Abstract
Top-down mass spectrometry is an emerging approach for the analysis of intact proteins. The term was coined as a contrast with the better-established, bottom-up strategy for analysis of peptide fragments derived from digestion, either enzymatically or chemically, of intact proteins. Although the term top-down originates from proteomics, it can also be applied to mass spectrometric analysis of intact large biomolecules that are constituents of protein assemblies or complexes. Traditionally, mass spectrometry has usually started with intact molecules, and in this regard, top-down approaches reflect the spirit of mass spectrometry. This article provides an overview of the methodologies in top-down mass spectrometry and then reviews applications covering protein posttranslational modifications, protein biophysics, DNAs/RNAs, and protein assemblies. Finally, challenges and future directions are discussed.
1. Introduction
Mass spectrometry (MS) has become the essential tool in proteomics research1. In the well-established “bottom-up” approach, protein samples are digested enzymatically or chemically to give smaller peptides that are easily characterized by mass spectrometers2. Following the development of electron-capture dissociation (ECD)3, and, more recently, electron-transfer dissociation (ETD)4, a new approach is emerging for analysis of intact protein samples by MS; this approach has been given the term “top-down” to distinguish it from “bottom-up”.
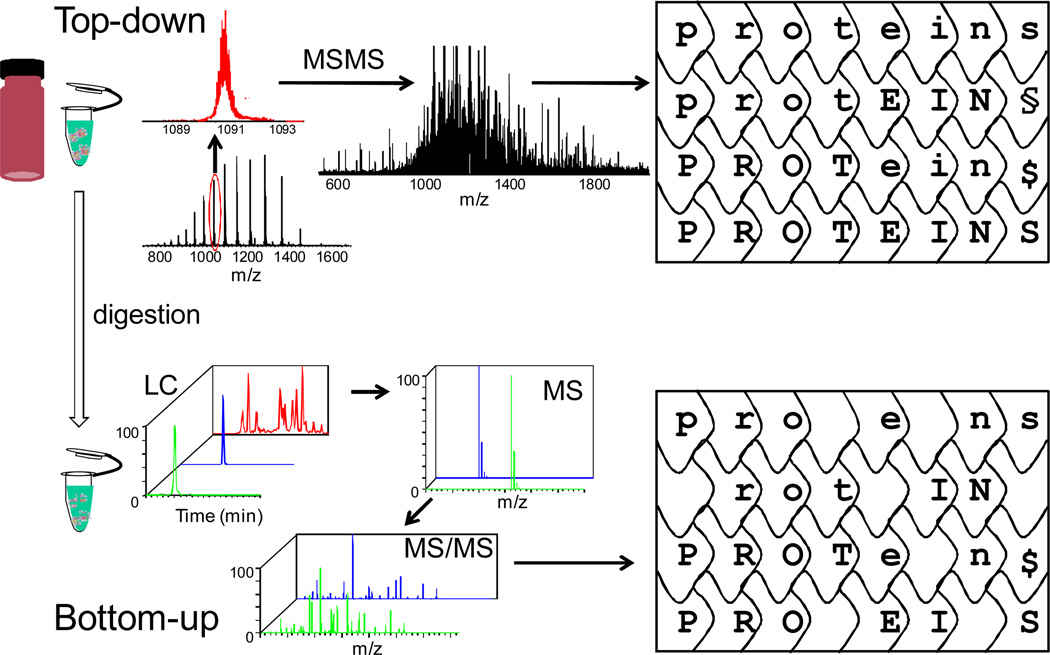
The workflows of top-down and bottom-up approaches for protein analysis are complementary (Figure 1). Although the bottom-up approach offers swift identification of a protein by utilizing a combination of tandem mass spectrometric (MS/MS) data and database searching, there are drawbacks for this approach5. Peptides from protein digestion cannot be completely recovered for MS measurements, leading to information loss. Correlations between peptides and their sources when dealing with mixtures of proteins including proteins containing various post-translational modifications are also lost. Furthermore, losses of peptides containing modifications can be particularly frustrating when seeking to locate post-translational modifications. Moreover, for complex protein mixtures, isomeric peptides are possible to be present in the digest, causing confusion as to the protein source of a peptide, which is vital for the protein identification. Top-down sequencing can overcome these information-loss problems. Measurement of the intact proteins gives information for possible protein isoforms provided they have sufficiently different masses so that the isoforms can be separated by the spectrometer. Given that the analyst has selected the protein of interest according to its m/z prior to activation, top-down sequencing, as an MS/MS method, always makes the connection between the intact protein and the fragments.
Figure 1.
Comparison of top-down and bottom-up workflows.
In contrast to the mature bottom-up approach, top-down sequencing is still in its growing stage, and the number of practitioners is much less, although quickly growing. An important factor is that a Fourier transform ion cyclotron resonance (FTICR) mass spectrometer is the most effective tool for sequencing proteins via election-capture dissociation. Unfortunately FTICR instruments are more costly and require more expertise than other mass spectrometers. The use of orbitraps in top-down may be an alternative. Although electron-transfer dissociation is an alternative to electron-capture dissociation and is suitable for ion-trap instruments, its application to top-down is not yet as effective because the mass resolving power of ion traps is insufficient to permit top-down studies.
The majority of the current top-down research is focused on protein identification and characterization of associated post-translational modifications. The latter application can be successful because electron-capture (transfer) dissociation retains labile modifying groups in proteins during fragmentation. Given that several recent review articles addressed the technical developments of top-down MS for protein analysis6–9, we will focus in this review on the applications of top-down sequencing for protein biophysics and protein conformational studies. Further, we consider nucleic acids and macromolecular assemblies. This review provides a brief survey of recent top-down applications and is not comprehensive.
2. Methodologies
Sample Preparation
To conduct top-down analysis by FTICR MS, it is desirable but not essential to have pure samples or simple mixtures. Although one might deal with complex mixtures and separate the proteins by using a quadrupole mass analyzer that is concatenated with the FTICR or double resonance strategies in the FTICR instrument rather than HPLC, the dispersion of signals during ionization and the suppression of individual proteins reduce the signal-to-noise ratio. Therefore, sample purification is usually an important step. Furthermore, because electron-capture dissociation cleaves the backbone N-Cα bond in a highly but not completely nonspecific manner, there are many reaction channels producing many fragments. Spectral averaging over a number of scans is usually required to improve the signal-to-noise ratio; this approach means that larger quantities of samples are needed compared to bottom-up approaches. Each protein is a unique case in top-down analysis, unlike the analysis of peptides in a bottom-up approach. For bottom-up peptide identification in proteomics, for example, the fragmentation conditions are established beforehand and not changed during the LC-MS/MS analysis. For proteins, optimum results, however, are achieved by tuning the conditions to fragment each. Thus, top-down is difficult to conduct with on-line HPLC separation. This poses some difficulties for high throughput analysis wherein coupling of sample separation and FTICR with ECD is necessary but sometimes incompatible with the shorter time scales of LC separation and the longer times of FTICR ECD spectral averaging. Therefore, fraction collection is very often spliced between protein separation and FTICR measurement. Despite the difficulties, there is strong motivation to develop further top-down analysis by increasing the speed and mass resolving power of current instrument platforms. These improvements are needed to increase throughput to augment bottom-up approaches that are inadequate for analyzing proteins with multiple modifications.
For bottom-up approaches, peptide separation is now a high-performance and mature technology. For top-down, on-line separation of intact proteins by either capillary HPLC10 or isoelectric focusing11 may be chosen. Similar to 2-D gel electrophoresis, which incorporates separation based on two properties of proteins (i.e., isoelectric point and molecular mass), on- or off-line HPLC can be combined with capillary isoelectric focusing12, chromatofocusing13, or ion exchange14–15 to achieve also 2-D separation16 of proteins for on-line MS measurement or for off-line fraction collection followed by MS analysis. Hydrophilic interaction liquid chromatography (HILIC)17 can successfully separate both peptides and proteins including histones18. Meng et al.19 combined gel electrophoresis using an acid-labile surfactant with RPLC to separate intact proteins within a ~5 kDa range for MS analysis. Newer technologies include gel-eluted liquid fraction entrapment electrophoresis (GELFrEE) to separate proteins over a broad MW range and then to utilize top-down FTICR experiments on the fractions that contain 5–100 kDa proteins20.
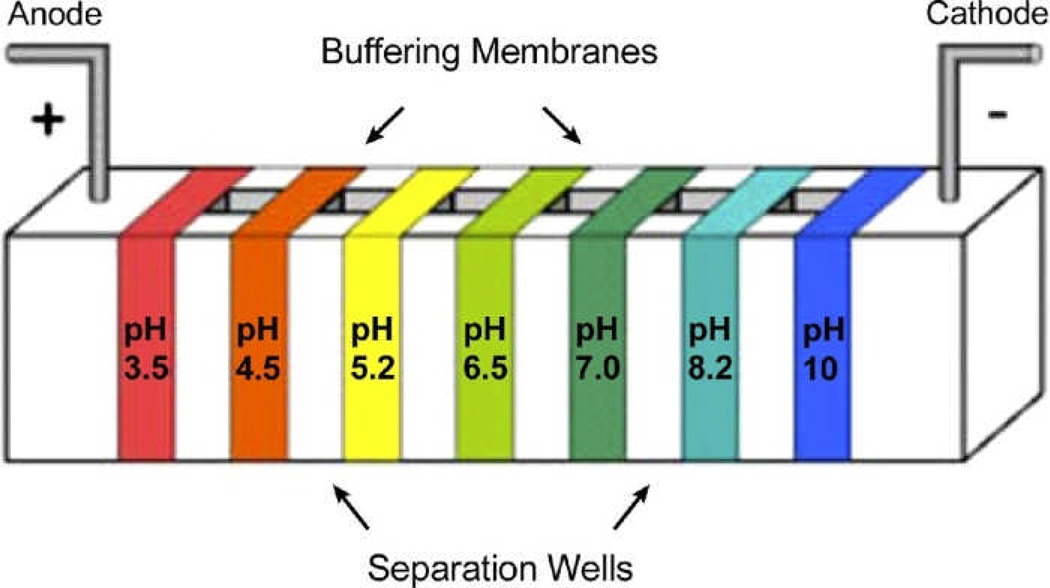
Similar to GELFrEE, an approach MSWIFT21 (membrane-separated wells for isoelectric focusing and trapping, Figure 2.) can also be utilized for sample clean-up in bottom-up proteomics22 and for removal of neutral buffering materials in protein sample23. The big advantage of MSWIFT is that peptides/proteins of different pI values are bracketed by membranes of fixed pH values, and the sample can be directly used for MS analysis unless the sample mixture is too complicated and needs further separation. This is well suited for top-down analysis.
Figure 2.
MSWIFT device for isoelectric point-based separation. Reprinted from ref. 22, J. Am. Soc. Mass Spectrom., 2010, 9, 1612–1619, with permission of the American Society for Mass Spectrometry.
Whitelegge24 developed methods to separate membrane proteins by using HPLC and choosing appropriate solvent systems. A compromise strategy, known as “middle-down”, was adopted in this research.25 Here, proteins are digested under mild conditions to produce large peptides that are then fragmented in a top-down manner. In this way, peptides that are smaller than the whole protein are easily separated by chromatography and submitted to productive top-down analysis. Moreover, the sequence coverage can be better than that afforded by bottom-up sequencing of digested peptides.
Higher charge states of the precursor ion are beneficial for fragmentation. Evan Williams26 first reported that glycerol and m-nitrobenzyl alcohol (mNBA) can be included in the spray media to achieve “supercharging” of electrosprayed protein ions. The proposed mechanism is that addition of the reagent into the sample solution causes the surface tension of the droplets to increase during evaporation of the solvent. Increased surface tension allows the droplets to accommodate more charges before reaching the Rayleigh limit27. Loo’s group28 screened sulfolane and other reagents for supercharging of native proteins and noncovalent protein complexes. The effect was also demonstrated on large 800 kDa GroEL by van Duijn29. Emmett30 identified some reagents, dimethylformamide, thiodiglycol, dimethylacetamide, dimethylsulfoxide, and N-methylpyrrolidone, that not only increase the charge of proteins with masses of 78 kDa when using ESI, but also improve the liquid chromatography performance when added in the LC solvents.
Mass spectrometry instrumentation
Although the early mass spectrometric analysis of intact proteins was performed by using triple quadrupole instruments31, the use of high mass resolving power FTICR instrumentation32 afforded resolution of overlapping fragment ions for more confident assignment33. Following recognition of the need for improved mass resolving power, FTICR instruments with higher magnetic fields and actively shielded magnets became available. A significant breakthrough was made with electron-capture dissociation, an activation method that can fragment protein ions and, therefore, be utilized for analysis of large biomolecules34. Thus, most top-down analyses to date have been done with FTICR-based instruments.
Other types of instruments (e.g., ion traps and time-of-flight mass spectrometers) can also be applied to analyze intact proteins, but to date they are less effective than FTICRs, and hybrid linear ion trap (LTQ) - orbitraps35 or LTQ-FTICRs36. In fact, the new-design orbitrap from Thermo Fisher utilizes a higher field orbitrap analyzer and improved ion-trap, providing another opportunity for top-down proteomics. The combination gives improved resolving power over older designs and higher speed, making it competitive with FTICR-based instruments37. Fragmentation so far is by ETD and others instead of ECD, but the relative effectiveness of ETD remains unknown for large proteins. Nevertheless, a significant advantage of orbitraps compared to FTICRs is that mass resolving power decreases as the square root of m/z not as its first power, making resolving power on orbitraps less sensitive to m/z. Less competitive than FTICR and orbitraps is MALDI TOF, and several groups utilized this approach for top-down analysis by employing the strategy of in-source decay (ISD)38–39.29 30
Fragmentation methods
Although collisionally activated dissociation (CAD)40–41 is mainly used for protein identification in top-down MS, detailed characterization of the proteins requires electron-capture dissociation3. The advantage of the latter is that the labile posttranslational modifications (e.g., phosphorylation and glycosylation) are retained during protein fragmentation. These modifying groups are usually lost as part of small neutral molecules (e.g., phosphoric acid) during the CAD processes. For large proteins, owing to intramolecular interactions of the residues, the fragments cleaved by ECD sometimes do not separate from each other. To solve this problem, activated-ion electron-capture dissociation (AI-ECD)42 was developed. Protein ions are first preheated by collisions with inert gas molecules or atoms or by irradiation with infrared laser light43 with energies less than the threshold of dissociation, and then ECD is applied. This pre-heating of protein ions leads to unfolding of the protein structure, breaking intramolecular interactions. As a result, the fragments produced by ECD separate more readily from the protein than in the absence of “heating” and are detected. Proteins with masses up to 200 kDa have been analyzed in part by this approach44.
In addition to the prevalent electron-capture dissociation approach, other electron-induced fragmentation methods are available for fragmenting peptides; they include electron-transfer dissociation (ETD)4,35, electron-detachment dissociation (EDD)45 and electron-ionization dissociation (EID)46. Negative-ion ETD can also fragment peptide anions by interaction with either the Xe radical cation47 or fluoranthene (C16H10+•) radical cation48. EDD, for negative ions, promotes cleavage of the C-C bond of the protein backbone. The application of most of these methods for structural analysis of intact proteins is in its infancy, and the rules and mechanisms of fragmentation need to be better elucidated. The first demonstration of EDD to a large protein was to ubiquitin49, whereas EDD can be applied to the analysis of RNAs up to 61 nucleotides50.
A complementary approach to high-resolving-power mass measurements in FT instruments is charge reduction by ion-ion reactions51; here the number of charges of the product ions are decreased so that their m/z’s shift to higher values, affording more confident assignments. This can be a useful approach, although the details are beyond the scope of this article. Interested readers are referred to the papers by the S. McLuckey group. Some applications of ion-ion chemistry are mentioned in the following section.
Software for data analysis
The Kelleher group has spearheaded the development of software for the top-down technology. Their algorithm for data processing, ProsightPTM52, is so far the most successful software package for analysis of top-down data of proteins with or without modifications (http://groups.molbiosci.northwestern.edu/kelleher/software.html). The software has been commercialized by Thermo Fisher. Another software platform is Biotools, from Bruker. Boston University makes available a protein-identifier software package, BUPID-top-down53, for analysis of top-down data. This package is capable of assigning internal fragments as well as sequence ions.
Applications
The first application of top-down mass spectrometry was the speciation of cytochrome c proteins from nine animal sources; this was accomplished by comparing the CAD fragments produced in a triple quadrupole instrument54–55, before the term “top-down” had been coined in MS56. In a similar way, Feng57 fragmented ESI-produced ions of a 150 kDa antibody. With the recent technological developments, top-down MS has found a number of application areas in biomacromolecular research; the most common is intact-protein characterization, DNA/RNA analysis, protein conformational characterization, protein-ligand binding-site location, and the very recent protein-protein assembly characterization. Potential clinical applications of top-down mass spectrometry are illustrated using transthyretin and hemoglobin as models53.
A. Intact-protein characterization
Posttranslational modifications (PTMs)
Given that ECD/ETD does not cause losses of labile modifications, it is especially useful for characterization of protein PTMs and mutations. The Kelleher group58 has been developing the workflow for streamlined characterization of proteins by top-down approaches by using histone H4 as a model for shotgun annotation of acetylation, methylation, and phosphorylation and for the characterization of isoforms. They studied four histone forms H2A59, H2B60, H361–62, and H463–64 to obtain comprehensive information of modifications and progressive profiles at different cell-cycle phases. They also demonstrated sample-separation methods and found that mild performic acid oxidation65 enhances chromatographic and top-down mass spectrometric analyses of histones.
A strategy for large scale top-down analysis using lysate from Saccharomyces cerevisiae was developed by Meng et al.66. An on-line, metal-free, weak cation exchange-hydrophilic interaction LC/RPLC system can be used to characterize core histone mixtures at the intact protein level15, demonstrating significant improvement in sensitivity, several orders of magnitude reduction in sample requirements, and a reduction in overall analysis time. The Garcia group67 characterized the high mobility group (HMG) protein A1a, a small heterochromatin-associated nuclear protein whose PTMs are partially linked to apoptosis and cancer. Taking a combined MS effort including bottom-up, middle-down and top-down MS, they found methylation, acetylation, and phosphorylation at unexpected sites. Bottom-up MS is capable of identifying low-level modification sites, whereas top- and middle-down MS can pinpoint the commonly occurring combinatorially modified forms (i.e., the occupancy pattern of multiple modifications on residues along the sequence). These workers, by using top-down methods, found that relatively few combinatorially modified forms dominate the population.
Glycosylation is common and critical in biological systems. Because glycosylation is heterogeneous in its sites and forms, detailed characterization is not a trivial task. It usually requires harvest of the glycans and glycosylated peptides cleaved by enzymes followed by chromatographic separation and analysis by various tools. Top-down MS can be clearly used to characterize glycosylation in different systems. Fridriksson et al.68 characterized the heterogeneous glycosylation of immunoglobulin E constructs of the Fc segment of IgE, the Fcε(3-4) (52 kDa) and Fcε(2-3-4)2 (76 kDa) disulfide-bonded homodimers. Secreted proteins of Mycobacterium tuberculosis are potential diagnostic and vaccine candidates. Previously unidentified species were discovered that contain extensive post-translational modifications69, including loss of a signal sequence, loss of the N-terminal residue, degradation by proteolysis, oxidation, and glycosylation. For example, seven hexose attachments were found in a 9 kDa protein, and 20 attachments in each of two 20 kDa proteins. Schirm et al.70 identified unusual bacterial glycosylation by tandem mass spectrometry analyses of intact flagellin from four bacterial pathogens (Campylobacter jejuni, Helicobacter pylori, Aeromonas caviae, and Listeria monocytogenes). The top-down approach provided Information on the extent of glycosylation, the molecular masses, and the identity of oligosaccharide residues on the flagellin. Whitelegge et al.71 studied apolipoproteins that are from high-density lipoproteins from dog (Canis lupus familiaris). The experimental molecular masses of apoA-II proapoA-I, apoA-I, apoC-I are in excellent agreement with the corresponding MWs calculated from genomic data. Top-down sequencing of apoC-I was performed.
Phosphorylation of proteins plays extremely important roles in biological function. A phosphate group can be added to or removed from serine, threonine, or tyrosine by a kinase or phosphotase. Characterization of both the sites of occupancy and the relative extent of phosphorylation of a protein sequence is crucial for understanding the functions a protein performs. In the case of limited number of phosphorylation sites, top-down MS can provide a full view of phosphorylation. For proteins containing many phosphorylation sites with partial occupancy of some, top-down results can be very complicated, and interpretation can be challenging. By minimizing the further cleavage of primary product ions and by denaturing the tertiary noncovalent bonding of the molecular ions under a variety of activation conditions (AI-ECD), 250 out of the 258 backbone cleavages of carbonic anhydrase can be produced72. Amazingly, posttranslational modifications can be identified to within one residue of 235 residues in the protein, and 24 of a possible 34 phosphate substitution sites can be located. The Ge group characterized phosphorylation of various cardiac proteins, and they showed that cTnI is regulated by protein kinase A and protein kinase C at five sites, Ser22/Ser23, Ser42/44, and Thr143. This information comes primarily from in vitro phosphorylation assays by the specific kinases. It was unambiguously shown that Ser22/23 are the only two sites basally phosphorylated in wild-type mouse cTnI73. To confirm these identifications, one can substitute Ser with Ala. All the phosphorylation sites in the truncated (28–115 kDa) and full-length forms of cardiac myosin binding protein C (cMyBP-C, 142 kDa) were also identified74, and sequential phosphorylations were characterized by using a combination of top-down and middle-down approaches. Remarkably, it was discovered that truncations in recombinant proteins dramatically alter the phosphorylation state. The significance of this work is that the usual use of recombination technology affords truncated and full-length recombinant proteins with alterations in the post-translational state, leading to variations in structure and function. Besides being able to provide unique insights into the global state of post-translational modification of rat cTn subunits, high resolving power, top-down MS readily revealed naturally occurring single amino acid sequence variants including a genetic polymorphism at residue 7 in cTnI, and an alternative splice isoform that affects a putative hinge region around residue 192 of cTnT, all of which co-exist within a single rat heart36. By the same method, the Ge group75 localized, for the first time, a single amino acid polymorphism at V116 of swine cardiac troponin I.
Relative Quantitation
Quantitative information in proteomics is often required especially in differential experiments where comparisons are made between test (disease) and control (healthy) states. In the bottom-up approach, isotope-coded affinity tags (ICAT), isobaric tag for relative and absolute quantitation (iTRAQ), stable isotope labeling by amino acids of cell culture (SILAC), and related isotope-based labeling methods can be used to label peptides for MS measurements1. Another strategy, called “label-free”, involves quantitation by counting the spectra taken for a given analyte (the assumption is that abundant analytes will be characterized by a larger number of spectra than will low-level analytes1. Because bottom-up measures peptides, information about protein isoforms is usually missing. Top-down analysis provides quantitative data on different isoforms including individual and post-translationally modified forms. Furthermore, given that a protein modification gives several site-specific isoforms, a crude measure of the abundance of one isoform can be made by comparing the intensities of the peaks corresponding to unmodified and modified peptides76.
Zabrouskov77 characterized sequentially the relative stepwise deamidation of bovine ribonuclease A at five sites: Asn67, Asn71, Asn94, Asn 34, and Gln4. In more detail, Ying Ge’s group78 considered the possible change of fragmentation efficiency owing to addition of a modification group to the protein and the variation of the modification sites. She introduced a quantitation scheme by using the fragments from the unmodified protein as yardsticks to normalize those unmodified fragments from the modified protein. Then the abundances of the modified fragments from the modified protein can be compared with those of unmodified fragments obtained in the same experiment. Hendrickson and coworkers79 used ETD in an Orbitrap to analyze quantitatively apolipoproteins from human high density lipoproteins (HDL). This paper is the first demonstration of differential mass spectrometry (dMS80–81) in a top-down manner to deal with two sets of samples that differ by 5-fold. In another timely application, Muddimann and coworkers82 showed that top-down quantitation of stem-cell proteins can be accomplished by SILAC labeling.
Membrane Proteins
Membrane proteins are important, but less-studied than other cellular proteins in proteomics research. Their intrinsic structures, interaction with the lipid bilayer, and function for either transporting or signaling are of great interest. Since crystallization of membrane proteins became possible, over 180 unique proteins have been studied at atomic resolution by X-ray crystallography. It was estimated that approximately 1,700 membrane-protein structures comprise each structural family83. Although the vast majority of MS and chemical labeling studies for membrane proteins, are by the bottom-up approach84, the top-down protocol is emerging. For example, Whitelegge and coworkers85–86 developed various solvent systems to collect integral membrane proteins for top-down analysis. Recently, this group used high-resolving power, top-down MS to characterize 11 integral and 5 peripheral subunits of the 750 kDa photosystem II complex from the eukaryotic red alga, Galdieria sulphuraria87. Both collisionally activated and electron-capture dissociation are capable for confirming the presence of 11 small subunits; PsbE, PsbF, PsbH, PsbI, PsbJ, PsbK, PsbL, PsbM, PsbT, PsbX and PsbZ, and with a mass accuracy that is better than 5 ppm. All subunits showed covalent modifications that fall into three classes including retention of initiating formyl-methionine, removal of methionine at the N-terminus with or without acetylation, and removal of a long, N-terminal peptide. Peripheral subunits identified by top-down analysis included oxygen-evolving complex subunits PsbO, PsbU, PsbV, as well as Psb28 (PsbW) and Psb27 (‘‘PsbZ-like’’). Granvogl et al.88 identified 13 one-helix less than 10 kDa integral membrane proteins in photosystem II subcomplexes of the thylakoid membrane in chloroplasts. Whitelegge et al.89 carried out top-down analysis of the small subunits of the cytochrome b6f complex from Arabidopsis thylakoids. The results revealed novel post-transcriptional/translational modifications including the presence at PetL position 2 of glutamic acid instead of the proline predicted from the gene sequence. Post-translationally modified integral membrane proteins with polyhelix bundles, transmembrane porin motifs, and molecular masses up to 35 kDa can now be investigated by CAD top-down high mass resolving power Fourier transform MS90.
B. Nucleic acids
Tandem MS of large pieces of DNA/RNA has also become an active application area where top-down protocols may bear fruit. The strengths of MS in this area lie in the ability to identify modifications and in the speed and sensitivity for analysis, compared with conventional biochemical methods. The McLafferty group91 studied the fragmentation of DNAs up to 108-mers and obtained complete sequence information for a 50-mer, by using mostly internal fragments92. This work builds on earlier work begun by McLuckey and coworkers in the early 90s93–94. The field has matured to the point that McLuckey proposed a nomenclature scheme to denote fragment ions. Structural characterization of nucleic acids by MS can clarify the modifications taking place in nature or produced artificially for therapeutic purposes. Furthermore, MS can contribute to the understanding of metabolic pathways of nucleic acids. Peptide nucleic acids95 and locked nucleic acids96 are two examples of modified DNAs that have been already studied by MS. Introduction of modification groups to an entity usually results in stability changes, which are vital to its function. In this regard, gas-phase fragmentation mechanisms of nucleic acid ions are both analytically and fundamentally important; in a fundamental sense, they provide insight to solvation by providing a benchmark of their intrinsic stability in the gas phase.
New classes of RNA97–98, (e.g., small interfering RNAs, microRNAs, transfer RNA, and ribosomal RNAs with nucleotide (nt) numbers varying from dozens to hundrends) do not act as messenger RNA to produce proteins, but perform other important biological functions. Several recent articles about top-down analysis of RNAs show the potential in this area. For example, the McLuckey group showed that highly modified transfer RNAs99 can now be approached with MS. Breuker100 completely sequenced a 34 nt RNA by CAD and EDD by carefully minimizing the internal energy of the products from the primary backbone cleavage. They also proposed a mechanism for backbone cleavage by EDD. Her group50 recently extended the strategy to a 61 nt RNA. Using IRMPD101 and SORI-CAD102, a few groups studied non-covalent RNA/drug complexes to infer the properties of the stem-loop structure with 20–30 nt RNAs as model systems. Interruptions in the sequence coverage indicate binding sites between RNA and drug molecules. If such interruptions are general, top-down MS will play an important role in locating binding sites in these systems.
C. Protein conformations
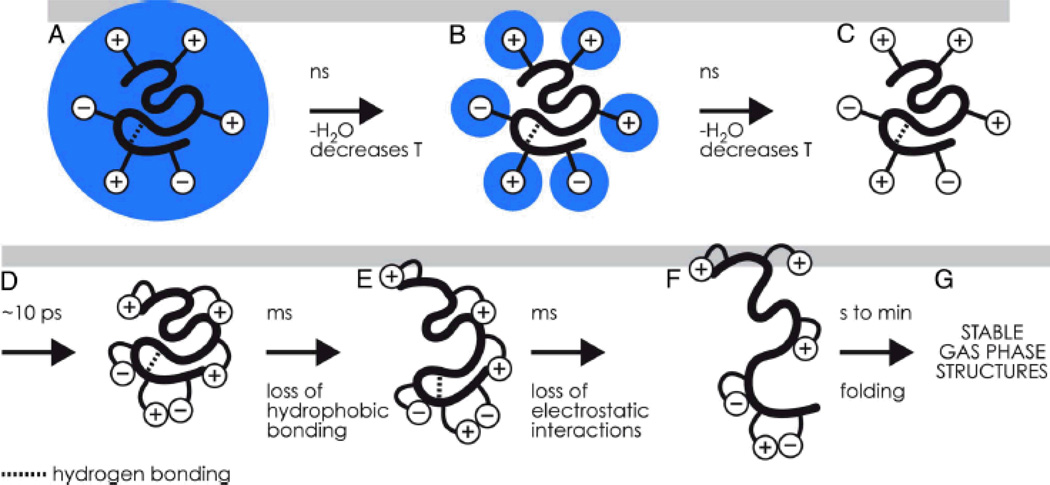
After the invention of ECD for MS, McLafferty immediately applied it to study not only the detailed folding and unfolding of the protein ubiquitin103, but also the folding kinetic intermediates of cytochrome c104, and the reductive unfolding pathways of RNase B isoforms105. Breuker106 discovered that gentle collisional activation of the homodimer ions of cytochrome c can generate regular ECD fragments without irradiating the protein with electrons. This technique was given a term of “native ECD” and applied to probe the thermal unfolding of native cytochrome c in the transition from solution to gas phase107. Based on considerable evidence from experiment and simulation studies, the authors devised a picture of the temporal evolution, from 10−12 to 102 s, of protein structure upon electrospray into gas phase108 (Figure 3); the picture clarifies some important questions regarding the preservation of native protein structure upon removal of hydrophilic solvent, a transition during which the relative importance of hydrophobic and electrostatic interactions changes considerably.
Figure 3.
Temporal structure evolution of globular proteins after ESI - ref. 108 Proc. Natl. Acad. Sci. U.S.A., 2008, 105, 18145–18152, copyright of the National Academy of Sciences.
Gas-phase electrostatic interactions can be revealed by ECD for various protein ions. An example is a 10 kDa, three-helix bundle protein with charges of 7+ to 16+ that was submitted to ECD109. The yield of each observed c or z ion was plotted versus the charge numbers of the protein ion, and a site-specific transition charge (“melting charge”) value was assigned to be 50% of the total yield. The transition charge value of each of the three helices was obtained by similarly plotting the integrated c/z ion yields versus the protein charges. The transition charge values increase as we go from the N- to C-terminus, indicating the three helices are more and more stable from N- to C-terminus, in agreement with NMR data. The ECD cleavage sites between the helices, have site-specific transition charge values that are close to those of the nearby helix ends. The native structure of this protein shows one, three, and six intrahelix salt-bridges in the three helices from N-to C-terminus, respectively. The density of salt-bridges (number of salt-bridges/number of residues) correlates well with the transition charge values, suggesting the protein structure is mainly stabilized by the salt-bridges. This work sheds light on how to analyze ECD data in terms of details of protein structures.
For protein conformations and dynamics, the bottom-up strategy combined with footprinting by deuterium exchange110 or other chemical reagents111 is very powerful. The top-down approach in this area is just emerging. One can take advantage of the dependence of protein folding on charge-state of electrosprayed protein ions; for example, highly charged ions tend to represent unfolded states112. Konermann’s group applied hydrogen/deuterium exchange (H/DX) coupled with ECD top-down mass spectrometry to study static113 and intermediate protein structures114. Another example of this approach is the work of Kaltashov, who investigated ETD top-down of deuterated proteins115 and indicated that the prospects for this approach are significant116. Interestingly, supercharging reagents (e.g., m-NBA) do not cause dramatic structure changes, as demonstrated by ETD in a top-down experiment of deuterated ubiquitin117. This overall approach may become useful for probing the real-time kinetics of proteins under physiological conditions, if more examples are demonstrated.
The Gross group has combined top-down and bottom-up approaches118 with footprinting by fast photochemical oxidation of proteins (FPOP119) to characterize protein conformations. To draw conclusions, the amino acid residues modified by footprinting reagents must be identified. According to Garcia and coworkers67, bottom-up strategies have the largest dynamic range (i.e., can detect the modifications at low levels) whereas top-down approaches can be used to assign regions undergoing the greatest modifications and distinguish forms with combinatorial modifications.
D. Non-covalent protein complexes
With the development of modern MS technology, most cellular proteins can be analyzed to show their identity. Attention is now needed to understand protein interactions with other cellular species. Identification of protein components in complexes became possible when ESI was invented and added to high performance mass spectrometers. Following Loo’s highly-cited review120, other reviews of this field appeared29,121–124. The protein interactions to be addressed involve those either with small ligands or other proteins, and there is early demonstrations that top-down MS can be successfully applied to these investigations. Although cross-linking and MS have been successfully applied to probe the structures of native protein complexes125, the approach is not covered in this article because reports of analysis by top-down MS are sparse.
Protein-ligand complexes
ECD can still fragment protein backbone in the presence of ligands interacting noncovalently, indicating that these interactions survive the fragmentation processes126. The Loo group127 also showed this to be the case for the ECD of the protein-ligand complex of the 140-residue, 14.5 kDa alpha-synuclein protein and one molecule of polycationic spermine (202 Da). From an analysis of the product ions, the spermine binding can be localized to residues 106–138, demonstrating good agreement with results from previous solution studies by NMR. The Loo group128 also determined the adenosine triphosphate (ATP)-binding site on chicken adenylate kinase by the same method. Although CAD of the protein-nucleotide triphosphate complexes yields products from the dissociation of a covalent phosphate bond of the nucleotide with subsequent release of the nucleotide monophosphate, the intrinsic stability of electrostatic interactions in the gas phase allows the diphosphate group to remain noncovalently bound to the protein. Using ECD, they found that the product ions bearing mono- and diphosphate groups were mapped to the region of the adenylate kinase-ATP complex, in agreement with NMR and X-ray crystalloagraphic identifications of the ATP-binding pocket. More evidence for success of this approach is from Erales et al.129, who mapped the copper-binding site of the small CP12 chloroplastic protein of Chlamydomonas reinhardtii by using top-down MS and site-directed mutagenesis. Protein-peptide interface can also be mapped out by this approach as demonstrated by the Langridge-Smith group130 using a complex of the 21kDa p53-inhibitor protein anterior gradient-2 and its hexapeptide binding ligand (PTTIYY). The results are supported by a solution-phase peptide-competition ELISA assay.
If fragmentation yields are insufficient, the application of supercharging by adding sulfolane or m-nitrobenzyl alcohol to increase the charge of a protein-ligand complex causes concomitant increases in fragmentation yields by CAD and ECD, all still retaining the ligands in the fragments, as shown in recent work by the Loo group131. However, it can not be excluded that rearrangement of the ligand in the supercharged whole complex may occur132. Other aspects of protein-ligand complexes studied by MS were already covered in other review articles133–134.
Protein-Protein complexes
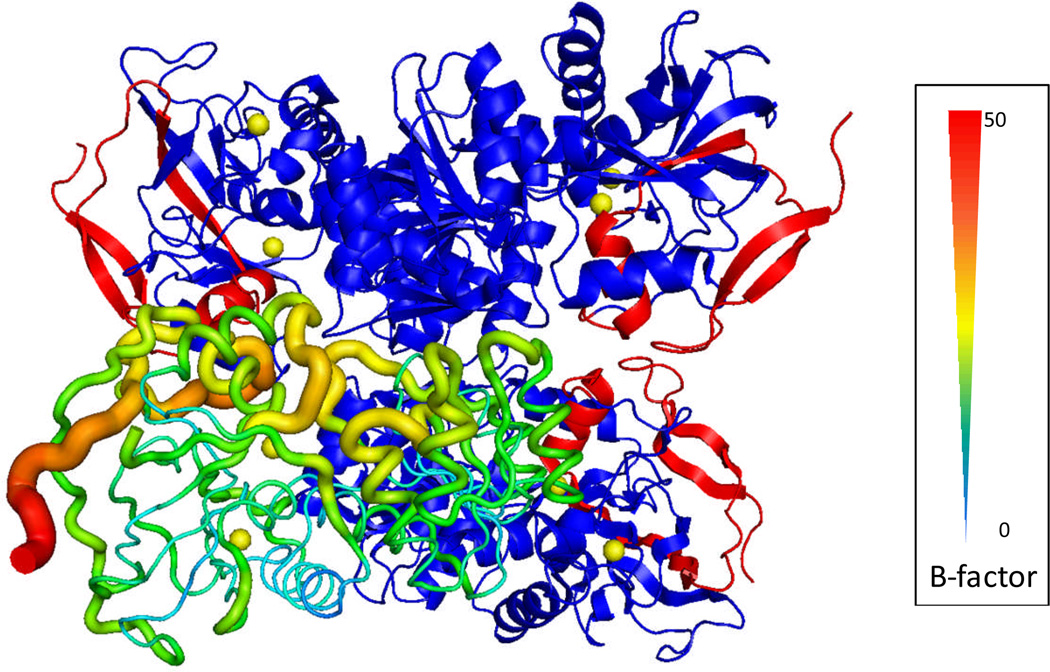
Robinson and coworkers have been studying intact macromolecular assemblies by native electrospray, MS/MS and ion mobility with Q-TOF instruments modified to go to relatively high m/z of 32,400. Her group recently extended their technology to membrane complexes that are protected in micelles135. The constituents can be identified by bottom-up proteomics after the whole assembly is denatured and digested by enzymes124. CAD of the intact complex generated by native electrospray produces sub-complexes or single subunits carrying more charges than expected by statistical considerations owing to asymmetric charge partitioning. The output is indicative of how the subunits interact with each other and with the assembly of the subunits 123. Obtaining sequence information of the components directly from the intact complex, however, has been limited when using CAD, IRMPD, SID, BIRD, and ECD. The discovery by the Gross group136 that ECD is capable of providing top-down sequencing of the exposed region of the subunit of 147 kDa yeast alcohol dehydrogenase tetramer in an FTICR mass spectrometer offers encouragement for development in this area. For example, they showed that sequence information is generated directly from the intact assembly and moreover that the sequence information is from the exposed region of the constituent component. The latter observation has implications for providing structural insights of the assembly. The observed ECD fragments correlate well with the B-factor, a temperature-factor in X-ray crystallography as shown in Figure 4137. The B-factor is a measure of the flexibility of certain region of the protein sequence when changes in temperature or crystal imperfection occur. The MSMS fragments from top-down mass spectrometry and the B-factors from X-ray crystallography build a connection between these two analytical tools. We think this opens up a new top-down approach to characterize macromolecular assemblies by obtaining both proteomics and structural-biology information in one experiment.
Figure 4.
Crystal structure of yeast ADH tetramer (PDB code 2HCY). The lower left subunit is shown in B-factor scale where the N-terminus has the highest B-factor value. In each of the other three subunits, the sequenced N-terminal region up to the 55th residue by ECD is highlighted in red – ref 137.
4. Perspectives
In the first ECD paper3 in 1998, the term “top down” was invoked. The first full article56 describing “top-down” MS appeared in 1999, and since then, more and more laboratories are adopting the approach. Its strength is to utilize electron-induced fragmentation to determine post-translational modifications and other changes (e.g., amino-acid substitutions) in proteins. One obstacle standing in the way of widespread use is the lack of high throughput analysis on the proteome scale, and improvements may be in the offing with the new orbitrap Elite, discussed earlier. The Kelleher group is spearheading the needed technological developments to realize the hope that top-down MS can be applied to solve problems in systems biology.
Another obstacle is the lower sensitivity of top-down approach for detection of PTMs than that of bottom-up. Technological advances are highly desired to address these issues. For example, it is clear that electrophoresis-based protein separation when coupled with top-down mass spectrometry will be a step forward to characterize therapeutic proteins. Another approach is a “middle-down” (“middle-out”) strategy whereby large proteins are digested to afford larger peptides than does trypsin digestion (discussed earlier). Promising in this area is the use of OmpT, and enzyme known to cleave principally at dibasic sites and give peptide fragments that can be 10 times larger in size than those from trypsin digestion138. As this enzyme becomes available, we expect to see new applications in proteomics.
On another front, construction of high-field FTICR instruments for improved performance is now going forward with a 21 T magnet139. In the meantime, the top-down technology will coexist with bottom-up and middle-down approaches for application to identification and biophysical investigation of proteins and other biomolecular entities.
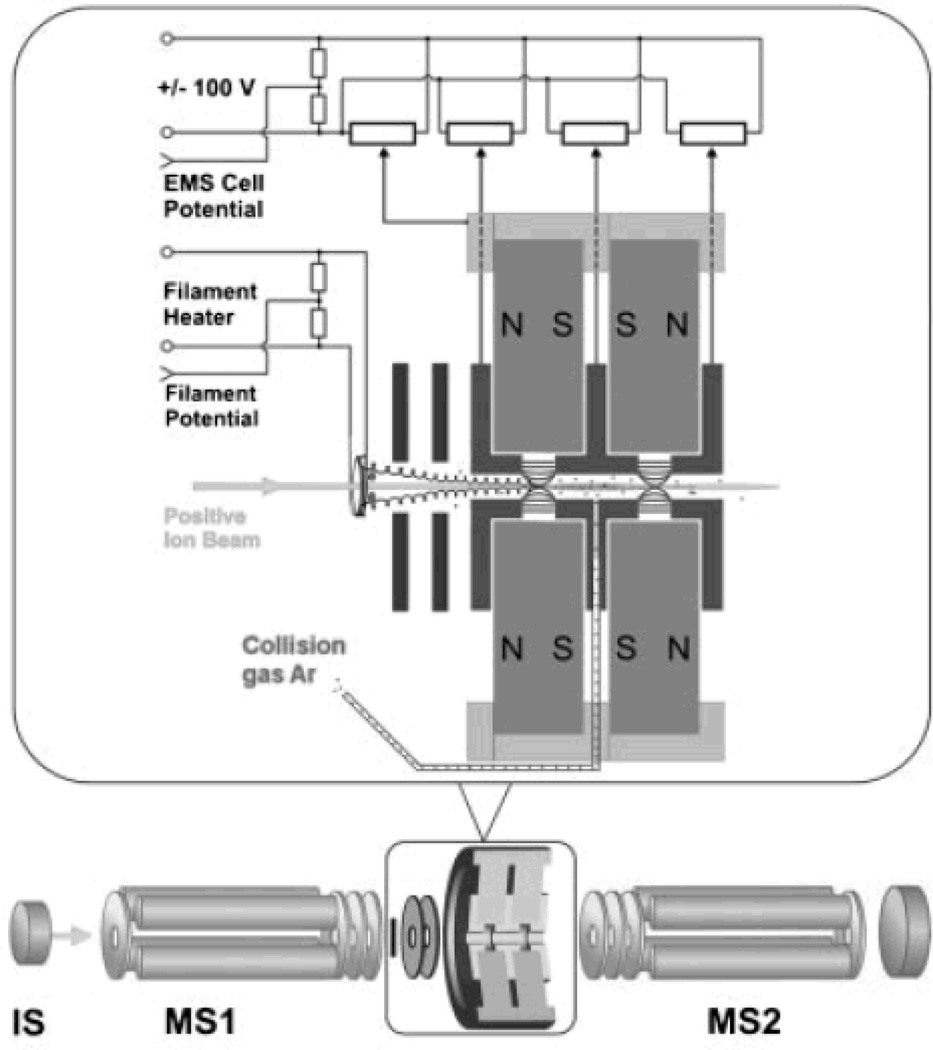
The extension of ECD to other non-FTICR types of mass spectrometers should have advantages in expanding the field. Recently, Voinov and coworkers140 built a radiofrequency-free magnetic cell in which ECD products were generated and detected in a triple quadrupole instrument by replacing the second quadrupole with this cell. This advance brings ECD into the realm of beam-type instruments. By simply adjusting the voltages, ECD and CAD can be performed sequentially in the cell141 (Figure 5). The coupling with Q-TOF instruments with their shorter duty-cycle may provide a more convenient means to achieve high-throughput ECD top-down analysis. In–source atmospheric pressure (AP)-ECD is another potentially interesting choice142.
Figure 5.
Triple quadrupole instrument with the 2nd Q replaced with a RF-free magnetic cell for ECD and CAD – ref 141, Rapid Commun. Mass Spectrom., 2009, 23, 3028–3030, copyright of John Wiley & Sons.
Given that top-down MS is well-suited for target-compound analysis, research in protein biophysics will grow in combination with hydrogen/deuterium exchange and other chemical labeling strategies to probe protein conformation and dynamics. In structural biology, determination of the compositions and stoichiometry of macromolecular assemblies is of great importance because many biological functions are performed by proteins in complexes rather than by isolated proteins. Our observation that ECD of an intact tetrameric complex of yeast alcohol dehydrogenase in FTICR provides both identification and structure information holds significant promise in this direction. Incorporation of ETD/ECD into the Q-TOF/ion mobility platform is also promising for the study of protein complexes; this combination gives cross sections, their changes for ETD/ECD fragments, and possibly CAD fragments to help infer structure.
Acknowledgements
Funding for the MS Resource at Washington University in St. Louis from the National Center for Research Resources of the National Institute of Health (grant P41RR000954 and grant 1S10025101 to Michael L. Gross) is greatly appreciated. Weidong Cui thanks the Deans of the Schools of Arts & Sciences and Medicine at Washington University for their support. This work is also supported by a grant from the National Science Foundation (IBDR 0964199).
References
- 1.JRR Yates CI, Nakorchevsky A. Annu. Rev. Biomed. Eng. 2009;11:49–79. doi: 10.1146/annurev-bioeng-061008-124934. [DOI] [PubMed] [Google Scholar]
- 2.SL Matallana-Surget B, Wattiez R. Expert Rev. Proteomics. 2010;7:5–7. doi: 10.1586/epr.09.101. [DOI] [PubMed] [Google Scholar]
- 3.Zubarev RA, Kelleher NL, McLafferty FW. J. Am. Chem. Soc. 1998;120:3265–3266. [Google Scholar]
- 4.Syka JEP, Coon JJ, Schroeder MJ, Shabanowitz J, Hunt DF. Proc. Natl. Acad. Sci. USA. 2004;101:9528–9533. doi: 10.1073/pnas.0402700101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chait BT. Science. 2006;314:65–66. doi: 10.1126/science.1133987. [DOI] [PubMed] [Google Scholar]
- 6.Armirotti A, Damonte G. Proteomics. 2010;10:3566–3576. doi: 10.1002/pmic.201000245. [DOI] [PubMed] [Google Scholar]
- 7.Garcia BA. J. Am. Soc. Mass Spectrom. 2010;21:193–202. doi: 10.1016/j.jasms.2009.10.014. [DOI] [PubMed] [Google Scholar]
- 8.Kellie JF, Tran JC, Lee JC, Ahlf DR, Thomas HM, Ntai I, Catherman AD, Durbin KR, Zamdborg L, Vellaichamy A, Thomas PM, Kelleher NL. Mol. Biosys. 2010;6:1532–1539. doi: 10.1039/c000896f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Scherperel GE, Reid G. Analyst. 2007;132:500–506. doi: 10.1039/b618499p. [DOI] [PubMed] [Google Scholar]
- 10.Li W, Hendrickson CL, Emmett MR, Marshall AG. Anal. Chem. 1999;71:4397–4402. doi: 10.1021/ac990011e. [DOI] [PubMed] [Google Scholar]
- 11.Jensen PK, Pasa-Tolic L, Anderson GA, Horner JA, Lipton MS, Bruce JE, Smith RD. Anal. Chem. 1999;71:2076–2084. doi: 10.1021/ac990196p. [DOI] [PubMed] [Google Scholar]
- 12.Zhou F, Johnson MV. Electrophoresis. 2005;26:1383–1388. doi: 10.1002/elps.200410125. [DOI] [PubMed] [Google Scholar]
- 13.Chong BE, yan F, Lubman DM, Miller FR. Rapid Commun. Mass Spectrom. 2001;15:291–296. doi: 10.1002/rcm.227. [DOI] [PubMed] [Google Scholar]
- 14.Liu H, Berger SJ, Chakraborty AB, Plumb RS, Cohen SA. J. Chromatogr. B. 2002;782:267–289. doi: 10.1016/s1570-0232(02)00554-8. [DOI] [PubMed] [Google Scholar]
- 15.Tian Z, Zhao R, Tolic N, Moore RJ, Stenojen DL, Robinson EW, Smith RD, Pasa-Tolic L. Proteomics. 2010;10:3610–3620. doi: 10.1002/pmic.201000367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lubman DM, Kachman MT, Wang H, Gong S, Yan F, Hamler RL, O'Neil KA, Zhu K, Buchanan NS, Barder TJ. J. Chromatogr. B. 2002;782:183–196. doi: 10.1016/s1570-0232(02)00551-2. [DOI] [PubMed] [Google Scholar]
- 17.Boersema PJ, Mohammed S, Heck AJ. Anal. Bioanal. Chem. 2008;391:151–159. doi: 10.1007/s00216-008-1865-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lindner H, Sarg B, Meraner C, Hellinger W. J. Chromatogr. A. 1996;743:137–144. doi: 10.1016/0021-9673(96)00131-8. [DOI] [PubMed] [Google Scholar]
- 19.Meng F, Cargile BJ, Patrie SM, Johnson JR, McLoughlin SM, Kelleher NL. Anal. Chem. 2002;74:2923–2929. doi: 10.1021/ac020049i. [DOI] [PubMed] [Google Scholar]
- 20.JL Lee JF, Tran JC, et al. J. Am. Soc. Mass Spectrom. 2009;20:2183–2191. doi: 10.1016/j.jasms.2009.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lim P, North R, Vigh G. Electrophoresis. 2007;28:1851–1859. doi: 10.1002/elps.200600846. [DOI] [PubMed] [Google Scholar]
- 22.Cologna SM, Russell WK, Lim PJ, Vigh G, Russell DH. J. Am. Soc. Mass Spectrom. 2010;21:1612–1619. doi: 10.1016/j.jasms.2010.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pai P, Cologna SM, Russell WK, Vigh G, Russell DH. Anal. Chem. 2011;83:2814–2818. doi: 10.1021/ac1029743. [DOI] [PubMed] [Google Scholar]
- 24.Whitelegge JP, le Coutre J, Lee JC, Engel CK, Prive GG, Faull KF, Kaback HR. Proc. Natl. Acad. Sci. U.S.A. 1999;96:10695–10698. doi: 10.1073/pnas.96.19.10695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Garcia BA, Siuti N, Thomas CE, Mizzen CA, Kelleher NL. Int. J. Mass Spectrom. 2007;259:184–196. [Google Scholar]
- 26.Iavarone AT, Jurchen JC, Williams ER. Anal. Chem. 2001;73:1455–1460. doi: 10.1021/ac001251t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Iavarone AT, Williams ER. J. Am. Chem. Soc. 2003;125:2319–2327. doi: 10.1021/ja021202t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lomeli SH, Peng IX, Yin S, Loo RRO, Loo JA. J. Am. Soc. Mass Spectrom. 2010;21:127–131. doi: 10.1016/j.jasms.2009.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.van Duijn E. J Am Soc Mass Spectrom. 2010;21:971–978. doi: 10.1016/j.jasms.2009.12.010. [DOI] [PubMed] [Google Scholar]
- 30.Valaja SG, Tipton JD, Emmett MR, Marshall AG. Anal. Chem. 2010;82:7515–7519. doi: 10.1021/ac1016858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Loo JA, CGaSEdmonds RR. Science. 1990;248:201–204. [Google Scholar]
- 32.Henry KD, Quinn JP, McLafferty FW. J. Am. Chem. Soc. 1991;113:5447–5449. [Google Scholar]
- 33.Loo JA, Quinn JP, Steven IR, Henry KD, Senko MW, McLafferty FW. Proc. Natl. Acad. Sci. USA. 1992;89:286–289. doi: 10.1073/pnas.89.1.286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cooper HJ, Hakansson K, Marshall AG. Mass Spectrom. Rev. 2005;24:201–222. doi: 10.1002/mas.20014. [DOI] [PubMed] [Google Scholar]
- 35.Macek B, Waanders LF, Olsen JV, Mann M. Mol. Cell. Proteomics. 2006;5:949–958. doi: 10.1074/mcp.T500042-MCP200. [DOI] [PubMed] [Google Scholar]
- 36.Sancho SR, Ge Y, Walker JW. J. Muscle Res. and Cell Motility. 2008;29:203–212. doi: 10.1007/s10974-009-9168-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Compton P, Damoc E, Denisov E, Tran JC, Ahlf DR, Wieghaus A, Senko MW, Horning SR, Makarov A, Kelleher NL. Proceedings of the 59th ASMS Conference on Mass Spectrometry and Allied Topics. Denver, Colorado: 2011. [Google Scholar]
- 38.Reiber DC, Grover TA, Brown RS. Anal. Chem. 1998;70:673–683. doi: 10.1021/ac971157l. [DOI] [PubMed] [Google Scholar]
- 39.Resemann A, Wunderlich D, Rothbauer U, Warscheid B, Leonhardt H, Fuchser J, Kuhlmann K, Suckau D. Anal. Chem. 2010;82:3283–3292. doi: 10.1021/ac1000515. [DOI] [PubMed] [Google Scholar]
- 40.McLafferty FW, Bente PF, Kornfeld R, Tsai SC, Howe I. J. Am. Chem. Soc. 1973;95:2120–2129. [Google Scholar]
- 41.Laskin J, Futrell JH. Mass Spectrom. Rev. 2003;22:158–181. doi: 10.1002/mas.10041. [DOI] [PubMed] [Google Scholar]
- 42.Horn DM, Ge Y, McLafferty FW. Anal. Chem. 2000;72:4778–4784. doi: 10.1021/ac000494i. [DOI] [PubMed] [Google Scholar]
- 43.Little DP, Speir JP, Senko MW, O'Connor PB, McLafferty FW. Anal. Chem. 1994;66:2809–2815. doi: 10.1021/ac00090a004. [DOI] [PubMed] [Google Scholar]
- 44.XJ Han M, Breuker K, McLafferty FW. Science. 2006;314:109–112. doi: 10.1126/science.1128868. [DOI] [PubMed] [Google Scholar]
- 45.Budnik BA, Haselmann KF, Zubarev RA. Chem. Phys. Lett. 2001;342:299–302. [Google Scholar]
- 46.Fung YME, Adams CM, Zubarev RA. J. Am. Chem. Soc. 2009;131:9977–9985. doi: 10.1021/ja8087407. [DOI] [PubMed] [Google Scholar]
- 47.Coon JJ, Shabanowitz J, Hunt DF, Syka JEP. J. Am. Soc. Mass Spectrom. 2005;16:880–882. doi: 10.1016/j.jasms.2005.01.015. [DOI] [PubMed] [Google Scholar]
- 48.Huzarska M, Ugalde I, Kaplan DA, Hartmer R, Easterling ML, Polfer NC. Anal. Chem. 2010;82:2873–2878. doi: 10.1021/ac9028592. [DOI] [PubMed] [Google Scholar]
- 49.Ganisl B, Valovka T, Hartl M, Taucher M, Bister K, Breuker K. Chem. - A Eur. J. 2011;17:4460–4469. doi: 10.1002/chem.201003709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Taucher M, Breuker K. J. Am. Soc. Mass Spectrom. 2010;21:918–929. doi: 10.1016/j.jasms.2010.02.025. [DOI] [PubMed] [Google Scholar]
- 51.McLuckey SC, Huang T. Anal. Chem. 2009;81:8669–8676. doi: 10.1021/ac9014935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.RDT LeDuc GK, Kim Y, Januszyk TE, Bynum LH, Sola JV, Garavelli JS, Kelleher LN. Nucleic Acids Research. 2004;32:W340–W345. doi: 10.1093/nar/gkh447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Theberge R, et al. Int. J. Mass Spectrom. 2010 [Google Scholar]
- 54.Smith RD, Loo JA, Barinaga CJ, Edmonds CG, Udseth HR. J. Am. Soc. Mass Spectrom. 1990;1:53–65. doi: 10.1016/1044-0305(90)80006-9. [DOI] [PubMed] [Google Scholar]
- 55.Reid GE, McLuckey SA. J. Mass Spectrom. 2002;37:63–75. doi: 10.1002/jms.346. [DOI] [PubMed] [Google Scholar]
- 56.Kelleher NL, Lin HY, Valaskovic GA, Aaserud DJ, Fridriksson EK, McLafferty FW. J. Am. Chem. Soc. 1999;121:806–812. [Google Scholar]
- 57.Feng R, Konishi Y. Anal. Chem. 1993;65:645–649. [Google Scholar]
- 58.Pesavento JJ, Kim Y, Taylor GK, Kelleher NL. J. Am. Chem. Soc. 2004;126:3386–3387. doi: 10.1021/ja039748i. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Boyne MTI, Pesavento JJ, Mizzen CA, Kelleher NL. J. Proteome Res. 2006;5:248–253. doi: 10.1021/pr050269n. [DOI] [PubMed] [Google Scholar]
- 60.Siuti N, Roth MJ, Mizzen CA, Kelleher NL, Pesavento JJ. J. Proteome Res. 2006;5:233–239. doi: 10.1021/pr050268v. [DOI] [PubMed] [Google Scholar]
- 61.Thomas CE, Kelleher NL, Mizzen CA. J. Proteome Res. 2006;5:240–247. doi: 10.1021/pr050266a. [DOI] [PubMed] [Google Scholar]
- 62.Jiang L, Smith JN, Anderson SL, Ma P, Mizzen CA, Kelleher NL. J. Biol. Chem. 2007;282:27923–27934. doi: 10.1074/jbc.M704194200. [DOI] [PubMed] [Google Scholar]
- 63.Pesavento JJ, Bullock CR, LeDuc RD, Mizzen CA, Kelleher NL. J. Biol. Chem. 2008;283:14927–14937. doi: 10.1074/jbc.M709796200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Pesavento JJ, Yang H, Kelleher NL, Mizzen CA. Mol. Cell. Biol. 2008;28:468–486. doi: 10.1128/MCB.01517-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Pesavento JJ, Garcia BA, Streeky JA, Kelleher NL, Mizzen CA. Mol. Cell. Proteomics. 2007;6:1510–1526. doi: 10.1074/mcp.M600404-MCP200. [DOI] [PubMed] [Google Scholar]
- 66.Meng F, Du Y, Miller LM, Patrie SM, Robinson DE, Kelleher NL. Anal. Chem. 2004;76:2852–2858. doi: 10.1021/ac0354903. [DOI] [PubMed] [Google Scholar]
- 67.Young NL, Plazas-Mayorca MD, DiMaggio PA, Flaniken IZ, Beltran AJ, Mishra N, LeRoy G, Floudas CA, Garcia BA. J. Am. Soc. Mass Spectrom. 2010;21:960–970. doi: 10.1016/j.jasms.2010.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Fridriksson EK, Beavil A, Holowka D, Gould HJ, Baird B, McLafferty FW. Biochem. 2000;39:3369–3376. doi: 10.1021/bi9919091. [DOI] [PubMed] [Google Scholar]
- 69.Ge Y, ElNaggar M, Sze SK, Oh H, Begley TP, McLafferty FW, Boshoff H, Barry CE. J. Am. Soc. Mass Spectrom. 2003;14:253–261. doi: 10.1016/s1044-0305(02)00913-3. [DOI] [PubMed] [Google Scholar]
- 70.Schirm M, Schoenhofen IC, Logan SM, Waldron KC, Thibault P. Anal. Chem. 2005;77:7774–7782. doi: 10.1021/ac051316y. [DOI] [PubMed] [Google Scholar]
- 71.Puppione DL, Bassilian S, Souda P, MacDonald MH, Hagland F, Whitelegge JP. Comparative Biochem Physiol, Part D: Genomics and Proteomics. 2008;3D:290–296. doi: 10.1016/j.cbd.2008.08.002. [DOI] [PubMed] [Google Scholar]
- 72.Sze SK, Ge Y, Oh H, McLafferty FW. Proc. Natl. Acad. Sci. USA. 2002;99:1774–1779. doi: 10.1073/pnas.251691898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Avaz-Guner S, Zhang J, Li L, Walker JW, Ge Y. Biochem. 2009;48:8161–8170. doi: 10.1021/bi900739f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ge Y, Rybakova IN, Xu Q, Moss RL. Proc. Natl. Acad. Sci. USA. 2009;106:12658–12663. doi: 10.1073/pnas.0813369106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zhang J, Dong X, Hacker TA, Ge Y. J. Am. Soc. Mass Spectrom. 2010;21:940–948. doi: 10.1016/j.jasms.2010.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Pesavento JJ, Mizzen CA, Kelleher NL. Anal. Chem. 2006;78:4271–4280. doi: 10.1021/ac0600050. [DOI] [PubMed] [Google Scholar]
- 77.Zabrouskov V, Han X, Welker E, Zhai H, Lin C, van Wijk KJ, Scheraga HA, McLafferty FW. Biochem. 2006;45:987–992. doi: 10.1021/bi0517584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Zabrouskov V, Ge Y, Schwartz J, Walker JW. Mol. Cell. Proteomics. 2008;7:1838–1849. doi: 10.1074/mcp.M700524-MCP200. [DOI] [PubMed] [Google Scholar]
- 79.Mazur MT, Cardasis HL, Spellman DS, Liaw A, Yates NA, Hendrickson RC. Proc. Natl. Acad. Sci. USA. 2010;107:7728–7733. doi: 10.1073/pnas.0910776107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Wiener MC, Sachs JR, Deyanova EG, Yates NA. Anal. Chem. 2004;76:6085–6096. doi: 10.1021/ac0493875. [DOI] [PubMed] [Google Scholar]
- 81.Meng F, Wiener MC, Sachs JR, Burns C, Verma P, Paweletz CP, Mazur MT, Deyanova EG, Yates NA, Hendrickson RC. J. Am. Soc. Mass Spectrom. 2007;18:226–233. doi: 10.1016/j.jasms.2006.09.014. [DOI] [PubMed] [Google Scholar]
- 82.Collier TS, Sarkar P, Rao B, Muddiman DC. J. Am. Soc. Mass Spectrom. 2010;21:879–889. doi: 10.1016/j.jasms.2010.01.031. [DOI] [PubMed] [Google Scholar]
- 83.White SH. Nature. 2009;459:344–346. doi: 10.1038/nature08142. and the other 345 articles in the same issue. [DOI] [PubMed] [Google Scholar]
- 84.Pan Y, Konermann L. Analyst. 2010;135:1191–1200. doi: 10.1039/b924805f. [DOI] [PubMed] [Google Scholar]
- 85.Whitelegge J, Halgand F, Souda P, Zabrouskov V. Expert Rev. Proteomics. 2006;3:585–596. doi: 10.1586/14789450.3.6.585. [DOI] [PubMed] [Google Scholar]
- 86.Whitelegge JP. Comprehensive Anal. Chem. 2009;52:179–196. [Google Scholar]
- 87.Thangaraj B, Ryan CM, Souda P, Krause K, Faull KF, Weber APM, Fromme P, Whitelegge JP. Proteomics. 2010;10:1–13. doi: 10.1002/pmic.201000190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Granvogl B, Zoryan M, Ploscher M, Eichacker LA. Anal. Biochem. 2008;383:279–288. doi: 10.1016/j.ab.2008.08.038. [DOI] [PubMed] [Google Scholar]
- 89.Whitelegge JP, Laganowsky A, Souda NJP, Zhang H, Cramer WA. J. Exp. Botany. 2006;57:1515–1522. doi: 10.1093/jxb/erj163. [DOI] [PubMed] [Google Scholar]
- 90.Ryan CM, Souda P, Bassilian S, Ujwal R, Zhang J, Abramson J, Ping P, al e. Mol. Cell. Proteomics. 2010;9:791–803. doi: 10.1074/mcp.M900516-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Little DP, Chorush RA, Speir JP, Senko MW, Kelleher NL, McLafferty FW. J. Am. Chem. Soc. 1994;116:4893–4897. [Google Scholar]
- 92.Little DP, Aaserud DJ, Valaskovic GA, McLafferty FW. J. Am. Chem. Soc. 1996;118:9352–9359. [Google Scholar]
- 93.McLuckey SC, Vanberkel GJ, Glish GL. J. Am. Soc. Mass Spectrom. 1992;3:60–70. doi: 10.1016/1044-0305(92)85019-G. [DOI] [PubMed] [Google Scholar]
- 94.McLuckey SC, Habibigoudarzi S. J. Am. Chem. Soc. 1993;115:12085–12095. [Google Scholar]
- 95.Flora JW, Muddiman DC. Rapid Commun. Mass Spectrom. 1998;12:759–762. [Google Scholar]
- 96.Huang T, Kharlamova A, McLuckey SA. J. Am. Soc. Mass Spectrom. 2010;21:144–153. doi: 10.1016/j.jasms.2009.09.020. [DOI] [PubMed] [Google Scholar]
- 97.Eddy SR. Nature Reviews Genetics. 2001;2:919–929. doi: 10.1038/35103511. [DOI] [PubMed] [Google Scholar]
- 98.Mercer TR, Dinger ME, Mattick JS. Nature Reviews Genetics. 2009;10:155–159. doi: 10.1038/nrg2521. [DOI] [PubMed] [Google Scholar]
- 99.T Huang LJ, McLuckey SC. J. Am. Soc. Mass Spectrom. 2010;21:890–898. doi: 10.1016/j.jasms.2009.12.007. [DOI] [PubMed] [Google Scholar]
- 100.Taucher M, Rieder U, Breuker K. J. Am. Soc. Mass Spectrom. 2010;21:278–285. doi: 10.1016/j.jasms.2009.10.010. [DOI] [PubMed] [Google Scholar]
- 101.Grifffey RH, Hofstadler SA, Sannes-Lowery KA, Ecker DJ, Crooke ST. Proc. Natl. Acad. Sci. USA. 1999;96:10129–10133. doi: 10.1073/pnas.96.18.10129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Turner KB, Hagan NA, Kohlway AS, Fabris D. J. Am. Soc. Mass Spectrom. 2006;17:1401–1411. doi: 10.1016/j.jasms.2006.06.009. [DOI] [PubMed] [Google Scholar]
- 103.Breuker K, Oh H, Horn DM, Cerda BA, McLafferty FW. J. Am. Chem. Soc. 2002;124:6407–6420. doi: 10.1021/ja012267j. [DOI] [PubMed] [Google Scholar]
- 104.Horn DM, Breuker K, Frank AJ, McLafferty FW. J. Am. Chem. Soc. 2001;123:9792–9799. doi: 10.1021/ja003143u. [DOI] [PubMed] [Google Scholar]
- 105.Xu G, Zhai H, Naravan M, McLafferty FW, Scheraga HA. Chem., Biol. 2004;11:517–524. doi: 10.1016/j.chembiol.2004.03.020. [DOI] [PubMed] [Google Scholar]
- 106.Breuker K, McLafferty FW. Angew. Chem. Int. Ed. 2003;42:4900–4904. doi: 10.1002/anie.200351705. [DOI] [PubMed] [Google Scholar]
- 107.Breuker K, McLafferty FW. Angew. Chem. Int. Ed. 2005;44:4911–4914. doi: 10.1002/anie.200500668. [DOI] [PubMed] [Google Scholar]
- 108.Breuker K, McLafferty FW. Proc. Natl. Acad. Sci. U.S.A. 2008;105:18145–18152. doi: 10.1073/pnas.0807005105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Breuker K, Bruschweiler S, Tollinger M. Angew. Chem. Int. Ed. 2011;50:873–877. doi: 10.1002/anie.201005112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Engen JR. Anal. Chem. 2009;81:7870–7875. doi: 10.1021/ac901154s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Mendoza VL, Vachet RW. Mass Spectrom. Rev. 2009;28:785–815. doi: 10.1002/mas.20203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Kaltashov IA, Bobst CE, Abzalimov RR, Berkowitz SA, Houde D. J. Am. Soc. Mass Spectrom. 2010;21:323–327. doi: 10.1016/j.jasms.2009.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Pan J, Han J, Borchers CH, Konermann L. J. Am. Chem. Soc. 2008;130:11574. doi: 10.1021/ja802871c. [DOI] [PubMed] [Google Scholar]
- 114.Pan J, Han J, Borchers CH, Konermann L. Anal. Chem. 2010;82:8591–8597. doi: 10.1021/ac101679j. [DOI] [PubMed] [Google Scholar]
- 115.RRK Abzalimov DA, Easterling ML, Kaltashov IA. J. Am. Soc. Mass Spectrom. 2009;20:1514–1517. doi: 10.1016/j.jasms.2009.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Kaltashov IA, Bobst CE, Abzalimov RR. Anal. Chem. 2009;81:7892–7899. doi: 10.1021/ac901366n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Sterling HJ, williams ER. Anal. Chem. 2010;82:9050–9057. doi: 10.1021/ac101957x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Chen J, Cui W, Giblin D, Gross ML. Manuscript in preparation. [Google Scholar]
- 119.Chen J, Rempel DL, Gross ML. J. Am. Chem. Soc. 2010;132:15502–15504. doi: 10.1021/ja106518d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Loo JA. Mass Spectrom Rev. 1997;16:1–23. doi: 10.1002/(SICI)1098-2787(1997)16:1<1::AID-MAS1>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- 121.Heck AJ, Van Den Heuvel RH. Mass Spectrom Rev. 2004;23:368–389. doi: 10.1002/mas.10081. [DOI] [PubMed] [Google Scholar]
- 122.Benesch JL, Ruotolo BT, Simmons DA, Robinson CV. Chem Rev. 2007;107:3544–3567. doi: 10.1021/cr068289b. [DOI] [PubMed] [Google Scholar]
- 123.Benesch JL. J Am Soc Mass Spectrom. 2009;20:341–348. doi: 10.1016/j.jasms.2008.11.014. [DOI] [PubMed] [Google Scholar]
- 124.Sharon M. J. Am. Soc. Mass Spectrom. 2010;21:487–500. doi: 10.1016/j.jasms.2009.12.017. [DOI] [PubMed] [Google Scholar]
- 125.Leitner A, Walzthoeni T, Kahraman A, Herzog F, Rinner O, Beck M, Aebersold R. Mol. Cell. Proteomics. 2010;9:1634–1649. doi: 10.1074/mcp.R000001-MCP201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Haselmann KF, Jorgensen TJ, Budnik BA, Jensen F, Zubarev RA. Rapid Commun. Mass Spectrom. 2002;16:2260–2265. doi: 10.1002/rcm.853. [DOI] [PubMed] [Google Scholar]
- 127.YZ Xie J, Yin S, Loo JA. J. Am. Chem. Soc. 2006;128:14432–14433. doi: 10.1021/ja063197p. [DOI] [PubMed] [Google Scholar]
- 128.Yin S, Loo JA. J Am Soc Mass Spectrom. 2010;21:899–907. doi: 10.1016/j.jasms.2010.01.002. [DOI] [PubMed] [Google Scholar]
- 129.Erales J, Brigitte G, Whitelegge J, Halgand F. Biochem. J. 2009;419:75–82. doi: 10.1042/BJ20082004. [DOI] [PubMed] [Google Scholar]
- 130.Clarke DJ, Murray E, Hupp T, Mackay CL, Langridge-Smith PRR. J. Am. Soc. Mass Spectrom. 2011 doi: 10.1007/s13361-011-0155-3. [DOI] [PubMed] [Google Scholar]
- 131.Yin S, Loo JA. Int. J. Mass Spectrom. 2010 doi: 10.1016/j.ijms.2010.06.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Sterling HJ, Cassou CA, Trnka MJ, Burlingame AL, Krantz BA, Williams ER. Phys. Chem. Chem. Phys. 2011 doi: 10.1039/c1cp20277d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Breuker K. Int. J. Mass Spectrom. 2004;239:33–41. [Google Scholar]
- 134.Schermann SM, Simmons DA, Konermann L. Expert Rev. Proteomics. 2005;2:475–485. doi: 10.1586/14789450.2.4.475. [DOI] [PubMed] [Google Scholar]
- 135.Barrera NP, Di Bartolo N, Booth PJ, Robinson CV. Science. 2008;321:243–246. doi: 10.1126/science.1159292. [DOI] [PubMed] [Google Scholar]
- 136.Zhang H, Cui W, Wen J, Blankenship RE, Gross ML. J Am Soc Mass Spectrom. 2010;21:1966–1968. doi: 10.1016/j.jasms.2010.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Zhang H, Cui W, Wen J, Blankenship RE, Gross ML. Anal. Chem. 2011 doi: 10.1021/ac200695d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Varadarajan N, Rodriguez S, Hwang B, Georgious G, Iverson BL. Nature Chem. Biol. 2008;4:290–294. doi: 10.1038/nchembio.80. and private communication with Neil Kelleher. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Arnaud CH. Chem. Eng. News. 2010 Jun 21;:10–15. [Google Scholar]
- 140.Voinov VG, Deinzer ML, Barofsky DF. Anal. Chem. 2009;81:1238–1243. doi: 10.1021/ac802084w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Voinov VG, Beckman JS, Deinzer ML, Barofsky DF. Rapid Commun. Mass Spectrom. 2009;23:3028–3030. doi: 10.1002/rcm.4209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Robb DB, Rogalski JC, Kast J, Blades MW. Rapid Commun. Mass Spectrom. 2010;24:3303–3308. doi: 10.1002/rcm.4773. [DOI] [PubMed] [Google Scholar]