Abstract
We have investigated the self-assembly of fluorenylmethoxycarbonyl-conjugated dialanine (Fmoc-AA) molecules using combined computational and experimental approaches. Fmoc-AA gels were characterized using TEM, circular dichroism, FTIR, and WAXS. Computationally, we simulated the assembly of Fmoc-AA using molecular dynamics techniques. All simulations converged to a condensed fibril structure in which the Fmoc groups stack mostly within in the center of the fibril. However, the Fmoc groups are partially exposed to water, creating an amphiphilic surface, which may be responsible for aggregation of fibrils into nano-scale fibers observed in TEM. From the fibril models, radial distribution calculations agree with d-spacings observed in WAXS for the fibril diameter and π-stacking interactions. Our analyses show that dialanine, despite its short length, adopts a mainly extended polyproline II conformation. In contrast to previous hypotheses, these results indicate that β-sheet-like hydrogen bonding is not prevalent. Rather, stacking of Fmoc groups, inter-residue hydrogen bonding and hydrogen bonding with water play the important roles in stabilizing the fibril structure of supramolecular assemblies of short conjugated peptides.
Keywords: Fmoc-dipeptide, supramolecular gel, self-assembly, replica-exchange molecular dynamics, nanofiber, molecular dynamics
Introduction
Molecular self-assembly continues to be explored as a powerful approach for fabricating functional nanostructured materials.1–4 Artificial self-assembly methods offer a “bottom-up” route to fabricating novel complex structures with nanometer precision. Numerous examples of the self-assembly of low molecular weight organic molecules leading to hydrogel formation can be found in literature,5–7 and of these, hydrogels composed of self-assembling, peptide-based small molecules have recently gained interest for their potential uses in biomedical applications such as tissue engineering and drug delivery.8 In contrast to traditional hydrogels composed of chemically cross-linked high molecular weight polymers, gel materials based on self-assembling peptide conjugates consist of low molecular weight components that associate based on non-covalent interactions, such as hydrophobic forces, π-stacking and hydrogen bonding. These materials may therefore be more sensitive to in vivo chemical and physical stimuli than polymeric hydrogels, and more easily degraded by the body and cleared by the kidneys.5 Also unlike many polymer hydrogel systems, these molecules self-assemble via a pH or temperature trigger and can therefore be delivered via transdermal injection and gelled in situ, minimizing the invasiveness of the procedure required to place them. Being peptide-derived, these materials are easily customized with bioactive sequences that may serve to enhance cell adhesion or induce specific signaling pathways.
Notable among self-assembling peptide-based gel materials are those created by Stupp and co-workers. 9 These engineered peptide amphihiles (PAs) consist of a hydrocarbon tail conjugated to a short (8–12 residue) peptide sequence, with the inner residues having a high propensity to form β-sheet type hydrogen bonding interactions with neighboring molecules.10 The putative supramolecular arrangement of these molecules is a cylindrical fibril, stabilized by hydrophobic interactions among the hydrocarbon tails within the core of a fibril and by hydrogen bonding between the β-sheet forming segments. A recent molecular dynamics study of the proposed assembly confirmed this general structure, demonstrating the water- and ion-excluding properties of the hydrophobic core as well as the extensive hydrogen bonding between adjacent peptide segments.11
Recently it has been shown that much shorter (2–5 residues) peptides conjugated to aromatic N-terminal protecting groups (such as fluorenylmethoxycarbonyl, naphthalene, pyrene, and even DNA nucleotide bases) can also self-assemble to form gels.7,8,11–17 Of these, the fluorenylmethoxycarbonyl (Fmoc) derivatives were the first such molecules that were shown to gel in part because Fmoc-protected peptides and dipeptides are widely available for use in solid phase peptide synthesis. Their gelation ability was likely discovered serendipitously during attempts to drive aqueous dissolution; gelation of certain Fmoc-peptides has been demonstrated by raising then lowering the solution temperature, adding an excess of water to a miscible organic solvent containing the peptide conjugate, or by raising then lowering the solution pH.8
Like PAs, these conjugate peptides are thought to form nanometer diameter fibril structures stabilized by noncovalent interactions between aromatic protecting groups and β-sheet-like hydrogen bonding between amide bonds on adjacent molecules. However, the proposed supramolecular arrangement of the conjugated peptides is markedly different from that of the PA system. Researchers observed, using Fourier-transform infrared spectroscopy (FTIR), circular dichroism (CD) spectroscopy, and x-ray diffraction (XRD) techniques, that these gels contain hydrogen-bonding patterns, molecular configurations, and intermolecular dimensions indicative of antiparallel β-sheet structures.12,18 Smith et al.18 proposed a model in which pairs of Fmoc-diphenylalanine molecules are arranged in a helix around the long axis of a tubular fibril in an anti-parallel manner, with the aromatic rings of the Fmoc group on one molecule located near the peptide carboxyl terminus of the adjacent molecule. The authors offer a computational/theoretical confirmation of the energetic feasibility of this structure, involving a potential energy minimization using the AMBER force field; however, potential energy minimization typically leads to local minima and excludes the contribution of thermal energy and entropic effects.
Considering the potential conformational changes during the time evolution in self-assembly, molecular dynamics (MD) simulations are more favorable than simple energy minimization to study the self-assembly of biomolecules.19,20 Besides providing atomistic information on kinetics, thermodynamics, and time evolution of molecular conformations, MD can also remove unfavorable interactions that remain after energy minimization. Therefore, MD can overcome the minimization’s inadequacy in predicting the entropically favored conformations and therefore has been widely used in other self-assembly studies in recent years.11,21–23 In one example, MD simulations were used to study the self-assembly of Fmoc-tetrapeptides based on the RGD cell adhesion sequence.24 However, the simulations were relatively short (10–20 ns), consisted of assemblies of only 21 molecules, and there was no clear evidence that the structures had converged. Furthermore, the study concerned Fmoc-tetrapeptides, which have two more peptide bonds available for hydrogen bonding than do Fmoc-dipeptides and is therefore an arguably more “peptide-like” system.
In our preliminary molecular dynamics study (see Supporting Information), the Fmoc-dipeptide fibril model proposed by Smith et al.18 did not maintain its hollow tube structure; instead, it shrunk into a more compact aggregate with water molecules excluded from the center. We thus focus on models in which the aromatic protecting groups are concentrated toward the center of the fibril with the attached peptide chains radiating outward, as is the case for PAs.
In this work, we sought to answer two questions about the self-assembly of Fmoc-dipeptides: 1) What are the major driving forces for self-assembly? and 2) How are individual molecules arranged within the gel-forming fibril structures? To this end we performed experimental validation of past results along with the first demonstration of the use of extensive molecular dynamics simulations to gain insight into the supramolecular structure of the Fmoc-dipeptide assembly. By answering these questions we will improve our understanding of the nano-scale properties of these materials and can therefore assess the feasibility of using self-assembling short peptide conjugates for in vivo biomedical applications. Our system and approach may also be extended to investigate the dependence of side chain identity and sequence on self-assembly25,26 in order to move toward rational design of peptide-based small molecule hydrogelators.
Materials and Methods
Experimental
Preparation of Fmoc-AA gels
Fmoc-Ala-Ala-OH was purchased from Bachem, Inc. (Torrance, CA) and used without further purification. To form gels via a pH trigger, Fmoc-AA was added to deionized water, vortexed, and sonicated for several minutes. 1 eq 0.5 M NaOH was added and mixture was vortexed until completely clear. The mixture was then added quickly to a vial containing 1 eq 0.1 M HCl, to trigger gelation.
Wide angle X-ray scattering
Fmoc-AA gel samples were prepared at 10 mg/ml (final pH = 2.6–2.8) by a pH trigger method (1 eq 0.5 M NaOH added to dissolve Fmoc-AA, then 1 eq 0.1 M HCl added to induce gelation, as described above) and were spread out onto a quartz slide and allowed to dry for two days. Samples were run on a Scintag X1 theta-theta powder diffractometer with a Cu x-ray source. Peak analysis was performed using Jade v9.1.1 (Materials Data Inc.) software, and d-spacings were calculated assuming an x-ray wavelength of 1.54059Å (Cu K-α1).
Transmission Electron Microscopy
Gels for TEM analysis were prepared at a final concentration of 5 mg/ml (final pH = 3.3–3.4). Approximately 10 μl of gel was placed on the shiny side of Formvar/Carbon coated 300 mesh nickel grids (Electron Microcopy Sciences, Hatfield, PA) and was allowed to adsorb for 2 minutes. Excess solvent was removed using a Kimwipe, and the grid was further allowed to air dry for 1 minute, after which the grid was washed with 10 μl deionized water for 1 minute and dried with a Kimwipe. Finally, the grids were placed on 10 μl drops of 2% uranyl acetate for 1.5 minutes, then dried with a Kimwipe and allowed to air dry before placing in the instrument. Samples were imaged using a FEI Tecnai instrument operating at 80 kV, and the associated AMT Advantage HR 1kX1k digital camera was used both for sample navigation and image capture.
Fourier Transform Infrared spectroscopy
Gels were prepared at 5 mg/ml (final pH = 3.3–3.4) as described above, except D2O was used in place of deionized water in all solutions. For pH triggered gels, NaOH and HCl solutions were diluted to the concentrations described above using D2O. Gel samples were sandwiched tightly between two calcium fluoride (CaF2) windows (25 mm wide by 2 mm thick) and placed in line with the beam in a transmission configuration within the instrument. A Thermo-Mattson Infinity Gold FTIR instrument was used to collect spectra, which were averaged over 200 scans.
Circular Dichroism spectroscopy
Fmoc-AA gels for circular dichroism were prepared at a concentration of 5 mg/ml. ~2.5 eq of Glucono-delta-lactone (GdL), a slowly-hydrolyzing lactone that ring-opens to form gluconic acid, was used to acidify and hence gel a solution of Fmoc-AA and 1 eq 0.5 M NaOH in deionized water, as described previously.27 Prior to GdL addition, a background spectrum was taken using deionized water with 1 eq 0.5 M NaOH added. After GdL addition and dissolution the Fmoc-AA solution was pipetted into a cylindrical demountable quartz cuvette with a 0.1 mm pathlength. Spectra were measured using a Jasco J-810 CD Spectrometer every 10 minutes after GdL addition. The instrument was operated with a data pitch of 0.5 nm, scan speed of 200 nm/min, and response time of 1 second. Three spectra were taken for each time point then averaged. After the final time point we checked for linear dichroism (LD) artifacts by performing the scan again with the cuvette rotated 90, 180, and 270 degrees from its starting position, but we observed no significant change in the CD signal and thus concluded that LD effects were minimal. The final pH of GdL-formed gels after 24 hours was in all cases between 3.5 and 3.7.
Molecular models and dynamics simulations
Initial structures preparations
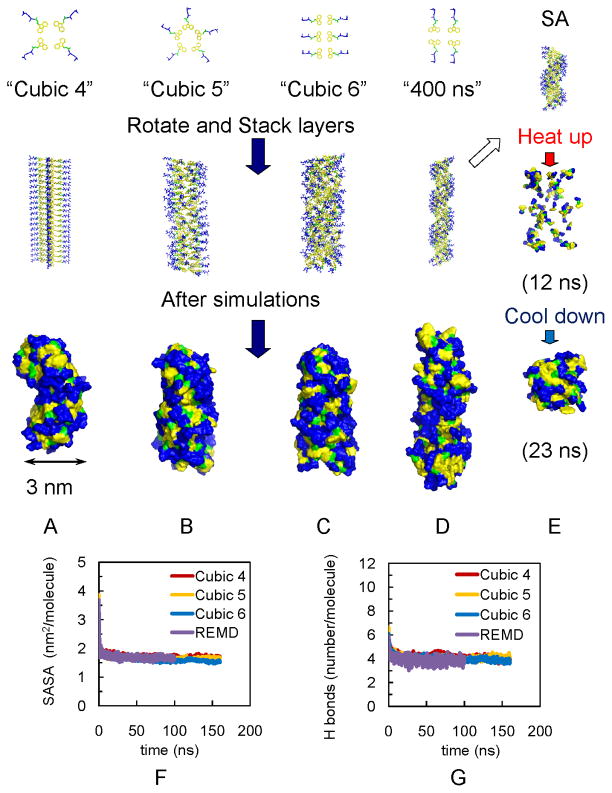
Fmoc-AA molecules were built in TINKER.28 As shown in Figure 1, all the fibrils are cylindrical nanofibrils, “Cubic 4” has 4 molecules placed as a “cross” on each layer; “Cubic 5” has 5 molecules with the alanines radiating outward; “Cubic 6” and “400 ns” has parallel placed Fmoc-AAs also with alanines radiating outward. Except Cubic 4’s fibril, the plate layers are rotated 35.9 degrees relative to the adjacent layer. The distance between each layer is 4.5 Å for SA and 400 ns models, and 5 Å for all Cubic models. A lower density of molecules on each layer (like 2 or 3 Fmoc-AA molecules on each layer) will result in an unstable fibril. Specific geometrical details are listed in Table S1of supporting information.
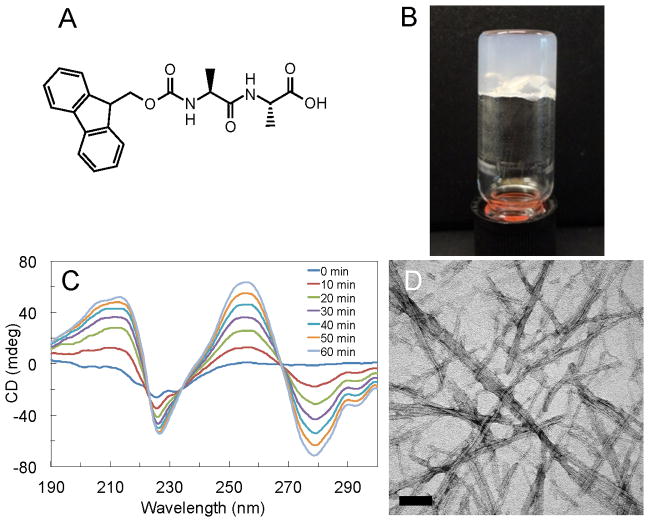
Figure 1.
Experimental analysis of Fmoc-AA gels. (A) Fmoc-AA chemical structure; (B) Vial inversion test confirms that self-assembly leads to formation of self-supporting gel. (C) CD of Fmoc-AA gels formed via slow acidification with GdL shows increase in dichroism over time due to handedness induced by self-assembly. Spectra shown were taken from 0 to 60 minutes at 10 minute intervals; (D) TEM shows nanoscale “ribbon” like features from 10–50 nm in width, similar to those seen in previous studies. Scale bar = 100 nm.
Details of atomistic simulations
Individual Fmoc-AA molecules were built and parameterized using TINKER and GROMACS (version 4.5.4) 29. For the Fmoc residue, an OPLS-AA-type molecular mechanics model was developed in both TINKER and GROMACS. The atomic charges for new atom types in the Fmoc residue were derived by quantum mechanical (QM) calculations; the bond, angle and vdW parameters were transferred from OPLS-AA, OPLS-AA/L30–32 and MM333 force fields; the torsion parameters were derived by fitting the molecular mechanics (MM) calculations to the QM potential surface (details about parameters are provided in supporting information).
Some of our molecular dynamics simulations were performed on the Texas Advanced Computing Center (TACC) Ranger (3,936 16-way SMP compute, 15,744 AMD Opteron™ processors for a total of 62,976 compute cores, 579 TFLops Peak) and TACC Lonestar (Dell PowerEdge 1955 Blade, 55.5 TFlops Peak, 1300 nodes, 2 Dual-Core Xeon 5100 at 2.66 GHz, InfiniBand interconnection).
All simulations and calculations were performed using GROMACS 4.5.4 and 4.5.329 with OPLS-AA force field34,35, and applied periodic boxes with explicit water molecules. All starting structures for MD simulation, except simulated annealing and replica exchange molecular dynamics (REMD), were subjected to steepest descent energy minimizations with 5000 maximum steps to remove high-energy contacts. Before the productive MD, in order to keep the system stable at 300 K and 1 bar, a 100 ps NVT and a 100 ps NPT equilibration were applied respectively. During the productive MD, all bonds were constrained using LINCS algorithm36,37; for every 10 fs, neighbors were searched in grid cells with 1 nm as cutoff values for short-range neighborlist, electrostatic and van der Waals; long-range electrostatics were treated with the particle mesh Ewald method38 with a grid spacing of 0.16 nm; constant temperature and pressure were maintained by coupling the system to an external bath at 300 K and 1 bar, using velocity rescaling39 and Parrinello–Rahman40, respectively. A 2 ps pressure coupling time was applied and the isothermal compressibility of water is 4.5×10−5 bar−1. A leap-frog integrator with the integration time step of 2 fs was used.41 GROMACS OPLS-AA force field with our fitted parameters for Fmoc group was used for the Fmoc-AA, and a TIP3P model was applied for water.42 In SA of Fmoc-AA molecules self-assembly, the initial fibril structure was the first 15 layers of the 400 ns model. The system temperature was firstly heated up from 288 to 300 K in 0.5 ns, then linearly and gradually cooled down to 0 K in the following 39.5 ns. We randomly extracted 120 frames during the temperature dropping from 700 to 290 K in SA that covered the conformational space broadly, and those frames were then used in REMD as initial structures; a set of temperatures ranging from 280 to 600 K was estimated and preset in order to maintain the success rates of exchange within 20–30%.
Results and Discussion
Our model gelator, Fmoc-Ala-Ala (or Fmoc-AA, Figure 1A), is a simple molecule known to self-assemble in aqueous solution to form a self-supporting gel.12,26 We confirm that Fmoc-AA does indeed form a self-supporting gel at 0.5% w/v by vial inversion (Figure 1B) with circular dichroism (CD) experiments (Figure 1B) and transmission electron microscopy (TEM) (Figure 1D) showing chiral and nanostructural evidence of self-assembly, respectively.
Circular dichroism measurements confirm the assembly of Fmoc-AA. Figure 1C illustrates the increase in dichroism over time when glucono-δ-lactone (GdL), a ring molecule that slowly hydrolyzes to gluconic acid, is used to slowly lower the pH and hence slowly gel a solution of Fmoc-AA (see Methods section for experimental details). The increase in dichroism can be attributed to an increase in the apparent chirality of the sample; this increase in “nanoscale chirality” is a known hallmark of the self-assembly process.43 Therefore, the data in Figure 1C corresponds to the progression of self-assembly over the course of an hour, and the concomitant tendency of individual Fmoc-AA molecules to adopt preferred orientations, giving rise to an overall “handedness” of the assembly. We confirmed that the hydrolysis of GdL itself does not contribute to the increase in dichroism over time (see Supporting Information). Within the data there are several important features: a broad positive peak centered at ~210 nm, a negative peak at 228 nm, and peaks at higher wavelengths (> 260 nm). The broad peak at ~210 nm is roughly 10 nm higher than a characteristic β-sheet peak typically between 195 nm and 202 nm.44 The negative peak at 228 nm is similar to what has been suggested as a “β-sheet peak” in previous studies,12,18,45 although it is also shifted about 10 nm higher than typical characteristic β-sheet-type peaks. It is important to note that there are published CD data for other Fmoc-dipeptide hydrogelators that do not contain this negative peak.27 The high wavelength peaks likely arise due to the induced chirality of aromatic residues (specifically, the Fmoc group) placed within the environment of a supramolecular assembly.44 Though slight wavelength variations exist, the CD data presented here are generally in good agreement with published data for Fmoc-dipeptides. Interestingly, the peaks in our CD align well with those reported for Fmoc-Leu-Gly by Adams et al.27 but are opposite in sign, indicating similar supramolecular structural features that give rise to nanoscale chirality, but with reversed handedness (e.g. left- vs. right-hand helix). Given the wavelength discrepancies with protein data and the fact that not all Fmoc-dipeptide gelators exhibit β-sheet-like peaks in CD, we hesitate to use CD to make specific conclusions regarding the structural arrangement of molecules within a fibril assembly. Rather, these data serve as confirmation that Fmoc-AA self-assembles in a manner similar to other Fmoc-dipeptide systems.
TEM analysis was also performed to confirm that the nanoscale morphological features of Fmoc-AA gels were similar to those reported for other Fmoc-dipeptide gels. Figures 1C and D clearly show the ribbon-like morphology seen in previous studies of various Fmoc-dipeptide gelators. The ribbon width ranges from 10 to 50 nm, dimensions also noted in previous studies.12,18,27,46,47
Molecular simulations of self-assembly and stability analysis
Next we carried out molecular dynamics simulations to investigate the self-assembly of Fmoc-AA fibrils. We performed several simulations on different initial fibril structures consisting of loosely stacked “layers” with four, five, or six fully protonated Fmoc-AA molecules, as is shown in Figure 2. These potential fibril structures were inspired by previous work on PAs and the duplex structure of DNA. Each fibril model was named according to the number of Fmoc-AA molecules in a single plate and the geometry of the water boundary, i.e. “Cubic 4” refers to the model in which the initial structure is a 20-layer containing 4 Fmoc-AA molecules each, and this structure is placed in a cubic box of water molecules for the simulation. The rationale for running several different initial configurations was to determine what effect initial conditions might have on the final assembled structures. Each system was subjected to 160 ns room-temperature MD simulations or more.
Figure 2.
Models of fibrils in simulations show independence of final assembly of initial structures. (A) Fibril with 4 Fmoc-AA soaked in cubic box (“Cubic 4”). (B) Fibril with 5 Fmoc-AA soaked in cubic box (“Cubic 5”). (C) Fibril with 6 Fmoc-AA soaked in cubic box (“Cubic 6”). (D) Fibril with 4 Fmoc-AA soaked in long rectangular box with 400 ns simulation (“400 ns”). For A–C, all simulations were 160 ns. (E) Simulated annealing of 15 layers from model in (D); also used as initial structures for REMD simulations. Note: yellow indicates fluorenyl group of Fmoc, green is the carbamate linker, and blue is the peptide chain. For A–D, Top: the arrangement of Fmoc-AA molecules (hydrogens are not shown) on each layer. Middle: initial structures for simulations. Bottom: snapshots of the solvent accessible surfaces from the last frames of simulations show self-assembled fibril structures. The convergence of SASA and hydrogen bonding calculations over time indicates structural stability: (F) SASA per molecule for Cubic 4, Cubic 5, Cubic 6 and REMD ensemble at 299 K (“REMD”); all converge to ~1.7 nm2/molecule, and (G) Number of hydrogen bonds per molecule for Cubic 4 to 6 and REMD simulations (~ 4).
Additionally, we attempted “ab initio” predictions using simulated annealing (SA)48 to investigate the assembly process without well-defined initial structures. SA is a global optimization algorithm designed to search for an optimal configuration. In SA, the system is “heated up” to yield random configurations and slowly “cooled back” down to a low temperature using molecular dynamics simulations. As shown in Figure 2E, during the SA procedure the initial fibril is heated up and the structure falls apart; Fmoc-AA molecules are randomly distributed in the water box without any aggregation. As the system is cooled, Fmoc-AA molecules begin to assemble together into a fibril. However, with finite simulation time length, SA cannot guarantee to output the optimal structure (at a global energy minimum). Therefore we performed further simulations using replica exchange molecular dynamics (REMD), a technique that utilizes the faster kinetics at the high temperature to accelerate the sampling at room temperature.49 REMD has been utilized to successfully fold small proteins 50–53 and study self-assembly. 54–56 In our REMD, simulations are performed in parallel on 120 replicas of the molecular system, each at different temperatures from 280 to 600 K for 100 ns. The structures in the neighboring replicas are allowed to exchange with a success rate greater than 0.18. In this work, the initial structures for REMD are a collection of structures at different temperatures extracted from the SA process. Due to the computational cost, we used a smaller system with 60 Fmoc-AA molecules in a 396 nm3 rectangular water-containing box.
Snapshots of the last frame of each simulation (Figure 2A to D) show that each model has been compacted and reorganized during the simulation but all maintain a fibril shape. The REMD ensemble at 299 K also lead to a similar fibril shape to the SA assembled fibrils in Figure 2E. It is also apparent from these snapshots that the hydrophobic fluorenyl rings are mostly buried but not completely inaccessible to water, and this feature may help to explain why lateral aggregation of fibrils into larger fiber structures is observed in TEM.
We also assessed the stability of each model’s assembly by calculating the solvent accessible surface area (SASA) per molecule over the course of the simulation. Results of this analysis are displayed in Figure 2F. SASA represents quantitatively the contact between solute and the solvent and therefore depicts the changes and rearrangements of hydrophobic surfaces during the molecular assembly and aggregation.57 During the assembly process, the space between Fmoc-AA plate layers lessens and the structure becomes more compact. Water molecules initially located in the hydrophobic “hollow” between the Fmoc rings in the starting structures are expelled out of the fibrils during the simulations, and therefore the SASA per molecule decreases, suggesting the process of the aggregation of molecules. Additionally, hydrogen bonding between water and Fmoc-AA would also provide information about assembly.
Figure 2F and G show, respectively, the convergence of the SASA of each fibril model and the number of hydrogen bonds between water and Fmoc-AA during the simulations. During the first few of nanoseconds of the simulation, SASA and hydrogen bond numbers drop dramatically, suggesting the rapid aggregation and assembly of Fmoc-AA molecules. After 50ns, all systems become stable; the constant values of both SASA (~1.7 nm2/molecule) and hydrogen bonds (~4/molecule) per molecule between simulations with different starting structures suggest similar self-assembled fibrils are formed regardless of the nature of the initial starting structures.
In the 299 K REMD ensemble, the initial structure was taken from the simulated annealing at a high temperature, and thus the Fmoc-AA units are not in a fibril form, but randomly distributed in the water box (as in Figure 2E the middle, the snapshot at 12 ns). However, during the exchanges of replicas between different temperature conditions, the stacked fibril-like configurations are quickly exchanged to and stayed in the 299 K ensemble. As a result, the REMD 299K ensemble gives remarkably similar results as the “traditional” MD results as shown in Figure 2F and G. These MD simulations confirm that the self-assembly is a rather robust phenomenon and the fibril structure is energetically very favorable.
In our simulations, we also observed that the fibril composed of completely deprotonated Fmoc-AA molecules would fall apart very quickly. This observation is confirmed by experiment: at high pH (wherein Fmoc-AA is likely totally deprotonated), the solution was free flowing and not gel-like, indicating that long, entangled fibrils were not present.
Experimental WAXS results confirm the molecular dimensions derived from simulations
We further validated our computational simulations by comparing molecular dimensions calculated from Radial Distribution Functions (RDF) with actual d-spacings obtained from Wide Angle X-ray Scattering (WAXS) experiments on dried films of Fmoc-AA gels triggered by a pH change. RDF describes the variation of the density of an atom (or group of atoms) with the distance from a reference particle,58 providing dimensional information that can be experimentally determined using X-ray scattering techniques. WAXS can give information about local order in non-crystalline polymers,59 and therefore is employed in this study to compare with RDF computational results. We recognize that drying gel films on quartz single crystal slides yields results likely unrepresentative of the gel phase;60 however, for the sake of comparison with previous studies we opted to use a similar sample preparation method.
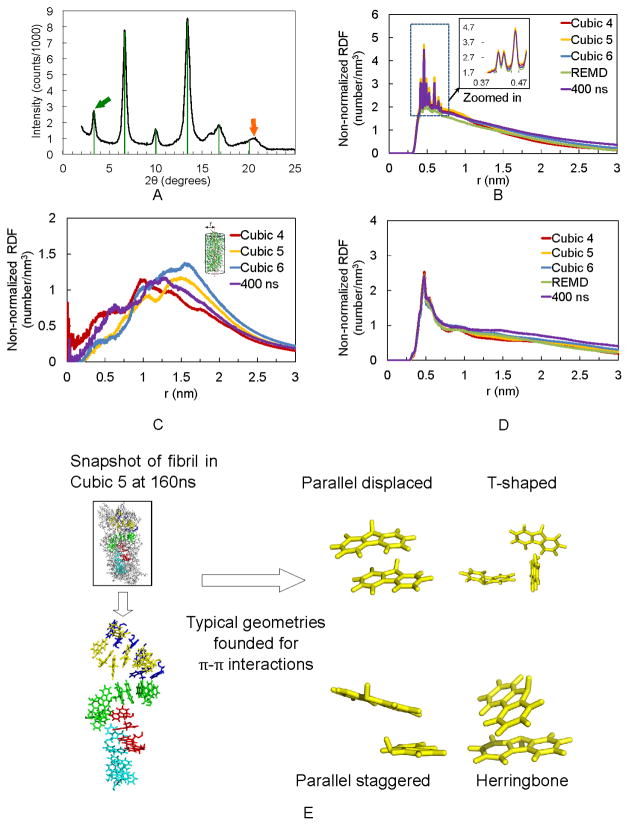
Our WAXS analysis of dried Fmoc-AA gel films gives molecular dimensions similar to those reported previously. The diffraction pattern is shown in Figure 3A. Similar to Smith et al.,18 we found a reflection corresponding to a d-spacing of 26.3 Å (green arrow in Figure 3A), with subsequent higher order reflections up to n=6, indicated by green lines. Distinct from the higher order reflections is a peak corresponding to a dimension of 4.35 Å (orange arrow). This dimension is also similar to one observed by Smith et al.18 and is seen in other self-assembling dipeptide-conjugate gel systems25,47 in which the dimension has frequently been attributed to a β-sheet like spacing between adjacent molecules within the fibril assembly.
Figure 3.
WAXS experiments confirm non-normalized RDF calculations. (A) WAXS diffraction pattern for Fmoc-AA gel film shows strong reflection (green arrow) and higher order reflections (green lines) corresponding to a d-spacing of 26.3 Å (n=1). Orange arrow indicates a reflection with d=4.35 Å. (B) Last 50 ns non-normalized RDF plot of distance between fluorenyl rings of Cubic 4, 5, and 6, REMD and the 400 ns simulations. (C) Last 50 ns non-normalized RDF plot of distance between terminal alanine’s hydroxyl hydrogen and fibril axis. (D) Last 50 ns non-normalized RDF plot of Fmoc-AAs’ strand-to-strand distance, showing predominance of a 0.46–0.48 nm (4.6–4.8 Å) distance between adjacent dipeptides. (E) Representative snapshot taken from Cubic 5 MD simulation displays the π-π stacking in fibril after 160 ns simulation.
The obtained characteristic dimensions from WAXS are consistent with our radial distribution analyses of simulated fibrils. Note that due to the various simulation box sizes and concentrations of Fmoc-AA, it is more meaningful to compare the non-normalized RDF, e.g. without dividing the probability density by the average density in the whole box. Otherwise, doubling the box size without increasing the number of peptides, reduces the RDF intensity by factor of two since the final fibril structure remains the same. The positions of peaks are not affected by the normalization. During self-assembly simulation, the fluorenyl rings (colored yellow in Figure 2) are stacked in an ordered way, as seen within differently-colored Fmoc-AA pairs highlighted in Figure 3E. The non-normalized RDF between fluorenyl rings is shown in Figure 3B. All atoms in the fluorenyl rings were used for the RDF calculation. The high peaks correspond to distances of around 0.41 to 0.5 nm, indicating an ordered stacking patterning between the fluorenyl rings within the Fmoc group. A similar distance at d= 4.35 Å also appears in the WAXS in Figure 3A, suggesting that this distance may be characteristic of the π-π stacking between fluorenyl rings, rather than a specific β-sheet-like spacing between peptide bonds on adjacent molecules. This interpretation was offered by Braun and Cardoso,61 who argued that reflections of this dimension likely arise due to spacing of the aromatic groups (in this case, Fmoc) along the fibril axis. Furthermore, by inspecting the fluorenyl rings in the simulated structures, we observed a few well-recognized π-π stacking geometries (Figure 3E right),62–64 which likely explain the range of distances seen in the non-normalized RDF of fluorenyl rings.
By calculating the non-normalized RDF between the terminal alanine’s hydroxyl hydrogen and the center of the fibril (the “center” here is defined by a set of dummy atoms along the z-axis of the fibril, as shown Figure 3C, inset), the radius of the fibril can be estimated as a range between 1 to 1.5 nm as shown in Figure 3C. This radius range is also consistent with the experimental results obtained from our WAXS in Figure 3A, which shows a pronounced peak at 26.3 Å. Smith et al.18 observed a very similar value (26.0 Å) in their WAXS experiments using dried gel films of Fmoc-diphenylalanine.
Figure 3D shows the non-normalized RDF of the distance between adjacent peptide backbones (including α-carbons, amide nitrogens and amide carbons) along the axis of the assembled fiber. The predominant value is 0.46–0.48 nm, which corresponds well with typical antiparallel β-sheet chain-to-chain distances.65,66 However, we do not see a distinct reflection corresponding to this dimension in our WAXS results (Figure 3A). Given the broadness of the peak corresponding to d=4.35 Å in WAXS and the simulated non-normalized RDF of fluorenyl suggesting multiple preferred interaction distances from 4.1 – 5.0 Å, it is possible that a reflection corresponding to d=4.8 Å is present but convoluted with reflections arising from π-π stacking dimensions. While it is not present in our experimental data, the presence of this dimension in our non-normalized RDF calculations may serve to explain why others have observed a similar d-spacing and ascribed it to the presence of β-sheets. We further explored the secondary structures using FTIR experiments and conformational analysis of the simulations, as discussed below.
Analysis of hydrogen bonding patterns within assembled structure
Table 1 shows the hydrogen bonds distribution between the Fmoc, middle alanine (“Ala-mid”), and terminal alanine (“Ala-term”) residues of adjacent Fmoc-AA molecules during the last 50 ns of the simulations. We found that the carbonyl oxygen of the Fmoc carbamate (-OC(O)NH-) group is particularly active in forming hydrogen bonds with alanine residues, especially the terminal alanines on adjacent molecules. The high degree of interaction between the Fmoc carbamate and the terminal alanine residue may help explain why single residue Fmoc-protected amino acids with non-aromatic side chain groups have not, to our knowledge, been shown to form stable gels. The only examples of gel-forming single residue Fmoc-protected amino acids in literature are phenylalanine, tyrosine, and derivatives of the two67–69—all of which have side chain groups capable of π-π stacking interactions that likely contribute to fibril stability.
Table 1.
Hydrogen bonds distribution between individual residues on adjacent Fmoc-AA molecules. Each matrix adds to 100%.
| Cubic 4 | Fmoc | Ala-mid | Ala-term |
|---|---|---|---|
| Fmoc | 0 % | 28 % | 30 % |
| Ala-mid | 15 % | 22 % | |
| Ala-term | 5 % | ||
|
| |||
| Cubic 5 | Fmoc | Ala-mid | Ala-term |
|
| |||
| Fmoc | 0 % | 31 % | 26 % |
| Ala-mid | 19 % | 20 % | |
| Ala-term | 4 % | ||
|
| |||
| Cubic 6 | Fmoc | Ala-mid | Ala-term |
|
| |||
| Fmoc | 0 % | 17 % | 37 % |
| Ala-mid | 15 % | 22 % | |
| Ala-term | 9 % | ||
The number of hydrogen bonds is determined with a donor-acceptor distance ≤ 3.5 Å and interaction angle ≤ 30°. The middle and terminal alanines are named “Ala-mid” and “Ala-term”, respectively.
We also calculated the fraction of hydrogen bonds formed between Fmoc-AAs and the solvent (water) molecules, as shown in Table 2. The Fmoc group’s carbonyl oxygen is almost as active as the terminal alanine carboxyl group in making hydrogen bonds with the surrounding water molecules. These data suggest, different from Stupp’s proposed structure for the PA assembly,11 that in the fibril composed of short peptide conjugates like Fmoc-AA, the hydrophobic Fmoc groups are not completely buried within the core of the fiber. Instead, our simulations indicate that, although the Fmoc groups are mostly concentrated toward the core of the fiber, both the Fmoc carbamate and the aromatic fluorenyl groups are somewhat accessible to water molecules, making the fibril surface amphiphilic. This observation becomes quite apparent looking at Figure 2 (bottom illustrations); the highly hydrophobic fluorenyl rings (yellow) are exposed to the polar solvent molecules, as well as the hydrophilic alanine residues (blue), and thus the fibril surfaces are composed of both hydrophobic and hydrophilic components, as suggested in part by Smith et al.18 This property may also explain why fibrils can further assemble into fibers; interactions between hydrophobic regions on adjacent fibrils may force their aggregation into ribbon-like fibers.
Table 2.
Distribution of total hydrogen bonds formed between hydrogen bond-capable groups on Fmoc-AA molecules and water molecules. Each row adds to 100%.
| Fmoc-O | Ala-mid-O | Ala-mid-NH | Ala-term-O | Ala-term-NH | Ala-term-OH | |
|---|---|---|---|---|---|---|
| Cubic 4 | 25 % | 17 % | 6 % | 17 % | 10 % | 25 % |
| Cubic 5 | 25 % | 17 % | 5 % | 17 % | 11 % | 25 % |
| Cubic 6 | 27 % | 19 % | 7 % | 14 % | 11 % | 22 % |
Backbone conformational and secondary-structure analysis by FTIR and Ramachandran plots
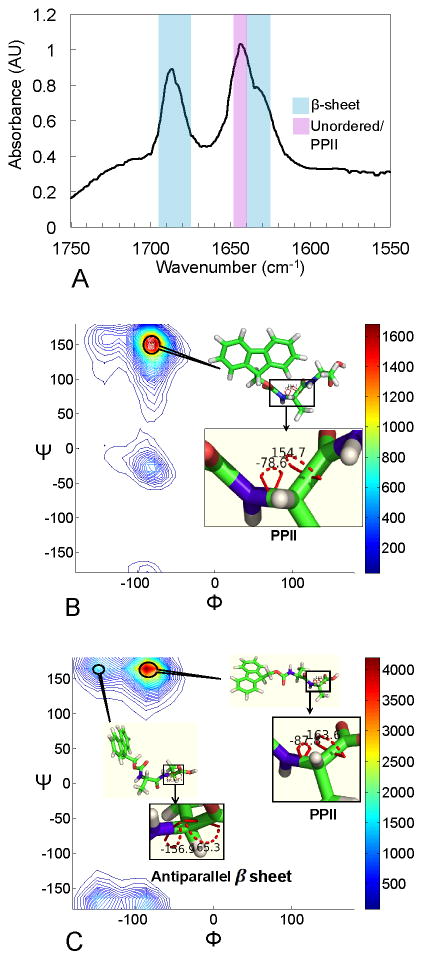
In transmission FTIR spectroscopy, Fmoc-AA gels formed from D2O show pronounced shifts in the amide I absorption region (~1600–1700 cm−1). Protein secondary structures involve specific hydrogen bonding interactions between amide bonds that shift the amide C=O stretch frequency resonance in an often-characteristic manner.70 FTIR can therefore be used to elucidate which secondary structures are most prevalent in a protein of unknown structure. Naturally, this technique has been applied to peptide-based self-assembled gel materials to probe for interactions that might point to certain secondary structures. Figure 4A shows the amide I absorption spectrum for an Fmoc-AA gel, with clear peaks centered at 1686 cm−1, 1644 cm−1, and a shoulder peak at 1634 cm−1. The 1686 cm−1 peak and 1634 cm−1 shoulder are within characteristic absorption frequency ranges seen in proteins with β-sheet content (blue overlay in Figure 4A).70 These peak locations and relative intensities are also quite consistent with previous FTIR data given for Fmoc-dipeptides.8,12,18,46 The large absorption at 1644 cm−1, similarly observed in related naphthalene-dipeptide gelator systems,47 lies outside the typical range of frequencies for β-sheet structures; instead, it falls within a frequency range indicative of hydrogen bonding within unordered protein structures (purple overlay)70 but the maximum absorption value itself is characteristic of polyproline-like hydrogen bonding interactions within collagen that are shown to decrease upon heating.71 The spectroscopic data alone would seem to indicate that hydrogen bonding interactions indicative of polyproline-like or random coil structure are present in numbers similar to or possibly even greater than those typical of β-sheet structures. However, the above analysis of hydrogen bonding patterns in our computational simulations showed a lack of significant β-sheet-like interactions (hydrogen bonding between amide groups on adjacent Fmoc-AA molecules).
Figure 4.
Experimental FTIR results and backbone conformational analysis (Ramachandran plot) of simulated assemblies show preferred conformations for Fmoc-AA molecules within fibril assembly. (A) FTIR amide I absorbance data give a peak at 1686 cm−1 and a shoulder at 1634 cm−1, similar to that seen in predominantly β-sheet proteins. Another strong absorbance at 1644 cm−1 correlates to random coil- or polyproline-like shift in amide C=O stretch frequency. (B) Backbone conformational analysis of middle alanine in Cubic 5 model during the last 50 ns of simulation; PPII-like peak is at φ,ψ = (−78°, 152°), β-sheet-like peak is at φ,ψ = (−78°, −23°). (C) Backbone conformational analysis of terminal alanine in Cubic 5 model during the last 50 ns of simulation, PPII-like peak is at φ,ψ = (−86°, 164°), β-sheet-like peak is at φ,ψ = (−148°, 164°). Results are representative of all simulations performed. In B and C, the color bar represents the population of residues in a range of angles defined by the contour lines.
Therefore, while the spectroscopic data suggest it is reasonable to construct a supramolecular model based on the assumption of β-sheet-like hydrogen bonding between amide bonds on adjacent Fmoc-AA molecules, computational data suggest that other significant interactions may be present and may point to an alternate model for the assembly. Given this contradiction and given that conjugate peptides are structurally quite distinct from long peptides and proteins, we comment that it may not always be appropriate to extrapolate from peptide/protein-level spectroscopic data to make structural conclusions about these conjugate peptide gel systems.
Analysis of the predominant peptide torsion angles indicative of specific secondary structures present in the simulated assemblies shows a strong preference for polyproline II (PPII) and antiparallel β-sheet-like conformations. The φ,ψ angles of each alanine residue for the population of Fmoc-AA molecules in a given simulation are illustrated with a Ramachandran plot for backbone conformational analysis, a classical approach to investigate the secondary structure of amino acids. In Figure 4B and C, Ramachandran plots of the middle and terminal alanines of our systems are calculated for the Cubic 5 model during the last 50 ns (results are representative of all fibril geometries simulated). Figure 4B demonstrates that most of the middle alanine (“Ala-mid”) residues are likely to adopt a PPII conformation: 64.1% of residues have φ,ψ angles in the range φ,ψ = (−100° to 0°, 100° to 180°) and a peak at φ,ψ = (−78°, 152°), whereas 10.6% of residues have backbone torsions in the range φ,ψ = (−180° to −100°, 100° to 180°) indicative of β-sheet conformation. For terminal alanine (“Ala-term”) residues, within the same φ,ψ ranges 54.7% have mostly PPII-like conformation, and 16.3% have β-sheet conformation. Figure 4C shows that the PPII conformation is preferred among the terminal alanine (“Ala-term”) residues. It is known that within the PPII structure, amide linkages preferably form hydrogen bonds with water rather than with other amide groups.72 Since our FTIR analysis showed distinct PPII character, we counted the hydrogen bonds of residues on Fmoc-AA (Fmoc, Ala-mid and Ala-term) with other Fmoc-AA versus with water (Table 3). We indeed observed that hydrogen-bond capable residues of Fmoc-AA all prefer to form hydrogen bonds with water rather than with other Fmoc-AA molecules even in the assembled form, further corroborating our computational observation of the alanine residues’ preference for adopting a PPII-like conformation.
Table 3.
Average number of hydrogen bonds between each residue and water, versus each residue and adjacent Fmoc-AA (Unit: Hydrogen bonds per residue)
| Fmoc···Fmoc-AA | Fmoc···water | Ala-mid···Fmoc-AA | Ala-mid···water | Ala-term···Fmoc-AA | Ala-term···water | |
|---|---|---|---|---|---|---|
| Cubic 4 | 0.59 | 1.00 | 0.66 | 1.06 | 0.58 | 2.23 |
| Cubic 5 | 0.58 | 1.00 | 0.71 | 1.03 | 0.51 | 2.28 |
| Cubic 6 | 0.51 | 1.05 | 0.51 | 1.09 | 0.66 | 1.95 |
“···” refers to the hydrogen bonding between the two objects/residues.
Besides the PPII conformation, the terminal alanine also displays a notable population at φ,ψ = (−148°, 164°), which is close to typical antiparallel β-sheet’s φ,ψ angles around (−140°, 135°). In Figure 3D, the distance between Fmoc-AA strands is presented using non-normalized RDF, and the peak at around 0.46–0.48 nm also suggest an β-sheet like conformation according to previous X-ray diffraction patterns studies of protein secondary structures 65,66. We did not observe a pronounced peak around 0.48 nm in our WAXS results in Figure 3A, but in our FTIR experiments we have observed absorbance shifts indicative both of β-sheet-like interactions between Fmoc-AA molecules and of PPII-like interactions with the solvent. While we observe β-sheet-like character in FTIR and in backbone conformational analysis of the simulated assemblies, our hydrogen bonding analysis shows that hydrogen bonding between peptides on adjacent strands (the mode of interaction for β-sheet structures) is not the predominant interaction. Rather, the Fmoc carbonyl oxygen and terminal alanine form hydrogen bonds most readily, both with each other and the surrounding water molecules. Clearly, hydrogen bonding plays a role in stabilizing the fibril structure; however, based on our data it becomes apparent that the importance of the β-sheet forming ability of these Fmoc-dipeptides in stabilizing the fiber assembly may have been previously overstated.
Conclusions
In this paper, we presented a computational molecular dynamics simulations alongside experimental analyses to gain new insight into the structure of the Fmoc-dipeptide supramolecular assembly. We have shown in our simulations that Fmoc-AA molecules assemble into well-defined fibril structures independent of starting conditions. Furthermore, we show that alternate molecular dynamics methods (REMD and SA) give nearly identical results, further confirming the convergence of the computational data. Our experimental data show that Fmoc-AA indeed assembles into gel-forming fiber structures. Computational analysis of characteristic dimensions for fibril diameter and π-π stacking interactions are consistent with results from WAXS experiments, and comparison of FTIR spectra with typical protein FTIR data suggests the presence of β-sheet- and polyproline II-like features. However, assigning typical secondary structure motifs to these short conjugated peptides is challenging. The alanine residues adopt mostly PPII conformations with limited β-sheet-like torsion angles, according to the simulations. From the analysis of hydrogen bonding patterns we find that the majority of hydrogen bonding interactions are between the Fmoc carbonyl oxygen and either the terminal alanine hydroxyl group or the surrounding water, potentially explaining why single Fmoc-protected amino acids with non-aromatic side chain groups have not been shown to form stable gels. It appears also that hydrogen bonding between peptide bonds on adjacent Fmoc-AA molecules is far less common, and therefore we argue that the putative β-sheet character observed in Fmoc-AA gels arises due to the preferential conformation of Fmoc-AA molecules within the assembled fibril and the chemical environment of the fibril, rather than from “typical” interactions specific to β-sheet strands. It is important to remember that no single spectroscopic analytical tool can provide information about both of the two main characteristics of protein secondary structure: hydrogen bonding and conformation (i.e. backbone torsion angles). Here, FTIR gives information about bond stretching affected by hydrogen bonding and CD gives information about sample chirality resulting from preferred molecular conformations. In the case of proteins and polypeptides with repeated secondary structural patterns, the hydrogen bonding effects on bond stretching and the sample chirality resulting from preferred backbone dihedrals are well characterized and can be correlated with secondary structure features thought or known to be present in the protein based on its primary sequence. However, Fmoc-dipeptides are far shorter, less complex, and contain non-native conjugate groups; therefore, they likely interact with each other in a manner distinct from peptide bonds within classical protein secondary structures. For this reason, it is our belief that a) researchers should exercise caution when extrapolating spectroscopic and other analytical data from pure protein solutions to small, conjugated peptide systems such as these, and b) computational tools can help us understand these systems where experimental analytical tools are unable to provide explicit details. By combining computational studies and experiments, we obtained missing critical insights into the supramolecular structure and mechanism of Fmoc-dipeptide self-assembly.
Finally, we show that hydrophobic groups, while mainly concentrated toward the center of the fibril, are partially exposed to the solvent and may therefore aid in aggregating fibrils into larger fibers seen in TEM, although the exact mechanism is unclear. It was reported by Tang et al.46 that there is a dependence on gel pH on fibril aggregation. However, current analytical techniques are unable to provide detailed supramolecular information that would give insight into this process. This “gap” may be addressed using computational models beyond atomic resolutions.
Fmoc-AA was chosen for comparison with the only other proposed structural model to date (also an Fmoc-dipeptide) and because its side chain uniformity and simplicity decrease the complexity of computational calculations performed during MD simulations. However, the knowledge and approaches established in this study will enable us to explore and rationally design novel self-assembling peptide-based materials that have bioactive sequences and biological functionality for biomedical applications.
Supplementary Material
Acknowledgments
This work was supported by the National Institutes of Health grant (R21 HL102806) and the National Science Foundation Teragrid MCB100057. We acknowledge L. Ricles for assistance with TEM and Dr. S. Swinnea for assistance with WAXS and helpful discussions. K.M.E. acknowledges the National Science Foundation for financial support through the Graduate Research Fellowship Program. X.M. is grateful to Dr. J. Zhang, Y. Shi, Dr. J. Fonner, Z. Xia, Dr. M. J. Schnieders and Dr. C. Yan for invaluable discussion and suggestions.
Footnotes
Author Contributions
P.R., L.J.S., and M.N. designed research. K.M.E. carried out experimental work, and X.M. carried out computational work. K.M.E. and X.M. wrote the manuscript. All authors have given approval to the final version of the manuscript.
Included in the Supporting Information are preliminary MD simulations of a previously proposed supramolecular structure for Fmoc-diphenylalanine, additional CD evidence of self-assembly, detailed information about simulation models, quantum mechanical characterization of the Fmoc group, and PDB files for the “Cubic 5” and topology files for GROMACS 4.5.4. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Whitesides GM, Mathias JP, Seto CT. Science. 1991;254:1312–1319. doi: 10.1126/science.1962191. [DOI] [PubMed] [Google Scholar]
- 2.Adams DJ, Topham PD. Soft Matter. 2010;6:3707–3721. [Google Scholar]
- 3.Aggeli A, Nyrkova IA, Bell M, Harding R, Carrick L, McLeish TCB, Semenov AN, Boden N. Proc Natl Acad Sci U S A. 2001;98:11857–11862. doi: 10.1073/pnas.191250198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Douglas SM, Dietz H, Liedl T, Hogberg B, Graf F, Shih WM. Nature. 2009;459:414–418. doi: 10.1038/nature08016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Estroff LA, Hamilton AD. Chem Rev. 2004;104:1201–1218. doi: 10.1021/cr0302049. [DOI] [PubMed] [Google Scholar]
- 6.Zhang S, Marini DM, Hwang W, Santoso S. Curr Opin Chemi Biol. 2002;6:865–871. doi: 10.1016/s1367-5931(02)00391-5. [DOI] [PubMed] [Google Scholar]
- 7.Jayawarna V, Richardson SM, Hirst AR, Hodson NW, Saiani A, Gough JE, Ulijn RV. Acta Biomater. 2009;5:934–943. doi: 10.1016/j.actbio.2009.01.006. [DOI] [PubMed] [Google Scholar]
- 8.Adams DJ. Macromol Biosci. 2011;11:160–173. doi: 10.1002/mabi.201000316. [DOI] [PubMed] [Google Scholar]
- 9.Hartgerink JD, Beniash E, Stupp SI. Science. 2001;294:1684–1688. doi: 10.1126/science.1063187. [DOI] [PubMed] [Google Scholar]
- 10.Cui H, Webber MJ, Stupp SI. Pept Sci. 2010;94:1–18. doi: 10.1002/bip.21328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lee OS, Stupp SI, Schatz GC. J Am Chem Soc. 2011;133:3677–3683. doi: 10.1021/ja110966y. [DOI] [PubMed] [Google Scholar]
- 12.Jayawarna V, Ali M, Jowitt TA, Miller AF, Saiani A, Gough JE, Ulijn RV. Adv Mater. 2006;18:611–614. [Google Scholar]
- 13.Li X, Kuang Y, Xu B. Soft Matter. 2012;8:2801–2806. doi: 10.1039/C2SM06920B. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Orbach R, Adler-Abramovich L, Zigerson S, Mironi-Harpaz I, Seliktar D, Gazit E. Biomacromolecules. 2009;10:2646–2651. doi: 10.1021/bm900584m. [DOI] [PubMed] [Google Scholar]
- 15.Adhikari B, Banerjee A. Soft Matter. 2011;7:9259–9266. [Google Scholar]
- 16.Adhikari B, Banerjee A. Chem – Eur J. 2010;16:13698–13705. doi: 10.1002/chem.201001240. [DOI] [PubMed] [Google Scholar]
- 17.Zhang Y, Kuang Y, Gao Y, Xu B. Langmuir. 2011;27:529–537. doi: 10.1021/la1020324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Smith AM, Williams RJ, Tang C, Coppo P, Collins RF, Turner ML, Saiani A, Ulijn RV. Adv Mater. 2008;20:37–41. [Google Scholar]
- 19.McCammon J, Gelin B, Karplus M. Nature. 1977;267:585–590. doi: 10.1038/267585a0. [DOI] [PubMed] [Google Scholar]
- 20.Karplus M, McCammon JA. Nat Struct Mol Biol. 2002;9:646–652. doi: 10.1038/nsb0902-646. [DOI] [PubMed] [Google Scholar]
- 21.Johnson RR, Johnson ATC, Klein ML. Nano Lett. 2007;8:69–75. doi: 10.1021/nl071909j. [DOI] [PubMed] [Google Scholar]
- 22.Sun S, Wong JTY, Zhang TY. Soft Matter. 2011;7:9307–9310. [Google Scholar]
- 23.Saha B, Shindo S, Irle S, Morokuma K. ACS Nano. 2009;3:2241–2257. doi: 10.1021/nn900494s. [DOI] [PubMed] [Google Scholar]
- 24.Castelletto V, Moulton CM, Cheng G, Hamley IW, Hicks MR, Rodger A, López-Pérez DE, Revilla-López G, Alemán C. Soft Matter. 2011;7:11405–11415. [Google Scholar]
- 25.Adams DJ, Morris K, Chen L, Serpell LC, Bacsa J, Day GM. Soft Matter. 2010;6:4144–4156. [Google Scholar]
- 26.Adams DJ, Mullen LM, Berta M, Chen L, Frith WJ. Soft Matter. 2010;6:1971–1980. [Google Scholar]
- 27.Adams DJ, Butler MF, Frith WJ, Kirkland M, Mullen L, Sanderson P. Soft Matter. 2009;5:1856. [Google Scholar]
- 28.Ponder JW. TINKER Version 6.0. Washington University; St. Louis: 2012. [Google Scholar]
- 29.Hess B, Kutzner C, van der Spoel D, Lindahl E. J Chem Theory Comput. 2008;4:435–447. doi: 10.1021/ct700301q. [DOI] [PubMed] [Google Scholar]
- 30.Jorgensen WL, Maxwell DS, Tirado-Rives J. J Am Chem Soc. 1996;118:11225–11236. [Google Scholar]
- 31.Jorgensen WL, Tirado-Rives J. Proc Natl Acad Sci USA. 2005;102:6665–6670. doi: 10.1073/pnas.0408037102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang J, King M, Suggs L, Ren P. Biomacromolecules. 2007;8:3015–3024. doi: 10.1021/bm070244c. [DOI] [PubMed] [Google Scholar]
- 33.Allinger NL, Yuh YH, Lii JH. J Am Chem Soc. 1989;111:8551–8566. [Google Scholar]
- 34.Jorgensen WL, Maxwell DS, Tirado-Rives J. J Am Chem Soc. 1996;118:11225–11236. [Google Scholar]
- 35.Kaminski GA, Friesner RA, Tirado-Rives J, Jorgensen WL. J Phys Chem B. 2001;105:6474–6487. [Google Scholar]
- 36.Hess B, Bekker H, Berendsen HJC, Fraaije JGEM. J Comput Chem. 1997;18:1463–1472. [Google Scholar]
- 37.Hess B. J Chem Theory Comput. 2007;4:116–122. doi: 10.1021/ct700200b. [DOI] [PubMed] [Google Scholar]
- 38.Darden T, York D, Pedersen L. J Chem Phys. 1993;98:10089–10092. [Google Scholar]
- 39.Bussi G, Donadio D, Parrinello M. J Chem Phys. 2007;126:014101–7. doi: 10.1063/1.2408420. [DOI] [PubMed] [Google Scholar]
- 40.Parrinello M, Rahman A. J Appl Phys. 1981;52:7182–7190. [Google Scholar]
- 41.Van Gunsteren WF, Berendsen HJC. Mol Simul. 1988;1:173–185. [Google Scholar]
- 42.Jorgensen WL, Chandrasekhar J, Madura JD, Impey RW, Klein ML. J Chem Phys. 1983;79:926–935. [Google Scholar]
- 43.Smith DK. Chem Soc Rev. 2009;38:684–694. doi: 10.1039/b800409a. [DOI] [PubMed] [Google Scholar]
- 44.Nordén B, Rodger A, Dafforn T. Linear Dichroism and Circular Dichroism: A Textbook on Polarized-Light Spectroscopy. Royal Society of Chemistry; 2010. [Google Scholar]
- 45.Zhang Y, Gu H, Yang Z, Xu B. J Am Chem Soc. 2003;125:13680–13681. doi: 10.1021/ja036817k. [DOI] [PubMed] [Google Scholar]
- 46.Kirkpatrick S, Gelatt CD, Vecchi MP. Science. 1983;220:671–680. doi: 10.1126/science.220.4598.671. [DOI] [PubMed] [Google Scholar]
- 47.Sugita Y, Okamoto Y. Chem Phys Lett. 1999;314:141–151. [Google Scholar]
- 48.García AE, Onuchic JN. Proc Natl Acad Sci USA. 2003;100:13898–13903. doi: 10.1073/pnas.2335541100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nymeyer H, Gnanakaran S, García AE. Numerical Computer Methods, Part D. Vol. 383. Academic Press; 2004. pp. 119–149. [DOI] [PubMed] [Google Scholar]
- 50.Zhou R. AIP Conference Proceedings. 2003;690:406–408. [Google Scholar]
- 51.Zhou R. Protein Folding Protocols. 2006;350:205–223. [Google Scholar]
- 52.Johnson RR, Kohlmeyer A, Johnson ATC, Klein ML. Nano Lett. 2009;9:537–541. doi: 10.1021/nl802645d. [DOI] [PubMed] [Google Scholar]
- 53.Wolf MG, Jongejan JA, Laman JD, de Leeuw SW. J Phys Chem B. 2008;112:13493–13498. doi: 10.1021/jp804285e. [DOI] [PubMed] [Google Scholar]
- 54.Teplow DB, Lazo ND, Bitan G, Bernstein S, Wyttenbach T, Bowers MT, Baumketner A, Shea JE, Urbanc B, Cruz L, Borreguero J, Stanley HE. Acc Chem Res. 2006;39:635–645. doi: 10.1021/ar050063s. [DOI] [PubMed] [Google Scholar]
- 55.Richmond TJ. J Mol Biol. 1984;178:63–89. doi: 10.1016/0022-2836(84)90231-6. [DOI] [PubMed] [Google Scholar]
- 56.Ren P, Ponder JW. J Phys Chem B. 2003;107:5933–5947. [Google Scholar]
- 57.Windle AH. Pure Appl Chem. 1985;57:1627–1638. [Google Scholar]
- 58.Houton KA, Morris KL, Chen L, Schmidtmann M, Jones JTA, Serpell LC, Lloyd GO, Adams DJ. Langmuir. 2012;28:9797–9806. doi: 10.1021/la301371q. [DOI] [PubMed] [Google Scholar]
- 59.Chen L, Morris K, Laybourn A, Elias D, Hicks MR, Rodger A, Serpell L, Adams DJ. Langmuir. 2009;26:5232–5242. doi: 10.1021/la903694a. [DOI] [PubMed] [Google Scholar]
- 60.Braun HG, Cardoso AZ. Colloids Surf, B. 2012;97:43–50. doi: 10.1016/j.colsurfb.2012.03.028. [DOI] [PubMed] [Google Scholar]
- 61.Gazit E. FASEB J. 2002;16:77–83. doi: 10.1096/fj.01-0442hyp. [DOI] [PubMed] [Google Scholar]
- 62.McGaughey GB, Gagné M, Rappé AK. J Biol Chem. 1998;273:15458–15463. doi: 10.1074/jbc.273.25.15458. [DOI] [PubMed] [Google Scholar]
- 63.Sun S, Bernstein ER. J Phys Chem. 1996;100:13348–13366. [Google Scholar]
- 64.Tycko R. Biochemistry. 2003;42:3151–3159. doi: 10.1021/bi027378p. [DOI] [PubMed] [Google Scholar]
- 65.Chan JCC, Oyler NA, Yau WM, Tycko R. Biochemistry. 2005;44:10669–10680. doi: 10.1021/bi050724t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ryan DM, Anderson SB, Nilsson BL. Soft Matter. 2010;6:3220–3231. [Google Scholar]
- 67.Yang Z, Gu H, Fu D, Gao P, Lam JK, Xu B. Adv Mater. 2004;16:1440–1444. [Google Scholar]
- 68.Sutton S, Campbell NL, Cooper AI, Kirkland M, Frith WJ, Adams DJ. Langmuir. 2009;25:10285–10291. doi: 10.1021/la9011058. [DOI] [PubMed] [Google Scholar]
- 69.Jackson M, Mantsch HH. Crit Rev Biochem Mol Biol. 1995;30:95–120. doi: 10.3109/10409239509085140. [DOI] [PubMed] [Google Scholar]
- 70.Tang C, Smith AM, Collins RF, Ulijn RV, Saiani A. Langmuir. 2009;25:9447–9453. doi: 10.1021/la900653q. [DOI] [PubMed] [Google Scholar]
- 71.Bryan MA, Brauner JW, Anderle G, Flach CR, Brodsky B, Mendelsohn R. J Am Chem Soc. 2007;129:7877–7884. doi: 10.1021/ja071154i. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Lakshminarayanan R, Fan D, Du C, Moradian-Oldak J. Biophys J. 2007;93:3664–3674. doi: 10.1529/biophysj.107.113936. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.