Abstract
We investigate the diffusion and the drift motion of λ DNA molecules near solid-state nanopores prior to their translocation though the nanopores using fluorescence microscopy. The radial dependence of the electric field near a nanopore generated by an applied voltage in ionic solution can be estimated quantitatively in 3D by analyzing the motion of negatively charged DNA molecules. We find that the electric field is approximately spherically symmetric around the nanopore under the conditions investigated. In addition, DNA clogging at the nanopore was directly observed. Surprisingly, the probability of the clogging event increases with increasing external bias voltage. We also find that DNA molecules clogging the nanopore reduce the electric field amplitude at the nanopore membrane surface. To better understand these experimental results, analytical method with Ohm’s law and computer simulation with Poisson and Nernst-Planck (PNP) equations are used to calculate the electric field near the nanopore. These results are of great interest in both experimental and theoretical considerations of the motion of DNA molecules near voltage-biased nanopores. These findings will also contribute to the development of solid-state nanopore based DNA sensing devices.
Keywords: single-molecule, sensing, nanopore, DNA
Nanometer scale pores have emerged as a promising single molecule detection tool since their invention.1 A nanopore offers a highly confined space that allows the analysis of single DNA molecules. Because of the promise of developing single molecule DNA sequencer for next generation high speed genome analysis, interest continues to increase in nanopore related research fields. As a result, excellent review articles describing the current progress and challenges towards the single DNA sequencing technique have recently been published.2–3
By applying an external voltage across a nanopore fabricated in a silicon nitride membrane submerged in ionic solution, negatively charged DNA molecules can electrophoretically thread through the nanopore. The ionic current decreases when an individual DNA molecule goes into the nanopore because the presence of the DNA molecule physically blocks a certain portion of the ionic current flow in the pore. The magnitude and duration of the current decrease depends on the molecule's radius and length.4–7 Various fabrication methods have been introduced to make nanometer scale pores and these pores have all shown similar results regarding DNA translocation.8–15
Recently, solid-state nanopore experiments have focused on improving its accuracy by measuring the same DNA molecule many times16 or on slowing down DNA translocation speed by additional techniques using nano-analytical instruments (for example, AFM,17 optical tweezers18 or magnetic tweezers19) to manipulate an individual DNA molecule. On the other hand, theoretical investigations have focused on elucidating the entire mechanism of DNA translocation processes with ion transport including the motion of DNA molecules before translocation near the nanopore mouth. Theoretical models of evaluating the DNA capture rate into a nanopore have been compared to the experimentally measured capture rate. 20–23 To provide further experimental information to theoreticians who study DNA’s electrophoretic motion before threading of DNA molecules through the nanopore, visualization the motion of DNA molecules near the nanopore is essential. Here, we present the direct observation of the motion of fluorescently dyed DNA molecules near nanopores by a fluorescence microscope.
The motions of DNA molecules are Brownian in solution far away from the nanopore that its electrophoretic motion is negligible. Upon randomly approaching the nanopore, when the electrophoretic force is significant, the DNA molecules are captured and pulled by the electric field produced by the voltage difference across the nanopore membrane. Previously, this capture process was optically imaged by Chen et al. in 2004.24 Chen et al. observed an evacuated region of DNA molecules within a few micrometers of the nanopore caused by the capture and translocation of DNA molecules. Unfortunately, because of the limitations of frame rate and spatial resolution of optical microscopy, the electric field strength inducing the electrophoretic drift motion of DNA was not quantified in their study. To extend their DNA observation approach and to estimate the field strength quantitatively for a better understanding of the mechanism of DNA translocation, we used nanopores with wider diameters near 100 nm to increase the field strength since the cross sectional area of the pore is expected to be proportional to the field strength smearing out from the nanopore mouth.25 Recently, Gershaw et al. recaptured DNA molecules right after the molecules finished their translocation by flipping the polarity of the external voltage. To evaluate the recapturing rates of the DNA molecules, they assumed the spherically symmetric electric field decreasing in strength as 1/r2, where r was the radial distance from the location of nanopore mouth, and they quantitatively estimated the strength of the field for the DNA molecules to drift back into nanopore.16 In our study, the electric field profile around a nanopore in 3D can be experimentally determined by direct observation of the motion of DNA molecules near the nanopore. We estimate the electric field profiles under various salt concentrations and bias voltages in this paper. Our results indicate that the electric fields are approximately spherically symmetric and the strength decreases as 1/r2 for the far field of r greater than 2 µm. A plot of r2E(r) vs theta graph shows an almost constant also support this conclusion (Supporting Information).
In addition, we will discuss the issue of DNA clogging of nanopores by taking advantage of our direct observation methods. Discrete reductions in current followed by a reduced DNA capture rate have been observed in DNA translocation experiments and are often end the nanopore experiment after several hours. One plausible explanation is that at least one DNA molecule is sticking to the nanopore wall and preventing another DNA molecule from translocating smoothly.4, 26 We have carefully measured the chance of this DNA clogging event under various external bias voltages and have investigated the influence of the clogged DNA molecules at nanopore on the motions of the other DNA molecules in ionic solutions. Surprisingly, higher bias voltages increase the chance of the DNA clogging. Additionally, the clogged DNA molecules significantly reduce the strength of the electric field near the nanopore, therefore reducing the DNA capture rate.
RESULTS AND DISCUSSION
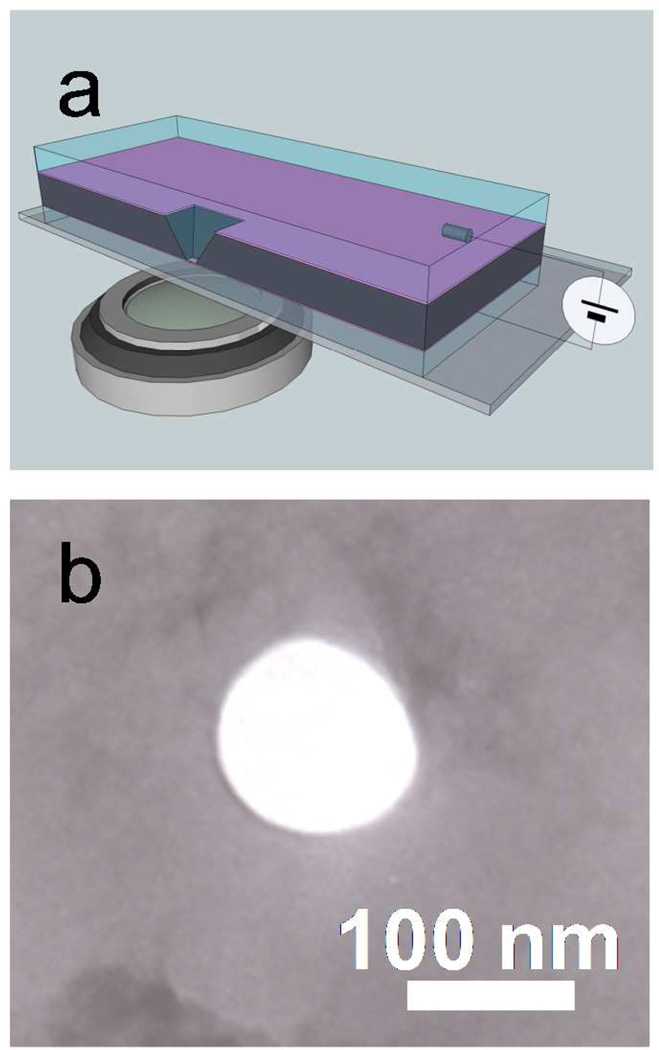
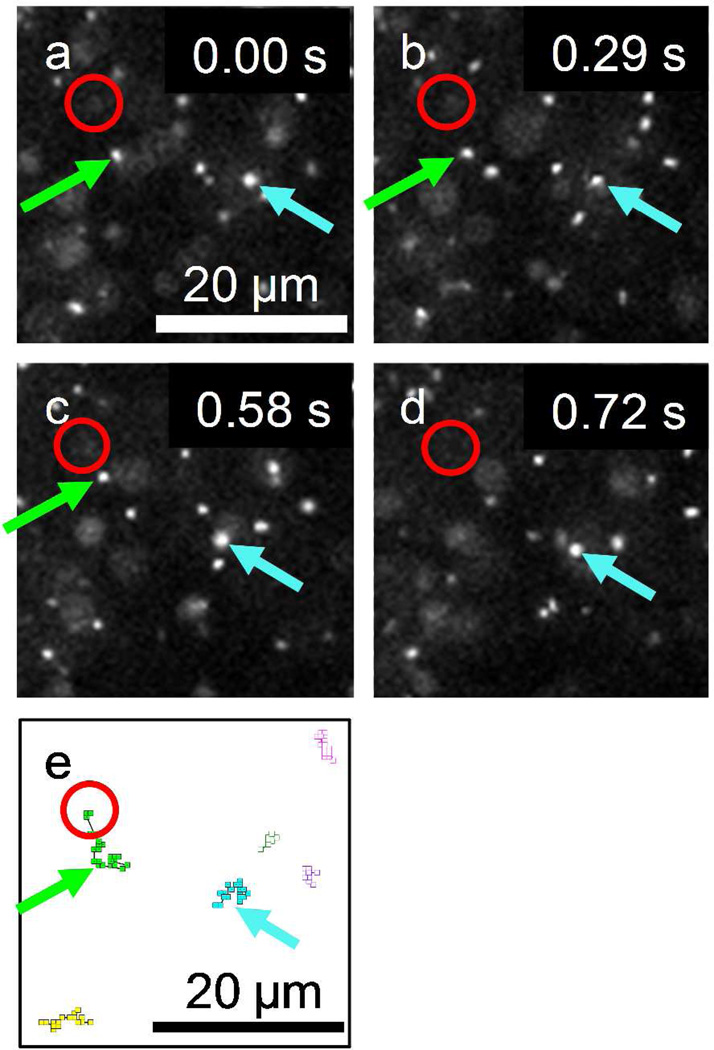
Figure 1 shows the solid-state nanopore setup and a TEM image of a typical 100 nm diameter pore. Examples of fluorescence microscope images of DNA molecules on membrane surface (height, z = 0) observed from this setup are shown in Fig. 2(a)–(d). These excerpts from successive images taken at a 14 Hz frame rate show the motion of DNA molecules near a 300 mV biased nanopore, marked by a red circle, in 0.02M KCl solution. A green arrow in each image depicts a fluorescently labeled DNA molecule moving toward the pore while other DNA molecules more than 10 µm away from the nanopore (i.e., the blue arrowed DNA molecule) are randomly diffusing. For example, to illustrate how the DNA trajectory data were taken, six DNA molecules are selected and their trajectories are shown in Fig. 2(e). Except for the green arrowed DNA molecule, the anticipated Brownian motion of λ DNA molecules in free solution were confirmed by observing that their mean squared displacements were proportional to time and the proportional coefficients were equal to the diffusion coefficient of λ DNA in free solution reported previously.27 For the green arrowed one, the drifting motion toward the nanopore is likely caused by the electric field produced by the applied voltage which exerts a force on the negatively charged DNA molecules.16, 27–28,29
Figure 1.
(a) Solid state nanopore setup. Schematic illustration shows Si chip containing a freestanding 200 nm SiN membrane in which a 100 nm pore was milled by a FIB. This membrane is immersed in aqueous solution on top of an optical microscope for the direct observation of DNA translocation. A home-build PDMS cell (not shown in this schematic) is used to seal between the silicon chip and the glass coverslip. Ionic current through the nanopore is measured using two Ag/AgCl electrodes immersed in the cis and trans chambers in the cell. (b) A TEM image of a 100 nm diameter nanopore.
Figure 2.
Example of time resolved fluorescence images focused on membrane surface (z = 0) showing the motion of fluorescently labeled DNA molecules in 0.02M KCl at 0.3V bias voltage. Images, (a) – (d) are extracted at t = 0.00, 0.29, 0.58 and 0.72 sec from a sequence of 600 frames recorded at 14 frames per second. One DNA molecule (blue arrow) shows a typical Brownian motion while another DNA molecule (green arrow) shows a drifting motion toward the nanopore located at the center of a drawn red circle in the images. From (c) to (d), the green arrowed DNA molecule disappears as this molecule translocates through the nanopore. (e) DNA molecule trajectories in the sequential frames. The trajectory of the molecule near the nanopore (green arrow) indicates the drifting motion.
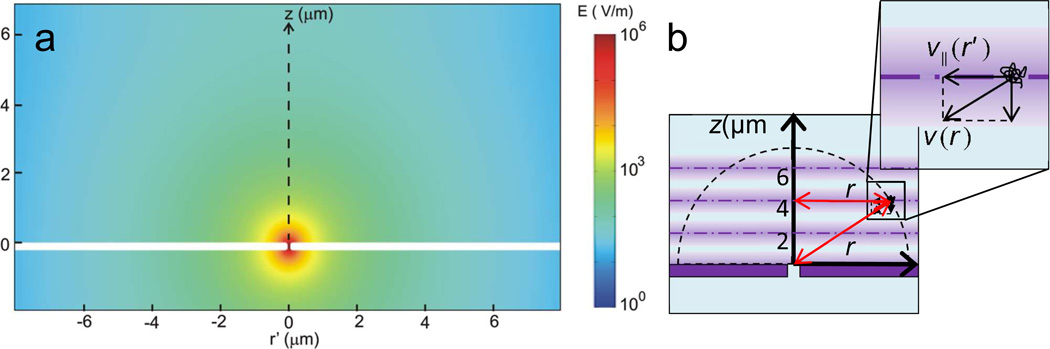
The velocity of individual DNA molecules are estimated by the following procedure: 1. Find the displacement and v‖ (velocity component parallel to the membrane surface) of DNA between consecutive frames at heights, z = 0, 2, 4 and 6 µm from the nanopore membrane surface. 2. Confirm that v‖ parallel to the membrane surface is a function of r’ only. 3. Take <v‖(r’)> as a component of v(r) by assuming a spherically symmetric electrophoretic motion toward nanopore. To better understand the experimental results in Figure 2, the electric field magnitude produced by the voltage across a nanopore membrane was simulated in 3D using a finite element analysis software (COMSOL, Multiphysics). A 2D cross-section along the nanopore is plotted in Fig. 3(a). The figure shows the electric field strength produced by a 100 nm size pore in a 200 nm thick membrane, biased by a voltage of 0.3 V in 0.02 M KCl solution as used by Figure 2. This simulation shows the electric field strength is about 10V/m or 10−5V/µm at 10Mm away from the pore, indeed very weak. The details of the simulation are explained in the supporting information.
Figure 3.
(a) A simulation of electric field magnitude around a nanopore. A 100 nm size pore in a 200 nm thick membrane is biased by a 0.3 V voltage in 0.02 M KCl solution. The white region corresponds to the membrane. (b) A schematic of a possible electrophoretic DNA motion caused by the electric field near a nanopore.
To analyze the DNA electrophoretic motion caused by the electric field near a nanopore in 3D, the trajectories of the DNA molecules in 2D were plotted at different heights, z = 0, 2, 4 and 6 µm from the nanopore membrane surface as depicted in Fig. 3(b). The locations of the DNA molecules as r’ from the z axis in cylindrical coordinates, are plotted while DNA molecules diffuse. Although the field depth of an optical microscopy at high magnifications was as thin as 1 µm in z direction, the average number of frames for one sequential trajectory of single DNA molecule is approximately 7 frames where the drift motion by electrophoretic force is not dominant, estimated to be about <z2> = 2Dt ~ 0.5 µm during the time of taking 7 frames.30 This frame number became low as DNA molecules were close to a nanopore where the electrophoretic motion became dominant, for example, the number of frames to observe one sequential trajectory of the DNA became approximately 3 at (r, z) = (6 µm, 6 µm), 45° of slant angle from surface with 0.7 V. This limited the number of locations where DNA can be tracked. However, below 0.7 V or below 45° the number of frames for sequential trajectory increased and on surface (z = 0, angle 0°) this number was near 7 frames since the direction of the electrophoretic force was parallel to the surface. As we discuss later in detail, a DNA molecule occasionally stacks and clogs around a nanopore during the above observations and the clogged DNA molecules can influence on the motion of the other DNA molecules near the nanopore. To exclude this effect, we did not use the data of the DNA trajectories after DNA was found to be clogging at a nanopore.
To estimate the electric driving force on DNA molecules toward a nanopore mouth, the stochastic Langevin equation was applied. We assume the charge distribution of a DNA molecule is symmetrical around its center of mass and can be treated as a point particle. Therefore, the Langevin equation can be expressed as
| (1) |
where f(t) is the Langevin Gaussian noise term replicating the effect of thermal fluctuations. ζ is the friction constant of a λ DNA molecule in the ionic solution. v and q are the velocity and the effective charge of a λ DNA molecule. Inertia effects are neglected because of the highly damped DNA motion in aqueous solution. The Gaussian noise term f(t) is presumably 0 by taking a time average, < f(t)> of more than 300 of the time sequential DNA’s motions, because of its random thermal fluctuation nature. Therefore, the electric field, E, near a pore can be quantitatively estimated if the velocities, v of each DNA molecule at various locations around the pore are measured. If a spherically symmetric field is assumed, and the equation can be expressed in spherical polar coordinates and the v and the E are functions of only r from the center of the nanopore mouth as the origin, r = 0.16 Further assuming on average that the electric driving force is balanced with the drag force thus F=0 in equation (1), one can write.
| (2) |
where μ is the electrophoretic mobility of the λ DNA molecule.27–28
In order to estimate E(r) from v(r), v(r) in a polar coordinate must be found by experimentally measured v‖ in a cylindrical coordinate as schematically depicted in Fig. 3(b). We consider that the velocity v‖ is a function of r’ only by assuming an azimuthal symmetric field around nanopore and tentatively v‖(r’) is a component of v(r) parallel to the nanopore membrane surface, and then E(r) can be calculated from the equation.
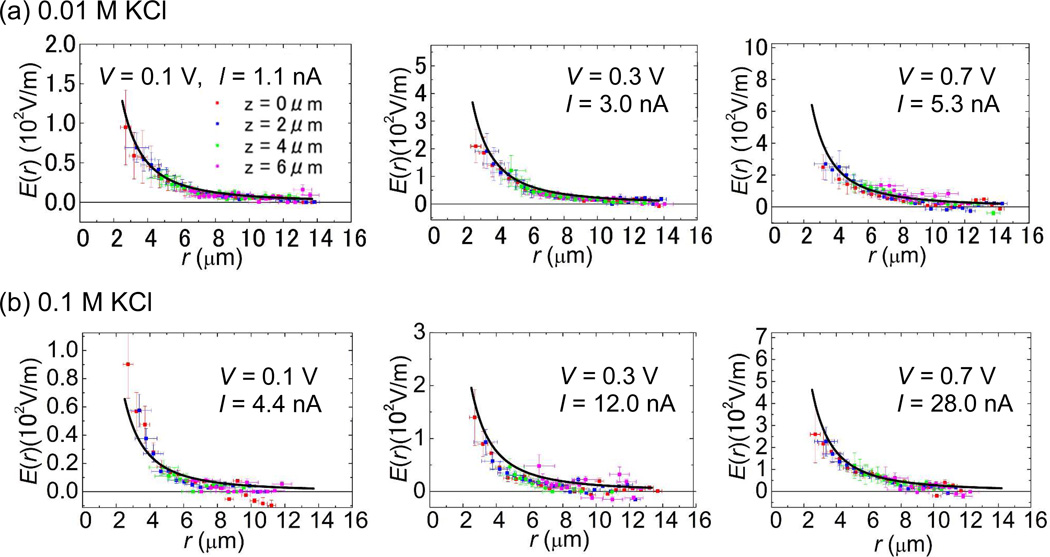
The magnitude of the electric field, E(r) = | E(r) | under various applied voltages is plotted in Fig. 4a and 4b for 0.01M and 0.1 M KCl respectively. E(r) at various heights, 0, 2, 4 and 6 µm from the membrane surface was calculated as a function of r, where r is the polar coordinate. We avoid plotting E(r) for r < 2 µm because the average displacements of DNA toward nanopore mouth exceeds 1 µm per frame there and this would underestimate v(r) since significant numbers of DNA molecules disappearing at the following frame indicates that some DNA molecules have already translocated through the nanopore.
Figure 4.
The strength of the electric field E(r) at bias voltages, 0.1, 0.3 and 0.7 V as a function of r at various heights, 0, 2, 4 and 6 µm from the nanopore membrane surface. The salt concentrations are 0.01M for (a) and 0.1 M KCl for (b). Each solid curve in each graph is plotted based on the simple theoretical model by Ohm’s law in equation (3), E(r) = |J(r)|/σ with the experimentally measured ionic currents through nanopore at given bias voltages. The y axis scales in each Fig. 4 plots are normalized by the proportionality constants, I/2πσ of 1/r2. The same figure was also plotted in the same scale for better comparisons in Figure S2 of the Supporting Information.
Next, we compare our experimental estimation of the electric fields near the nanopores with predictions by theoretical models. Using the relation between an ionic current density J and an electric field E given by Ohm’s law, one can write
| (3) |
where |r| = r, σ is the electrical conductivity of the ionic solutions and I is the ionic current measured through the nanopore under an applied voltage.16 The electrical conductivities for both 0.01M and 0.1 M KCl salt concentrations can be estimated by measured linear I–V characteristics of the nanopore with its pore diameter estimated by TEM observation. These values of the conductivities are close to those found previously.31-30 By using the experimental value of I at given bias voltages, E(r) = |J(r)|/σ is plotted (solid curves) into each graph of Fig. 4a and 4b. These plots show that the experimental data fit well with the solid curves that were plotted based on the Ohm’s law in equation (3). This result confirms that the electric field around the location of a nanopore mouth is approximately spherically symmetric as its center at z = 0 µm, or E(r) ~ r−2. This result indicates that the theoretical model16 is also valid on relatively larger electric fields at higher voltages through 100 nm diameter nanopore used in our experiment. However, our measured E(r) data in Fig.4 do show small divagations from Ohm’s law.
To better understand the electric behavior of the solid-state nanopores at approximately the experimental conditions, including the nanopore geometry, salt concentration, and surface charge, which simple Ohm’s law is incapable of, we also simulated electric field strength at various heights of 0, 2, 4 and 6 µm from membrane surface with Poisson-Nernst-Planck (PNP) equations (using Multiphysics from COMSOL). The simulation results are comparable to the experimental results and the Ohm’s law prediction of equation (3) within experimental errors (see supporting information Figure S2). The simulation results predict that the magnitude of electric field at higher z height is a little greater than at lower height when the polar coordinate length r is the same and this prediction matches well with the experimental results. The simulated 2D electric field strength at each condition of Fig.4 is shown in Fig. S3 (supporting information).
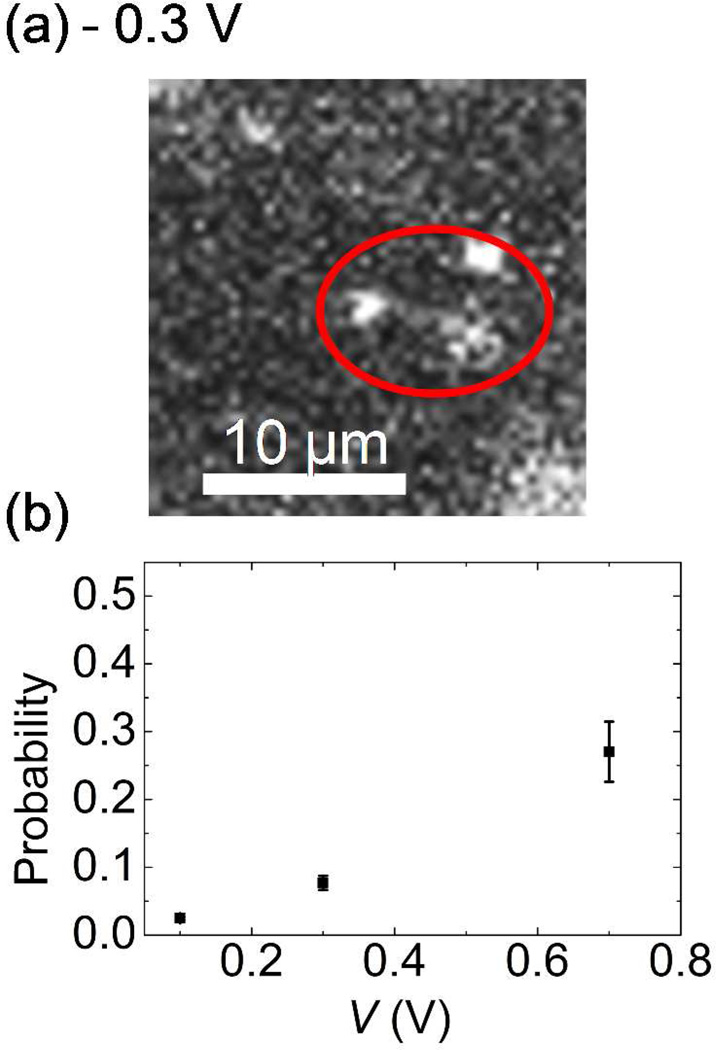
In addition, one interesting phenomenon was observed directly from our fluorescent images of DNA translocation. Single DNA molecules occasionally sticks and then clog nanopores. Either increasing or reversing the polarity of the external bias voltages did not remove the DNA sticking to a nanopore. Occasionally, DNA molecules stretching from the pore mouth were observed when the magnitude of the reversed bias voltages was increased above 0.3 V. A typical example is shown in Fig. 5. This indicates the clogging DNA cannot be removed only by the electrophoretic force. To investigate the origin of the DNA clogging, we plot the probability of this DNA clogging for each DNA capture at clean pores under various external bias voltages for DNA translocation in Fig. 5. Interestingly, the clogging probability rises from 0.024 to 0.26 as the external voltage increases from 0.1 V to 0.7 V. One of the possible explanations is that the probability of this DNA clogging is associated with the initial configurations such as the hairpin configuration of a single DNA molecule entering the nanopore mouth. Theoretically, a single λ DNA molecule forms a ~1 µm diameter sphere in a free solution that has to unwind itself before entering a small pore meaning it has to overcome a free energy barrier for the conformational change due to the confinement inside the nanopore.22 A higher bias voltage could allow a DNA molecule with more complicated configurations such as multiple folded DNA to enter a nanopore mouth then to stick there before its conformation changes.24 Since our nanopore diameter is larger than the nanopores used in the most of the DNA translocation experiments by other groups,4–5, 15 the chance of these multiple fold DNA molecules to occur and to be stuck may increase. Furthermore, the multiple fold DNA tends to stick to the channel wall of the pore better because the area for the interaction between a DNA molecule and the channel wall increases.29 This would increase the DNA clogging rate as the external voltage increases. To confirm this argument quantitatively, numerical simulation will be necessary. However, we believe this higher probability of the DNA clogging at higher bias voltage is general for DNA translocation experiments using solid state nanopores for any diameter of nanopore wide enough for the translocation of folded DNA molecules.
Figure 5.
(a) A DNA molecule stretching from the nanopore mouth by switching the polarity of bias voltage. The clogged DNA at pore is rarely removed only by electrophoretic force. (b) Probability of the DNA clogging at nanopore. The probability increases with increasing the external bias voltage.
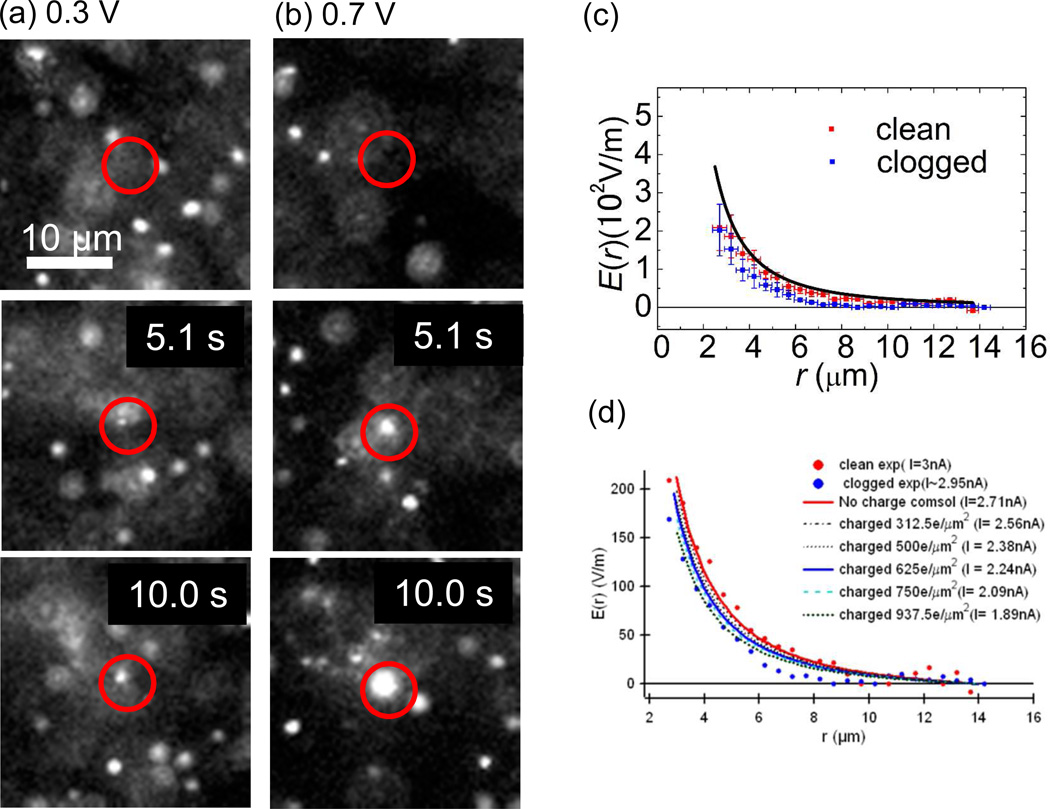
We have also investigated the DNA clogging rate after DNA clogging event had occurred at a nanopore. The probability of the DNA clogging on the pre-DNA-clogged pore increases as the number of clogging DNA molecules increases. Examples of these multiple clogging events are depicted in the images of Fig. 6 (a) and (b). The top images are a frame before the first DNA clogs at the pores and the middle and the bottom images are 5.1 sec and 10.0 sec after the top images with the bias voltages at 0.3 V (a) and 0.7 V (b). It is difficult to estimate the precise quantitative values of the DNA clogging probability for the pre-DNA-clogged pore since the presence of multiple clogging DNA molecules seen as a white spot at the location of the nanopore prevents from counting exact rate as the DNA molecules arriving from the top and the clogged ones is indistinguishable as the images in Fig. 5 indicate. However, the probability is appeared to be close to 1 because the apparent size of the white spot at the nanopore increases as single DNA molecule enters the white spot.
Figure 6.
Fluorescence images showing the DNA clogging into a nanopore with bias voltage 0.3 V for (a) and 0.7 V for (b). Images are extracted at 5.1 sec and 10.0 sec after the first DNA molecule is clogged. A nanopore is located at the center of a drawn red circle in the images. One and three DNA molecules were clogging while two and five molecules entered nanopore at 5.1 sec and 10.0 sec for (a). Near 10 and 20 DNA molecules are clogged by 5.1 sec and 10.0 sec for (b). (c) The magnitude of the electric fields near the clean nanopore and DNA clogged nanopore on the nanopore membrane surface. The magnitudes of the fields are reduced for the DNA clogged pore. (d) Computer simulation shows that a charged nanopore surface can reduce the electric field near a nanopore.
It is well-known that YOYO-1 dye molecules for our observation could increase the contour length of a DNA molecule up to about 35 % at a saturating dye concentration, 4:1 for the number of base pair to YOYO-dye molecule ratio.32,33,34Although a low dye content has been chosen to minimize such effects, this dye intercalating DNA molecules may have increased the clogging probability well.
Finally, we estimate the electric field at 0.3 V bias near the DNA clogged nanopore and compare the magnitudes with the field near a clean nanopore at z = 0 in 0.01M KCl solutions as shown in Fig. 6(c). The magnitudes are reduced and almost zero at more than 6 µm away from the nanopore. Although these values must depend on the number of DNA molecules clogging the nanopore, this reduction of the field is noteworthy and most likely decreases the capture rate of DNA translocation.
By simulating the effect of surface charge added by DNA coating the nanopore using COMSOL, we have endeavored to explain the reduction of the electric field after the nanopore is clogged with DNA in Fig. 6(d). This simulation was based on two possible nanopore property changes after negatively charged large λ DNA molecules, ~1um in 3D, physically stick to the nearby surface of a nanopore: 1) It would decrease the pore volume both inside and nearby outside. Which would increase both the pore and access resistance, causing a decrease in the electric field strength E(r) as observed. 2) It would increase the pore surface charge. This would result in: A) a repulsion of the incoming negatively charged DNA molecules or a smaller E(r) observed, and B) an increase in local ion concentration or pore current which could be canceled out by the decrease in pore current due to the increase in pore resistance in part 1).
The dashed and solid curves in Fig. 6(d) are the electric field magnitude after charge is added to the surface ~3.8 µm2, an area of radius 1.1 µm, as well as a reduction of pore current due to an increase in pore resistance. The charged surface region is shown in Fig. S1 in the Supporting information. This simulation implies the possibility of a DNA-clogged-pore as a nanopore with some negative surface charge would reduce the electric field strength near a nanopore. Alternatively, a DNA-clogged-pore can be considered as a smaller diameter pore or larger resistance due to some volume of the pore being occupied by the clogging DNA molecules, a smaller pore would also have reduced electric field strength. Further experimental studies of this phenomenon are necessary to compare with the result of numerical simulations.
CONCLUSIONS
Our direct observation of DNA molecules near ~100 nm diameter pores by fluorescent microscopy has revealed the motion of DNA before its translocation through the nanopores. We have determined the drifting motion of DNA where the electrophoretic driving force on DNA molecules toward nanopore exceeds the thermal fluctuation force. Careful analysis of the DNA motion near the nanopores at various heights from the nanopore membrane surface suggests the electric field around the nanopore mouth is spherically symmetric. In addition, the strength of the field E decreases as 1/r2 and quantitatively fits with a theoretical model based on simple Ohm’s law. With our direct observation method, DNA clogging into nanopore is observed first time and surprisingly the chance for DNA clogging during a translocation experiment increases with increasing external bias voltage. We have also demonstrated that reversing the polarity of the bias voltage does not remove DNA molecules clogged into the nanopore. The probability of DNA clogging increases as the number of molecules clogged into the pore increases. Finally, the reduction of the magnitudes of the electric field near a nanopore is revealed for the DNA clogged nanopore. These findings by our direct observation technique will be of great interest in both experimental and theoretical considerations regarding the motion of DNA molecules as they approach the nanopore.
METHODS
Fabrication of nanopore
A 100-nm-diameter nanopore was fabricated in a 40 µm × 40 µm, 200 nm thick freestanding silicon nitride membrane using conventional photolithography. The membranes were supported on 500 µm thick silicon (100) substrate. A focused ion beam (FIB) was used to mill pores on the nanometer scale. To determine the diameter and the shape of individual nanopores, transmission electron microscopy (TEM) was used. The measurement of ionic current though individual nanopores in ionic solutions confirmed the depth of the membrane and the diameter of the nanopores.4, 15
DNA observation
YOYO-1 dye (Molecular Probes) was used to stain the DNA with a dye to base pair ratio near 1:10. The final DNA concentration was 1 ng/mL in 0.1 M and 0.01M KCl solution containing 10 mM Tris-HCl (pH 8.0) and 1 mM EDTA.
A home-build nanopore translocation setup is placed on the stage of fluorescence microscope. A schematic drawing in Fig. 1 shows our measurement setup. The silicon chip containing a nanopore was placed on a coverslip on the upright microscope (TE2000 Nikon) where the solutions containing fluorescently tagged DNA molecules were injected between the coverslip and the silicon chip (cis side). A voltage difference between the cis and trans chambers was applied via Ag /AgCl electrodes. The stained DNA molecules were illuminated by a 100 W mercury arc lamp and observed with using the microscope. Sequential images were acquired using an intensified charge coupled device camera (ORCA-ER Hamamatsu Photonics) down to 36 ms time intervals. Since the field depth of the optical microscopy at high magnification (i.e. 100k× oil immersion objective) was as thin as 1 µm, the motions of the dyed DNA molecules at specific heights from the membrane surface were plotted by varying the z focal depths. This measurements at the different z focal depths were made sequentially (i.e. the objective was fixed at z = 2 µm, measurements were made for a long period of time, then the objective was moved to different height). The heights at z = 0, 2, 4 and 6 µm were chosen. To trace the DNA motion, the conformation of the DNA molecule was ignored. More than 300 of DNA molecules were randomly chosen to trace their trajectories. Because the effect of DNA clogging on the motion of other DNA molecules, we have repeatedly performed nanopore cleaning to remove the clogging DNA by rinsing Si chip with a dilute bleach solution. Multiple nanopores were used especially for the 0.7 V bias observations since the cleaning processes describe above occasionally degrade nanopores for example, cracking nanopore membrane. Since our finding of the magnitude of electric fields is expected proportional to the value of ionic current though nanopore, the current value were carefully adjusted to be the same for the measurements for the same measurements. In order to do this, the bias voltage was actually deviated for each pore although the pores selected for the deviations to be fewer than 5%.
Supplementary Material
Acknowledgements
T.M. acknowledges support from Grant-in-Aid for Young Scientists (B) of Ministry of Education, Culture, Sports, Science and Technology (MEXT). J.L. acknowledges support from NHGRI R21HG004776. The Authors thank Ryan Rollings for making comments and suggestions to improve the manuscript.
Footnotes
Supporting Information Available: Computational simulation near the region of solid-state nanopores is available free of charge via the Internet at http://pubs.acs.org.
REFERENCES AND NOTES
- 1.Li J, Stein D, McMullan C, Branton D, Aziz MJ, Golovchenko JA. Ion-Beam Sculpting at Nanometre Length Scales. Nature. 2001;412:166–169. doi: 10.1038/35084037. [DOI] [PubMed] [Google Scholar]
- 2.Branton D, Deamer DW, Marziali A, Bayley H, Benner SA, Butler T, Di Ventra M, Garaj S, Hibbs A, Huang X, et al. The Potential and Challenges of Nanopore Sequencing. Nat Biotechnol. 2008;26:1146–1153. doi: 10.1038/nbt.1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dekker C. Solid-State Nanopores. Nat Nanotechnol. 2007;2:209–215. doi: 10.1038/nnano.2007.27. [DOI] [PubMed] [Google Scholar]
- 4.Li J, Gershow M, Stein D, Brandin E, Golovchenko JA. DNA Molecules and Configurations in a Solid-State Nanopore Microscope. Nat. Mater. 2003;2:611–615. doi: 10.1038/nmat965. [DOI] [PubMed] [Google Scholar]
- 5.Storm AJ, Storm C, Chen J, Zandbergen H, Joanny JF, Dekker C. Fast DNA Translocation through a Solid-State Nanopore. Nano Lett. 2005;5:1193–1197. doi: 10.1021/nl048030d. [DOI] [PubMed] [Google Scholar]
- 6.Aksimentiev A, Schulten K. Imaging α-Hemolysin with Molecular Dynamics: Ionic Conductance, Osmotic Permeability, and the Electrostatic Potential Map. Biophys. J. 2005;88:3745–3761. doi: 10.1529/biophysj.104.058727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fologea D, Gershow M, Ledden B, McNabb DS, Golovchenko JA, Li J. Detecting Single Stranded DNA with a Solid State Nanopore. Nano Lett. 2005;5:1905–1909. doi: 10.1021/nl051199m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mitsui T, Stein D, Kim YR, Hoogerheide D, Golovchenko JA. Nanoscale Volcanoes: Accretion of Matter at Ion-Sculpted Nanopores. Phys. Rev. Lett. 2006;96 doi: 10.1103/PhysRevLett.96.036102. 036102. [DOI] [PubMed] [Google Scholar]
- 9.Ashkenasy N, Sanchez-Quesada J, Bayley H, Ghadiri MR. Recognizing a Single Base in an Individual DNA Strand: A Step toward DNA Sequencing in Nanopores. Angew. Chem. Int. Ed. 2005;44:1401–1404. doi: 10.1002/anie.200462114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tobkes N, Wallace BA, Bayley H. Secondary Structure and Assembly Mechanism of an Oligomeric Channel Protein. Biochemistry. 1985;24:1915–1920. doi: 10.1021/bi00329a017. [DOI] [PubMed] [Google Scholar]
- 11.Yemini M, Hadad B, Liebes Y, Goldner A, Ashkenasy N. The Controlled Fabrication of Nanopores by Focused Electron-Beam-Induced Etching. Nanotechnology. 2009;20 doi: 10.1088/0957-4484/20/24/245302. 245302. [DOI] [PubMed] [Google Scholar]
- 12.Garaj S, Hubbard W, Reina A, Kong J, Branton D, Golovchenko JA. Graphene as a Subnanometre Trans-Electrode Membrane. Nature. 2010;467:190–193. doi: 10.1038/nature09379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stein D, Li J, Golovchenko JA. Ion-Beam Sculpting Time Scales. Phys. Rev. Lett. 2002;89 doi: 10.1103/PhysRevLett.89.276106. 276106. [DOI] [PubMed] [Google Scholar]
- 14.Storm AJ, Chen JH, Zandbergen HW, Dekker C. Translocation of Double-Strand DNA through a Silicon Oxide Nanopore. Phys. Rev. E. 2005;71 doi: 10.1103/PhysRevE.71.051903. 051903. [DOI] [PubMed] [Google Scholar]
- 15.Chen P, Mitsui T, Farmer DB, Golovchenko J, Gordon RG, Branton D. Atomic Layer Deposition to Fine-Tune the Surface Properties and Diameters of Fabricated Nanopores. Nano Lett. 2004;4:1333–1337. doi: 10.1021/nl0494001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gershow M, Golovchenko JA. Recapturing and Trapping Single Molecules with a Solid-State Nanopore. Nat. Nanotechnol. 2007;2:775–779. doi: 10.1038/nnano.2007.381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.King GM, Golovchenko JA. Probing Nanotube-Nanopore Interactions. Phys. Rev. Lett. 2005;95 doi: 10.1103/PhysRevLett.95.216103. 216103. [DOI] [PubMed] [Google Scholar]
- 18.Hall AR, van Dorp S, Lemay SG, Dekker C. Electrophoretic Force on a Protein-Coated DNA Molecule in a Solid-State Nanopore. Nano Lett. 2009;9:4441–4445. doi: 10.1021/nl9027318. [DOI] [PubMed] [Google Scholar]
- 19.Peng H, Ling XS. Reverse DNA Translocation through a Solid-State Nanopore by Magnetic Tweezers. Nanotechnology. 2009;20 doi: 10.1088/0957-4484/20/18/185101. 185101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Muthukumar M, Kong CY. Simulation of Polymer Translocation through Protein Channels. Proc. Natl. Acad. Sci. USA. 2006;103:5273–5278. doi: 10.1073/pnas.0510725103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Muthukumar M, Wong CTA. Polymer Capture by Electro-Osmotic Flow of Oppositely Charged Nanopores. J. Chem. Phys. 2007;126 doi: 10.1063/1.2723088. 164903. [DOI] [PubMed] [Google Scholar]
- 22.Muthukumar M. Theory of Capture Rate in Polymer Translocation. J. Chem. Phys. 2010;132 doi: 10.1063/1.3429882. 195101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hatlo MM, Panja D, van Roij R. Translocation of DNA Molecules through Nanopores with Salt Gradients: The Role of Osmotic Flow. Phys. Rev. Lett. 2011;107 doi: 10.1103/PhysRevLett.107.068101. 068101. [DOI] [PubMed] [Google Scholar]
- 24.Chen P, Gu J, Brandin E, Kim Y-R, Wang Q, Branton D. Probing Single DNA Molecule Transport Using Fabricated Nanopores. Nano Lett. 2004;4:2293–2298. doi: 10.1021/nl048654j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hyun C, Rollings R, Li J. Probing Access Resistance of Solid-State Nanopores with a Scanning Probe Microscope Tip. Small. 2012;8:385–392. doi: 10.1002/smll.201101337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lu B, Albertorio F, Hoogerheide DP, Golovchenko JA. Origins and Consequences of Velocity Fluctuations During DNA Passage through a Nanopore. Biophys J. 2011;101:70–79. doi: 10.1016/j.bpj.2011.05.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nkodo AE, Garnier JM, Tinland B, Ren H, Desruisseaux C, McCormick LC, Drouin G, Slater GW. Diffusion Coefficient of DNA Molecules During Free Solution Electrophoresis. Electrophoresis. 2001;22:2424–2432. doi: 10.1002/1522-2683(200107)22:12<2424::AID-ELPS2424>3.0.CO;2-1. [DOI] [PubMed] [Google Scholar]
- 28.Stellwagen NC, Gelfi C, Righetti PG. The Free Solution Mobility of DNA. Biopolymers. 1997;42:687–703. doi: 10.1002/(SICI)1097-0282(199711)42:6<687::AID-BIP7>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- 29.Bonthuis DJ, Meyer C, Stein D, Dekker C. Conformation and Dynamics of DNA Confined in Slitlike Nanofluidic Channels. Phys. Rev. Lett. 2008;101 doi: 10.1103/PhysRevLett.101.108303. 108303. [DOI] [PubMed] [Google Scholar]
- 30.Tang J, Levy SL, Trahan DW, Jones JJ, Craighead HG, Doyle PS. Revisiting the Conformation and Dynamics of DNA in Slitlike Confinement. Macromolecules. 2010;43:7368–7377. [Google Scholar]
- 31.Klein SD, Bates RG. Conductance of Tris(Hydroxymethyl)-Aminomethane Hydrochloride (Tris·Hcl) in Water at 25 and 37°C. J. Solution Chem. 1980;9:289–292. [Google Scholar]
- 32.Reisner W, Morton KJ, Riehn R, Wang YM, Yu Z, Rosen M, Sturm JC, Chou SY, Frey E, Austin RH. Statics and Dynamics of Single DNA Molecules Confined in Nanochannels. Phys. Rev. Lett. 2005;94 doi: 10.1103/PhysRevLett.94.196101. 196101. [DOI] [PubMed] [Google Scholar]
- 33.Zhang C, Zhang F, van Kan JA, van der Maarel JR. Effects of Electrostatic Screening on the Conformation of Single DNA Molecules Confined in a Nanochannel. J Chem Phys. 2008;128 doi: 10.1063/1.2937441. 225109. [DOI] [PubMed] [Google Scholar]
- 34.Gunther K, Mertig M, Seidel R. Mechanical and Structural Properties of Yoyo-1 Complexed DNA. Nucleic Acids Res. 2010;38:6526–6532. doi: 10.1093/nar/gkq434. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.