Abstract
There are potentially several ways Mg2+ might promote formation of an RNA tertiary structure: by causing a general “collapse” of the unfolded ensemble to more compact conformations, by favoring a re-organization of structure within a domain to a form with specific tertiary contacts, and by enhancing cooperative linkages between different sets of tertiary contacts. To distinguish these different modes of action, we have studied Mg2+ interactions with the adenine riboswitch, in which a set of tertiary interactions that forms around a purine-binding pocket is thermodynamically linked to the tertiary “docking” of two hairpin loops in another part of the molecule. Each of four RNA forms with different extents of tertiary structure were characterized by small-angle x-ray scattering. The free energy of interconversion between different conformations in the absence of Mg2+ and the free energy of Mg2+ interaction with each form have been estimated, yielding a complete picture of the folding energy landscape as a function of Mg2+ concentration. At 1 mM Mg2+ (50 mM K+), the overall free energy of stabilization by Mg2+ is large, −9.8 kcal/mol, and about equally divided between its effect on RNA collapse to a partially folded structure and on organization of the binding pocket. A strong cooperative linkage between the two sets of tertiary contacts is intrinsic to the RNA. This quantitation of the effects of Mg2+ on an RNA with two distinct sets of tertiary interactions suggests ways Mg2+ may work to stabilize larger and more complex RNA structures.
The stabilization of RNA tertiary structure by low concentrations of Mg2+ ions was first described in early studies of tRNA folding (1–4). Much attention has been focused on those factors which promote strong Mg2+ interactions with native structures: a high charge density causes accumulation of Mg2+ in an “ion atmosphere” (5), and the tertiary fold of an RNA may create special environments with unusually strong ion interactions (6, 7). To understand how Mg2+ stabilizes RNA structures, it is also important to consider partially unfolded forms of the RNA, which also interact strongly with Mg2+ (8–10). Particularly intriguing is the ability of Mg2+ to cause the “collapse” of RNAs from extended forms to compact intermediates that lack fully formed tertiary contacts (11–15).
A relatively simple RNA with which to explore the varied effects of Mg2+ on the formation of both a native RNA structure and its folding intermediates is the aptamer domain of riboswitches that sense purine ligands (16–18). These RNAs have a simple topology with two distinct sets of tertiary hydrogen bonds (Figure 1A): the two hairpin loops dock with each other, and a binding pocket becomes organized around the bound purine ligand. From FRET and NMR studies of several purine riboswitch RNAs, it is known that Mg2+ favors the docked conformation in the absence of ligand (19–21). Furthermore, docking of the loops strengthens the ligand affinity of the binding pocket (22). The purine riboswitch is thus a particularly simple case in which Mg2+ - dependent formation of a compact but not fully native intermediate is coupled to formation of a core functional tertiary structure.
Figure 1.
The adenine riboswitch aptamer domain (A-riboswitch) used in these studies. A, Secondary structure of the RNA. The sequence is that of the add switch (18) with modification of the 5'-3' base pairs to aid transcription (52). Arrows denote 5'-3' backbone connectivity, horizontal black bars represent canonical Watson-Crick base pairing, black dots represent non-canonical pairs, and thick red bars represent base-base tertiary interactions. The large green A denotes the ligand. Yellow boxes enclose two distinct sets of tertiary contacts. Bases in blue were mutated to disrupt the docked loops tertiary structure. B, schematic drawings of potential forms of the RNA with different combinations of the two sets of tertiary contacts: N and Idock, docked loops formed; N and ILBP, binding pocket organized around bound ligand.
In this work, we ask how Mg2+ alters the folding landscape of an adenine-specific riboswitch (A-riboswitch, Figure 1A) by measuring the free energy of Mg2+ interaction with the native structure and each of several partially folded forms that are potentially part of the equilibrium folding pathway. We use a titration method that monitors the thermodynamic activity of Mg2+ in the presence of RNA and yields the extent of charge neutralization by Mg2+ (the “excess” Mg2+) for a particular RNA conformation. An advantage of this approach for our purposes is that the effect of Mg2+ on the free energy of individual RNA forms at different positions in the folding landscape (e.g., extended, collapsed, or native conformations) can be examined over a wide range of Mg2+ concentrations (8). Together with structural characterization of the RNA forms by small angle x-ray scattering (SAXS) and hydroxyl radical probing, we have developed a quantitative picture of the folding transitions taking place in the A-riboswitch in the absence of Mg2+ (50 mM K+), and the ways these folding equilibria shift as Mg2+ is titrated. Mg2+ strongly stabilizes both the docked and binding pocket tertiary structures, and because of a large cooperative linkage between these structures (−5.3 kcal/mol) the ion may enhance the ligand binding affinity of the RNA by as much as −9.8 kcal/mol (1 mM Mg2+, 50 mM K+). In the Discussion, we note some parallels between the A-riboswitch folding intermediates and collapsed intermediates seen in larger RNAs.
Materials & Methods
Materials and solution preparation
Preparation of plasmid DNA encoding the A-riboswitch RNA, its transcription with T7 RNA polymerase, and purification of the RNA have been described (10). Plasmids encoding RNAs with the mutations C60G or G38C were prepared by the same methods. Buffers and salts were high purity (> 99%, Fluka). Standard buffer conditions included 20 mM MOPS (pH 6.8) or MES (pH 6.0) buffer adjusted to the desired pH with KOH; sufficient KCl was added to bring the total concentration of K+ to 50 mM in all experiments.
Spectroscopic measurements
RNA melting curves and isothermal titrations of RNA with MgCl2 were both monitored by UV absorption in a Cary 400 spectrophotometer as described (10). Measurement of Γ2+ using the fluorescent dye 8-hydroxyquinoline sulfonic acid (HQS) to monitor Mg2+ activity has been described in detail (10, 23). Both UV and HQS-monitored Mg2+ titrations were carried out at 20 °C. Three or four data sets were collected at each of two pH values (pH 6.0 with MES, and pH 6.8 with MOPS) for each condition. Changing pH alters the affinity of the dye for Mg2+ and allows the measurement of Γ2+ over a wider range of bulk Mg2+ concentration (C2+). No pH dependence of the stability of the A-riboswitch tertiary structure has been observed by UV melting analysis. To merge all the data sets, data collected at a single pH were first compared and the data range with the lowest scatter in Γ2+ was selected. For pH 6.8, the range was logC2+ between −7.0 and −3.7; at pH 6.0, the corresponding range was −4.0 to −2.8. All selected data were combined and fit to a sixth-order polynomial (in logC2+) with no linear term (to force a slope of zero at the x-axis) using the software proFit from Quantum Soft. The reported error bars were calculated by taking the square root of the sum of the squares of the residuals over a window of nine points.
X-ray scattering
RNA samples (at 1 – 2 mg/mL) for small-angle X-ray scattering (SAXS) measurements were extensively exchanged into standard MOPS-KCl buffer (50 mM K+) supplemented with the indicated MgCl2 and ligand concentrations. The samples were allowed to equilibrate at room temperature for an hour prior to the experiment. SAXS measurements were performed at beamlines 12-ID and 18-ID of the Advanced Photon Source (APS) at Argonne National Laboratory. The wavelength of the incident X-ray radiation was set to 1.033 Å (which provides a beam energy of 12 keV), the exposure time was set to 0.15 seconds, and samples were moved through an X-ray flow cell while in the path of the beam to minimize radiation damage. The ambient temperature of the experiment was ~29 °C. Twenty data sets were collected for each sample or buffer solution in order to obtain good statistics. Calculation of Rg and distance distribution functions from the data have been described (24). Structural models were computed from the scattering profiles using DAMMIN (25). For each model shown herein, the program DAMMIN was run in jagged mode, on scattering data up to q = 0.3 Å−1. Twenty converged solutions were computed, superimposed using SUPCOMB, and then averaged using DAMAVER (26) to obtain the final solution.
Hydroxyl radical probing
Hydroxyl radical footprinting experiments were carried out with 5' 32P-labeled RNA as described (10). Cleavage products were quantitated using the software SAFA (27).
Results
Characterization of A-riboswitch loop mutants
The goal of this study is to examine Mg2+ interactions with A-riboswitch RNAs in each of the four possible combinations of two distinct sets of tertiary structures, the docked loops and binding pocket (Figure 1A). Besides the native structure (N) and the RNA without any tertiary structure (the extended state, E), there are two intermediate RNA forms: one with docked loops but a disordered binding pocket, Idock, and another with bound ligand but undocked loops, ILBP (Figure 1B). The binding pocket is easily switched between native and disordered forms by the presence or absence of ligand, but a mutant sequence is needed to enforce an undocked loop conformation. Following work on the similar pbuE A-riboswitch (19), we prepared two variants that disrupt a base pair between the loops, C60G and G38C (Figure 1A).
All experiments reported here were carried out with a background of 50 mM K+ (the anions are Cl− and the sulfonate forms of either MOPS or MES buffer). UV-monitored isothermal titrations of the RNAs with Mg2+ in the presence of ligand (either 2, 6 diaminopurine, DAP, or adenine) showed that both mutations strongly affect the ability of the RNA to fold to the native conformation (Figure 2). [DAP is known to bind the A-riboswitch with higher affinity than adenine (17), and has been useful for studying other purine riboswitch variants with weak ligand binding affinity (22). In comparisons of wild type RNA titrated in the presence of either 82 μM adenine or 11 μM DAP (10), we found that DAP binds 34 fold more tightly over a wide range of Mg2+ concentrations.] Comparing titrations of the wild type RNA (in the presence of 82 μM adenine) and C60G RNA (82 μM DAP), the midpoint of the C60G RNA titration is shifted to higher Mg2+ concentration and the hypochromic change is less than half (Figure 2). The extent of folding of the two RNAs can be directly compared at 0.1 mM Mg2+ (Figure 2); taking into account the 34 fold tighter affinity of DAP over adenine, C60G RNA has ~570 fold weaker affinity for ligand than does wild type RNA at this Mg2+ concentration. Similar experiments with G38C RNA failed to observe any reproducible folding transition at Mg2+ concentrations up to 1 mM. A trend of increasing stability for G38C, C60G, and wild type RNAs was found in UV melting experiments in the presence of 1 mM Mg2+ and ligand, and a very weak affinity of G38C RNA for DAP (~26 mM−1) was estimated from melting experiments carried out in 10 mM Mg2+ (Figure S1 of Supporting Information). Overall, these titration and melting experiments imply that the mutation G38C severely weakens the docked loop conformation while C60G has a substantial but less drastic effect, consistent with ligand affinities reported for analogous mutations in other purine riboswitch sequences (19, 28). In subsequent sections we present evidence that C60G RNA bound to ligand is in the desired ILBP form, with native-like tertiary structure formed around the ligand but undocked hairpin loops.
Figure 2.
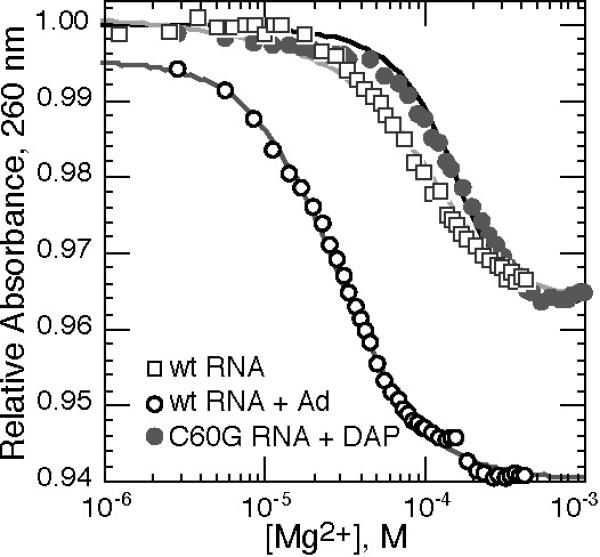
Titration of wild type A-riboswitch (open symbols) or C60G RNA (gray circles) with MgCl2 in buffer containing 50 mM K+ (20 °C), monitored by absorbance at 260 nm. Mg2+-dependent baselines have been subtracted from the data sets (see Materials and Methods). Wild type A-riboswitch was titrated in the presence of 82 μM adenine (open circles), or no ligand (open squares); the C60G RNA titration included 82 μM DAP (gray circles). Titration of the wild type RNA with ligand has been fit to the Hill equation (midpoint 29.0 μM Mg2+, n = 1.58). The other two curves are optimum fits of a Mg2+-dependent two-state equilibrium between E and Idock forms (wild type RNA) or E and ILBP forms (C60G RNA) (see Results for further explanation).
As previously noted in melting experiments with the Figure 1A A-riboswitch sequence (10), Mg2+ may induce tertiary structure formation in the absence of ligand. The Mg2+-dependence of this folding is seen by Mg2+ titration of the wild type RNA (Figure 2). We argue below that the RNA conformation adopted at 1 mM Mg2+ is the desired Idock form.
RNA dimensions from small angle X-ray scattering
We used small-angle X-ray scattering (SAXS) to monitor docking of the hairpin loops by changes in the overall solution dimensions of the RNAs. In these experiments, wild-type A-riboswitch in the presence of both ligand and Mg2+ has the same radius of gyration (Rg) as calculated from the crystal structure (PDB 1Y26, 20.3 Å). In the absence of either Mg2+ or ligand (DAP), the RNA adopts a much more extended conformation (Rg = 24.9 Å). Titration with Mg2+ in the absence of ligand gradually brings Rg close to that of the native form (Figure 3A). In contrast to the wild type RNA, C60G RNA dimensions in the absence of ligand are much less sensitive to the presence of Mg2+ (Figure 3A); even conditions that saturate C60G RNA with ligand (1 mM Mg2+, 250 μM DAP) leave the RNA in an extended conformation (Figure 3A).
Figure 3.
SAXS studies of wild type and variant A-riboswitch RNAs in standard buffer; ligand is 250 μM DAP, where present. Samples were equilibrated with the indicated bulk Mg2+ concentrations (C2+). A, Rg as a function of bulk Mg2+ concentration for wild type RNA with (blue) or without (red) ligand, or C60G RNA with (green) or without (orange) ligand. The solid red line is the best fit of a Mg2+-dependent two-state equilibrium between E and Idock forms of the RNA (see text). B, distance distribution function P(r) for RNAs in the absence of Mg2+: wild type RNA with (blue) or without (red) ligand, or C60G RNA without ligand (orange). C, P(r) for RNAs with 1 mM bulk Mg2+ concentration: wild type RNA with (blue) or without (red) ligand, and C60G RNA with (green) or without (orange) ligand. D, reconstruction from SAXS data of the wild type A-riboswitch bound to ligand in the presence of Mg2+. The RNA backbone from the corresponding crystal structure (1Y26) has been aligned with the calculated density; the bound adenine ligand is shown as a space-filling model. The two views differ by 90° rotation. E, reconstruction from SAXS data of the C60G (green) and G38C (yellow) RNAs in the absence of ligand or Mg2+. The two structures are superimposed to show their similar overall shape.
The distance distribution function derived from the scattering data, P(r), contains a more detailed description of macromolecular conformation than reported by the single parameter Rg. Comparing P(r) for wild type and C60G RNAs, it is clear that the extended forms found in the absence of Mg2+ and ligand are not identical; the wild type A-riboswitch has a slightly more compact form than C60G RNA (Figure 3B). Addition of 1 mM Mg2+ has very little effect on the P(r) function of C60G RNA in the absence of ligand (Figure 3B and C), unlike the strong effect of Mg2+ on the wild-type RNA.
Another point of consequence is that P(r) curves for wild type RNA in 1 mM Mg2+ are nearly identical whether or not ligand is bound (Figure 3C). Thus, any organization of the binding pocket that accompanies ligand binding does not affect the dimensions of the RNA in a way that is detectable by SAXS; the experiment is primarily sensitive to the disposition of the two loops.
We attempted to calculate low resolution models of the wild type and variant A-riboswitch RNAs that satisfy the constraints of the various scattering data sets (see Materials and Methods). The shape found for the wild type RNA in the presence of Mg2+ and ligand corresponds to the overall dimensions of the crystal structure (Figure 3D). Similar shapes were calculated for wild type RNA in 1 or 10 mM Mg2+ without ligand (not shown). Reconstructions of C60G and G38C RNAs in the absence of Mg2+ gave similar extended structures suggestive of undocked hairpin loops (Figure 3E). It is ambiguous which of the three arms of this T-shaped structure might be the 5'–3' terminal helix. A proposed model of ligand-free purine riboswitch based on FRET measurements (21) has the 3' 20 nucleotides of helices P1 and P3 (A64–C83) remain co-axially stacked as they are in the native crystal structure (18), and the P2 helix protruding at right angles. However, the P1–P3 helices span ~70 Å, while the extended state RNA reconstruction has a long dimension of ~100 Å; it seems unlikely that the sequence in question could be stretched enough to cover this distance. Alternatively, the 29 nucleotides of backbone extending between the P2 and P3 hairpins (A35–U63) could form a stacked structure of about the right dimensions. Although the identity of the three helical segments in the E state structure is not crucial for the present work, the nature of the three helix junction structure that enforces the extended conformation is an interesting question for further investigation.
Calculations with scattering data collected on either the wild type A-riboswitch without ligand or Mg2+, or C60G RNA saturated with ligand at 1 mM Mg2+, did not yield unique structures, a possible indication that an ensemble of disparate structures is present in solution (29).
Hydroxyl radical probing of tertiary structure in C60G RNA
To further delineate conformational differences between wild type and C60G RNAs, we measured the reactivity of the two sequences towards hydroxyl radical, a probe sensitive to ribose exposure to solvent (30). Loop nucleotides G37 and G38 were protected in the native structure of the wild type RNA (relative to unfolded RNA in the absence of both Mg2+ and ligand), but neither residue changed reactivity in response to high Mg2+ concentration and saturating ligand concentration in the C60G RNA (Figure 4). Six different residues within the binding pocket became protected in the wild-type RNA in the presence of ligand; the same residues were protected to about the same extent in C60G RNA bound to ligand (Figure 4). Disruption of the docked loops apparently does not affect the structure of the RNA around the bound ligand, at least at a level detectable by hydroxyl radical reactivity.
Figure 4.
Hydroxyl radical footprinting results for the wild-type and C60G variant A-riboswitch (data analysis was as described in Materials & Methods). Experiments were performed in standard buffer at 0 °C, with the indicated amount of Mg2+ and 250 μM DAP present in all but the unfolded control samples. The difference in normalized reactivity between the control unfolded form (RNA in the absence of Mg2+ and DAP) and the sample RNA (with DAP and Mg2+) is plotted; a positive number represents reduced reactivity in the sample RNA. Open column, wild type RNA with 5 mM Mg2+; gray – black columns, C60G RNA with increasing Mg2+ concentrations of 0.1, 0.2, 0.5 1.0, and 5.0 mM. Residue numbering is the same as Figure 1A. Relative reactivities of nucleotides 32–75 are in Supplementary Information (Figure S2).
Direct measurement of excess Mg2+
The excess Mg2+ accumulated by different conformations of the wild type and mutant RNAs was measured using a fluorescent chelator that monitors the effect of an RNA on the thermodynamic activity of Mg2+ ions (8, 23). The results are reported in terms of the interaction parameter Γ2+, which is the excess number of Mg2+ ions present in compensation for the RNA negative charge at a given level of bulk ion concentration C2+. It is essentially the same as the ions accumulated in a dialysis experiment in response to the presence of an RNA, where C2+ is the ion concentration in the solution external to the RNA dialysis solution (31). Similar interaction parameters describe the excess monovalent ion (Γ+) and excluded anion (Γ−), though they are not measured here.
The wild type RNA was titrated with Mg2+ either in the absence of ligand or in the presence of a high enough concentration of DAP that the RNA was in the native structure without Mg2+ present (10) (Figure 5A). As expected, the more compact native form accumulated more Mg2+ than the unfolded form of the RNA. The difference between these two curves, ΔΓ2+, is the uptake of Mg2+ upon folding. Excess Mg2+ for the native RNA structure is 15.5 ± 0.2 ions/RNA at 1 mM Mg2+ (Table 2, Supplementary Information), which corresponds to 43% of the RNA phosphate charge neutralized by Mg2+. The data shown here extend our previous measurements of Γ2+ with this RNA to much higher Mg2+ concentrations (10).
Figure 5.
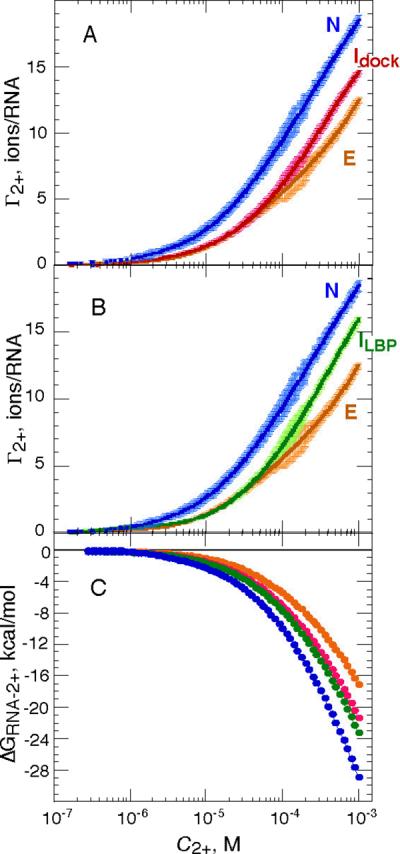
Excess Mg2+ (Γ2+) as a function of bulk Mg2+ concentration (C2+) for wild type and C60G RNAs in standard conditions (50 mM K+, 20 °C), with and without the ligand DAP present (250 μM). Six or seven independent data sets were merged to obtain each titration curve; errors are shown in the plots as lighter color bars (see Materials and Methods). A, wild type RNA in the presence (blue) or absence (magenta) of ligand. C60G RNA in the absence of ligand is shown for comparison (orange). B, C60G RNA in the presence (green) or absence (orange) of ligand. Wild type RNA in the presence of ligand is shown for comparison (blue). C, Free energies of Mg2+-RNA interaction (ΔGRNA-2+) calculated from Γ2+ curves in panels A and B (see Materials and Methods). Blue, native RNA (N); orange, C60G RNA in the absence of ligand (E); green, the Idock conformation, extrapolated from wild type RNA at 1 mM Mg2+; magenta, ILBP conformation, extrapolated from C60G RNA bound to ligand at 1 mM Mg2+.
Because the C60G RNA tertiary structure does not form tertiary structure in the absence of Mg2+, even at very high ligand concentrations, the Γ2+ curves obtained with this RNA in the presence or absence of DAP were initially identical to each other (Figure 5B) and to the titration curve of unfolded wild type RNA (Figure 5A). The two C60G RNA curves diverge significantly when C2+ exceeds ~50 μM Mg2+; based on UV-monitored titrations (Figure 2 and data not shown), the midpoint of ligand binding is expected at ~90 μM Mg2+ at the ligand concentration of the experiment. Notably, the ligand-bound form of C60G RNA, which is essentially the only form present above C2+ ≈ 0.6 mM, did not reach the same level of Γ2+ as the wild type RNA (at 1 mM Mg2+, Γ2+ was 13.4 ± 0.2 ions/RNA for C60G RNA). This observation is consistent with SAXS measurements that showed a more extended structure for ligand-bound C60G RNA (Figure 3A). In the absence of ligand, the wild type and C60G RNA Γ2+ curves matched at low Mg2+ concentrations but diverged above ~ 0.1 mM (Figure 5A), again implying a different, probably more extended structure for the mutant.
An important point is that the uptake of Mg2+ upon ligand binding (ΔΓ2+, as calculated from Figure 5 data) is about the same for C60G RNA (2.6 ± 0.3 ions/RNA) as wild type (3.2 ± 0.3 ions) when evaluated at 1 mM Mg2+ (Table S2). However, ΔΓ2+ only appears insensitive to the mutation because Mg2+ interactions with both the ligand-bound and unbound states of the RNA have been altered to comparable extents, presumably because C60G RNA loops remain undocked whether or not ligand is bound.
Free energies of Mg2+-RNA interactions
If no conformational changes take place in an RNA during its titration with Mg2+, the Γ2+ curve may be integrated to give the total free energy of Mg2+-RNA interaction (ΔGRNA-2+) as a function of bulk Mg2+ concentration (8, 31). Based on scattering measurements (Figure 3A), neither the native, ligand-bound structure (labeled N in Figure 5A) nor the ligand-free form of C60G RNA (E in Figure 5B) is affected by the presence or absence of Mg2+; ΔGN-2+ and ΔGE-2+ have been calculated from these respective Γ2+ curves (Fig 5C). The other two forms of the RNA observed at 1 mM Mg2+, Idock for wild type RNA in the absence of ligand and ILBP for C60G RNA saturated with ligand (Figure 5A and 5B), were achieved only when Mg2+ was present. We calculated ΔGRNA-2+ for these two RNA forms by noting that the ΔGN-2+ and ΔGE-2+ curves differ by a uniform factor (1.95±0.19 fold) independent of Mg2+ concentration (Figure S3, Supplementary Information). We assume that corresponding curves for the two intermediate forms will also have similar shapes and differ from ΔGN-2+ or ΔGE-2+ by a constant factor over the entire range of Mg2+ concentrations. The free energies ΔGIdock-2+ and ΔGILBP-2+ (shown in Figure 5C) have therefore been scaled from an average of the ΔGN-2+ and ΔGE-2+ curves; the interaction free energies found at 1 mM bulk Mg2+ (Table S1) were used to calculate the scaling factor.
Intrinsic free energies of formation of intermediates from extended state RNA
To fully evaluate the effects of Mg2+ on the A-riboswitch folding landscape, it would be useful to know the initial free energy differences between the four RNA forms (Figure 1B) before Mg2+ is added. Equilibria between the extended form of the RNA and the two intermediates, Idock and ILBP, strongly favor the extended form in the absence of Mg2+ and are therefore difficult to measure directly. However, using SAXS or UV absorbance, we have observed these equilibria shift towards the intermediate form as Mg2+ is titrated (Figures 2 and 3A). The Mg2+-dependence of the observed equilibrium between an extended (E) and intermediate (I) form is derived as
| (1) |
where the ΔG values are the known free energies of Mg2+-RNA interaction with either RNA form (Figure 5C) and R is the gas constant. KE-I is the Mg2+-dependent, observed equilibrium constant for interconverting the extended and intermediate forms. is the intrinsic equilibrium constant in the absence of Mg2+, and the only unknown in the equation. A two-state equilibrium between E and I is assumed; potential limitations imposed by the assumption are considered below. In deriving the equation (see Supplementary Information), the reference state is defined as our standard experimental conditions (50 mM K+) in the absence of Mg2+. The free energy associated with the conformational equilibrium in the absence of Mg2+, , is therefore a standard state free energy, ΔG°E-I. The corresponding free energies in the presence of Mg2+ are denoted by ΔGE-I, which varies with the Mg2+ concentration.
For the docking reaction, E ⇌ Idock, we compare eq (1) with the Mg2+-dependences of UV absorption (Figure 2) and Rg (Figure 3A) measurements with wild type RNA in the absence of ligand. When was systematically varied, the best fit values were 0.12 (ΔG°E-Idock = 1.3 kcal/mol) or 0.065 (ΔG°E-Idock = 1.6 kcal/mol) for the UV and SAXS data, respectively. The Mg2+-dependence of ligand binding to C60G RNA (Figure 2) corresponds to the equilibrium E ⇌ ILBP; we obtain the intrinsic equilibrium constant for the binding of DAP. Given that adenine binds the wild type RNA 34 fold more weakly than DAP, we estimate ΔG°E-ILBP = −0.7 kcal/mol as the standard state affinity of C60G RNA for adenine in the absence of Mg2+.
There is reason to think that the docking reaction, E ⇌ Idock, might not be strictly two-state: single-molecule FRET experiments with a similar A-riboswitch sequence as used here detected a population of molecules with hairpin loops in an intermediate conformation between docked and extended (19). We have therefore considered the folding pathway
in which the intermediate I' is assigned an Rg half-way between those of the E and Idock forms, and various Mg2+ interaction energies between ΔGE-2+ and ΔGIdock-2+. Although it is possible to find combinations of the intrinsic equilibrium constants for the E ⇌ I' and I' ⇌ Idock reactions that give slightly better fits to the scattering data than the two-state model (Figure 3A), the product of the two equilibrium constants is always within 10% of ; the maximum population of the I' state in these simulations was less than 10%. Although intermediate states between the fully extended and docked RNA forms may well exist, we conclude that the overall free energy estimated for the E ⇌ Idock reaction is reasonable.
Discussion
Γ2+ and Rg define four forms of the A-riboswitch RNA
To determine the ways Mg2+ might affect folding of the A-riboswitch, we used the complementary methods of X-ray scattering and excess Mg2+ ion (Γ2+) measurements to characterize the A-riboswitch – ligand complex and its various partially unfolded forms. Four species have been studied, each with a distinct combination of size (Rg) and excess Mg2+. We first summarize essential characteristics of the four RNA forms and their interconversions, and in the following section consider the different ways Mg2+ stabilizes the RNA tertiary structure and enhances ligand binding affinity.
The form of the RNA with the highest value of Γ2+ is the native structure N, the ligand complex formed with the wild type RNA (Figure 6A). The wild type RNA in the absence of ligand adopts a docked-loop conformation at 1 mM Mg2+, as anticipated from FRET and NMR studies of related purine riboswitch sequences (19–21). This form of the RNA, Idock, has similar dimensions and shape as the native form (Figure 3C), but accumulates Mg2+ to a lesser extent than the native structure (Figure 6A); the smaller Γ2+ must be due to structural differences in the binding pocket when ligand is absent.
Figure 6.
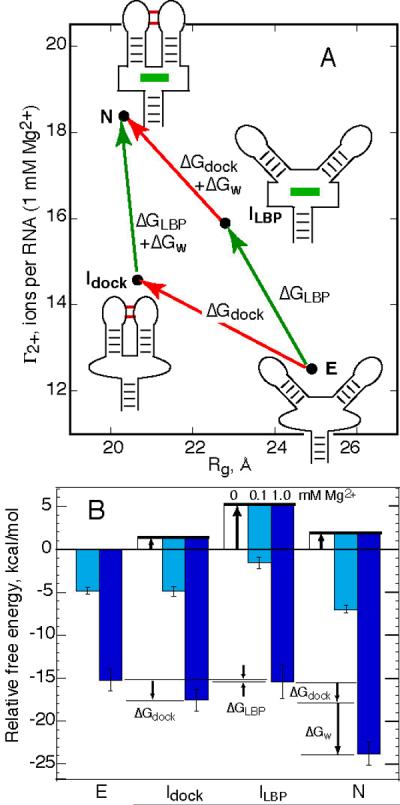
Effects of Mg2+ on the A-riboswitch folding landscape. A, four states of the A- riboswitch, distinguished by their dimensions (Rg) and excess Mg2+ (Γ2+ reported at 1 mM Mg2+). See Discussion for definitions of the four states. Arrows connecting the RNA forms define a thermodynamic cycle with three free energies, all of which are potentially Mg2+-dependent. The number of ions taken up (ΔΓ2+) in the reaction defined by each arrow is indicated. B, Stabilities of each of the RNA forms from panel A, relative to the extended form (E) in its standard state (50 mM K+ buffer without Mg2+). Stabilities of the ligand-bound RNA forms (N and ILBP) are calculated for an adenine concentration of 10 μM. Thick arrows, free energies of the two-state equilibria between E and each of the three other forms; cyan and blue bars, stabilization of each form by 0.1 or 1.0 mM bulk Mg2+ concentration, respectively. Calculations of the three ΔG values defined in panel A are illustrated for 1 mM Mg2+ (arrows labeled by ΔG). Error bars for the N and E state Mg2+-RNA interaction free energies are based on the reproducibility of the Γ2+ data that were integrated to find ΔGRNA-2+ (Figure 5); errors for the other two forms include an additional estimated uncertainty from the method used to interpolate the free energies.
The C60G riboswitch variant maintains an extended structure with dimensions that are insensitive to Mg2+ in the absence of ligand (E in Figure 6A). The convergence of reconstructions on a unique structure with plausible dimensions (Figure 3E) argues that the RNA adopts a single conformation, within the resolution of SAXS experiments. C60G RNA can be saturated with ligand at 1 mM Mg2+, but based on hydroxyl radical probing and scattering measurements (Figure 3C and Figure 4), the loops remain undocked with an average angle between helices P2 and P3 that must be intermediate between the docked and fully extended structures (ILBP in Figure 6A). The binding pocket, however, probably retains the same tertiary structure around the bound ligand as the native RNA (Figure 4).
The free energy differences between an extended E state and each of the other three forms in the absence of Mg2+ can be estimated, if two-state equilibria are assumed. (As described in Results, we use ΔG° to refer to our standard buffer conditions, 50 mM K+ without Mg2+. ΔG° for formation of the two ligand-bound states, N and ILBP, includes the standard free energy of adenine binding.) Two of these ΔG° values were calculated by extrapolating the equilibria observed over a range of Mg2+ concentrations to zero Mg2+ (see Results). We can also estimate ΔG° for a third transition, E → N. Upon melting of the wild type RNA in the presence of 11 μM DAP without Mg2+, RNA unfolding from the native, ligand-bound conformation to an equilibrium mixture of E and Idock states was observed with a Tm of 20.9 °C (31). After correcting for the small amount of Idock calculated to be present and for the 34 fold weaker binding affinity of adenine, ΔG° for E → N is estimated as −4.7 kcal/mol (20 °C).
We combine our ΔG° estimates for interconversion between different RNA forms with the Mg2+ - RNA interaction energies of each form (ΔGRNA-2+) to construct a free energy landscape for A-riboswitch folding.1 The relative free energies of the four RNA forms are shown in Figure 6B; the free energies of the two ligand-bound species have been adjusted to reflect 10 μM adenine rather than the standard state 1 M, as an order-of-magnitude approximation of the in vivo adenine concentration (32, 33). In the absence of Mg2+, the extended E state is the most stable RNA conformation; as Mg2+ is added the relative stabilities change until the ligand-bound N state becomes by far the most stable conformation.
As a caution about applying the data in Figure 6B to physiological conditions, note that the free energies determined in 1 mM Mg2+ are probably larger than those attained in vivo. The K+ activity in our standard 50 mM buffer is about three-fold lower than the K+ activity present in bacteria under typical growth conditions (34). K+ competes with Mg2+ for interactions with any RNA; thus, the higher cellular levels of K+ would shift our Γ2+ curves to a higher Mg2+concentration range. An increase in Γ2+ by one ion is coupled to a decrease in Γ+ by ~1.8 K+ ions (35, 36); therefore a three-fold increase in K+ activity should cause a ~6 fold (31.8) increase in the Mg2+ concentration needed to attain a given Γ2+ value. Estimates of the effective Mg2+concentration in vivo span 0.5 to ~2 mM (37–39); in our experiments, the same accumulation of excess Mg2+ as occurs with an RNA in vivo would be expected in the range 83 μM – 0.33 mM.
How Mg2+ affects the A-riboswitch energy landscape
Having defined four A-riboswitch conformations with different combinations of tertiary interactions, we are now able to quantitatively parse the Mg2+ - dependence of RNA folding and ligand binding into several components. We consider all four conformations in Figure 6A as structures that are accessible to the wild type sequence and capable of interconversion; the diagram then becomes a four-sided thermodynamic cycle that is defined, in the simplest possible model, by three ΔG values. There is a free energy associated with docking of the hairpin loops (ΔGdock), a free energy for ligand-binding pocket formation (ΔGLBP, which includes interactions with the ligand and therefore depends on the concentration of ligand present), and a coupling factor that links loop docking and ligand binding (ΔGw). Each of these three free energies can be deduced from the relative free energy values of the four RNA forms (Figure 6B). The Mg2+-dependence of each Δ represents a different potential mechanism for translating Mg2+ - RNA interactions into an enhancement of ligand binding affinity; each is considered in turn below.
First, the binding of ligand to an RNA with undocked loops (Δ°LBP) is intrinsically very weak but strongly enhanced by Mg2+. It is known from structure mapping [(22), and Figure 4], NMR (20), and fluorescent tag (40) studies that ligand binding induces distinctive structural changes in the binding pocket; presumably the organization of the binding pocket needed to accommodate ligand is energetically costly. Mg2+ strongly favors the structural rearrangements that accompany binding; ΔGLBP is enhanced −5.1 kcal/mol by 1 mM Mg2+. Backbone surrounding the bound ligand forms two sharp bends which bring phosphate oxygens as close as 3.6 Å (18); such regions of high charge density could be responsible for the sensitivity of the binding pocket structure to Mg2+ (40) and the accumulation of excess Mg2+.
Second, docking of the hairpin loops (ΔGdock) is also an intrinsically unfavorable reaction but strongly favored by Mg2+, as expected from other studies (20, 21, 41). Here we find that 1 mM Mg2+ stabilizes the docked conformation by ~ −3.5 kcal/mol. However, the docking reaction will influence ligand binding only to the degree that loop docking and formation of the binding pocket are favorably linked (ΔGw < 0, Figure 6A). From studies of a purine riboswitch RNA with both hairpins deleted, this linkage has been estimated as −3.3 kcal/mol in the presence of 10 mM Mg2+ (125 mM K+) (22); it has not been known whether a favorable ΔGW depends on Mg2+. From our estimations of ΔG°dock and ΔG°LBP, we deduce ΔG°W ≈ −5.3 kcal/mol (see arrow in Figure 6B). This value becomes more negative by −0.5 kcal/mol in 0.1 mM Mg2+, or −1.2 kcal/mol in 1 mM Mg2+. (These free energies, because they are small differences between large ΔGRNA-2+, have uncertainties comparable to their size.) The increase in Γ2+ that accompanies loop docking also appears somewhat larger when the binding pocket is occupied (2.1 ions/RNA, at 1 mM Mg2+) than unoccupied (1.8 ions/RNA), though the difference of 0.3 ions is comparable to the measurement error. We conclude that a strong linkage between the two tertiary structures is inherent to the RNA structure, but the linkage may be enhanced by Mg2+ to a small degree.
The value of ΔG°w we calculate assumes that the DAP complex with C60G RNA mimics a ligand-bound state that is accessible to the wild type riboswitch sequence. The G38C RNA is more difficult to saturate (ΔG°LBP more positive) and would therefore yield a somewhat more negative value of ΔG°w if used as a mimic of the ligand-bound intermediate. The conclusion that there is strong coupling between the two sets of tertiary structure in the absence of Mg2+ nevertheless remains.
Overall, we conclude that Mg2+ enhances the A-riboswitch affinity for adenine principally by two mechanisms of nearly equal effectiveness: stabilization of the docked loop structure, and stabilization of the ligand binding pocket (cf. ΔGIdock-E2+ and ΔGILBP, Figure 5C). The Mg2+ interaction free energies are large compared to the intrinsic stability differences between the various RNA folds, and above ~ 0.1 mM Mg2+ the relative stabilities of the RNA forms reflect, to a large degree, differences in the strengths of their interactions with Mg2+.
Parallels to Mg2+ -induced collapse in larger RNAs
Mg2+-induced “collapse” of an RNA to compact, non-native structures has been documented in several larger sequences (~200–400 nt), with implications for the kinetic and equilibrium pathways by which tertiary structures form in response to Mg2+ (11, 15, 42, 43). There is a structural parallel between these larger RNAs and the A-riboswitch RNA, in that large RNAs tend to use pseudoknot, tetraloop receptor (44), and A-minor (45) motifs to link peripheral elements together structurally. Such tertiary crosslinks presumably restrain the RNA dimensions and the organization of the central core of the structure [see, for instance, the structures of RNase P (46) and Azoarcus group I intron (47)]. In a similar way, a peripheral crosslink in the A-riboswitch (the docked loops) is linked to formation of a core tertiary structure (the ligand binding site). Are there also parallels between the Mg2+-induced collapse phenomenon and the response of the A-riboswitch RNA to Mg2+?
Although specific characteristics of the collapse phenomenon vary between RNAs, usually two distinct types of collapse can be distinguished. Partial collapse takes place at lower Mg2+ concentrations, does not depend on tertiary contacts (11, 15, 48), and is promoted by a variety of ions, presumably because it relies on simple screening of the electrostatic repulsion between phosphates (48, 49). At higher Mg2+ concentrations, further collapse takes place to a form with nearly the same dimensions as the native structure. This second collapse phase depends on the potential of the RNA to form specific tertiary contacts (11, 15, 48). However, chemical probing of the near-native collapsed form does not uncover any fully-formed tertiary contacts (15, 48, 50). Among several tertiary interactions mutated in one study of a group I intron, only disruption of a peripheral tetraloop-receptor contact destabilized the compact intermediate in addition to the native structure (15). These near-native intermediates apparently sample peripheral contact(s) without fully forming either peripheral or central core tertiary structures. The thermodynamic linkages between peripheral contacts and the central core are therefore important parameters that determine the cooperativity of the folding pathway in response to titration with Mg2+.
The Mg2+-dependent equilibrium folding pathway of the A-riboswitch is similar to that of larger RNAs in that it is predicted to proceed through a compact intermediate with docked loops. Based on calculations with the free energy landscape, the Idock RNA form may reach 30% of the total RNA at some ligand and Mg2+ concentrations. In the case of the A-riboswitch, we were able to estimate the cooperative free energy that links the peripheral and core structures; it is large (−5.3 kcal/mol) and only weakly dependent on Mg2+. In the A-riboswitch, at least, the core and peripheral structures take advantage of Mg2+ for stability, while the linkage between them is largely built into the RNA structure.
In contrast to the two collapse phases seen with larger RNAs, we do not observe the first, electrostatic collapse that is independent of peripheral tertiary contacts; the extended E state of the A-riboswitch appears completely insensitive to Mg2+. Screening of the RNA negative charge by ions must not be able to alter the intrinsic stiffness of the unliganded junction in this RNA. A similar finding has been made in a study of the S domain of RNase P, which concluded that a Mg2+-independent, extended conformation of a folding intermediate is maintained because of a specific, non-native structure within a three-helix junction, and that electrostatic repulsion plays a minor role in enforcing the extended conformation (51). Modulation of the stiffness vs. flexibility of folding intermediates is another way RNA sequence may adjust the stability of native structures in the presence Mg2+.
Concluding remarks
Γ2+ measurements provide a useful perspective for considering the effects of Mg2+ on RNA folding. As illustrated with the A-riboswitch RNA in this work, a knowledge of Γ2+ as a function of Mg2+ concentration can be used to extract the intrinsic (Mg2+-independent) free energy of folding reactions which might not be directly observable. Γ2+ measurements also reveal the strong Mg2+-dependences of ion uptake (ΔΓ2+) and Mg2+ - RNA interaction free energies (ΔGRNA-2+), which are usually not apparent from folding reaction analyses based on the Hill equation (10). Lastly, Γ2+ provides benchmarks for computational efforts to quantitatively model Mg2+ interactions with known RNA conformations (9).
Supplementary Material
Acknowledgments
We thank Drs. Xiaobing Zuo and Sonke Seifert for help with collection and analysis of the scattering data, and Robert Trachman for providing Mg2+ titration data for the A-riboswitch in the absence of ligand.
Abbreviations used
- A-riboswitch
the adenine-binding domain of an adenine riboswitch RNA
- DAP
2,6 diaminopurine
- HQS
8-hydroxy-quinoline-5-sulfonic acid
- SAXS
small angle X-ray scattering
- Rg
radius of gyration
Footnotes
This work was supported by National Institutes of Health Grants GM58545 (to D.E.D.) and T32 GM008403 (Program in Molecular Biophysics).
We assume that the extended states of wild type and C60G RNAs have the same standard state chemical potential, set to zero in Figure 6B, so that free energy changes measured with each RNA may be compared. This assumption implies that the two RNA E forms have similar structures and interaction with ions. Although the C60G mutation may extend the P2 helix by one base pair, we found no significant difference in Γ2+ between the two RNAs up to ~30 μM Mg2+ in the absence of ligand (Figure 5A), and suggest that the assumption is a reasonable approximation within the errors of the Figure 6B free energy landscape.
Supporting Information Available Three figures showing melting curves of wild type, C60G, and G38C RNAs, the hydroxyl radical reactivity of C60G RNA, and the correspondence of normalized Mg2+ – RNA interaction free enrgies for the native A– riboswitch and extended C60G RNAs; two tables list Rg values for the three RNAs under various conditions and Γ2+ and GRNA–2+ values evaluated at 1 mM Mg2+ for wild type and C60G RNAs; a complete derivation of equation (1). This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Lynch DC, Schimmel PR. Cooperative Binding of Magnesium to Transfer Ribonucleic Acid Studied by a Fluorescent Probe. Biochemistry. 1974;13:1841–1852. doi: 10.1021/bi00706a012. [DOI] [PubMed] [Google Scholar]
- 2.Römer R, Hach R. tRNA conformation and magnesium binding. A study of a yeast phenylalanine-specific tRNA by a fluorescent indicator and differential melting curves. Eur J Biochem. 1975;55:271–284. doi: 10.1111/j.1432-1033.1975.tb02160.x. [DOI] [PubMed] [Google Scholar]
- 3.Stein A, Crothers DM. Conformational Changes of Transfer RNA. The Role of Magnesium(II) Biochemistry. 1976;15:160–167. doi: 10.1021/bi00646a025. [DOI] [PubMed] [Google Scholar]
- 4.Leroy JL, Guéron M, Thomas G, Favre A. Role of divalent ions in folding of tRNA. Eur J Biochem. 1977;74:567–574. doi: 10.1111/j.1432-1033.1977.tb11426.x. [DOI] [PubMed] [Google Scholar]
- 5.Misra VK, Draper DE. Mg(2+) binding to tRNA revisited: the nonlinear Poisson-Boltzmann model. J Mol Biol. 2000;299:813–25. doi: 10.1006/jmbi.2000.3769. [DOI] [PubMed] [Google Scholar]
- 6.Cate JH, Hanna RL, Doudna JA. A magnesium ion core at the heart of a ribozyme domain. Nat Struct Biol. 1997;4:553–8. doi: 10.1038/nsb0797-553. [DOI] [PubMed] [Google Scholar]
- 7.Misra VK, Draper DE. A thermodynamic framework for Mg2+ binding to RNA. Proc Natl Acad Sci U S A. 2001;98:12456–12461. doi: 10.1073/pnas.221234598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Grilley D, Soto AM, Draper DE. Mg2+-RNA interaction free energies and their relationship to the folding of RNA tertiary structures. Proc Natl Acad Sci U S A. 2006;103:14003–14008. doi: 10.1073/pnas.0606409103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Soto AM, Misra V, Draper DE. Tertiary structure of an RNA pseudoknot is stabilized by “diffuse” Mg(2+) ions. Biochemistry. 2007;46:2973–2983. doi: 10.1021/bi0616753. [DOI] [PubMed] [Google Scholar]
- 10.Leipply D, Draper DE. The dependence of RNA tertiary structure stabilityon Mg2+ concentration: interpretation of the Hill equation and coefficient. Biochemistry. 2010;49:1843–1853. doi: 10.1021/bi902036j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Buchmueller KL, Webb AE, Richardson DA, Weeks KM. A collapsed non-native RNA folding state. Nat Struct Biol. 2000;7:362–366. doi: 10.1038/75125. [DOI] [PubMed] [Google Scholar]
- 12.Russell R, Millett IS, Doniach S, Herschlag D. Small angle X-ray scattering reveals a compact intermediate in RNA folding. Nat Struct Biol. 2000;7:367–370. doi: 10.1038/75132. [DOI] [PubMed] [Google Scholar]
- 13.Rangan P, Masquida B, Westhof E, Woodson SA. Assembly of core helices and rapid tertiary folding of a small bacterial group I ribozyme. Proc Natl Acad Sci U S A. 2003;100:1574–1579. doi: 10.1073/pnas.0337743100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Perez-Salas UA, Rangan P, Krueger S, Briber RM, Thirumalai D, Woodson SA. Compaction of a bacterial group I ribozyme coincides with the assembly of core helices. Biochemistry. 2004;43:1746–1753. doi: 10.1021/bi035642o. [DOI] [PubMed] [Google Scholar]
- 15.Chauhan S, Caliskan G, Briber RM, Perez-Salas U, Rangan P, Thirumalai D, Woodson SA. RNA tertiary interactions mediate native collapse of a bacterial group I ribozyme. J Mol Biol. 2005;353:1199–1209. doi: 10.1016/j.jmb.2005.09.015. [DOI] [PubMed] [Google Scholar]
- 16.Mandal M, Boese B, Barrick JE, Winkler WC, Breaker RR. Riboswitches control fundamental biochemical pathways in Bacillus subtilis and other bacteria. Cell. 2003;113:577–586. doi: 10.1016/s0092-8674(03)00391-x. [DOI] [PubMed] [Google Scholar]
- 17.Mandal M, Breaker RR. Adenine riboswitches and gene activation by disruption of a transcription terminator. Nat Struct Mol Biol. 2004;11:29–35. doi: 10.1038/nsmb710. [DOI] [PubMed] [Google Scholar]
- 18.Serganov A, Yuan YR, Pikovskaya O, Polonskaia A, Malinina L, Phan AT, Hobartner C, Micura R, Breaker RR, Patel DJ. Structural basis for discriminative regulation of gene expression by adenine- and guanine-sensing mRNAs. Chem Biol. 2004;11:1729–1741. doi: 10.1016/j.chembiol.2004.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lemay JF, Penedo JC, Tremblay R, Lilley DM, Lafontaine DA. Folding of the adenine riboswitch. Chem Biol. 2006;13:857–868. doi: 10.1016/j.chembiol.2006.06.010. [DOI] [PubMed] [Google Scholar]
- 20.Noeske J, Schwalbe H, Wohnert J. Metal-ion binding and metal-ion induced folding of the adenine-sensing riboswitch aptamer domain. Nucleic Acids Res. 2007;35:5262–5273. doi: 10.1093/nar/gkm565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Brenner MD, Scanlan MS, Nahas MK, Ha T, Silverman SK. Multivector fluorescence analysis of the xpt guanine riboswitch aptamer domain and the conformational role of guanine. Biochemistry. 2010;49:1596–1605. doi: 10.1021/bi9019912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stoddard CD, Gilbert SD, Batey RT. Ligand-dependent folding of the three-way junction in the purine riboswitch. RNA. 2008;14:675–684. doi: 10.1261/rna.736908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Grilley D, Soto AM, Draper DE. Direct quantitation of Mg2+ - RNA interactions by use of a fluorescent dye. Methods in Enzymology. 2009;455:71–94. doi: 10.1016/S0076-6879(08)04203-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lambert D, Leipply D, Draper DE. The Osmolyte TMAO Stabilizes Native RNA Tertiary Structures in the Absence of Mg(2+): Evidence for a Large Barrier to Folding from Phosphate Dehydration. J Mol Biol. 2010;404:138–157. doi: 10.1016/j.jmb.2010.09.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Petoukhov MV, Konarev PV, Kikhney AG, Svergun DI. ATSAS 2.1 – towards automated and web-supported small-angle scattering data analysis. Journal of Applied Crystallography. 2007;40:s223–s228. [Google Scholar]
- 26.Kozin MB, Svergun DI. Automated matching of high- and low-resolution structural models. Journal of Applied Crystallography. 2001;34:33–41. [Google Scholar]
- 27.Das R, Laederach A, Pearlman SM, Herschlag D, Altman RB. SAFA: semi-automated footprinting analysis software for high-throughput quantification of nucleic acid footprinting experiments. RNA. 2005;11:344–354. doi: 10.1261/rna.7214405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gilbert SD, Love CE, Edwards AL, Batey RT. Mutational analysis of the purine riboswitch aptamer domain. Biochemistry. 2007;46:13297–13309. doi: 10.1021/bi700410g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rambo RP, Tainer JA. Improving small-angle X-ray scattering data for structural analyses of the RNA world. Rna. 2010;16:638–46. doi: 10.1261/rna.1946310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Latham JA, Cech TR. Defining the Inside and Outside of a Catalytic RNA Molecule. Science. 1989;245:276–245. doi: 10.1126/science.2501870. [DOI] [PubMed] [Google Scholar]
- 31.Leipply D, Lambert D, Draper DE. Ion-RNA Interactions: Thermodynamic Analysis of the Effects of Mono- and Divalent Ions on RNA Conformational Equilibria. Methods in Enzymology. 2009;469:433–463. doi: 10.1016/S0076-6879(09)69021-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Burton K. Adenine transport in Escherichia coli. Proc Biol Sci. 1994;255:153–157. doi: 10.1098/rspb.1994.0022. [DOI] [PubMed] [Google Scholar]
- 33.Nygaard P, Saxild HH. The purine efflux pump PbuE in Bacillus subtilis modulates expression of the PurR and G-box (XptR) regulons by adjusting the purine base pool size. J Bacteriol. 2005;187:791–794. doi: 10.1128/JB.187.2.791-794.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cayley S, Lewis BA, Guttman HJ, Record MT., Jr. Characterization of the cytoplasm of Escherichia coli K-12 as a function of external osmolarity. Implications for protein-DNA interactions in vivo. J Mol Biol. 1991;222:281–300. doi: 10.1016/0022-2836(91)90212-o. [DOI] [PubMed] [Google Scholar]
- 35.Record MT, Jr., deHaseth PL, Lohman TM. Interpretation of monovalent and divalent cation effects on the lac repressor-operator interaction. Biochemistry. 1977;16:4791–4796. doi: 10.1021/bi00641a005. [DOI] [PubMed] [Google Scholar]
- 36.Misra VK, Draper DE. The interpretation of Mg2+ binding isotherms for nucleic acids using Poisson-Boltzmann theory. J Mol Biol. 1999;294:1135–47. doi: 10.1006/jmbi.1999.3334. [DOI] [PubMed] [Google Scholar]
- 37.Alatossava T, Jutte H, Kuhn A, Kellenberger E. Manipulation of intracellular magnesium content in polymyxin B nonapeptide-sensitized Escherichia coli by ionophore A23187. J Bacteriol. 1985;162:413–419. doi: 10.1128/jb.162.1.413-419.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.London RE. Methods for measurement of intracellular magnesium: NMR and fluorescence. Annu Rev Physiol. 1991;53:241–258. doi: 10.1146/annurev.ph.53.030191.001325. [DOI] [PubMed] [Google Scholar]
- 39.Froschauer EM, Kolisek M, Dieterich F, Schweigel M, Schweyen RJ. Fluorescence measurements of free [Mg2+] by use of mag-fura 2 in Salmonella enterica. FEMS Microbiol Lett. 2004;237:49–55. doi: 10.1016/j.femsle.2004.06.013. [DOI] [PubMed] [Google Scholar]
- 40.Rieder R, Lang K, Graber D, Micura R. Ligand-Induced Folding of the Adenosine Deaminase A-Riboswitch and Implications on Riboswitch Translational Control. Chembiochem. 2007;8:896–902. doi: 10.1002/cbic.200700057. [DOI] [PubMed] [Google Scholar]
- 41.Lemay JF, Lafontaine DA. Core requirements of the adenine riboswitch aptamer for ligand binding. RNA. 2007;13:339–350. doi: 10.1261/rna.142007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fang X, Littrell K, Yang XJ, Henderson SJ, Siefert S, Thiyagarajan P, Pan T, Sosnick TR. Mg2+-dependent compaction and folding of yeast tRNAPhe and the catalytic domain of the B. subtilis RNase P RNA determined by small-angle X-ray scattering. Biochemistry. 2000;39:11107–11113. doi: 10.1021/bi000724n. [DOI] [PubMed] [Google Scholar]
- 43.Russell R, Millett IS, Tate MW, Kwok LW, Nakatani B, Gruner SM, Mochrie SG, Pande V, Doniach S, Herschlag D, Pollack L. Rapid compaction during RNA folding. Proc Natl Acad Sci U S A. 2002;99:4266–4271. doi: 10.1073/pnas.072589599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Costa M, Michel F. Frequent use of the same tertiary motif by self-folding RNAs. Embo J. 1995;14:1276–1285. doi: 10.1002/j.1460-2075.1995.tb07111.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nissen P, Ippolito JA, Ban N, Moore PB, Steitz TA. RNA tertiary interactions in the large ribosomal subunit: the A-minor motif. Proc Natl Acad Sci U S A. 2001;98:4899–4903. doi: 10.1073/pnas.081082398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kazantsev AV, Krivenko AA, Harrington DJ, Holbrook SR, Adams PD, Pace NR. Crystal structure of a bacterial ribonuclease P RNA. Proc Natl Acad Sci U S A. 2005;102:13392–13397. doi: 10.1073/pnas.0506662102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Adams PL, Stahley MR, Kosek AB, Wang J, Strobel SA. Crystal structure of a self-splicing group I intron with both exons. Nature. 2004;430:45–50. doi: 10.1038/nature02642. [DOI] [PubMed] [Google Scholar]
- 48.Das R, Kwok LW, Millett IS, Bai Y, Mills TT, Jacob J, Maskel GS, Seifert S, Mochrie SG, Thiyagarajan P, Doniach S, Pollack L, Herschlag D. The fastest global events in RNA folding: electrostatic relaxation and tertiary collapse of the Tetrahymena ribozyme. J Mol Biol. 2003;332:311–319. doi: 10.1016/s0022-2836(03)00854-4. [DOI] [PubMed] [Google Scholar]
- 49.Moghaddam S, Caliskan G, Chauhan S, Hyeon C, Briber RM, Thirumalai D, Woodson SA. Metal ion dependence of cooperative collapse transitions in RNA. J Mol Biol. 2009;393:753–764. doi: 10.1016/j.jmb.2009.08.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Buchmueller KL, Weeks KM. Near native structure in an RNA collapsed state. Biochemistry. 2003;42:13869–13878. doi: 10.1021/bi035476k. [DOI] [PubMed] [Google Scholar]
- 51.Baird NJ, Gong H, Zaheer SS, Freed KF, Pan T, Sosnick TR. Extended structures in RNA folding intermediates are due to nonnative interactions rather than electrostatic repulsion. J Mol Biol. 2010;397:1298–1306. doi: 10.1016/j.jmb.2010.02.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lambert D, Draper DE. Effects of Osmolytes on RNA Secondary and Tertiary Structure Stabilities and RNA-Mg2+ Interactions. J Mol Biol. 2007;370:993–1005. doi: 10.1016/j.jmb.2007.03.080. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.





