Abstract
The ventrolateral dermomyotome gives rise to all muscles of the limbs through the delamination and migration of cells into the limb buds. These cells proliferate and form myoblasts, withdraw from the cell cycle and become terminally differentiated. The myogenic lineage colonizes pre-patterned regions to form muscle anlagen as muscle fibers are assembled. The regulatory mechanisms that control the later steps of this myogenic program are not well understood. The homeodomain transcription factor Pitx2 is expressed in the muscle lineage from the migration of precursors to adult muscle. Ablation of Pitx2 results in distortion, rather than loss, of limb muscle anlagen, suggesting that its function becomes critical during the colonization of, and/or fiber assembly in, the anlagen. Gene expression arrays were used to identify changes in gene expression in flow-sorted migratory muscle precursors, labeled by Lbx1EGFP, which resulted from the loss of Pitx2. Target genes of Pitx2 were clustered using the “David Bioinformatics Functional Annotation Tool” to bin genes according to enrichment of gene ontology keywords. This provided a way to both narrow the target genes and identify potential gene families regulated by Pitx2. Representative target genes in the most enriched bins were analyzed for the presence and evolutionary conservation of Pitx2 consensus binding sequence, TAATCY, on the −20kb, intronic, and coding regions of the genes. Fifteen Pitx2 target genes were selected based on the above analysis and were identified as having functions involving cytoskeleton organization, tissue specification, and transcription factors. Data from these studies suggest that Pitx2 acts to regulate cell motility and expression of muscle specific genes in the muscle precursors during forelimb muscle development. This work provides a framework to develop the gene network leading to skeletal muscle development, growth and regeneration.
Keywords: Homeobox, Pitx2, Development, Myoblast, Gene Expression Analysis, Bioinformatics
1. Introduction
The forelimb muscles originate from the hypaxial dermomyotome of the interlimb somites during embryonic development. Inductive cues from the lateral plate mesoderm synergistically induce the expression of Lbx1 within the ventrolateral Pax3 expression domain of the dermomyotome (Tremblay et al., 1998). These cells delaminate from the dermomyotome and migrate into the developing limb bud (Bladt et al., 1995; Dietrich et al., 1999; Hayashi and Ozawa, 1995). The dorsal and ventral muscle masses of mouse limb bud consist of Lbx1+/Pax3+ limb muscle progenitor cells at E10.5 and this gene expression persists until E12.5 (Gross et al., 2000). At E11-E12.5 muscle masses enlarge, shape and position themselves with respect to bone anlagen. Muscle progenitor cells increase their numbers through proliferation, undergo withdrawal from the cell cycle and become terminally differentiated myocytes. Pax3 and Lbx1 have generally been placed at the beginning of myogenic progression and activation of the Muscle Regulatory Factors (MRF) in the embryonic limb because they are expressed earlier and their mutation leads to a loss of migratory precursors before MRFs are normally expressed (Bober et al., 1994; Goulding et al., 1994; Gross et al., 2000; Mennerich et al., 1998; Schafer and Braun, 1999). These myocytes fuse with each other to form multinucleated myotubes and then muscle fibers. The precise regulatory mechanisms that control each step of the myogenic program are not well understood to date.
The bicoid–related homeobox gene Pitx2 is expressed in muscle anlagen in all stages of myogenic progression (Shih et al., 2007a, b). Pitx2 contributes to the establishment of network kernels that specify pre-myogenic progenitors for extraocular and mastication muscles (Shih et al., 2008). Ablation of Pitx2 causes lethality in the mouse at E10.5–E14.5 with axial malformations, open body wall, heart defects, and arrest of organ development (Gage et al., 1999; Kitamura et al., 1999; Lin et al., 1999; Lu et al., 1999). Pitx2 is positioned downstream of both Wnt and growth factor signaling pathways in skeletal myogenesis and promotes muscle progenitor proliferation by direct regulation of the expression of a number of cyclin-dependent kinases (Kioussi et al., 2002). Analysis of Lbx1+ muscle progenitor cells isolated from E12.5 Pitx2 mutant forelimbs has revealed that a small group of cytoskeletal, adhesion, and signaling genes are potential targets for regulation by Pitx2. Muscle progenitors of Pitx2 mutants were smaller, more symmetrical, had increased actin bundling, and decreased motility preventing myogenic cells from filling limb bud anlagen (Campbell et al., 2012).
In this study we utilized gene expression data from microarray experiments in combination with online gene ontology databases and in house scripts to predict the presence of Cis-Regulatory Modules (CRM). The gene expression profile of Lbx1+ muscle precursors isolated from forelimb tissue at E12.5 from Pitx2 WT, HET, and MUT mice was obtained using Affymetrix Mouse Genome 430 2.0 arrays, with RMA normalization and analyzed with SAMExcel (Kioussi and Gross, 2008; Tusher et al., 2001). This resulted in a total of 772 probe sets, representing 688 unique genes that were significantly differentially expressed. Genes were placed into a total of 175 bins based on putative functional annotations using DAVID Bioinformatics Functional Annotation Tool (Dennis et al., 2003; Huang da et al., 2009). The first 10 bins had similar enrichment scores reported and were split with GOTERMs referring to cytoskeletal or transcription factor functions. These genes were analyzed for predicted Pitx2 binding sites within the genomic gene sequence and the −20kb upstream region. The top 2 genes from each bin that contained Pitx2 binding sites conserved in at least 4 species gave us a representative pool of 20 genes. A predicted network model was constructed using BioTapestry version 5.0.2 to visually link Pitx2 with its target genes.
1. Material and methods
1.1 Mice
ICR Pitx2LacZ/+ mouse embryos (HET) (Lin et al., 1999), Lbx1EGFP/+ (Gross et al., 2000) were used. Pitx2LacZ/+ mice were bred with Pitx2LacZ/+|Lbx1EGFP/+ to generate Lbx1EGFP/+|Pitx2LacZ/LacZ (MUT), Lbx1EGFP/+|Pitx2LacZ/+ (HET) and Lbx1EGFP/+|Pitx2+/+ (WT) mice. Tail genomic DNA was extracted and used for PCR genotyping (Gross et al., 2000; Lin et al., 1999). For cell flow sorting, embryos were rapidly genotyped under a fluorescent microscope to identify Lbx1 HET mice. Positive identification of Lbx1 HET embryos were followed up with X-gal staining to determine Pitx2 genotype.
1.2 RNA Preparation and Microarray Analysis
We analyzed microarrays from E12.5 mouse forelimb tissue enriched for Lbx1+ muscle progenitor cells (Campbell et al., 2012). These data have been deposited in NCBI’s Gene Expression Omnibus (Edgar et al., 2002) and are accessible through GEO Series accession number GSE31945 (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE31945). The differentially expressed genes determined to be significantly altered above the 1.2 fold cutoff were analyzed with DAVID Bioinformatics Functional Annotation Tool (Dennis et al., 2003; Huang da et al., 2009). The DAVID Functional Annotation Clustering application analyzed and sorted the genes into functional clusters that we then individually searched for the presence of Pitx2 binding sites.
2. Pitx2 Binding Site Analysis and BioTapestry
The genomic sequence with the −20kb upstream sequence of each gene was downloaded from MGI’s database to the mouse reference genome (NCBI v37, mm9). The data was then processed with a script generated in our lab, binding_site_search.pl, to determine the location of Pitx2 binding sites, TAATCY, relative to the start of the gene. The level of evolutionary conservation of the identified binding sites were determined by the use of our script, binding_site_compare.pl, by comparing the site to gene entry locations in the UCSC Genome browser (Eng et al., 2010). Cytoscape v2.8.2 was utilized to compose visualizations of gene expression data and presence of binding sites (Shannon et al., 2003). BioTapestry v5.0.2 was utilized to compose the predicted gene network in the developing forelimb muscles (Longabaugh et al., 2005).
3. Results and Discussion
3.1 Pitx2 Target Gene Functional Clusters in Forelimb
The DAVID Functional Annotation Clustering tool was applied to the genes identified as differentially regulated greater than a 1.2 fold cutoff. A total of 175 bins covered 668 unique genes. The first 10 bins showed enrichment scores of 3 or greater (Table 1). Key gene ontology terms (GOTERMS) such as cell differentiation, cytoskeleton, transcription factor, etc. were ranked according to their frequency of occurrence in the Pitx2 target genes compared to their occurrence in the mouse genome as a whole. The Ptix2 target genes were arranged according to their putative function in the developing forelimb muscles. The top ten clusters enrichment score ranged from 4.9 to 3.0 with the top scoring clusters being genes involved in cell differentiation and morphogenesis, cytoskeleton binding, and sequence specific transcription factors (SSTFs) (Table 1). These genes represent the regulatory priorities of the cells at E12.5 of limb muscle development. These 10 categories can be divided into two main groups the structural genes (Bin 1, 2, 4, 8, 9, 10) and sequence specific transcription factors (SSTF) (Bin 3, 5, 6, 7).
Table 1.
David Functional Annotation Categories of Pitx2 Target Genes in Forelimb Muscle Progenitor Cells
| Annotation Bin | Enrichment Score | GOTERM | # of Genes | P- Value | Benjamini Corrected P-value | Average # Pitx2 Binding Sites |
|---|---|---|---|---|---|---|
| 1 | 4.9 | Differentiation/Morphogenesis | 26 | 1.7E-7 | 3.9E-4 | 13 |
| 2 | 4.2 | Cytoskeleton Binding | 37 | 1.5E-6 | 2.8E-4 | 7 |
| 3 | 4.2 | Sequence Specific DNA Binding | 47 | 2.1E-7 | 1.2E-4 | 13 |
| 4 | 4.0 | Cytoskeleton Organization | 31 | 2.2E-6 | 7.5E-4 | 6 |
| 5 | 3.6 | Negative Regulation of Transcription | 32 | 7.4E-5 | 8.0E-3 | 17 |
| 6 | 3.4 | Transcription Factor Activity | 58 | 3.8E-7 | 1.1E-4 | 13 |
| 7 | 3.4 | Regulation of Transcription | 46 | 4.4E-6 | 1.3E-3 | 15 |
| 8 | 3.2 | Cytoskeleton | 67 | 1.4E-5 | 4.8E-3 | 9 |
| 9 | 3.0 | Cell Adhesion | 21 | 3.3E-4 | 1.6E-2 | 16 |
| 10 | 3.0 | Cell Motility | 34 | 1.1E-6 | 1.4E-3 | 15 |
3.2 Evolutionary Conserved Pitx2 Binding Sites
The identification of Pitx2 binding sites in the promoter, coding, and intronic regions of the target genes was accomplished through comparative genome analysis. We analyzed the −20kb region of the transcriptional start site along with the genomic sequence of each gene in bins 1 to 10 (Table 1). We searched for the presence of the TAATCY consensus sequence for evidence of regulation by Pitx2. The average number of binding sites identified ranged from 17 to 6 per gene (Table 1). If genes were directly regulated by Pitx2, then the bin with the highest enrichment score should correlate to a higher number of Pitx2 binding sites. Our data show that the difference between the bins, either the average number of binding sites and enrichment scores has similar values compared to each other. This non-linear correlation between binding site and enrichment score may be due indirect regulation through co-factor binding complexes. To address this possibility, we chose to focus on those genes that may be directly regulated by Pitx2 based on predicted binding sites. We have chosen to focus on the top 20 genes between all 10 functional bins because they contained a higher than average number of Pitx2 binding sites for further analysis for evolutionary conserved binding sites (Table 2). The use of a Perl script that was developed in our lab allowed us to align the output for each gene from the UCSC Genome Browser Assembly and identify the absolute location and species conservation of that site (Kent et al., 2002).
Table 2.
Pitx2 Target Genes with Conserved Binding sites
| Gene | Gene Location | Number of Pitx2 binding sites | Species | Function | Bibliography |
|---|---|---|---|---|---|
|
Sox6 SRY-box containing gene 6 |
Chr7: 122,594,858- 123,138,600 | 89 (18) | M, R, H, O, D, Ho, Op, Ch | Skeletal muscle differentiation | (An et al., 2011) |
|
Zfpm2 Zinc finger protein, multitype 2 |
Chr15: 40,466,588- 40,936,138 | 85 (22) | M, R, H, O, D, Ho, Op, Ch | Cardiac development | (Svensson et al., 2000; Tevosian et al., 2000) |
|
Prkg1 Protein kinase, cGMP-dependent type 1 |
Chr19: 30,622,041- 31,839,523 | 78 (0) | M, R, H, O, D, Ho, Op, Ch | Intracellular cGMP signaling | (Hofmann et al., 2009) |
|
Nfib Nuclear factor 1/B |
Chr4: 81,916,077- 82,151,212 | 59 (16) | M, R, H, O, D, Ho, Op, Ch | Adipocyte differentiation | (Waki et al., 2011) |
|
Foxp2 Forkhead box P2 |
Chr6: 155,115,506- 15,391,977 | 53 (25) | M, R, H, O, D, Ho, Op, Ch | Smooth muscle differentiation | (Shu et al., 2007) |
|
Zfhx3 Zinc finger homeobox 3 |
Chr8: 111,218,544- 111,485,536 | 44 (3) | M, R, H, O, D, Ho, Op, Ch | Muscle differentiation | (Berry et al., 2001) |
|
Mid1 Midline 1 |
ChrX: 166,103,179- 166,428,729 | 42 (2) | M, R, H, O, D, Ho, Op, Ch | E3 ligase | (Liu et al., 2011) |
|
Meis2 Meis homeobox 2 |
Chr2: 115,667,000- 115,890,794 | 42 (12) | M, R, H, O, D, Ho, Op, Ch | Limb patterning | (Capdevila et al., 1999; Salsi et al., 2008) |
|
Trps1 Trichorhinophalangeal syndrome 1 |
Chr15: 50,466,305- 50,721,587 | 41 (4) | M, R, H, O, D, Ho, Op, Ch | Cartilage, skeleton, lung, and trachea development | (Kunath et al., 2002; Gai et al., 2011) |
|
Dscaml1 Down syndrome cell adhesion molecule like 1 |
Chr9: 45,218,376- 45,561,796 | 37 (6) | M, R, H, O, D, Ho, Op, Ch | Cell adhesion, Cell type self avoidance | (Agarwala et al., 2001; Fuerst et al., 2009) |
|
Tle4 Transducin-like enhancer of split 4 |
Chr19: 14,502,562- 14,672,473 | 33 (2) | M, R, H, O, D, Ho, Op, Ch | Mesoderm specification | (Van Hateren et al., 2005; Murai et al., 2007) |
|
Palld Palladin, cytoskeletal associated protein |
Chr8: 63,972,041- 64,381,487 | 29 (0) | M, R, H, O, D, Ho, Op | Cytoskeleton associated protein, Muscle differentiation | (Jin et al., 2009; Jin et al., 2010) |
|
Met Met proto-oncogene |
Chr6: 17,393,957- 17,523,980 | 29 (1) | M, R, H, O, D, Ho, Op | Limb muscle development | (Christ and Brand- Saberi, 2002) |
|
Ank2 Ankyrin 2 |
Chr3: 126,630,030- 127,111,949 | 28 (0) | M, R, H, O, D, Ho, Op | Ca2+ channel membrane targeting and stability | (Cunha et al., 2011) |
|
Dclk1 Doublecortin-like kinase 1 |
Chr3: 55,026,448- 55,342,990 | 27 (1) | M, R, H, O, D, Ho, Op, Ch | Microtubule associated protein | (Lin et al., 2000; Sureban et al., 2011) |
M, mouse; R, rat; H, human; O, orangutans; D, dog; Ho, horse; Op, opossum; Ch, chicken
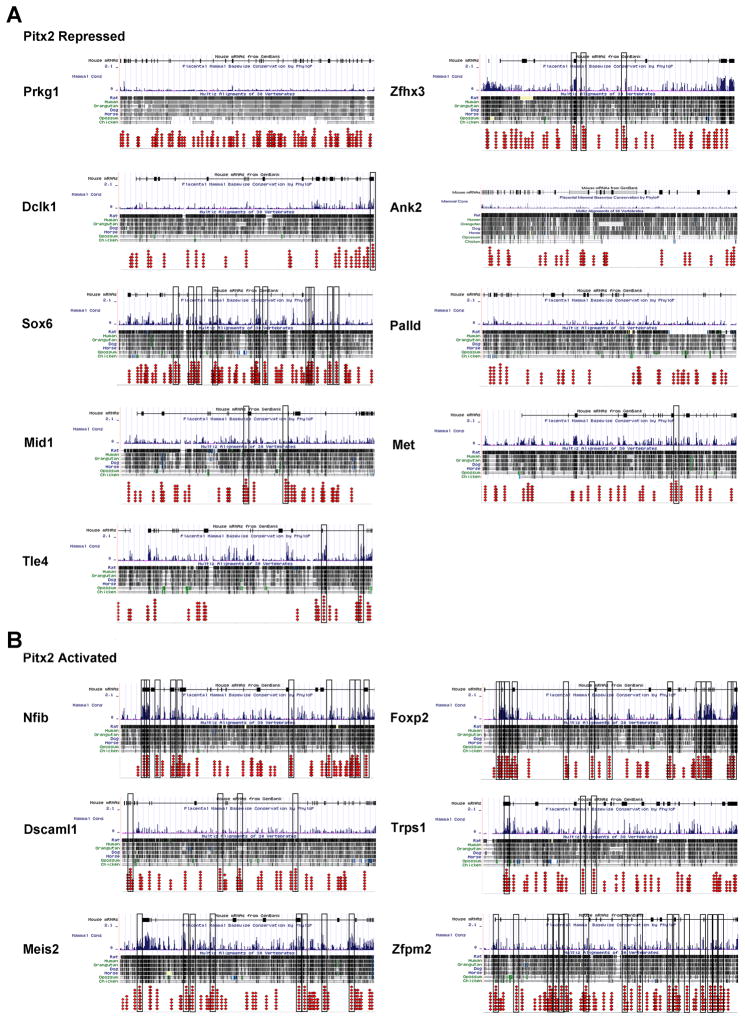
The Pitx2 binding sites on target genes were selected based on their conservation between a minimum of 4 species (mouse, human, plus 2 others) and a maximum of 8 (mouse, human, rat, orangutan, dog, horse, opossum, chicken). Within the top 20 genes, 5 were eliminated because of the presence of an additional gene in the −20kb region. The top 15 genes all contained a number of Pitx2 binding sites conserved through the minimum number of species (Table 2), when the minimum number of species was increased to the maximum of 8, the number of conserved Pitx2 binding sites reduced significantly to an average of 7 Pitx2 binding sites per gene (Fig 1, boxed regions). Almost all of the Pitx2 binding sites conserved in all 8 species were located within intronic regions of the gene rather than the proximal promoter sequence (Fig 1).
Figure 1. Evolutionary Conserved Pitx2 Binding Sites.
Comparative analysis of Pitx2 consensus sequence binding sites in Pitx2 target genes. The gene and the −20kb region upstream of the transcriptional start site were mapped against the aligned sequences in the top 15 Pitx2 target genes. Columns of red dots indicate a predicted Pitx2 binding site and the number of species the motif was conserved in, for all sites with conservation in at least 4 species. Binding sites with 3 or less conserved species not shown for clarity. Black boxes outline the binding sites that were found to be conserved in all 8 of the species.
3.3 Pitx2 Target Gene Function
The order of Pitx2 target genes based on the absolute fold change of gene expression from the microarray analysis was different that the order based on the number of conserved Pitx2 binding sites (Table 3). The genes with fold changes of 1.5 or greater were Doublecortin like kinase 1 (Dclk1), Met pro-oncogene (c-Met), Forkhead box P2 (Foxp2), and Down syndrome cell adhesion molecule like 1 (Dscaml1). The gene Dclk1 encodes for a protein that binds and regulates microtubule polymerization and dynamics (Lin et al., 2000). The expression of Dclk1 is highly enriched in the developing brain; a low level of expression can be detected by northern blot in skeletal muscle of developing mice (E7–E11) (Sossey-Alaoui and Srivastava, 1999). Microtubule dynamics are important for migration of the cell as they provide stability to the cell body, allowing for actin cytoskeleton bundling to form protrusions of the cell membrane and allow the cell to elongate and form attachments to the extracellular matrix. Met encodes a tyrosine kinase receptor, is expressed in the lateral dermomyotome of all somites and is involved in the development of hypaxial musculature. Mice deficient for the c-Met receptor, develop myogenic precursors but these precursors fail to delaminate from the dermomyotome and migrate to eventually populate the limb bud (Dietrich et al., 1999). The Foxp family members of SSTFs regulate gene expression in a multiple of developmental process including lung, heart, and cerebral development. Foxp1/2/4 regulate smooth muscle differentiation and proliferation (Shu et al., 2007). The Dscaml1 protein is a member of the Ig superfamily of cell adhesion molecules. Dscaml1 is highly expressed in the adult brain with lower levels during embryogenesis. Members of the Ig-superfamily are known to mediate cell adhesion and same cell type recognition through homophilic interaction between neighboring cells, similar mechanism for same cell sorting have been demonstrated for NCAM (Agarwala et al., 2001). Dclk1, Met were repressed by Pitx2 whereas Foxp2 and Dscaml1 were activated by Pitx2 in the Lbx1+ migratory muscle precursors. All four genes are involved in regulating cell motility and expression of surface receptors to allow for homophilic cell recognition of the muscle precursors. We have shown that Pitx2 regulates cytoskeletal, adhesion, and signaling genes in migratory muscle precursor cells (Campbell et al., 2012). Proper regulation of the cytoskeleton allows migratory cells to form protrusions in directions of positive signals in order to explore the extracellular environment. This is followed by the formation of nascent adhesions to the extracellular matrix, transmission of positive signals stabilizes the these attachments allowing for anchoring of the cell body and providing traction points to allow the cell to propel itself forward. Once muscle cells reach their intended destination, cell-cell contacts also act to provide intracellular signaling cues that promote myogenic differentiation and fusion (Abmayr et al., 2003).
Table 3.
Highly Regulated Pitx2 Target Genes in Muscle Precursor Cells
| Gene | WT | HT | MT | Fold Change |
|---|---|---|---|---|
| Dclk1 | 150 ± 24 | 160 ± 3 | 217 ± 63 | 1.5 |
| Met | 1438 ± 273 | 1605 ± 79 | 2093 ± 119 | 1.5 |
| Zfhx3 | 365 ± 12 | 531 ± 124 | 515 ± 101 | 1.4 |
| Mid1 | 873 ± 83 | 947 ± 197 | 1132 ± 448 | 1.3 |
| Tle4 | 146 ± 5 | 153 ± 28 | 188 ± 24 | 1.3 |
| Palld | 1033 ± 1 | 1045 ± 65 | 1240 ± 39 | 1.2 |
| Sox6 | 147 ± 16 | 185 ± 47 | 171 ± 87 | 1.2 |
| Ank2 | 238 ± 79 | 236 ± 40 | 253 ± 49 | 1.2 |
| Zfpm2 | 187 ± 13 | 160 ± 22 | 222 ± 53 | 1.2 |
| Prkg1 | 441 ± 36 | 425 ± 28 | 327 ± 13 | −1.3 |
| Meis2 | 873 ± 202 | 433 ± 182 | 689 ± 110 | −1.3 |
| Trps1 | 473 ± 101 | 418 ± 61 | 328 ± 115 | −1.4 |
| Nfib | 758 ± 112 | 523 ± 60 | 553 ± 67 | −1.4 |
| Foxp2 | 225 ± 76 | 133 ± 33 | 147 ± 45 | −1.5 |
| Dscaml1 | 278 ± 10 | 269 ± 8 | 144 ± 9 | −1.8 |
The next group of genes with a fold change 1.4 - 1.3 were the Zinc finger homeobox 3 (Zfhx3), Midline 1 (Mid1), Transducin-like enhancer of split 4 (Tle4), Trichorhinophalangeal syndrome 1 (Trps1), Nuclear factor I/B (Nfib), Meis homeobox 2 (Meis2), and Protein kinase, cGMP-dependent, type 1 (Prkg1). The SSTF Zfhx3 contains both a homeodomain and zinc finger motifs. Zfhx3-A isoform inhibits myogenic differentiation and expression of MRFs, while the B variant promotes myogenic differentiation and expression of MRFs (Berry et al., 2001). Mid1 is an E3 ubiquitin ligase protein, which targets the microtubule associated protein phosphatase 2A (PP2A-C) for degradation. Mutations in the Mid1 result in reduced activity or loss of function and cause accumulation of PP2A-C, disrupts the downstream mTORC1 signaling which controls a number of cellular processes such as growth, autophagy, cell motility, cell cytoskeleton (Liu et al., 2011). The transducin-like enhancer gene (Groucho-related genes or Grgs) encodes for a member of a closely related family of proteins that mediate Notch signaling through repression of the Hairy-related transcription factors (Hes genes) and the Lef/TCF transcription factors that mediate Wnt signaling. The isoform Tle4 is expressed in the posterior of the fore and hind limb buds (Van Hateren et al., 2005). The SSTF Trps1 recognizes GATA consensus to regulate transcription. Trps1 represses prostate-specific antigen (PSA), runt-related transcription factor 2 (Runx2), osteocalcin (Bglap), signal transducer and activator of transcription (Stat3), and parathyroid hormone related protein (Pthrp). Trps1 activates the expression of Wnt inhibitors Wif1, Apcdd1, and Dkk4 during hair follicle development (Fantauzzo and Christiano, 2012). Trps1 null mice have shown that Trps1 acts downstream of BMP7 and functions in the development of bone, kidney, and hair follicles (Gai et al., 2011). The SSTF Nfib is expressed in adipose tissue and brain in adult mice. Nfib induces differentiation through the induction of adipogenic transcription factors PPAR-gamma and C/EBP-alpha (Waki et al., 2011). The SSTF Meis2 is expressed in the trunk of the embryo prior to limb bud induction (E9.0) and in the mesenchyme of the early limb bud (E10.0). As the limb elongates Meis2 becomes restricted to the proximal region of the limb bud (E11.0). This restriction of Meis2 to the proximal region of the limb is important, since Meis2 represses Fgf8 at the apical ectodermal ridge and Shh, Tbx2, Bmp, and Hox genes in the mesenchyme of the limb bud. This restriction sets up zones to allow for proximal to distal patterning of the limb bud (Capdevila et al., 1999). Prkg1 encodes for two isozymes, cGKI-alpha and cGKI-beta, which are expressed in smooth muscle, platelets, and purkinje cells, hippocampal neurons, and lateral amygdale. In the cardiovascular system small signaling molecules nitric oxide (NO) and natriuretic peptides lead to the elevation of cGMP which in turn activates cGKI to interact with IRAG to reduce intracellular Ca2+ concentrations and activate myosin light chain phosphatase (MLC-P), leading to reduced contractility and vasodilatation. Signaling through NO/cGMP can induce cGKI-dependent switching between a proliferative/migratory cell and a differentiated contractile cell. Although the exact mechanisms are unclear, in response to smooth muscle injury vascular smooth muscle cells (VSMCs) de-differentiate by down regulating cGKI expression which in turn causes decreased expression of other smooth-muscle specific genes. Then the VSMCs migrate to the injured area, then re-differentiate by upregulation of cGKI and downstream target genes and switch to a contractile cell state to repair the damage (Hofmann et al., 2009). The genes identified in this group collectively all share a similar function as regulating cell type specification and/or proliferation, primarily through the Wnt pathway. Pitx2 is positioned downstream of both Wnt and growth factor signaling pathways in skeletal myogenesis and promotes muscle progenitor proliferation by direct regulation of the expression of a number of cyclin-dependent kinases (Kioussi et al., 2002).
The third group of genes with fold change of 1.2 was Palladin cytoskeleton associated protein (Palld), SRY-box containing gene 6 (Sox6), Ankyrin 2 (Ank2), and Zinc finger protein, multitype 2 (Zfpm2). Palladin is associated with actin stress fibers, focal adhesions, and Z-discs were it acts as a scaffolding protein to allow for binding of other actin binding proteins and adhesion signaling proteins. Cells lacking Palladin have disrupted actin cytoskeleton and knockout of Palladin in mice shows embryonic lethality at E15.5. The exact mechanism of how Palladin knockout leads to embryonic lethality is unclear, but overexpression analysis of Palladin in cell culture revealed that Palladin is able to induce the expression of smooth muscle differentiation genes (Jin et al., 2010). Sox6 is a member of the Sox SSTF family. Due to the lack of a regulatory domain, the Sox6 protein is completely dependent on cofactors to specify which genes to regulate. In developing skeletal muscle, activation of Sox6 expression leads to the inhibition of cardiac and embryonic muscle myosin isoforms, the inhibition of slow twitch fiber specification genes, the activation of fast twitch fiber specification genes (An et al., 2011). The ankyrin-2 protein is an adaptor protein for stabilizing L-type calcium channel 1.3 (Cav1.3) to the surface of cardiac myocytes. Individuals with loss of function studies mutations of the ankryin-2 protein develop early onset atrial fibrillation (AF). In cell culture the loss of ankyrin-2 protein results in the reduction of Cav1.3 at the surface of atrial myocytes leading to shortened action potential duration (APD) a clinical sign of AF (Cunha et al., 2011). The zinc finger SSTF, multitype 2 encodes a GATA interacting protein called Friend of GATA 2 (Fog-2). The expression of Fog-2 is observed in the developing heart at E13.5–15.5 and in the adult expression is seen in heart, brain, testis, liver and lung. This pattern is very similar to the expression of GATA-4/-5/-6 suggesting they may serve as binding partners for Fog-2 to regulate target genes essential for cardiac development. Knockout mice for Fog-2 die at embryonic stage E12–15.5 due to abnormal coronary vessel and gonadal development (Cantor and Orkin, 2005). The genes represented in this group encode for proteins required for proper expression of cell types specific genes required for organogenesis.
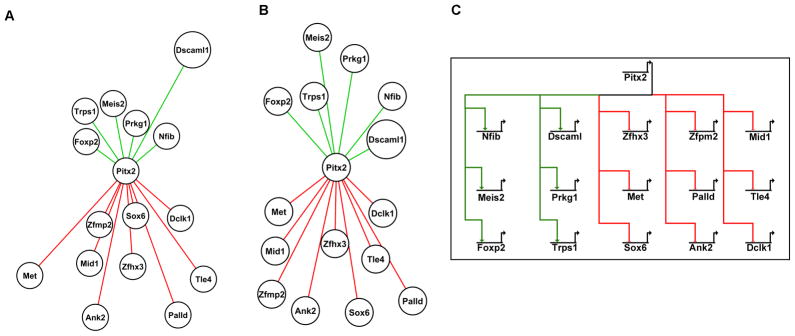
The interaction of Pitx2 with its target gene is illustrated in Fig 2 in two ways. The first network was presented with respect to the distance of the target gene to Pitx2 by how many Pitx2 binding sites were found through analysis of the genomic and −20kb sequences of the gene; the more binding sites present the closer the gene is to Pitx2 (Fig 2A). The second network was presented with respect to the fold change of the target gene expression by microarray analysis; the greater the fold change the closer the gene was placed to Pitx2 (Fig 2B). Finally, a proposed Biotapestry (Longabaugh et al., 2005) diagram of the role of Pitx2 in forelimb development, with red lines representing Pitx2 acting as a repressor and green lines representing Pitx2 acting as an activator on the target gene (Fig 2C). The target genes Meis2, Prkg1, Nfib, Zmp2, Sox6 and Foxp2 were placed closer to the Pitx2 core with respect to the number of binding sites found in the genomic plus −20kb sequences. When these target genes were mapped with respect to fold change we observed that these genes were placed further away from Pitx2 but as a group they had similar levels of significant fold changes. The target genes Dscaml1 and Met were interesting because both genes had relatively low number of Pitx2 sites, but both exhibited the largest fold changes; the inverse relationship of Pitx2 binding sites within the target gene and expression may be due to multiple signaling pathways important in forelimb development regulating these genes. Furthermore, this in silico analysis does not take into account indirect and cofactor binding which is known to play a role in the mechanism of Pitx2 regulation of downstream genes.
Figure 2. Pitx2 Target Genes Visualized with Cytoscape and Biotapestry.
Individual target genes are represented by a white circle with Pitx2 as the core regulator. Lines colored red represent targets of Pitx2 and lines colored green represent targets of activation by Pitx2. The distance of the gene from Pitx2 represents the strength of their respective relationship. (A) Representation of target gene relationship based on the sum of evolutionary conserved binding sites from Table 2. Genes with greater number of binding sites are mapped closer to Pitx2; genes with fewer binding sites are further away. (B) Representation of target gene relationship based on the fold changes observed by microarray analysis. The greater the fold change the closer the gene is mapped to Pitx2. It appears that Pitx2 acts as a repressor for the majority of our target genes. (C) BioTapestry was used to generate a model of the Pitx2 regulatory network in the limb. Red links represent genes repressed by Pitx2, while green links represent genes activated by Pitx2. The mechanism of activation or repression is not shown.
4. Conclusion
This brief study used SAMExcel to analyze microarray data from migratory muscle precursors isolated from Pitx2 MUT, HET and WT forelimb tissue for alterations in gene expression. Using DAVID Functional Annotation Clustering Tool we identified genes grouped according to GOTERMS. This provided a lead to identify Pitx2 dependent cellular/biological processes in the developing forelimb muscles. Our data suggest that Pitx2 regulates genes involved in cytoskeletal organization and gene transcription, which are involved in cell motility, organogenesis, and myogenesis. Pitx2 also regulates SSTFs and members of the Wnt signaling involved in cell lineage specification and organ formation. These studies define Pitx2 as an essential node in the transcriptional network required for skeletal muscle development of the forelimbs.
Future work will begin with validation of the target genes through qPCR. DNA from the same cell populations used for microarray analysis will be collected and analyzed (Hilton et. al. 2010). This can be used to confirm the changes in gene expression due to the loss of Pitx2 in the Lbx1+ muscle progenitor cells. We expect that the genes will show differential expression in the same direction, but not necessarily in the same relative magnitude, much like our other works (Hilton et al 2010, Eng et al 2012). The predicted binding sites will be further validated by ChIP-qpcr. Interaction of Pitx2 with co-activators and/or co-repressors will be also investigated in the Pitx2 specific cis-regulatory elements. Ultimately, with validated binding sites and the inspection of a small subset of genes of interest, a genome wide approach (ChIP seq) will identify the Pitx2 binding sites on the muscle progenitor cell population. This approach will be determined in adult myoblasts and the collectively these data will determine how the network state changes over time.
Acknowledgments
We thank Hsiao-Yen Ma for assisting in animal husbandry, Anne-Marie Girard for microarray processing and Roy Brown for Perl code development. This research was supported by NIH-NIAMS grant AR054406 to CK.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abmayr SM, Balagopalan L, Galletta BJ, Hong SJ. Cell and molecular biology of myoblast fusion. Int Rev Cytol. 2003;225:33–89. doi: 10.1016/s0074-7696(05)25002-7. [DOI] [PubMed] [Google Scholar]
- Agarwala KL, Ganesh S, Tsutsumi Y, Suzuki T, Amano K, Yamakawa K. Cloning and functional characterization of DSCAML1, a novel DSCAM-like cell adhesion molecule that mediates homophilic intercellular adhesion. Biochem Biophys Res Commun. 2001;285:760–772. doi: 10.1006/bbrc.2001.5214. [DOI] [PubMed] [Google Scholar]
- An CI, Dong Y, Hagiwara N. Genome-wide mapping of Sox6 binding sites in skeletal muscle reveals both direct and indirect regulation of muscle terminal differentiation by Sox6. BMC Dev Biol. 2011;11:59. doi: 10.1186/1471-213X-11-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berry FB, Miura Y, Mihara K, Kaspar P, Sakata N, Hashimoto-Tamaoki T, Tamaoki T. Positive and negative regulation of myogenic differentiation of C2C12 cells by isoforms of the multiple homeodomain zinc finger transcription factor ATBF1. J Biol Chem. 2001;276:25057–25065. doi: 10.1074/jbc.M010378200. [DOI] [PubMed] [Google Scholar]
- Bladt F, Riethmacher D, Isenmann S, Aguzzi A, Birchmeier C. Essential role for the c-met receptor in the migration of myogenic precursor cells into the limb bud. Nature. 1995;376:768–771. doi: 10.1038/376768a0. [DOI] [PubMed] [Google Scholar]
- Bober E, Franz T, Arnold HH, Gruss P, Tremblay P. Pax-3 is required for the development of limb muscles: a possible role for the migration of dermomyotomal muscle progenitor cells. Development. 1994;120:603–612. doi: 10.1242/dev.120.3.603. [DOI] [PubMed] [Google Scholar]
- Campbell AL, Shih HP, Xu J, Gross MK, Kioussi C. Regulation of Motility of Myogenic Cells in Filling Limb Muscle Anlagen by Pitx2. PloS one. 2012;7:e35822. doi: 10.1371/journal.pone.0035822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantor AB, Orkin SH. Coregulation of GATA factors by the Friend of GATA (FOG) family of multitype zinc finger proteins. Semin Cell Dev Biol. 2005;16:117–128. doi: 10.1016/j.semcdb.2004.10.006. [DOI] [PubMed] [Google Scholar]
- Capdevila J, Tsukui T, Rodriquez Esteban C, Zappavigna V, Izpisua Belmonte JC. Control of vertebrate limb outgrowth by the proximal factor Meis2 and distal antagonism of BMPs by Gremlin. Mol Cell. 1999;4:839–849. doi: 10.1016/s1097-2765(00)80393-7. [DOI] [PubMed] [Google Scholar]
- Christ B, Brand-Saberi B. Limb muscle development. Int J Dev Biol. 2002;46:905–914. [PubMed] [Google Scholar]
- Cunha SR, Hund TJ, Hashemi S, Voigt N, Li N, Wright P, Koval O, Li J, Gudmundsson H, Gumina RJ, et al. Defects in ankyrin-based membrane protein targeting pathways underlie atrial fibrillation. Circulation. 2011;124:1212–1222. doi: 10.1161/CIRCULATIONAHA.111.023986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dennis G, Jr, Sherman BT, Hosack DA, Yang J, Gao W, Lane HC, Lempicki RA. DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol. 2003;4:P3. [PubMed] [Google Scholar]
- Dietrich S, Abou-Rebyeh F, Brohmann H, Bladt F, Sonnenberg-Riethmacher E, Yamaai T, Lumsden A, Brand-Saberi B, Birchmeier C. The role of SF/HGF and c-Met in the development of skeletal muscle. Development. 1999;126:1621–1629. doi: 10.1242/dev.126.8.1621. [DOI] [PubMed] [Google Scholar]
- Edgar R, Domrachev M, Lash AE. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002;30:207–210. doi: 10.1093/nar/30.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eng D, Campbell A, Hilton T, Leid M, Gross MK, Kioussi C. Prediction of regulatory networks in mouse abdominal wall. Gene. 2010;469:1–8. doi: 10.1016/j.gene.2010.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fantauzzo KA, Christiano AM. Trps1 activates a network of secreted Wnt inhibitors and transcription factors crucial to vibrissa follicle morphogenesis. Development. 2012;139:203–214. doi: 10.1242/dev.069971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuerst PG, Bruce F, Tian M, Wei W, Elstrott J, Feller MB, Erskine L, Singer JH, Burgess RW. DSCAM and DSCAML1 function in self-avoidance in multiple cell types in the developing mouse retina. Neuron. 2009;64:484–497. doi: 10.1016/j.neuron.2009.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gage PJ, Suh H, Camper SA. Dosage requirement of Pitx2 for development of multiple organs. Development. 1999;126:4643–4651. doi: 10.1242/dev.126.20.4643. [DOI] [PubMed] [Google Scholar]
- Gai Z, Gui T, Muragaki Y. The function of TRPS1 in the development and differentiation of bone, kidney, and hair follicles. Histol Histopathol. 2011;26:915–921. doi: 10.14670/HH-26.915. [DOI] [PubMed] [Google Scholar]
- Goulding M, Lumsden A, Paquette AJ. Regulation of Pax-3 expression in the dermomyotome and its role in muscle development. Development. 1994;120:957–971. doi: 10.1242/dev.120.4.957. [DOI] [PubMed] [Google Scholar]
- Gross MK, Moran-Rivard L, Velasquez T, Nakatsu MN, Jagla K, Goulding M. Lbx1 is required for muscle precursor migration along a lateral pathway into the limb. Development. 2000;127:413–424. doi: 10.1242/dev.127.2.413. [DOI] [PubMed] [Google Scholar]
- Hayashi K, Ozawa E. Myogenic cell migration from somites is induced by tissue contact with medial region of the presumptive limb mesoderm in chick embryos. Development. 1995;121:661–669. doi: 10.1242/dev.121.3.661. [DOI] [PubMed] [Google Scholar]
- Hofmann F, Bernhard D, Lukowski R, Weinmeister P. cGMP regulated protein kinases (cGK) Handb Exp Pharmacol. 2009:137–162. doi: 10.1007/978-3-540-68964-5_8. [DOI] [PubMed] [Google Scholar]
- Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- Jin L, Gan Q, Zieba BJ, Goicoechea SM, Owens GK, Otey CA, Somlyo AV. The actin associated protein palladin is important for the early smooth muscle cell differentiation. PloS one. 2010;5:e12823. doi: 10.1371/journal.pone.0012823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin L, Yoshida T, Ho R, Owens GK, Somlyo AV. The actin-associated protein Palladin is required for development of normal contractile properties of smooth muscle cells derived from embryoid bodies. J Biol Chem. 2009;284:2121–2130. doi: 10.1074/jbc.M806095200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent WJ, Sugnet CW, Furey TS, Roskin KM, Pringle TH, Zahler AM, Haussler D. The human genome browser at UCSC. Genome Res. 2002;12:996–1006. doi: 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kioussi C, Briata P, Baek SH, Rose DW, Hamblet NS, Herman T, Ohgi KA, Lin C, Gleiberman A, Wang J, et al. Identification of a Wnt/Dvl/beta-Catenin --> Pitx2 pathway mediating cell-type-specific proliferation during development. Cell. 2002;111:673–685. doi: 10.1016/s0092-8674(02)01084-x. [DOI] [PubMed] [Google Scholar]
- Kioussi C, Gross MK. How to build transcriptional network models of mammalian pattern formation. PloS one. 2008;3:e2179. doi: 10.1371/journal.pone.0002179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitamura K, Miura H, Miyagawa-Tomita S, Yanazawa M, Katoh-Fukui Y, Suzuki R, Ohuchi H, Suehiro A, Motegi Y, Nakahara Y, et al. Mouse Pitx2 deficiency leads to anomalies of the ventral body wall, heart, extra- and periocular mesoderm and right pulmonary isomerism. Development. 1999;126:5749–5758. doi: 10.1242/dev.126.24.5749. [DOI] [PubMed] [Google Scholar]
- Kunath M, Ludecke HJ, Vortkamp A. Expression of Trps1 during mouse embryonic development. Gene Expr Patterns. 2002;2:119–122. doi: 10.1016/s0925-4773(02)00300-3. [DOI] [PubMed] [Google Scholar]
- Lin CR, Kioussi C, O’Connell S, Briata P, Szeto D, Liu F, Izpisua-Belmonte JC, Rosenfeld MG. Pitx2 regulates lung asymmetry, cardiac positioning and pituitary and tooth morphogenesis. Nature. 1999;401:279–282. doi: 10.1038/45803. [DOI] [PubMed] [Google Scholar]
- Lin PT, Gleeson JG, Corbo JC, Flanagan L, Walsh CA. DCAMKL1 encodes a protein kinase with homology to doublecortin that regulates microtubule polymerization. J Neurosci. 2000;20:9152–9161. doi: 10.1523/JNEUROSCI.20-24-09152.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu E, Knutzen CA, Krauss S, Schweiger S, Chiang GG. Control of mTORC1 signaling by the Opitz syndrome protein MID1. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:8680–8685. doi: 10.1073/pnas.1100131108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longabaugh WJ, Davidson EH, Bolouri H. Computational representation of developmental genetic regulatory networks. Dev Biol. 2005;283:1–16. doi: 10.1016/j.ydbio.2005.04.023. [DOI] [PubMed] [Google Scholar]
- Lu MF, Pressman C, Dyer R, Johnson RL, Martin JF. Function of Rieger syndrome gene in left-right asymmetry and craniofacial development. Nature. 1999;401:276–278. doi: 10.1038/45797. [DOI] [PubMed] [Google Scholar]
- Mennerich D, Schafer K, Braun T. Pax-3 is necessary but not sufficient for lbx1 expression in myogenic precursor cells of the limb. Mech Dev. 1998;73:147–158. doi: 10.1016/s0925-4773(98)00046-x. [DOI] [PubMed] [Google Scholar]
- Murai K, Vernon AE, Philpott A, Jones P. Hes6 is required for MyoD induction during gastrulation. Dev Biol. 2007;312:61–76. doi: 10.1016/j.ydbio.2007.09.011. [DOI] [PubMed] [Google Scholar]
- Salsi V, Vigano MA, Cocchiarella F, Mantovani R, Zappavigna V. Hoxd13 binds in vivo and regulates the expression of genes acting in key pathways for early limb and skeletal patterning. Dev Biol. 2008;317:497–507. doi: 10.1016/j.ydbio.2008.02.048. [DOI] [PubMed] [Google Scholar]
- Schafer K, Braun T. Early specification of limb muscle precursor cells by the homeobox gene Lbx1h. Nature genetics. 1999;23:213–216. doi: 10.1038/13843. [DOI] [PubMed] [Google Scholar]
- Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shih HP, Gross MK, Kioussi C. Cranial muscle defects of Pitx2 mutants result from specification defects in the first branchial arch. Proceedings of the National Academy of Sciences of the United States of America. 2007a;104:5907–5912. doi: 10.1073/pnas.0701122104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shih HP, Gross MK, Kioussi C. Expression pattern of the homeodomain transcription factor Pitx2 during muscle development. Gene Expr Patterns. 2007b;7:441–451. doi: 10.1016/j.modgep.2006.11.004. [DOI] [PubMed] [Google Scholar]
- Shih HP, Gross MK, Kioussi C. Muscle development: forming the head and trunk muscles. Acta Histochem. 2008;110:97–108. doi: 10.1016/j.acthis.2007.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shu W, Lu MM, Zhang Y, Tucker PW, Zhou D, Morrisey EE. Foxp2 and Foxp1 cooperatively regulate lung and esophagus development. Development. 2007;134:1991–2000. doi: 10.1242/dev.02846. [DOI] [PubMed] [Google Scholar]
- Sossey-Alaoui K, Srivastava AK. DCAMKL1, a brain-specific transmembrane protein on 13q12.3 that is similar to doublecortin (DCX) Genomics. 1999;56:121–126. doi: 10.1006/geno.1998.5718. [DOI] [PubMed] [Google Scholar]
- Sureban SM, May R, Lightfoot SA, Hoskins AB, Lerner M, Brackett DJ, Postier RG, Ramanujam R, Mohammed A, Rao CV, et al. DCAMKL-1 regulates epithelial-mesenchymal transition in human pancreatic cells through a miR-200a-dependent mechanism. Cancer Res. 2011;71:2328–2338. doi: 10.1158/0008-5472.CAN-10-2738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svensson EC, Huggins GS, Lin H, Clendenin C, Jiang F, Tufts R, Dardik FB, Leiden JM. A syndrome of tricuspid atresia in mice with a targeted mutation of the gene encoding Fog-2. Nat Genet. 2000;25:353–356. doi: 10.1038/77146. [DOI] [PubMed] [Google Scholar]
- Tevosian SG, Deconinck AE, Tanaka M, Schinke M, Litovsky SH, Izumo S, Fujiwara Y, Orkin SH. FOG-2, a cofactor for GATA transcription factors, is essential for heart morphogenesis and development of coronary vessels from epicardium. Cell. 2000;101:729–739. doi: 10.1016/s0092-8674(00)80885-5. [DOI] [PubMed] [Google Scholar]
- Tremblay P, Dietrich S, Mericskay M, Schubert FR, Li Z, Paulin D. A crucial role for Pax3 in the development of the hypaxial musculature and the long-range migration of muscle precursors. Developmental biology. 1998;203:49–61. doi: 10.1006/dbio.1998.9041. [DOI] [PubMed] [Google Scholar]
- Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proceedings of the National Academy of Sciences of the United States of America. 2001;98:5116–5121. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Hateren N, Belsham A, Randall V, Borycki AG. Expression of avian Groucho-related genes (Grgs) during embryonic development. Gene Expr Patterns. 2005;5:817–823. doi: 10.1016/j.modgep.2005.03.011. [DOI] [PubMed] [Google Scholar]
- Waki H, Nakamura M, Yamauchi T, Wakabayashi K, Yu J, Hirose-Yotsuya L, Take K, Sun W, Iwabu M, Okada-Iwabu M, et al. Global mapping of cell type-specific open chromatin by FAIRE-seq reveals the regulatory role of the NFI family in adipocyte differentiation. PLoS Genet. 2011;7:e1002311. doi: 10.1371/journal.pgen.1002311. [DOI] [PMC free article] [PubMed] [Google Scholar]