Abstract
A recently discovered interaction of rod transducin α subunit (Gαt1) with UNC119a is thought to be important for transducin trafficking in photoreceptors. In this study, we analyzed the subcellular distribution of UNC119a under different conditions of illumination in vivo. Analyses by immunofluorescence and Western blotting of retina serial tangential sections demonstrated that UNC119a resides predominantly in the rod inner segment, with a small fraction of UNC119a also appearing to infiltrate the rod outer segment. Such a distribution is consistent with the proposed role of UNC119a in facilitating transducin transport from the rod inner segment to the outer segment in the dark. In addition, UNC119a was present in smaller amounts in the cell body and synaptic region of rods. The profile of UNC119a subcellular distribution remained largely unchanged under all tested conditions of illumination, and correlated with the profile of Gαt1 following its light-dependent translocation. Quantification by Western blotting suggested that mouse retina contains ~17 pmol of UNC119a, giving a ~1 to 4 molar ratio of UNC119a to Gαt1. Hence, light-translocated Gαt1 can serve as a major partner of UNC119a. Supporting this role, the levels of UNC119a were downregulated by about 2-fold in mouse retina lacking Gαt1. As a dominant partner, Gαt1 may potentially modulate function of other known UNC119a-interacting proteins involved in photoreceptor ciliary trafficking and synaptic regulation, in light-dependent manner.
Keywords: retina, rods, UNC119, transducin
1. Introduction
Rod photoreceptors are polarized cells with a specialized phototransduction compartment called the outer segment, which is linked to the rest of the cell by a slender connecting cilium. Phototransduction proteins synthesized in the rod inner segment are trafficked to the rod outer segment, whereas proteins regulating synaptic transmission to downstream bipolar neurons are delivered to the synaptic terminal [1, 2]. Transducin (Gt1 or Gαt1β1γ1), the key visual G-protein, is one of the few photoreceptor proteins that translocates in both directions between the outer segment and the inner compartments depending on the dark/light conditions [3–6]. In dark transducin traffics to the outer segment, whereas in relatively intense light, it translocates from the outer segment to the inner segment and synaptic terminal. The light-dependent translocation of transducin is thought to play a role in light-adaptation and neuroprotection [6, 7], but the significance of this phenomenon is still poorly understood. The mechanism of the light-induced outer segment to inner segment transport of transducin has received significant attention, and a consensus has emerged that the translocation occurs by diffusion [3–6]. In a simple diffusion model, when the rate of transducin activation by photoexcited rhodopsin (R*) exceeds the rate of its inactivation by the rod RGS9 GAP complex, dissociated Gαt1GTP and Gβ1γ1 are able to diffuse into the inner segment and synaptic terminal [3–6, 8]. Much less is known about the inner segment to outer segment transport of transducin in the dark [4]. The large quantities of transducin returning to the outer segment in the dark on a time–scale of several hours, and the potential energy independence of the process [9] lend support to the involvement of diffusion. As there is no Gt activation in the inner segment, transducin subunits are presumed to form a heterotrimeric complex on the inner segment membranes [4]. Yet, the heterotrimer with two lipid moieties, the Gαt1 N-acyl and the Gγ1 farnesyl, appears to lack the capacity to dissociate by itself from the membranes. Gt dissociates from the disc membrane in the dark only in hypotonic extracts in vitro [10]. Surprisingly, studies of transducin diffusion using Fluorescence Recovery After Photobleaching, reveal significant interdisc transfer of Gt [11, 12]. The two lipids might not provide sufficiently strong membrane affinity to block dissociation entirely, particularly if they form a single membrane insertion site and/or have little additive effect [13]. Alternatively, Gt1 dissociation from the membrane may be facilitated by interaction with protein partners and sequestration of one or both of the lipid moieties [4].
Recently, UNC119, a mammalian ortholog of C. elegans unc-119 [14], also known as Retina Gene 4 protein (RG4) [15] has been identified as a novel protein partner of Gαt1 [16, 17]. UNC119 (isoform a) is abundantly expressed in the rod inner segment and synaptic terminal [18], and, to a much lesser extent in several other tissues [19, 20]. A second isoform of UNC119, UNC119b, exists in vertebrates, but its presence in photoreceptor cells has not been investigated [16]. UNC119 shares significant sequence and structural homology with the prenyl-binding protein, PrBP/δ [16, 21, 22]. PrBP/δ has been implicated in the inner segment to outer segment transport of several prenylated proteins, apparently through sequestration of the prenyl groups [23]. Remarkably, the lipid specificity of UNC119a is very different from that of PrBP/δ [16, 17]. UNC119a was shown to interact with the acylated (myristoylated) N-terminus of the GTP-bound Gαt1 [16]. Moreover, UNC119a can interact with heterotrimeric Gt1, promote dissociation of Gαt1 from Gβ1γ1, and release them from the membrane [17]. The finding that the return of Gt1 to the outer segment in the dark was impaired in UNC119a knockout mice indicates the role of UNC119a in the inner segment to outer segment transport of transducin [16]. Although UNC119a did not appreciably bind recoverin [15, 16], other myristoylated proteins in photoreceptors are potential partners for UNC119a [24]. Recently, UNC119a has been shown to interact with the renal celiopathy nephronophthisis (NPHP) protein nephrocystin-3 (NPHP3) and ciliary protein cystin in a myristoyl-dependent manner [24].
Besides myristoyl-dependent partners, UNC119a has several known lipid-independent partners. In the synaptic terminal, UNC119a interacts with CaBP4 [25], a Ca2+-binding modulator of the voltage-gated Ca2+ channel (Cav 1.4) [26], and with RIBEYE, a major component of the synaptic ribbons [27]. In the inner segment and the connecting cilium, UNC119a binds monomeric Arf-like GTPases ARL2 [28] and ARL3 [29] that are involved in microtubule-dependent processes and protein trafficking to and through the connecting cilium [30, 31]. Interestingly, ARL2 and ARL3 are also binding partners for PrBP/δ [32, 33]. Unc119a and PrBP/δ preferentially interact with the GTP-bound ARL2 and ARL3 [29, 32]. Furthermore, ARL3•GTP binding facilitated release of myristoylated cargo from UNC119a [24] and farnesylated cargo from PrBP/δ [34]. Thus, UNC119a and PrBP/δ appear to serve as ARL3 effectors in ciliary transport of myristoylated and prenylated proteins, respectively. Mutations in UNC119-binding proteins have been linked to ciliopathies and visual disorders [24, 35], while an UNC119a null mutation causes a late-onset dominant cone dystrophy in a human patient [36]. To better understand the role of the Gαt1-Unc119a interaction in transducin trafficking and in the overall network of UNC119a interactions, it is essential to develop a quantitative account of UNC119a distribution in photoreceptor cells under different conditions of illumination in vivo. In this study, we qualitatively and quantitatively examined the subcellular distribution of UNC119a in mouse and rat rods under variable light conditions, and compared the localization of UNC119a and Gαt1. Furthermore, we assessed the overall retinal abundance of UNC119a and its dependence on transducin expression.
2. Materials and Methods
2.1 Immunofluorescence
Mice were dark-adapted overnight. For light adaptation, the pupils were dilated by applying a drop of 1% tropicamide, followed by a drop of 2.5% phenylephrine hydrochloride. The mice were then kept in the dark for 30 min before light exposure. During light exposure, mice were kept in a Styrofoam box (20 × 15 × 15 cm) covered with semi-transparent polyethylene film. Light from white fluorescent bulbs in the ceiling was used for strong light exposure of ~500 lux for 45 min. The mice were then euthanized with CO2. Mouse eyeballs were enucleated, poked through the cornea with a 21-gauge needle, and fixed in 4% paraformaldehyde in phosphate-buffered saline (PBS) for 1 hr in the dark at 25 °C. For dark-adapted mice, euthanasia, enucleation of eyeballs and initial fixation were all performed under dim red light. After this initial fixation, the eyeballs were cut open to remove the cornea and lens, and the eyecups were fixed in 4% paraformaldehyde in PBS for an additional 3 hrs at 25 °C. The eyecups were then transferred to a 1:1 solution of 30% sucrose in PBS and 4% paraformaldehyde in PBS for 1 hr at 25 °C. Finally, the eyecups were submersed in a 30% sucrose solution in PBS and left at 4°C overnight. The next day, eyecups were embedded in tissue freezing medium (Triangle Biomedical Sciences) and frozen into cryo blocks on dry ice. Cryo blocks were subsequently sectioned (10 μm, Microm HM 505E) and stored at −80 °C until use. Before staining, sections were warmed up to room temperature and were first incubated in 0.1% Triton X-100 in PBS for 30 min, followed by a 1 hr incubation with either a rabbit anti-rod Gαt1 antibody K-20 (1:1000) (Santa Cruz Biotechnology, Santa Cruz, CA), or a monoclonal anti-UNC119 antibody (1:50) (a kind gift from Dr. Francoise Haeseleer, University of Washington). The sections were then stained with either goat anti-rabbit AlexaFluor 546 or goat anti-mouse AlexaFluor 488 secondary antibody (Molecular Probes) (1:1000) for 1 hr. All sections were subsequently counterstained with TO-PRO3 nuclear stain and mounted using Vectashield mounting medium (Vector Laboratories). The stained sections were visualized under a Zeiss LSM 510 confocal microscope.
2.2 Serial tangential sectioning of the retina
Tangential serial sectioning of the rat retina was carried according to a previously described procedure [37]. Adult Long Evans rats (Charles River) were dark-adapted overnight and then subjected to controlled diffused white illumination while free running in a cage placed inside a large Styrofoam box. At the end of this light conditioning rats were euthanized and their eyes promptly harvested and dissected in Dulbecco’s modified Eagle’s medium (DMEM) medium under dim red light using a Stemi 2000-C stereomicroscope (Zeiss), equipped with OWL Gen 3+ intensifiers (B.E. Meyers and Co. Inc). Fragments of the retinas were flattened between two glass plates separated by 0.5 mm spacers, and frozen on dry ice. From this point, all tissue manipulations were carried out under ambient light. Frozen retina fragments were tangentially cut in a Leica CM1850 Cryostat. Serial 10 μm sections, spanning the entire retina, were collected, and each one transferred into 50 μl of buffer containing 6 M urea, 125 mM Tris-HCl, pH 6.8, 4% SDS, bromphenol blue, and 10 mg/ml dithiothreitol. The resulting set of extracts was analyzed by immunoblotting. In each set of serial sections, proteins of interest, UNC119 and Gαt1, were detected along with four protein markers, including rod outer segment marker (rom-1), rod inner segment and mitochondrial marker (cytochrome c oxidase subunit I, COX I), general cellular marker (β-tubulin) and general photoreceptor marker (phosducin). Quantification of the specific bands was performed on an Odyssey Infrared Imaging System (LI-COR Biosciences). To generate protein profiles, the fluorescence value of each band was plotted as a percentage of the combined fluorescence in all sections.
We used commercial primary antibodies against rod transducin α subunit (Gαt1) (sc-389, Santa Cruz Biotechnology), β-tubulin (T0198, Sigma Aldrich), and cytochrome c oxidase subunit I, COX I (MS404, MitoSciences). A monoclonal antibody against UNC119a was a kind gift from Dr. Francoise Haeseleer (University of Washington). Antibody against rod outer segment membrane protein, rom-1, was generously provided by Dr. Andrew Goldberg (Oakland University). Antibodies against phosducin were described previously [38].
2.3 Expression and quantification of mouse UNC119a protein standard
Recombinant His6-tagged mouse UNC119a was designed as a protein standard for the quantification of UNC119 in mouse retina. The UNC119a coding sequence was PCR amplified from the pCMV6-mouse unc119 plasmid (Origene), and cloned into the pET15b vector using NdeI/XhoI sites. Protein expression in BL21-codon plus E. coli cells was induced by adding 40 μM IPTG for 5 h at 30°C. The His6-tagged unc119 protein was initially purified on an Ni-NTA resin (Novagen) followed by further purification by gel filtration on a Superdex 75 sepharose column. The protein was then dialyzed against hypotonic buffer containing 50% glycerol, and stored at −20°C in aliquots. The concentration of the UNC119a standard was determined by quantifying UNC119a bands along with serial dilutions of two protein standards of known concentration, BSA (A8531, Sigma) and carbonic anhydrase (C7025, Sigma), on Coomassie-stained gels. The concentrations of the BSA and carbonic anhydrase standards were determined by UV absorption using extinction coefficients ε280 of 43824 M−1cm−1 and 43430 M−1cm−1, respectively. The calibration curves were plotted based on the integrated densities of the protein bands on the Coomassie-stained gels measured using ImageJ.
2.4 Analysis of UNC119a protein levels in mouse retina
To estimate the quantity of UNC119a protein in mouse retina, we first obtained a hypotonic retina extract. Two mouse retinas were homogenized in 100 μl of hypotonic buffer (5 mM Tris, pH 7.5, 1 mM MgSO4, 1 M DTT and complete protease inhibitor mixture (Roche)) with a pestle and brief sonication. The retinal homogenates were then separated into soluble and membrane fractions by centrifuging at 100,000g for 45 min, and analyzed by Western blotting with an anti-UNC119a antibody [25], along with the His6-tagged UNC119a protein standard.
For comparison of UNC119a protein levels in control and Gαt1-knockout mice, total mouse retinal homogenates were obtained by solubilization of two retinas in 100 μl of 10% SDS-Na using brief sonication. Protein concentrations were determined using the DC Protein Assay (Bio-Rad) with bovine serum albumin dissolved in 10% SDS-Na serving as the standard. Samples of retinal homogenates were probed using standard Western blotting procedure and primary antibodies against UNC119a (1:260) [25] and RGS9 (1: 1000) (Elmira Biologicals, Iowa City).
2.5 Quantitative Real Time PCR analysis
Total RNA was isolated from single retinas of wild-type, Tg(Δ1−83PhLP+/−), and Gα t1−/− mice. The Absolutely RNA Miniprep Kit (Agilent Technologies) was used including two separate DNAse treatments, and following the manufacturer’s modified protocol for small samples. Using a two-step qRT-PCR process, 5 ng of each total RNA sample was reverse transcribed using oligo(dT) primers and the AffinityScript QPCR cDNA Synthesis Kit (Agilent Technologies). In addition, the following negative controls were also prepared: no reverse transcriptase and no RNA template, and these were run through the same reverse transcription parameters. 0.5 ng of each subsequent cDNA was amplified in triplicate using the Brilliant II SYBR Green QPCR Master Mix (Agilent Technologies), including reference dye and 200 nM of each forward and reverse primer. The PCR reaction was run under the following conditions: denaturation at 95°C 10 minutes, followed by 32 cycles at 95°C for 30 seconds, 55°C for 1 minute, and 72°C for 30 seconds using a Stratagene Mx3000P real-time PCR system and the comparative quantitation setting. A melting curve analysis was also added at the end to verify a single product from each reaction, and the fluorescence recorded during every qPCR cycle at the annealing step (55°C) was used for data analysis. Primers were designed using GenBank mouse mRNA sequences and PrimerQuest software, and selected so that one primer would cross an exon-exon junction, where possible. The primers were synthesized and HPLC-purified by Integrated DNA Technologies (IDT) (Coralville, IA), and the primer concentrations optimized for each reaction before beginning. Primers for UNC119a (forward, 5′-CGC TTT GTT CGC TAC CAG TTC ACA -3′; reverse 5′-ATG CAG AAG CCA AAG TGG AAG TCG - and primers for 3′), and primers for hypoxanthine phosphorybosyl transferase (HPRT) housekeeping gene (forward, 5′-CAG GCC AGA CTT TGT TGG AT -′; reverse, 5′-GGA CGC AGC AAC TGA CAT T -3′). Final amplification plot data was averaged for all samples within a group and their replicates, and the MxPro-MX300P QPCR software version 4.10 (Stratagene) calculated the threshold cycle values (Ct) by its default amplification-based threshold settings. The software then calculated the Ct difference between the sample groups, and corrected it by the difference in the Cts for the normalizer, HPRT, in these samples. A relative quantity chart was prepared to show the fold-differences +/− SEM of UNC119 gene expression between the different types of mice, with the 1054 negatives serving as the calibrators or controls.
3. Results
3.1 Analysis of the subcellular distribution of UNC119a and Gαt1 under different illumination
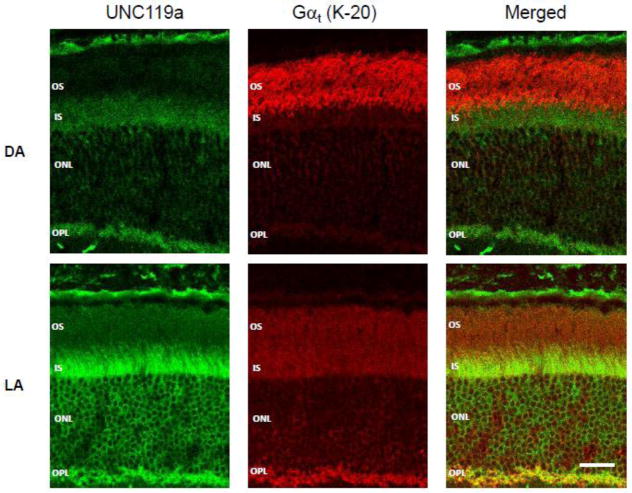
The distribution of UNC119 and Gαt1 in dark- and light-adapted mouse retinas was first examined qualitatively by immunofluorescence using monoclonal anti-UNC119 antibody [25]. Among the two known UNC119 isoforms, UNC119a and UNC119b, this antibody specifically recognizes UNC119a. Therefore, all the analyses described below pertain to UNC119a. Retina cross-sections were obtained from wild type (C57BL/6) mice either dark-adapted overnight or exposed to 500 lux light for 45 min. In dark-adapted (DA) animals the strongest UNC119a immunostaining was observed in the inner segment and synapses (outer plexiform layer), whereas Gαt1 was almost exclusively localized to the outer segment (OS) (Fig. 1, DA). This is illustrated in the merged image showing no co-localization of UNC119a and Gαt1. Following light-adaptation, the overall distribution of UNC119a remained largely unchanged, however a major fraction of Gαt1 underwent translocation from the outer segment to the other rod cellular compartments, where it co-localized with UNC119a (Fig. 1, LA).
Figure 1. Gαt1 and UNC119a co-localize following light-dependent translocation of transducin.
Cryosections of mouse retinas were obtained from dark- (DA) and light-adapted (LA: 45 min, 500 lux) Gαt1+/− mice. The sections were stained with a monoclonal anti-UNC119a antibody [25] (green) and a rabbit anti-Gαt1 antibody K-20 (SCBT) (red). The staining was visualized using goat anti-rabbit AlexaFluor 568 and goat anti-mouse AlexaFluor 488 secondary antibodies under a Zeiss LSM 510 confocal microscope. Bar – 20 μm. OS – outer segment, IS – inner segment, ONL – outer nuclear layer, OPL – outer plexiform layer.
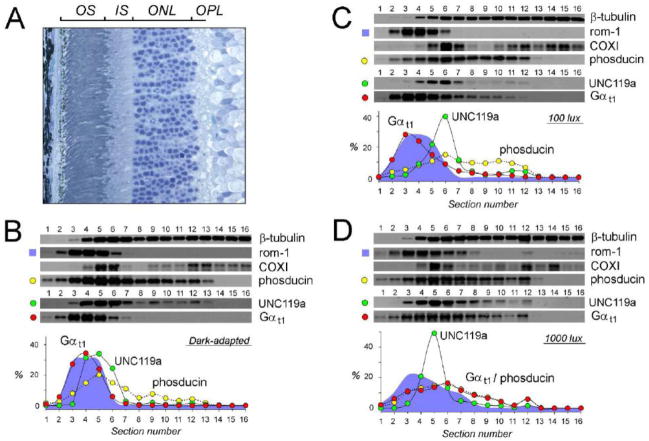
While conducting immunofluorescence analyses, we made an observation that the overall intensity of the UNC119a signal in the retina sections obtained from mice exposed to 500 lux light was appreciably higher than in those from their dark-adapted counterparts (Fig. 1). Because the total amount of UNC119a in the retinas does not change significantly during the course of the light conditioning procedure (data not shown), the observed variation of the immunofluorescence signal could perhaps be explained in terms of epitope masking affecting binding of the antibody to UNC119a in a light-dependent manner. This observation prompted us to repeat our measurements using an alternative technique based on Western blotting and not sensitive to epitope masking effects [6, 37]. This technique takes advantage of the highly ordered arrangement of rod photoreceptors with their major cellular compartments present in paralleled retina layers (Fig. 2A). Owing to such an alignment, the average distribution of any protein in an individual rod can be determined by measuring the distribution of this protein in serial sections tangential to the plane of retina layers. For these experiments we choose to use rats, as their retinas are better suited for the tangential sectioning due to their bigger size and smaller curvature compared to mouse retina. Animals were dark-adapted overnight, and then exposed to 100 lux and 1000 lux light for 45 min. Following light conditioning rats were euthanized and their retinas harvested and serially sectioned. The amount of UNC119a and Gαt1 in each section was determined by Western blotting, along with the following reference proteins. Rod outer segment membrane protein, rom-1, was used as a marker of the outer segment. A mitochondrial marker, COXI, was used to visualize the distal part of the inner segment adjacent to the outer segment and containing most of the photoreceptors’ mitochondria. A photoreceptor specific soluble protein, phosducin, was used to identify the photoreceptor boundaries, while ubiquitous β-tubulin was used as a loading control to ensure even distribution of cellular material among sections, with the exception of the outer segment, which contains decreasing amounts of microtubules. Three representative analyses, for each type of illumination used, are illustrated in Fig. 2B–D.
Figure 2. Analysis of the distribution of UNC119a and Gαt1 by Western blotting of serial sections of the retina.
(A) Cross-section of rat retina stained with toluidine blue, retina layers abbreviated as in Fig. 1. (B–D) Retinas were obtained from either dark adapted rat (B) or rat exposed to 100 lux (C), or 1000 lux (D) light for 45 min, flat mounted, frozen and serially 10 μm sectioned (1–16). Each section was dissolved in SDS sample buffer, and analyzed by Western blotting with antibodies against the proteins indicated next to each blot. Plots show quantification of UNC119a (green), and Gαt1 (red), phosducin (yellow), and rom-1 (cyan) distribution in the blots: the fluorescence value of a specific band in each section on the corresponding blot was plotted as a percentage of the total fluorescence value of all bands in all sections on the blots.
Our results, obtained by the serial sectioning technique, were generally consistent with the immunofluorescence data. As such we confirmed that, while UNC119a could be detected in any cellular compartment, it is predominantly localized to the rod inner segment. This subcellular distribution of UNC119a remained essentially the same under all tested levels of illumination. As calculated from the shown blots, 24–32% of total UNC119a is present in the outer segment (sections 1–4), 51–62% in the inner segment (sections 6 and 7), and 9–16% is evenly distributed through the outer nuclear and outer plexiform layers (sections 8–13). The absence of any significant subcellular translocation of UNC119a was really noticeable when compared to Gαt1, whose compartmentalization shows a strong dependency on the history of illumination. During prolong dark adaptation Gαt1 accumulates in the outer segment, as evident from the near identical distribution of rom-1 and Gαt1 under this condition (Fig. 2B). Under a dim 100 lux light, some translocation of Gαt1 from the outer segment is already taking place (Fig. 2C), however only bright 1000 lux light is triggering a full scale event (Fig. 2D). Interestingly, the profile of light-dispersed Gαt1 closely resembles that of phosducin (Fig. 2D).
3.2 Quantification of UNC119a in mouse retina
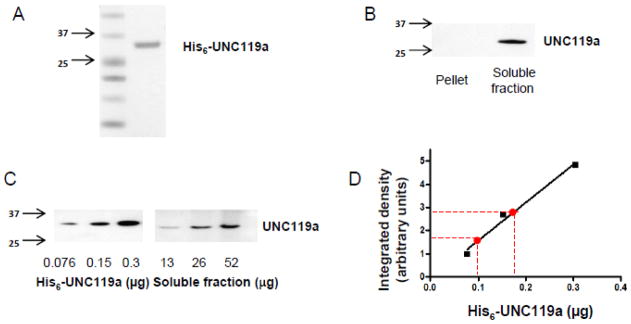
The interaction of UNC119a with Gαt1 [16, 17] and their protein distributions in light-adapted retinas (Figs 1 and 2) support the complex formation between UNC119a and Gαt1 in the rod inner compartments. Transducin is a major photoreceptor protein, and its mouse retina content is known [39]. However, the relative abundance of UNC119a and its molar ratio to Gαt1 in rods is unknown. To quantify UNC119a in a mouse retina, we first expressed and purified the His6-tagged mouse UNC119a to use as a protein standard. After purification, no additional bands were seen in the UNC119a standard on a Coomassie-stained gel (Fig. 3A). UNC119a contains no Trp residues, and therefore, its protein concentration measurement by UV absorption at 280 nm is highly unreliable. Instead, we determined the UNC119a standard concentration by quantifying UNC119a bands along with serial dilutions of two protein standards of known concentration, BSA and carbonic anhydrase, on Coomassie-stained gels. The use of both BSA and carbonic anhydrase yielded linear calibration curves, and produced nearly identical estimates of the concentration of UNC119a standard. To determine the UNC119a mouse retina content, we first analyzed the protein partitioning between the soluble and membrane fractions. This analysis revealed that UNC119 is fully solubilized by retina extraction with hypotonic buffer (Fig. 3B). The soluble retina fraction was utilized in the quantification of UNC119a by Western blotting since this allowed us to minimize the effects of protein masking. According to our analyses, mouse retina contains 0.47 ± 0.06 μg or ~17 pmol of UNC119a (Fig. 3C & D).
Figure 3. Quantitative immunoblot analysis of UNC119a in mouse retina.
A. Coomassie blue-stained UNC119a standard. For use as a standard, mouse UNC119a was cloned into the pET15b vector, and the His6-tagged protein was purified as described in Section 2.3
B. Mouse retinas were homogenized in hypotonic buffer by brief sonication, separated into soluble and membrane fractions (100,000g, 45 min), and analyzed by western blotting with an anti-UNC119a antibody [25].
C. Soluble fractions from the mouse retinal homogenates and His6-tagged mouse UNC119a standards were analyzed by western blot using an anti-UNC119a antibody.
D. Integrated densities of the individual His6-tagged UNC119a bands in the western blot were measured using ImageJ, and a linear fit curve was obtained by plotting these values. Using this standard curve, the amount of UNC119a in a single mouse retina was estimated to be ~0.46 μg. Shown here is a result from a representative experiment. From six similar experiments, a single mouse retina is estimated to contain 0.47 ± 0.06 μg (Mean ± SE) or ~17 pmol of UNC119a.
3.3 Gαt1 is increasing the protein level of UNC119a in rods
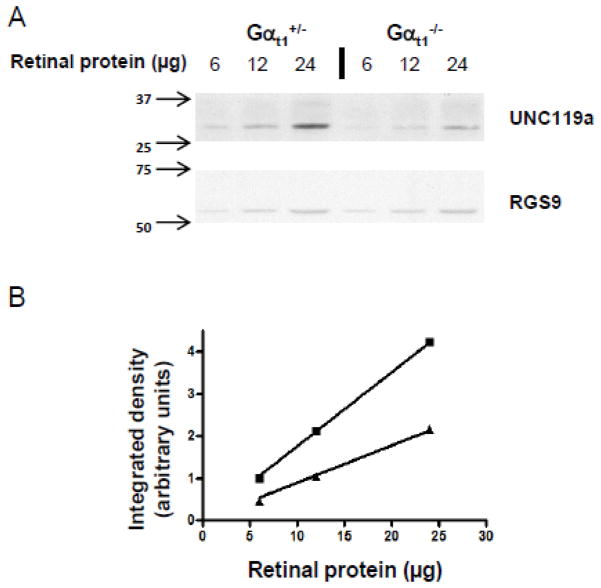
It is not unusual for interacting proteins to regulate each other’s expression in the cell. To test whether the expression of UNC119a and Gαt1 is co-dependent, we determined the protein level of UNC119a in the retinas of Gαt1 knockout mice (Gαt1−/−) using their heterozygous littermates (Gαt1+/−) that express normal levels of Gαt1 as controls [40]. We found that the level of UNC119a in the Gαt1−/− retina was on average reduced by two fold (Fig. 4A & B). Because knockout of the Gαt1 gene neither negatively affects photoreceptor viability nor the expression of other visual signaling proteins [40], the down-regulation of UNC119a thus appears to be specifically related to the absence of Gαt1 in these cells. Furthermore, we previously documented an even stronger four fold reduction of UNC119a in the retinas of transgenic mice expressing truncated phosducin-like protein, [41]. Like Gαt1−/− mice, these mice express no detectable Gαt1 in the retina [41], which further supports the notion that the expression of UNC119a depends on Gαt1. To explore a common molecular mechanism underlying this effect, we quantified the levels of UNC119a transcript in both Gαt1−/− and Tg(Δ1−83PhLP+/−) mice by real-time RT-PCR. We found no evidence of any significant reduction of the UNC119a transcript due to the absence of Gαt1 in each model (Fig. 5). Thus, in all probability, Gαt1 is increasing the level of UNC119a in photoreceptors by a posttranslational mechanism, such as protein stabilization via complex formation.
Figure 4. UNC119a protein levels are reduced by ~2-fold in the retina of Gαt1 knockout mice.
A. Retinal homogenates were obtained by solubilizing two retinas in 100 μl of 10% SDS. Total homogenates from control (Gαt1+/−) and transducin-α knockout (Gαt1−/−) mice were analyzed by western blotting with monoclonal anti-UNC119a and anti-RGS9 antibodies. RGS9 served as a loading control.
B. The UNC119a protein levels in each sample were quantified by measuring the integrated densities of the bands using ImageJ, and linear fit curves were obtained for both control and transducin-α knockout (Gαt1−/−) mice. From these curves, UNC119a protein levels were found to be reduced by 2.04-fold in retinas of Gαt1 knockout mice. Shown here is one representative experiment. From three similar experiments, the fold change in UNC119a protein levels in control vs transducin-α knockout (Gαt1−/−) mice was determined to be 2.223 ± 0.138 (Mean ± SE).
Figure 5. Normal levels of UNC119 transcript in the retinas lacking Gαt1 expression.
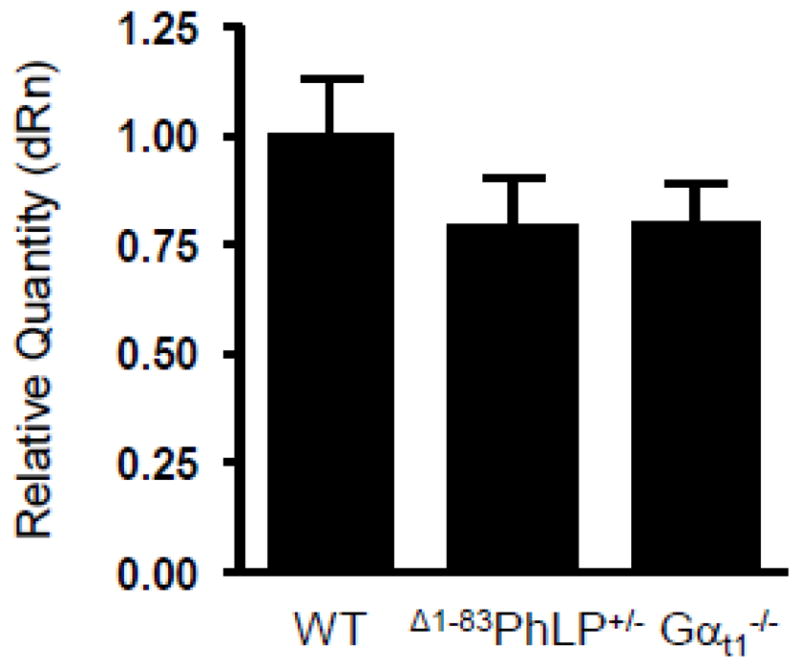
The levels of UNC119a transcript in the retinas of 10 days-old Δ1−83PhLP−/− (WT), Δ1−83PhLP+/−, and Gαt1−/− mice were determined by quantitative real-time PCR. The mean of three independent measurements in Δ1−83PhLP+/− and Gαt1−/− mice was normalized to that in Δ1−83 PhLP−/− mice regarded as a wild-type control.
4. Discussion
UNC119a is an abundant photoreceptor protein with multifaceted, yet poorly understood functions in protein trafficking and synaptic regulation in photoreceptors [15–18]. The recently discovered interaction of UNC119a with the Gαt1 subunit of the visual heterotrimeric G protein, transducin, and its potential role in transducin trafficking has renewed interest in understanding the significance of UNC119a for photoreceptor cell function {15, 16]. Most intriguing is the finding of the unique selectivity on UNC119a for the N-acyl lipid modification, which is different from that of a homologous prenyl-binding protein PrBP/δ [16, 17]. This feature makes UNC119a well-suited to facilitate subcellular trafficking of Gαt1 by acting as its myristoyl sheath.
To understand this cellular function of UNC119a further, here we qualitatively and quantitatively analyzed UNC119a expression in rod photoreceptors. We found that UNC119a is specifically enriched in the inner segment of rods, although smaller amounts of this protein could be detected essentially everywhere (Fig. 1, 2). There is no significant co-localization of UNC119a with Gαt1 in dark-adapted rods. However, exposure of animals to saturating levels of illumination triggers transducin translocation from the rod outer segment, whereas subcellular localization of UNC119a is unaffected by light. This results in substantial, but not complete, co-localization of UNC119a and Gαt1 in the inner segment. Our conclusion that UNC119a is compartmentalized primarily in the inner segment contrasts with earlier assessments that UNC119 is predominantly a synaptic protein with lesser presence in the inner segment [18, 25]. The difference between the UNC119 localization observed here and previously [18, 25] is likely due to the application of the serial sectioning/Western blotting technique that is insensitive to the epitope masking effects [6, 37]. Furthermore, we found that while UNC119a is largely excluded from the apical outer segment, a fraction of the protein is present in the basal portion of the outer segment (Fig. 2). It is plausible that such an asymmetric distribution of UNC119a reflects its function in the trafficking of transducin in the inner segment→outer segment direction in the dark. Indeed, recent analyses of UNC119a-null mice revealed that UNC119a has no effect on the transducin outer segment to inner segment translocation in light [16]. In agreement, we found no evidence of any light-evoked redistribution of UNC119a concurrent with the movement of Gαt1. Also, since UNC119a has been shown to promote transducin partitioning from the membranes and block phototransduction [17], its exclusion from the outer segment is necessary to preserve signaling and prevent uncontrolled outflow of transducin from this compartment. On the contrary, this property of UNC119a may be beneficial in the inner segment, where UNC119a could promote dissociation of transducin from the inner segment membranes. Thus, by keeping light-dispersed transducin moveable in the inner segment, UNC119a is facilitating its return to the outer segment in the dark, as shown experimentally [16]. The targeting of the Unc119a/Gαt1 complex to the connecting cilium and release of Gαt1 into the outer segment are probably regulated by ciliary ARL3•GTP, as it was shown for NPHP3 [24]. The mechanism of ARL•GTP-dependent dissociation of Gαt1 from UNC119a may be similar to the model of allosteric release of farnesylated cargo from PrBP/δ [34]. Subsequent disruption the ARL•GTP/UNC119a complex requires GTP hydrolysis and the activity of the ARL3-specific GAP, retinitis pigmentosa protein 2 (RP2) [24, 42]. RP2 is not confined to the cilium, rather it is localized to the plasma membrane including the outer segment membrane [43]. Such distribution of RP2 may be responsible for the presence of UNC119a in the base of the outer segment observed in this study.
Overall, our findings on UNC119a compartmentalization in rods are consistent with the phenotype of the UNC119a knockout mouse [44]. This mouse model revealed a retinal degeneration phenotype linked to a dysfunction in the distal inner segment/outer segment compartments of rods rather than in the synapse [44]. Although our results indicate that UNC119a is significantly less abundant in the synaptic terminal compared to the inner segment, its concentration in the synaptic terminal might be relatively high considering the lower cytoplasmic volume of a rod spherule [45]. The fraction of UNC119a in the synaptic terminal may support synaptic roles of the protein suggested by several studies [18, 25, 27].
Knowing the relative abundance of UNC119a and its molar ratio to Gαt1 in rods can provide additional clues to the significance of UNC119a for transducin trafficking. Our measurements demonstrate that a mouse retina contains ~17 pmol of UNC119a. Based on two different measurement approaches, Western blotting [39] and binding of a nonhydrolysable GTP analog GTPγS [46], and a widely accepted ~1:8 transducin to rhodopsin molar ratio [47], the amount of Gαt1 in adult mouse retina containing ~500 pmol of rhodopsin [48] could be estimated as ~60 pmol. Thus, the most conservative estimation of the relative UNC119a abundance yields a molar ratio of one molecule of UNC119a per four molecules of Gαt1. Depending on the light-exposure and the extent of transducin translocation, the relative concentrations of UNC119a and Gαt1 in the inner rod compartments may range from nearly stoichiometric to a 3–4-fold excess of Gαt1, suggesting that Gαt1 may serve as a major partner of UNC119a. This notion is also supported by down-regulation of UNC119a in Gαt1-null rods (Fig. 4). A similar down-regulation of UNC119a was previously observed in several other models - most recently in transgenic Δ1−83PhLP+/− mice, where it was attributed to the suppression of the chaperonin containing TCP-1 [41]. These mutant mice, however, also lack any detectable Gαt1 expression, which therefore makes it possible that the reduction of UNC119a in this model was due to the loss of Gαt1. A reduction of UNC119a was also observed in mouse photoreceptors lacking CaBP4 [25]. CaBP4 is one of the binding partners for UNC119a that lacks N-terminal myristoylation [25]. As a major UNC119a interacting partner, Gαt1 can outcompete myristoylated cargo for UNC119a, or form ternary complexes with UNC119a and its lipid-independent partners. UNC119a partners have been linked to regulation of ciliary function and synaptic transmission in photoreceptor cells [25, 27–29]. Our study highlights a novel potential role for the light-induced translocation of transducin in rods, i. e. the possibility that translocated transducin, via its interaction with UNC119a, modulates ciliary trafficking and synaptic transmission in photoreceptors.
5. Conclusion
Our study demonstrates that UNC119a is enriched in the inner segment of rod photoreceptor cells, whereas its amounts in the synaptic terminal are significantly lower. We also found that a fraction of UNC119a resides in the basal portion of the rod outer segment. The intracellular distribution of UNC119 is light-independent and consistent with its proposed role in transducin trafficking to the outer segment. The estimated 4:1 molar ratio of transducin to UNC119a and downregulation of UNC119a in rods lacking Gαt1 suggest that light-dispersed transducin is a major binding partner for UNC119a in the inner compartments of the rod.
Highlights.
UNC119 is enriched in the inner segment of rod photoreceptors.
UNC119a distribution in rods is light-independent and consistent with its role in transducin trafficking.
The estimated molar ratio of UNC119a to transducin is ~1 to 4.
UNC119a is downregulated in the absence of transducin-α.
Light-translocated transducin-α is apparently a major binding partner for UNC119a.
Acknowledgments
We thank K. Boyd (University of Iowa) for excellent technical assistance. We thank Drs. F. Haeseleer (University of Washington) and A. Goldberg (Oakland University) for providing antibodies for this research. This work was supported by National Institutes of Health Grants EY-12682 to N.O.A., and EY019665 to M.S., and an unrestricted Research to Prevent Blindness Grant awarded to West Virginia University Eye Institute.
Abbreviations
- Gαt1
rod transducin α subunit
- UNC119a
uncoordinated 119a protein
- PrBP/δ
prenyl-binding protein/PDE delta subunit
- BSA
Bovine Serum Albumin
- DTT
dithiothreitol
- IPTG
isopropyl β-D-1-thiogalactopyranoside
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Burns ME, Arshavsky VY. Neuron. 2005;48:387–401. doi: 10.1016/j.neuron.2005.10.014. [DOI] [PubMed] [Google Scholar]
- 2.Karan S, Zhang H, Li S, Frederick JM, Baehr W. Vision Research. 2008;48:442–452. doi: 10.1016/j.visres.2007.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Calvert PD, Strissel KJ, Schiesser WE, Pugh EN, Jr, Arshavsky VY. Trends in Cell Biology. 2006;16:560–568. doi: 10.1016/j.tcb.2006.09.001. [DOI] [PubMed] [Google Scholar]
- 4.Artemyev NO. Molecular Neurobiology. 2008;37:44–51. doi: 10.1007/s12035-008-8015-2. [DOI] [PubMed] [Google Scholar]
- 5.Slepak VZ, Hurley JB. IUBMB Life. 2008;60:2–9. doi: 10.1002/iub.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sokolov M, Lyubarsky AL, Strissel KJ, Savchenko AB, Govardovskii VI, Pugh EN, Jr, Arshavsky VY. Neuron. 2002;34:95–106. doi: 10.1016/s0896-6273(02)00636-0. [DOI] [PubMed] [Google Scholar]
- 7.Fain GL. BioEssays : news and reviews in molecular, cellular and developmental biology. 2006;28:344–354. doi: 10.1002/bies.20382. [DOI] [PubMed] [Google Scholar]
- 8.Lobanova ES, Finkelstein S, Song H, Tsang SH, Chen CK, Sokolov M, Skiba NP, Arshavsky VY. The Journal of Neuroscience : the official journal of the Society for Neuroscience. 2007;27:1151–1160. doi: 10.1523/JNEUROSCI.5010-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nair KS, Hanson SM, Mendez A, Gurevich EV, Kennedy MJ, Shestopalov VI, Vishnivetskiy SA, Chen J, Hurley JB, Gurevich VV, Slepak VZ. Neuron. 2005;46:555–567. doi: 10.1016/j.neuron.2005.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Goc A, Angel TE, Jastrzebska B, Wang B, Wintrode PL, Palczewski K. Biochemistry. 2008;47:12409–12419. doi: 10.1021/bi8015444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang Q, Zhang X, Zhang L, He F, Zhang G, Jamrich M, Wensel TG. The Journal of Biological Chemistry. 2008;283:30015–30024. doi: 10.1074/jbc.M803953200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kerov V, Artemyev NO. Molecular and Cellular Neurosciences. 2011;46:340–346. doi: 10.1016/j.mcn.2010.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Seitz HR, Heck M, Hofmann KP, Alt T, Pellaud J, Seelig A. Biochemistry. 1999;38:7950–7960. doi: 10.1021/bi990298+. [DOI] [PubMed] [Google Scholar]
- 14.Maduro M, Pilgrim D. Genetics. 1995;141:977–988. doi: 10.1093/genetics/141.3.977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Higashide T, Murakami A, McLaren MJ, Inana G. The Journal of Biological Chemistry. 1996;271:1797–1804. doi: 10.1074/jbc.271.3.1797. [DOI] [PubMed] [Google Scholar]
- 16.Zhang H, Constantine R, Vorobiev S, Chen Y, Seetharaman J, Huang YJ, Xiao R, Montelione GT, Gerstner CD, Davis MW, Inana G, Whitby FG, Jorgensen EM, Hill CP, Tong L, Baehr W. Nature Neuroscience. 2011;14:874–880. doi: 10.1038/nn.2835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gopalakrishna KN, Doddapuneni K, Boyd KK, Masuho I, Martemyanov KA, Artemyev NO. The Journal of Biological Chemistry. 2011;286:28954–28962. doi: 10.1074/jbc.M111.268821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Higashide T, McLaren MJ, Inana G. Investigative Ophthalmology & Visual Science. 1998;39:690–698. [PubMed] [Google Scholar]
- 19.Swanson DA, Chang JT, Campochiaro PA, Zack DJ, Valle D. Investigative Ophthalmology & Visual Science. 1998;39:2085–2094. [PubMed] [Google Scholar]
- 20.Karim Z, Vepachedu R, Gorska M, Alam R. Cellular Signalling. 2010;22:128–137. doi: 10.1016/j.cellsig.2009.09.022. [DOI] [PubMed] [Google Scholar]
- 21.Gillespie PG, Prusti RK, Apel ED, Beavo JA. The Journal of Biological Chemistry. 1989;264:12187–12193. [PubMed] [Google Scholar]
- 22.Norton AW, Hosier S, Terew JM, Li N, Dhingra A, Vardi N, Baehr W, Cote RH. The Journal of Biological Chemistry. 2005;280:1248–1256. doi: 10.1074/jbc.M410475200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang H, Li S, Doan T, Rieke F, Detwiler PB, Frederick JM, Baehr W. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:8857–8862. doi: 10.1073/pnas.0701681104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wright KJ, Baye LM, Olivier-Mason A, Mukhopadhyay S, Sang L, Kwong M, Wang W, Pretorius PR, Sheffield VC, Sengupta P, Slusarski DC, Jackson PK. Genes & Development. 2011;25:2347–2360. doi: 10.1101/gad.173443.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Haeseleer F. Investigative Ophthalmology & Visual Science. 2008;49:2366–2375. doi: 10.1167/iovs.07-1166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Haeseleer F, Imanishi Y, Maeda T, Possin DE, Maeda A, Lee A, Rieke F, Palczewski K. Nature Neuroscience. 2004;7:1079–1087. doi: 10.1038/nn1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Alpadi K, Magupalli VG, Kappel S, Koblitz L, Schwarz K, Seigel GM, Sung CH, Schmitz F. The Journal of Biological Chemistry. 2008;283:26461–26467. doi: 10.1074/jbc.M801625200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kobayashi A, Kubota S, Mori N, McLaren MJ, Inana G. FEBS Letters. 2003;534:26–32. doi: 10.1016/s0014-5793(02)03766-3. [DOI] [PubMed] [Google Scholar]
- 29.Veltel S, Kravchenko A, Ismail S, Wittinghofer A. FEBS Letters. 2008;582:2501–2507. doi: 10.1016/j.febslet.2008.05.053. [DOI] [PubMed] [Google Scholar]
- 30.Kahn RA, Volpicelli-Daley L, Bowzard B, Shrivastava-Ranjan P, Li Y, Zhou C, Cunningham L. Biochemical Society Transactions. 2005;33:1269–1272. doi: 10.1042/BST0331269. [DOI] [PubMed] [Google Scholar]
- 31.Schrick JJ, Vogel P, Abuin A, Hampton B, Rice DS. The American Journal of Pathology. 2006;168:1288–1298. doi: 10.2353/ajpath.2006.050941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hanzal-Bayer M, Renault L, Roversi P, Wittinghofer A, Hillig RC. The EMBO Journal. 2002;21:2095–2106. doi: 10.1093/emboj/21.9.2095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Linari M, Hanzal-Bayer M, Becker J. FEBS Letters. 1999;458:55–59. doi: 10.1016/s0014-5793(99)01117-5. [DOI] [PubMed] [Google Scholar]
- 34.Ismail SA, Chen YX, Rusinova A, Chandra A, Bierbaum M, Gremer L, Triola G, Waldmann H, Bastiaens PI, Wittinghofer A. Nature Chemical Biology. 2011;7:942–949. doi: 10.1038/nchembio.686. [DOI] [PubMed] [Google Scholar]
- 35.Zeitz C, Kloeckener-Gruissem B, Forster U, Kohl S, Magyar I, Wissinger B, Matyas G, Borruat FX, Schorderet DF, Zrenner E, Munier FL, Berger W. American Journal of Human Genetics. 2006;79:657–667. doi: 10.1086/508067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kobayashi A, Higashide T, Hamasaki D, Kubota S, Sakuma H, An W, Fujimaki T, McLaren MJ, Weleber RG, Inana G. Investigative Ophthalmology & Visual Science. 2000;41:3268–3277. [PubMed] [Google Scholar]
- 37.Song H, Belcastro M, Young EJ, Sokolov M. The Journal of Biological Chemistry. 2007;282:23613–23621. doi: 10.1074/jbc.M701974200. [DOI] [PubMed] [Google Scholar]
- 38.Sokolov M, Strissel KJ, Leskov IB, Michaud NA, Govardovskii VI, Arshavsky VY. The Journal of Biological Chemistry. 2004;279:19149–19156. doi: 10.1074/jbc.M311058200. [DOI] [PubMed] [Google Scholar]
- 39.Kerov VS, Natochin M, Artemyev NO. Molecular and Cellular Neurosciences. 2005;28:485–495. doi: 10.1016/j.mcn.2004.10.010. [DOI] [PubMed] [Google Scholar]
- 40.Calvert PD, Krasnoperova NV, Lyubarsky AL, Isayama T, Nicolo M, Kosaras B, Wong G, Gannon KS, Margolskee RF, Sidman RL, Pugh EN, Jr, Makino CL, Lem J. Proceedings of the National Academy of Sciences of the United States of America. 2000;97:13913–13918. doi: 10.1073/pnas.250478897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Posokhova E, Song H, Belcastro M, Higgins L, Bigley LR, Michaud NA, Martemyanov KA, Sokolov M. Molecular & Cellular Proteomics : MCP. 2011;10:M110 000570. doi: 10.1074/mcp.M110.000570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Veltel S, Gasper R, Eisenacher E, Wittinghofer A. Nature Structural and Molecular Biology. 2008;15:373–380. doi: 10.1038/nsmb.1396. [DOI] [PubMed] [Google Scholar]
- 43.Grayson C, Bartolini F, Chapple JP, Willison KR, Bhamidipati A, Lewis SA, Luthert PJ, Hardcastle AJ, Cowan NJ, Cheetham ME. Human Molecular Genetics. 2002;11:3065–3074. doi: 10.1093/hmg/11.24.3065. [DOI] [PubMed] [Google Scholar]
- 44.Ishiba Y, Higashide T, Mori N, Kobayashi A, Kubota S, McLaren MJ, Satoh H, Wong F, Inana G. Experimental Eye Research. 2007;84:473–485. doi: 10.1016/j.exer.2006.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zampighi GA, Schietroma C, Zampighi LM, Woodruff M, Wright EM, Brecha NC. PLoS One. 2011;6:e16944. doi: 10.1371/journal.pone.0016944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tsang SH, Burns ME, Calvert PD, Gouras P, Baylor DA, Goff SP, Arshavsky VY. Science. 1998;282:117–121. doi: 10.1126/science.282.5386.117. [DOI] [PubMed] [Google Scholar]
- 47.Pugh EN, Jr, Lamb TD. Biochimica et Biophysica Acta. 1993;1141:111–149. doi: 10.1016/0005-2728(93)90038-h. [DOI] [PubMed] [Google Scholar]
- 48.Wenzel A, Reme CE, Williams TP, Hafezi F, Grimm C. The Journal of Neuroscience : the official journal of the Society for Neuroscience. 2001;21:53–58. doi: 10.1523/JNEUROSCI.21-01-00053.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]