Abstract
Cell-free microRNAs (miRNAs) that circulate in the blood are promising surrogate biomarkers of disease and physiological processes. The ease of quantifying specific miRNA species using made-to-order approaches based on Taq-polymerase has led to numerous studies that have identified changes in the abundance of circulating cell-free miRNA species that correlate with pathology or other events. The growing interest in developing miRNAs as blood biomarkers necessitates the careful consideration of the unique properties of such body fluids that can make the reproducible and quantitative assessment of RNA abundance challenging. For example, enzymes involved in the amplification and analysis of RNA can be affected by blood components that copurify with miRNA. Thus, if miRNAs are to be effectively utilized as biomarkers, it is important to establish standardized protocols for blood collection and miRNA analysis to ensure accurate quantitation. Here we outline several considerations, including the type of collection tube used in sampling, the influence of added anticoagulants and stabilizers, sample processing, enrichment of vesicular and other miRNA species, RNA extraction approaches and enzyme selection, that affect quantitation of miRNA isolated from plasma and should be considered in order to achieve reproducible, sensitive and accurate quantitation.
1. INTRODUCTION
MicroRNAs (miRNAs) are non-coding RNA molecules of 19–22 nucleotides that are deregulated in cancer [1, 2] neurodegeneration [3], and are temporally over- or underrepresented in physiological conditions including pregnancy [4], aging [5], and longevity [6]. Cell free miRNAs that circulate in blood serum and other body fluids have emerged as promising markers of disease and physiological processes [1, 7–10]. The validation of circulating miRNAs as biomarkers requires methods to accurately identify and quantify miRNAs in complex samples collected from patients. The present protocol provides a standard operating procedure for collection and analysis of blood-derived miRNAs. This protocol was developed based on rigorous testing of the effects of variables on the accuracy and sensitivity of circulating miRNA quantitation, including vacutainer choice, plasma/serum preparation/fractionation, plasma quality control, RNA extraction, and the use of a Taq polymerase mix resistant to endogenous inhibitors of polymerases of plasma [11, 12].
1.1. Quantitative RT-PCR
A common method used to quantify miRNAs is quantitative reverse-transcriptase polymerase chain reaction (qRT-PCR) [13]. qRT-PCR is a preferred approach for the identification and quantitation of blood biomarkers, because alternative approaches including array platforms do not yet match the sensitivity of PCR-based approaches [14]. Using a qRT-PCR approach, changes in plasma and serum miRNA profiles have been reported to reflect various physiological and pathological conditions, including diagnostic and prognostic value for colorectal cancer, breast cancer, gastric cancer, leukemia, lung cancer, lymphoma, oral cancer, ovarian cancer, pancreatic cancer, prostate cancer (reviewed in [1, 7–10]) and other diseases or conditions [15–18]. The rising interest in quantifying cell-free circulating miRNA for diagnostic and prognostic purposes requires assurance that the measured concentration accurately represents the actual amounts in the samples. A major consideration in the processing of RNA from plasma for analysis by qRT-PCR is the sufficient removal of endogenous inhibitors of polymerases, which include hemoglobin [19], lactoferrin [20], immunoglobulin G [21], and calcium, [22] which can co-purify with nucleic acids [23].
1.2. Blood Samples
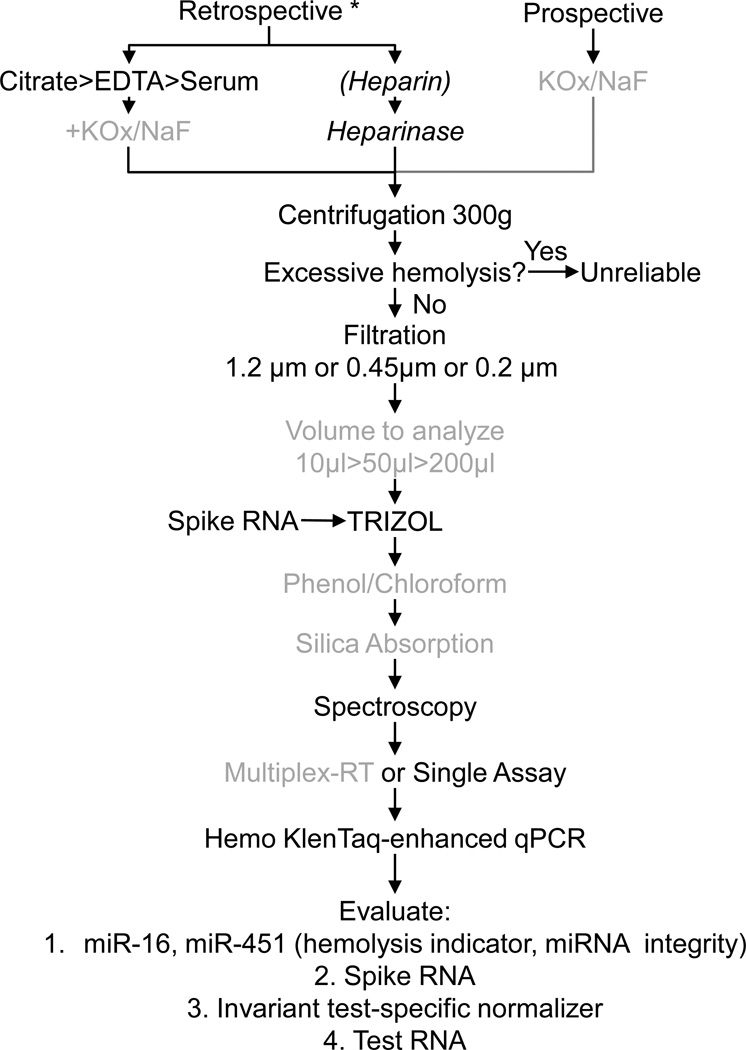
Currently, retrospective studies (using existing samples and data) to quantify cell-free circulating miRNAs frequently rely on plasma collected in commonly used EDTA-tubes (i.e. BD Vacutainer® tubes containing EDTA (7.2 mg, 4.0 ml) or blood serum collected in the absence of anti-coagulants (i.e. BD SST™, BD Vacutainers®), both of which can produce satisfactory results. Another common plasma collection method, which uses heparin as an anticoagulant, is undesirable, as heparin strongly interferes with the quantitation of miRNAs and other RNA species using PCR-based approaches [24]. This inhibition can be alleviated by digesting heparin with lyase heparinase I prior to qRT-PCR [24, 25], though this alternative encumbers a high cost and low efficiency [11]. For prospective studies (de novo design of study and collection), we have found that collection of blood into citrate (sodium citrate, 0.105 M, 4.5ml), or sodium fluoride and potassium oxalate (sodium fluoride/potassium oxalate, 5mg/4mg, 2ml) can provide superior miRNA quantitation and should be considered [11] (Figure 1).
Figure 1. Plasma Collection and Processing for miRNA Quantitation.
For retrospective studies, plasma collected into citrate or EDTA, or blood collected in the absence of anti4 coagulants (serum) provide acceptable data. The use of heparin as an anticoagulant strongly interferes with miRNA quantitation. This inhibition can be partially overcome by treating the samples with heparinase. For prospective studies, collection into KOx/NaF provides high reproducibility and sensitivity in miRNA quantitation [11]. Plasma/serum cleared of most cells by centrifugation at 300g are analyzed for signs of hemolysis [27, 31, 32], and should be interpreted with caution or eliminated from the study if excessive hemolysis is present. To remove all cells and most cell debris from plasma, samples should be filtered. The choice of cut-off sizes determines which of the known miRNA-containing plasma complexes are included in the quantitation. For some collections, especially heparin13 containing plasma, the best volume for purification and quantitation should be evaluated to identify the highest miRNA: interfering contaminant ratio in the RNA preparation. A spiked RNA is added during TRIzol extraction to assess yield and amplification efficiency for each RNA preparation. An additional phenol/chloroform extraction and differential silica-absorption can increase purity and quantitation efficiency several-fold. Spectroscopy is used to quantify RNA yield and purity. Single-assay, or previously validated multiplex RT analyses using looped-primers specifically designed to 3’ ends of mature miRNAs are used for cDNA production. Quantitative PCR with the addition of Hemo KlenTaq, which is much less sensitive to blood-borne inhibitors of Taq polymerase, provides high sensitivity in miRNA quantitation. MiRNAs to evaluate for quality control and yield include miR-16, miR-451 and spiked RNA. Assay-specific normalizing miRNAs or other RNAs should be quantified along with the miRNAs of interest.
* If cell pellet present in frozen plasma/serum, physically separate cell free portion from cell pellet before thaw by cutting cores. Italizised steps: not recommended if alternatives exist. Grey steps: Should be empirically validated for benefits in each study.
1.3. Proper Serum and Plasma Handling
An important consideration in working with cell-free plasma is to avoid contamination with cells and cell lysate during processing of the sample, as the contribution of cellular miRNAs [26] and other components will mask or confound the plasma miRNA profile [27]. Common starting material is cell-free plasma or serum in which the cell-free phase was promptly separated from the cell pellet after phlebotomy to prevent loss of components [28] or hemolysis [29]. In these cases, the cellular and cell-free proportion is typically frozen separately, which minimizes artifactual cell pellet contribution to the plasma.
Alternatively, and especially appropriate in multi-center trials where identical sample preparation procedures must be maintained, the cell mass may be pelleted from plasma by centrifugation, yet not separated prior to freezing. In this case, it is important to separate the plasma from the cell pellet before thawing in order to avoid hemolysate contribution to the cell-free portion of the plasma. One approach is to separate the cell-free plasma from the pellet by cutting the vacutainer using a guillotine or “saw before thaw”. Regardless of the approach, the supernatant contains some cells, which can be reduced using medium speed centrifugation (300–800g). Additionally, filtration through 1.2 µm-cut off filters will remove necrotic or apoptotic debris, as well platelets, if elimination of these miRNA-containing particles is desired.
1.4. Sample Quality Control
Recent recommendations on proper plasma handling include quality control to ensure the starting material is not contaminated with cellular components that arise from cell lysis during blood draw, storage, sample thaw, or extraction. For most applications, excessive abundance of hemoglobin is a quantifiable measure to assess the quality of the plasma. While blood cells may shed up to 20% of their hemoglobin content [30], excessive abundance of hemoglobin in plasma is indicative of improper sample handling or disease, and will interfere with both proper polymerase function [19], as well as interpretation of results [27, 31]. Quantitation of hemolysis visually [27], and by spectroscopy [31, 32] are effective methods by which to select high-quality samples (Figure 1). In addition, excessive miR-16 and miR-451 concentrations, two miRNAs that are common in blood cells, can be indicative of hemolysis [27], if not disease [12]. Alternatively, detecting low or no miR-16 can be indicative of sample degeneration or interfering substances [11], putting into question the utility of the plasma for quantifying cell-free circulating miRNAs (Figure 1).
1.5. Nature of Cell-Free miRNA Complexes in Plasma
MiRNAs in the plasma can be associated with vesicles or protein complexes. MiRNAs are associated with a variety of microvesicles [33], including well-characterized exosomes [34]. Alternatively, miRNAs have been found to be associated with non-vesicular lipoprotein complexes [35], which co-purify with microvesicles enriched by ultracentrifugation [36, 37], other non-vesicular RNA-binding proteins [38], or miRNA processing proteins [39, 40]. The majority of miRNAs detected in blood plasma are thought to be associated with protein complexes rather than vesicles [39, 40], yet alternative findings have been reported [41]. Focusing analyses on specific miRNA subpopulations may reveal signatures specific to tissue or cell of origin, or allow the quantitation of changes in miRNA concentrations that are otherwise concealed in the high total miRNA concentration of plasma. However, it is important to consider that particles and vesicles change in the blood which includes the loss of surface antigens for enrichment [42]. Protocols and tools for the isolation of plasma exosomes, and lipoprotein complexes are available [35, 37, 43], and several commercial kits are offered. It is not yet clear whether protein-associated miRNAs are of value or an impediment in diagnosis or prognosis [39, 40]. Most studies that aim to identify plasma biomarkers do not distinguish among the various plasma complexes that contain miRNA. It is also important to note that during disease progression, the miRNAs that change the plasma signatures above or below homeostatic levels may be of different origin, and may include platelets [44] early in disease, liver origin during drug-administration [45], and in the case of cancer, tumor-origin later in the disease. If microvesicles (100–1000 nm), or exosomes (50–100 nm), or lipoprotein and other miRNA-protein complexes (<50 nm) are of interest [46], different filters (1.2 µm, 0.45 µm, 0.2 µm, or 0.1 µm) may aid in enrichment of desired miRNA population. Alternative approaches such as gel exclusion chromatography or gradients have been developed for larger volumes in order to further purify the particles, lipoprotein complexes and vesicles [39, 40], yet whether the yield is sufficient to analyze typically available plasma volumes of 100 µl-5 ml is not clear.
1.6. Extraction Methods
Extraction and purification of miRNAs from plasma must be sufficient to remove endogenous inhibitors of polymerases, [19–22] which can co-purify with nucleic acids [11, 23]. A common approach is to use TRIzol reagent (Invitrogen) for extraction of RNA [47], including miRNAs. Alternative approaches should be compared side-by side [11, 48]. A recent report highlighted the importance of reagent choice, as different reagents extract specific miRNA species selectively, especially at low RNA concentrations {Kim, 2012 #85}.
Importantly, the nature of the complex containing the miRNAs may determine the yield and enrichment of specific RNA subspecies. For example, the extraction method used to purify RNA from exosomes, including the choice of organic solvent for removal of lipids and proteins, and the use of silica membranes to enrich for RNA species affects the enrichment of some miRNA species over others as well as the RNA yield and purity [48]. Some of the purest miRNA preparations from whole plasma are produced using differential silica-adsorption. However, this approach increases the cost of preparation [11], and may be unsuitable in situations where other RNA species need to be quantified. The use of ribonuclease inhibitors may help in maintaining RNA stability [49], however, we have found no benefit in using RNAsin [11].
RNA yield can be established using spectroscopy or an Agilent Bioanalyzer (Figure 1). However, for plasma-derived samples, it is not unusual to find high A320nm readings, suggestive of low purity. The required purity for proper analyses should be established empirically for miRNAs to quantify before undertaking large-scale investments into both expensive purification and quantitation approaches on precious samples.
1.7. Volumes Analyzed
Typically, 200 µl of plasma is sufficient for analyses by qRT-PCR. Indeed using less volume, as low as 10 µl may provide superior amplification efficiency over larger volumes for some blood collections, in part due to a balance between RNA yield and co-purification of substances that interfere with quantitation [11]. However, the volume required depends on the number and abundance of miRNAs to be quantified, whether the reverse transcriptase reaction can be multiplexed, and the purification and subfractionation requirements of the blood plasma.
1.8. Normalization
In order to achieve accurate and reproducible quantitation of miRNAs from blood plasma, several normalizing parameters should be recorded in order to establish a base-line for comparison across samples. Choices include total original plasma/serum volume, total RNA yield, endogenous invariant miRNA standards, other RNAs or spiked RNAs. An advantage of normalizing to endogenous miRNAs is that their origin, processing and amplification are very similar to the test-miRNA, and thus representative across multiple variables. Which RNA species represent the best normalizing miRNAs or other noncoding RNAs is a matter of debate, and may depend upon the specific application [27, 50–52]. The use of total RNA as a normalizer, while very useful in cell-culture and tissue studies, is more problematic for RNA of plasma origin, as reliable quantitation is highly dependent on the purity of the sample [13]. Identical concentrations of synthetic C. elegans miR-39 miRNA mimic RNA oligonucleotides, or other synthetic RNA sequences [11, 13] spiked during preparation has the advantage of detecting loss of sample, the presence of inhibitors of detection, and other issues involving miRNA preparation and analysis. For full benefits of using spiked RNA, care must be taken that it is used in the femtomolar-to attomolar range, the concentration range of most plasma miRNAs, because more abundant RNA species are less susceptible to inhibition by endogenous inhibitors of polymerases [11].
Relative miRNA abundance can be measured using common approaches [53], or with correction for amplification efficiency based on an exponential model of PCR [54, 55]. However, miRNA abundance is best measured by computing moles based on comparing CT values of samples to dilutions of a synthetic DNA corresponding to the cDNA produced by RT for each miRNA measured [56] to make a standard curve, or by using an RNA oligonucleotide as a standard. Furthermore, the calculation of the amplification efficiency using the equation (T2/T1)(1/(CT2 ave-CT1 ave))-1 can detect the presence of some inhibitors, as they may significantly reduce the amplification efficiency below 1 [55].
1.9. Taq Poymerase
Intact Taq polymerase activity is sensitive to blood-borne inhibitors [57]. An alternative approach to avoid interference from such inhibitors of PCR is to use different polymerases [20, 22, 23, 57, 58]. One enzyme in particular, Hemo KlenTaq, has a 100-fold lower sensitivity to blood inhibitors than wild-type Taq [57]. However, reduced proofreading [59] limits the utility of Hemo KlenTaq in quantitative PCR (qPCR). This limitation can be overcome by the use of a Taq polymerase cocktail of normal and Hemo KlenTaq which improves sensitivity up to 30-fold [11], thus reducing the need of stringent purification (Figure 1).
Whatever the quantitative approach, it is important to go beyond “blind-faith” when one considers TaqMan approaches, and to optimize the qPCR approach empirically to produce a single band of the correct size by native PAGE in order to confirm the specificity and purity of amplified and quantitated products [11, 12].
2. Methodological Overview
The following protocol is optimized for quantitation of six miRNA species in frozen or fresh cell-free blood plasma of human or murine origin [11, 12] (Figure 1). The protocol includes the following steps: 1) Blood collection, 2) Removal of debris from plasma using a combination of low-speed centrifugation and filtration; 3) separation of protein-complex associated miRNAs from microvesicle and lipoprotein-complex associated miRNAs; 4) extraction of RNA from fractions; 5) multiplex reverse-transcription of miRNA with stem-loops, and 6) amplification of cDNA using a polymerase cocktail resistant to plasma-inhibitors. At each point, empirical evaluation of additional steps that we have determined to affect quantitation are advised to the user in cases where the outcome is not satisfactory.
3. Protocol
3.1. Physical separation of tasks
To achieve the sensitivity necessary to measure attomolar RNA concentrations faithfully, physical separation of each procedural step is advisable to avoid cross-contamination of samples, synthetic oligonucleotides, reagents, cDNA or environmental components. It is strongly advised that the different steps in the protocol are carried out at different benches, hoods or rooms. For example, Area 1 is restricted for use in preparing and separating plasma and is equipped with a hood and bench-top, and dedicated pipettors. Area 2 includes 3 separate areas dedicated to different operations of the project. Area 2.1 is a bench-top used for aliquotting primers, standards and control oligonucleotides. Area 2.2 is a hood dedicated for RT-reaction set-up. Area 2.3 is a bench-top used for the sole purpose of setting up microwell plate PCR-reactions. Area 3 is used for RT, PCR, RNA gels, RNA quantitation and other support for the project.
3.1. Blood collection
Plasma and serum samples are collected by venipuncture using BD Vacutainer® tubes containing sodium fluoride/potassium oxalate, 5mg/4mg, 2ml, (BD, 367921) for human samples, or Capiject T-MPS tubes (Terumo, Somerset, NJ) for mouse samples. Collection as serum, citrate, or EDTA plasma produces different efficiencies and specificities of miRNA quantitation and should not be interchanged [11]. Tubes are inverted several times for anticoagulants to be evenly distributed, and incubated for 15 min at room temperature (r.t.).
Cell-free plasma is obtained by centrifugation of blood samples at 200×g for 15 min at 4°C. Supernatants are removed and collected in 15 ml (or smaller as appropriate) polypropylene tubes. All steps from here on are carried out on ice (or alternatively chilled on sterilized Bath Armor Beads, (Lab Armor, 42370) Cornelius, OR), unless otherwise specified. The plasma is further centrifuged twice at 800×g for 15 min at 4°C to obtain cell-free plasma.
Remaining cell debris is removed by passing through detergent-free filters. For small volumes (≤500 µl) use Millipore Durapore© Centrifugal Filter 0.22 µm cut-off (UFC30GV00) 0.45 µm cut-off (UFC30HV00), or 0.65 µm cut-off (UFC30DV00) and centrifuge for 4 min at 12,000×g at 4°C. Alternatively, use 1.2 µm, 0.45 µm, or 0.2 µm pore-size cut-off syringe filters (Supor® Membrane, PALL, Port Washington, NY 4656, 4654, or 4652 respectively) depending on the plasma component to be quantified, as outlined in section 1.4.
Plasma is inspected visually for any pink hue, which is indicative of hemolysis, or, if sample volumes are generous, by spectrophotometry [60].
Plasma is preferably processed immediately, especially if sub-fractionated. Alternatively, plasma is flash-frozen in liquid nitrogen for future processing. It is advisable to store samples in the vapor phase of liquid nitrogen in the case of vial rupture, which would contaminate the contents of the ruptured vial, as well as raise the possibility that the spilled content can contaminate other vials in storage if stored in the liquid phase.
3.2. Plasma processing
If starting with frozen plasma supernatant, with no further processing, thaw plasma slowly on ice. Process 500 µl if available. Centrifuge at 4°C for 15 min at 300×g, and filter through Durapore© Filter 0.45 µm cut off (Millipore, UFC30HV00) for 4 min at 12,000×g at 4°C, or as outlined in 3.1.3. Transfer 450 µl to a fresh tube (VWR, 87003-294) and process.
EMPERICAL consideration: Addition of 1.0 µg NaF/0.8 µg KOx per µl of centrifuged and filtered serum/plasma, (NaF (Sigma-Aldrich Corp, S-6776;) and KOx (Aqua Solutions, Deer Park, TX, P5311)) can improve detection of miRNAs up to 2.4-fold for serum and 3.6-fold for EDTA plasma when quantifying by standard SYBR Green o r TaqMan approaches [11].
3.3. Separation of particulate from soluble miRNA complexes and extraction
EMPERICAL consideration: The addition of a single acidic phenol-chloroform (Amresco, Solon, OH; 0966) extraction, followed by absorption, and differential elution from a silica resin (Invitrogen PureLink miRNA Isolation Kit, K1570-01) can raise sensitivity about 4 fold, but will nearly double the cost of extraction [11].
If quantifying total cell-free plasma, replace steps 1–3 (of section 3.3. below), by the addition of 500 µl TRIzol (Invitrogen, 15596-018) reagent directly to the plasma and incubation for 10 min at r.t.
If vesicular/lipoprotein associated miRNAs are to be quantified separately from soluble protein-complexed miRNAs, transfer each sample into appropriate SW60 tubes (Beckman, 328874) or smaller that are designed for ultracentrifugation.
Centrifuge samples for 2 hours at 100,000×g in a swinging-bucket rotor (Beckman, SW 60Ti rotor). Remove 400 µl of supernatant promptly into a fresh VWR tube (VWR, 87003-294) labeled “S100” containing 500 µl TRIzol (Invitrogen, 15596-018) reagent. Swinging buckets are preferred over fixed-angle rotors, as pellets can easily dislodge from the side of tubes.
Remove the rest of the supernatant VERY CAREFULLY from the pellet (most likely invisible) and discard. Add 500 µl TRIzol reagent directly onto the pellet in the SW60 tube and mix by pipetting. Transfer the pellet/TRIzol mix to an appropriate VWR tube labeled “P100”. Incubate at r.t. for 10 min.
To both tubes, add 100 µl chloroform (J.T. Baker, 9175-02) and shake vigorously by hand for 15 s. Do NOT vortex. Incubate at room temperature for 3 min. Centrifuge the samples at 300×g for 15 min at 4° C. Transfer as much as is possible of the clear, upper, aqueous phase without disturbing the interphase, and transfer into new, labeled 1.7 ml microfuge tube (Costar, 3620) or other low RNA-binding tubes are used for extracted RNA). Leave about 50 µl of aqueous solution behind as not to disturb the interphase. Transfer the same volume from each sample within a study, which is usually between 250 µl – 350 µl. Save the lower, pink, organic phase for later protein or DNA extraction if desired, and store at −80°C.
-
Add 750 µl (1.5 × volume of TRIzol used) of isopropanol to each aqueous sample. Mix well by shaking. Add 1 µl glycoblue (Ambion, AM9515) to each sample. Mix well by shaking. Incubate overnight at −80°. Centrifuge at 4°C for 40 min at 21,000−g (or maximum speed on bench-top microfuge). Locate the RNA pellets (blue) and very carefully remove the supernatant. Leave about 50 µl so as to not disturb the pellet. Add 1 ml of FRESHLY PREPARED high-grade 75% EtOH (OmniPur, 200 proof, 4450) in GIBCO water (10977) or other high-purity water. Centrifuge at max speed for 5 min on the bench-top microfuge and very carefully remove as much of the supernatant as possible without disturbing the blue pellet. Centrifuge again at maximum speed for 1 min on the bench top and very carefully remove any remaining supernatant with a P20 pipettor, or similar size.
Note: We have observed that RNA isolated from serum and plasma has variable stability when stored at −80° C in water. Thus, long-term storage of RNA in isopropanol or ethanol is advisable.
Invert tubes on a Kimwipe (Kimtech, 34155) to dry. Be careful not to over-dry, as completely dried pellets can be difficult to resuspend. Drying should take as little as 3–5’. Take care with dried pellets and gloves, as static from gloves can cause pellets to “jump” out of tubes.
Resuspend in 20 µl GIBCO water or other high purity water. Measure 2 µl on a biophotometer (Eppendorf 22331, fitted with a Hellma, 105.800 UVS Tray Cell, and 0.2 mm and/or 1 mm cap) or other low-volume spectrophotometer. The typical yield for the pellet is 2 µg/ml of plasma, and for the supernatant is 5 µg/ml of plasma. It is best to RT immediately. Alternatively store in −80°C ultra-low freezer.
3.4. Reverse transcription (RT)
EMPERICAL consideration: 1) Multiplexing the RT reactions with several reverse primers has the advantages of conserving precious RNA sample, and can provide more direct correlation among the different miRNAs that are reverse transcribed, by eliminating tube-to-tube variation. However, it is imperative to exclude the possibilities of primer-dimers, false-priming, synergistic amplification or other artifacts that cannot be corrected for in retrospect. Therefore, proper multiplex mixes, including the addition of spiked RNA need to be confirmed by trial on control samples. Start by analyzing the sequences of the TaqMan RT-primers [56] for base-pair overlaps that might lead to primer-dimers. Other alternatives include the use of the LNA system, which work on the basis of poly-A tailed RNA.
2) The inability to detect specific miRNAs in plasma or serum, in many cases, reflects the low abundance of particular miRNAs in the circulation. Alternatively, absence of detection may reflect that release of some miRNAs from cells into blood is limited or selective [12, 61, 62]
Remove TaqMan MicroRNA Reverse Transcription Kit components from freezer and thaw on ice.
Prepare the RT master mix (ABI 4366596) on ice in a Costar microcentrifuge tube, per sample: 100 mM dNTPs (with dTTP), 0.15 µl; MultiScribe Reverse Transcriptase, 50 U/µl, 1.0 µl; 10X Reverse Transcription Buffer, 1.5 µl; RNase Inhibitor, 20 U/µl; 0.19 µl, Gibco dH2O, 4.16 µl; RT Primer (20×) 0.75 µl; Total Volume, 10 µl. (All ABI) Note: In the case of RT primer multiplexing with more than 2 primers, it is necessary to purchase the medium size TaqMan assays (ABI, 4440887) in order to receive 20X rather than 5X RT Primer concentrations that are provided with the small size TaqMan assay (ABI, 4427975).
Mix master mix gently and centrifuge briefly to bring solution to bottom of tube. Keep master mix on ice until ready to use. Label PCR strip tubes (VWR, 53509-304) and add 10 µl of master mix to each tube. Keep on ice (PCR plate cold racks (Isofreeze, 5640-T4) work well for this). Add 5 µl of RNA (on average, 0.4 µg total RNA) to each labeled tube, then mix by pipetting up and down. Cap tubes and centrifuge briefly to collect solution down to bottom of tube.
Incubate reactions on ice for 5 min. Load One Step Program on thermal cycler (16°C, 30 min, 42°C, 30 min, 85°C, 5 min, 4°C ∞) with reaction volume set to 15 µl. Load tubes into thermal cycler and start. The reaction takes a little over an hour.
3.5. Quantitative PCR (qPCR)
Remove TaqMan MicroRNA Assay kit and cDNA from freezer and thaw on Bath Armor Beads or ice. Produce a master mix composed of TaqMan 2× Universal PCR MM, no UNG, 10.0 µl; Gibco dH2O 7.07 µl; TaqMan MicroRNA Assay (20×) 1.0 µl, Hemo KlenTaq™ [57] (New England BioLabs, Ipswich, MA, M0332), 0.6 µl; cDNA 0.19 µl in a total volume of 20 µl per reaction. It is recommended to measure each sample/primer combination in triplicate. Note: If not multiplexing, leave out cDNA from the mastermix, and add to appropriate well on the qPCR plate.
-
Centrifuge plate for 5 min at 300g. Run TaqMan program on the qPCR machine (95°C, 10 min (1×); [95°C, 15 s, 60°C, 60 sec (40×)].
Note: Commercial ABI PCR primers can only amplify properly cDNA from miRNA produced with specific RT stem-loop primers that match the ABI PCR primers. This is because the PCR primers are designed to hybridize to portions of both the RT primer as well as sequences that are specific to the 3’ end of the miRNA [56] [41]. Therefore, cDNA templates produced by other means, including tailing or linkering lack sequences required for efficient and specific binding of the 3’ primer.
4. Pitfalls
Assurance of validity of plasma quantitation includes 1) elimination of sources of cross-over material contamination, 2) a balance of purity and yield, 3) removal of all organic material from extracted RNA, 4) confirmation of the plasma origin of TaqMan qRT-PCR products.
More specifically:
As outlined in section 3.1, handling and storage of plasma, RNA, DNA (primers), cDNA, and reagents in mutually exclusive areas of the lab is required to prevent detrimental carry6 over of material into the test sample during the time ranging from storage to the final PCR plate, and conversely into reagents, stocks and standards.
It is important to consider the cost, extra time and handling of material, as each step carries a risk of degradation and contamination [11].
The effective exclusion of phenol, chloroform of other organic phases from the aqueous RNA in section 3.3.4. is imperative for purity (A260/A280, A320), and to avoid interference with RT and/or PCR. It is advisable for researchers with little experience in effective phase separation to add absorption methods (such as PureLink (Invitrogen, K1570-01), which can more definitively remove phenolic compounds from the interrogated RNA.
The ease of TaqMan or LNA technology, the pervasive use of which has enabled biomarker discovery by qPCR the day after serum/plasma collection, should not lead the investigator, who observes detectable gaps in the Ct curves of control and test sample, to the conclusion that they represent the true product of interest. Initially, all qRT-PCR products should be validated by separation of the PCR products by native PAGE [11, 12], a process that takes 2 hours. If more than one band, or no band is visible, further work is required to obtain specific qRT-PCR amplification.
5. Concluding Remarks
The current method addresses aspects that limit miRNA quantitation in blood, including vacutainers, hemolysis, insufficient RNA purification, limited volume, normalization procedure, quantitation, and the use of polymerase cocktails that are not inhibited by material that co-purifies with RNA. Following the protocol outlined above will circumvent these limitations and allows the quantitation of miRNAs that could not otherwise be accurately quantitated.
As the technology to quantify miRNA with greater sensitivity develops, enrichment of cell-free plasma miRNA subpopulations may become valuable to quantify miRNA changes that are otherwise hidden in the vast sea of plasma miRNAs. It is important to recognize that the nature of the complex circulating miRNAs are associated with [12, 34, 35, 38, 39, 63–66], may affect their stability [39] and determine how amenable these miRNA subpopulations are to particular purification methods [48], or amplification by polymerases. The improvements outlined here allow the quantitation of miRNAs with very low abundance, where usual techniques fail. The use of these approaches is expected to increase the repertoire of miRNAs that can be analyzed as potential biomarkers of disease. This protocol is considered one step towards assuring that the measured concentration represents the actual amounts in the samples [67]. The applicability of any one method to another study should be independently verified [68, 69]. Recent developments [70, 71], using LNA approaches [14], and alternative qRT-PCR approaches [72] are expected to improve sensitivity.
ACKNOWLEDGEMENTS
This work was funded by the US Army Medical Research and Materiel Command under W81XWH-08-1-0641, an American Cancer Society of Illinois Research Grant #189903, a Schweppe Research Foundation Fellowship, and Rosalind Franklin University of Medicine and Science start-up funds to DMD, and a National Institutes of Health grant R01NS069759 to M.L.H. We thank the IU Simon Cancer Center, and the Susan G Komen for the Cure tissue bank (https://komentissuebank.iu.edu/), and Dr. Jeff Martin of the University of California at San Francisco for making the blood samples available. We thank Alicia Case and Mallory Havens for critical reading discussing the manuscript.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
BIBLIOGRAPHY
- 1.Almeida MI, Reis RM, Calin GA. Mutation research. 2011;717:1–8. doi: 10.1016/j.mrfmmm.2011.03.009. [DOI] [PubMed] [Google Scholar]
- 2.He X, He L, Hannon GJ. Cancer Res. 2007;67:11099–11101. doi: 10.1158/0008-5472.CAN-07-2672. [DOI] [PubMed] [Google Scholar]
- 3.Gascon E, Gao FB. Frontiers in neuroscience. 2012;6:48. doi: 10.3389/fnins.2012.00048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Morales Prieto DM, Markert UR. Journal of reproductive immunology. 2011;88:106–111. doi: 10.1016/j.jri.2011.01.004. [DOI] [PubMed] [Google Scholar]
- 5.Smith-Vikos T, Slack FJ. Journal of cell science. 2012;125:7–17. doi: 10.1242/jcs.099200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Elsharawy A, Keller A, Flachsbart F, Wendschlag A, Jacobs G, Kefer N, Brefort T, Leidinger P, Backes C, Meese E, Schreiber S, Rosenstiel P, Franke A, Nebel A. Aging cell. 2012 doi: 10.1111/j.1474-9726.2012.00824.x. [Epub ahead of print]. [DOI] [PubMed] [Google Scholar]
- 7.Corcoran C, Friel AM, Duffy MJ, Crown J, O'Driscoll L. Clinical chemistry. 2011;57:18–32. doi: 10.1373/clinchem.2010.150730. [DOI] [PubMed] [Google Scholar]
- 8.Brase JC, Wuttig D, Kuner R, Sultmann H. Mol Cancer. 2010;9:306. doi: 10.1186/1476-4598-9-306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zen K, Zhang CY. Med Res Rev. 2010 doi: 10.1002/med.20215. [Epub ahead of print]. [DOI] [PubMed] [Google Scholar]
- 10.Kosaka N, Iguchi H, Ochiya T. Cancer Sci. 2010;101:2087–2092. doi: 10.1111/j.1349-7006.2010.01650.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kim DJ, Linnstaedt S, Palma J, Park JC, Ntrivalas E, Kwak-Kim JY, Gilman-Sachs A, Beaman K, Hastings ML, Martin JN, Duelli DM. The Journal of molecular diagnostics : JMD. 2012;14:71–80. doi: 10.1016/j.jmoldx.2011.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pigati L, Yaddanapudi SC, Iyengar R, Kim DJ, Hearn SA, Danforth D, Hastings ML, Duelli DM. PloS one. 2010;5:e13515. doi: 10.1371/journal.pone.0013515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kroh EM, Parkin RK, Mitchell PS, Tewari M. Methods. 2010;50:298–301. doi: 10.1016/j.ymeth.2010.01.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jensen SG, Lamy P, Rasmussen MH, Ostenfeld MS, Dyrskjot L, Orntoft TF, Andersen CL. BMC genomics. 2011;12:435. doi: 10.1186/1471-2164-12-435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen X, Ba Y, Ma L, Cai X, Yin Y, Wang K, Guo J, Zhang Y, Chen J, Guo X, Li Q, Li X, Wang W, Wang J, Jiang X, Xiang Y, Xu C, Zheng P, Zhang J, Li R, Zhang H, Shang X, Gong T, Ning G, Zen K, Zhang CY. Cell Res. 2008;18:997–1006. doi: 10.1038/cr.2008.282. [DOI] [PubMed] [Google Scholar]
- 16.Go AT, Visser A, van Dijk M, Mulders MA, Eijk P, Ylstra B, Blankenstein MA, van Vugt JM, Oudejans CB. Methods Mol Biol. 2008;444:291–302. doi: 10.1007/978-1-59745-066-9_23. [DOI] [PubMed] [Google Scholar]
- 17.Zhang Y, Jia Y, Zheng R, Guo Y, Wang Y, Guo H, Fei M, Sun S. Clin Chem. 2010;56:1830–1838. doi: 10.1373/clinchem.2010.147850. [DOI] [PubMed] [Google Scholar]
- 18.Hung EC, Chiu RW, Lo YM. J Clin Pathol. 2009;62:308–313. doi: 10.1136/jcp.2007.048470. [DOI] [PubMed] [Google Scholar]
- 19.Akane A, Matsubara K, Nakamura H, Takahashi S, Kimura K. J Forensic Sci. 1994;39:362–372. [PubMed] [Google Scholar]
- 20.Al-Soud WA, Radstrom P. J Clin Microbiol. 2001;39:485–493. doi: 10.1128/JCM.39.2.485-493.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Al-Soud WA, Jonsson LJ, Radstrom P. J Clin Microbiol. 2000;38:345–350. doi: 10.1128/jcm.38.1.345-350.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Eilert KD, Foran DR. J Forensic Sci. 2009;54:1001–1007. doi: 10.1111/j.1556-4029.2009.01116.x. [DOI] [PubMed] [Google Scholar]
- 23.de Franchis R, Cross NC, Foulkes NS, Cox TM. Nucleic Acids Res. 1988;16:10355. doi: 10.1093/nar/16.21.10355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Johnson ML, Navanukraw C, Grazul-Bilska AT, Reynolds LP, Redmer DA. Biotechniques. 2003;35:1140–1142. doi: 10.2144/03356bm03. 1144. [DOI] [PubMed] [Google Scholar]
- 25.Tsai M, Miyamoto M, Tam SY, Wang ZS, Galli SJ. Am J Pathol. 1995;146:335–343. [PMC free article] [PubMed] [Google Scholar]
- 26.Duttagupta R, Jiang R, Gollub J, Getts RC, Jones KW. PLoS One. 2011;6:e20769. doi: 10.1371/journal.pone.0020769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kirschner MB, Kao SC, Edelman JJ, Armstrong NJ, Vallely MP, van Zandwijk N, Reid G. PLoS One. 2011;6:e24145. doi: 10.1371/journal.pone.0024145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Waring WS, Evans LE, Kirkpatrick CT. J Clin Pathol. 2007;60:820–823. doi: 10.1136/jcp.2006.039925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Oremek GM, Gerstmeier F, Sauer-Eppel H, Sapoutzis N, Wechsel HW. Anticancer Res. 2003;23:1127–1130. [PubMed] [Google Scholar]
- 30.Willekens FL, Bosch FH, Roerdinkholder-Stoelwinder B, Groenen-Dopp YA, Werre JM. European journal of haematology. 1997;58:246–250. doi: 10.1111/j.1600-0609.1997.tb01662.x. [DOI] [PubMed] [Google Scholar]
- 31.Pritchard CC, Kroh E, Wood B, Arroyo JD, Dougherty KJ, Miyaji MM, Tait JF, Tewari M. Cancer Prev Res (Phila) 2012 doi: 10.1158/1940-6207.CAPR-11-0370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sanderink GJ, van Rijn HJ. Clin Chim Acta. 1985;146:65–73. doi: 10.1016/0009-8981(85)90124-x. [DOI] [PubMed] [Google Scholar]
- 33.Orozco AF, Lewis DE. Cytometry. Part A : the journal of the International Society for Analytical Cytology. 2010;77:502–514. doi: 10.1002/cyto.a.20886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Taylor DD, Gercel-Taylor C. Gynecol Oncol. 2008;110:13–21. doi: 10.1016/j.ygyno.2008.04.033. [DOI] [PubMed] [Google Scholar]
- 35.Vickers KC, Palmisano BT, Shoucri BM, Shamburek RD, Remaley AT. Nat Cell Biol. 2011;13:423–433. doi: 10.1038/ncb2210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Looze C, Yui D, Leung L, Ingham M, Kaler M, Yao X, Wu WW, Shen RF, Daniels MP, Levine SJ. Biochem Biophys Res Commun. 2009;378:433–438. doi: 10.1016/j.bbrc.2008.11.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang J, Hawari FI, Shamburek RD, Adamik B, Kaler M, Islam A, Liao DW, Rouhani FN, Ingham M, Levine SJ. Biochem Biophys Res Commun. 2008;366:579–584. doi: 10.1016/j.bbrc.2007.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang K, Zhang S, Weber J, Baxter D, Galas DJ. Nucleic acids research. 2010;38:7248–7259. doi: 10.1093/nar/gkq601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Arroyo JD, Chevillet JR, Kroh EM, Ruf IK, Pritchard CC, Gibson DF, Mitchell PS, Bennett CF, Pogosova-Agadjanyan EL, Stirewalt DL, Tait JF, Tewari M. Proc Natl Acad Sci U S A. 2011;108:5003–5008. doi: 10.1073/pnas.1019055108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Turchinovich A, Weiz L, Langheinz A, Burwinkel B. Nucleic Acids Res. 2011;39:7223–7233. doi: 10.1093/nar/gkr254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gallo A, Tandon M, Alevizos I, Illei GG. PLoS One. 2012;7:e30679. doi: 10.1371/journal.pone.0030679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rupp AK, Rupp C, Keller S, Brase JC, Ehehalt R, Fogel M, Moldenhauer G, Marme F, Sultmann H, Altevogt P. Gynecol Oncol. 2011;122:437–446. doi: 10.1016/j.ygyno.2011.04.035. [DOI] [PubMed] [Google Scholar]
- 43.Han X. Biochimica et biophysica acta. 2010;1801:774–783. doi: 10.1016/j.bbalip.2010.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Stratz C, Nuhrenberg TG, Binder H, Valina CM, Trenk D, Hochholzer W, Neumann FJ, Fiebich BL. Thrombosis and haemostasis. 2012;107:634–641. doi: 10.1160/TH11-10-0742. [DOI] [PubMed] [Google Scholar]
- 45.Wang K, Zhang S, Marzolf B, Troisch P, Brightman A, Hu Z, Hood LE, Galas DJ. Proc Natl Acad Sci U S A. 2009;106:4402–4407. doi: 10.1073/pnas.0813371106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Palma J, Yaddanapudi SC, Pigati L, Havens MA, Jeong S, Weiner GA, Weimer KM, Stern B, Hastings ML, Duelli DM. Nucleic acids research. 2012 doi: 10.1093/nar/gks656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rio DC, Ares M, Jr, Hannon GJ, Nilsen TW. Cold Spring Harbor protocols. 2010;2010 doi: 10.1101/pdb.prot5439. pdb prot5439. [DOI] [PubMed] [Google Scholar]
- 48.Eldh M, Lotvall J, Malmhall C, Ekstrom K. Molecular immunology. 2012;50:278–286. doi: 10.1016/j.molimm.2012.02.001. [DOI] [PubMed] [Google Scholar]
- 49.Tzimagiorgis G, Michailidou EZ, Kritis A, Markopoulos AK, Kouidou S. Cancer epidemiology. 2011;35:580–589. doi: 10.1016/j.canep.2011.02.016. [DOI] [PubMed] [Google Scholar]
- 50.Zhu HT, Dong QZ, Wang G, Zhou HJ, Ren N, Jia HL, Ye QH, Qin LX. Molecular biotechnology. 2012;50:49–56. doi: 10.1007/s12033-011-9414-6. [DOI] [PubMed] [Google Scholar]
- 51.Appaiah HN, Goswami CP, Mina LA, Badve S, Sledge GW, Jr, Liu Y, Nakshatri H. Breast Cancer Res. 2011;13:R86. doi: 10.1186/bcr2943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hu Z, Dong J, Wang LE, Ma H, Liu J, Zhao Y, Tang J, Chen X, Dai J, Wei Q, Zhang C, Shen H. Carcinogenesis. 2012;33:828–834. doi: 10.1093/carcin/bgs030. [DOI] [PubMed] [Google Scholar]
- 53.Livak KJ, Schmittgen TD. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 54.Liu W, Saint DA. Anal Biochem. 2002;302:52–59. doi: 10.1006/abio.2001.5530. [DOI] [PubMed] [Google Scholar]
- 55.Cikos S, Bukovska A, Koppel J. BMC Mol Biol. 2007;8:113. doi: 10.1186/1471-2199-8-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Schmittgen TD, Lee EJ, Jiang J, Sarkar A, Yang L, Elton TS, Chen C. Methods. 2008;44:31–38. doi: 10.1016/j.ymeth.2007.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kermekchiev MB, Kirilova LI, Vail EE, Barnes WM. Nucleic Acids Res. 2009;37:e40. doi: 10.1093/nar/gkn1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Abu Al-Soud W, Radstrom P. Appl Environ Microbiol. 1998;64:3748–3753. doi: 10.1128/aem.64.10.3748-3753.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Barnes WM. Gene. 1992;112:29–35. doi: 10.1016/0378-1119(92)90299-5. [DOI] [PubMed] [Google Scholar]
- 60.Noe DA, Weedn V, Bell WR. Clinical chemistry. 1984;30:627–630. [PubMed] [Google Scholar]
- 61.Ohshima K, Inoue K, Fujiwara A, Hatakeyama K, Kanto K, Watanabe Y, Muramatsu K, Fukuda Y, Ogura S, Yamaguchi K, Mochizuki T. PLoS One. 2010;5:e13247. doi: 10.1371/journal.pone.0013247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Collino F, Deregibus MC, Bruno S, Sterpone L, Aghemo G, Viltono L, Tetta C, Camussi G. PLoS One. 2010;5:e11803. doi: 10.1371/journal.pone.0011803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Pegtel DM, Cosmopoulos K, Thorley-Lawson DA, van Eijndhoven MA, Hopmans ES, Lindenberg JL, de Gruijl TD, Wurdinger T, Middeldorp JM. Proc Natl Acad Sci U S A. 2010;107:6328–6333. doi: 10.1073/pnas.0914843107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Valadi H, Ekstrom K, Bossios A, Sjostrand M, Lee JJ, Lotvall JO. Nat Cell Biol. 2007;9:654–659. doi: 10.1038/ncb1596. [DOI] [PubMed] [Google Scholar]
- 65.Skog J, Wurdinger T, van Rijn S, Meijer DH, Gainche L, Curry WT, Jr, Carter BS, Krichevsky AM, Breakefield XO. Nat Cell Biol. 2008;10:1470–1476. doi: 10.1038/ncb1800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Tsui NB, Ng EK, Lo YM. Clin Chem. 2002;48:1647–1653. [PubMed] [Google Scholar]
- 67.Bustin SA, Nolan T. J Biomol Tech. 2004;15:155–166. [PMC free article] [PubMed] [Google Scholar]
- 68.Moltzahn F, Olshen AB, Baehner L, Peek A, Fong L, Stoppler H, Simko J, Hilton JF, Carroll P, Blelloch R. Cancer Res. 2011;71:550–560. doi: 10.1158/0008-5472.CAN-10-1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Nolan T, Hands RE, Bustin SA. Nat Protoc. 2006;1:1559–1582. doi: 10.1038/nprot.2006.236. [DOI] [PubMed] [Google Scholar]
- 70.Andreasen D, Fog JU, Biggs W, Salomon J, Dahslveen IK, Baker A, Mouritzen P. Methods. 2010;50:S6–S9. doi: 10.1016/j.ymeth.2010.01.006. [DOI] [PubMed] [Google Scholar]
- 71.Tzimagiorgis G, Michailidou EZ, Kritis A, Markopoulos AK, Kouidou S. Cancer Epidemiol. 2011 doi: 10.1016/j.canep.2011.02.016. [DOI] [PubMed] [Google Scholar]
- 72.Yao B, Li J, Huang H, Sun C, Wang Z, Fan Y, Chang Q, Li S, Xi J. RNA. 2009;15:1787–1794. doi: 10.1261/rna.1555209. [DOI] [PMC free article] [PubMed] [Google Scholar]