Abstract
An efficient screening method was developed for functionalized DNA-targeted platinum-containing hybrid anticancer agents based on metal-mediated amine-to-nitrile addition, a form of “click” chemistry. The goal of the study was to generate platinum–acridine agents for their use as cytotoxic “warheads” in targeted and multifunctional therapies. This was achieved by introducing hydroxyl, carboxylic acid, and azide functionalities in the acridine linker moiety and by varying the nonleaving groups attached to platinum. The assay, which was based on microscale reactions between 6 platinum–nitrile complexes and 10 acridine derivatives, yielded a small library of 60 platinum–acridines. Reactions were monitored and product mixtures were quantitatively analyzed by automated in-line high-performance liquid chromatography– electrospray mass spectrometry (LC-ESMS) analysis and subjected to cell viability screening using a non-radioactive cell proliferation assay. The new prescreening methodology proves to be a powerful tool for establishing structure–activity relationships and for identifying target compounds.
INTRODUCTION
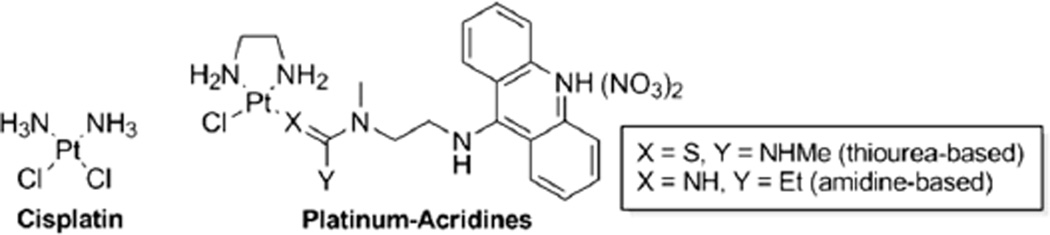
Platinum-based anticancer drugs continue to be the cornerstone of many chemotherapy regimens despite their clinical drawbacks and the recent advent of more tolerable molecularly targeted therapies.1, 2 DNA-targeted agents acting by novel mechanisms at the molecular level show considerable promise as potential treatments for intractable tumors.3 In particular, we and others have demonstrated that DNA adducts that do not mimic the cytotoxic lesions produced by the anticancer agent cisplatin (cis-diamminedichloridoplatinum(II), Chart 1) may overcome tumor resistance to the clinical platinum drugs.4–6 Platinum–acridine agents, represented by the prototypical structures in Chart 1, unlike cisplatin, do not induce DNA cross-links but form monofunctional–intercalative adducts.7 The unique structural and thermodynamic features of this dual binding mode and the rapid formation of the hybrid adducts in cellular DNA result in an up to 500-fold higher cytotoxic potency compared to cisplatin in aggressive, rapidly proliferating cancers.8–12 While the cytotoxic properties of the platinum–acridines translate into promising tumor growth inhibition in vivo, the dose-limiting systemic toxicity observed in mice suggests that modifications of these agents are necessary to improve their pharmacological properties.12 Specifically, we are interested in attaching the platinum–acridines as cytotoxic “warheads” to receptor- or tumor tissue-targeted vehicles. To achieve this goal, it is necessary to install chemically or enzymatically reversible linkers in the original structures. This is not a trivial task as many modifications made to a cytotoxic agent for the purpose of delivering it may compromise its target interactions and decrease its potency.13
Chart 1.
To accelerate the preclinical development of our technology, we have developed new methodology based on efficient platinum-mediated click chemistry to generate and screen a modular library of functionalized, conjugatable platinum–acridines. Fragment-based approaches using simple condensation and click chemistries have demonstrated great utility for the design of enzyme-targeted inhibitors.14, 15 Here, we demonstrate, for the first time, that modular library screening can be applied to DNA-interacting anticancer agents. Proof-of-concept data is presented demonstrating that the new assay provides a powerful tool for establishing structure– activity relationships (SAR) and identifying candidates for the desired biological application.
RESULTS AND DISCUSSION
To generate the desired compound library, sets of suitably modified platinum complexes and 9-aminoacridine derivatives, the two building blocks of the hybrid agents, were synthesized. We then used highly efficient amine addition to platinum-coordinated nitrile ligand, a classical metal-catalyzed reaction for forming CN bonds,16 to generate the amidine-linked hybrids (based on the prototype with X = NH in Chart 1). The clean conversion to hybrid agent without the formation of stoichiometric by-products, which is reminiscent of organic click chemistry,17 provides an ideal platform for generating diversity within the library of platinum–acridines (for details see the Experimental Section).
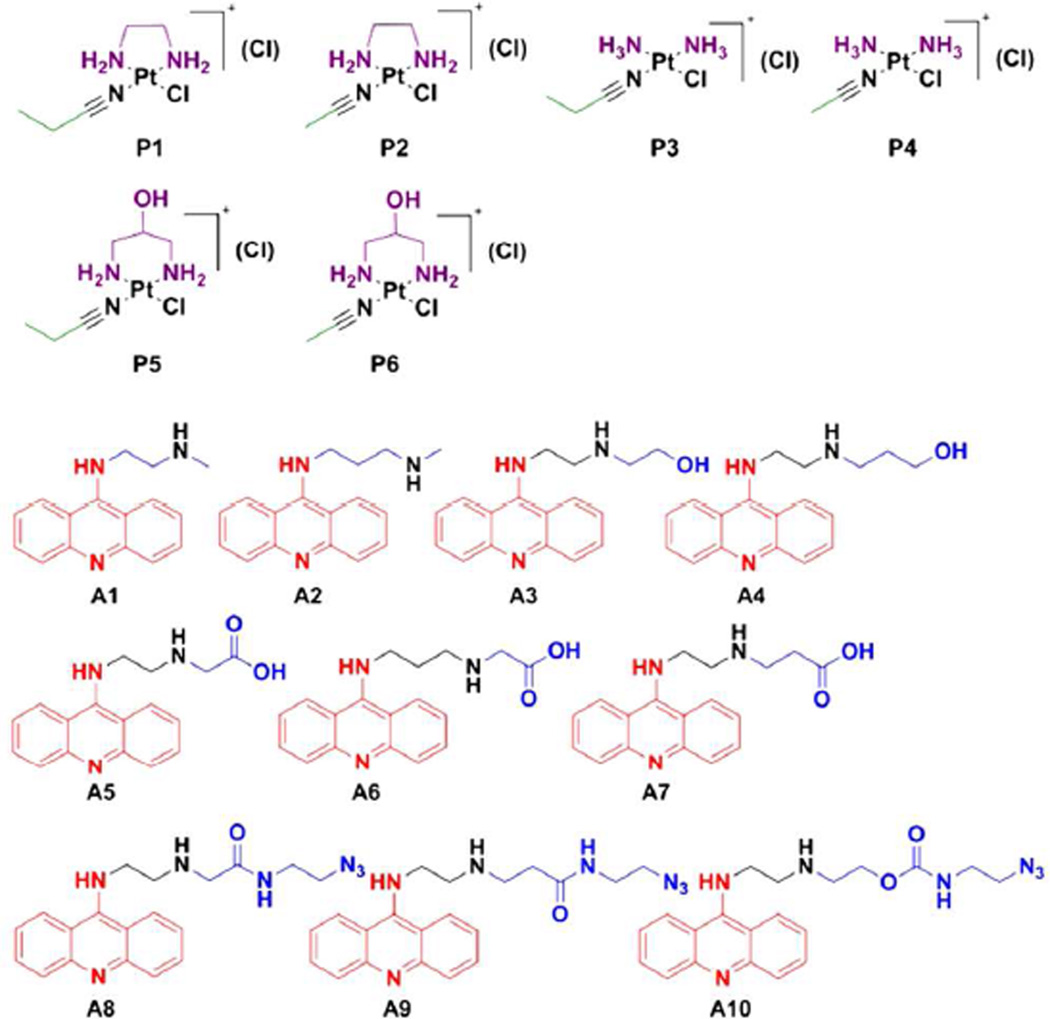
A total of 6 nitrile-modified platinum complexes and 10 acridines were synthesized as building blocks for the modular click assembly of 60 hybrids (Chart 2). Structural diversity in this small library was achieved by varying the nonleaving group(s) (R1) and nitrile ligands (R2) in the platinum moieties, as well as the geometry of the acridine side chain (R3). Several of the unmodified parent compounds studied previously were also generated. Conjugatable functional groups were placed strategically within the platinum and acridine moieties as attachment points for targeted carriers. These include hydroxyl (in P5, P6, A3, and A4) and carboxylic acid (in A5–A7) groups, which lend themselves as coupling partners in (reversible) ester, carbamate, and amide linkages.18 In addition, terminal azide groups were installed in extended side chains using amide (A8, A9) and carbamate (A10) coupling chemistry. These derivatives are also of interest for their use as imaging tools in bioorthogonal reactions19 with alkyne-modified fluorophores to study the subcellular distribution of the hybrid agents.
Chart 2.
Library of Platinum (P) and Acridine (A) Building Blocks with Design Elements Highlighted.
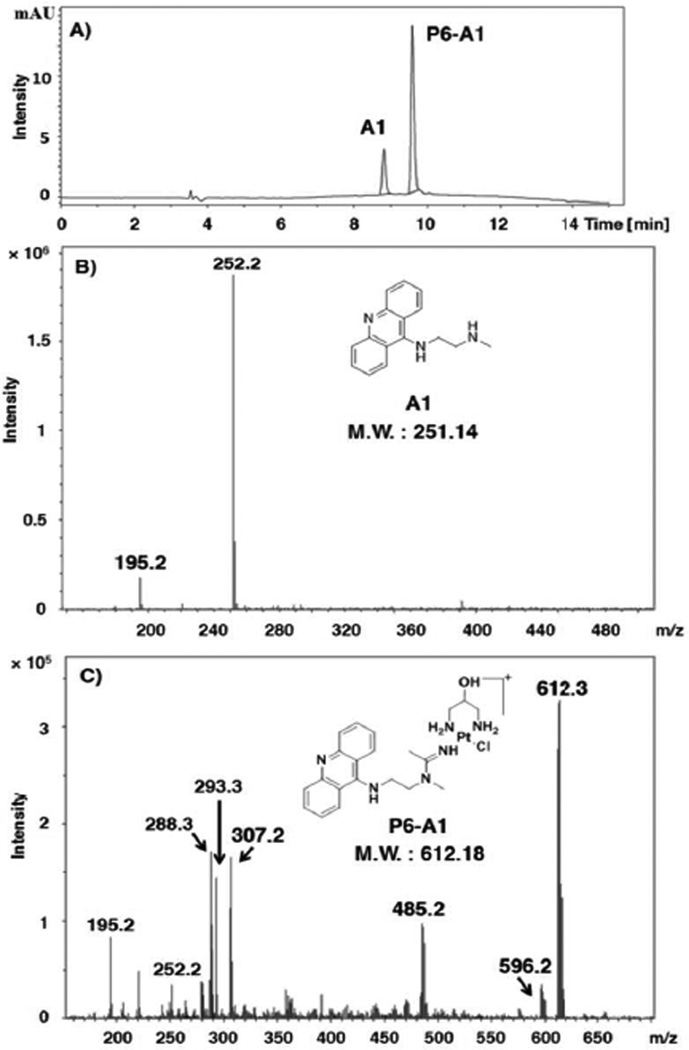
To demonstrate the utility of this approach, 60 micro-scale reactions were assembled containing stoichiometric amounts of platinum (P) and acridine (A) and allowed to incubate in DMF under conditions that produce hybrid agent without any detectable side products (see experimental details and the Supporting Information). Automated in-line high-performance liquid chromatography–electrospray mass spectrometry (LC-ESMS) was used to monitor the progress of the reactions and to calculate the yield of each hybrid. This was possible by integrating the HPLC traces recorded at an acridine-specific wavelength assuming the chromophores in the acridine percursors (A) and in the newly formed hybrids show the same absorptivity. An example of an LC profile along with ESMS characterization of the reaction mixture is given in Figure 1 for hybrid P6-A1 (for a complete set of 60 LC-MS profiles see the Supporting Information). Note the absence of side products and high yield (80%) of the conversion. Baseline separation of the two fractions corresponding to unreacted acridine and product platinum–acridines was observed for all 60 samples analysed, which greatly facilitated quantification and mass spectrometric characterization of the hybrids.
Figure 1.
High-throughput LC-ESMS analysis of “click” reaction mixtures. (A) Reverse-phase HPLC trace for the reaction of platinum precursor P6 with acridine A1. (B) ESMS spectrum of A1 (HPLC fraction with retention time 8.8 min) recorded in positive-ion mode. Characteristic molecular and fragment ions: [M+H]+ (m/z 252.2) (C) ESMS spectrum of P6-A1 (HPLC fraction with retention time 9.6 min) recorded in positive-ion mode. Characteristic molecular and fragment ions: [M]+ (m/z 613.2), [M+H]2+ (m/z 307.1, z = 2), [M-OH+H]+ (m/z 596.2), [M-{PtCl(pn2-OH)}]+ (free acridine–amidine ligand, m/z 293.3) [M-2H-Cl]+ (m/z 575.3), [M-H-Cl]2+ (m/z 288.3, z = 2), [M-pn2-OH-Cl]+ (m/z 485.2). The structures of A1 and P6-A1 are shown as insets in (B) and (C).
Species in solution were identified by their molecular-ion peaks and fragments generated by in-source collision-induced dissociation (CID). The reaction mixtures were diluted with appropriate amounts of media so that each sample contained the same concentration of hybrid agent prior to incubations with cancer cells. NCI-H460 cells were then exposed to a fixed concentration of 50 nM of each platinum–acridine and the viability of the cancer cells relative to untreated control was assessed after 72 h of incubation using a colorimetric cell proliferation assay. The assay was also performed with the platinum and acridine precursors, which proved to be at least two orders of magnitude less cytotoxic than the hybrids. These control experiments were necessary to demonstrate that unreacted precursors in reactions that did not go to completion did not significantly contribute to the inhibition of cell proliferation (see the Supporting Information for an example). Finally, to further validate the pre-screening approach, selected compounds of interest were resynthesized and their IC50 values determined in the same cell line (Chart 2).
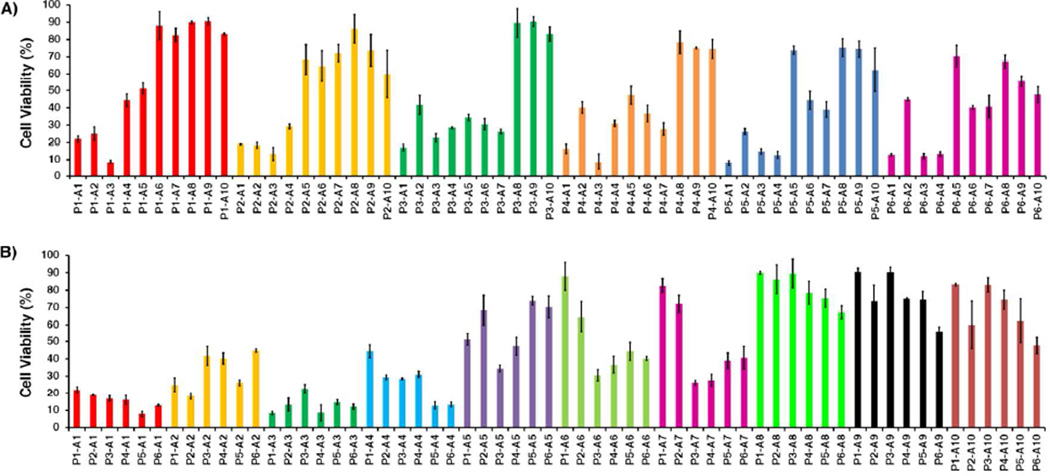
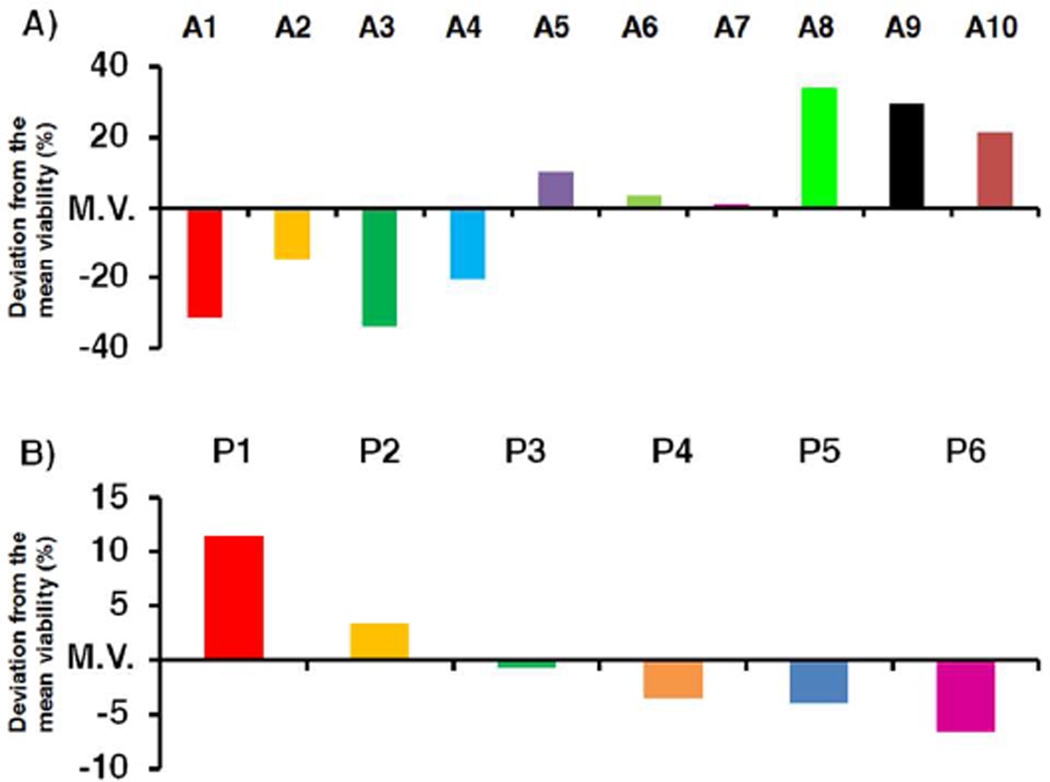
To validate our combinatorial approach as a tool for establishing SAR in platinum–acridines, the data set generated was examined by plotting cell viabilities for the 60 samples (P1-A1 to P6-A10). To gain insight into the relative importance of the fragments P and A in each compound, samples were numerically sorted into groups of hybrids containing the same platinum moiety and groups sharing the same acridine ligand (Figure 2). The former alignment demonstrates that for hybrids containing the same platinum moiety (P1–P6) the biological activity usually decreases significantly with increasing degree of modification and length of the (functionalized) 9-aminoacridine side chain (Figure 2A). Analysis of the global activity profiles for P1–P6 also shows that variation of the nitrile ligand (R2 = Et in P1, P3, and P5 vs. R2 = Me in P2, P4, and P6) leads to little variation in activity and produces pairs of most similar compounds. By contrast, replacement of the ethane-1,2-diaminoethane (en) non-leaving group with ammine (NH3) ligands (P1/P2 vs. P3/P4) enhances the cytotoxicity of hybrids containing acridines A5–A7. For several of the less active derivatives introduction of rac-1,3-diaminopropan-2-ol (pn2-OH) as a nonleaving group (in P5 and P6) resulted in improved cancer cell kill (average cell viabilities in Figure 2A were 55% for P1/P2, 45% for P3/P4, and 42% for P5/P6, relative to control cells). Likewise, a plot of cell viabilities for the 10 types of hybrids defined by common acridine moieties (Figure 2B) gives important clues about SAR in this library of compounds. In general, the derivatives containing ethylene groups in the acridine side chain performed better than those containing extended propylene chains, based on average cell viabilities of 15% and 30% calculated for the pairs A1/A3 and A2/A4, respectively. Derivatives A6 and A7, which contain extended, carboxylic acid-modified side chains that only differ in the positioning of the secondary amino function (Chart 2) produce an average reduction in cell viability of merely 50% while showing similar activity profiles. Extension of the side chains to contain azide functionalities (A8–A10) follows the same trend and leads to a further decrease in cytotoxicity of the corresponding hybrids, which form a cluster of highly similar profiles (Figure 2B). On the basis of mean cell viabilities calculated across the entire set of 60 library members, hybrids containing A1, A3, and P6 resulted in the strongest cell kill effect (Figure 3).
Figure 2.
Biological activity profiles for the 60 compounds based on the viabilities of treated cells relative to untreated control determined in a colorimetric cell proliferation assay. The test compounds are sorted and color-coded by common platinum moieties in (A) and by common acridine moieties in (B). Error bars indicate ± standard deviations for sets of three data points for each compound.
Figure 3.
Relative potencies of acridine (A) and platinum (B) fragments as building blocks in hybrid agents expressed as ± % deviations from the mean cell viability (M.V.) determined across the entire set of 60 library members.
To assess the utility of the library screen as a tool for target compound identification, a structurally diverse subset of four analogues of interest was resynthesized and tested in NCIH460 cells. IC50 values were calculated from the corresponding dose–response curves (see in the Supporting Information) and are summarized along with data acquired previously for the parent compounds in Table 1. Of the hybrids chosen, the azide-functionalized compound P1-A8 required the highest concentration (110 ± 8 nM) to inhibit cell proliferation by 50%, consistent with its relatively poor performance in the prescreen. (It should be noted, however, that this derivative is still an order of magnitude more potent than cisplatin in this cell line.9) Compounds that reduced cell viability to levels of ≤20% of control in the high-throughput assay resulted in significantly lower IC50 values. The hydroxy-modified hybrids P4-A3 and P6-A1 showed the highest cell-kill potential with the latter reaching cytotoxicity levels in the low-nanomolar concentration range similar to the unmodified platinum–acridines P1-A1 and P2-A1.10, 12
Table 1.
Summary of structural elements and biological activity for selected library members.
The pre-screening method provides insight into the relative importance of the two functionally interdependent components, based on the similarity of activity profiles and clustering of individual library members. Because the method is based on testing reaction mixtures without purification steps at an arbitrarily fixed concentration of test compound, the cell viabilities extracted from it do not correlate with IC50 values, which have to be calculated from the corresponding drug–response curves. Potential complications in evaluating the biological activity of library members may arise in cases in which incomplete conversion of the building blocks to hybrid is observed. Under such circumstances, unreacted platinum or acridine precursors may contribute to the cell kill observed in the pre-screen. Furthermore, synergistic effects between multiple components, which might lead to enhanced cell kill, cannot be completely ruled out. Generally these mixtures resulted in poor inhibition of cancer cell proliferation, indicating that both the precursors and the hybrid agent were only marginally cytotoxic. However, in some cases, such as P3-A7, which showed only 50% conversion but reduced cell viability by more than 70%, it had to be confirmed that the hybrid and not unreacted precursor caused the cell kill, which proved to be the case (see the Supporting Information). Finally, in addition to demonstrating the feasibility of our library design and its validity as a prescreening tool, we have identified promising hydroxyl-modified hybrids P4-A3 and P6-A1 as potential “warheads” for the desired application of targeted prodrug delivery.
CONCLUSION
In summary, we have demonstrated the potential of library screening for delineating SAR in a class of DNA-targeted hybrid agents and for lead discovery. The modular click approach in conjunction with the facile preparation of building blocks provides a powerful tool for efficient screening of second-generation derivatives of these promising cytotoxics. The use of metalbased coupling chemistry as an efficient screening tool for the discovery of metal-containing (hybrid) pharmacophores is a previously unexplored approach. Here, we provided proof-ofconcept data by generating 60 hybrid agents and testing them in one cancer cell line. We envision that this assay can easily be extended to multi-step library synthesis, for instance by combining the platinum-mediated additions with other forms of click or coupling chemistries. Likewise, parallel testing in multiple cell lines would add another dimension to this assay. Such an approach would ultimately lend itself to cluster analysis of extended databases with the ultimate goal of designing personalized oncology drugs that can be targeted to specific cancer types.
EXPERIMENTAL SECTION
Reagents and Instrumentation
All reagents were used as obtained from commercial sources without further purification unless indicated otherwise. 1H NMR spectra of the target compounds and intermediates were recorded on a Bruker Advance 300 MHz instrument. Proton-decoupled 13C NMR spectra were recorded on a Bruker Advance 300 instrument operating at 75 MHz. Chemical shifts (δ) are given in parts per million (ppm) relative to tetramethylsilane (TMS). 1H NMR data are reported in the conventional form including chemical shifts (δ, ppm), multiplicities (s = singlet, d = doublet, t = triplet, q = quartet, m = multiplet, br = broad), coupling constants (Hz), and integral intensities). 13C{1H} NMR data are reported as chemical shifts (δ, ppm). HPLC-grade solvents were used for all HPLC and mass spectrometry experiments. LC-ESMS analysis was performed on an Agilent 1100LC/MSD ion trap mass spectrometer equipped with an atmospheric pressure electrospray ionization source. Eluent nebulization was achieved with a N2 pressure of 50 psi, and solvent evaporation was assisted by a flow of N2 drying gas (350 °C). Positive-ion mass spectra were recorded with a capillary voltage of 2800 V and a mass-to-charge scan range of 150 to 2200 m/z. To establish the purity of target compounds, samples were diluted in methanol containing 0.1 % formic acid and analyzed using a 4.6 mm × 150 mm reverse-phase Agilent ZORBAX SB-C18 (5 Um) analytical column at 25 °C. The following solvent system was used for the characterization of “click” reactions: solvent A, optima water, and solvent B, methanol/0.1% formic acid, with a flow rate of 0.5 mL/min and a gradient of 95% A to 5% A over 12 minutes. The purity of the resynthesized compounds (> 95%) was confirmed using the following solvent system: solvent A, optima water, and solvent B, methanol/0.1% formic acid, with a flow rate of 0.5 mL/min and a gradient of 95% A to 5% A over 25 minutes. A wavelength range of 363–463 nm was used to monitor the reactions and to determine reaction yields. Peak integration was done using the Agilent LC/MSD Trap Control 4.0 data analysis software.
Experimental procedure for “click” reactions
Stock solutions of the platinum–nitrile complexes P1–P6 (20 mM) and acridine derivatives A1–A10 (20 mM) were prepared in anhydrous DMF. Concentrations of A1–A10 were determined spectrophotometrically (λmax = 413 nm, ε = 10,000 M−1 cm−1). The reactions between platinum complexes and acridine derivatives were carried out in 1.5-mL Eppendorf tubes by mixing equal volumes (100 μL) of platinum–nitrile complex and acridine derivative. The reaction mixtures were placed in a shaker and incubated at 4 °C for 5 days. To detect and characterize the hybrid agents and to determine conversion yields, 1-μL samples were removed from each reaction and diluted with 1000 µL of methanol containing 0.1% formic acid prior to in-line LC-ESMS analysis. Sample injections into the LC unit were accomplished via a thermostatted (4 °C) autosampler. Chromatographic separations were performed with a 4.6 mm × 150 mm reverse-phase Agilent ZORBAX SB-C18 (5 Um) analytical column with the column temperature maintained at 25 °C. The binary mobile phase consisted of: solvent A, optima water, and solvent B, methanol/0.1% formic acid delivered at a gradient of 95% A to 5% A over 12 minutes and a flow rate of 0.5 mL/min. The formation and the extent of conversion of the platinum–acridines was monitored in the corresponding chromatograms using the LC/MSD Trap Control 4.0 data analysis software.
Typical Procedure for the Preparation of Platinum–Acridine Derivatives: Compound P1-A3
(For details of the synthesis and characterization of all complexes and precursor building blocks see the Supporting Information.) Platinum complex P1 (381 mg, 1.00 mmol) was converted to its nitrate salt by reaction with AgNO3 (162 mg, 0.950 mmol) in 7 mL of anhydrous DMF. AgCl was removed by syringe filtration, and the filtrate was cooled to −10 °C. Acridine precursor A3 (282 mg, 0.100 mmol) was added to the solution, and the suspension was stirred at −10 °C for 24 h. After treatment with activated carbon, the reaction mixture was added into 300 mL of vigorously stirred diethyl ether. The yellow precipitate was stirred for approximately 30 min and then recovered by membrane filtration and dried in a vacuum overnight. The solid was dissolved in anhydrous methanol containing one equivalent of HNO3 (1 M) and stirred at room temperature for 30 min, and the crude dinitrate salt was precipitated with 300 mL of anhydrous diethyl ether. The product was further purified by recrystallization from hot ethanol to give 606 mg of the dinitrate salt as a yellow microcrystalline solid (Yield: 82%). 1H NMR (MeOH-d4) δ 8.42 (d, J = 8.4 Hz, 2 H), 7.88 (t d, J = 6.9, 1.2 Hz, 2 H), 7.74 (d, J = 8.4 Hz, 2 H), 7.51 (t d, J = 7.2, 1.2 Hz, 2H 1 H), 6.12 (br s, 1 H), 5.32 (s, 2 H), 5.11 (s, 2 H), 4.32 (t, J = 6.7 Hz, 2 H), 3.89 (t, J = 6.7 Hz, 2H), 3.63 (t, J = 5.0 Hz, 2 H), 3.47 (t, J = 5.1 Hz, 2 H), 2.94 (q, J = 7.2 Hz, 1 H), 2.47 (m, 4 H), 1.18 (t, J = 7.2 Hz, 3 H). 13C NMR (MeOH-d4) δ 171.73, 160.21, 141.38, 136.63, 126.48, 125.35, 119.73, 114.13, 60.70, 29.26, 11.85. MS (ESI, positive-ion mode): m/z for C22H32ClN6OPt ([M]+), 627.06; found: 627.3.
Cell proliferation assay
Cell viability was assessed using the CellTiter 96 Aqueous Non-Radioactive Cell Proliferation Assay (Promega, Madison, WI, USA) as described previously.9, 10 Briefly, NCI-H460 cell suspensions were harvested and seeded into 96-well microplates at a density of 750 cells/well. The cells were preincubated at 37 °C overnight and then treated with 50 nM compound from the library reactions or serial dilutions of selected purified compounds (the DMF content in the mixtures was less than 1% at the highest incubation concentration), as well as control samples containing platinum and acridine precursors and samples containing media/DMF. After an incubation period of 72 h, 20 μL of MTS/PMS solution was added to each well and incubated at 37 °C for 4 h. The absorbance of tetrazolium dye was measured at 490 nm using a plate reader. The fraction of viable cells was calculated as a percentage of untreated control and is reported as the mean ± standard deviation for 3 incubations of each compound. IC50 values were calculated from non-linear curve fits using a sigmoidal dose–response equation in GraphPad Prism (GraphPad Software, La Jolla, CA) and are averages of two individual experiments performed in triplicate.
Supplementary Material
Scheme 1.
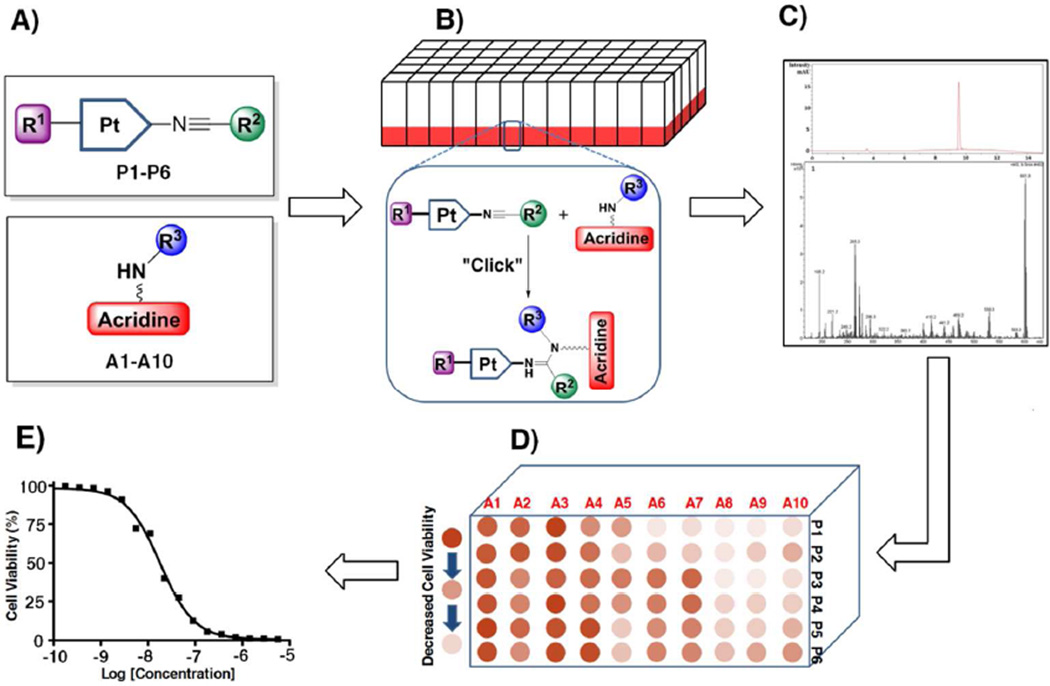
Design of the Combinatorial Screening Assaya
a (A) Synthesis of fragment libraries containing variable and functionalizable groups R1–R3. For the color-coding of residues, Chart 2. (B) Microscale coupling (“click”) reactions. (C) Qualitative and quantitative LC-ESMS analysis. (D) High-throughput screening in cancer cells on 96-well plates. (E) SAR data analysis and hit identification.
ACKNOWLEDGMENTS
Xin Qiao gratefully acknowledges support from the China Scholarship Council (CSC, grant #2011694010).
Funding Sources
This work was supported by the National Institutes of Health (grant CA101880).
ABBREVIATIONS
- LC–MS
high-performance liquid chromatography–mass spectrometry
- MTS
3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium
- NSCLC
non-small cell lung cancer
Footnotes
ASSOCIATED CONTENT
SUPPORTING INFORMATION
Experimental procedures, details of product characterization and purity, LC-ESMS analysis of “click” reactions. This material is available free of charge via the Internet at http://pubs.acs.org.
Notes
The authors declare no competing financial interest.
REFERENCES
- 1.Kelland L. The resurgence of platinum-based cancer chemotherapy. Nat. Rev. Cancer. 2007;7:573–584. doi: 10.1038/nrc2167. [DOI] [PubMed] [Google Scholar]
- 2.Graham J, Mushin M, Kirkpatrick P. Oxaliplatin. Nat. Rev. Drug. Discov. 2004;3:11–12. doi: 10.1038/nrd1287. [DOI] [PubMed] [Google Scholar]
- 3.Momekov G, Bakalova A, Karaivanova M. Novel approaches towards development of non-classical platinum-based antineoplastic agents: Design of platinum complexes characterized by an alternative DNA-binding pattern and/or tumor-targeted cytotoxicity. Curr. Med. Chem. 2005;12:2177–2191. doi: 10.2174/0929867054864877. [DOI] [PubMed] [Google Scholar]
- 4.Guddneppanavar R, Bierbach U. Adenine-N3 in the DNA minor groove - an emerging target for platinum containing anticancer pharmacophores. Anticancer Agents Med. Chem. 2007;7:125–138. doi: 10.2174/187152007779313991. [DOI] [PubMed] [Google Scholar]
- 5.Komeda S, Casini A. Next-generation anticancer metallodrugs. Curr. Top. Med. Chem. 2012;12:219–235. doi: 10.2174/156802612799078964. [DOI] [PubMed] [Google Scholar]
- 6.Komeda S. Unique platinum-DNA interactions may lead to more effective platinum-based antitumor drugs. Metallomics. 2011;3:650–655. doi: 10.1039/c1mt00012h. [DOI] [PubMed] [Google Scholar]
- 7.Baruah H, Wright MW, Bierbach U. Solution structural study of a DNA duplex containing the guanine-N7 adduct formed by a cytotoxic platinum-acridine hybrid agent. Biochemistry. 2005;44:6059–6070. doi: 10.1021/bi050021b. [DOI] [PubMed] [Google Scholar]
- 8.Qiao X, Zeitany AE, Wright MW, Essader AS, Levine KE, Kucera GL, Bierbach U. Analysis of the DNA damage produced by a platinum-acridine antitumor agent and its effects in NCI-H460 lung cancer cells. Metallomics. 2012;4:645–652. doi: 10.1039/c2mt20031g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Smyre CL, Saluta G, Kute TE, Kucera GL, Bierbach U. Inhibition of DNA Synthesis by a Platinum-Acridine Hybrid Agent Leads to Potent Cell Kill in Non-Small Cell Lung Cancer. ACS Med. Chem. Lett. 2011;2:870–874. doi: 10.1021/ml2001888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Graham LA, Wilson GM, West TK, Day CS, Kucera GL, Bierbach U. Unusual Reactivity of a Potent Platinum-Acridine Hybrid Antitumor Agent. ACS Med. Chem. Lett. 2011;2:687–691. doi: 10.1021/ml200104h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Choudhury JR, Rao L, Bierbach U. Rates of intercalator-driven platination of DNA determined by a restriction enzyme cleavage inhibition assay. J. Biol. Inorg. Chem. 2011;16:373–380. doi: 10.1007/s00775-010-0733-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ma Z, Choudhury JR, Wright MW, Day CS, Saluta G, Kucera GL, Bierbach U. A non-cross-linking platinum-acridine agent with potent activity in non-small-cell lung cancer. J. Med. Chem. 2008;51:7574–7580. doi: 10.1021/jm800900g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Alley SC, Okeley NM, Senter PD. Antibody-drug conjugates: targeted drug delivery for cancer. Curr. Opin. Chem. Biol. 2010;14:529–537. doi: 10.1016/j.cbpa.2010.06.170. [DOI] [PubMed] [Google Scholar]
- 14.Srinivasan R, Li J, Ng SL, Kalesh KA, Yao SQ. Methods of using click chemistry in the discovery of enzyme inhibitors. Nat. Protocols. 2007;2:2655–2664. doi: 10.1038/nprot.2007.323. [DOI] [PubMed] [Google Scholar]
- 15.Bahta M, Liu F, Kim SE, Stephen AG, Fisher RJ, Burke TR., Jr Oxime-based linker libraries as a general approach for the rapid generation and screening of multidentate inhibitors. Nat. Protocols. 2012;7:686–702. doi: 10.1038/nprot.2012.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kukushkin VY, Pombeiro AJ. Additions to metal-activated organonitriles. Chem. Rev. 2002;102:1771–1802. doi: 10.1021/cr0103266. [DOI] [PubMed] [Google Scholar]
- 17.Baskin JM, Prescher JA, Laughlin ST, Agard NJ, Chang PV, Miller IA, Lo A, Codelli JA, Bertozzi CR. Copper-free click chemistry for dynamic in vivo imaging. Proc. Natl. Acad. Sci. U S A. 2007;104:16793–16797. doi: 10.1073/pnas.0707090104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang RE, Costanza F, Niu Y, Wu H, Hu Y, Hang W, Sun Y, Cai J. Development of self-immolative dendrimers for drug delivery and sensing. J. Controlled Release. 2012;159:154–163. doi: 10.1016/j.jconrel.2011.11.032. [DOI] [PubMed] [Google Scholar]
- 19.Karver MR, Weissleder R, Hilderbrand SA. Bioorthogonal reaction pairs enable simultaneous, selective, multi-target imaging. Angew. Chem. Int. Ed. Engl. 2012;51:920–922. doi: 10.1002/anie.201104389. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.