Summary
T lymphocytes make use of their major integrin LFA-1 to migrate on surfaces that express ICAM-1 such as blood vessels and inflamed tissue sites. How the adhesions are turned over in order to supply traction for this migration has not been extensively investigated. By following the fate of biotinylated membrane LFA-1 on T lymphocytes, we show in this study that LFA-1 internalization and re-exposure on the plasma membrane are linked to migration. Previously we demonstrated the GTPase Rap2 to be a regulator of LFA-1-mediated migration. SiRNA knockdown of this GTPase inhibits both LFA-1 internalization and also its ability to be re-exposed, indicating that Rap2 participates in recycling of LFA-1 and influences its complete endocytosis–exocytosis cycle. Confocal microscopy images reveal that the intracellular distribution of Rap2 overlaps with endosomal recycling vesicles. Although the homologous GTPase Rap1 is also found on intracellular vesicles and associated with LFA-1 activation, these two homologous GTPases do not co-localize. Little is known about the conformation of the LFA-1 that is recycled. We show that the extended form of LFA-1 is internalized and in Rap2 siRNA-treated T lymphocytes the trafficking of this LFA-1 conformation is disrupted resulting in its intracellular accumulation. Thus LFA-1-mediated migration of T lymphocytes requires Rap2-expressing vesicles to recycle the extended form of LFA-1 that we have previously found to control migration at the leading edge.
Key words: Rap2, LFA-1, Integrin, Migration, Recycling
Introduction
As cells move forward they make use of integrins to create the adhesions that allow migration to proceed. A migrating cell needs to attach at the front to move the leading edge membrane forward, then release at the rear and re-establish its adhesions (Vicente-Manzanares et al., 2009). Immune cells are the most dynamic cells in the body in terms of motility having the ability to migrate rapidly on the luminal walls of blood vessels as well as in extra-vascular tissues (Ley et al., 2007). By using the β2 integrin LFA-1 (CD11a/CD18; αLβ2) to attach to intercellular adhesion molecule-1 (ICAM-1)-expressing surfaces, the cells are able to migrate both randomly as well as in a directed fashion towards chemoattractants (Evans et al., 2009).
An integrin such as LFA-1 has three basic conformations reflecting increasing affinity for binding ligand ICAM-1 that are characterized as bent, extended/closed and extended/open (Hogg et al., 2011; Springer and Dustin, 2012). These forms can be distinguished through the use of conformation-specific monoclonal antibodies. For example mAb KIM127 detects extended LFA-1 with the open form considered to bind ICAM-1 with higher affinity than the closed form. MAb 24 exclusively binds the extended/open high affinity conformation. In our previous work we found the extended/closed form of LFA-1 to be distributed chiefly at the leading edge of migrating T cells where there is dynamic adhesion turnover, whereas the extended/open form localized further back with highest concentration in the more firmly bound lamellar region (Evans et al., 2011; Smith et al., 2005; Stanley et al., 2008).
There have been various ideas as to how turnover of LFA-1 adhesions on migrating T cells might occur. One possibility is of a regulated cycle of LFA-1 adhesion and de-adhesion, potentially aided by the protease calpain (Franco and Huttenlocher, 2005; Hogg et al., 2011). Alternatively it is now well-established that other integrins undergo endocytosis and re-exposure on the plasma membrane, a process creating fresh attachments that allow migration to be ongoing (Caswell et al., 2009; Lawson and Maxfield, 1995; Pellinen and Ivaska, 2006). In support of the latter option, LFA-1 was found to be internalized in CHO cells and neutrophils (Fabbri et al., 2005; Fabbri et al., 1999).
The generation of active LFA-1 able to bind ICAM-1 requires delivery of the GTPase Rap1 to the cell membrane, a key element in the signaling originating from both chemokine receptors and the T cell receptor (Dustin et al., 2004; Hogg et al., 2011; Kinashi, 2005). Mechanistically Rap1 activity is dependent on the effector RAPL that transports the integrin to the leading edge of migrating T cells (Katagiri et al., 2006; Katagiri et al., 2003). There is increasing evidence that LFA-1 and Rap1 are components of intracellular vesicles that have the characteristics of recycling endosomes (Bivona et al., 2004; Katagiri et al., 2006; Mor et al., 2009; Raab et al., 2010).
The functions of the Rap1 homologue, GTPase Rap2, have been less extensively investigated. Rap2 has a role in integrin-mediated adhesion and in migration of B cells (McLeod et al., 2002). Similarly we found Rap2 to regulate LFA-1-mediated migration of T cells (Miertzschke et al., 2007). In this study we have investigated the recycling of LFA-1 in T cells and find that this GTPase is involved in the turnover of LFA-1 that drives T cell migration.
Results
An association between LFA-1 recycling and T lymphocyte migration
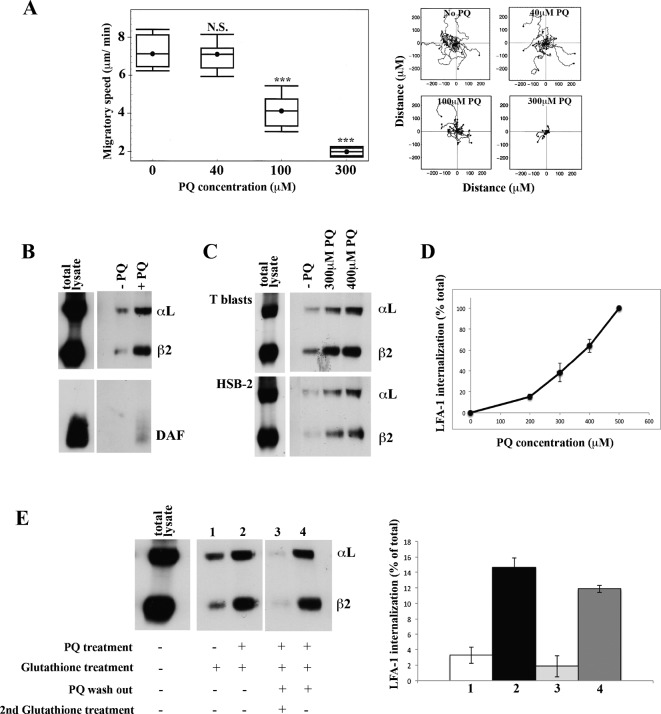
We investigated whether intracellular vesicle transport in the T cell is essential for LFA-1-mediated migration. Primaquine (PQ) is a lysosomotrophic amine that slows recycling by blocking membrane fusion of exocytic vesicles and has been used to assess a requirement for vesicular recycling in various T cell activities (Pathak and Blum, 2000; Reid and Watts, 1990; Roberts et al., 2002; Roy et al., 2008). Following 5 min pre-treatment with PQ, both the random migration and average speed of HSB2 T cells were normal at 40 µM, reduced by 40% at 100 µM and by 70% at 300 µM PQ with no impact on cell viability (Fig. 1A). Thus prevention of docking of intracellular transport vesicles at the cell membrane reduced the ability of T cells to migrate.
Fig. 1. LFA-1 is internalized and re-expressed by T cells migrating on ICAM-1.
(A) Speed of T lymphoblasts treated with increasing concentrations of primaquine (PQ) migrating on immobilized ICAM-1 showing cell trajectories tracked by video microscopy. Mean speed ± s.e.m. of three independent experiments (left) and the migratory tracks in a typical experiment (right) are shown; n = 20 cells per condition, ***P<0.001. (B) Western blots of immunoprecipitated internalized LFA-1 and DAF following biotinylation of T lymphoblast surface membranes followed by 30 min migration on ICAM-1: total cell lysate immunoprecipitated for LFA-1 and DAF (sample diluted 2×) and internalized protein ± 300 µM PQ revealed by removal of biotin from cell membrane receptors with glutathione. (C) Western blots comparing internalized LFA-1 in T lymphoblasts and HSB2 T cell line: total biotinylated LFA-1 (total lysate at 2× dilution) and similar amounts of internalized LFA-1 ± PQ. (D) Internalized T cell LFA-1 after 30 min on ICAM-1 in the presence of increasing amounts of PQ ± s.d. from n = 3 experiments. (E) Total biotinylated LFA-1 (total lysate at 2× dilution). Lanes 1 and 2: LFA-1 internalized after 30 min ± 300 µM PQ. LFA-1 re-exposure on the membrane following PQ washout is demonstrated by lack of intracellular LFA-1 in lane 3 (treated with glutathione to remove detection of membrane LFA-1) and lane 4 (no glutathione treatment allowing membrane and intracellular LFA-1 to be detected). Left: typical experiment. Right: mean ± s.d. of n = 3 experiments.
To investigate whether recycling of LFA-1 might be involved, we examined integrin internalization and re-exposure by biotinylating T cell membrane receptors and then allowing the cells to migrate on ICAM-1 in the presence of PQ. By subsequent removal of biotin from LFA-1 remaining on the cell membrane using glutathione, the LFA-1 that had become internalized could be assessed (Fabbri et al., 2005; Fabbri et al., 1999; Reid and Watts, 1990). This internalized LFA-1 was detected in cell lysates by anti-LFA-1 immunoprecipitation and anti-biotin blotting using conditions where all biotinylated LFA-1 was detected (data not shown). After 30 min of T lymphoblast adhesion to ICAM-1, a proportion of the LFA-1 was located intracellularly and this was increased in the presence of PQ (Fig. 1B). To show that the internalization was selective, we compared LFA-1 with DAF, a glycophosphatidylinositol-linked lipid raft protein that is poorly endocytosed. Unlike LFA-1, little DAF was located intracellularly either with or without addition of PQ (Fig. 1B). LFA-1 was endocytosed in an equivalent manner in T lymphoblasts and the T cell line HSB2 and retained in increased amounts at PQ concentrations that halted migration (Fig. 1C). The amount of intracellular LFA-1 correlated closely with the level of PQ with which the T cells were treated, consistent with the ability of PQ to block re-exposure of LFA-1 on the surface membrane (Fig. 1D).
An issue concerned the fate of the internalized LFA-1 and whether it was degraded or, alternatively, returned to the plasma membrane. We first allowed LFA-1 to internalize over 30 min ± PQ. In the PQ-treated samples where exocytosis was blocked, ∼15% of total LFA-1 accumulated inside the T cell over this time period with a much lower level when the endocytosis–exocytosis cycle was allowed to proceed normally (Fig. 1E, lanes 1 and 2). To evaluate the fate of this internalized LFA-1, we concentrated on the T cells that had been pre-treated with PQ. The PQ was washed out and then the cells were incubated for 20 min before assessing the LFA-1 compartmentalization. The majority of the internalized LFA-1 left the intracellular compartment during this time period and re-appeared on the plasma membrane as glutathione sensitive LFA-1 (Fig. 1E, lanes 3 and 4). Thus LFA-1 is not degraded when it is internalized, but is returned to the plasma membrane.
Rap2 regulates endocytosis of LFA-1 in T cells migrating on ICAM-1
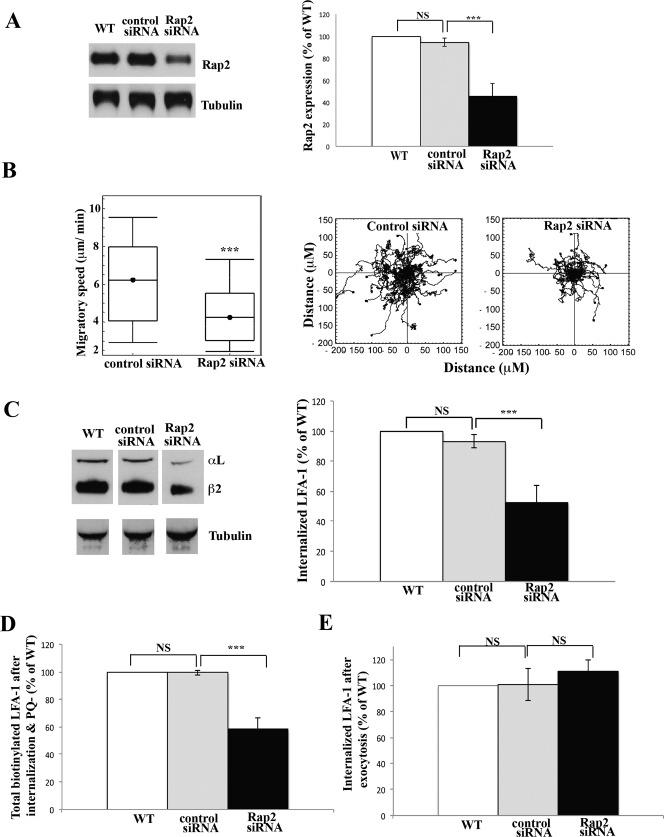
We previously demonstrated an essential role for the GTPase Rap2B in T cell migration as assessed by Rap2 siRNA knockdown and reconstitution with wild type Rap2B cDNA (Miertzschke et al., 2007). We therefore wondered whether the GTPase might be affecting migration by influencing LFA-1 internalization. Treatment of HSB2 T cells with Rap2 siRNA for 72 h reduced expression of Rap2 to 45.4±11.5% of control T cells (Fig. 2A). Rap2 knockdown had no effect on the level of the homologous GTPase Rap1 (data not shown; Miertzschke et al., 2007). This reduced level of Rap2 was sufficient to inhibit the speed and randomness in direction of T cell migration by ∼35% and could be reversed by transfection with WT Rap2B as previously described (Fig. 2B) (Miertzschke et al., 2007).
Fig. 2. Rap2 regulates internalization of T cell LFA-1 during migration.
(A) HSB2 T cells were either not electroporated as control WT or electroporated with control or Rap2 siRNAs. Western blots were probed for Rap2 after 72 h and α-tubulin as a loading sample control. Left: typical experiment showing Rap2 siRNA knockdown compared with non-electroporated (WT) or control siRNA-treated T cell controls. Right: quantification of siRNA knockdown compared with controls, mean ± s.d. of n = 5 experiments, ***P<0.001. (B) HSB2 T cells electroporated with control or Rap2 siRNAs migrating on ICAM-1 for 30 min. Left: mean speed ± s.d. of n = 3 experiments. Right: migratory tracks of individual cells; n = 40 cells per condition, ***P<0.001. (C) Biotinylated LFA-1 internalization + PQ in WT, control siRNA- and Rap2 siRNA-treated HSB2 T cells. LFA-1 internalization following Rap2 siRNA compared with controls, showing (left) Western blot of a typical experiment and (right) quantification of 3 experiments, mean ± s.d. ***P<0.001. (D) Using T cells treated as in C, total biotinylated LFA-1 in control and Rap2 siRNA-treated T cells is shown following PQ washout and a 20 min incubation. No glutathione treatment allows assessment of internal and re-exposed LFA-1; mean ± s.d. of 3 experiments, ***P<0.001. (E) Re-expression of LFA-1 in control and Rap2 siRNA-treated T cells following PQ washout and 20 min incubation. LFA-1 remaining inside the cells revealed following removal of glutathione sensitive membrane LFA-1; mean ± s.d. of 3 experiments, ***P<0.001.
We next investigated the level of internalized LFA-1 in Rap2 siRNA-treated T cells and found a consistent reduction to 47.5±11.2% of that found in control siRNA-treated T cells (Fig. 2C). Thus the internalized LFA-1 correlated with the level of Rap2 knockdown. The next question was whether the LFA-1 internalized in the Rap2 siRNA-treated T cells was capable of being re-exposed at the membrane. To address this issue, we washed out the PQ from this same set of T cells and incubated them for 20 min to allow re-exposure on the cell membrane of previously internalized LFA-1. We then re-assessed the total amount of biotin-labeled LFA-1 in control and Rap2 siRNA-treated T cells including both intracellular and re-exposed populations (Fig. 2D). The first point was that the proportions of total biotin-labeled LFA-1 recovered in each sample were identical to the quantities of LFA-1 internalized in Fig. 2C, indicating that this initially endocytosed LFA-1 was not degraded (Fig. 2D). Secondly following removal of glutathione-sensitive LFA-1 on the cell membrane, the proportion of intracellular LFA-1 remaining in the Rap2 siRNA-treated cells was observed to be increased compared with that in the control siRNA-treated cells (Fig. 2E). Thus it was concluded that Rap2 siRNA-treated T cells were deficient in promoting subsequent membrane re-exposure of LFA-1, implying that Rap2 is a controlling factor not only in the endocytosis of LFA-1, but is also in recycling LFA-1 back to the membrane.
Rap2 is located in recycling vesicles
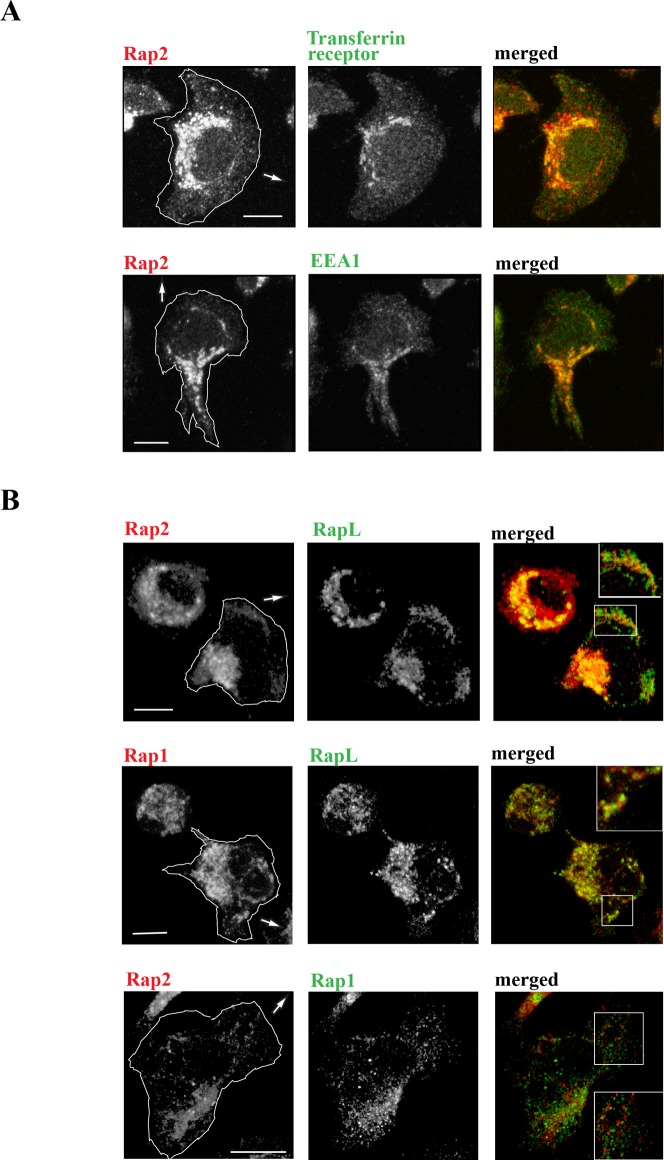
We next used confocal microscopy to determine the intracellular location of Rap2 and its relationship with markers of endosomal vesicles. Rap2 staining was punctate and chiefly concentrated in the juxta-nuclear region but with scattered labeling towards the leading edge (Fig. 3A,B). In terms of vesicle markers, Rap2 co-localized with early endosomal protein, EEA-1, a key Rab5 effector protein (Christoforidis et al., 1999) and with transferrin receptor that labels recycling vesicles (Fig. 3A). To quantify overlap in expression between Rap2 with EEA1 and transferrin receptor respectively, we performed a pixel-by-pixel analysis to calculate Manders' coefficient (Manders' coefficient for Rap2/EEA1 = 61.90±0.08%; Rap2/transferring receptor = 66.33±0.11%; n = 5 cells each). Rap2 did not notably overlap with Rab11 that also marks a subset of endosomal vesicles (data not shown).
Fig. 3. Expression of Rap2, Rap1 and recycling vesicles in migrating T lymphoblasts.
(A) Confocal microscopy images showing the co-distribution of Rap2 with intracellular vesicle markers EEA1 and transferrin receptor at the interface with ICAM-1. (B) Rap2 and Rap1 each co-localize with effector RAPL, but there is a lack of overlap between these two GTPases; insets show detail at leading edge. Arrows show direction of T cell migration. Scale bar = 5 µm.
The closely homologous GTPase, Rap1, is also found in intracellular vesicles in T cells (Bivona et al., 2004; Katagiri et al., 2006; Mor et al., 2009; Raab et al., 2010). However, we observed little overlap in expression of Rap2 with Rap1, indicating that they must be chiefly present in separate intracellular compartments (Manders' overlap coefficient for Rap2 with Rap1 was 24.3±0.06%; n = 5 cells) (Fig. 3B). RAPL is a downstream effector of Rap1, necessary for intracellular transport of LFA-1 to the leading edge of T cells (Katagiri et al., 2003). In our previous study we found that RAPL bound Rap2 with greater stability than it bound Rap1, suggesting that it had a role in the functioning of both these GTPases (Miertzschke et al., 2007). When viewed by confocal microscopy, both Rap2 and Rap1 were observed to co-localize with RAPL (Manders' coefficient for Rap2/RAPL = 60.30±0.15%; Rap1/RAPL = 0.68±0.05%; n = 5 cells each) (Fig. 3B). Thus Rap1 and Rap2 appear to be present on different endosomal vesicles with both subsets however co-expressing RAPL.
The effect of Rap2 knockdown on the conformation of internalized LFA-1
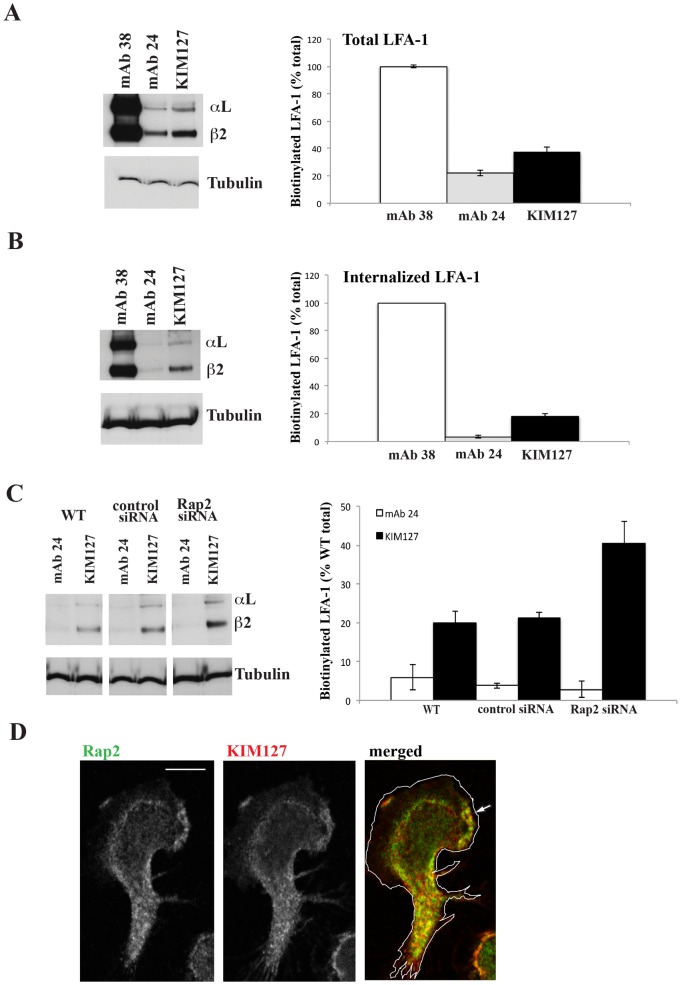
The major conformations of bent, extended and extended/high affinity LFA-1 can be distinguished through the use of mAbs that detect conformation-specific epitopes (Hogg et al., 2011; Springer and Dustin, 2012). To gain insight into the conformations of LFA-1 that might be recycled, we immunoprecipitated LFA-1 using pan-LFA-1 mAb 38, mAb KIM127 that detects both forms of extended LFA-1 and mAb 24 that recognizes only the extended/open (high affinity) conformation. T cells expressed 37.4±3.6% extended LFA-1 and 22.2±2.1% high affinity LFA-1 compared with total LFA-1 (Fig. 4A). However, examination of internalized LFA-1 following removal of glutathione sensitive membrane LFA-1 revealed 17.9±1.9% extended LFA-1 and essentially no high affinity LFA-1 compared with the total LFA-1 (Fig. 4B). Thus at least a proportion of the intracellular LFA-1 was in the extended, but apparently not high affinity (extended/open) conformation.
Fig. 4. Rap2 regulates internalization of the extended conformation of LFA-1.
MAbs specific for pan-LFA-1 (38), high affinity (24) and extended (KIM127) LFA-1 used to immunoprecipitate (A) total LFA-1 and (B) internalized LFA-1 from T cells. (A,B) Left: typical western blot experiment. Right: mean ± s.d. of n = 4 experiments. (C) Internalization of extended but not high affinity conformations of LFA-1 are increased following Rap2 siRNA knockdown compared with control siRNA and WT treatment of T cells. Left: typical western blot experiment. Right: mean ± s.d. of n = 3 experiments. (D) Confocal microscopy image showing overlap of Rap2 and KIM127-expressing LFA-1 concentrated in the lamella with some lamellipodial distribution in the direction of T lymphoblast migration indicated by an arrow. Scale bar = 5 µm.
Following Rap2 siRNA knockdown, there was a two-fold increase in the amount of internalized, extended KIM127-expressing LFA-1 compared with control siRNA-treated and WT T cells, but there was no impact on the poorly internalized high affinity LFA-1 (Fig. 4C). As a control we showed that the total cellular levels for each of extended and high affinity LFA-1 expressed by Rap2 knockdown, siRNA control and WT T cells were identical indicating that there was no loss of LFA-1 in the Rap2 knockdown samples due to intracellular degradation (supplementary material Fig. S1). Therefore the difference in the amount of extended LFA-1 internalized by Rap2 siRNA-treated cells was a matter of compartmentalization and not caused by a difference in total LFA-1 expression level.
Finally we further confirmed the association between Rap2 and the extended conformation of LFA-1 using confocal microscopy by showing co-localization of KIM127-positive LFA-1 with Rap2 vesicles in T lymphoblasts (Manders' coefficient for Rap2/KIM127 = 77.0±0.06%; n = 5 cells) (Fig. 4D).
Discussion
Previously we showed that the GTPase Rap2B was a controlling factor in LFA-1-mediated migration of T cells, but we did not explore where it might be having its influence. In this study we demonstrate that recycling of LFA-1 is important for migration and it is this step that is regulated by the GTPase Rap2. There is increasing evidence that recycling of integrin is essential for the successful migration of leukocytes. For example, the presence of α5β1 (Lawson and Maxfield, 1995; Pierini et al., 2000) and LFA-1 (Fabbri et al., 2005) in recycling vesicles drives neutrophil migration.
LFA-1 is turned over on T cells through endocytosis and re-exposure without being degraded. This finding helps explain the stability of LFA-1 expression on leukocyte membrane that is maintained without extensive new synthesis (Dustin et al., 1989). The internalization of LFA-1 over 30 min occurs whether the T cells are migrating on ICAM-1 or maintained in suspension (data not shown) indicating that the recycling may be constitutive. Similarly, LFA-1 recycling in neutrophils is considered to occur constitutively (Fabbri et al., 2005).
Rap2 immunostaining that co-localized with LFA-1 was punctate and concentrated in the juxta-nuclear region with some spread toward the leading edge, a pattern resembling that of endosomal vesicles. Association of Rap2 with recycling endosomes in COS-1 cells displays a similar staining pattern (Uechi et al., 2009). Additionally Rap2 co-localized with EEA1, a downstream effector of Rab5 associated with early endosomal vesicles (Christoforidis et al., 1999), in keeping with a role for Rap2 in LFA-1 internalization. Rap2 also co-localized with the transferrin receptor, similarly to other reports linking intracellular integrins to transferrin receptor-expressing recycling vesicles (Laukaitis et al., 2001; Pellinen et al., 2006). Together the immunoprecipitation and immunostaining data indicated that recycling of at least a proportion of the T cell membrane LFA-1 was controlled by Rap2 expressed on endosomal vesicles associated with recycling.
There was, however, a lack of Rap2 immunostaining with its homologous GTPase, Rap1. This is of relevance because LFA-1 recycling has been associated with Rap1 that is found on vesicles expressing EEA1, Rab5 and Rab11, but not transferrin receptor (Katagiri et al., 2006) or, alternatively, on vesicles with limited overlap between EEA1 and Rab7 (Raab et al., 2010). Thus there must be heterogeneity in LFA-1-transporting vesicles with Rap2 characterizing one set and Rap1, a separate set. The implication is that each vesicle type is involved in a distinctive set of events contributing to LFA-1 recycling. Some evidence for this comes from previous findings where Rap1 was found to dominate in adhesive interactions, whereas Rap2 was restricted to migration of T cells (Miertzschke et al., 2007). A more complete characterization is needed of the LFA-1 recycling mediated by Rap2 compared with Rap1.
It was unexpected that the extended conformation of LFA-1 was being recycled. However, other examples of the recycling of an active integrin conformation are those of α2β1 in MDA breast cancer cells, where an integrin activation epitope was also associated with transferrin receptor-expressing vesicles (Pellinen et al., 2006) and α5β1 in several cancer cell lines (Arjonen et al., 2012). We showed previously that the KIM127-expressing LFA-1 conformation was linked to α-actinin at the leading edge of the migrating T cell (Stanley et al., 2008). As siRNA knockdown of α-actinin did not affect LFA-1 internalization (data not shown), the implication is that migrating T cells appear to be recycling active LFA-1 that is not only ligand unoccupied but disengaged from the cytoskeleton. As suggested by others (Caswell et al., 2009; Laukaitis et al., 2001), this recycling may be ongoing locally at membrane level, providing fresh integrin, in this case LFA-1, for new adhesions that are formed as the T cell advances.
Reduction in Rap2 expression through siRNA knockdown disrupts LFA-1 intracellular trafficking causing a decrease in the pool of internalized LFA-1, but without disturbing the total level of LFA-1. As the majority of this reduced level of internalized LFA-1 is neither in extended nor high affinity conformation and detected with a pan-LFA-1 mAb, it must by default be in the bent, inactive conformation of LFA-1. However, Rap2 also controls the correct trafficking of a subset of LFA-1 that is in the extended conformation and this LFA-1 forms an increased proportion of intracellular pool of LFA-1 when the level of Rap2 is reduced. Thus by slowing down LFA-1 recycling, Rap2 knockdown causes both accumulation of the extended form of LFA-1, but also a decrease in total intracellular LFA-1. An explanation is that Rap2 is involved in two recycling pathways, dealing separately with the intracellular transport of inactive, bent LFA-1 and extended KIM127-expressing LFA-1. Distinct recycling of active and inactive α1 integrins has been observed in carcinoma cell lines (Arjonen et al., 2012). It is also possible that Rap2 is responsible for the deactivation of LFA-1, mediating its conversion from extended to inactive bent form.
In conclusion we have highlighted a role for the GTPase Rap2 in regulating the recycling of at least two conformations of LFA-1 and that this activity is important for the random migration of T cells on ICAM-1. As both Rap2 and Rap1 have apparently separate roles in LFA-1 recycling, future effort should be directed towards characterizing the full heterogeneity of the endosomal vesicles involved in LFA-1 recycling and uncovering their distinct functions. The fact that the extended conformation of LFA-1, but not the high affinity form, was found intracellularly suggests that the turnover by T cells of these two LFA-1 conformations differs mechanistically. Thus the findings imply that the regulation of other forms of LFA-1 might occur potentially by alternative forms of recycling.
Materials and Methods
Monoclonal antibodies and other reagents
The following mAbs were used: Rap2, Rap1 and EEA1 mAbs (BD Transduction Labs, Oxford Science Park, Oxford, UK); pan-LFA-1 mAb 38 (CD11a), LFA-1 activation mAbs KIM127 (CD18 extension reporter) and 24 (CD18 activation reporter) (Stanley et al., 2008), mAb 67 (DAF, CD55) (Leitinger and Hogg, 2002) all prepared at CR UK, LRI; DM1A (α-tubulin) (Sigma–Aldrich Ltd, Gillingham, Kent, UK); H68.4 (transferrin receptor) (Cambridge BioScience, Cambridge, UK); RAPL mAb B6.4 was a generous gift from Dr Tatsuo Kinashi, Kansai Medical School, Japan. Secondary antibodies used were Zenon AlexaFluor488 anti-mouse IgG2a, Zenon AlexaFluor546 anti-mouse IgG1, AlexaFluor488 anti-rat IgG and AlexaFluor546 anti-mouse IgG (Molecular Probes/Life Technologies Ltd, Paisley, UK). ICAM-1Fc was prepared as previously described (Smith et al., 2005; Stanley et al., 2008).
T cell culture and transfection
Peripheral blood mononuclear cells were prepared from single donor leukocyte buffy coats (National Blood Service, Tooting, UK) and dealt with according to the local CR UK Ethics Committee regulations. T cells were expanded as previously described and used between days 10 and 14 (Smith et al., 2003). The human T lymphoblast CD3− T cell line, HSB2 isolated from an acute lymphoblastic leukemia source (ATCC number CCL-120.1, known as CCRF-HSB-2 or HSB2) was maintained in RPMI 1640/10% FCS (Wright et al., 1994).
HSB2 T cells were washed in OptiMEM + GlutaMAX (Life Technologies, Paisley, UK) and 2×107 cells were electroporated using a Gene Pulser with Capacitance Extender (Bio-Rad UK, Hemel Hempstead, UK) set at 960 µF and 260 mV. Rap2 knockdown siRNAs for Rap2A, Rap2B and Rap2C (Dharmacon smart pools for Rap2A, gene ID 5911, Rap2B, gene ID 5912 and Rap2C, gene ID 57826) and control siRNA, siCONTROL Non-targeting smart pool, (Dharmacon, Inc., ThermoFisher Scientific, Loughborough, UK) were all used at 200 nmol. Transfected cells were maintained for 72 h in RPMI 1640 with 10% FCS. The efficiency of siRNA knockdown was evaluated by Western blotting using Rap2 mAb and α-tubulin mAb serving as a sample loading control.
LFA-1 internalization and re-exposure assay
The protocol used was adapted from Fabbri et al. (Fabbri et al., 1999). Glass coverslips (32 mm) were coated with 3 µg/ml ICAM-1Fc in PBS at 4°C overnight then blocked with 2.5% BSA. To biotinylate membrane proteins, washed T cells were re-suspended in 0.5 mg/ml EZ-link sulpho-NHS-SS-biotin (21331, Pierce, ThermoFisher Scientific, Loughborough, UK) at 2.5×107 cells/ml and incubated on ice for 1 h. After washing, 4×106 T cells in HBSS buffer were added to each ICAM-1-coated coverslip. Primaquine diphosphate (PQ) (160393, Sigma–Aldrich Ltd) at 40–500 µM was added and the cells incubated for 30 min at 37°C to allow adhesion and internalization of receptors. To remove membrane bound biotin, freshly made cold reduced glutathione buffer (46 mM glutathione, 75 mM NaCl, 1 mM EDTA, 1% BSA, 75 mM NaOH) was added and the cells incubated on ice for 30 min. Controls for biotinylation of total LFA-1 were incubated in PBS. To investigate re-exposure of LFA-1 on the cell surface, after reduction and removal of membrane biotin with glutathione buffer, the T cells were placed onto new ICAM-1-coated coverslips and incubated at 37°C for 20 min and then treated on ice for 30 min with cold PBS to detect total biotinylated LFA-1 or with glutathione buffer as above to detect only intracellular biotinylated LFA-1.
To detect biotinylated LFA-1, T cells were lysed with a standard cell lysis buffer containing 0.2% NP40 and protease inhibitors for 20 min. Following centrifugation, mAbs were added to the supernatant and incubated at 4°C for 2 h followed by addition of protein A sepharose (Sigma–Aldrich Ltd) and further incubation at 4°C for 1.5 h. The bound protein samples were then washed 5 times and non-reducing gel sample buffer added prior to SDS-PAGE on 4–12% NuPAGE gels (Life Technologies Ltd). Proteins were transferred to nitrocellulose (GE Healthcare, Chalfont St Giles, UK) and blocked with 5% milk powder in PBS/0.1% Tween 20. To reveal biotinylated LFA-1, the blot was incubated with ECL streptavidin-HRP conjugate (RPN1231, GE Healthcare) in PBS/0.1% Tween 20 for 1 h, washed 3× and treated with ECL reagent (GE Healthcare) before exposure to film. A set of samples was probed with α-tubulin mAb (Sigma–Aldrich Ltd) followed by anti-mouse IgG-HRP Ab (GE Healthcare) to check for equivalent cell loading between samples. Films were scanned and the band densities analyzed using ImageJ software.
Video microscopy
Thirty-five mm glass bottom microwell dishes (MatTek Corp., Ashland, Mass, USA) were coated at 4°C overnight with 3 µg/ml ICAM-1Fc, then blocked with 2.5% BSA. 4×105 HSB-2 T cells per dish were allowed to settle for 10 min before filming. PQ was added immediately before imaging. Images were taken at intervals of 15 sec using a Nikon Diaphot 300 microscope and AQM2001 Kinetic Acquisition Manager software (Kinetic Imaging Ltd., Bromborough, UK). Cells were tracked over 240–480 frames corresponding to 40 min using Motion Analysis software (Kinetic Imaging Ltd.) and the data analyzed using a Mathematica notebook (Wolfram Research Europe Ltd, Long Hanborough, UK) developed by Daniel Zicha (CR UK, London). The migration data are depicted using box-and-whisker plots.
Confocal microscopy
Glass coverslips (13 mm) were coated with ICAM-1Fc as for video microscopy. Washed T cells were added to coated coverslips (2×105 cells/coverslip) for 30 min. Adherent cells were fixed with 3% paraformaldehyde (Sigma–Aldrich Ltd) in Pipes buffer (pH 8) for 5 min at RT and then in 3% paraformaldehyde in sodium tetraborate (pH 11, Sigma–Aldrich Ltd) for 10 min. Cells were permeabilized with 0.5% Triton X-100 in PBS for 5 min on ice. Autofluorescence was quenched using 1 mg/ml sodium tetraborate (pH 8) for 15 min and the cells were blocked with 5% BSA in PBS for 45 min. Coverslips were incubated with primary mAbs for 30 min, followed by AlexaFluor488-goat anti-mouse IgG or AlexaFluor546-goat anti-rat IgG (Life Technologies Ltd) for 20 min. Alternatively coverslips were incubated for 30 min with Zenon AlexaFluor anti-mouse IgG1 or IgG2a labeled primary mAbs and then fixed for 15 min with 4% paraformaldehyde in PBS. Images were acquired using a Zeiss Laser Scanning LSM 710 Microscope.
The extent of co-localization of different markers was analyzed using Image J software and JACoB analysis. Co-localization was measured using the Manders' coefficient to evaluate the overlap in fluorescence of Rap2 with other markers (Bolte and Cordelières, 2006).
Statistical analysis
The migration assays are presented as the mean ± s.e.m. The unpaired Student's t test was performed using GraphPad Prism software version 4 for Macintosh computers. The following significant differences are as indicated: *P<0.05, **P<0.01 and ***P<0.001.
Supplementary Material
Acknowledgments
This work was supported by Cancer Research UK.
Footnotes
Competing interests: The authors have no competing interests to declare.
References
- Arjonen A., Alanko J., Veltel S., Ivaska J. (2012). Distinct recycling of active and inactive β1 integrins. Traffic 13, 610–625 10.1111/j.1600-0854.2012.01327.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bivona T. G., Wiener H. H., Ahearn I. M., Silletti J., Chiu V. K., Philips M. R. (2004). Rap1 up-regulation and activation on plasma membrane regulates T cell adhesion. J. Cell Biol. 164, 461–470 10.1083/jcb.200311093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolte S., Cordelières F. P. (2006). A guided tour into subcellular colocalization analysis in light microscopy. J. Microsc. 224, 213–232 10.1111/j.1365-2818.2006.01706.x [DOI] [PubMed] [Google Scholar]
- Caswell P. T., Vadrevu S., Norman J. C. (2009). Integrins: masters and slaves of endocytic transport. Nat. Rev. Mol. Cell Biol. 10, 843–853 10.1038/nrm2799 [DOI] [PubMed] [Google Scholar]
- Christoforidis S., McBride H. M., Burgoyne R. D., Zerial M. (1999). The Rab5 effector EEA1 is a core component of endosome docking. Nature 397, 621–625 10.1038/17618 [DOI] [PubMed] [Google Scholar]
- Dustin M. L., Garcia–Aguilar J., Hibbs M. L., Larson R. S., Stacker S. A., Staunton D. E., Wardlaw A. J., Springer T. A. (1989). Structure and regulation of the leukocyte adhesion receptor LFA-1 and its counterreceptors, ICAM-1 and ICAM-2. Cold Spring Harb. Symp. Quant. Biol. 54, 753–765 10.1101/SQB.1989.054.01.089 [DOI] [PubMed] [Google Scholar]
- Dustin M. L., Bivona T. G., Philips M. R. (2004). Membranes as messengers in T cell adhesion signaling. Nat. Immunol. 5, 363–372 10.1038/ni1057 [DOI] [PubMed] [Google Scholar]
- Evans R., Patzak I., Svensson L., De Filippo K., Jones K., McDowall A., Hogg N. (2009). Integrins in immunity. J. Cell Sci. 122, 215–225 10.1242/jcs.019117 [DOI] [PubMed] [Google Scholar]
- Evans R., Lellouch A. C., Svensson L., McDowall A., Hogg N. (2011). The integrin LFA-1 signals through ZAP-70 to regulate expression of high-affinity LFA-1 on T lymphocytes. Blood 117, 3331–3342 10.1182/blood-2010-06-289140 [DOI] [PubMed] [Google Scholar]
- Fabbri M., Fumagalli L., Bossi G., Bianchi E., Bender J. R., Pardi R. (1999). A tyrosine-based sorting signal in the β2 integrin cytoplasmic domain mediates its recycling to the plasma membrane and is required for ligand-supported migration. EMBO J. 18, 4915–4925 10.1093/emboj/18.18.4915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabbri M., Di Meglio S., Gagliani M. C., Consonni E., Molteni R., Bender J. R., Tacchetti C., Pardi R. (2005). Dynamic partitioning into lipid rafts controls the endo-exocytic cycle of the αL/β2 integrin, LFA-1, during leukocyte chemotaxis. Mol. Biol. Cell 16, 5793–5803 10.1091/mbc.E05-05-0413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franco S. J., Huttenlocher A. (2005). Regulating cell migration: calpains make the cut. J. Cell Sci. 118, 3829–3838 10.1242/jcs.02562 [DOI] [PubMed] [Google Scholar]
- Hogg N., Patzak I., Willenbrock F. (2011). The insider's guide to leukocyte integrin signalling and function. Nat. Rev. Immunol. 11, 416–426 10.1038/nri2986 [DOI] [PubMed] [Google Scholar]
- Katagiri K., Maeda A., Shimonaka M., Kinashi T. (2003). RAPL, a Rap1-binding molecule that mediates Rap1-induced adhesion through spatial regulation of LFA-1. Nat. Immunol. 4, 741–748 10.1038/ni950 [DOI] [PubMed] [Google Scholar]
- Katagiri K., Imamura M., Kinashi T. (2006). Spatiotemporal regulation of the kinase Mst1 by binding protein RAPL is critical for lymphocyte polarity and adhesion. Nat. Immunol. 7, 919–928 10.1038/ni1374 [DOI] [PubMed] [Google Scholar]
- Kinashi T. (2005). Intracellular signalling controlling integrin activation in lymphocytes. Nat. Rev. Immunol. 5, 546–559 10.1038/nri1646 [DOI] [PubMed] [Google Scholar]
- Laukaitis C. M., Webb D. J., Donais K., Horwitz A. F. (2001). Differential dynamics of α5 integrin, paxillin, and α-actinin during formation and disassembly of adhesions in migrating cells. J. Cell Biol. 153, 1427–1440 10.1083/jcb.153.7.1427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawson M. A., Maxfield F. R. (1995). Ca2+- and calcineurin-dependent recycling of an integrin to the front of migrating neutrophils. Nature 377, 75–79 10.1038/377075a0 [DOI] [PubMed] [Google Scholar]
- Leitinger B., Hogg N. (2002). The involvement of lipid rafts in the regulation of integrin function. J. Cell Sci. 115, 963–972. [DOI] [PubMed] [Google Scholar]
- Ley K., Laudanna C., Cybulsky M. I., Nourshargh S. (2007). Getting to the site of inflammation: the leukocyte adhesion cascade updated. Nat. Rev. Immunol. 7, 678–689 10.1038/nri2156 [DOI] [PubMed] [Google Scholar]
- McLeod S. J., Li A. H. Y., Lee R. L., Burgess A. E., Gold M. R. (2002). The Rap GTPases regulate B cell migration toward the chemokine stromal cell-derived factor-1 (CXCL12): potential role for Rap2 in promoting B cell migration. J. Immunol. 169, 1365–1371. [DOI] [PubMed] [Google Scholar]
- Miertzschke M., Stanley P., Bunney T. D., Rodrigues–Lima F., Hogg N., Katan M. (2007). Characterization of interactions of adapter protein RAPL/Nore1B with RAP GTPases and their role in T cell migration. J. Biol. Chem. 282, 30629–30642 10.1074/jbc.M704361200 [DOI] [PubMed] [Google Scholar]
- Mor A., Wynne J. P., Ahearn I. M., Dustin M. L., Du G., Philips M. R. (2009). Phospholipase D1 regulates lymphocyte adhesion via upregulation of Rap1 at the plasma membrane. Mol. Cell. Biol. 29, 3297–3306 10.1128/MCB.00366-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pathak S. S., Blum J. S. (2000). Endocytic recycling is required for the presentation of an exogenous peptide via MHC class II molecules. Traffic 1, 561–569 10.1034/j.1600-0854.2000.010706.x [DOI] [PubMed] [Google Scholar]
- Pellinen T., Ivaska J. (2006). Integrin traffic. J. Cell Sci. 119, 3723–3731 10.1242/jcs.03216 [DOI] [PubMed] [Google Scholar]
- Pellinen T., Arjonen A., Vuoriluoto K., Kallio K., Fransen J. A., Ivaska J. (2006). Small GTPase Rab21 regulates cell adhesion and controls endosomal traffic of β1-integrins. J. Cell Biol. 173, 767–780 10.1083/jcb.200509019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierini L. M., Lawson M. A., Eddy R. J., Hendey B., Maxfield F. R. (2000). Oriented endocytic recycling of α5β1 in motile neutrophils. Blood 95, 2471–2480. [PubMed] [Google Scholar]
- Raab M., Wang H., Lu Y., Smith X., Wu Z., Strebhardt K., Ladbury J. E., Rudd C. E. (2010). T cell receptor “inside-out” pathway via signaling module SKAP1-RapL regulates T cell motility and interactions in lymph nodes. Immunity 32, 541–556 10.1016/j.immuni.2010.03.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reid P. A., Watts C. (1990). Cycling of cell-surface MHC glycoproteins through primaquine-sensitive intracellular compartments. Nature 346, 655–657 10.1038/346655a0 [DOI] [PubMed] [Google Scholar]
- Roberts T. J., Sriram V., Spence P. M., Gui M., Hayakawa K., Bacik I., Bennink J. R., Yewdell J. W., Brutkiewicz R. R. (2002). Recycling CD1d1 molecules present endogenous antigens processed in an endocytic compartment to NKT cells. J. Immunol. 168, 5409–5414. [DOI] [PubMed] [Google Scholar]
- Roy K. C., Maricic I., Khurana A., Smith T. R. F., Halder R. C., Kumar V. (2008). Involvement of secretory and endosomal compartments in presentation of an exogenous self-glycolipid to type II NKT cells. J. Immunol. 180, 2942–2950. [DOI] [PubMed] [Google Scholar]
- Smith A., Bracke M., Leitinger B., Porter J. C., Hogg N. (2003). LFA-1-induced T cell migration on ICAM-1 involves regulation of MLCK-mediated attachment and ROCK-dependent detachment. J. Cell Sci. 116, 3123–3133 10.1242/jcs.00606 [DOI] [PubMed] [Google Scholar]
- Smith A., Carrasco Y. R., Stanley P., Kieffer N., Batista F. D., Hogg N. (2005). A talin-dependent LFA-1 focal zone is formed by rapidly migrating T lymphocytes. J. Cell Biol. 170, 141–151 10.1083/jcb.200412032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Springer T. A., Dustin M. L. (2012). Integrin inside-out signaling and the immunological synapse. Curr. Opin. Cell Biol. 24, 107–115 10.1016/j.ceb.2011.10.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanley P., Smith A., McDowall A., Nicol A., Zicha D., Hogg N. (2008). Intermediate-affinity LFA-1 binds α-actinin-1 to control migration at the leading edge of the T cell. EMBO J. 27, 62–75 10.1038/sj.emboj.7601959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uechi Y., Bayarjargal M., Umikawa M., Oshiro M., Takei K., Yamashiro Y., Asato T., Endo S., Misaki R., Taguchi T.et al. (2009). Rap2 function requires palmitoylation and recycling endosome localization. Biochem. Biophys. Res. Commun. 378, 732–737 10.1016/j.bbrc.2008.11.107 [DOI] [PubMed] [Google Scholar]
- Vicente–Manzanares M., Choi C. K., Horwitz A. R. (2009). Integrins in cell migration – the actin connection. J. Cell Sci. 122, 199–206 10.1242/jcs.018564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright D. D., Sefton B. M., Kamps M. P. (1994). Oncogenic activation of the Lck protein accompanies translocation of the LCK gene in the human HSB2 T-cell leukemia. Mol. Cell. Biol. 14, 2429–2437 10.1128/MCB.14.4.2429 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.