Abstract
Xylan-1,4-β-xylosidase (β-xylosidase) hydrolyses xylo-oligomers at their non-reducing ends into individual xylose units. Recently, XylC, a β-xylosidase from Thermoanaerobacterium saccharolyticum JW/SL-YS485, was found to be structurally different from corresponding glycosyl hydrolases in the CAZy database (http://www.cazy.org/), and was subsequently classified as the first member of a novel family of glycoside hydrolases (GH120). In the present paper, we report three crystal structures of XylC in complex with Tris, xylobiose and xylose at 1.48–2.05 Å (1 Å=0.1 nm) resolution. XylC assembles into a tetramer, and each monomer comprises two distinct domains. The core domain is a right-handed parallel β-helix (residues 1–75 and 201–638) and the flanking region (residues 76–200) folds into a β-sandwich domain. The enzyme contains an open carbohydrate-binding cleft, allowing accommodation of longer xylo-oligosaccharides. On the basis of the crystal structures and in agreement with previous kinetic data, we propose that XylC cleaves the glycosidic bond by the retaining mechanism using two acidic residues Asp382 (nucleophile) and Glu405 (general acid/base). In addition to the active site, nine other xylose-binding sites were consistently observed in each of the four monomers, providing a possible reason for the high tolerance of product inhibition.
Keywords: crystal structure, glycoside hydrolase, synchrotron radiation, Thermoanaerobacterium saccharolyticum, xylan, β-xylosidase
Abbreviations: AmHyp2, Acaryochloris marina hypothetical protein; BaXylB, Bifidobacterium adolescentis XylB; BfHyp1, Butyrivibrio fibrisolvens hypothetical protein; BlXyl, Bifidobacterium longum xylosidase; GH, glycoside hydrolase; MIR, multiple isomorphous replacement; MR, molecular replacement; pNP, p-nitrophenyl; RiHyp3, Roseburia intestinalis hypothetical protein; TsXylC, Thermoanaerobacterium saccharolyticum XylC
INTRODUCTION
Xylans, the most abundant hemicellulosic heteropolysaccharides in plant cell wall, have to be efficiently decomposed into constituent sugars before they can be utilized as a biofuel substrate and other feedstock commodities [1]. The enzymatic strategies for complete destruction of xylan require a number of xylanolytic enzymes with different activities because of the structural complexity of xylan. These xylan-degrading enzymes, including endo-1,4-β-xylanases (EC 3.2.1.8), acetylxylan esterases (EC 3.1.1.72), feruloyl esterases (EC 3.1.1.73), α-L-arabinofuranosidases (EC 3.2.1.55) and α-glucuronidases (EC 3.2.1.139), act synergistically to efficiently release the substituted group, such as ferulic acid, acetic acid, arabinose and glucuronic acid, from a repeating β-1,4-linked xylose backbone to produce short xylo-oligosaccharides. Eventually, the xylo-oligosaccharides are hydrolysed by β-D-xylosidase (EC 3.2.1.37) to yield xylose units [2]. To date, 102 β-xylosidases have been characterized. There are 48, 4, 9, 33, 6, 1 and 1 β-xylosidases belonging to GH (glycoside hydrolase) family 3, 30, 39, 43, 52, 54 and 116 respectively (see CAZy, the carbohydrate-active enzymes database, at http://www.cazy.org/Glycoside-Hydrolases.html, accessed July 2012) [3]. Nowadays, most β-xylosidases suitable for industrial application and processes are thermophilic enzymes because of their unique thermostability features. Nevertheless, only a few β-xylosidases from anaerobic thermophilic bacteria have been reported [4].
Thermoanaerobacterium saccharolyticum JW/SL-YS485 is a highly xylanolytic anaerobic thermophile that grows within a broad pH range (3.85–6.35) and over a broad temperature range (30–66°C), and can use xylan as the sole carbon and energy source. Several hemicellulolytic enzymes from this bacterial species have been purified and characterized, including one large and cell-associated endoxylanase [5], one glucuronidase [6], two acetylxylan esterases [7] and three xylosidases (XylA, XylB and XylC) [4,8]. However, XylA cannot be classified to any GH family as the gene encoding XylA has yet to be sequenced and cloned [4]. XylB is composed of a 500-residue mature protein and shows 91% and 35% protein sequence identity with XynB and XylB from T. saccharolyticum and Caldicellosiruptor saccharolyticus respectively [8]. XylB was assigned to the GH39 family. XylC has drawn much attention since its discovery because of several aspects. First, no similar xylosidase sequences has been identified, whereas BlastP searches of the XylC protein sequence against the non-redundant protein sequences database have suggested that its novel structural features indicate that it is a new glycosyl hydrolase. Thus XylC has been classified recently as the first member of the new glycohydrolase family GH120 [4]. Secondly, XylC appears to be less sensitive to product inhibition by xylose and may well be functional in the presence of high product concentrations that are usually found in industrial applications. To investigate the substrate specificity of XylC, a series of oligosaccharides were tested for the enzyme's activity, including xylobiose, xylotriose, pNP-xyloside (pNP is p-nitrophenyl), pNP-α-L-arabinofuranoside, pNP-α-D-xylopyranoside, pNP-α-D-glucopyranoside, oat spelt xylan, birchwood xylan and carboxymethyl cellulose. The results indicate that only xylobiose, xylotriose and pNP-xyloside are preferred substrates [4]. As mentioned above, XylC showed low product inhibition, with approximately 70% of XylC activity retained at a xylose concentration of 200 mM by using the artificial substrate pNP-xyloside [4]. Taken together, these properties render XylC a potent tool for industrial applications. Solving the crystal structure not only will reveal the catalytic mechanism of XylC, but also can provide a basis for the rational engineering of this novel GH120 family enzyme.
MATERIALS AND METHODS
Protein expression, purification, crystallization and data collection
The expression and purification methods employed for the protein have been described previously [4,9]. The wild-type XylC crystals were grown in 0.2 M sodium citrate (pH 5.6) and 15–17% (w/v) poly(ethylene glycol) 3350 [9]. To prepare heavy-atom derivatized crystals, 15 different mercury compounds of the Heavy Atom Screen Hg kit (Hampton Research) were dissolved (final concentration 2 mM) in the cryoprotectant solution [0.2 M sodium citrate (pH 5.6), 18% (w/v) poly(ethylene glycol) 3350 and 15% (w/v) glycerol]. The crystals were soaked in the mercury-containing cryoprotectant for at least 1 h. Seven mercury datasets were collected for solving phase using the MIR (multiple isomorphous replacement) method. The XylC–xylose and XylC–xylobiose crystals were obtained by soaking the wild-type crystals with the cryoprotectant solution which contained 200 mM xylose and 50 mM xylobiose respectively.
The X-ray diffraction datasets from the mercury-derivatized crystals and three complex crystals (with Tris, xylose and xylobiose), were collected to 1.48–2.5 Å (1 Å=0.1 nm) resolution, at the beamline BL13B1 of the National Synchrotron Radiation Research Center (NSRRC, Taiwan) and at the beamline BL17U of the Shanghai Synchrotron Radiation Facility (SSRF, Shanghai, China). The data were processed using the HKL2000 program [10]. Before structural refinements, 5% randomly selected reflections were set aside for calculating Rfree as a monitor [11].
Structural determination and refinement
The XylC crystal structure was solved using a modified MIR method, which we have used in solving other structures [12–15]. Basically, the crystals were soaked with several mercury-containing compounds which can bind to cysteine residues and they turned out to be sufficient in providing good phasing power. The MIR datasets of mercury-containing derivatives were collected at wavelengths of 1.0000 Å (BL13B1) and 0.9793 Å (BL17U). When any four of the seven mercury datasets were combined with the native dataset for phase calculation, up to 2213 amino acids (total 2552 amino acids) were auto-built with the FOM (figure of merit) ranging from 0.19 to 0.41 and the Z-scores from 6.32 to 61.53 using SOLVE and RESOLVE [16–19]. The best results were obtained using the datasets of mersalyl acid, mercury acetate, phenylmercury acetate and tetrakis(acetoxymercuri)methane.
The electron density was improved further and the polypeptide tracings became continuous after preliminary refinement using Refmac5 [20], and a complete model with virtually all side chains was built using ARP/wARP [21]. The complex structures (XylC–Tris, XylC–xylobiose and XylC–xylose) were determined by using the MR (molecular replacement) method with PHASER [22]. The 2Fo−Fc difference Fourier map showed clear electron densities for most amino acid residues. Subsequent refinements by incorporating ligands and water molecules were according to a 1.0σ map level. The structural refinements were finalized using Coot [23] and CNS [24]. Some data collection and refinement statistics of these crystals are summarized in Table 1 and Supplementary Table S1 http://www.BiochemJ.org/bj/448/bj4480401add.htm. All Figures were prepared using PyMOL (http://www.pymol.org).
Table 1. Summary of data processing and refinement statistics.
Values in parentheses are for the highest resolution shell. Rmerge=ΣhklΣi|Ii(hkl)−<I(hkl)>|ΣhklΣiIi(hkl).
| Parameter | XylC–Tris (PDB code 3VST) | XylC–xylobiose (PDB code 3VSU) | XylC–xylose (PDB code 3VSV) |
|---|---|---|---|
| Data collection | |||
| Wavelength (Å) | 1.00 | 1.00 | 1.00 |
| Resolution (Å) | 25–1.75 (1.81–1.75) | 25–2.05 (2.12–2.05) | 25–1.48 (1.53–1.48) |
| Space group | P21 | ||
| Unit cell | |||
| a/b/c (Å) | 88.64/202.24/99.92 | 88.68/202.31/100.40 | 88.78/201.95/100.11 |
| β (o) | 99.186 | 99.141 | 99.177 |
| Number of reflections | |||
| Measured | 1471142 (145181) | 902385 (68612) | 2097975 (200889) |
| Unique | 346368 (34567) | 212277 (19059) | 574432 (57397) |
| Completeness (%) | 99.8 (99.9) | 98.2 (88.4) | 100.0 (100.0) |
| Rmerge (%) | 6.2 (37.5) | 14.0 (49.0) | 7.1 (49.6) |
| Mean I/σ(I) | 19.1 (2.5) | 8.5 (1.6) | 20.9 (3.3) |
| Multiplicity | 4.2 (4.2) | 4.3 (3.6) | 3.7 (3.5) |
| Refinement | |||
| Number of reflections used | 332107 (30562) | 197345 (14891) | 548128 (49577) |
| Rwork (%) | 15.1 (21.6) | 15.3 (24.0) | 15.0 (21.7) |
| Rfree (%) | 17.6 (23.7) | 19.4 (27.6) | 16.8 (22.8) |
| Geometry deviations | |||
| Bond lengths (Å) | 0.015 | 0.015 | 0.015 |
| Bond angles (o) | 1.77 | 1.78 | 1.74 |
| Number of atoms/mean B-values (Å2) | |||
| Protein atoms | 20588/16.6 | 20588/17.3 | 20588/13.7 |
| Tris atoms | 32/13.6 | – | – |
| Xylose atoms | – | – | 350/34.1 |
| Xylobiose atoms | – | 76/38.3 | – |
| Water molecules | 3164/32.9 | 2518/32.8 | 3614/34.0 |
| Ramachandran plot (%) | |||
| Most favoured | 86.9 | 86.2 | 87.4 |
| Additionally allowed | 12.7 | 13.4 | 12.3 |
| Disallowed | 0.4 | 0.4 | 0.4 |
Mutant production and specific activity measurement
Site-directed mutagenesis was performed by inverse PCR and circulation of PCR products using the wild-type xylosidase gene in the expression plasmid pHsh-xylC [4]. Xylosidase activity is given in units, the amount of enzyme that releases 1 μmol of product per min, and the specific activity is defined as the number of enzyme units per mg of protein. Protein concentrations were determined by absorbance at 280 nm, and an absorption coefficient of 124025 M−1·cm−1 for XylC was obtained by the ProtParam tool from the Expasy server (http://web.expasy.org/protparam/). The β-xylosidase activity was determined by assaying the amount of p-nitrophenol released from the artificial substrate pNP-xyloside as described previously [4]. The reaction mixture contained 10 μl of properly diluted enzyme (approximately 0.01 unit of XylC and its mutants), 180 μl of 0.1 M potassium phthalate buffer (pH 6.0) and 10 μl of 40 mM pNP-β-D-xylopyranoside. After incubation at 65°C for 5 min, the reaction was stopped by adding 0.6 ml of 1 M Na2CO3, and the A405 was read. A standard curve was prepared using p-nitrophenol.
RESULTS AND DISCUSSION
The overall structure
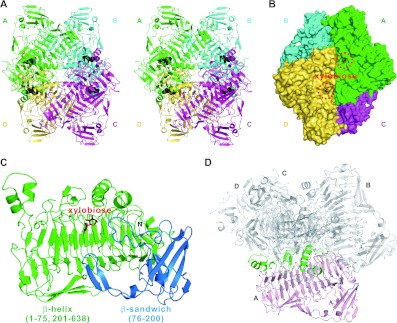
There are four XylC monomers in an asymmetric unit, forming a tetramer. The contact interface encompasses six protein segments, including residues 53–59, 177–179, 276–305, 449–466, 509–519 and 584–596 (Figures 1A and 1D), that buries a total surface area of 4330 Å2 on each monomer. A PDBePISA analysis shows a CSS (Complex Formation Significance Score) (which ranges from 0 to 1) of 0.978, also indicating that the tetramer formation is not a result of crystal packing [25].
Figure 1. The overall structure of XylC—xylobiose.
(A) Stereo view of the tetrameric XylC–xylobiose structure. The four monomers are depicted in green, cyan, magenta and yellow; the xylobiose molecules are shown in black sphere representation. (B) Surface representation of XylC. (C) Two different domains (parallel β-helix in green and β-sandwich domain in blue) of a XylC monomer. (D) The regions from the parallel β-helix domain in green (residues 53–59, 276–305, 449–466, 509–519 and 584–596) and one small loop from the β-sandwich domain in blue (residues 177–179) are involved in the tetramer interactions.
We present three XylC complex structures (XylC–Tris, XylC–xylobiose and XylC–xylose). Initially, a Tris ion was observed in each monomer in the ‘native’ structure, occupying the active site. It appeared to mimic the substrate/product oligosaccharide as seen in a previous case of β-glucanase [26]. To obtain an apo-form structure, the Tris buffer was changed to either Hepes or citrate buffer. However, a Hepes or citrate ion still bound to the same site as Tris did (results not shown). The protein molecules in XylC–xylobiose and XylC–xylose complexes, solved by MR using XylC–Tris as a search model, also adopt the closed conformation as does XylC–Tris, and all three structures superimpose very well, with RMSDs (root mean square deviations) ranging from 0.108 to 0.145 Å for all Cα atoms.
XylC, like the other β-xylosidases (e.g. in the GH39 and GH43 families), has an open oligosaccharide-binding cleft (Figure 1B). The space of that active site seems to allow only two xylose units to enter and bind (Supplementary Figure S1 at http://www.BiochemJ.org/bj/448/bj4480401add.htm). Xylo-oligosaccharides longer than two sugar units will be exposed to the bulk solvent that surrounds the enzyme. The xylobiose-binding site constitutes the −1 and +1 subsites, which appear to be necessary and sufficient for XylC catalysis. The kinetic results also indicate that longer substrates (e.g. xylotriose) did not have a lower Km (results not shown).
The crystal structures show that the XylC protein is folded into two domains (Figure 1C). The major domain is a long parallel β-helix (shown in green) that comprises residues 1–75 and 201–636. This β-helix domain contains six β-helix repeats, and it is surrounded by seven α-helices and five additional β-strands. According to the original definition of a β-helical structure, the first five and the last one repeats are not complete because these repeats at the N- and C-terminus respectively all lack the PB1 strands [27].
The second domain of XylC is a β-sandwich (coloured blue; residues 76–200) composed of two antiparallel β-sheets, a β-hairpin and three short α-helices (Figure 1C). Interestingly, this β-sandwich domain shows an Ig-like fold according to the structural homology search using DALI [28], having the highest Z-score of 3.8 (a Z-score above 2 means significant similarities and these two structures usually correspond to similar folds). These two domains are both involved in the active-site formation and provide interactions for substrate binding.
Some novel features by structural comparison
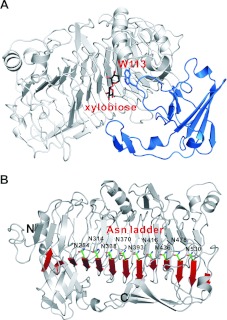
As mentioned above, XylC is composed of two domains. When XylC is superimposed with other related parallel β-helix proteins (PelC from Erwinia chrysanthemi and RGases from Aspergillus aculeatus), an extra β-sandwich domain in XylC becomes prominent (Supplementary Figures S2A and S2B at http://www.BiochemJ.org/bj/448/bj4480401add.htm). Interestingly, this β-sandwich domain is not involved in any protein oligomerization interaction and it is located in a distal side of the enzyme (coloured blue in Supplementary Figure S2C). Nevertheless, this domain is indispensable for the enzyme activity, because it contains the −1 sugar-stacking residue Trp113, located in a loop here (Figure 2A).
Figure 2. Structure features.
(A) The β-sandwich domain (in blue) is indispensable for XylC, because Trp113 in the loop is important for substrate binding by providing stacking interaction with the xylose unit at the subsite −1. (B) XylC has the longest asparagine ladder (nine asparagine residues) of all asparagine ladders observed from parallel β-helix structures solved to date.
As a common feature of the right-handed parallel β-helix domain, the internal amino acids with hydrophobic and polar groups constitute the aliphatic, aromatic and polar stacks within the inner core that maintain the structural stability [29]. Inside the internal of β-helix, an asparagine ladder formed by three or four aspargine residues was often observed in some other structures [27,30,31]. The asparagine ladder is characteristic of parallel β-helix structure, but is not consistently seen in all proteins with this fold. Interestingly, here, the XylC structure contains a much longer asparagine ladder (of nine asparagine residues: Asn254, Asn314, Asn338, Asn370, Asn393, Asn416, Asn436, Asn478 and Asn530) than all others that have been reported (Figure 2B).
To understand further whether all GH120 xylosidase enzymes have a longer asparagine ladder or not, sequence alignments of some GH120 enzymes have been carried out. There are 33 proteins that have now been classified to GH120 from the CAZy website. None of these proteins has been characterized, except for T. saccharolyticum XylC (TsXylC) and Bifidobacterium adolescentis XylB (BaXylB) proteins. The sequences of TsXylC, BaXylB, Bifidobacterium longum xylosidase (BlXyl), Butyrivibrio fibrisolvens hypothetical protein (BfHyp1), Acaryochloris marina hypothetical protein (AmHyp2) and Roseburia intestinalis hypothetical protein (RiHyp3), which all belong to the GH120 family, were aligned. TsXylC shares 48%, 48%, 40%, 30% and 26% protein identity with BaXylB, BlXyl, BfHyp1, AmHyp2 and RiHyp3 respectively (Supplementary Figure S3 at http://www.BiochemJ.org/bj/448/bj4480401add.htm). Interestingly, there are indeed longer asparagine ladders (from three to nine asparagine residues) observed among these protein sequences.
XylC shows a higher optimal temperature (65°C) than for other β-xylosidases in the GH3, GH39 and GH43 families (25–45°C) [32]. Recently, a new GH120 β-xylosidase B (XylB) having 48% sequence identity with XylC was purified from B. adolescentis, and it was found to have an optimal temperature of 60°C [33]. The higher thermostability of XylC and BaXylB (with eight asparagine ladder residues) might be attributed not only to the larger β-helix domain, but also to the extensive tetramer interface. The longer asparagine ladder may provide intrinsic interactions to stabilize the β-helix fold and increase the thermostability. Because only two GH120 β-xylosidase enzymes have been characterized to date, more studies are warranted to understand the structure–function relationship in the future.
Active-site interactions
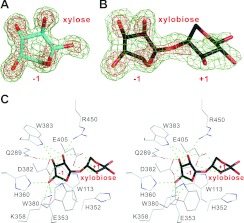
The electron-density maps of the Tris ion and the bound xylobiose and xylose, are all very clear in the −1 subsite (Figures 3A and 3B and Supplementary Figure S4A at http://www.BiochemJ.org/bj/448/bj4480401add.htm). The Tris ion superimposes very well with the xylose unit bound to the −1 subsite and is stabilized by nine and two hydrogen bonds between the Tris ion and seven protein residues, and the Tris ion and two water molecules respectively (Supplementary Figures S4B and S4C). However, the xylose unit of xylobiose in the +1 site is not as clearly seen as in the −1 site because of fewer interactions with the enzyme (Figures 3B and 3C).
Figure 3. Electron-density maps and the active site.
The 2Fo−Fc electron-density maps of xylose (A) and xylobiose (B) are contoured at 1.0σ and 2.5σ levels (in green and red respectively). (C) Stereo view of the detailed interaction networks of XylC active-site residues with xylobiose. There are 14 hydrogen bonds (green broken lines) between the active-site residues with the xylose unit at the −1 subsite, but only two hydrogen bonds with the xylose unit at the +1 subsite.
From the XylC–xylobiose structure, there are 14 hydrogen bonds formed between the −1 xylose unit and eight amino acid residues, Trp113, Gln289, Glu353, Lys358, His360, Asp382, Trp383 and Arg450. In contrast, only two hydrogen bonds between the +1 xylose unit and Glu405 are observed (Figure 3C). Besides, Trp113 and His352 provide stacking interactions with the −1 and +1 xylopyranose rings respectively.
To elucidate further the importance of active-site residues, a few amino acids involved in binding xylobiose were mutated and the specific activities were measured (Table 2). The measurement was performed with pNP-xylose as an artificial substrate that structurally mimics xylobiose, whereas the hydrolysed product of p-nitrophenol (and xylose) is easier to monitor. The results show that the residues involved in the hydrogen bonds with xylobiose are important for catalysis. The mutants E353A, K358A, W380A, D382A, W383A and E405A lost activity completely, but H360A and R450A retained 23.3% and 37.3% activity respectively (Table 2). The stacking interactions with the xylopyranose rings are also investigated. The mutant W113A lost all of its enzymatic activity, but the activity was restored when the tryptophan residue was replaced with an alternative aromatic residue (43.8% activity for W113Y and 97.6% for W113F). This result indicates that the stacking interaction between Trp113 and the −1 xylopyranose ring is essential for substrate binding. Regarding the stacking interaction of H352 with the +1 xylopyranose ring, the mutant H352A retained 30.5% activity (Table 2). We tried to obtain XylC complex structures with longer substrates (e.g. xylotriose) by co-crystallization and soaking methods, but it turned out that only xylobiose density was observed in the active site (results not shown). It is possible that the xylotriose had been hydrolysed to xylobiose. Another, perhaps more likely, possibility is that the third xylose unit was too flexible to be observed, because of the lack of specific interaction for this sugar.
Table 2. Activities of wild-type and mutant XylC.
ND, not detectable.
| Name | Activity (units/mg) | Relative activity (%) |
|---|---|---|
| Wild-type | 24.9 | 100 |
| W113A | ND | – |
| W113Y | 10.9 | 43.8 |
| W113F | 24.3 | 97.6 |
| H352A | 7.6 | 30.5 |
| E353A | ND | – |
| K358A | ND | – |
| H360A | 5.8 | 23.3 |
| W380A | ND | – |
| D382A | ND | – |
| W383A | ND | – |
| E405A | ND | – |
| R450A | 9.3 | 37.3 |
Possible mechanisms for catalysis and low product inhibition
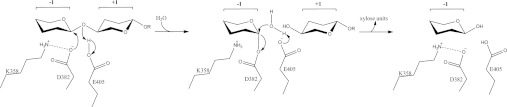
All β-xylosidases characterized hydrolyse xylo-oligosaccharides by using two carboxylate residues, such as aspartate and glutamate (GH3 and GH43), glutamate and glutamate (GH30 and GH39), and glutamate and aspartate (GH52 and GH116) as nucleophile and general acid respectively. They are classified according to the two-step retaining and the single-step inverting mechanisms (http://www.cazy.org/Glycoside-Hydrolases.html). At present, only seven β-xylosidase crystal structures of two GH families (GH39 and GH42) were determined and the PDB codes of the structures are 1W91, 2BFG, 2BS9, 1PX8 and 1UHV (GH39), and 1YRZ and 1YIF (GH43). It is not feasible to propose the catalytic mechanism comparing the XylC structure with the available β-xylosidase structures because the active sites are significantly different between GH39 [(β/α)8], GH43 (5-fold β-propeller) and GH120 (right-handed parallel β-helix). In XylC, the side chains of Glu353, Asp382 and Glu405 are all located within an appropriate distance from the xylobiose. However, further analysis shows that Glu353 is too far from the anomeric carbon of the −1 xylose (C1B) and the bridging oxygen to the +1 xylose (O4A). Thus Glu353 is probably not a catalytic residue, but only involved in substrate binding. On the other hand, Asp382 is more likely to serve as the nucleophile because it is properly oriented toward the −1 xylose, with the oxygen atoms at 3.1 Å and 2.8 Å from the C1 (C1B) and O2 (O2B) atoms. In addition, hydrogen bonding to the adjacent Lys358 side chain ensures that the carboxy oxygen near the C1 atom is negatively charged and ready for nucleophilic attack. After the nucleophilic attack, an acylxylose intermediate is formed, which in turn is hydrolysed by an incoming water molecule, supposed to take over the original place of the O4 (O4A) atom of the +1 sugar. In this step, Glu405 probably serves as a general acid to subtract a proton from the water. Moreover, the short distance between the Asp382 and Glu405 of 4.9 Å also suggests that XylC cleaves the glycosidic bond via the retaining pathway rather than the inverting mechanism (Scheme 1). This retaining mechanism is also supported by the results of stereochemical analysis, which showed that XylC produced β-xylose from β-xylopyranosides [4]. From the sequence alignment results, the active-site residues are highly conserved among all these sequences, especially for the two proposed catalytic residues (Asp382 and Glu405) (Supplementary Figure S3).
Scheme 1. The proposed catalytic mechanism of XylC.
R=xylose.
β-Xylosidase is an important enzyme in the final steps of xylan hydrolysis to xylose. Most of the xylosidases known to date exhibit a strong product inhibition by xylose, although it is not so strong for a few recently discovered β-xylosidases from fungi and bacteria, which might possibly be beneficial for industrial use. Examples are the fungal β-xylosidases from Scytalidium thermophilum (0% inhibition at 200 mM xylose), Humicola grisea var. thermoidea (0% inhibition at 10 mM) and Aspergillus nidulans (44% inhibition at 25 mM) [4]. On the other hand, bacterial β-xylosidases usually exhibit lower Ki values ranging from 2 to 10 mM xylose [34]. Interestingly, with 70% activity retained in the presence of 200 mM xylose, XylC has by far the highest xylose tolerance among all the known bacterial β-xylosidases [4].
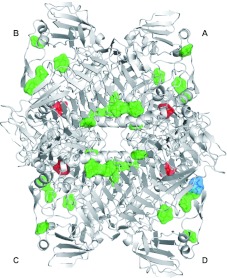
The XylC–xylose complex crystal (with 200 mM xylose) shows, in addition to the active-site bound xylose molecules (coloured red), 31 other molecules (six observed at the dimer interface) of bound xylose (coloured green), which correspond to nine binding sites consistently present in each monomer (plus one bound to a different site in monomer D, coloured blue) (Figure 4). Despite the diverse interactions in the different sites (Supplementary Figure S5 at http://www.BiochemJ.org/bj/448/bj4480401add.htm), these well-defined symmetrically distributed binding sites in the XylC tetramer are probably specific for xylose. These findings suggest a working hypothesis for future investigation, that the presence of ‘allosteric’ xylose-binding sites on XylC may attract free xylose to reduce the local product concentration and it can be related to the observed high tolerance against product inhibition. This hypothesis, however, still needs to be verified by further studies.
Figure 4. Xylose-binding sites from the XylC–xylose structure.
Four xyloses in the active site are shown in red and 31 reproducible xyloses bound to the other additional sites are shown in green. The non-specific site is shown in blue.
Online data
ACKNOWLEDGEMENTS
The synchrotron data collection was conducted at beamline BL13B1 of NSRRC (National Synchrotron Radiation Research Center, Taiwan) supported by the National Science Council of Taiwan, and at beamline BL17U of the Shanghai Synchrotron Radiation Facility (SSRF, Shanghai, China).
FUNDING
This work was supported by the National High Technology Research and Development Program of China [grant number 2012AA022200 (to R.-T.G.)], the National Basic Research Program of China [grant number 2011CB710800 (to R.-T.G.)] and the National Natural Science Foundation of China [grant number 30970062 (to W.S.)].
References
- 1.Saha B. C. Hemicellulose bioconversion. J. Ind. Microbiol. Biotechnol. 2003;30:279–291. doi: 10.1007/s10295-003-0049-x. [DOI] [PubMed] [Google Scholar]
- 2.Biely P. Microbial xylanolytic systems. Trends Biotechnol. 1985;3:286–290. [Google Scholar]
- 3.Cantarel B. L., Coutinho P. M., Rancurel C., Bernard T., Lombard V., Henrissat B. The Carbohydrate-Active EnZymes database (CAZy): an expert resource for glycogenomics. Nucleic Acids Res. 2009;37:D233–D238. doi: 10.1093/nar/gkn663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shao W., Xue Y., Wu A., Kataeva I., Pei J., Wu H., Wiegel J. Characterization of a novel β-xylosidase, XylC, from Thermoanaerobacterium saccharolyticum JW/SL-YS485. Appl. Environ. Microbiol. 2011;77:719–726. doi: 10.1128/AEM.01511-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shao W., Deblois S., Wiegel J. A high-molecular-weight, cell-associated xylanase isolated from exponentially growing Thermoanaerobacterium sp. strain JW/SL-YS485. Appl. Environ. Microbiol. 1995;61:937–940. doi: 10.1128/aem.61.3.937-940.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shao W., Obi S., Puls J., Wiegel J. Purification and characterization of the α-glucuronidase from Thermoanaerobacterium sp. strain JW/SL-YS485, an important enzyme for the utilization of substituted xylans. Appl. Environ. Microbiol. 1995;61:1077–1081. doi: 10.1128/aem.61.3.1077-1081.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shao W., Wiegel J. Purification and characterization of two thermostable acetyl xylan esterases from Thermoanaerobacterium sp. strain JW/SL-YS485. Appl. Environ. Microbiol. 1995;61:729–733. doi: 10.1128/aem.61.2.729-733.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lorenz W. W., Wiegel J. Isolation, analysis, and expression of two genes from Thermoanaerobacterium sp. strain JW/SL YS485: a β-xylosidase and a novel acetyl xylan esterase with cephalosporin C deacetylase activity. J. Bacteriol. 1997;179:5436–5441. doi: 10.1128/jb.179.17.5436-5441.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu W., Sun Y., Ko T. P., Wiegel J., Shao W., Lu F., Guo R. T., Huang C. H. Crystallization and preliminary X-ray diffraction analysis of a novel GH120 β-xylosidase (XylC) from Thermoanaerobacterium saccharolyticum JW/SL-YS485. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 2012;68:914–916. doi: 10.1107/S1744309112025328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Otwinowski Z., Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 11.Brunger A. T. Assessment of phase accuracy by cross validation: the free R value: methods and applications. Acta Crystallogr. Sect. D Biol. Crystallogr. 1993;49:24–36. doi: 10.1107/S0907444992007352. [DOI] [PubMed] [Google Scholar]
- 12.Guo R. T., Kuo C. J., Chou C. C., Ko T. P., Shr H. L., Liang P. H., Wang A. H. Crystal structure of octaprenyl pyrophosphate synthase from hyperthermophilic Thermotoga maritima and mechanism of product chain length determination. J. Biol. Chem. 2004;279:4903–4912. doi: 10.1074/jbc.M310161200. [DOI] [PubMed] [Google Scholar]
- 13.Sun H. Y., Ko T. P., Kuo C. J., Guo R. T., Chou C. C., Liang P. H., Wang A. H. Homodimeric hexaprenyl pyrophosphate synthase from the thermoacidophilic crenarchaeon Sulfolobus solfataricus displays asymmetric subunit structures. J. Bacteriol. 2005;187:8137–8148. doi: 10.1128/JB.187.23.8137-8148.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cheng Y. S., Ko T. P., Wu T. H., Ma Y., Huang C. H., Lai H. L., Wang A. H., Liu J. R., Guo R. T. Crystal structure and substrate-binding mode of cellulase 12A from Thermotoga maritima. Proteins. 2011;79:1193–1204. doi: 10.1002/prot.22953. [DOI] [PubMed] [Google Scholar]
- 15.Ren F., Ko T. P., Feng X., Huang C. H., Chan H. C., Hu Y., Wang K., Ma Y., Liang P. H., Wang A. H., et al. Insights into the mechanism of the antibiotic-synthesizing enzyme MoeO5 from crystal structures of different complexes. Angew. Chem. Int. Ed. 2012;51:4157–4160. doi: 10.1002/anie.201108002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Terwilliger T. C., Berendzen J. Automated MAD and MIR structure solution. Acta Crystallogr. Sect. D Biol. Crystallogr. 1999;55:849–861. doi: 10.1107/S0907444999000839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Terwilliger T. C. Maximum-likelihood density modification. Acta Crystallogr. Sect. D Biol. Crystallogr. 2000;56:965–972. doi: 10.1107/S0907444900005072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Terwilliger T. C. Automated main-chain model building by template matching and iterative fragment extension. Acta Crystallogr. Sect. D Biol. Crystallogr. 2003;59:38–44. doi: 10.1107/S0907444902018036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Terwilliger T. C. SOLVE and RESOLVE: automated structure solution and density modification. Methods Enzymol. 2003;374:22–37. doi: 10.1016/S0076-6879(03)74002-6. [DOI] [PubMed] [Google Scholar]
- 20.Murshudov G. N., Vagin A. A., Dodson E. J. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. Sect. D Biol. Crystallogr. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 21.Perrakis A., Morris R., Lamzin V. S. Automated protein model building combined with iterative structure refinement. Nat. Struct. Biol. 1999;6:458–463. doi: 10.1038/8263. [DOI] [PubMed] [Google Scholar]
- 22.McCoy A. J., Grosse-Kunstleve R. W., Adams P. D., Winn M. D., Storoni L. C., Read R. J. Phaser crystallographic software. J. Appl. Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Emsley P., Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr. Sect. D Biol. Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 24.Brunger A. T., Adams P. D., Clore G. M., DeLano W. L., Gros P., Grosse-Kunstleve R. W., Jiang J. S., Kuszewski J., Nilges M., Pannu N. S., et al. Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. Sect. D Biol. Crystallogr. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 25.Krissinel E., Henrick K. Inference of macromolecular assemblies from crystalline state. J. Mol. Biol. 2007;372:774–797. doi: 10.1016/j.jmb.2007.05.022. [DOI] [PubMed] [Google Scholar]
- 26.Huang J. W., Cheng Y. S., Ko T. P., Lin C. Y., Lai H. L., Chen C. C., Ma Y., Zheng Y., Huang C. H., Zou P., et al. Rational design to improve thermostability and specific activity of the truncated Fibrobacter succinogenes 1,3-1,4-β-D-glucanase. Appl. Microbiol. Biotechnol. 2012;94:111–121. doi: 10.1007/s00253-011-3586-7. [DOI] [PubMed] [Google Scholar]
- 27.Yoder M. D., Lietzke S. E., Jurnak F. Unusual structural features in the parallel β-helix in pectate lyases. Structure. 1993;1:241–251. doi: 10.1016/0969-2126(93)90013-7. [DOI] [PubMed] [Google Scholar]
- 28.Dietmann S., Park J., Notredame C., Heger A., Lappe M., Holm L. A fully automatic evolutionary classification of protein folds: Dali Domain Dictionary version 3. Nucleic Acids Res. 2001;29:55–57. doi: 10.1093/nar/29.1.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jenkins J., Pickersgill R. The architecture of parallel β-helices and related folds. Prog. Biophys. Mol. Biol. 2001;77:111–175. doi: 10.1016/s0079-6107(01)00013-x. [DOI] [PubMed] [Google Scholar]
- 30.Kobe B., Deisenhofer J. Proteins with leucine-rich repeats. Curr. Opin. Struct. Biol. 1995;5:409–416. doi: 10.1016/0959-440x(95)80105-7. [DOI] [PubMed] [Google Scholar]
- 31.Zheng Y., Huang C. H., Liu W., Ko T. P., Xue Y., Zhou C., Guo R. T., Ma Y. Crystal structure and substrate-binding mode of a novel pectate lyase from alkaliphilic Bacillus sp. N16-5. Biochem. Biophys. Res. Commun. 2012;420:269–274. doi: 10.1016/j.bbrc.2012.02.148. [DOI] [PubMed] [Google Scholar]
- 32.Jordan D. B., Wagschal K. Properties and applications of microbial β-D-xylosidases featuring the catalytically efficient enzyme from Selenomonas ruminantium. Appl. Microbiol. Biotechnol. 2010;86:1647–1658. doi: 10.1007/s00253-010-2538-y. [DOI] [PubMed] [Google Scholar]
- 33.Lagaert S., Pollet A., Delcour J. A., Lavigne R., Courtin C. M., Volckaert G. Characterization of two β-xylosidases from Bifidobacterium adolescentis and their contribution to the hydrolysis of prebiotic xylooligosaccharides. Appl. Microbiol. Biotechnol. 2011;92:1179–1185. doi: 10.1007/s00253-011-3396-y. [DOI] [PubMed] [Google Scholar]
- 34.Yan Q. J., Wang L., Jiang Z. Q., Yang S. Q., Zhu H. F., Li L. T. A xylose-tolerant β-xylosidase from Paecilomyces thermophila: characterization and its co-action with the endogenous xylanase. Bioresour. Technol. 2008;99:5402–5410. doi: 10.1016/j.biortech.2007.11.033. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.