Abstract
Nrf2, a master regulator of intracellular redox homeostasis, is indicated to participate in fatty acid metabolism in liver. However, its role in diet-induced obesity remains controversial. In the current study, genetically engineered Nrf2-null, wild-type (WT), and Nrf2-activated, Keap1-knockdown (K1-KD) mice were fed either a control or a high-fat western diet (HFD) for 12 weeks. The results indicate that the absence or enhancement of Nrf2 activity did not prevent diet-induced obesity, had limited effects on lipid metabolism, but affected blood glucose homeostasis. Whereas the Nrf2-null mice were resistant to HFD-induced glucose intolerance, the Nrf2-activated K1-KD mice exhibited prolonged elevation of circulating glucose during a glucose tolerance test even on the control diet. Feeding a HFD did not activate the Nrf2 signaling pathway in mouse livers. Fibroblast growth factor 21 (Fgf21) is a liver-derived anti-diabetic hormone that exerts glucose- and lipid-lowering effects. Fgf21 mRNA and protein were both elevated in livers of Nrf2-null mice, and Fgf21 protein was lower in K1-KD mice than WT mice. The inverse correlation between Nrf2 activity and hepatic expression of Fgf21 might explain the improved glucose tolerance in Nrf2-null mice. Furthermore, a more oxidative cellular environment in Nrf2-null mice could affect insulin signaling in liver. For example, mRNA of insulin-like growth factor binding protein 1, a gene repressed by insulin in hepatocytes, was markedly elevated in livers of Nrf2-null mice. In conclusion, genetic alteration of Nrf2 does not prevent diet-induced obesity in mice, but deficiency of Nrf2 improves glucose homeostasis, possibly through its effects on Fgf21 and/or insulin signaling.
Keywords: Nrf2, high-fat diet, obesity, Fgf21, glucose intolerance, redox signaling
Introduction
The nuclear factor erythoid 2-related factor 2 (Nrf2), a member of the cap‘n’collar (CNC) family of transcription factors, is a crucial molecule in the maintenance of redox homeostasis. Inactive Nrf2 is retained in the cytoplasm by association with an actin-binding protein, Kelch-like ECH-associated protein 1 (Keap1), which functions as an adapter for Cul3/Rbx1-mediated ubiquitination and degradation of Nrf2. Keap1 senses the cellular oxidative stress and releases Nrf2. Nrf2 then translocates to the nucleus, where it forms a heterodimer with small Maf (Katsuoka et al., 2005) or Jun (Jeyapaul and Jaiswal, 2000), and activates transcription of genes encoding detoxifying and antioxidant enzymes as well as membrane transporters through binding to the antioxidant-responsive elements in their promoters. Nrf2 target genes include NAD(P)H:quinone oxidoreductase 1 (Nqo1) (Itoh et al., 1997; Ishii et al., 2002), glutathione-S-transferases (Gsts) (Itoh et al., 1997), glutamate cysteine ligase catalytic subunit (Gclc) (Chan and Kwong, 2000), multidrug resistance-associated proteins (Hayashi et al., 2003; Klaassen and Slitt, 2005; Maher et al., 2007), etc. Multiple cellular protective functions of Nrf2 during hepatic toxicity, inflammation, aging, and chemotherapy, as well as mechanisms of activating the Nrf2-Keap1 signaling pathway have been thoroughly reviewed (Aleksunes and Manautou, 2007; Kensler et al., 2007; Giudice et al., 2010; Klaassen and Reisman, 2010).
In addition to its well-recognized roles in detoxification and cytoprotection, Nrf2 signaling has recently been associated with nutrient disposition, especially lipid metabolism. A microarray study first revealed that either genetic or chemical activation of Nrf2 decreased the mRNA levels of a large number of lipid metabolism genes, especially those involved in lipogenesis, in mouse livers (Yates et al., 2009). An independent proteomic analysis further demonstrated that many proteins involved in lipogenesis were upregulated in livers of Nrf2-null mice (Kitteringham et al., 2010). These two studies suggested a negative regulation of lipogenesis by Nrf2 in mouse livers. However, studies using diet-induced obesity mouse models have reached conflicting conclusions. Whereas chronic activation of Nrf2 by administering agonists, CDDO-Im (Shin et al., 2009) or oltipraz (Yu et al., 2011), was reported to prevent high-fat diet-induced obesity in mice, others found that deficiency of Nrf2 protected mice from diet-induced obesity, due to impaired adipogenesis (Pi et al., 2010), or by increasing fibroblast growth factor 21 (Fgf21), a liver-derived pro-lipolytic hormone (Chartoumpekis et al., 2011).
In order to elucidate the impact of Nrf2 on diet-induced obesity, the current study employed not only Nrf2 deficient (Nrf2-null) mice, but also mice with genetically enhanced Nrf2 activity (Keap1-knockdown, K1-KD). Nrf2-null, WT, and K1-KD mice were fed either a high-fat Western diet (HFD) or a control diet for 12 weeks. Application of the Nrf2-null and K1-KD models during intake of HFD allows us to assess the effect of the Nrf2 “gene dose response” while avoiding the potential off-target effects of chemical activators of Nrf2. By comparing various metabolic parameters in Nrf2-null, wild-type (WT), and K1-KD mice fed either a control diet or a HFD, putative roles of Nrf2 in glucose and lipid homeostasis were determined.
Materials and Methods
Animals and experimental design
Nrf2-null (from Dr. Jefferson Chan, University of California, Irvine, CA) (Chan et al., 1996) and K1-KD mice (from Dr. Masayukin Yamamoto, Tohoku University, Aoba-ku, Sendai, Japan) (Okada et al., 2008) were bred in the Laboratory Animal Facility at the University of Kansas Medical Center. Both strains were backcrossed to achieve a >99% congenic C57BL/6 background, which was verified by the Jackson Laboratory (Bar Harbor, ME). Wild type (WT) C57BL/6 mice were purchased from Charles River Laboratories, Inc. (Wilmington, MA). At 3 months of age, male Nrf2-null, WT, and K1-KD mice (n=6) were fed either a control diet (5SXC; catalog # 1813907; 12.1% fat-derived calorie) or a high-fat Western diet (HFD) (5SXB; catalog# 1813906; 39.7% fat-derived calorie) that were purchased from TestDiet (Richmond, IN) for 12 weeks. To mimic the Western diet, both control diet and HFD contained fat from a variety of sources, including lard, hydrogenated vegetable oil, butter fat, corn oil, and soy oil. The feed preservative t-butylhydroquinone was removed from both diets because it is a known Nrf2 agonist (Imhoff and Hansen, 2010). All animals were fed ad libitum and had free access to water. Body weights were monitored weekly. All mice were housed in an Association for Assessment and Accreditation of Laboratory Animal Care-accredited animal care facility in a temperature-, light-, and humidity-controlled environment. The University of Kansas Medical Center Institutional Animal Care and Use Committee approved the studies.
Glucose/insulin tolerance test
Glucose tolerance test (GTT) was conducted after 10 weeks of feeding, and insulin tolerance test (ITT) was performed after 11 weeks of feeding the control or Western diets. The feed was removed for 6 h before the GTT or ITT. For the GTT, a single dose of D-glucose (20% solution in water; 10 ml/kg) was injected i.p.. For the ITT, a single dose of insulin (Humulin N, purchased from a CVS Pharmacy, Roeland Park, KS) (0.75 U/kg; 5 ml/kg in saline) was injected i.p.. Blood was taken from tails of mice at 0, 30, 60, 90, and 120 min thereafter, and glucose concentrations were determined using a ReliOn Ultima glucose monitor (Arkray USA, Inc., Minneapolis, MN).
Serum analysis
All mice were sacrificed in the morning after being fed either a control diet or a HFD for 12 weeks. Right before mice were sacrificed, the glucose concentrations were determined with a ReliOn Ultima glucose monitor using blood from the tail as described above. Blood was collected from these mice without pre-fasting. Concentrations of triglycerides, nonesterified fatty acids (NEFAs), and cholesterol in plasma were measured using kits from Wako Diagnostics (Richmond, VA). Plasma insulin was quantified using an enzyme-linked immunosorbent assay kit from Millipore (Billerica, MA). Beta-hydroxybutyrate was determined using a kit from Cayman Chemical Company (Ann Arbor, MI). All assays were performed according to the manufacturers' protocols.
Histopathology
Liver tissues were fixed in 10% formalin for 48 h, transferred to 70% ethanol for 48 h, and embedded in paraffin blocks for sectioning. Liver sections (5 μm) were stained with hematoxylin and eosin using standard protocols.
Liver biochemistry
Liver lipids were extracted as described (McGrath and Elliott, 1990), and determined with the same protocol as serum lipids. GSH concentrations in livers were quantified by UPLC-MS/MS as described previously (Wu et al., 2011). NADH concentrations in livers were determined by using a kit from BioVision Inc. (Milpitas, CA) per their protocol. Lipid peroxidation was monitored by quantifying thiobarbituric acid reactive substances (TBARS) using the OXItek TBARS kit (ZeptoMetrix, Buffalo, NY) as previously described (Zhang et al., 2010).
Glycogen content assays
Glycogen was prepared from mouse livers as reported (Passonneau and Lauderdale, 1974). Briefly, 20 mg of frozen liver tissue was homogenized in 0.03 N HCl. The homogenates were heated at 100°C for 5 min, followed by centrifugation at 12,000 g for 5 min. The supernatant was transferred to a new tube and appropriate dilutions were made. Amyloglucosidase from Aspergillus niger (A-7420) for glycogen hydrolysis, and the glucose assay kit (GOGA-20) were both purchased from Sigma-Aldrich (St. Louis, MO). The assay was carried out according to the manufacturer's protocol with slight modifications.
RNA isolation
Total RNA was extracted from livers using RNA-Bee reagent (Tel-Test, Inc., Friendswood, TX) per the manufacturer's protocol. RNA was dissolved in diethyl pyrocarbonate-treated deionized water, and RNA concentrations were determined with a NanoDrop Spectrophotometer ND-1000.
Messenger RNA quantification
Total RNA was reverse-transcribed into first-strand cDNA using multiscript reverse transcriptase from a High Capacity RT kit (Applied Biosystems, Foster City, CA) according to the manufacturer's protocol. Messenger RNA of genes of interest was determined with quantitative real time-PCR performed on an Applied Biosystems Prism 7900HT sequence detection system. The reaction system contains 2 ng of cDNA, 150 nM of each primer, and 5 μl of Power SYBR Green PCR Master Mix (Applied Biosystems, Foster City, CA) in a total volume of 10 μl. The specific primers used to quantify gene expression are listed in Table 1. The relative mRNA levels were calculated by cycle threshold (Ct) values, which were normalized to the internal control glyceraldehyde 3-phosphate dehydrogenase (Gapdh) mRNA.
Table 1.
Primers used for quantitative real time-PCR.
| Gene Name | Forward | Reverse |
|---|---|---|
| Acaca | 5′-agaaacccgaacagtggaact-3′ | 5′-aggtagcccttcacggttaaa-3′ |
| CAT | 5′-cctcgttcaggatgtggttt-3′ | 5′-tctggtgatatcgtgggtga-3′ |
| CD36 | 5′-catattggtcaagccagctag-3′ | 5′-agcaacaaacatcaccactcc-3′ |
| ChREBP | 5′-cctcacttcactgtgcctca-3′ | 5′-acaggggttgttgtctctgg-3′ |
| Cyp4a10 | 5′-cacaccctgatcaccaacag-3′ | 5′-tccttgatgcacattgtggt-3′ |
| Fasn | 5′-gaccttcatggacacaatgct-3′ | 5′-ataccaccagagaccgttatg-3′ |
| Fgf21 | 5′-ctgggggtctaccaagcata-3′ | 5′-atcctccctgatctccaggt-3′ |
| G6pase | 5′-atgactttgggatccagtcg-3′ | 5′-tggaaccagatgggaaagag-3′ |
| Gclc | 5′-tggccactatctgcccaatt-3′ | 5′-gtctgacacgtagcctcggtaa-3′ |
| Gstm l | 5′-ctcccgactttgacagaagc-3′ | 5′-ttgctctgggtgatcttgtg-3′ |
| IGF-1 | 5′-ctggaccagagaccctttgc-3′ | 5′-ggacggggacttctgagtctt-3′ |
| lgfbp-1 | 5′-ctgccaaactgcaacaagaatg-3′ | 5′-ggtcccctctagtctccaga-3′ |
| Me1 | 5′-gggattgctcacttggttgt-3′ | 5′-agtgggtgaaccctcacaag-3′ |
| Nqo1 | 5′-tatccttccgagtcatctctagca-3′ | 5′-tctgcagcttccagcttcttg-3′ |
| Nrf2 | 5′-cgagatatacgcaggagaggtaaga-3′ | 5′-gctcgacaatgttctccagctt-3′ |
| Pklr | 5′-ctggtgattgtggtgacagg-3′ | 5′-atggggtgcaactaggtcag-3′ |
| PPARα | 5′-atgaagagggctgagcgtag-3′ | 5′-aaacgcaacgtagagtgctgt-3′ |
| SREBP-1c | 5′-taggtcaccgtttctttgtgg-3′ | 5′-atccaagggcatctgagaact-3′ |
Western blot analysis
Liver protein was extracted with NE-PER nuclear extraction kit (Pierce Biotechnology, Rockford, IL) according to the manufacturer's manual. Protein concentrations were determined using the Bradford assay (Bio-Rad Laboratories, Hercules, CA). Approximately 30 μg of nuclear protein extracts and 60 μg of cytosolic protein were used for immunoblotting of proteins of interest. The primary antibodies used in this study include Nrf2 (sc-13032), PPARα (sc-9000), and histone 3 (H3; sc-8654-R) from Santa Cruz Biotechnology (Santa Cruz, CA); ChREBP (NB400-135) from Novus Biologicals, LLC (Littleton, CO); Nqo1 (Ab2346) and β-actin (Ab8227) from Abcam (Cambridge, MA); and Fgf21 (RD281108100) from BioVendor R&D (Modrice, Czech Republic). Secondary antibodies were purchased from Sigma-Aldrich (St. Louis, MO). Protein-antibody complexes were detected using an enhanced chemiluminescent kit (Peirce Biotechnology, Rockford, IL) and exposed to HyBlot CL autoradiography film (Denville Scientific Inc., Metuchen, NJ). Density of individual blots was quantified by Quantity One 1-D Analysis Software (Bio-Rad Laboratories, Hercules, CA). The blot densities of proteins of interest were normalized to respective loading controls.
Statistical analysis
Differences among individual groups were evaluated by two-way analysis of variance (ANOVA) with genotype and diet as the two variables, followed by Student-Newman-Keuls comparisons. Differences were considered statistically significant at p < 0.05.
Results
Gross characteristics of mice with low to high Nrf2 activities fed a HFD
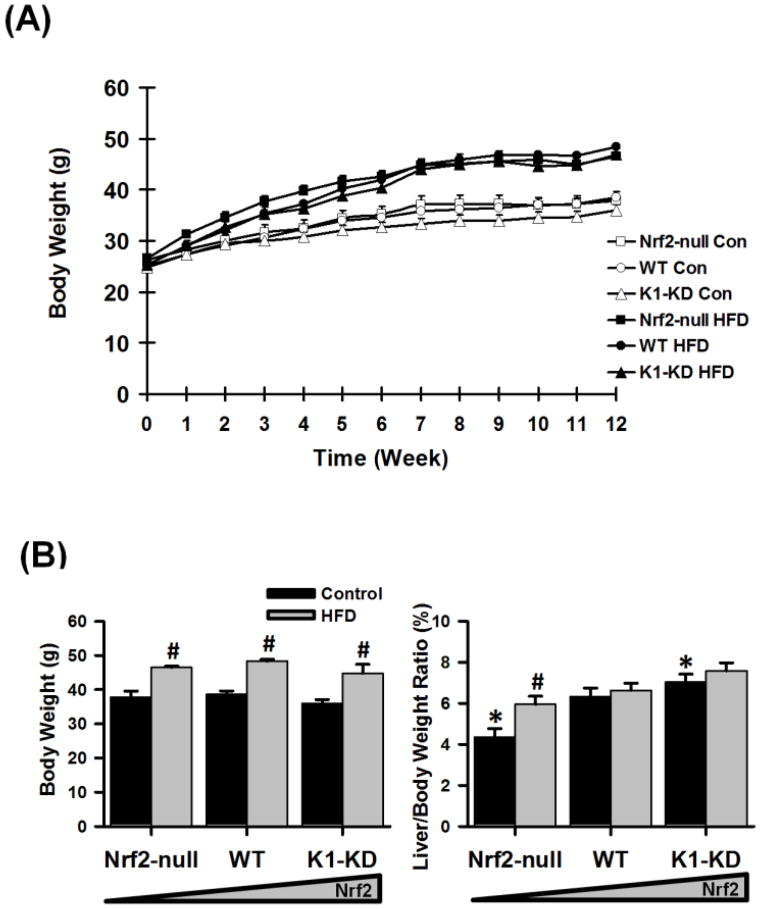
Starting at 3 months of age, Nrf2-null (absent of Nrf2 expression), wild-type (WT; with regular Nrf2 activity), and Keap1-knockdown (K1-KD; with enhanced Nrf2 activity) mice were fed either a control diet (12.1% calories derived from fat) or a high-fat western diet (HFD; 39.7% fat-derived calories) for 12 continuous weeks. Fat in both diets were from a variety of sources, instead of a single animal- or plant-origin. The growth curves of the three genotypes of mice were similar throughout the 12-week feeding period on either the control diet or the HFD (Fig. 1A). Mice fed the HFD for 12 weeks gained approximately 10 g more weight than those fed the control diet, regardless of their genotype (Fig. 1B). Although having similar body size, Nrf2-null mice had smaller livers, whereas K1-KD mice had larger livers than WT mice. Feeding the HFD increased the liver-to-body weight ratio only in Nrf2-null mice, but not in the other two genotypes.
Figure 1. Body weight and liver weight.
(A) Growth curves of Nrf2-null, wild-type (WT), and Keap-1 knockdown (K1-KD) mice fed with either a high-fat Western Diet (HFD) or a control diet over a 12-week period. (B) End point body weight as well as liver/body weight ratio. Data are presented as mean ± S.E. (n=6). Two-way ANOVA was performed with genotype and treatment as main factors. Significant main effects and interactions were followed by Student-Newman-Keuls comparisons to assess the differences between groups. Asterisks (*) represent statistical differences (p < 0.05) between mice with altered Nrf2 activity and WT mice on the same diet. Pounds (#) represent statistical differences (p < 0.05) between HFD-treated and control diet-treated mice of the same genotype.
Liver histology
Hematoxylin and eosin staining showed that the microscopic structures of livers of mice with different levels of Nrf2 activity were similar on each diet (data not shown).
Blood and liver chemistry
Blood chemistries were determined in serum collected from mice in the morning following overnight ad libitum access to food and water. Blood glucose concentrations were higher in K1-KD mice than in Nrf2-null and WT mice when either control or HFD was fed (Table 2). The HFD did not affect blood glucose concentrations in any of the 3 genotypes of mice. The concentrations of circulating triglycerides were comparable among the 3 genotypes of mice fed the control diet. The HFD increased blood triglycerides in WT and K1-KD mice, but not in Nrf2-null mice. Blood concentrations of non-esterified free fatty acids (NEFAs) were similar among the 3 genotypes of mice fed the control diet. The HFD tended to increase circulating NEFAs in all 3 genotypes, but was statistically significant only in the K1-KD mice. The blood cholesterol concentrations in mice fed the control diet were not statistically different among genotypes. When the HFD was given, the circulating cholesterol increased in all 3 genotypes, but tended to be higher in K1-KD mice (K1-KD vs. WT, p=0.076). The concentrations of β-hydroxybutyrate were not affected by either the Nrf2 genotype or the fat content of diets.
Table 2.
Blood and liver chemistry.
| Control | HFD | |||||
|---|---|---|---|---|---|---|
| Nrf2-null | WT | K1-KD | Nrf2-null | WT | K1-KD | |
| Blood | ||||||
| Glucose (Fed, mg/dL) | 182 ± 10.2 | 194 ± 10.2 | 242 ± 9.5* | 180 7 ± 9.5 | 177 ± 8.9 | 220 + 9.5 * |
| Triglyceride (mg/dL) | 33.9 ± 9 | 20.2 ± 3.2 | 30.1 ± 2.5 | 41.3 ± 4.3 * | 58.6 ± 3.9 # | 63.8 ± 12.1 # |
| NEFA (mg/dL) | 8.6 ± 1.0 | 8.6 ± 1.6 | 7.0 ± 0.7 | 9.7 ± 1.2 | 12.7 ± 0.9 | 11.6 ± 1.7 # |
| Cholesterol (mg/dL) | 111,0 ± 27.9 | 175.4 ± 28.3 | 172.4 ± 17.4 | 183.4 ± 22.4 # | 224,8 ± 11.7 | 2650 ± 24.5 # |
| β-Hydroxybutyrate (mM) | 0.23 ± 0.03 | 0.24 ± 0.01 | 0.22 ± 0.01 | 0.26 ± 0.02 | 0.22 ± 0.01 | 0.23 ± 0.02 |
| Liver | ||||||
| Glycogen (mg/g liver) | 81 ± 11 | 78 ± 2 | 75 ± 4 | 51 ± 3 # | 68 ± 6 | 69 ± 7 |
| Triglyceride (mg/g liver) | 14.4 ± 2.46 | 14.2 ± 1.5 | 12.7 ± 2.06 | 25 ± 2.13 # | 25.9 ± 1.53 # | 21.5 ±2.14 # |
| NEFA (mg/g liver) | 10.6 ± 1.24 | 10.6 ± 0.45 | 10.7 ± 0.38 | 14.6 ± 1.34 # | 13.4 ± 0.48 # | 14.4 ± 0.8 # |
| GSH (μM/g liver) | 5.5 ± 0.35 * | 6.8 ± 0.39 | 7.5 ± 0.32 | 6.4 ± 0.32 * | 7.7 ± 0.32 | 7.3 ± 0.32 |
| NADH (nM/mg protein) | 0.81 ± 0.1 * | 1.22± 0.1 | 1.09 ± 0.09 | 0.76 ± 0.1 | 1.04 ± 0.09 | 0.86 ± 0.09 |
| MDA (μM/g liver) | 14.1 ± 2.6 | 8.3 ± 0.9 | 9.9 ± 2.0 | 11.9 ± 2.9 | 10.6 ± 0.7 | 10.2 ± 0.8 |
Statistical significance is established at p < 0.05:
different from WT on the same diet;
different from mice of the same genotype on the control diet.
Glycogen concentrations were similar in livers of mice possessing various Nrf2 activities when they were on the control diet (Table 2). The HFD decreased glycogen content in livers of Nrf2-null mice, but not in WT and K1-KD mice. Liver triglyceride concentrations were similar in mice with graded Nrf2 activity when the control diet was given. The HFD nearly doubled the concentration of triglycerides in livers of all 3 genotypes of mice. The concentrations of NEFAs increased approximately 40% in livers of all three genotypes of mice fed the HFD, but no genotype differences were observed in mice fed either the control or the HFD. Concentrations of the two reducing sources in hepatocytes, glutathione (GSH) and nicotinamide adenine dinucleotide (NADH), were not affected by HFD, but were lower or tended to be lower in livers of Nrf2-null mice than WT mice, regardless of the fat content of diets. Concentrations of malondialdehyde (MDA), a marker of oxidative stress, were not affected by either Nrf2 genotype or fat content of diets in the liver.
Glucose and insulin tolerance tests
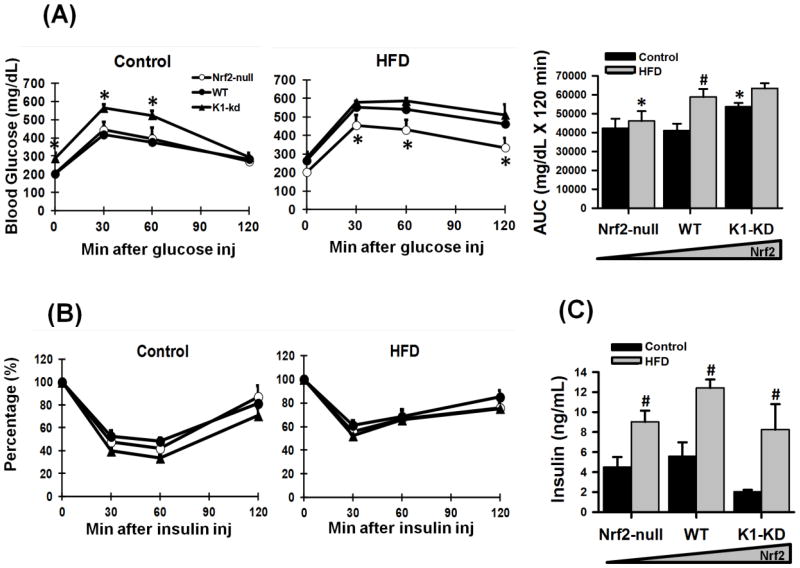
To evaluate the impact of Nrf2 on whole-body glucose disposition, an intraperitoneal glucose tolerance test (GTT) was conducted in the 10th week of feeding. When the control diet was fed, K1-KD mice exhibited higher blood glucose concentrations (Fig. 2A) and decreased glucose clearance. The HFD increased the blood glucose concentration and reduced the glucose clearance in WT mice to a similar level as K1-KD mice. In contrast, Nrf2-null mice were able to maintain low blood glucose concentrations and normal glucose clearance even when a HFD was fed. Thus, the deficiency of Nrf2 seems to protect mice from HFD-induced hyperglycemia. An i.p. insulin tolerance test (ITT) was performed in the 11th week of feeding. Intraperitoneal insulin injection was equally efficient in lowering blood glucose concentrations in all 3 genotypes of mice on either a control or a HFD (Fig. 2B), indicating that insulin sensitivity was not affected by Nrf2. Feeding the HFD increased the circulating insulin concentrations in all 3 genotypes of mice (Fig. 2C), and no apparent differences in circulating insulin concentrations were observed among the 3 genotypes.
Figure 2. Glucose and insulin parameters in Nrf2-null, WT, K1-KD mice fed a control diet or HFD.
(A) Glucose tolerance test (GTT) as well as area under the curves (AUCs) of serial glucose concentrations in the GTT. Asterisks (*) represent statistically significant differences (p < 0.05) compared with WT on the same diet at that time point. (B) Insulin tolerance test (ITT). (C) Blood insulin concentrations. For GTT, a bolus of intraperitoneal injection of 20% D-glucose was given after the basal blood glucose (time 0) was taken. Serial glucose levels were taken at indicated time points thereafter. The procedure of ITT resembled that of GTT except a single dose of Humulin N (0.75 U/kg) was given at time 0. The blood glucose levels at indicated time points in ITT were presented as percentages of time 0. Blood insulin concentrations were determined after euthanasia via a bolus of pentobarbital. Asterisks (*) represent statistical differences (p < 0.05) between mice with altered Nrf2 activity and WT mice on the same diet. Pounds (#) represent statistical differences (p < 0.05) between HFD-treated and control diet-treated mice of the same genotype.
Hepatic expression of Nrf2 and Nrf2-target genes
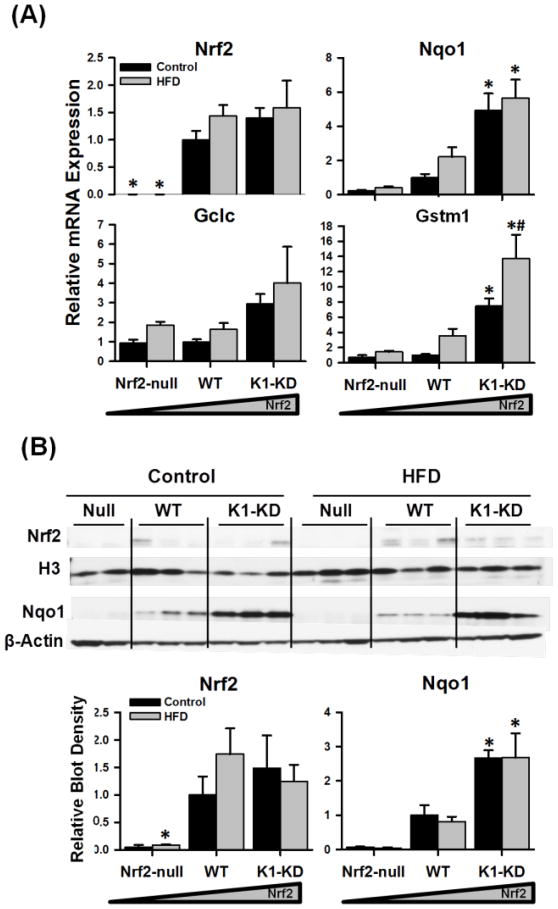
To explore the in vivo effects of an overload of dietary fatty acids on the Nrf2-Keap1 signaling pathway, the mRNA expression of Nrf2 and its prototypical target genes in mouse livers was determined (Fig. 3A). Nrf2 mRNA was absent in Nrf2-null mouse livers as expected. The HFD did not affect the mRNA expression of Nrf2 in WT or K1-KD mouse livers. Graded expression of mRNA of well-characterized Nrf2-regulated genes, including Nqo1, glutathione S-transferase mu 1 (Gstm1), and Gclc, was observed in both control- and HFD-fed mice that possess none (Nrf2-null) to high levels (K1-KD) of Nrf2 activity. However, the HFD had negligible effects on the mRNA levels of these Nrf2-target genes, except a slight induction of Gstm1 in K1-KD mice. Furthermore, the nuclear accumulation of Nrf2 protein was not significantly increased in the liver by feeding a HFD (Fig. 3B). Accordingly, the amount of Nqo1 protein, one of the most sensitive Nrf2-target genes, was not increased in the HFD-fed mice compared with the control-fed mice in all three genotypes. Consistent with mRNA expression pattern, Nqo1 protein was much higher in livers of K1-KD mice than the other genotypes. Taken together, both mRNA and protein data indicate that dietary fatty acids do not alter the Nrf2 signaling pathway in mouse livers.
Figure 3. Hepatic mRNA and protein expression of Nrf2 and Nrf2-target genes.
(A) Messenger RNA of Nrf2, Nqo1, Gstm1, and Gclc were quantified by RT-qPCR, normalized to Gapdh, and presented as a ratio to WT control. The mRNA expression of Gapdh in mouse livers was not affected by either Nrf2 genotype or diet. (B) Western blots were performed with nuclear (Nrf2) or cytosol (Nqo1) fraction from control- and HFD-treated Nrf2-null, WT, and K1-KD mouse livers. Histone 3 (H3) and β-actin were used as nuclear and cytosol loading control, respectively. Density of blots was determined by Quantity One 1D Analiysis software, normalized to respective loading control, and presented as a ratio to wild-type control. Asterisks (*) represent statistical differences (p < 0.05) between mice with altered Nrf2 activity and WT mice on the same diet. Pounds (#) represent statistical differences (p < 0.05) between HFD-treated and control diet-treated mice of the same genotype.
Expression of hepatic genes involved in glucose metabolism
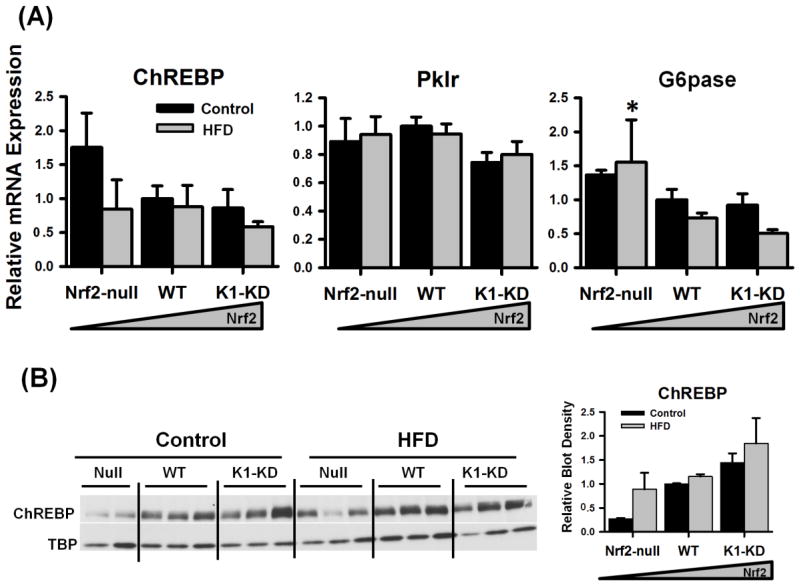
In liver, the carbohydrate responsive element-binding protein (ChREBP) regulates a series of genes involved in glycolysis, gluconeogenesis, and fatty acid synthesis to convert excess carbohydrates into triglycerides through de novo lipogenesis. When the control diet was fed, the mRNA of ChREBP tended to be higher in livers of Nrf2-null than Nrf2-expressing mice, but was not statistically significant (Fig. 4A). In contrast, the nuclear accumulation of ChREBP protein tended to be lower in livers of Nrf2-null mice than WT and K1-KD mice when a control diet was fed (Fig. 4B). Liver-type pyruvate kinase (Pklr), the key enzyme for glycolysis in liver, is a prototypical target gene of ChREBP. No differences were observed in the mRNA expression of Pklr among mice possessing different Nrf2 activities (Fig. 4A). Glucose-6-phosphatase (G6pase) catalyzes the final step of gluconeogenesis in hepatocytes, resulting in the formation of free glucose. ChREBP also regulates the transcription of G6pase. Nrf2-null mice expressed more G6pase mRNA than the other two genotypes when the HFD was fed (Fig. 4A). Moreover, the mRNA of another key enzyme for glycolysis, glucokinase (Gck), tended to be higher in Nrf2-null mice when a HFD was fed (Nrf2 vs. WT, p=0.084), and phosphoenolpyruvate carboxykinase (Pepck), another rate-limiting enzyme for gluconeogenesis was higher in livers of Nrf2-null mice when control diet was fed (data not shown). In general, the HFD did not markedly influence the mRNA expression of glucose metabolism-related genes in mouse livers.
Figure 4. Quantification of genes involved in glucose metabolism in the liver.
(A) Messenger RNA of genes involved in hepatic glucose metabolism. Messenger RNA were determined by RT-qPCR, normalized to Gapdh, and presented as a ratio to WT control. (B) Nuclear accumulation of ChREBP protein. Density of ChREBP blots was normalized to the loading control TBP and presented as a ratio to wild-type control. Asterisks (*) represent statistical differences (p < 0.05) between mice with altered Nrf2 activity and WT mice on the same diet. Pounds (#) represent statistical differences (p < 0.05) between HFD-treated and control diet-treated mice of the same genotype.
Hepatic expression of genes involved in fatty acid metabolism
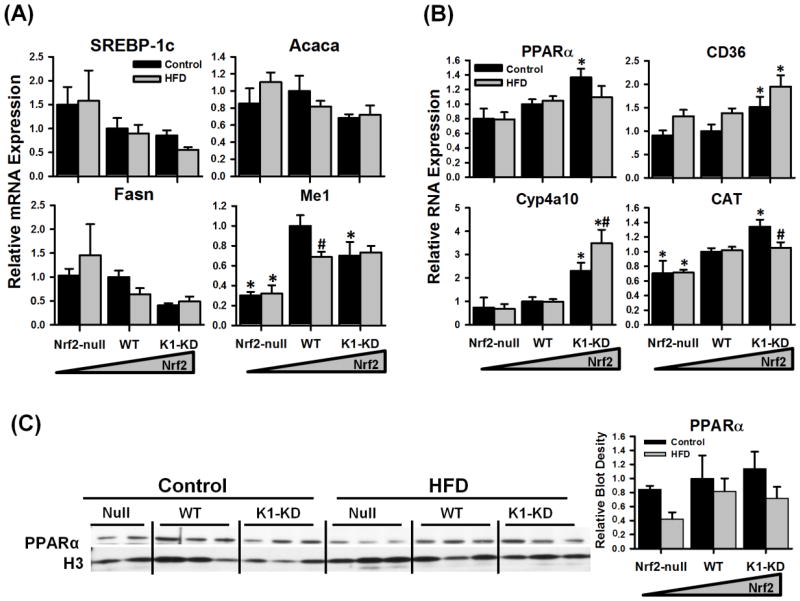
In liver, sterol regulatory element-binding protein 1c (SREBP-1c) is a critical transcription factor that mediates the insulin signal in regulating fatty acid synthesis. Acetyl-CoA carboxylase 1 (Acaca) and fatty acid synthase (Fasn) are two key enzymes for hepatic fatty acid synthesis, and are both transcriptionally regulated by SREBP-1c. Messenger RNA of SREBP-1c, Acaca, and Fasn was not statistically different among genotypes on either diet but had a trend of being higher in livers of Nrf2-null mice or lower in K1-KD mice than in WT mice, on both the control and HFD (Fig. 5A). However, the differences were not statistically significant. Malic enzyme (Me1) is an enzyme involved in pyruvate cycling that transfers reducing equivalents from NADH to NADPH, which is important for the reductive reactions of FA synthesis. The transcription of Me1 is regulated by SREBP-1c. Me1 mRNA was expressed lower in livers of Nrf2-null mice than in WT mice. In addition, Acaca, Fasn, and Me1 are also transcriptionally regulated by ChREBP. Similar to glucose metabolism-genes, FA synthesis genes in liver were not significantly affected by the HFD or Nrf2 activity.
Figure 5. Hepatic mRNA and protein expression of fatty acid metabolism-related genes.
(A) Messenger RNA of genes involved in fatty acid biosynthesis. (B) Messenger RNA expression of genes involved in fatty acid catabolism. (C) Immunoblots of PPARα protein using nuclear fraction prepared from livers of mice with various Nrf2 activity. Density of PPARα protein blots was normalized to the loading control H3 and presented as a ratio to wild-type control. Asterisks (*) represent statistical differences (p < 0.05) between mice with altered Nrf2 activity and WT mice on the same diet. Pounds (#) represent statistical differences (p < 0.05) between HFD-treated and control diet-treated mice of the same genotype.
Peroxisome proliferator-activated receptor α (PPARα) regulates the transcription of a broad range of genes encoding enzymes and transporters involved in fatty acid catabolism. In mice fed the control diet, the mRNA of PPARα was higher in livers of K1-KD than the other two genotypes (Fig. 5B), although the nuclear accumulation of PPARα protein was not statistically different between genotypes (Fig. 5C). Messenger RNA of several genes involved in FA oxidation was expressed more in K1-KD mouse livers (Fig. 5B). The fatty acid translocase CD36 is an important membrane transporter that mediates the uptake of fatty acids in macrophages, endothelium, smooth muscle cells, as well as hepatocytes (He et al., 2011). Interestingly, CD36 was recently identified on mitochondrial membranes, and appears to participate in β-oxidation by moving FA across the mitochondrial membrane (Bonen et al., 2004). The mRNA of CD36 was higher in K1 -KD mouse livers than the other two genotypes when mice were fed the control or the HFD. Cytochrome P450 4a10 (Cyp4a10), an enzyme involved in FA ω-oxidation in peroxisomes, was expressed higher in livers of K1-KD mice than Nrf2-null or WT mice when the control diet was given. Cyp4a10 mRNA was further increased by feeding the HFD only in K1-KD mice. Catalase (CAT) decomposes hydrogen peroxide, a reactive byproduct of fatty acid β-oxidation in peroxisomes. Compared with WT mice, CAT mRNA was lower in Nrf2-null mouse livers fed a control diet or HFD, but higher in K1-KD mouse livers when control diet was given. CAT mRNA was slightly decreased by the HFD in K1-KD mouse livers. In summary, the Nrf2-activated K1-KD mice appear to have higher expression of fatty acid catabolism-related enzymes.
Hepatic expression of liver-derived hormones
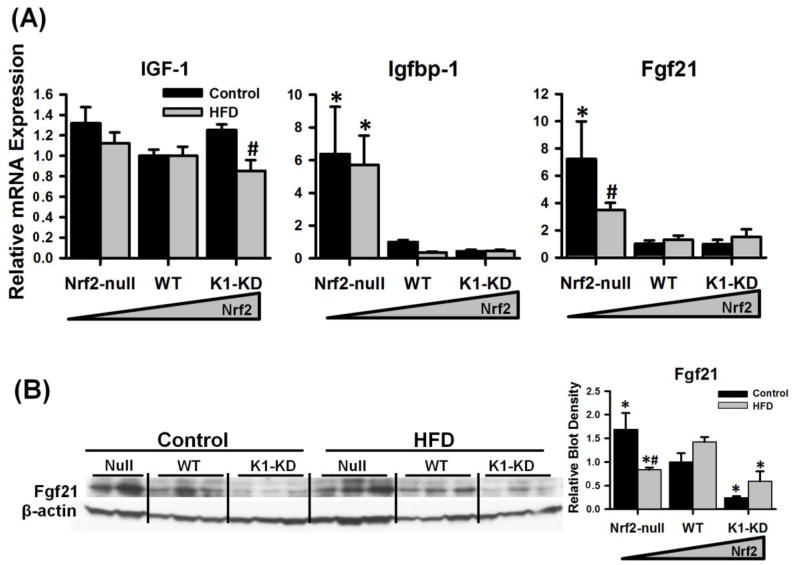
Insulin-like growth factor 1 (IGF-1) is a hormone primarily produced by liver. IGF-1 stimulates cell growth and proliferation in almost every organ in the body. Under normal physiological conditions, IGF-1 protein (approximately 98%) is bound to one of the 6 binding proteins (Igfbps) in the blood. Igfbp-1, which is predominantly synthesized in liver, is considered the key Igfbp that regulates the bioactivity of IGF-1 (Lewitt and Baxter, 1991). As shown in Fig. 6A, hepatic expression of IGF-1 mRNA was not affected by the genetic alteration of Nrf2 activity, and was slightly decreased by the HFD only in K1-KD mice. However, the mRNA of Igfbp-1 was markedly higher in livers of Nrf2-null mice, compared with mice expressing Nrf2. The HFD did not affect the mRNA expression of Igfbp-1 in any of the three genotypes. Fibroblast growth factor 21 (Fgf21) is a newly identified peptide hormone that is synthesized by the liver, but promotes glucose uptake and lipolysis in white adipose tissue. Both mRNA (Fig. 6A) and protein (Fig. 6B) of Fgf21 were much higher in livers of Nrf2-null mice compared with Nrf2-expressing mice when the control diet was fed, and were both reduced by the HFD in Nrf2-null mice. In comparison, Fgf21 protein was expressed less in livers of K1-KD mice than WT mice on either control or HFD. These data suggest that Nrf2 activity negatively regulates the hepatic expression of Igfbp-1 and Fgf21.
Figure 6. Hepatic mRNA expression of liver-derived endocrine hormones.
(A) Messenger RNA expression of insulin-like growth factor-1 (IGF-1), IGF-1 binding protein-1 (Igfbp-1), and Fgf21. (B) Immunoblots of Fgf21 in mouse livers with graded activation of Nrf2. Density of Fgf21 protein blots was normalized to the loading control β-actin and presented as a ratio to wild-type control. Asterisks (*) represent statistical differences (p < 0.05) between mice with altered Nrf2 activity and WT mice on the same diet. Pounds (#) represent statistical differences (p < 0.05) between HFD-treated and control diet-treated mice of the same genotype.
Discussion
The prevalence of obesity has increased significantly in the world. Health consequences of obesity include type 2 diabetes, metabolic syndrome, hypertension, cardiovascular disease, cancer, liver disease, etc (Ginter and Simko, 2008). Nrf2, a master regulator of antioxidative and detoxification enzymes, has recently been suggested to participate in the regulation of lipid metabolism. However, the exact function of Nrf2 in energy metabolism remains controversial. Data from the current study demonstrate that Nrf2 activity has minimal influence on HFD-induced obesity, but rather adversely affects glucose homeostasis in mice.
In the literature, both activation (Shin et al., 2009; Yu et al., 2011) and deficiency of Nrf2 (Pi et al., 2010; Chartoumpekis et al., 2011) have been implicated in preventing diet-induced obesity in mice. The present study, however, shows that genetically modified mice with graded Nrf2 activity were equally susceptible to HFD-induced obesity (Fig. 1), and exhibited similar blood and hepatic lipid profiles when a HFD was fed (Table 2). The discrepancies on the role of Nrf2 during diet-induced obesity in these studies could be due to differences in experimental design. Fat sources of the diet (Buettner et al., 2006), length of HFD feeding (VanSaun et al., 2009), gender, genetic background (Zhu et al., 2009), and age of the mice, as well as the mechanism of activating Nrf2 may all contribute to the outcome of the HFD studies.
The current study fed a Western-style HFD to genetically modified mice that have none, normal, and enhanced Nrf2 activity. In order to mimic the diets of humans, the Western HFD utilized in the present study contained 40% calories from various fat sources. All mice used in the present study were male, on the C57BL/6 background, and fed a control diet or HFD for 12 weeks. In comparison, some of the other studies used female mice (Shin et al., 2009), some used mice on a C57BL6/129SV mixed background (Pi et al, 2010); some fed mice diets containing very high amounts of fat (60% of fat calorie) (Shin et al., 2009; Chartoumpekis et al., 2011); and, some employed diets using soybean oil as the only fat source (45% calorie from fat) (Yu et al., 2011) or without indication of fat sources (Shin et al., 2009; Pi et al., 2010; Chartoumpekis et al., 2011). The longest periods of feeding the HFD in these previous studies ranged from 12 (Pi et al., 2010) to 28 weeks (Yu et al., 2011). Moreover, most of the previous studies started feeding the HFD to juvenile mice (3-6 weeks of age), whereas the present study and another study (Chartoumpekis et al., 2011) started feeding the HFD to adult mice (3-month old) to eliminate potential effects of fluctuations of hormones during development.
In addition to the aforementioned reasons, the mechanism of activating Nrf2 may also impact the experimental results. The current study used K1-KD mice, which express lower levels of Keap1, the Nrf2 repressing protein, as the Nrf2-activated model. Using a genetic model can avoid potential off-target effects of chemicals that are used to activate Nrf2. For example, a daily oral gavage of CDDO-Im inhibited food intake in mice (Shin et al., 2009). In conclusion, the aspects discussed above could all influence the outcome of a diet-induced obesity study. The current study indicates that activation of Nrf2 itself does not affect the development of obesity in mice fed a Western HFD.
The present study shows that dietary fatty acids do not activate Nrf2 signaling in mouse livers. This conclusion is supported by the fact that neither Nrf2 protein accumulation in the nucleus nor the expression of prototypical Nrf2 target genes was markedly increased by feeding a HFD in any of the 3 genotypes of mice (Fig. 3). Moreover, hepatic GSH and MDA were not affected by Western HFD in the current study (Table 2), indicating that no oxidative stress was induced in mouse livers. The lack of oxidative stress could also explain why Nrf2 signaling was not activated by feeding the Western HFD.
To further explore the role of Nrf2 in lipid metabolism, mRNA of the main regulators of fatty acid synthesis (SREBP-1c and ChREBP) and fatty acid oxidation (PPARα), as well as several key enzymes for fatty acid metabolism that are known to be regulated by these transcription factors were quantified in mouse livers. As shown in Figs. 4A and 5A, genes involved in fatty acid synthesis tended to be expressed higher in Nrf2-null mice, but the differences were not statistically significant; whereas genes involved in lipid catabolism (Fig. 5B) seemed to be expressed slightly higher in livers of the Nrf2-activated K1-KD mice. From these data, one would expect a positive correlation between Nrf2-activity and fatty acid catabolism in mouse livers. Caution is required in interpreting these data because the activity of these transcription factors is subjected to post-translational modifications that await examination.
An interesting phenotype observed in the current study is that genetic activation of Nrf2 aggravates glucose intolerance in control-fed mice, and the deficiency of Nrf2 improves glucose intolerance in mice fed a HFD. Glucose is the key substrate for the central nervous system and red blood cells. The liver plays an important role in maintaining glucose homeostasis by converting dietary carbohydrates into glycogen or triglycerides for storage after food intake, and producing glucose through glycogenolysis and gluconeogenesis under fasting conditions. The glycogen and triglyceride contents in livers of mice with graded Nrf2 activity were similar regardless of diet, suggesting that the energy storage function of liver is not affected by Nrf2. Messenger RNA of the glucose transporter 2 (data not shown), rate-limiting enzymes for glycolysis (Pklr, Fig4A; and hexokinase, data not shown) and gluconeogenesis (G6pase, Fig 4A; and phosphoenolpyruvate carboxykinase, data not shown) were either unaffected by Nrf2 activity, or slightly elevated when Nrf2 is absent. These data suggest that the capability of the liver in handling glucose is not markedly impacted by the absence or activation of Nrf2, at least at the gene transcription level.
Insulin and glucagon are the major hormones that control the transcription of glycolytic and lipogenic genes in liver. Because neither circulating insulin concentrations nor insulin tolerance was affected by the absence or activation of Nrf2 (Fig.2), the pancreatic function and insulin sensitivity in peripheral tissues appear to be normal in mice with various Nrf2 activities. In liver, insulin exerts its transcriptional regulation on lipogenic genes mainly through the transcription factor SREBP-1c. Messenger RNA data from this study (Fig. 5A) suggest that Nrf2-null mice might have better hepatic insulin sensitivity because various lipogenic genes tended to be expressed slightly more in Nrf2-null mice and slightly less in K1-KD mice. Therefore, enhanced lipogenesis from carbohydrates could be one of the reasons that Nrf2-null mice had lower blood glucose concentrations.
In addition to its function in processing nutrients, the liver also synthesizes hormones that help maintain whole-body energy homeostasis. One of them is IGF-1, which can lower blood glucose levels by promoting glucose uptake in peripheral tissues and inhibiting hepatic glucose production (Sjogren et al., 2002). The biological access of IGF-1 to target tissues is tightly regulated by a family of IGF-binding proteins. Of the six known Igfbps, Igfbp-1 is predominantly synthesized by liver and is considered the major regulator of the immunoreactive IGF-1 in the circulation (Lewitt and Baxter, 1991). Increased Igfbp-1 protein in vivo is associated with reduced free IGF-1 and hyperglycemia in rodents (Lewitt et al., 1991; Murphy, 2000). In the current study, IGF-1 mRNA was comparable in livers of mice with graded Nrf2 activity, which is consistent with a previous study demonstrating that the circulating concentration of total IGF-1 was not affected by Nrf2 deficiency (Beyer et al., 2007). Nevertheless, the mRNA of Igfbp-1 was markedly higher in livers of Nrf2-null mice (Fig. 6). The transcription of Igfbp-1 in liver is repressed by insulin, and this repression is impaired by oxidative stress due to the interruption of insulin-dependent phosphorylation of mammalian target of rapamycin (Kimura et al., 2010). Therefore, the increased mRNA of Igfbp-1 in Nrf2-null mice could be due to the more oxidative cellular environment in their hepatocytes. However, the elevated Igfbp-1 mRNA did not seem to result in higher circulating Igfbp-1 protein in Nrf2-null mice, because the blood glucose concentration is lower instead of higher in these mice compared with WT and K1-KD mice. This suggests additional regulation of Igfbp-1 at steps of post-transcription, post-translation, as well as protein secretion.
The current study and previous reports (Beyer et al., 2007; Zhang et al., 2010; Wu et al., 2011) demonstrate that enhanced Nrf2 activity is correlated with increased reducing equivalents (i.e., GSH, NADH, and NADPH) and decreased oxidative stress in liver. In addition to scavenging oxidative stress, these reducing equivalents are also important for glucose and lipid metabolism. In addition, growing evidence indicates that reactive oxygen species are not just inert by-products of mitochondrial respiration, but rather function as signaling molecules in mammalian cells. For example, H2O2 is a signal for glucose-stimulated insulin secretion in the pancreas (Pi et al., 2007). The absence (Nrf2-null) or activation (K1-KD) of Nrf2 can alter the intracellular redox balance in hepatocytes or other types of cells, and thus interfere with cell metabolism pathways that require reactive oxygen species as signaling molecules.
Fgf21 is a newly identified endocrine hormone that is abundantly expressed in liver, released into the blood, and promotes glucose uptake and lipolysis in white adipose tissue (Kharitonenkov et al., 2005; Inagaki et al., 2007). Over-expression or administration of recombinant Fgf21 prevents weight gain, promotes weight loss, improves hepatic and peripheral insulin sensitivity, as well as decreases plasma glucose and triglycerides in multiple obesity/diabetes models (Kharitonenkov et al., 2005; Kharitonenkov et al., 2007; Xu et al., 2009). The expression of Fgf21 in liver is transcriptionally regulated by PPARα in response to fasting (Inagaki et al., 2007). Enhanced expression of Fgf21 mRNA in livers of Nrf2-null mice was first described in our previous study (Zhang et al., 2010). Recently, another group reported that Nrf2 deficiency correlated with higher hepatic Fgf21 mRNA and increased plasma Fgf21, which they suggested partially protected mice from HFD-induced obesity and insulin intolerance (Chartoumpekis et al., 2011). Moreover, they demonstrated that Nrf2 directly repressed the transcription of Fgf21 in ST-2 cells. Consistent with these previous reports, the current study shows that Fgf21 protein was expressed more in livers of Nrf2-null mice and less in Nrf2-activated K1-KD mice. Therefore, the improved glucose tolerance of Nrf2-null mice could be the result of elevated expression of the anti-diabetic hormone Fgf21 in liver. However, the enhanced expression of Fgf21 in Nrf2-null mice was not sufficient to prevent HFD-induced weight gain within a 12-week period in either the current study or a previous report (Chartoumpekis et al., 2011). It has been shown that i.p. administration of recombinant Fgf21 prevents weight gain only at higher doses (>1 mg/kg/day), but was able to lower blood glucose at a much lower dose (0.1 mg/kg/day) in HFD-fed mice (Xu et al., 2009). Thus, the increase of Fgf21 in Nrf2-null mice should be sufficient to improve glucose tolerance, but not enough to prevent weight gain in mice fed a HFD.
In conclusion, by using mice with genetically altered Nrf2 activity, the current study demonstrates that Nrf2 does not prevent the development of obesity induced by a Western HFD in mice, nor directly regulates fatty acid metabolism in liver. Nrf2 deficiency improves glucose tolerance in mice fed a HFD, whereas Nrf2 activation worsens glucose tolerance in mice fed either a control diet or a HFD. The effect of Nrf2 on glucose homeostasis appears to be mediated by its negative regulation of the anti-diabetic hormone Fgf21 in liver, as well as its effects on cell metabolism pathways that interact with reactive oxygen species.
Highlights.
Nrf2 deficiency improves glucose tolerance in mice fed a high-fat diet.
The anti-diabetic hormone, Fgf21, is highly expressed in livers of Nrf2-null mice.
The absence of Nrf2 increases the insulin-regulated Igfbp-1 mRNA in liver.
Nrf2 deficiency improves glucose tolerance by influencing Fgf21 and insulin signaling.
Acknowledgments
The authors would like to thank colleagues in Dr. Klaassen's laboratory for helping in collecting tissues and critically reviewing the manuscript. This research was supported by NIH grants: DK-081461, ES-019487, and ES-009649.
Abbreviations
- Acaca
acetyl-CoA carboxylase 1
- CAT
catalase
- CD36
cluster of differentiation 36
- ChREBP
carbohydrate responsive element-binding protein
- Cyp4a10
cytochrome P450 4a10
- Fasn
fatty acid synthase
- Fgf21
fibroblast growth factor 21
- G-6-P
glucose-6-phosphatase
- Gapdh
glyceraldehyde 3-phosphate dehydrogenase
- Gclc
glutamate cysteine ligase catalytic subunit
- Gst
glutathione S-transferase
- GTT
glucose tolerance test
- HFD
high-fat diet
- IGF1
insulin-like growth factor 1
- Igfbp-1
insulin-like growth factor binding protein
- ITT
insulin tolerance test
- Keap1
Kelch-like ECH-associated protein 1
- K1-KD
Keap1-knockdown
- MDA
malondialdehyde
- Me1
malic enzyme
- Nqo1
NAD(P)H:quinione oxidoreductase 1
- Nrf2
nuclear factor erythoid 2-related factor 2
- Pklr
liver-type pyruvate kinase
- PPARα
peroxisome proliferator-activated receptor alpha
- SREBP-1c
sterol regulatory element binding protein 1c
- WT
wild-type
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Aleksunes LM, Manautou JE. Emerging role of Nrf2 in protecting against hepatic and gastrointestinal disease. Toxicol Pathol. 2007;35:459–473. doi: 10.1080/01926230701311344. [DOI] [PubMed] [Google Scholar]
- Beyer TA, Xu W, Teupser D, Auf dem Keller U, Bugnon P, Hildt E, Thiery J, Kan YW, Werner S. Impaired liver regeneration in Nrf2 knockout mice: role of ROS-mediated insulin/IGF-1 resistance. Embo J. 2007;27:212–223. doi: 10.1038/sj.emboj.7601950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonen A, Campbell SE, Benton CR, Chabowski A, Coort SL, Han XX, Koonen DP, Glatz JF, Luiken JJ. Regulation of fatty acid transport by fatty acid translocase/CD36. Proc Nutr Soc. 2004;63:245–249. doi: 10.1079/PNS2004331. [DOI] [PubMed] [Google Scholar]
- Buettner R, Parhofer KG, Woenckhaus M, Wrede CE, Kunz-Schughart LA, Scholmerich J, Bollheimer LC. Defining high-fat-diet rat models:metabolic and molecular effects of different fat types. J Mol Endocrinol. 2006;36:485–501. doi: 10.1677/jme.1.01909. [DOI] [PubMed] [Google Scholar]
- Chan JY, Kwong M. Impaired expression of glutathione synthetic enzyme genes in mice with targeted deletion of the Nrf2 basic-leucine zipper protein. Biochim Biophys Acta. 2000;1517:19–26. doi: 10.1016/s0167-4781(00)00238-4. [DOI] [PubMed] [Google Scholar]
- Chan K, Lu R, Chang JC, Kan YW. NRF2, a member of the NFE2 family of transcription factors, is not essential for murine erythropoiesis, growth, and development. Proc Natl Acad Sci U S A. 1996;93:13943–13948. doi: 10.1073/pnas.93.24.13943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chartoumpekis DV, Ziros PG, Psyrogiannis AI, Papavassiliou AG, Kyriazopoulou VE, Sykiotis GP, Habeos IG. Nrf2 represses FGF21 during long-term high-fat diet-induced obesity in mice. Diabetes. 2011;60:2465–2473. doi: 10.2337/db11-0112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ginter E, Simko V. Adult obesity at the beginning of the 21st century: epidemiology, pathophysiology and health risk. Bratisl Lek Listy. 2008;109:224–230. [PubMed] [Google Scholar]
- Giudice A, Arra C, Turco MC. Review of molecular mechanisms involved in the activation of the Nrf2-ARE signaling pathway by chemopreventive agents. Methods Mol Biol. 2010;647:37–74. doi: 10.1007/978-1-60761-738-9_3. [DOI] [PubMed] [Google Scholar]
- Hayashi A, Suzuki H, Itoh K, Yamamoto M, Sugiyama Y. Transcription factor Nrf2 is required for the constitutive and inducible expression of multidrug resistance-associated protein 1 in mouse embryo fibroblasts. Biochem Biophys Res Commun. 2003;310:824–829. doi: 10.1016/j.bbrc.2003.09.086. [DOI] [PubMed] [Google Scholar]
- He J, Lee JH, Febbraio M, Xie W. The emerging roles of fatty acid translocase/CD36 and the aryl hydrocarbon receptor in fatty liver disease. Exp Biol Med (Maywood) 2011;236:1116–1121. doi: 10.1258/ebm.2011.011128. [DOI] [PubMed] [Google Scholar]
- Imhoff B, Hansen J. Tert-butylhydroquinone induces mitochondrial oxidative stress causing Nrf2 activation. Cell Biol Toxicol. 2010;26:541–551. doi: 10.1007/s10565-010-9162-6. [DOI] [PubMed] [Google Scholar]
- Inagaki T, Dutchak P, Zhao G, Ding X, Gautron L, Parameswara V, Li Y, Goetz R, Mohammadi M, Esser V, Elmquist JK, Gerard RD, Burgess SC, Hammer RE, Mangelsdorf DJ, Kliewer SA. Endocrine regulation of the fasting response by PPARα-mediated induction of fibroblast growth factor 21. Cell Metab. 2007;5:415–425. doi: 10.1016/j.cmet.2007.05.003. [DOI] [PubMed] [Google Scholar]
- Ishii T, Itoh K, Yamamoto M. Roles of Nrf2 in activation of antioxidant enzyme genes via antioxidant responsive elements. Methods Enzymol. 2002;348:182–190. doi: 10.1016/s0076-6879(02)48637-5. [DOI] [PubMed] [Google Scholar]
- Itoh K, Chiba T, Takahashi S, Ishii T, Igarashi K, Katoh Y, Oyake T, Hayashi N, Satoh K, Hatayama I, Yamamoto M, Nabeshima Y. An Nrf2/small Mafheterodimer mediates the induction of phase II detoxifying enzyme genes through antioxidant response elements. Biochem Biophys Res Commun. 1997;236:313–322. doi: 10.1006/bbrc.1997.6943. [DOI] [PubMed] [Google Scholar]
- Jeyapaul J, Jaiswal AK. Nrf2 and c-Jun regulation of antioxidant response element (ARE)-mediated expression and induction of gamma-glutamylcysteine synthetase heavy subunit gene. Biochem Pharmacol. 2000;59:1433–1439. doi: 10.1016/s0006-2952(00)00256-2. [DOI] [PubMed] [Google Scholar]
- Katsuoka F, Motohashi H, Ishii T, Aburatani H, Engel JD, Yamamoto M. Genetic evidence that small maf proteins are essential for the activation of antioxidant response element-dependent genes. Mol Cell Biol. 2005;25:8044–8051. doi: 10.1128/MCB.25.18.8044-8051.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kensler TW, Wakabayashi N, Biswal S. Cell survival responses to environmental stresses via the Keap1-Nrf2-ARE pathway. Annu Rev Pharmacol Toxicol. 2007;47:89–116. doi: 10.1146/annurev.pharmtox.46.120604.141046. [DOI] [PubMed] [Google Scholar]
- Kharitonenkov A, Shiyanova TL, Koester A, Ford AM, Micanovic R, Galbreath EJ, Sandusky GE, Hammond LJ, Moyers JS, Owens RA, Gromada J, Brozinick JT, Hawkins ED, Wroblewski VJ, Li DS, Mehrbod F, Jaskunas SR, Shanafelt AB. FGF-21 as a novel metabolic regulator. J Clin Invest. 2005;115:1627–1635. doi: 10.1172/JCI23606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kharitonenkov A, Wroblewski VJ, Koester A, Chen YF, Clutinger CK, Tigno XT, Hansen BC, Shanafelt AB, Etgen GJ. The metabolic state of diabetic monkeys is regulated by fibroblast growth factor-21. Endocrinology. 2007;148:774–781. doi: 10.1210/en.2006-1168. [DOI] [PubMed] [Google Scholar]
- Kimura K, Katsumata Y, Ozawa T, Tawara S, Igarashi K, Cho Y, Shibata N, Hakuno F, Takahashi S, Takenaka A. Effect of paraquat-induced oxidative stress on insulin regulation of insulin-like growth factor-binding protein-1 gene expression. J Clin Biochem Nutr. 2010;46:157–167. doi: 10.3164/jcbn.09-97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitteringham NR, Abdullah A, Walsh J, Randle L, Jenkins RE, Sison R, G CEP, Sanderson C, Williams S, Higgins L, Yamamoto M, Hayes J, Park BK. Proteomic analysis of Nrf2 deficient transgenic mice reveals cellular defence and lipid metabolism as primary Nrf2-dependent pathways in the liver. J Proteomics. 2010;73:1612–1631. doi: 10.1016/j.jprot.2010.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klaassen CD, Reisman SA. Nrf2 the rescue: effects of the antioxidative/electrophilic response on the liver. Toxicol Appl Pharmacol. 2010;244:57–65. doi: 10.1016/j.taap.2010.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klaassen CD, Slitt AL. Regulation of hepatic transporters by xenobiotic receptors. Curr Drug Metab. 2005;6:309–328. doi: 10.2174/1389200054633826. [DOI] [PubMed] [Google Scholar]
- Lewitt MS, Baxter RC. Insulin-like growth factor-binding protein-1: a role in glucose counterregulation? Mol Cell Endocrinol. 1991;79:C147–152. doi: 10.1016/0303-7207(91)90086-8. [DOI] [PubMed] [Google Scholar]
- Lewitt MS, Denyer GS, Cooney GJ, Baxter RC. Insulin-like growth factor-binding protein-1 modulates blood glucose levels. Endocrinology. 1991;129:2254–2256. doi: 10.1210/endo-129-4-2254. [DOI] [PubMed] [Google Scholar]
- Maher JM, Dieter MZ, Aleksunes LM, Slitt AL, Guo G, Tanaka Y, Scheffer GL, Chan JY, Manautou JE, Chen Y, Dalton TP, Yamamoto M, Klaassen CD. Oxidative and electrophilic stress induces multidrug resistance-associated protein transporters via the nuclear factor-E2-related factor-2 transcriptional pathway. Hepatology. 2007;46:1597–1610. doi: 10.1002/hep.21831. [DOI] [PubMed] [Google Scholar]
- McGrath LT, Elliott RJ. Lipid analysis and fatty acid profiles of individual arterial atherosclerotic plaques. Anal Biochem. 1990;187:273–276. doi: 10.1016/0003-2697(90)90456-j. [DOI] [PubMed] [Google Scholar]
- Murphy LJ. Overexpression of insulin-like growth factor binding protein-1 in transgenic mice. Pediatr Nephrol. 2000;14:567–571. doi: 10.1007/s004670000347. [DOI] [PubMed] [Google Scholar]
- Okada K, Shoda J, Taguchi K, Maher JM, Ishizaki K, Inoue Y, Ohtsuki M, Goto N, Takeda K, Utsunomiya H, Oda K, Warabi E, Ishii T, Osaka K, Hyodo I, Yamamoto M. Ursodeoxycholic acid stimulates Nrf2-mediated hepatocellular transport, detoxification, and antioxidative stress systems in mice. Am J Physiol Gastrointest Liver Physiol. 2008;295:G735–747. doi: 10.1152/ajpgi.90321.2008. [DOI] [PubMed] [Google Scholar]
- Passonneau JV, Lauderdale VR. A comparison of three methods of glycogen measurement in tissues. Anal Biochem. 1974;60:405–412. doi: 10.1016/0003-2697(74)90248-6. [DOI] [PubMed] [Google Scholar]
- Pi J, Bai Y, Zhang Q, Wong V, Floering LM, Daniel K, Reece JM, Deeney JT, Andersen ME, Corkey BE, Collins S. Reactive oxygen species as a signal in glucose-stimulated insulin secretion. Diabetes. 2007;56:1783–1791. doi: 10.2337/db06-1601. [DOI] [PubMed] [Google Scholar]
- Pi J, Leung L, Xue P, Wang W, Hou Y, Liu D, Yehuda-Shnaidman E, Lee C, Lau J, Kurtz TW, Chan JY. Deficiency in the nuclear factor E2-related factor-2 transcription factor results in impaired adipogenesis and protects against diet-induced obesity. J Biol Chem. 2010;285:9292–9300. doi: 10.1074/jbc.M109.093955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin S, Wakabayashi J, Yates MS, Wakabayashi N, Dolan PM, Aja S, Liby KT, Sporn MB, Yamamoto M, Kensler TW. Role of Nrf2 in prevention of high-fat diet-induced obesity by synthetic triterpenoid CDDO-imidazolide. Eur J Pharmacol. 2009;620:138–144. doi: 10.1016/j.ejphar.2009.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sjogren K, Jansson JO, Isaksson OG, Ohlsson C. A model for tissue-specific inducible insulin-like growth factor-I (IGF-I) inactivation to determine the physiological role of liver-derived IGF-I. Endocrine. 2002;19:249–256. doi: 10.1385/ENDO:19:3:249. [DOI] [PubMed] [Google Scholar]
- VanSaun MN, Lee IK, Washington MK, Matrisian L, Gorden DL. High fat diet induced hepatic steatosis establishes a permissive microenvironment for colorectal metastases and promotes primary dysplasia in a murine model. Am J Pathol. 2009;175:355–364. doi: 10.2353/ajpath.2009.080703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu KC, Cui JY, Klaassen CD. Beneficial role of Nrf2 in regulating NADPH generation and consumption. Toxicol Sci. 2011;123:590–600. doi: 10.1093/toxsci/kfr183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J, Lloyd DJ, Hale C, Stanislaus S, Chen M, Sivits G, Vonderfecht S, Hecht R, Li YS, Lindberg RA, Chen JL, Young Jung D, Zhang Z, Ko HJ, Kim JK, Véniant MM. Fibroblast growth factor 21 reverses hepatic steatosis, increases energy expenditure, and improves insulin sensitivity in diet-induced obese mice. Diabetes. 2009;58:250–259. doi: 10.2337/db08-0392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yates MS, Tran QT, Dolan PM, Osburn WO, Shin S, McCulloch CC, Silkworth JB, Taguchi K, Yamamoto M, Williams CR, Liby KT, Sporn MB, Sutter TR, Kensler TW. Genetic versus chemoprotective activation of Nrf2 signaling: overlapping yet distinct gene expression profiles between Keap1 knockout and triterpenoid-treated mice. Carcinogenesis. 2009;30:1024–1031. doi: 10.1093/carcin/bgp100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Z, Shao W, Chiang Y, Foltz W, Zhang Z, Ling W, Fantus I, Jin T. Oltipraz upregulates the nuclear respiratory factor 2 alpha subunit (NRF2) antioxidant system and prevents insulin resistance and obesity induced by a high-fat diet in C57BL/6J mice. Diabetologia. 2011;54:922–934. doi: 10.1007/s00125-010-2001-8. [DOI] [PubMed] [Google Scholar]
- Zhang YKJ, Yeager RL, Tanaka Y, Klaassen CD. Enhanced expression of Nrf2 in mice attenuates the fatty liver produced by a methionine- and choline-deficient diet. Toxicol Appl Pharmacol. 2010;245:326–334. doi: 10.1016/j.taap.2010.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu M, Ji G, Jin G, Yuan Z. Different responsiveness to a high-fat/cholesterol diet in two inbred mice and underlying genetic factors: a whole genome microarray analysis. Nutr Metab. 2009;6:43. doi: 10.1186/1743-7075-6-43. [DOI] [PMC free article] [PubMed] [Google Scholar]