Abstract
The processes of cellular growth regulation and cellular metabolism are closely inter-related. The c-Myc oncogene is a “master regulator” which controls many aspects of both of these processes. The metabolic changes which occur in transformed cells, many of which are driven by c-Myc overexpression, are necessary to support the increased need for nucleic acids, proteins and lipids necessary for rapid cellular proliferation. At the same time, c-Myc overexpression results in coordinated changes in level of expression of gene families which result in increased cellular proliferation. This interesting duality of c-Myc effects places it in the mainstream of transformational changes and gives it a very important role in regulating the “transformed phenotype”. The effects induced by c-Myc can occur either as a “primary oncogene” which is activated by amplification or translocation; or as a downstream effect of other activated oncogenes. In either case, it appears that c-Myc plays a central role in sustaining the changes which occur with transformation. Although efforts to utilize c-Myc as a therapeutic target have been quite frustrating, it appears that this may change in the next few years.
Keywords: c-Myc, Glycolysis, Cancer Metabolism, Metabolic Transformation
Introduction
It has become increasingly clear over the past two decades that the metabolic changes that accompany transformation are intimately related to the growth abnormalities of malignant cells and that these metabolic changes are necessary in order to provide the energy required for rapid cell division. It has also become clear that the multifaceted oncogene, c-Myc, plays important regulatory roles in many aspects of transformation1–4. Although c-Myc may play a primary oncogenic role in tumors such as Burkitt’s lymphoma in which it is translocated under the promoter regions of the heavy- or light-chain immunoglobulin genes5, it is more commonly a downstream “early-response” gene, which responds to activation of many diverse signaling pathways. It remains unclear whether c-Myc overexpression is primarily responsible for the metabolic changes induced by transformation, or whether its common overexpression may be a result of the complex metabolic changes which occur when cells become malignant.
Although our detailed understanding of cancer metabolism and growth regulation have evolved independently6, these two areas of investigation have now merged. For a period of time extending into the 1970’s, cellular metabolism was regarded as a collection of metabolic pathways which largely served for energy production. We now know that malignant transformation induces changes in almost every aspect of metabolism, allowing transformed cells to fill the huge demand for proteins, lipids and nucleic acids necessary to support rapid cell division. The changes in glycolysis which accompany transformation, known as the Warburg effect7, 8, which have been recognized for more than eighty years are “the tip of the iceberg” in terms of the many metabolic changes now known to occur. Our current understanding of the Warburg effect and its implications for the regulation of cancer cell growth has been refined by several generations of tumor biologists and has had increasing clarity as the molecular biology of the malignant phenotype has been elucidated. The Warburg effect9, has been observed in many tumor types and is now being used clinically to detect tumors by fluorodeoxyglucose positron emission tomography (FDG-PET)10.
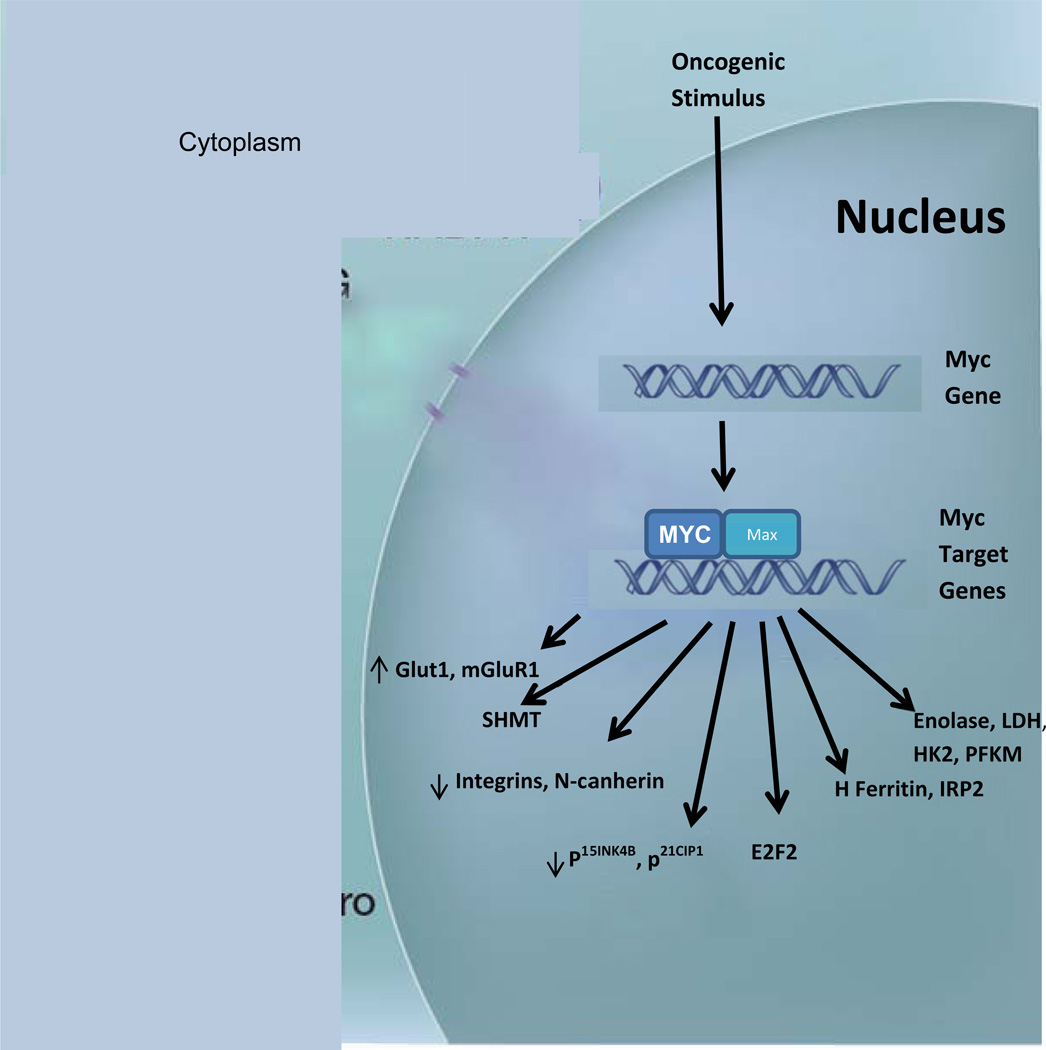
Since the discovery of transforming oncogenes in the late 1970’s, the biochemical study of cancer metabolism has been overshadowed by efforts to identify the mutations that contribute to cancer initiation and progression. Recently, however, it has been demonstrated that all of the key elements of “metabolic transformation” (the Warburg effect) — increased glucose consumption, decreased oxidative phosphorylation, and accompanying lactate production — are induced by oncogene activation. Induced overexpression of the c-Myc gene is responsible for many of the changes that induce malignant changes (see Figure 1). These changes support the production of intermediates for cell growth and division, and are regulated by both oncogenes and tumor suppressor genes in a number of key cancer-producing pathways. Metabolic transformation is the result of complex interactions between a generally hypoxic tumor microenvironment and multiple oncogenic mutations which drive the alterations in cellular metabolism which occur in transformed cells. The metabolic changes which accompany malignant transformation represent one of a relatively few “hallmarks” of malignancy11, 12.
Figure 1.
Diagram of Myc effects in transformed cells. There are a wide variety of downstream pathways which are both positively and negatively regulated by Myc expression.
Regulation of c-Myc Expression
In normal (nontransformed) cells, c-Myc expression and function is tightly regulated by developmental or mitogenic signals. c-Myc mRNA is very short-lived and in the absence of positive regulatory signals, c-Myc transcription decreases and c-Myc protein levels are low, providing no proliferative drive. In tumor cells, on the other hand, c-Myc function is almost always increased, sometimes by mutations in the gene itself, but more commonly through the induction of c-Myc expression via upstream oncogenic pathways. The oncogenic properties of c-Myc are counterbalanced by its ability to induce apoptosis through several pathways13, 14. This dichotomy most likely explains why c-Myc is not commonly the driving oncogene in early tumors.
Another safeguard mechanism by which c-Myc expression is tightly controlled is stability of the c-Mmyc protein. The short half-life of c-Myc in proliferating cells (approximately 30 minutes)15 make this a particularly effective mechanism of gene regulation. c-Myc has been shown to undergo ubiquitylation and degradation by the proteasome16, 17. This includes phosphorylation-dependent degradation of c-Myc18.
On the other hand, c-Myc overexpression may also occur as a result of post-translational modifications. For example, mutations in the coding region of c-Myc are commonly found in human lymphomas, particularly in the Thr58 phosphorylation site. This mutation has been shown to enhance the transforming activity of c-Myc by cuasing inefficient ubuiquitination and decreased proteasome-mediated protein turnover19
MicroRNAs (miRNAs) are intertwined in the c-Myc regulatory network, both as targets of c-myc and as regulators of c-Myc expression. The application of microarray technology has revealed both c-Myc-induced and c-Myc-repressed miRNAs. A miRNA wich is consistently repressed by c-Myc in multiple tumors is miR-26a20. On the other hand, overespression of the let-7a miRNA causes dramatic decreases in c-Myc gene expression and its target genes, as well as antiproliferative activity in lymphoma cells21. More recent work has also shown that miR-33b22miR-143 and miR-145 repress ERK5/c-Myc signaling23.
c-Myc Target Genes
Thousands of c-Myc target genes have been identified by one or more differential expression screens including SAGE24, DNA microarray25, and subtractive hybridization3, 26. The list of c-Myc-responsive genes27 includes genes involved in almost every important cellular function (Table 1). In addition to those genes which are positively regulated by c-Myc, the transcriptional activity of other genes, including cyclin D1 and carboxypedidase D is repressed by the c-Myc-Max complex or c-Myc alone28, 29. Interestingly, it has recently been shown that a network of c-Myc-induced genes accounts for similarities between the transcriptional programs of embryonic stem cells and transformed cells30.
Table 1.
| Target Gene | Regulation | Pathway | Functional Relevance |
|---|---|---|---|
| Cyclin A2, Cyclin D2, Cyclin E1 | Up | Growth Factor Response | Response to mitogenic stimuli |
| Enolase, LDH-A | Up | Glycolysis | Metabolic Transformation |
| Serine hydroxymethyl transferase | Up | C1 Metabolism | Anaplerosis |
| EIF4E, Ribosomal Proteins L3, L6, S15A | Up | Translation Initiation | Global increase in translation |
| Ornithine Decarboxylase, prothymosin-α, HMG1/Y | Up | Transformation | Anchorage-independent growth |
| Iron-regulatory protein-2 H-ferritin, transferrin receptor | Up | Iron metabolism | Required for Myc-induced proliferation |
| Nucleolin, NM23, Nucleophosmin | Up | Cellular proliferation | Required for RNA and DNA synthesis |
| p21CIP1 | Down | DNA damage response | Differentiation |
| p15INK4B | Down | TGFβ pathway | Resistance to growth arrest |
| N-cadherin, Integrins | Down | Cell adhesion | Metastatic Potential |
c-Myc and Transformation
The human c-Myc gene was discovered as a result of early studies of very aggressive chicken tumors which led to the identification of the v-myc oncogene as the cause of myelocytomatosis (leukemia and sarcoma)31, 32. The discovery that human c-Myc is consistently altered by balanced chromosomal translocation in Burkitt’s lymphoma marked it as a bona fide human oncogene33. Myc is frequently translocated in multiple myeloma34 and is one of the most highly amplified oncogenes among many different human cancers35. Myc is downstream of the deregulated Notch signaling pathways found in T cell leukemia36. Hence, c-Myc overexpression may reflect amplification or physiologic overexpression as a downstream member of a signaling pathway.
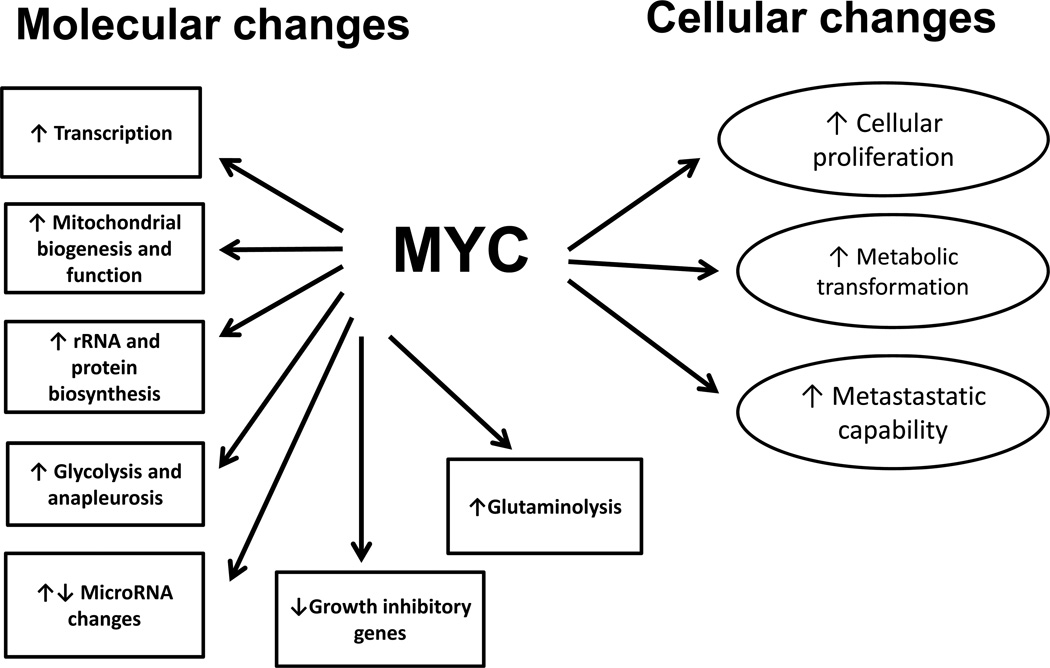
The c-Myc oncogene is overexpressed in the majority of human cancers and contributes to the cause of at least 40% of tumors37. It encodes a helix-loop-helix leucine zipper transcription factor that dimerizes with its partner protein, Max, to transactivate gene expression. The c-Myc heterodimer can also repress gene expression via binding to the transcription factor Miz138, 39. The other members of the myc gene family (l-Myc and n-Myc) also encode essential transcription regulators whose expression is altered in a wide variety of tumor types. c-Myc binds to the promoters of thousands of genes, although only a fraction of these respond with transcriptional changes40, 41. c-Myc regulates several large gene families resulting in coordinated changes in cell proliferation and cellular metabolism. c-Myc stimulates genes involved in protein biosynthesis, cancer metabolism, transcription factors and cell cycle genes and some microRNAs, while inhibiting expression of other microRNAs and some tumor suppressor genes38. The pleiotropic effects of c-Myc expression occur at the molecular and cellular level (Figure 2) and affect almost every activity of cell life.
Figure 2.
Pleiotropic Effects of c-Myc Expression. The c-myc gene has a variety of molecular and cellular effects (which are closely related). These effects result from myc-mediated changes in large gene families which drive cellular functions. Microarray studies have shown that these changes occur in concert and have major effects on cellular function.
c-Myc Drives Cell Proliferation
c-Myc is critically involved in the regulation of many growth promoting signal transduction pathways and is an immediate early response gene which is downstream of many ligand-membrane receptor complexes42, 43. c-Myc expression is tightly regulated, its level of expression is influenced at the transcriptional level by a number of transcriptional regulatory motifs within its proximal promoter region4, 44, 45. Although the c-Myc gene product has been most widely characterized as a driver of cell proliferation38, 46, it also stimulates glycolysis47, 48 and has been shown to upregulate expression of a broad variety of metabolic enzymes49.
Normal proliferating cells express low levels of c-Myc RNA and protein in response to a broad variety of mitogenic stimuli. Almost all cancer-associated genetic changes in c-Myc are associated with noncoding regulatory regions rather than protein coding sequences. This reflects the fact that deregulation of c-Myc expression, rather than expression of a mutated protein is what drives the c-Myc effect. Although almost all of the abnormalities of metabolic transformation can be related to c-Myc overexpression, as noted above, it is not always clear whether c-Myc overexpression is a primary or secondary effect in transformed cells
Myc Stimulates Glycolysis
The first suggestion that c-Myc played an important role in regulation of glycolysis was the observation that lactate dehydrogenase A (LDHA), which converts pyruvate to lactate as part of the glycolytic pathway, was one of twenty putative c-Myc target genes26, 50, 51. Subsequent work has shown that many other glucose metabolism genes are directly regulated by c-Myc, as well37. These genes include glucose transporter GLUT1, hexokinase 2 (HK2), phosphofructokinase (PFKM), and enolase 1 (ENO1)52–54. Through the up-regulation of these genes, c-Myc contributes directly to the Warburg effect (aerobic glycolysis) and the ability of transformed cells to convert glucose to pyruvate even under adequate oxygen tension. Interestingly, ENO1 has been shown to give rise to an alternative translation initiation product, MBP-1, which is a negative regulator of c-Myc expression55. This provides a negative feedback look which is modulated by hypoxia56.
The direct effects of c-Myc expression on glycolytic activity have been confirmed in studies with transgenic animals57. Mice which overexpress c-Myc in the liver demonstrate increased glycolytic enzyme activity in the liver and overproduce lactic acid. On the other hand, stably transfected rodent fibroblasts overexpressing LDH-A alone, or those transformed by c-Myc, overproduce lactate. This suggests that LDH-A, which is a downstream target of c-Myc, is able to induce the Warburg effect. Soft agar clonogenicity of Burkitt’s Lymphoma cells is markedly decreased by inhibiting expression of LDH-A50.
Other cancer-related genes also play a role in regulating glycolysis. For example, phosphoinositol-3-kinase (PI3K) and its downstream effector AKT have a direct role in stimulating glucose uptake and metabolism, rendering the transformed cell addicted to glucose for the maintenance of survival. More recent studies have linked Ras, VHL, and mutations of isocitrate dehydrogenase 1 (IDH1)58, succinate dehydrogenase (SDH), and fumarate hydratase (FH) to the activation of glycolysis through HIF-1. This results in increased glycolytic enzyme gene expression50, 59–61. On the other hand, the Akt oncogene was shown to stimulate glycolysis post-transcriptionally, and the p53 tumor suppressor emerged as another regulator of mitochondrial function and glycolysis, demonstrating that loss of p53 is associated with enhanced glycolysis59. The signaling molecule Ras, which becomes a powerful oncogene when mutated, stimulates glycolysis51, 62, 63. Akt kinase, a well-characterized downstream effector of insulin signaling, reprises its role in glucose uptake and utilization in the cancer setting64. Other work also suggests that p53-mediated regulation of glucose metabolism may be dependent on the transcription factor NF-κB65. The M2 splice isoform of pyruvate kinase (PKM2) is a key regulator of aerobic glycolysis in cancer cells66.
c-Myc Stimulates Mitochondrial Biogenesis
In addition to its important role in regulating cellular metabolism by altering expression of genes involved in metabolic pathways, c-Myc also plays an important role in mitochondrial biogenesis. Large scale studies of gene expression in rat and human systems first suggested that c-Myc overexpression can induce nuclearly encoded mitochondrial genes24, 25, 29. In addition, c-Myc has been shown to bind to the promoters of genes encoding progeins involved in mitochondrial function24, 67. Using an inducible c-Myc-dependent human B cell model of cell proliferation it was shown that mitochondrial biogenesis is completely dependent on c-Myc expression68. Moreover, the genes involved with mitochondrial biogenesis were among the c-Myc target genes most highly induced.
In addition to its role in generation of functional mitochondria, c-Myc appears to increase mitochondrial function. It has been shown that c-Myc increases mitochondrial synthesis of acetyl-CoA which, in turn, contributes to significant increases in histone acetylation and fatty acid biosynthesis in rapidly dividing cells69, 70. The ability of c-Myc to induce mitochondrial biogenesis in proliferating cells while inhibiting mitochondrial respiration is very important, because mitochondria not only provide a means for efficient production of ATP in the presence of oxygen, but they also play a role in generating substrates for macromolecular synthesis in dividing cells. These components include pyrimidines, whose synthesis is directly linked to the electron transport chain, the carbon backbone for amino acids, as well as citrate which is converted to acetyl-CoA for lipid biosynthesis. These functions complement the stimulation of glucose uptake and metabolism by c-Myc, which provides carbon backbones for critical cellular components, including ribose for nucleotide biosynthesis and NADPH through the pentose phosphate pathway for redox homeostasis, triglycerides and ATP through glycolysis.
c-Myc Regulates Glutamine Metabolism
In addition to their use of glucose, mammalian cells obtain energy for growth and proliferation through the catabolism of glutamine48, 71, 72. Induced c-Myc overexpression coordinates the expression of genes necessary for cells to engage in glutamine catabolism that exceeds the cellular requirement for protein and nucleotide biosynthesis. A consequence of this c-Myc-dependent glutaminolysis is the reprogramming of mitochondrial metabolism to depend on glutamine catabolism to sustain cellular viability and tricarboxcylic acid (TCA) cycle anapleurosis. Some human tumors have been reported to consume so much glutamine that they decrease circulating plasma glutamine levels73, 74.
Glutamine is used as a source of energy and nitrogen for biosynthesis, and a carbon substrate for anabolic processes in cancer cells75, 76, but the regulation of glutamine metabolism is not well understood75, 77, 78. In contrast to most other metabolites that are taken up by proliferating cells which are not catabolized, but instead are used as substrates for anabolic macromolecular synthesis, glutamine is a very important energy source72. Glutamine metabolism is an important mitochondrial function in cancer cells, specifically enzymatic glutaminolysis that catabolizes glutamine to generate ATP and lactate77.
Recent studies have shown that cancer-related alterations in glucose and glutamine metabolism are strongly influenced by c-Myc expression79. In particular, the demonstration of persistent, c-Myc-dependent hypoxic metabolism of glutamine, even in the absence of glucose suggests that this is a major influence of c-Myc expression. In fact, in transformed cells, overexpression of c-Myc results in the concurrent conversion of glucose to lactate and the oxidation of glutamine via the TCA cycle79. Under hypoxic conditions with high c-Myc, a substantial fraction of the glucose consumed was converted to excreted lactate, and glutamine continued to be utilized by the TCA cycle, which was used for cell survival. This study also found that a glutamine-dependent and glucose independent TCA cycle may operate under both aerobic and hypoxic conditions under glucose-depleted culture conditions. Moreover, they observed an enhanced conversion of glutamine to glutathione under hypoxia; glutathione is an important reducing agent for controlling the accumulation of mitochondrial reactive oxygen species (ROS). They also demonstrated that inhibition of glutaminase effectively kills hypoxic cancer cells in vitro and delays tumor xenograft growth. The essential role of glutamine metabolism in cell survival and proliferation under hypoxia and glucose deficiency makes cells susceptible to the glutaminase inhibitor bis-2-(5-phenylacetamido-1,2,4-thiadiazol-2-yl)ethyl sulfide (BPTES) and hence could be targeted for cancer therapy (as described below).
Interestingly, c-Myc appears to alter glutamine metabolism by transcriptionally repressing the miRNAs, miR-23a and miR-23b80, 81. This results in increased expression of their target protein, mitochondrial glutaminase (GLS). This leads, in turn, to up-regulation of glutamine catabolism82, resulting in increased glutamate which is further metabolized through the TCA cycle or serves as a substrate for glutathione synthesis. This unexpected mechanism of gene regulation connects myc regulation of miRNAs, glutamine metabolism and ROS homeostasis.
c-Myc is a Therapeutic Target
Because of its ubiquitous role in human tumors, c-Myc is an attractive therapeutic target. Blocking the metabolic pathways which are “driven” by c-Myc or restoring these altered pathways could lead to a new approach in cancer treatment. Inhibition of lactate dehydrogenase A has been shown to inhibit tumor progression83. Other groups have targeted c-Myc transcription by interfering with chromatin-dependent-signal-transduction84. Using a potent, selective small-molecule inhibitor of (BET) bromodomain proteins, proteins which associate with acetylated chromatin and activate transcription by increasing the concentration of attracted transcriptional activators85, 86. Using this approach, growth inhibitory activity was seen in three murine models of multiple myeloma87. Histone deacetylase inhibitors, including several which are in early phase clinical trials, have also shown marked downregulation of c-Myc88–91. Another interesting approach has been the development of a dominant negative c-Myc construct, Omomyc, which is a c-Myc-derived bHLHZip domain which forms heterodimers with wild type c-Myc, but interferes with the formation of c-Myc/Max dimers and suppresses binding to E-box elements92–94. More recent work94 has demonstrated that this interesting c-Myc inhibitor effects some, but not all, c-Myc functions. In addition, work which targets closely related pathways, such as the HIF-1 pathway, may have dramatic effects on c-Myc function95.
Other groups have focused on transcriptional inhibition of the c-Myc gene. Preliminary evidence from experiments using c-Myc antisense oligonucleotides has been encouraging, but has not translated into effective clinical treatments96. Using the Pu27 quadruplex-forming sequence present in the c-Myc promoter, quadruplex stabilizing compounds have been shown to decrease c-Myc expression levels97. Recent work has shown that treating cells with oligonucleotides encoding the genomic c-Myc promoter quadruplex-forming sequence, Pu27, results in leukemic cell death98. New approaches, such as genome-scale metabolic modeling hold promise for the identification of novel drug targets and biomarkers99
Summary
The c-Myc gene serves as a “master regulator” of cellular metabolism and proliferation. Since it is activated by a large number of oncogenic pathways and, in turn, stimulates many of the metabolic changes that result in malignant transformation, it is truly “both the chicken and the egg”. Under normal circumstances c-Myc is dependent on mitogenic stimulation for its expression and function. c-Myc is a multifunctional transcription factor which drives the multiple synthetic functions necessary for rapid cell division while at the same time inhibiting expression of genes with antiproliferative functions. Because of its propensity to induce apoptosis, its expression is tightly regulated. It influences expression of a wide variety of gene families which contribute to the abnormal growth abilities of transformed cells. It is quite clear that this central role in regulating cell function represents a unique opportunity to develop novel cancer therapies.
Acknowledgments
Dr. Miller is funded by NIH Grant 3P20RR18733
Footnotes
Dr. Miller is on the Board of Directors of Advanced Cancer Therapeutics; All other authors declare no potential conflict of interest.
References
- 1.Morrish F, Neretti N, Sedivy JM, Hockenbery DM. The oncogene c-Myc coordinates regulation of metabolic networks to enable rapid cell cycle entry. Cell Cycle. 2008;7:1054–1066. doi: 10.4161/cc.7.8.5739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wolfer A, Ramaswamy S. MYC and metastasis. Cancer Res. 2011;71:2034–2037. doi: 10.1158/0008-5472.CAN-10-3776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Meyer N, Penn LZ. Reflecting on 25 years with MYC. Nat Rev Cancer. 2008;8:976–990. doi: 10.1038/nrc2231. [DOI] [PubMed] [Google Scholar]
- 4.Levens D. You Don't Muck with MYC. Genes Cancer. 2010;1:547–554. doi: 10.1177/1947601910377492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lombardi L, Newcomb EW, Dalla-Favera R. Pathogenesis of Burkitt lymphoma: expression of an activated c-myc oncogene causes the tumorigenic conversion of EBV-infected human B lymphoblasts. Cell. 1987;49:161–170. doi: 10.1016/0092-8674(87)90556-3. [DOI] [PubMed] [Google Scholar]
- 6.Teicher BA, Linehan WM, Helman LJ. Targeting cancer metabolism. Clin Cancer Res. 2012;18 doi: 10.1158/1078-0432.CCR-12-2587. xx-xx. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Warburg O. On the origin of cancer cells. Science. 1956;123:309–314. doi: 10.1126/science.123.3191.309. [DOI] [PubMed] [Google Scholar]
- 8.Warburg O. On respiratory impairment in cancer cells. Science. 1956;124:269–270. [PubMed] [Google Scholar]
- 9.Koppenol WH, Bounds PL, Dang CV. Otto Warburg's contributions to current concepts of cancer metabolism. Nat Rev Cancer. 2011;11:325–337. doi: 10.1038/nrc3038. [DOI] [PubMed] [Google Scholar]
- 10.Fletcher JW, Djulbegovic B, Soares HP, Siegel BA, Lowe VJ, Lyman GH, et al. Recommendations on the use of 18F-FDG PET in oncology. J Nucl Med. 2008;49:480–508. doi: 10.2967/jnumed.107.047787. [DOI] [PubMed] [Google Scholar]
- 11.Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000;100:57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- 12.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 13.Pelengaris S, Khan M, Evan G. c-MYC: more than just a matter of life and death. Nat Rev Cancer. 2002;2:764–776. doi: 10.1038/nrc904. [DOI] [PubMed] [Google Scholar]
- 14.Kelly GL, Rickinson AB. Burkitt lymphoma: revisiting the pathogenesis of a virus-associated malignancy. Hematology Am Soc Hematol Educ Program. 2007:277–284. doi: 10.1182/asheducation-2007.1.277. [DOI] [PubMed] [Google Scholar]
- 15.Hann SR, Eisenman RN. Proteins encoded by the human c-myc oncogene: differential expression in neoplastic cells. Mol Cell Biol. 1984;4:2486–2497. doi: 10.1128/mcb.4.11.2486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ciechanover A, DiGiuseppe JA, Schwartz AL, Brodeur GM. Degradation of MYCN oncoprotein by the ubiquitin system. Prog Clin Biol Res. 1991;366:37–43. [PubMed] [Google Scholar]
- 17.Salghetti SE, Kim SY, Tansey WP. Destruction of Myc by ubiquitin-mediated proteolysis: cancer-associated and transforming mutations stabilize Myc. Embo J. 1999;18:717–726. doi: 10.1093/emboj/18.3.717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yada M, Hatakeyama S, Kamura T, Nishiyama M, Tsunematsu R, Imaki H, et al. Phosphorylation-dependent degradation of c-Myc is mediated by the F-box protein Fbw7. Embo J. 2004;23:2116–2125. doi: 10.1038/sj.emboj.7600217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bahram F, von der Lehr N, Cetinkaya C, Larsson LG. c-Myc hot spot mutations in lymphomas result in inefficient ubiquitination and decreased proteasome-mediated turnover. Blood. 2000;95:2104–2110. [PubMed] [Google Scholar]
- 20.Sander S, Bullinger L, Wirth T. Repressing the repressor: a new mode of MYC action in lymphomagenesis. Cell Cycle. 2009;8:556–559. doi: 10.4161/cc.8.4.7599. [DOI] [PubMed] [Google Scholar]
- 21.Sampson VB, Rong NH, Han J, Yang Q, Aris V, Soteropoulos P, et al. MicroRNA let-7a down-regulates MYC and reverts MYC-induced growth in Burkitt lymphoma cells. Cancer Res. 2007;67:9762–9770. doi: 10.1158/0008-5472.CAN-07-2462. [DOI] [PubMed] [Google Scholar]
- 22.Takwi AA, Li Y, Becker Buscaglia LE, Zhang J, Choudhury S, Park AK, et al. A statin-regulated microRNA represses human c-Myc expression and function. EMBO Mol Med. 2012 doi: 10.1002/emmm.201101045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Takaoka Y, Shimizu Y, Hasegawa H, Ouchi Y, Qiao S, Nagahara M, et al. Forced Expression of miR-143 Represses ERK5/c-Myc and p68/p72 Signaling in Concert with miR-145 in Gut Tumors of Apc(Min) Mice. PLoS One. 2012;7:e42137. doi: 10.1371/journal.pone.0042137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Menssen A, Hermeking H. Characterization of the c-MYC-regulated transcriptome by SAGE: identification and analysis of c-MYC target genes. Proc Natl Acad Sci U S A. 2002;99:6274–6279. doi: 10.1073/pnas.082005599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Coller HA, Grandori C, Tamayo P, Colbert T, Lander ES, Eisenman RN, et al. Expression analysis with oligonucleotide microarrays reveals that MYC regulates genes involved in growth, cell cycle, signaling, and adhesion. Proc Natl Acad Sci U S A. 2000;97:3260–3265. doi: 10.1073/pnas.97.7.3260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lewis BC, Shim H, Li Q, Wu CS, Lee LA, Maity A, et al. Identification of putative c-Myc-responsive genes: characterization of rcl, a novel growth-related gene. Mol Cell Biol. 1997;17:4967–4978. doi: 10.1128/mcb.17.9.4967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zeller KI, Jegga AG, Aronow BJ, O'Donnell KA, Dang CV. An integrated database of genes responsive to the Myc oncogenic transcription factor: identification of direct genomic targets. Genome Biol. 2003;4:R69. doi: 10.1186/gb-2003-4-10-r69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Philipp A, Schneider A, Vasrik I, Finke K, Xiong Y, Beach D, et al. Repression of cyclin D1: a novel function of MYC. Mol Cell Biol. 1994;14:4032–4043. doi: 10.1128/mcb.14.6.4032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Guo QM, Malek RL, Kim S, Chiao C, He M, Ruffy M, et al. Identification of c-myc responsive genes using rat cDNA microarray. Cancer Res. 2000;60:5922–5928. [PubMed] [Google Scholar]
- 30.Kim J, Woo AJ, Chu J, Snow JW, Fujiwara Y, Kim CG, et al. A Myc network accounts for similarities between embryonic stem and cancer cell transcription programs. Cell. 2010;143:313–324. doi: 10.1016/j.cell.2010.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Duesberg PH, Vogt PK. Avian acute leukemia viruses MC29 and MH2 share specific RNA sequences: evidence for a second class of transforming genes. Proc Natl Acad Sci U S A. 1979;76:1633–1637. doi: 10.1073/pnas.76.4.1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sheiness D, Bishop JM. DNA and RNA from uninfected vertebrate cells contain nucleotide sequences related to the putative transforming gene of avian myelocytomatosis virus. J Virol. 1979;31:514–521. doi: 10.1128/jvi.31.2.514-521.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dalla-Favera R, Bregni M, Erikson J, Patterson D, Gallo RC, Croce CM. Human c-myc onc gene is located on the region of chromosome 8 that is translocated in Burkitt lymphoma cells. Proc Natl Acad Sci U S A. 1982;79:7824–7827. doi: 10.1073/pnas.79.24.7824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shou Y, Martelli ML, Gabrea A, Qi Y, Brents LA, Roschke A, et al. Diverse karyotypic abnormalities of the c-myc locus associated with c-myc dysregulation and tumor progression in multiple myeloma. Proc Natl Acad Sci U S A. 2000;97:228–233. doi: 10.1073/pnas.97.1.228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Beroukhim R, Mermel CH, Porter D, Wei G, Raychaudhuri S, Donovan J, et al. The landscape of somatic copy-number alteration across human cancers. Nature. 2010;463:899–905. doi: 10.1038/nature08822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Palomero T, Lim WK, Odom DT, Sulis ML, Real PJ, Margolin A, et al. NOTCH1 directly regulates c-MYC and activates a feed-forward-loop transcriptional network promoting leukemic cell growth. Proc Natl Acad Sci U S A. 2006;103:18261–18266. doi: 10.1073/pnas.0606108103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dang CV, Le A, Gao P. MYC-induced cancer cell energy metabolism and therapeutic opportunities. Clin Cancer Res. 2009;15:6479–6483. doi: 10.1158/1078-0432.CCR-09-0889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Eilers M, Eisenman RN. Myc's broad reach. Genes Dev. 2008;22:2755–2766. doi: 10.1101/gad.1712408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Herkert B, Eilers M. Transcriptional repression: the dark side of myc. Genes Cancer. 2010;1:580–586. doi: 10.1177/1947601910379012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zeller KI, Zhao X, Lee CW, Chiu KP, Yao F, Yustein JT, et al. Global mapping of c-Myc binding sites and target gene networks in human B cells. Proc Natl Acad Sci U S A. 2006;103:17834–17839. doi: 10.1073/pnas.0604129103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Patel JH, Loboda AP, Showe MK, Showe LC, McMahon SB. Analysis of genomic targets reveals complex functions of MYC. Nat Rev Cancer. 2004;4:562–568. doi: 10.1038/nrc1393. [DOI] [PubMed] [Google Scholar]
- 42.Armelin HA, Armelin MC, Kelly K, Stewart T, Leder P, Cochran BH, et al. Functional role for c-myc in mitogenic response to platelet-derived growth factor. Nature. 1984;310:655–660. doi: 10.1038/310655a0. [DOI] [PubMed] [Google Scholar]
- 43.Kelly K, Cochran BH, Stiles CD, Leder P. Cell-specific regulation of the c-myc gene by lymphocyte mitogens and platelet-derived growth factor. Cell. 1983;35:603–610. doi: 10.1016/0092-8674(83)90092-2. [DOI] [PubMed] [Google Scholar]
- 44.Brooks TA, Hurley LH. Targeting MYC Expression through G-Quadruplexes. Genes Cancer. 2010;1:641–649. doi: 10.1177/1947601910377493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hurley LH, Von Hoff DD, Siddiqui-Jain A, Yang D. Drug targeting of the c-MYC promoter to repress gene expression via a G-quadruplex silencer element. Semin Oncol. 2006;33:498–512. doi: 10.1053/j.seminoncol.2006.04.012. [DOI] [PubMed] [Google Scholar]
- 46.Freie BW, Eisenman RN. Ratcheting Myc. Cancer Cell. 2008;14:425–426. doi: 10.1016/j.ccr.2008.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dang CV, O'Donnell KA, Juopperi T. The great MYC escape in tumorigenesis. Cancer Cell. 2005;8:177–178. doi: 10.1016/j.ccr.2005.08.005. [DOI] [PubMed] [Google Scholar]
- 48.Dang CV. Rethinking the Warburg effect with Myc micromanaging glutamine metabolism. Cancer Res. 2010;70:859–862. doi: 10.1158/0008-5472.CAN-09-3556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gordan JD, Thompson CB, Simon MC. HIF and c-Myc: sibling rivals for control of cancer cell metabolism and proliferation. Cancer Cell. 2007;12:108–113. doi: 10.1016/j.ccr.2007.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shim H, Dolde C, Lewis BC, Wu CS, Dang G, Jungmann RA, et al. c-Myc transactivation of LDH-A: implications for tumor metabolism and growth. Proc Natl Acad Sci U S A. 1997;94:6658–6663. doi: 10.1073/pnas.94.13.6658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ramanathan A, Wang C, Schreiber SL. Perturbational profiling of a cell-line model of tumorigenesis by using metabolic measurements. Proc Natl Acad Sci U S A. 2005;102:5992–5997. doi: 10.1073/pnas.0502267102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kim JW, Gao P, Liu YC, Semenza GL, Dang CV. Hypoxia-inducible factor 1 and dysregulated c-Myc cooperatively induce vascular endothelial growth factor and metabolic switches hexokinase 2 and pyruvate dehydrogenase kinase 1. Mol Cell Biol. 2007;27:7381–7393. doi: 10.1128/MCB.00440-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Osthus RC, Shim H, Kim S, Li Q, Reddy R, Mukherjee M, et al. Deregulation of glucose transporter 1 and glycolytic gene expression by c-Myc. J Biol Chem. 2000;275:21797–21800. doi: 10.1074/jbc.C000023200. [DOI] [PubMed] [Google Scholar]
- 54.Kim JW, Tchernyshyov I, Semenza GL, Dang CV. HIF-1-mediated expression of pyruvate dehydrogenase kinase: a metabolic switch required for cellular adaptation to hypoxia. Cell Metab. 2006;3:177–185. doi: 10.1016/j.cmet.2006.02.002. [DOI] [PubMed] [Google Scholar]
- 55.Subramanian A, Miller DM. Structural analysis of alpha-enolase. Mapping the functional domains involved in down-regulation of the c-myc protooncogene. J Biol Chem. 2000;275:5958–5965. doi: 10.1074/jbc.275.8.5958. [DOI] [PubMed] [Google Scholar]
- 56.Sedoris KC, Thomas SD, Miller DM. Hypoxia induces differential translation of enolase/MBP-1. BMC Cancer. 2010;10:157. doi: 10.1186/1471-2407-10-157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Valera A, Pujol A, Gregori X, Riu E, Visa J, Bosch F. Evidence from transgenic mice that myc regulates hepatic glycolysis. Faseb J. 1995;9:1067–1078. doi: 10.1096/fasebj.9.11.7649406. [DOI] [PubMed] [Google Scholar]
- 58.Yang H, Ye D, Guan K, Xiong Y. IDH1 and IDH2 mutations in tumorigenesis: mechanistic insights and clinical perspectives. Clin Cancer Res. 2012;18 doi: 10.1158/1078-0432.CCR-12-1773. xx-xx. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kim JW, Dang CV. Cancer's molecular sweet tooth and the Warburg effect. Cancer Res. 2006;66:8927–8930. doi: 10.1158/0008-5472.CAN-06-1501. [DOI] [PubMed] [Google Scholar]
- 60.Parsons DW, Jones S, Zhang X, Lin JC, Leary RJ, Angenendt P, et al. An integrated genomic analysis of human glioblastoma multiforme. Science. 2008;321:1807–1812. doi: 10.1126/science.1164382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Flier JS, Mueckler MM, Usher P, Lodish HF. Elevated levels of glucose transport and transporter messenger RNA are induced by ras or src oncogenes. Science. 1987;235:1492–1495. doi: 10.1126/science.3103217. [DOI] [PubMed] [Google Scholar]
- 62.Gaglio D, Metallo CM, Gameiro PA, Hiller K, Danna LS, Balestrieri C, et al. Oncogenic K-Ras decouples glucose and glutamine metabolism to support cancer cell growth. Mol Syst Biol. 2011;7:523. doi: 10.1038/msb.2011.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Chesney J, Telang S. Regulation of Glycolytic and Mitochondrial Metabolism by Ras. Curr Pharm Biotechnol. 2012 doi: 10.2174/1389201011314030002. [DOI] [PubMed] [Google Scholar]
- 64.Manning BD, Cantley LC. AKT/PKB signaling: navigating downstream. Cell. 2007;129:1261–1274. doi: 10.1016/j.cell.2007.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kawauchi K, Araki K, Tobiume K, Tanaka N. p53 regulates glucose metabolism through an IKK-NF-kappaB pathway and inhibits cell transformation. Nat Cell Biol. 2008;10:611–618. doi: 10.1038/ncb1724. [DOI] [PubMed] [Google Scholar]
- 66.Tamada M, Suematsu M, Saya H. Pyruvate kinase M2: multiple faces for conferring benefits on cancer cells. Clin Cancer Res. 2012;18 doi: 10.1158/1078-0432.CCR-12-0859. xx-xx. [DOI] [PubMed] [Google Scholar]
- 67.Fernandez PC, Frank SR, Wang L, Schroeder M, Liu S, Greene J, et al. Genomic targets of the human c-Myc protein. Genes Dev. 2003;17:1115–1129. doi: 10.1101/gad.1067003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Li F, Wang Y, Zeller KI, Potter JJ, Wonsey DR, O'Donnell KA, et al. Myc stimulates nuclearly encoded mitochondrial genes and mitochondrial biogenesis. Mol Cell Biol. 2005;25:6225–6234. doi: 10.1128/MCB.25.14.6225-6234.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Morrish F, Noonan J, Perez-Olsen C, Gafken PR, Fitzgibbon M, Kelleher J, et al. Myc-dependent mitochondrial generation of acetyl-CoA contributes to fatty acid biosynthesis and histone acetylation during cell cycle entry. J Biol Chem. 2010;285:36267–36274. doi: 10.1074/jbc.M110.141606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Morrish F, Hockenbery D. Myc's mastery of mitochondrial mischief. Cell Cycle. 2003;2:11–13. doi: 10.4161/cc.2.1.275. [DOI] [PubMed] [Google Scholar]
- 71.Kaadige MR, Elgort MG, Ayer DE. Coordination of glucose and glutamine utilization by an expanded Myc network. Transcription. 2010;1:36–40. doi: 10.4161/trns.1.1.12142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wise DR, DeBerardinis RJ, Mancuso A, Sayed N, Zhang XY, Pfeiffer HK, et al. Myc regulates a transcriptional program that stimulates mitochondrial glutaminolysis and leads to glutamine addiction. Proc Natl Acad Sci U S A. 2008;105:18782–18787. doi: 10.1073/pnas.0810199105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Klimberg VS, McClellan JL, Claude H, Organ Honorary Lectureship. Glutamine, cancer, and its therapy. Am J Surg. 1996;172:418–424. doi: 10.1016/s0002-9610(96)00217-6. [DOI] [PubMed] [Google Scholar]
- 74.Chen MK, Espat NJ, Bland KI, Copeland EM, 3rd, Souba WW. Influence of progressive tumor growth on glutamine metabolism in skeletal muscle and kidney. Ann Surg. 1993;217:655–666. doi: 10.1097/00000658-199306000-00007. discussion 66-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Reitzer LJ, Wice BM, Kennell D. Evidence that glutamine, not sugar, is the major energy source for cultured HeLa cells. J Biol Chem. 1979;254:2669–2676. [PubMed] [Google Scholar]
- 76.Gallagher FA, Kettunen MI, Day SE, Lerche M, Brindle KM. 13C MR spectroscopy measurements of glutaminase activity in human hepatocellular carcinoma cells using hyperpolarized 13C-labeled glutamine. Magn Reson Med. 2008;60:253–257. doi: 10.1002/mrm.21650. [DOI] [PubMed] [Google Scholar]
- 77.Deberardinis RJ, Sayed N, Ditsworth D, Thompson CB. Brick by brick: metabolism and tumor cell growth. Curr Opin Genet Dev. 2008;18:54–61. doi: 10.1016/j.gde.2008.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Curthoys NP, Watford M. Regulation of glutaminase activity and glutamine metabolism. Annu Rev Nutr. 1995;15:133–159. doi: 10.1146/annurev.nu.15.070195.001025. [DOI] [PubMed] [Google Scholar]
- 79.Le A, Lane AN, Hamaker M, Bose S, Gouw A, Barbi J, et al. Glucose-independent glutamine metabolism via TCA cycling for proliferation and survival in B cells. Cell Metab. 2012;15:110–121. doi: 10.1016/j.cmet.2011.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.O'Donnell KA, Wentzel EA, Zeller KI, Dang CV, Mendell JT. c-Myc-regulated microRNAs modulate E2F1 expression. Nature. 2005;435:839–843. doi: 10.1038/nature03677. [DOI] [PubMed] [Google Scholar]
- 81.Gao P, Tchernyshyov I, Chang TC, Lee YS, Kita K, Ochi T, et al. c-Myc suppression of miR-23a/b enhances mitochondrial glutaminase expression and glutamine metabolism. Nature. 2009;458:762–765. doi: 10.1038/nature07823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Yuneva M, Zamboni N, Oefner P, Sachidanandam R, Lazebnik Y. Deficiency in glutamine but not glucose induces MYC-dependent apoptosis in human cells. J Cell Biol. 2007;178:93–105. doi: 10.1083/jcb.200703099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Le A, Cooper CR, Gouw AM, Dinavahi R, Maitra A, Deck LM, et al. Inhibition of lactate dehydrogenase A induces oxidative stress and inhibits tumor progression. Proc Natl Acad Sci U S A. 2010;107:2037–2042. doi: 10.1073/pnas.0914433107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Schreiber SL, Bernstein BE. Signaling network model of chromatin. Cell. 2002;111:771–778. doi: 10.1016/s0092-8674(02)01196-0. [DOI] [PubMed] [Google Scholar]
- 85.Jang MK, Mochizuki K, Zhou M, Jeong HS, Brady JN, Ozato K. The bromodomain protein Brd4 is a positive regulatory component of P-TEFb and stimulates RNA polymerase II-dependent transcription. Mol Cell. 2005;19:523–534. doi: 10.1016/j.molcel.2005.06.027. [DOI] [PubMed] [Google Scholar]
- 86.Rahman S, Sowa ME, Ottinger M, Smith JA, Shi Y, Harper JW, et al. The Brd4 extraterminal domain confers transcription activation independent of pTEFb by recruiting multiple proteins, including NSD3. Mol Cell Biol. 2011;31:2641–2652. doi: 10.1128/MCB.01341-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Delmore JE, Issa GC, Lemieux ME, Rahl PB, Shi J, Jacobs HM, et al. BET bromodomain inhibition as a therapeutic strategy to target c-Myc. Cell. 2011;146:904–917. doi: 10.1016/j.cell.2011.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Gui CY, Ngo L, Xu WS, Richon VM, Marks PA. Histone deacetylase (HDAC) inhibitor activation of p21WAF1 involves changes in promoter-associated proteins, including HDAC1. Proc Natl Acad Sci U S A. 2004;101:1241–1246. doi: 10.1073/pnas.0307708100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Peart MJ, Smyth GK, van Laar RK, Bowtell DD, Richon VM, Marks PA, et al. Identification and functional significance of genes regulated by structurally different histone deacetylase inhibitors. Proc Natl Acad Sci U S A. 2005;102:3697–3702. doi: 10.1073/pnas.0500369102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Seo SK, Jin HO, Woo SH, Kim YS, An S, Lee JH, et al. Histone deacetylase inhibitors sensitize human non-small cell lung cancer cells to ionizing radiation through acetyl p53-mediated c-myc down-regulation. J Thorac Oncol. 2011;6:1313–1319. doi: 10.1097/JTO.0b013e318220caff. [DOI] [PubMed] [Google Scholar]
- 91.Kretzner L, Scuto A, Dino PM, Kowolik CM, Wu J, Ventura P, et al. Combining histone deacetylase inhibitor vorinostat with aurora kinase inhibitors enhances lymphoma cell killing with repression of c-Myc, hTERT, and microRNA levels. Cancer Res. 2011;71:3912–3920. doi: 10.1158/0008-5472.CAN-10-2259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Soucek L, Jucker R, Panacchia L, Ricordy R, Tato F, Nasi S. Omomyc, a potential Myc dominant negative, enhances Myc-induced apoptosis. Cancer Res. 2002;62:3507–3510. [PubMed] [Google Scholar]
- 93.Soucek L, Helmer-Citterich M, Sacco A, Jucker R, Cesareni G, Nasi S. Design and properties of a Myc derivative that efficiently homodimerizes. Oncogene. 1998;17:2463–2472. doi: 10.1038/sj.onc.1202199. [DOI] [PubMed] [Google Scholar]
- 94.Savino M, Annibali D, Carucci N, Favuzzi E, Cole MD, Evan GI, et al. The action mechanism of the Myc inhibitor termed Omomyc may give clues on how to target Myc for cancer therapy. PLoS One. 2011;6:e22284. doi: 10.1371/journal.pone.0022284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Meijer TW, Kaanders JH, Span PN, Bussink J. Targeting hypoxia, HIF-1 and tumor glucose metabolism to improve radiotherapy efficacy. Clin Cancer Res. 2012;18 doi: 10.1158/1078-0432.CCR-12-0858. xx-xx. [DOI] [PubMed] [Google Scholar]
- 96.Sekhon HS, London CA, Sekhon M, Iversen PL, Devi GR. c-MYC antisense phosphosphorodiamidate morpholino oligomer inhibits lung metastasis in a murine tumor model. Lung Cancer. 2008;60:347–354. doi: 10.1016/j.lungcan.2007.10.028. [DOI] [PubMed] [Google Scholar]
- 97.Brown RV, Danford FL, Gokhale V, Hurley LH, Brooks TA. Demonstration that drug-targeted down-regulation of MYC in non-Hodgkins lymphoma is directly mediated through the promoter G-quadruplex. J Biol Chem. 2011;286:41018–41027. doi: 10.1074/jbc.M111.274720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Sedoris KC, Thomas SD, Clarkson CR, Muench D, Islam A, Singh R, et al. Genomic c-Myc quadruplex DNA selectively kills leukemia. Mol Cancer Ther. 2012;11:66–76. doi: 10.1158/1535-7163.MCT-11-0515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Jerby L, Ruppin E. Predicting drug-targets and biomarkers of cancer via genome-scale metabolic modeling. Clin Cancer Res. 2012;18 doi: 10.1158/1078-0432.CCR-12-1856. xx-xx. [DOI] [PubMed] [Google Scholar]
- 100.Adhikary S, Eilers M. Transcriptional regulation and transformation by Myc proteins. Nat Rev Mol Cell Biol. 2005;6:635–645. doi: 10.1038/nrm1703. [DOI] [PubMed] [Google Scholar]