Abstract
As HIV-1 evolves over the course of infection, resistance against antiretrovirals may arise in the absence of drug pressure, especially against receptor and fusion blockers because of the extensive changes observed in the envelope glycoprotein. Here we show that viruses from the chronic phase of disease are significantly less sensitive to CCR5 receptor and fusion blockers compared to early infection variants. Differences in susceptibility to CCR5 antagonists were observed in spite of no demonstrable CXCR4 receptor utilization. No significant sensitivity differences were observed to another entry blocker, soluble CD4, or to reverse transcriptase, protease, or integrase inhibitors. Chronic as compared to early phase variants demonstrated greater replication when passaged in the presence of subinhibitory concentrations of fusion but not CCR5 receptor inhibitors. Fusion antagonist resistance, however, emerged from only one chronic phase virus culture. Because sensitivity to receptor and fusion antagonists is correlated with receptor affinity and fusion capacity, respectively, changes that occur in the envelope glycoprotein over the course of infection confer greater ability to use the CCR5 receptor and increased fusion ability. Our in vitro passage studies suggest that these evolving phenotypes increase the likelihood of resistance against fusion but not CCR5 receptor blockers.
Introduction
It has been hypothesized that as HIV-1 evolves within an infected host, resistant variants can emerge in the absence of antiretroviral pressure.1 Indeed, studies have observed polymorphisms at sites that confer resistance among subjects who have not received antiretroviral drugs.2–7 Modifications that confer antiretroviral resistance often impart replicative fitness costs, and as a result, viruses with such changes may not persist in the absence of antiretroviral pressure.8–12 However, a number of antiretroviral resistance mutations confer minimal replicative disadvantage, which provides a basis for their persistence in the absence of drug.13 The presence of low-frequency variants with drug resistance polymorphisms has been associated with greater risk of treatment failure.14
Minority drug-resistant variants are more likely to exist during the chronic as opposed to acute phase of infection. Most newly infected subjects harbor a limited number of viral species immediately after HIV-1 acquisition.15 Thus, if drug-resistant variants are not acquired, it is unlikely they will be circulating during acute infection. Over the course of infection, HIV-1 diversifies into a swarm of variants, termed quasispecies, which may contain low-frequency drug-resistant isolates.1,16 As a result, the potential for treatment failure theoretically increases if therapy is instituted during chronic as opposed to early phase disease because of the de novo evolution of drug-resistant variants. This premise, however, has never been directly tested because in most subjects, treatment is initiated without knowledge about the duration of infection.
Other phenotypic changes may also predispose to higher risk for treatment failure during the chronic as compared to the early stage of HIV-1 infection. For instance, during the acute phase of disease, most viruses utilize the CCR5 receptor and over time, the virus can evolve to use the CXCR4 receptor.17–20 Thus, the presence of CXCR4 using viruses will render CCR5 receptor blockers mostly ineffective during the chronic phase of disease. In addition to receptor usage changes, HIV-1's sensitivity to various antiretroviral drugs changes over time. We and others have shown that over the course of infection, envelopes have decreased sensitivity to CCR5 receptor and fusion inhibitors,21–25 although this has not been a universal observation in all subjects.26 Given the extensive changes observed in both the envelope and polymerase gene, it remains unclear if the sensitivity changes are observed only for entry blockers or also against other drugs from different antiretroviral classes.2,27
Drug resistance may also emerge more frequently among chronic as opposed to early infection variants because longitudinal sensitivity changes may affect replication in the presence of subtherapeutic drug concentrations. In vivo, resistance has been documented against all current antiretroviral drugs.28 All anti-HIV-1 drugs, except coreceptor antagonists, can become ineffective as a result of specific single point mutations within the viral genome. In vivo resistance against the CCR5 receptor inhibitors requires a set of genetic modifications, rather than a single canonical change.29,30 Although single point mutations can emerge due to random reverse transcriptase errors, de novo drug resistance primarily arises because of continued replication while on antiretroviral therapy. As a result, replication capacity in the presence of drug serves as a potential surrogate marker for future drug resistance evolution. Thus, in addition to the increasing viral diversity, longitudinal sensitivity changes may be another factor important in the emergence of drug resistance.
In this study, we compared sensitivity to a diverse panel of drugs among early and chronic infection variants from subjects with well-defined duration of infection. We also examined replication capacity and emergence of drug resistance among early and chronic infection variants passaged in the presence of subtherapeutic drug concentrations. We found that over time, HIV-1 becomes less sensitive to fusion and CCR5 receptor blockers. This decreased sensitivity impacts replication in the presence of fusion but not receptor blockers. Our results suggest that longitudinal sensitivity changes have the potential to impact treatment success with fusion but not necessarily CCR5 receptor blockers.
Materials and Methods
Subjects
All subjects examined were from the AIDS Linked to the IntraVenous Experience (ALIVE) cohort, which follows HIV-1-uninfected and HIV-1-infected injection drug users in Baltimore, Maryland through semiannual visits.31 HIV-1 seroconverters were identified through serological testing of longitudinal samples. The seroconversion date was estimated as the midpoint between the last HIV-1-seronegative visit and the day the first HIV-1-seropositive sample was obtained. We identified 30 antiretroviral-naive subjects in whom peripheral blood mononuclear cells (PBMCs) were available from around 3 months and around 2–3 years after estimated HIV-1 seroconversion.
The study was approved by human subjects review boards at Johns Hopkins University, Bloomberg School of Public Health, and Brigham and Women's Hospital; all participants provided informed written consent.
Viruses
Each subject's early and chronic infection PBMCs were cocultured with HIV-1-negative PBMCs. Viral supernatants were collected within 10 days after infection. DNA was isolated from each PBMC sample using the QIAmp DNA Blood Midi Kit (Qiagen). Bulk PCRs were used to amplify full-length envelope genes from both the early and chronic infection PBMC DNA sample as described previously.29,32 Amplified products from a minimum of four independent PCRs were combined to minimize resampling bias. A modified yeast gap-repair homologous recombination method was used to incorporate the envelope products within the NL4-3 clone as previously described with minor modifications.21,33,34 First, the full-length HIV-1 clone NL4-3 from pNL4-3 was incorporated into the multiple cloning site (MCS) of pRS315 (New England Biolabs).
Briefly, oligonucleotides 5′- CTATAGGGCGAATTGGAG CTCTACTTACACCAGGAAAGGCGCTACTTCTAGATGT ACT-3′ and 5′- GGAACAAAAGCTGGGTACCGTGCAAC CTCTACCTCCTGGGCGTACATCTAGAAGTAGC-3′ were used as templates in overlap PCR with primers 5′-CTATA GGGCGAATTGGAGCT-3′ and 5′-GGAACAAAAGCTGGG TACCG-3′. The resulting PCR product was inserted into XbaI-digested pRS315 using yeast gap-repair homologous recombination. Transformed yeast were selected in leucine dropout media. This newly engineered pRS315 was digested with XbaI and combined with AatII-digested pNL4-3 to transform and select yeast in leucine dropout media. Plasmids with NL4-3 within the MCS of pRS315 (pRS315-NL4-3) were confirmed by restriction digestion.
Second, the URA3 gene was amplified from pRS316 (New England Biolabs) using primers AGTCCCTGTTCGGGCGC CAGGTATTTCACACCGCAGGG and ACGACTCACTATA GGGCGAAAGATTGTACTGAGAGTGCAC. The underlined segments highlight portions homologous to primer binding site (PBS) and sequences immediately upstream from the 5′ long terminal repeat (LTR), respectively. The NL4-3 5′ LTR to PBS sequence was replaced with this URA3 PCR product by transforming yeast with linearized pRS315-NL4-3 and the URA3 PCR product, and selecting on CMM plates lacking leucine and uracil to generate pRS315-NL4-3Δ5′ end. The CMV immediate early promoter was amplified from pCDNA3 (Invitrogen) using primers AGTCCCTGTTCGG GCGCCACTCGGTACCAAGCTTGGGTC and ACGACTCA CTATAGGGCGAACGATGTACGGGCCAGATATAC; the underlined portions overlap with PBS and pRS315 sequences as above. Yeast were transformed with linearized pRS315-NL4-3Δ5′ end and the CMV promoter PCR product and selected on plates containing 5-fluoro-1,2,3,6-tetrahydro-2,6-dioxo-4-pyrimidine carboxylic acid (FOA) but lacking leucine to generate a plasmid incorporating NL4-3 sequences from PBS to the 3′ LTR and CMV promoter immediately upstream of PBS, pCMV-NL4-3-PBS→LTR. Within pCMV-NL4-3-PBS→LTR, the NL4-3 envelope gene was replaced with URA3 PCR product, which was amplified with the primers AGAAAGAGCAGAAGACAGTGGCAATGATTAATTAAA CCACCTTTTCAATTCATC and TTTTGACCACTTGCCAC CCATGGTATTTCACACCGCAGGG from pRS316. The underlined portions correspond to primer sequences used to amplify full-length envelopes from subject samples, and the sequences in italics contain a PacI restriction site. Different full-length envelopes were shuttled into NL4-3 plasmid by transforming yeast with pCMV-NL4-3-PBS→LTRΔGp160 linearized with PacI and amplified full-length envelope PCR products from subject PBMC samples. All recombinant NL4-3 clones containing a subject's envelope genes were pooled to generate a library of envelopes (pCMV-NL4-3-PBS→LTR+Envs).
PCR product containing yeast centromere sequence (CEN6), an autonomously replicating sequence (ARSH4), and the LEU2 gene was amplified from pRS315 as described previously.33 This PCR product was inserted into the MCS of pcDNA3 (Invitrogen) to generate pCDNA3-Leu. The HIV-1 NL4-3 fragment from 5′ LTR to around 700 gag base pairs was generated using primers GGTAGGCGTGTACGGTGGGAG GTCTATATAAGCAGAGCTCGTTTAGTGAACCGACAAG AAATCCTTGATCTG and GGCTGATCAGCGAGCTCTAGC ATTTAGGTGACACTATAGAATAGTGCTATGTCACTTCC CCTTGGTTCTCTC; the underlined portions correspond to pCDNA3 sequences. The italic portion of the primer is homologous to the beginning of the NL4-3 5′ LTR and gag sequences, respectively. Yeast were transformed with linearlized pCNA-Leu and the NL4-3-gag PCR product and selected on leucine media to generate CMV-NL4-3-LTR→Gag4. All yeast transformation were done with 33% PEG3350, 100 mM lithium acetate, and 0.28 μg/μl salmon sperm DNA. TOP10F E. coli (Invitrogen) were transformed with plasmids rescued from yeast to generate larger quantities of the plasmid. Incorporation of the different envelopes within NL4-3 was confirmed by sequence analysis. Replication-competent recombinant viruses were generated by cotransfecting 293T cells with equivalent quantities of CMV-NL4-3-LTR→Gag4 and the library of recombinant NL4-3 with the subject's envelopes (pCMV-NL4-3-PBS→LTR+Envs). Supernatants were collected 48 to 72 h after transfection, and the number of infectious particles (IP) was assessed on TZM-bl cells as previously described.21
Inhibitor sensitivity
TZM-bl, U87/CD4/CXCR4 and U87/CD4/CCR5 cells, zidovudine, lopinavir, efavirenz, raltegravir, TAK779, T-20, maraviroc, and soluble CD4 were obtained through the NIH AIDS Research and Reference Reagent Program.35,36 Infection of TZM-bl cells in the absence and presence of 2-fold serial dilution of the inhibitor was used to estimate the 50% inhibitory concentration (IC50) as previously described.21 Coreceptor usage was determined by monitoring p24 production in U87/CD4/CXCR4 and U87/CD4/CCR5 cells infected with 500 IP of each virus supernatant.
In vitro passaging in the presence and absence of drug
Viruses were passaged on PBMCs from HIV-1-negative donors in the presence of increasing concentrations of maraviroc or T-20. PBMC cultures at a concentration of 2×106 cells per milliliter were maintained in RPMI 1640 media containing glutamine, 10% fetal calf serum (FCS), and interleukin-2 (IL-2). About 5000 IP of each virus was adsorbed to 10×106 cells for 2 h prior to adding additional culture media. Drug was added after infections were established, generally 2 days after infection. Initial maraviroc and T-20 concentrations were around 1 nM and 0.005 μg/ml, respectively. Culture drug concentration was doubled approximately twice a week. Cultures were monitored for viral replication twice a week using an in house p24 antigen assay as previously described.37 The cultures were fed weekly with fresh phytohemagglutinin (PHA)-stimulated donor PBMCs. Cultures were also maintained without any drug to monitor virus replication on PBMCs. Days since infection were plotted against p24 values and the area under the curve was estimated in Prism5 (version 5.02).
Statistical analysis
Antiretroviral sensitivity, replication, and resistance characteristics of early infection viruses were compared to the chronic phase variants using the Wilcoxon matched-pairs signed-rank test. All p-values were based on a two-sided test. All statistical analyses were done with Intercooled Stata version 8.0 (Stata Corporation, College Station, TX).
Results
Sensitivity to various antiretrovirals
Virus was successfully expanded from 35 of the 60 PBMC samples, but both the early and the chronic infection PBMC sample yielded infectious virus stocks in only 12 subjects. HIV-1 virus copy numbers were significantly higher in the corresponding plasma for the PBMC samples that yielded successful cultures (median 3.8×104, range 9.8×102 −2.2×106 copies/ml) compared to the PBMCs from which virus could not be expanded (median 3.6×103, range 4.0×102 −6.5×104 copies/ml, p=0.0002, Wilcoxon rank-sum test). In the 12 subjects with chronic and early infection virus stocks, PBMCs were collected a median of 4.1 months (range 1.1–5.0 months) and 27.1 months (range 18–28.9 months) after estimated seroconversion, respectively (Table 1). In these 12 subjects, CD4 counts decreased from a median of 803.5 cells/μl at early infection to a median 508 cells/μl during the chronic phase. HIV-1 plasma levels were also lower for the chronic (median 3.2×104 copies per ml) as compared to the early (median 9.3×104 copies per ml) infection sample. All assays were performed with the coculture supernatants that demonstrated infectious virus on TZM-bl cells within 10 days of culture. Early phase infection virus supernatant titers (median 46, range 10–2150 IP/μl) were not significantly different from the chronic stage virus stock titers (median 196, range 10–2203 IP/μl, p=0.9).
Table 1.
Subject Demographics
| |
Early infection |
Chronic infection |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| |
|
|
|
U87b |
|
|
|
U87 |
||
| Subject | Interval (months)a | Virus level (copies/ml) | CD4 count (cells/μl) | CCR5 | CXCR4 | Interval (months)a | Virus level (copies/ml) | CD4 count (cells/μl) | CCR5 | CXCR4 |
| A1 | 4.2 | 94,947 | 439 | 1143 | <25 | 21.0 | 89,047 | 475 | 1124 | <25 |
| A2 | 3.0 | 85,439 | 142 | 3428 | <25 | 21.4 | 30,996 | 144 | 3771 | <25 |
| A4 | 5.0 | 2,158,680 | 342 | 2476 | 4999 | 24.5 | 79,094 | 293 | 2987 | 4976 |
| A5 | 4.1 | 279,745 | 895 | 998 | <25 | 27.2 | 137,661 | 698 | 766 | <25 |
| A8 | 4.1 | 346,923 | 780 | 1024 | <25 | 28.9 | 35,219 | 293 | 998 | <25 |
| A10 | 4.7 | 53,998 | 827 | 1240 | <25 | 28.1 | 2,709 | 1103 | 1085 | <25 |
| A17 | 4.9 | 40,634 | 1253 | 1098 | <25 | 20.9 | 20,995 | 541 | 995 | <25 |
| A18 | 3.2 | 188,258 | 1231 | 847 | <25 | 27.0 | 12,299 | 574 | 956 | <25 |
| A22 | 4.2 | 51,696 | 775 | 1987 | <25 | 27.7 | 32,334 | 448 | 2365 | <25 |
| A23 | 4.9 | 56,189 | 1006 | 855 | <25 | 27.7 | 79,490 | 407 | 925 | <25 |
| A26 | 1.1 | 90,967 | 1022 | 996 | <25 | 18.0 | 2,060 | 725 | 998 | <25 |
| A27 | 4.1 | 152,472 | 480 | 887 | <25 | 27.1 | 17,668 | 548 | 989 | <25 |
Interval between the estimated seroconversion date and day of sample collection.
Level of p24 antigen (pg/ml) in supernatants after 4 days of infection in either U87/CD4/CCR5 or U87/CD4/CXCR4 cells. Reference X4 virus (NL4-3) generated 4909 and <12.5 pg/ml p24 in U87/CD4/CXCR4 and U87/CD4/CCR5 cells, respectively. Reference R5 virus (YU-2) generated <12.5 and 3789 pg/ml p24 in U87/CD4/CXCR4 and U87/CD4/CCR5 cells, respectively. In the p24 assay, the limit of detection was around 25 pg/ml.
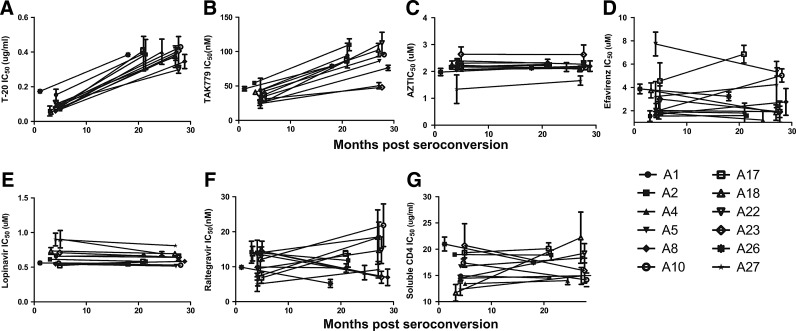
Early infection viruses demonstrated approximately 5-fold lower IC50s (median fold change 4.8, range 2.2–7.0) to a fusion inhibitor, T-20, compared to the chronic infection viruses (p=0.002) (Fig. 1A). Early infection viruses were also around two times more sensitive (median fold change 2.2, range 1.5–4.7) to CCR5 antagonist, TAK779, compared to HIV isolates from the chronic phase of infection (p=0.003) (Fig. 1B). Early and chronic infection virus supernatants, however, displayed no significant difference in their sensitivity to a representative compound from each of the four other different antiretroviral classes (nucleoside reverse transcriptase, nonnucleoside reverse transcriptase, protease, and integrase inhibitors) (Fig. 1C–F). CCR5 and fusion inhibitors IC50s demonstrated a significant monotonic association (r=0.7, p=0.0002, Spearman rank correlation). Significant sensitivity differences among early and chronic infection variants were observed with drugs that prevent virus entry into cells but not with compounds that inhibit reverse transcription, integration, or viral protein cleavage. Similar differences, however, were not evident among early and chronic infection variants with another entry inhibitor, soluble CD4, which prevents the virus from binding the CD4 receptor on target cells (Fig. 1G).
FIG. 1.
Antiretroviral sensitivity changes among longitudinally collected viruses. Graph shows longitudinal change in sensitivity to T-20 (A), TAK779 (B), AZT (C), efavirenz (D), lopinavir (E), raltegravir (F), and soluble CD4 (G). The y-axis shows the IC50s, which represent mean values from two or more independent experiments with the error bars showing the standard deviation. The x-axis shows the interval of time from the estimated date of seroconversion.
Coreceptor usage
The presence of a higher percentage of viruses that can use the CXCR4 receptor could be one potential explanation for the observed significant differences in the sensitivity to TAK779 among early and chronic infection viruses because the differential presence of CXCR4 utilizing variants could increase the CCR5 inhibitor IC50s. All subjects' viruses produced high p24 levels in U87/CD4/CCR5 cells and none, except subject A4 early and chronic infection variants, was able to replicate in U87/CD4/CXCR4 cells (Table 1). Thus, coreceptor usage variation does not account for the differences in sensitivity to CCR5 inhibitors among early and chronic phase infection variants. Because subject A4 viruses were able to utilize the CXCR4 receptor, they were insensitive to the CCR5 antagonist, TAK779, even at high concentrations.
Generation of replication-competent recombinant viruses
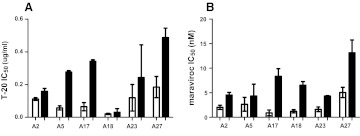
Virus stocks derived from PBMC passages may change the proportion and types of viruses present in the original subject sample.38 To examine inhibitor sensitivity changes over the course of infection in the absence of in vitro passaging, we constructed replication-competent recombinant viruses that incorporated subjects' early and chronic infection full-length envelope glycoproteins. For six subjects' early and chronic infection envelopes incorporated into NL4-3, the 293T-derived virus stocks demonstrated relatively high virus titer of greater than 10 IP/ml. In the remaining individuals, PBMC passages were required to generate workable titers from the 293T transfections from one of the time points. Among the 293T-derived viruses, the chronic (median 0.26 μg/ml, range 0.03–0.49 μg/ml) as compared to early (median 0.09 μl/ml, range 0.02–0.18 μg/ml, p=0.03) infection envelopes displayed significantly lower sensitivity to T-20 (Fig. 2A). In addition, viruses with the chronic (median 5.3 nM, range 4.3–13.1 nM) as compared to early (median 1.8 nM, range 0.9–5.1 nM, p=0.03) infection envelopes had significantly lower IC50 to another CCR5 inhibitor, maraviroc (Fig. 2B). Thus, the NL4-3-based, nonpassaged recombinant viruses displayed longitudinal sensitivity changes to CCR5 receptor and fusion blockers similar to that observed among the PBMC coculture virus stocks.
FIG. 2.
Sensitivity of replication competent recombinant viruses to CCR5 and fusion inhibitors. Figures show IC50s to T-20 (A) and maraviroc (B) among recombinant viruses with early infection (white bars) and chronic phase (black bars) envelopes. Subject IDs are on the x-axis. All IC50s represent mean values from two or more independent experiments with the error bars showing the standard deviation.
In vitro passage in the presence of subinhibitory drug
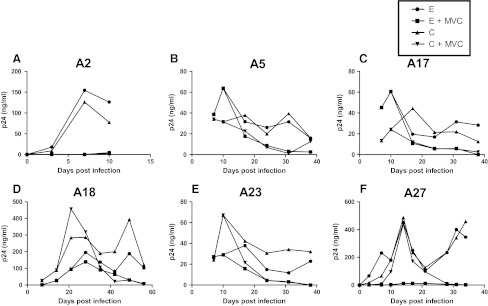
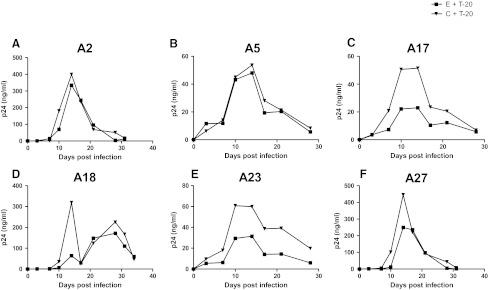
Decreasing sensitivity to CCR5 and fusion blockers among chronic as compared to early phase variants suggests that as the virus evolves over the course of infection, it may have greater likelihood of developing resistance. To examine this issue, we passaged the replication-competent recombinant viruses in the presence of subinhibitory concentrations of either maraviroc or T-20 (Figs. 3 and 4). Area under the p24 curve (AUC) was used to estimate replication over the entire culture period. Compared to replication in the absence of drug (AUC median 1128.5, range 526.4–10721) (shown in Fig. 3 only), viruses replicated significantly less efficiently in the presence of maraviroc (AUC median 621.1, range 5.2–7359, p=0.002) (Fig. 3) and showed a trend toward lower replication in the presence of T-20 (AUC median 1444.0, range 319.6–3310, p=0.07) (Fig. 4).
FIG. 3.
Replication in the presence and absence of subinhibitory concentrations of maraviroc. Figures (A–F) show the replication of recombinant 293T derived nonpassaged virus with early (E) and chronic (C) phase envelopes in the presence (+MVC) or absence of maraviroc. Subject IDs are above each graph. The x-axis shows the days since infection. The y-axis shows the p24 concentration and a line connects points from different times. The area under the curve (AUC) was estimated using GraphPad Prism5.
FIG. 4.
Replication in the presence of subinhibitory concentrations of T-20. Figures (A–F) show the replication of recombinant 293T-derived nonpassaged virus with early (E) and chronic (C) phase envelopes in the presence of T-20 (+T-20). Subject IDs are above each graph. The x-axis shows the days since infection. The y-axis shows the p24 concentration and a line connects points from different times. The area under the curve (AUC) was estimated using GraphPad Prism5.
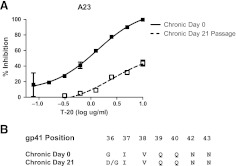
Maraviroc-resistant viruses did not emerge from the in vitro passaging of either the early or the chronic phase viruses by the time cultures were stopped due to insufficient virus (Fig. 3). In subject A5, chronic phase virus appears to break through around day 38 of culture, but no replication-competent virus was detected after this point (data not shown). In the absence of the emergence of resistant virus, we assessed whether early versus chronic phase viruses replicated more efficiently in the presence of maraviroc. As assessed by AUC, replication was not significantly different between early (AUC median 1069, range 793.2–7219) and chronic (AUC median 1191.5, range 526.4–10721, p=0.9) envelope recombinant virus in the absence of drug. In addition, AUC was also not significantly different among chronic (AUC median 621.1, range 9.1–7359) versus early (AUC median 585.1, range 5.2–3169, p=0.3) envelope recombinant viruses in the presence of maraviroc. In contrast to maraviroc, chronic envelope variants (AUC median 2067.0, range 660.2–3310.0) replicated significantly better in the presence of subinhibitory T-20 compared to the early phase viruses (AUC median 1274.6, range 319.6–2695.0, p=0.03) (Fig. 4). In addition, in subject A23, T-20-resistant virus emerged from the chronic but not early phase envelope recombinant viruses. By day 21, the chronic phase in vitro-passaged virus demonstrated minimal inhibition in the presence of 10 μg/ml of T-20, which was significantly different from the original stock (Fig. 5A). In the remaining five individuals, no resistant virus emerged from either the early or the chronic in vitro-passage cultures. Sequence analysis demonstrated that by day 21, A23 virus had acquired mutation, which has been associated with high level T-20 resistance (Fig. 5B).
FIG. 5.
Emergence of T-20-resistant virus. (A) T-20 inhibition of chronic phase day 0, nonpassaged (filled squares) versus post-day 21 passaged virus (hollow squares). The x-axis shows the amount of input T-20 in log μg/ml, and the y-axis shows the percent inhibition relative to infection without any inhibitor. Each point represents an average of at least two independent experiments performed in triplicate and is the mean percent inhibition with standard errors of mean. Nonlinear regression was used to estimate a fitted curve in GraphPad Prism5. (B) Predicated amino acids of gp41 envelope positions associated with resistance to T-20. Sequences are shown of bulk PCR envelopes amplified from day 0 and day 21 virus supernatants.
Discussion
In this study, we show that virus sensitivity to some HIV-1 antiretroviral agents changes over time, which influences replication capacity and potentially emergence of resistance. We found that viruses from around 2–3 years after acquisition were significantly less sensitive to coreceptor and fusion inhibitors compared to variants present during the first 6 months of infection. In contrast, we observed no significant differences among the viruses from the two infection time points in their sensitivity to CD4 receptor, reverse transcriptase, protease, and integrase inhibitors. Chronic as compared to early phase variants replicated more in the presence of subinhibitory T-20 but not maraviroc, although virus resistant to fusion antagonist emerged in only one subject. Our results suggest that the envelope as opposed to polymerase gene changes over the course of infection impact drug sensitivity, but the envelope glycoprotein evolution likely has minimal impact on the effectiveness of most entry inhibitors except potentially the fusion blockers.
A previous study showed that CCR5 and fusion inhibitors were equally potent against acute and chronic infection isolates, although the variants from the two different time points were not obtained from longitudinally collected samples.26 In contrast, other investigations of longitudinally followed HIV-1-infected subjects demonstrate that sensitivity to coreceptor and fusion antagonists decreases from the chronic to the late phase of disease.22–25 One of the major differences with our study and these previous publications is that we examined variants from the early and chronic phase of infection in a highly characterized seroincident cohort with well-defined dates of estimated seroconversion as opposed to comparing variants from the chronic and late stage of disease. In addition, we observed no significant differences in both PBMC-passaged and 293T-derived nonpassaged recombinant viruses, suggesting that sensitivity changes to CCR5 and fusion blockers were not an artifact from in vitro selection. Collectively, the results suggest that susceptibility to coreceptor and fusion inhibitors gradually decreases from acute infection to the chronic and late disease stages. It should be noted, however, that there may be an abrupt emergence of hypersensitive variants just prior to coreceptor switching as has been documented in one subject previously.39
Currently, entry inhibitors, such as maraviroc and T-20, are primarily used during the chronic or late periods of infection as salvage therapy. Usage of these entry inhibitors, especially the CCR5 receptor antagonists, may be more preferential in early disease stages because circulating variants have higher sensitivity to these drugs, and there is lower likelihood of CXCR4 using variants.17–20 Studies also suggest that variants with higher IC50s are the intermediate forms in the pathway to resistance against coreceptor and fusion inhibitors,35,40,41 and thus chronic as opposed to early infection isolates are potentially more likely to develop clinically relevant drug resistance under T-20 or maraviroc pressure. Therefore, use of these entry inhibitors during the chronic phase of infection may predispose to a relatively higher likelihood of failure, which could compromise the efficacy of the entire regimen.
To examine this issue, we passaged early and chronic phase variants in the presence of subinhibitory concentrations of T-20 and maraviroc. In one case, resistant virus emerged from chronic phase cultures, and in all subjects, the chronic as compared to early infection variants replicated at higher levels in the presence of low T-20 concentrations. Similar observations were not observed in the presence of subinhibitory maraviroc levels. This suggests that envelope modifications over the course of infection that confer decreased sensitivity to CCR5 receptor and fusion blockers could potentially increase the likelihood of eliciting resistance to the fusion but not the CCR5 receptor antagonists. Although chronic as compared to early phase variants are less sensitive to both CCR5 and fusion inhibitors, the likelihood of resistance may increase only against T-20 as opposed to maraviroc potentially because of the differences in the barrier to escape. In general, single amino acid mutations in the heptad repeat region 1 (HR1) can confer high level T-20 resistance while viruses generally require multiple mutations within the envelope V3 loop to replicate efficiently in the presence of CCR5 inhibitors.29,42–48 Thus, decreases in sensitivity alone may favor chronic phase viruses to develop T-20 resistance more efficiently, but this may be insufficient for the emergence of resistance against CCR5 receptor antagonists. Our observations reinforce the limited clinical utility of fusion inhibitors, especially with this theoretical disadvantage during the chronic and late phases of disease. On the other hand, in the absence of X4 variants, disease stage likely does not impact maraviroc efficacy.
As opposed to coreceptor and fusion entry inhibitors, chronic infection viruses were not more resistant to CD4 receptor blocker, soluble CD4. These observations suggest that the selection forces on the envelope glycoprotein affect its CCR5 interaction and fusion capacity but not CD4 engagement. During infection, increased production of CCR5 ligands, such as RANTES, MIP-1α, and MIP-1β, may drive this evolution.40,49 The presence of these chemokines decreases CCR5 receptor expression on target cells,50 which potentially forces viruses to acquire an ability to infect cells that have low CCR5 densities. The ability to replicate in cells with low CCR5 expression correlates with inhibition by CCR5 blockers, which is further related to T-20 susceptibility and fusion capacity.21,22,42,51 In aggregate, the presence of natural CCR5 ligands may drive the emergence of variants with a greater ability to use the CCR5 receptor and higher fusion capacity. Interestingly, the longitudinal difference in sensitivity to CCR5 antagonists relative to the change in T-20 susceptibility was fairly similar among the majority of subjects, which suggests that modifications that affect CCR5 use have similar consequences for fusion kinetics among fairly diverse envelopes. This implies that changes in CCR5 and fusion inhibitor susceptibility are potentially influenced by sequence modifications in one portion and less likely by simultaneous changes in different parts of the envelope gene.
To our knowledge, this is the first study that has examined the phenotypic susceptibility of variants isolated during different phases of infection to nucleoside and nonnucleoside reverse transcriptase, protease, and integrase inhibitors. Interestingly, no significant differences were observed, even though previous studies have demonstrated increasing polymorphisms at drug resistance sites in the polymerase gene over the course of infection.2 One caveat with our study is that we mostly used cocultures to generate virus stocks, and in vitro cultures and bulk PCR amplification fail to recapitulate the diversity present in the original subject sample.38,52 Even though the methods used in this study do not generate the minor species present in each subject's sample, we can conclude that sensitivity to the reverse transcriptase, protease, and integrase inhibitors remains relatively unchanged for the major variants present during the chronic as compared to the initial phase of disease. Our results imply that the duration of infection is unlikely to influence sensitivity to most antiretroviral drugs, except the CCR5 receptor and fusion inhibitors. Longitudinal decreased sensitivity, however, potentially facilitates the emergence of drug resistance against the fusion but not the CCR5 receptor inhibitors.
Acknowledgments
We thank all the subjects who have contributed samples for these studies as part of the ALIVE cohort. We thank Daniel Kuritzkes for insightful comments on the manuscript. This study was supported by the Doris Duke Charitable Foundation (M.S.), and from NIH Grants AI077473 (M.S.), DA04334 (G.K.), and DA12568 (S.M.). Nikolaos Chatziandreou and Ana Belen Arauz contributed equally to this publication.
Author Disclosure Statement
No competing financial interests exist.
References
- 1.Coffin JM. HIV population dynamics in vivo: Implications for genetic variation, pathogenesis, and therapy. Science. 1995;267:483–489. doi: 10.1126/science.7824947. [DOI] [PubMed] [Google Scholar]
- 2.Kearney M. Palmer S. Maldarelli F, et al. Frequent polymorphism at drug resistance sites in HIV-1 protease and reverse transcriptase. AIDS. 2008;22:497–501. doi: 10.1097/QAD.0b013e3282f29478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cornelissen M. van den Burg R. Zorgdrager F. Lukashov V. Goudsmit J. pol gene diversity of five human immunodeficiency virus type 1 subtypes: Evidence for naturally occurring mutations that contribute to drug resistance, limited recombination patterns, and common ancestry for subtypes B and D. J Virol. 1997;71:6348–6358. doi: 10.1128/jvi.71.9.6348-6358.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kozal MJ. Shah N. Shen N, et al. Extensive polymorphisms observed in HIV-1 clade B protease gene using high-density oligonucleotide arrays. Nat Med. 1996;2:753–759. doi: 10.1038/nm0796-753. [DOI] [PubMed] [Google Scholar]
- 5.Lech WJ. Wang G. Yang YL, et al. In vivo sequence diversity of the protease of human immunodeficiency virus type 1: Presence of protease inhibitor-resistant variants in untreated subjects. J Virol. 1996;70:2038–2043. doi: 10.1128/jvi.70.3.2038-2043.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mohri H. Singh MK. Ching WT. Ho DD. Quantitation of zidovudine-resistant human immunodeficiency virus type 1 in the blood of treated and untreated patients. Proc Natl Acad Sci USA. 1993;90:25–29. doi: 10.1073/pnas.90.1.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Najera I. Holguin A. Quinones-Mateu ME, et al. Pol gene quasispecies of human immunodeficiency virus: mutations associated with drug resistance in virus from patients undergoing no drug therapy. J Virol. 1995;69:23–31. doi: 10.1128/jvi.69.1.23-31.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Back NK. Nijhuis M. Keulen W, et al. Reduced replication of 3TC-resistant HIV-1 variants in primary cells due to a processivity defect of the reverse transcriptase enzyme. EMBO J. 1996;15:4040–4049. [PMC free article] [PubMed] [Google Scholar]
- 9.Croteau G. Doyon L. Thibeault D. McKercher G. Pilote L. Lamarre D. Impaired fitness of human immunodeficiency virus type 1 variants with high-level resistance to protease inhibitors. J Virol. 1997;71:1089–1096. doi: 10.1128/jvi.71.2.1089-1096.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gerondelis P. Archer RH. Palaniappan C, et al. The P236L delavirdine-resistant human immunodeficiency virus type 1 mutant is replication defective and demonstrates alterations in both RNA 5′-end- and DNA 3′-end-directed RNase H activities. J Virol. 1999;73:5803–5813. doi: 10.1128/jvi.73.7.5803-5813.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Archer RH. Dykes C. Gerondelis P, et al. Mutants of human immunodeficiency virus type 1 (HIV-1) reverse transcriptase resistant to nonnucleoside reverse transcriptase inhibitors demonstrate altered rates of RNase H cleavage that correlate with HIV-1 replication fitness in cell culture. J Virol. 2000;74:8390–8401. doi: 10.1128/jvi.74.18.8390-8401.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Martinez-Picado J. Savara AV. Sutton L. D'Aquila RT. Replicative fitness of protease inhibitor-resistant mutants of human immunodeficiency virus type 1. J Virol. 1999;73:3744–3752. doi: 10.1128/jvi.73.5.3744-3752.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Martinez-Picado J. Martinez MA. HIV-1 reverse transcriptase inhibitor resistance mutations and fitness: A view from the clinic and ex vivo. Virus Res. 2008;134:104–123. doi: 10.1016/j.virusres.2007.12.021. [DOI] [PubMed] [Google Scholar]
- 14.Li JZ. Paredes R. Ribaudo HJ, et al. Low-frequency HIV-1 drug resistance mutations and risk of NNRTI-based antiretroviral treatment failure: A systematic review and pooled analysis. JAMA. 2011;305:1327–1335. doi: 10.1001/jama.2011.375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sagar M. HIV-1 transmission biology: Selection and characteristics of infecting viruses. J Infect Dis. 2010;202(Suppl 2):S289–296. doi: 10.1086/655656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shankarappa R. Margolick JB. Gange SJ, et al. Consistent viral evolutionary changes associated with the progression of human immunodeficiency virus type 1 infection. J Virol. 1999;73:10489–10502. doi: 10.1128/jvi.73.12.10489-10502.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Scarlatti G. Tresoldi E. Bjorndal A, et al. In vivo evolution of HIV-1 co-receptor usage and sensitivity to chemokine-mediated suppression. Nat Med. 1997;3:1259–1265. doi: 10.1038/nm1197-1259. [DOI] [PubMed] [Google Scholar]
- 18.Connor RI. Sheridan KE. Ceradini D. Choe S. Landau NR. Change in coreceptor use correlates with disease progression in HIV-1-infected individuals. J Exp Med. 1997;185:621–628. doi: 10.1084/jem.185.4.621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Brumme ZL. Goodrich J. Mayer HB, et al. Molecular and clinical epidemiology of CXCR4-using HIV-1 in a large population of antiretroviral-naive individuals. J Infect Dis. 2005;192:466–474. doi: 10.1086/431519. [DOI] [PubMed] [Google Scholar]
- 20.Moyle GJ. Wildfire A. Mandalia S, et al. Epidemiology and predictive factors for chemokine receptor use in HIV-1 infection. J Infect Dis. 2005;191:866–872. doi: 10.1086/428096. [DOI] [PubMed] [Google Scholar]
- 21.Etemad B. Fellows A. Kwambana B, et al. Human immunodeficiency virus type 1 V1-to-V5 envelope variants from the chronic phase of infection use CCR5 and fuse more efficiently than those from early after infection. J Virol. 2009;83:9694–9708. doi: 10.1128/JVI.00925-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Repits J. Oberg M. Esbjornsson J, et al. Selection of human immunodeficiency virus type 1 R5 variants with augmented replicative capacity and reduced sensitivity to entry inhibitors during severe immunodeficiency. J Gen Virol. 2005;86:2859–2869. doi: 10.1099/vir.0.81111-0. [DOI] [PubMed] [Google Scholar]
- 23.Koning FA. Kwa D. Boeser-Nunnink B, et al. Decreasing sensitivity to RANTES (regulated on activation, normally T cell-expressed and -secreted) neutralization of CC chemokine receptor 5-using, non-syncytium-inducing virus variants in the course of human immunodeficiency virus type 1 infection. J Infect Dis. 2003;188:864–872. doi: 10.1086/377105. [DOI] [PubMed] [Google Scholar]
- 24.Jansson M. Popovic M. Karlsson A, et al. Sensitivity to inhibition by beta-chemokines correlates with biological phenotypes of primary HIV-1 isolates. Proc Natl Acad Sci USA. 1996;93:15382–15387. doi: 10.1073/pnas.93.26.15382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Koning FA. Koevoets C. van der Vorst TJ. Schuitemaker H. Sensitivity of primary R5 HTV-1 to inhibition by RANTES correlates with sensitivity to small-molecule R5 inhibitors. Antivir Ther. 2005;10:231–237. [PubMed] [Google Scholar]
- 26.Rusert P. Kuster H. Joos B, et al. Virus isolates during acute and chronic human immunodeficiency virus type 1 infection show distinct patterns of sensitivity to entry inhibitors. J Virol. 2005;79:8454–8469. doi: 10.1128/JVI.79.13.8454-8469.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sagar M. Kirkegaard E. Lavreys L. Overbaugh J. Diversity in HIV-1 envelope V1–V3 sequences early in infection reflects sequence diversity throughout the HIV-1 genome but does not predict the extent of sequence diversity during chronic infection. AIDS Res Hum Retroviruses. 2006;22:430–437. doi: 10.1089/aid.2006.22.430. [DOI] [PubMed] [Google Scholar]
- 28.Johnson VA. Brun-Vezinet F. Clotet B, et al. Update of the drug resistance mutations in HIV-1. Top HIV Med. 2008;16:138–145. [PubMed] [Google Scholar]
- 29.Tsibris AM. Sagar M. Gulick RM, et al. In vivo emergence of vicriviroc resistance in a human immunodeficiency virus type 1 subtype C-infected subject. J Virol. 2008;82:8210–8214. doi: 10.1128/JVI.00444-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Henrich TP. Tsibris A. Lewine N, et al. Evolution of CCR5 antagonist resistance in an HIV-1 subtype C clinical isolate. J Acquir Immune Defic. 2010;55:420–427. doi: 10.1097/QAI.0b013e3181f25574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vlahov D. Graham N. Hoover D, et al. Prognostic indicators for AIDS and infectious disease death in HIV-infected injection drug users: Plasma viral load and CD4+ cell count. JAMA. 1998;279:35–40. doi: 10.1001/jama.279.1.35. [DOI] [PubMed] [Google Scholar]
- 32.Keele BF. Giorgi EE. Salazar-Gonzalez JF, et al. Identification and characterization of transmitted and early founder virus envelopes in primary HIV-1 infection. Proc Natl Acad Sci USA. 2008;105:7552–7557. doi: 10.1073/pnas.0802203105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Marozsan AJ. Arts EJ. Development of a yeast-based recombination cloning/system for the analysis of gene products from diverse human immunodeficiency virus type 1 isolates. J Virol Methods. 2003;111:111–120. doi: 10.1016/s0166-0934(03)00166-6. [DOI] [PubMed] [Google Scholar]
- 34.Dudley DM. Gao Y. Nelson KN, et al. A novel yeast-based recombination method to clone and propagate diverse HIV-1 isolates. Biotechniques. 2009;46:458–467. doi: 10.2144/000113119. [DOI] [PubMed] [Google Scholar]
- 35.Wei X. Decker JM. Liu H, et al. Emergence of resistant human immunodeficiency virus type 1 in patients receiving fusion inhibitor (T-20) monotherapy. Antimicrob Agents Chemother. 2002;46:1896–1905. doi: 10.1128/AAC.46.6.1896-1905.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bjorndal A. Deng H. Jansson M, et al. Coreceptor usage of primary human immunodeficiency virus type 1 isolates varies according to biological phenotype. J Virol. 1997;71:7478–7487. doi: 10.1128/jvi.71.10.7478-7487.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hatch SC. Archer J. Gummuluru S. Glycosphingolipid composition of human immunodeficiency virus type 1 (HIV-1) particles is a crucial determinant for dendritic cell-mediated HIV-1 trans-infection. J Virol. 2009;83:3496–3506. doi: 10.1128/JVI.02249-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Voronin Y. Chohan B. Emerman M. Overbaugh J. Primary isolates of human immunodeficiency virus type 1 are usually dominated by the major variants found in blood. J Virol. 2007;81:10232–10241. doi: 10.1128/JVI.01035-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Coetzer M. Nedellec R. Salkowitz J, et al. Evolution of CCR5 use before and during coreceptor switching. J Virol. 2008;82:11758–11766. doi: 10.1128/JVI.01141-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Trkola A. Kuhmann SE. Strizki JM, et al. HIV-1 escape from a small molecule, CCR5-specific entry inhibitor does not involve CXCR4 use. Proc Natl Acad Sci USA. 2002;99:395–400. doi: 10.1073/pnas.012519099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kuhmann SE. Pugach P. Kunstman KJ, et al. Genetic and phenotypic analyses of human immunodeficiency virus type 1 escape from a small-molecule CCR5 inhibitor. J Virol. 2004;78:2790–2807. doi: 10.1128/JVI.78.6.2790-2807.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Reeves JD. Gallo SA. Ahmad N, et al. Sensitivity of HIV-1 to entry inhibitors correlates with envelope/coreceptor affinity, receptor density, and fusion kinetics. Proc Natl Acad Sci USA. 2002;99:16249–16254. doi: 10.1073/pnas.252469399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Reeves JD. Miamidian JL. Biscone MJ, et al. Impact of mutations in the coreceptor binding site on human immunodeficiency virus type 1 fusion, infection, and entry inhibitor sensitivity. J Virol. 2004;78:5476–5485. doi: 10.1128/JVI.78.10.5476-5485.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Xu L. Pozniak A. Wildfire A, et al. Emergence and evolution of enfuvirtide resistance following long-term therapy involves heptad repeat 2 mutations within gp41. Antimicrob Agents Chemother. 2005;49:1113–1119. doi: 10.1128/AAC.49.3.1113-1119.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Moore JP. Kuritzkes DR. A piece de resistance: How HIV-1 escapes small molecule CCR5 inhibitors. Curr Opin HIV AIDS. 2009;4:118–124. doi: 10.1097/COH.0b013e3283223d46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pugach P. Marozsan AJ. Ketas TJ. Landes EL. Moore JP. Kuhmann SE. HIV-1 clones resistant to a small molecule CCR5 inhibitor use the inhibitor-bound form of CCR5 for entry. Virology. 2007;361:212–228. doi: 10.1016/j.virol.2006.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ogert RA. Wojcik L. Buontempo C, et al. Mapping resistance to the CCR5 co-receptor antagonist vicriviroc using heterologous chimeric HIV-1 envelope genes reveals key determinants in the C2-V5 domain of gp120. Virology. 2008;373:387–399. doi: 10.1016/j.virol.2007.12.009. [DOI] [PubMed] [Google Scholar]
- 48.Westby M. Smith-Burchnell C. Mori J, et al. Reduced maximal inhibition in phenotypic susceptibility assays indicates that viral strains resistant to the CCR5 antagonist maraviroc utilize inhibitor-bound receptor for entry. J Virol. 2007;81:2359–2371. doi: 10.1128/JVI.02006-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Alkhatib G. Locati M. Kennedy PE. Murphy PM. Berger EA. HIV-1 coreceptor activity of CCR5 and its inhibition by chemokines: Independence from G protein signaling and importance of coreceptor downmodulation. Virology. 1997;234:340–348. doi: 10.1006/viro.1997.8673. [DOI] [PubMed] [Google Scholar]
- 50.Pastore C. Picchio GR. Galimi F, et al. Two mechanisms for human immunodeficiency virus type 1 inhibition by N-terminal modifications of RANTES. Antimicrob Agents Chemother. 2003;47:509–517. doi: 10.1128/AAC.47.2.509-517.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lobritz MA. Marozsan AJ. Troyer RM. Arts EJ. Natural variation in the V3 crown of human immunodeficiency virus type 1 affects replicative fitness and entry inhibitor sensitivity. J Virol. 2007;81:8258–8269. doi: 10.1128/JVI.02739-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Salazar-Gonzalez JF. Bailes E. Pham KT, et al. Deciphering human immunodeficiency virus type 1 transmission and early envelope diversification by single-genome amplification and sequencing. J Virol. 2008;82:3952–3970. doi: 10.1128/JVI.02660-07. [DOI] [PMC free article] [PubMed] [Google Scholar]