Abstract
Cells of the adaptive immune response undergo dynamic epigenetic changes as they develop and respond to immune challenge. Plasticity is a necessary prerequisite for the chromosomal dynamics of lineage specification, development, and the immune effector function of the mature cell types. The alterations in DNA methylation and histone modification that characterize activation may be integral to the generation of immunologic memory, thereby providing an advantage on secondary exposure to pathogens. While the immune system benefits from the dynamic nature of the epigenome, such benefit comes at a cost – increased likelihood of disease-causing mutation.
Keywords: T cells, B cells, Adaptive Immunity, Memory, Secondary Immune Response, Epigenetics, Histone Modifications, DNA Methylation, DNMT
1. Introduction
Eukaryotic gene expression is regulated at multiple levels including the influence of local properties of chromatin on regulatory DNA. Covalent modifications to DNA and the protein constituents of the chromatin fiber constitute instructional information for the genome and are unique for each cell type. Elucidation of the mechanisms by which this biologically essential information, the epigenome, is established and maintained through development and differentiation represents a major challenge to the current generation of biologists.
1.1 The Epigenome
1.1.1 DNA Methylation
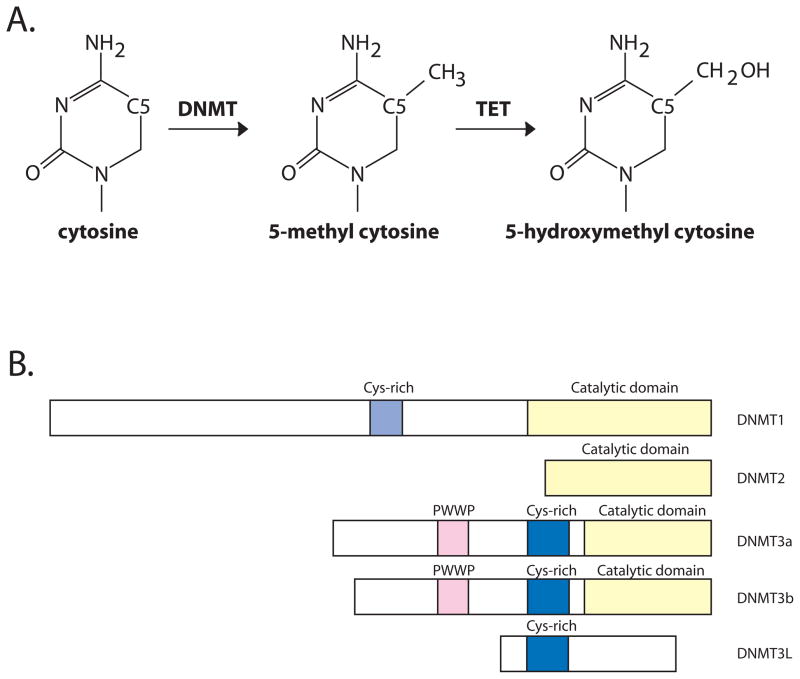
DNA in mammals is subject to covalent modification that alters the chemical information content displayed in the major groove. The most abundant DNA modification found in mammals is methylation of cytosine, first described in calf thymus DNA in the 1940’s (Hotchkiss, 1948). A methyl group is deposited at the C5 position of the pyrimidine ring (Figure 1A). In mammals, an overwhelming majority of methylated cytosine is found in the context of a simple sequence palindrome, 5′-CG-3′, where the modification is placed symmetrically on both strands of the DNA duplex.
Figure 1. DNA methylation and DNA methyltransferases.
A. The figure depicts enzymatic methylation at C5 of cytosine by DNA methyltransferases. 5-methylcytosine serves as a substrate for TET family enzymes which oxidize the methyl carbon to generate 5-hydroxymethyl cytosine.
B. The mammalian DNMT family members are depicted in cartoon fashion. Domains are indicated where relevant.
DNA methylation in mammals is classically associated with functions in genome defense and in genomic imprinting (Jurkowska et al., 2011). Mammalian genomes are packed with transposable elements (and their relics) that, in general, are densely methylated. The advent of high throughput sequencing has provided further insight into localization of methylation in mammalian genomes. On the order of 60 to 80 % of all CpG sites are highly methylated in human cells with some variation by cell type (Hodges et al., 2011; Lister et al., 2009). Surprisingly, approximately 25% of methylated cytosine in pluripotent cells is found in sequence contexts other than the CpG dinucleotide. Methylation density shows large variations throughout human chromosomes, with sub-telomeric DNA frequently showing higher methylation density than chromosome arms (Lister et al., 2009). Regions in which methylation levels at individual cytosine residues fall under 70% are also common in human chromosomes – they constitute roughly 1/3 of autosomal regions and 80% of the X chromosome in females. These regions tend to be enriched in genes with cell type specific expression patterns that are not expressed in that cell type (Lister et al., 2009) consistent with longstanding observations that DNA methylation levels within gene bodies are generally higher in expressed genes than in silent genes (Hellman and Chess, 2007).
At the level of individual genes, DNA methylation patterns differ with cell type and reflect transcriptional output. Differentially methylated regions that distinguish developmental states frequently correlate with protein binding and with histone marks characteristic of enhancers (Hodges et al., 2011; Lister et al., 2009; Meissner et al., 2008). Hypomethylated regions are characteristic of core promoters, the size of the hypomethylated region relative to transcription start correlates with expression level and varies by cell type (Hodges et al., 2011).
Methylation of DNA in mammals is catalyzed by a well characterized family of enzymes, the DNA methyltransferases (Figure 1B). These enzymes utilize an activated methyl donor, S-adenosyl methionine and an elegant reaction mechanism involving eversion of the methylated base from the double helix (Klimasauskas et al., 1994). Mammals have multiple sequence homologs of cytosine methyltransferase enzymes. Of these, four catalyze cytosine methylation reactions and one, DNMT3L, lacks the conserved residues necessary for catalysis.
DNMT1 was the first mammalian DNA methyltransferase enzyme characterized at the biochemical (Bestor and Ingram, 1983) and molecular level (Bestor et al., 1988). This enzyme displays a marked preference for substrate DNA with methylation of only one strand – hemimethylated DNA. Further, it localizes to replication forks during S phase (Jurkowska et al., 2011), making it an ideal candidate for restoration of symmetric methylation of CG dinucleotides following DNA replication. Consistent with this hypothesis, DNMT1 is essential for normal development in mice (Li et al., 1992).
The DNMT3 family of proteins consists of two catalysis proficient cytosine methyltransferases, DNMT3a and DNMT3b, as well as a non-catalytic protein, DNMT3L. The DNMT3a and DNMT3b enzymes are essential for life and are involved in the establishment of global DNA methylation patterns in early development and in germ cells (Jurkowska et al., 2011). DNMT3L is a regulatory component of a multimeric holoenzyme along with DNMT3a that functions in genomic imprinting during gametogenesis (Cheng and Blumenthal, 2008). The expression and function of these enzymes in adult somatic cells is not well understood.
DNMT2 has considerable sequence similarity to the mammalian DNA methyltransferases and adopts a similar structure (Cheng and Blumenthal, 2008). However, this enzyme does not methylate DNA - it methylates a cytosine residue in a specific transfer RNA (Goll et al., 2006). The biological outcome of this RNA modification is currently unknown.
As a covalent modification, DNA methylation is stable and mechanisms for rapid reversal of this epigenetic mark have been a topic of intense interest. That cellular mechanisms permitting rapid remodeling of DNA methylation must exist is exemplified by the dynamics of this mark during development and also during cellular reprogramming. Recently, two enzyme families, the Ten-eleven translocation (TET) proteins and AID/APOBEC deaminases, have been proposed as active DNA demethylases (Bhutani et al., 2011). The TET family enzymes catalyze α-ketoglutarate dependent oxidation of 5-methylcytosine, generating progressively oxidized forms of this base (Wu and Zhang, 2011). The recent discovery of readily detectable levels of 5-hydroxymethyl cytosine, the product of TET-dependent oxidation of 5-methyl cytosine (Figure 1A), in somatic and embryonic stem cells has led to formulation of models depicting this modification as an intermediate in the process of active demethylation (Bhutani et al., 2011; Wu and Zhang, 2011).
1.1.2 Histone modifications
A cell’s epigenome is also comprised of post-translational histone modifications that decorate nucleosomes, the basic unit of chromatin. The nucleosomes is composed of approximately 150 bp of DNA wrapped around a histone octamer containing two copies of each of the four histone proteins (H2A, H2B, H3 and H4). Histones, in turn have a structured core domain bounded by flexible amino termini. Conserved residues within the core histones are important sites of post-translational modification. These modifications have been proposed to function as information that can act in a combinatorial fashion (Rando, 2012). A subset of modifications (including most acetylation events and methylation of lysine 4 of histone H3) are associated with gene activation. A different subset (including methylation of lysines 9 and 27 of histone H3) are associated with gene silencing.
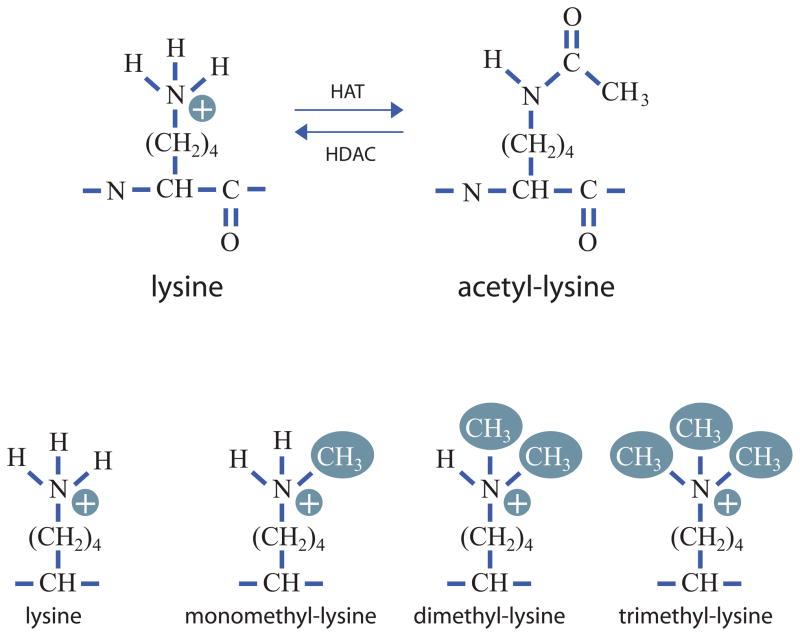
More than 100 different modification events on core histone residues have been biochemically characterized, many with well described functions (Rando, 2012). One of the earliest modifications to be discovered is acetylation of lysine residues. Lysine acetylation is a reversible modification that results in the formation of an amide bond at the ε-amino group of lysine (Figure 2A). Acetylation is carried out by acetyltransferases which use acetyl-CoA as an activated acetate donor. Deacetylation is carried out by histone deacetylases that add a water molecule across the amide bond to regenerate the free amine plus acetate. As acetylation neutralizes the positive charge of lysine, it has long been thought to weaken histone-DNA contacts (Hayes and Hansen, 2001). Densely acetylated regions are, in general, associated with regions of active transcription.
Figure 2. Lysine modification as an epigenetic mark.
The figure depicts lysine acetylation (top) and methylation (bottom). The epsilon amino group of lysine is acetylated by histone acetyltransferase enzymes (HATs). Acetyl-lysine is deacetylated by histone deacetylases (HDACs). Lysine can also be modified by the addition of one, two or three methyl groups. Methylation of lysine retains the positive charge, unlike acetylation which neutralizes it.
A second commonly found histone modification is methylation of lysine residues. Lysine residues can bear one, two or three methyl groups (Figure 2B). Histone methyltransferases catalyze methylation of lysine residues using S-adenosyl methionine as an activated methyl donor. Like acetylation, lysine methylation is reversible and removal of methyl groups is catalyzed by multiple lysine demethylases. The different classes of histone demethylases utilize distinct reaction mechanisms to remove these functional groups. Unlike acetylation, lysine methylation can be associated with either gene activity or with gene repression depending on the amino acid modified. Specificity and function of lysine methylation are determined by ‘reader’ proteins that recognize the modified lysine residue along with adjacent amino acids. Importantly, lysine methylation does not neutralize charge. Instead, it generates ammonium ions with increasing hydrophobicity that provide binding sites for reader proteins that dictate downstream function.
1.1.3 Inheritance of the Epigenome
The epigenome, like the genome, must be faithfully copied during cell division. DNA methylation patterns are widely believed to be copied by DNMT1, the enzyme with a strong preference for hemimethylated DNA. The inheritance of histone modifications, especially in mammalian cells, is an active field of research. Passage of the replication fork requires complete dismantling of the chromatin fiber and removal of histones and non-histone proteins from DNA. Subsequently, parental and newly synthesized histones are reassembled onto the daughter DNA duplexes. The precise mechanisms by which histone modification and non-histone protein association are restored are not completely understood. Acetylation marks distinguishing newly synthesized histone must be removed (Polo and Almouzni, 2006), candidate enzymes for this process have been identified (Bhaskara et al., 2010). Re-establishment of methylation marks appears to be somewhat slower and may not be complete until the G1 phase of the subsequent cell cycle (Zee et al., 2012). How the complex structures characterizing euchromatin and heterochromatin are formed is currently understudied, although ATP dependent chromatin remodeling enzymes have been implicated in this process (Polo and Almouzni, 2006; Sims and Wade, 2011).
1.2 The Immune System as a Model to Study Epigenetics
The immune system has been a useful and powerful system to study how the epigenome impacts gene expression. In large part, this results from the fact that immune cells develop in a precisely orchestrated fashion that has been the topic of intense study for decades. This has resulted in a detailed characterization of how immune cells develop and in the generation of elegant molecular and genetic tools to study this process.
Immune cells arise from a common progenitor, the hematopoietic stem cell (HSC), which is found within bone marrow. As the HSCs differentiate into multiple lineages and ultimately different cell types, the intermediate stages can be separated by combinations of surfaces markers. This useful system allows for the study of primary cells from a single individual for analysis of the dynamic changes in gene expression and chromatin as a cell differentiates. A small stem cell pool is maintained in the bone marrow, a continuous fraction differentiates into three main lineages to produce red blood cells, innate immune cells, and adaptive immune cells (Seita and Weissman, 2010). While many of the transcription factors are shared between the innate and adaptive immune cells, each lineage has a stepwise development. At each step the epigenome is remodeled to restrict gene expression from other lineages and to promote the gene expression of the appropriate lineage.
When an organism is infected by a foreign pathogen, the two arms of the immune system are activated. Innate immune cells are at the front line and immediately respond to an infection. These cells are typically short lived and are responsible for the efficient recruitment and activation of adaptive immune cells. Adaptive immune cells - B and T cells - express a unique class of antigen surface receptors. The B and T cell receptors (BCRs and TCRs) are encoded by multiple gene segments that are assembled by a process called V(D)J recombination. The combinatorial and imprecise joining of the gene segments in each individual B and T cell creates, in the population, a vast repertoire of BCRs and TCRs. B and T cells develop from a common lymphoid progenitor (CLP) population in the bone marrow and thymus, respectively, and produce mature naïve cells that circulate through the body. During an immune response only the naïve B and T cells specific for the pathogen are stimulated to proliferate, differentiate, and respond. After an infection is resolved the activated innate cells die and are cleared, but not all of the activated B and T cells are removed. The immune system has evolved to retain B and T cells for a specific pathogen by creating memory B and T cells. These memory cells are able to proliferate, differentiate and respond in a shorter time frame the next time the organism is infected with the same pathogen; thus controlling and clearing the pathogen with potentially less damage to the organism.
2 T Lymphocyte development and epigenetic regulation
2.1 T cell development and differentiation
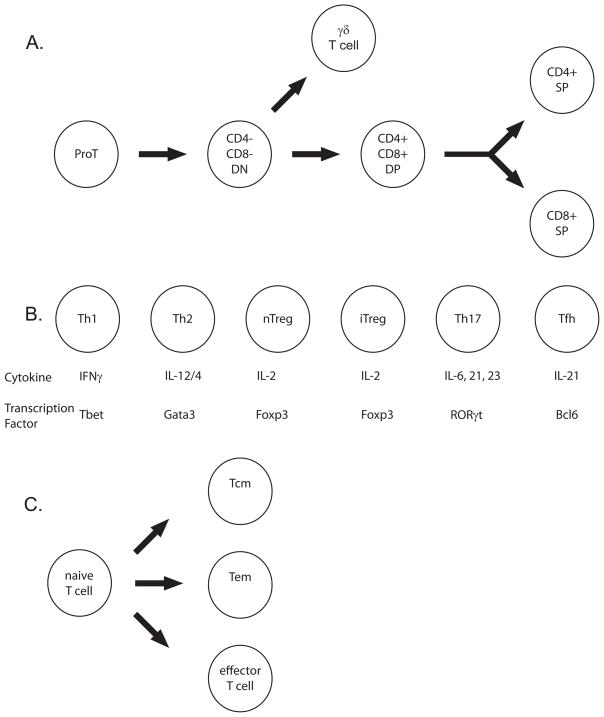
Initially, T cells develop (Figure 3A) from HSCs in the bone marrow until T progenitor cells exit this niche and migrate to the thymus (Rothenberg and Taghon, 2005). In the thymus, developing T cells rearrange the gene segments (Figure 4A) found within the T cell receptor loci (Cobb et al., 2006). After successful generation of a complete TCR, most T cells are then subjected to positive and negative selection. They are positively selected for their ability to recognize antigen in the context of MHC. In addition, they are negatively selected against auto-antigens to prevent development of self-reactive T cells. During positive selection, T cells express both co-stimulatory molecules (Figure 3A), CD4 and CD8, necessary for TCR signaling (Xiong and Bosselut, 2012). The exact mechanism by which a T cell ‘decides’ to be CD4 or CD8 is still currently under investigation (Adoro et al., 2012), but it is believed that signaling cascades establishes the expression of key transcription factors that maintain the CD4 or CD8 fate (Xiong and Bosselut, 2012).
Figure 3. T cell development in the thymus and differentiation in the periphery.
A. This figure depicts T cell development in the thymus.
B. This figure depicts the various Th subtypes. The Th subtypes (top), the key cytokine responsible for initiating differentiation (middle), and the key transcription factors responsible for maintaining differentiation (bottom).
C. This figure depicts T cell differentiation into effector, effector memory (Tem) and central memory (Tcm) cells.
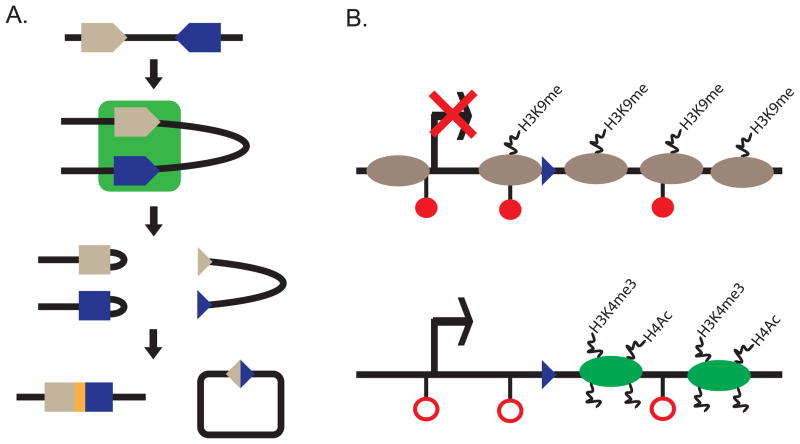
Figure 4. V(D)J recombination.
A. Schematic diagram of V(D)J recombination. A 12 (gray triangle) and 23 (blue triangle) RSS are bound by the same RAG1/2 complex (green square). Couple cleavage occurs to generate the two products of the reaction hairpin sealed coding ends (left) and blunt signal ends (right). The hairpins are opened by Artemis, repaired and ligated by the NHEJ pathway that creates a novel coding junction (yellow rectangle) between the two coding gene segments. The signal ends are repaired by the NHEJ pathway.
B. Schematic of an inaccessible or accessible RSS. Inacessible RSS chromatin (top) has been correlated with DNA methylation (filled red circles), high nucleosome occupancy (gray ovals), H3K9me and a lack of transcription (crossed out arrow). Accessible RSS chromatin (bottom) has been correlated with loss of DNA methylation (open red circles), nucleosome loss or repositioning (green ovals), H3K4me3, H3 and H4 acetylation, and active transcription (arrow).
CD8+ T cells, known as cytotoxic T cells, recognize antigen presented in the context of MHC class I, which is present on all cell types. Once activated, they produce cytokines to further activate immune cells and cytotoxic molecules to directly kill infected cells. CD4+T cells, known as T helper (Th) cells, recognize antigen presented in the context of MHC class II whose expression is restricted to immune cells such as dendritic cells and B cells. Upon activation, CD4+ T cells can differentiate into at least five sub types: Th1, Th2, Treg, Th17, and Tfh (Figure 3B) depending upon the cytokine signaling environment in which they are activated (Zygmunt and Veldhoen, 2011). Memory T cells can be produced from CD8 or CD4 T cells (Figure 3C) as central memory (TCM) or effector memory (TEM) (Masopust and Picker, 2012). Upon secondary challenge with the same pathogen, memory T cells quickly proliferate and differentiate into effector cells.
2.2 Epigenetics of T cell biology
2.2.1 V(D)J Recombination
As mentioned above, BCRs and TCRs are assembled by a process known as V(D)J recombination (Figure 4A). Variable (V), Diversity (D) and Joining (J) gene segments are flanked by recombination signal sequences (RSSs) which are the substrate for the Recombinase Activating Gene 1 and 2 (RAG1/2) complex (Cobb et al., 2006). RSSs are comprised of a conserved nonamer and hexamer sequence separated by either 12 or 23 nucleotides known as 12 or 23 RSSs, respectively (Schatz and Spanopoulou, 2005; Schatz and Swanson, 2011). The reaction occurs as a coupled cleavage event where double strand breaks are generated at the 12 and 23 RSS at the same time within a larger RAG1/2 complex. Due to the generation of double strand breaks which could be potentially genotoxic, V(D)J recombination is a highly regulated reaction to restrict rearrangement to the appropriate antigen receptor locus during the correct developmental stage. The first level of regulation is the restriction of RAG1/2 protein expression to developing B and T lymphocytes (Oettinger et al., 1990). The second level of regulation is the epigenetic changes at the antigen receptor loci to make RSSs accessible to the recombinase in addition to alter global locus conformation to facilitate coupled cleavage of distal V gene segments (Cobb et al., 2006).
RSSs are made accessible in a lineage specific manner so that Tcr loci recombine in developing T lymphocytes while Ig loci recombine in developing B lymphocytes (Cobb et al., 2006). In addition the individual Tcr loci are further regulated so Tcrb, Tcrd and Tcrg gene segments are accessible in CD4 and CD8 double negative (DN) thymocytes, while Tcra gene segments are accessible (Figure 3A) in CD4 and CD8 double positive (DP) thymocytes (Krangel, 2009). A parallel activation is found during B lymphocyte development when the heavy chain locus is activated prior to the light chain loci (Degner-Leisso and Feeney, 2010). Observational studies (Figure 4B) correlated RSS accessibility to RAG1/2 with transcription, loss of DNA methylation, sensitivity to DNaseI and endonuclease digestion, histone H3 and H4 acetylation, histone H3 lysine 4 di- and tri- methylation (H3K4me2 and H3K4me3), and alterations in nucleosome positioning and occupancy (Spicuglia et al., 2010). This lineage and stage specific regulation is enforced by the activation of cis elements that recruit histone modifying and nucleosome remodeling complexes alter the chromatin landscape to activate transcription.
In particular, epigenetic regulation of the Tcrb locus has been carefully characterized and investigated because while the locus rearranges in DN thymocytes, recombination must be inhibited when RAG1/2 is re-expressed in DP thymocytes, a process known as feedback inhibition (Jackson and Krangel, 2006). Feedback inhibition specifically inhibits further Vβ to DJβ recombination events. While the epigenetic state of the D and J gene segments is indistinguishable between DN and DP thymocytes (Mathieu et al., 2000; McMurry and Krangel, 2000; Spicuglia et al., 2002; Tripathi et al., 2002), the Vβ gene segments lose a permissive chromatin structure by having reduced transcription, reduced active histone modifications, gain of DNA methylation and reduced sensitivity to DNase I and endonuclease digestion in DP as compared to DN thymocytes (McMurry and Krangel, 2000; Tripathi et al., 2002). In addition, the Tcrb locus undergoes large scale chromatin fiber changes where the locus adopts a contracted conformation in DN thymocytes bringing the V and DJ gene segments closer together and a decontracted conformation in DP thymocytes (Skok et al., 2007). Changes in locus conformation has been characterized at most of the antigen receptor loci (Fuxa et al., 2004; Jhunjhunwala et al., 2008; Jhunjhunwala et al., 2009; Roldan et al., 2005; Shih et al., 2011; Skok et al., 2007). When Vβ chromatin accessibility was maintained by introducing enhancer elements active in DP, the modified allele failed to further recombine Vβ to DJβ gene segments (Jackson et al., 2005). Only when a recombination substrate was introduced addition to the enhancer element that Vβ to DJβ gene segments recombined in DP thymocytes (Kondilis-Mangum et al., 2011). These findings support the growing body of evidence that V(D)J recombination is regulated by multiple mechanisms and that RSSs must have accessible chromatin in addition to being brought closer together in 3 dimensional space. While the cis elements that regulate transcription and histone modifications have been characterized (see below), the analysis of potential regulators locus configuration are just beginning. CTCF and cohesin are two proteins known to be involved in creating loops on mammalian chromosomes and are found at the antigen receptor loci (Seitan et al., 2012). Deletion of cohesin during T cell development does perturb Tcra recombination (Seitan et al., 2011). In addition transcription factors such as Pax5 have been implicated in regulating locus conformation (Fuxa et al., 2004). At the Igh locus, motif analysis found CTCF sites to be near Pax5 and E2A binding sites (Ebert et al., 2011). This suggests CTCF binding may be a regulated event that may involve changes in the chromatin and methylation state since CTCF binding can be regulated by DNA methylation (Phillips and Corces, 2009).
Deletion studies of various cis elements within the antigen receptor loci provide insight as to how the epigenetic state permissible for recombination is established. For example, deletion of the known TCR enhancers, Eβ and Eα, abolishes recombination by reducing or eliminating accessible histone modification, reducing transcription and altering nucleosome occupancy (Hawwari and Krangel, 2005; Mathieu et al., 2000; Sleckman et al., 1997; Spicuglia et al., 2002). While the activity of Eβ is restricted to the most 3′ end of the Tcrb locus, the activity of Eα regulates up to 1.5 Mb 5′ of the element to include the Jα and 1/3 of the Vα/δ gene segments. Promoters also are an integral part of creating a permissive chromatin state. Deletion of promoters (Hawwari et al., 2005; Whitehurst et al., 1999) or termination (Abarrategui and Krangel, 2009) of transcription prior to RNA Pol II transcribing through an RSS specifically reduces or abolishes H3K4me3 (Abarrategui and Krangel, 2009) and alters nucleosome occupancy (Kondilis-Mangum et al., 2010). One mechanism by which the enhancer regulates promoter activity is through direct interaction between protein complexes loaded onto each cis element. One example of this is the physical interaction detected by 3C between Eβ and the Dβ1 associated promoter, PDβ1, which is mediated by RUNX1/3. RUNX1/3 binds Eβ and in required for the physical interaction with and transcription from PDβ1 (Oestreich et al., 2006). Moreover, cis elements recruit nucleosome modifying enzymes and nucleosome remodeling complexes (Morshead et al., 2003). The activity of PDβ1 can be replaced by the SWI/SNF complex (Osipovich et al., 2007). Conversely, artificial recruitment of G9a, an H3 lycine9 methyltransferase, to PDβ1 suppresses recombination (Osipovich et al., 2004).
The epigenetic modifications established by cis regulatory elements not only establish an active chromatin state, but are important in the recruitment and stable binding of the RAG1/2 complex. In addition to the RSS sequence specificity directed by RAG1, the RAG proteins contain histone modification reader (RAG2 PHD finger that binds H3K4me3 (Liu et al., 2007; Matthews et al., 2007) and writer (RAG1 RING domain that monoubiquitylates histone H3 (Grazini et al., 2010)) domains that restrict RAG binding to accessible RSSs (Ji et al., 2010a; Ji et al., 2010b) and protect the genome from RAG1/2 mediated double strand breaks.
2.2.2 Th subtypes
CD4+ T helper (Th) cells exit the thymus as Th0 cells and have evolved to further differentiate in the periphery after antigen stimulation (Zygmunt and Veldhoen, 2011). Cytokine secreted by immune and other local cells during an immune response lead to differentiation into five subtypes Th1, Th2, Treg, Th17, and Tfh by activating master transcription factors that alter the epigenetic and transcriptional profiles (Figure 3B). The newly acquired epigenetic and transcriptional state allows for the different Th cell subsets to perform specific functions and to modulate the immune response.
How epigenetic modifications regulate transcription and Th cell function have been extensively analyzed at the IFNγ and Th2 cytokine loci (as reviewed in (Lee et al., 2006)). The Th2 cytokine locus contains three important cytokines (IL-4, IL-5 and IL-13) involved in Th2 cell differentiation and function. Like the antigen receptor loci, the IFNγ and Th2 cytokine loci undergo dynamic epigenetic changes as Th0 cells differentiate into either Th1 or Th2 cells (Avni et al., 2002; Fields et al., 2002; Messi et al., 2003; Winders et al., 2004; Yamashita et al., 2002; Yano et al., 2003). Comparative analysis of Th0, Th1 and Th2 cells lead to a general model that the IFNγ and Th2 cytokine loci are unmodified in Th0. In Th1 cells IFNγ gains active histone modifications, while the Th2 cytokine locus gains repressive histone modifications and DNA methylation. Conversely, in Th2 cells, the Th2 cytokine locus gains active histone modifications while the IFNγ locus gains repressive histone modifications.
Many cis elements have been identified within the IFNγ and Th2 cytokine loci (Ansel et al., 2006; Aune et al., 2009) that include promoters, locus control regions and cell type specific enhancers. Each cis elements contributes to proper IFNγ, IL-4, IL-5 and IL-13 expression in various T cell populations in addition to other immune cells (as reviewed in (Ansel et al., 2006)). A vast literature exists dissecting the contribution of each cis element in addition to the key transcription factors that bind to each element. Some interesting observations about lineage decision and locus priming have emerged from these studies. One finding is that some of the regulatory elements physically interact in cis within the Th2 or IFNγ locus (intrachromosomal interactions) and in trans between the two loci (interchromosomal interactions) (as reviewed in (Amsen et al., 2009)). Moreover, the intrachromosomal interactions are different between an inactive and active Th2 cytokine locus (as reviewed in (Lee et al., 2006)). The intra- and interchromosomal interactions may facilitate coordinate gene expression within a large locus and can be applied to other multiple gene immune loci. It may be possible that these regulatory element interactions play a role in establishing the proper chromatin environment for transcription, which is still an open question in the field.
Wei et al (Wei et al., 2009) performed H3K4m3 and H3K27m3 ChIP-Seq characterization in addition to gene expression analysis from Th0, Th1, Th2, Th17, iTreg and nTreg ex vivo stimulated cells. Like in the limited analysis on Th1 and Th2 cells, lineage specific effector genes such as IFNγ, IL-17 and IL-4 were only expressed in the proper cell type and repressed in the others. In addition, the chromatin status in Th0 cells of lineage specific genes reviled some non-transcribing genes lack H3K4m3 or H3K27m3 modifications while others have both histone modifications. These promoters that have both active and repressive histone modifications are thought to be poised for activation. Upon differentiation, these poised genes lose H3K27m3 and maintain H3K4m3. It remains unclear if the chromatin structure of poised genes in Th0 is necessary for proper gene expression or repression in the subsequent cell types following antigen stimulation. An additional open question is if and how these new chromatin signatures for effector Th1, Th2, Th17, iTreg and nTreg are important in memory CD4+ cells.
2.2.3 DNA Methylation and T cell Memory
The role of DNA methylation has been addressed in naïve T cells and CD8+ T cell memory. In general, cytokine loci (e.g. IFNγ, Th2 and IL-2 loci) lose DNA methylation in activated T-cells (Agarwal and Rao, 1998; Bird et al., 1998; Fitzpatrick et al., 1998; Gett and Hodgkin, 1998; Hu-Li et al., 2001; Lee et al., 2001; Reiner and Seder, 1999; Richter et al., 1999), thus suggesting a potential role for DNA methylation. To experimentally test whether DNA methylation is necessary for proper T cell function, the conditional DNMT1 allele was bred with T cell specific Cre recombinase transgenes. Early deletion of DNMT1, using an Lck-Cre transgene, blocks early T cell development from DN to DP thymocytes and generates few peripheral CD8+ or CD4+ T cells (Lee et al., 2001). When CD4-Cre was used, deleting DNMT1 later in T cell development, naïve CD4+ and CD8+ DNMT1 conditionally deleted T cells were present in the periphery (Lee et al., 2001; Makar and Wilson, 2004). While DNMT1 deficient T cells were able to be activated during an immune response, they had a reduced ability to proliferate and produce memory (Lee et al., 2001). Upon Th1 or Th2 skewing conditions, DNMT1 deficient T cells miss-expressed lineage specific cytokines without affecting the expression of Th1 or Th2 master regulatory transcription factors (Makar and Wilson, 2004). There exists a strong interdependence between CD4+ and CD8+ T cells, and therefore the effects of DNMT1 deletion in one population may have an adverse effect on the other. To more clearly address the role of DNMT1 deletion in activated CD8+ T cells, the conditional DNMT1 allele was bred with a Granzyme B-Cre to specifically delete in activated CD8 T+ cells (Chappell et al., 2006). DNMT1 deletion reduced the proliferation of activated CD8+ T cells and decreased the number of memory CD8 T cells generated. The DNMT1 deficient memory T cells upon secondary challenge had reduced cytokine production, reduced proliferation and increased cell death. Together these studies indicate proper inheritance of the methylome is key for T cell activation, proliferation, memory cell formation and memory cell activation. It remains unclear is how the methylome is impacting these various stages of differentiation or how histone modifications together with the methylome regulate the inducibility of effector genes when reactivating T cells. Moreover, the defect in proliferation seen with all three Cre transgenes is not fully elucidated. The defect could be due to perturbations in the cell cycle or in cell survival. For early thymocyte deletion of DNMT1, the proliferation defect was partially rescued by expression of the pro-survival factor Bcl-XL (Lee et al., 2001) which would argue for defects in cell survival. DNMT3A and DNMT3B have been shown in other cell types to compensate for the loss of DNMT1, but are insufficient for T cell proliferation and differentiation.
3. Development, differentiation and epigenetic regulation in B lymphocytes
3.1 B Cell Development and the Germinal Center Reaction
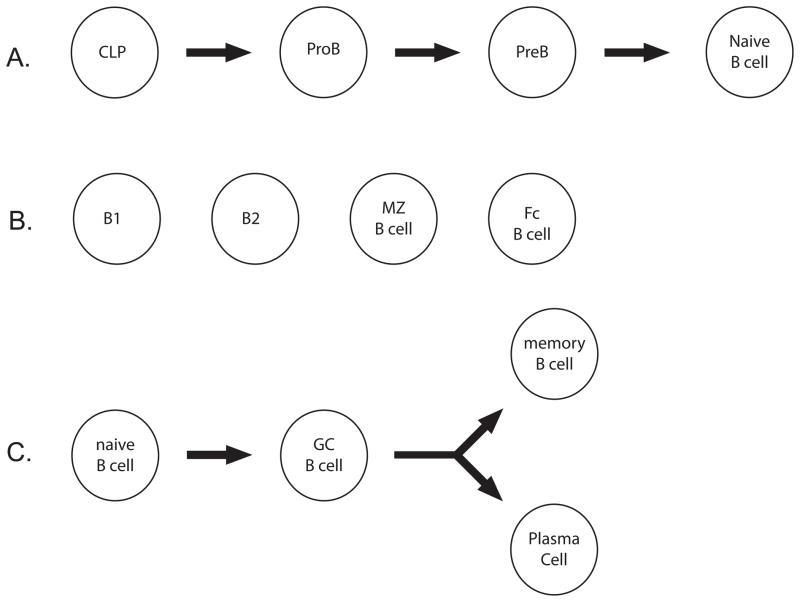
B lymphocytes develop and recombine their antigen receptor loci, the immunoglobulin (Ig) heavy and light chains, in the bone marrow (Figure 5A). Their maturation is dependent upon the expression of key transcription factors such as E47, HEB, and Pax5 (Jones and Zhuang, 2009). Without these factors the antigen receptor loci fail to recombine and the cells cannot further differentiate into naïve B cells. If an Ig receptor is successfully generated, the naïve B cell can leave the bone marrow to the spleen. Four main sub groups of naïve B-cells exist in the periphery: B1, B2, marginal zone and follicular B cells (Figure 5B).
Figure 5. B cell development in the bone marrow and differentiation in the periphery.
A. The figure depicts B cell development in the bone marrow.
B. The figure depicts the various mature B cell populations found in the periphery.
C. The figure depicts B cell maturation during a germinal center reaction.
B1, marginal zone and follicular B cells are typically stimulated during an immune response to produce short lived, low affinity IgM+ plasma cells (PC) and memory B cells. B2 cells can also undergo activation via a complex mechanism that requires TH function (Figure 5C). The activation of B cells in this fashion generates a novel structure within the follicle of secondary lymphoid tissue, the germinal center. Within the germinal center two regions are created - the light and dark zones. Activated germinal center (GC) B cells enter the germinal center reaction and cycle between the two zones. In the dark zone GC B cells, also known as centroblasts, rapidly divide and express activation-induced cytidine deaminase, AID, the protein responsible for class switch recombination and somatic hypermutation. In the light zone GC B cells, known as centrocytes, are selected for the production of higher affinity antibodies by follicular dendritic cells and TFH cells. BCL6, the key transcription factor for GC B cells, suppresses the DNA damage response and stimulates cell cycle progression. To exit the germinal center reaction, cells express PRDM1 to directly inhibit Bcl6 expression. Follicular activation in this fashion produces memory B cells with various antibody isotypes (e.g. IgG or IgA). Moreover, long lived PCs are generated that home to the bone marrow and secrete protective antibodies.
3.2 B cell epigenetics
3.2.1 The methylome of B lymphocytes
Elegant genetic methods have been used to study the role of DNA methylation in development and differentiation of the B cell lineage in mice (Broske et al., 2009). Hematopoietic stem cells with reduced dosage of DNMT1 skew towards myeloerythroid lineages, suggesting that DNA methylation is an important requisite for lymphoid development. Indeed, analysis of the transcriptome of such cells indicates enrichment for myeloerythroid transcription factors at the expense of the corresponding lymphoid factors. Forced expression of a master regulator of lymphoid fate, Ebf1, could suppress this defect, leading to the conclusion that HSC’s with deficient DNA methylation cannot silence myeloerythroid specific transcription factors and activate the corresponding lymphoid factors (Broske et al., 2009). Following commitment at the HSC stage, naïve resting B lymphocytes could survive at normal levels with reduced dosage of DNMT1.
Similar methodology has recently been applied to B lymphocyte activation in the germinal center reaction (Shaknovich et al., 2011). Mice with reduced levels of DNMT1 form smaller germinal centers than their wild type counterparts and treatment of animals with DNA methylation inhibitors completely inhibits the formation of germinal centers. Mechanistically, genetic manipulation of DNMT1 levels in GC B lymphocytes resulted in increased DNA damage, suggesting that this enzyme is critical to maintenance of genome integrity in activated, GC B lymphocytes (Shaknovich et al., 2011).
DNA methylation levels have been measured by multiple techniques in adult human B cell populations. Melnick and colleagues sampled the B cell methylome in sorted cell populations from adult human tonsil. The assay utilized for these experiments measures methylation status at restriction enzyme sites enriched in CpG islands (Shaknovich et al., 2011). Their analyses indicate that GC B cells have reduced methylation levels at the roughly 50,000 sites surveyed when compared to naïve counterparts. However, direct chemical determination of global DNA methylation found little to no difference between GC and resting B cells, suggesting the differences measured are specific for genomic regions enriched in the restriction sites utilized. These restriction sites are enriched in CpG dense promoter regions (Shaknovich et al., 2011). Hannon and colleagues have performed genomic shotgun bisulfite sequencing of sorted peripheral B lymphocytes pooled from female human volunteers. They find hypomethylated regions associated with promoters and in intergenic regions. In the latter category, enrichment of binding sites for B cell specific transcription factors is notable (Hodges et al., 2011).
3.2.2 Transcription factors direct epigenetic lineage determinants in B lymphocytes
The specification of the B cell lineage is similar in concept to that of any specialized cell type. Beginning with multipotent progenitor cells (in this case hematopoietic stem cells), development involves activation of lineage specific genes, repression of genes integral to competing lineages, and in the case of lymphocytes, appropriate regulation of recombination at antigen receptor loci. In the case of B cell development in bone marrow, a concerted and sequential action of a network of transcription factors regulate the transcriptional program and the antigen receptor loci (Ramirez et al., 2010). These include the zinc finger protein Ikaros which is required for lymphocyte development (Georgopoulos et al., 1994). During B cell development, Ikaros function is required to recruit histone acetylation to regulatory DNA at the loci encoding the V(D)J recombinases, RAG1/2 as well as for the establishment of open chromatin necessary for recombination at the immunoglobulin heavy chain loci (Reynaud et al., 2008).
Pax5 is also required for commitment to the B cell lineage where it maintains B cell identity through activation of B cell specific transcripts and repression of genes from other lineages (Ramirez et al., 2010). Regulation of Pax5 itself is complex, involves a tissue specific enhancer, requires the presence of acetylated H3 lysine 9 and methylation of H3 lysine 4 and the absence of H3K27 trimethylation (Decker et al., 2009). Interestingly, the generation of induced pluripotent stem cells (iPS cells) from murine B cells required silencing of Pax5 via RNA interference or expression of a myeloid specific transcription factor (Hanna et al., 2008), suggesting this transcription factor directs the epigenome to maintain B cell identity.
The exit of B lymphocytes from the germinal center is orchestrated by the expression of the transcriptional repressor PRDM1, also known as Blimp1. PRDM1 mediates repression of the transcription factors that drive GC B cell fate, including BCL6 and Pax5. It does so through recruitment of the histone methyltransferase G9a to establish locally repressive chromatin architecture (Gyory et al., 2004).
3.2.3 B lymphocyte activation and DNA methylation
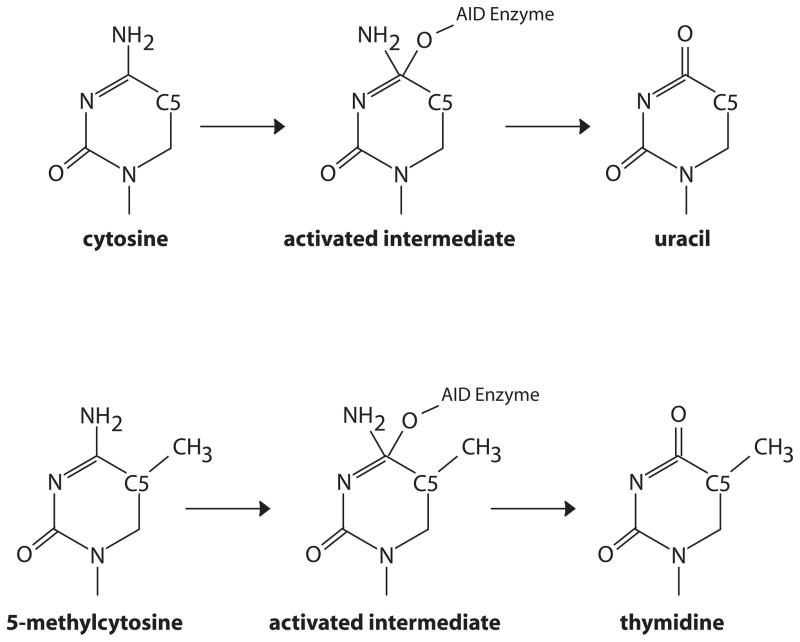
Activated B cells in the germinal center express extremely high levels of AID, the enzyme directly responsible for somatic hypermutation and class switch recombination of antibody chains. AID, in turn, has been implicated in DNA demethylation in other contexts (Bhutani et al., 2011). These findings raise the question of whether AID action results in remodeling of the DNA methylome in the activated B cell. The deamination reaction catalyzed by AID (Figure 6) converts cytosine to uracil and 5-methyl cytosine to thymidine – each resulting in a mismatch. Subsequent resolution of the mismatch lesion by DNA repair pathways would result in the loss of the methyl mark (Bhutani et al., 2011).
Figure 6. Deamination of cytosine by AID.
The proposed mechanism for enzymatic deamination of cytosine by AID/APOBEC enzymes involves production of an activated intermediate where the enzyme is covalently linked to the pyrimidine base through an oxygen atom. The intermediate is resolved by attack of a water molecule, regenerating the active enzyme and uracil. The reaction mechanism proceeds unaltered in the presence of a methyl carbon at the 5 position of the ring. However, the resulting product is thymidine, rather than uracil.
The AID enzyme requires intimate association with the nucleotide surfaces required for Watson-Crick base pairing, resulting in an obligate requirement for single-stranded DNA as a substrate. AID also preferentially acts at a consensus sequence motif – RGYW (R=purine, Y=pyrimidine, W=A or T). Given these properties, it seems likely that one consequence of AID action would be loss of methylation – albeit in limited regions of the genome. AID is known to act at the immunoglobulin loci, in addition it is also found at most transcriptional start sites in the B cell genome (Yamane et al., 2011) and this enzyme has been proposed as a mediator of major alterations in the B cell methylome (Shaknovich et al., 2011).
4 Concluding Remarks
Dynamic epigenetic changes occur during the development of lymphocytes and during immune responses. These alterations in the instructions for the genome impact both primary effector populations as well as memory. One cost of the epigenetic plasticity that characterizes the immune system is a propensity to disease. Indeed, proteins that write, read and erase epigenetic marks are frequent targets of mutation in lymphoma and leukemia. DNMT3A (Yan et al., 2011), DNMT3B (Amara et al., 2010), and the MLL histone modifying proteins (Pasqualucci et al., 2011) are found to be mutated in B cell lymphomas. Mutations in TET2 have recently been identified in lymphoma (Quivoron et al., 2011). TET2 and DNMT3A mutations have been seen together in human T cell lymphomas (Couronne et al., 2012). Inappropriate recombination during antibody class switching is proposed to be a major mechanism in lymphomagenesis (Gu et al., 2012). These findings suggest that a more detailed understanding of the epigenetic plasticity of lymphocyte development and activation may inform on oncogenic events in immune cells.
Acknowledgments
This work was supported by the Intramural Research Program of the National Institute of Environmental Health Sciences, NIH (ES101965 to P.A.W.). We thank the members of the Wade laboratory for useful discussions throughout the course of preparation of this manuscript. We apologize to our many colleagues whose work could not be cited here due to space constraints.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abarrategui I, Krangel MS. Germline transcription: a key regulator of accessibility and recombination. Adv Exp Med Biol. 2009;650:93–102. doi: 10.1007/978-1-4419-0296-2_8. [DOI] [PubMed] [Google Scholar]
- Adoro S, Park JH, Singer A. Coreceptor gene “imprinting:” A genetic solution to a developmental dilemma in T cells. Cell Cycle. 2012:11. doi: 10.4161/cc.11.5.19596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agarwal S, Rao A. Modulation of chromatin structure regulates cytokine gene expression during T cell differentiation. Immunity. 1998;9:765–775. doi: 10.1016/s1074-7613(00)80642-1. [DOI] [PubMed] [Google Scholar]
- Amara K, Ziadi S, Hachana M, Soltani N, Korbi S, Trimeche M. DNA methyltransferase DNMT3b protein overexpression as a prognostic factor in patients with diffuse large B-cell lymphomas. Cancer Sci. 2010;101:1722–1730. doi: 10.1111/j.1349-7006.2010.01569.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amsen D, Spilianakis CG, Flavell RA. How are T(H)1 and T(H)2 effector cells made? Curr Opin Immunol. 2009;21:153–160. doi: 10.1016/j.coi.2009.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ansel KM, Djuretic I, Tanasa B, Rao A. Regulation of Th2 differentiation and Il4 locus accessibility. Annu Rev Immunol. 2006;24:607–656. doi: 10.1146/annurev.immunol.23.021704.115821. [DOI] [PubMed] [Google Scholar]
- Aune TM, Collins PL, Chang S. Epigenetics and T helper 1 differentiation. Immunology. 2009;126:299–305. doi: 10.1111/j.1365-2567.2008.03026.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avni O, Lee D, Macian F, Szabo SJ, Glimcher LH, Rao A. T(H) cell differentiation is accompanied by dynamic changes in histone acetylation of cytokine genes. Nature immunology. 2002;3:643–651. doi: 10.1038/ni808. [DOI] [PubMed] [Google Scholar]
- Bestor T, Laudano A, Mattaliano R, Ingram V. Cloning and sequencing of a cDNA encoding DNA methyltransferase of mouse cells. The carboxyl-terminal domain of the mammalian enzymes is related to bacterial restriction methyltransferases. Journal of molecular biology. 1988;203:971–983. doi: 10.1016/0022-2836(88)90122-2. [DOI] [PubMed] [Google Scholar]
- Bestor TH, Ingram VM. Two DNA methyltransferases from murine erythroleukemia cells: purification, sequence specificity, and mode of interaction with DNA. Proc Natl Acad Sci U S A. 1983;80:5559–5563. doi: 10.1073/pnas.80.18.5559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhaskara S, Knutson SK, Jiang G, Chandrasekharan MB, Wilson AJ, Zheng S, Yenamandra A, Locke K, Yuan JL, Bonine-Summers AR, et al. Hdac3 is essential for the maintenance of chromatin structure and genome stability. Cancer Cell. 2010;18:436–447. doi: 10.1016/j.ccr.2010.10.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhutani N, Burns DM, Blau HM. DNA demethylation dynamics. Cell. 2011;146:866–872. doi: 10.1016/j.cell.2011.08.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bird JJ, Brown DR, Mullen AC, Moskowitz NH, Mahowald MA, Sider JR, Gajewski TF, Wang CR, Reiner SL. Helper T cell differentiation is controlled by the cell cycle. Immunity. 1998;9:229–237. doi: 10.1016/s1074-7613(00)80605-6. [DOI] [PubMed] [Google Scholar]
- Broske AM, Vockentanz L, Kharazi S, Huska MR, Mancini E, Scheller M, Kuhl C, Enns A, Prinz M, Jaenisch R, et al. DNA methylation protects hematopoietic stem cell multipotency from myeloerythroid restriction. Nat Genet. 2009;41:1207–1215. doi: 10.1038/ng.463. [DOI] [PubMed] [Google Scholar]
- Chappell C, Beard C, Altman J, Jaenisch R, Jacob J. DNA methylation by DNA methyltransferase 1 is critical for effector CD8 T cell expansion. J Immunol. 2006;176:4562–4572. doi: 10.4049/jimmunol.176.8.4562. [DOI] [PubMed] [Google Scholar]
- Cheng X, Blumenthal RM. Mammalian DNA methyltransferases: a structural perspective. Structure. 2008;16:341–350. doi: 10.1016/j.str.2008.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cobb RM, Oestreich KJ, Osipovich OA, Oltz EM. Accessibility control of V(D)J recombination. Adv Immunol. 2006;91:45–109. doi: 10.1016/S0065-2776(06)91002-5. [DOI] [PubMed] [Google Scholar]
- Couronne L, Bastard C, Bernard OA. TET2 and DNMT3A mutations in human T-cell lymphoma. N Engl J Med. 2012;366:95–96. doi: 10.1056/NEJMc1111708. [DOI] [PubMed] [Google Scholar]
- Decker T, Pasca di Magliano M, McManus S, Sun Q, Bonifer C, Tagoh H, Busslinger M. Stepwise activation of enhancer and promoter regions of the B cell commitment gene Pax5 in early lymphopoiesis. Immunity. 2009;30:508–520. doi: 10.1016/j.immuni.2009.01.012. [DOI] [PubMed] [Google Scholar]
- Degner-Leisso SC, Feeney AJ. Epigenetic and 3-dimensional regulation of V(D)J rearrangement of immunoglobulin genes. Seminars in immunology. 2010;22:346–352. doi: 10.1016/j.smim.2010.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebert A, McManus S, Tagoh H, Medvedovic J, Salvagiotto G, Novatchkova M, Tamir I, Sommer A, Jaritz M, Busslinger M. The distal V(H) gene cluster of the Igh locus contains distinct regulatory elements with Pax5 transcription factor-dependent activity in pro-B cells. Immunity. 2011;34:175–187. doi: 10.1016/j.immuni.2011.02.005. [DOI] [PubMed] [Google Scholar]
- Fields PE, Kim ST, Flavell RA. Cutting edge: changes in histone acetylation at the IL-4 and IFN-gamma loci accompany Th1/Th2 differentiation. J Immunol. 2002;169:647–650. doi: 10.4049/jimmunol.169.2.647. [DOI] [PubMed] [Google Scholar]
- Fitzpatrick DR, Shirley KM, McDonald LE, Bielefeldt-Ohmann H, Kay GF, Kelso A. Distinct methylation of the interferon gamma (IFN-gamma) and interleukin 3 (IL-3) genes in newly activated primary CD8+ T lymphocytes: regional IFN-gamma promoter demethylation and mRNA expression are heritable in CD44(high)CD8+ T cells. J Exp Med. 1998;188:103–117. doi: 10.1084/jem.188.1.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuxa M, Skok J, Souabni A, Salvagiotto G, Roldan E, Busslinger M. Pax5 induces V-to-DJ rearrangements and locus contraction of the immunoglobulin heavy-chain gene. Genes & development. 2004;18:411–422. doi: 10.1101/gad.291504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Georgopoulos K, Bigby M, Wang JH, Molnar A, Wu P, Winandy S, Sharpe A. The Ikaros gene is required for the development of all lymphoid lineages. Cell. 1994;79:143–156. doi: 10.1016/0092-8674(94)90407-3. [DOI] [PubMed] [Google Scholar]
- Gett AV, Hodgkin PD. Cell division regulates the T cell cytokine repertoire, revealing a mechanism underlying immune class regulation. Proc Natl Acad Sci U S A. 1998;95:9488–9493. doi: 10.1073/pnas.95.16.9488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goll MG, Kirpekar F, Maggert KA, Yoder JA, Hsieh CL, Zhang X, Golic KG, Jacobsen SE, Bestor TH. Methylation of tRNAAsp by the DNA methyltransferase homolog Dnmt2. Science. 2006;311:395–398. doi: 10.1126/science.1120976. [DOI] [PubMed] [Google Scholar]
- Grazini U, Zanardi F, Citterio E, Casola S, Goding CR, McBlane F. The RING domain of RAG1 ubiquitylates histone H3: a novel activity in chromatin-mediated regulation of V(D)J joining. Molecular cell. 2010;37:282–293. doi: 10.1016/j.molcel.2009.12.035. [DOI] [PubMed] [Google Scholar]
- Gu X, Shivarov V, Strout MP. The role of activation-induced cytidine deaminase in lymphomagenesis. Current opinion in hematology. 2012;19:292–298. doi: 10.1097/MOH.0b013e328353da3a. [DOI] [PubMed] [Google Scholar]
- Gyory I, Wu J, Fejer G, Seto E, Wright KL. PRDI-BF1 recruits the histone H3 methyltransferase G9a in transcriptional silencing. Nat Immunol. 2004;5:299–308. doi: 10.1038/ni1046. [DOI] [PubMed] [Google Scholar]
- Hanna J, Markoulaki S, Schorderet P, Carey BW, Beard C, Wernig M, Creyghton MP, Steine EJ, Cassady JP, Foreman R, et al. Direct reprogramming of terminally differentiated mature B lymphocytes to pluripotency. Cell. 2008;133:250–264. doi: 10.1016/j.cell.2008.03.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawwari A, Bock C, Krangel MS. Regulation of T cell receptor alpha gene assembly by a complex hierarchy of germline Jalpha promoters. Nat Immunol. 2005;6:481–489. doi: 10.1038/ni1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawwari A, Krangel MS. Regulation of TCR delta and alpha repertoires by local and long-distance control of variable gene segment chromatin structure. J Exp Med. 2005;202:467–472. doi: 10.1084/jem.20050680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayes JJ, Hansen JC. Nucleosomes and the chromatin fiber. Current opinion in genetics & development. 2001;11:124–129. doi: 10.1016/s0959-437x(00)00168-4. [DOI] [PubMed] [Google Scholar]
- Hellman A, Chess A. Gene body-specific methylation on the active X chromosome. Science. 2007;315:1141–1143. doi: 10.1126/science.1136352. [DOI] [PubMed] [Google Scholar]
- Hodges E, Molaro A, Dos Santos CO, Thekkat P, Song Q, Uren PJ, Park J, Butler J, Rafii S, McCombie WR, et al. Directional DNA methylation changes and complex intermediate states accompany lineage specificity in the adult hematopoietic compartment. Molecular cell. 2011;44:17–28. doi: 10.1016/j.molcel.2011.08.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hotchkiss RD. The quantitative separation of purines, pyrimidines, and nucleosides by paper chromatography. J Biol Chem. 1948;175:315–332. [PubMed] [Google Scholar]
- Hu-Li J, Pannetier C, Guo L, Lohning M, Gu H, Watson C, Assenmacher M, Radbruch A, Paul WE. Regulation of expression of IL-4 alleles: analysis using a chimeric GFP/IL-4 gene. Immunity. 2001;14:1–11. doi: 10.1016/s1074-7613(01)00084-x. [DOI] [PubMed] [Google Scholar]
- Jackson A, Kondilis HD, Khor B, Sleckman BP, Krangel MS. Regulation of T cell receptor beta allelic exclusion at a level beyond accessibility. Nature immunology. 2005;6:189–197. doi: 10.1038/ni1157. [DOI] [PubMed] [Google Scholar]
- Jackson AM, Krangel MS. Turning T-cell receptor beta recombination on and off: more questions than answers. Immunol Rev. 2006;209:129–141. doi: 10.1111/j.0105-2896.2006.00342.x. [DOI] [PubMed] [Google Scholar]
- Jhunjhunwala S, van Zelm MC, Peak MM, Cutchin S, Riblet R, van Dongen JJ, Grosveld FG, Knoch TA, Murre C. The 3D structure of the immunoglobulin heavy-chain locus: implications for long-range genomic interactions. Cell. 2008;133:265–279. doi: 10.1016/j.cell.2008.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jhunjhunwala S, van Zelm MC, Peak MM, Murre C. Chromatin architecture and the generation of antigen receptor diversity. Cell. 2009;138:435–448. doi: 10.1016/j.cell.2009.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji Y, Little AJ, Banerjee JK, Hao B, Oltz EM, Krangel MS, Schatz DG. Promoters, enhancers, and transcription target RAG1 binding during V(D)J recombination. J Exp Med. 2010a;207:2809–2816. doi: 10.1084/jem.20101136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji Y, Resch W, Corbett E, Yamane A, Casellas R, Schatz DG. The in vivo pattern of binding of RAG1 and RAG2 to antigen receptor loci. Cell. 2010b;141:419–431. doi: 10.1016/j.cell.2010.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones ME, Zhuang Y. Regulation of V(D)J recombination by E-protein transcription factors. Advances in experimental medicine and biology. 2009;650:148–156. doi: 10.1007/978-1-4419-0296-2_12. [DOI] [PubMed] [Google Scholar]
- Jurkowska RZ, Jurkowski TP, Jeltsch A. Structure and function of mammalian DNA methyltransferases. Chembiochem. 2011;12:206–222. doi: 10.1002/cbic.201000195. [DOI] [PubMed] [Google Scholar]
- Klimasauskas S, Kumar S, Roberts RJ, Cheng X. HhaI methyltransferase flips its target base out of the DNA helix. Cell. 1994;76:357–369. doi: 10.1016/0092-8674(94)90342-5. [DOI] [PubMed] [Google Scholar]
- Kondilis-Mangum HD, Cobb RM, Osipovich O, Srivatsan S, Oltz EM, Krangel MS. Transcription-dependent mobilization of nucleosomes at accessible TCR gene segments in vivo. J Immunol. 2010;184:6970–6977. doi: 10.4049/jimmunol.0903923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondilis-Mangum HD, Shih HY, Mahowald G, Sleckman BP, Krangel MS. Regulation of TCRbeta allelic exclusion by gene segment proximity and accessibility. J Immunol. 2011;187:6374–6381. doi: 10.4049/jimmunol.1102611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krangel MS. Mechanics of T cell receptor gene rearrangement. Curr Opin Immunol. 2009;21:133–139. doi: 10.1016/j.coi.2009.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee GR, Kim ST, Spilianakis CG, Fields PE, Flavell RA. T helper cell differentiation: regulation by cis elements and epigenetics. Immunity. 2006;24:369–379. doi: 10.1016/j.immuni.2006.03.007. [DOI] [PubMed] [Google Scholar]
- Lee PP, Fitzpatrick DR, Beard C, Jessup HK, Lehar S, Makar KW, Perez-Melgosa M, Sweetser MT, Schlissel MS, Nguyen S, et al. A critical role for Dnmt1 and DNA methylation in T cell development, function, and survival. Immunity. 2001;15:763–774. doi: 10.1016/s1074-7613(01)00227-8. [DOI] [PubMed] [Google Scholar]
- Li E, Bestor TH, Jaenisch R. Targeted mutation of the DNA methyltransferase gene results in embryonic lethality. Cell. 1992;69:915–926. doi: 10.1016/0092-8674(92)90611-f. [DOI] [PubMed] [Google Scholar]
- Lister R, Pelizzola M, Dowen RH, Hawkins RD, Hon G, Tonti-Filippini J, Nery JR, Lee L, Ye Z, Ngo QM, et al. Human DNA methylomes at base resolution show widespread epigenomic differences. Nature. 2009;462:315–322. doi: 10.1038/nature08514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Subrahmanyam R, Chakraborty T, Sen R, Desiderio S. A plant homeodomain in RAG-2 that binds Hypermethylated lysine 4 of histone H3 is necessary for efficient antigen-receptor-gene rearrangement. Immunity. 2007;27:561–571. doi: 10.1016/j.immuni.2007.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makar KW, Wilson CB. DNA methylation is a nonredundant repressor of the Th2 effector program. J Immunol. 2004;173:4402–4406. doi: 10.4049/jimmunol.173.7.4402. [DOI] [PubMed] [Google Scholar]
- Masopust D, Picker LJ. Hidden memories: frontline memory T cells and early pathogen interception. J Immunol. 2012;188:5811–5817. doi: 10.4049/jimmunol.1102695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathieu N, Hempel WM, Spicuglia S, Verthuy C, Ferrier P. Chromatin remodeling by the T cell receptor (TCR)-beta gene enhancer during early T cell development: Implications for the control of TCR-beta locus recombination. J Exp Med. 2000;192:625–636. doi: 10.1084/jem.192.5.625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthews AG, Kuo AJ, Ramon-Maiques S, Han S, Champagne KS, Ivanov D, Gallardo M, Carney D, Cheung P, Ciccone DN, et al. RAG2 PHD finger couples histone H3 lysine 4 trimethylation with V(D)J recombination. Nature. 2007;450:1106–1110. doi: 10.1038/nature06431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMurry MT, Krangel MS. A role for histone acetylation in the developmental regulation of VDJ recombination. Science. 2000;287:495–498. doi: 10.1126/science.287.5452.495. [DOI] [PubMed] [Google Scholar]
- Meissner A, Mikkelsen TS, Gu H, Wernig M, Hanna J, Sivachenko A, Zhang X, Bernstein BE, Nusbaum C, Jaffe DB, et al. Genome-scale DNA methylation maps of pluripotent and differentiated cells. Nature. 2008;454:766–770. doi: 10.1038/nature07107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messi M, Giacchetto I, Nagata K, Lanzavecchia A, Natoli G, Sallusto F. Memory and flexibility of cytokine gene expression as separable properties of human T(H)1 and T(H)2 lymphocytes. Nature immunology. 2003;4:78–86. doi: 10.1038/ni872. [DOI] [PubMed] [Google Scholar]
- Morshead KB, Ciccone DN, Taverna SD, Allis CD, Oettinger MA. Antigen receptor loci poised for V(D)J rearrangement are broadly associated with BRG1 and flanked by peaks of histone H3 dimethylated at lysine 4. Proc Natl Acad Sci U S A. 2003;100:11577–11582. doi: 10.1073/pnas.1932643100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oestreich KJ, Cobb RM, Pierce S, Chen J, Ferrier P, Oltz EM. Regulation of TCRbeta gene assembly by a promoter/enhancer holocomplex. Immunity. 2006;24:381–391. doi: 10.1016/j.immuni.2006.02.009. [DOI] [PubMed] [Google Scholar]
- Oettinger MA, Schatz DG, Gorka C, Baltimore D. RAG-1 and RAG-2, adjacent genes that synergistically activate V(D)J recombination. Science. 1990;248:1517–1523. doi: 10.1126/science.2360047. [DOI] [PubMed] [Google Scholar]
- Osipovich O, Cobb RM, Oestreich KJ, Pierce S, Ferrier P, Oltz EM. Essential function for SWI-SNF chromatin-remodeling complexes in the promoter-directed assembly of Tcrb genes. Nature immunology. 2007;8:809–816. doi: 10.1038/ni1481. [DOI] [PubMed] [Google Scholar]
- Osipovich O, Milley R, Meade A, Tachibana M, Shinkai Y, Krangel MS, Oltz EM. Targeted inhibition of V(D)J recombination by a histone methyltransferase. Nature immunology. 2004;5:309–316. doi: 10.1038/ni1042. [DOI] [PubMed] [Google Scholar]
- Pasqualucci L, Dominguez-Sola D, Chiarenza A, Fabbri G, Grunn A, Trifonov V, Kasper LH, Lerach S, Tang H, Ma J, et al. Inactivating mutations of acetyltransferase genes in B-cell lymphoma. Nature. 2011;471:189–195. doi: 10.1038/nature09730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips JE, Corces VG. CTCF: master weaver of the genome. Cell. 2009;137:1194–1211. doi: 10.1016/j.cell.2009.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polo SE, Almouzni G. Chromatin assembly: a basic recipe with various flavours. Current opinion in genetics & development. 2006;16:104–111. doi: 10.1016/j.gde.2006.02.011. [DOI] [PubMed] [Google Scholar]
- Quivoron C, Couronne L, Della Valle V, Lopez CK, Plo I, Wagner-Ballon O, Do Cruzeiro M, Delhommeau F, Arnulf B, Stern MH, et al. TET2 inactivation results in pleiotropic hematopoietic abnormalities in mouse and is a recurrent event during human lymphomagenesis. Cancer Cell. 2011;20:25–38. doi: 10.1016/j.ccr.2011.06.003. [DOI] [PubMed] [Google Scholar]
- Ramirez J, Lukin K, Hagman J. From hematopoietic progenitors to B cells: mechanisms of lineage restriction and commitment. Curr Opin Immunol. 2010;22:177–184. doi: 10.1016/j.coi.2010.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rando OJ. Combinatorial complexity in chromatin structure and function: revisiting the histone code. Current opinion in genetics & development. 2012;22:148–155. doi: 10.1016/j.gde.2012.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiner SL, Seder RA. Dealing from the evolutionary pawnshop: how lymphocytes make decisions. Immunity. 1999;11:1–10. doi: 10.1016/s1074-7613(00)80076-x. [DOI] [PubMed] [Google Scholar]
- Reynaud D, Demarco IA, Reddy KL, Schjerven H, Bertolino E, Chen Z, Smale ST, Winandy S, Singh H. Regulation of B cell fate commitment and immunoglobulin heavy-chain gene rearrangements by Ikaros. Nat Immunol. 2008;9:927–936. doi: 10.1038/ni.1626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richter A, Lohning M, Radbruch A. Instruction for cytokine expression in T helper lymphocytes in relation to proliferation and cell cycle progression. J Exp Med. 1999;190:1439–1450. doi: 10.1084/jem.190.10.1439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roldan E, Fuxa M, Chong W, Martinez D, Novatchkova M, Busslinger M, Skok JA. Locus ‘decontraction’ and centromeric recruitment contribute to allelic exclusion of the immunoglobulin heavy-chain gene. Nature immunology. 2005;6:31–41. doi: 10.1038/ni1150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothenberg EV, Taghon T. Molecular genetics of T cell development. Annu Rev Immunol. 2005;23:601–649. doi: 10.1146/annurev.immunol.23.021704.115737. [DOI] [PubMed] [Google Scholar]
- Schatz DG, Spanopoulou E. Biochemistry of V(D)J recombination. Curr Top Microbiol Immunol. 2005;290:49–85. doi: 10.1007/3-540-26363-2_4. [DOI] [PubMed] [Google Scholar]
- Schatz DG, Swanson PC. V(D)J recombination: mechanisms of initiation. Annu Rev Genet. 2011;45:167–202. doi: 10.1146/annurev-genet-110410-132552. [DOI] [PubMed] [Google Scholar]
- Seita J, Weissman IL. Hematopoietic stem cell: self-renewal versus differentiation. Wiley interdisciplinary reviews. 2010;2:640–653. doi: 10.1002/wsbm.86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seitan VC, Hao B, Tachibana-Konwalski K, Lavagnolli T, Mira-Bontenbal H, Brown KE, Teng G, Carroll T, Terry A, Horan K, et al. A role for cohesin in T-cell-receptor rearrangement and thymocyte differentiation. Nature. 2011;476:467–471. doi: 10.1038/nature10312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seitan VC, Krangel MS, Merkenschlager M. Cohesin, CTCF and lymphocyte antigen receptor locus rearrangement. Trends Immunol. 2012;33:153–159. doi: 10.1016/j.it.2012.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaknovich R, Cerchietti L, Tsikitas L, Kormaksson M, De S, Figueroa ME, Ballon G, Yang SN, Weinhold N, Reimers M, et al. DNA methyltransferase 1 and DNA methylation patterning contribute to germinal center B-cell differentiation. Blood. 2011;118:3559–3569. doi: 10.1182/blood-2011-06-357996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shih HY, Hao B, Krangel MS. Orchestrating T-cell receptor alpha gene assembly through changes in chromatin structure and organization. Immunol Res. 2011;49:192–201. doi: 10.1007/s12026-010-8181-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sims JK, Wade PA. Mi-2/NuRD complex function is required for normal S phase progression and assembly of pericentric heterochromatin. Molecular biology of the cell. 2011;22:3094–3102. doi: 10.1091/mbc.E11-03-0258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skok JA, Gisler R, Novatchkova M, Farmer D, de Laat W, Busslinger M. Reversible contraction by looping of the Tcra and Tcrb loci in rearranging thymocytes. Nature immunology. 2007;8:378–387. doi: 10.1038/ni1448. [DOI] [PubMed] [Google Scholar]
- Sleckman BP, Bardon CG, Ferrini R, Davidson L, Alt FW. Function of the TCR alpha enhancer in alphabeta and gammadelta T cells. Immunity. 1997;7:505–515. doi: 10.1016/s1074-7613(00)80372-6. [DOI] [PubMed] [Google Scholar]
- Spicuglia S, Kumar S, Yeh JH, Vachez E, Chasson L, Gorbatch S, Cautres J, Ferrier P. Promoter activation by enhancer-dependent and -independent loading of activator and coactivator complexes. Molecular cell. 2002;10:1479–1487. doi: 10.1016/s1097-2765(02)00791-8. [DOI] [PubMed] [Google Scholar]
- Spicuglia S, Zacarias-Cabeza J, Pekowska A, Ferrier P. Epigenetic regulation of antigen receptor gene rearrangement. F1000 Biol Rep. 2010:2. doi: 10.3410/B2-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tripathi R, Jackson A, Krangel MS. A change in the structure of Vbeta chromatin associated with TCR beta allelic exclusion. J Immunol. 2002;168:2316–2324. doi: 10.4049/jimmunol.168.5.2316. [DOI] [PubMed] [Google Scholar]
- Wei G, Wei L, Zhu J, Zang C, Hu-Li J, Yao Z, Cui K, Kanno Y, Roh TY, Watford WT, et al. Global mapping of H3K4me3 and H3K27me3 reveals specificity and plasticity in lineage fate determination of differentiating CD4+ T cells. Immunity. 2009;30:155–167. doi: 10.1016/j.immuni.2008.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitehurst CE, Chattopadhyay S, Chen J. Control of V(D)J recombinational accessibility of the D beta 1 gene segment at the TCR beta locus by a germline promoter. Immunity. 1999;10:313–322. doi: 10.1016/s1074-7613(00)80031-x. [DOI] [PubMed] [Google Scholar]
- Winders BR, Schwartz RH, Bruniquel D. A distinct region of the murine IFN-gamma promoter is hypomethylated from early T cell development through mature naive and Th1 cell differentiation, but is hypermethylated in Th2 cells. J Immunol. 2004;173:7377–7384. doi: 10.4049/jimmunol.173.12.7377. [DOI] [PubMed] [Google Scholar]
- Wu H, Zhang Y. Mechanisms and functions of Tet protein-mediated 5-methylcytosine oxidation. Genes & development. 2011;25:2436–2452. doi: 10.1101/gad.179184.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong Y, Bosselut R. CD4-CD8 differentiation in the thymus: connecting circuits and building memories. Curr Opin Immunol. 2012;24:139–145. doi: 10.1016/j.coi.2012.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamane A, Resch W, Kuo N, Kuchen S, Li Z, Sun HW, Robbiani DF, McBride K, Nussenzweig MC, Casellas R. Deep-sequencing identification of the genomic targets of the cytidine deaminase AID and its cofactor RPA in B lymphocytes. Nature immunology. 2011;12:62–69. doi: 10.1038/ni.1964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashita M, Ukai-Tadenuma M, Kimura M, Omori M, Inami M, Taniguchi M, Nakayama T. Identification of a conserved GATA3 response element upstream proximal from the interleukin-13 gene locus. J Biol Chem. 2002;277:42399–42408. doi: 10.1074/jbc.M205876200. [DOI] [PubMed] [Google Scholar]
- Yan XJ, Xu J, Gu ZH, Pan CM, Lu G, Shen Y, Shi JY, Zhu YM, Tang L, Zhang XW, et al. Exome sequencing identifies somatic mutations of DNA methyltransferase gene DNMT3A in acute monocytic leukemia. Nat Genet. 2011;43:309–315. doi: 10.1038/ng.788. [DOI] [PubMed] [Google Scholar]
- Yano S, Ghosh P, Kusaba H, Buchholz M, Longo DL. Effect of promoter methylation on the regulation of IFN-gamma gene during in vitro differentiation of human peripheral blood T cells into a Th2 population. J Immunol. 2003;171:2510–2516. doi: 10.4049/jimmunol.171.5.2510. [DOI] [PubMed] [Google Scholar]
- Zee BM, Britton LM, Wolle D, Haberman DM, Garcia BA. Origins and Formation of Histone Methylation across the Human Cell Cycle. Mol Cell Biol. 2012 doi: 10.1128/MCB.06673-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zygmunt B, Veldhoen M. T helper cell differentiation more than just cytokines. Adv Immunol. 2011;109:159–196. doi: 10.1016/B978-0-12-387664-5.00005-4. [DOI] [PubMed] [Google Scholar]