Abstract
K5 lyase A (KflA) is a tailspike protein from the K5A phage that catalyzes the degradation of the capsule polysaccharide of K5 strains of Escherichia coli. The K5 E. coli capsule polysaccharide, also known as heparosan, is composed of the disaccharide repeating unit of [-4)-GlcA-β(1,4)-GlcNAc-α(1-] and therefore identical to the biological precursor of heparin and heparan sulfate (HS). KflA could supplement the heparin lyases for heparin and HS analysis. The first part of this study aimed to clarify ambiguity resulting from the revision of the KflA amino acid sequence in 2010 from that published in 2000. We found that only the expression of the updated sequence gave a soluble active enzyme, which produced heparosan degradation products similar to those of previous studies. Next, we examined the specificity of KflA toward heparosan oligosaccharides of varying sizes, all containing a single N-sulfated glucosamine (GlcNS) residue. The presence of GlcNS in an octasaccharide and a nonasaccharide chain directed cleavage by KflA to a single position at the reducing end of the substrate. However, an N-sulfated decasaccharide exhibited extensive cleavage at the nonreducing end of the chain, illustrating a distinct change in the cleavage pattern of KflA toward substrates of differing sizes. Because KflA is able to cleave a substrate containing isolated GlcNS residues, this enzyme could be used for the analysis of low-sulfate content HS domains.
Keywords: heparan sulfate, heparin, heparosan, lyase, oligosaccharide
Introduction
K5 lyase A (KflA) is a viral tailspike protein from the K5A bacteriophage that infects the K5 strain of Escherichia coli (Hänfling et al. 1996). The K5 strain of E. coli possesses a polysaccharide capsule that is composed of heparosan, a glycosaminoglycan with the repeating disaccharide unit: [-4)-GlcA-β(1,4)-GlcNAc-α(1-] (Roberts 1996). This polysaccharide capsule is recognized by the K5A phage and allows for host specificity, but the capsule must be penetrated for the phage to access the entry receptor on the cell surface. To accomplish this, KflA molecules on the phage tailspike bind and depolymerize the heparosan capsule and expose the viral entry receptor (Clarke et al. 2000; Leiman et al. 2007). KflA follows the general mechanism of the large class of polysaccharide-degrading enzymes called polysaccharide lyases (Garron and Cygler 2010). It cleaves heparosan by catalyzing a β-elimination reaction at C-4 of glucoronic acid (GlcA), which breaks the glycosidic bond and yields a Δ4,5 unsaturated GlcA at the nonreducing terminus of the oligosaccharide product (Clarke et al. 2000). Although it is believed to follow a catalytic mechanism like that of other polysaccharide lyases, the structure of KflA most closely resembles that of glycoside hydrolases in the GH90 family, many of which are also phage tailspike proteins serving an analogous function in viral entry (Garron and Cygler 2010). These tailspike proteins, including KflA, all feature homotrimeric parallel β-helix architecture (Barbirz et al. 2008; Thompson et al. 2010). It has been proposed that the intensive structural similarity of the phage tailspike proteins indicates an evolutionary relationship in spite of their lack of significant sequence homology (Jenkins et al. 1998; Barbirz et al. 2008).
The enzyme KflA and K5 strain of E. coli have been of interest in the field of heparin and heparan sulfate (HS) glycobiology, because the K5 capsule polysaccharide is structurally identical to the biosynthetic precursor of heparin and HS. Heparin and HS function in a wide variety of processes in mammals and have roles that include blood coagulation, cell proliferation and development (Lindhardt 2003). During synthesis by the cell, heparosan is assembled by glycosyltransferases and enzymatically modified in several ways: deacetylation and subsequent N-sulfation of GlcNAc, epimerization of GlcA to iduronic acid (IdoA), sulfation of the 2-OH on GlcA or IdoA and sulfation of 6- and 3-OH groups on N-acetylglucosamine (GlcNAc) or N-sulfated glucosamine (GlcNS; Lindhardt 2003). This results in a highly variable, heterogenous pattern of sulfation and epimerization in the chain. The tendency of polysaccharide lyases to cleave these complex substrates in a predictable manner has made them important tools in glycosaminoglycan research (Michaud et al. 2003; Sasisekharan et al. 2006). The bacterial polysaccharide lyases include three heparin lyases (heparinases I, II and III) that have been integral to many advances in the understanding of heparin and HS biology, such as the identification of the antithrombin-binding site on heparin that is responsible for its anticoagulant properties (Lindahl et al. 1979). More recently, KflA has been used to propose a structure for HS consisting of highly sulfated regions interspersed with unmodified domains possessing the heparosan-like structure and domains with intermediate levels of sulfation (Murphy et al. 2004). KflA has also been used to identify vascular endothelial growth factor-binding sites in HS (Robinson et al. 2006). Both of these experiments exploit the specificity of KflA for heparosan that has not been modified by sulfation or epimerization. The potential for further applications of KflA in heparin and HS research make it an attractive target for investigation.
There were two primary goals in this study. The first originated with the revision of the amino acid sequence of KflA (GenBank accession number CAA71133.2) that accompanied the publication of its crystal structure in 2010 (Thompson et al. 2010). In 2000, the protein sequence CAA71133.1 was published alongside a study that identified the gene for KflA within the K5 phage genome and described the expression of recombinant KflA (Clarke et al. 2000). Figure 1 shows a partial alignment of the two sequences of the regions with the most consecutive mismatches. There are a total of 42 mismatched amino acid residues between the two sequences. In the period after the first sequence was published, but before it was revised in 2010, a number of fundamental biochemical studies were done with KflA. It was not completely clear that the sequence was used for these studies, which included experiments on the specificity of KflA. Therefore, before beginning further experiments on the specificity of KflA, we wanted to be certain that the same protein sequence had been used throughout the earlier studies on KflA and that there was simply a mistake in the sequence data.
Fig. 1.
(A) There are a total of 42 mismatched amino acids in the two published sequences for KflA (CAA71133.1 from 2000 and CAA71133.2 from 2010). Shown here are 38 of the mismatches that are mostly concentrated in three rows of the alignment of the two sequences. (B) Expression of CAA71133.2 gave soluble, active protein with the expected molecular mass (56.3 kDa theoretical for post-translationally cleaved protein).
Our second goal was to assess the substrate specificity of KflA using synthetic oligosaccharide substrates. Products from treating the substrates with KflA were characterized by mass spectrometry. A similar strategy has been successfully employed by other groups on heparinases I and II (Ernst et al. 1998; Rhomberg et al. 1998). Previous studies on the specificity of KflA have focused on polymeric glycosaminoglycans and have shown that KflA does not degrade chondroitin sulfate, heparin, hyaluronic acid, N-sulfo heparosan or chemically desulfated N-acetylated heparin (Murphy et al. 2004; Rek et al. 2007). HS showed modest amounts of degradation by KflA, which was correlated with cleavage at regions possessing the heparosan-like structure (Murphy et al. 2004). Earlier work has also investigated the minimum size of substrates required for cleavage by KflA, and it was reported that heparosan decasaccharide was fully degraded, while octasaccharide and hexasaccharide left 50 and 90% undigested, respectively (Murphy et al. 2004). The heparosan-degrading activity of KflA was first observed in whole K5 phage particles, where it was reported to cleave the substrate with a random, endolytic mode of action (Hänfling et al. 1996). However, the cleavage of heparosan with K5 phage particles also gave larger-sized oligosaccharide products (decasaccharides to hexasaccharides) than the mixture of octasaccharides to disaccharides observed when heparosan is cleaved with the recombinant KflA enzyme (Hänfling et al. 1996; Murphy et al. 2004; Blundell et al. 2009). The reason for this difference is not clear.
Here, we have set to understand the specificity of KflA, especially with regard to isolated GlcNS residues and the substrate size. Up to this time, KflA has not been studied using structurally homogeneous sulfated oligosaccharides, so we expect this approach should move toward a more consistent understanding of its specificity and insight into its mode of action. We synthesized a series of oligosaccharide substrates for KflA based on the structure of heparosan and established the effect of a single GlcNS in the oligosaccharide chain. In the N-sulfated octasaccharide (Octa-4) substrate, no cleavage was seen at the [GlcNS-GlcA] bond; however, in an N-trifluoroacetylated octasaccharide (Octa-3), the [N-trifluoroacetyl glucosamine (GlcNTFA)-GlcA] bond was cleaved. Additionally, Octa-4 was only cleaved on the reducing side of the GlcNS. We elongated the Octa-4 substrate at the nonreducing end one sugar residue at a time, which allowed the GlcNS to be retained in the same relative position. When the substrate was extended to Deca-6, cleavage was observed on both the nonreducing and reducing sides of the GlcNS residue. Our study advances the understanding of the substrate specificity of KflA, providing an additional tool to analyze the structures of HS.
Results
Sequence CAA71133.2 gives a soluble and active enzyme
A recent paper in 2010 reports substantial changes in the amino acid sequence of KflA compared with the gene for KflA that was first identified in 2000 (Thompson et al. 2010). In the 10-year period between 2000 and 2010, a number of studies were done characterizing the biochemistry of KflA. To verify the sequence, we expressed and purified KflA protein based on the Genbank sequences for both CAA71133.1 (the sequence published in 2000) and CAA71133.2 (the sequence published in 2010). Both sequences were expressed in E. coli as (His)6-tagged fusion proteins and purified on a nickel-Sepharose column. Only the sequence CAA71133.2 yielded a protein with measurable activity and gave a band near the 56.3-kDa size as determined with sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) (Figure 1). While the estimated molecular weight (70.9 kDa) is larger than the apparent value, a post-translational protease cleavage is speculated. It is believed that the C-terminal end of KflA from Ser505 is autocatalytically cleaved to remove a 14.6-kDa autochaperone fragment from the initial 70.9-kDa protein (Thompson et al. 2010). To verify that the CAA771133.1 was being expressed, but in an insoluble form, we included a control where the CAA771133.1-containing vector was present but was not induced with isopropyl-β-d-thiogalactopyranoside. The 65-kDa band was only seen in the sample where expression was induced, but this band was not visible in the supernatant from the centrifugation step that followed cell lysis (Supplementary data, Figure S1). The lack of solubility for CAA771133.1 could be indicative of the full amino acid sequence that has not undergone the post-translational autocatalytic cleavage that removes a 14.6-kDa C-terminal fragment from the protein, yielding an insoluble form of protein. The C-terminal region from Ser505 contains 17 of the 42 total amino acid mismatches, possibly destabilizing the autochaperone and autocatalytic function that has been proposed to be associated with this region of the sequence (Thompson et al. 2010).
N-sulfo heparosan and 6-O-sulfo heparosan are not substrates for KflA
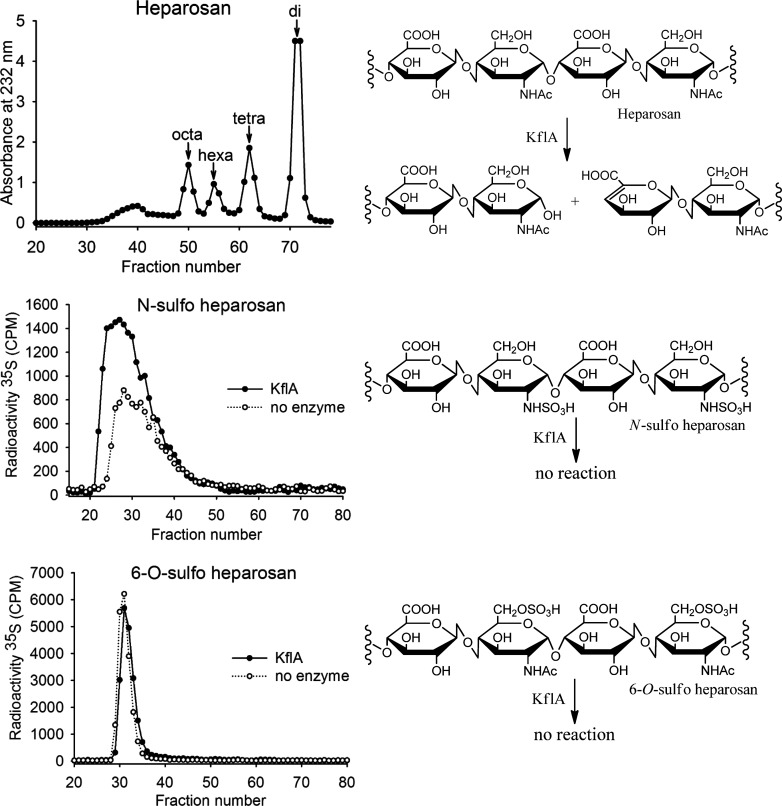
To verify that sequence CAA71133.2 encodes an active lyase, we evaluated the oligosaccharide products formed when heparosan was incubated with KflA. The elution profile of KflA-degraded heparosan from a BioGel P-10 column is shown in Figure 2A. The absorbance at 232 nm, which corresponds to the Δ4,5 unsaturated glucuronic acid product of KflA cleavage (as depicted in Figure 2D), was used to monitor the presence of product in fractions. Electrospray ionization mass spectrometry (ESI-MS) confirmed the identity of the peaks that are labeled in Figure 2A. Both the elution profile and indentifies of the products were largely consistent with those published in earlier work by other groups, namely the products are mixtures of oligosaccharides ranging from di- to octasaccharides (Murphy et al. 2004; Blundell et al. 2009).
Fig. 2.
Heparosan depolymerization by KflA gives a mixture of octasaccharides through disaccharides. These were identified by P-10 fractionation of the depolymerization products followed by scanning fractions at 232 nm and confirming peaks with ESI-MS. N-[35S]sulfo heparosan and 6-O-[35S]sulfo heparosan were treated with KflA or with vehicle. P-10 fractions show experimental and control samples elute in the same range and indicate that the enzyme was nonreactive with these polymers.
Next we set out to assess whether N-sulfo heparosan or 6-O-sulfo heparosan could act as a substrate for KflA. Both substrates were 35S-radiolabeled as described in “Experimental Procedures”. N-[35S]-sulfo heparosan and 6-O-[35S]sulfo heparosan were incubated with KflA overnight. After incubation, the reaction mixture was separated on a BioGel P-10 column, and the plots of counts per minute (CPM) vs. fraction were compared with the plot of a control consisting of the respective sulfated heparosan incubated without the enzyme. As illustrated in Figure 2B and C, N-sulfo heparosan and 6-O-sulfo heparosan treated with KflA were eluted the same as those of undigested polysaccharide substrates, suggesting that neither served as a substrate for KflA. With respect to N-sulfated heparosan, our result here confirmed that of others, which showed N-sulfated heparosan is not cleaved (Murphy et al. 2004). The effect of 6-O-sulfation on KflA substrate had not been assessed previously.
Minimum size of the KflA substrate
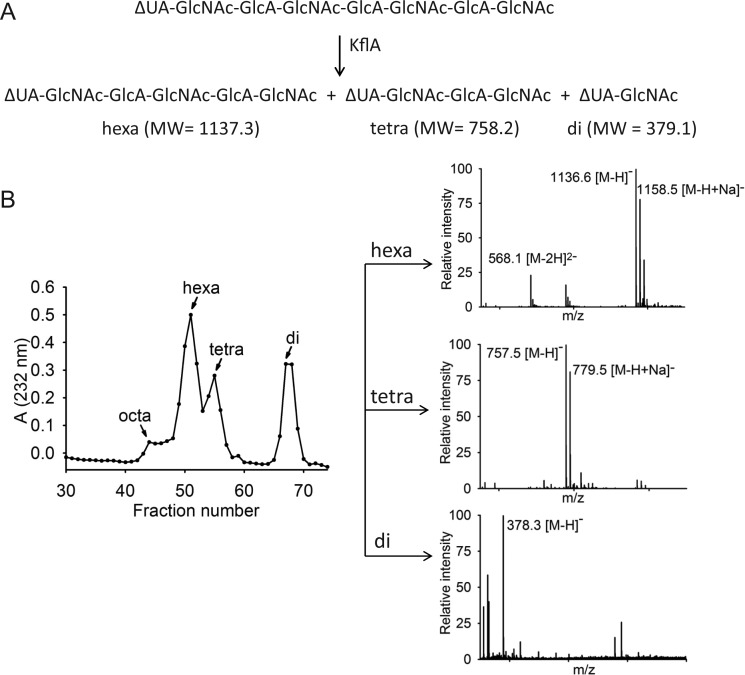
To determine the minimum length of substrate required for cleavage by KflA, we exposed the lyase to hexa- and octasaccharides. Both oligosaccharide substrates were isolated from KflA-degraded heparosan, and their structures were confirmed by ESI-MS as shown in Table I. The digestion reaction was carried out by incubating Δ4,5 hexasaccharide (Hexa-1) and Δ4,5 octasaccharide (Octa-2) with KflA, and the products were analyzed by the BioGel P-2 column coupled with ESI-MS (Figure 3). Octa-2 was almost fully degraded to yield di-, tetra- and hexasaccharides (Figure 3A and B). For example, the analysis of fraction 51 revealed a strong signal at the m/z value of 1136.6, along with 1158.5 m/z and 568.1 m/z. Taking these values as the [M−H]−, [M−H+Na]− and [M−2H]2− fragments, respectively, these indicate an oligosaccharide product with a molecular weight of 1137.7 ± 0.4 (Figure 3B). This measurement is very close to the calculated molecular weight of 1137.3 for a hexasaccharide with a structure of ΔUA-GlcNAc-GlcA-GlcNAc-GlcA-GlcNAc. The exposure of Hexa-1 to KflA resulted in mostly (undigested) hexasaccharide along with only a small amount of disaccharide, suggesting that Hexa-1 was resistant to digestion (data not shown). Our data suggest that octasaccharide is the minimum size for KflA digestion, which is consistent with the previous publication (Murphy et al. 2004). Based on this conclusion, our subsequent studies were focused on using substrates at least as large as octasaccharides to further investigate the substrate specificity of KflA.
Table I.
Summary of oligosaccharide substrates
| Abbreviated structures | Calculated MW | Measured MW | |
|---|---|---|---|
| Hexa-1 | ΔUA-GlcNAc-GlcA-GlcNAc-GlcA-GlcNAc | 1137.3 | 1138.2 ± 0.5 |
| Octa-2 | ΔUA-GlcNAc-GlcA-GlcNAc-GlcA-GlcNAc-GlcA-GlcNAc | 1516.4 | 1517.1 ± 0.4 |
| Octa-3 | GlcNAc-GlcA-GlcNTFA-GlcA-GlcNAc-GlcA-GlcNAc-GlcA-pNP | 1709.4 | 1710.2 ± 0.1 |
| Octa-4 | GlcNAc-GlcA-GlcNS-GlcA-GlcNAc-GlcA-GlcNAc-GlcA-pNP | 1693.4 | 1694.4 ± 0.1 |
| Nona-5 | GlcA-GlcNAc-GlcA-GlcNS-GlcA-GlcNAc-GlcA-GlcNAc-GlcA-pNP | 1869.5 | 1870.4 ± 0.1 |
| Deca-6 | GlcNAc-GlcA-GlcNAc-GlcA-GlcNS-GlcA-GlcNAc-GlcA-GlcNAc-GlcA-pNP | 2072.5 | 2073.4 ± 0.1 |
Fig. 3.
(A) The possible products from KflA catalyzed degradation of the Δ4,5 octasaccharide, Octa-2. The theoretical molecular weights of each of the fragments are also shown. (B) BioGel P-2 fraction absorbance profile from KflA cleavage of Octa-2. ESI-MS confirmed the identity of the peak fractions.
Effect of a single N-sulfation on cleavage of octasaccharide substrate
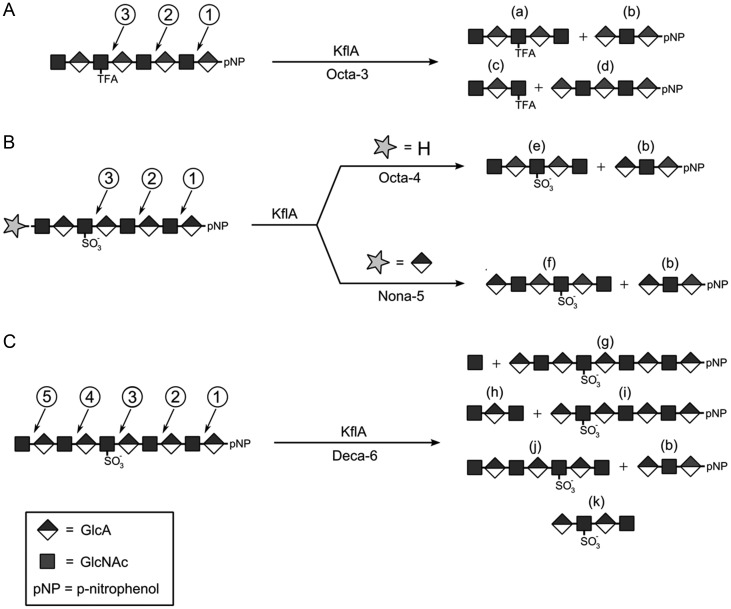
We determined the contribution of N-sulfation to the susceptibility to degradation by KflA using a series of synthetic oligosaccharides. To this end, a total of four oligosaccharides, ranging from octa- to decasaccharides, were synthesized (Octa-3 to Deca-6, Table I). The detailed procedures for the synthesis of these oliogsaccharides are described in “Experimental Procedures”. The results from ESI-MS analysis confirmed the identity of the anticipated products (Table II–IV). To first determine the sites cleaved by KflA for substrate lacking N-sulfation, we began with the octasaccharide shown in Figure 4A. In Octa-3, the third sugar residue from the nonreducing end is a GlcNTFA residue. The GlcNTFA residue is a bioisostere of GlcNAc, and it can be readily converted to a GlcNS via chemoenzymatic methods (Masuko et al. 2012). In addition, a para-nitrophenol (pNP) group is attached to the reducing end of the substrate, which is used as a means for ultra violet detection (monitored at 310 nm). The substrates were incubated with KflA, and the products were analyzed by BioGel P-2 and ESI-MS. The plots of A310 vs. fraction number illustrate the relative amounts of the pNP-tagged species and are shown in Supplementary data, Figure S2. The summary of these analyses is shown in Tables II–IV. For Octa-3 substrate, degradation with KflA yielded four products (Figure 4A), indicative of two sets of cleavages by KflA (Table II). One cleavage occurred at position 2, yielding pentasaccharide(a) and trisaccharide(b); another cleavage occurred at position 3, yielding trisaccharide(c) and pentasaccharide(d). Our data also confirmed that neither the pNP group nor the TFA substitution on the glucosamine residue affects the susceptibility to degradation by KflA.
Table II.
ESI-MS and BioGel P-2 analysis of KflA digested Octa-3
| Substrates | Abbreviated structures | Fraction# from BioGel P2 | Calculated MW | Measured MW |
|---|---|---|---|---|
| Octa-3 | GlcNAc-GlcA-GlcNTFA-GlcA-GlcNAc-GlcA-GlcNAc-GlcA-pNP | – | – | – |
| Products | ||||
| a | GlcNAc-GlcA-GlcNTFA-GlcA-GlcNAc | 35 | 1033.3 | 1033.5 |
| b | ΔUA-GlcNAc-GlcA-pNP | 44 | 676.2 | 676.4 ± 0.1 |
| c | GlcNAc-GlcA-GlcNTFA | 41 | 654.2 | 654.4 |
| d | ΔUA-GlcNAc-GlcA-GlcNAc-GlcA-pNP | 36 | 1055.3 | 1055.4 ± 0.04 |
Table III.
ESI-MS and BioGel P-2 analysis of KflA digested Octa-4 and Nona-5
| Substrates | Abbreviated structures | Fraction# from BioGel P2 | Calculated MW | Measured MW |
|---|---|---|---|---|
| Octa-4 | GlcNAc-GlcA-GlcNS-GlcA-GlcNAc-GlcA-GlcNAc-GlcA-pNP | – | – | – |
| Products | ||||
| e | GlcNAc-GlcA-GlcNS-GlcA-GlcNAc | 35 | 1017.3 | 1017.4 |
| b | ΔUA-GlcNAc-GlcA-pNP | 42 | 676.2 | 676.4 ± 0.1 |
| Substrate | ||||
| Nona-5 | GlcA-GlcNAc-GlcA-GlcNS-GlcA-GlcNAc-GlcA-GlcNAc-GlcA-pNP | |||
| Products | ||||
| f | GlcA-GlcNAc-GlcA-GlcNS-GlcA-GlcNAc | 35 | 1193.3 | 1194.1 ± 0.6 |
| b | ΔUA-GlcNAc-GlcA-pNP | 47 | 676.2 | 676. 4 ± 0.1 |
Table IV.
ESI-MS and BioGel P-2 analysis of KflA digested Deca-6
| Substrates | Abbreviated structures | Fraction# from BioGel P2 | Calculated MW | Measured MW |
|---|---|---|---|---|
| Deca-6 | GlcNAc-GlcA-GlcNAc-GlcA-GlcNS-GlcA-GlcNAc-GlcA-GlcNAc-GlcA-pNP | – | – | – |
| Products | ||||
| b | ΔUA-GlcNAc-GlcA-pNP | 49 | 676.5 | 676.4 ± 0.1 |
| g | ΔUA-GlcNAc-GlcA-GlcNS-GlcA-GlcNAc-GlcA-GlcNAc-GlcA-pNP | 31 | 1852.5 | 1852.4 ± 0.6 |
| h | GlcNAc-GlcA-GlcNAc | 41 | 600.5 | 600.5 ± 0.1 |
| i | ΔUA-GlcNS-GlcA-GlcNAc-GlcA-GlcNAc-GlcA-pNP | 34 | 1473.2 | 1473.4 ± 0.2 |
| j | GlcNAc-GlcA-GlcNAc-GlcA-GlcNS-GlcA-GlcNAc | 32 | 1397.2 | 1397.6 ± 0.1 |
| k | ΔUA-GlcNS-GlcA-GlcNAc | 37 | 796.7 | 796.4 |
Fig. 4.
The cleavage of synthetic N-trifluoroacetylated or N-sulfated oligosaccharides by KflA. (A) The third sugar residue from the Octa-3 substrate at the nonreducing end is a GlcNAc bioisostere, GlcNTFA. The products identified by ESI-MS correspond to cleavage at two positions at the reducing side of the GlcNTFA. (B) Replacement of the trifluoroacetyl group with a sulfate eliminates cleavage at the [GlcNS-GlcA] bond, a result observed in all N-sulfated substrates. The N-sulfated substrates Octa-4 and Nona-5 are both cleaved at position 2. (C) The N-sulfo decassacharide Deca-6 was lysed at positions 2, 4 and 5, with two sites on the nonreducing side of GlcNS. The presence of fragment (k) indicates that some chains were cleaved multiple times.
After establishing the pattern of cleavage for Octa-3, the TFA group was exchanged with an N-sulfate to give Octa-4. The digestion of Octa-4 with KflA only yielded two products (Figure 4B), suggesting that only a single cleavage occurred on this substrate. Analysis with BioGel P-2 and ESI-MS revealed that the cleavage site is located at position 2. The lack of products corresponding to cleavage at position 3 demonstrates that KflA is unable to cleave the linkage consisting of –GlcNS-GlcA–.
The effect of the GlcNS residue on the KflA cleavage pattern was further investigated by extending the length of the oligosaccharide substrates. The addition of one GlcA to the nonreducing end of Octa-4 resulted in Nona-5. The digestion pattern of Nona-5 to KflA is very similar to that observed for Octa-4 (Figure 4B and Table III), namely only single cleavage was observed. The addition of another GlcNAc residue to Nona-5 yielded Deca-6. Incubation of Deca-6 with KflA resulted in a more complex mixture of products. The results from product analysis revealed that KflA cleaved on three sites in Deca-6 (Figure 4C): the first scenario is to cleave at position 5 to yield a nonasaccharide(g) and a monosaccharide product. Indeed, we observed the presence of nonasaccharide(g) in the product by ESI-MS analysis. The second scenario is to cleave at position 4, yielding trisaccharide(h) and hexasaccharide(i). Both trisaccharide(h) and hexasaccharide(i) were observed by ESI-MS (Table IV). The third scenario is to cleave at position 2, resulting in trisaccharide(b) and hexasaccharide(j). In addition, hexasaccharide(j) was further degraded to tetrasaccharide(k). All these oligosaccharide products were observed by ESI-MS (Table IV). As expected, no cleavage at position 3 was found.
Discussion
Polysaccharide lyases are valuable tools for glycosaminoglycan research, and KflA itself has already been shown to be useful in the study of the structure of HS. Understanding its specificity toward single sulfations of the heparosan substrate could further extend the applications of KflA in heparin and HS analysis. Here, we demonstrate that only Genbank sequence CAA71133.2 produced a soluble and active KflA enzyme. The degradation of polymeric heparosan with CAA71133.2 gave a product profile similar to that seen using KflA before the sequence was corrected in 2010 (Murphy et al. 2004). Through comparing our results with those published by other groups, it seems likely that CAA71133.1 was never used in the previous studies and was reported as the result of errors in gene sequencing. However, there were anomalies in the work that first identified and sequenced the KflA gene that led us to question, which protein sequence was used for this study. Their SDS–PAGE result shows a band at approximately 67 kDa, which is much closer to the 70.9 kDa expected for the full peptide than 56.3 kDa for the post-translationally cleaved product (Clarke et al. 2000). Our SDS–PAGE experiment gave a band positioned near 56 kDa, which corresponds to the removal of the 14.6-kDa segment. The crystal structure of KflA verifies that this segment is excised in the mature protein (Thompson et al. 2010). It is unclear why CAA71133.1 failed to give a soluble product. There are 17 of the 42 total mismatched amino acids from the sequence alignment clustered at the C-terminal end of CAA771133.1 (from Ser505 to the C-terminus), which could disrupt the autochaperone and autocatalytic functions associated with this region (Thompson et al. 2010).
In this article, we used defined oligosaccharides ranging from oligosaccharides to decasaccharides to show that KflA is sensitive to the presence of GlcNS residues. We have also shown that the glycosidic bond consisting of [GlcNS-GlcA] is resistant to degradation by KflA. Replacing the GlcNS residue with the GlcNAc bioisostere, GlcNTFA renders this bond susceptible to cleavage. Although the oligosaccharides were not fully digested, the extent of cleavage can be enhance by adding more KflA enzyme or prolonging the incubation time (Rek et al. 2007).
Our data show that heparosan substrates smaller than an octasaccharide are no longer susceptible to degradation. At the same time, incubation of KflA with polymeric heparosan results in mostly disaccharide products (Figure 2). Using a model of random endolytic cleavage by KflA (Hänfling et al. 1996), one would expect the majority of the products to be larger than hexasaccharides. A model of processive cleavage by polymeric heparosan KflA could explain the predominance of tetrasaccharides and disaccharides that is observed. Indeed, a processive cleavage model for the K1-5 phage has been proposed (Leiman et al. 2007).
It is apparent that the size of the substrate has effects on the cleavage pattern of KflA. All three of the synthetic substrates have a common cleavage site at position 2. However, when the substrate is extended to Deca-6, there is a dramatic change in the cleavage pattern by KflA (Figure 4). For instance, position 4 is not susceptible for cleavage in Nona-5, but is cleaved in Deca-6. The basis for this is unclear based on our understanding of the crystal structure of KflA. This work does show that KflA is able to recognize and cleave domains containing isolated GlcNS residues. Therefore, it may be possible to use KflA for illuminating the detailed structure of HS domains with low-sulfate content. However, the variable cleavage patterns of KflA due to substrate size effects should be accounted for when using KflA for HS structural analysis.
Materials and methods
Expression and purification of (His)6-tagged KflA
The cDNA for KflA with NCBI GenBank accession numbers CAA71133.2 and CAA71133.1 were purchased from GenScript optimized for expression in E. coli, in plasmid pUC57. The same protocol was used for both CAA71133.2 and CAA71133.1. The primers 5′-AGTACATATGATGGCAAAACTGACCAAACCG-3′ and 5′- AGGAGGATCCTTACTTCGGCAGGGCGGCCAG-3′ were synthesized (Invitrogen, NY). The restriction sites for Nde1 and BamH1, respectively, are underlined. Amplification via polymerase chain reaction consisted of 5 min at 95°C, then 25 cycles of the following: 30 s at 94°C, 30 s at 52°C, 2.5 min at 68°C and a final period of 10 min at 68°C. The vector pET-15b and the cDNA were treated with Nde1 and BamH1 and ligated using the manufacturer's protocol from the DNA rapid ligation kit (Roche, IN). E. coli DH5α (Invitrogen) was transformed with the ligation product, colonies from the transformation were shaken overnight in 3 mL Luria-Bertani (LB) media with carbenicillin (50 μg/mL for all LB media), and plasmid was extracted following the manufacturer's protocol with the QIAprep Spin Miniprep Kit (Qiagen, Hilden, Germany). Samples of purified plasmid were treated successively with Nde1 and BamH1 and resolved on 5% agarose gel to identify correctly ligated plasmids for sequencing (Genome Analysis Core Facility, University of North Carolina, CA). Plasmids (KflA in pET-15b) were transformed into BL21* E. coli (Invitrogen), and colonies were shaken in 3 mL LB media overnight (37°C), added to 100 mL LB media and grown to A600 between 0.6 and 0.8 at 37°C, then cooled to 22°C and induced with isopropyl-β-d-thiogalactopyranoside (20 mM). The induced cells were shaken at 22°C for 18 h and harvested by centrifugation (4000 × g, 12 min, 5°C). The pellet was suspended in ice cold buffer A (25 mM Tris–HCl, 500 mM NaCl, 40 mM imidazole, pH 8.5) and lysed with 3 × 30 s rounds of sonication (Branson, 45% duty cycle, power output 9), followed by centrifugation (15,000 × g, 30 min, 5°C). Supernatant was filtered (Millipore, 45 μm) and loaded on a nickel sepharose column (GE Healthcare, Uppsala, Sweden, 700 μL bed volume; Bio-Rad 0.8 × 4 cm, 0.5 mL/min). The column was washed with 75 mL buffer A and eluted with 1 mL buffer B (25 mM Tris–HCl, 500 mM NaCl, 40 mM imidazole, pH 8.5). The quantity of protein was estimated by A280, with 0.72 for sequence CAA71133.2 and 0.14 for CAA71133.1. Each eluent was tested for activity by adding 1 μL eluent to 3 μg of heparosan in 100 μL (Tris–HCl 25 mM, pH 8.5) and monitoring A232 and A280, with only CAA71133.2 showing significant (42%) increase in A232 over a 1-min period. Analyzed with 12% SDS–PAGE, eluent from protein CAA71133.2 ran at purity judged to be ≥90%, while no significant band was visible for eluent from sequence CAA71133.1. To test CAA71133.1 cells for the expression of insoluble protein, approximately 10 μL of pellet from the postlysis centrifugation of CAA71133.1 was added to 20 μL water and 20 μL Laemelli buffer (5% β-mercaptoethanol), then boiled for 10 min to give a strong band at 65 kDa. Postlysis supernatant from CAA71133.1 was also used for. Finally, a 100 μL sample of cells for CAA71133.1 taken directly after induction and incubation, harvested by tabletop centrifuge (5300 × g, 3 min), decanted, suspended in 20 μL water and 20 μL Laemlli buffer (5% β-mercaptoethanol) and boiled for 10 min prior to 12% SDS–PAGE.
Expression and purification of sulfotransferases and glycosyltransferases
The synthesis of the N-sulfated and N-trifluoroacetylated oligosaccharides and sulfated heparosan required the enzymes N-sulfotransferase (NST), 6-O-sulfotransferase isoforms 1 and 3 (6-OST-1, 6-OST-3), N-acetylglucosaminyltransferase (KfiA) and Pasteurella multocida heparosan synthase (pmHS2). The expression and purification of these enzymes has been described before (Chen et al. 2005, 2007). Briefly, the enzymes were expressed in E. coli as fusion proteins and purified accordingly, with NST tagged to a glutathione S-transferase and purified on a glutathione sepharose column (GE Healthcare), while 6-OST-1 and 6-OST-3 were expressed as maltose-binding protein fusions and purified on an amylose column (GE Healthcare). Both KfiA and pmHS2 were expressed as (His)6 -tag fusions and purified on a nickel sepharose column (GE Healthcare).
Heparosan incubation with KflA
A solution of 5 mg/mL heparosan (molecular weight ∼30 kDa; Volpi 2004) was incubated with 9.0 μg of KflA in a total volume of 1 mL (25 mM HCl, pH 8.5) for 2 h at 37°C. The method used for the production of heparosan is described in section “Synthesis of heparosan”. The crude reaction mixture was mixed with 1 μL 0.5% phenol red and separated on a BioGel P-10 column (BioRad, 0.75 × 200 cm) with an elution buffer containing 20 mM Tris–HCl and 1 mM NaCl (pH 7.5) at a flow rate of 3 mL/h. The absorbance of each fraction was measured at 232 nm. The absorbance vs. fraction number plot shown in Figure 2 was constructed, and the three fractions with the highest absorbance for each of the four prominent peaks were pooled. Prior to analysis with mass spectrometry, these were either desalted by dialysis or BioGel P-2. The pooled fractions corresponding to the peak eluting last were assumed to be disaccharide. We desalted a 0.5-mL sample of these pooled fractions (with 1 μL 0.5% phenol red) with BioGel P-2 (Biorad) in 0.1 M ammonium bicarbonate with a flow rate of 3 mL/h. The other pooled fractions from the P-10 separation were dialyzed against 20 mM ammonium bicarbonate using molecular weight cutoff (MWCO) 3500 membrane and dried. Each sample was identified by ESI-MS analysis.
N-[35S]-sulfo heparosan and 6-O-[35S]-sulfo heparosan incubation with KflA
N-[35S]-sulfo heparosan (10,000 CPM, 0.4 μg in 30 μL water) and KflA in buffer B (7 μg in 20 μL; 25 mM HCl, 500 mM NaCl, 250 mM imidazole, pH 8.5) were added to 100 μL buffer (25 mM Tris–HCl, pH 8.5) and kept at 37°C for 24 h. For the negative control sample, 20 μL buffer B with no enzyme was added. The reaction mixture was resolved on BioGel P-10, and 35S CPM for each fraction was measured (Packard Tri-Carb 2500 TR) using 100 μL of eluent in 2 mL scintillation fluid (Econo-Safe, Research Products International, IL). The 6-O-[35S]-sulfo heparosan (3 × 105 CPM, 2.5 μg in 10 μL water) and KflA in buffer B (7 μg in 20 μL; 25 mM HCl, 500 mM NaCl, 250 mM imidazole, pH 8.5) were mixed in 200 μL buffer (25 mM Tris, pH 8.5) at 37°C for 24 h, followed by P-10 purification and 35S counting as for the N-[35S]sulfo heparosan sample.
Minimum substrate size requirement of KflA
The dried Hexa-1 and Octa-2 were redissolved in water (see “Heparosan incubation with KflA”). Each of the two reactions contained approximately 250 μg of Hexa-1 (in 66 µL H2O) or Octa-2 (in 40 µL H2O), and KflA (9 μg in 25 µL water) diluted to a total volume of 525 μL (25 mM Tris–HCl, pH 8.5). After 18 h incubation at 37°C, phenol red (5 μL of 0.5%) was added and the individual mixtures were separated on a BioGel P-2 column. The fractions were scanned at 232 nm, and fractions corresponding to peaks in the absorbance vs. fraction plot were analyzed with ESI-MS.
Preparation of heparosan
Both polysaccharides were prepared from heparosan, which is derived from E. coli K5 and based on a previously reported method (Vann et al. 1981). E. coli K5 (Bi 8337-41) (50 μL stock) is added to 100 mL LB media and grown 8 h (37°C). The culture is expanded with 15 mL of this culture added per liter of LB media and incubated for 16–18 h (37°C). This method uses 6 L of culture. The cells are pelleted (4000 × g, 30 min), and the supernatant is filtered and adjusted to pH 5.0. This is diluted 1:1 with buffer A (20 mM sodium acetate, 50 mM NaCl, pH 4.0) and loaded onto a column containing 100 mL diethylaminoethyl cellulose (DEAE) per 12 L of the diluted supernatant (flow rate 10 mL/min). After loading, the column is washed with 1 L buffer A and eluted with 300–350 mL buffer B (20 mM sodium acetate, 1 M NaCl, pH 4.0). Ethanol (100%) is added to the eluent and a precipitate is formed. The eluent is kept overnight at −20°C and centrifuged (4000 × g, 30 min, 5°C). The pellet is resuspended in 25 mL H2O. A solution of 4 M ammonium sulfate is added to give a 1:1 mixture. This is cooled on ice for 15 min, then centrifuged (3500 × g, 30 min). The gel-like precipitate is saved and the supernatant is removed and precipitated again with the addition of ammonium sulfate to 60%. This is centrifuged (3500 × g, 30 min) and the precipitate is combined with the previous precipitate and resuspended in 10 mL H2O. This is dialyzed overnight against H2O with MWCO 12,000–14,000. The average MW of heparosan isolated from E. coli K5 strain is about 30 kDa (Vann et al. 1981).
35S labeled N-[35S]-sulfo heparosan and 6-O-[35S]-sulfo heparosan
The synthesis of 35S-labeled 6-O-[35S]-sulfo heparosan and N-[35S]-sulfo heparosan has been discussed (Peterson and Liu 2010). To prepare N-[35S]-sulfo heparosan, the heparosan was first deacetylated under basic conditions and subsequently dialyzed. The heparosan or chemically deacetylated heparosan was sulfated with three molar equivalents of [35S] 3′-phosphoadenosine-5′-phosphosulfate (PAPS) and its respective sulfotransferase enzyme (6-OST-1 and -3 isoforms, or NST). The reaction mixtures were purified by a DEAE column. A bed volume of 0.2 mL DEAE was used for approximately 10 µg of substrate. The column was washed with 1 mL H2O (2×), 1 mL 1 M NaCl (2×), 1 mL 250 mM NaCl (2×) and equilibrated with 1 mL DEAE buffer (2 × 1 mL) (50 mM sodium acetate, 150 mM NaCl, 3 M urea, 2 mM ethylenediaminetetraacetic acid, 0.01% Triton X-100). The reaction mixture is loaded onto the column, which is then washed with DEAE buffer (4×) and 250 mM NaCl (4×). The sulfated polysaccharide is eluted with 1 mL of 1 M NaCl, dialyzed and dried.
Synthesis of Octa-3, Octa-4, Nona-5, Deca-6
A detailed description of the general method for the synthesis of heparin-like oligosaccharides has been given before (Liu et al. 2010). The starting compound GlcA-pNP (Sigma, MO) was mixed with roughly equimolar amounts of UDP-GlcNAc and 0.1 g/L KfiA and incubated overnight at 37°C. High performance liquid chromatography (HPLC) (YMC polyamine II column) was used to insure that the reaction had run to ≥95% completion. After completion, UDP-GlcA (1.1 equimolar) and pmHS2 (0.1 g/L) were added to the reaction mixture, incubated (37°C) and again monitored with HPLC. The products of the reaction were purified with a BioGel P-10 column, and the trisaccharide was dialyzed (MWCO 1000), dried and elongated with two additional cycles. GlcNTFA residue was incorporated into the chain by substituting UDP-GlcNAc with UDP-GlcNTFA in the third cycle. A final addition of GlcNAc followed by P-10 purification and drying gave Octa-3, which was the basis of the other three synthetic substrates. The position of the GlcNTFA group was selectively N-sulfated at the position of the GlcNTFA. This was done by first hydrolyzing the GlcNTFA to the free amine (triethylamine in methanol/water), followed by drying. The resulting de-N-trifluoroacetylated octasaccharide was sulfated with 3 M equivalents of PAPS in the presence of NST (overnight, 37°C), the reaction mixture was purified on a BioGel P-2 column and the product was dried. To obtain Nona-5, a sample of free amine octasaccharide was extended once with pmHS2 catalyzed addition of UDP-GlcA, purified on a BioGel P-2 column and dried. The resulting nonasaccharide was sulfated (PAPS and NST), purified on P-2 and dried. Deca-6 was obtained from Nona-5 by adding UDP-GlcNAc (KfiA), followed by P-2 purification and drying. The identity of each of the final products was confirmed by ESI-MS.
Synthesis of GlcNTFA and PAPS
A chemoenzymatic method was used to synthesize GlcNTFA (Liu et al. 2010; Zhou et al. 2011). The starting material GlcNH2 1-phosphate was reacted with S-ethyl trifluorothioacetate to give GlcNTFA 1-phosphate, which was condensed with uridine diphosphate with glucosamine-1-phosphate acetyl-transferase/N-acetylglucosamine-1-phosphate uridyltransferase (GlmU). GlmU is expressed in E. coli and purified with a nickel sepharose column. The sulfotranferase co-factor PAPS is also synthesized by a chemoenzymatic method, starting from adenosine triphosphate (ATP) (Zhou et al. 2011). In a one pot reaction, ATP is mixed with organic sulfate and is phosphorylated at the 3′ position by adenosine 5′-phosphosulfate (APS) kinase, while ATP sulfurylase hydrolytically cleaves pyrophosphate and replaces it with sulfate to give PAPS. The reaction is carried out in the presence of pyrophosphatase to drive the reaction toward product. PAPS is purified from the reaction mixture using a DEAE column. The three enzymes used are expressed in E. coli as (His)6-tagged fusions and purified with a nickel column.
Cleavage of synthetic oligosaccharides
All reactions were carried out in 525 μL (Tris–HCl 25 mM, pH 8.5) total volume containing 9 μg of KflA (in 25 μL buffer: 25 mM Tris–HCl, 500 mM NaCl, 250 mM imidazole, pH 8.5) at 37°C for 18 h. Approximately 100–300 μg of substrate was contained in each reaction. For Octa-3 and Octa-4, 100 μg of substrate was added; 300 μg of Nona-5; 200 μg of Deca-6 was used. The reaction mixture was separated on BioGel P-2 and fractions were scanned at 232 and 310 nm. A sample of each fraction was analyzed with ESI-MS.
Electrospray ionization mass spectrometry analysis
All mass spectrometry was done using electrospray negative ionization on a Thermo LCQ Deca. Eluent from P-2 column or from dialyzed P-10 fractions (1 μL for heparosan digest, 15 μL for all other oligosaccharides) was added to 200 μL in 8:2 water/methanol. The sample was injected by a syringe pump (Hamilton, NV) at 35 μL/min. Nonsulfated oligosaccharides were ionized at 5 kV potential and 275°C, with sulfated oligosaccharides at 3 kV and 175°C. Scans were averaged over 1 min and data were analyzed with Xcalibur software.
Supplementary data
Supplementary data for this article is available online at http://glycob.oxfordjournals.org/.
Funding
This work was supported by the National Institutes of Health grants 5R25GM055336 (to support Tim O'Leary's PhD studies) and HL094463.
Abbreviations
APS, adenosine 5′-phosphosulfate; ATP, adenosine triphosphate; CPM, counts per minute; DEAE, diethylaminocellulose; EDTA, ethylenediaminetetraacetic acid; ESI-MS, Electrospray ionization mass spectrometry; GlcA, glucoronic acid; GlcNAc, N-acetylglucosamine; GlcNS, N-sulfated glucosamine; GlcNTFA, N-trifluoroacetyl glucosamine; GlmU, glucosamine-1-phosphate acetyl-transferase/N-acetylglucosamine-1-phosphate uridyltransferase; HPLC, high performance liquid chromatography; HS, heparan sulfate; Hexa-1, Δ4,5 hexasaccharide; IdoA, iduronic acid; KflA, K5 lyase A; LB, Luria-Bertani; MS, mass spectrometry; MWCO, molecular weight cutoff; NST, N-sulfotransferase; Octa-2, Δ4,5 octasaccharide; 6-OST-1 and -3, 6-O-sulfotransferase 1 and 3; PAPS, [35S] 3′-phosphoadenosine-5′-phosphosulfate; PCR, polymerase chain reaction; pmHS2, Pasteurella multocida heparosan synthase; pNP, para-nitrophenol; SDS–PAGE, sodium dodecylsulfate polyacrylamide gel electrophoresis; UDP, uridine diphosphate.
Supplementary Material
References
- Barbirz S, Müller JJ, Uetrecht C, Clark AJ, Heinemann U, Seckler R. Crystal structure of Escherichia coli phage HK620 tailspike: Podoviral tailspike endoglycosidase modules are evolutionarily related. Mol Microbiol. 2008;69:303–316. doi: 10.1111/j.1365-2958.2008.06311.x. doi:10.1111/j.1365-2958.2008.06311.x. [DOI] [PubMed] [Google Scholar]
- Blundell CD, Roberts IS, Sheehan JK, Almond A. Investigating the molecular basis for the virulence of Escherichia coli K5 by nuclear magnetic resonance analysis of the capsule polysaccharide. J Mol Microbiol Biotechnol. 2009;17:71–82. doi: 10.1159/000215933. doi:10.1159/000215933. [DOI] [PubMed] [Google Scholar]
- Chen J, Avci FY, Muñoz EM, McDowell LM, Chen M, Pedersen LC, Zhang L, Linhardt RJ, Liu J. Enzymatically redesigning of biologically active heparan sulfate. J Biol Chem. 2005;280:42817–42825. doi: 10.1074/jbc.M504338200. doi:10.1074/jbc.M504338200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Jones CL, Liu J. Using an enzymatic combinatorial approach to identify anticoagulant heparan sulfate structures. Chem Biol. 2007;14:986–993. doi: 10.1016/j.chembiol.2007.07.015. doi:10.1016/j.chembiol.2007.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke BR, Fred E, Roberts IS. Cloning, expression, and purification of the K5 capsular polysaccharide lyase (KflA) from coliphage K5A: Evidence for two distinct K5 lyase enzymes. J Bacteriol. 2000;182:3761–3766. doi: 10.1128/jb.182.13.3761-3766.2000. doi:10.1128/JB.182.13.3761-3766.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ernst S, Rhomberg AJ, Biemann K, Sasisekharan R. Direct evidence for a predominantly exolytic processive mechanism for depolymerization of heparin-like glycosaminoglycans by heparinase I. Proc Natl Acad Sci USA. 1998;95:4182–4187. doi: 10.1073/pnas.95.8.4182. doi:10.1073/pnas.95.8.4182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garron M-L, Cygler M. Structural and mechanistic classification of uronic acid-containing polysaccharide lyases. Glycobiology. 2010;20:1547–1573. doi: 10.1093/glycob/cwq122. doi:10.1093/glycob/cwq122. [DOI] [PubMed] [Google Scholar]
- Hanfling P, Shashkov AS, Jann B, Jann K. Analysis of the enzymatic cleavage (β elimination) of the capsular K5 polysaccharide of Escherichia coli by the K5-specific coliphage: A reexamination. J Bact. 1996;178:4747–4750. doi: 10.1128/jb.178.15.4747-4750.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenkins J, Mayans O, Pickersgill R. Structure and evolution of parallel β-helix proteins. J Struct Biol. 1998;122:236–246. doi: 10.1006/jsbi.1998.3985. doi:10.1006/jsbi.1998.3985. [DOI] [PubMed] [Google Scholar]
- Leiman PG, Battisti AJ, Valorie D, Bowman1 KD, Mühlenhoff M, Gerardy-Schahn R, Scholl D, Molineux IJ. The structures of bacteriophages K1E and K1–5 explain processive degradation of polysaccharide capsules and evolution of new host specificities. J Mol Biol. 2007;371:836–849. doi: 10.1016/j.jmb.2007.05.083. doi:10.1016/j.jmb.2007.05.083. [DOI] [PubMed] [Google Scholar]
- Lindahl U, Backström G, Höök M, Thunberg L, Fransson LA, Linker A. Structure of the antithrombin binding site in heparin. Proc Natl Acad Sci USA. 1979;76:1218–1222. doi: 10.1073/pnas.76.7.3198. doi:10.1073/pnas.76.3.1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindhardt RJ. 2003 Claude S. Hudson Award address in carbohydrate chemistry. heparin: structure and activity. J Med Chem. 2003;46:2551–2564. doi: 10.1021/jm030176m. doi:10.1021/jm030176m. [DOI] [PubMed] [Google Scholar]
- Liu R, Xu Y, Chen M, Weïwer M, Zhou X, Bridges AS, DeAngelis PL, Zhang Q, Lindhardt RJ, Liu J. Chemoenzymatic design of heparan sulfate oligosaccharides. J Biol Chem. 2010;285:34240–34249. doi: 10.1074/jbc.M110.159152. doi:10.1074/jbc.M110.159152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masuko S, Bera S, Dixy GE, Michel W, Liu J, DeAngelis PL, Linhardt RJ. Chemoenzymatic synthesis of uridine diphosphate-GlcNAc and uridine diphosphate-GalNAc analogs for the preparation of unnatural glycosaminoglycans. J Org Chem. 2012;77:1449–1456. doi: 10.1021/jo202322k. doi:10.1021/jo202322k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaud P, Da Costa A, Courtois B, Courtois J. Polysaccharide lyases: Recent developments as biotechnological tools. Crit Rev Biotech. 2003;23:233–266. doi: 10.1080/07388550390447043. doi:10.1080/07388550390447043. [DOI] [PubMed] [Google Scholar]
- Murphy KJ, Merry CLR, Lyon M, Thompson JE, Roberts IS, Gallagher JT. A new model for the domain structure of heparan sulfate based on the novel specificity of K5 lyase. J Biol Chem. 2004;279:27239–27245. doi: 10.1074/jbc.M401774200. doi:10.1074/jbc.M401774200. [DOI] [PubMed] [Google Scholar]
- Peterson SB, Liu J. Unraveling the specificity of heparanase utilizing synthetic substrates. J Biol Chem. 2010;285:14504–14513. doi: 10.1074/jbc.M110.104166. doi:10.1074/jbc.M110.104166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rek A, Thompson J, Roberts IS, Kungl AJ. Biophysical investigation of recombinant K5 lyase: Structural implications of substrate binding and processing. Biochim Biophys Acta. 2007;1774:72–77. doi: 10.1016/j.bbapap.2006.10.017. doi:10.1016/j.bbapap.2006.10.017. [DOI] [PubMed] [Google Scholar]
- Rhomberg AJ, Shriver Z, Biemann K, Sasisekharan R. Mass spectrometric evidence for the enzymatic mechanism of the depolymerization of heparin-like glycosaminoglycans by heparinase II. Proc Natl Acad Sci USA. 1998;95:12232–12237. doi: 10.1073/pnas.95.21.12232. doi:10.1073/pnas.95.21.12232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts IS. The biochemistry and genetics of capsular polysaccharide production in bacteria. Annu Rev Microbiol. 1996;50:285–315. doi: 10.1146/annurev.micro.50.1.285. doi:10.1146/annurev.micro.50.1.285. [DOI] [PubMed] [Google Scholar]
- Robinson CJ, Mulloy B, Gallagher JT, Stringer SE. VEGF165-binding sites within heparan sulfate encompass two highly sulfated domains and can be liberated by K5 lyase. J Biol Chem. 2006;281:1731–1740. doi: 10.1074/jbc.M510760200. doi:10.1074/jbc.M510760200. [DOI] [PubMed] [Google Scholar]
- Sasisekharan R, Raman R, Prabhakar V. Glycomics approaches to structure function relationships of glycosaminoglycans. Annu Rev Biomed Eng. 2006;8:181–231. doi: 10.1146/annurev.bioeng.8.061505.095745. doi:10.1146/annurev.bioeng.8.061505.095745. [DOI] [PubMed] [Google Scholar]
- Thompson JE, Pourhossein M, Waterhouse A, Hudson T, Goldrick M, Derrick JP, Roberts IS. The K5 lyase KflA combines a viral tail spike structure with a bacterial polysaccharide lyase mechanism. J Biol Chem. 2010;285:23963–23969. doi: 10.1074/jbc.M110.127571. doi:10.1074/jbc.M110.127571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vann WF, Schmidt MA, Jann B, Jann K. The structure of the capsular polysaccharide (K5 antigen) of urinary-tract-infective Escherichia coli 010:K5:H4: A polymer similar to desulfo-heparin. Eur J Biochem. 1981;116:359–364. doi: 10.1111/j.1432-1033.1981.tb05343.x. doi:10.1111/j.1432-1033.1981.tb05343.x. [DOI] [PubMed] [Google Scholar]
- Volpi N. Purification of the Escherichia coli K5 capsular polysaccharide and use of high-performance capillary electrophoresis to qualitative and quantitative monitor the process. Electrophoresis. 2004;25:3307–3312. doi: 10.1002/elps.200305856. doi:10.1002/elps.200305856. [DOI] [PubMed] [Google Scholar]
- Zhou X, Chandarajoti K, Truong QP, Liu R, Liu J. Expression of heparan sulfate sulfotransferases in Kluyveromyces lactis and preparation of 3′-phosphoadenosine-5′-phosphosulfate. Glycobiology. 2011;21:771–780. doi: 10.1093/glycob/cwr001. doi:10.1093/glycob/cwr001. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.