Abstract
Herein proposed is a simple system to realize hands-free labeling and simultaneous detection of two human cell lines within a microfluidic device. This system was realized by novel covalent immobilization of pH-responsive poly(methacrylic acid) microgels onto the inner glass surface of an assembled polydimethylsiloxane/glass microfluidic channel. Afterwards, selected thiophene labeled monoclonal antibodies, specific for recognition of CD4 antigens on T helper/inducer cells and CD19 antigens on B lymphocytes cell lines, were encapsulated in their active state by the immobilized microgels. When the lymphocytes suspension, containing the two target subpopulations, was flowed through the microchannel, the physiological pH of the cellular suspension induced the release of the labeled antibodies from the microgels and thus the selective cellular staining. The selective pH-triggered staining of the CD4- and CD19-positive cells was investigated in this preliminary experimental study by laser scanning confocal microscopy. This approach represents an interesting and versatile tool to realize cellular staining in a defined module of lab-on-a-chip devices for subsequent detection and counting.
INTRODUCTION
The simultaneous labeling and detection of two or more different markers are the basic steps for multiparametric cellular analysis (MA). MA is an invaluable tool in cell biology and clinical diagnostics to analyze cell subpopulations and to assess their function in both normal and disease processes.
Currently, MA involves extensive and time-consuming sample preparation for multicolor cell staining.1 Improper stimuli during sample processing could also alter the original immunophenotype of the cells, hence inducing sample preparation artifacts.2, 3, 4 In this context, the fully integration of multiple cellular staining with counting and characterization of cells in lab-on-a-chip devices (LOCs) is highly desirable.5, 6 Additionally, microfluidic LOCs offer the advantages of high-throughput assays, multistage automation, parallel processing of multiple analytes, and portability.7, 8, 9
On the other hand the integration of cellular staining on-chip is not a trivial task as it involves complex and time-consuming sample pre-treatments. To this aim, different approaches including micro-mixers, membranes, and microfluidic arrays among others have been developed.10, 11, 12, 13, 14, 15, 16, 17 However, these systems do not meet the requirements for applications like point-of-care diagnostics since they require injections of both cell suspension and staining reagents within the microdevice as well as the transport of labile staining reagents where the analysis takes place.
Hence, the development of reagent storage systems within the microfluidic device may be very appealing to reduce demands on end-users and to facilitate the employment in lower-resource settings.
To this aim, Yager et al. propose a fibrous pad containing lyophilized fluorescent proteins where the functional biomolecules are rehydrated with phosphate-buffered saline to perform immunoassay.18 In an earlier strategy, a glass chamber coated with a dried mixture of gelatin and fluorochrome conjugates antibodies has been proposed to retain the functional biomolecules in hydrated state for cell staining.19 However, both these systems are disposable, and they cannot be reloaded with protein cargo after the staining process.
Accordingly there is the need to develop innovative strategies to retain the functional biomolecules within the microdevice and to release them in response to a proper stimulus to stain target cells in a specific section of microfluidic device.
In this paper, the in situ cell staining was achieved by the use of a novel microfluidic platform functionalized with poly(methacrylic acid) (PMAA) microgels.
Owing to their pH-responsiveness, PMAA microparticles were able to encapsulate labeled antibodies in their active state and release them upon appropriate pH variations. The intrinsic properties (high water content, biocompatibility, and chemical/mechanical stability) of stimuli-responsive microgels make them ideal candidates for the uptake and release of biomolecules in cellular analyses.20, 21, 22, 23, 24 Indeed, the hydrophilic network of microgels provides the encapsulated biomolecules with a protective environment and inhibits their degradation. Particles of micrometer size can also fit into microfluidic devices and provide large surface to volume ratio and short response time.20, 21, 22, 23, 24
Finally, microgels may fulfill the need of a more controlled microenvironment during the cell staining process, thus reducing the potential for alterations of the target cell.
For microfluidic applications, microgels need to be fixed within the active regions of the device channels and not flow out during the analysis.25 Hence, a robust and reliable method, easy to manufacture in miniaturized systems leading to place them in a microfluidic device, should be properly developed. Further, this system, thanks to the employment of smart microgel, can be reloaded and reused several times making this flexible approach applicable for a wide range of lab-on-a-chip applications.
In this work the inner glass wall of an assembled polydimethylsiloxane (PDMS)/glass microfluidic channel was covalently functionalized with pH-responsive PMAA microgels by exploiting the functional groups existing on the microparticles and the chemically modified glass surface.
The leukocytes superfamily is composed of many subtypes of white blood cells such as B and T cells, whose variations in concentration are major data for diagnostics of many pathologies.26
In this study, in order to mimic biological samples, two different human cell lines were used, namely SUP-T1 T lymphocytes and TK6 B lymphocytes. Anti-human CD4 and CD19 monoclonal antibodies (MAbs) were chosen for the detection of T and B lymphocytes, respectively.27, 28 These selected monoclonal antibodies were previously labeled with thiophene fluorophores (TFs). TFs are fluorescent probes characterized by high optical stability, bright fluorescence, and large Stokes shifts, particularly well-suited for multiplexing analyses.28, 29 Labeled MAbs are retained in the hydrated state within the microchannel where the sample is pumped and their release, to accomplish cellular staining, is induced only from the passage of cell suspension.
EXPERIMENTAL
Materials
All chemicals and solvents were purchased from Sigma Aldrich, unless otherwise stated. The 4-nitrophenyl methacrylate monomer (NPMA) was obtained from monomer and polymer, whereas the O,O′-Bis (2-aminoethyl) poly(ethylene glycol)(diamino-PEG) 2000 was purchased from Rapp Polymere. All reagents were used as received. The PMAA microgels were synthesized in solution according to the procedure reported in Appendix 1 of the supplementary material.30 TF1-conjugated anti-human CD4 (TF1-antiCD4) and TF2-conjugated anti-human CD19 (TF2-antiCD19) antibodies were obtained from Mediteknology srl. The chemical structures of the two employed thiophene fluorophores 2,5-dioxopyrrolidin-1-yl-5′-(7-(thiophen-2-yl) benzo[c][1,2,5]thiadiazol-4-yl)2-2′bithiophene-5-carboxylate (TF1) and 2,2′:5′,2″-terthiophene-5-carboxylic acid 4-sulfo-2,3,5,6-tetrafluorophenyl ester, sodium salt (TF2), together with the absorption and photoluminescence spectra of TF1 and TF2 conjugated MAbs, are shown in Appendix 2 of the supplementary material.30 The human B lymphoblastoid and T lymphoblastoid cells, TK6 and SUP-T1, respectively, were purchased from Health Protection Agency Culture Collections (Porton Down, Salisbury, UK). Both the cell lines were grown according to the supplier's instructions. Deionized water with a resistivity of ≥18.0 MΩ/cm was employed in all experiments. A commercial PDMS elastomer kit (Sylgard 184) composed of an elastomer base and a curing agent was purchased from Dow Corning. Unless specified, the experiments and measurements were run at room temperature (RT). The smart surface was realized on the top of a glass substrate whose dimensions are 1 cm in width and 1 cm in depth.
Microgels functionalized platforms
Smart platforms were realized by covalent immobilization of PMAA microgels onto pretreated glass substrates. Briefly, glass substrates were pre-cleaned by sonication in acetone and isopropylic alcohol and then treated with oxygen plasmas (RIE IONVAC, PGF 600 RF HUTTER) to increase the hydroxyl groups content on their surfaces. The oxygen flow rate was set to 20 sccm, at power of 30 W and pressure of 30 mTorr for 3 s. Subsequently, the glass surfaces were silylated with 1% (v/v) 3-triethoxysilyl-propyl isocyanate (TESPI) in toluene overnight. TESPI reacted with hydroxyl groups of the glass surfaces, thus leaving free isocyanate terminals for further reactions with a nucleophilic group. The glass samples were thoroughly rinsed with toluene and dried by nitrogen flow. Immediately after the silanization step, the diamino-PEG spacers were linked by nucleophilic attack of the amino groups on the isocyanate terminals. A solution of diamino-PEG (0.8% w/v) in tetrahydrofuran (THF) was added to the isocyanate-functionalized glass substrates and kept with gentle stirring for 1 h to induce the formation of the ureidic bond. Then the solution was removed, and the glass substrates were rinsed abundantly with THF and dried under nitrogen stream.
Smart platforms were realized by covalent immobilization of PMAA microgels, synthesized in a separate flask, onto the pretreated glass substrates by carbodiimide chemistry (the carboxylic groups of the PMAA microgels were activated with 1-Ethyl-3 (3-dimethylaminopropyl) carbodiimide hydrochloride (EDC)) by optimizing the procedure reported in our previous work. In particular, the reaction solution consisting of PMAA microgels (2.5 mg/ml) and 6.0 mM EDC in MES buffer (1.0 mM) was adjusted to pH 4.0 and incubated at RT for 1 h. Then glass substrates were rinsed carefully with distilled water and sonicated, in order to remove the non-covalently bonded microgels.
The experimental conditions for the uptake of single labeled MAb (either the TF1-antiCD4 or the TF2-antiCD19) into the PMAA microgels, immobilized on glass surfaces, were the following. The optimal concentration of the labeled antibody was found to be 5.0 μg/ml (data not shown). The TF1-antiCD4 antibody solution was diluted with MES buffer (1.0 mM, pH 5.3) to a final concentration of 5.0 μg/ml and incubated with the microgel-functionalized platform on a rotary mixer overnight.
To induce the simultaneous uptake of TF1-antiCD4 and TF2-antiCD19 MAbs, a solution consisting of both the labeled MAbs at the concentration of 2.5 μg/ml (thus the total concentration of proteins was 5.0 μg/ml) in MES buffer (1.0 mM, pH 5.3) was incubated with the microgel-based platform at RT overnight. Soon after, the loaded smart platforms with the labeled antibody was rinsed thoroughly with distilled water, to remove the free labeled antibodies, and finally dried under a nitrogen flow.
After the uptake process, the smart platforms were analyzed by laser scanning confocal microscopy (LSCM). LSCM was selected in this preliminary study for convenience and simplicity. Conversely counting and detection of labeled cells in the integrated smart platforms on-chip will be performed by developed systems of detection for flow cytometry on-chip. LSCM measurements were performed by using a FV-1000 Olympus microscope in epilayer configuration. The samples were excited by a diode laser (λ = 405 nm) through an objective lens 40 × with a numerical aperture of 0.85.
The release process was performed by adding MES buffer (1.0 mM, pH 7.8) onto the previously loaded platforms and incubating overnight on a rotary mixer. Afterwards, the glass substrates were rinsed thoroughly with distilled water to remove the free labeled antibodies and finally dried under nitrogen flow. The uptake and release processes were characterized by LSCM to test the ability of the immobilized microgels to encapsulate and release either the TF1-antiCD4 or the TF2-antiCD19 or both under pH variations.
Cell staining experiments were run by incubating the smart platforms loaded with one or two labeled MAbs with cell suspension of either TK6 or SUP-T1 cells, or both simultaneously at an estimated concentration of 1.5 × 106 cells/ml at pH 7.8 overnight. The physiological pH of the cell suspension allowed the in situ cell staining, thanks to the release of fluorescent monoclonal antibodies. After overnight incubation, the system consisting of tested cells onto the smart surfaces was analyzed by LSCM. The cell staining experiments, including the labeled antibodies and the cell lines, are summarized in Table TABLE I..
TABLE I.
List of the cell staining experiments using different combinations of TF-labeled antibodies and lymphoblastoid cells lines, together with the experimental results obtained by LSCM.
| TF1-antiCD4 | TF2-antiCD19 | SUP-T1 (CD4+) | TK6 (CD19+) | Results | |
|---|---|---|---|---|---|
| 1 | − | + | − | + | Single stain (TF2) |
| 2 | − | + | + | − | No stain |
| 3 | − | + | + | + | Single stain (TF2) |
| 4 | + | − | + | + | Single stain (TF1) |
| 5 | + | − | − | + | No stain |
| 6 | + | − | + | + | Single stain (TF1) |
| 7 | + | + | + | − | Single stain (TF1) |
| 8 | + | + | − | + | Single stain (TF2) |
| 9 | + | + | + | + | Multiple stain (TF1+TF2) |
The binding of labeled conjugates by cell surface CD4 and CD19 antigens was assessed by analyzing the fluorescence signals in a spectral window of 25 nm around 580 and 490 nm for TF1 and TF2, respectively. The emission spectra were recorded by the internal spectral unit of the microscope with a spectral resolution of 2 nm. TK6 and SUP-T1 T cells were analyzed cell-by-cell and autofluorescence was excluded by spectral analysis of individual cells. Cell treatment procedures are reported in Appendix 3 of the supplementary material.30
Microgels functionalized microchannel
The assembled PDMS/glass microfluidic device with one inlet and one exit was constructed as follows: glass substrates were pre-cleaned, treated with oxygen plasma, and functionalized with TESPI and diaminoPEG, according to the procedure reported in Sec. 2B.
Separately, linear microchannels were fabricated via soft lithography technique (Appendix 4 of the supplementary material30) by mixing PDMS prepolymer and a curing agent in a weight ratio of 10:1, respectively.31, 32, 33
The microfluidic device had the dimension of 1.7 cm in width, 3 mm in height, and 2.3 cm in depth. The microchannel within it had the dimensions of 500 μm in width, 25 μm in height, and 1 cm in depth.
After deposition of the PDMS channel on the diaminoPEG-functionalized glass substrates, the PMAA microgels were covalently attached onto the inner glass surface of the microchannel. To this aim, a diluted suspension of microgels in 6.0 mM EDC solution (MES buffer 1.0 mM), at the ratio of 1:50 was injected by a syringe pump (flow rate of 2 ml/h) and incubated for 2 h within the microfluidic channel. Afterwards, the microchannel surface was cleaned with distilled water using the syringe pump at a flow rate 0.5 ml/h, 2 h, to remove the non-covalently bonded microgels. The PMAA functionalized microchannel was loaded with both the TF1 and TF2 antibodies: the uptake solution consisting of both the labeled MAbs was diluted at ratio of 1:2 and was flushed into the microchannel by a syringe pump (flow rate of 0.5 ml/h) for 1 h. Any remaining free labeled antibodies were removed by flushing distilled water (1 ml/h, 1 h).
The cell staining experiments were run by injection of the diluted cell suspension, at 1:10 ratio, with a syringe in a pulsed way within the loaded microchannel.34 After 30 min, the tested cell suspension, within the functionalized microchannel, was analyzed by confocal microscopy to check the cell staining induced by release of both labeled MAbs. To definitely demonstrate the specificity of the immunostaining within the double loaded microgels functionalized microchannel, control experiments were carried out with single CD4+ SUP-T1 and CD19+ TK6 cells.
RESULTS AND DISCUSSION
Smart PMAA microgels surfaces for uptake and release of TF-labeled antibodies
In our previous work, we verified that PMAA microgels preserve their pH-sensitivity even after covalent immobilization on glass substrates. In particular, the atomic force microscopy (AFM) analysis indicated that immobilized microgels were able to swell from 0.5 to 1.2 μm upon switching from pH 5.3 to pH 8.0. Owing to their pH-responsiveness, the immobilized PMAA microgels were exploited to induce the pH-controlled uptake and release of one labeled antibody for single cell staining of T cells.35, 36, 37
In the current work, we initially improved the covalent immobilization process of the PMAA microgels onto glass substrates, and then we assessed their capability to retain up to two different labeled antibodies and to release them simultaneously under physiological pH. The primary aim was to show that the labeled MAbs were retained in their active state and then released in response to the pH-environment thus inducing the double labeling of B and T cell subpopulations.
Afterwards, PMAA microgels were covalently immobilized onto the inner glass wall of an assembled PDMS/glass microfluidic device, and the double cell staining was performed on-chip.
For cell staining experiments, two human lymphocyte lines, TK6 (B cells) and SUP-T1 (T cells) were used. Indeed, the T cell/B cell ratio has diagnostic relevance and plays a crucial role in immune response and many diseases. Considering that T cells and B cells express CD4 and CD19 surface antigens, respectively, two monoclonal anti-CD4 and anti-CD19 antibodies labeled with two different thiophene fluorophores were employed in the following experiments. They are indicated as TF1-antiCD4 and TF2-antiCD19 MAbs.26, 27, 28 Thiophene fluorophores provide MAbs with attractive features for spectroscopic characterization.28, 29
First, the platforms were incubated with each single TF-labeled antibody at pH 5.0, to ensure that the proteins would be loaded via ion exchange mechanisms. Indeed, at this pH the proteins have a net positive charge (pI 5.9) whereas the microgels are negatively charged (Appendix 5 of the supplementary material30). The optimal concentration of the labeled antibody was found to be 5.0 μg/ml (data not shown). In second step, the microgels functionalized surfaces were loaded with both labeled antibodies for the simultaneous cell staining of B and T cells. The concurrent uptake of TF1-antiCD4 and TF2-antiCD19 MAbs was run under the same conditions described for the single uptake process. However, this time the concentration of each antibody was 2.5 μg/ml, so that their total concentration was kept to 5.0 μg/ml. This ensures that the microgels would be exposed to the same concentration of bioactive molecules either simultaneous or single uptake experiments.
LSCM has been used to study the simultaneous uptake of TF1-antiCD4 and TF2-antiCD19 MAbs into the covalently immobilized microgels (Fig. 1). Specifically, TF1 and TF2 were visualized in real time using a diode laser (λ = 405 nm), while their emission spectra were recorded by the internal spectral unit of the microscope (spectral resolution of 2 nm) and collected without switching the excitation light source.
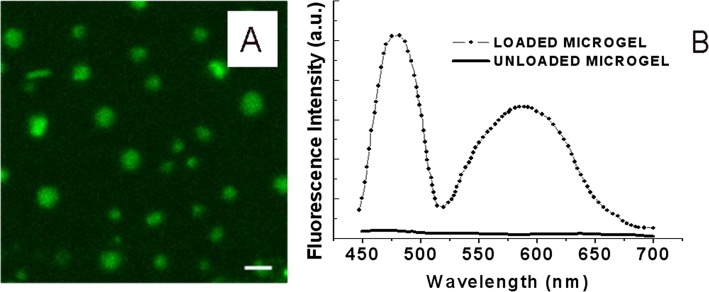
Figure 1.
(a) The uptake of TF1-antiCD4 and TF2-antiCD19 antibodies into the covalently immobilized microgels onto smart platform was analyzed by detecting the fluorescence arising from the two TFs fluorophores. (b) Fluorescence spectra of the immobilized microgels after the uptake process (dotted line), assessing the location of both fluorescent biomolecules inside the immobilized microparticles, and after release of labeled antibodies (straight line). The scale bar was 5 μm.
According to the LSCM analysis, the fluorescence spectra of TF1 and TF2 were highly defined, thus suggesting that thiophene molecules were well-spaced within the hydrophilic matrix of the microgels and did not interfere with each other upon nonspecific binding. To assess the concentrations of the encapsulated labeled antibodies, calibration curves were prepared for both the TF1-antiCD4 and the TF2-antiCD19. The estimated values of encapsulated labeled MAbs into the PMAA microgels were found to be 0.22 μg/ml for TF1-antiCD4 and 0.25 μg/ml for TF2-antiCD19 (Appendix 6 of the supplementary material30). Therefore, the covalently attached PMAA microgels were able to encapsulate approximately the same amount of the labeled MAbs, thus indicating that either the specificity of thiophenes or the antigen did not affect the uptake process.
The water environment of the PMAA microgels could also minimize biomolecules denaturing, thereby keeping them in their active state during storage. Afterwards, the release of the loaded MAbs was induced by incubating the smart surface into fresh buffer with a pH value of 7.8 overnight. In these conditions, labeled antibodies were released into the external solution as a result of the increased permeability of the microgels network, as well as the electrostatic repulsions between the proteins and the hydrogel. The presence of fluorescent signal within the microparticles after the uptake, as well its decrease after the release, clearly demonstrated that the immobilized PMAA microgels were able to encapsulate and deliver TF1-antiCD4 and TF2-antiCD19 simultaneously under pH variations (Fig. 1b). As reported in Figure 1, the release of both labeled antibodies from immobilized microgels was almost complete.
Cellular staining experiments with smart PMAA microgels surfaces
Cell staining experiments were run by incubating the loaded smart platforms with cell suspension of either TK6 or SUP-T1 cells, or both simultaneously at an estimated concentration of 1.5 × 106 cells/ml at pH 7.8. This physiological pH allowed the release of fluorescent antibodies through the enlarged meshes of the microgel network and the in situ cell staining.
Cytotoxic effects of PMAA microgels, tested by the 3-(4,5-dimethyl-2-thiazolyl)-2, 5-diphenyl-2H-tetrazolium bromide (MTT) assay, have been reported in our previous paper, and the microgels were found to be nontoxic up to the highest tested concentration.36
After overnight incubation the system consisting of cell suspension and the smart surfaces was analyzed by LSCM. The cell staining experiments, including the selected labeled antibodies and the cell lines used are summarized in Table TABLE I..
The experiments were initially carried out incubating individual TF1-antiCD4 and TF2-antiCD19 antibodies with a suspension of either T or B cells, or both (Table TABLE I., Expts. 1-6) to realize positive and negative controls.
The positive controls showed that only TK6 B lymphocytes, within a mixture of T and B cells, were stained after incubation with the single loaded TF2-antiCD19 platform (Table TABLE I., Expts. 1-3). Similarly, the release of single TF1-antiCD4 determines only the staining of CD4+ SUP-T1 lymphocytes within a mixture of both target cells (Table TABLE I., Expts. 4-6).
The negative controls were run by incubating the single loaded TF1-antiCD4 or TF2-antiCD19 MAbs platforms with single CD19+ TK6 and CD4+ SUP-T1 cells, respectively (Table TABLE I., Expts. 2 and 5). In both cases cellular staining did not occur, thus assessing the absence of nonspecific binding.
Then the microgels functionalized surfaces loaded with both TF1-antiCD4 and TF2-antiCD19 antibodies were incubated with the CD4 positive SUP-T1 (Table TABLE I., Expt. 7), with the CD19 positive TK6 cells (Table TABLE I., Expt. 8) and with a mixture of SUP-T1 and TK6 cells in a 50/50 ratio (Table TABLE I., Expt. 9).
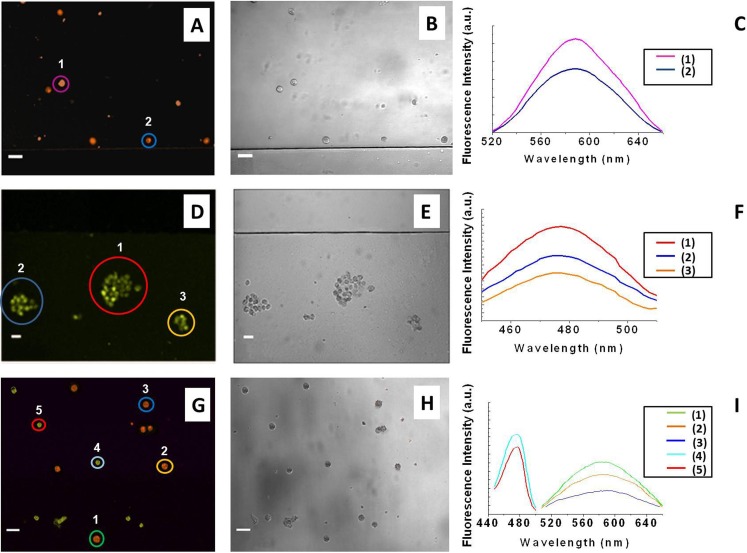
As shown in Figures 2a, 2b, 2c, only the TF1 fluorescence spectrum was recorded in Expt. 7; thus, the TF1-antiCD4 MAb exclusively interacted with specific cell surface antigens, whereas the TF2-antiCD19 MAbs were released in solution. Conversely, in Expt. 8 only the TF2-antiCD19 staining was observed (Figs. 2d, 2e, 2f). These findings revealed that the cell-antibody interaction was highly selective via the T or B cell receptor complex, and any nonspecific binding was detectable.
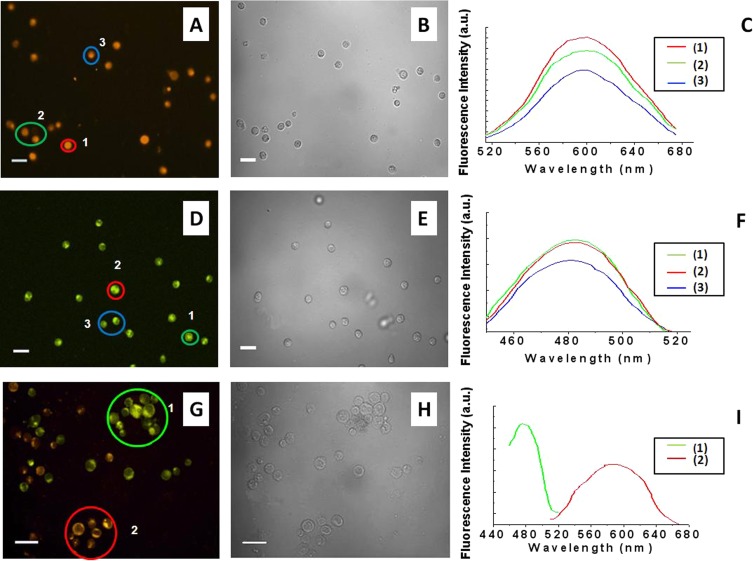
Figure 2.
Confocal microscopy images of (a) SUP-T1 T cells, (d) TK6 B cells, and (g) SUP-T1 T and TK6 B cells after the incubation onto loaded smart platforms with the two labeled MAbs. The double cell staining was shown in the latter experiment. It was observed that TF1-antiCD4 antibody positively stained SUP-T1 cells whereas TF2-antiCD19 positively stained TK6 cells. The corresponding images (b), (e), (h) were acquired under bright field exposure. For the sake of clarity, only selected cells are shown. CD4-positive cells are false-colored orange and CD19-positive cells are false-colored green. The scale bar was 25 μm.
Finally, the dual-labeled smart platform was incubated with the mixture of CD4+ SUP-T1 and CD19+ TK6 cells. Under these conditions, both the TF1 and TF2 fluorescence spectra were recorded by LSCM, thus indicating that the simultaneous double staining on the tested cells occurred (Figs. 2g, 2h, 2i). The ratio of labeled cells to total tested cells was estimated to be 50% ± 5.% for CD19+ TK6 cells and 50% ± 5% for CD4+ SUP-T1. These results represent the mean and standard deviation of three independent measurements, each performed with different smart platforms.
Functionalized PMAA microgels microchannel for double cell staining
After the development of microgels functionalized surfaces onto glass substrates, the inner glass surface of microfluidic channel was functionalized with PMAA microgels to exploit the benefits of miniaturization and to realize hands-free cellular labeling and on-chip detection (Fig. 3).38, 39, 40, 41, 42, 43
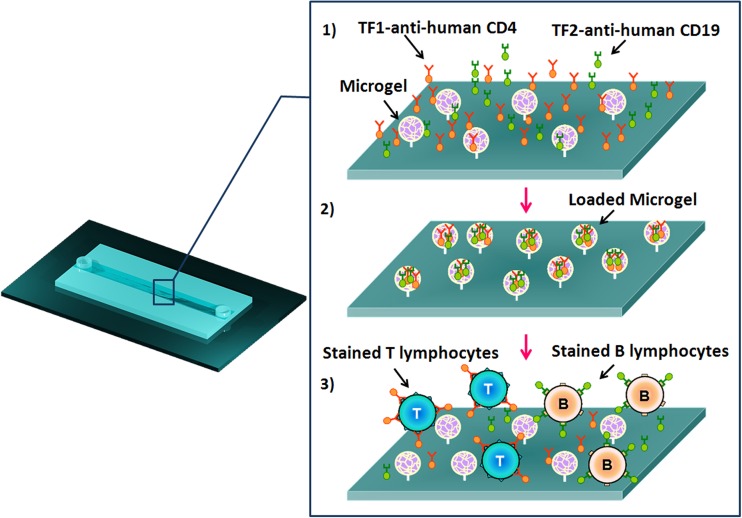
Figure 3.
Schematic representation of double cell staining within microgels functionalized microchannel. (1) Uptake of TF-labeled MAbs into the covalently immobilized PMAA microgels within the microchannel specific for the detection of B and T cells. (2) PMAA microgels microchannel loaded with TF1-antiCD4 and TF2-antiCD19 antibodies. (3) A defined cell mixture of two human lymphoblastoid cells (TK6 and SUP-T1 cells) was incubated within the microchannel. The physiological pH of cells suspension induces the pH-triggered cell staining.
Miniaturized systems are particularly attractive for the integration of on-chip cells staining, thanks to their dimensions and volume handling capacities.44 Specifically, miniaturization allows reduction of the sample volume and reaction times.45, 46 In addition, microfluidics technologies are particularly appealing for the automation of routine assay and sample preparation, thanks to the advantages of high-throughput assays, multistage automation, parallel processing of multiple analytes, and reduced error rate.44 Finally, miniaturized technology will lead to increased portability, paving the way for on-site analysis with rapid results also in remote settings. PDMS is an attractive material for microfluidics devices and possesses many advantages such as, biocompatibility, high optical transparency, mechanical compliance, and flexibility.47, 48, 49
Accordingly, we have fabricated a PDMS/glass microdevice and developed a novel procedure to immobilize PMAA microgels onto the inner glass wall of the microchannel.
Before assembling the microdevice, the glass substrate was chemically treated with TESPI and diamino-PEG, in order to achieve primary amines as pendant groups on its surface. At this stage, the PDMS replica was sealed to the previously functionalized glass substrate. Finally, PMAA microgels activated by carbodiimide chemistry were flushed by syringe pump into the microdevice. To remove nonspecifically bound microgels, the microchannel was thoroughly flushed with bidistilled water, and then analyzed by optical microscopy. The covalent attachment onto the inner glass wall was found to occur with a surface dense coverage of approximately 9.27 × 10−4 microgels/μm2. This result is representative of at least five independent measurements performed on separate microchips.
The microgel functionalized microchannel was also demonstrated to be able to encapsulate functional molecules and to release them at the passage of pretreated cells.
In particular, the uptake experiment was run by injecting both a solution of TF1-antiCD4 and TF2-antiCD19 MAbs diluted at a ratio of 1:2 into the microchannel.
The simultaneous uptake of both labeled MAbs into the immobilized microgels within the microchannel was demonstrated by LSCM. The highly defined fluorescence signals of both the conjugates are reported in Fig. 4.
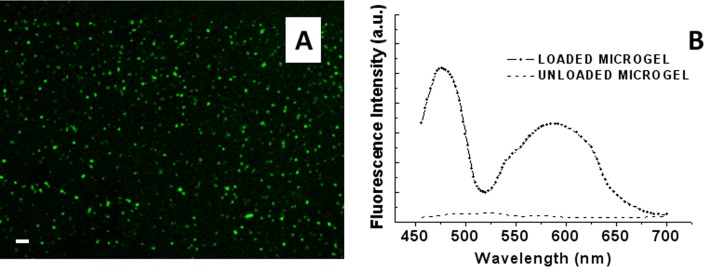
Figure 4.
Confocal microscopy image of the PMAA microgels microchannel loaded with TF1-antiCD4 and TF2-antiCD19 antibodies. (a) Fluorescence image of the double loaded microgels functionalized microchannel. (b) Fluorescence spectra of the immobilized microgels after the uptake (dotted line) and after the release process of labeled antibodies (straight line). The scale bar was 10 μm.
The encapsulation efficiency (EE) was determined by fluorescence data according to Eq. 1:
| (1) |
where FII is the fluorescence intensity of the initial protein solution before the uptake experiment and FIF is the fluorescence intensity of final protein solution after the uptake process within the functionalized microchannel.50, 51
Accordingly, the encapsulation efficiencies of TF1-antiCD4 and TF2-antiCD19 were found to be 51.4 ± 4.3% and 57.0 ± 4.1%, respectively.
Further, the release of labeled MAbs from microgels was approximately complete at the passage of MES buffer (1.0 mM) at physiological pH (pH 7.8) or cellular suspension, as demonstrated by fluorescence emission of thiophene fluorophores (Fig. 4b). After the release process, no loaded microgel particles were evident in any of the samples analyzed by LSCM (Fig. 5).
Figure 5.
Confocal microscopy images of the unloaded PMAA microgels microchannel after the release process of TF1-antiCD4 and TF2-antiCD19 antibodies induced by the passage of MES buffer (1.0 mM) at physiological pH (pH 7.8) within it. (a) Fluorescence image of the empty microgels functionalized microchannel. (b) The corresponding image acquired under bright field exposure. The scale bar was 20 μm.
The cell staining experiments were run by injection of a diluted (1/10) cell suspension using a syringe in a pulsed way within the loaded microchannel. The injection in a pulsed way prevents the cell settling to the bottom of the syringe during the experiment.34
After 30 min, the system consisting of double loaded smart microchannel, and cell suspension was analyzed by LSCM. The presence of both TF1 and TF2 fluorescence signals by LSCM proved the concurrent release of both labeled MAbs from the smart microchannel and therefore the double cell staining (Figs. 6g, 6i). Similarly to the results on smart platforms, the ratio of stained to total cells within the microchannel was 50% ± 4% for CD19+ TK6 cells and 50% ± 6% for CD4+ SUP-T1.
Figure 6.
Confocal microscopy images of (a) SUP-T1 T cells, (d) TK6 B cells, and (g) SUP-T1 T and TK6 B cells after the incubation within the loaded functionalized microchannel with the two labeled MAbs. The double cell staining was shown in the latter experiment. It was observed that TF1-antiCD4 antibody positively stained SUP-T1 T cells whereas TF2-antiCD19 positively stained TK6 B cells. The corresponding images (b), (e), and (h) were acquired under bright field exposure. For the sake of clarity, only selected cells are shown. All panels are false-color confocal images of cells stained with TF1-antiCD4 (orange) or with TF2-anti CD19 (green). The scale bar was 25 μm.
To definitely demonstrate the specificity of the immunostaining within the double loaded microgels functionalized microchannel, control experiments were run with single CD4+ SUP-T1 and CD19+ TK6 cell suspension. In particular, the single signal of TF1 and TF2 fluorescence was recorded using either CD4+ SUP-T1 or CD19+ TK6 cells, respectively, after their incubation within the loaded microchannel (Figs. 6a, 6b, 6c and 6d, 6e, 6f).
Finally the double cell staining was shown within the functionalized microchannel: both the fluorescence signals were recorded by LSCM on the tested cells after the release of TF1-antiCD4 and TF2-antiCD19 antibodies (Figs. 6g, 6i).
It is noteworthy that after release, the unloaded microparticles do not interfere with cell detection within the microfluidic channel. Indeed PMAA microgels have no intrinsic fluorescence properties; their size (3–6 μm) is lower than those one of tested cells (10–15 μm). The free unbound antibodies released within the microchannel do not interfere with the analysis of cellular suspension as shown by control staining experiments.
This assembled PDMS/glass microchannel functionalized with PMAA microgels can be easily integrated with a wide range of microdevices which require on-chip staining, since it does not involve complex design and extremely time-consuming procedures for its realization.
Although the fabrication process of PDMS replicas is simple and inexpensive and easily prototyped, there are limitations associated with the fabrication of a master which typically requires clean room facilities and equipment, which increase process time and costs. However, this is problematic only when the device is in the early development stage, especially when only a few prototype devices are needed.52
Indeed, the covalent attachment of PMAA microgels, the uptake of the labeled MAbs as well as the double cell staining were improved through miniaturization. In particular the immobilization of PMAA microgels within the microchannel required a diluted reagent solution (at ratio of 1:50) compared to microgel-functionalized platform. Similarly, the uptake of labeled monoclonal antibodies within the microfluidic system was carried out with a 1:5 diluted solution and reduced time (1 h instead of overnight). Finally, the on-chip double cell staining requires less sample (the cell suspension was diluted in buffer at ratio of 1:10 in buffer) and reduced time (the incubation time was 30 min instead of overnight).
The so-realized microchannel is particularly advantageous with respect to the traditional flow cytometry even though it employs comparable incubation times. Typical immunofluorescence protocols for conventional flow cytometers require a sample consisting of 0.5–1.6 × 106 cells/ml, staining reagents concentrations in the range of 10–20 μg/ml; conversely, the reported system involves a solution of labeled antibodies of 2.5 μg/ml and cell sample of 1.5 × 105 cells/ml to perform the staining process.14
Additionally, it can be cleaned and reused, thus paving the way to repeated load/release of labeled biomolecules, dyes, or drugs, depending on the analysis being carried out.
The water environment ensured by PMAA microgels should also minimize biomolecules denaturing within the chip, thus keeping them in their active state during storage.
This microgels functionalized microdevice represented a proof-of-concept prototype to realize the on-chip cellular staining between the sample pretreatment module and cell counting/detection. The counting and detection of stained cells in LOCs will be successively carried out by current systems for on-chip cell detection.
CONCLUSIONS
In conclusion, we propose a simple hands-free approach to integrate double staining and detection of two human lymphoblastoid cell lines on-chip. The microgels functionalized microchannel combines the advantages of realizing the pH-triggered in situ cell staining together with minute amounts of sample and reagents and reduced incubation time. Further, this system is cheap, reusable, and may allow in future the detection of unlimited cell surface antigens, preserving the proper labeled antibodies in their hydrated and folded state. Nevertheless, the great optical stability of thiophene fluorophores allowed the easy monitoring of labeled cells and prolonged experiment times. According to all these advantages, the proposed smart microfluidic system would represent a significant tool to fully integrate cell staining in LOCs for multiparametric cellular analysis and flow cytometry.
ACKNOWLEDGMENTS
The authors are grateful to Dr. I. E. Palamà and Dr. M. R. Musarò for having provided SUPT1 and TK6 cells, and E. Perrone and S. D'Amone for their expert technical help. Mediteknology srl is also acknowledged for providing the labeled antibodies.
References
- Kelliher A. S., Parent D. W., Anderson D. C., Dorn M. E., Hahn J. L., Eapen S., and Preffer F. I., Cytometry, B 66B(1), 40–45 (2005). 10.1002/cyto.b.20055 [DOI] [PubMed] [Google Scholar]
- Alyassin M. A., Moon S. J., Keles H. O., Manzur F., Lin R. L., Haeggstrom E., Kuritzkes D. R., and Demirci U., Lab Chip. 9(23), 3364–3369 (2009). 10.1039/b911882a [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piuri V. and Scotti F., in IEEE International Conference on Computational Intelligence for Measurement Systems and Applications CIMSA, 2004, pp. 103–108.
- Toner M. and Irimia D., Annu. Rev. Biomed. Eng. 7, 77–103 (2005). 10.1146/annurev.bioeng.7.011205.135108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weigl B., Domingo G., La Barre P., and Gerlach J., Lab Chip. 8(12), 1999–2014 (2008). 10.1039/b811314a [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srinivasan V., Pamula V. K., and Fair R. B., Lab Chip 4(4), 310–315 (2004). 10.1039/b403341h [DOI] [PubMed] [Google Scholar]
- Knight J., Nature 418(6897), 474–475 (2002). 10.1038/418474a [DOI] [PubMed] [Google Scholar]
- Li P. C. H., de Camprieu L., Cai J., and Sangar M., Lab Chip 4(3), 174–180 (2004). 10.1039/b400770k [DOI] [PubMed] [Google Scholar]
- Walt D. R., Science 308(5719), 217–219 (2005). 10.1126/science.1108161 [DOI] [PubMed] [Google Scholar]
- Liu R. H., Yang J., Pindera M. Z., Athavale M., and Grodzinski P. Lab Chip 2(3), 151–157 (2002). 10.1039/b201952c [DOI] [PubMed] [Google Scholar]
- Lin H. K., Zheng S., Williams A. J., Balic M., Groshen S., Scher H. I., Fleisher M., Stadler W., Datar R. H., Tai Y. C., and Cote R. J., Clin. Cancer Res. 16(20) 5011–5018 (2010). 10.1158/1078-0432.CCR-10-1105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi N., Torii M., Uebayashi Y., and Nasu M., Appl. Environ. Microbiol. 77(4), 1536–1539 (2011). 10.1128/AEM.01765-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan S. D. H., Luedke G., Valer M., Buhlmann C., and Preckel T., Cytometry, A 55A(2), 119–125 (2003). 10.1002/cyto.a.10070 [DOI] [PubMed] [Google Scholar]
- Sims C. E. and Allbritton N. L., Lab Chip 7(4), 423–440 (2007). 10.1039/b615235j [DOI] [PubMed] [Google Scholar]
- Buhlmann C., Preckel T., Chan S., Luedke G., and Valer M., J. Biomol. Tech. 14(2), 119–127 (2003). [PMC free article] [PubMed] [Google Scholar]
- Lancaster C., Kokoris M., Nabavi M., Clemmens J., Maloney P., Capadanno J., Gerdes J., and Battrell C. F., Methods 37(1), 120–127 (2005). 10.1016/j.ymeth.2005.07.003 [DOI] [PubMed] [Google Scholar]
- Song H., Chen T., Zhang B., Ma Y., and Wang Z., Biomicrofluidics 4(1), 044104 (2010). 10.1063/1.3497376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens D. Y., Petri C. R., Osborn J. L., Mihalic P. S., McKenzie K. G., and Yager P., Lab Chip 8, 2038–2045 (2008). 10.1039/b811158h [DOI] [PubMed] [Google Scholar]
- Beck M., Brockhuis S., Velde N., Breukers C., Greve J., and Terstappen L. W. M. M., Lab Chip 12(1), 167–173 (2012). 10.1039/c1lc20565j [DOI] [PubMed] [Google Scholar]
- Nolan C. M., Reyes C. D., Debord J. D., Garcia A. J., and Lyon L. A., Biomacromolecules 6(4), 2032–2039 (2005). 10.1021/bm0500087 [DOI] [PubMed] [Google Scholar]
- Peppas N. A. and Langer R., Science 263(5154), 1715–1720 (1994). 10.1126/science.8134835 [DOI] [PubMed] [Google Scholar]
- Kharlampieva E., Erel-Unal I., and Sukhishvili S. A., Langmuir 23(1), 175–181 (2007). 10.1021/la061652p [DOI] [PubMed] [Google Scholar]
- Guschin D., Yershov G., Zaslavsky A., Gemmell A., Shick V., Proudnikov D., Arenkov P., and Mirzabekov A., Anal. Biochem. 250(2), 203–211 (1997). 10.1006/abio.1997.2209 [DOI] [PubMed] [Google Scholar]
- Verpoorte E., Lab Chip 3(2), 60–68 (2003). 10.1039/b313217j [DOI] [PubMed] [Google Scholar]
- Peterson D. S., Lab Chip 5(2), 132–139 (2005). 10.1039/b405311g [DOI] [PubMed] [Google Scholar]
- Murthy S. K., Sin A., Tompkins R. G., and Toner M., Langmuir 20(26), 11649–11655 (2004). 10.1021/la048047b [DOI] [PubMed] [Google Scholar]
- Park J. and Han K., Ann. Lab Med. 32, 171–176 (2012). 10.3343/alm.2012.32.3.171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zambianchi M., Di Maria F., Cazzato A., Gigli G., Piacenza M., Sala F. D., and Barbarella G., J. Am. Chem. Soc. 131(31), 10892–10900 (2009). 10.1021/ja902416s [DOI] [PubMed] [Google Scholar]
- Quarta A., Di Corato R., Manna L., Argentiere S., Cingolani R., Barbarella G., and Pellegrino T., J. Am. Chem. Soc. 130(32), 10545–10555 (2008). 10.1021/ja800102v [DOI] [PubMed] [Google Scholar]
- See supplementary material at http://dx.doi.org/10.1063/1.4763560 for the synthetic procedure for PMAA microgels, the molecular structures of TF1 and TF2 and the absorption and photoluminescence spectra of TF1 and TF2 conjugated MAbs, the cell treatment procedures, the realization of PDMS/glass microfluidic device, the cell staining experiments with the smart platforms loaded with one labeled antibody, the evaluation of thiophenes (TFs) labeled monoclonal antibodies (MAbs) encapsulated in PMAA microgels.
- Duffy D. C., McDonald J. C., Schueller O. J. A., and Whitesides G. M., Anal. Chem. 70(23), 4974–4984 (1998). 10.1021/ac980656z [DOI] [PubMed] [Google Scholar]
- Berdichevsky Y., Sabolek H., Levine J. B., Staleyb K. J., and Yarmush M. L., J. Neurosci. Meth. 178(1), 59–64 (2009). 10.1016/j.jneumeth.2008.11.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villa-Diaz L. G., Torisawa Y. S., Uchida T., Ding J., Nogueira-de-Souza N. C., O'Shea K. S., Takayama S., and Smith G. D., Lab Chip 9(12), 1749–1755 (2009). 10.1039/b820380f [DOI] [PMC free article] [PubMed] [Google Scholar]
- See http://www.rsc.org/Publishing/Journals/lc/Chips_and_Tips/suspension_injection.asp for lab on a chip, chips and tips (last accessed June 2011).
- Argentiere S., Blasi L., Ciccarella G., Cazzato A., Barbarella G., Cingolani R., and Gigli G., Soft Matter 5(21), 4101–4103 (2009). 10.1039/b914277k [DOI] [Google Scholar]
- Blasi L., Argentiere S., Morello G., Palama I., Barbarella G., Cingolani R., and Gigli G., Acta Biomater. 6(6), 2148–2156 (2010). 10.1016/j.actbio.2009.12.031 [DOI] [PubMed] [Google Scholar]
- Eichenbaum G. M., Kiser P. F., Simon S. A., and Needham D., Macromolecules 31(15), 5084–5093 (1998). 10.1021/ma970897t [DOI] [PubMed] [Google Scholar]
- Daw R. and Finkelstein J., Nature 442(7101), 367–418 (2006). 10.1038/442367a [DOI] [Google Scholar]
- Whitesides G. M., Nature 442(7101), 368–373 (2006). 10.1038/nature05058 [DOI] [PubMed] [Google Scholar]
- Janasek D., Franzke J., and Manz A., Nature 442(7101), 374–380 (2006). 10.1038/nature05059 [DOI] [PubMed] [Google Scholar]
- El-Ali J., Sorger P. K., and Jensen K. F., Nature 442(7101), 403–411 (2006). 10.1038/nature05063 [DOI] [PubMed] [Google Scholar]
- Yager P., Edwards T., Fu E., Helton K., Nelson K., Tam M. R., and Weigl B. H., Nature 442(7101), 412–418 (2006). 10.1038/nature05064 [DOI] [PubMed] [Google Scholar]
- Mark D., Haeberle S., Roth G., von Stetten F., and Zengerle R., Chem. Soc. Rev. 39(3), 1153–1182 (2010). 10.1039/b820557b [DOI] [PubMed] [Google Scholar]
- Kim S. M., Lee S. H., and Suh K. Y., Lab Chip 8, 1015–1023 (2008). 10.1039/b800835c [DOI] [PubMed] [Google Scholar]
- Beebe D. J., Mensing G. A., and Walker G. M., Annu. Rev. Biomed. Eng. 4, 261–286 (2002). 10.1146/annurev.bioeng.4.112601.125916 [DOI] [PubMed] [Google Scholar]
- Dittrich P. S. and Manz A., Nature 5, 210–218 (2006). 10.1038/nrd1985 [DOI] [PubMed] [Google Scholar]
- McDonald J. C. and Whitesides G. M., Acc. Chem. Res. 35(7), 491–499 (2002). 10.1021/ar010110q [DOI] [PubMed] [Google Scholar]
- Zhao Y., Lim C. C., Sawyer D. B., Liao R., and Zhang X., J. Micromech. Microeng. 15(9), 1649–1656 (2005). 10.1088/0960-1317/15/9/006 [DOI] [Google Scholar]
- Gadre A., Kastantin M., Li S., and Ghodssi R., in Proceedings of International Semiconductor Device Research Symposium, Washington DC, 2001, pp. 186–189.
- O'Neil C. P., Suzuki T., Demurtas D., Finka A., and Hubbell J. A., Langmuir 25(16), 9025–9029 (2009). 10.1021/la900779t [DOI] [PubMed] [Google Scholar]
- Szebeni J., Di Iorio E. E., Hauser H., and Winterhalter K. H., Biochemistry 24(12), 2827–2832 (1985). 10.1021/bi00333a003 [DOI] [PubMed] [Google Scholar]
- Bhagat A. A. S., Jothimuthu P., and Papautsky I., Lab Chip 7(9), 1192–1197 (2007). 10.1039/b704946c [DOI] [PubMed] [Google Scholar]