Abstract
Retroviral overexpression of reprogramming factors (Oct4, Sox2, Klf4, c-Myc) generates induced pluripotent stem cells (iPSCs). However, the integration of foreign DNA could induce genomic dysregulation. Cell-permeant proteins (CPPs) could overcome this limitation. To date this approach has proved exceedingly inefficient. We discovered a striking difference in the pattern of gene expression induced by viral versus CPP-based delivery of the reprogramming factors, suggesting that a signaling pathway required for efficient nuclear reprogramming was activated by the retroviral, but not CPP approach. In gain- and loss-of function studies, we find that the toll-like receptor 3 (TLR3) pathway enables efficient induction of pluripotency by viral or mmRNA approaches. Stimulation of TLR3 causes rapid and global changes in the expression of epigenetic modifiers to enhance chromatin remodeling and nuclear reprogramming. Activation of inflammatory pathways are required for efficient nuclear reprogramming in the induction of pluripotency.
Introduction
Seminal studies by Yamanaka and colleagues revealed that ectopic expression of four transcriptional factors (Oct4, Sox2, Klf4 and c-Myc; OSKM) induces pluripotency in somatic cells (Takahashi et al., 2007; Takahashi and Yamanaka, 2006). However, much remains to be understood about the underlying mechanisms of reprogramming of somatic cells to iPSCs. Furthermore, retroviral overexpression of the reprogramming factors raises concerns that the integration of foreign DNA into the host genome could silence or induce dysregulation of indispensable genes (Maherali et al., 2007; Okita et al., 2007; Takahashi et al., 2007; Takahashi and Yamanaka, 2006; Wernig et al., 2007). A protein-based approach for iPSC generation avoids all concerns for integration of foreign DNA, and theoretically provides for greater control over the concentration, timing, and sequence of transcription factor stimulation (Yang et al., 2009; Zhou et al., 2009). Successful reprogramming to pluripotency has been achieved by using purified recombinant proteins, or cell extracts containing these proteins, but is exceedingly inefficient (Cho et al., 2010; Kim et al., 2009; Zhou et al., 2009) (~0.001%) by comparison to reprogramming with retroviral vectors (0.1%–1%) (Stadtfeld and Hochedlinger, 2010). In seeking to develop effective reprogramming protocols with CPPs, we fortuitously observed an intriguing difference in the pattern of gene expression induced by viral as opposed to CPP-based methods. The accelerated and sustained induction of gene expression that is observed with retroviral methodology is recapitulated by combining CPPs with retroviral particles encoding non-relevant genes, or more practically by combining CPPs with agonists of toll-like receptors (TLRs). We further observed that the induction of TLR3-mediated signaling promotes epigenetic remodeling that is generally required for efficient reprogramming. Recognition of the role of innate immunity in nuclear reprogramming, and its directed manipulation, provides key insights that may facilitate our understanding of both innate immunity and reprogramming and increase our understanding of the genetic and epigenetic pathways that function in induced pluripotency.
Results
Different patterns of gene expression induced by reprogramming factors delivered as viral constructs versus cell-permeant peptides
The CPPs are fusion peptides, each with a reprogramming factor, a linker, and a cell transduction domain (Yang et al., 2009). These CPPs exhibit cognate DNA-binding activity, rapidly translocate across the plasma and nuclear membranes, uniformly transduce nearly all cells in the treated wells, and exert transcriptional control on known downstream target genes. Nevertheless, after multiple attempts with a variety of experimental protocols, we initially failed to generate iPSC lines from human fibroblasts using CPPs generated by our group or by commercial vendors.
In an effort to understand and overcome this hurdle, we examined the temporal sequence of target gene expression in response to a retroviral construct (pMX-Sox2) versus the corresponding CPP (CPP-SOX2). We focused on validated downstream targets such as Jarid2, Zic2, and bMyb for Sox2-activated genes, as well as, downstream genes known to function in nuclear reprogramming, i.e. Nanog, Sox2 and Oct4 (Boyer et al., 2006; Li et al., 2009; Sharov et al., 2008; Tarasov et al., 2008). Human fibroblasts were synchronized by serum starvation and then subjected to either a single infection with pMX-Sox2 or daily treatments of CPP-SOX2, and gene expression was assayed up to 20 days. We used a daily dose of CPP-SOX2 (200nM) that we had shown could rescue human iPSCs treated with shRNA against Sox2 (d.n.s).
An intriguing difference in the pattern of gene expression was observed. As early as Day 1 of transfection with pMX-Sox2, human BJ fibroblasts manifested increased expression of the pluripotency (e.g. Nanog) and target genes (Figures 1A, 1B and Figure S1). By contrast, despite its rapid entry into the cytoplasm and nucleus of treated cells (within 2h; d.n.s.), CPP-SOX2 did not show a similar increase in target gene expression in the first 48h (Figure 1A). Because the temporal pattern of expression of the selected genes (Jarid2, Zic2, bMyb, Oct4, Sox2 and Nanog) was remarkably similar for each treatment condition, their change in fold-expression over time is shown as an average in Figure 1B. The observed difference in target gene expression occurred despite high transfection efficiency with the CPP (over 80%; d.n.s.).
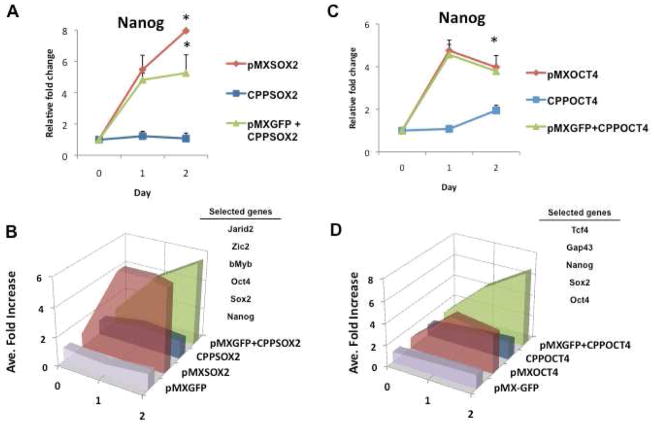
Figure 1. Irrelevant retroviral vector accelerates CPP-induced gene expression.
Changes in expression of pluripotency genes were assessed after exposure of human fibroblasts to the retroviral vectors encoding Sox 2 or Oct 4, or the cell permeant peptides of Sox2 or Oct 4. The addition of an irrelevant retroviral vector encoding GFP (pMX-GFP) enhanced the response to the cell permeant peptides. (A) Relative fold change in gene expression levels of Nanog after exposure to pMX-Sox2 (red line), CPP-SOX2 (blue line) or pMX-GFP + CPP-SOX2 (green line). Relative fold change in gene expression was determined following treatment with 200nM CPP-SOX2 daily or after a single pMX-Sox2 infection on Day 0. All data represented as mean ± s.d., n=3, *P <0.005. (B) Summary figure showing the average fold-change in the selected genes (i.e. Jarid2, Zic2, bMyb, Oct4, Sox2, and Nanog) over time for each condition. When Sox2 is given in the form of a CPP, activation of the downstream target genes is delayed. However, in the presence of an irrelevant retroviral vector, target gene expression increases rapidly, and mimics that of pMX-Sox2. (C) Relative fold change in gene expression levels of Nanog following pMX-Oct4, CPP-OCT4 or pMX-GFP + CPP-OCT4 treatments. (D) Summary figure showing the average fold-change in the selected genes (i.e. Tcf4, Gap43, Nanog, Sox2 and Oct4) over time for each condition. All data represented as mean ± s.d., n=3, *P <0.005.
To exclude the possibility that the delay in target gene expression was a function of the design of a single CPP, we repeated these experiments comparing a CPP versus viral vector for Oct4 (pMX-Oct4), assessing the effect on downstream targets (Tcf4 and GAP43) and pluripotency genes. We observed a similar pattern of delayed gene expression in cells treated with CPP-OCT4 compared to those transfected with pMX-Oct4 (Figures 1C and 1D). Again, because the temporal pattern of expression of the selected genes was remarkably similar for each treatment condition, their change in fold-expression over time is shown as a group average in Figure 1D. Thus, the pattern of delayed expression of downstream target genes appeared to be an attribute of CPP reprogramming factors. To determine whether this difference in gene expression was attributable to the temporal differences in the expression levels of the reprogramming factors between the two delivery methods, we quantified the nuclear expression of Oct4 following CPP or retroviral delivery of Oct4 (d.n.s.). Both of the delivery methods showed equal expression of Oct4 at 24 hr post-treatment.
To further determine the role of CPPs on induction of pluripotency, we checked the gene expression of pluripotency factors at later time points (days 4–20). Retroviral Oct4 induced an exponential increase in pluripotency gene expression at later time points (Figure S2A–C). In contrast, after an initial delay, CPP-OCT4 induced only a modest increase in pluripotency gene expression (“low and slow”).
Viral particles accelerate and augment CPP-induced expression of target genes
We hypothesized that an intrinsic feature of viral particles, independent of the genes encoded, might influence the reprogramming process. To test this hypothesis, we assessed the effect of the CPPs alone or in the presence of a retroviral particle encoding a gene not involved in reprogramming. The pMX-GFP control vector did not affect target gene expression (Figure S1C). However, when the pMX-GFP vector was combined with CPP-SOX2, the expression of the SOX2 target genes was enhanced substantially. In fact, the combination reproduced the pattern of gene expression induced by the retrovirus expression vector pMX-Sox2 (Figures 1A, 1B and Figures S1A–B). We repeated these studies with CPP-OCT4 (Figures 1C, 1D, and Figures S1D–E). When the pMX-GFP vector was combined with CPP-OCT4, again the temporal expression of the downstream genes was accelerated, mimicking the effect observed with the viral vector pMX-Oct4 (Figures 1C–D). These studies suggested that some intrinsic feature of the viral particle contributed to nuclear reprogramming.
Mechanism by which viral particles enable CPP-induced expression of target genes
To gain more insight into the mechanisms by which a retroviral particle could accelerate nuclear reprogramming, we substituted a non-integrating mutant of pMX-GFP by introducing a frameshift mutation near the pol coding region of the retrovirus (Hagino-Yamagishi et al., 1987). Notably, the pMX-GFP non-integrating mutant was fully capable of accelerating the target gene expression induced by CPP-OCT4 (Figure S1F–J). Accordingly, integration of foreign DNA into the host genome is not required for the difference in gene expression between the CPPs and their corresponding viral vectors.
Viral infection activates innate immunity, by virtue of interaction with Toll-like receptors (TLRs) (McWhirter et al., 2004). The TLRs recognize pathogen-associated molecular patterns (PAMPs) associated with viral protein, lipopolysaccharides, DNA or RNA (Akira and Takeda, 2004; Beutler, 2004; Kato et al., 2005). We hypothesized that activation of innate immunity through TLRs may be involved in the difference in gene expression. Indeed, we observed that our retroviral vectors (but not the CPPs) activated inflammatory (innate immune response) genes, including the toll-like receptor 3 (TLR3), NF-κB, IFN-β, Stat1 and Stat2 (d.n.s.).
TLR3 signaling enables efficient induction of pluripotency genes by retroviral Oct4
The TLR-signaling pathway consists of two distinct pathways: a myeloid differentiation primary response gene (MyD) 88-dependent pathway, and a MyD88-independent pathway (Kawai and Akira, 2010). The MyD88-dependent pathway is common to all TLRs, except TLR3 (Adachi et al., 1998; Kaisho and Akira, 2006). To distinguish which TLR signaling pathway might be involved in nuclear reprogramming with viral vectors, we used inhibitory peptides or shRNA knockdown directed against the MyD88-dependent and -independent pathways.
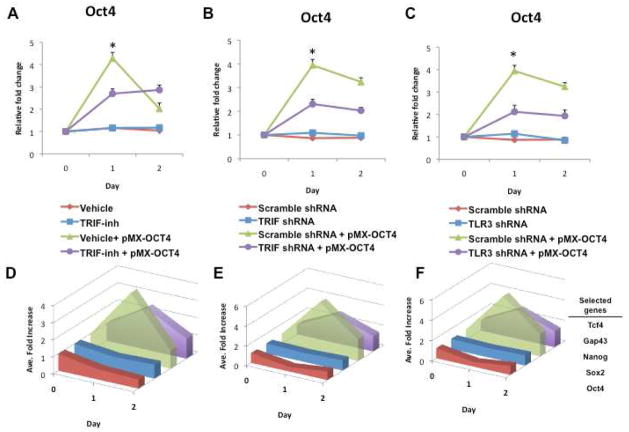
The TLR3 pathway is activated by viral dsRNA, and is independent of MyD88. The adaptor for TLR3 is TRIF (for TIR-domain-containing adapter-inducing interferon-β) (Yamamoto et al., 2003). To explore the role of this pathway in the action of retroviral reprogramming constructs, we knocked down TLR3 or TRIF. In addition, we employed a cell permeable peptide inhibitor of TRIF. As expected, peptide inhibition of the TRIF adaptor molecule or knockdown by shRNA of TRIF or TLR3 reduced the activation by pMX-Oct4 of immune response genes (Figure S2D–F). Notably, these knockdowns of TLR3 signaling also decreased the target and pluripotency gene expression induced by pMX-Oct4. The peptide inhibitor of TRIF (TRIF-Inh) attenuated the effect of pMX-Oct4 to induce Oct4 expression (Figure 2A), as well as expression of the other target genes (Figure 2D). Similarly, shRNA knockdown of TRIF (Figures 2B and 2E), as well as shRNA knockdown of TLR3 (Figures 2C and 2F) also attenuated the effect of pMX-Oct 4 to induce expression of the target genes.
Figure 2. Knockdown of TLR3 pathway inhibits action of retroviral vector encoding Oct4.
(A) Gene expression of Oct4 following retroviral-Oct4 (pMX-Oct4) infection is reduced in fibroblasts treated with the TRIF-inhibitory peptide (TRIF-inh). The lower panel shows a summary diagram of the average fold-changes over time in the selected pluripotency genes (Oct4, Sox2 and Nanog) in the four conditions. (B) Gene expression of Oct4 following retroviral-Oct4 (pMX-Oct4) infection is reduced in TRIF shRNA-knockdown fibroblasts. The lower panel shows a summary diagram of the average fold-changes over time in the selected pluripotency genes (Oct4, Sox2 and Nanog) in the four conditions. (C) Gene expression of Oct4 following retroviral-Oct4 (pMX-Oct4) infection is reduced in TLR3 shRNA-knockdown fibroblasts. The lower panel shows a summary diagram of the average fold-changes over time in the selected pluripotency genes (Oct4, Sox2 and Nanog) in the four conditions. All data represented as mean ± s.d., n=3, *P <0.005.
By contrast, inhibition of MyD88 by an inhibitory peptide (Figure S2G), or by a stable shRNA knockdown (Figure S2H) had no effect on the target and pluripotency gene expression induced by pMX-Oct4. Together, these studies indicate that TLR3, but not the other TLR pathways, are required for full induction of target gene expression by the retrovirus expression vector.
TLR3 signaling enables efficient generation of human iPSCs by retroviral vectors
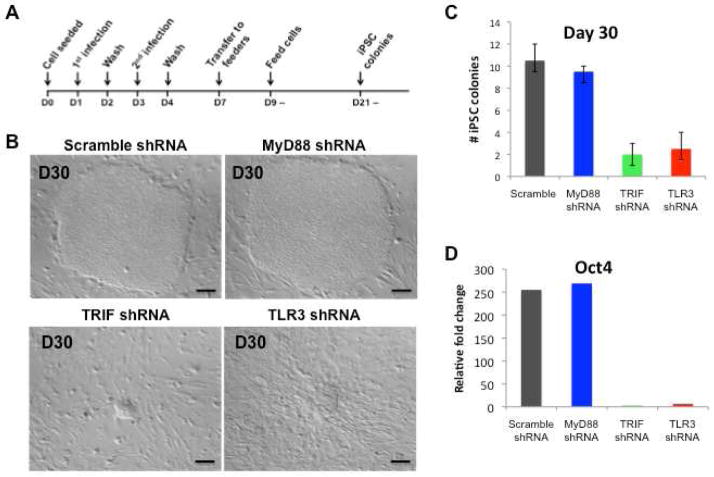
To determine if TLR3 signaling was necessary for efficient generation of human iPSCs, we assessed retroviral reprogramming in BJ fibroblasts previously treated with scrambled shRNA or shRNA to knockdown (KD) the expression of TLR3, TRIF, or MyD88 (Figure 3A). Six days following transduction, the cells were seeded on mitomycin C-treated mouse embryonic fibroblasts (MEFs) and the following day, the medium was replaced with iPSC medium. Around day 25 we observed small colonies in the Scramble- and MyD88-KD cells; by contrast, no colonies were observed in the TRIF- or TLR3-KD cells. By day 30 distinct colonies with typical iPSC colony morphology were noted in dishes containing the scramble- and MyD88-KD cells (Figure 3B). At this time, the TRIF- and TLR3-KD cells manifested only small granulated colonies. It took another 9 days for the TRIF- and TLR3-KD cells to yield morphologically distinct iPSC colonies (d.n.s.). We manually counted each distinct colony with typical morphological features, as well as those smaller granulated colonies, appearing from days 30–39 (in two independent experiments by an observer blinded to the treatment group). As seen in Figure 3C, at early time points the TRIF- and TLR3-KD cells generated fewer colonies by comparison to scramble- and MyD88-KD cells. Furthermore, we compared the gene expression values between these colonies at day 30. The expression of Oct4 (Figure 3D), Sox2 and Nanog (d.n.s) were upregulated more than 10-fold when compared to the TRIF- and TLR3-KD iPSCs. These findings provided the first evidence that TLR3 activation is necessary for efficient induction of pluripotency genes and generation of human iPSC colonies using the approach first described by Yamanaka.
Figure 3. TLR3 or TRIF knockdown fibroblasts exhibit impaired nuclear reprogramming.
(A) Protocol for iPSC generation using the reprogramming factors, delivered as retroviral vectors. (B) Representative images of iPSCs on day 30 after initiation of retroviral nuclear reprogramming for scramble, MyD88, TRIF and TLR3 shRNA knockdown fibroblasts. In fibroblasts where the TLR3 signaling pathway was knocked down (i.e. TRIF shRNA or TLR3 shRNA cell lines), the development of human iPSC colonies was markedly delayed. By contrast, in fibroblasts where the adaptor for all other TLRs was knocked down (MyD88 shRNA), no delay in hiPSC development was noted. (C) Total number of hiPSC colonies on day 30 in scramble, MyD88, TRIF and TLR3 shRNA knockdown fibroblast cell lines transduced by the reprogramming factors delivered by retroviral transfection. The yield of hiPSC colonies was reduced by knocking down the TLR3 signaling pathway. All data represented as mean ± s.d., *P<0.05; scramble compared to TRIF or TLR3 shRNA knockdown fibroblasts. (D) Fold change in Oct4 gene expression in scramble, MyD88, TRIF and TLR3 shRNA knockdown fibroblasts at day 35.
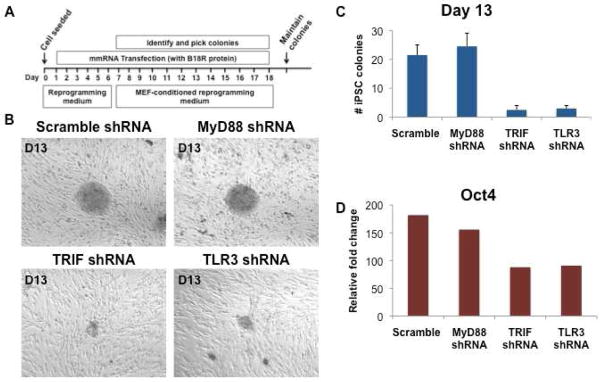
TLR3 signaling enables efficient generation of human iPSCs by mmRNA encoding reprogramming factors
To determine if TLR signaling was required for other methods of reprogramming, we assessed the effects of TLR knockdowns on mmRNA based reprogramming. The scramble or KD human BJ fibroblasts were treated with modified messenger RNA (mmRNA) encoding OSKM (Figure 4A). We observed that when the TLR3 pathway is knocked down (using shRNA against TLR3 or TRIF), expression of the pluripotency genes in single colonies is reduced (Figure 4D). Furthermore, we find that TRIF- and TLR3 KD cells generated fewer colonies by comparison to scramble- and MyD88-KD cells (Figures 4B and 4C). These observations indicate that TLR3 signaling enables efficient induction of pluripotency using mmRNA, as well as retroviral vectors, encoding the reprogramming factors.
Figure 4. Nuclear reprogramming using mmRNA is inhibited by TLR3 or TRIF knockdown.
(A) Protocol for iPSC generation using the reprogramming factors, delivered as mmRNA. (B) Representative images of human iPSCs on day 13 after initiation of mmRNA nuclear reprogramming in the scramble, MyD88, TRIF and TLR3 shRNA knockdown fibroblasts. In fibroblasts where the TLR3 signaling pathway was knocked down (ie. TRIF shRNA or TLR3 shRNA cell lines), the development of human iPSC colonies was markedly delayed. By contrast, in fibroblasts where the adaptor for other TLRs was knocked down (MyD88 shRNA), no delay in hiPSC development was noted. (C) Total number of hiPSC colonies on day 13 in scramble, MyD88, TRIF and TLR3 shRNA knockdown fibroblast cell lines transduced by the mmRNA reprogramming factors. The yield of hiPSC colonies was reduced by knocking down the TLR3 signaling pathway. All data represented as mean ± s.d., *P<0.05; scramble compared to TRIF or TLR3 shRNA knockdown fibroblasts. (D) Fold change in Oct4 gene expression in individual colonies derived from scramble, MyD88, TRIF and TLR3 shRNA knockdown fibroblasts at day 13 of their mmRNA reprogramming.
TLR3 agonist accelerates CPP-induced target gene expression
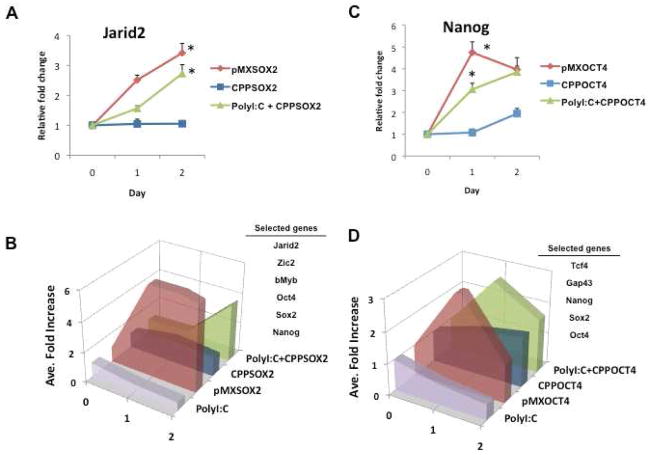
If TLR3 activation plays a role in the efficiency of viral-based reprogramming, then the addition of a TLR3 agonist would be predicted to enhance CPP-induced reprogramming. Polyinosinic-polycytidylic acid (Poly I:C) is a synthetic analog of dsRNA that is recognized specifically by TLR3 and which induces the expression of genes involved in innate immunity (d.n.s.) (Alexopoulou et al., 2001). Accordingly, we assessed the effect of the CPPs alone or in the presence of poly I:C. The expression of target genes was unaffected by poly I:C alone. However, when poly I:C (300ng/ml) was combined with CPP-SOX2, the expression of the downstream genes was accelerated, reproducing the time course of gene expression induced by pMX-Sox2 (Figures 5A–B). We repeated these studies with CPP-OCT4. When poly I:C was combined with CPP-OCT4, again the temporal expression of the downstream genes was accelerated, mimicking the effect observed with the viral vector pMX-Oct4 (Figures 5C–D). Furthermore, as seen in Figure S2A–C, the effect of poly I:C to enhance gene expression of CPP-OCT4 was maintained at later time points. These studies supported the hypothesis that TLR3 signaling is required for efficient induction of the target genes of the reprogramming factors.
Figure 5. Poly I:C accelerates CPP-induced target gene expression.
(A) Relative fold change in gene expression levels of Jarid2 following pMX-Sox2 (red line), CPP-SOX2 (blue line) or poly I:C + CPP-SOX2 (green line) treatments. (B) Summary figure of these selected genes (i.e. Jarid2, Zic2, bMyb, Oct4, Sox2, and Nanog) exhibiting the temporal pattern of average gene expression following each treatment. Poly I:C markedly enhances the expression of downstream genes by CPP-SOX2. (C) Relative fold change in gene expression levels of Nanog following pMX-Oct4, CPP-OCT4 or Poly I:C + CPP-OCT4 treatments. (D) Summary figure of these selected genes (i.e. Tcf4, Gap43, Nanog, Sox2 and Oct4) exhibiting the temporal pattern of gene expression following each treatment. Poly I:C markedly enhances the expression of downstream genes by CPP-Oct4. All data represented as mean ± s.d., n=3, *P <0.005.
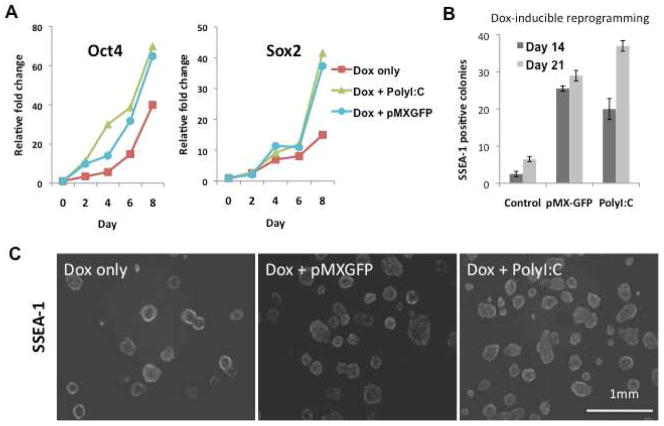
TLR3 activation enhances efficiency of a doxycycline-inducible system for generating iPSCs
To further test the hypothesis that TLR3 activation was required for efficient reprogramming, we isolated MEFs from murine embryos expressing a doxycycline (Dox)-inducible polycistronic transgene construct encoding the four reprogramming factors (Wernig et al., 2008). To generate iPSCs, 105 MEFs/per well in 6-well plates were treated with Dox. In some wells, poly I:C was also added for the initial 6 days of the reprogramming process. In other wells, cells were infected with pMX-GFP on the first day of Dox treatment. Poly I:C or pMX-GFP each accelerated the dox-induced expression of Oct4 and Sox2 (Figure 6A). Poly I:C as well as pMX-GFP accelerated changes in the morphology of the MEFs with small, compact rounded cells aggregating in the wells at 3 days (d.n.s.). Similarly, infection with pMX-GFP accelerated colony formation, as a number of small colonies were observed by day 7 in the viral particle infected group (d.n.s.). By day 14, typical mES-like colonies appeared, many of which expressed SSEA-1. At this time point, the number of typical SSEA-1+ colonies were increased by 7–8 fold in wells exposed to viral particles or poly I:C (Figure 6B). Colony number increased further by day 21–28 (Figures 6B and 6C). These studies strengthened the hypothesis that TLR3 activation enhances nuclear reprogramming to pluripotency.
Figure 6. TLR3 activation enhances reprogramming in a doxycycline-inducible system.
MEFS containing the Dox-inducible polycistronic transgene construct encoding the four reprogramming factors were stimulated by doxycycline, in the absence or presence of TLR3 activation with Poly I:C or the irrelevant retroviral vector pMX-GFP. (A) Gene expression of Oct4 and Sox2 was accelerated by co-administration of poly I:C, or a retroviral construct encoding GFP. (B) Histogram showing SSEA-1+ colonies at 2 and 3 weeks in primary plates. Co-administration of poly I:C, or a retroviral construct encoding GFP, markedly increased the yield of doxycycline-induced reprogramming. (C) SSEA-1 live staining showing iPSC colonies derived from Dox-inducible MEFs 4 wks after exposure to doxycycline. In some wells, MEFs were also exposed to a retroviral construct encoding GFP, or to poly I:C.
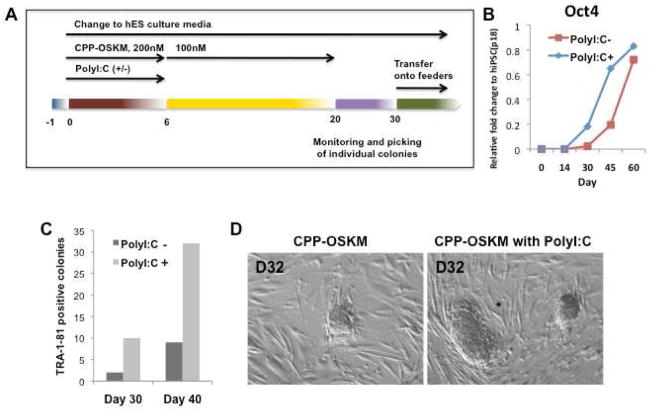
TLR3 activation enhances CPP-induced generation of human iPSCs
It is known that persistent expression (about 2 wks) of the reprogramming factors is required using viral vectors to generate mouse iPSCs (Brambrink et al., 2008; Wernig et al., 2008). We hypothesized that activation of the TLR3 pathway might facilitate epigenetic alterations required for full transcriptional effect of the CPPs. Accordingly, we exposed human fibroblasts to the four CPP-transcription factors (Oct4-R11, Sox2-R11, Klf4-R11 and cMyc-R11) (at a dose of 200nM for days 1–6, and a dose of 100nM for days 7–21), in the presence or absence of poly I:C (300ng/ml) for days 1–6 (Figure 7A). The cultures were transferred to feeder cells (inactivated MEFs) at day 26. In the presence of poly I:C, Oct4 expression was accelerated (Figure 7B). Furthermore, poly I:C accelerated iPSC generation (Figures 7C–D). Small colonies were observed by day 21 and ES-like colonies appeared by day 30, many of which expressed TRA-1-81 as indicated by live cell staining. By contrast, in human fibroblasts not exposed to poly I:C, colonies were not observed until day 30. By day 40, colony number was increased by more than 4-fold in cells exposed to poly I:C (Figure 7C). Human iPSCs induced by CPP alone (piPSC) or in combination with poly I:C (ppiPSCs) manifested characteristic ESC morphology, immunoreactivity for SSEA3 and OCT4, and transcriptional expression (i.e. POU5f1, SOX2, UTF1, ZFP42 and NANOG), and differentiated into all three embryonic germ layers in vitro and in teratomas (Figure S3). Thus, CPP-induced reprogramming to pluripotency of human fibroblasts was enhanced and accelerated by stimulation of TLR signaling.
Figure 7. TLR3 activation stimulates CPP-induced reprogramming of human fibroblasts.
(A) Protocol for human iPSC generation using four CPP-TFs (OCT4-R11, SOX2-R11, KLF4-R11 and cMYC-R11). (B) Gene expression of Oct4 was increased by co-administration of poly I:C, by day 30–45. (C) TRA-1-81 positive colonies counted at day 30 and day 40 in the presence and absence of poly I:C. Co-administration of poly I:C markedly increased the yield of CPP-induced reprogramming. (D) ES-like colony formation at day 32 of CPP-induced transactivation (10×).
TLR3 activation enables epigenetic changes that favor reprogramming
We hypothesized that TLR3 activation might enhance early transcriptional activation by inducing an open chromatin state, permitting the reprogramming factors to induce an ESC-specific gene expression pattern (Niwa, 2007a, b). Accordingly, we performed chromatin immunoprecipitation followed by PCR analysis (ChIP-PCR) to detect trimethylation of histone H3 at lysine 4 (H3K4me3). This epigenetic modification marks transcriptionally active genes. Human fibroblasts were treated with pMX-Sox2, or with CPP-Sox2 in the presence of poly I:C or pMX-GFP. By day 2 of treatment, pMX-Sox2 induced H3K4 trimethylation at the Oct4 promoter (Figure S4A). By contrast, CPP-Sox2 had no effect at day 2, except in the presence of viral vector or poly I:C (Figure S4A). [At later time points, CPP-Sox2 alone could induce H3K4 trimethylation, a time lag that reflected its delayed effects on target gene expression (d.n.s)]. Similarly, poly I:C, or the retroviral vector encoding GFP, accelerated H3K4 trimethylation at the Sox2 promoter in CPP-treated fibroblasts (Figure S5A).
In a similar fashion, we assessed histone H3 at lysine 9 (H3K9me3) in the Oct4 and Sox2 promoters. This epigenetic modification marks transcriptionally silenced genes. By day 2 of treatment, pMXSox2 but not CPPSox2 alone, fully reversed H3K9 trimethylation at the Oct4 promoter (Figure S4B). Poly I:C or pMX-GFP enhanced the reversal of H3K9 trimethylation induced by CPP-Sox2 on day 2 (Figure S4B). [At later time points, CPP-Sox2 alone could fully reverse H3K9 trimethylation at the Oct4 promoter, after a time lag that reflected its delayed effects on target gene expression (d.n.s)]. Similarly, poly I:C, or the retroviral vector encoding GFP, accelerated the loss of H3K9 trimethylation at the Sox2 promoter in CPP-treated fibroblasts (Figure S5B). These studies provided an epigenetic mechanism to explain the effect of TLR3 activation to enhance nuclear reprogramming.
TLR3 activation regulates epigenetic machinery
Histone acetylation status influences the accessibility of DNA to the transcriptional machinery for gene expression. Histone de-acetylation is generally associated with a closed chromatin state, and inhibitors of histone de-acetylase (HDAC) such as valproic acid are employed to enhance nuclear reprogramming. Therefore it is notable that poly I:C downregulated the expression of a suite of HDAC genes in CPP treated human fibroblasts (Supplementary Table S1). The downregulation of HDAC1 expression by poly I:C or by pMX-GFP was confirmed by Western analysis (Figure S4C and S4D). Similar downregulation of the HDAC family by poly I:C was noted in the dox-inducible MEFs described above and in Figure 5. To determine if the effect of poly I:C on epigenetic modifiers was dependent upon TLR3 signaling, we repeated these studies in the TLR knockdown lines. As anticipated, poly I:C reduced HDAC1 expression in the scrambled or Myd88 KD lines, but failed to downregulate HDAC1 expression in the TRIF- and TLR3-KD lines (Figure S5C). These studies indicated that activation of TLR3 signaling causes changes in epigenetic modifiers that favor reprogramming.
Poly I:C affected the expression of other epigenetic modifiers (Table S1). In addition, the changes in methylation status of the Oct4 and Sox2 promoters that were accelerated by TLR3 activation were also associated with an accelerated redistribution of heterochromatin protein 1 (HP1; d.n.s.). HP1 bound to methylated H3K9 recruits the methylase Suv39h, leading to further methylation of H3K9, so as to consolidate a repressed state (Bannister et al., 2001; Lachner et al., 2001). The redistribution of HP1 induced by poly I:C is consistent with genome-wide epigenetic alterations promoted by TLR3 activation, and directed by the reprogramming factors.
Role of NF-κB in TLR-mediated regulation of epigenetic machinery
Histone acetylation favors an open chromatin state, maintained by proteins containing histone acetyltransferase (HAT) domains, such as p300 and CBP. NF-κB is a transcriptional effector of TLR3 activation (Hayden and Ghosh, 2004; Hayden et al., 2006), and interacts with CBP/p300 to positively regulate gene expression (Farlik et al., 2010; Li and Verma, 2002). We used a luciferase reporter assay system to document that poly I:C, but not the CPPs alone, induced NF-κB activation (Figure S6A). This effect was mediated by TLR3, as shRNA knockdown of TLR3 or its adaptor protein TRIF reduced the effect of poly I:C on NF-κB activation (Figure S6A). Similarly, the retroviral constructs, as well as poly I:C, induced a sustained increase in the expression of NF-κB and TLR3, whereas CPP-SOX2 did not (Figure S6A). These studies suggest that the effect of TLR3 activation to induce changes in gene expression of the epigenetic machinery might be mediated in part by NF-κB.
To further elucidate the elements of TLR3 signaling involved in reprogramming, we inhibited the action of downstream effectors IKKε, NF-κB and IRF3. As before, we found that poly I:C enhanced reprogramming of doxycycline-inducible MEFs. This effect of poly I:C to induce NF-κB translocation to the nucleus (Figure S6B); and to increase colony formation (Figure S6D); was markedly reduced by the addition of p65 decoy. In additional experiments, shRNA knockdown of IRF3 or IKK (Figure S6C) also impaired iPSC generation (Figure S6D). These studies indicated that TLR3-induced activation of both NF-κB and IRF3 were required for efficient reprogramming.
To further document a role for these transcriptional factors in TLR3-facilitated chromatin modification, we performed ChIP assay for H3K4me3 and H3K27me3. We observed that the inhibition of NF-κB, IRF3 or IKKε fully eliminated H3K4 trimethylation at the Oct4 promoter after doxycycline stimulation in the presence of poly I:C (Figure S7A). Furthermore, H3K27 trimethylation of the Oct4 promoter was increased upon inhibition of the innate immune pathway (Figure S7B).
In the reprogramming dox-inducible MEFs, the addition of poly I:C induced changes in the expression in a set of enzymes involved in chromatin modification (Figure S7C). We observed that poly I:C upregulated histone acetyltransferase (i.e. HAT1 and Esco2), histone methyltransferase (SMYD) as well as downregulated the histone deacetylase family (i.e. HDAC 5, 8, 9 and 10), H3K4 histone methyltransferase (Ash1l) and H3K79 histone methyltransferase (Dot1L). Inhibition of the latter methyltransferase is critical in somatic cell reprogramming (Onder et al., 2012). Notably, the effects of poly I:C on these epigenetic modifiers was reversed by decoy oligonucleotide inhibition of NF-κB (Figure S7D).
Discussion
This report is the first to posit a direct link between nuclear reprogramming efficiency and inflammatory pathways in the induction of pluripotency. In seeking to induce pluripotency while employing cell permeant peptides (CPPs), we serendipitously discovered a role for innate immunity signaling in effective nuclear reprogramming. Our salient observations are: 1.) A consistent difference in the temporal characteristics of gene expression is observed between cells exposed to the reprogramming factors in the form of retroviral vectors versus CPPs; 2.) This difference is reversed by combining CPPs with TLR 3 agonists; 3.) TLR3 knockdown inhibits the activation of downstream pluripotency genes when using retroviral vectors or mmRNA to overexpress the reprogramming factors, and reduces the efficiency and yield of human iPSC generation; 4.) TLR3 activation enhances the efficiency and yield of miPSC generation in a dox-inducible system; and enhances the efficiency and yield of human iPSC generation when using the reprogramming factors in the form of CPPs, and 5.) TLR3 activation enables epigenetic alterations, including changes in methylation status of the Oct4 and Sox2 promoters, as well as changes in the expression of epigenetic effectors, that promote an open chromatin configuration. The knowledge that the activation of innate immune response affects nuclear reprogramming permitted us to enhance the efficiency and yield of human iPSCs using reprogramming factors in the form of CPPs.
An unappreciated role for TLR3 activity in reprogramming
TLR3 recognizes double-stranded RNA (dsRNA) generated by retroviruses (Agger et al., 2007; De Santa et al., 2007). The importance of TLR signaling for effective nuclear reprogramming has not been appreciated. We show that the efficiency and yield of human iPSC generation, using retroviral vectors, is reduced by knockdown of the pathway with peptide inhibitors or shRNA knockdown of TLR3 or its adaptor protein TRIF. This effect of TLR3 knockdown is also observed when using mmRNA to express the reprogramming factors.
We confirmed the importance of TLR signaling in a virus-free system using murine embryonic fibroblasts that were genetically engineered to express a doxycycline-inducible cassette encoding the reprogramming factors. In this system, the co-administration of the TLR3 agonist poly I:C increased the efficiency and yield of murine iPSCs (Figure 6). Notably, the same effect was observed with co-administration of the retrovirus encoding GFP, which retrovirus would be expected to activate TLR3 without otherwise affecting the nuclear reprogramming process. Our work indicates that the retroviral vectors used for inducing pluripotency are more than vehicles for delivering the reprogramming factors, and actively contribute to the reprogramming process.
It is worth noting that there seems to be an optimal level of TLR3 activation. Because retroviral vectors activate the TLR3 pathway, further activation does not seem to be necessary during reprogramming with retroviral vectors, and may even be counterproductive. Higher levels of activation are associated with less colony formation, probably due to cell death (d.n.s.)
TLR3 and epigenetic modification
The effect of TLR3 activation to enhance the yield and efficiency of human iPSC generation is due in part to its regulation of the expression and/or distribution of epigenetic modifiers (Table S1; Figures 7, S4C, S4D and S5C). Associated with the changes in expression of epigenetic modifiers, we observed histone modifications consistent with an open chromatin configuration on the promoter regions of Oct4 and Sox2 (Figures S4 and S5). Notably however, the increase in H3K4 trimethylation and the decrease in H3K9 trimethylation of these promoters were not observed with TLR3 activation alone. Only in the presence of the CPPs did poly I:C induce changes in these methylation marks. This observation supports the concept that, although TLR3 activation causes widespread changes in the expression of epigenetic modifiers that might promote the open chromatin configuration of pluripotency genes, the reprogramming proteins are likely necessary to direct the epigenetic modifiers to the appropriate promoter sequences. This notion is also supported by the confocal images of HP1α distribution (d.n.s.). Heterochromatin protein1 (HP1) is associated with the closed conformation of chromatin. Although we did not see changes in the expression levels of HP1 expression (Figures S4C–D), we observed marked changes in its distribution when CPP-SOX2 was co-administered with poly I:C or with the retroviral vector encoding GFP. However, in the absence of the CPP, there was no observable redistribution of HP1 induced by poly I:C or the retroviral vector alone.
Any of five classes of pathogen recognition receptors (PRRs) have been shown to signal in ways that are comparable to TLR3 (Roy and Mocarski, 2007), and might be expected to accelerate nuclear reprogramming. The fact that the current work implicated TRIF (Choe et al., 2005) signaling aligns with the fact that the retrovirus RNA, or the mmRNA, triggers TLR3 signaling to engage NF-κB, IRF3 and IFNβ. Other TLR or PRRs drive a similar inflammatory response converging on NF-κB, IRF-3 and IFNβ (Akira and Takeda, 2004).
Type 1 interferons (e.g. IFNβ) are elaborated during exposure to mmRNA or viral vectors, and we have not ruled out their role in reprogramming efficiency. In addition, silencing of endogenous retrotransposons is characteristic of the fully reprogrammed pluripotent state (Brambrink et al., 2008; Stadtfeld et al., 2008; Wernig et al., 2008). Notably, IRF3 induces expression of the tripartite motif (TRIM) protein family, which is implicated in silencing of endogenous retroelements in ES cells (Wolf and Goff, 2007). We observed that poly I:C induces the expression of the TRIM family (Table S1), and this effect of TLR3 activation may play a role in the reprogramming process by silencing endogenous retrotransposons.
In any event, our data suggest that activation of inflammatory pathways enhance nuclear reprogramming. Innate immunity signaling appears to favor an open chromatin state, which increases cell plasticity in response to a pathogen. We speculate that this cell state may enhance induction of pluripotentiality, transdifferentiation, or even malignant transformation, and is characterized by global changes in epigenetic modifiers in a process we term ‘transflammation’.
Conclusion
Our observations highlight a previously unrecognized role for innate immunity activation in nuclear reprogramming. The viral vectors or mmRNA constructs used to induce pluripotency are more than mere vehicles for the reprogramming factors. Innate immune activation causes striking changes in epigenetic modifiers that favor an open chromatin configuration. These changes enable a fluidity of cell phenotype that contributes to successful nuclear reprogramming.
Experimental Procedures
Cells
BJ human fibroblast cells (Stemgent) were cultured in DMEM with 10% FBS and 1% penicillin/streptomycin (5% CO2, 37°C). For isolation of secondary dox-inducible M EFs, chimeric embryos were obtained from transgenic R26rtTA;Col1a12lox-4F2A mice expressing the loxP-flanked, dox-inducible polycistronic 4F2A cassette (Oct4, Sox2, Klf4, c-Myc; Jackson Laboratory). Secondary MEFS were isolated as previously described (Wernig et al., 2008), and expanded for two passages before freezing.
Viral preparation and infection
HEK293FT cells were plated at 6 × 106 cells per T225 flask and incubated o.n. Cells were transfected with 10 μg of VSV-G (envelope protein), 15 μg of pUMVC (packaging plasmid) and 10 μg of gene of interest (Sox2 or Oct4) with Lipofectamine. 48 hours after transfection, the supernatant of transfectant was collected and filtered (0.45 μm filter). Following spinning at 17,100 rpm for 2.3hrs, the viral pellet was resuspended to make 100× stock solutions. Human fibroblasts (5 × 104 cells/well) were seeded a day before transduction. The medium was replaced with virus-containing supernatant supplemented with 8 μg/ml polybrene, and incubated for 24 hr.
Treatments
At 60–70% confluency, BJ fibroblast cells were serum-starved (1% serum) to induce G1 cell cycle arrest, and then subjected to either a single infection with retroviral constructs or daily treatments with 200 nM CPPs (CPP-SOX2 or CPP-OCT4). Poly I:C (300 ng/ml) was added to the cells simultaneously with the CPPs. For experiments involving peptide inhibitors, cells were pretreated for 6 hrs at 40uM with either MyD88 inhibitory peptide (Pepinh-MyD) or TRIF inhibitory peptide (Pepinh-TRIF) followed by CPP treatments.
Gene Expression and Microarray Analyses
RNA was isolated (RNeasy), first-strand cDNA primed with oligo(dT) primers, and qPCR performed (primer sets from Applied Biosystems). RNA probes were prepared and hybridized to Illumina HumanHT-12 v4 Expression BeadChip microarrays.
Short Hairpin RNA Design
Short hairpin RNA was obtained from Invivogen. Target sequences: MyD88 shRNA, AACTGGAACAGACAAACTATC; TRIF shRNA, AAGACCAGACGCCACTCCAAC and TLR3 shRNA, GCTTGGCTTCCACAACTAGAA
Chromatin Immunoprecipitation and ChIP-qPCR
qChIP was performed (Lim et al., 2009; Peng et al., 2009). For qChIP and qRT-PCR, error estimates are standard deviations. Recovery of genomic DNA as the percentage input was calculated as the ratio of copy numbers in the immunoprecipitate to the input control. Primers of Oct4 and Sox2 promoters were purchased from Cell Signaling.
Generation of iPSCs
Retroviral-iPSCs
Human fibroblasts previously treated with MyD88, TRIF, TLR3 or Scramble shRNA were transduced with pMX-Oct4, Sox2, Klf4, and cMyc retroviruses (Takahashi et al., 2007; Takahashi and Yamanaka, 2006), and were cultured in iPSC medium on mitomycin-treated MEFs. Colonies were counted over time, and were harvested for RNA isolation qPCR analysis for pluripotency gene expression.
Protein-iPSCs
Recombinant Oct4, Sox2, Klf4, and cMyc human proteins (CPPs) contained an eleven-arginine membrane penetration domain at the C terminus (Stemgent). Human fibroblasts were treated with CPP-Oct4, CPP-Sox2, CPP-Klf4 and CPP-Myc daily for 6 days (200 nM) CPPs, then daily from days 7–20 (100nM). Poly I:C (300ng/ml) or vehicle was added to the cell simultaneously up to day 6. The cells were passed onto MEF feeders at day 30. After 20 days of CPP treatments, wells were scanned for colonies.
Doxycycline-induced iPSCs
4 × 104/well secondary MEFs (passage #4) were plated and treated with doxycycline (2μg/mL) ± poly I:C (300ng/ml). The generation of iPSC colonies was scored at days 14 and 21.
Induction of pluripotency using mmRNA
Knockdown human fibroblasts were transduced with mmRNA encoding OSKM and were cultured in iPSC medium on mitomycin-treated MEFs (Yakubov et al., 2010). Colonies were counted by day 13, and single colonies were harvested for RNA isolation and qPCR analysis for pluripotency gene expression.
NF-κB Luciferase assay
BJ fibroblasts (3 × 105/well) were subjected to either pMX-GFP infection, CPP-SOX2 treatment with or without poly IC (300 ng/ml). Cells were transfected with pNF-κB-Luc and pFC-MEKK as a positive control plasmid using Lipofectamine 2000, and after 24h, cells were studied using the Bright-Glo™ Luciferase Assay System and a luminometer.
Immunostaining of Live Cells
For the detection of SSEA-1 or TRA-1-81 in live cells, the primary Ab (anti-mouse SSEA-1, anti-human TRA-1-81, Stemgent) was diluted to a final concentration of 2.5 to 5 μg/ml in cell culture medium and incubated with cells for 30 min (37°C, 5 % CO2). After gentle washing the cells were examined under a fluorescent microscope.
Western Blotting
Proteins were extracted from BJ fibroblasts by solubilizing the cells in RIPA buffer containing 1× protease inhibitor cocktail. 25 μg of total protein was loaded and resolved on SDS-polyacrylamide gels, transferred to PVDF membranes and probed with Abs against HP1α, HDAC (Cell Signaling), and β-actin (Sigma, A5441). Immunoblots were developed with enhanced chemiluminescence reagents (Amersham).
Supplementary Material
Acknowledgments
We thank Drs. Joanna Wysocka, Karla Kirkegaard, and Vittorio Sebastiano for their guidance, Mr. Liqun Zhu for technical assistance, Dr. Yaso Natkunam for histological studies. This work was supported by grants to J.P.C from National Institutes of Health (U01HL100397, RC2HL103400). J.L was supported by grant from the Tobacco Related Disease Research Program of the University of California. N.S was supported by NRSA grant (HL098049-01A1).
Footnotes
Disclosures: JPC, NS and JL are inventors of the intellectual property, assigned to Stanford University, which was generated by this research.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Adachi O, Kawai T, Takeda K, Matsumoto M, Tsutsui H, Sakagami M, Nakanishi K, Akira S. Targeted disruption of the MyD88 gene results in loss of IL-1- and IL-18-mediated function. Immunity. 1998;9:143–150. doi: 10.1016/s1074-7613(00)80596-8. [DOI] [PubMed] [Google Scholar]
- Agger K, Cloos PA, Christensen J, Pasini D, Rose S, Rappsilber J, Issaeva I, Canaani E, Salcini AE, Helin K. UTX and JMJD3 are histone H3K27 demethylases involved in HOX gene regulation and development. Nature. 2007;449:731–734. doi: 10.1038/nature06145. [DOI] [PubMed] [Google Scholar]
- Akira S, Takeda K. Toll-like receptor signalling. Nat Rev Immunol. 2004;4:499–511. doi: 10.1038/nri1391. [DOI] [PubMed] [Google Scholar]
- Alexopoulou L, Holt AC, Medzhitov R, Flavell RA. Recognition of double-stranded RNA and activation of NF-kappaB by Toll-like receptor 3. Nature. 2001;413:732–738. doi: 10.1038/35099560. [DOI] [PubMed] [Google Scholar]
- Bannister AJ, Zegerman P, Partridge JF, Miska EA, Thomas JO, Allshire RC, Kouzarides T. Selective recognition of methylated lysine 9 on histone H3 by the HP1 chromo domain. Nature. 2001;410:120–124. doi: 10.1038/35065138. [DOI] [PubMed] [Google Scholar]
- Beutler B. Inferences, questions and possibilities in Toll-like receptor signalling. Nature. 2004;430:257–263. doi: 10.1038/nature02761. [DOI] [PubMed] [Google Scholar]
- Boyer LA, Mathur D, Jaenisch R. Molecular control of pluripotency. Current opinion in genetics & development. 2006;16:455–462. doi: 10.1016/j.gde.2006.08.009. [DOI] [PubMed] [Google Scholar]
- Brambrink T, Foreman R, Welstead GG, Lengner CJ, Wernig M, Suh H, Jaenisch R. Sequential expression of pluripotency markers during direct reprogramming of mouse somatic cells. Cell Stem Cell. 2008;2:151–159. doi: 10.1016/j.stem.2008.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho HJ, Lee CS, Kwon YW, Paek JS, Lee SH, Hur J, Lee EJ, Roh TY, Chu IS, Leem SH, et al. Induction of pluripotent stem cells from adult somatic cells by protein-based reprogramming without genetic manipulation. Blood. 2010;116:386–395. doi: 10.1182/blood-2010-02-269589. [DOI] [PubMed] [Google Scholar]
- Choe J, Kelker MS, Wilson IA. Crystal structure of human toll-like receptor 3 (TLR3) ectodomain. Science (New York, NY. 2005;309:581–585. doi: 10.1126/science.1115253. [DOI] [PubMed] [Google Scholar]
- De Santa F, Totaro MG, Prosperini E, Notarbartolo S, Testa G, Natoli G. The histone H3 lysine–27 demethylase Jmjd3 links inflammation to inhibition of polycomb-mediated gene silencing. Cell. 2007;130:1083–1094. doi: 10.1016/j.cell.2007.08.019. [DOI] [PubMed] [Google Scholar]
- Farlik M, Reutterer B, Schindler C, Greten F, Vogl C, Muller M, Decker T. Nonconventional initiation complex assembly by STAT and NF-kappaB transcription factors regulates nitric oxide synthase expression. Immunity. 2010;33:25–34. doi: 10.1016/j.immuni.2010.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagino-Yamagishi K, Donehower LA, Varmus HE. Retroviral DNA integrated during infection by an integration-deficient mutant of murine leukemia virus is oligomeric. J Virol. 1987;61:1964–1971. doi: 10.1128/jvi.61.6.1964-1971.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayden MS, Ghosh S. Signaling to NF-kappaB. Genes & development. 2004;18:2195–2224. doi: 10.1101/gad.1228704. [DOI] [PubMed] [Google Scholar]
- Hayden MS, West AP, Ghosh S. NF-kappaB and the immune response. Oncogene. 2006;25:6758–6780. doi: 10.1038/sj.onc.1209943. [DOI] [PubMed] [Google Scholar]
- Kaisho T, Akira S. Toll-like receptor function and signaling. J Allergy Clin Immunol. 2006;117:979–987. doi: 10.1016/j.jaci.2006.02.023. quiz 988. [DOI] [PubMed] [Google Scholar]
- Kato H, Sato S, Yoneyama M, Yamamoto M, Uematsu S, Matsui K, Tsujimura T, Takeda K, Fujita T, Takeuchi O, et al. Cell type-specific involvement of RIG-I in antiviral response. Immunity. 2005;23:19–28. doi: 10.1016/j.immuni.2005.04.010. [DOI] [PubMed] [Google Scholar]
- Kawai T, Akira S. The role of pattern-recognition receptors in innate immunity: update on Toll-like receptors. Nat Immunol. 2010;11:373–384. doi: 10.1038/ni.1863. [DOI] [PubMed] [Google Scholar]
- Kim D, Kim CH, Moon JI, Chung YG, Chang MY, Han BS, Ko S, Yang E, Cha KY, Lanza R, et al. Generation of Human Induced Pluripotent Stem Cells by Direct Delivery of Reprogramming Proteins. Cell Stem Cells. 2009;4:472–476. doi: 10.1016/j.stem.2009.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lachner M, O’Carroll D, Rea S, Mechtler K, Jenuwein T. Methylation of histone H3 lysine 9 creates a binding site for HP1 proteins. Nature. 2001;410:116–120. doi: 10.1038/35065132. [DOI] [PubMed] [Google Scholar]
- Li G, Margueron R, Ku M, Chambon P, Bernstein BE, Reinberg D. Jarid2 and PRC2, partners in regulating gene expression. Genes & development. 2009;24:368–380. doi: 10.1101/gad.1886410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q, Verma IM. NF-kappaB regulation in the immune system. Nat Rev Immunol. 2002;2:725–734. doi: 10.1038/nri910. [DOI] [PubMed] [Google Scholar]
- Lim DA, Huang YC, Swigut T, Mirick AL, Garcia-Verdugo JM, Wysocka J, Ernst P, Alvarez-Buylla A. Chromatin remodelling factor Mll1 is essential for neurogenesis from postnatal neural stem cells. Nature. 2009;458:529–533. doi: 10.1038/nature07726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maherali N, Sridharan R, Xei W, Utikal J, Eminli S, Arnold K, Stadtfeld M, Yachechko R, Tchieu J, Jaenisch R, et al. Directly reprogrammed fibroblasts show global epigenetic reprogramming and widespread tissue contribution. Cell Stem Cell. 2007;1:55–70. doi: 10.1016/j.stem.2007.05.014. [DOI] [PubMed] [Google Scholar]
- McWhirter SM, Fitzgerald KA, Rosains J, Rowe DC, Golenbock DT, Maniatis T. IFN-regulatory factor 3-dependent gene expression is defective in Tbk1-deficient mouse embryonic fibroblasts. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:233–238. doi: 10.1073/pnas.2237236100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niwa H. How is pluripotency determined and maintained? Development (Cambridge, England) 2007a;134:635–646. doi: 10.1242/dev.02787. [DOI] [PubMed] [Google Scholar]
- Niwa H. Open conformation chromatin and pluripotency. Genes & development. 2007b;21:2671–2676. doi: 10.1101/gad.1615707. [DOI] [PubMed] [Google Scholar]
- Okita K, Ichisaka T, Yamanaka S. Generation of germline-competent induced pluripotent stem cells. Nature. 2007;448:313–317. doi: 10.1038/nature05934. [DOI] [PubMed] [Google Scholar]
- Onder TT, Kara N, Cherry A, Sinha AU, Zhu N, Bernt KM, Cahan P, Marcarci BO, Unternaehrer J, Gupta PB, et al. Chromatin-modifying enzymes as modulators of reprogramming. Nature. 2012;483:598–602. doi: 10.1038/nature10953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng JC, Valouev A, Swigut T, Zhang J, Zhao Y, Sidow A, Wysocka J. Jarid2/Jumonji coordinates control of PRC2 enzymatic activity and target gene occupancy in pluripotent cells. Cell. 2009;139:1290–1302. doi: 10.1016/j.cell.2009.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy CR, Mocarski ES. Pathogen subversion of cell-intrinsic innate immunity. Nat Immunol. 2007;8:1179–1187. doi: 10.1038/ni1528. [DOI] [PubMed] [Google Scholar]
- Sharov AA, Masui S, Sharova LV, Piao Y, Aiba K, Matoba R, Xin L, Niwa H, Ko MS. Identification of Pou5f1, Sox2, and Nanog downstream target genes with statistical confidence by applying a novel algorithm to time course microarray and genome-wide chromatin immunoprecipitation data. BMC Genomics. 2008;9:269. doi: 10.1186/1471-2164-9-269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stadtfeld M, Hochedlinger K. Induced pluripotency: history, mechanisms, and applications. Genes & development. 2010;24:2239–2263. doi: 10.1101/gad.1963910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stadtfeld M, Maherali N, Breault DT, Hochedlinger K. Defining molecular cornerstones during fibroblast to iPS cell reprogramming in mouse. Cell Stem Cell. 2008;2:230–240. doi: 10.1016/j.stem.2008.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, Tomoda K, Yamanaka S. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- Tarasov KV, Tarasova YS, Tam WL, Riordon DR, Elliott ST, Kania G, Li J, Yamanaka S, Crider DG, Testa G, et al. B-MYB is essential for normal cell cycle progression and chromosomal stability of embryonic stem cells. PLoS ONE. 2008;3:e2478. doi: 10.1371/journal.pone.0002478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wernig M, Lengner CJ, Hanna J, Lodato MA, Steine E, Foreman R, Staerk J, Markoulaki S, Jaenisch R. A drug-inducible transgenic system for direct reprogramming of multiple somatic cell types. Nat Biotechnol. 2008;26:916–924. doi: 10.1038/nbt1483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wernig M, Meissner A, Foreman R, Brambrink T, Ku M, Hochedlinger K, Bernstein BE, Jaenisch R. In vitro reprogramming of fibroblasts into a pluripotent ES-cell-like state. Nature. 2007;448:318–324. doi: 10.1038/nature05944. [DOI] [PubMed] [Google Scholar]
- Wolf D, Goff SP. TRIM28 mediates primer binding site-targeted silencing of murine leukemia virus in embryonic cells. Cell. 2007;131:46–57. doi: 10.1016/j.cell.2007.07.026. [DOI] [PubMed] [Google Scholar]
- Yakubov E, Rechavi G, Rozenblatt S, Givol D. Reprogramming of human fibroblasts to pluripotent stem cells using mRNA of four transcription factors. Biochemical and biophysical research communications. 2010;394:189–193. doi: 10.1016/j.bbrc.2010.02.150. [DOI] [PubMed] [Google Scholar]
- Yamamoto M, Sato S, Hemmi H, Hoshino K, Kaisho T, Sanjo H, Takeuchi O, Sugiyama M, Okabe M, Takeda K, et al. Role of adaptor TRIF in the MyD88-independent toll-like receptor signaling pathway. Science. 2003;301:640–643. doi: 10.1126/science.1087262. [DOI] [PubMed] [Google Scholar]
- Yang WC, Patel KG, Lee J, Ghebremariam YT, Wong HE, Cooke JP, Swartz JR. Cell-free production of transducible transcription factors for nuclear reprogramming. Biotechnology and bioengineering. 2009;104:1047–1058. doi: 10.1002/bit.22517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou H, Wu S, Joo J, Zhu S, Han D, Lin T, Trauger S, Bien G, Yao S, Zhu Y, et al. Generation of induced pluripotent stem cells using recombinant proteins. Cell Stem Cell. 2009;4:381–384. doi: 10.1016/j.stem.2009.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.