Abstract
LacI/GalR transcription regulators have extensive, non-conserved interfaces between their regulatory domains and the 18 amino acids that serve as ‘linkers’ to their DNA-binding domains. These non-conserved interfaces might contribute to functional differences between paralogs. Previously, two chimeras created by domain recombination displayed novel functional properties. Here, we present a synthetic protein family, which was created by joining the LacI DNA-binding domain/linker to seven additional regulatory domains. Despite ‘mismatched’ interfaces, chimeras maintained allosteric response to their cognate effectors. Therefore, allostery in many LacI/GalR proteins does not require interfaces with precisely matched interactions. Nevertheless, the chimeric interfaces were not silent to mutagenesis, and preliminary comparisons suggest that the chimeras provide an ideal context for systematically exploring functional contributions of non-conserved positions. DNA looping experiments revealed higher order (dimer–dimer) oligomerization in several chimeras, which might be possible for the natural paralogs. Finally, the biological significance of repression differences was determined by measuring bacterial growth rates on lactose minimal media. Unexpectedly, moderate and strong repressors showed an apparent induction phase, even though inducers were not provided; therefore, an unknown mechanism might contribute to regulation of the lac operon. Nevertheless, altered growth correlated with altered repression, which indicates that observed functional modifications are significant.
INTRODUCTION
Domain recombination commonly occurs in nature and is exploited in biotechnology as a means for combinatorial discovery of new protein functions (1–5). On a ‘gross’ level, the functions of the independent domains are expected to be unchanged by recombination. For example, a DNA-binding domain is expected to retain binding to a specific sequence of DNA. On a ‘fine’ level, domain recombination can result in unanticipated functional modification. Even when exchanged domains are homologous to each other, functional modification can arise when inter-domain contacts include amino acids that are not conserved between family members. In corollary, amino acid substitutions at non-conserved positions provide opportunity to fine-tune the functions of both engineered and natural proteins.
Our long-term goal is to experimentally demonstrate whether a family-wide set of ‘rules’ can be defined for functional modification through domain recombination and/or substitution of functionally important, non-conserved positions. As a model family, we are using the LacI/GalR transcription regulatory proteins, which are found in a wide range of bacteria and mediate responses to a wide range of environmental and metabolic changes. These proteins have an N-terminal DNA-binding domain that is connected to a regulatory domain by a linker of ∼18 amino acids [Figure 1; reviewed in (6)]. When these repressors bind DNA, the linker and the regulatory domain form an interface that comprises a large number of non-conserved side chains, including 8–10 non-conserved linker positions [the number varies for different homologs, Figure 1A, green side chains, (7–11)]. Therefore, we hypothesized that domain recombination would alter the functions of LacI/GalR homologs, as was indeed shown with two previously studied chimeras.
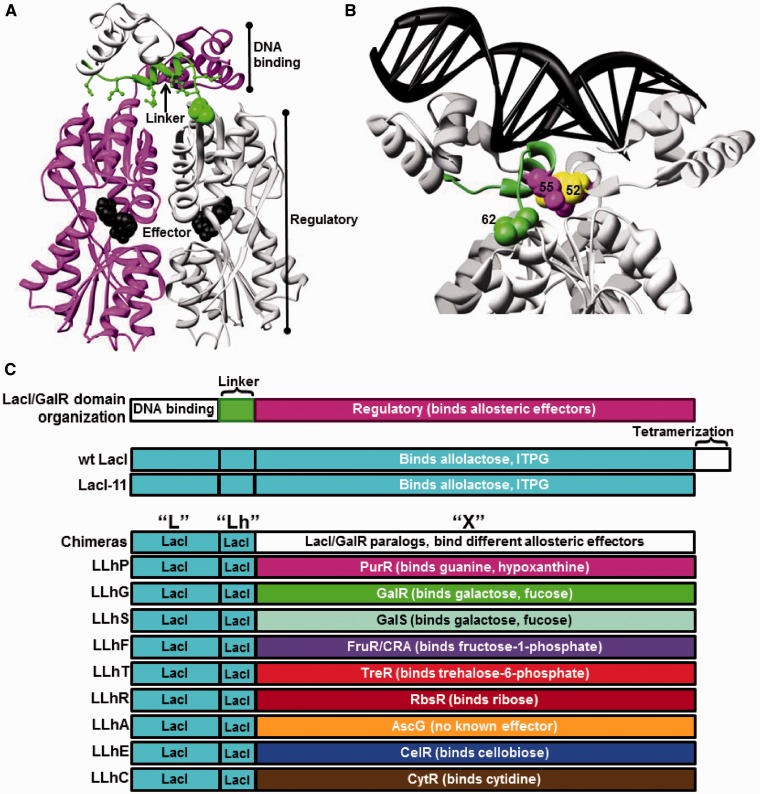
Figure 1.
Structure of a representative LacI/GalR protein and schematic of chimeric homologs. Dimeric LacI (8) (pdb 1efa) illustrates the structure common to LacI/GalR homologs. (A) The protein is oriented to show the two monomers (magenta and light gray) and one linker (green). DNA-binding domains are at the top of the structure, and effector ligand (black spheres) is bound in the clefts of the regulatory domains. Non-conserved linker positions that interact with the regulatory domains are shown with green ball-and-stick. Position 62 is highlighted with green spheres. (B) The protein structure is rotated and zoomed to show bound DNA (black ladder) and the side chains of positions 52 (yellow), 55 (magenta) and 62 (green). Structure depictions were created using UCSF Chimera (89). (C) Cartoon depiction of domain compositions for LacI/GalR homologs and chimeras. Chimeric proteins comprise the DNA-binding domain and linker of LacI joined to the regulatory domains of nine natural homologs (Table 1). LLhP was previously reported in (12). LLhG and LLhG/E62K were previously reported in (13). A C-terminal, 11 amino acid deletion of the tetramerization domain in wild-type LacI results in a dimeric version (23,90).
The first of these chimeras—‘LLhP’—was created from the DNA-binding domain of LacI, the 18-amino acid linker encompassing the LacI hinge helix, and the regulatory domain of PurR (Figure 1 and Table 1) (12). The second chimera—‘LLhG’—was created from the DNA-binding domain and linker of LacI and the regulatory domain of GalR (13). At a gross level, DNA binding is intact in both chimeras; both LLhP and LLhG bind the lac operator. Nevertheless, the functions of LLhP and LLhG are not identical to each other or to wild-type LacI—the three proteins repress the lac promoter to different extents (12–14). They have different binding affinities for lac operator DNA sequences (15–17). LLhP does not discriminate between variant operators as effectively as wild-type LacI (16). Both LLhP and LLhG have unexpected in vivo phenotypes, suggesting that the LacI DNA-binding domain has acquired specificity for other regions of the Escherichia coli genome (12,13). In addition, mutating non-conserved linker positions altered LLhP and LLhG repression and DNA binding (12–14), which confirms that these positions provide opportunities to fine-tune the functions of both engineered and natural LacI/GalR proteins.
Table 1.
Natural LacI/GalR homologs
| Natural proteina | ‘X’ abbreviation | Allosteric response | Position numbersb | Effector ligand |
|---|---|---|---|---|
| LacI | – | Induction | 1–360 | Allolactose; IPTG (18,56) |
| PurR | P | Co-repression | 60–341 | Hypoxanthine; guanine (38,39) |
| GalR | G | Induction | 60–343 | Galactose; fucose (82) |
| GalS | S | Induction | 60–346 | Galactose; fucose (45) |
| FruR (CRA) | F | Induction | 62–334 | Fructose-1-phosphate (83,84) |
| TreR | T | Induction | 63–315 | Trehalose-6-phosphate (85) |
| RbsR | R | Induction | 60–330 | Ribose (86) |
| CytR | C | Induction | 57–330 | Cytidine (87) |
| CelRc | E | Induction | 65–340 | Cellobiose (88) |
| AscG | A | None known | 60–334 | Transcription occurs when IS186 is inserted into ascG (44,61) |
aLacI, Lactose repressor protein; PurR, Purine repressor protein; GalR, Galactose repressor protein; GalS, Galactose isorepressor protein; FruR, Fructose repressor protein, which is also called the ‘catabolite repressor/activator’ (CRA); TreR, Trehalose repressor protein; RbsR, Ribose repressor protein; CytR, Cytidine repressor protein; CelR, Cellobiose repressor protein; AscG, Cryptic asc operon repressor. bThe total numbers of amino acids are indicated for LacI. For chimeras, the numbers correspond to the amino acids of the parent regulatory domains that are fused to the LacI DNA-binding domain/linker (positions 1–62). cFrom Thermobifida fusca. All other proteins are from E. coli.
Toward our goal of demonstrating family-wide ‘rules’ for functional modification through domain recombination and substitution of non-conserved amino acids, we were motivated (i) to create a family of synthetic LacI/GalR repressors and (ii) to determine how much change in repression is required to have a biological impact on the host organism, E. coli. Here, we report the creation of seven additional chimeras, using the LacI DNA-binding domain and linker fused to the regulatory domains from E. coli FruR/CRA, GalS, TreR, RbsR, CytR, AscG and Thermobifida fusca CelR. Functional attributes for the parent proteins are listed in Table 1 and chimera compositions are illustrated in Figure 1.
Chimera characterization included assessment of (i) in vivo transcription repression, (ii) preliminary mutagenesis of non-conserved linker positions, (iii) allosteric response and (iv) ability to simultaneously bind two DNA-binding sites, with looping of the intervening DNA. We also designed an in vivo assay to determine a biological threshold for ‘significant’ functional change. This assay took advantage of the fact that the synthetic chimeras regulate the lac operon but are no longer induced by the natural effector of LacI, which is allolactose (18). Chimeric repressors were transformed into E. coli, which were then grown in lactose minimal media. Cultures transformed with strong repressors should produce only small quantities of the lac metabolic enzymes and thus grow slowly. In contrast, cultures expressing weak repressors should express large quantities of the metabolic enzymes and use lactose for rapid growth. By correlating growth and repression levels, we determined how much change in repression was required to alter bacterial growth.
MATERIALS AND METHODS
Modification of the vector plasmid pHG165
In vivo β-galactosidase assays of transcription repression are most reproducible when the repressor protein is encoded by the low copy plasmid pHG165 (19,20); previous experiments with LLhP variants utilized this vector (12). For experiments with LLhG variants, the endogenous lacO1-binding site of pHG165 was disabled by mutation to create the plasmid pHG165a, in case this sequence allowed auto-repression of the chimera-coding regions (13). We subsequently showed that repressor expression and function via the two plasmids were equivalent (14), and both plasmids were used in the current experiments. The pHG165 plasmid also contains residual LacI C-terminal codons; these were removed to create plasmid pHG165c, allowing mutagenesis of full-length lacI when cloned into this version of the plasmid. To create a ‘no repressor’ control plasmid (‘DEL’), the LLhG coding region on pHG165a was interrupted with a single base-pair deletion that caused a frameshift and created a stop signal at codon 4.
Chimera construction and mutagenesis
The nomenclature of the novel chimeras follows the convention described in the ‘Introduction’ section—the first letter indicates the source of the DNA-binding domain, the second letter (followed by an ‘h’ representing the hinge helix) is the linker and the third is the regulatory domain (Table 1 and Figure 1). As a group, we refer to the chimeras in this article as the ‘LLhX’ proteins.
To create the LLhX repressors, the LacI-encoding plasmid pLS1 (21) was the source for codons 1–61. The coding regions for the regulatory domains of six E. coli repressors (GalS, TreR, RbsR, FruR/CRA, AscG and CytR) were amplified from DH5α (PCR Master Mix, Promega, Madison, WI, USA; or Easy-A High-Fidelity PCR Cloning Enzyme, Stratagene/Agilent Technologies, Santa Clara, CA, USA). The coding region for the regulatory domain from T. fusca CelR was cloned from the plasmid pNS2, a generous gift from Dr David B. Wilson (Cornell University). The sequences of these parent proteins are listed in Supplementary Table S1, along with details of chimera construction. The primers used to amplify the coding regions are listed in Supplementary Table S2.
Similar to the construction of LLhG (13), LLhS creation required the ‘E230K’ mutation of the GalS regulatory domain to alleviate toxicity in E. coli (Supplementary Table S1). In GalR (the parent protein of LLhG), the E230K mutation diminishes ‘repressosome’ formation (22). The necessity of the E230K mutation in LLhG and LLhS thus suggests that these chimeras have potential to form tetramers that can simultaneously bind two DNA operators, ‘looping’ the intervening DNA. For these and various other chimeras, additional random mutations in the linker region were created using a modified version of the QuikChange protocol [Stratagene, La Jolla, CA, USA; (13)]; the primers are listed in Supplementary Table S3. Site-specific mutations were generated using the QuikChange protocol.
Construction of LacI variants
The coding regions for full-length LacI and the LacI-11 variant were subcloned from the genome of DH5α E. coli to the plasmid pHG165c. LacI-11 has a stop codon that is inserted 11 amino acids from the 3′-end of the gene, which disrupts the tetramerization domain, and only dimeric protein is created (23). Historically, two polymorphisms of LacI have been used, with either threonine or alanine at position 109. These have been presumed equivalent because 12 other amino acid substitutions were classified as having no effect on in vivo repression by LacI (24). However, the prior study is low resolution, with ‘wild-type’ activity defined as <200-fold change in repression. In the absence of quantitative characterization, we chose the threonine polymorphism, which is the primary sequence used for myriad thermodynamic studies by the Matthews Lab [e.g. (17,21,23,25,26)].
Pull-down assays of LLhX protein expression
Expression of active, soluble protein was confirmed for all chimera variants as previously described (13,15). In brief, biotinylated operator DNA variants were immobilized to Streptavidin magnetic beads (New England Biolabs, Ipswich, MA, USA). Two of the DNA sequences (lacO1 and lacO2) are natural operators of the lac operon (27); lacOsym is a symmetrized version of lacO1 that binds most variants of LacI and LLhP more tightly than lacO1(16,17,25,28). A non-specific sequence, Onon, (25) was also used. Bead-immobilized operator was incubated with lysate from E. coli strain 3.300 cells (E. coli Genetic Stock Center, Yale University) expressing chimeras. Beads were magnetically separated from the lysate and washed. Proteins that adhered to the DNA beads were visualized with sodium dodecyl sulfate–polyacrylamide gel electrophoresis. Most variants were expressed to high levels of repressor protein (e.g. Supplementary Figure S1), corresponding to ≥2500 repressors per cell [calculations are detailed in (14,15)]. Exceptions are noted in the text below.
Quantification of LLhX transcription repression
Transcription repression was monitored by transforming pHG165-chimera plasmids into E. coli 3.300 cells, which are lacI- but have normal coding regions for lacZYA. To monitor repression of lacZYA, β-galactosidase assays were carried out on agar plates [using both Luria borth (LB) and MOPS minimal media (see below)] and in liquid culture (MOPS minimal medium) in the presence and absence of effector ligands. The β-galactosidase substrate in plate assays was X-gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside; Gold Biotechnology, St Louis, MO, USA), whereas the substrate in liquid culture assays was ONPG (o-nitrophenyl-β-d-galactopyranoside; Research Products International, Mt. Prospect, IL, USA). For each potential allosteric effector, liquid culture assays were determined for the ‘no repressor’ control ‘DEL’ plasmid (Supplementary Figure S2). MOPS minimal medium (Teknova, Hollister, CA, USA; 40 mM morpholinopropanesulfonic acid, 10 mM NH4Cl, 4 mM tricine, 50 mM NaCl and other trace metals listed for product number M2101) was supplemented with 0.04% or 0.8% glycerol, 1.32 mM dibasic potassium phosphate, 10 mM NaHCO3, 0.2% casamino acids (CAA), 0.0025% thiamine and 100 μg/ml ampicillin (19,20).
For moderately and weakly repressing chimeras with blue plate phenotypes, the liquid culture assay used the high-throughput, 96-well format previously described (13). Current data analysis included empirically determined, A420 scattering constants to correct for cell debris, for each row of the 96-well plate (1.35 to 1.15×A540), which tightly correlated with the lag time between shaking the plate and the read-time for each row. These values are lower than the value typically used for 1-cm path-length cuvettes, [1.75×A540] (29).
For tightly repressing chimeras with white plate phenotypes, the assay required further modification in order for results to agree with plate and low-throughput (10 ml culture) liquid culture assays. Cultures were grown overnight in 48-well blocks in 2.5 ml of overnight MOPS media (0.04% glycerol). Cultures were then transferred to new 48-well blocks containing 2.5 ml of growth MOPS media (0.8% glycerol) and grown to OD600 ∼ 0.4 in either the absence or presence of effector (Table 2). Blocks were centrifuged for 10 min at 1200g and the supernatant was discarded. Pellets were resuspended in 500 µl of working buffer (61 mM Na2HPO4, 40 mM NaH2PO4, pH 7.0, 10 mM KCl, 1 mM MgSO4, 0.4 mM DTT); 100 µl was removed to determine OD600. To the remaining 400 µl of resuspended culture, 10 µl of polymixin B was added and incubated at room temperature for 10 min. The assay of enzymatic activity proceeded in 96-well plates as previously described (13). For extremely tight repressors, which required overnight reaction times to generate measurable reaction product, 1 mM TCEP (Tris[2-carboxyethyl] phosphine hydrochloride; Gold Biotechnology, St Louis, MO, USA) was added to maintain reducing conditions. Control experiments with weak repressors showed no difference ± TCEP. The scattering constant for cell debris from 48-well growth at 420 nm was determined empirically to be [1.4×A540].
Table 2.
Known and potential effectors added to the β-galactosidase assays
| Effector | Assay concentration (mM) |
|---|---|
| d-ribose | 5 |
| Trehalose | 10 |
| Cellobiose | 7.6 |
| 2, d-deoxyribose | 20 |
| d-arabinose | 60 |
| IPTG | 1 |
| d-fructose | 20 |
| d-fucose | 20 |
| d-fructose-1,6-bisphosphate | 20 |
| d-fructose-6-phosphate | 20 |
| d-xylose | 20 |
| l-rhamnose | 20 |
| Sucrose | 20 |
| d-mannose | 20 |
| Maltose | 20 |
| l-arabinose | 10 |
| Melibiose | 2 |
| d-lyxose | 16.6 |
| Adenine | 0.19 |
| Cytidine | 2 |
| d-glucose | 28 |
For each chimera variant, a phenotype was determined in the absence and presence of effector (i) once on LB plate media, (ii) once on minimal plate media and (iii) for at least two separate colonies (four determinations each) in liquid culture. In most cases, the three assays agreed. Deviations between LB and minimal plate media are discussed in the ‘Results’ section. Two instances of apparent allosteric regulation on plate assays did not hold in liquid culture assays. These were (i) LLhG + IPTG ‘anti-induction’ and (ii) LacI ‘induction’ on minimal media/glycerol plates. Published experiments with purified LacI and LacI-11 show that glycerol is a ‘neutral’ ligand; it binds to the regulatory domain but does not elicit an allosteric change in DNA binding (26,30). We speculate that glycerol in plate assays somehow facilitates air oxidation of LacI, which inactivates the repressor [(31) K. S. Matthews, personal communication]. Similarly, high-affinity DNA binding by LLhG variants is affected by air oxidation (15). If IPTG is a neutral ligand for LLhG, ligand binding could protect from air oxidation.
In liquid culture, repression was sometimes sporadically lost for a colony, leading to high values. This happened for ∼5% of all colonies. When this occurred, assays were performed for one to three additional colonies. Using all values, an average and standard deviation were determined. If the values for the sporadic colony fell outside of the range, they were excluded from reported results. The reported values more closely agreed with the plate phenotypes than the sporadically high values.
One final difference from previous studies is the normalization scale used for liquid culture assays. Previously, LLhP and LLhG repression were normalized to different standard conditions. Since this study expands the synthetic family, we herein report un-normalized activity. Values for LLhP and LLhG variants are un-normalized in this article.
Looping assays
Looping assays were performed using modified versions of strains developed by Maher, Becker and colleagues (32). The original system comprised multiple E. coli strains with an episome carrying the wild-type lacI gene and a lacZ reporter gene, under control of the lacUV5 promoter flanked by two lac operators—lacOsym and lacO2. The various strains differ in spacing between the two operators, which results in a periodicity of repression if two repressor dimer–operator complexes associate with each other. For the chimeras, we chose four diagnostic operator spacings for which LacI showed dramatic repression differences—with 70.5, 72.5, 77.5 and 75.5 bp between the centers of the lacOsym and lacO2 operators. These spacings sample a well-documented (32) unstable twisted DNA loop (70.5 and 72.5 bp) and a very stable relaxed loop (75.5 and 77.5 bp). Measuring repression for these four diagnostic reporters is therefore sufficient to assess whether repressor tetramerization—which produces bidentate proteins required for DNA looping—occurs for any of the chimeras.
To create a lacI-genetic background in the four looping strains, wild-type lacI was disabled with the Y282D mutation; the resulting monomer is incapable of tight DNA binding or in vivo repression (33–35). This mutation was created on looping plasmids pJ976, pJ977, pJ962, pJ978 (32) using the QuikChange Lightning Site-Directed Mutagenesis Kit. Mutagenic primers are listed in Supplementary Table S3. The LacI Y282D mutant looping constructs were placed on the single-copy F128 episome by homologous recombination. Bacterial conjugation and phenotypic selections were carried out as described (36). After mating and selection, correct strain recombinants were confirmed by PCR amplification to detect the inactivated internal lacZ lacO2 sequence that naturally occurs in this gene (32). The Y282D mutation was confirmed by PCR amplification followed by product sequencing.
Using the four looping strains, β-galactosidase activity was determined using the 48-well assay, since unrepressed β-galactosidase activity is lower in the looping strains than in 3.300 cells. Control experiments with the DEL (no repressor) plasmid showed a small spacing dependence, with similar magnitude to the one operator/position control experiments described in reference (32). Thus, RNA polymerase might be modestly sensitive to the operator position. To account for this, repression values for LacI and the chimeras were normalized relative to DEL values. The presence of pHG165 plasmid also reduced the level of unrepressed β-galactosidase activity relative to previous experiments, which had no exogenous plasmid (32). A final difference is that the repressor protein expression levels are several orders of magnitude higher in the current study. Taking these differences into account, the current values for wild-type LacI were quite similar to those reported by Becker, Maher and colleagues (32).
Potential looping behavior was assayed for one tightly repressing variant of each chimera other than LLhC. At least three colonies with consistent phenotypes were assayed in liquid culture (four values each). Results for colonies that sporadically lost repression were treated as above.
Growth assays
Growth assays were carried out with E. coli 3.300 cells. Cultures transformed with various LLhX chimeras were grown overnight with 100 µg/ml ampicillin in 2×YT media at 37°C. For each variant, a 100 µl sample of an overnight culture was pelleted and washed in 1 ml MOPS minimal growth medium without sugar and resuspended in 1 ml MOPS medium with 100 µg/ml ampicillin. Unless otherwise indicated, the media contained 0.2% CAA. These culture conditions corresponded to those of β-galactosidase repression assays; the only difference is that repression assays included glycerol as a carbon source, whereas lactose or glucose was substituted during growth assays. Purified protein and thermodynamic-binding assays are available for LLhP and the LLhG/E62K variant (15,16); these binding assays were used to verify that lactose does not induce either protein (data not shown).
The OD600 was determined for the resuspended cell pellet and a normalized volume of cells (∼100 µl resuspension) was used to inoculate 20 ml of pre-warmed MOPS media + sugar. This media contained 2 ml of sugar stock solution, so that the final concentration was either 0.2% glucose (Fisher Scientific) or 0.2% lactose (Sigma Chemical Company). A ‘no sugar’ control substituted 2 ml ddH2O. The OD600 of time zero was determined for each culture. If indicated, non-metabolizable inducer d-fucose (Acros Organics, NJ, USA) was added to a final concentration of 20 mM at various time points.
Cultures were grown in baffled 250 ml Erlenmeyer flasks at 37°C with shaking. Samples (0.5 ml) were taken every 1 or 2 h and an OD600 determined. For cultures that took >10 h to reach stationary phase, the experiment was paused by transferring the culture to a sterile test tube and centrifuging at 480 g for 2 min. Pelleted cells were stored in the remaining growth media overnight at 4°C. The next morning, the culture was slowly warmed to room temperature, gently resuspended, transferred to a clean, warm Erlenmeyer flask and growth was resumed at 37°C. As long as arrests occurred prior to OD600 = 1.0, values before and after pausing were equivalent. The duration of the growth assays ranged from ∼500 to 1500 min. At the end of the assay for one slow-growing lactose culture, pH of the media was tested and found to be close to the starting pH. Time to OD600 = 1 was determined by linear regression of the data points that fall in the linear region of the last growth phase. Growth curves in lactose and glucose minimal media were repeated at least three times or performed as part of a time series (d-fucose addition) or concentration series (CAA concentration).
For each chimera variant, after the completion of one replicate of the growth assay, cells were pelleted and plasmid DNA was isolated with a QIAprep Spin Miniprep Kit. The full coding region was sequenced for each repressor variant and found to be intact in all cases. Expression of LLhX proteins was verified at the end of a growth assay using a DNA pull-down assay.
To monitor transcription of the lac operon during the growth assay, β-galactosidase (lacZ) activity was assayed at various times. To that end, 1 ml of culture was mixed with 50–100 µl of 20 mg/ml 4-methylumbelliferyl-β-d-galactopyranoside (MUG) dissolved in DMSO (21,29). Cultures were viewed under UV light to detect the fluorescent product of MUG that results from β-galactosidase activity.
Graphs and correlation analyses were created using GraphPad Prism 5 (GraphPad Software, Inc, La Jolla, CA, USA).
RESULTS
We have created and characterized a synthetic protein family comprising chimeric repressors created via domain recombination of LacI/GalR homologs (Figure 1 and Table 1). Two chimeric proteins (LLhP and LLhG) were created and reported earlier (12,13); seven new chimeras are reported herein. The ‘LLhX’ nomenclature of the chimeras reflects their domain composition: The first ‘L’ indicates that LacI DNA-binding domain, ‘Lh’ represents the LacI linker; and ‘X’ indicates the parent protein of the regulatory domain (listed in Table 1). Note that all position numbers mentioned in this manuscript correspond to numbering of the E. coli LacI protein. Upon their creation, all LLhX variants were assayed for in vivo protein expression (Supplementary Figure S1). Almost all showed high levels; two exceptions are described in Supplementary Figure S1.
Repressor function of the chimeras
Natural LacI/GalR proteins use the two N-terminal domains of a homodimer to bind operator DNA (Figure 1), most often resulting in repression of downstream genes [reviewed in (6,37)]. The repressor–DNA interaction is modulated when repressor binds to effector molecules in a cleft at the center of each regulatory domain (allosteric response): Of the chimera parent proteins discussed in this work, seven are induced upon binding metabolites (Table 1), so that DNA binding is diminished and transcription is increased. Three parent proteins use different allosteric variations: PurR represses more strongly upon binding purines (38,39). CytR binding to cytidine induces RNA transcription, but the mechanism is via altered CytR–CRP interactions rather than diminished DNA-binding affinity (40–43). AscG has no known effector and must be mutated for induction to occur (44).
Chimera function was assayed by monitoring in vivo repression of the wild-type, genomic E. coli lacZYA operon. Activity of the lacZ reporter gene was assayed by monitoring β-galactosidase activity on plates and in liquid culture (12–14), with low enzyme activity (little color) corresponding to tight repression. Plate assays were performed in parallel, using LB and MOPS minimal media + glycerol; phenotypes under these two conditions were the same for most chimeras and showed a range of repression abilities for the chimeras (data not shown). However, both LLhF and LLhS did not repress as well (colonies were more blue) on LB relative to minimal media (data not shown). Consistent with the plate assays, liquid culture β-galactosidase assays for the different chimeras showed different repression of the lac operon (Figure 2A), from strong repression to none at all.
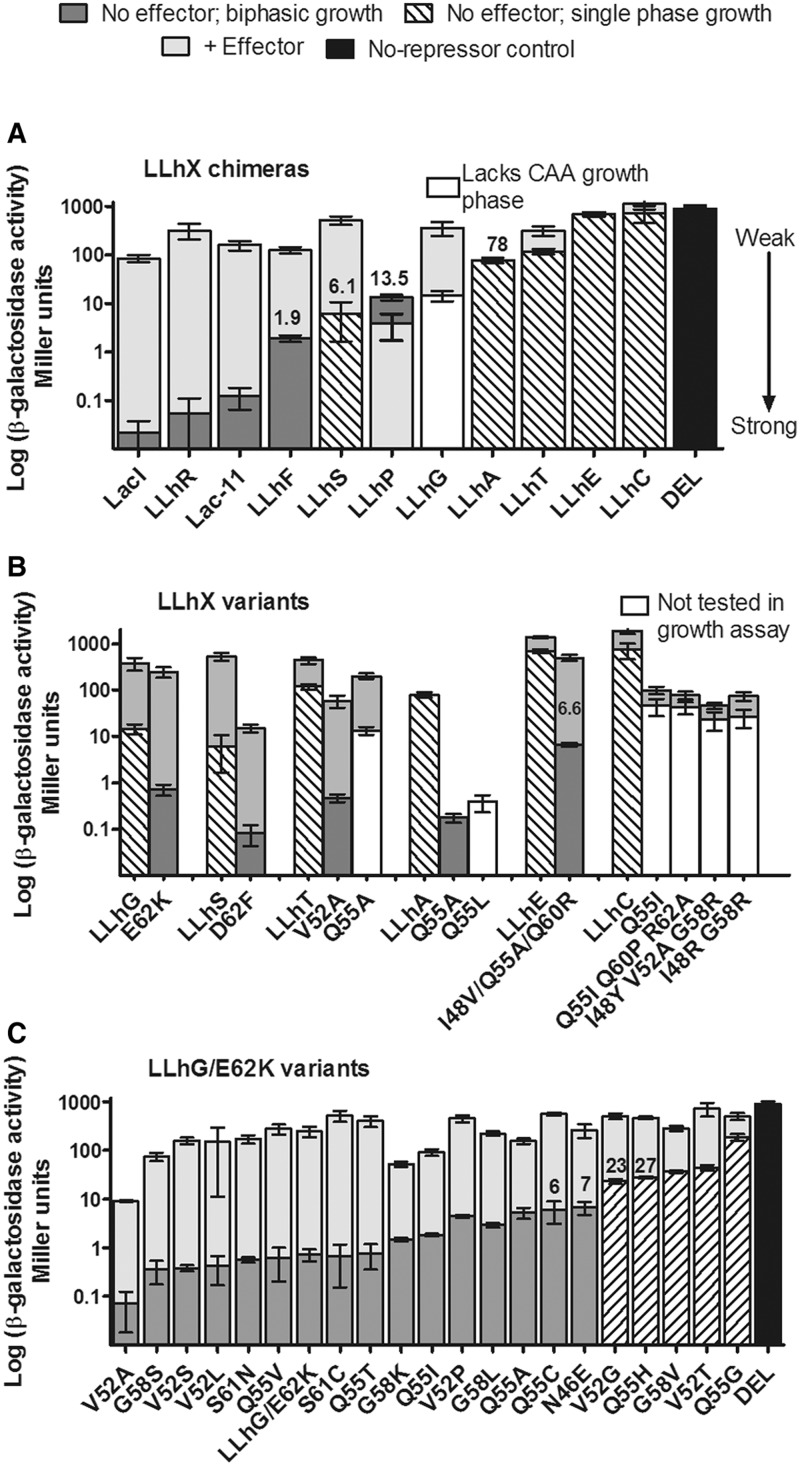
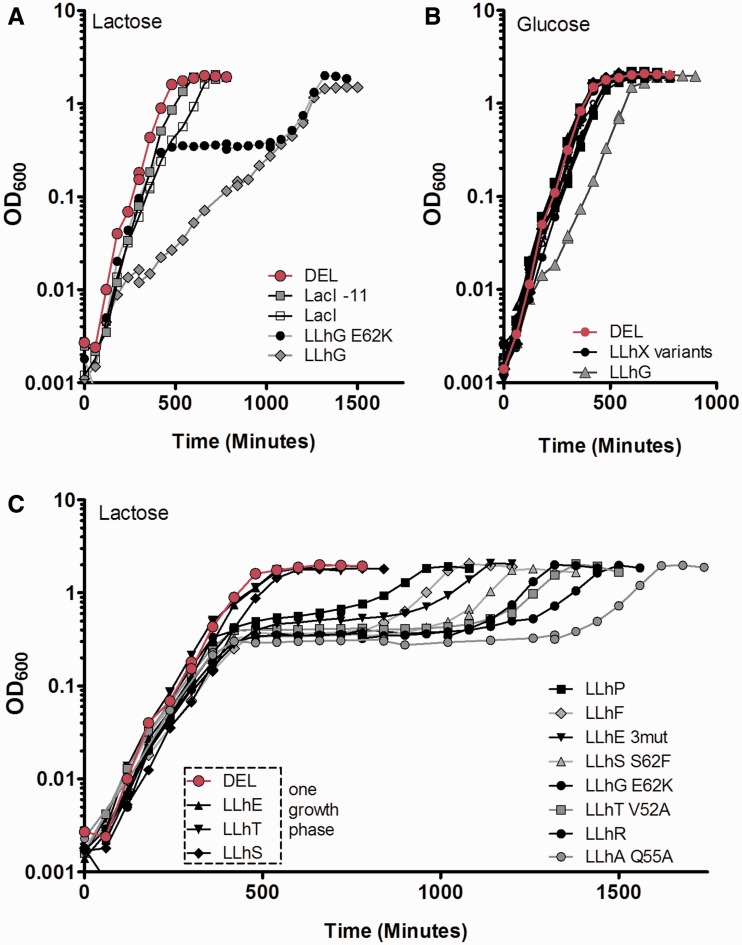
Figure 2.
Repression of the lac operon by the (A) LLhX chimeras, (B) tight-binding LLhX variants and (C) LLhG/E62K variants. Transcription repression is inversely proportional to β-galactosidase activity; low bars correspond to stronger repression. In all panels, the dark gray, striped, and white bars show repression in the absence of effector. Dark gray and striped bars, respectively, correspond to the upper (biphasic growth) and lower (single phase growth) trends shown in Figure 6. For variants near the correlation gap in Figure 6, repression values are printed above the relevant bars. White bars are explained in the legends to panels A and B. The light gray bars depict repression in the presence of effector (Tables 1, 2 and Figure 1). For most chimeras, effector was added directly to the media; an upstream metabolite was used for LLhP (adenine), LLhF (fructose) and LLhT (trehalose). Most of the chimeras are induced by effector (repression is alleviated), but LLhP is co-repressed and repression is tighter in the presence of effector. LLhA has no known inducer. In panel B, note that none of the LLhC variants showed statistically significant induction or co-repression in the presence of cytidine when the effects of cytidine were taken into account (Supplementary Figure S2). The black bars show the ‘DEL’ β-galactosidase activity in the absence of repressor. The truncated LacI-11 variant is a dimer (23,90,91) and showed an expected decrease in in vivo repression relative to wild-type, tetrameric LacI. In panel C, many LLhG/E62K repression values were reported earlier (13) and are here shown on the same scale as panels A/B; new values are reported for N46E/E62K, Q55C/E62K, Q55H/E62K, Q55T/E62K, Q55V/E62K and S61C/E62K. All error bars indicate one standard deviation from the mean.
We previously found that single mutations at non-conserved linker positions could greatly enhance repression by versions of LLhP and LLhG (13,14). To determine whether the non-conserved linker positions were functionally important in the new chimeras, we used various mutagenesis strategies to strengthen repression of the weaker chimeras—LLhS, LLhT, LLhE, LLhC and LLhA (Figure 2B). The first candidate was position 62, since this was the site of domain fusion and might be the site of a side chain clash between the LacI linker and the ‘X’ regulatory domain. Like prior results for LLhG (13), random mutagenesis of LLhS at position 62 produced variants with repression enhanced up to two orders of magnitude (Figure 2B and Supplementary Figure S3). However, mutagenesis of position 62 did not identify tightly-repressing variants of the other weak chimeras.
Therefore, other non-conserved linker positions were randomly mutated in order to identify variants with stronger repression. For LLhT, random mutations at eleven non-conserved positions identified two mutations at positions 52 and 55 that enhanced repression. For LLhA, mutagenesis of position 55 created the highly repressing Q55A and Q55L substitutions. For LLhE, single-mutant variants did not restore repression; however, combinatorial random mutagenesis resulted in double and triple mutations that enhanced repression: Q55A/D62T and I48V/Q55A/Q60R. For LLhC, simultaneous random mutagenesis of 12 non-conserved positions yielded four ‘light blue’ variants: Q55I, Q55I/Q60P/R62A, I48Y/V52A/G58R and I48R/G58R. In liquid culture, LLhC repression was enhanced 10- to 20-fold by these mutations.
Together, mutagenesis results show that the non-conserved linker positions make important contributions to repressor function. The fact that each tightly repressing variant comprised different amino acid substitutions suggests that each LLhX chimera is distinct from the others, despite otherwise identical LacI DNA-binding domains and linkers.
Allosteric regulation is preserved in most of the chimeras
Chimera construction should result in a ‘mismatched’ interface between the linker and regulatory domain, which might disrupt allosteric regulation of DNA binding [The DNA-binding and regulatory domains do not directly contact each other; all contacts are mediated through the linker (10)]. Nevertheless, in the previously studied LLhP and LLhG chimeras, allostery was intact and dictated by the regulatory domain. The regulatory domain dictated not only the type of effector ligand (Table 1 and Figure 1), but also the direction of the allosteric response: LLhG induction paralleled GalR induction by fucose (13), whereas LLhP co-repression paralleled PurR co-repression by the adenine metabolite hypoxanthine (12). The ∼2-fold magnitude of LLhP co-repression was also quite similar to that of PurR (39).
Similarly, allosteric regulation was preserved in the new chimeras that had measurable repression (Figure 2A). The repression-competent variant of LLhE also showed allosteric regulation (Figure 2B). The exception was LLhC. Even with enhanced repression, none of the four LLhC variants showed significant induction or co-repression when effector cytidine was added to the assay (Figure 2B). Correcting for enhanced β-galactosidase activity in the presence of cytidine (Supplementary Figure S2) did not result in statistically significant induction or co-repression by the LLhC variants.
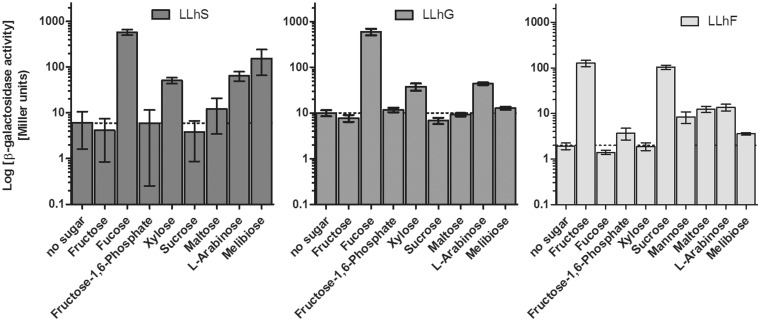
As noted above, both LLhF and LLhS did not repress as well on LB plates, which suggested that these chimeras might be induced by ‘gratuitous’ effectors in addition to their known effectors (Table 1). Thus, we screened these and other chimeras for in vivo induction by various sugars (Table 2), to determine whether gratuitous effectors could be identified. Both LLhS and LLhG were induced by xylose and l-arabinose (Figure 3); the similar behaviors might be expected because the parent proteins GalS and GalR are ‘iso-repressors’ with the same natural inducer (45), (although note that LLhG was ‘not’ induced on LB plates). The allosteric response of LLhS was larger than that of LLhG, and LLhS was also induced by melibiose and maltose. The enhanced allosteric responses of LLhS might be related to the fact that GalS requires 15-fold less galactose to release operator DNA than does GalR (46). The variant LLhG/E62K responded to the same inducers as LLhG (data not shown). Since LLhG/E62K has been purified and is amenable to thermodynamic DNA-binding assays (15), we used this variant to confirm the novel inducer behaviors in vitro (Supplementary Figure S4). LLhF was also induced by several sugars (Figure 3). However, since metabolism of many sugars generates the fructose metabolite that is the natural inducer of the parent protein (FruR) this result might be trivial.
Figure 3.
Additional effectors for LLhS, LLhG and LLhF. β-galactosidase activity was monitored in the presence of potential, gratuitous inducers (Table 2). Horizontal, dashed lines are included to aid comparison to the ‘no sugar’ condition. Error bars indicate one standard deviation from the mean.
Several chimeras show evidence of DNA looping in vivo
Repressor dimerization is required for high-affinity binding to cognate DNA operators [reviewed in (6,37)]. If two dimers interact with each other while binding to two operators on a contiguous piece of DNA, repression can be enhanced (47). To that end, wild-type LacI forms a ‘dimer of dimers’ through an additional C-terminal domain. This domain is absent in most other LacI/GalR proteins. However, LacI/GalR homologs can form tetramers through other means. For example, two dimers of GalR interact with each other when they are bound to two appropriately-spaced DNA-binding sites, creating a ‘repressosome’ (22,48). Hence, the chimeras have potential to form tetramers. Indeed, LLhG is toxic unless it contains the E230K mutation that diminishes GalR repressosome formation [see the ‘Materials and Methods’ section and (13)]. Thus, we decided to assess the chimeric proteins for potential tetramerization/looping.
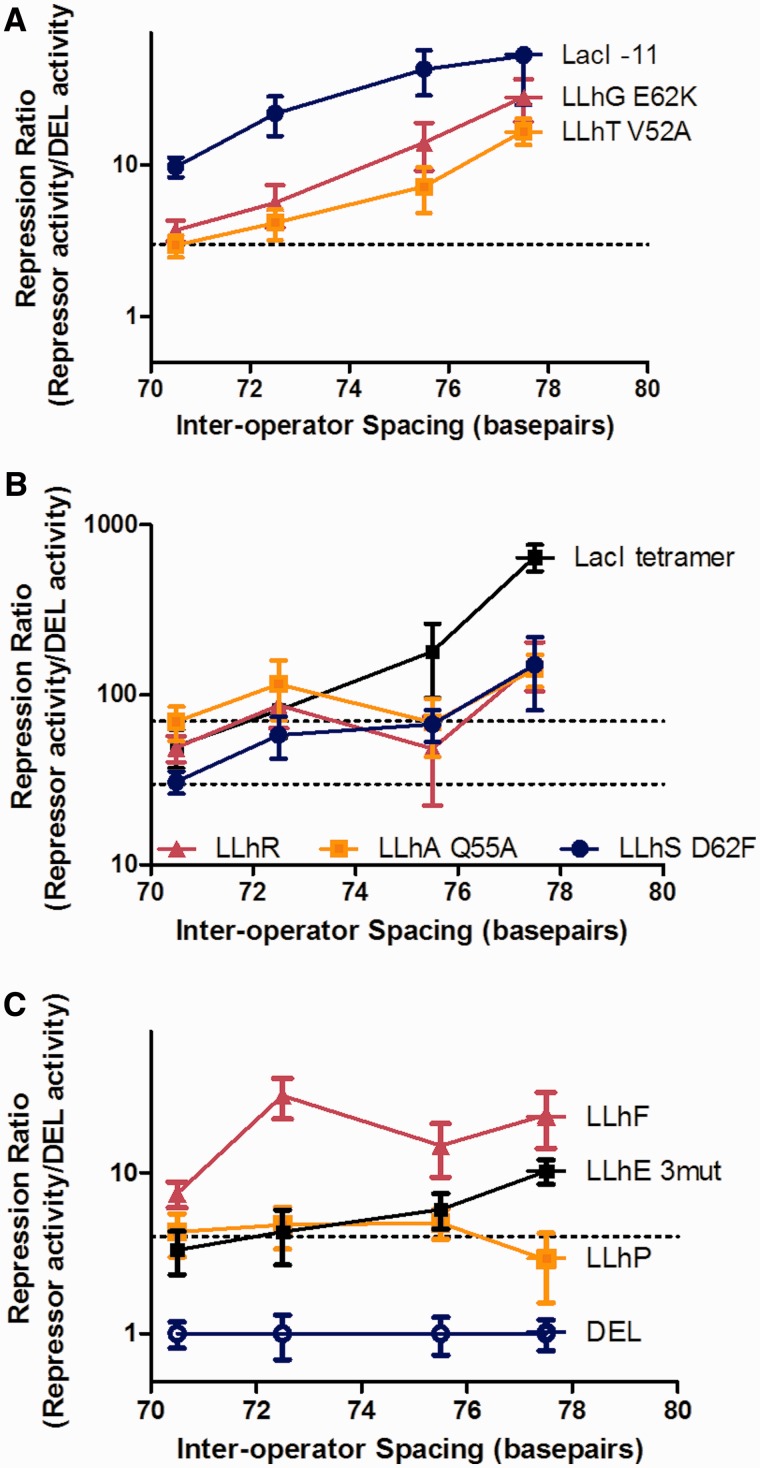
When two DNA-bound repressor dimers interact with each other, the intervening DNA sequence forms a ‘loop’ that can adopt various topologies (32). Under these conditions, repression is sensitive to the juxtapositions of the two operators (32,47,49–52), which can be altered by increasing the spacing between them, so that one operator rotates to a different face of the DNA helix. Operator pairs with optimal spacing for untwisted DNA loops should exhibit tighter repression than those for which simultaneous dimer–dimer binding requires unfavorable DNA twisting. We assayed for potential dimer–dimer interactions using four diagnostic strains of E. coli that carry a lacZ reporter gene regulated by two lac operators—lacOsym and lacO2 (32). The four strains efficiently sample two unfavorable and two favorable DNA loop lengths, as based on prior studies (32). Repression of these four constructs by wild-type LacI tetramer differed by up to 10-fold. Repression measurements for these four diagnostic constructs therefore provide a rapid assessment of tetramerization. Results for tightly repressing chimera variants are shown in Figure 4 (Note that data are normalized to the ‘DEL’ control plasmid, so that high values correspond to tight repression).
Figure 4.
Repression by several chimeras shows dependence on the spacing between two operator-binding sites. β-galactosidase activity was measured in four strains of E. coli that express lacZ under regulation by the lacOsym and lacO2 operators. Because spacing between the two operators varies (X axis), chimeras that form dimer–dimer interactions are expected to have altered repression in the different strains. Repression was normalized to the ‘DEL’ no repressor condition; larger values correspond to tighter repression. Error bars show the standard deviation of the average. Dotted lines are to aid visual inspection of the data. (A) LacI-11, LLhG/E62K, and LLhT/V52A. (B) Wild-type LacI, LLhR, LLhA/Q55A, and LLhS/D62F. (C) LLhP, LLhF, and LLhE ‘3mut’ (I48V/Q55A/Q60R).
As described in the ‘Materials and Methods’ section, measured values for wild-type LacI are in good agreement with published values (32). The LacI-11 dimeric variant showed weaker spacing-dependent repression (Figure 4A), consistent with the work of Müller-Hill and colleagues for the same inter-operator spacings (53). The molecular basis of this result is unclear, but may reflect weak protein–protein interactions that diminish as the operator spacing increased, consistent with diminished local concentration of repressor (53).
Several chimeric repressors clearly showed spacing-dependent repression, consistent with DNA looping supported by repressor tetramerization. Like its parent protein GalR, LLhG/E62K showed spacing-dependence even though the chimera contains the ‘E230K’ mutation that diminished GalR repressosome formation by ∼3-fold (22). Repression by the LLhT/V52A showed similar behavior (Figure 4A). LLhS/D62F showed the same direction in its spacing dependence, although with smaller magnitude (Figure 4B). In contrast, LLhR, LLhA/Q55A, and a tight-binding variant of LLhE showed <2-fold difference between the four strains (Figure 4B and C). LLhP showed no significant difference between the four strains. LLhF had inconclusive results (Figure 4C); the fold-change between the four strains was comparable to LLhG/E62K and LLhT/V52A, but the pattern of change suggests a different orientation requirement for the two operator-binding sites.
Growth curves for cultures expressing LacI/GalR synthetic homologs
Both domain recombination and amino acid substitutions can be used to modify protein function, but for biological significance, the change must be large enough to alter the biology of a host organism. To determine which repressor variants were biologically distinct, E. coli 3.300 cultures expressing LLhX variants were grown in lactose minimal media. A key feature of experimental design was that the chimeras are not induced by the allolactose effector of wild-type LacI; thus the fast induction seen for LacI variants grown on lactose (Figure 5A) does not occur for the chimeras. This allowed us to directly monitor how altered repression affects bacterial growth rates. We expected that cultures expressing stronger repressors would show impeded growth on lactose relative to those with weaker repressors, which would manifest as altered growth rates (slopes of the growth curve). As a control, each culture was also grown on glucose minimal media. For most variants, glucose cultures were essentially identical to the control plasmids (Figure 5B); two exceptions are noted below.
Figure 5.
LLhX growth curves in lactose and glucose minimal media. (A) Growth on lactose minimal media for E. coli cultures with the ‘DEL’ control (magenta), wild-type LacI, dimeric LacI-11, LLhG and LLhG/E62K. Note the biphasic growth curve for LLhG/E62K; the first phase is due to diauxic growth on CAA (Supplementary Figure S5). Cultures expressing LLhG (filled triangles) do not utilize CAA as a distinct carbon source. (B) Growth curves in glucose minimal media for ‘DEL’ (magenta) and LLhX chimera variants. LLhG growth on glucose (gray triangles) is slowed relative to the other cultures. (C) Growth curves in lactose minimal media for cultures expressing LLhX variants. LLhE ‘3mut’ comprises the triple mutant I48V/Q55A/Q60R. The ‘DEL’ control plasmid is shown with magenta diamonds.
For weak LLhX repressors on lactose minimal media (LLhE, LLhT and LLhS), the growth curves showed a single growth phase, as expected (Figure 5C). However, for even moderately strong LLhX repressors, the growth curves on lactose minimal media were biphasic (Figure 5A and C). These cultures exhibited a common growth phase with a plateau at ∼500 min, followed by a variable lag phase preceding a second, exponential growth phase. The first growth phase has been observed in other studies, but the source was not known (54); we have shown that it arises from metabolism of CAA (Supplementary Figure S5), which was included in growth assays to match the conditions of repression assays as closely as possible. Surprisingly, two repressor variants were not able to use CAA as a carbon source (described later; Figure 5A).
For the second growth phase of the biphasic curves, the slopes were very similar to each other. Instead, the primary difference between cultures was the duration of the lag time (Figure 5C). For strong repressors (longer lag phases), the slopes of the lag phases were near zero. For growth curves with the shortest lag times (e.g. LLhP and LLhE/I48V/Q55A/Q60R in Figure 5C), the ‘lag’ phase showed a low but non-zero growth rate (slope) prior to the second exponential growth phase.
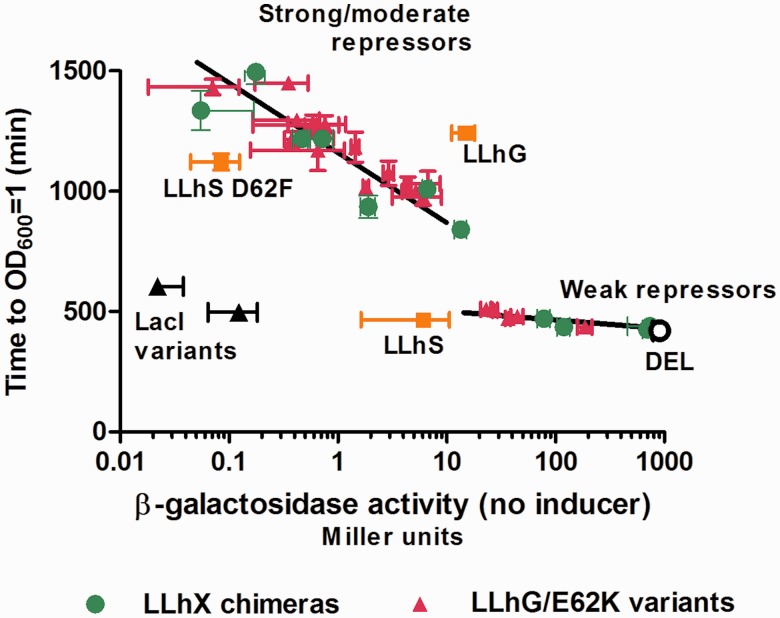
Thus, to answer our initial question, ‘How much change in repression is required to alter bacterial growth?’ we faced the challenge of simultaneously analyzing disparate data. To that end, we chose to plot the time to OD600 = 1 versus repression values (Figure 6). At this OD, all of the cultures were clearly in their last growth phase, and all cultures presumably had the same access to various carbon and metabolic sources, albeit at different times and perhaps used in different manners. For the LLhX chimeras and their variants, plots of growth time versus β-galactosidase activity (inverse repression) suggested a hyperbolic correlation, but the data were too sparse to draw conclusions. To increase the number of data points, we chose 21 previously studied mutational variants of LLhG/E62K (Figure 2C) that showed a wide-range of repression values (13). When results for these variants were added to Figure 6, the resulting plot shows a robust correlation.
Figure 6.
Culture growth times correlate with repressor strength. Time to OD600 = 1 are plotted against log [β-galactosidase activity] for LLhG/E62K (magenta squares) and LLhX (green circles) variants. Data for LLhG, LLhS and LLhS D62F are indicated with orange squares. Results for wild-type LacI and dimeric LacI-11 are shown with black triangles; as expected, these repressors are quickly induced when lactose is converted to allolactose. Growth times are the average of at least three replicate experiments; error bars show the standard deviations. Black lines were drawn to aid visual inspection of the data.
Notably, the data in Figure 6 segregate into two series, with an abrupt threshold separating the series at ∼t = 500 min. Most variants of the LLhX and LLhG/E62K series follow the same two trends (exceptions are noted below). After initial experiments, additional repressors were selected based on repression values that should fill the gap. All attempts were unsuccessful, even though repression values at the ends of the two correlation series were within 2-fold (Figure 2). All of the repressors in the upper trend showed the two separate growth phases, whereas all repressors in the lower series had only a single growth phase. The abrupt break might be related to the fact that the cells with strong repressors have metabolized CAA and now must synthesize amino acids in order to grow.
Upregulation of lac operon transcription leads to the second growth phase
The biphasic curves seen in lactose minimal media for moderate/strong repressors were reminiscent of classic diauxie, when one carbon source is preferred over another (55). When wild-type E. coli is grown on a mixture of glucose and lactose, the glucose is utilized first, followed by a lag phase. A second growth phase occurs via a positive feedback mechanism: some of the lactose is converted to allolactose, binds to LacI, and decreases LacI operator-binding so that transcription of lacZYA is allowed, which in turn produces more allolactose (18,56). However, as noted above, the chimeras should not be induced by allolactose and the various LLhX inducers were not expected to be present in the growth assays. Furthermore, no inducers are known for either PurR (the parent protein to LLhP) or AscG (LLhA) (Table 1).
Nevertheless, when exogenous inducer was added at various times during growth assays, the lag phase was shortened and was followed by a growth phase that was similar to the second growth phase (Supplementary Figure S6). This suggested that, even in the probable absence of chimera inducers, the second growth phase occurred via a mechanism that displaced the chimeric repressors from the operator. To test whether induction of the lac operon was occurring during the second growth phase, bacterial cultures were assayed just prior to and after completion of the second growth phase for β-galactosidase activity. Experiments used the substrate MUG, which is hydrolyzed to a fluorescent product (Supplementary Figure S7). For cultures grown on lactose, the cells showed no detectable hydrolysis of MUG at the end of the lag phase (after CAA growth), but did hydrolyze MUG at the end of the second growth phase. Cultures grown on glucose showed no detectable hydrolysis of MUG, including when they were maintained at stationary phase for long times to match the end of the second lactose growth phase. Thus, the second growth phase on lactose correlated with increased β-galactosidase activity, even in the probable absence of inducer.
To rule out the next obvious source of lost repression—mutated repressor proteins—we verified that the chimeras were un-mutated and expressed at high levels. At the end of the growth assay, the plasmids expressing the repressor variants were isolated and the coding regions were sequenced. No spontaneous mutations were found for any repressor in the study. Second, for a subset of repressors, the presence of active protein at the completion of a growth curve was verified with DNA ‘pull-down’ experiments (see the ‘Materials and Methods’ section). Again, results showed the presence of repressor protein expressed at high levels and capable of binding DNA. We previously showed that this high level of expression has the effect of ‘buffering’ small changes in protein expression levels (15). Thus, upregulation of the lac operon was not due to loss of repressor protein.
A few variants did not follow the global trends
Of the tightly repressing chimeras, LLhG (lacking the E62K polymorphism) lacked the first growth phase. Thus, LLhG must somehow repress the use of CAA as a carbon source (Figure 5A), perhaps due to acquired repression for some other part of the E. coli genome. The effect on metabolism might be global, since LLhG also grew significantly slower on glucose than the other chimeras and LLhG/E62K variants (Figure 5B), both with and without CAA (data not shown). Therefore, the fact that this chimera does not follow the trend of the other repressors in Figure 6 is not surprising (orange square above the trend line). Note that LLhG/E62K does follow the trend line. The other exceptions to the correlations were variants of LLhS (Table 1). Based on the correlations in Figure 6, cultures with LLhS repressors grew more quickly than expected (orange squares below the trendline). Since the parent protein GalS requires 15-fold less galactose to release operator DNA than does GalR (46), one possibility is that galactose is created from lactose at sufficient levels to induce LLhS in the growth assays.
DISCUSSION
Chimera characterization illuminated features of the LacI/GalR protein family
Characterization of the chimeras identified features of allostery and oligomerization that probably apply to the naturally-occurring LacI/GalR homologs.
1. Allosteric regulation does not require precise, ‘jigsaw’ interactions between domains
Logically, allosteric communication between the regulatory domain and the DNA-binding domain of the LacI/GalR homologs should involve the interface that is formed between the regulatory domain and the linker. Since both sides of the interface comprise several non-conserved amino acids, allosteric communication could be interrupted by chimera formation. However, most of the chimeras had intact allosteric responses that were dictated by the regulatory domains. Thus, despite extensive interactions between the regulatory domain and linker (10), allosteric communication does not require a precise ‘jig-saw’ pattern of interactions.
The LLhC chimeras created from CytR appear to lack allosteric regulation (Figure 3). Escherichia coli CytR differs from all the other parent proteins in Table 1, in that it lacks a functionally important ‘YPAL’ motif in its linker (11) and that cytidine binding modulates CytR–CRP interactions instead of DNA-binding affinity [e.g. (40–43)]. Thus, two scenarios might explain the lack of allosteric regulation in LLhC. One option is that the CytR regulatory domain is performing as in wild-type CytR, with no change in DNA binding upon cytidine binding. This would suggest that CytR allosteric change only alters its CRP-binding site without rearranging the N-subdomain, as occurs for YPAL homologs LacI, PurR, and CcpA (9,57). Alternatively, although detailed interactions are not required for allostery when domains are recombined from YPAL homologs, a YPAL linker ‘round peg’ might not fit into the ‘square hole’ of the non-YPAL CytR regulatory domain.
2. Several LacI/GalR homologs have potential to form dimer–dimer interactions
Using four diagnostic constructs that comprised the lacZ reporter gene under control of ‘looping’ promoters, several chimeras showed a dependence on the spacing between the two operator DNA-binding sites (Figure 4), which suggests that dimer–dimer interactions occur. These results are easily explained for LLhG: dimer–dimer interactions via the regulatory domains are well documented for GalR [e.g. (22)]; RegulonDB (58) shows the positions of the two operator-binding sites in the regulatory regions of the galETKM and galP. Note that both GalR and GalS can bind to the regulatory regions of these operons (45,46). No evidence was previously known for GalS looping, but the necessity of the ‘E230K’ mutation in LLhS—which diminishes repressosome formation in GalR—suggests that GalS might be capable of tetramerization when expressed at the high levels used in the current experiments.
Tetramerization has not been directly assessed for many of the other parent proteins. Therefore, to determine the potential of the natural homolog dimers to associate, we examined the number of DNA-binding sites in the promoter regions of the naturally-regulated genes using RegulonDB (58). Indeed, the treBC operon that is regulated by TreR (the parent protein of LLhT) contains two operator-binding sites (58), and LLhT/V52A shows behavior consistent with looping. The master regulator FruR (parent to LLhF) probably acts at >150 promoters (59); at least one operon—fruBKA—has two FruR-binding sites annotated in its regulatory region, whereas other operons have only one annotated site (58). The purR and purA genes each have two PurR-binding sites in their regulatory regions, whereas many of the other ∼53 genes regulated by PurR (parent to LLhP) have only one annotated site (58,60). The ascFB genes regulated by AscG has two binding sites (61), which contrasts with LLhA results from the current experiments. However, the rbsDACBKR operon that is regulated by RbsR has only one annotated operator-binding site (58), and the chimera LLhR shows no spacing dependence.
Thus, repeated DNA-binding sites that mediate dimer–dimer interactions may be a common feature of transcription regulation by even the dimeric LacI/GalR homologs. Although the homologs are not permanently tethered with a tetramerization domain like wild-type LacI, this arrangement could give them flexibility to regulate either as a dimer, as two separate dimers, or as an associated dimer–dimer at different genes.
Mutagenesis of non-conserved positions suggests predictable, albeit limited, trends in functional outcomes
The current studies include preliminary results for the parallel mutagenesis studies that are needed to understand whether family-wide ‘rules’ can be identified for functional modification via mutagenesis of non-conserved positions. Functional outcomes for substitutions at position 55 and 52 are compared in Table 3. Notably, position 55 was consistently important for restoring repression to weak chimeras, although the effects of individual amino acids differed among the chimeras. For example, the Q55A substitution enhanced repression in some chimeras and diminished repression in others. Nevertheless, most of the chimeras had at least one substitution at position 55 that enhanced repression [current results and previously published in (13); the exception is discussed in the footnotes of Table 3]. The V52A substitution enhanced repression in all four chimeras tested (Table 3); it will be interesting to determine whether this trend is maintained in additional chimeras. Finally, mutagenesis of position 62 strengthened repression for both LLhG and LLhS, but again via different amino acid substitutions (Supplementary Figure S3). Thus, results suggest that the overall role of a non-conserved amino acid position can be preserved between homologs, but that the outcomes of individual amino acids show significant epistasis.
Table 3.
Comparison of changes in in vivo repression compared to unmodified chimeras
| LacI | LLhGa | LLhG/E62Ka | LLhPb | LLhT | LLhA | LLhC | LLhE | |
|---|---|---|---|---|---|---|---|---|
| Q55A | ↓ | ↓ | ↓ | ↑ | ↑ | ↑ as 2mut or 3mutc | ||
| Q55L | = | ↓ | ↑ | |||||
| Q55I | ↑ | ↑ | ||||||
| V52A | ↑d | ↑ | ↑ | ↑as 3mute |
aPreviously reported in (13). bPreviously reported in (12); LLhP and several of its variants showed bacterial toxicity, and thus the mutagenesis/selection protocol might have missed variants that enhance repression (e.g. at position 55). cThe Q55A mutation only enhanced repression in the presence of one or two additional substitutions (see ‘Results’ section). Q55A alone did not enhance repression. dPreviously reported in (17). eIn the presence of two additional substitutions (see the ‘Results’ section).
Functional differences between chimera variants are biologically significant
The chimeras and their variants show a range of repression values that span more than three orders of magnitude (Figure 2). However, even though the effect of a mutation can be measured in the laboratory, a change might not be large enough to alter the life cycle of the host organism. For example, even though we can reproducibly measure a 3-fold change in repression, bacterial growth might not be altered unless a 10-fold change occurs. Moreover, the necessary threshold for change might manifest as a continuous change, as a step function, or as some behavior in between. By correlating repression with bacterial growth, we aimed to detect both the magnitude of the required repression change and the nature of the switch.
In addition, these correlation studies might provide a means for reconciling discrepancies between experimental and computational predictions about important non-conserved amino acids. A huge dataset of LacI mutational variants and their phenotypes is available (24), and the natural LacI/GalR proteins have been used in the development of myriad computational analyses (62–77). However, mutagenesis results for LLhP and LLhG suggested that various analyses under-estimate the number of functionally-important positions (11). One possible source of the discrepancy between experiment and prediction is that we previously over-estimated the biological significance of a repression ‘change’ in LLhP and LLhG.
Experiments to determine how altered transcription impacts growth of E. coli cultures returned a two-part answer—for tight and moderate repressors, the correlation is so steep that essentially any change in repression alters growth (Figure 6, upper trend). Note that the upper trend likely encompasses the up to 200-fold change that was classified as ‘wild-type’ (‘+’) repression for LacI variants (24), which might bias computational studies that use this dataset. Once repression reaches a certain weak threshold, further loss of repression has little impact on culture growth (Figure 6, lower trend). The threshold occurred near 13 Miller units, which provides a point of comparison with the published mutational studies [(12–14); data from earlier manuscripts have been converted to the current un-normalized scale]. These studies examined 5–15 amino acids per position in LLhG, LLhG/E62K and LLhP. In LLhP, most of the linker variants diminished repression below the threshold. However, many LLhP variants are toxic to several strains of E. coli (16); tighter repressors might be more toxic and thus selected against in the random mutagenesis protocol. For variants of LLhG/E62K, all of the positions had one or more amino acids that were tighter repressors than the threshold. In the current work, mutagenesis of non-conserved positions 52, 55 and 62 created several chimera variants with significant repression changes. Therefore, all non-conserved linker positions are functionally important in the LacI/GalR family.
Growth assays identified unexpected bacterial response
Finally, the growth assays suggested a mystery about regulation of the lac operon: what process triggered upregulation of the lac operon, facilitating the second growth phase? Control experiments directly ruled out the possibilities that the repressors are mutated or that repressor expression is turned off. One possibility is that the cells synthesized the various inducers for each of the chimeras. Although this might have occurred for LLhS variants (described earlier), this possibility is not satisfactory for other chimeras. For example, the cellobiose inducer for LLhE variants should not be made in E. coli, and neither LLhP nor LLhA have known inducers (Table 1). Furthermore, the inducers for LLhR, LLhT and LLhF (Figure 1) would be synthesized by different metabolic pathways, probably at different rates under the nutrient-limited conditions; if direct induction was occurring for these repressors, the different chimeras should not fall on the same trend-line. Thus, we ruled out induction via metabolites.
A second possibility is that the cells could be mutated to up-regulate transcription. However, the repeatability of the growth curves, which were generated from separate transformation of repressor-encoding plasmids, argues against that process which should be random. Third, the cells could turn on a generalized stress response. However, this explanation is unsatisfactory because cultures with strong repressors should be more stressed than cultures with weak repressors. An increased stress response should compress growth rates to a common threshold for strong repressors (the opposite of what is shown in Figure 6). A fourth possible source of upregulation is the CRP transcription activator (78), which binds to and activates the lac promoter in the presence of increased cAMP (which arises from decreased glucose). To test this hypothesis, growth experiments could be repeated in a crp- strain; however, the same counter-argument about more stress (reflected in increased cAMP levels) in cultures with strong repressors should still apply.
Finally, upregulation might be explained by competition between repressor and the activating CRP-cAMP complex or between repressor and RNA polymerase, which has been documented for wild-type LacI (79,80). We previously showed for both LLhP (16) and LLhG/E62K (15) variants that repression strength correlates with DNA-binding affinity. By extension, the growth rates must also correlate with DNA-binding affinities, and we expect similar behavior for other LLhX chimeras. Binding affinities reflect equilibrium conditions—even very strong DNA-binding repressors must occasionally vacate the lac operator, providing CRP-cAMP opportunity to activate or RNA polymerase opportunity to transcribe the operon.
Competition between transcription factors provides a satisfactory explanation at the level of a single cell; and multiple situations are found in which the effect in a single cell persists for many generations (depending on the half-lives of lacZ mRNA and protein). However, the shape of the second growth phase is consistent with a positive feedback mechanism, which suggests an initiating event. This contrasts with the originally-expected behavior of variable growth rate slopes that were expected from the variable expression of the lac operon (β-galactosidase activity assays). Therefore, perhaps a novel initiating event is at play in regulation of the lac operon.
CONCLUSION
Characterization of the synthetic LLhX protein family led to several discoveries about function in the LacI/GalR family. First, despite ‘mismatched’ interfaces, chimeras maintained allosteric response to their cognate effectors. Therefore, allostery in many LacI/GalR proteins does not require interfaces with precisely matched interactions. Looping experiments suggest that dimer–dimer interactions could be common among the LacI/GalR homologs. In addition, the chimeric interfaces were sensitive to mutagenesis of non-conserved positions; preliminary comparisons between the synthetic homologs suggest that the chimeras provide an ideal context for systematically exploring the sequence/function relationship for non-conserved amino acids. When the biological significance of functional modification was explored, experiments identified unexpected bacterial behaviors. Nevertheless, experiments showed a strong correlation between repression strength and bacterial growth, which demonstrates that most changes arising from domain recombination or mutations of LacI/GalR linkers are significant.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Tables 1–3, Supplementary Figures 1–7, and Supplementary References [12,13,15,81].
FUNDING
The National Institutes of Health [GM079423 to L.S.K.; GM075965 to L.J.M. and P20 RR17708 from the Institutional Development Award program of the National Center for Research Resources to L.S.K.]; Mayo Foundation (to L.J.M.). Funding for open access charge: NIH.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The coding region for CelR was a generous gift from Dr David B. Wilson (Cornell University). The authors thank Dr Susan Egan (KU-Lawrence), Dr Christina Hester (KUMC), and Dr Barry Hall (Bellingham Research Institute) for their helpful discussions about growth assays; and Reggie Nguyen (KUMC and Rockhurst University) for performing preliminary growth assay experiments. The authors thank Daniel J. Parente (KUMC) for assistance with data analysis and comments on this article. The authors thank Sudheer Tungtur (KUMC) for validating in vivo sugar responses with thermodynamic-binding assays. And the authors thank Dr Egan, Dr Sarah Bondos (Texas A&M Health Science Center), and Dr Kathleen Matthews (Rice University) for helpful comments on the manuscript.
REFERENCES
- 1.Bashton M, Chothia C. The generation of new protein functions by the combination of domains. Structure. 2007;15:85–99. doi: 10.1016/j.str.2006.11.009. [DOI] [PubMed] [Google Scholar]
- 2.Lim WA. The modular logic of signaling proteins: building allosteric switches from simple binding domains. Curr. Opin. Struct. Biol. 2002;12:61–68. doi: 10.1016/s0959-440x(02)00290-7. [DOI] [PubMed] [Google Scholar]
- 3.Guntas G, Mitchell SF, Ostermeier M. A molecular switch created by in vitro recombination of nonhomologous genes. Chem. Biol. 2004;11:1483–1487. doi: 10.1016/j.chembiol.2004.08.020. [DOI] [PubMed] [Google Scholar]
- 4.Wong-Deyrup SW, Prasannan C, Dupureur CM, Franklin SJ. DNA targeting and cleavage by an engineered metalloprotein dimer. J. Biol. Inorg. Chem. 2011;17:387–398. doi: 10.1007/s00775-011-0861-0. [DOI] [PubMed] [Google Scholar]
- 5.Goyal R, Salahudeen AA, Jansen M. Engineering a prokaryotic Cys-loop receptor with a third functional domain. J. Biol. Chem. 2011;286:34635–34642. doi: 10.1074/jbc.M111.269647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Swint-Kruse L, Matthews KS. Allostery in the LacI/GalR family: variations on a theme. Curr. Opin. Microbiol. 2009;12:129–137. doi: 10.1016/j.mib.2009.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schumacher MA, Glasfeld A, Zalkin H, Brennan RG. The X-ray structure of the PurR-guanine-purF operator complex reveals the contributions of complementary electrostatic surfaces and a water-mediated hydrogen bond to corepressor specificity and binding affinity. J. Biol. Chem. 1997;272:22648–22653. doi: 10.1074/jbc.272.36.22648. [DOI] [PubMed] [Google Scholar]
- 8.Bell CE, Lewis M. A closer view of the conformation of the Lac repressor bound to operator. Nat. Struct. Biol. 2000;7:209–214. doi: 10.1038/73317. [DOI] [PubMed] [Google Scholar]
- 9.Schumacher MA, Allen GS, Diel M, Seidel G, Hillen W, Brennan RG. Structural basis for allosteric control of the transcription regulator CcpA by the phosphoprotein HPr-Ser46-P. Cell. 2004;118:731–741. doi: 10.1016/j.cell.2004.08.027. [DOI] [PubMed] [Google Scholar]
- 10.Swint-Kruse L, Larson C, Pettitt BM, Matthews KS. Fine-tuning function: correlation of hinge domain interactions with functional distinctions between LacI and PurR. Protein Sci. 2002;11:778–794. doi: 10.1110/ps.4050102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tungtur S, Parente DJ, Swint-Kruse L. Functionally important positions can comprise the majority of a protein's architecture. Prot. Struct. Func. Bioinf. 2011;79:1589–1608. doi: 10.1002/prot.22985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tungtur S, Egan SM, Swint-Kruse L. Functional consequences of exchanging domains between LacI and PurR are mediated by the intervening linker sequence. Proteins. 2007;68:375–388. doi: 10.1002/prot.21412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Meinhardt S, Swint-Kruse L. Experimental identification of specificity determinants in the domain linker of a LacI/GalR protein: bioinformatics-based predictions generate true positives and false negatives. Proteins. 2008;73:941–957. doi: 10.1002/prot.22121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tungtur S, Meinhardt S, Swint-Kruse L. Comparing the functional roles of nonconserved sequence positions in homologous transcription repressors: implications for sequence/function analyses. J. Mol. Biol. 2010;395:785–802. doi: 10.1016/j.jmb.2009.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tungtur S, Skinner H, Zhan H, Swint-Kruse L, Beckett D. In vivo tests of thermodynamic models of transcription repressor function. Biophys. Chem. 2011;159:142–151. doi: 10.1016/j.bpc.2011.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhan H, Taraban M, Trewhella J, Swint-Kruse L. Subdividing repressor function: DNA binding affinity, selectivity, and allostery can be altered by amino acid substitution of nonconserved residues in a LacI/GalR homologue. Biochemistry. 2008;47:8058–8069. doi: 10.1021/bi800443k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhan H, Swint-Kruse L, Matthews KS. Extrinsic interactions dominate helical propensity in coupled binding and folding of the lactose repressor protein hinge helix. Biochemistry. 2006;45:5896–5906. doi: 10.1021/bi052619p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jobe A, Bourgeois S. lac Repressor-operator interaction. VI. The natural inducer of the lac operon. J. Mol. Biol. 1972;69:397–408. doi: 10.1016/0022-2836(72)90253-7. [DOI] [PubMed] [Google Scholar]
- 19.Neidhardt FC, Bloch PL, Smith DF. Culture medium for enterobacteria. J. Bacteriol. 1974;119:736–747. doi: 10.1128/jb.119.3.736-747.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bhende PM, Egan SM. Amino acid-DNA contacts by RhaS: an AraC family transcription activator. J. Bacteriol. 1999;181:5185–5192. doi: 10.1128/jb.181.17.5185-5192.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Swint-Kruse L, Zhan H, Fairbanks BM, Maheshwari A, Matthews KS. Perturbation from a distance: mutations that alter LacI function through long-range effects. Biochemistry. 2003;42:14004–14016. doi: 10.1021/bi035116x. [DOI] [PubMed] [Google Scholar]
- 22.Geanacopoulos M, Adhya S. Genetic analysis of GalR tetramerization in DNA looping during repressosome assembly. J. Biol. Chem. 2002;277:33148–33152. doi: 10.1074/jbc.M202445200. [DOI] [PubMed] [Google Scholar]
- 23.Chen J, Matthews KS. Subunit dissociation affects DNA binding in a dimeric lac repressor produced by C-terminal deletion. Biochemistry. 1994;33:8728–8735. doi: 10.1021/bi00195a014. [DOI] [PubMed] [Google Scholar]
- 24.Suckow J, Markiewicz P, Kleina LG, Miller J, Kisters-Woike B, Müller-Hill B. Genetic studies of the Lac repressor. XV: 4000 single amino acid substitutions and analysis of the resulting phenotypes on the basis of the protein structure. J. Mol. Biol. 1996;261:509–523. doi: 10.1006/jmbi.1996.0479. [DOI] [PubMed] [Google Scholar]
- 25.Falcon CM, Matthews KS. Engineered disulfide linking the hinge regions within lactose repressor dimer increases operator affinity, decreases sequence selectivity, and alters allostery. Biochemistry. 2001;40:15650–15659. doi: 10.1021/bi0114067. [DOI] [PubMed] [Google Scholar]
- 26.Swint-Kruse L, Zhan H, Matthews KS. Integrated insights from simulation, experiment, and mutational analysis yield new details of LacI function. Biochemistry. 2005;44:11201–11213. doi: 10.1021/bi050404+. [DOI] [PubMed] [Google Scholar]
- 27.Oehler S, Eismann ER, Kramer H, Müller-Hill B. The three operators of the lac operon cooperate in repression. EMBO J. 1990;9:973–979. doi: 10.1002/j.1460-2075.1990.tb08199.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sadler JR, Sasmor H, Betz JL. A perfectly symmetric lac operator binds the lac repressor very tightly. Proc. Natl Acad. Sci. USA. 1983;80:6785–6789. doi: 10.1073/pnas.80.22.6785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Miller JH. A Short Course in Bacterial Genetics: A Laboratory Handbook for Escherichia Coli and Related Bacteria. Plainview, NY: Cold Spring Harbor Laboratory Press; 1992. [Google Scholar]
- 30.Riggs AD, Newby RF, Bourgeois S. lac repressor–operator interaction. II. Effect of galactosides and other ligands. J. Mol. Biol. 1970;51:303–314. doi: 10.1016/0022-2836(70)90144-0. [DOI] [PubMed] [Google Scholar]
- 31.Manly SP, Matthews KS. Activity changes in lac repressor with cysteine oxidation. J. Biol. Chem. 1979;254:3341–3347. [PubMed] [Google Scholar]
- 32.Becker NA, Kahn JD, Maher LJ., 3rd Bacterial repression loops require enhanced DNA flexibility. J. Mol. Biol. 2005;349:716–730. doi: 10.1016/j.jmb.2005.04.035. [DOI] [PubMed] [Google Scholar]
- 33.Chen J, Matthews KS. T41 mutation in lac repressor is Tyr282—Asp. Gene. 1992;111:145–146. doi: 10.1016/0378-1119(92)90618-y. [DOI] [PubMed] [Google Scholar]
- 34.Chakerian AE, Matthews KS. Characterization of mutations in oligomerization domain of Lac repressor protein. J. Biol. Chem. 1991;266:22206–22214. [PubMed] [Google Scholar]
- 35.Schmitz A, Schmeissner U, Miller JH. Mutations affecting the quaternary structure of the lac repressor. J. Biol. Chem. 1976;251:3359–3366. [PubMed] [Google Scholar]
- 36.Whipple FW. Genetic analysis of prokaryotic and eukaryotic DNA-binding proteins in Escherichia coli. Nucleic Acids Res. 1998;26:3700–3706. doi: 10.1093/nar/26.16.3700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Weickert MJ, Adhya S. A family of bacterial regulators homologous to Gal and Lac repressors. J. Biol. Chem. 1992;267:15869–15874. [PubMed] [Google Scholar]
- 38.Choi KY, Zalkin H. Structural characterization and corepressor binding of the Escherichia coli purine repressor. J. Bacteriol. 1992;174:6207–6214. doi: 10.1128/jb.174.19.6207-6214.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Meng LM, Nygaard P. Identification of hypoxanthine and guanine as the co-repressors for the purine regulon genes of Escherichia coli. Mol. Microbiol. 1990;4:2187–2192. doi: 10.1111/j.1365-2958.1990.tb00580.x. [DOI] [PubMed] [Google Scholar]
- 40.Gavigan SA, Nguyen T, Nguyen N, Senear DF. Role of multiple CytR binding sites on cooperativity, competition, and induction at the Escherichia coli udp promoter. J. Biol. Chem. 1999;274:16010–16019. doi: 10.1074/jbc.274.23.16010. [DOI] [PubMed] [Google Scholar]
- 41.Pedersen H, Sogaard-Andersen L, Holst B, Valentin-Hansen P. Heterologous cooperativity in Escherichia coli. The CytR repressor both contacts DNA and the cAMP receptor protein when binding to the deoP2 promoter. J. Biol. Chem. 1991;266:17804–17808. [PubMed] [Google Scholar]
- 42.Tretyachenko-Ladokhina V, Cocco MJ, Senear DF. Flexibility and adaptability in binding of E. coli cytidine repressor to different operators suggests a role in differential gene regulation. J. Mol. Biol. 2006;362:271–286. doi: 10.1016/j.jmb.2006.06.085. [DOI] [PubMed] [Google Scholar]
- 43.Kallipolitis BH, Norregaard-Madsen M, Valentin-Hansen P. Protein-protein communication: structural model of the repression complex formed by CytR and the global regulator CRP. Cell. 1997;89:1101–1109. doi: 10.1016/s0092-8674(00)80297-4. [DOI] [PubMed] [Google Scholar]
- 44.Hall BG, Xu L. Nucleotide sequence, function, activation, and evolution of the cryptic asc operon of Escherichia coli K12. Mol. Biol. Evol. 1992;9:688–706. doi: 10.1093/oxfordjournals.molbev.a040753. [DOI] [PubMed] [Google Scholar]
- 45.Weickert MJ, Adhya S. Isorepressor of the gal regulon in Escherichia coli. J. Mol. Biol. 1992;226:69–83. doi: 10.1016/0022-2836(92)90125-4. [DOI] [PubMed] [Google Scholar]
- 46.Geanacopoulos M, Adhya S. Functional characterization of roles of GalR and GalS as regulators of the gal regulon. J. Bacteriol. 1997;179:228–234. doi: 10.1128/jb.179.1.228-234.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mossing MC, Record MT., Jr Upstream operators enhance repression of the lac promoter. Science. 1986;233:889–892. doi: 10.1126/science.3090685. [DOI] [PubMed] [Google Scholar]
- 48.Semsey S, Virnik K, Adhya S. Three-stage regulation of the Amphibolic gal operon: from repressosome to GalR-free DNA. J. Mol. Biol. 2006;358:355–363. doi: 10.1016/j.jmb.2006.02.022. [DOI] [PubMed] [Google Scholar]
- 49.Krämer H, Niemöller M, Amouyal M, Revet B, von Wilcken-Bergmann B, Müller-Hill B. lac repressor forms loops with linear DNA carrying two suitably spaced lac operators. EMBO J. 1987;6:1481–1491. doi: 10.1002/j.1460-2075.1987.tb02390.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Müller J, Oehler S, Müller-Hill B. Repression of lac promoter as a function of distance, phase, and quality of an auxilary lac operator. J. Mol. Biol. 1996;257:21–29. doi: 10.1006/jmbi.1996.0143. [DOI] [PubMed] [Google Scholar]
- 51.Matthews KS. DNA looping. Microbiol. Rev. 1992;56:123–136. doi: 10.1128/mr.56.1.123-136.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Eismann E, von Wilcken-Bergmann B, Müller-Hill B. Specific destruction of the second lac operator decreases repression of the lac operon in Escherichia coli fivefold. J. Mol. Biol. 1987;195:949–952. doi: 10.1016/0022-2836(87)90499-2. [DOI] [PubMed] [Google Scholar]
- 53.Müller J, Barker A, Oehler S, Müller-Hill B. Dimeric lac repressors exhibit phase-dependent co-operativity. J. Mol. Biol. 1998;284:851–857. doi: 10.1006/jmbi.1998.2253. [DOI] [PubMed] [Google Scholar]
- 54.Poelwijk FJ, Heyning PD, de Vos MG, Kiviet DJ, Tans SJ. Optimality and evolution of transcriptionally regulated gene expression. BMC Syst. Biol. 2011;5:128. doi: 10.1186/1752-0509-5-128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Monod J. The phenomenon of enzymatic adaptation and its bearings on problems of genetics and cellular differentiation. Growth. 1947;11:223–289. [Google Scholar]
- 56.Wilson CJ, Zhan H, Swint-Kruse L, Matthews KS. The lactose repressor system: paradigms for regulation, allosteric behavior and protein folding. Cell Mol. Life Sci. 2007;64:3–16. doi: 10.1007/s00018-006-6296-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mowbray SL, Björkman AJ. Conformational changes of ribose-binding protein and two related repressors are tailored to fit the functional need. J. Mol. Biol. 1999;294:487–499. doi: 10.1006/jmbi.1999.3271. [DOI] [PubMed] [Google Scholar]
- 58.Gama-Castro S, Salgado H, Peralta-Gil M, Santos-Zavaleta A, Muniz-Rascado L, Solano-Lira H, Jimenez-Jacinto V, Weiss V, Garcia-Sotelo JS, Lopez-Fuentes A, et al. RegulonDB version 7.0: transcriptional regulation of Escherichia coli K-12 integrated within genetic sensory response units (Gensor Units) Nucleic Acids Res. 2011;39:D98–D105. doi: 10.1093/nar/gkq1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Shimada T, Yamamoto K, Ishihama A. Novel members of the Cra regulon involved in carbon metabolism in Escherichia coli. J. Bacteriol. 2011;193:649–659. doi: 10.1128/JB.01214-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Cho BK, Federowicz SA, Embree M, Park YS, Kim D, Palsson BO. The PurR regulon in Escherichia coli K-12 MG1655. Nucleic Acids Res. 2011;39:6456–6464. doi: 10.1093/nar/gkr307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ishida Y, Kori A, Ishihama A. Participation of regulator AscG of the beta-glucoside utilization operon in regulation of the propionate catabolism operon. J. Bacteriol. 2009;191:6136–6144. doi: 10.1128/JB.00663-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Pei J, Cai W, Kinch LN, Grishin NV. Prediction of functional specificity determinants from protein sequences using log-likelihood ratios. Bioinformatics. 2006;22:164–171. doi: 10.1093/bioinformatics/bti766. [DOI] [PubMed] [Google Scholar]
- 63.Kalinina OV, Novichkov PS, Mironov AA, Gelfand MS, Rakhmaninova AB. SDPpred: a tool for prediction of amino acid residues that determine differences in functional specificity of homologous proteins. Nucleic Acids Res. 2004;32:424–428. doi: 10.1093/nar/gkh391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ye K, Feenstra AK, Heringa J, Ijzerman AP, Marchiori E. Multi-RELIEF: a method to recognize specificity determining residues from multiple sequence alignments using a machine-learning approach for feature weighting. Bioinformatics. 2008;24:18–25. doi: 10.1093/bioinformatics/btm537. [DOI] [PubMed] [Google Scholar]
- 65.Ye K, Vriend G, Ijzerman AP. Tracing evolutionary pressure. Bioinformatics. 2008;24:908–915. doi: 10.1093/bioinformatics/btn057. [DOI] [PubMed] [Google Scholar]
- 66.Binkley J, Karra K, Kirby A, Hosobuchi M, Stone EA, Sidow A. ProPhylER: A curated online resource for protein function and structure based on evolutionary constraint analyses. Genome Res. 2010;20:142–154. doi: 10.1101/gr.097121.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Cai ZH, Tsung EF, Marinescu VD, Ramoni MF, Riva A, Kohane IS. Bayesian approach to discovering pathogenic SNPs in conserved protein domains. Hum. Mutat. 2004;24:178–184. doi: 10.1002/humu.20063. [DOI] [PubMed] [Google Scholar]
- 68.Chasman D, Adams RM. Predicting the functional consequences of non-synonymous single nucleotide polymorphisms: structure-based assessment of amino acid variation. J. Mol. Biol. 2001;307:683–706. doi: 10.1006/jmbi.2001.4510. [DOI] [PubMed] [Google Scholar]
- 69.Chen HL, Zhou HX. Prediction of solvent accessibility and sites of deleterious mutations from protein sequence. Nucleic Acids Res. 2005;33:3193–3199. doi: 10.1093/nar/gki633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Chiu HC, Chang CA, Hu YJ. Prediction of orthologous relationship by functionally important sites. Comput. Meth. Programs Biomed. 2005;78:209–222. doi: 10.1016/j.cmpb.2005.03.002. [DOI] [PubMed] [Google Scholar]
- 71.Jiang R, Yang H, Sun F, Chen T. Searching for interpretable rules for disease mutations: a simulated annealing bump hunting strategy. BMC Bioinformatics. 2006;7:417. doi: 10.1186/1471-2105-7-417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Krishnan VG, Westhead DR. A comparative study of machine-learning methods to predict the effects of single nucleotide polymorphisms on protein function. Bioinformatics. 2003;19:2199–2209. doi: 10.1093/bioinformatics/btg297. [DOI] [PubMed] [Google Scholar]
- 73.Lau AY, Chasman DI. Functional classification of proteins and protein variants. Proc. Natl Acad. Sci. USA. 2004;101:6576–6581. doi: 10.1073/pnas.0305043101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Lee W, Zhang Y, Mukhyala K, Lazarus RA, Zhang Z. Bi-directional SIFT predicts a subset of activating mutations. PloS One. 2009;4:e8311. doi: 10.1371/journal.pone.0008311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Marini NJ, Thomas PD, Rine J. The use of orthologous sequences to predict the impact of amino acid substitutions on protein function. PloS Genet. 2010;6:e1000968. doi: 10.1371/journal.pgen.1000968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Needham CJ, Bradford JR, Bulpitt AJ, Care MA, Westhead DR. Predicting the effect of missense mutations on protein function: analysis with Bayesian networks. BMC Bioinformatics. 2006;7:405. doi: 10.1186/1471-2105-7-405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Ng PC, Henikoff S. Predicting deleterious amino acid substitutions. Genome Res. 2001;11:863–874. doi: 10.1101/gr.176601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.De Crombrugghe B, Chen B, Anderson W, Nissley P, Gottesman M, Pastan I, Perlman R. Lac DNA, RNA polymerase and cyclic AMP receptor protein, cyclic AMP, lac repressor and inducer are the essential elements for controlled lac transcription. Nat. New Biol. 1971;231: 139–142. doi: 10.1038/newbio231139a0. [DOI] [PubMed] [Google Scholar]
- 79.Schlax PJ, Capp MW, Record TM., Jr Inhibition of transcription initiation by IacRepressor. J. Mol. Biol. 1995;245:331–350. doi: 10.1006/jmbi.1994.0028. [DOI] [PubMed] [Google Scholar]
- 80.Sanchez A, Osborne ML, Friedman LJ, Kondev J, Gelles J. Mechanism of transcriptional repression at a bacterial promoter by analysis of single molecules. EMBO J. 2011;30:3940–3946. doi: 10.1038/emboj.2011.273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Jones DH. PCR mutagenesis and recombination in vivo. PCR Methods Appl. 1994;3:S141–S148. doi: 10.1101/gr.3.6.s141. [DOI] [PubMed] [Google Scholar]
- 82.Majumdar A, Rudikoff S, Adhya S. Purification and properties of Gal repressor:pL-galR fusion in pKC31 plasmid vector. J. Biol. Chem. 1987;262:2326–2331. [PubMed] [Google Scholar]
- 83.Saier MH, Jr, Ramseier TM. The catabolite repressor/activator (Cra) protein of enteric bacteria. J. Bacteriol. 1996;178:3411–3417. doi: 10.1128/jb.178.12.3411-3417.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Ramseier TM, Negre D, Cortay JC, Scarabel M, Cozzone AJ, Saier MH., Jr In vitro binding of the pleiotropic transcriptional regulatory protein, FruR, to the fru, pps, ace, pts and icd operons of Escherichia coli and Salmonella typhimurium. J. Mol. Biol. 1993;234:28–44. doi: 10.1006/jmbi.1993.1561. [DOI] [PubMed] [Google Scholar]
- 85.Horlacher R, Boos W. Characterization of TreR, the major regulator of the Escherichia coli trehalose system. J. Biol. Chem. 1997;272:13026–13032. doi: 10.1074/jbc.272.20.13026. [DOI] [PubMed] [Google Scholar]
- 86.Mauzy CA, Hermodson MA. Structural homology between rbs repressor and ribose binding protein implies functional similarity. Protein Sci. 1992;1:843–849. doi: 10.1002/pro.5560010702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Barbier CS, Short SA, Senear DF. Allosteric mechanism of induction of CytR-regulated gene expression. Cytr repressor-cytidine interaction. J. Biol. Chem. 1997;272:16962–16971. doi: 10.1074/jbc.272.27.16962. [DOI] [PubMed] [Google Scholar]
- 88.Spiridonov NA, Wilson DB. Characterization and cloning of celR, a transcriptional regulator of cellulase genes from Thermomonospora fusca. J. Biol. Chem. 1999;274:13127–13132. doi: 10.1074/jbc.274.19.13127. [DOI] [PubMed] [Google Scholar]
- 89.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. UCSF Chimera–a visualization system for exploratory research and analysis. J. Comput. Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 90.Alberti S, Oehler S, von Wilcken-Bergmann B, Krämer H, Müller-Hill B. Dimer-to-tetramer assembly of lac repressor involves a leucine heptad repeat. New Biologist. 1991;3:57–62. [PubMed] [Google Scholar]
- 91.Taraban M, Zhan H, Whitten AE, Langley DB, Matthews KS, Swint-Kruse L, Trewhella J. Ligand-induced conformational changes and conformational dynamics in the solution structure of the lactose repressor protein. J. Mol. Biol. 2008;376:466–481. doi: 10.1016/j.jmb.2007.11.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.