression of the human MYCN cDNA was targeted to the neural crest using a derivatized construct in which stabilized expression of rat tyrosine hydroxylase (TH), a relatively weak promotor [24], was achieved by incorporating a rabbit beta globin intron element [21]. TH expression in mice is confined to relatively differentiated, PHOX2B positive neuronal precursors of sympathoadrenal origin, a potential cell-of-origin for the initiation of NB [25]. In addition to investigating the specific role of MYCN in driving NB tumorigenesis, this modeling approach also tested the general hypothesis that aberrant stimulation of proliferation, blockade of apoptosis or impaired differentiation within this target cell population could initiate and maintain NB.
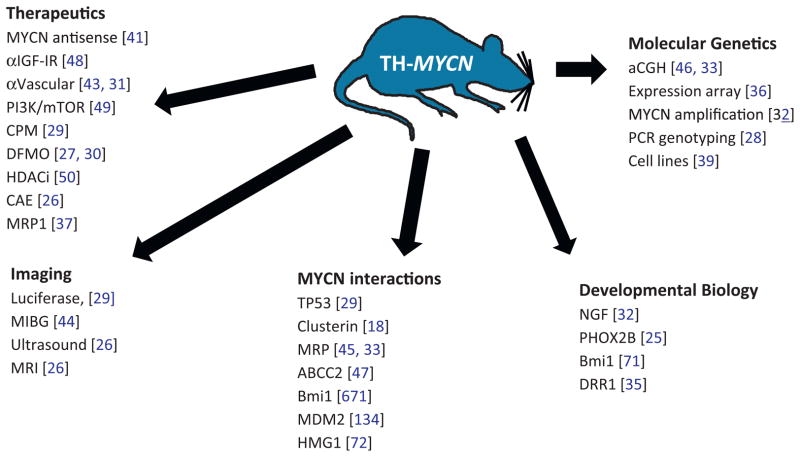
The diverse research originating from the use of this model illustrates the power of GEMM approaches to elucidate molecular genetics and therapeutic sensitivity in pediatric cancers (Fig. 1). TH-MYCN recapitulates most major genetic and clinical aspects of high-risk MYCN-amplified disease, and the model has been widely used for a multitude of basic biology and applied therapeutics studies [18,25–46]. Imaging characteristics are quite similar to that of human disease using variety of modalities, including ultrasound, luciferase (in E2F1-Luc/TH-MYCN double transgenic animals), MRI, PET and 131I-MIBG [26,29,44]. Of most relevance, TH-MYCN tumors arise spontaneously and to high penetrance within their native tissue of origin (sympathetic paraspinal, celiac and periadrenal sympathetic ganglia), replicate many major genetic changes of human MYCN-amplified disease [33,39,46], including amplification of the MYCN transgene [32,33], retain native tumor–stromal interactions and vasculature, and progress in a fashion reminiscent of MYCN-amplified primary human tumors, such that murine staging systems developed for TH-MYCN tumors relate very closely to INSS (surgical) staging regimens based on gross pathology and imaging data [26,29,31]. That they are to some extent “addicted” to continuous expression of MYCN is implied by the ability of antisense RNA to MYCN to block the formation and progression of tumors in the model [41]. Micrometastases are present in the model although do not accurately represent the clinical spectrum seen in clinical disease [21]. Although the specific cell of origin for tumors in this model is still undefined, as mentioned above, tumors appear to arise from PHOX2B, TH positive cells localized to hyperproliferative microfoci within periadrenal and paraspinal sympathetic ganglia [25].
Fig. 1.
Relevant publications arising from the TH-MYCN mouse model. TH-MYCN has had numerous uses in multiple areas of NB research, and some key publications are grouped above. The main areas of impact include analysis of secondary genomic changes, elucidation and evaluation of MYCN modifiers, developmental studies on the role of MYCN in neural crest development, and pre-clinical studies assessing the efficacy of emerging therapeutics in treatment of high-risk MYCN-driven NB. [×], reference number, CAE, cyclophosphamide, adriamycin, etoposide, CPM, cyclophosphamide.
These lesions express high levels of MYCN, fail to undergo apoptosis in response to NGF withdrawal, and generate tumors characterized by PHOX2B positive neural derivatives with high expression of MYCN protein [32]. Several useful cell lines have been derived from the model [39]. A variety of therapeutic studies have been conducted (discussed below) which collectively reflect therapeutic sensitivity to conventional single-agent chemotherapeutics or combinations commonly in use for treatment of human NB (cyclophosphamide, platinum compounds, adriamycin, VP16, irinotecan, and vincristine), and in some cases, acquisition of therapeutic resistance to these agents [26,29,37]. TH-MYCN is increasingly used for validation of novel small-molecule therapeutics (NDGA, DFMO, TNP470, HDAC, and reversan) either alone or in combination with retrieval chemotherapy regimens [27,30,31,37,43,47–50].
4. Modeling NB genetics – MYCN modifiers
One of the obvious strengths of GEMM is in the identification of genetic modifiers through forward genetic screens. Cellular expression of MYCN induces a potent oncogenic stimulus that requires significant remodeling of metabolism, cell cycle checkpoint function, apoptotic potential and DNA damage repair mechanisms [51–53]. A critical question for understanding the role of MYCN as a target is how overexpression of MYCN, and MYC genes in general, is tolerated by cells [54–56]. The TH-MYCN model shows significant strain-dependent differences in the ability of MYCN to drive tumorigenesis, with high versus absent penetrance in 129sv/Jx1 and FvBN/N strains, respectively [21,33]. Ongoing efforts are focused at determining discrete secondary genomic changes in modifier genes (single nucleotide polymorphisms, expression changes, epigenetic alterations) that co-segregate with susceptibility to neuroblastoma in this model [36,42]. Additional efforts are directed at chemical (ENU [57]) and insertional (transposon [58]) mediated mutagenesis of the TH-MYCN and other GEMM models as strategies to identify modifier loci, and will undoubtedly identify additional targets critical to the function of MYCN and other initiator genes in NB.
5. Modeling MYCN-targets and cooperating genes
Simple genetic intercrosses (Fig. 2) provide an additional, candidate-driven means to assess cooperativity between an oncogene and additional gene targets in the genesis of a particular tumor. This approach can also help to clarify the oncogenically relevant targets, which in the case of MYC is particularly useful since the gene-set regulated by this enigmatic family of transcription factors encompasses thousands of targets regulating ribosomal and protein biosynthesis, metabolism and proliferation [59–63]. Adding to the confusion, MYC genes regulate a variety of micro-RNA and epigenetic targets that regulate global stability of chromatin [19,64–67]. Genetic intercrosses of MYC-driven models against mice with modulated expression of MYC interacting genes such as p53, MDM2, clusterin, HMG1 and Bmi1 have established definitive roles for these genes in modulation of MYC-driven oncogenesis, and are leading to the identification of new druggable targets and therapy approaches for MYC blockade [18,29,34,68–72]. Deletion or silencing of caspase-8 occurs frequently in high-risk disease [73–76], and the potential for this mutation to block MYCN-driven tumorigenesis in NB is of great interest. One less prominent feature of the TH-MYCN model that has escaped modeling is the relatively reduced propensity of these mice to develop bulky metastasis. Whether the lack of bulky metastases represents an artifact of murine physiology or a definitive difference between TH-MYCN tumorigenesis and human counterparts is as yet unclear. Nevertheless, rational candidate genes that might enhance metastasis in the TH-MYCN GEMM include TrkB, nm23, c-Myb, slug1, DKK1, NCAM, integrins and selectins [18,77–81]. Specific micro-RNAs targeted by MYC genes mediate critical oncogenic functions, as revealed in GEMM in which miR’s were conditionally deleted. Homozygous deletion of the miR-17 complex in mice revealed an absolute requirement for expression of miR-19a and -19b in lymphoma driven by MYC [82]. Mechanistically, sustained overexpression of these miR’s suppressed MYC-driven apoptosis and accelerated tumorigenesis; and discrete secondary miR targets were identified that mediated the prosurvival function of this miR cluster, which could represent new therapeutic targets.
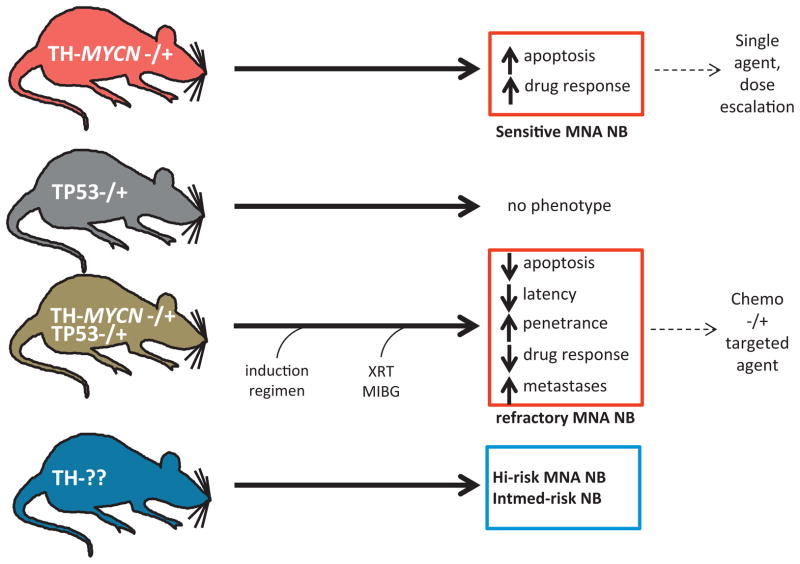
Fig. 2.
Utilizing TH-driven models of NB to study MYCN-driven oncogenesis and modifier genes. Simple genetic crosses elucidate the functional interdependence of two genes in mice. One example involves the contribution of p53 to the genesis or progression of MYCN-driven tumors. In the crosses above, haploinsufficiency of p53 accelerates NB in the TH-MYCN model. These tumors display defective apoptosis and are resistant to cyclophosphamide. This approach can establish whether specific MYCN targets or cooperating genes are suitable for indirect therapeutic inhibition of MYCN in NB.
6. Modeling the role of p53 pathway
Trp53 is important in NB, since MYC-driven cancers are characterized by coordinate induction of proliferation and apoptosis, with both ARF and p53 pathways implicated [68,83–86]. Concurrent deletion of p53 enhances tumorigenesis by MYC in a wide range of tumor models [87,88]. The advent of GEMM with reactivatable p53 transgenes has provided an additional opportunity to study the impact of p53 modulation in the context of MYCN misexpression. Mutations in p53 occur commonly in adult malignancies driven by MYC and p53 pathway deficiencies (mutations in p53, MDM2, INK4A/ARF). Perhaps uniquely in NB, these mutations are not present at diagnosis, are acquired in 50% of heavily pre-treated NB patients, and herald the emergence of chemoresistance. TH-MYCN mice doubly transgenic for MYCN and an inactive allele of p53 (MYCN-+/p53-+) display enhanced penetrance and shortened latency to NB development [29]. Interestingly, these mice exhibit resistance to cyclophosphamide, a first-line chemotherapeutic utilized in most NB high-risk induction regimens, and reduced apoptosis [29]. This and derivative models should ultimately serve as platforms to dissect the role of the p53 pathway in chemoresistance to conventional therapies and in studies of novel p53-targeted agents.
7. Beyond MYCN modeling – ALK and PHOX2B
That relatively few mutations drive the majority of NB cases is clear from recent genome-wide studies. MYCN is a definitive marker of 40% of high-risk NB, however the majority of high-risk cases are not defined by known single gene alterations [89]. The recognition that distinct ALK mutations are differentially present within the germline and somatic tumor tissue of NB patients, and underlie the majority of hereditary NB, led to the elegant validation of Knudson’s two-hit hypothesis (describing sequential loss of suppressor oncogene alleles in cancer) in NB [90–94]. A minor fraction of remaining cases also relate to loss of function mutations in PHOX2B [95]. Both genes probably play critical roles in the early specification of neural crest so that elegant GEMM strategies will be required to elucidate the way in which these genes function during early development. To date, targeted deletion in GEMM indicates no obvious anatomic phenotype in ALK null mice, although such mice appear to have neurological performance deficiencies [96].
Homozygous PHOX2B knockout is embryonic lethal and animals lack peripheral sympathetic neural derivatives, whereas heterozygous deficiency is associated with hypoventilation [97,98]. No models have yet been reported assessing overexpression of either gene in the neural crest. Various outcomes are conceivable for each, such as impaired development of neural crest, loss of sympathetic neuronal cell lineages or tumorigenesis. However, it remains to be seen whether overexpression of these genes in either wild-type or mutated form could induce NB in the absence of additional mutations, such as a second ALK mutation or amplification of MYCN, is unclear. The differential distribution of such mutations (R1275Q and F1174L) in germline versus somatic tissue, and in MYCN-independent versus MYCN-amplified NB, respectively, implies that these mutations may play distinct roles in the initiation and maintenance of tumors, during early neurogenesis [92,93,99]. It therefore remains to be seen whether targeted overexpression of either mutation to neural crest induces NB of distinct phenotypes.
Accurate modeling of the role of ALK mutations will require sophisticated combinatorial approaches to GEMM. For example, ALK-R1275Q is present almost exclusively as a germline mutation in patients with hereditary NB and is a weak kinase (perhaps explaining how this mutation is tolerated in the germline), whereas ALK-F1174L is among the most potent ALK mutations yet identified and localizes primarily to tumor tissue, with some data suggesting preferential association with high-risk MYCN-amplified tumors. This supports a sequential loss model, and prompts several questions. Is ALK-R1275Q functionally distinct and does it play a discrete role during neural crest maturation, or is its tolerance in the germline simply a function of the level of expressed wild-type ALK kinase activity? Is overexpression of ALK alone capable of driving tumorigenesis? Do distinct ALK mutations have equivalent capacity to drive tumorigenesis? Does this require abnormal expression of MYCN or other alterations? A simple approach that would accurately model sequential loss could be envisioned. Mice carrying a constitutively driven, heritable transgenic TH-ALK-R1275Q construct would be bred against mice carrying an inducible, knock-in allele of ALK-F1174L permitting temporally manipulable induction of ALK-F1174L expression. Finally, for the development of improved ALK-targeted small-molecule inhibitors, basic questions relate to how these mutations transduce signals in vivo. Of course, such models would also be vital to assess the efficacy of novel ALK-targeted therapeutics in NB.
PHOX2B is a homeodomain-containing transcription factor that regulates the commitment and differentiation of sympathoadrenergic precursor cells, together with ALK and dHAND2 [97,100,101]. Loss-of-function mutations in the homeodomain of PHOX2B occur in a minority (<10%) of hereditary NBs [95,102], and the incidence of these mutations in sporadic tumors is unclear but appears to be very uncommon, although most NB tumors appear to express and are composed of rapidly proliferating PHOX2B positive neuroblasts (making PHOX2B expression analysis an ideal assay for detection of minimal residual disease) [103–106]. Enforced expression of wild-type PHOX2B and patient-derived loss-of-function mutations appears to drive proliferation but not differentiation in vitro, implying a suppressive role in NB. As discussed above, murine modeling demonstrates that PHOX2B deletion is associated with failure to develop neural crest derivatives and is embryonically lethal. It would therefore seem that the role of PHOX2B is in early specification and differentiation of sympathetic derivatives and in NB tumors its loss is growth-promoting (methylation and silencing of PHOX2B also occurs with some prevalence in NB). GEMM approaches are therefore of great interest to clarify the role of this gene in neurodevelopment and NB tumorigenesis. Alternatively, PHOX2B may serve as an ideal promotor for neural-crest specific overexpression strategies seeking to define the role of additional mutations in future NB GEMMs.
8. Beyond MYCN modeling – additional targets
Since aberrant control of MYCN and ALK accounts for only a minority of high-risk patients, additional models are needed both for MYCN-independent high-risk disease, and for intermediate-risk patients. Genome-wide-association-studies are rapidly defining the spectrum of nucleotide polymorphisms, copy number variations, expression changes and epigenetic events that account for a predisposition to both sporadic and familial NB and which appear to selectively associate with discrete tumor phenotypes. Recent sequencing of NB genomes, transcriptomes or expressed transcripts (RNA-seq), combined with SNP and aCGH studies confirm that aberrations in MYCN (20%), ALK (10–15%), LMO1 (12%) are generally the only major single-gene alterations that occur with any frequency in NB, in addition to four additional loci at which copy number variations are found (1p36, 3p, 11q, and 17q). Additional uncommonly mutated genes with sporadic SNP or CNV associations in NB and associated with high-risk disease included PTPN11, NF1, FLJ22536, and BARD1 [107–112]. Strikingly however, the relative frequency and number of mutated genes identified through such approaches has been surprisingly low, implying that NB genomes are relatively simple, or that key drivers are somehow being overlooked systematically. These additional loci identified to date in sporadic NB are all gain- or loss-of-function mutations amenable to GEM modeling approaches.
RAS-MAPK signaling abnormalities probably also play a prominent role in NB, as somatic mutations and silencing of multiple pathway components have been described [113]. Activating mutations in PTPN11 occur in 3% of NB and are frequently associated with Noonan syndrome, an autosomal dominant hereditary dysmorphic syndrome associated with mutations in the NF1-RAS pathway, and a propensity to malignancy, and uncommonly NB. SHP-2, the protein product of PTPN11 is a tyrosine phosphatase that augments signaling through the RAS-MEK pathway [114]. Somatic NF1 mutations occur infrequently in NB [108]. Decreased expression of NF1 (typically in the absence of mutational inactivation) is strongly correlated with poor outcome [108]. MAPK signaling negatively regulates the transcription factor ZNF423 [108], critical to the ability of neural crest precursors and NB tumor cell lines to differentiate in response to retinoids. Patients with aggressive tumors characterized by low NF1 expression have hyperactive RAS-MEK signaling, a potential biomarker for retinoid resistance. Interestingly, treatment of such cell lines with MAPK pathway (MEK) inhibitors (now in early clinical trials) may reactivate ZNF423, enabling retinoid responsiveness.
Rare germline polymorphisms of BARD1 and LMO1, detected in recent GWAS [109–111,115], are associated with high-risk disease and occur, in sporadic tumors at both loci in recent sequencing analyses of NB genomes. LMO1 is a (LIM domain) transcriptional regulator that exhibits both germline SNP and somatic copy number gain in advanced NB [109,112]. LIM proteins are implicated in predisposition to B-ALL and lymphoma, and are expressed in neural tissues. Enforced expression of LMO1 drives proliferation in NB cell lines. Murine modeling of LMO2 translocations generate T-ALL. BARD1 (BRCA1 associated ring domain-1 protein) binds BRCA1, is mutated in breast cancer and fails to dimerize with BRCA1-mutated proteins in that disease. This complex is likely to function as an ubiquitin ligase [116]. Finally, the recent finding that the high-risk NB patient cohort lacking MYCN gene-amplification is characterized by frequent upregulation of MYCC transcriptional targets provides a powerful rationale for murine modeling of MYCC over-expression [117]. The possibility that MYCC could be a significant driver of high-risk disease is therefore of significant interest. Collectively, murine modeling of these candidate driver genes should inform our understanding of disparate NB phenotypes; but nevertheless would still fail to provide models for the majority of tumors, in which mutated genetic loci have not been specifically identified. As discussed below, expression profiling datasets are providing a new array of candidates that will provide important opportunities for modeling of non-MYCN-driven and other forms of NB.
9. Modeling of additional expression and genomic alterations
Cross-platform, integrative genomic screens that analyze copy number abnormalities (CNAs) suggest that distinct patient populations are defined by global patterns that cluster alterations in DNA ploidy and focal-segmental copy number (numerical or focal segmental copy number changes) [118–120]. These include the commonly observed regions of focal gain and loss (1p36, 11q, 13, 14, gain of 17p), which are being incorporated into clinical risk-assignment systems. With the routine introduction of Cre-recombinase and associated DNA modifying enzymes into GEM modeling approaches, it is possible, although technically challenging, to model focal segmental subchromosomal copy number losses and large scale chromosomal alterations in mice [121–123]. Of note, the gain or loss of a chromosomal region is likely associated with altered expression in a number of genes. For example, approximately 10 candidate genes map to a region of chromosome 1p36 that shows common loss in NB and include CHD5, a member of the chromodomain helicase DNA-binding domain family of SWI/SNF-like ATP dependent-chromatin remodeling proteins, proposed to regulate chromatin dynamics [124–126]. CAMTA1, a calmodulin binding transcription activator that is clonogenic when suppressed in NB cell lines and oncogenic in xenograft studies [127], CASZ1, a zinc-finger transcription factor that regulates neural cell fate specification [128], and mir-34A [129–132], a micro-RNA and candidate tumor suppressor that regulates expression of MYCN and other targets.
The significant role of micro-RNAs in the development of the neural crest precursors and malignancies makes modeling of these targets a high priority. Micro-RNAs involved in reciprocal regulatory loops with MYCN or with expression changes that segregate with MYCN amplification include let7b, miR-34A, miR-17-92, miR-181a, and mir-181b and others (reviewed in [66,133,134]). Both knockout and transgenic overexpression of micro-RNAs and RNA-interference has been achieved in mice (reviewed in [17]), and presents no special technologic issues.
One area of distinct difficulty in murine models involves the role of telomere alterations, since regulation of telomere length in mice does not perfectly model that of human telomeres (reviewed in [135]). Given that telomere length in NB correlates with prognosis, and telomerase expression segregates benign NB histology (including stage 4S) from malignant disease forms, this is an important area of biology requiring murine models. Recent reports have assessed the differential penetrance and progression of particular cancers in the setting of short or long telomere length utilizing Terc (telomerase) knockout murine models [136], so that this may yet be a possibility in NB.
That specific mutational associations do not account for a majority of sporadic NBs argues strongly for the role of epigenetic modifications in NB tumorigenesis. Because the basis of methylation changes in NB and other cancers is not clearly driven by a specific driver, methylation changes have not been adequately modeled utilizing GEMM approaches. However candidate-driven and more recently genome wide profiling approaches have identified widespread modulation of gene expression in NB through hypermethylation of promotors, yielding new gene candidates with modulated expression that correlates closely with unfavorable prognosis (reviewed in [137]). Historically, caspase 8 and RASSF1A were the first silenced genes identified using candidate approaches, and are both targets of interest for GEMM strategies [74,138]. In any event, modeling of gene silencing via methylation is essentially equivalent to simple gene-knockout strategies and presents no special issues for GEMM approaches.
10. Modeling NB as a deficiency of neural crest development
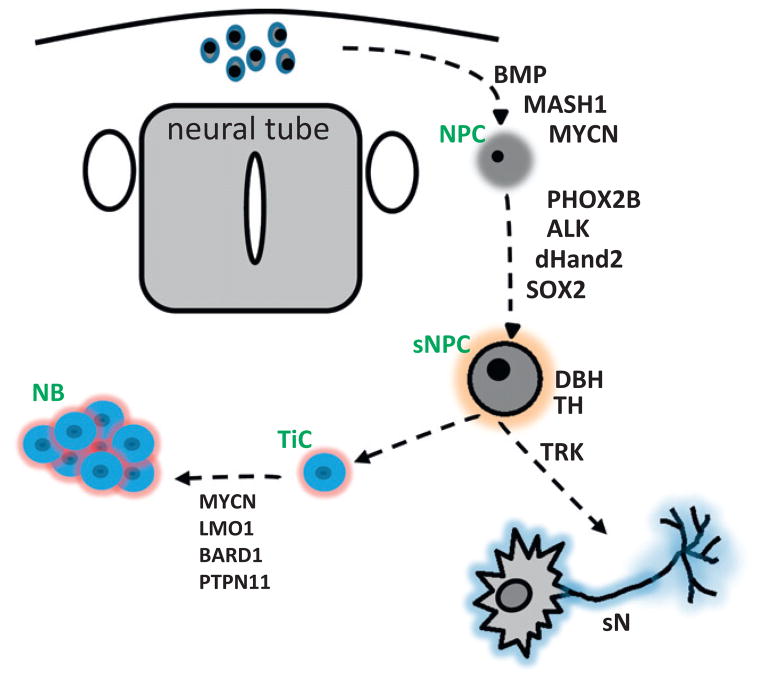
NB is one of several neurocristopathies associated with disordered development of the autonomic nervous system, including central hypoventilation syndrome, Hirschsprung disease, pheochromocytoma and neurofibromatosis [139]. The high degree of clinical diversity and specificity of age distribution reinforces the hypothesis that specific NB phenotypes are likely to originate from distinct aberrations that occur at specific points within the neural crest fate-specification pathway. The use of GEMM for understanding the developmental nature of neuronal pediatric tumors has been extensively reviewed [140]. Of interest, NB exhibits the highest frequency of spontaneous regression in cancer and approximately 10% of cases display spontaneous regression of metastases and primary lesions characterized by delayed differentiation, impaired apoptotic function and aberrant expression of telomerase. Regression has been observed in TH-MYCN mice although no true entity representing 4S (4M/INRG) NB has been observed [26,32]. Modeling stage 4S NB represents a challenge, given the lack of definable genetic associations, and the uncertainty that 4S represents a true clonal entity (as opposed to a metastable developmental intermediate). Recent advances in developmental biology may help in this regard, as the genetic expression changes that occur in the progression of neural precursors to differentiated sympathetic neurons is being defined (recently reviewed in [141]). Ex vivo maintenance of neural crest precursors and recapitulation of the neural crest differentiation program (utilizing both embryonally stem cells and murine and human iPS cell-programming) has provided starting materials for identifying specific fate decisions that dictate maturation of neural crest precursors [142,143]. Complementary animal models are lacking, but conceptually should be quite easily constructed. Fig. 3 below provides some relevant genes and their expression sequence at critical stages of commitment and specification of neural crest (adapted from [25]). The promoters from each of these genes could conceivably be used to drive particular oncogenes-of-interest (such as ALK, MYC or MYCN) at varied stages of neural crest development. Such comparative models could further illuminate relationships between developmental and cancer biology in NB. Finally, the specific ability to fully reverse and image very early proliferation and hyperplasia in these models is a critical feature of such approaches to model tumor regression and/or differentiation, as discussed below.
Fig. 3.
Gene expression in the developing neural crest. The sequence of expressed genes activated during development of the neural crest has largely been determined. Modulation of these genes utilizing conditional and regulable GEMM approaches will help to elucidate the specific role of each gene in fatespecification and commitment to lineage restricted (sympathetic and parasympathetic) derivatives. These genes are also ideal for targeting of NB specific initiator mutations to the appropriate tissue-compartment and precursor cell that may be required for induction of NB. TiC, tumor initiating cell, sNPC, sympathetic neuronal precursor cell, NPC, neuronal precursor cell, sN, sympathetic neuron.
11. Technologies for current and future modeling strategies
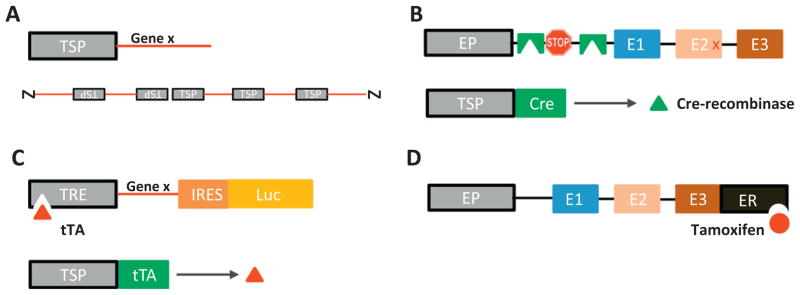
The TH-MYCN model utilized comparatively simple, non-targeted tissue-specific mis-expression (a conventional transgenic approach, to establish that targeted expression of an initiating oncogene could drive NB (Fig. 4A). Given the unique biology of this disease as a derivative of aberrant neural crest differentiation and its propensity to regress in an age- and time-dependent manner, future modeling strategies should incorporate technologies allowing for inducible or fully reversible expression of mutations. Specifically with the identification of ALK as a contributor to both inherited and sporadic NB, improved modeling of two-hit allele losses will require a combination of conventional knock-out approach with a targeted, inducible knock-in or knock-out strategy (Fig. 4B) to properly recapitulate the sequential loss of endogenous alleles and acquisition of mutated copies. Some additional possibilities for inducible and regulable expression are outlined in Fig. 4.
Fig. 4.
GEM modeling approaches. A summary of major non-conditional, conditional and gene-regulable modeling approaches in mice. A. classical transgenic, non-targeted, tissue specific constitutive overexpression of a gene utilizing a tissue defined promotor, integrated randomly into the genome in multiple repeats. B. Targeted Cre-recombinase based gene activation replaces an endogenous allele with a construct containing a lox-stop-lox target site. Excision of loxP sites flanking a stop codon is activated in tissue-specific manner by a genetic cross to an animal expressing Cre recombinase protein under control of a TSP. C. Tetracycline/doxycycline based in which animals expressing a tetracycline ransactivator protein under control of a TSP and a tetracycline-response element controlling expression of one or two genes are intercrossed to provide fully-reversible tissue-specific gene expression dependent on administration of doxycycline to mice. D. Tamoxifen-dependent targeted system in which an endogenous allele is replaced with a construct expressing a fusion protein containing the tamoxifen-responsive estrogen receptor ligand binding site. TSP, tissue specific promotor, EP, endogenous promotor, STOP, stop codon, tTA, tetracycline transactivator protein, TRE, tetracycline response element, ER, estrogen response element.
The most common strategy for inducibility has utilized “Cre-loxP” technology, which harnesses the ability of bacteriophage derived (Cre) recombinases to excise genetic sequences delimited by loxP sites (loxP-geneX-loxP) [144–147]. Tissue-targeted knockout of a murine gene is possible if the endogenous allele is replaced with a “floxed” (flanked by loxP sites) copy and bred against a mouse engineered to express Cre under control of a tissue specific promotor (TSP-Cre). Activation of a mutated or wild-type allele is possible using Cre-mediated excision of loxP-stop-loxP sites engineered into transgenic or knock-in constructs (Fig. 4B). In this case, activation is determined by the expression characteristics of the TSP. Time-dependent inducibility of Cre-driven systems can be achieved through the use of TSP-Cre-ER fusion constructs, in which Cre is constitutively inactivated through fusion with an estrogen receptor construct that activates in response to Tamoxifen treatment. More recently, a second recombinase approach Flp-FRT, using a yeast-derived recombinase (Flp) has been used in combination with Flippase recognition sites (Frt) to drive inducible recombination strategies that allow more fine-control over this process [148,149]. A disadvantage of these approaches is the irreversibility of the Cre or Flp-driven recombination event.
A third and increasingly popular (and fully reversible) GEMM approach utilizes Escherichia coli derived tetracycline operator (TetO) sequences fused to a minimal cytomegalovirus (CMV) promotor to produce a tetracycline responsive promotor (TRE, tetracycline response element) that enables tissue-specific, time-dependent and fully reversible control over transgene expression (without excision of genetic sequences) [150–152]. Expression of the TRE (and of the gene of interest) is responsive to binding of tetra-cycline transactivating proteins (fusions of tetracycline repressor (TetR) proteins with HSV-derived VP16) that either induce (rtTA, TET-on) or repress (tTA, TET-off) transcription when bound to the TRE (Fig. 4C). This technology is in third-generation development and over 250 GEM strains are available utilizing TRE variants. Oral administration of doxycycline induces virtually complete induction or suppression of transcription within 0–24 h in vivo. The disadvantages include some degree of baseline leakage of transcription (tTA > rtTA based systems) and concerns over the administration of (potentially mutagenic) tetracyclines to pregnant mice. The full-reversibility of these systems underlies their extensive use to model oncogene addiction in a variety of cancers, and the impact of suppression of genes such as MYC on cancer suppression, emergence of genetic and therapeutic resistance in lymphoma, breast cancer and other malignancies involving aberrant control of MYC expression. These issues have made TRE-based approaches of great interest for study of NB tumor regression, spontaneous differentiation, and particularly to assess the impact of time-dependent expression of developmentally important genes, such as MYCN, PHOX2B, and ALK. A variety of modeling efforts are currently underway.
A final strategy permitting direct modulation of function at the protein level involves estrogen receptor (ER) fusion protein driven GEMM utilizing transgene constructs in which DNA encoding a Tamoxifen-responsive estrogen receptor (ER) element is added to the c-terminus of the gene-of-interest [87,153]. ER-fusion proteins are cytoplasmically localized so that proteins with nuclear-dependent functions are inactivated but can be restored following subcutaneous administration of Tamoxifen (Fig. 4D). The Myc-ER and p53-ER GEMM [87,88] have successfully utilized these approaches, which provide for very rapid restoration of protein function, however maintained expression requires continuous administration of Tamoxifen, and complete restoration of wild-type function can be variable.
12. GEMM-based pre-clinical drug development
A potential strength of GEMM is their applicability to pre-clinical drug development, but efficient utilization for this purpose has not yet been fully realized. A significant deficiency of tumor implantation modeling has been lack of predictive power for clinical response, most likely related to altered behavior of implanted human cell lines in the subcutaneous murine microenvironment. It is becoming apparent that in certain trial designs, GEMM may provide superior modeling of drug response as compared to exogenous implantation models [154,155].
A specific advantage of GEM tumors compared to exogenous implants is that they arise naturally, in immunocompetent hosts, within the appropriate tissue and developmental microenvironment, thereby retaining native tumor–stromal interactions and tumor-derived vasculature. For particular applications, such as validation of targeted antibodies, anti-vascular and anti-angiogenic agents, and immunosuppressives, these advantages are most readily apparent. The most frequent criticism of the use of GEMM models in the pre-clinical arena is that genetically primed models do not accurately model the natural clonal evolution of human tumors, resulting in tumors with exaggerated sensitivity to both conventional chemotherapeutics and novel small-molecules that target transgene expression. A second criticism is that GEMM are not practically useful for routine pre-clinical screening of novel therapeutics, due to issues of cost, patents, long latencies to tumor development, low tumor penetrance or an inability to monitor tumor development and therapeutic response (reviewed in [156]).
For TH-MYCN and an increasing number of GEMM, these criticisms are being challenged as pre-clinical experience increases (and with it the body of data that describes the concordance between pre-clinical and clinical response). In our experience, the most appropriate rules for generation of high-quality and predictive pre-clinical data in GEMM are to: (1) use GEMM that closely model a discrete subset of clinical tumors in which both tumor biology and clinical outcome are powerfully associated with aberrant expression of a single gene (i.e. gene-addicted tumors), (2) use GEMM with high penetrance and short latency to tumor formation, and (3) deploy a validated pre-clinical imaging modality that is practically useful for assessment of tumor burden. Clearly, the systematic use of GEMM for pre-clinical drug development must also be underpinned by case-specific evidence for each GEMM, establishing that the growth, progression, imaging characteristics and chemoresponsiveness of murine tumors closely recapitulates human counterparts. “Credentialing” of several GEMM have been completed in this manner (reviewed in [157]). The use of highly validated models within the framework of a “mouse-hospital” in which rigorously controlled trial designs are employed using dedicated pathology and imaging-based response assessment, has led to several examples in which the accuracy of GEMM-based pre-clinical data supersedes that of transplantation models.
In the NB field, experience with credentialing of the TH-MYCN model spans 15 years, and from multiple studies it is clear that this GEMM is a useful and valid indicator of therapy response at least for high-risk NB patients defined by amplification of MYCN. Several trial designs have been reported, including intervention (single agent debulking trial), survival (single agents alone and in combination with chemotherapeutics) and prevention (empirical treatment at conventional or low doses initiated prior to onset of detectable tumors). Practical protocols for the use of multiple imaging modalities have been reported, including MRI, ultrasound and 18FDG-PET. A variety of therapeutics have been assessed, including front-line chemotherapeutics (cyclophosphamide, cis-platinum, adriamycin, irinotecan, temozolomide, and etoposide) [26,29,30,37,47], steroids (dexamethasone), anti-vascular/anti-angiogenic agents and novel small-molecules [27,30,31,37,43,48]. TH-MYCN closely models the response of chemotherapeutics in humans. Recent work extends these efforts to include modeling of relapse and therapeutic resistance [29]. The latter issue is particularly important, since chemoresistant relapse and metastasis is the major source of morbidity and death in the high-risk patient population with MYCN gene-amplification. As referred to above, an important issue to solve with this model is the lack of bulky metastatic deposits.
13. Conclusions
GEMM are continuing to define the role of clinically important mutations in the genesis of the major adult cancers. The use of GEMM-based approaches is increasing in pediatric cancer, and will play an important role in the development of novel therapeutics targeted at specific oncogenes and pathways. NB can be a paradigm for the rational development of GEMM that define all discrete clinical cohorts of this distinctive cancer.
Footnotes
Conflict of interest
None.
References
- 1.Greaves M. Darwinian medicine: a case for cancer. Nat Rev Cancer. 2007;7:213–21. doi: 10.1038/nrc2071. [DOI] [PubMed] [Google Scholar]
- 2.Stratton MR. Exploring the genomes of cancer cells: progress and promise. Science. 2011;331:1553–8. doi: 10.1126/science.1204040. [DOI] [PubMed] [Google Scholar]
- 3.Evan GI, Christophorou M, Lawlor EA, Ringshausen I, Prescott J, Dansen T, et al. Oncogene-dependent tumor suppression: using the dark side of the force for cancer therapy. Cold Spring Harb Symp Quant Biol. 2005;70:263–73. doi: 10.1101/sqb.2005.70.054. [DOI] [PubMed] [Google Scholar]
- 4.Grimmer MR, Weiss WA. Childhood tumors of the nervous system as disorders of normal development. Curr Opin Pediatr. 2006;18:634–8. doi: 10.1097/MOP.0b013e32801080fe. [DOI] [PubMed] [Google Scholar]
- 5.Hayward WS, Neel BG, Astrin SM. Activation of a cellular onc gene by promoter insertion in ALV-induced lymphoid leukosis. Nature. 1981;290:475–80. doi: 10.1038/290475a0. [DOI] [PubMed] [Google Scholar]
- 6.Payne GS, Bishop JM, Varmus HE. Multiple arrangements of viral DNA and an activated host oncogene in bursal lymphomas. Nature. 1982;295:209–14. doi: 10.1038/295209a0. [DOI] [PubMed] [Google Scholar]
- 7.Schwab M, Alitalo K, Klempnauer KH, Varmus HE, Bishop JM, Gilbert F, et al. Amplified DNA with limited homology to myc cellular oncogene is shared by human neuroblastoma cell lines and a neuroblastoma tumour. Nature. 1983;305:245–8. doi: 10.1038/305245a0. [DOI] [PubMed] [Google Scholar]
- 8.Zimmerman KA, Yancopoulos GD, Collum RG, Smith RK, Kohl NE, Denis KA, et al. Differential expression of myc family genes during murine development. Nature. 1986;319:780–3. doi: 10.1038/319780a0. [DOI] [PubMed] [Google Scholar]
- 9.Sejersen T, Bjorklund H, Sumegi J, Ringertz NR. N-myc and c-src genes are differentially regulated in PCC7 embryonal carcinoma cells undergoing neuronal differentiation. J Cell Physiol. 1986;127:274–80. doi: 10.1002/jcp.1041270213. [DOI] [PubMed] [Google Scholar]
- 10.Ruppert C, Goldowitz D, Wille W. Proto-oncogene c-myc is expressed in cerebellar neurons at different developmental stages. EMBO J. 1986;5:1897–901. doi: 10.1002/j.1460-2075.1986.tb04442.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Charron J, Malynn BA, Fisher P, Stewart V, Jeannotte L, Goff SP, et al. Embryonic lethality in mice homozygous for a targeted disruption of the N-myc gene. Gene Dev. 1992;6:2248–57. doi: 10.1101/gad.6.12a.2248. [DOI] [PubMed] [Google Scholar]
- 12.Moens CB, Stanton BR, Parada LF, Rossant J. Defects in heart and lung development in compound heterozygotes for two different targeted mutations at the N-myc locus. Development (Cambridge, England) 1993;119:485–99. doi: 10.1242/dev.119.2.485. [DOI] [PubMed] [Google Scholar]
- 13.Stanton BR, Perkins AS, Tessarollo L, Sassoon DA, Parada LF. Loss of N-myc function results in embryonic lethality and failure of the epithelial component of the embryo to develop. Gene Dev. 1992;6:2235–47. doi: 10.1101/gad.6.12a.2235. [DOI] [PubMed] [Google Scholar]
- 14.Knoepfler PS, Cheng PF, Eisenman RN. N-myc is essential during neurogenesis for the rapid expansion of progenitor cell populations and the inhibition of neuronal differentiation. Gene Dev. 2002;16:2699–712. doi: 10.1101/gad.1021202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wey A, Martinez Cerdeno V, Pleasure D, Knoepfler PS. c- and N-myc regulate neural precursor cell fate, cell cycle, and metabolism to direct cerebellar development. Cerebellum. 2010;9:537–47. doi: 10.1007/s12311-010-0190-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Malynn BA, de Alboran IM, O’Hagan RC, Bronson R, Davidson L, DePinho RA, et al. N-myc can functionally replace c-myc in murine development, cellular growth, and differentiation. Gene Dev. 2000;14:1390–9. [PMC free article] [PubMed] [Google Scholar]
- 17.Premsrirut PK, Dow LE, Kim SY, Camiolo M, Malone CD, Miething C, et al. A rapid and scalable system for studying gene function in mice using conditional RNA interference. Cell. 2011;145:145–58. doi: 10.1016/j.cell.2011.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chayka O, Corvetta D, Dews M, Caccamo AE, Piotrowska I, Santilli G, et al. Clusterin, a haploinsufficient tumor suppressor gene in neuroblastomas. J Natl Cancer Inst. 2009;101:663–77. doi: 10.1093/jnci/djp063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schulte JH, Horn S, Otto T, Samans B, Heukamp LC, Eilers UC, et al. MYCN regulates oncogenic MicroRNAs in neuroblastoma. Int J Cancer. 2008;122:699–704. doi: 10.1002/ijc.23153. [DOI] [PubMed] [Google Scholar]
- 20.Dews M, Homayouni A, Yu D, Murphy D, Sevignani C, Wentzel E, et al. Augmentation of tumor angiogenesis by a Myc-activated microRNA cluster. Nat Genet. 2006;38:1060–5. doi: 10.1038/ng1855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Weiss WA, Aldape K, Mohapatra G, Feuerstein BG, Bishop JM. Targeted expression of MYCN causes neuroblastoma in transgenic mice. Embo J. 1997;16:2985–95. doi: 10.1093/emboj/16.11.2985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Iwakura H, Ariyasu H, Kanamoto N, Hosoda K, Nakao K, Kangawa K, et al. Establishment of a novel neuroblastoma mouse model. Int J Oncol. 2008;33:1195–9. [PubMed] [Google Scholar]
- 23.Hattori Y, Kanamoto N, Kawano K, Iwakura H, Sone M, Miura M, et al. Molecular characterization of tumors from a transgenic mouse adrenal tumor model: comparison with human pheochromocytoma. Int J Oncol. 2010;37:695–705. doi: 10.3892/ijo_00000719. [DOI] [PubMed] [Google Scholar]
- 24.Patankar S, Lazaroff M, Yoon SO, Chikaraishi DM. A novel basal promoter element is required for expression of the rat tyrosine hydroxylase gene. J Neurosci. 1997;17:4076–86. doi: 10.1523/JNEUROSCI.17-11-04076.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Alam G, Cui H, Shi H, Yang L, Ding J, Mao L, et al. MYCN promotes the expansion of Phox2B-positive neuronal progenitors to drive neuroblastoma development. Am J Pathol. 2009;175:856–66. doi: 10.2353/ajpath.2009.090019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Teitz T, Stanke JJ, Federico S, Bradley CL, Brennan R, Zhang J, et al. Preclinical models for neuroblastoma: establishing a baseline for treatment. PLoS ONE. 2011;6:e19133. doi: 10.1371/journal.pone.0019133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rounbehler RJ, Li W, Hall MA, Yang C, Fallahi M, Cleveland JL. Targeting ornithine decarboxylase impairs development of MYCN-amplified neuroblastoma. Cancer Res. 2009;69:547–53. doi: 10.1158/0008-5472.CAN-08-2968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Haraguchi S, Nakagawara A. A simple PCR method for rapid genotype analysis of the TH-MYCN transgenic mouse. PLoS ONE. 2009;4:e6902. doi: 10.1371/journal.pone.0006902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chesler L, Goldenberg DD, Collins R, Grimmer M, Kim GE, Tihan T, et al. Chemotherapy-induced apoptosis in a transgenic model of neuroblastoma proceeds through p53 induction. Neoplasia. 2008;10:1268–74. doi: 10.1593/neo.08778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hogarty MD, Norris MD, Davis K, Liu X, Evageliou NF, Hayes CS, et al. ODC1 is a critical determinant of MYCN oncogenesis and a therapeutic target in neuroblastoma. Cancer Res. 2008;68:9735–45. doi: 10.1158/0008-5472.CAN-07-6866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chesler L, Goldenberg DD, Seales IT, Satchi-Fainaro R, Grimmer M, Collins R, et al. Malignant progression and blockade of angiogenesis in a murine transgenic model of neuroblastoma. Cancer Res. 2007;67:9435–42. doi: 10.1158/0008-5472.CAN-07-1316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hansford LM, Thomas WD, Keating JM, Burkhart CA, Peaston AE, Norris MD, et al. Mechanisms of embryonal tumor initiation: distinct roles for MycN expression and MYCN amplification. Proc Natl Acad Sci U S A. 2004;101:12664–9. doi: 10.1073/pnas.0401083101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hackett CS, Hodgson JG, Law ME, Fridlyand J, Osoegawa K, de Jong PJ, et al. Genome-wide array CGH analysis of murine neuroblastoma reveals distinct genomic aberrations which parallel those in human tumors. Cancer Res. 2003;63:5266–73. [PubMed] [Google Scholar]
- 34.Chen Z, Lin Y, Barbieri E, Burlingame S, Hicks J, Ludwig A, et al. Mdm2 deficiency suppresses MYCN-Driven neuroblastoma tumorigenesis in vivo. Neoplasia. 2009;11:753–62. doi: 10.1593/neo.09466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Asano Y, Kishida S, Mu P, Sakamoto K, Murohara T, Kadomatsu K. DRR1 is expressed in the developing nervous system and downregulated during neuroblastoma carcinogenesis. Biochem Biophys Res Commun. 2010;394:829–35. doi: 10.1016/j.bbrc.2010.03.085. [DOI] [PubMed] [Google Scholar]
- 36.Balamuth NJ, Wood A, Wang Q, Jagannathan J, Mayes P, Zhang Z, et al. Serial transcriptome analysis and cross-species integration identifies centromere-associated protein E as a novel neuroblastoma target. Cancer Res. 2010;70:2749–58. doi: 10.1158/0008-5472.CAN-09-3844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Burkhart CA, Watt F, Murray J, Pajic M, Prokvolit A, Xue C, et al. Small-molecule multidrug resistance-associated protein 1 inhibitor reversan increases the therapeutic index of chemotherapy in mouse models of neuroblastoma. Cancer Res. 2009;69:6573–80. doi: 10.1158/0008-5472.CAN-09-1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Moore HC, Wood KM, Jackson MS, Lastowska MA, Hall D, Imrie H, et al. Histological profile of tumours from MYCN transgenic mice. J Clin Pathol. 2008;61:1098–103. doi: 10.1136/jcp.2007.054627. [DOI] [PubMed] [Google Scholar]
- 39.Cheng AJ, Cheng NC, Ford J, Smith J, Murray JE, Flemming C, et al. Cell lines from MYCN transgenic murine tumours reflect the molecular and biological characteristics of human neuroblastoma. Eur J Cancer. 2007;43:1467–75. doi: 10.1016/j.ejca.2007.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lastowska M, Chung YJ, Cheng Ching N, Haber M, Norris MD, Kees UR, et al. Regions syntenic to human 17q are gained in mouse and rat neuroblastoma. Gene Chromosome Cancer. 2004;40:158–63. doi: 10.1002/gcc.20031. [DOI] [PubMed] [Google Scholar]
- 41.Burkhart CA, Cheng AJ, Madafiglio J, Kavallaris M, Mili M, Marshall GM, et al. Effects of MYCN antisense oligonucleotide administration on tumorigenesis in a murine model of neuroblastoma. J Natl Cancer Inst. 2003;95:1394–403. doi: 10.1093/jnci/djg045. [DOI] [PubMed] [Google Scholar]
- 42.Dam V, Morgan BT, Mazanek P, Hogarty MD. Mutations in PIK3CA are infrequent in neuroblastoma. BMC Cancer. 2006;6:177. doi: 10.1186/1471-2407-6-177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Morowitz MJ, Barr R, Wang Q, King R, Rhodin N, Pawel B, et al. Methionine aminopeptidase 2 inhibition is an effective treatment strategy for neuroblastoma in preclinical models. Clin Cancer Res. 2005;11:2680–5. doi: 10.1158/1078-0432.CCR-04-1917. [DOI] [PubMed] [Google Scholar]
- 44.Accorsi R, Morowitz MJ, Charron M, Maris JM. Pinhole imaging of 131I-metaiodobenzylguanidine (131I-MIBG) in an animal model of neuroblastoma. Pediatr Radiol. 2003;33:688–92. doi: 10.1007/s00247-003-1006-6. [DOI] [PubMed] [Google Scholar]
- 45.Norris MD, Burkhart CA, Marshall GM, Weiss WA, Haber M. Expression of N-myc and MRP genes and their relationship to N-myc gene dosage and tumor formation in a murine neuroblastoma model. Med Pediatr Oncol. 2000;35:585–9. doi: 10.1002/1096-911x(20001201)35:6<585::aid-mpo20>3.0.co;2-p. [DOI] [PubMed] [Google Scholar]
- 46.Weiss WA, Godfrey T, Francisco C, Bishop JM. Genome-wide screen for allelic imbalance in a mouse model for neuroblastoma. Cancer Res. 2000;60:2483–7. [PubMed] [Google Scholar]
- 47.Henderson MJ, Haber M, Porro A, Munoz MA, Iraci N, Xue C, et al. ABCC Multidrug transporters in childhood neuroblastoma: clinical and biological effects independent of cytotoxic drug efflux. J Natl Cancer Inst. 2011;103:1236–51. doi: 10.1093/jnci/djr256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Meyer GE, Chesler L, Liu D, Gable K, Maddux BA, Goldenberg DD, et al. Nordi-hydroguaiaretic acid inhibits insulin-like growth factor signaling, growth, and survival in human neuroblastoma cells. J Cell Biochem. 2007;102:1529–41. doi: 10.1002/jcb.21373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chesler L, Schlieve C, Goldenberg DD, Kenney A, Kim G, McMillan A, et al. Inhibition of phosphatidylinositol 3-kinase destabilizes Mycn protein and blocks malignant progression in neuroblastoma. Cancer Res. 2006;66:8139–46. doi: 10.1158/0008-5472.CAN-05-2769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Liu T, Tee AE, Porro A, Smith SA, Dwarte T, Liu PY, et al. Activation of tissue transglutaminase transcription by histone deacetylase inhibition as a therapeutic approach for Myc oncogenesis. Proc Natl Acad Sci U S A. 2007;104:18682–7. doi: 10.1073/pnas.0705524104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Vita M, Henriksson M. The Myc oncoprotein as a therapeutic target for human cancer. Semin Cancer Biol. 2006;16:318–30. doi: 10.1016/j.semcancer.2006.07.015. [DOI] [PubMed] [Google Scholar]
- 52.Mo H, Vita M, Crespin M, Henriksson M. Myc overexpression enhances apoptosis induced by small molecules. Cell Cycle. 2006;5:2191–4. doi: 10.4161/cc.5.19.3320. [DOI] [PubMed] [Google Scholar]
- 53.Henriksson M, Luscher B. Proteins of the Myc network: essential regulators of cell growth and differentiation. Adv Cancer Res. 1996;68:109–82. doi: 10.1016/s0065-230x(08)60353-x. [DOI] [PubMed] [Google Scholar]
- 54.Eilers M, Eisenman RN. Myc’s broad reach. Gene Dev. 2008;22:2755–66. doi: 10.1101/gad.1712408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Soucek L, Evan GI. The ups and downs of Myc biology. Curr Opin Genet Dev. 2010;20:91–5. doi: 10.1016/j.gde.2009.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Secombe J, Pierce SB, Eisenman RN. Myc: a weapon of mass destruction. Cell. 2004;117:153–6. doi: 10.1016/s0092-8674(04)00336-8. [DOI] [PubMed] [Google Scholar]
- 57.Gondo Y, Fukumura R, Murata T, Makino S. ENU-based gene-driven mutagenesis in the mouse: a next-generation gene-targeting system. Exp Anim. 2010;59:537–48. doi: 10.1538/expanim.59.537. [DOI] [PubMed] [Google Scholar]
- 58.Dupuy AJ, Akagi K, Largaespada DA, Copeland NG, Jenkins NA. Mammalian mutagenesis using a highly mobile somatic Sleeping Beauty transposon system. Nature. 2005;436:221–6. doi: 10.1038/nature03691. [DOI] [PubMed] [Google Scholar]
- 59.Lawlor ER, Soucek L, Brown-Swigart L, Shchors K, Bialucha CU, Evan GI. Reversible kinetic analysis of Myc targets in vivo provides novel insights into Myc-mediated tumorigenesis. Cancer Res. 2006;66:4591–601. doi: 10.1158/0008-5472.CAN-05-3826. [DOI] [PubMed] [Google Scholar]
- 60.Boon K, Caron HN, van Asperen R, Valentijn L, Hermus MC, van Sluis P, et al. N-myc enhances the expression of a large set of genes functioning in ribosome biogenesis and protein synthesis. Embo J. 2001;20:1383–93. doi: 10.1093/emboj/20.6.1383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Mac SM, D’Cunha CA, Farnham PJ. Direct recruitment of N-myc to target gene promoters. Mol Carcinogen. 2000;29:76–86. [PubMed] [Google Scholar]
- 62.Grandori C, Cowley SM, James LP, Eisenman RN. The Myc/Max/Mad network and the transcriptional control of cell behavior. Annu Rev Cell Dev Biol. 2000;16:653–99. doi: 10.1146/annurev.cellbio.16.1.653. [DOI] [PubMed] [Google Scholar]
- 63.Murphy DM, Buckley PG, Bryan K, Das S, Alcock L, Foley NH, et al. Global MYCN transcription factor binding analysis in neuroblastoma reveals association with distinct E-box motifs and regions of DNA hypermethylation. PLoS ONE. 2009;4:e8154. doi: 10.1371/journal.pone.0008154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lin CH, Jackson AL, Guo J, Linsley PS, Eisenman RN. Myc-regulated microRNAs attenuate embryonic stem cell differentiation. EMBO J. 2009;28:3157–70. doi: 10.1038/emboj.2009.254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.O’Donnell KA, Wentzel EA, Zeller KI, Dang CV, Mendell JT. c-Myc-regulated microRNAs modulate E2F1 expression. Nature. 2005;435:839–43. doi: 10.1038/nature03677. [DOI] [PubMed] [Google Scholar]
- 66.Bray I, Bryan K, Prenter S, Buckley PG, Foley NH, Murphy DM, et al. Widespread dysregulation of MiRNAs by MYCN amplification and chromosomal imbalances in neuroblastoma: association of miRNA expression with survival. PLoS ONE. 2009;4:e7850. doi: 10.1371/journal.pone.0007850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Shohet JM, Ghosh R, Coarfa C, Ludwig A, Benham AL, Chen Z, et al. A genome-wide search for promoters that respond to increased MYCN reveals both new oncogenic and tumor suppressor microRNAs associated with aggressive neuroblastoma. Cancer Res. 2011;71:3841–51. doi: 10.1158/0008-5472.CAN-10-4391. [DOI] [PubMed] [Google Scholar]
- 68.Martins CP, Brown-Swigart L, Evan GI. Modeling the therapeutic efficacy of p53 restoration in tumors. Cell. 2006;127:1323–34. doi: 10.1016/j.cell.2006.12.007. [DOI] [PubMed] [Google Scholar]
- 69.Hemann MT, Bric A, Teruya-Feldstein J, Herbst A, Nilsson JA, Cordon-Cardo C, et al. Evasion of the p53 tumour surveillance network by tumour-derived MYC mutants. Nature. 2005;436:807–11. doi: 10.1038/nature03845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Jacobs JJ, Scheijen B, Voncken JW, Kieboom K, Berns A, van Lohuizen M. Bmi-1 collaborates with c-Myc in tumorigenesis by inhibiting c-Myc-induced apoptosis via INK4a/ARF. Gene Dev. 1999;13:2678–90. doi: 10.1101/gad.13.20.2678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Cui H, Hu B, Li T, Ma J, Alam G, Gunning WT, et al. Bmi-1 is essential for the tumorigenicity of neuroblastoma cells. Am J Pathol. 2007;170:1370–8. doi: 10.2353/ajpath.2007.060754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Giannini G, Cerignoli F, Mellone M, Massimi I, Ambrosi C, Rinaldi C, et al. High mobility group A1 is a molecular target for MYCN in human neuroblastoma. Cancer Res. 2005;65:8308–16. doi: 10.1158/0008-5472.CAN-05-0607. [DOI] [PubMed] [Google Scholar]
- 73.Eggert A, Grotzer MA, Zuzak TJ, Wiewrodt BR, Ikegaki N, Brodeur GM. Resistance to TRAIL-induced apoptosis in neuroblastoma cells correlates with a loss of caspase-8 expression. Med Pediatr Oncol. 2000;35:603–7. doi: 10.1002/1096-911x(20001201)35:6<603::aid-mpo24>3.0.co;2-1. [DOI] [PubMed] [Google Scholar]
- 74.Teitz T, Wei T, Valentine MB, Vanin EF, Grenet J, Valentine VA, et al. Caspase 8 is deleted or silenced preferentially in childhood neuroblastomas with amplification of MYCN. Nat Med. 2000;6:529–35. doi: 10.1038/75007. [DOI] [PubMed] [Google Scholar]
- 75.Hopkins-Donaldson S, Bodmer JL, Bourloud KB, Brognara CB, Tschopp J, Gross N. Loss of caspase-8 expression in neuroblastoma is related to malignancy and resistance to TRAIL-induced apoptosis. Med Pediatr Oncol. 2000;35:608–11. doi: 10.1002/1096-911x(20001201)35:6<608::aid-mpo25>3.0.co;2-u. [DOI] [PubMed] [Google Scholar]
- 76.Hopkins-Donaldson S, Bodmer JL, Bourloud KB, Brognara CB, Tschopp J, Gross N. Loss of caspase-8 expression in highly malignant human neuroblastoma cells correlates with resistance to tumor necrosis factor-related apoptosis-inducing ligand-induced apoptosis. Cancer Res. 2000;60:4315–9. [PubMed] [Google Scholar]
- 77.Tanaka N, Fukuzawa M. MYCN downregulates integrin alpha1 to promote invasion of human neuroblastoma cells. Int J Oncol. 2008;33:815–21. [PubMed] [Google Scholar]
- 78.Vitali R, Mancini C, Cesi V, Tanno B, Mancuso M, Bossi G, et al. Slug (SNAI2) down-regulation by RNA interference facilitates apoptosis and inhibits invasive growth in neuroblastoma preclinical models. Clin Cancer Res. 2008;14:4622–30. doi: 10.1158/1078-0432.CCR-07-5210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Airoldi I, Cocco C, Morandi F, Prigione I, Pistoia V. CXCR5 may be involved in the attraction of human metastatic neuroblastoma cells to the bone marrow. Cancer Immunol Immunother. 2008;57:541–8. doi: 10.1007/s00262-007-0392-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Zhang L, Yeger H, Das B, Irwin MS, Baruchel S. Tissue microenvironment modulates CXCR4 expression and tumor metastasis in neuroblastoma. Neoplasia. 2007;9:36–46. doi: 10.1593/neo.06670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Leone A, Seeger RC, Hong CM, Hu YY, Arboleda MJ, Brodeur GM, et al. Evidence for nm23 RNA overexpression, DNA amplification and mutation in aggressive childhood neuroblastomas. Oncogene. 1993;8:855–65. [PubMed] [Google Scholar]
- 82.Olive V, Bennett MJ, Walker JC, Ma C, Jiang I, Cordon-Cardo C, et al. miR-19 is a key oncogenic component of mir-17-92. Gene Dev. 2009;23:2839–49. doi: 10.1101/gad.1861409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Tweddle DA, Pearson AD, Haber M, Norris MD, Xue C, Flemming C, et al. The p53 pathway and its inactivation in neuroblastoma. Cancer Lett. 2003;197:93–8. doi: 10.1016/s0304-3835(03)00088-0. [DOI] [PubMed] [Google Scholar]
- 84.Carr-Wilkinson J, O’Toole K, Wood KM, Challen CC, Baker AG, Board JR, et al. High frequency of p53/MDM2/p14ARF pathway abnormalities in relapsed neuroblastoma. Clin Cancer Res: Off J Am Assoc Cancer Res. 2010;16:1108–18. doi: 10.1158/1078-0432.CCR-09-1865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Chen L, Iraci N, Gherardi S, Gamble LD, Wood KM, Perini G, et al. p53 is a direct transcriptional target of MYCN in neuroblastoma. Cancer Res. 2010;70:1377–88. doi: 10.1158/0008-5472.CAN-09-2598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Keshelava N, Zuo JJ, Waidyaratne NS, Triche TJ, Reynolds CP. p53 mutations and loss of p53 function confer multidrug resistance in neuroblastoma. Med Pediatr Oncol. 2000;35:563–8. doi: 10.1002/1096-911x(20001201)35:6<563::aid-mpo15>3.0.co;2-j. [DOI] [PubMed] [Google Scholar]
- 87.Christophorou MA, Martin-Zanca D, Soucek L, Lawlor ER, Brown-Swigart L, Verschuren EW, et al. Temporal dissection of p53 function in vitro and in vivo. Nat Genet. 2005;37:718–26. doi: 10.1038/ng1572. [DOI] [PubMed] [Google Scholar]
- 88.Amaravadi RK, Yu D, Lum JJ, Bui T, Christophorou MA, Evan GI, et al. Autophagy inhibition enhances therapy-induced apoptosis in a Myc-induced model of lymphoma. J Clin Invest. 2007;117:326–36. doi: 10.1172/JCI28833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Pugh T. Exome sequencing of 81 neuroblastomas identifies a wide diversity of somatic mutation. Orlando, FL, USA: American Association for Cancer Research; 2011. [Google Scholar]
- 90.Chen Y, Takita J, Choi YL, Kato M, Ohira M, Sanada M, et al. Oncogenic mutations of ALK kinase in neuroblastoma. Nature. 2008;455:971–4. doi: 10.1038/nature07399. [DOI] [PubMed] [Google Scholar]
- 91.Mosse YP, Laudenslager M, Longo L, Cole KA, Wood A, Attiyeh EF, et al. Identification of ALK as a major familial neuroblastoma predisposition gene. Nature. 2008;455:930–5. doi: 10.1038/nature07261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Janoueix-Lerosey I, Lequin D, Brugieres L, Ribeiro A, de Pontual L, Combaret V, et al. Somatic and germline activating mutations of the ALK kinase receptor in neuroblastoma. Nature. 2008;455:967–70. doi: 10.1038/nature07398. [DOI] [PubMed] [Google Scholar]
- 93.George RE, Sanda T, Hanna M, Frohling S, Luther W, 2nd, Zhang J, et al. Activating mutations in ALK provide a therapeutic target in neuroblastoma. Nature. 2008;455:975–8. doi: 10.1038/nature07397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.George RE, Attiyeh EF, Li S, Moreau LA, Neuberg D, Li C, et al. Genome-wide analysis of neuroblastomas using high-density single nucleotide polymorphism arrays. PLoS ONE. 2007;2:e255. doi: 10.1371/journal.pone.0000255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Mosse YP, Laudenslager M, Khazi D, Carlisle AJ, Winter CL, Rappaport E, et al. Germline PHOX2B mutation in hereditary neuroblastoma. Am J Hum Genet. 2004;75:727–30. doi: 10.1086/424530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Bilsland JG, Wheeldon A, Mead A, Znamenskiy P, Almond S, Waters KA, et al. Behavioral and neurochemical alterations in mice deficient in anaplastic lymphoma kinase suggest therapeutic potential for psychiatric indications. Neuropsychopharmacol: Off Publ Am Coll Neuropsychopharmacol. 2008;33:685–700. doi: 10.1038/sj.npp.1301446. [DOI] [PubMed] [Google Scholar]
- 97.Pattyn A, Morin X, Cremer H, Goridis C, Brunet JF. The homeobox gene Phox2b is essential for the development of autonomic neural crest derivatives. Nature. 1999;399:366–70. doi: 10.1038/20700. [DOI] [PubMed] [Google Scholar]
- 98.Durand E, Dauger S, Pattyn A, Gaultier C, Goridis C, Gallego J. Sleep-disordered breathing in newborn mice heterozygous for the transcription factor Phox2b. Am J Resp Crit Care Med. 2005;172:238–43. doi: 10.1164/rccm.200411-1528OC. [DOI] [PubMed] [Google Scholar]
- 99.De Brouwer S, De Preter K, Kumps C, Zabrocki P, Porcu M, Westerhout EM, et al. Meta-analysis of neuroblastomas reveals a skewed ALK mutation spectrum in tumors with MYCN amplification. Clin Cancer Res: Off J Am Assoc Cancer Res. 2010;16:4353–62. doi: 10.1158/1078-0432.CCR-09-2660. [DOI] [PubMed] [Google Scholar]
- 100.Bachetti T, Di Paolo D, Di Lascio S, Mirisola V, Brignole C, Bellotti M, et al. PHOX2B-mediated regulation of ALK expression: in vitro identification of a functional relationship between two genes involved in neuroblastoma. PLoS ONE. 2010:5. doi: 10.1371/journal.pone.0013108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Stanke M, Junghans D, Geissen M, Goridis C, Ernsberger U, Rohrer H. The Phox2 homeodomain proteins are sufficient to promote the development of sympathetic neurons. Development (Cambridge, England) 1999;126:4087–94. doi: 10.1242/dev.126.18.4087. [DOI] [PubMed] [Google Scholar]
- 102.Raabe EH, Laudenslager M, Winter C, Wasserman N, Cole K, LaQuaglia M, et al. Prevalence and functional consequence of PHOX2B mutations in neuroblastoma. Oncogene. 2008;27:469–76. doi: 10.1038/sj.onc.1210659. [DOI] [PubMed] [Google Scholar]
- 103.Stutterheim J, Gerritsen A, Zappeij-Kannegieter L, Kleijn I, Dee R, Hooft L, et al. PHOX2B is a novel and specific marker for minimal residual disease testing in neuroblastoma. J Clin Oncol. 2008;26:5443–9. doi: 10.1200/JCO.2007.13.6531. [DOI] [PubMed] [Google Scholar]
- 104.Longo L, Borghini S, Schena F, Parodi S, Albino D, Bachetti T, et al. PHOX2A and PHOX2B genes are highly co-expressed in human neuroblastoma. Int J Oncol. 2008;33:985–91. [PubMed] [Google Scholar]
- 105.Beiske K, Ambros PF, Burchill SA, Cheung IY, Swerts K. Detecting minimal residual disease in neuroblastoma patients-the present state of the art. Cancer Lett. 2005;228:229–40. doi: 10.1016/j.canlet.2005.02.053. [DOI] [PubMed] [Google Scholar]
- 106.Burchill SA, Kinsey SE, Picton S, Roberts P, Pinkerton CR, Selby P, et al. Minimal residual disease at the time of peripheral blood stem cell harvest in patients with advanced neuroblastoma. Med Pediatr Oncol. 2001;36:213–9. doi: 10.1002/1096-911X(20010101)36:1<213::AID-MPO1052>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- 107.Bentires-Alj M, Paez JG, David FS, Keilhack H, Halmos B, Naoki K, et al. Activating mutations of the noonan syndrome-associated SHP2/PTPN11 gene in human solid tumors and adult acute myelogenous leukemia. Cancer Res. 2004;64:8816–20. doi: 10.1158/0008-5472.CAN-04-1923. [DOI] [PubMed] [Google Scholar]
- 108.Holzel M, Huang S, Koster J, Ora I, Lakeman A, Caron H, et al. NF1 is a tumor suppressor in neuroblastoma that determines retinoic acid response and disease outcome. Cell. 2010;142:218–29. doi: 10.1016/j.cell.2010.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Nguyenle B, Diskin SJ, Capasso M, Wang K, Diamond MA, Glessner J, et al. Phenotype restricted genome-wide association study using a gene-centric approach identifies three low-risk neuroblastoma susceptibility Loci. PLoS Genet. 2011;7:e1002026. doi: 10.1371/journal.pgen.1002026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Capasso M, Diskin SJ. Genetics and genomics of neuroblastoma. Cancer Treat Res. 2010;155:65–84. doi: 10.1007/978-1-4419-6033-7_4. [DOI] [PubMed] [Google Scholar]
- 111.Capasso M, Devoto M, Hou C, Asgharzadeh S, Glessner JT, Attiyeh EF, et al. Common variations in BARD1 influence susceptibility to high-risk neuroblastoma. Nat Genet. 2009;41:718–23. doi: 10.1038/ng.374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Maris JM, Mosse YP, Bradfield JP, Hou C, Monni S, Scott RH, et al. Chromosome 6p22 locus associated with clinically aggressive neuroblastoma. N Engl J Med. 2008;358:2585–93. doi: 10.1056/NEJMoa0708698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Kamp DW, Shacter E, Weitzman SA. Chronic inflammation and cancer: the role of the mitochondria. Oncology. 2011;25:400–10. 13. [PubMed] [Google Scholar]
- 114.Martinelli S, Carta C, Flex E, Binni F, Cordisco EL, Moretti S, et al. Activating PTPN11 mutations play a minor role in pediatric and adult solid tumors. Cancer Genet Cytogenet. 2006;166:124–9. doi: 10.1016/j.cancergencyto.2005.10.003. [DOI] [PubMed] [Google Scholar]
- 115.Wang K, Diskin SJ, Zhang H, Attiyeh EF, Winter C, Hou C, et al. Integrative genomics identifies LMO1 as a neuroblastoma oncogene. Nature. 2011;469:216–20. doi: 10.1038/nature09609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Wu JY, Vlastos AT, Pelte MF, Caligo MA, Bianco A, Krause KH, et al. Aberrant expression of BARD1 in breast and ovarian cancers with poor prognosis. Journal international du cancer. 2006;118:1215–26. doi: 10.1002/ijc.21428. [DOI] [PubMed] [Google Scholar]
- 117.Fredlund E, Ringner M, Maris JM, Pahlman S. High Myc pathway activity and low stage of neuronal differentiation associate with poor outcome in neuroblastoma. Proc Natl Acad Sci U S A. 2008;105:14094–9. doi: 10.1073/pnas.0804455105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Schleiermacher G, Janoueix-Lerosey I, Ribeiro A, Klijanienko J, Couturier J, Pierron G, et al. Accumulation of segmental alterations determines progression in neuroblastoma. J Clin Oncol: Off J Am Soc Clin Oncol. 2010;28:3122–30. doi: 10.1200/JCO.2009.26.7955. [DOI] [PubMed] [Google Scholar]
- 119.Janoueix-Lerosey I, Schleiermacher G, Delattre O. Molecular pathogenesis of peripheral neuroblastic tumors. Oncogene. 2010;29:1566–79. doi: 10.1038/onc.2009.518. [DOI] [PubMed] [Google Scholar]
- 120.Schleiermacher G, Michon J, Huon I, d’Enghien CD, Klijanienko J, Brisse H, et al. Chromosomal CGH identifies patients with a higher risk of relapse in neuroblastoma without MYCN amplification. Br J Cancer. 2007;97:238–46. doi: 10.1038/sj.bjc.6603820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Liu P, Jenkins NA, Copeland NG. A highly efficient recombineering-based method for generating conditional knockout mutations. Genome Res. 2003;13:476–84. doi: 10.1101/gr.749203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Bunting M, Bernstein KE, Greer JM, Capecchi MR, Thomas KR. Targeting genes for self-excision in the germ line. Gene Dev. 1999;13:1524–8. doi: 10.1101/gad.13.12.1524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Brault V, Pereira P, Duchon A, Herault Y. Modeling chromosomes in mouse to explore the function of genes, genomic disorders, and chromosomal organization. PLoS Genet. 2006;2:e86. doi: 10.1371/journal.pgen.0020086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Fujita T, Igarashi J, Okawa ER, Gotoh T, Manne J, Kolla V, et al. CHD5, a tumor suppressor gene deleted from 1p36. 31 in neuroblastomas. J Natl Cancer Inst. 2008;100:940–9. doi: 10.1093/jnci/djn176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.White PS, Thompson PM, Gotoh T, Okawa ER, Igarashi J, Kok M, et al. Definition and characterization of a region of 1p36. 3 consistently deleted in neuroblastoma. Oncogene. 2005;24:2684–94. doi: 10.1038/sj.onc.1208306. [DOI] [PubMed] [Google Scholar]
- 126.Thompson PM, Gotoh T, Kok M, White PS, Brodeur GM. CHD5, a new member of the chromodomain gene family, is preferentially expressed in the nervous system. Oncogene. 2003;22:1002–11. doi: 10.1038/sj.onc.1206211. [DOI] [PubMed] [Google Scholar]
- 127.Okawa ER, Gotoh T, Manne J, Igarashi J, Fujita T, Silverman KA, et al. Expression and sequence analysis of candidates for the 1p36. 31 tumor suppressor gene deleted in neuroblastomas. Oncogene. 2008;27:803–10. doi: 10.1038/sj.onc.1210675. [DOI] [PubMed] [Google Scholar]
- 128.Liu Z, Yang X, Li Z, McMahon C, Sizer C, Barenboim-Stapleton L, et al. CASZ1, a candidate tumor-suppressor gene, suppresses neuroblastoma tumor growth through reprogramming gene expression. Cell Death Differ. 2011;18:1174–83. doi: 10.1038/cdd.2010.187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Tivnan A, Tracey L, Buckley PG, Alcock LC, Davidoff AM, Stallings RL. MicroRNA-34a is a potent tumor suppressor molecule in vivo in neuroblastoma. BMC Cancer. 2011;11:33. doi: 10.1186/1471-2407-11-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Cole KA, Attiyeh EF, Mosse YP, Laquaglia MJ, Diskin SJ, Brodeur GM, et al. A functional screen identifies miR-34a as a candidate neuroblastoma tumor suppressor gene. Mol Cancer Res: MCR. 2008;6:735–42. doi: 10.1158/1541-7786.MCR-07-2102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Wei JS, Song YK, Durinck S, Chen QR, Cheuk AT, Tsang P, et al. The MYCN oncogene is a direct target of miR-34a. Oncogene. 2008;27:5204–13. doi: 10.1038/onc.2008.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Welch C, Chen Y, Stallings RL. MicroRNA-34a functions as a potential tumor suppressor by inducing apoptosis in neuroblastoma cells. Oncogene. 2007;26:5017–22. doi: 10.1038/sj.onc.1210293. [DOI] [PubMed] [Google Scholar]
- 133.Stallings RL, Foley NH, Bryan K, Buckley PG, Bray I. Therapeutic targeting of miRNAs in neuroblastoma. Expert Opin Ther Targets. 2010;14:951–62. doi: 10.1517/14728222.2010.510136. [DOI] [PubMed] [Google Scholar]
- 134.Chen Y, Stallings RL. Differential patterns of microRNA expression in neuroblastoma are correlated with prognosis, differentiation, and apoptosis. Cancer Res. 2007;67:976–83. doi: 10.1158/0008-5472.CAN-06-3667. [DOI] [PubMed] [Google Scholar]
- 135.Sahin E, Depinho RA. Linking functional decline of telomeres, mitochondria and stem cells during ageing. Nature. 2010;464:520–8. doi: 10.1038/nature08982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Blasco MA, Lee HW, Rizen M, Hanahan D, DePinho R, Greider CW. Mouse models for the study of telomerase. Ciba Found Symp. 1997;211:160–70. doi: 10.1002/9780470515433.ch11. discussion 70-6. [DOI] [PubMed] [Google Scholar]
- 137.Decock A, Ongenaert M, Vandesompele J, Speleman F. Neuroblastoma epigenetics: from candidate gene approaches to genome-wide screenings. Epigenetics. 2011:6. doi: 10.4161/epi.6.8.16516. [DOI] [PubMed] [Google Scholar]
- 138.Agathanggelou A, Bieche I, Ahmed-Choudhury J, Nicke B, Dammann R, Baksh S, et al. Identification of novel gene expression targets for the Ras association domain family 1 (RASSF1A) tumor suppressor gene in non-small cell lung cancer and neuroblastoma. Cancer Res. 2003;63:5344–51. [PMC free article] [PubMed] [Google Scholar]
- 139.Pizzo PA, Poplack DG. Principles and practice of pediatric oncology. 6. Philadelphia: Wolters Kluwer/Lippincott Williams & Wilkins Health; 2011. [Google Scholar]
- 140.Dyer MA. Mouse models of childhood cancer of the nervous system. J Clin Pathol. 2004;57:561–76. doi: 10.1136/jcp.2003.009910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Jiang M, Stanke J, Lahti JM. The connections between neural crest development and neuroblastoma. Curr Top Dev Biol. 2011;94:77–127. doi: 10.1016/B978-0-12-380916-2.00004-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Kawaguchi J, Nichols J, Gierl MS, Faial T, Smith A. Isolation and propagation of enteric neural crest progenitor cells from mouse embryonic stem cells and embryos. Development (Cambridge, England) 2010;137:693–704. [Google Scholar]
- 143.Lee G, Chambers SM, Tomishima MJ, Studer L. Derivation of neural crest cells from human pluripotent stem cells. Nat Protoc. 2010;5:688–701. doi: 10.1038/nprot.2010.35. [DOI] [PubMed] [Google Scholar]
- 144.Sauer B, Henderson N. Site-specific DNA recombination in mammalian cells by the Cre recombinase of bacteriophage P1. Proc Natl Acad Sci U S A. 1988;85:5166–70. doi: 10.1073/pnas.85.14.5166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Tuveson DA, Jacks T. Technologically advanced cancer modeling in mice. Curr Opin Genet Dev. 2002;12:105–10. doi: 10.1016/s0959-437x(01)00272-6. [DOI] [PubMed] [Google Scholar]
- 146.Tuveson DA, Jacks T. Modeling human lung cancer in mice: similarities and shortcomings. Oncogene. 1999;18:5318–24. doi: 10.1038/sj.onc.1203107. [DOI] [PubMed] [Google Scholar]
- 147.Jacks T. Modeling cancer in the mouse. Harvey Lect. 2005;101:1–19. [PubMed] [Google Scholar]
- 148.Dymecki SM. Flp recombinase promotes site-specific DNA recombination in embryonic stem cells and transgenic mice. Proc Natl Acad Sci U S A. 1996;93:6191–6. doi: 10.1073/pnas.93.12.6191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Vooijs M, van der Valk M, te Riele H, Berns A. Flp-mediated tissue-specific inactivation of the retinoblastoma tumor suppressor gene in the mouse. Oncogene. 1998;17:1–12. doi: 10.1038/sj.onc.1202169. [DOI] [PubMed] [Google Scholar]
- 150.Urlinger S, Baron U, Thellmann M, Hasan MT, Bujard H, Hillen W. Exploring the sequence space for tetracycline-dependent transcriptional activators: novel mutations yield expanded range and sensitivity. Proc Natl Acad Sci U S A. 2000;97:7963–8. doi: 10.1073/pnas.130192197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Baron U, Bujard H. Tet repressor-based system for regulated gene expression in eukaryotic cells: principles and advances. Methods Enzymol. 2000;327:401–21. doi: 10.1016/s0076-6879(00)27292-3. [DOI] [PubMed] [Google Scholar]
- 152.Gossen M, Bujard H. Tight control of gene expression in mammalian cells by tetracycline-responsive promoters. Proc Natl Acad Sci U S A. 1992;89:5547–51. doi: 10.1073/pnas.89.12.5547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Littlewood TD, Hancock DC, Danielian PS, Parker MG, Evan GI. A modified oestrogen receptor ligand-binding domain as an improved switch for the regulation of heterologous proteins. Nucleic Acid Res. 1995;23:1686–90. doi: 10.1093/nar/23.10.1686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Kraman M, Bambrough PJ, Arnold JN, Roberts EW, Magiera L, Jones JO, et al. Suppression of antitumor immunity by stromal cells expressing fibroblast activation protein-alpha. Science. 2010;330:827–30. doi: 10.1126/science.1195300. [DOI] [PubMed] [Google Scholar]
- 155.Olive KP, Jacobetz MA, Davidson CJ, Gopinathan A, McIntyre D, Honess D, et al. Inhibition of Hedgehog signaling enhances delivery of chemotherapy in a mouse model of pancreatic cancer. Science. 2009;324:1457–61. doi: 10.1126/science.1171362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Frese KK, Tuveson DA. Maximizing mouse cancer models. Nat Rev Cancer. 2007;7:645–58. doi: 10.1038/nrc2192. [DOI] [PubMed] [Google Scholar]
- 157.Nishijo K, Chen QR, Zhang L, McCleish AT, Rodriguez A, Cho MJ, et al. Credentialing a preclinical mouse model of alveolar rhabdomyosarcoma. Cancer Res. 2009;69:2902–11. doi: 10.1158/0008-5472.CAN-08-3723. [DOI] [PMC free article] [PubMed] [Google Scholar]