Abstract
Pot1 is a single-stranded-DNA-binding protein that recognizes telomeric G-strand DNA. It is essential for telomere capping in Saccharomyces pombe and regulates telomere length in humans. Human Pot1 also interacts with proteins that bind the duplex region of the telomeric tract. Thus, like Cdc13 from S. cerevisiae, Pot 1 may have multiple roles at the telomere. We show here that endogenous chicken Pot1 (cPot1) is present at telomeres during periods of the cell cycle when t loops are thought to be present. Since cPot1 can bind internal loops and directly adjacent DNA-binding sites, it is likely to fully coat and protect both G-strand overhangs and the displaced G strand of a t loop. The minimum binding site of cPot1 is double that of the S. pombe DNA-binding domain. Although cPot can self associate, dimerization is not required for DNA binding and hence does not explain the binding-site duplication. Instead, the DNA-binding domain appears to be extended to contain a second binding motif in addition to the conserved oligonucleotide-oligosaccharide (OB) fold present in other G-strand-binding proteins. This second motif could be another OB fold. Although dimerization is inefficient in vitro, it may be regulated in vivo and could promote association with other telomere proteins and/or telomere compaction.
Telomeric DNA is composed of a length of duplex repeated sequence that terminates in a single-strand overhang on the 3′ G-rich strand. It is packaged by proteins that bind to the single- and double-strand regions of the telomeric tract (33) to form a complex that acts as a protective cap over the end of the chromosome. This cap hides the DNA terminus from nucleases and the DNA repair activities that would otherwise detect the terminus as a double-strand break and promote chromosome fusions or signal cell cycle arrest (6, 8, 14). However, the cap must also allow regulated access to the DNA terminus by telomerase, the specialized reverse transcriptase that adds new repeats onto the telomeric G strand, and to other activities that are needed to maintain the telomeric DNA.
This regulated access seems to be achieved both via the active recruitment of telomerase and other replication enzymes to the telomere (16) and by the telomere cycling between closed and open states (6). The closed, protective structure is thought to render the DNA terminus inaccessible during much of the cell cycle, while the open structure would make it available to telomerase during S phase. In some organisms, the closed structure seems to involve folding of the DNA to form a lariat-like structure (termed a t loop) on the chromosome end (21). T loops result from the 3′ overhang invading the duplex region of the telomeric tract, an event that is promoted by the duplex telomere-binding protein TRF2 (43).
3′ Overhangs range in length from 14 nucleotides (nt) in ciliates to 150 to 350 nt in mammals and are present at both chromosome ends (49). The overhangs are an important aspect of telomere structure because they allow telomerase to maintain both telomeres and because they provide a substrate for the specialized G-strand-binding proteins that are an essential part of the protective cap. These G-strand-binding proteins include Cdc13 from budding yeast, TEBP (telomere end-binding protein) from the ciliates Oxytricha and Euplotes, and Pot1 (protection of telomeres), a protein that is present in vertebrates, plants, and fungi (49). TEBP, Cdc13, and Pot1 are functional and, to a certain extent, structural homologs that are each required for telomere end protection (45).
The Oxytricha TEBP is a dimeric protein that can bind DNA either as an αβ heterodimer or as an α2 homodimer (18, 38). In the crystal structure of the αβ-DNA complex, the terminal 12 nt of telomeric DNA lie in a cleft between the α and β subunits, which explains why the protein is so effective at protecting the DNA terminus (23). The DNA-binding surface is made up of a series of oligonucleotide-oligosaccharide (OB)-binding folds (11, 23, 35, 36). The α subunit contains three such folds, while the β subunit has one. The two N-terminal folds from the α subunit cooperate to make the DNA-binding surface in the α2 homodimer, while these same folds interact with the β subunit fold to form the binding surface in the αβ heterodimer. The OB fold seems to be a structurally conserved motif that is generally used by G-overhang-binding proteins. The only significant region of sequence identity between the Oxytricha TEBP and the Pot1 protein family corresponds to the N-terminal fold of the TEBP α subunit (3). In Saccharomyces pombe Pot1, this conserved region serves as the DNA-binding domain and the X-ray crystal structure indicates that it exists as an OB fold (29, 30). Although Cdc13 shares little sequence identity with TEBP or Pot1, the DNA-binding domain is also comprised of an OB fold that is structurally similar to the N-terminal fold of the TEBP α subunit (34, 45).
In vivo studies with S. cerevisiae have revealed that Cdc13 is an essential protein that is required for telomere protection, with loss of Cdc13 resulting in extensive degradation of the telomeric C strand (19). Cdc13 also functions in telomerase recruitment, telomerase repression, and coordination of G- and C-strand synthesis (9, 37). It achieves these multiple functions by binding the G-strand overhang and acting as a landing pad that recruits a series of unique protein complexes that each perform a different task (32). Although the Pot1 protein family has not been as extensively characterized as Cdc13, these proteins also function in telomere protection and/or length regulation. Deletion of the S. pombe Pot1 gene results in a cell division defect leading to elongated cells that fail to divide further and a high incidence of chromosome missegregation (3). Those cells that survive undergo rapid loss of telomeric and subtelomeric sequences, followed by chromosome circularization. In mammalian cells, Pot1 localizes to telomeres and loss of the G-strand overhang causes a reduction in Pot1 binding (4, 31). Overexpression of either the full-length protein or a C-terminal fragment lacking the DNA-binding domain can cause rapid telomere lengthening, indicating that Pot1 somehow regulates telomerase access to the DNA terminus (12, 31).
To learn more about how Pot1 functions in telomere protection and length regulation in vertebrates, we have identified and characterized chicken Pot1 (cPot1). We chose to study the chicken protein because the chicken DT40 cell line provides an excellent system for making gene disruptions, due its high level of homologous recombination. Thus, we may be able to correlate future studies of Pot1 knockout cells with information from the present biochemical analysis. The work described here examines the subcellular location of cPot1 throughout the cell cycle and its binding specificity and binding-domain architecture. Our studies indicate that cPot1 is likely to bind both to G-strand overhangs when they are exposed during S phase and to the internal regions of single-stranded G-strand DNA that are thought to be present during much of the cell cycle as a result of t-loop formation. We also show that cPot1 requires two full and directly adjacent telomeric repeats for binding. Although the protein can self associate to form dimers or multimers, the DNA-binding surface is unlikely to be composed of OB folds from two separate subunits because dimerization is inefficient and is not required for DNA binding. We speculate that the binding surface may instead be composed of two tandem OB folds that cooperate to form one long binding groove.
MATERIALS AND METHODS
Library screening and 5′ RACE.
The cPot1 gene was isolated from a λZAPII chicken embryonic fibroblast cDNA library as recommended by the manufacturer (Strategene) by using the human Pot1 (hPot1) DNA-binding domain as a probe (3). cPot1 genomic DNA was isolated from a λFix II library containing DNA from the DT40 cell line. Sequence alignments were performed by using the GCG Wisconsin package (Accelrys, San Diego, Calif.). RNA for 5′ rapid amplification of cDNA ends (RACE) was obtained by lysing DT40 cells in guanidinium thiocyanate followed by extraction with acidic phenol. 5′ RACE was performed with the GeneRacer kit (Invitrogen) by using a cPot1 gene-specific reverse primer, cPot1-R1 (5′-CATCCACTTTTGCCTTCCCTAC-3′), and a universal 5′ nested primer (5′-GGACACTGACATGGACTGAAGGAGTA-3′). The products were cloned with a TOPO cloning kit (Invitrogen) and sequenced.
Antibody production and cPot1 detection.
A 5′ fragment of the cPot1 cDNA encoding the extra N-terminal domain (END) and DNA-binding domain (amino acids 1 to 253) was cloned into pQE30 (QIAGEN) and expressed in Escherichia coli M15 (QIAGEN). His-tagged cPot1 protein was purified by using Ni2+-charged chelating Sepharose (Amersham Pharmacia). Polyclonal antibody was generated in rabbits (Covance) and purified by using a cPot1 affinity column. The cTRF1 antibody was also generated in rabbits by using purified cTRF1 as previously described (15). Nuclei were released from DT40 cells (ATCC CRL-2111) by homogenization in 10 mM HEPES (pH 7.9)-10 mM KCl-1.5 mM MgCl2-20% glycerol-1 mM dithiothreitol-0.5 mM phenylmethylsulfonyl fluoride-104 μM AEBSF (4-[2-aminoethyl]benzeuesulfonyl fluoride hydrochloride)-0.08 μM aprotinin-2.1 μ M leupeptin-3.6 μM bestatin-1.5 μM pepstatin A-1.4 μM E-64 and collected by centrifugation. The isolated nuclei were resuspended in sodium dodecyl sulfate (SDS) sample buffer and sonicated prior to loading on an SDS gel.
Indirect immunofluorescence.
Subcellular localization of cPot1 was performed by using chicken DT40 and LMH (ATCC CRL-2117) cells as previously described (42). In brief, DT40 cells were treated with RSB buffer (10 mM Tris [pH 7.4], 10 mM NaCl, and 5 mM MgCl2), fixed with 3.7% formaldehyde for 10 min, and then centrifuged onto Alcian Blue-treated coverslips. The LMH cells were grown and fixed directly on collagen-coated coverslips. Cells were then permeabilized with 0.5% IGEPAL CA-630 (NP-40) for 20 min. To prepare metaphase chromosome spreads, DT40 cells were treated with colcimid (0.1 μg/ml) for 90 min prior to the addition of RSB buffer. Endogenous cPot1 and cTRF1 were detected with affinity-purified polyclonal cPot1 or cTRF1 antibody and fluorescein isothiocyanate-conjugated secondary antibody (Jackson ImmunoResearch). DNA was stained with DAPI (4′,6′-diamidino-2-phenylindole) (0.2 μg/ml). Cells were photographed with a SPOT digital camera. To examine cPot1-TRF1 colocalization, DT40 and LMH cells were transfected with pTET-Flag-hTRF1 or pCMV-Flag-hTRF1. Expressed hTRF1 was detected with mouse anti-Flag antibody (M2) and Cy5-conjugated secondary antibody (Jackson ImmunoResearch) by using a Zeiss LSM 510 confocal microscope. hTRF1 was used instead of cTRF1 because the chicken cDNA may be missing a few bases from the 5′ end (15).
To examine whether Pot1 is present at telomeres during early or late S phase and G2/M phase, DT40 cells were cultured in the presence of aphidicolin (5 μg/ml) for 12 h. The cells were then washed to remove the drug, grown for 0 to 4 h under normal culture conditions (RPMI supplemented with 10% fetal calf serum, 1% [vol/vol] chicken serum, 50 μM 2-mercaptoethanol, 100 U of penicillin/ml, 50 μg of streptomycin/ml, and 2 mM glutamine), and fixed as described above. Fluorescence-activated cell sorter analyses were performed to determine the synchrony of the culture.
Yeast two-hybrid analysis.
Full-length and truncated cPot1 cDNAs were cloned into yeast two-hybrid vectors pACT2 (Clontech) and/or pBTM116 (2) to generate either a Gal4 activation domain (GAD) fusion (GAD-cPot1) or a LexA DNA-binding domain fusion (LexA-cPot1). Full-length cTRF1, cTRF2, cRap1, cKu70, and the ankyrin domain of chicken tankyrase 1 (amino acids 370 to 736) were also cloned into the pACT2 and pMTM116 vectors to generate GAD and LexA fusion proteins. Constructs were transformed into the S. cerevisiae strain L40 (22) and selected on SD plates without leucine or tryptophan (−Leu/−Trp). To screen for interactions, yeast cells containing the relevant plasmids were grown on SD plates −Leu/−Trp/−His with 30 mM 3-aminotriazole (3-AT). β-Galactosidase activities were measured by liquid assay as previously described (15) by using o-nitrophenol-β-d-galactopyranoside as the substrate. Constructs that failed to give a positive result in the two-hybrid assay were checked for protein expression by Western blotting of yeast extracts.
Protein expression and glutathione S-transferase (GST) pull downs.
Full-length cPot1 was cloned into the baculovirus expression vector pFastBac1 (Invitrogen) and expressed in insect Sf21 cells. His-cPot1 protein was purified by using Ni2+-charged Sepharose. To prepare GST-cPot1 fusion, full-length cPot1 was cloned into the vector pGEX4T2 (Amersham Pharmacia) in frame with GST. The recombinant GST-cPot1 was expressed in E. coli BL21 (DE3) pLysS and allowed to attach to glutathione Sepharose (Amersham Pharmacia). The beads were washed and then resuspended in pull-down buffer (phosphate-buffered saline, 10% glycerol, 1 mM dithiothreitol, 0.1% Triton X, 0.5 mM phenylmethylsulfonyl fluoride, 104 μM AEBSF, 0.08 μM aprotinin, 2.1 μM leupeptin, 3.6 μM bestatin, 1.5 μM pepstatin A, 1.4 μM E-64). For GST pull-down analysis, 20 pmol of purified His-cPot1 was incubated with 40 pmol of Sepharose-bound GST-cPot1, Sepharose-bound GST, or protein-free glutathione Sepharose beads for 2 h at 4°C. The beads were washed with pull-down buffer and resuspended in SDS sample buffer before being loaded on SDS gels.
DNA-binding assays.
Mobility shift assays were performed as previously described (39). In brief, oligonucleotides were 5′ end labeled, and the relative labeling efficiency was determined by DE81 assay or PhosphorImaging. Each sample was then adjusted to a standard counts per minute per picomole by addition of cold oligonucleotide. 32P-labeled oligonucleotide (0.2 pmol) was incubated with ∼10 pmol of purified His-tagged cPot1 for 60 min at 37°C in 10 mM Tris [pH 8.0]-15 mM EGTA-100 mM NaCl-1 μg sonicated calf thymus DNA, and the sample was then loaded on 4 to 20% or 8% acrylamide gels. In competition experiments, the cold competitor was either mixed with labeled TelG oligonucleotide prior to addition of cPot1 or the cold competitor was incubated with cPot1 for 15 min prior to addition of the labeled oligonucleotide.
To determine the fraction of protein able to bind DNA, 2.3 pmol of cPot1 (the protein concentration was estimated by comparison to bovine serum albumin standards) was incubated with 0.1 to 3.5 pmol of 32P-labeled minimum binding site (MBS) oligonucleotide, and the products were separated in 4 to 20% gels and quantified by PhosphorImager. To convert the PhosphorImager values to molar concentrations, a standard curve was generated for each experiment by loading known quantities of 32P-labeled MBS oligonucleotide on gels and exposing the gels to the same PhosphorImager cassette as the gels containing the DNA/protein complex and free DNA.
RESULTS
Identification of cPot1.
The cPot1 gene was isolated by screening a chicken cDNA library with a probe corresponding to the DNA-binding domain of the hPot1 protein (3). cDNAs were isolated from several positive plaques, and the longest was found to contain a 2,334-bp open reading frame (ORF), 144-bp 5′ untranslated region (UTR), and 1,572-bp 3′ UTR. To ensure that the first ATG in the cDNA was the true start codon, we performed 5′ RACE by using a primer located downstream of the putative DNA-binding domain (see below). Sequencing of the products identified an additional 43 bp of 5′ UTR. Although two ATGs were located within the resulting 187-nt 5′ UTR, they were both followed by stop codons.
The open reading frame (ORF) encodes a protein of 778 amino acids that is clearly recognizable as a Pot1 homolog, as it has 61% overall sequence identity with hPot1 and 75% identity within the region corresponding to the DNA-binding domain (Fig. 1A and B) (3, 4). The main difference between the chicken, human, and mouse Pot1 proteins is that cPot1 has a 144-amino-acid extension at the N terminus, termed the END. This extra domain is unlikely to be an artifact created by template switching during cDNA synthesis because it was found in three independent cDNA clones and was present in the 5′ RACE products. It also occurs in a cPot1 expressed sequence tag that contains the adjacent DNA-binding domain and is present on genomic DNA restriction fragments that contain the DNA-binding domain (data not shown).
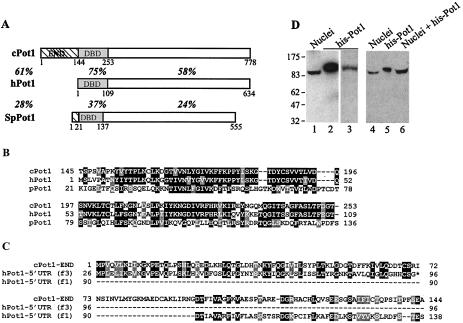
FIG. 1.
Identification of cPot1. (A) Domain structure of chicken, human, and S. pombe Pot1. The box marked with diagonal lines is the END, the shaded box represents the DNA-binding domain (DBD), and the open box is the C-terminal domain. The overall amino acid sequence identity between the human and chicken or human and S. pombe proteins is shown to the left, and the identity between domains is shown below each domain. (B) Sequence alignment of the S. pombe DNA-binding domain with the equivalent regions of human and chicken Pot1. (C) Comparison of the cPot1 END to conceptually translated regions of the hPot1 5′ UTR. Numbers beside the cPot1 sequence indicate the number of amino acids from the initiating methionine. Numbers beside each hPot1 sequence represent the number of codons from the 5′ end of the UTR. The upper alignment is with the 71-codon ORF translated in reading frame 3; the lower alignment is with the adjacent region of 5′ UTR translated in reading frame 1. The star represents a stop codon. (D) Lanes 1, 2, and 4 to 6, Western blots with cPot1 antibody showing endogenous and recombinant cPot1; lanes 1 and 4, nuclei isolated from chicken DT40 cells; lanes 2 and 5, purified His-tagged cPot1 expressed in baculovirus; lane 6, mix of nuclear proteins and recombinant Pot1; lane 3, purified His-tagged cPot1 stained with Coomassie blue.
The cPot1 gene resembles hPot1 in that the 5′ UTR contains multiple ATGs that are followed by in-frame stop codons (4). However, the cPot1 gene has only two such ATGs, while the hPot1 gene has nine, so the start codon that gives rise to full-length hPot1 is preceded by a series of short ORFs. Conceptual translation of the longest (71-codon) ORF and the adjacent region of the hPot1 5′ UTR revealed a surprising degree of sequence similarity to portions of the chicken END. The ORF has 38% identity and 49% similarity, while the adjacent region of 5′ UTR has 24% identity and 42% similarity (Fig. 1C). Overall, these two regions of the hPot1 5′ UTR align with a sequence encoding ∼120 out of 144 amino acids in the cPot1 END. This finding suggests that the cPot1 END has been lost from mammalian Pot1 during evolution. Translation of the hPot1 5′ UTR to generate an END would require complex frame-shifting events, and there is no evidence that this occurs (4). Moreover, the mouse Pot1 gene has a much shorter 5′ UTR that lacks obvious sequence identity to the cPot1 END. Thus, loss of the domain from hPot1 appears to be the result of mutations that prevent it from being translated, while in mice the END has been completely removed from the gene, probably by a chromosomal rearrangement (41).
Although hPot1 transcripts are subject to alternative splicing reactions that generate mRNAs encoding variants that have quite different sizes and DNA binding affinities (4), we did not see any evidence of alternative splicing in the three cPot1 cDNA clones that were sequenced. We were also unable to detect splice variants when we examined the endogenous cPot1 protein by Western blotting. Antibody was generated against a fragment of recombinant cPot1 that included both the END and the DNA-binding domain, so it should be able to detect translation products equivalent to those generated by four out of the five human splice variants. When affinity-purified antibody was used to perform Western blotting with either whole cells or nuclei from chicken DT40 cells, only a single band was observed. Thus, splice variants must be absent from, or expressed at very low levels in, this cell line (Fig. 1D, lanes 1 and 4 and data not shown). Although purified recombinant Pot1 migrated more slowly than the endogenous protein, this difference in mobility was probably caused by a combination of the His tag and the higher concentration of total protein in the nuclear preparation containing endogenous Pot1. When the purified protein was mixed with total nuclear proteins, the recombinant and endogenous proteins migrated together (Fig. 1D, lanes 4 to 6), indicating that the antibody is specific for cPot1.
Subcellular localization of endogenous cPot1 during the cell cycle.
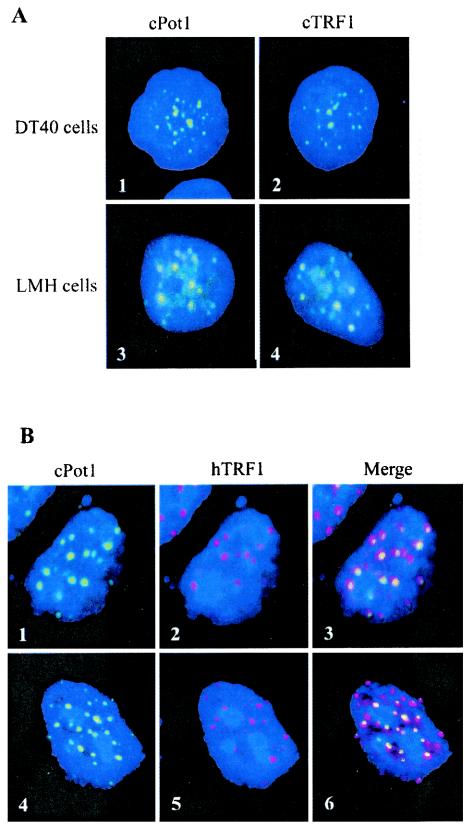
Since transiently transfected hPot1 had been show to localize to telomeres (4), we next checked whether the endogenous chicken protein also had a telomeric location. Both the DT40 B-cell line and LMH hepatocytes were fixed and incubated with antibody to either cPot1 or the duplex telomeric DNA-binding protein cTRF1 (15). As shown in Fig. 2A, the two antibodies gave rise to a similar punctate pattern of nuclear staining, suggesting that cPot1 was also present at telomeres. To further examine the localization of cPot1, we transiently expressed Flag-tagged hTRF1 in DT40 and LMH cells and determined the degree of Pot1 and TRF1 colocalization by using a mouse monoclonal antibody to the TRF1 Flag tag and the rabbit polyclonal cPot1 antibody. When the Pot1 and TRF1 staining patterns were overlaid, it was apparent that much of the cPot1 colocalized with TRF1 (Fig. 2B and data not shown), indicating that cPot1 is indeed present at telomeres.
FIG. 2.
cPot1 colocalizes with TRF1 at telomeres. (A) Panels 1 and 2, chicken DT40 cells; panels 3 and 4, LMH cells stained with antibody to cPot1 (1 and 3) or cTRF1 (2 and 4). (B) Confocal microscopic images showing hTRF1 and cPot1 colocalization. Chicken LMH cells were transiently transfected with a Flag-tagged hTRF1 expression construct and then were fixed and stained with antibody to cPot1 (1 and 4) or the Flag tag (2 and 5). Panels 3 and 6 show an overlay of the cPot1 and hTRF1 staining.
Although human and yeast Pot1 have the appropriate substrate specificity to bind telomeric G-strand overhangs, the likelihood that the G overhangs are sequestered into t loops for much of the cell cycle has raised questions about the timing of Pot1 association (3). T-loop formation should render an overhang inaccessible to Pot1, but invasion of the overhang into the duplex region of the telomeric tract appears to cause displacement of a more internal region of the G strand. Since it was unclear whether this displacement loop would also be a substrate for Pot1 binding, we set out to determine whether cPot1 binds to telomeres only during S phase, when t loops become dissociated, or whether it is also present at telomeres during the rest of the cell cycle. To achieve this, we first stained unsynchronized cells with antibody to cPot1 or cTRF1 and counted the number of cells with punctate (i.e., telomeric) signals. There was very little difference in the number of cells with punctate cPot1 and cTRF1 staining (96 versus 94%) (Fig. 3A), suggesting that there was little cell cycle-related difference in the relative location of the two proteins.
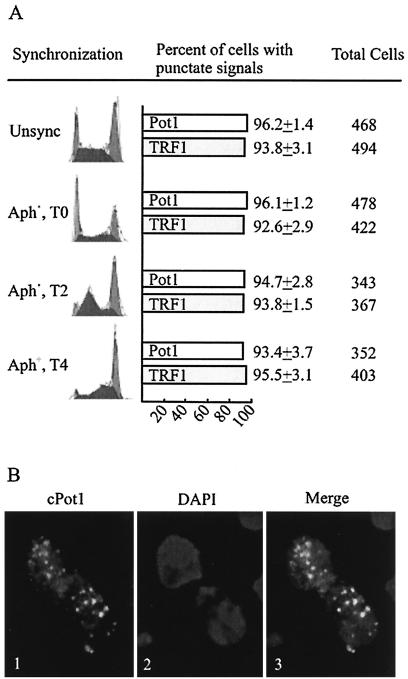
FIG. 3.
cPot1 is present at telomeres throughout interphase. (A) Histogram showing the percentage of cells with punctate cPot1 and cTRF1 staining. Cells were synchronized with aphidicolin, removed from the drug, and isolated 0, 2, or 4 h later. Synchronized and unsynchronized (unsync) cells were then fixed and stained with antibody to cPot1 or cTRF1. The fluorescence-activated cell sorter analysis for each population of cells is shown to the left. The total number of cells counted for each sample is shown to the right. Aph+, aphidicolin treated; T0, time zero; T2, and T4, 2 and 4 h, respectively, after aphidicolin removal. (B) cPot1 staining in G1 cells. Panel 1, cPot1 antibody; panel 2, DAPI; panel 3, overlay of cPot1 and DAPI.
To look more closely at cells in S phase and G2, cultures were blocked in early S phase with aphidicolin, released, and harvested 0, 2, or 4 h later. When the cells were processed for indirect immunofluorescence, there was again little difference in the fraction of cells that exhibited punctate cPot1 and cTRF1 staining regardless of whether they were in early or mid-S phase or late S/G2. Moreover, when the pattern of cPot1 staining was compared between the unsynchronized and various S phase/G2-enriched cultures, no significant difference was observed in the number of cells with punctate staining (Fig. 3A). Since telomeres appear to be replicated throughout S phase (50), our analysis would not detect the transient release of cPot1 while individual telomeres are being replicated. Nonetheless, our results indicate that cPot1 is present at telomeres throughout most of S and G2 of the cell cycle.
To determine whether Pot1 is also present at telomeres during metaphase and early G1, as opposed to the G1/S boundary (the stage of the aphidicolin block), cells at these stages in the cell cycle were identified by microscopy and examined for telomeric staining. Newly divided G1 cells exhibited a punctate staining pattern similar to that of other interphase cells (Fig. 3B), indicating that Pot1 remains associated with telomeric DNA throughout interphase. Thus, cPot1 is present at telomeres during stages of the cell cycle when t loops are also thought to be present. Although cPot1 staining could sometimes be seen in metaphase cells, the signal was frequently weak, and staining was absent from many of the cells even when we tried to maximize chromatin decondensation by swelling the cells prior to fixation or digesting them briefly with micrococcal nuclease (data not shown). It is possible that the reason for the decreased cPot1 staining is that compaction of the chromosomes during metaphase renders the protein inaccessible to the antibody. However, since cTRF1 staining was not noticeably decreased at this time (data not shown), it is also possible that some of the Pot1 is released from the chromosome.
cPot1 self association.
Telomere-binding proteins frequently interact with themselves and/or other telomere proteins to form large multiprotein complexes (15, 17, 37, 40, 52). To determine whether this is also the case for cPot1, we used a yeast two-hybrid analysis to look for self association and/or interaction with cTRF1, cTRF2, cRap1, cKu70, or the ankyrin repeat domain of tankyrase 1. The various telomere proteins were expressed both as GAD and LexA DNA-binding-domain fusions, and interactions were detected by growth on selection plates. Growth was observed in cells expressing the cPot1 activation- and binding-domain fusions (data not shown), suggesting that the protein can form homodimers or multimers. In contrast, no interaction was detected between Pot1 and any of the other telomere proteins (data not shown). As hPot1 has recently been shown to interact with the TRF1/tankyrase/Tin2 complex (31), the lack of association between cPot1 and cTRF1 or tankyrase 1 is somewhat unexpected. However, the native interaction could be between cPot1 and cTin2 (the chicken Tin2 gene has not yet been isolated) or with a region of tankyrase outside the ankyrin domain.
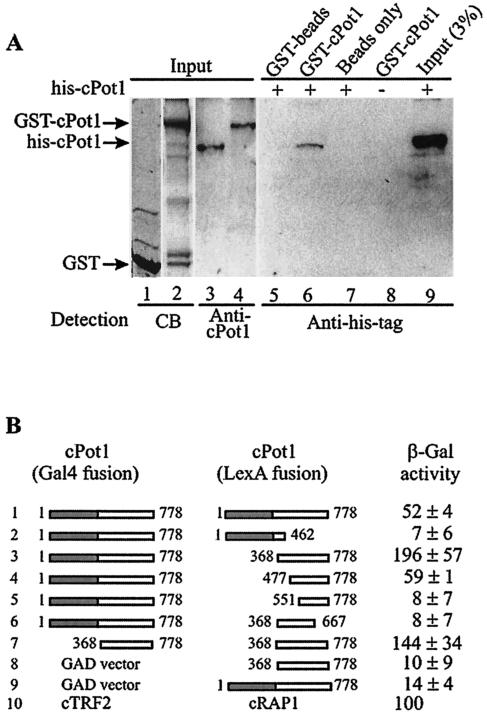
To confirm the cPot1 self association, we performed pull-down experiments with recombinant His-tagged Pot1 and a GST-Pot1 fusion protein. When His-tagged Pot1 was incubated with the GST-cPot1 bound to glutathione beads, the His-tagged protein copurified with the beads (Fig. 4A, lane 6), which did not occur when the His-tagged protein was incubated with GST bound to beads or the beads alone (Fig. 4A, lanes 5 and 7), indicating that the Pot1-Pot1 self association was specific.
FIG. 4.
cPot1 self associates via the C-terminal domain. (A) Western blots showing association between His-tagged cPot1 and a GST-cPot1 fusion protein. Lanes 1 and 2, Coomassie blue stain of protein released from the GST and GST-cPot1 beads; lanes 3 to 4 and 9, input His-tagged Pot1 or GST-Pot1 fusion protein; lanes 5 to 8, His-tagged cPot precipitated by glutathione beads coupled to GST, GST-cPot1 fusion protein, or beads alone. Detection was with Pot1 antibody (lanes 3 to 4) or His-tagged antibody (lanes 5 to 9). (B) Two-hybrid assay for interactions between different regions of cPot1. The pairs of DNA-binding domain and activation domain constructs are illustrated on the left, and the β-galactosidase activity is indicated at the right. The Pot1 END plus DNA-binding domain is shaded gray, and the C-terminal domain is unshaded. The values expressed are relative to that of the cTRF2/cRap1 control (construct set 10) and represent the average of the results obtained with two independent transformants.
We next used the two-hybrid assay to map the domain responsible for cPot1 self association. We generated a series of LexA-cPot1 fusion constructs lacking various portions of Pot1 and tested for their association with the full-length Gal4-cPot1 fusion protein (Fig. 4B). Interactions were detected between an ∼400-amino-acid C-terminal Pot1 fragment and the full-length protein (construct set 3). Interaction was also detected when the same C-terminal fragment was expressed from both the GAD and LexA vectors (set 7). Interestingly, this interaction was consistently stronger than that observed with the full-length protein, suggesting that the N-terminal portion of the protein may exert an inhibitory effect. The interaction between C-terminal fragments was reduced by removal of a further ∼100 N-terminal amino acids and abolished by removal of the C-terminal ∼100 amino acids (sets 4 and 6). No interaction was observed between the full-length protein and a fragment containing the END plus the DNA-binding domain (set 2). Thus, it appears that cPot1 self associates through portions of the C-terminal domain. This domain is fairly conserved in the vertebrate Pot1 proteins but is diverged in the related ciliate TEBPs.
cPot1 DNA-binding specificity.
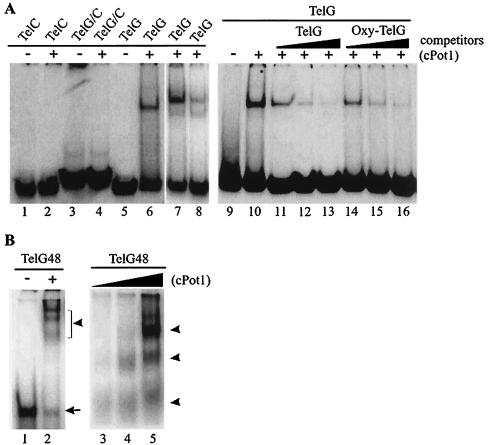
To examine the binding specificity of cPot1, purified protein (Fig. 1D, lane 3) was incubated with a variety of telomeric and nontelomeric substrate oligonucleotides, and binding was monitored by mobility shift assay. The protein-bound oligonucleotides corresponded to extended stretches of the chicken telomeric G strand (T2AG3) but not to the telomeric C strand, duplex telomeric DNA, or nontelomeric DNA (Fig. 5A, Table 1, and data not shown). Although binding was specific for G-strand DNA, the protein did not display an absolute preference for the vertebrate telomeric sequence (T2AG3) because binding was competed by an excess of cold T4G4 oligonucleotide corresponding to Oxytricha telomeric DNA (Fig. 5A, lanes 14 to 16). Thus, cPot1 resembles hPot1 and the Oxytricha TEBP in that it binds preferentially to its own telomeric DNA sequence but will bind variations of this sequence with somewhat reduced affinity (3, 47). Interestingly, binding to the labeled TelG oligonucleotide was not completely abolished by 1,000× the competitor of cold TelG even when the cold competitor was added prior to the hot oligonucleotide (data not shown), probably because the increase in total amount of TelG drives more of the cPot1 to bind DNA. Thus, while the ratio of hot to cold TelG present in Pot1-DNA complexes decreases with increasing cold competitor, the overall increase in the amount of complex means a significant amount of labeled DNA is still present.
FIG. 5.
cPot1 binds to G-strand telomeric DNA with low cooperativity. Mobility shift gels with various telomeric oligonucleotides incubated with (+) or without (−) purified cPot1 are shown. (A) Lanes 1 and 2, telomeric C-strand DNA (TelC); lanes 3 and 4, DNA duplex formed from telomeric G- and C-strand DNA (TelG/C); lanes 5 to 16, telomeric G strand DNA (TelG); lanes 9 to 16, binding to labeled TelG in the presence of 100-, 500-, and 1,000-fold excesses of cold TelG (lanes 11 to 13) or Oxytricha telomeric DNA (OxyTelG, lanes 14 to 16). The binding reactions shown in lanes 1 to 6 and 9 to 16 contained 0.25 pmol of hot oligonucleotide and 10 pmol of cPot1; those in lanes 7 and 8 contained 0.4 and 6.4 pmol of TelG, respectively, and 2 pmol cPot1. (B) A constant amount of TelG48 was incubated with increasing amounts of cPot1. Lanes 1 and 2, a 4 to 20% gradient gel with electrophoresis time limited to retain the unbound DNA on the gel; lanes 3 to 5, an 8% gel with electrophoresis time extended to separate the DNA-protein complexes. The arrow marks unbound oligonucleotide, and the arrowhead marks DNA-protein complexes. The binding reaction shown in 1ane 2 contained 0.2 pmol of TelG48 and 2.5 pmol of cPot1; those shown in lanes 3 to 5 contained 0.4 pmol of TelG48 and 0.3, 0.6, or 3 pmol of cPot1, respectively.
TABLE 1.
Oligonucleotides used in electrophoretic mobility shift assays
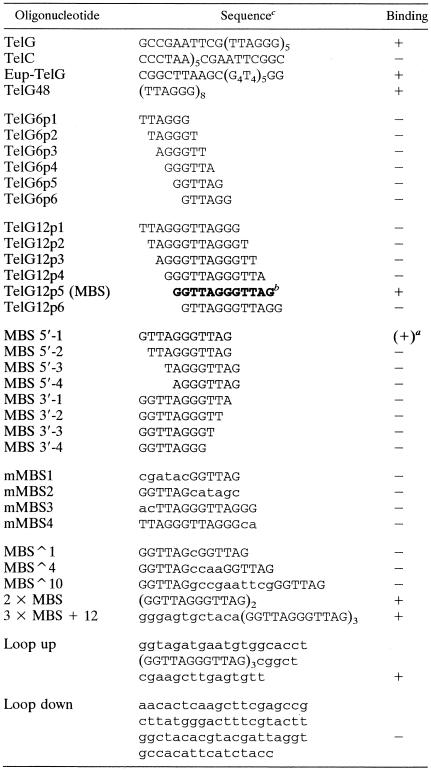
(+), weak binding.
The minimum binding site.
Lowercase letters indicate nontelomeric sequences.
The purified DNA-binding domain from S. pombe Pot1 binds G-strand DNA in a highly cooperative manner, so only one shifted band is observed when the protein is incubated with an oligonucleotide that has multiple Pot1 binding sites (29). In contrast, binding of the full-length chicken protein to a G-strand oligonucleotide with five T2AG3 repeats gave rise to either one or two DNA protein complexes, depending on the concentrations of DNA and protein used in the binding reaction (Fig. 5A, lanes 6 to 8). Since the cPot1 was largely full length (Fig. 1D), this observation suggested that binding of cPot1 might be noncooperative. To test this possibility, increasing amounts of cPot1 were incubated with a constant amount of G-strand oligonucleotide that contained eight T2AG3 repeats (Table 1, TelG48). As shown in Fig. 5B, three to four different DNA-protein complexes could be seen, indicating that the number of Pot1 molecules bound to each oligonucleotide molecule was variable. Thus, unlike the purified S. pombe DNA-binding domain, binding of full-length cPot1 is largely noncooperative.
Size of the minimum DNA-binding site.
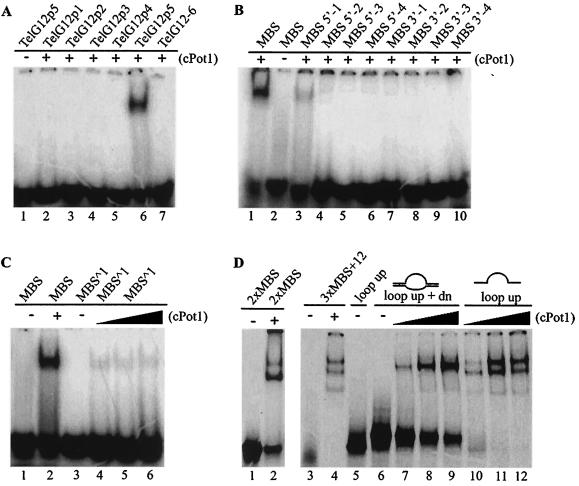
Although the S. pombe Pot1 DNA-binding domain binds tightly to a substrate of only six nucleotides (29), the binding site of the full-length cPot1 protein appeared to be longer because a maximum of two different DNA protein complexes were visible after binding to the G-strand oligonucleotide that contained five T2AG3 repeats (Fig. 5A, lanes 7 and 8, TelG). We therefore tested whether cPot1 could bind to G-strand oligonucleotides that contained all six possible permutations of a single or double telomeric repeat (Table 1). No binding was observed with any permutation of the single repeat (data not shown) or with five of the six permutations of the double repeat (Fig. 6A). However, cPot1 did bind to the double repeat corresponding to the sequence GGTTAGGGTTAG (Fig. 6A, lane 6, TelG12p5). Thus, the binding site of full-length cPot1 appears to be approximately double that of the purified S. pombe DNA-binding domain. Double-stranded-DNA-binding proteins commonly use dimerization as a strategy to double the size of their binding sites, and some OB fold-containing single-stranded-DNA-binding proteins also use this approach to extend their DNA-binding surfaces (46). Thus, since the DNA-binding domain of S. pombe Pot1 forms a single OB fold (30), it seemed likely that the extended binding site of cPot1 might be the result of homodimerization placing the OB folds from two separate subunits over adjacent 6-nt repeats. If this were the case, the isolated cPot1 DNA-binding domain might also be able to bind a single telomeric repeat. Unfortunately, we were unable to test this possibility because multiple attempts to express a region containing the predicted DNA-binding domain yielded only insoluble protein. We therefore examined whether two complete GGTTAG repeats are required for cPot1 binding, as might be expected if the full binding domain is made up of two copies of a single DNA-binding motif. Since the permutation of the repeat was clearly important (Fig. 6A), we used oligonucleotides that had from 1 to 4 nt removed from either the 5′ end (MBS 5′ series) or the 3′ end (MBS 3′ series) of the GGTTAGGGTTAG double repeat but maintained the correct permutation at the other end. Binding was reduced ∼80% by removal of only one nucleotide from the 5′ end (Fig. 6B, lane 3) and completely eliminated by removal of two or more nucleotides from the 5′ end or one or more nucleotides from the 3′ end (Fig. 6B, lanes 4 to 10). Binding was also eliminated if the final length of the oligonucleotide was maintained at 12 nt but 2 or 6 nt from one of the two telomeric repeats was changed to a nontelomeric sequence (Table 1, mMBS1 to MBS4). Thus, the minimum binding site (MBS) for cPot1 is composed of two complete GGTTAG repeats, supporting the idea that the full DNA-binding domain is composed of a duplicated DNA-binding motif.
FIG. 6.
cPot1 binding-site size and spacing. (A and B) The cPot1 minimum binding site is 12 nt. (A) Mobility shift gel with the six different permutations of the 12-nt double repeat (TelG12p1 to TelG12p6) incubated with (+) or without (−) cPot1. (B) MBS oligonucleotides were incubated with (+) or without (−) cPot1. MBS is the same as TelG12p5, MBS 5′-1 to MBS 5′-4 have 1 to 4 nucleotides removed from the 5′ end of the MBS double repeat, and MBS 3′-1 to MBS 3′-4 have 1 to 4 nucleotides removed from the 3′ end. Binding reactions shown in panels A and B contained 0.2 pmol of labeled oligonucleotide and 10 pmol of cPot1. (C and D) cPot1 can bind adjacent MBSs and DNA loops but not split half sites. (C) Mobility shift gels showing reduced cPot1 binding to oligonucleotides containing the MBS, with one nucleotide inserted between the two GGTTAG half sites. Binding reactions contained 0.2 pmol of labeled oligonucleotide and 9.2 pmol (lanes 2 and 5), 4.6 pmol (lane 4), or 18.4 pmol (lane 6) of cPot1. (D) Binding of cPot1 to oligonucleotides containing two adjacent MBSs (lanes 1 and 2, 2× MBS), three adjacent MBSs (lanes 3 and 4, 3× MBS plus 12), the looped substrate containing three MBSs in the loop (lanes 6 to 9, loop up plus down [dn]), or the upper strand of the loop (lanes 5 and 10 to 12, loop up). Binding reactions contained 0.2 pmol of labeled oligonucleotide and 10 pmol (lanes 2, 4, 8, and 11), 5 pmol (lanes 7 and 10), or 15 pmol (lanes 9 and 12) of cPot1.
To probe the architecture of the cPot1 DNA-binding domain, we next examined whether the two GGTTAG repeats have to lie directly adjacent to each other or whether they can be separated by one or more nucleotides. Although most homodimeric double-stranded-DNA-binding proteins recognize a palindromic sequence, a few resemble cPot1 in that they bind direct repeats (5, 26). Studies of these proteins have revealed that half-site spacing requirements can be indicative of the way in which this is achieved. For example, the telomere protein TRF1 has a flexible linker between the DNA-binding and the dimerization domains, so the DNA-binding domain can swivel relative to the dimerization domain (5). This flexibility in the linker region allows binding to the nonpalindromic telomeric sequence, but it leads to a corresponding flexibility in half-site spacing on the DNA such that the two half sites can be separated by a variable number of nucleotides. In contrast, the transcription factor HapI has an extensive and highly asymmetric dimerization interface that places the two DNA-binding surfaces a defined distance apart so that no variability is tolerated in half-site spacing (26).
To test for flexibility between the two halves of the cPot1 binding motif, oligonucleotides that had either 4 or 10 nt of nontelomeric sequence inserted between the two GGTTAG repeats (Table 1, MBŜ4 and MBŜ10) were used to compete for binding to the 12-nt MBS oligonucleotide. As no competition was detected (data not shown), we performed a direct binding assay with four times the standard concentration of cPot1 to evaluate binding to both the oligonucleotide with the 4-nt insertion and one with a 1-nt insertion (MBŜ1). Again binding was undetectable with the 4-nt insertion, and it was reduced by >95% with the 1-nt insertion (Fig. 6C and data not shown). This result indicates that the two halves of the cPot1 DNA-binding domain must be anchored together in a rigid manner to form one long, inflexible DNA-binding surface. This rigidity in the DNA-binding domain could be achieved either by having a single extended DNA-binding surface or through formation of a rigid but asymmetric dimer or multimer that brings the DNA-binding domains of two subunits into close proximity.
cPot1 dimerization is not required for DNA binding.
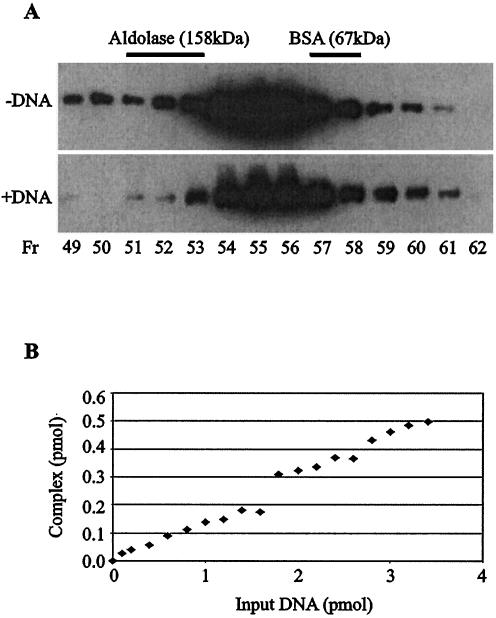
To address more directly whether cPot1 binds DNA as a monomer or a dimer, we used gel filtration to examine the extent of dimerization in the presence and absence of DNA. As predicted from the two-hybrid and pull-down assays, we were able to detect cPot1 in fractions that would be expected to contain 172-kDa dimers. However, the dimeric form of cPot1 represented only a few percent total protein (Fig. 7A). This was true even when the cPot1 was prebound to a 10- to 15-fold molar excess of MBS oligonucleotide, suggesting that dimerization was not driven by DNA binding. As the small amount of dimer could result from a high rate of subunit exchange with resultant complex dissociation during the course of the experiment, we next tried to detect cPot1 dimerization by cross-linking with bis(sulfosuccinimidyl)suberate (BS3). The cross-linking yielded some large cPot1 complexes, but again the fraction of dimers (or multimers) to monomers was low and was not changed significantly by binding the protein to the MBS oligonucleotide prior to cross-linking (data not shown). We therefore conclude that cPot1 self association is quite inefficient even in the presence of telomeric DNA. This result is consistent with our observations in the GST pull-down experiments, where less than 1% of the input His-tagged cPot1 copurified with the GST-cPot1 beads (Fig. 4).
FIG. 7.
cPot1 can bind DNA as a monomer. (A) Fractionation of cPot1 on a Superdex 200 gel filtration column. Upper panel, purified His-tagged cPot1; lower panel, purified cPot1 that had been incubated with a 12-fold molar excess of MBS oligonucleotide in one-half times PBS. cPot1 was detected by Western blotting with cPot1 antibody. Peak fractions containing the marker proteins bovine serum albumin and aldolase are indicated with bars. Fr, fraction. (B) Titration curve for cPot1 binding to the MBS oligonucleotide. The plot shows the quantity of complex-formed DNA versus the amount of input DNA that accumulated when increasing amounts of labeled MBS oligonucleotide were incubated with a fixed amount (2.3 pmol) of cPot1.
Although the above results indicated that our binding reactions contained only a low concentration of cPot1 dimers, it remained possible that this concentration was sufficient to provide the observed cPot1 DNA-binding activity. If cPot1 resembles Cdc13 in having a dissociation constant in the 0.1- to 10-nM range (1, 25), less than 1.5% of the input protein would have to form dimers in order to shift 50% of the MBS oligonucleotide. To determine the fraction of protein molecules that were active for DNA binding compared to the fraction that formed dimers, we performed a series of titrations to measure how much of the input protein could bind DNA when increasing amounts of 32P-labeled MBS oligonucleotide were added to a set (200 ng or 2.3 pmol) amount of purified cPot1. The cPot1/MBS complexes were separated from free DNA in mobility shift gels, and bound oligonucleotide was quantified by using a PhosphorImager. As cPot1 binds the MBS oligonucleotide at a 1:1 molar ratio (only one band shift is observed in mobility shift assays), the moles of bound DNA equal the moles of bound cPot1. Although we were not able to obtain full saturation of the cPot 1 because the apparent Kd is quite high (see below), we were routinely able to titrate 10 to 20% of the protein into a complex with the MBS oligonucleotide. As these experiments were performed with protein preparations where only a few percent molecules existed as dimers (Fig. 7A), this result means that most of the cPot1 must have bound to DNA as a monomer.
Although it is possible to determine the Kd of a protein from titration curves plotting the amount of DNA-protein complex formed as a function of DNA concentration (the concentration of free DNA at half saturation is equal to the Kd), we were unable to determine an accurate Kd for cPot1 because of our inability to attain full saturation. When data from the beginning of each binding curve were fit to the Hill equation by using a two-parameter fit, we obtained Kds that were substantially in excess of 100 nM, the approximate Kd for SpPot1 (3, 29). However, these values are likely to overestimate the true Kd because there was a loss of total labeled DNA due to smearing of the signal in the mobility shift gels, which is a known problem with gel shift assays that can lead to aberrantly low apparent affinities (10).
Binding to adjacent binding sites and internal loops.
As cPot1 is proposed to bind and protect G-strand DNA either at G-strand overhangs or at t loops (4, 12, 31), we next tested how closely cPot1 molecules can pack along a length of G-strand DNA and whether the protein can bind to stretches of GGTTAG repeats that are part of a structure designed to mimic a t loop. To examine the spacing of cPot1 binding, we determined whether multiple protein molecules could bind an oligonucleotide that had two or three MBSs directly adjacent to each other. As shown in Fig. 6D, assays with the 2× MBS oligonucleotide gave rise to bands corresponding to both one and two protein molecules per DNA molecule (Fig. 6D, lanes 1 and 2), while a 3× MBS oligonucleotide that had an additional 12 nt of nontelomeric DNA at the 5′ end gave rise to three bands (Fig. 6D, lanes 3 and 4). These results indicate that the protein can bind to directly adjacent binding sites.
The substrate mimicking a t loop was made by annealing two partially complementary oligonucleotides to generate a loop that contained three minimum binding sites on the upper strand, nontelomeric DNA on the lower strand, and 20-bp nontelomeric duplex DNA on either side of the loop (Table 1 and Fig. 6D, diagram). A 10-fold excess of lower-strand oligonucleotide was used to ensure complete hybridization of the upper MBS-containing oligonucleotide, and the efficiency of duplex formation was monitored by looking for a decrease in mobility in the gel shift assay (Fig. 6D, compare lanes 5 and 6). As shown in Fig. 6D, lanes 7 to 9, cPot1 was clearly able to bind to the loop substrate, although the efficiency of binding was reduced relative to that of the upper-strand oligonucleotide. Thus, while binding is enhanced by the presence of single-stranded DNA at either the 5′ or 3′ end, it is not required for the protein to load onto an internal stretch of GGTTAG repeats. Our results therefore indicate that although cPot1 binding lacks cooperativity, the protein has the capacity to fully coat and hence protect any pair of GGTTAG repeats regardless of whether they are present at the G-strand overhang or on the displaced strand of a t loop.
DISCUSSION
Although the members of the Pot1 protein family are required for telomere end protection and appear to be functional homologs of Cdc13, we presently have very limited information about how these proteins exert their protective function or whether they play additional roles at telomeres. To broaden our understanding of Pot1 function, we have identified the chicken protein and have examined its distribution throughout the cell cycle and its DNA-binding specificity and binding-domain organization. As has been observed for other telomere proteins, we have found that the domain structure and overall amino acid sequence of Pot1 is quite well conserved between chickens and mammals (15, 27, 44), indicating that protein function is also likely to be similar. The main difference between the mammalian and avian proteins is that cPot1 has an extra 144 amino acids at the N terminus. This N-terminal extension appears to have been lost from mammalian Pot1 during evolution, because its remnants still exist in the 5′ UTR of the human gene.
One question we have examined concerns the timing of Pot1 binding to the telomere. Because the telomeres of vertebrate cells appear to cycle between a closed conformation where the G overhang is incorporated into a t loop and an open structure where the overhang is more accessible, it was unclear whether Pot1 would bind to telomeres throughout the cell cycle. Our localization studies indicate that cPot1 is present at telomeres throughout most, if not all, of interphase. This means that Pot1 is bound to telomeres during stages of the cell cycle when the G-strand overhangs are thought to be incorporated into t loops. Consequently, cPot1 is unlikely to function only during DNA replication when t loops are dismantled. Since we have also shown that cPot1 binds telomeric repeats at internal loops as well as at DNA termini and can fully coat a molecule bearing multiple binding sites, our results indicate that cPot1 is likely to protect not only G-strand overhangs but also the G-strand DNA that is displaced when t loops are formed.
Although cPot1 clearly binds single-stranded telomeric DNA with high specificity, several aspects of the immunofluorescence-staining pattern were unexpected for a protein that binds only to G-strand overhangs or displaced G-strand DNA. First, the intensity of staining was only slightly less than that seen with the TRF1 antibody (Fig. 2A). As TRF1 binds along the length of the 5- to 9-kb duplex telomeric tract, much-weaker staining would be anticipated for a protein that binds solely to the short G-strand overhangs (24, 48). Second, the degree of overlap between the cPot1 and hTRF1 staining was surprisingly high, because although TRF1 would be expected to bind the many large blocks of interstitial T2AG3 repeats that are present on chicken chromosomes (28, 48), this would not be expected for a G-overhang-binding protein. These two observations suggested that Pot1 might not be localized only at the telomeric G-strand overhang or on displaced G-strand DNA but that it may also be present along duplex regions of T2AG3 repeats. This possibility is supported by a recent report indicating that human Pot1 interacts with proteins that associate with the duplex region of the telomeric tract (the TRF1/Tin2/tankyrase complex) and that a hPot1 mutant that is missing the DNA-binding domain is still able to localize to telomeres (31). As neither the human nor the chicken Pot1 proteins can bind directly to duplex telomeric DNA, Pot1 is unlikely to be directly responsible for telomere-end protection when present at duplex regions of the telomeric tract. Instead, Pot1 may resemble Cdc13 in that it plays different roles in telomere maintenance, depending on its location or binding partners, which might explain the reduced intensity of cPot1 staining at metaphase. The protein associated with the duplex region of the telomeric tract might be preferentially released from the telomere or assume a very different conformation at this time so that it is rendered inaccessible to the Pot1 antibody.
Our finding that cPot1 needs a full 12 nt for binding was unexpected because the purified DNA-binding domain from S. pombe Pot1 requires only a single 6-nt telomeric repeat (29). The much larger binding-site size of the chicken protein could reflect the presence of a more extended DNA-binding surface on each cPot1 subunit, or it could result from homodimer or multimer formation positioning the DNA-binding motifs from two separate subunits over adjacent GGTTAG repeats. The ability of cPot1 to self associate together with the perfect doubling of the binding site compared to that of the S. pombe DNA-binding domain initially favored the dimerization-multimerization model. However, when we examined the extent of cPot multimerization, it became clear that most of the protein exists as a monomer and that there was insufficient dimer or multimer to give the level of binding observed in our assays. Thus, cPot1 can bind DNA as a monomer and hence must have an expanded DNA-binding surface relative to the isolated S. pombe DNA-binding domain.
One way the size of the DNA-binding surface of an OB fold can be expanded is by extending the length of one or more of the loops that connect the five β sheets that form the core of the fold (46). This strategy is observed in Cdc13, where a 30-amino-acid loop connecting β2-β3 provides much of the surface used to recognize the 11-nt binding site (34). However, it is unlikely to be the cause of the large cPot1 DNA-binding surface, because sequence alignment indicates that the conserved region predicted to form the OB fold in the S. pombe protein lacks amino acid insertions in the chicken protein (Fig. 1B). Thus, the expanded DNA-binding surface of the chicken protein is likely to be formed from portions of the protein that lie outside the main region of sequence homology between the Oxytricha TEBP and the yeast and vertebrate Pot1 proteins.
While the structural elements that extend the cPot1 DNA-binding surface remain to be identified, one interesting possibility is that the protein contains a second OB fold that cooperates with the OB fold in the conserved region to make a single larger DNA-binding surface. Such tandem OB folds are seen in the Oxytricha TEBP, human replication protein A, and BRCA2, where they cooperate to form a single, long, DNA-binding groove (7, 11, 23, 51). The existence of tandem OB folds in cPot1 would fit with our finding that the protein binds two directly adjacent telomeric repeats, and it could explain why attempts to express portions of the protein predicted to contain only the first (conserved) OB fold were unsuccessful. However, structural analysis will be needed to test this hypothesis, because the family of OB folds involved in nucleic acid binding lack discernible sequence similarity and hence cannot be identified on the basis of primary sequence (35, 46).
Given that dimerization is not a requirement for DNA binding, the function of the cPot1 self association is presently unclear. However, such self association is not without precedent among G-overhang-binding proteins, because the α subunit of the Oxytricha TEBP forms α2 dimers in which the two DNA-binding domains remain independent and spatially separate (36). Formation of the α2 dimer is thought to be a step in a specific assembly pathway that results in formation of the α/β/DNA ternary complex (11). Positioning of the DNA in the binding groove of the α/β heterodimer requires cofolding of the protein subunits with the DNA, and the α2 dimer is thought to promote β subunit association and subsequent cofolding. Since cPot1 is likely to interact with a variety of telomere proteins (31, 32), it is possible that dimerization serves a similar chaperone-like function to promote these associations. An alternative but not mutually exclusive possibility is that dimerization is important for telomere compaction or clustering. cPot1 might compact regions of G-strand DNA within a telomere in a manner analogous to the compaction of the duplex telomeric tract by TRF1 (20). Alternatively, like the S. pombe telomere protein Taz1, cPot1 might be required to bring together the termini of sister chromatids or even separate chromosomes (13). Given that cPot1 dimerization is so inefficient in vitro and appears to be inhibited by the N-terminal domain, in vivo dimerization and hence the resultant downstream events may well be subject to some form of regulation.
Acknowledgments
We thank Ming Tan for his help with the baculovirus system, Saugata Ray for assistance with the two hybrid screens, and Mark Rance for help with the binding analysis. We are grateful to Yolanda Sanchez, Carol Caperelli, Iain Cartwright, and members of the Price lab for helpful comments. pTET-Flag-hTRF1 and pCMV-Flag-hTRF1 were kindly provided by Titia deLange and Dominique Broccoli, respectively.
This work was supported by NIH grant AG17212 to C.M.P.
REFERENCES
- 1.Anderson, E. M., W. A. Halsey, and D. S. Wuttke. 2002. Delineation of the high-affinity single-stranded telomeric DNA-binding domain of Saccharomyces cerevisiae Cdc13. Nucleic Acids Res. 30:4305-4313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bartel, P., C. T. Chien, R. Sternglanz, and S. Fields. 1993. Elimination of false positives that arise in using the two-hybrid system. BioTechniques 14:920-924. [PubMed] [Google Scholar]
- 3.Baumann, P., and T. R. Cech. 2001. Pot1, the putative telomere end-binding protein in fission yeast and humans. Science 292:1171-1175. [DOI] [PubMed] [Google Scholar]
- 4.Baumann, P., E. Podell, and T. R. Cech. 2002. Human Pot1 (protection of telomeres) protein: cytolocalization, gene structure, and alternative splicing. Mol. Cell. Biol. 22:8079-8087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bianchi, A., R. M. Stansel, L. Fairall, J. D. Griffith, D. Rhodes, and T. de Lange. 1999. TRF1 binds a bipartite telomeric site with extreme spatial flexibility. EMBO J. 18:5735-5744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Blackburn, E. H. 2001. Switching and signaling at the telomere. Cell 106:661-673. [DOI] [PubMed] [Google Scholar]
- 7.Bochkarev, A., R. A. Pfuetzner, A. M. Edwards, and L. Frappier. 1997. Structure of the single-stranded-DNA-binding domain of replication protein A bound to DNA. Nature 385:176-181. [DOI] [PubMed] [Google Scholar]
- 8.Chan, S. W., and E. H. Blackburn. 2002. New ways not to make ends meet: telomerase, DNA damage proteins and heterochromatin. Oncogene 21:553-563. [DOI] [PubMed] [Google Scholar]
- 9.Chandra, A., T. R. Hughes, C. I. Nugent, and V. Lundblad. 2001. Cdc13 both positively and negatively regulates telomere replication. Genes Dev. 15:404-414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Classen, S., D. Lyons, T. R. Cech, and S. C. Schultz. 2003. Sequence-specific and 3′-end selective single-strand DNA binding by the Oxytricha nova telomere end binding protein alpha subunit. Biochemistry 42:9269-9277. [DOI] [PubMed] [Google Scholar]
- 11.Classen, S., J. A. Ruggles, and S. C. Schultz. 2001. Crystal structure of the N-terminal domain of Oxytricha nova telomere end-binding protein alpha subunit both uncomplexed and complexed with telomeric ssDNA. J. Mol. Bio. 314:1113-1125. [DOI] [PubMed] [Google Scholar]
- 12.Colgin, L. M., K. Baran, P. Baumann, T. R. Cech, and R. R. Reddel. 2003. Human POT1 facilitates telomere elongation by telomerase. Curr. Biol. 13:942-946. [DOI] [PubMed] [Google Scholar]
- 13.Cooper, J. P., Y. Watanabe, and P. Nurse. 1998. Fission yeast Taz1 protein is required for meiotic telomere clustering and recombination. Nature 392:828-831. [DOI] [PubMed] [Google Scholar]
- 14.de Lange, T. 2002. Protection of mammalian telomeres. Oncogene 21:532-540. [DOI] [PubMed] [Google Scholar]
- 15.De Rycker, M., R. N. Venkatesan, C. Wei, and C. M. Price. 2003. Vertebrate tankyrase domain structure and sterile alpha motif (SAM)-mediated multimerization. Biochem. J. 372:87-96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Evans, S. K., and V. Lundblad. 2000. Positive and negative regulation of telomerase access to the telomere. J. Cell Sci. 113:3357-3364. [DOI] [PubMed] [Google Scholar]
- 17.Fairall, L., L. Chapman, H. Moss, T. de Lange, and D. Rhodes. 2001. Structure of the TRFH dimerization domain of the human telomeric proteins TRF1 and TRF2. Mol. Cell 8:351-361. [DOI] [PubMed] [Google Scholar]
- 18.Fang, G., and T. R. Cech. 1995. Telomere proteins, p. 69-107. In E. H. Blackburn and C. W. Greider (ed.), Telomeres. Cold Spring Harbor Press, Cold Spring Harbor, N.Y.
- 19.Garvik, B., M. Carson, and L. Hartwell. 1995. Single-stranded DNA arising at telomeres in cdc13 mutants may constitute a specific signal for the RAD9 checkpoint. Mol. Cell. Biol. 15:6128-6138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Griffith, J., A. Bianchi, and T. de Lange. 1998. TRF1 promotes parallel pairing of telomeric tracts in vitro. J. Mol. Biol. 278:79-88. [DOI] [PubMed] [Google Scholar]
- 21.Griffith, J. D., L. Comeau, S. Rosenfield, R. M. Stansel, A. Bianchi, H. Moss, and T. de Lange. 1999. Mammalian telomeres end in a large duplex loop. Cell 97:503-514. [DOI] [PubMed] [Google Scholar]
- 22.Hollenberg, S. M., R. Sternglanz, P. F. Cheng, and H. Weintraub. 1995. Identification of a new family of tissue-specific basic helix-loop-helix proteins with a two-hybrid system. Mol. Cell. Biol. 15:3813-3822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Horvath, M. P., V. L. Schweiker, J. M. Bevilacqua, J. A. Ruggles, and S. C. Schultz. 1998. Crystal structure of the Oxytricha nova telomere end binding protein complexed with single strand DNA. Cell 95:963-974. [DOI] [PubMed] [Google Scholar]
- 24.Huffman, K. E., S. D. Levene, V. M. Tesmer, J. W. Shay, and W. E. Wright. 2000. Telomere shortening is proportional to the size of the 3′ G-rich telomeric overhang. J. Biol. Chem. 275:19719-19722. [DOI] [PubMed] [Google Scholar]
- 25.Hughes, T. R., R. G. Weilbaecher, M. Walterscheid, and V. Lundblad. 2000. Identification of the single-strand telomeric DNA binding domain of the Saccharomyces cerevisiae Cdc13 protein. Proc. Natl. Acad. Sci. USA 97:6457-6462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.King, D. A., L. Zhang, L. Guarente, and R. Marmorstein. 1999. Structure of a HAP1-DNA complex reveals dramatically asymmetric DNA binding by a homodimeric protein. Nat. Struct. Biol. 6:64-71. [DOI] [PubMed] [Google Scholar]
- 27.Konrad, J. P., W. Mills, D. J. Easty, and C. J. Farr. 1999. Cloning and characterisation of the chicken gene encoding the telomeric protein TRF2. Gene 239:81-90. [DOI] [PubMed] [Google Scholar]
- 28.Krutilina, R. I., S. Oei, G. Buchlow, P. M. Yau, A. O. Zalensky, I. A. Zalenskaya, E. M. Bradbury, and N. V. Tomilin. 2001. A negative regulator of telomere-length protein TRF1 is associated with interstitial (TTAGGG)n blocks in immortal Chinese hamster ovary cells. Biochem. Biophys. Res. Commun. 280:471-475. [DOI] [PubMed] [Google Scholar]
- 29.Lei, M., P. Baumann, and T. R. Cech. 2002. Cooperative binding of single-stranded telomeric DNA by the Pot1 protein of Schizosaccharomyces pombe. Biochemistry 41:14560-14568. [DOI] [PubMed] [Google Scholar]
- 30.Lei, M., E. R. Podell, P. Baumann, and T. R. Cech. 2003. DNA self-recognition in the structure of Pot1 bound to telomeric single-stranded DNA. Nature 426:198-203. [DOI] [PubMed] [Google Scholar]
- 31.Loayza, D., and T. De Lange. 2003. POT1 as a terminal transducer of TRF1 telomere length control. Nature 25:25. [DOI] [PubMed] [Google Scholar]
- 32.Lustig, A. J. 2001. Cdc13 subcomplexes regulate multiple telomere functions. Nat. Struct. Biol. 8:297-299. [DOI] [PubMed] [Google Scholar]
- 33.McEachern, M. J., A. Krauskopf, and E. H. Blackburn. 2000. Telomeres and their control. Annu. Rev. Genet. 34:331-358. [DOI] [PubMed] [Google Scholar]
- 34.Mitton-Fry, R. M., E. M. Anderson, T. R. Hughes, V. Lundblad, and D. S. Wuttke. 2002. Conserved structure for single-stranded telomeric DNA recognition. Science 296:145-147. [DOI] [PubMed] [Google Scholar]
- 35.Murzin, A. G. 1993. OB(oligonucleotide/oligosaccharide binding)-fold: common structural and functional solution for non-homologous sequences. EMBO J. 12:861-867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Peersen, O. B., J. A. Ruggles, and S. C. Schultz. 2002. Dimeric structure of the Oxytricha nova telomere end-binding protein alpha-subunit bound to ssDNA. Nat. Struct. Biol. 9:182-187. [DOI] [PubMed] [Google Scholar]
- 37.Pennock, E., K. Buckley, and V. Lundblad. 2001. Cdc13 delivers separate complexes to the telomere for end protection and replication. Cell 104:387-396. [DOI] [PubMed] [Google Scholar]
- 38.Price, C. M. (ed.). 1995. Telomere-binding proteins of ciliated protozoa, vol. 9. Springer Verlag, Berlin, Germany.
- 39.Price, C. M., R. Skopp, J. Krueger, and D. Williams. 1992. DNA recognition and binding by the Euplotes telomere protein. Biochemistry 31:10835-10843. [DOI] [PubMed] [Google Scholar]
- 40.Seimiya, H., and S. Smith. 2002. The telomeric poly(ADP-ribose) polymerase, tankyrase 1, contains multiple binding sites for telomeric repeat binding factor 1 (TRF1) and a novel acceptor, 182-kDa tankyrase-binding protein (TAB182). J. Biol. Chem. 277:14116-14126. [DOI] [PubMed] [Google Scholar]
- 41.Smith, J., I. R. Paton, F. Murray, R. P. Crooijmans, M. A. Groenen, and D. W. Burt. 2002. Comparative mapping of human Chromosome 19 with the chicken shows conserved synteny and gives an insight into chromosomal evolution. Mamm. Genome 13:310-315. [DOI] [PubMed] [Google Scholar]
- 42.Smith, S., I. Giriat, A. Schmitt, and T. de Lange. 1998. Tankyrase, a poly(ADP-ribose) polymerase at human telomeres. Science 282:1484-1487. [DOI] [PubMed] [Google Scholar]
- 43.Stansel, R. M., T. de Lange, and J. D. Griffith. 2001. T-loop assembly in vitro involves binding of TRF2 near the 3′ telomeric overhang. EMBO J. 20:5532-5540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tan, M., C. Wei, and C. Price. 2003. The telomeric protein Rap1 is conserved in vertebrates and is expressed from a bidirectional promoter positioned between the Rap1 and KARS genes. Gene 323:1-10. [DOI] [PubMed] [Google Scholar]
- 45.Theobald, D. L., R. B. Cervantes, V. Lundblad, and D. S. Wuttke. 2003. Homology among telomeric end-protection proteins. Structure 11:1049-1050. [DOI] [PubMed] [Google Scholar]
- 46.Theobald, D. L., R. M. Mitton-Fry, and D. S. Wuttke. 2003. Nucleic acid recognition by OB-fold proteins. Annu. Rev. Biophys. Biomol. Struct. 32:115-133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Theobald, D. L., and S. C. Schultz. 2003. Nucleotide shuffling and ssDNA recognition in Oxytricha nova telomere end-binding protein complexes. EMBO J. 22:4314-4324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Venkatesan, R. N., and C. Price. 1998. Telomerase expression in chickens: constitutive activity in somatic tissues and down-regulation in culture. Proc. Natl. Acad. Sci. USA 95:14763-14768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wei, C., and C. M. Price. 2003. Protecting the terminus: T-loops and telomere end-binding proteins. Cell. Mol. Life Sci. 60:2283-2294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wright, W. E., V. M. Tesmer, M. L. Liao, and J. W. Shay. 1999. Normal human telomeres are not late replicating. Exp. Cell Res. 251:492-499. [DOI] [PubMed] [Google Scholar]
- 51.Yang, H., P. D. Jeffrey, J. Miller, E. Kinnucan, Y. Sun, N. H. Thoma, N. Zheng, P. L. Chen, W. H. Lee, and N. P. Pavletich. 2002. BRCA2 function in DNA binding and recombination from a BRCA2-DSS1-ssDNA structure. Science 297:1837-1848. [DOI] [PubMed] [Google Scholar]
- 52.Zhu, X. D., B. Kuster, M. Mann, J. H. Petrini, and T. Lange. 2000. Cell-cycle-regulated association of RAD50/MRE11/NBS1 with TRF2 and human telomeres. Nat. Genet. 25:347-352. [DOI] [PubMed] [Google Scholar]