Abstract
Autotaxin (ATX, nucleotide pyrophosphate/phosphodiesterase-2, NPP2) is an autocrine motility factor initially characterized from A2058 melanoma cell conditioned medium. ATX is known to contribute to cancer cell survival, growth, and invasion. Recently ATX was shown to be responsible for the lysophospholipase D activity that generates lysophosphatidic acid (LPA). Production of LPA is sufficient to explain the effects of ATX on tumor cells. Cyclic phosphatidic acid (cPA) is a naturally occurring analog of LPA in which the sn-2 hydroxy group forms a 5-membered ring with the sn-3 phosphate. Cellular responses to cPA generally oppose those of LPA despite activation of apparently overlapping receptor populations, suggesting that cPA also activates cellular targets distinct from LPA receptors. cPA has previously been shown to inhibit tumor cell invasion in vitro and cancer cell metastasis in vivo. However, the mechanism governing this effect remains unresolved. Here we show that 3-carba analogs of cPA lack significant agonist activity at LPA receptors yet are potent inhibitors of ATX activity, LPA production, and A2058 melanoma cell invasion in vitro and B16F10 melanoma cell metastasis in vivo.
INTRODUCTION
Autotaxin (ATX‡, nucleotide pyrophosphate/ phosphodiesterase-2, NPP2) was initially purified and characterized as an autocrine motility factor from A2058 melanoma cell conditioned medium nearly 15 years ago (1). ATX has been shown to be an important mediator in cancer cell survival, growth, migration, invasion, and metastasis (2–6). Until recently, ATX was merely thought to be a nucleotide phosphodiesterase (7); however, the apparent central role of ATX in cancer cell biology, owing to its numerous effects on tumor cells, was difficult to reconcile with this singular activity. In 2002, two independent groups showed that ATX was responsible for the long elusive lysophospholipase D activity that generates phosphatidic acid (LPA) (6,8). It is now fully appreciated that ATX is a multifunctional phosphodiesterase that hydrolyzes both nucleotides and lysophospholipids (6,8,9) using a single active site (10).
LPA (1-acyl-2-hydroxy-sn-glycero-3-phosphate) is the prototype member of a family of phospholipid growth factors. LPA elicits numerous biological effects including the promotion of cellular survival, mitogenesis, angiogenesis, migration, and invasion that are mediated by cell surface G protein-coupled receptors (GPCR) (11–14) and the intracellular nuclear hormone receptor PPARγ (15). LPA has long been associated with ovarian and breast cancers (16) but its production by ATX means it likely plays some role in many, if not all, metastatic cancers (17). ATX expression positively correlates with the metastatic and invasive properties of human tumors including melanoma (1,18), breast cancer (19,20), renal cell cancer (21), lung cancer (22,23), neuroblastoma (24), hepatocellular carcinoma (25), and glioblastoma multiforme (26). Euer et al. reported compatible results, using Affymetrix gene chip assays, in which ATX was among the 40 most upregulated genes associated with highly metastatic cancers (27).
Although many questions were answered when the biology associated with ATX was linked to the generation of LPA; one significant question remained, namely how are plasma LPA levels maintained in the low nanomolar range (28)? ATX is a ubiquitous plasma protein (8,29) and its substrate, lysophosphatidylcholine (LPC), is abundantly present in the 100–200 µM range in plasma (30,31), a concentration equivalent to the Km for ATX hydrolysis (9,32). Therefore, why are plasma LPA levels only ~100 nM? van Meeteren and colleagues recently showed that ATX is subject to feedback inhibition by its hydrolysis products LPA and sphingosine-1-phosphate (32). This phenomenon is the likely explanation for low plasma LPA levels and has provided insight into modulation of ATX activity.
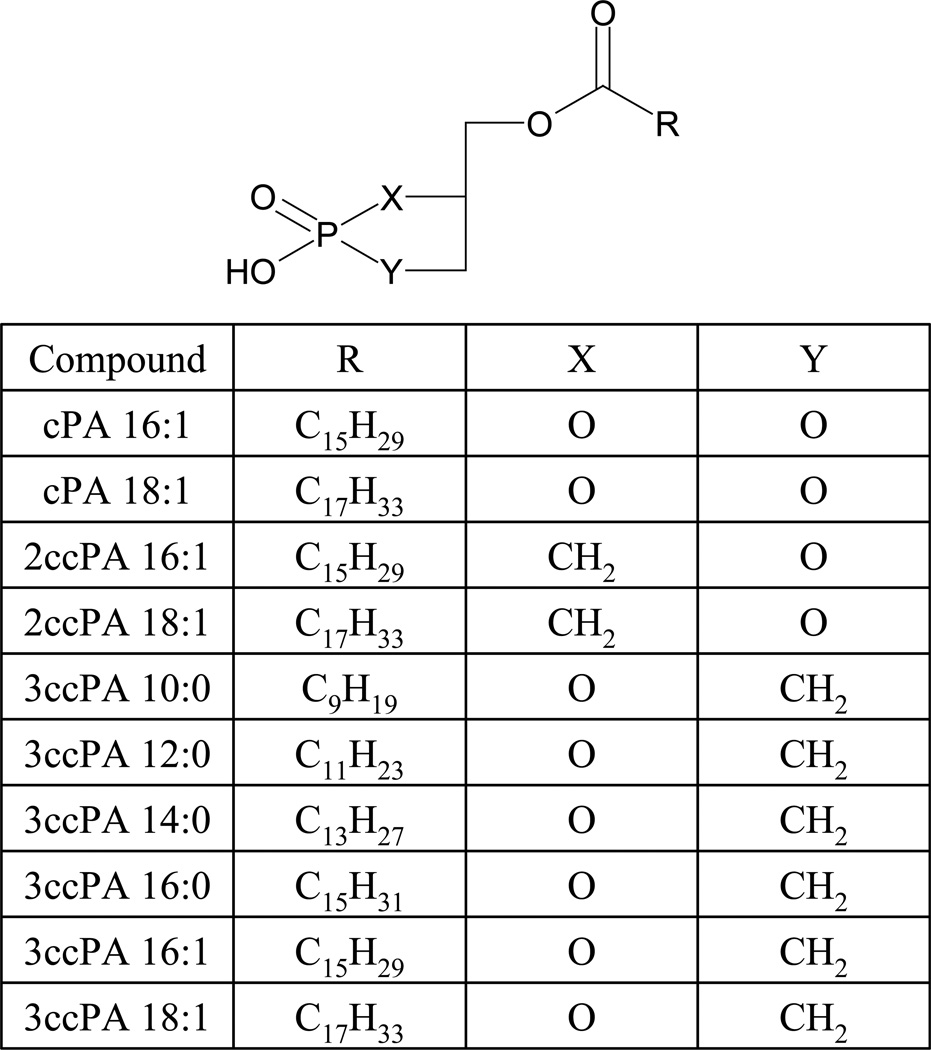
Cyclic phosphatidic acid (1-acyl-sn-glycero-2,3-cyclic-phosphate, cPA) was initially isolated as a eukaryotic DNA polymerase inhibitor from slime mold (33). cPA is known to be generated via phospholipase D catalyzed transphosphatidylation of LPC (34) and has been shown to be present in mammalian plasma bound to albumin (35). cPA partially desensitizes LPA responses in NIH3T3 cells and Xenopus oocytes, suggesting activation of at least partially overlapping receptor populations (36,37). Despite this apparent overlap in receptor targets, cPA and LPA elicit very distinct cellular effects. Unlike LPA, cPA is antimitogenic (36,38) and prevents cancer cell invasion in vitro (39) and metastasis in vivo (40,41). Analogs of cPA, in which a methylene (CH2, carba) group replaces either the sn-1 or sn-2 oxygen within the cyclic phosphate have been prepared by total chemical synthesis (See Figure 1 for structures) (42) and were shown to be more potent than the parent compounds as inhibitors of tumor cell invasion in vitro (41).
Figure 1.
Structures of the cPA analogs tested.
Here we show that carba analogs of cPA are (A) poor activators of LPA GPCR, (B) highly potent inhibitors of ATX activity and LPA production and (C) inhibitors of ATX-mediated invasion of melanoma cells in vitro and in vivo.
EXPERIMENTAL PROCEDURES
Materials
1-Oleoyl-2-hydroxy-sn-glycero-3-phosphocholine (LPC 18:1) was obtained from Avanti Polar Lipids (Alabaster, AL). 1-Palmitoleoyl-sn-glycero-2,3-cyclic-phosphate (cPA 16:1, Fig.1), 1-oleoyl-sn-glycero-2,3-cyclic-phosphate (cPA 18:1) and stabilized cPA analogs, in which the phosphate oxygen was replaced with a methylene group at either the sn-2 (2ccPA 16:1 and 2ccPA 18:1) or the sn-3 position (3ccPA 10:0, 12:0, 14:0, 16:0, 16:1 and 18:1) position were synthesized as described previously (42,43). Bis-(p-nitrophenyl) phosphate (BPNPP) and FS-3 were purchased from Sigma-Aldrich (St Louis, MO) and Echelon Biosciences (Salt Lake City, UT) respectively.
Spectrophotometric ATX Inhibition Assay
This assay utilizes BPNPP as substrate and recombinant ATX expressed in conditioned media isolated from transiently transfected HEK293T cells (32). Briefly, 4 × 106 HEK293 cells were plated in 10-cm dishes and allowed to attach and recover overnight. On day 2, the cells were transfected with an expression plasmid generated by Clair and colleagues (9) using Effectene (Qiagen, Valencia, CA) according to the manufacturer’s protocol. After 24 h, the transfection medium was replaced with serum-free DMEM and the cells were cultured at 37° C and 5% CO2 for an additional 30 h (32). Conditioned medium was collected and centrifuged at 3000×g for 10 min at 4° C. The clarified medium was stored at −20° C in aliquots until used. For analysis, 50 µl of conditioned medium was added to 100 µl of BPNPP, 1 mM final concentration in assay buffer (Tris 50 mM, pH 8.0, NaCl 150 mM, KCl 5mM, CaCl2 1 mM, MgCl2 1 mM) and 50 µl of test compound in assay buffer containing 1:1 bovine serum albumin in 96-well plates. The absorbance at 405 nm was measured at time zero and after 4 hours of incubation at 37° C using a Molecular Devices Versamax (Sunnyvale, CA) plate reader. Data were normalized to the corresponding vehicle control and the mean and standard deviations of triplicate wells were expressed as percent ATX inhibition.
Spectrofluorometric ATX Inhibition Assay
This assay utilizes FS-3 as substrate and recombinant ATX-HA as enzyme. For analysis, 0.5 µl of purified ATX-HA was added to 75 µl of FS-3, 1 µM final concentration in assay buffer (Tris 50 mM, pH 8.0, NaCl 150 mM, KCl 5mM, CaCl2 1 mM, MgCl2 1 mM) and 25 µl of test compound in assay buffer containing 1:1 bovine serum albumin in 96-well plates. The fluorescence (excitation 485 nm, emission 538 nm) was measured at time zero and after 2 hours of incubation at 37° C using a FLEXstation fluorescence plate reader (Molecular Devices, Sunnyvale, CA). Data were normalized to the corresponding vehicle control and the mean and standard deviations of triplicate wells were expressed as percent ATX activity (% of control).
Purification of Recombinant ATX
ATX-HIS6 (generously provided by Drs. Mary Stracke and Tim Clair, NCI), lacking the 31 N-terminal amino acids of ATX but containing the honeybee melittin secretion sequence at the N-terminus and a HIS6 epitope tag at the C-terminus, was purified by Con-A and Ni-NTA chromatography from media conditioned by stably expressing HighFive cells as previously described (9).
Full-length ATX cDNA possessing a C-terminal HA epitope tag (ATX-HA) was generated and cloned into pcDNA3.1(+) (Invitrogen, Carlsbad, CA). ATX-HA was immuno-purified by anti-HA affinity matrix (Covance Research Products, Berkeley, CA) from media conditioned by stably expressing COS-1 cells. A detailed description of ATX-HA production and purification will be published elsewhere (Bandle et al., manuscript in preparation).
Synthesis of fluorescent LPC analog (ADMAN-LPC)
The fluorescent LPC analog 1-(7’-(dimethylamino)-3’-(pentadecanoyl)-1-naphthyl)-2-O-methyl-sn-glycero-3-phosphocholine (3-acyl-7-dimethylamino-naphthyl-1-LPC (ADMAN-LPC), for structure see Figure 3) was synthesized in 9 steps using 3-pentadecanoyl-7-(dimethylamino)-1-naphthol and (S)-3-iodo-2-O-methyl-3-(O-tert-butyldiphenylsilyl) propane-1,2-diol as the key intermediates. After deprotection of the silyl group from the resulting coupling product, a phosphocholine moiety was incorporated at the sn-3 position as described previously (44) to generate the desired reagent. A detailed description of the synthesis and characterization of ADMAN-LPC will be published elsewhere (Byun et al., manuscript in preparation). This compound was readily visualized using long wavelength UV for detection following TLC as described below.
Figure 3.
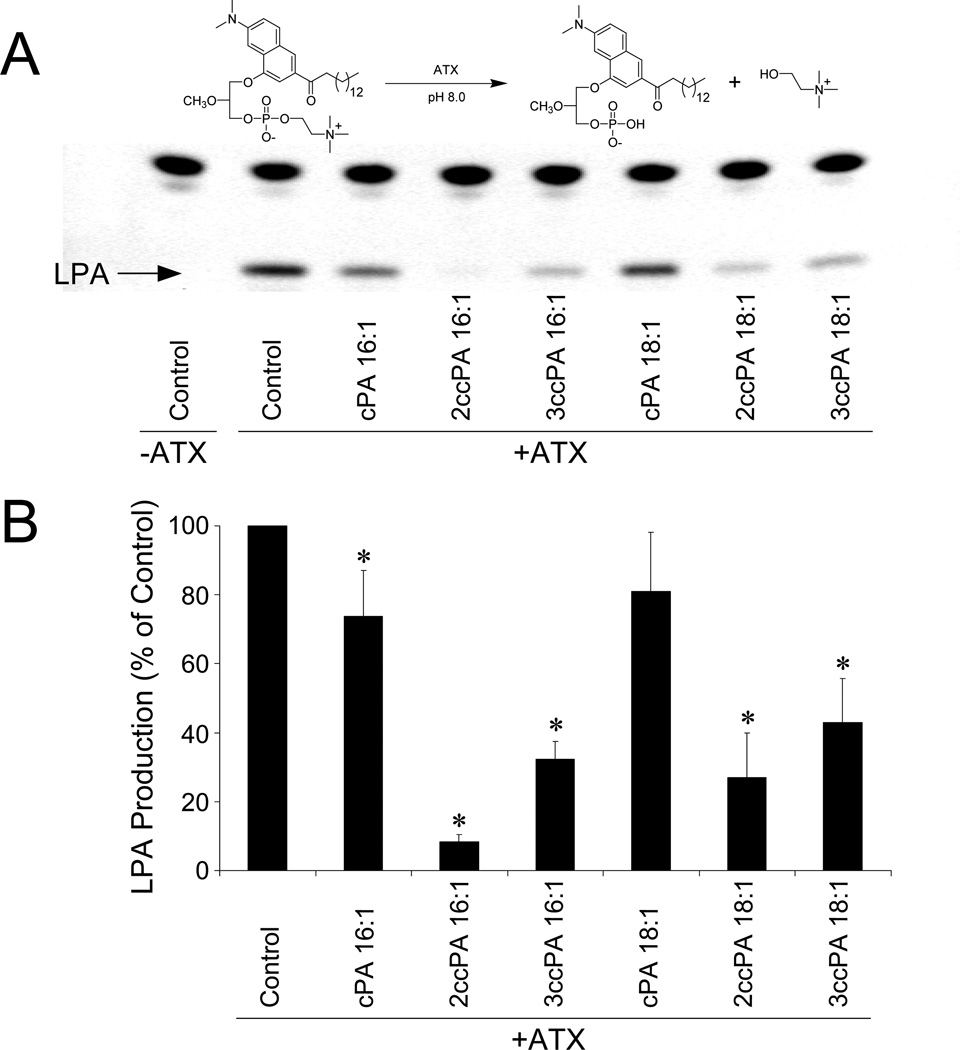
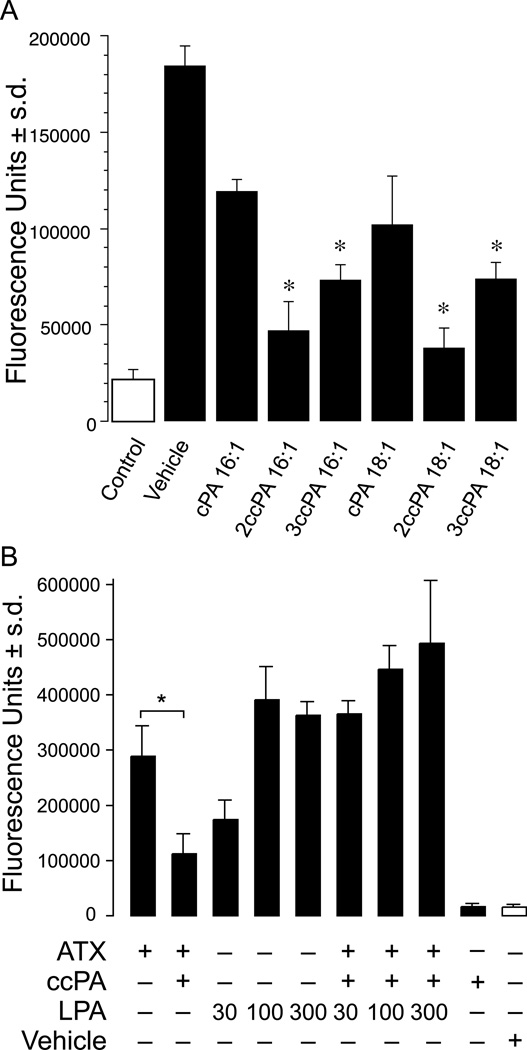
Inhibition of ATX-mediated LPA production by cPA analogs. Purified, recombinant ATX-HA was incubated with 2.5 µM ADMAN-LPC, a synthetic, fluorescent LPC analog in the absence or presence of 1 µM individual cPA analogs for 4 h. Lipids were extracted using a modified Bligh-Dyer protocol, separated by TLC (chloroform/ methanol/ ammonium hydroxide, 65:30:8 v/v/v), and visualized by UV illumination. (A) Representative TLC separation (colors are inverted). (B) Quantitative analysis of four independent experiments (* p<0.05, T-test).
Fluorescent ATX Inhibition Assay
This method is a modification of that reported by van Meteeren et al. (32). ADMAN-LPC (2.5 µM) was dried and resuspended by sonication in assay buffer (Tris 50 mM, pH 8.0, NaCl 150 mM, KCl 5mM, CaCl2 1 mM, MgCl2 1 mM) containing 2 mg/ml BSA in the presence and absence of the individual cPA analogs (1µM). Recombinant ATX-HA (1 µl) was added and the reaction was allowed to proceed for 4 h at 37° C. Lipids were extracted using a modification of the Bligh and Dyer protocol that utilized 3.5 volumes of citrate phosphate buffer (pH 4) prior to chloroform and methanol addition. Extracted lipids were dried in vacuo, resuspended in 25 µl of chloroform/methanol (1:1 v/v), and separated by TLC on silica gel using chloroform/ methanol/ammonium hydroxide (60:35:8 v/v) (45). Fluorescent LPC and LPA species were visualized by UV trans-illumination.
In vitro Invasion Assay
A-2058 melanoma cells (a generous gift from Dr. Tim Clair, NCI) were maintained in DMEM + 10% FBS. Invasion of these cells across a Matrigel™-coated membrane was assessed using Fluoroblok™ 24-well plates (BD Biosciences, Oxford, UK, 8-µm pore size) according to the manufacturer’s protocol. Briefly, cell suspensions (1×105 cells/ml) were prepared by trypsinizing and resuspending A2058 cells in DMEM containing 0.1% BSA. The plate inserts were prepared by rehydrating the matrix coating with PBS for 2 h at 37°C. The PBS was then carefully removed and 0.75 ml of DMEM/BSA containing chemoattractant (recombinant ATX-HIS6 + 1 µM LPC 18:1) in the presence or absence of cPA analogs was added to the lower well, and 0.5 ml of the cell suspension in DMEM + 0.1% BSA was added to each upper well insert. Plates were incubated for 22 h at 37°C. Following incubation, the medium was removed from the upper chamber and the entire insert was transferred to a 24-well plate containing 2 µg calcein AM (Molecular Probes, Eugene, OR) in 0.5 ml Hank’s balanced salt solution (Na2HPO4 0.34 mM, KH2PO4 0.44 mM, pH 7.4, NaCl 138 mM, KCl 5.3 mM, NaHCO3 4.17 mM, D-glucose 5.56 mM). The plates were incubated for 1 h at 37°C and bottom read in a FLEXstation fluorescence plate reader (Molecular Devices, Sunnyvale, CA) at excitation and emission wavelengths of 485 and 530 nm, respectively. Data are presented as the mean and standard deviation of triplicate wells.
LPA GPCR Activation Assay
This method measures real-time, transient increases in intracellular Ca2+ to determine LPA GPCR activation. The McArtl rat hepatoma (RH7777) cell lines used here stably express LPA1/2/3 G protein-coupled receptors and have been used extensively to examine the pharamacology of natural LPA isoforms and synthetic LPA analogs (46–48). CHO cells that stably express LPA4 were a generous gift of Dr. Takao Shimizu (Tokyo University). The procedure of cell labeling and monitoring has been described previously (49,50). The mean ± standard deviation of triplicate wells was determined. Curves were fitted and EC50 values were calculated using KaleidaGraph (Synergy Software, Reading, PA).
In vivo Metastasis Assay
This model utilizes highly metastatic B16F10 melanoma cells to examine the therapeutic effect of metastasis blockers in vivo. PBS (vehicle control), 3ccPA 16:1 or 3ccPA 18:1 were delivered as intraperitoneal injections (250 µg/kg, 5 µg per dose) 15 min and 48 h after the intravenous injection of 5 × 105 melanoma cells per mouse via the tail vein. Three weeks post inoculation, animals were euthanized, the lungs were washed, fixed with formalin, and the number of lung metastases was determined. No tumors were noted in tissues other than the lungs in animals treated with ccPA.
RESULTS
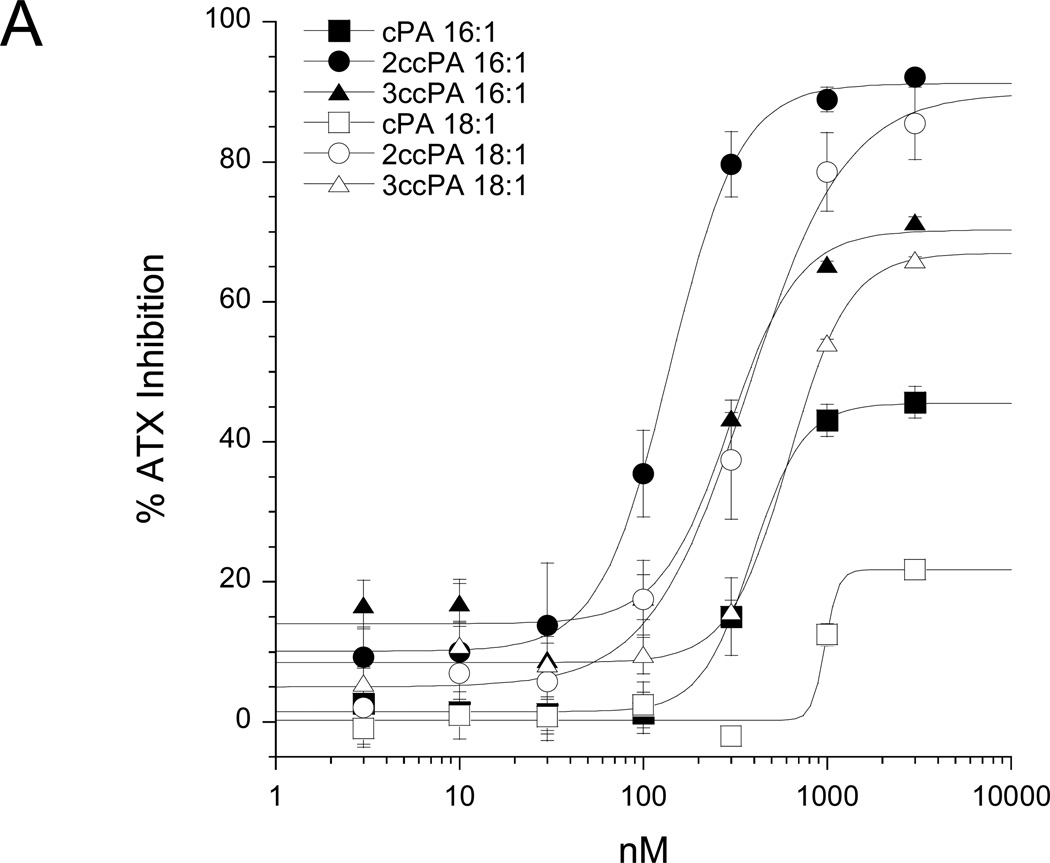
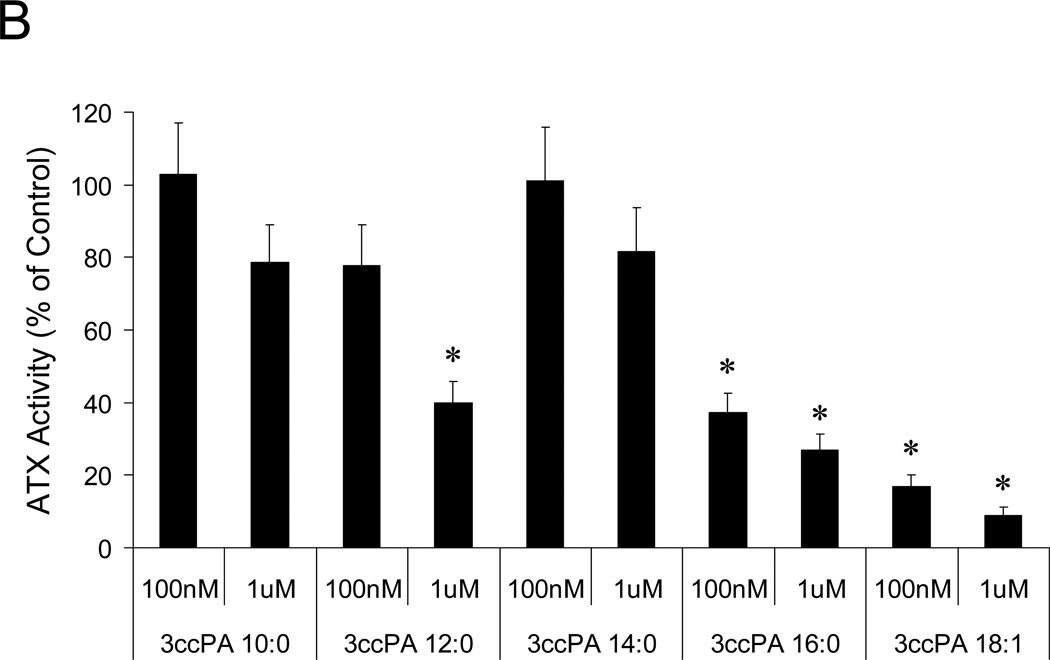
cPA has previously been shown to block the invasion and migration of cancer cells in vitro (39) and in vivo (40); however, the cellular targets and mechanisms governing this effect remain unresolved. Recently, van Meeteren et al. showed that ATX is subject to feedback inhibition by LPA and S1P, the products of ATX-mediated hydrolysis of lysophosphatidylcholine and sphingosyl-phosphorylcholine, respectively (32). These results led us to hypothesize that cyclic phosphatidic acids, naturally occurring analogs of LPA, might achieve their anti-invasive effect through inhibition of ATX-mediated LPA production. In this study we examined ten cPA analogs (for structures see Figure 1). First, we examined the effect of these analogs on ATX activity by measuring ATX-mediated hydrolysis of BPNPP spectrophotometrically (Figure 2A) and FS-3 spectrofluorometrically (Figure 2B). The cPA analogs containing a natural 2,3-cyclic phosphate group (cPA 16:1 and cPA 18:1) were poor inhibitors of ATX; cPA 16:1 and 18:1 showed maximum ATX inhibitions of 45% and 22% respectively (Figure 2A). Conversely, stabilized cPA analogs containing an isosteric replacement of the sn-2 oxygen with a methylene group (2ccPA 16:1 and 2ccPA 18:1) were highly effective ATX inhibitors (Figure 2A). 2ccPA 16:1 blocked ATX activity by 91% with an IC50 of 140 nM, whereas 2ccPA 18:1 blocked ATX activity by 90% with an IC50 of 370 nM. Stabilized cPA analogs containing a methylene unit in place of the sn-3 oxygen (3ccPA 16:1 and 3ccPA 18:1) were intermediately potent inhibitors of ATX. 3ccPA 18:1 blocked ATX activity by 70% with an IC50 of 294 nM, whereas 3ccPA 16:1 blocked 67% of ATX activity with an IC50 of 620 nM. Analogs of 3ccPA containing 16 and 18 carbon fatty acyl groups were superior to those containing shorter hydrocarbon chains (Figure 2B).
Figure 2.
Inhibition of ATX by cPA analogs. (A) Conditioned media from HEK293 cells transiently transfected with ATX-HIS6 was incubated with 1 mM BPNPP in presence of increasing amounts of individual cPA analogs complexed 1:1 with BSA. Data were normalized to the appropriate BSA control and are reported as percent ATX inhibition (n=3 for each data point (mean ± standard deviation), results are representative of two independent experiments). (B) Recombinant ATX-HA was incubated with 1 µM FS-3 in the presence of 3ccPA analogs complexed 1:1 with BSA (n=3 for each bar (mean ± standard deviation), results are representative of two independent experiments, * p<0.05, T-test).
Following initial screening using BPNPP and FS-3 as ATX substrates, we conducted secondary screening of the effect of 1 µM cPA (16:1 and 18:1 analogs) on ATX-mediated production of LPA via the hydrolysis of a synthetic LPC substrate (Figure 3). The synthetic, fluorescent LPC analog ADMAN-LPC was hydrolyzed to a fluorescent analog of LPA by ATX (compare lanes 1 and 2). Both cPA 16:1 and 18:1 modestly inhibited ATX-mediated LPC hydrolysis (compare lanes 3 and 6 to lane 2). In contrast, 2ccPA 16:1 was a highly effective inhibitor of LPA production (compare lanes 4 and 2). 3ccPA 16:1, 2ccPA 18:1 and 3ccPA 18:1 were intermediate in their ability to inhibit ATX-mediated LPA production (compare lanes 5, 7 and 8 to lane 2). Similar results were obtained when a combination of 1 µM LPC 18:1 and [14C]-LPC 16:0 were used as substrate (data not shown).
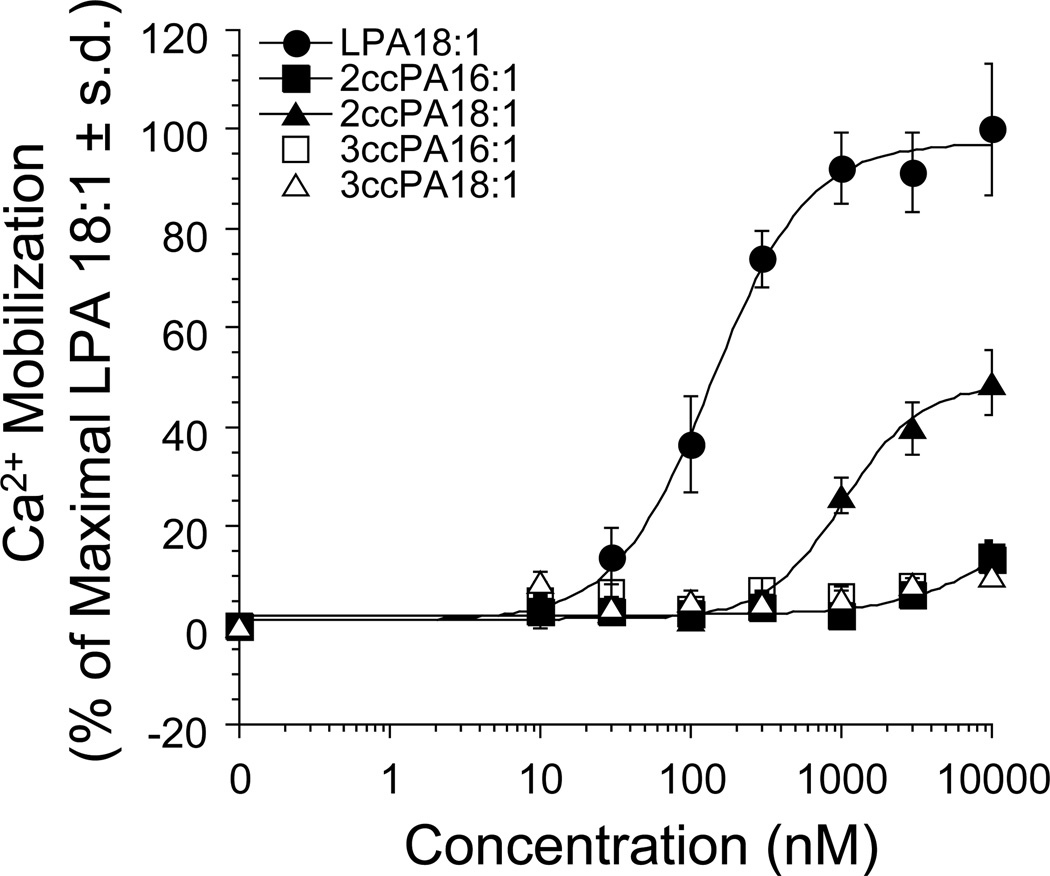
We have previously described the pharmacology of natural cPA analogs with respect to activation of the LPA1, LPA2 and LPA3 receptors (51). Here we expand our understanding of this structure-activity relationship to include eight 2ccPA and 3ccPA analogs. Knowledge of the activation profiles of these reagents is critically important given the role of LPA GPCR in cancer cell migration associated with LPA produced via ATX (5). Table 1 summarizes the results of the pharmacological screening of the ten cPA analogs with respect to activation of all four known LPA GPCR. cPA 16:1 activates LPA1/2/4 and cPA 18:1 activates LPA1/2/3/4 both with significantly higher (>10-fold) EC50 concentrations than LPA 18:1. The 2ccPA 16:1 analog is a weak agonist (13% and 14% maximal efficacy as compared to LPA 18:1 on LPA1 and LPA3 respectively). Likewise, the 2ccPA 18:1 analog is a partial agonist of both LPA1 (54% efficacy, 1.7 µM EC50) and LPA3 (46% efficacy, 0.17 µM EC50). 2ccPA also activated LPA2 (68% efficacy) and LPA4 (90% efficacy) in a nonsaturable fashion over the concentration range tested. In contrast, all 3ccPA analogs tested failed to elicit significant activation LPA1/2/3/4. In order to highlight the difference in LPA GPCR activation between the 2ccPA and 3ccPA analogs, full dose response curves are shown for the activation of LPA1 by LPA 18:1 (positive control), and the 16:1 and 18:1 analogs of 2ccPA and 3ccPA (Figure 4).
Table 1.
Summary of pharmacological evaluation of cPA analogs in RH7777 cells stably expressing individual LPA GPCR.
| Compound | EC50 [µM] (Efficacy)§ | |||
|---|---|---|---|---|
| LPA1 | LPA2 | LPA3 | LPA4 | |
| LPA 18:1 | 0.20 ± 0.068 (100%) |
0.007 ± 0.001 (100%) |
0.26 ± 0.023 (100%) |
0.50 ± 0.027 (100%) |
| cPA 16:1 | NS¶ (99%) |
0.18 ± 0.04 (121%) |
NS (19%) |
NS (70%) |
| cPA 18:1 |
1.7 ± 0.23 (66%) |
0.085 ± 0.011 (140%) |
1.0 ± 0.62 (61%) |
NS (100%) |
| 2ccPA 16:1 | NS (13%) |
NA‖ | NS (14%) |
NA |
| 2ccPA 18:1 | 1.7 ± 0.89 (54%) |
NS (68%) |
0.17 ± 0.073 (46%) |
NS (90%) |
| 3ccPA 10:0 | NA | NA | NA | NA |
| 3ccPA 12:0 | NA | NA | NA | NA |
| 3ccPA 14:0 | NA | NA | NA | NA |
| 3ccPA 16:0 | NA | NA | NA | NA |
| 3ccPA 16:1 | NA | NA | NA | NA |
| 3ccPA 18:1 | NA | NA | NS (15%) |
NA |
Normalized to the maximum LPA 18:1 effect
onsaturable activation up to the highest concentration tested (10 µM for LPA1/3/4 and 3 µM for LPA2)
No activation up to the highest concentration tested (10 µM for LPA1/3/4 and 3 µM for LPA2)
Figure 4.
The effect of increasing amounts of ccPA analogs on intracellular Ca2+ mobilization was determined in RH7777 cells stably expressing the LPA1 receptor. 100% represents maximal Ca2+ mobilization elicited by LPA 18:1. Each data point represents the mean ± standard deviation of triplicate wells and are representative of at least 3 independent experiments.
In order to characterize the functional effect of the cPA analogs on an ATX-mediated cellular endpoint, we examined the invasion of A2058 melanoma cells using a modified Boyden chamber assay in the presence and absence of the test compounds. ATX, LPC, and cPA analogs were all applied in the lower chamber, whereas cells in DMEM + 0.1% BSA were plated in the upper chamber containing a Matrigel-coated membrane bearing 8-µm pores. First, we examined assay conditions to optimize concentrations of recombinant ATX, LPC 18:1, and cPA analogs (Supplemental Figure 1). In our hands, purified recombinant ATX-HIS6 (100-fold dilution), in the presence of 1 µM LPC, elicited a 5.6-fold increase in the number of invading A2058 cells as compared to cells exposed to LPC in the absence of ATX. Exposure of cells to ATX without LPC resulted in a modest (1.6-fold) increase in invading cells. Increasing the dilution of recombinant ATX to 300 fold significantly reduced the number of invading cells in both the presence and absence of LPC, demonstrating the importance of ATX activity in this effect. In this assay, invasion of A2058 cells was significantly attenuated (81%) by co-application of 3 µM 3ccPA 16:1. Next, the effect of the 16:1 and 18:1 cPA analogs was examined using a 100-fold dilution of recombinant ATX-HIS6, 1 µM exogenous LPC 18:1, and 3 µM of the test compound (Figure 5A). Treatment with ATX and LPC without the test lipid resulted in an 8-fold increase in the number of A2058 cells that invaded the lower chamber. Co-administration of 3 µM cPA 16:1 or cPA 18:1 with ATX and LPC resulted in modest 40 and 51% decreases in the number of invading cells, respectively. In contrast, co-application of 2ccPA 16:1 or 2ccPA 18:1 with ATX and LPC significantly attenuated the number of invading cells by 84 and 90%, respectively. Co-treatment with 3ccPA 16:1 or 3ccPA 18:1 plus ATX and LPC resulted in intermediate inhibitions of ~68% each.
Figure 5.
Panel A. Inhibition of cell invasion by cPA analogs. Optimized conditions were used to assess the inhibition of individual cPA analogs on A2058 melanoma cell invasion. The data are means ± standard deviations of triplicate wells and are representative of 3 separate experiments (* p<0.01, T-test).
Panel B. Exogenously added LPA bypasses the inhibition by 3ccPA 16:1. 1 µM LPC in 0.1% BSA was present in the lower chamber of the invasion chamber and recombinant purified ATX, LPA 18:1, 3-ccPA 16:1 (3 µM), or their combination was included. LPA 18:1 dose dependently enhanced invasion. In contrast, 3ccPA 16:1 inhibited ATX-induced migration. Exogenously added LPA 18:1 (30–300 nM) bypassed the inhibitory effect of 3ccPA 16:1. Vehicle (0.1 % BSA without ATX) did not induce invasion when added with 1 µM LPC, and 3ccPA 16:1 (3 µM) in the absence of ATX had no effect on basal A2058 cell invasion. The data are means ± standard deviations of triplicate wells and are representative of 3 separate experiments (* p<0.01, T-test).
We reasoned that if the inhibitory effect of the ccPA analogs on cell migration is due to selective inhibition of LPA production by ATX, than exogenous LPA added to the bottom of the invasion chambers should overcome this inhibition. LPA 18:1 alone added to the bottom chamber, showed a dose-dependent enhancement of A2058 invasion (Figure 5B). Indeed, 3ccPA 16:1 mediated inhibition of cell invasion was dose dependently bypassed by LPA 18:1. The 1 µM LPC substrate without exogenous ATX failed to elicit invasion and 3ccPA 16:1 did not have any detectable effect on this low level of A2058 cell invasion. Taken together these results provide strong support to the hypothesis that ccPA elicits its inhibitory effect on cancer cell invasion by targeting ATX and decreasing the production of the LPA.
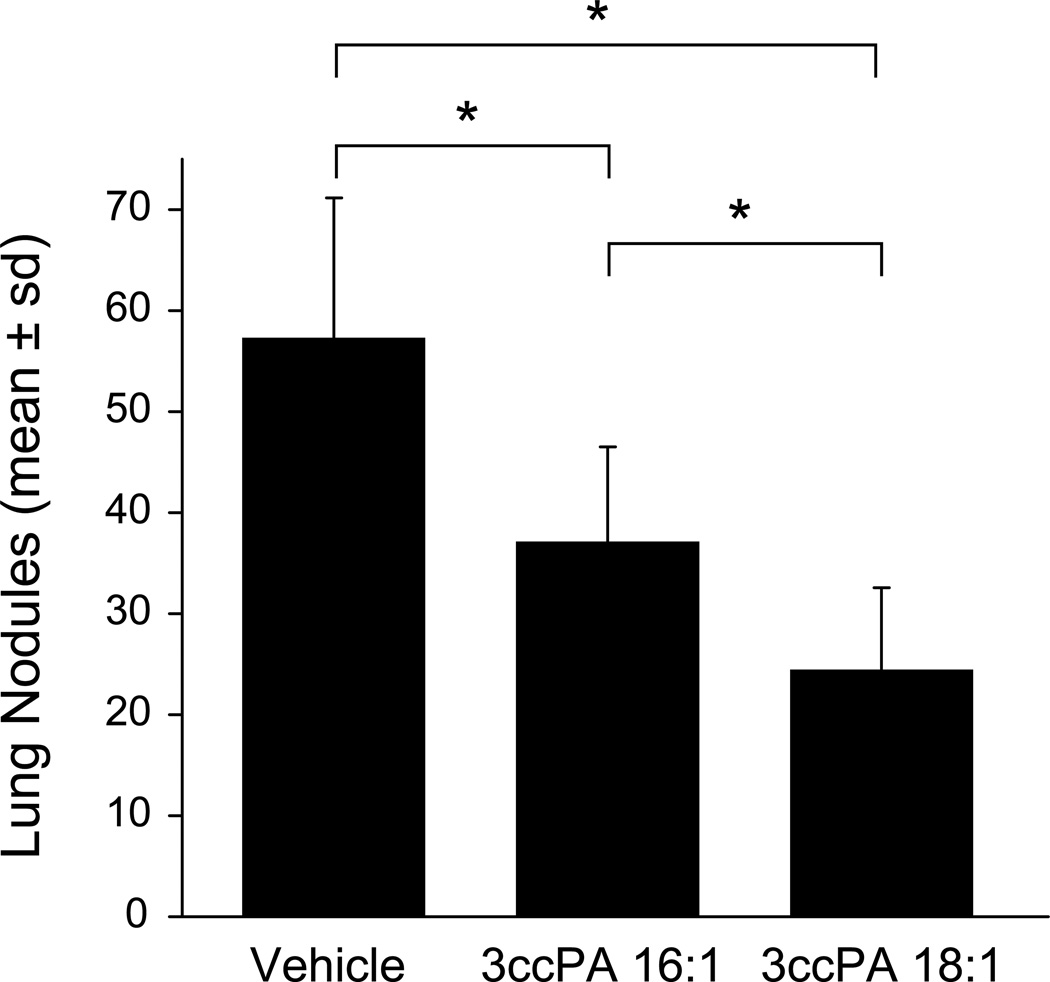
B16F10 mouse melanoma cells are widely used as a model to assess the therapeutic effect of anti-metastatic compounds in vivo (52,53). We used this model to examine the therapeutic effect of 3ccPA 16:1 and 3ccPA 18:1, delivered as intraperitoneal injections (250 µg/kg, 5 µg per dose), 15 min and 48 h after the intravenous injection of 5 × 105 cells per mouse. Three weeks post inoculation, animals were euthanized and the number of lung metastases was determined (Figure 6). Treatment with PBS (vehicle control) resulted in animals bearing 57 ± 14 tumors (range 44–80, n = 6). In contrast, animals treated with 3ccPA 16:1 contained 37 ± 10 tumors (range 27–50, n = 5, p < 0.01 ANOVA), whereas those treated with 3ccPA 18:1 had only 24 ± 8 tumors (range 16–40, n = 5, p < 0.01, ANOVA). 3ccPA 18:1 was significantly more effective at blocking lung metastasis than was 3ccPA 16:1 (p < 0.01).
Figure 6.
Inhibition of melanoma cell metastasis in vivo. B16F10 melanoma cells (5 × 105) were injected in the tail vein of C57BL/6 mice, followed by i.p. injections of PBS (vehicle control) or 3ccPA 16:1 or 3ccPA 18:1 (250 µg/kg, 5 µg per dose) 15 min and 48 h after inoculation. Animals were sacrificed 3 weeks later and lungs were washed, fixed in formalin, and lung nodules were counted. The data are means ± standard deviations from groups of 6 (PBS) or 5 (3ccPA 16:1 and 3ccPA 18:1) animals (* p < 0.01, ANOVA).
DISCUSSION
Here we have shown that carba-cPA analogs, in particular the 3ccPA (16:1 and 18:1) forms, are potent inhibitors of ATX that lack the ability to activate LPA GPCR. This ATX inhibitory activity was correlated with the inhibition of LPA-dependent invasion of A2058 cells and the inhibition of pulmonary metastasis of B16 melanoma in mice. This is the second report describing ATX inhibition mediated by naturally occurring lysophospholipids. van Meeteren et al. first showed that LPA and S1P blocked ATX activity with nanomolar potency (32). In our hands, LPA and S1P blocked ATX activity with IC50 values of 2.2 µM and 280 nM, respectively. These concentrations are significantly higher than the previously reported values (32). The reasons for these differences are not clear but could be the result of differences in the ATX construct, substrate, or assay conditions used. Nonetheless, under identical assay conditions, the 2ccPA 16:1 and 2ccPA 18:1 analogs were 15-and 6-fold more potent inhibitors of ATX than was LPA. We have previously shown that cPA 18:1 is stable in neutral buffered aqueous medium for up to 24h using LC-MS (51). The modest drop in cPA concentration over this time frame was not accompanied by a significant increase in LPA levels (51). This suggests that cPA analogs may be more effective ATX inhibitors in vivo due to their increased stability. Detailed analysis of the pharmacokinetics of cPA analogs is currently ongoing. The findings that 3ccPA analogs show a significantly lower IC50 for ATX inhibition than does LPA, and that cPA analogs are stable under assay conditions ((51) and Figure 3, lane 1), suggest that ccPA analogs are themselves novel, potent ATX inhibitors with promising in vivo activity.
Previous reports on the anti-invasive properties of cPA implicated increased cellular cAMP levels (39,54) and subsequent RhoA inactivation (39) in this effect. This implicated cell surface GPCR activation in the response. We have previously shown that natural cPA isoforms are poor activators of the LPA1, LPA2, and LPA3 receptors (51). In the course of this study, we have shown that natural cPA analogs are also poor activators of LPA4 (Table 1), a recently identified LPA GPCR that shares sequence homology with the purinergic receptor family (50). We have now extended the pharmacological knowledge of this structural family to include the 16:1 and 18:1 forms of 2ccPA and the 10:0, 12:0, 14:0, 16:0, 16:1 and 18:1 forms of 3ccPA. The 3ccPA analogs were all poor activators of LPA1/2/3/4 (Table 1). On the other hand, 2ccPA 16:1 and 2ccPA 18:1 were partial agonists of this receptor family (Table 1). Interestingly, whereas dodecyl LPC is the best substrate of ATX, the longer chain unsaturated ccPA analogs were the more effective inhibitors. This observation hints that the substrate binding site and the product feedback regulatory site show different acyl chain preference, hence are likely to be distinct. It is worth noting that the invasion of PC3 prostate cancer cells was blocked by 2ccPA 16:1, 3ccPA 16:1, and 3ccPA 18:1 but was unaffected by 2ccPA 18:1 (data not shown). This lack of effect by 2ccPA 18:1 in PC3 cells is in stark contrast to the significant inhibition of A2058 cell invasion (Figure 5). 2ccPA 18:1 is the only ccPA analog that possesses significant agonist activity, albeit as a significantly less potent, partial agonist as compared with LPA 18:1, at LPA1 (Table 1). PC3 cells express significant levels of LPA1 (~125 copies/ GAPDH × 103), whereas A2058 express 10-fold lower levels (~12 copies/GAPDH × 103) (5). These results suggest that GPCR activation is not likely to explain the anti-invasive properties of cPA analogs. Likewise, the cPA analogs tested did not antagonize LPA-mediated GPCR activation (51 and data not shown). Therefore, cPA does not seem to inhibit invasion via direct LPA GPCR effects. Furthermore, our results indicate that exogenously added LPA can bypass the inhibitory effect of ccPA on ATX-induced cell invasion, lending strong support to the hypothesis that ccPA inhibits invasion through ATX. These data also show the importance of designing/ identifying ATX inhibitors that do not activate LPA GPCR.
Two previous studies reported the anti-metastatic potential of cPA in vivo using injected B16 melanoma cells (41) and azoxymethane-induced intestinal tumors (40) in mice. In the first study cPA analogs were delivered as single doses intravenously (4 or 8 µg) mixed with 1×106 tumor cells (41). This protocol resulted in an 88% decrease in B16 derived lung metastases at the highest concentration (41). The second study delivered cPA analogs (3 and 6 mg/kg) subcutaneously in olive oil every other day for 30 weeks (40). cPA blocked bombesin-enhanced metastasis by up to 95% in this model (40). Here we show that 3ccPA 16:1 and 3ccPA 18:1 were able to block B16F10 tumor cell-derived lung metastasis by 35 and 57%, respectively, following only two 5 µg doses (250 µg/kg) 15 min and 48 h after inoculation. This decrease in metastatic potential resulted from intraperitoneal injections applied during the earliest phase of the metastatic process, suggesting that ccPA acted by inhibiting the formation of metastatic foci. We are currently working to optimize delivery and dosing of these reagents in an effort to maximize the anti-metastatic effect.
In this study we have described a novel role of cPA analogs as potent, natural ATX inhibitors. These are the most potent inhibitors we have identified to date. The 3ccPA 16:1 and 3ccPA 18:1 analogs are attractive lead compounds since they block ATX at nanomolar levels, yet they fail to significantly activate LPA GPCR at these concentrations. These compounds blocked cancer cell invasion in vitro (Figure 5) and tumor cell metastasis in vivo (Figure 6) without evidence of toxicity. Our results further validate ATX activity as a viable target of future cancer chemotherapeutics. These results also show proof of principle that compounds can be designed that selectively block LPA affects, via inhibition of LPA production, without the need for specific subtype selective LPA receptor antagonists.
Supplementary Material
ACKNOWLEDGEMENTS
This research was supported, in part, by an American Heart Association Award 0535195N and a University of Tennessee Cancer Institute Pilot Grant (DLB); the Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research (RWB); PO1 CA64602 and DAMD 17-02-1-0694 (GBM); and USPHS grants CA92160, and HL61469 (GT).
Footnotes
‡ABBREVIATIONS
ATX, autotaxin; cPA, 1-acyl-sn-glycero-2,3-cyclic-phosphate; 2ccPA, 2-carba-cyclic phosphatidic acid, 3ccPA 3-carba-cyclic phosphatidic acid; GPCR, G protein-coupled receptor; LPA, 1-acyl-2-hydroxy-sn-glycero-3-phosphate; LPA1, LPA receptor type 1 (EDG2); LPA2, LPA receptor type 2 (EDG4); LPA3, LPA receptor type 3 (EDG7); LPA4, LPA receptor type 4 (P2Y9).
REFERENCES
- 1.Stracke ML, Krutzsch HC, Unsworth EJ, Arestad A, Cioce V, Schiffmann E, Liotta LA. J Biol Chem. 1992;267:2524–2529. [PubMed] [Google Scholar]
- 2.Murata J, Lee HY, Clair T, Krutzsch HC, Arestad AA, Sobel ME, Liotta LA, Stracke ML. J Biol Chem. 1994;269:30479–30484. [PubMed] [Google Scholar]
- 3.Nam SW, Clair T, Campo CK, Lee HY, Liotta LA, Stracke ML. Oncogene. 2000;19:241–247. doi: 10.1038/sj.onc.1203263. [DOI] [PubMed] [Google Scholar]
- 4.Nam SW, Clair T, Kim YS, McMarlin A, Schiffmann E, Liotta LA, Stracke ML. Cancer Res. 2001;61:6938–6944. [PubMed] [Google Scholar]
- 5.Hama K, Aoki J, Fukaya M, Kishi Y, Sakai T, Suzuki R, Ohta H, Yamori T, Watanabe M, Chun J, Arai H. J Biol Chem. 2004;279:17634–17639. doi: 10.1074/jbc.M313927200. [DOI] [PubMed] [Google Scholar]
- 6.Umezu-Goto M, Kishi Y, Taira A, Hama K, Dohmae N, Takio K, Yamori T, Mills GB, Inoue K, Aoki J, Arai H. J Cell Biol. 2002;158:227–233. doi: 10.1083/jcb.200204026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Clair T, Lee HY, Liotta LA, Stracke ML. J Biol Chem. 1997;272:996–1001. doi: 10.1074/jbc.272.2.996. [DOI] [PubMed] [Google Scholar]
- 8.Tokumura A, Majima E, Kariya Y, Tominaga K, Kogure K, Yasuda K, Fukuzawa K. J Biol Chem. 2002;277:39436–39442. doi: 10.1074/jbc.M205623200. [DOI] [PubMed] [Google Scholar]
- 9.Clair T, Aoki J, Koh E, Bandle RW, Nam SW, Ptaszynska MM, Mills GB, Schiffmann E, Liotta LA, Stracke ML. Cancer Res. 2003;63:5446–5453. [PubMed] [Google Scholar]
- 10.Gijsbers R, Aoki J, Arai H, Bollen M. FEBS Lett. 2003;538:60–64. doi: 10.1016/s0014-5793(03)00133-9. [DOI] [PubMed] [Google Scholar]
- 11.Contos JJ, Ishii I, Chun J. Mol Pharmacol. 2000;58:1188–1196. doi: 10.1124/mol.58.6.1188. [DOI] [PubMed] [Google Scholar]
- 12.Anliker B, Chun J. Semin Cell Dev Biol. 2004;15:457–465. doi: 10.1016/j.semcdb.2004.05.005. [DOI] [PubMed] [Google Scholar]
- 13.Tigyi G, Parrill AL. Prog Lipid Res. 2003;42:498–526. doi: 10.1016/s0163-7827(03)00035-3. [DOI] [PubMed] [Google Scholar]
- 14.Tigyi G. Prostaglandins Other Lipid Mediat. 2001;64:47–62. doi: 10.1016/s0090-6980(01)00107-1. [DOI] [PubMed] [Google Scholar]
- 15.McIntyre TM, Pontsler AV, Silva AR, St Hilaire A, Xu Y, Hinshaw JC, Zimmerman GA, Hama K, Aoki J, Arai H, Prestwich GD. Proc Natl Acad Sci U S A. 2003;100:131–136. doi: 10.1073/pnas.0135855100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fang X, Schummer M, Mao M, Yu S, Tabassam FH, Swaby R, Hasegawa Y, Tanyi JL, LaPushin R, Eder A, Jaffe R, Erickson J, Mills GB. Biochim Biophys Acta. 2002;1582:257–264. doi: 10.1016/s1388-1981(02)00179-8. [DOI] [PubMed] [Google Scholar]
- 17.Mills GB, Moolenaar WH. Nat Rev Cancer. 2003;3:582–591. doi: 10.1038/nrc1143. [DOI] [PubMed] [Google Scholar]
- 18.Kohn EC, Hollister GH, DiPersio JD, Wahl S, Liotta LA, Schiffmann E. Int J Cancer. 1993;53:968–972. doi: 10.1002/ijc.2910530618. [DOI] [PubMed] [Google Scholar]
- 19.Debies MT, Welch DR. J Mammary Gland Biol Neoplasia. 2001;6:441–451. doi: 10.1023/a:1014739131690. [DOI] [PubMed] [Google Scholar]
- 20.Yang SY, Lee J, Park CG, Kim S, Hong S, Chung HC, Min SK, Han JW, Lee HW, Lee HY. Clin Exp Metastasis. 2002;19:603–608. doi: 10.1023/a:1020950420196. [DOI] [PubMed] [Google Scholar]
- 21.Stassar MJ, Devitt G, Brosius M, Rinnab L, Prang J, Schradin T, Simon J, Petersen S, Kopp-Schneider A, Zoller M. Br J Cancer. 2001;85:1372–1382. doi: 10.1054/bjoc.2001.2074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yang Y, Mou L, Liu N, Tsao MS. Am J Respir Cell Mol Biol. 1999;21:216–222. doi: 10.1165/ajrcmb.21.2.3667. [DOI] [PubMed] [Google Scholar]
- 23.Klominek J, Robert KH, Bergh J, Hjerpe A, Gahrton G, Sundqvist KG. Anticancer Res. 1998;18:759–767. [PubMed] [Google Scholar]
- 24.Deissler H, Blass-Kampmann S, Bruyneel E, Mareel M, Rajewsky MF. Faseb J. 1999;13:657–666. doi: 10.1096/fasebj.13.6.657. [DOI] [PubMed] [Google Scholar]
- 25.Zhang G, Zhao Z, Xu S, Ni L, Wang X. Chin Med J (Engl) 1999;112:330–332. [PubMed] [Google Scholar]
- 26.Hoelzinger DB, Mariani L, Weis J, Woyke T, Berens TJ, McDonough WS, Sloan A, Coons SW, Berens ME. Neoplasia. 2005;7:7–16. doi: 10.1593/neo.04535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Euer N, Schwirzke M, Evtimova V, Burtscher H, Jarsch M, Tarin D, Weidle UH. Anticancer Res. 2002;22:733–740. [PubMed] [Google Scholar]
- 28.Sano T, Baker D, Virag T, Wada A, Yatomi Y, Kobayashi T, Igarashi Y, Tigyi G. J Biol Chem. 2002;277:21197–21206. doi: 10.1074/jbc.M201289200. [DOI] [PubMed] [Google Scholar]
- 29.Tokumura A, Yamano S, Aono T, Fukuzawa K. Ann N Y Acad Sci. 2000;905:347–350. doi: 10.1111/j.1749-6632.2000.tb06576.x. [DOI] [PubMed] [Google Scholar]
- 30.Okita M, Gaudette DC, Mills GB, Holub BJ. Int J Cancer. 1997;71:31–34. doi: 10.1002/(sici)1097-0215(19970328)71:1<31::aid-ijc7>3.0.co;2-4. [DOI] [PubMed] [Google Scholar]
- 31.Croset M, Brossard N, Polette A, Lagarde M. Biochem J. 2000;345(Pt 1):61–67. [PMC free article] [PubMed] [Google Scholar]
- 32.van Meeteren LA, Ruurs P, Christodoulou E, Goding JW, Takakusa H, Kikuchi K, Perrakis A, Nagano T, Moolenaar WH. J Biol Chem. 2005;280:21155–21161. doi: 10.1074/jbc.M413183200. [DOI] [PubMed] [Google Scholar]
- 33.Murakami-Murofushi K, Shioda M, Kaji K, Yoshida S, Murofushi H. J Biol Chem. 1992;267:21512–21517. [PubMed] [Google Scholar]
- 34.Friedman P, Haimovitz R, Markman O, Roberts MF, Shinitzky M. J Biol Chem. 1996;271:953–957. doi: 10.1074/jbc.271.2.953. [DOI] [PubMed] [Google Scholar]
- 35.Kobayashi T, Tanaka-Ishii R, Taguchi R, Ikezawa H, Murakami-Murofushi K. Life Sci. 1999;65:2185–2191. doi: 10.1016/s0024-3205(99)00483-x. [DOI] [PubMed] [Google Scholar]
- 36.Fischer DJ, Liliom K, Guo Z, Nusser N, Virag T, Murakami-Murofushi K, Kobayashi S, Erickson JR, Sun G, Miller DD, Tigyi G. Mol Pharmacol. 1998;54:979–988. doi: 10.1124/mol.54.6.979. [DOI] [PubMed] [Google Scholar]
- 37.Liliom K, Murakami-Murofushi K, Kobayashi S, Murofushi H, Tigyi G. Am J Physiol. 1996;270:C772–C777. doi: 10.1152/ajpcell.1996.270.3.C772. [DOI] [PubMed] [Google Scholar]
- 38.Murakami-Murofushi K, Kaji K, Kano K, Fukuda M, Shioda M, Murofushi H. Cell Struct Funct. 1993;18:363–370. doi: 10.1247/csf.18.363. [DOI] [PubMed] [Google Scholar]
- 39.Mukai M, Imamura F, Ayaki M, Shinkai K, Iwasaki T, Murakami-Murofushi K, Murofushi H, Kobayashi S, Yamamoto T, Nakamura H, Akedo H. Int J Cancer. 1999;81:918–922. doi: 10.1002/(sici)1097-0215(19990611)81:6<918::aid-ijc13>3.0.co;2-e. [DOI] [PubMed] [Google Scholar]
- 40.Ishihara R, Tatsuta M, Iishi H, Baba M, Uedo N, Higashino K, Mukai M, Ishiguro S, Kobayashi S, Murakami-Murofushi K. Int J Cancer. 2004;110:188–193. doi: 10.1002/ijc.20069. [DOI] [PubMed] [Google Scholar]
- 41.Murakami-Murofushi K, Uchiyama A, Fujiwara Y, Kobayashi T, Kobayashi S, Mukai M, Murofushi H, Tigyi G. Biochim Biophys Acta. 2002;1582:1–7. doi: 10.1016/s1388-1981(02)00131-2. [DOI] [PubMed] [Google Scholar]
- 42.Kobayashi S, Murofushi H, Murakami-Murofushi K. International Patent. 2004
- 43.Kobayashi S, Tokunoh R, Shibasaki M, Shinagawa R, Murakami-Murofushi K. Tetrahedron Letters. 1993;34:4047–4050. [Google Scholar]
- 44.Erukulla RK, Byun H-S, Bittman R. Tetrahedron Letters. 1994;35:5783–5784. [Google Scholar]
- 45.Tokumura A, Kanaya Y, Kitahara M, Miyake M, Yoshioka Y, Fukuzawa K. J Lipid Res. 2002;43:307–315. [PubMed] [Google Scholar]
- 46.Fischer DJ, Nusser N, Virag T, Yokoyama K, Wang D, Baker DL, Bautista D, Parrill AL, Tigyi G. Mol Pharmacol. 2001;60:776–784. [PubMed] [Google Scholar]
- 47.Ohta H, Sato K, Murata N, Damirin A, Malchinkhuu E, Kon J, Kimura T, Tobo M, Yamazaki Y, Watanabe T, Yagi M, Sato M, Suzuki R, Murooka H, Sakai T, Nishitoba T, Im DS, Nochi H, Tamoto K, Tomura H, Okajima F. Mol Pharmacol. 2003;64:994–1005. doi: 10.1124/mol.64.4.994. [DOI] [PubMed] [Google Scholar]
- 48.Virag T, Elrod DB, Liliom K, Sardar VM, Parrill AL, Yokoyama K, Durgam G, Deng W, Miller DD, Tigyi G. Mol Pharmacol. 2003;63:1032–1042. doi: 10.1124/mol.63.5.1032. [DOI] [PubMed] [Google Scholar]
- 49.Durgam GG, Virag T, Walker MD, Tsukahara R, Yasuda S, Liliom K, van Meeteren LA, Moolenaar WH, Wilke N, Siess W, Tigyi G, Miller DD. J Med Chem. 2005;48:4919–4930. doi: 10.1021/jm049609r. [DOI] [PubMed] [Google Scholar]
- 50.Noguchi K, Ishii S, Shimizu T. J Biol Chem. 2003;278:25600–25606. doi: 10.1074/jbc.M302648200. [DOI] [PubMed] [Google Scholar]
- 51.Fujiwara Y, Sardar V, Tokumura A, Baker DL, Murakami-Murofushi K, Parrill AL, Tigyi G. J Biol Chem. 2005 doi: 10.1074/jbc.M504351200. [DOI] [PubMed] [Google Scholar]
- 52.Qian F, Zhang ZC, Wu XF, Li YP, Xu Q. Biochem Biophys Res Commun. 2005;333:1269–1275. doi: 10.1016/j.bbrc.2005.06.039. [DOI] [PubMed] [Google Scholar]
- 53.Yamaguchi H, Kitayama J, Takuwa N, Arikawa K, Inoki I, Takehara K, Nagawa H, Takuwa Y. Biochem J. 2003;374:715–722. doi: 10.1042/BJ20030381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mukai M, Nakamura H, Tatsuta M, Iwasaki T, Togawa A, Imamura F, Akedo H. FEBS Lett. 2000;484:69–73. doi: 10.1016/s0014-5793(00)02129-3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.