Abstract
Life abounds with genetic variations writ in sequences that are often only a few hundred nucleotides long. Rapid detection of these variations for identification of genetic diseases, pathogens and organisms has become the mainstay of molecular science and medicine. This report describes a new, highly informative closed-tube polymerase chain reaction (PCR) strategy for analysis of both known and unknown sequence variations. It combines efficient quantitative amplification of single-stranded DNA targets through LATE-PCR with sets of Lights-On/Lights-Off probes that hybridize to their target sequences over a broad temperature range. Contiguous pairs of Lights-On/Lights-Off probes of the same fluorescent color are used to scan hundreds of nucleotides for the presence of mutations. Sets of probes in different colors can be combined in the same tube to analyze even longer single-stranded targets. Each set of hybridized Lights-On/Lights-Off probes generates a composite fluorescent contour, which is mathematically converted to a sequence-specific fluorescent signature. The versatility and broad utility of this new technology is illustrated in this report by characterization of variant sequences in three different DNA targets: the rpoB gene of Mycobacterium tuberculosis, a sequence in the mitochondrial cytochrome C oxidase subunit 1 gene of nematodes and the V3 hypervariable region of the bacterial 16 s ribosomal RNA gene. We anticipate widespread use of these technologies for diagnostics, species identification and basic research.
INTRODUCTION
Life abounds with genetic variations writ in sequences that are often only a few hundred nucleotides long. Rapid detection of these variations for identification of genetic diseases, pathogens and organisms has become the mainstay of molecular science and medicine. Variants can be single or multiple, known or unknown, in areas of high G/C or A/T content and can also cause or interrupt secondary structures into which single-stranded nucleic acids can fold. This immense range of possibilities creates technical challenges for detection methods based on probe/target hybridization. DNA sequencing, by either conventional or newer methods, may ultimately provide the greatest possible information about a selected target or a whole genome, but sequencing is currently impractical for routine diagnostic use. For these reasons, most molecular diagnostic assays in use today depend on closed-tube polymerase chain reaction (PCR) amplification of short stretches of double-stranded DNA. The detection of specific variants is usually determined by sequence-specific probes of different color (1). Analysis of multiple variants in the same reaction typically requires the use of several sequence-specific probes (2) or sloppy molecular beacons (3,4) of different colors. These approaches are limited, however, by the fact that most commercial fluorescent thermocyclers use a maximum of four to six fluorescent colors. High resolution melt analysis using DNA-binding fluorescent dyes overcomes these limitations by scanning stretches several hundred nucleotides long in just one color (5). But, DNA–binding dyes cannot be used to analyze several targets in the same tube, unless those targets melt at non-overlapping temperatures. Moreover, high-resolution melt analysis requires specialized equipment because sequence differences in whole amplicons cause only subtle changes in melt curves over a narrow range in temperature (6).
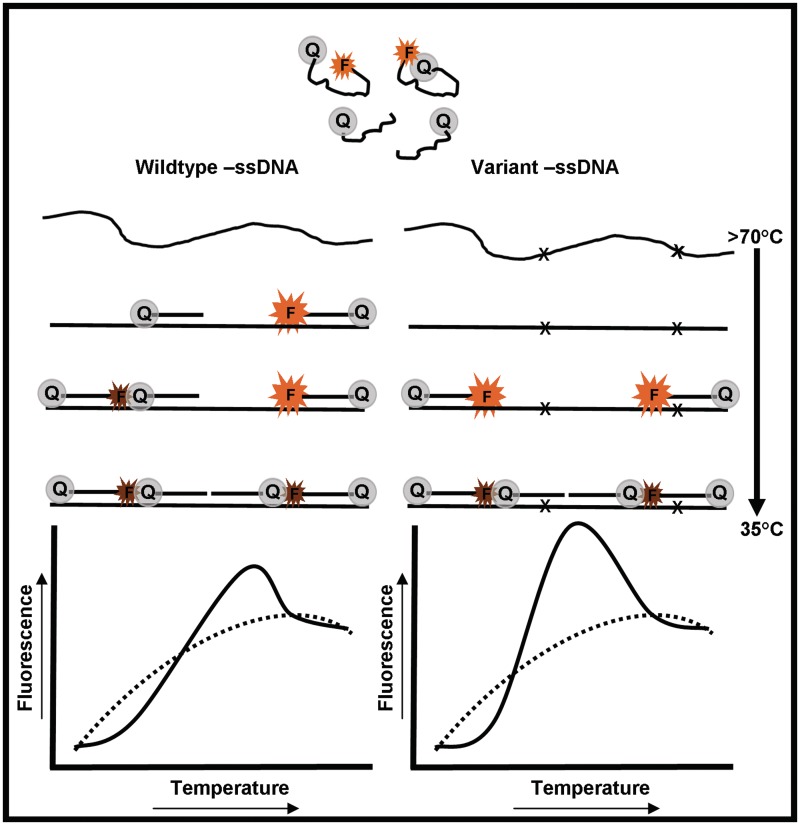
Here, we describe a new, highly informative PCR approach that overcomes these challenges by combining efficient quantitative amplification of single-stranded DNA, through LATE-PCR (7,8), with sets of Lights-On/Lights-Off probe pairs. Figure 1 illustrates this new technology for two pairs of Lights-On/Lights-Off probes and their temperature-dependent hybridization to two target strands that differ by the absence or presence of two mutations that decrease the melting temperature (Tm) of a single Lights-On probe and a single Lights-Off probe. We refer to the resulting temperature-dependent signals as fluorescent contours because they are a composite of all probe–target interactions (see the lower portion of Figure 1).
Figure 1.
A schematic description of the temperature-dependent hybridization of a set of Lights-On/Lights-Off probes to two variant targets and their resulting fluorescent contours.
LATE-PCR is an advanced form of non-symmetric PCR that uses pairs of limiting and excess primers. Together, these primers efficiently generate double-stranded DNA followed by the production of single-stranded amplicons. Depending on the number of target molecules present at the start of amplification and the number of thermal cycles used, there are 10–20 fold more single-stranded amplicons than double-strand amplicons at the end of amplification. When the temperature is lowered at end-point, the complementary double-stranded molecules are unavailable to interact with the Lights-On/Lights-Off probes, because Lights-On/Lights-Off probes are low-temperature probes with Tm’s that are at least 5°C below the Tm of the limiting primer. The accumulated single-stranded molecules, in contrast, are fully available to bind probes over a wide range of temperatures.
Each Lights-On probe is labeled with a fluorophore, for example, a molecular beacon with a two base pair self-complementary stem (Figure 1). A Lights-Off probe, in contrast, is labeled with only a quencher moiety, such as a black hole quencher. Each Lights-Off probe serves to absorb energy from the fluorophore of its adjacent Lights-On probe, when both are bound to a target. As each probe is relatively short and interrogates only a portion of the target, variants that differ by only a single base have distinct and consistently different fluorescent signatures in standard thermocyclers. As the signal from each Lights-On probe is extinguished, multiple pairs of probes of the same color or different colors can be used to analyze sequences several hundred nucleotides long.
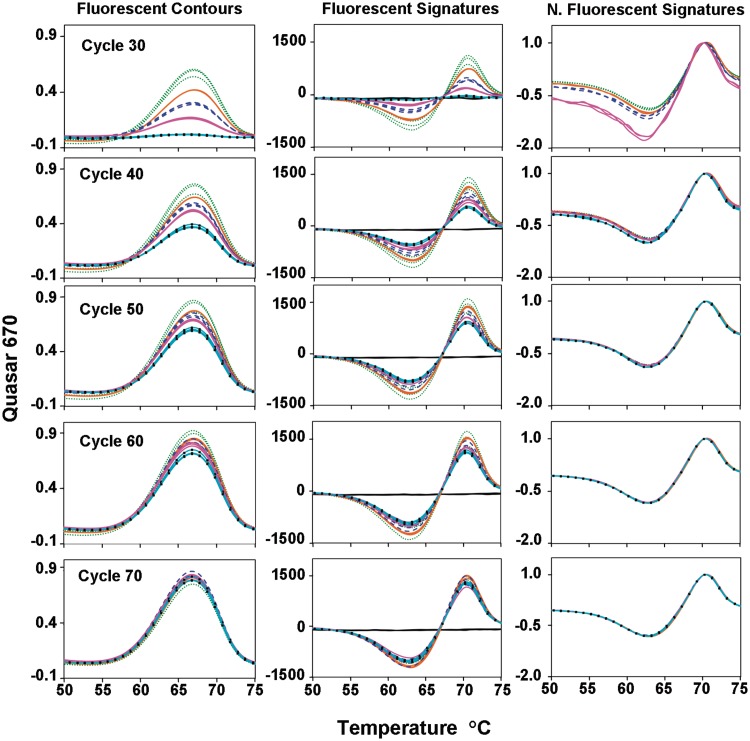
For convenience, we mathematically transform each fluorescent contour into its first derivative curve, which we refer to as a fluorescent signature. Once these fluorescent signatures are normalized, it is easy to compare different samples for the presence of variants. Fluorescent signatures using the same targets and probes are highly reproducible (Figure 2). When an unfamiliar fluorescent signature is observed, its sequence can be determined from an aliquot using a convenient Dilute-‘N’-Go protocol (9,10). Libraries of fluorescent signatures and their corresponding sequences can be mathematically coded and stored in a database. Subsequent samples need only to be sequenced if their signatures differ from all known signatures in the library.
Figure 2.
Reproducibility and detection of the RRDR region of the rpoB gene for reference strain H37Rv of M. tuberculosis using triplicate replicate reactions in a dilution series, each with two pairs of Lights-On/Lights-Off probes. Fluorescent contours, signatures and normalized signatures for 100 000 (dotted green), 10 000 (solid orange), 1000 (dashed blue) 100 (solid magenta) and 10 copies (solid light blue with black squares) across 70 cycles of amplification starting at cycle 30.
The aim of this report is to provide an overview of how to design and utilize LATE-PCR Lights-On/Lights-Off assays as well as our evidence for their reliability and versatility. These findings are illustrated using different DNA targets amplified in monoplex assays including (i) the RRDR portion of the rpoB gene responsible for rifampin resistance in Mycobacterium tuberculosis. This target was amplified with one pair of primers and was probed with two pairs of Lights-On/Lights-Off probes (Figure 2). Replicate reactions containing 10–100 000 copies illustrate the sensitivity and reproducibility of normalized fluorescent signatures; (ii) a portion of the mitochondrial cytochrome C oxidase subunit 1 (CO1) gene of Heterohabditis bacteriophora, a species of nematode (11). This target was amplified with one pair of primers and probed with five pairs of Lights-On/Lights-Off probes in three different fluorescent colors. This example illustrates that unique fluorescent signatures are generated regardless of whether the Tm of a Lights-On probe is higher or lower than the Tm of its adjacent Lights-Off probe, and that the overall fluorescent signature of a target can be comprised of differently colored sub-signatures; (iii) the RRDR portion of the rpoB gene responsible for rifampin resistance in M. tuberculosis was also probed with three pairs of Lights-On/Lights-Off probes. This example illustrates that the Lights-On/Lights-Off probes readily detect single base pair changes in a target sequence; (iv) the V3 region of the 16 s ribosomal RNA gene (12) was amplified using one pair of primers, and the intervening hypervariable sequences were probed with four pairs of Lights-On/Lights-Off probes in a single color. This example shows that Gram-positive and Gram-negative bacteria can be distinguished both as individual species and as classes of organisms, because the same probes hybridize in different portions of the detection temperature range.
MATERIALS AND METHODS
rpoB gene amplification and data analysis protocols
All LATE-PCR amplifications were performed in triplicate in 25 µl volume with 6 ng of human genomic DNA (Promega, WI, USA). Each reaction consisted of 1× PCR buffer 20 mM Tris–HCl (pH 8.0), 50 mM KCl (Invitrogen, CA, USA), 2 mM MgCl2, 200 nM deoxynucleotide triphosphates (dNTPs; Promega), 50 nM limiting primer, 1000 nM excess primer, 1.25 units of Platinum® Taq DNA polymerase (Invitrogen), 500 nM of probes 1, 3 and 6 and 200 nM of probes 2, 4 and 5 (Biosearch Technologies, CA, USA). In one experiment, a serial dilution of M. tuberculosis was used starting at 100 000–10 copies, whereas all other experiments contained ∼1000 genomes of each strain. The thermal profile conditions used on the Stratagene MxPro 3005P were as follows: 95°C/3 min for one cycle, followed by 98°C/10 s–75°C/40 s for 60 cycles, a hold at 75°C for 10 min followed by 10 min hold at 25°C. This was followed by a melt starting at 25°C with 1°C increments at 30 s intervals to 92°C. The fluorescence data analysis of the probe-target hybridizations following amplification was by melt-curve analysis using the first derivative for Quasar 670 fluorescence for temperatures between 40°C and 75°C. From this dataset, the highest fluorescent value was used to normalize the data to one.
COI gene amplification and data analysis protocols
The DNA of a single nematode H. bacteriophora was extracted by placing the individual worm into 25 µl volume of a lysis buffer containing 100 µg/ml proteinase K (Sigma-Aldrich, MO, USA); 10 mM Tris–Cl, pH 8.3 (Sigma-Aldrich) and 5 µM sodium-dodecyl-sulfate (Sigma-Aldrich); heating to 50°C for 30 min followed by 95°C for 10 min; then adding 25 µl of 10 mM Tris–Cl, pH 8.3, buffer prior to storage at −20°C. LATE-PCR amplifications were performed in triplicate in 25 µl volume with ∼10 000 mitochondrial genomes consisting of 1× PCR buffer (Invitrogen), 100 nM of each probe (Biosearch Technologies), 3 mM MgCl2, 250 nM dNTPs (Promega), 100 nM of each limiting primer, 1000 nM of excess primer, 1.25 units of Platinum® Taq DNA Polymerase (Invitrogen) and 1 µl of previously extracted nematode DNA. The thermal profile conditions on the Bio-Rad IQ5 were as follows: 95°C for 3 min followed by 95°C/5 s–55°C/10 s–72°C/45 s for five cycles followed by 95°C/5 s–64°C/10 s–72°C/45 s for 55 cycles followed by a melt starting at 25°C with 1°C increments at 30 s intervals to 95°C followed by an annealing starting at 95°C with 1°C decrements at 30 s intervals to 25°C. The fluorescence data analysis of the probe–target hybridizations following amplification were analyzed by anneal curve analysis using the first derivative for each fluorophore separately Cal Orange 560, Cal Red 610 and Quasar 670.
16s ribosomal RNA gene amplification and data analysis protocols
LATE-PCR amplifications were performed in triplicate in 25 µl volume with ∼10 000 genomes for 11 bacterial species (Table 1). These reactions used illustra PuReTaq Ready-To-Go™ PCR Beads (GE Healthcare, NJ, USA), consisting of 1× PCR buffer, 1.5 mM MgCl2 (supplemented with 1.5 mM MgCl2), 200 nM dNTPs, 1.25 units of Taq DNA Polymerase, 50 nM limiting primer, 1000 nM excess primer, 100 nM of probes 1–4 and 300 nM of probes 5–8 (Biosearch Technologies). The fluorescent signatures for the Gram-positive and Gram-negative targets were determined using the first derivative of the final anneal curve with background fluorescence of the no template control subtracted. The highest peaks of each target were then normalized to one.
Table 1.
Predicted probe Tm’s (°C) for Lights-On/Lights-Off probes within V3 region of 16 s ribosomal RNA gene
| Target | On Probe 1 | On Probe 2 | On Probe 3 | On Probe 4 | Off Probe 1 | Off Probe 2 | Off Probe 3 | Off Probe 4 |
|---|---|---|---|---|---|---|---|---|
| AB | 50.2 | 64.9 | 18.1 | 59.5 | 50.8 | 42.4 | 18.3 | 40.3 |
| EA | 69.5 | 58.6 | 11.6 | 69.7 | 61.5 | 54.5 | 18.3 | 42.1 |
| EC | 69.5 | 58.6 | 11.6 | 69.7 | 61.5 | 54.5 | 33 | 56.4 |
| ENFS | 24.1 | 59.7 | –2.8 | 70.6 | 7.8 | 34.6 | 20.6 | –3.2 |
| ENFM | 24.1 | 59.7 | –2.8 | 70.6 | 7.8 | 34.6 | 20.6 | –3.2 |
| KP | 70.6 | 66 | 13.8 | 70.6 | 62.6 | 64.5 | 35 | 57.7 |
| PA | 68.9 | 66 | –2.8 | 58 | 52 | 60 | 35 | 39.6 |
| SA | 54.5 | 53.8 | 63.9 | 34.7 | 36.8 | 39.3 | 20.6 | 30.3 |
| SE | 54.5 | 53.8 | 53.9 | 34.7 | 36.8 | 39.3 | 20.6 | 30.3 |
| SH | 54.5 | 53.8 | 50.1 | 34.7 | 36.8 | 38 | 20.6 | 30.3 |
Mixtures of known genomic DNA proportions were prepared with a total copy number of 10 000 genomes. A dilution series of each target of interest was first determined by real-time PCR with SYBR® Green (Invitrogen) to assess initial copy number. These dilutions were then adjusted to match CT-values ensuring that the starting concentrations prior to mixture were the same. From these concentrations, known proportions were made to assess the level of detection in mixtures. The following proportions of Staphylococcus epidermidis (SE) and Klebsiella pneumoniae (KP) were constructed; 100% SE, 90% SE;10% KP, 50% SE;50% KP, 10% SE;90% KP and 100% KP. The thermal profile on the Bio-Rad IQ5 were as follows: 95°C for 3 min followed by 95°C/10 s–65°C/15 s–72°C/45 s for 45 cycles followed by a melt starting at 25°C with 1°C/30 s increases up to 80°C followed by an annealing starting at 80°C with 1°C/30 s decreases down to 25°C. Fluorescence of the probe–target hybridizations following amplification was by anneal curve analysis for Quasar 670. The fluorescent signatures were determined using the first derivative of the final anneal curve with the temperature range restricted between 43°C and 75°C to emphasize the signatures of the mixtures.
A complete list of sequences for all primers and probes used can be found in the supplementary data.
RESULTS
Design options for Lights-On/Lights-Off probe pairs are very flexible
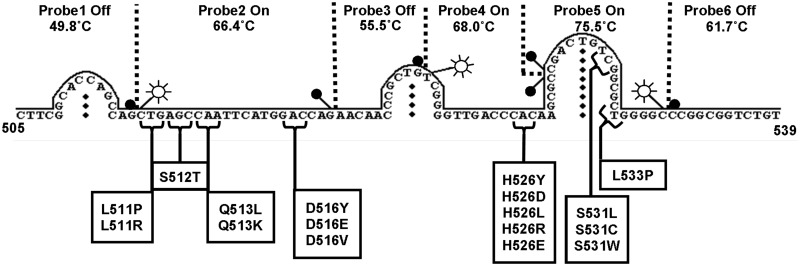
In the simplest case, each Lights-On probe has a Tm, TmOn, that is at least a few degrees (3–5°C) higher than the Tm of its paired Lights-Off probe, TmOff. Under these conditions when the temperature of the reaction is decreased at end-point, a fluorescent signal is observed above background and is then extinguished in accord with the number of degrees between TmOn and TmOff. This relationship can be extended to more than one pair of probes. For instance, in Figure 2, the rpoB gene target shown in Figure 4 was probed using two pairs of probes having the relationship Tm1On (75.5°C) > Tm2On (68.0°C) > Tm1Off (61.7°C) > Tm2Off (55.5°C). The combined fluorescent contour of the four probes rises and descends a single time as the detection temperature is lowered because the Lights-On probes bind sequentially followed by the two Lights-Off probes. Had the probes been arranged in the order: Tm1On (75.5°C) > Tm1Off (68.0°C) > Tm2On (61.7°C) > Tm2Off (55.5°C), the fluorescent contour would have gone up and down twice within the same temperature range. This alternative design was not possible due to the secondary structure of the single-stranded target (see Figure 4 and subsequent sections).
Figure 4.
A schematic diagram of the secondary structure for 101 nucleotides of the RRDR region between codons 505 and 539 for reference strain H37Rv of M. tuberculosis including the location and Tm's of the Lights-On/Lights-Off probe set, as well as a list and locations of known mutations that cause rifampin resistance.
The fluorescent contours displayed in the first column of Figure 2 illustrate another important feature of LATE-PCR. As the number of thermal cycles increases, single-stranded amplicons accumulate until these product strands reach their ‘terminal concentration’, at which point, synthesis stops because the single strands out compete their own primers for binding to the template strand. The number of thermal cycles required to reach terminal concentration depends on the efficiency of amplification (7) as well as the number of target molecules present at the start of the reaction. The more targets, the fewer cycles required to reach terminal concentration (8). Thus, in Figure 2, the replicate reactions begun with 100 000 copies of the target reached terminal concentration after 50 thermal cycles, whereas those begun with 10 copies of the target reached terminal concentration after 70 thermal cycles.
Figure 2, Column 2 demonstrates that each fluorescent contour can be converted to a fluorescent signature by plotting the first derivative of all values as a function of temperature. Just like the fluorescent contour, the amplitude of the fluorescent signature changes depending on target concentration and the number of cycles. But when each fluorescent signature is normalized to its highest value, Column 3, it becomes clear that this target and its probes have the same fluorescent signature as soon as a measurable number of single-stranded molecules has accumulated. This high degree of reproducibility (15-fold in this case) is characteristic of fluorescent signatures generated by these technologies.
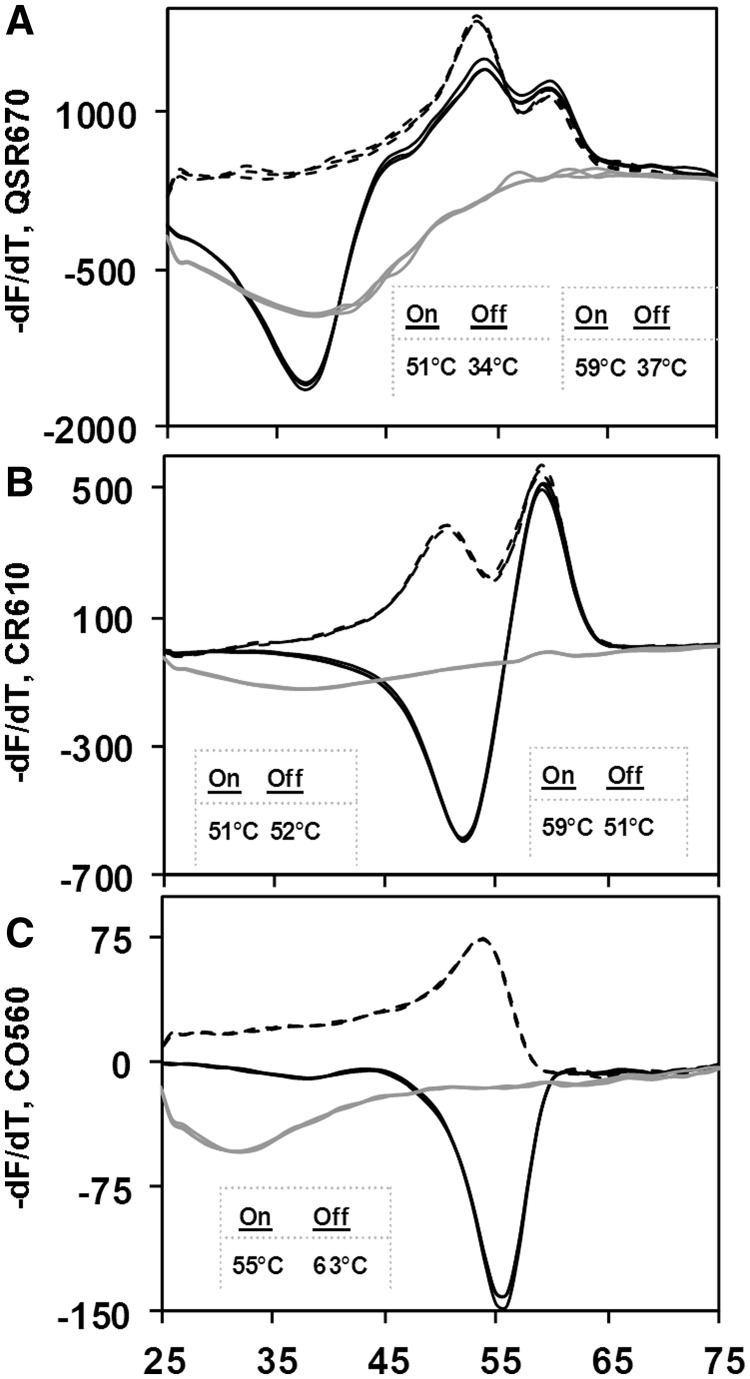
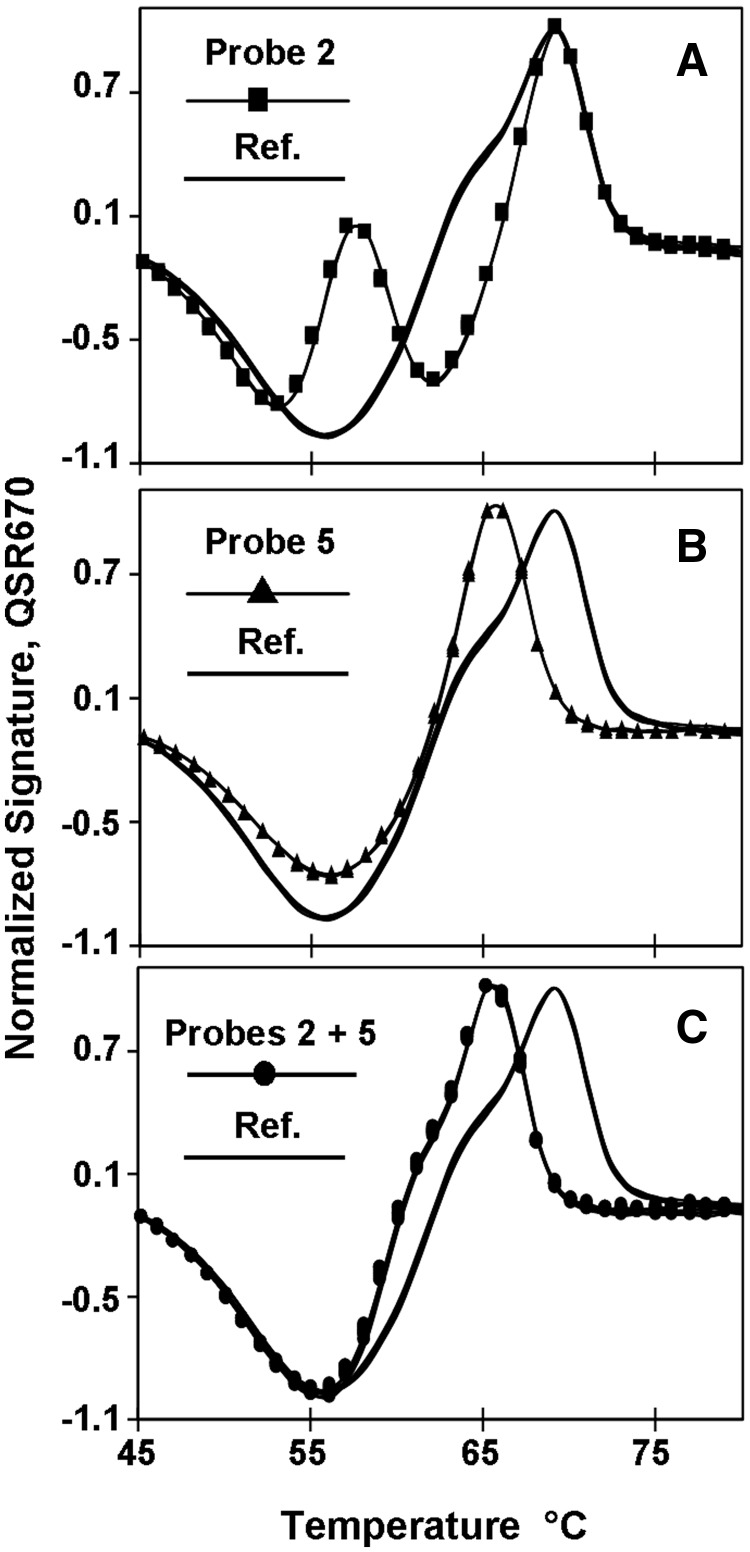
In Figure 3, a single pair of primers was used to generate a 460 base long single-stranded amplicon containing a portion of the mitochondrial CO1 gene in the nematode H. bacteriophora. The resulting target sequence was 388 bases long and was probed in sections using two Lights-On/Lights-Off probe pairs labeled in Quasar, Panel A, two Lights-On/Lights-Off probe pairs labeled in Cal Red, Panel B and one Lights-On/Lights-Off probe pair labeled in Cal Orange, Panel C. Each set of colored probes generated its own fluorescent sub-signature spanning the detection temperature (solid lines).
Figure 3.
Comparison of fluorescent signatures with Lights-On probes only and pairs of Lights-On/Lights-Off probes using a portion of the mitochondrial cytochrome oxidase I gene of H. bacteriophora (NC_008534). All reactions were performed in triplicate. Lights-On probes (dotted lines), Lights-On/Lights-Off probe pairs (solid lines) and negative controls (gray lines). (A) when Tm's for both Lights-On probes > Lights-Off probes. (B) when Tm of a single Lights-On probe = Lights-Off probe for one set in a pair. (C) when Tm of Lights-On probe < Lights-Off probe.
The results in Figure 3 also illustrate that the relative temperatures of probes in a pair or several pairs in a set is very flexible. One set of reactions was analyzed using just the Lights-On probes in each set (dotted lines), whereas a second set of reactions was analyzed using the Lights-On/Lights-Off pairs of probes. In Panel A, the two Lights-On probes had TmOn of 59°C and 51°C; and when they were used alone, the rate of fluorescence increased in two steps as the temperature was decreased. Addition of two Lights-Off probes, both with TmOff well below those of their adjacent Lights-On probes extinguished the fluorescent signals at low temperature. For this reason, the fluorescent signature reached its lowest negative value at ∼37°C.
Figure 3, Panel B illustrates a more complex case in which two Lights-On probes labeled in Cal Red were used to scan a different section of the target without or with addition of two Lights-Off probes. In this case, the Lights-On probes had TmOn of 59°C and 51°C, whereas the two Lights-Off probes have very similar TmOff of 51°C and 52°C. Once again in the absence of the Lights-Off probes, hybridization of the Lights-On probes generated two peaks in the rate of fluorescence. Addition of the Lights-Off probes extinguished the Lights-On probe with the 59°C TmOn as soon as it reached its highest rate of fluorescence and also suppressed any signal from the Lights-On probe with the 51°C TmOn.
The results shown in Figure 3, Panel C were obtained using just one pair of Lights-On/Lights-Off probes labeled in Cal Orange. In this case, the TmOff of the Lights-Off probe, 63°C, is actually higher than the TmOn of the Lights-On probe, 55°C. In the absence of the Lights-Off probe, the Lights-On probe generates a single melt peak at ∼54°C. But, in the presence of the Lights-Off probe, the pair of probes generates a distinctive melt valley at ∼56°C, because the Lights-Off probe quenches the fluorescence of the Lights-On probe as soon as it hybridizes to the target. Less light emanates from the fluorophore of the Lights-On probe when it is in close contact with the quencher of the adjacent Lights-Off probe on the target, than emanates from the same fluorophore on the unbound Lights-On probe. In solution, the probe is a vibrating random coil, and the interactions of fluorophore and the quencher moieties are not as stable (13).
Hairpin structures can impose constraints on probe design
A hairpin in a single-stranded target can interfere with probe binding if it has a high Tm. Therefore, if the target sequence and its variants are known in advance, the first step in designing probes is to establish in silico whether these sequences are likely to form hairpins as the detection temperature is decreased. For example, the RRDR sequence in the M. tuberculosis rpoB gene target has a high GC content and is predicted to form three hairpins whose Tm’s are 57.9°C, 69.7°C and 79.2°C (left to right in Figure 4). The hairpin with the highest Tm is also a sequence containing many of the mutations that result in rifampicin resistance. These features drove the design of the three pairs of Lights-On/Lights-Off probes used to coat this target. The two probes in the set having the highest TmOn, 75.5°C and 68.0°C, are Lights-On probes, which subdivide the hairpin and prevent its closure. Neither probe alone is sufficient to keep this hairpin open at lower temperatures (results not shown). Binding of the high Tm probes also prevents formation of the lower Tm hairpins. As soon as the optimal location of the high Tm probes was established, designing the locations and compositions of the other probes needed to cover the rest of the rifampin resistance mutations was straight forward. The six probes shown in Figure 4 comprise the single optimal set for this target.
Single nucleotide changes alter fluorescent signatures
Each probe within a set responds independently to mutations in its portion of the target sequence, but each fluorescent signature integrates the temperature-dependent signal of the entire set of probes. The set of six probes described in Figure 4 provided a tool with which to explore how sensitive this new analytic technology would be to single nucleotide changes within a target. As shown in Figure 5, solid line, the fluorescent signature of the reference target, has a high temperature peak at ∼69°C, which is due to the rapid rise in fluorescence that occurs when the two highest Tm Lights-On probes bind to the target. Once these two probes have saturated the target, the fluorescent signature begins to descend. Binding of the third Lights-On probe, at TmOn 66.4°C, is observed as a shoulder in the decreasing rate of fluorescence between 69°C and 56°C. The fluorescent signature descends below zero due to sequential binding of the three Lights-Off probes. These events result in the valley at ∼56°C. The rate of fluorescence then ascends back to zero at low temperature, reflecting gradual extinction of fluorescence by the Lights-Off probes.
Figure 5.
Fluorescent signatures for several mutations in the RRDR region of the rpoB gene of M. tuberculosis using triplicate replicate reactions. (A) A to G substitution in codon 516 beneath the Lights-On probe, Tm 66.4°C. (B) T to C substitution in codon 533 beneath Lights-On probe, Tm 75.5°C. (C) The same A to G substitution in codon 516 with the same T to C substitution in codon 533. See Figure 3 for location of probes.
Figure 5 also demonstrates that two different point mutations in the targets of two different Lights-On probes cause the Tm of each probe to decrease independently of the other probes in the set. Thus, the mutation in the target for Lights-On probe TmOn 66.4°C causes the 63°C shoulder to shift to the left, Panel A. The mutation in the target for Lights-On probe TmOn 75.5°C causes the initial rise in fluorescence to shift to a lower temperature, Panel B. Furthermore, because the Tm’s of all three Lights-On probes are now very similar, the fluorescent signature exhibits a single peak at 65°C. The fluorescent signature of the double mutation, Panel C, combines the changes observed in Panels A and B. The peak in the fluorescent signature once again is at 65°C, but a shoulder in the descending rate of fluorescence is now observed at ∼58°C.
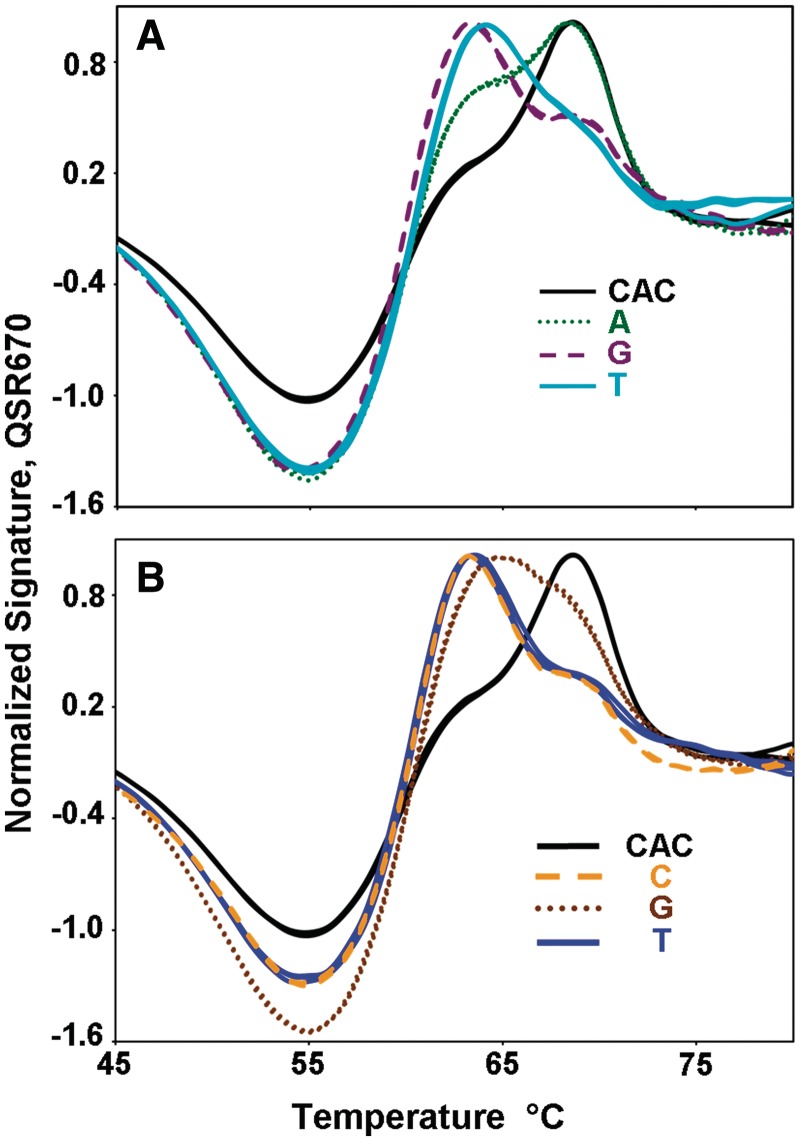
Figure 6 illustrates an even more rigorous test of the capacity of Lights-On/Lights-Off probes to resolve closely related sequence variants. In this case, all seven of the possible variants of the first and second nucleotides of codon 526 in the rpoB gene target were found to have unique fluorescent signatures. All four of the possible base changes at the first nucleotide of the codon, Panel A, are easily distinguished. Only two of the possible four variants at the second nucleotide, CCC and CTC, are very similar, but both of these are very different from the reference sequence, CAC, of the rifampin sensitive strain.
Figure 6.
Analysis of seven alleles of codon 526 in the RRDR region of the rpoB gene of M. tuberculosis using triplicate replicate reactions. (A) Normalized fluorescent signatures for each nucleotide in the first position, C: solid black, A: dotted green, G: dashed purple and T: solid light blue. (B) Normalized fluorescent signatures for each nucleotide in the second position, A: solid black, C: dashed orange, G: dotted brown and T: solid blue.
Analysis of variants for which there is no reference sequence
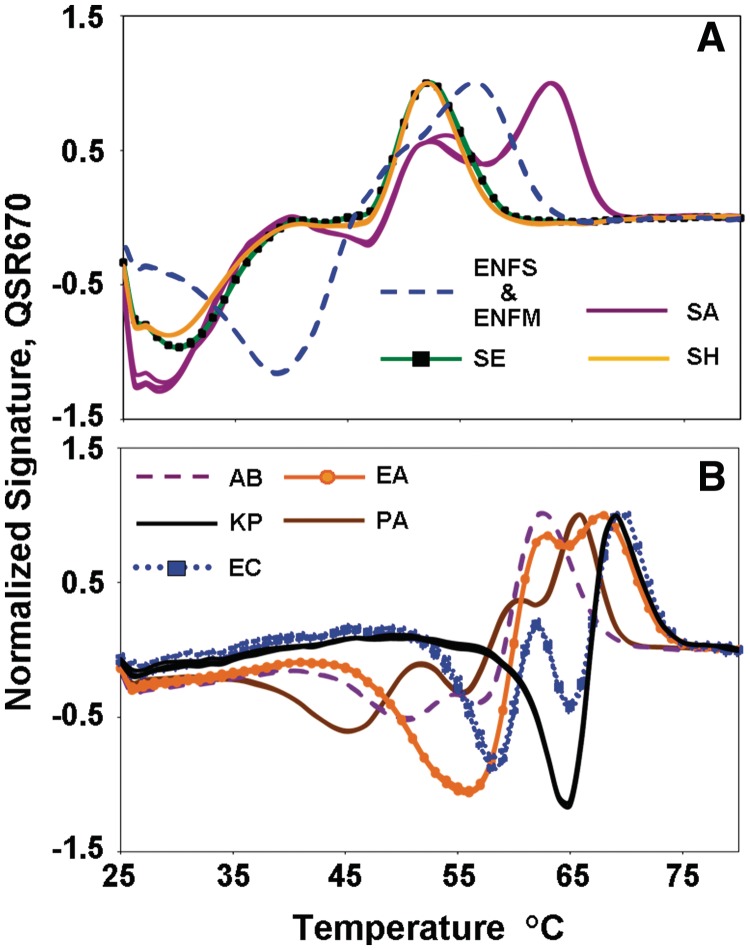
The 16s ribosomal RNA gene in bacteria is ideal for species genotyping because it contains three hypervariable sequences, V1, V2 and V3, each flanked by a pair of conserved sequences (12). But, the Michigan State Ribosomal Database Project [http://rdp.cme.msu.edu/] lists 1.9 million entries for bacterial 16s ribosomal RNA gene sequences, making it obvious that no sequence can a priori be regarded as the wild type nor can any variant be predicted in advance. Nevertheless, Lights-On/Lights-Off probes can be used to analyze these sequences. Figure 7 shows the fluorescent signatures for 11 species of bacteria using a single pair of primers to amplify the V3 target plus a set of four Lights-On probes labeled in Quasar 670 and four Lights-Off probes. All probes were designed as consensus probes, in as much as none was perfectly matched to any of these 11 targets (Table 1). The fluorescent signatures of the Gram-positive species as a group share similarities, Panel A, as do the fluorescent signatures of the Gram-negative species, Panel B. This observation is readily explained by the fact that the Gram-negative targets have a higher GC content than the Gram-positive targets. As a consequence, the four consensus Lights-On probes hybridize at higher Tm’s to the Gram-negative species than to the Gram-positive species.
Figure 7.
The fluorescent signatures of the 16s ribosomal RNA gene V3 region for 10 bacterial species using four pairs of Lights-On/Lights-Off probes in one color: (A) five Gram-positive species, E. faecalis (ENFS) and E. faecium (ENFM): dashed blue, S. aureus (SA): solid purple, S. epidermidis (SE): green with black square and S. haemolyticus (SH): solid orange (B) Six Gram-negative bacterial species. A. baumannii (AB): dashed purple, E. aerogenes (EA): solid orange with circle, K. pneumoniae (KP): solid black, P. aeruginosa (PA): solid brown and E. cloacae (EC): dashed blue with square. All reactions were performed in triplicates.
The fluorescent signatures of Enterococcus faecalis and Enterococcus faecium (Panel A, ENFS and ENFM) were identical to each other. Only three of the eight probes in the set actually hybridized to E. faecalis and E. faecium, because the Tm’s of the other five probes were too low and because the sequences to which the three probes bind in these species are identical. This ambiguity can be eliminated by increasing the length of the V3 amplicon to include a second highly variable region which is then probed using a second set of four pairs of consensus Lights-On/Lights-Off probes labeled in Cal Red. The complete reaction generates unique fluorescent signatures for each species comprised of two sub-signatures in two colors (Carver-Brown et al., in preparation).
Mixtures of targets having distinctly different fluorescent signatures
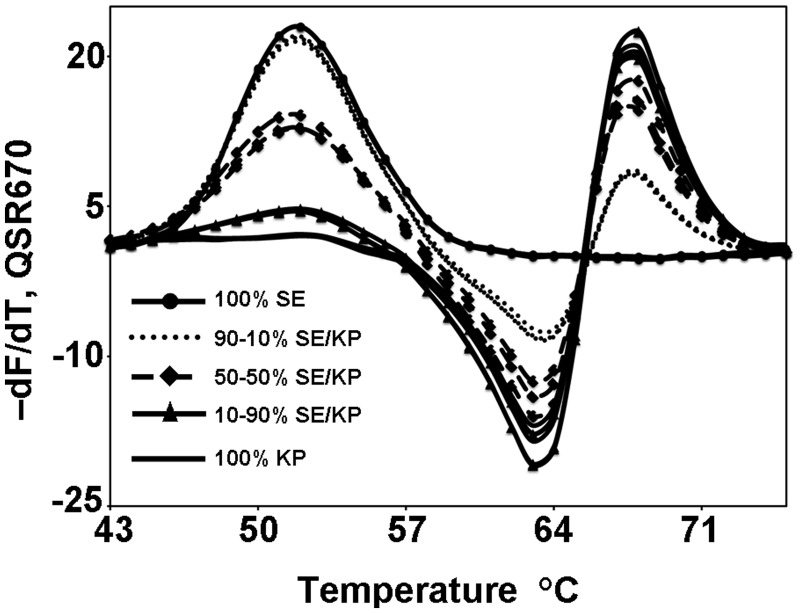
Mixtures of variants are notoriously difficult to analyze, and it is clear from Figures 6 and 7 that it would be virtually impossible to resolve target mixtures whose individual fluorescent signatures are very similar. However, the distinctly different fluorescent signatures of the Gram-positive and Gram-negative species shown in Figure 7 allowed us to measure the extent to which composites of two very dissimilar fluorescent signatures can be distinguished from their pure component signatures. Figure 8 shows that the fluorescent signature for a 10:90 mixture of SE:KP can readily be distinguished from that of pure KP, and the fluorescent signature of a 90:10 mixture of SE:KP can readily be distinguished from that of pure SE, provided analysis is based on the temperature ranges, 48–56°C and 66–71°C, in which the component signatures exhibit their greatest differences.
Figure 8.
The fluorescent signatures of different ratios in a mixture of S. epidermidis (SE) and K. pneumoniae (KP). 100% SE: solid line with circle, 90–10% SE/KP: dotted line, 50–50% SE/KP: dashed line with diamond, 10–90% SE/KP: solid line with triangle and 100% KP: solid line.
DISCUSSION
Lights-On/Lights-Off probes were initially conceived as a way to convert a symmetric PCR assay for mutations in the RRDR of the rpoB gene that requires five differently colored molecular beacons to analyze double-strand DNA (2) into a LATE-PCR assay that uses just one fluorescent color to analyze single-stranded DNA. But as this report shows, the combination of LATE-PCR with Lights-On/Lights-Off probes has emerged as a new and exciting platform for molecular diagnostics. The fundamental insight that we have drawn from the invention and exploration of Lights-On/Lights-Off probes in LATE-PCR assays is that closed-tube reactions are whole systems composed of enzymes, inorganic ions, nucleotide precursors and their by products, amplicons and oligonucleotide probes. The fluorescent contours measured at end-point are not traditional melt curves, and Tm values cannot be determined from the peaks of fluorescent signatures. This is because fluorescent contours describe the sum of numerous temperature-dependent dynamic equilibriums between all of the reaction components in all of their possible conformations, as well as all interactions of these components. It is therefore not surprising that alteration of a single component, such as the sequence of an amplified target, changes these equilibriums and results in a new, highly reproducible fluorescent signature.
This report does not provide a comprehensive description of the options for building diagnostic assays using this new technology, because Lights-On/Lights-Off probes have proven far more versatile and informative than was originally conceived. Thus far, every single base change in the rpoB gene target we have examined alters the fluorescent signature, probably because individual Lights-On/Lights-Off probes are relatively short. As we show here, sets of differently colored Lights-On/Lights-Off probes can be combined to examine adjacent sections of a long amplicon, such as the CO1 gene target. Additional experiments in our laboratory suggest that it will be possible to define sets of consensus primers and consensus probes that can distinguish a great many species within a genus or even a whole phylum of organisms (14). This closed-tube approach could be used to reduce the cost of cataloging all living species, because it will only be necessary to sequence new signatures that differ from those in an ever increasing library of fluorescent signatures (L. Rice, in preparation).
Sets of differently colored Lights-On/Lights-Off probes can also be used to analyze separate amplicons generated in the same closed-tube multiplexed LATE-PCR assay. For instance, in the case of multi-drug-resistant tuberculosis, a single-tube reaction can generate separate amplicons for the rpoB gene target, the mabA gene target and the katG gene target in order to simultaneously determine whether a particular strain is resistant or sensitive to either rifampin or isoniazid the two first line antibiotics (J. Rice et al., in preparation). Similarly, in the realm of cancer diagnostics, we have used a multicolored multiplexed approach to build a single-tube assay that scans for virtually all possible mutations in exons 18–21 of the epidermal growth factor receptor gene (15,16) (Tetrault et al., in preparation).
As Lights-On/Lights-Off probes are informative in the absence of prior knowledge of the target sequence, they can also be used to compare variants for which there is no reference sequence. In the case of the 16s ribosomal RNA V3 target, we estimate that a single pair of LATE-PCR primers will likely amplify ∼883 634 of the 1.9 million sequences listed in the Michigan State Ribosomal Database Project. The bacterial species encompassed in this data base are spread over many phyla. Nevertheless, computer modeling indicates that tens of thousands of these variants will have unique fluorescent signatures when analyzed with the two sets of differently colored Lights-On/Lights-Off probes describe here (Reis et al., in preparation).
Analysis of variable sequences using Lights-On/Lights-Off probes is more informative than analysis using multiple dual-labeled probes and is also a significant improvement over High Resolution Melt Analysis. HRMA analysis of homozygous mutations is a multistep process that involves symmetric PCR quantification of each target, mixing of targets in equimolar amounts and then amplification and analysis of the resulting heteroduplexes. A DNA-fluorescent dye is used to observe the subtle differences in the melt curvatures arising from heteroduplexes and homoduplexes. These steps require special reagents, special devices and sophisticated software (6).
Heteroduplexes do not play a part in Lights-On/Lights-Off analysis. Instead, one strand of the chosen target or its variant is interrogated using standard fluorescent thermocyclers with a set of probes that have been optimized in terms of Tm, composition and position to distinguish known alleles as well as unanticipated sequence changes. The resulting fluorescent signatures for each sequence are highly reliable as soon as single strands are generated. There is no need to quantify the number of targets in the sample and, in fact, the intensity of the signals prior to normalization provides an end-point measure of the number of initial targets (17). We anticipate widespread and diverse use of these new technologies.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Table 1.
FUNDING
Smiths Detection Diagnostics, Inc. (to L.J.W.) and NIH [1RC1EB010543-01 and 5RC1EB010643-02]. Lights-On/Lights-Off probes have been trademarked as ThermaLight™ probes. Funding for open access charge: Smiths Detection Diagnostics, Inc.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We are grateful to Drs Barry Kreiswirth and Natalia Kurepina of PHRI, Newark, NJ, for their encouragement and gift of M. tuberculosis genomic DNA.
REFERENCES
- 1.Holland PM, Abramson RD, Watson R, Gelfand DH. Detection of specific polymerase chain reaction product by utilizing the 5′-3′ exonuclease activity of Thermus aquaticus DNA polymerase. Proc. Natl Acad. Sci. USA. 1991;88:7276–7280. doi: 10.1073/pnas.88.16.7276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.El-Hajj HH, Marras SA, Tyagi S, Kramer FR, Alland D. Detection of rifampin resistance in Mycobacterium tuberculosis in a single tube with molecular beacons. J. Clin. Microbiol. 2001;39:4131–4137. doi: 10.1128/JCM.39.11.4131-4137.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Huang Q, Liu Z, Liao Y, Chen X, Zhang Y, Li Q. Multiplex fluorescence melting curve analysis for mutation detection with dual-labeled, self-quenched probes. PLoS One. 2011;6:e19206. doi: 10.1371/journal.pone.0019206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.El-Hajj HH, Marras SA, Tyagi S, Shashkina E, Kamboj M, Kiehn TE, Glickman MS, Kramer FR, Alland D. Use of sloppy molecular beacon probes for identification of mycobacterial species. J. Clin. Microbiol. 2009;47:1190–1198. doi: 10.1128/JCM.02043-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wittwer CT, Reed GH, Gundry CN, Vandersteen JG, Pryor RJ. High-resolution genotyping by amplicon melting analysis using LCGreen. Clin. Chem. 2003;49:853–860. doi: 10.1373/49.6.853. [DOI] [PubMed] [Google Scholar]
- 6.Herrmann MG, Durtschi JD, Bromley LK, Wittwer CT, Voelkerding KV. Amplicon DNA melting analysis for mutation scanning and genotyping: cross-platform comparison of instruments and dyes. Clin. Chem. 2006;52:494–503. doi: 10.1373/clinchem.2005.063438. [DOI] [PubMed] [Google Scholar]
- 7.Sanchez JA, Pierce KE, Rice JE, Wangh LJ. Linear-after-the-exponential (LATE)-PCR: an advanced method of asymmetric PCR and its uses in quantitative real-time analysis. Proc. Natl Acad. Sci. USA. 2004;101:1933–1938. doi: 10.1073/pnas.0305476101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pierce KE, Sanchez JA, Rice JE, Wangh LJ. Linear-After-The-Exponential (LATE)-PCR: primer design criteria for high yields of specific single-stranded DNA and improved real-time detection. Proc. Natl Acad. Sci. USA. 2005;102:8609–8614. doi: 10.1073/pnas.0501946102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rice JE, Sanchez JA, Pierce KE, Reis AH, Osborne A, Wangh LJ. Monoplex/multiplex linear-after-the-exponential-PCR assays combined with PrimeSafe and Dilute-'N'-Go sequencing. Nat. Protoc. 2007;2:2429–2438. doi: 10.1038/nprot.2007.362. [DOI] [PubMed] [Google Scholar]
- 10.Jia Y, Osborne A, Rice JE, Wangh LJ. Dilute-'N'-Go dideoxy sequencing of all DNA strands generated in multiplex LATE-PCR assays. Nucleic Acids Res. 2010;38:e119. doi: 10.1093/nar/gkq111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hebert PD, Cywinska A, Ball SL, deWaard JR. Biological identifications through DNA barcodes. Proc. Biol. Sci. 2003;270:313–321. doi: 10.1098/rspb.2002.2218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chakravorty S, Aladegbami B, Burday M, Levi M, Marras SA, Shah D, El-Hajj HH, Kramer FR, Alland D. Rapid universal identification of bacterial pathogens from clinical cultures by using a novel sloppy molecular beacon melting temperature signature technique. J. Clin. Microbiol. 2010;48:258–267. doi: 10.1128/JCM.01725-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Marras SA, Kramer FR, Tyagi S. Efficiencies of fluorescence re resonance energy transfer and contact-mediated quenching in oligonucleotide probes. Nucleic Acids Res. 2002;30:e122. doi: 10.1093/nar/gnf121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Luo A, Zhang A, Ho SY, Xu W, Zhang Y, Shi W, Cameron SL, Zhu C. Potential efficacy of mitochondrial genes for animal DNA barcoding: a case study using eutherian mammals. BMC Genomics. 2011;12:84. doi: 10.1186/1471-2164-12-84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lynch TJ, Bell DW, Sordella R, Gurubhagavatula S, Okimoto RA, Brannigan BW, Harris PL, Haserlat SM, Supko JG, Haluska FG, et al. Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib. N. Engl. J. Med. 2004;350:2129–2139. doi: 10.1056/NEJMoa040938. [DOI] [PubMed] [Google Scholar]
- 16.Paez JG, Jänne PA, Lee JC, Tracy S, Greulich H, Gabriel S, Herman P, Kaye FJ, Lindeman N, Boggon TJ, et al. EGFR mutations in lung cancer: correlation with clinical response to gefitinib therapy. Science. 2004;304:1497–1500. doi: 10.1126/science.1099314. [DOI] [PubMed] [Google Scholar]
- 17.Sanchez JA, Abramowitz JD, Salk JJ, Reis AH, Rice JE, Pierce KE, Wangh LJ. Two-temperature LATE-PCR endpoint genotyping. BMC Biotech. 2006;6:44. doi: 10.1186/1472-6750-6-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.