Abstract
Wip1 (PPM1D) is a stress responsive PP2C phosphatase that plays a key role in stress signaling. Although originally identified as a gene induced by p53 after genotoxic stress, we now know that Wip1 expression is additionally regulated by other mechanisms. Wip1 is not only a target of p53, but is also a target of other transcription factors, including Estrogen Receptor-alpha and NF-kappaB. Additionally, Wip1 expression is regulated by post-transcriptional mechanisms such as mRNA stabilization and alternative splicing. Upon induction, Wip1 dampens the stress response by dephosphorylating and inactivating proteins such as p53, p38 MAPK, and ATM, usually as part of a negative feedback loop. As a result, Wip1 functions to abrogate cell cycle checkpoints and inhibit senescence, apoptosis, DNA repair, and the production of inflammatory cytokines. Furthermore, Wip1 is overexpressed in several types of human cancers and has oncogenic functions. The regulation of Wip1, the role of Wip1 in stress signaling, and the cooperation of Wip1 with oncogenes in promoting tumorigenesis will be discussed in this review.
Keywords: Wip1, PPM1D, Stress Signaling, DNA Damage Response, Review
2. INTRODUCTION
Signal transduction, such as the transmission of a signal through protein modifications, is essential for the cell to cope with stress. A good example of stress signaling is the DNA damage response (DDR). Cells have adapted the DDR as a way for the normal functions, such as those involved in cell cycle progression, to temporarily shut down while stress-induced damage is repaired. Depending on the cell cycle phase of the cell at the time of damage, the cell can halt cell cycle progression by eliciting checkpoints (1, 2). However, if the damage is not repairable, then the cell may trigger apoptosis. Apoptosis and cell cycle arrest are crucial, since cells that have dysfunctional checkpoint and apoptotic responses are able to replicate despite persistent damage, potentially leading to mutations and tumor formation. Therefore, the existence of signaling proteins responsible for the initiation of cell cycle arrest during damage repair or apoptosis if the damage cannot be repaired implies the existence of mechanisms for suppressing these pathways upon the completion of repair to allow the resumption of normal cell cycle progression. The Wild-type p53 inducible protein 1 (Wip1) phosphatase, the product of the PPM1D gene, plays an important role in the latter process. In other words, Wip1 dampens stress signaling and facilitates the return of the cell to a homeostatic state once the damage is repaired.
Wip1 was originally identified as a nuclear phosphatase induced in a p53-dependent manner after IR exposure (3). It is a member of the PP2C family of serine/threonine phosphatases, which by definition means it is magnesium-dependent and okadaic acid-insensitive (3). Expression of Wip1 is induced by a variety of stresses, such as exposure to ionizing radiation (IR), ultraviolet (UV) radiation, anisomycin, hydrogen peroxide (H2O2), methyl methane sulfonate, and inflammatory cytokines (3–6). Once induced, Wip1 directly binds to and dephosphorylates several key signaling proteins involved in stress signaling. Some examples of Wip1 targets include p38 MAPK, p53, the phosphorylated form of the histone 2A variant H2AX (gamma-H2AX) and ataxia-telangiectasia mutated (ATM), which play important roles in apoptosis, cell cycle arrest, and DNA repair (4, 7–10).
Wip1 also plays an important role in tumorigenesis, which is evident by its overexpression in several types of human cancers such as breast cancer, ovarian cancer, and gastric carcinomas (11–14). Oncogenic stress triggers DNA damage-like signaling, which appears to act as a barrier to cellular transformation in response to oncogenic stress (15, 16), and inhibition of this signaling by Wip1 is most likely a tumor promoting mechanism. Additionally, although its overexpression alone does not promote tumorigenesis, Wip1 has been shown to cooperate with other oncogenes, including Erbb2 and HRas1, in promoting tumorigenesis (17, 18). This review will focus on the regulation of Wip1 expression after stress, the functional effects of Wip1 on signaling in the stress response, and the cooperation of Wip1 with oncogenes in promoting tumorigenesis.
3. REGULATION OF WIP1 EXPRESSION
Wip1 expression is induced by a variety of exogenous stresses and subsequently regulates stress signaling through its phosphatase activity. To date, the activity of Wip1 has not been shown to be regulated through post-translational modification; the major known modulation of Wip1 phosphatase activity is through the level of its expression. The following section will review the regulation of Wip1 expression, including transcriptional and post-transcriptional mechanisms.
3.1. Transcriptional regulation
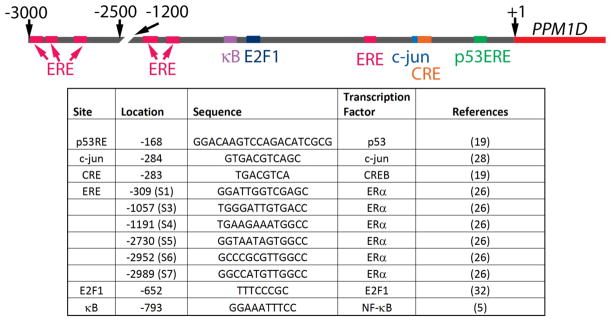
The promoter region of the PPM1D gene is GC-rich and contains binding motifs for numerous transcription factors, suggesting complex regulation during development and in modulating tissue-specific responses to stress. The transcription factors that have been experimentally validated, namely p53, CREB, NF-kappaB, ERalpha, c-jun, and E2F1, are important in the genotoxic and oncogenic stress responses, and the relative locations of their binding sites in the Wip1 promoter are shown in Figure 1. The regulation of the expression of Wip1 by these transcription factors is discussed in the sections below.
Figure 1.
Transcription factor binding sites in the PPM1D promoter. A schematic of the PPM1D promoter region shows the location of the transcription factor binding sites (based on the NCBI March 2006 human reference sequence Build 36.1). A list of the transcription factors and the location and the sequence of the respective binding site is shown.
3.1.1. p53
Wip1 was first identified as a gene upregulated after DNA damage in a p53-dependent manner (3). In particular, Rossi et al. analyzed the promoter region of Wip1 and confirmed Wip1 as a p53 transcriptional target (Figure 1 & Figure 2) (19). The authors identified two potential p53 response elements (p53RE) in the PPM1D promoter region, but showed that only the p53RE located in the 5′ untranslated region (5′UTR) conferred p53-responsiveness. By using reporter constructs, PCR, and chromatin immunoprecipitation (ChIP) analysis, they showed that p53 bound to the PPM1D promoter region after IR, and that the 5′UTR p53RE was responsible for the DNA damage-induced upregulation of Wip1 by p53 (19).
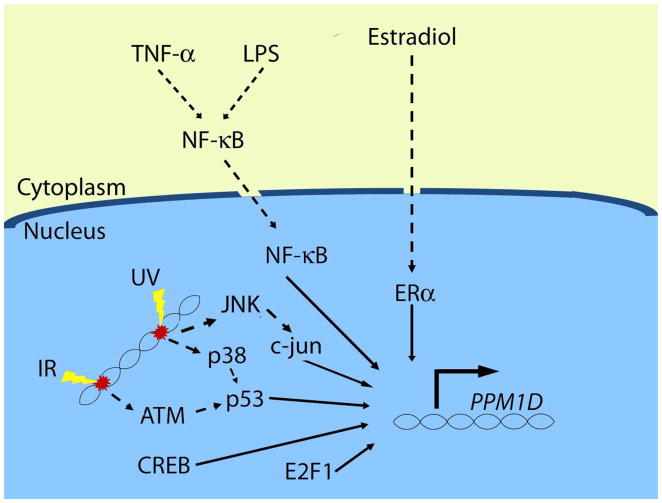
Figure 2.
Transcriptional regulation of PPM1D. The currently known transcription factors that enhance PPM1D transcription are p53, CREB, E2F1, c-jun, ERalpha, and NF-kappaB. The regulation by each of these factors depends on context, namely the type of stress and possibly the tissue type (see text for details).
Most types of DNA damage lead to activation of p53 and subsequent induction of Wip1, but the signaling pathways leading to p53 activation may differ, depending on the nature of the damage. A good example of this is the difference in signaling to p53 after UV radiation and IR exposure. Following exposure to IR, the PI3K-like kinase ATM becomes activated through autophosphorylation and subsequently phosphorylates p53 on Ser15, leading to its activation (20). This differs from p53 activation after UV radiation exposure, which involves phosphorylation of p53 on Ser33 and Ser46 by p38 MAPK (21). Wip1 expression is induced by UV radiation exposure (17), and not surprisingly, p38 is required for induction of Wip1 post-UV radiation (and not IR) exposure. This was shown by the fact that an inhibitor of p38 MAPK reduced Wip1 induction in a dose-dependent manner in UV radiation-exposed (and not IR-exposed) A549 cells (Figure 2) (4). Therefore, p53 transcriptionally activates PPM1D after genotoxic stress, and the upstream mechanism depends on the type of genotoxic stress.
3.1.2. Cyclic AMP response element binding protein (CREB)
A conserved cyclic AMP response element (CRE) was identified in the human and mouse PPM1D promoter regions (Figure 1), and the binding of the CRE binding protein (CREB) to the PPM1D promoter in HEK 293 cells and human hepatocytes was determined in a genome-wide association study (22, 23), suggesting that CREB regulates the expression of Wip1. The positive regulation of PPM1D transcription by CREB was confirmed by reporter assays and ChIP experiments (19). In contrast to p53 regulation of Wip1, which occurs in response to genotoxic stress, CREB also regulates basal PPM1D transcription in cultured cells (Figure 2). In the absence of exogenous stress, a luciferase reporter construct with a mutated CRE in the PPM1D promoter showed reduced luciferase activity levels (by over 40%) compared to the wild type PPM1D promoter in colon cancer HCT-116 cells. Additionally, ChIP analysis in HCT-116 wild type and p53−/− cells revealed that CREB was bound to the PPM1D promoter region in the absence of stress and independent of the presence of p53, which further supports a role of CREB in basal PPM1D transcriptional regulation (19). Like p53, CREB also regulates PPM1D transcription after IR exposure. This was illustrated by the finding that the amount of CREB bound to the PPM1D promoter region increased two hours after IR. Surprisingly, results from experiments with mutated reporter constructs suggested that even though both p53 and CREB regulate PPM1D, they appear to do so independently (19). Thus, CREB facilitates the transcription of PPM1D basally and after DNA damage independently of p53.
3.1.3. Nuclear Factor-kappa B (NF-kappaB)
The fact that NF-kappaB is activated in response to stress (24) and that there is a conserved kappaB site in the PPM1D promoter region (Figure 1) suggested that NF-kappaB regulates levels of PPM1D transcription, which was validated by our group (5). Inhibition of NF-kappaB chemically or by siRNA directed to the p65 subunit reduced basal levels of Wip1 mRNA and protein in MCF-7 breast cancer cells, which indicates that NF-kappaB is responsible, at least in part, for basal Wip1 expression. Reporter constructs and ChIP assays showed that the kappaB site is required for full basal PPM1D promoter activity and that p65 binds directly to the PPM1D promoter region (5). Additionally, as shown in Figure 2, stimulation with lipopolysaccharide (LPS) or the cytokine Tumor Necrosis Factor-alpha (TNF-alpha), both well-known NF-kappaB activators, increased PPM1D mRNA levels (5, 6). These studies showed that constitutively active NF-kappaB in cancer cells (such as in MCF-7 breast cancer cells) or cytokine-activated NF-kappaB induced Wip1 expression. However, as NF-kappaB is activated in response to many types of stress including genotoxic stress, NF-kappaB may regulate Wip1 expression after other types of stress as well.
3.1.4. Estrogen Receptor-alpha (ERalpha)
Several lines of evidence indicate that Wip1 may be regulated in a steroid-dependent manner. First, Wip1 is over-expressed in a number of cancers regulated by steroids such as breast cancer and ovarian cancer (11, 13, 25). Additionally, a large proportion of primary breast tumors that exhibit overexpression of Wip1 also have high expression of ERalpha (12). Indeed, Han et al. showed that Wip1 is regulated by ERalpha. They found that ERalpha binds to the promoter region of PPM1D and treatment of MCF-7 cells with 17beta-estradiol (E2) and ectopic expression of ERalpha increased Wip1 expression levels (Figure 1 and Figure 2) (26). Furthermore, Wip1 has been shown to increase the activity of several nuclear hormone receptors, most likely by enhancing their association with Nuclear Receptor Coactivator-1 (NCOA1), NCOA2 or NCOA3 (27). Recent work has demonstrated that Wip1 induction by E2 and ERalpha promotes cell cycle progression and cell proliferation, thus establishing a positive feedback loop whereby E2 stimulates ERalpha–dependent induction of Wip1, which then further promotes the upregulation of Estrogen-dependent genes to augment cell proliferation (26). Thus, Wip1 is positively regulated by ERalpha and then elicits pro-tumorigenic effects by enhancing ERalpha signaling in breast cancer cells (26).
3.1.5. c-jun
As discussed above, p38 MAPK/p53 signaling is important for Wip1 induction after UV radiation exposure (4). However, a recent report by Song et al. showed that JNK/c-jun signaling contributes to Wip1 induction in A549 cells (28). A sequence matching a c-jun binding motif is located 283 bp upstream from the translation start site in the PPM1D promoter region, overlapping the CRE (Figure 1). Overexpression of c-jun enhanced UV radiation-induced Wip1 promoter activity, and Electophoretic Mobility Shift Assays (EMSAs) indicated that c-jun directly bound to the consensus sequence in the PPM1D promoter region (28). ChIP assays demonstrated that p53 was bound to the Wip1 promoter a short time after UV radiation exposure, and JNK-activated c-jun was bound later (28). Therefore, the JNK/c-jun pathway may contribute to the UV-induced induction of Wip1 (Figure 2).
3.1.6. E2F1
E2F1 is a member of the E2F family of transcription factors that regulates genes important for cell growth, cell differentiation and apoptosis (29–31). As shown in Figure 1, an E2F family binding motif is present in the PPM1D promoter at −652 base pairs from the translation start site (32). Hershko et al. showed that PPM1D is a target of E2F1 by RT-PCR and ChIP analysis (Figure 2). Furthermore, E2F1 modulates PPM1D transcription independently of p53, since cells in which E2F1 is conditionally activated and that have depleted p53 by siRNA show a similar increase in Wip1 expression compared to cells that fully express p53. The fact that E2F1 upregulated Wip1 in a p53-deficient lung carcinoma cell line, H1299, also indicated that E2F1 induction of Wip1 is independent of p53 (32). It may be worth noting, however, that these experiments were done in cell systems in which E2F1 is conditionally activated upon the addition of a 4-hydroxytamoxifen rather than through its intrinsic regulation.
3.2. Post-transcriptional regulation
3.2.1. p53-dependent shift in transcription start site
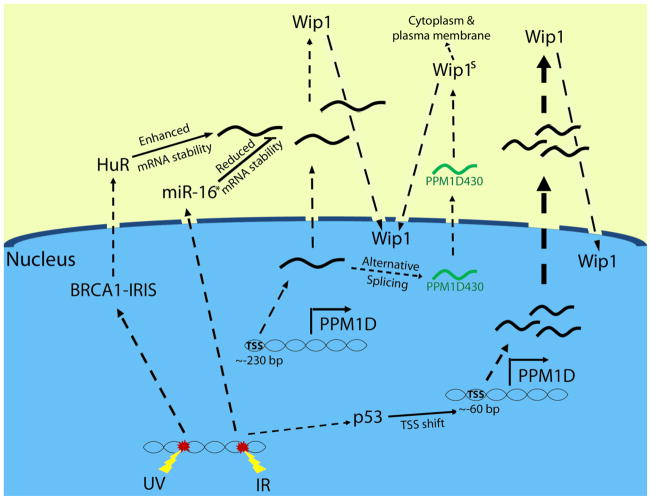
Detailed studies on p53-regulated Wip1 expression revealed that p53 also regulates Wip1 expression through post-transcriptional control (Figure 3). Rossi et al. identified two clusters of PPM1D transcription start sites, distinguished by their 5′UTR lengths (19). The longer 5′UTR transcripts had about 220 bases of highly structured mRNA preceding the translation initiation codon, whereas the shorter 5′UTR transcripts had about 65 bases before the translation initiation codon. The authors showed that CREB binding to the CRE site was associated with production of transcripts with longer 5′UTRs. However, in response to UV radiation or IR, p53 bound to the p53RE enhanced transcription of Wip1 transcripts with shorter 5′UTRs. In fact, RT-PCR-based analysis with specific primers showed that the majority of the PPM1D transcripts induced by either UV radiation or IR exposure were the shorter species. Since shorter transcripts may be more efficiently exported from the nucleus, this represents a post-transcriptional mechanism through which p53 enhances Wip1 expression (19).
Figure 3.
Regulation of Wip1 expression: emphasis on post-transcriptional mechanisms. The four mechanisms of post-transcriptional modulation of Wip1 expression are depicted. After DNA damage, BRCA1-IRIS enhances the expression of HuR, which stabilizes Wip1 mRNA leading to enhanced Wip1 expression. IR-induced miR-16 destabilizes Wip1 mRNA and decreases Wip1 expression. The Wip1 transcript is alternatively spliced to form a shorter variant (“PPM1D430”), which leads to an enhanced expression of a smaller Wip1 protein (“Wip1s”) that localizes to the nucleus, cytoplasm and plasma membrane. IR induces a p53-dependent shift in the transcriptional start site (“TSS”) of PPM1D, which produces a shorter transcript allowing for more efficient export from the nucleus.
3.2.2. Regulation by microRNA-16
Recent work has shown that microRNAs, including miR-16, are regulated by and play a role in the DDR and, furthermore, that p53 affects the level of specific microRNAs through transcription-dependent and independent mechanisms (33–35). Specifically, miR-16 expression was found to be induced following DNA damage through p53-dependent promotion of its processing and maturation (34). The mature form of miR-16 binds to the 3′UTR of Wip1 mRNA and promotes its degradation (Figure 3) (36). Expression of miR-16 decreased Wip1 mRNA levels through binding to a 12-base region of the 3′UTR, since the activity of a reporter construct with a mutation in this region was not affected by miR-16 expression. Furthermore, the authors showed that miR-16 inhibits Wip1 expression in a time-dependent manner after exposure to neocarzinostatin (NCS) (36). Thus, miR-16 may negatively regulate Wip1 expression by decreasing Wip1 mRNA stability.
Taken together, these studies describe a mechanism by which p53 inhibits Wip1 expression – through induction of miR-16. However, this is contrary to the previously demonstrated induction of Wip1 by p53 after DNA damage (3, 19). The differences may reflect cell type-specific differences in miR-16 expression levels or the complex temporal regulation of Wip1 expression. Indeed, Zhang et al. emphasized that miR-16 reduced Wip1 protein levels at early times after NCS addition, but later, miR-16 expression was reduced, and both p53 and Wip1 protein levels continued to increase (36). The p53-dependent inhibition of Wip1 expression through increased miR-16 levels may be an important mechanism that prevents tumorigenesis, since Wip1 promotes tumorigenesis and miR-16 has tumor suppressor properties (10, 18, 37–39).
3.2.3. BRCA1-IRIS enhances PPM1D mRNA stability
Another mechanism by which PPM1D mRNA stability is affected is through BRCA-IRIS. Unlike the tumor suppressor BRCA1, which is derived from the same locus, BRCA-IRIS is oncogenic and promotes cell cycle progression, cell survival and proliferation (40–42). In contrast to destabilization by miR-16, BRCA-IRIS enhances PPM1D mRNA stability. It does so through the enhancement of the RNA binding protein HuR, which stabilizes mRNA by binding to the 3′ untranslated region (43, 44). It was shown that, in breast cancer cells, overexpression or knock down of BRCA-IRIS leads, respectively, to increased or decreased Wip1 expression, as determined by immunoblotting and RT-PCR. Immunoblot analysis showed that BRCA-IRIS stabilized HuR and increased its cytoplasmic expression. This occurs presumably through NF-kappaB, since NF-kappaB is known to regulate HuR expression and overexpression of BRCA-IRIS leads to nuclear localization of the p65 subunit of NF-kappaB. HuR then binds to Wip1 mRNA to prevent its degradation, which leads to an enhanced Wip1 expression (Figure 3) (43). As BRCA1-IRIS, HuR, and Wip1 are all induced after UV radiation exposure, it is thought that this mechanism is activated after UV radiation and that Wip1 inhibits UV radiation-induced apoptosis through inhibition of p38 MAPK and p53, which will be discussed in Section 4 (43).
3.2.4. Alternative splicing – PPM1D430
As depicted in Figure 3, the final currently known post-transcriptional regulation of Wip1 expression to be discussed is alternative splicing. Alternative splicing of Wip1 was discovered by the cloning of a longer than predicted cDNA fragment of Wip1 that included a region beyond exon 5 that encoded a stop codon (45). Subsequently, two alternatively spliced products were found, one corresponding to the full length protein (PPM1D605) and the second to a shorter version (by 175 base pairs, PPM1D430). The two spliced products share the entire catalytic domain plus an additional 42 residues, suggesting that both proteins have phosphatase activity, which was confirmed by an in vitro phosphatase activity assay with purified recombinant proteins (45). Furthermore, PPM1D605 and PPM1D430 are similarly induced in MCF-7 cells after adriamycin exposure, and both are functional phosphatases, since either the specific knock down of PPM1D430 or the knock-down of both variants resulted in enhanced p53 phosphorylation and stabilization (45). On the other hand, other clues suggest functional differences between the two variants. For example, whereas PPM1D605 expression is ubiquitous, PPM1D430 is highly expressed in the testis and in leukocytes. Furthermore, in MCF-7 cells, both variants localize to the nucleus, whereas in T47D cells, PPM1D605 localizes to the nucleus but PPM1D430 was found in the cytoplasm, the plasma membrane, and the nucleus (Figure 3). Given the differences in tissue expression patterns and subcellular localization, the two variants may exhibit functional differences, but this is currently unknown.
4. WIP1 SUPPRESSES THE STRESS RESPONSE: UPDATE
Although Wip1 has been shown to be induced after a variety of stresses, most of the studies on this phosphatase have focused on its role in the DDR and in the response to oncogenic stress. Wip1 directly binds to, dephosphorylates, and inactivates a number of stress signaling proteins, leading to increased tumorigenesis and inhibition of the processes of DNA repair, cell cycle arrest, and apoptosis. Recent studies have shown that Wip1 also suppresses inflammatory signaling, facilitates senescence escape, and maintains stemness in stem cells. These aspects of Wip1 signaling will be reviewed below and are summarized in Figure 4.
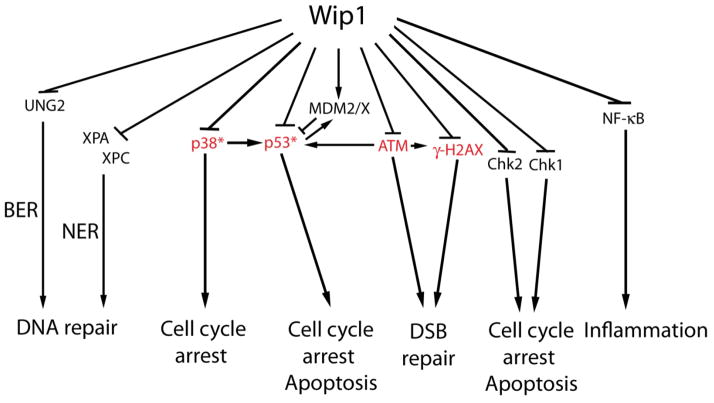
Figure 4.
The targets and functional consequences of Wip1 signaling. Wip1 signals through direct dephosphorylation of target proteins, which prevents apoptosis, inhibits DNA repair, reverses cell cycle arrest, and reduces inflammation. The proteins highlighted in red suppress tumorigenesis, and Wip1 inhibition of these proteins promotes tumorigenesis. *p53 and p38 MAPK enhance the tumor suppressor effects of p16Ink4a/p19Arf.
4.1. Wip1 targets stress-induced proteins
Extensive studies characterizing the substrate specificity of Wip1 have shown that it is capable of dephosphorylating phospho-serines or phospho-threonines residing in two different motifs. The first identified motif was the diphosphorylated peptide X−1pTX+1pYX+3, where X−1 can be any amino acid, X+1 is any aliphatic amino acid, and X+3 is any amino acid except proline (46). This motif is found in p38 MAPK and uracil-DNA glycosylase (UNG2), both of which are targeted by Wip1. In addition, the p(S/T)Q motif was identified as a Wip1 target sequence (47). The S/T in this motif is phosphorylated by the PI3K-like kinases such as ATM. By using phosphopeptides and recombinant Wip1 protein, Wip1 was shown to efficiently dephosphorylate p(S/T)Q peptides corresponding to several protein targets of the PI3K-like kinases, including p53 phosphorylated on Serine 15 and Chk1 phosphorylated on Serine 345 (47). Further biochemical analysis showed that the optimal p(S/T)Q phosphopeptide substrate for Wip1 is (D/E)(D/E)X−1p(S/T)QX+2, where X−1 is any amino acid and X+2 is any amino acid except basic amino acids or Proline (47). Indeed, many studies have characterized specific Wip1 targets exhibiting these motifs, which include p38, p53, ATM, gamma-H2AX, Chk1, Chk2, UNG2, Mdm2, MdmX, NF-kappaB, XPA, and XPC (Table 1).
Table 1.
Identified Wip1 targets
| Target protein | Phosphorylation site | Functional Consequence | Reference |
|---|---|---|---|
| p38 | T180 | Reduction of p38 activity | (4) |
| p53 | S15 | Reduction of p53 activity | (75) |
| UNG2 | T6 | Inhibition of BER | (109) |
| Chk1 | S345 | Reduction of Chk1 activity | (75) |
| Chk2 | T68, S19*, S33/S35* T432* | Reduction of Chk2 activity | (70) |
| ATM | S1981 | Reduction of ATM activity | (9, 73) |
| Mdm2 | S395 | Mdm2 stabilization and p53 destabilization | |
| MdmX | S403 | MdmX stabilization and p53 destabilization | (50) |
| gamma-H2AX | S139 | Inhibition of recruitment of repair factors to DNA breaks and DNA double strand break repair | (7, 8, 56) |
| NF-kappaB | S536 | Reduction of NF-kappaB activity | (6) |
| XPA | S196* | Inhibition of NER | (52) |
| XPC | S892* | Inhibition of NER | (52) |
indicates in vitro evidence only.
Some of these Wip1 targets and the subsequent biological consequences of their inactivation by Wip1 have been reviewed previously, including p38, p53, ATM, Chk1, Chk2, Mdm2 and UNG2 (25). However, since last reviewed, additional Wip1 targets have been identified. Therefore, the interaction of Wip1 with MdmX, gamma-H2AX, NF-kappaB, XPA, and XPC will be discussed below.
4.1.1. New Wip1 targets (MdmX, NF-kappaB, XPA, XPC)
MDMX, like Wip1, is a negative regulator of p53. Upregulation of MDMX leads to an autoregulatory negative feedback loop, since MDMX in complex with MDM2 enhances ubiquitination and degradation of the p53 protein (48, 49). Because of its negative regulation of p53, MDMX facilitates down-regulation of the DNA damage response after stress and is thought to promote tumorigenesis. The finding that Wip1 enhances the negative effects of MDMX on p53 signaling indicates that Wip1 and MDMX cooperate to reduce p53 activity and stability. Zhang et al. showed that Wip1 directly dephosphorylated MDMX on serine 403, a site that is phosphorylated by ATM after genotoxic stress (Table 1), and additionally inhibited phosphorylation of MDMX of S342 and S376 by an indirect mechanism (50). Overexpression of Wip1 reduced the levels of pS403-MDMX after NCS treatment, indicating that Wip1 inhibits MdmX phosphorylation. Co-immunoprecipitation experiments showed that Wip1 and MdmX physically interact, and recombinant Wip1 was able to dephosphorylate a phosphopeptide corresponding to MdmX (pS403). Furthermore, Wip1−/− mouse embryonic fibroblasts (MEFs) treated with NCS had reduced levels of MdmX compared to Wip1+/+ MEFs (which was dependent on ATM), and overexpression of Wip1 inhibited MdmX degradation after NCS. Additionally, Wip1 inhibits p53 through MdmX and enhances the interaction of MdmX with ubiquitin-specific peptidase 7 (USP7), which deubiquitinates and stabilizes MdmX (50). Therefore, Wip1 dephosphorylation of MdmX and enhancement of MdmX-dependent p53 degradation provides an additional mechanism by which Wip1 returns the cell to homeostasis after stress and promotes tumorigenesis (Figure 4).
As shown in Figure 4, NF-kappaB was identified as a Wip1 target by Chew et al. (6). By using an NF-kappaB reporter and by monitoring the expression levels of NF-kappaB targets, they showed that overexpression of Wip1 or knockdown of Wip1 with siRNA reduced or increased, respectively, NF-kappaB activation resulting from treatment with Interleukin-1 (IL-1) or TNF-alpha. This enhancement of NF-kappaB activity by Wip1 knockdown was not due to alterations in the expression of IkappaBalpha (an inhibitor of NF-kappaB), as determined by immunoblot, or in the DNA binding of the p50:p65 NF-kappaB heterodimeric complex after stimulation, as determined by EMSA. On the other hand, evaluation of the levels of phosphorylation of NF-kappaB p65 on Ser536 (pS536-p65) showed that overexpression of Wip1 or knockdown of Wip1 reduced or enhanced, respectively, the levels of pS536-p65 after stimulation by TNF-alpha. This effect was independent of p38 MAPK since the p38 inhibitor, SB202190, did not rescue the heightened pS536-p65 levels in Wip1-depleted cells. Furthermore, overexpression of Wip1 inhibited the TNF-alpha-induced binding of p65 to its transcriptional cofactor p300. Finally, recombinant Wip1 dephosphorylated immunopurified pS536-p65 in an in vitro phosphatase assay, indicating that Wip1 can directly dephosphorylate pS536 of p65 (Table 1 and Figure 4). The authors concluded that Wip1 reduces the expression of cytokines such as TNFalpha after stimulation through inhibition of NF-kappaB activity (6).
Xeroderma pigmentosum complementation group A and C (XPA and XPC) are critical proteins of the Nucleotide Excision Repair (NER) pathway, which resolves many of the UV radiation-induced DNA lesions such as cyclobutane pyrimidine dimers (CPDs) (51). XPA is recruited with other NER proteins and plays an important role in the assembly of repair proteins at the damage site. On the other hand, XPC facilitates the recognition of UV radiation-induced lesions specifically in transcriptionally inactive DNA, a process named global genome-NER (GG-NER) (51). As shown in Table 1 and Figure 4, a potential mechanism for the inhibition of NER by Wip1 is through dephosphorylation of XPA and XPC (52). XPA and XPC have p(S/T)Q motifs, which are consensus sequences for Wip1 that are targeted by PI3K-like kinases after DNA damage. Recombinant Wip1 protein efficiently dephosphorylated phosphopeptides corresponding to XPA S196 and XPC S892 (the PI3K-like kinase targets) in an in vitro phosphatase assay, and this was dependent on Wip1 phosphatase activity, since a phosphatase-dead mutant form of Wip1 (Wip1-D314A) was unable to dephosphorylate the phosphopeptides. The authors conclude that Wip1 may inhibit NER (see section 4.3.2.1 below) through dephosphorylation of XPA and XPC (52).
4.2. Wip1 inhibits DSB repair and cell cycle checkpoints through gamma-H2AX
gamma-H2AX is a critical component of the DDR and is important in maintaining genomic stability (53, 54). In the DDR, gamma-H2AX facilitates the recruitment of repair factors such as Mdc1, Rad51, and NBS1 to the sites of double strand breaks (DSBs), which is necessary for efficient repair (55). These repair factors, including gamma-H2AX, form foci when viewed with immunofluorescence microscopy that are removed once the repair is complete and DDR signaling is silenced. True to its role as a stress signaling silencer, Wip1 plays an important role in reversing gamma-H2AX levels through dephosphorylation. Three research groups (7, 56, 57), including ours, have shown that gamma-H2AX is a target of Wip1. Wip1 was found in the chromatin-bound subcellular fraction, and immunofluorescence analysis showed that Wip1 co-localized with foci formed by the DNA repair factors gamma-H2AX and Mdc1 (8). In vitro phosphatase assays showed that Wip1 dephosphorylated gamma-H2AX phosphopetides, and co-immunoprecipitation assays showed that Wip1 physically interacts with H2AX. Furthermore, deletion of Wip1 enhanced gamma-H2AX levels after genotoxic stress, whereas overexpression of Wip1 reduced gamma-H2AX levels, which was ATM-independent (7, 8, 56). Importantly, our group showed that formation of gamma-H2AX foci, per se, was not affected by Wip1 overexpression, since gamma-H2AX foci were observed in cells overexpressing Wip1 shortly after IR exposure (10 minutes) at levels comparable to those in control cells (7). Together, these studies showed that Wip1 is a gamma-H2AX phosphatase and is important for removal of gamma-H2AX after genotoxic stress (Table 1 and Figure 4).
The functional consequences of gamma-H2AX dephosphorylation by Wip1 include inhibition of cell cycle checkpoints and inhibition of DNA repair (Figure 4). Following exposure to the genotoxic agent doxorubicin, cells overexpressing Wip1 progressed into mitosis and exhibited an absence of gamma-H2AX or Mdc1 foci, indicating that overexpression of Wip1 prevents the G2/M cell cycle checkpoint after genotoxic stress through inhibition of gamma-H2AX (8). In addition, overexpression of Wip1 impaired foci formation of many repair factors including Nbs1, Rad50, and 53BP1 (7, 56). Impaired recruitment of DNA repair factors should also reduce DNA repair, and this was shown to be true in cells overexpressing Wip1. As measured by a neutral comet assay, IR-induced DSBs persisted in Wip1-overexpressing cells compared to similarly treated control cells (7). In a similar manner, site-specific DSBs initiated by the homing endonuclease, I-PpoI, exhibited impaired recruitment of γ-H2AX and were inefficiently repaired in control-treated MCF7 cells, which express high levels of Wip1, compared with Wip1 shRNA-treated cells, in which Wip1 was depleted (56). Therefore, accumulation of gamma-H2AX at DSBs is critical for cell cycle arrest and efficient DNA repair and overexpression of Wip1 inhibits both processes. Additionally, gamma-H2AX plays a role in tumorigenesis (58), and a role for Wip1 in removal of gamma-H2AX after oncogenic stress and in the absence of DNA damage will be discussed below in section 4.5.
4.3. Wip1 and the UV response
Signaling in response to genotoxic stress is specific to the type of stress and the nature of DNA damage. For example, the most lethal type of damage induced by IR exposure is DSBs, whereas UV radiation exposure induces CPDs and other types of DNA damage. Therefore, it is not surprising that regulation of Wip1 and Wip1 signaling after UV radiation exposure is different than after IR exposure. Several recent studies have focused on elucidating Wip1 signaling after UV radiation exposure, which will be discussed in this section.
4.3.1. UV-induced Wip1 expression
4.3.1.1. c-Jun vs. p53
As discussed in section 3, both c-jun and p53 have been shown to contribute to Wip1 induction after UV radiation exposure (4, 28). Song et al. showed by ChIP analysis that p53 binding to the PPM1D promoter peaks very soon after UV radiation exposure, after approximately 1 hour, whereas c-jun binding peaks at about 5 hours after UV radiation exposure in A549 cells. Additionally, JNK kinase activity upstream from c-jun is required for c-jun-mediated Wip1 induction, as indicated by reduced Wip1 expression in the presence of a JNK inhibitor. Therefore, in addition to the BRCA1-IRIS-dependent mechanism discussed in section 3.1.2, two additional signaling pathways lead to UV radiation-induced Wip1 expression – the JNK/c-jun pathway and the p38 MAPK/p53 pathway (28).
4.3.1.2. NER-dependence
Oh et al. showed that following UV radiation exposure, Wip1 is induced in a p53-dependent manner at later time points, and that Wip1 induction requires an intact NER system (59). The authors found that cells deficient in xeroderma pigmentosum complementation group B (XPB), a helicase that plays an essential role in NER (51), failed to induce Wip1 expression. Since NER is not functional in XPB cells, transcription-blocking UV radiation-induced DNA lesions persist at high levels. As the p53-dependent induction of large gene size targets, including Wip1, previously had been shown to be inhibited at high doses of UV radiation (60), the authors speculated that unrepaired lesions in the PPM1D gene in XPB cells prevented its transcription and expression (59). Therefore, NER is required for the repair of UV radiation-induced DNA lesions within the PPM1D gene and the subsequent p53-dependent induction at later time points after UV radiation exposure.
4.3.1.3. Dose-dependence
Induction of Wip1 after UV radiation also appears to be dose-dependent (60, 61). Low doses of Ultraviolet C (UVC, 240–290 nm) radiation, such as 5 J/m2, induce Wip1 expression as expected. In contrast, Wip1 is not induced and its expression actually decreases after a high dose of UVC radiation, such as 50 J/m2 (61). Using actinomycin D to inhibit transcription and MG132 to inhibit proteasome-dependent protein degradation, Xia et al. showed that Wip1 is down-regulated after high doses of UVC at the transcription and protein level. They determined that this is independent of p53, but did not elucidate the mechanism by which PPM1D transcription is inhibited and the Wip1 protein is destabilized. However, as described above, the lack of Wip1 induction may result from the presence of unrepaired DNA lesions at this high dose of UVC radiation and the consequent block of PPM1D transcription (59, 60). Additionally, the sustained phosphorylation of p53 on S15 and increased level of apoptosis that result from exposure to high doses of UVC radiation are consistent with the lack of Wip1 induction (61). The authors conclude that whereas Wip1 is induced in a p53-dependent manner in response to a low dose of UV radiation, high doses of UVC radiation suppress Wip1 induction in a p53-independent manner and disrupt the p53/Wip1 negative feedback loop (61).
4.3.2. Wip1 dampens the UV response
4.3.2.1. Inhibition of Nucleotide Excision Repair (NER)
Nguyen et al. were the first to show inhibition of NER by Wip1 (Figure 4) (52). Wip1−/− MEFs had almost a three-fold increase in NER activity as measured by the ability of the cells to repair a UV-irradiated luciferase reporter plasmid. Likewise, overexpression of wild type Wip1 (but not a nonfunctional mutant Wip1) inhibited NER by over 30%, and this was p53-independent, since overexpression of Wip1 in a p53−/− cell line, Saos2, similarly reduced NER activity by over 40%. As expected, Wip1 overexpression had no effect on the low level of reporter activity exhibited by cells deficient in xeroderma pigmentosum complementation group D (XPD), but did significantly reduce reporter activity in XPD cells complemented with an XPD expression vector. Similar effects of Wip1 on NER activity were also shown by measuring the levels of unrepaired CPDs (51, 52). Wip1−/− MEFs exhibited lower amounts of remaining CPDs after UV radiation compared to Wip1+/+ MEFs, whereas Wip1 overexpressing cells exhibited increased amounts of CPDs after UV radiation compared to the control cells. This effect was also shown in vivo by comparing CPD levels in the skin of UV irradiated Wip1−/− and Wip1+/+ mice. Wip1−/− MEFs and the skin from Wip1−/− mice showed a reduction in UV radiation-induced apoptosis, which was measured by the presence of cleaved Caspase-3 and PARP proteins as well as TUNEL staining (52). Thus, as shown in Figure 4, Wip1 negatively regulates NER after UV radiation (presumably through inactivation of XPA and XPC as discussed in section 4.1.1), and consequently enhances apoptosis through the higher levels of remaining CPDs.
4.3.2.2. Dampening of the DDR
Although not directly produced by UV radiation exposure, DSBs can be formed indirectly through, for example, the stalling of replication forks. This is evident by the measurement of DSBs by comet assays and the presence of phosphorylated DDR proteins such as gamma-H2AX, ATM, and NBS1 after UV radiation exposure (51, 59), which Oh et al. showed was independent of an intact NER system, as this occurred in cells deficient in the NER helicase, XPB (59). Since Wip1 is induced after UV radiation exposure in normal human fibroblasts, it most likely plays a role in dephosphorylating and inactivating the DDR proteins after UV radiation. As described in section 4.3.1.2, Wip1 was not induced by UV radiation exposure in XPB fibroblasts (59). Additionally, immunofluorescence assays revealed that foci corresponding to phosphorylated and activated DDR proteins such as gamma-H2AX and phospho-ATM were resolved 24 hours after UV radiation exposure when Wip1 expression was high. However, these foci persisted in the XPB cells. Since Wip1 was not induced in XPB cells and since DDR proteins such as gamma-H2AX and phospho-ATM are Wip1 targets, the lack of Wip1 expression is likely to contribute to persistent DDR activation in XPB cells. Likewise, UV radiation-induced Wip1 in normal cells is able to reverse DDR signaling, which is important for the cells to return to homeostasis (59).
4.4. Functional consequences of Wip1 after stress
4.4.1. Inhibition of apoptosis
Apoptosis, or programmed cell death, ensures that injured cells do not replicate and pass on genomic DNA damage to progeny cells. Therefore, the inability to proceed with apoptosis after severe damage is disadvantageous for an organism, since replication of lesion-containing DNA can lead to the immortalization of genomic mutations and, potentially, to tumor formation (62). Indeed, many tumors exhibit defects in proteins important in apoptosis; the p53 protein is a good example, since it functions in promoting apoptosis after stress and is one of the most frequently mutated genes in human tumors (63). On the other hand, the cell must also have proteins that negatively regulate apoptotic pathways in the event that stress-induced damage is efficiently repaired, and Wip1 is one of these proteins (Figure 4).
p53-dependent apoptosis is well characterized and involves numerous signaling proteins to induce the caspase-dependent and mitochondrial apoptotic pathways (reviewed in (64)). Since p53 is a central regulator of apoptosis, Wip1 inhibition of p53 by direct dephosphorylation and through the inactivation of upstream proteins promotes cell survival by suppressing apoptosis (Figure 4). For example, Wip1 suppresses myc-induced apoptosis through inhibition of the ATM-p53 apoptotic pathway and thereby promotes tumorigenesis. Myc-induced lymphoma tumors from Wip1−/− mice exhibited elevated levels of apoptosis compared to control mice, and the additional deletion of ATM or p53 lowered the level of apoptosis to values similar to those seen in tumors from ATM−/− or p53−/− mice (9). Thus, Wip1 inactivates ATM signaling to p53 and promotes cell survival when myc expression levels are high. Additionally, intestinal stem cells from Wip1−/− mice exhibited higher levels of apoptosis that correlated with higher levels of activated p53, thus demonstrating that Wip1 inhibits intestinal stem cell apoptosis through inhibition of p53 (65). Furthermore, Kong et al. showed that depletion of Wip1 in MCF-7 breast cancer cells lead to higher levels of apoptosis in response to doxorubicin. This most likely occurs through a p53-dependent mechanism, since these cells also showed higher levels of active p53 and Bax expression (66). Similarly, Parssinen et al. showed that Wip1 depletion by siRNA induced apoptosis in breast cancer cells that had functional p53. This study was done in the absence of exogenous stress, which indicates that Wip1 plays a role in breast cancer cell survival through the inhibition of p53 (67). Therefore, Wip1 inhibition of p53 appears to be a major mechanism by which Wip1 inhibits apoptosis.
The inhibition of apoptosis by Wip1 also reflects its negative feedback on the p38 MAPK/p53 pathway. Takekawa et al. showed that Wip1-overexpressing A549 cells (p53-proficient) exhibited a significant decrease in apoptosis after UV radiation compared to control cells, and this decrease was dependent on Wip1 phosphatase activity, since over-expression of a Wip1 phosphatase dead mutant had no effect (4). Furthermore, Schito et al. illustrated that the DP thymocyte population (double-positive for CD4 and CD8) and not the DN population (double-negative for CD4 and CD8) from Wip1−/− mice exhibited a higher rate of apoptosis (68). The relevance to Wip1 negative feedback on the p38 MAPK/p53 pathway resides in the fact that active p38 MAPK facilitates early thymocyte differentiation, but its inactivation (and consequent inactivation of p53) is necessary for thymocyte differentiation from DN to DP. Hence, persistent activation of p38 MAPK in mice deficient in Wip1 led to both defective T-cell maturation to the DP state and increased apoptosis in the DP population due to higher levels of p53 activation (68).
Wip1 also inhibits E2F1-induced and UV radiation-induced apoptosis, presumably through p38 MAPK (32, 43). Hershko et al. showed that active E2F1 induces the phosphorylation and activation of p38 MAPK through apoptosis signal-regulating kinase 1 (ASK1), which promotes apoptosis (32). As discussed in Section 3, the authors also identified Wip1 as a target of E2F1. E2F1-induced Wip1 is required for p38 MAPK dephosphorylation, since depletion of Wip1 by siRNA lead to prolonged p38 MAPK phosphorylation. This Wip1-dependent suppression of activated p38 MAPK led to suppression of E2F1-induced apoptosis, as indicated by an increase in the percentage of apoptotic cells when Wip1 is knocked down by siRNA (32). Additionally, Chock et al. showed that BRCA1-IRIS-induced Wip1 inhibited UV radiation-induced apoptosis through inhibition of p38 MAPK/p53 signaling (43). Overexpression of BRCA1-IRIS attenuates p38 MAPK and p53 activation and suppressed cell death after UV radiation exposure, whereas BRCA1-IRIS knock down by siRNA enhances p38 MAPK and p53 activation. Since BRCA1-IRIS upregulates Wip1, the authors conclude that the mechanism by which BRCA1-IRIS inhibits UV radiation-induced apoptosis is through Wip1 inhibition of the p38 MAPK/p53 pathway (43).
The effect of Wip1 over-expression on Chk2-dependent apoptosis has also been examined. Chk1 and Chk2 function in apoptosis-signaling pathways, as both proteins phosphorylate p53 and E2F1 and facilitate both p53-independent and p53-dependent apoptosis (69). Phosphorylation of T68 of Chk2 is important for activation of its kinase activity and induction of apoptosis: phosphorylation of this residue was reduced after IR in Wip1-overexpressing cells compared to control cells (70). As anticipated, overexpression of Wip1 inhibited apoptosis by decreasing phosphorylation of Chk2 on T68 and several other sites of phsophorylation following exposure to IR and, furthermore, that Chk2 and Wip1 interact through the (S/T)Q domain of Chk2 and the N-terminal domain of Wip1 (70, 71).
Finally, Xia et al. showed that Wip1 not only protects from DNA damage-induced apoptosis, but also protects the cell from apoptosis induced by various types of oxidative and ribotoxic stress (72). Wip1−/− MEFs exhibited higher levels of apoptosis (as measured by flow cytometry, nuclei morphology, and Caspase and PARP cleavage) after treatment with anisomycin, etoposide, H202, UV radiation, or staurosporine. Furthermore, Wip1−/− MEFs have higher levels of active p38 MAPK and JNK/c-jun pathways. Therefore, the authors concluded that Wip1 is a global regulator of apoptosis through the inhibition of p38 MAPK, JNK, and c-jun pathways (72).
4.4.2. Recovery and rescue from cell cycle arrest
Like apoptosis, cell cycle arrest is critical in preventing DNA replication of a damaged genome. Functional studies have confirmed the role of Wip1 in the G1/S, intra-S, and G2/M checkpoints (Figure 4). Oiva-Trastoy et al. showed that cells overexpressing Wip1 have a dampened G2 arrest (as measured by flow cytometry) after IR (73). Additionally, flow cytometry analysis of Wip1−/− and wild type MEFs revealed that MEFs deficient in Wip1 have a significantly higher G2:M ratio (52.7) compared to the wild type (17.4), indicating that Wip1 plays a role in the transition from the G2 cell cycle phase to mitosis (74). Wip1 also functions in the G1 checkpoint, since synchronized Wip1−/− MEFs exhibited an exaggerated G1 arrest after IR compared to wild type due to higher p53 activity (74). These data indicate that Wip1 is likely to participate in the reversal of signaling important for the maintenance, and not initiation, of the G1 checkpoint (through p53 inhibition) since there was no difference at earlier time points following IR (74). Finally, evidence suggests that Wip1 has a function in the intra S-phase cell cycle checkpoint after stress. Lu et al. showed that Wip1 plays a role in regulating the intra-S phase after IR and UV radiation in cells either overexpressing Wip1 or that have Wip1 knocked down by siRNA (75). Therefore, Wip1 helps to return the cell to normal cell cycle progression after stress by negatively regulating pathways involved G2/M, G1/S, and the intra-S phase checkpoints.
More recently, Lindqvist et al. showed that Wip1 is required for G2 checkpoint recovery competence (76). Cells were synchronized in G2, treated with Doxorubicin and then treated with ATM/ATR, Chk1/MAPKAP2, or p38 MAPK inhibitors to promote exit from G2 arrest. In this assay, cells with endogenous levels of Wip1 efficiently recovered from the G2 arrest, whereas cells with depleted Wip1 failed to do so (76). Furthermore, induction of Wip1 using a Tet-on system after DNA damage in the G2-arrested cells promoted checkpoint recovery. This was dependent on the phosphatase activity of Wip1, since expression of a phosphatase-dead mutant (D314A) failed to have this effect on G2 checkpoint recovery. The mechanism by which Wip1 maintains checkpoint recovery competence is through inhibition of p53, since failed G2 checkpoint recovery in Wip1 depleted cells was rescued by p53 depletion. Additionally, like HCT116 wild type cells, Wip1-depleted HCT116 p53−/− cells were able to recover from G2 arrest, indicating that activated p53 in Wip1-depleted cells caused the failure to recover from G2 arrest. Specifically, the authors showed that Wip1 inhibits the transcriptional repression of target genes, specifically Cyclin B1, by p53 (76). Therefore, Wip1 plays a critical role through p53 to facilitate checkpoint recovery. Furthermore, the authors concluded that this may be another mechanism by which Wip1 promotes tumorigenesis – by maintaining the ability of a cell to proliferate during oncogenic stress (76).
4.4.3. Function of Wip1 in stem cells
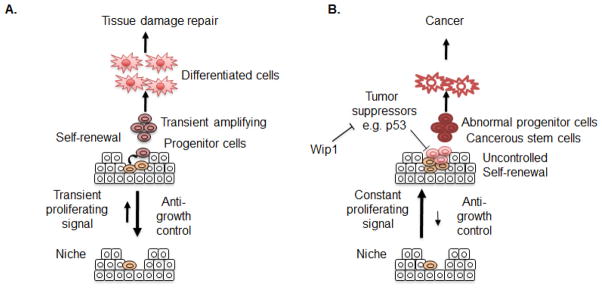
Adult stem cells act as a reservoir for the regeneration of damaged tissue. These stem cells normally reside in a quiescent state in order to minimize unnecessary replication to presumably prolong their life-span, and they undergo ‘self-renewal’ (e.g., a specific type of cellular proliferation, maintaining ‘stemness’) only when damaged tissue needs to be regenerated (77). In this circumstance, stem cells are transiently stimulated to undergo asymmetric self-renewal, which generates one identical stem cell and one progenitor cell. The generation of the identical stem cells is critical for maintaining the stem cell pool, whereas the new progenitor cells proliferate (hence their name ‘transient amplifying (TA) cells’) and differentiate to replace the damaged tissue (78) (Figure 5A). Maintaining a balance between sufficient numbers of progenitor cells and stem cells is critical for individual homeostasis (e.g., aging or cancer) (Figure 5). For example, recent studies have revealed that the reduced proliferative potential of stem cells and the subsequent depletion of the stem cell pool are correlated with age-related diseases such as rheumatoid arthritis, osteoarthritis, refractory hypertension (78, 79). More clearly, an animal model revealed that premature depletion of the stem cell pool by massive tissue damage and subsequent major repair is strongly associated with the premature aging phenotype (80). These data indicate that dysfunction of stem cells or depletion of an adequate pool of stem cells may be closely related to aging and/or aging related symptoms (such as degenerative diseases). On the other hand, uncontrolled proliferation of stem cells by constant stimulation or loss of tumor suppressor activity is suggested to be the origin of tumors such as intestinal cancer (81) and malignant astrocytoma (82). Therefore, well-defined tumor suppressors such as p16Ink4a, 19Arf and p53 have been extensively studied in a variety of stem cell systems and have a role in regulating the cellular fate of stem cells such as aging or senescence (83–86), tumorigenesis (87, 88) and stemness or self-renewality (89, 90).
Figure 5.
Model of stem cell self-renewal under normal and cancerous conditions. Stem cells normally remain quiescent due to strong anti-growth control from their surrounding microenvironment, termed a niche. A transient proliferation signal stimulates self-renewal to support tissue regeneration. Maintenance of stemness is promoted by Wip1. Under cancerous conditions, internal mutations and alteration of the microenvironment promote abnormal self-renewal of stem cells favoring growth. Unless tumor suppressors detect such an abnormality, cancerous stem cells (CSCs) or cancer initiating cells (TICs) may be generated from stem cells after a secondary genetic mutation. The proliferation of CSCs or TICs may be promoted by overexpression of Wip1.
Recently, Wip1 was found to be expressed in intestinal stem cells and to inhibit p53-mediated apoptosis, which promotes tumor initiation (65). Thus, abrogation of functionality of the tumor suppressor p53 by Wip1 in constantly proliferating intestinal stem cells of the APCmin mouse model was sufficient to induce cancerous stem cells or cancer initiating stem cells, which was demonstrated to be the origin of intestinal cancer (81) (Figure 5B). In contrast, continuous activation of the stress response in stem cells lacking Wip1 also had an effect on the life span of stem cells. For instance, the neural stem/progenitor cell (NPC) population in Wip1-deficient mice was significantly reduced in the subventricular zone (SVZ), which is where the NPCs normally reside. Consequently, the neural stemness (especially toward neurogenesis) was lowered, and this was shown to be dependent on p53 (91). On the other hand, human mesenchymal stem cells (hMSCs) undergo premature senescence in culture due to accumulated oxidative stress and/or constant DNA damage signaling (92, 93) (unpublished data). Stable expression of Wip1 in hMSCs lowers the stress response and overcomes premature senescence, which maintains the multi-differentiation potential (94). This suggests that modulation of the stress response by Wip1 expression in hMSCs is important for their stemness.
Dysregulation of Wip1 signaling may also play a role in aging. Wip1 expression is significantly reduced in aged mice (65, 95), and Wip1-deficient mice have a premature aging phenotype (74). The lack of Wip1 expression in aged cells results in hyperactivation of the stress response, for example through activation of p53 or p16Ink4a (96, 97). Such an accumulated stress response in stem cells by loss of Wip1 function and subsequent relief of stress signaling inhibition may also be responsible for the premature aging phenotype of Wip1 deficient mice (74).
4.4.4. Inhibition of inflammation
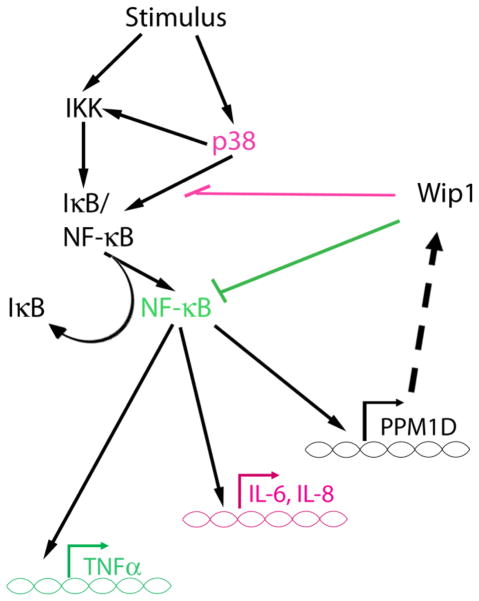
Analysis of the phenotype of Wip1−/− mice provided the first clue that Wip1 may play a role in inflammation. Wip1−/− mice are more susceptible to infection and have a higher frequency of ulcerated skin lesions, and furthermore, splenic lymphocytes harvested from Wip1−/− mice exhibited a dampened proliferative response after stimulation with phytohemagglutinin (PHA) and LPS compared to the wild type controls (74). Additionally, splenocytes from LPS-injected Wip1−/− mice showed a “hyperactivated” phenotype (6). Indeed, inhibition of inflammation was reported as an additional function of Wip1 by Chew et al. (6). Wip1 was shown to suppress the expression of targets of the transcription factor NF-kappaB after cytokine stimulation. Two mechanisms were identified – one through direct inhibition of NF-kappaB (described in section 4.1.1) and the other through inhibition of a previously identified Wip1 target, p38 MAPK. Inhibition of cytokine-induced p38 MAPK activity by Wip1 is presumed to occur through dephosphorylation of T180 of p38, its previously defined function. The functional consequence of Wip1 inhibition of cytokine-induced NF-kappaB and p38 activity was shown to be a reduction in the expression of NF-kappaB target genes such as interleukin-6 (IL-6), TNF-alpha, and IL-8. In particular, Wip1 reduction of TNF-alpha expression was shown to be through dephosphorylation of NF-kappaB, whereas Wip1 inhibition of IL-6 and IL-8 expression occurred through inhibition of p38 (Figure 6). Therefore, Wip1 appears to play a role in inflammation similar to that after genotoxic stress, i.e., Wip1 helps to turn off the inflammatory response (Figure 4 and Figure 6). Furthermore, taken together with the study from Lowe et al. (5), this describes another negative feedback loop involving Wip1. Wip1 is induced by NF-kappaB activated by inflammatory stimuli, and then Wip1 negatively regulates NF-kappaB signaling to reduce the inflammatory response (Figure 6).
Figure 6.
Wip1 negative feedback in inflammatory signaling. Schematic adapted from (6). Wip1 expression is induced by NF-kappaB. Wip1 inhibits production of TNFalpha by dephosphorylating and inhibiting the NF-kappaB subunit p65 (highlighted in green) and inhibits IL-6 and IL-8 production through p38 MAPK (highlighted in purple).
4.5. Wip1 attenuates the response to oncogenic stress and promotes tumorigenesis
4.5.1. Cooperation of Wip1 with oncogenes in tumorigenesis
Many human cancers exhibit amplification of the PPM1D gene or overexpression of the Wip1 protein. As shown in Table 2, these include breast cancer, ovarian clear cell adenocarcinoma, neuroblastoma, medulloblastoma, gastric carcinoma and pancreatic adenocarcinoma (11, 13, 14, 17, 98–106). Further work using mouse models validated Wip1 as an oncogene, and this has been reviewed by Lu et al. (25). For example, Bulavin et al. showed that deletion of PPM1D inhibited Erbb2 and Hras1 tumor formation through a p38 MAPK and p16/p19 mechanism (18). However, Wip1 is a proto-oncogene, since overexpression of Wip1 along with other oncogenes enhances in vitro oncogenic transformation and tumorigenesis whereas Wip1 overexpression alone has no effect (17, 37). In addition, several recent studies described below have investigated Wip1 as an oncogene.
Table 2.
Wip1 amplification and overexpression in human cancers
| Organ type | p53 mutation | PPM1D amplification | Wip1 overexpression | Associated prognosis | Reference |
|---|---|---|---|---|---|
| Neuroblastoma | 1/8 | 4/4* | 9/32** | Poorer | (102) |
| Medulloblastoma | 6/16 | 3/11** | Poorer | (103) | |
| 7/11 | 16/33** | (105) | |||
| 24/47 | 7/9, 148/168 | (104) | |||
| Gastric carcinoma | 39/53** | Poorer | (14) | ||
| Pancreatic adenocarcinoma | 13/31 | Poorer | (106) | ||
| Breast adenocarcinoma | 1/8 | 37/326 | 7/8 | Poorer | (17) |
| 1/10 | 11/117 | 10/11 | Poorer | (98) | |
| 5/10* | 5/5 | (99) | |||
| 3/5 | 3/3 | none | (11) | ||
| Ovarian clear cell carcinoma | 8/20 | 5/5 | Poorer | (101) | |
| 9/89 | *** | (13) | |||
| Prostate cancer | 0/3 | 3/3 | (11) |
In most cases, tumors or cell lines that showed PPM1D amplification were tested for Wip1 overexpression (the exception is indicated by **).
Cancer cell lines (not primary tumors) were tested.
Tumors were tested for Wip1 expression regardless of PPM1D amplification.
Specific numbers were not indicated, but there was a significant correlation of PPM1D amplification and Wip1 overexpression.
Several reports have characterized the incidence of PPM1D amplification and Wip1 overexpression in breast cancer (11, 13, 17, 98–100), and there is a positive correlation between PPM1D amplification and poor prognosis (98). More recently, Lambros et al. showed that Wip1 overexpression was more prevalent than PPM1D amplification, since Wip1 was overexpressed in 21% of the tumors, whereas PPM1D amplification was present in only 6% of the tumors (107). It should be noted, however, that the reported 6% PPM1D amplification in breast cancer is low compared to previous reports (11, 13, 17, 98–100), and the reason for this inconsistency is unclear. Lambros et al. also showed a significant correlation of Wip1 overexpression and tumor grade. Most of the tumors that exhibited Wip1 overexpression were also negative for p53 (107), which is consistent with other reports that p53 is frequently wild-type and not overexpressed, whereas mutant p53 frequently is overexpressed (98). On the other hand, and unlike the results of Rauta et al. (98), PPM1D amplification was not correlated with a poor prognosis (107). These results imply that, in addition to PPM1D amplification, up-regulation of Wip1 expression is important in breast cancer and suggest that Wip1 inhibition of p53 may be an important mechanism by which Wip1 promotes breast tumorigenesis.
In addition to breast cancer, studies of additional types of cancers, such as neuroendocrine tumors (NETs) and medulloblastomas, indicate that inhibition of p53 may be an important function for Wip1 in tumorigenesis. NETs rarely have mutated p53, but Hu et al. showed that the majority of pancreatic neuroendocrine tumors express higher levels of p53-negative regulators such as Wip1 (108). In this study, the authors showed that 51% of pancreatic NETs have amplified PPM1D, which correlates with a high level of Wip1 mRNA expression (108). Wip1 is also overexpressed in medulloblastomas (103, 104), and Castellino et al. showed that Wip1 is amplified in 7 out of 11 primary medulloblastoma tumors analyzed, which correlated with high Wip1 expression. Furthermore, Wip1 inhibits basal and etoposide-induced p53-mediated apoptosis in medulloblastoma cells (105), implicating Wip1 as a positive regulator of medulloblastoma cell survival.
Several recent in vitro studies have described a role for Wip1 in tumorigenesis. As mentioned above, Wip1 plays an important role in intestinal tumorigenesis through regulation of stem cell apoptosis, and furthermore, Zhang et al. showed that Wip1 also plays an important role in mammary tumor stem cell proliferation (36). Although Wip1 expression was slightly increased in primary tumor cells from MMTV-Erbb2 mice, Wip1 expression in mammary tumor stem cells isolated from these mice exhibited a 3-to-5 fold increase. On the other hand, expression of miR-16, a negative regulator of Wip1 (see section 3.1.1), was reduced by about 70-to-80% in mammary tumor stem cells whereas it was only slightly reduced in the whole population of primary tumor cells (36). These results suggest that inhibition of miR-16 leads to Wip1 overexpression in mammary tumor stem cells. Furthermore, overexpression of miR-16 or depletion of Wip1 inhibited mammary tumor stem cell proliferation. Wip1 overexpression also partially reversed the miR-16-dependent inhibition of proliferation, indicating that Wip1 facilitates mammary tumor stem cell proliferation (36). Although the mechanism is unknown, it would be interesting to test whether Wip1 inhibits p53-dependent apoptosis in mammary tumor stem cells, as is the case in intestinal stem cells (65).
In vitro studies from Chock et al. showed that Wip1 can cooperate with BRCA1-IRIS to transform human mammary epithelial (HME) cells. As described in section 3.2.2, BRCA1-IRIS enhances Wip1 expression through HuR-dependent stabilization of Wip1 mRNA. The authors also showed that expression of BRCA1-IRIS and Wip1 significantly increased cellular transformation of HME cells, as measured by the number of cells and colonies in soft agar assay after co-overexpresson of BRCA1-IRIS and Wip1 (43). In addition, co-overexpression of Ras and Wip1, which previously had been shown to cooperate in tumorigensis (17, 18), increased cell and colony numbers in the soft agar assay to the same extent as BRCA1-IRIS/Wip1 co-overexpression (43). Since overexpression of Wip1 in the mammary tissue of mice did not induce mammary tumorigenesis (37), this study indicates that Wip1, through an unknown mechanism, cooperates with BRCA1-IRIS to promote tumorigenesis.
4.5.2. Dampening of the oncogene-induced DDR response (gamma-H2AX)
Another way Wip1 may promote tumorigenesis is through inhibition of gamma-H2AX and the DDR. Through its role in the DDR, gamma-H2AX also plays an important role in tumorigenesis (58). The DDR acts as a barrier to oncogenic transformation and is triggered after oncogenic stress (15, 16). As discussed in section 4.2, Wip1 dephosphorylates gamma-H2AX after genotoxic stress. Our group also showed that Wip1 reduced gamma-H2AX levels in cells experiencing oncogenic stress (7). Wild-type MEFs transformed with E1A and Ras exhibited a limited number of gamma-H2AX foci. However, Wip1−/− MEFs transformed with E1A and Ras exhibited a much larger number of basal gamma-H2AX foci (7). Furthermore, gamma-H2AX levels remained high in Wip1−/− E1A/Ras MEFs compared to wild-type E1A/Ras MEFs after IR exposure. Wip1 dephosphorylation of gamma-H2AX, therefore, may be a mechanism by which Wip1 promotes genomic instability, facilitates cellular transformation, and promotes tumorigenesis.
5. CONCLUSION
The phosphatase Wip1 is induced by a variety of types of stress, and, although several mechanisms regulating Wip1 expression have been described recently, it is likely that many aspects of the regulation of Wip1 remain to be discovered. Although Wip1 was originally described as a p53-induced protein, recent studies have shown that p53-independent regulation of Wip1 also plays a critical role in the cellular response to stress. p53-independent mechanisms for Wip1 regulation may be especially important in tumor cells. Many of the tumors that overexpress Wip1 also lack over-expressed (mutant) p53, suggesting that they possess wild-type but functionally inactive p53. This is not surprising given that inhibition of p53, through direct dephosphorylation and stabilization of its negative regulators Mdm2 and MdmX, is an important role for Wip1. As described in this review, the stabilization of Wip1 mRNA by BRCA1-IRIS and transcriptional upregulation by CREB and NF-kappaB provide examples of p53-independent mechanisms leading to increased Wip1 levels.
Many aspects of Wip1 signaling in the stress response consist of feedback loops. For example, DNA damage activates p53, which induces Wip1 expression. Once expressed, Wip1 negatively regulates p53 either by dephosphorylation and subsequent inactivation or by destabilization of the p53 protein through Mdm2. The same is true for NF-kappaB and upstream regulators of Wip1 such as ATM and p38. Depending on the nature of the stress and the cellular context, NF-kappaB, ATM or p38 each can upregulate Wip1, and Wip1 then dephosphorylates and inactivates NF-kappaB, ATM or p38. These negative feedback loops in Wip1 signaling highlight its physiological role in response to stress, i.e. it dampens the stress response to help return the cell to a homeostatic state.
Given that Wip1 has a role in tumorigenesis, Wip1 is an attractive chemotherapeutic target. Therefore, full understanding of the regulation of Wip1 expression and characterization of the targets of Wip1 phosphatase activity in specific types of tumors may aid the development of more effective anti-tumor therapies. In conclusion, Wip1 negatively regulates stress responses, such as apoptosis, DNA repair, cell cycle arrest and inflammation, and as a consequence, cooperates with activated oncogenes in promoting tumorigenesis.
Acknowledgments
The authors would like to acknowledge support by NASA NNJ06ZSA001N (AJF) and NNX09AU95G (AJF), the NIEHS (JL) and NCI (EA and SJM) intramural programs, and the Korea Science and Engineering Foundation (KOSEF) (201131076) (HC and MOL) and the Priority Research Centers Program (2010-0028297) (HC and MOL).
References
- 1.Karlsson-Rosenthal C, Millar JB. Cdc25: mechanisms of checkpoint inhibition and recovery. Trends Cell Biol. 2006;16(6):285–92. doi: 10.1016/j.tcb.2006.04.002. S0962-8924(06)00099-7 [pii] [DOI] [PubMed] [Google Scholar]
- 2.Lavin MF, Kozlov S. ATM activation and DNA damage response. Cell Cycle. 2007;6(8):931–42. doi: 10.4161/cc.6.8.4180. 4180 [pii] [DOI] [PubMed] [Google Scholar]
- 3.Fiscella M, Zhang H, Fan S, Sakaguchi K, Shen S, Mercer WE, Vande Woude GF, O’Connor PM, Appella E. Wip1, a novel human protein phosphatase that is induced in response to ionizing radiation in a p53-dependent manner. Proc Natl Acad Sci U S A. 1997;94(12):6048–53. doi: 10.1073/pnas.94.12.6048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Takekawa M, Adachi M, Nakahata A, Nakayama I, Itoh F, Tsukuda H, Taya Y, Imai K. p53-inducible wip1 phosphatase mediates a negative feedback regulation of p38 MAPK-p53 signaling in response to UV radiation. EMBO J. 2000;19(23):6517–26. doi: 10.1093/emboj/19.23.6517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lowe JM, Cha H, Yang Q, Fornace AJ., Jr Nuclear factor-kappaB (NF-kappaB) is a novel positive transcriptional regulator of the oncogenic Wip1 phosphatase. J Biol Chem. 2010;285(8):5249–57. doi: 10.1074/jbc.M109.034579. M109.034579 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chew J, Biswas S, Shreeram S, Humaidi M, Wong ET, Dhillion MK, Teo H, Hazra A, Fang CC, Lopez-Collazo E, Bulavin DV, Tergaonkar V. WIP1 phosphatase is a negative regulator of NF-kappaB signalling. Nat Cell Biol. 2009;11(5):659–66. doi: 10.1038/ncb1873. ncb1873 [pii] [DOI] [PubMed] [Google Scholar]
- 7.Cha H, Lowe JM, Li H, Lee JS, Belova GI, Bulavin DV, Fornace AJ., Jr Wip1 directly dephosphorylates gamma-H2AX and attenuates the DNA damage response. Cancer Res. 2010;70(10):4112–22. doi: 10.1158/0008-5472.CAN-09-4244. 0008-5472.CAN-09-4244 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Macurek L, Lindqvist A, Voets O, Kool J, Vos HR, Medema RH. Wip1 phosphatase is associated with chromatin and dephosphorylates gammaH2AX to promote checkpoint inhibition. Oncogene. 2010;29(15):2281–91. doi: 10.1038/onc.2009.501. onc2009501 [pii] [DOI] [PubMed] [Google Scholar]
- 9.Shreeram S, Demidov ON, Hee WK, Yamaguchi H, Onishi N, Kek C, Timofeev ON, Dudgeon C, Fornace AJ, Anderson CW, Minami Y, Appella E, Bulavin DV. Wip1 phosphatase modulates ATM-dependent signaling pathways. Mol Cell. 2006;23(5):757–64. doi: 10.1016/j.molcel.2006.07.010. S1097-2765(06)00490-4 [pii] [DOI] [PubMed] [Google Scholar]
- 10.Shreeram S, Hee WK, Demidov ON, Kek C, Yamaguchi H, Fornace AJ, Jr, Anderson CW, Appella E, Bulavin DV. Regulation of ATM/p53-dependent suppression of myc-induced lymphomas by Wip1 phosphatase. J Exp Med. 2006;203(13):2793–9. doi: 10.1084/jem.20061563. jem.20061563 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li J, Yang Y, Peng Y, Austin RJ, van Eyndhoven WG, Nguyen KC, Gabriele T, McCurrach ME, Marks JR, Hoey T, Lowe SW, Powers S. Oncogenic properties of PPM1D located within a breast cancer amplification epicenter at 17q23. Nat Genet. 2002;31(2):133–4. doi: 10.1038/ng888. ng888 [pii] [DOI] [PubMed] [Google Scholar]
- 12.Yu E, Ahn YS, Jang SJ, Kim MJ, Yoon HS, Gong G, Choi J. Overexpression of the wip1 gene abrogates the p38 MAPK/p53/Wip1 pathway and silences p16 expression in human breast cancers. Breast Cancer Res Treat. 2007;101(3):269–78. doi: 10.1007/s10549-006-9304-y. [DOI] [PubMed] [Google Scholar]
- 13.Tan DS, Lambros MB, Rayter S, Natrajan R, Vatcheva R, Gao Q, Marchio C, Geyer FC, Savage K, Parry S, Fenwick K, Tamber N, Mackay A, Dexter T, Jameson C, McCluggage WG, Williams A, Graham A, Faratian D, El-Bahrawy M, Paige AJ, Gabra H, Gore ME, Zvelebil M, Lord CJ, Kaye SB, Ashworth A, Reis-Filho JS. PPM1D is a potential therapeutic target in ovarian clear cell carcinomas. Clin Cancer Res. 2009;15(7):2269–80. doi: 10.1158/1078-0432.CCR-08-2403. 1078-0432.CCR-08-2403 [pii] [DOI] [PubMed] [Google Scholar]
- 14.Fuku T, Semba S, Yutori H, Yokozaki H. Increased wild-type p53-induced phosphatase 1 (Wip1 or PPM1D) expression correlated with downregulation of checkpoint kinase 2 in human gastric carcinoma. Pathol Int. 2007;57(9):566–71. doi: 10.1111/j.1440-1827.2007.02140.x. PIN2140 [pii] [DOI] [PubMed] [Google Scholar]
- 15.Bartkova J, Horejsi Z, Koed K, Kramer A, Tort F, Zieger K, Guldberg P, Sehested M, Nesland JM, Lukas C, Orntoft T, Lukas J, Bartek J. DNA damage response as a candidate anti-cancer barrier in early human tumorigenesis. Nature. 2005;434(7035):864–70. doi: 10.1038/nature03482. nature03482 [pii] [DOI] [PubMed] [Google Scholar]
- 16.Mallette FA, Gaumont-Leclerc MF, Ferbeyre G. The DNA damage signaling pathway is a critical mediator of oncogene-induced senescence. Genes Dev. 2007;21(1):43–8. doi: 10.1101/gad.1487307. 21/1/43 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bulavin DV, Demidov ON, Saito S, Kauraniemi P, Phillips C, Amundson SA, Ambrosino C, Sauter G, Nebreda AR, Anderson CW, Kallioniemi A, Fornace AJ, Jr, Appella E. Amplification of PPM1D in human tumors abrogates p53 tumor-suppressor activity. Nat Genet. 2002;31(2):210–5. doi: 10.1038/ng894. ng894 [pii] [DOI] [PubMed] [Google Scholar]
- 18.Bulavin DV, Phillips C, Nannenga B, Timofeev O, Donehower LA, Anderson CW, Appella E, Fornace AJ., Jr Inactivation of the Wip1 phosphatase inhibits mammary tumorigenesis through p38 MAPK-mediated activation of the p16(Ink4a)-p19(Arf) pathway. Nat Genet. 2004;36(4):343–50. doi: 10.1038/ng1317. ng1317 [pii] [DOI] [PubMed] [Google Scholar]
- 19.Rossi M, Demidov ON, Anderson CW, Appella E, Mazur SJ. Induction of PPM1D following DNA-damaging treatments through a conserved p53 response element coincides with a shift in the use of transcription initiation sites. Nucleic Acids Res. 2008;36(22):7168–80. doi: 10.1093/nar/gkn888. gkn888 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Canman CE, Lim DS, Cimprich KA, Taya Y, Tamai K, Sakaguchi K, Appella E, Kastan MB, Siliciano JD. Activation of the ATM kinase by ionizing radiation and phosphorylation of p53. Science. 1998;281(5383):1677–9. doi: 10.1126/science.281.5383.1677. [DOI] [PubMed] [Google Scholar]
- 21.Bulavin DV, Saito S, Hollander MC, Sakaguchi K, Anderson CW, Appella E, Fornace AJ., Jr Phosphorylation of human p53 by p38 kinase coordinates N-terminal phosphorylation and apoptosis in response to UV radiation. EMBO J. 1999;18(23):6845–54. doi: 10.1093/emboj/18.23.6845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Conkright MD, Guzman E, Flechner L, Su AI, Hogenesch JB, Montminy M. Genome-wide analysis of CREB target genes reveals a core promoter requirement for cAMP responsiveness. Mol Cell. 2003;11(4):1101–8. doi: 10.1016/s1097-2765(03)00134-5. S1097276503001345 [pii] [DOI] [PubMed] [Google Scholar]
- 23.Zhang X, Odom DT, Koo SH, Conkright MD, Canettieri G, Best J, Chen H, Jenner R, Herbolsheimer E, Jacobsen E, Kadam S, Ecker JR, Emerson B, Hogenesch JB, Unterman T, Young RA, Montminy M. Genome-wide analysis of cAMP-response element binding protein occupancy, phosphorylation, and target gene activation in human tissues. Proc Natl Acad Sci U S A. 2005;102(12):4459–64. doi: 10.1073/pnas.0501076102. 0501076102 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hayden MS, Ghosh S. Signaling to NF-kappaB. Genes Dev. 2004;18(18):2195–224. doi: 10.1101/gad.1228704. 18/18/2195 [pii] [DOI] [PubMed] [Google Scholar]
- 25.Lu X, Nguyen TA, Moon SH, Darlington Y, Sommer M, Donehower LA. The type 2C phosphatase Wip1: an oncogenic regulator of tumor suppressor and DNA damage response pathways. Cancer Metastasis Rev. 2008;27(2):123–35. doi: 10.1007/s10555-008-9127-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Han HS, Yu E, Song JY, Park JY, Jang SJ, Choi J. The estrogen receptor alpha pathway induces oncogenic Wip1 phosphatase gene expression. Mol Cancer Res. 2009;7(5):713–23. doi: 10.1158/1541-7786.MCR-08-0247. 1541-7786.MCR-08-0247 [pii] [DOI] [PubMed] [Google Scholar]
- 27.Proia DA, Nannenga BW, Donehower LA, Weigel NL. Dual roles for the phosphatase PPM1D in regulating progesterone receptor function. J Biol Chem. 2006;281(11):7089–101. doi: 10.1074/jbc.M511839200. M511839200 [pii] [DOI] [PubMed] [Google Scholar]
- 28.Song JY, Han HS, Sabapathy K, Lee BM, Yu E, Choi J. Expression of a homeostatic regulator, Wip1 (wild-type p53-induced phosphatase), is temporally induced by c-Jun and p53 in response to UV irradiation. J Biol Chem. 2010;285(12):9067–76. doi: 10.1074/jbc.M109.070003. M109.070003 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.DeGregori J. The genetics of the E2F family of transcription factors: shared functions and unique roles. Biochim Biophys Acta. 2002;1602(2):131–50. doi: 10.1016/s0304-419x(02)00051-3. S0304419X02000513 [pii] [DOI] [PubMed] [Google Scholar]
- 30.McClellan KA, Slack RS. Specific in vivo roles for E2Fs in differentiation and development. Cell Cycle. 2007;6(23):2917–27. doi: 10.4161/cc.6.23.4997. 4997 [pii] [DOI] [PubMed] [Google Scholar]
- 31.Polager S, Ginsberg D. E2F - at the crossroads of life and death. Trends Cell Biol. 2008;18(11):528–35. doi: 10.1016/j.tcb.2008.08.003. S0962-8924(08)00214-6 [pii] [DOI] [PubMed] [Google Scholar]
- 32.Hershko T, Korotayev K, Polager S, Ginsberg D. E2F1 modulates p38 MAPK phosphorylation via transcriptional regulation of ASK1 and Wip1. J Biol Chem. 2006;281(42):31309–16. doi: 10.1074/jbc.M601758200. M601758200 [pii] [DOI] [PubMed] [Google Scholar]
- 33.Hu H, Gatti RA. MicroRNAs: new players in the DNA damage response. J Mol Cell Biol. 2010 doi: 10.1093/jmcb/mjq042. mjq042 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Suzuki HI, Yamagata K, Sugimoto K, Iwamoto T, Kato S, Miyazono K. Modulation of microRNA processing by p53. Nature. 2009;460(7254):529–33. doi: 10.1038/nature08199. nature08199 [pii] [DOI] [PubMed] [Google Scholar]
- 35.Boominathan L. The tumor suppressors p53, p63, and p73 are regulators of microRNA processing complex. PLoS One. 2010;5(5):e10615. doi: 10.1371/journal.pone.0010615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang X, Wan G, Mlotshwa S, Vance V, Berger FG, Chen H, Lu X. Oncogenic Wip1 phosphatase is inhibited by miR-16 in the DNA damage signaling pathway. Cancer Res. 2010;70(18):7176–86. doi: 10.1158/0008-5472.CAN-10-0697. 0008-5472.CAN-10-0697 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Demidov ON, Kek C, Shreeram S, Timofeev O, Fornace AJ, Appella E, Bulavin DV. The role of the MKK6/p38 MAPK pathway in Wip1-dependent regulation of ErbB2-driven mammary gland tumorigenesis. Oncogene. 2007;26(17):2502–6. doi: 10.1038/sj.onc.1210032. 1210032 [pii] [DOI] [PubMed] [Google Scholar]
- 38.Cittelly DM, Das PM, Salvo VA, Fonseca JP, Burow ME, Jones FE. Oncogenic HER2{Delta}16 suppresses miR-15a/16 and deregulates BCL-2 to promote endocrine resistance of breast tumors. Carcinogenesis. 2010;31(12):2049–57. doi: 10.1093/carcin/bgq192. bgq192 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gatt ME, Zhao JJ, Ebert MS, Zhang Y, Chu Z, Mani M, Gazit R, Carrasco DE, Dutta-Simmons J, Adamia S, Minvielle S, Tai YT, Munshi NC, Avet-Loiseau H, Anderson KC, Carrasco DR. MicroRNAs 15a/16-1 function as tumor suppressor genes in multiple myeloma. Blood. 2010 doi: 10.1182/blood-2009-11-253294. blood-2009-11-253294 [pii] [DOI] [PubMed] [Google Scholar]
- 40.ElShamy WM, Livingston DM. Identification of BRCA1-IRIS, a BRCA1 locus product. Nat Cell Biol. 2004;6(10):954–67. doi: 10.1038/ncb1171. ncb1171 [pii] [DOI] [PubMed] [Google Scholar]
- 41.Pettigrew CA, French JD, Saunus JM, Edwards SL, Sauer AV, Smart CE, Lundstrom T, Wiesner C, Spurdle AB, Rothnagel JA, Brown MA. Identification and functional analysis of novel BRCA1 transcripts, including mouse Brca1-Iris and human pseudo-BRCA1. Breast Cancer Res Treat. 2010;119(1):239–47. doi: 10.1007/s10549-008-0256-2. [DOI] [PubMed] [Google Scholar]
- 42.Nakuci E, Mahner S, Direnzo J, ElShamy WM. BRCA1-IRIS regulates cyclin D1 expression in breast cancer cells. Exp Cell Res. 2006;312(16):3120–31. doi: 10.1016/j.yexcr.2006.06.021. S0014-4827(06)00224-2 [pii] [DOI] [PubMed] [Google Scholar]
- 43.Chock K, Allison JM, Elshamy WM. BRCA1-IRIS overexpression abrogates UV-induced p38MAPK/p53 and promotes proliferation of damaged cells. Oncogene. 2010;29(38):5274–85. doi: 10.1038/onc.2010.262. onc2010262 [pii] [DOI] [PubMed] [Google Scholar]
- 44.Doller A, Pfeilschifter J, Eberhardt W. Signalling pathways regulating nucleo-cytoplasmic shuttling of the mRNA-binding protein HuR. Cell Signal. 2008;20(12):2165–73. doi: 10.1016/j.cellsig.2008.05.007. S0898-6568(08)00152-6 [pii] [DOI] [PubMed] [Google Scholar]
- 45.Chuman Y, Kurihashi W, Mizukami Y, Nashimoto T, Yagi H, Sakaguchi K. PPM1D430, a novel alternative splicing variant of the human PPM1D, can dephosphorylate p53 and exhibits specific tissue expression. J Biochem. 2009;145(1):1–12. doi: 10.1093/jb/mvn135. mvn135 [pii] [DOI] [PubMed] [Google Scholar]
- 46.Yamaguchi H, Minopoli G, Demidov ON, Chatterjee DK, Anderson CW, Durell SR, Appella E. Substrate specificity of the human protein phosphatase 2Cdelta, Wip1. Biochemistry. 2005;44(14):5285–94. doi: 10.1021/bi0476634. [DOI] [PubMed] [Google Scholar]
- 47.Yamaguchi H, Durell SR, Chatterjee DK, Anderson CW, Appella E. The Wip1 phosphatase PPM1D dephosphorylates SQ/TQ motifs in checkpoint substrates phosphorylated by PI3K-like kinases. Biochemistry. 2007;46(44):12594–603. doi: 10.1021/bi701096s. [DOI] [PubMed] [Google Scholar]
- 48.Batchelor E, Mock CS, Bhan I, Loewer A, Lahav G. Recurrent initiation: a mechanism for triggering p53 pulses in response to DNA damage. Mol Cell. 2008;30(3):277–89. doi: 10.1016/j.molcel.2008.03.016. S1097-2765(08)00240-2 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wu X, Bayle JH, Olson D, Levine AJ. The p53-mdm-2 autoregulatory feedback loop. Genes Dev. 1993;7(7A):1126–32. doi: 10.1101/gad.7.7a.1126. [DOI] [PubMed] [Google Scholar]
- 50.Zhang X, Lin L, Guo H, Yang J, Jones SN, Jochemsen A, Lu X. Phosphorylation and degradation of MdmX is inhibited by Wip1 phosphatase in the DNA damage response. Cancer Res. 2009;69(20):7960–8. doi: 10.1158/0008-5472.CAN-09-0634. 0008-5472.CAN-09-0634 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rastogi RP, Richa, Kumar A, Tyagi MB, Sinha RP. Molecular mechanisms of ultraviolet radiation-induced DNA damage and repair. J Nucleic Acids. 2010;2010:592980. doi: 10.4061/2010/592980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Nguyen TA, Slattery SD, Moon SH, Darlington YF, Lu X, Donehower LA. The oncogenic phosphatase WIP1 negatively regulates nucleotide excision repair. DNA Repair (Amst) 2010;9(7):813–23. doi: 10.1016/j.dnarep.2010.04.005. S1568-7864(10)00133-3 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.van Attikum H, Gasser SM. Crosstalk between histone modifications during the DNA damage response. Trends Cell Biol. 2009;19(5):207–17. doi: 10.1016/j.tcb.2009.03.001. S0962-8924(09)00060-9 [pii] [DOI] [PubMed] [Google Scholar]
- 54.Celeste A, Petersen S, Romanienko PJ, Fernandez-Capetillo O, Chen HT, Sedelnikova OA, Reina-San-Martin B, Coppola V, Meffre E, Difilippantonio MJ, Redon C, Pilch DR, Olaru A, Eckhaus M, Camerini-Otero RD, Tessarollo L, Livak F, Manova K, Bonner WM, Nussenzweig MC, Nussenzweig A. Genomic instability in mice lacking histone H2AX. Science. 2002;296(5569):922–7. doi: 10.1126/science.1069398. 1069398 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kinner A, Wu W, Staudt C, Iliakis G. Gamma-H2AX in recognition and signaling of DNA double-strand breaks in the context of chromatin. Nucleic Acids Res. 2008;36(17):5678–94. doi: 10.1093/nar/gkn550. gkn550 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Moon SH, Lin L, Zhang X, Nguyen TA, Darlington Y, Waldman AS, Lu X, Donehower LA. Wild-type p53-induced phosphatase 1 dephosphorylates histone variant gamma-H2AX and suppresses DNA double strand break repair. J Biol Chem. 2010;285(17):12935–47. doi: 10.1074/jbc.M109.071696. M109.071696 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Moon SH, Nguyen TA, Darlington Y, Lu X, Donehower LA. Dephosphorylation of gammaH2AX by WIP1: An important homeostatic regulatory event in DNA repair and cell cycle control. Cell Cycle. 2010;9(11) doi: 10.4161/cc.9.11.11810. 11810 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bonner WM, Redon CE, Dickey JS, Nakamura AJ, Sedelnikova OA, Solier S, Pommier Y. GammaH2AX and cancer. Nat Rev Cancer. 2008;8(12):957–67. doi: 10.1038/nrc2523. nrc2523 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Oh KS, Bustin M, Mazur SJ, Appella E, Kraemer KH. UV-induced histone H2AX phosphorylation and DNA damage related proteins accumulate and persist in nucleotide excision repair-deficient XP-B cells. DNA Repair (Amst) 2010 doi: 10.1016/j.dnarep.2010.09.004. S1568–7864(10)00309-5 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.McKay BC, Stubbert LJ, Fowler CC, Smith JM, Cardamore RA, Spronck JC. Regulation of ultraviolet light-induced gene expression by gene size. Proc Natl Acad Sci U S A. 2004;101(17):6582–6. doi: 10.1073/pnas.0308181101. 0308181101 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Xia Y, Yang Q, Gong X, Ye F, Liou YC. Dose-dependent mutual regulation between Wip1 and p53 following UVC irradiation*. Int J Biochem Cell Biol. 2010 doi: 10.1016/j.biocel.2010.12.009. S1357-2725(10)00421-8 [pii] [DOI] [PubMed] [Google Scholar]
- 62.Green DR, Evan GI. A matter of life and death. Cancer Cell. 2002;1(1):19–30. doi: 10.1016/s1535-6108(02)00024-7. S1535610802000247 [pii] [DOI] [PubMed] [Google Scholar]
- 63.Goh AM, Coffill CR, Lane DP. The role of mutant p53 in human cancer. J Pathol. 2011;223(2):116–26. doi: 10.1002/path.2784. [DOI] [PubMed] [Google Scholar]
- 64.Chipuk JE, Green DR. Dissecting p53-dependent apoptosis. Cell Death Differ. 2006;13(6):994–1002. doi: 10.1038/sj.cdd.4401908. 4401908 [pii] [DOI] [PubMed] [Google Scholar]
- 65.Demidov ON, Timofeev O, Lwin HN, Kek C, Appella E, Bulavin DV. Wip1 phosphatase regulates p53-dependent apoptosis of stem cells and tumorigenesis in the mouse intestine. Cell Stem Cell. 2007;1(2):180–90. doi: 10.1016/j.stem.2007.05.020. S1934-5909(07)00064-1 [pii] [DOI] [PubMed] [Google Scholar]
- 66.Kong W, Jiang X, Mercer WE. Downregulation of Wip-1 phosphatase expression in MCF-7 breast cancer cells enhances doxorubicin-induced apoptosis through p53-mediated transcriptional activation of Bax. Cancer Biol Ther. 2009;8(6) doi: 10.4161/cbt.8.6.7742. 7742 [pii] [DOI] [PubMed] [Google Scholar]
- 67.Parssinen J, Alarmo EL, Karhu R, Kallioniemi A. PPM1D silencing by RNA interference inhibits proliferation and induces apoptosis in breast cancer cell lines with wild-type p53. Cancer Genet Cytogenet. 2008;182(1):33–9. doi: 10.1016/j.cancergencyto.2007.12.013. S0165-4608(08)00005-8 [pii] [DOI] [PubMed] [Google Scholar]
- 68.Schito ML, Demidov ON, Saito S, Ashwell JD, Appella E. Wip1 phosphatase-deficient mice exhibit defective T cell maturation due to sustained p53 activation. J Immunol. 2006;176(8):4818–25. doi: 10.4049/jimmunol.176.8.4818. 176/8/4818 [pii] [DOI] [PubMed] [Google Scholar]
- 69.Roos WP, Kaina B. DNA damage-induced cell death by apoptosis. Trends Mol Med. 2006;12(9):440–50. doi: 10.1016/j.molmed.2006.07.007. S1471-4914(06)00168-7 [pii] [DOI] [PubMed] [Google Scholar]
- 70.Fujimoto H, Onishi N, Kato N, Takekawa M, Xu XZ, Kosugi A, Kondo T, Imamura M, Oishi I, Yoda A, Minami Y. Regulation of the antioncogenic Chk2 kinase by the oncogenic Wip1 phosphatase. Cell Death Differ. 2006;13(7):1170–80. doi: 10.1038/sj.cdd.4401801. 4401801 [pii] [DOI] [PubMed] [Google Scholar]
- 71.Yoda A, Xu XZ, Onishi N, Toyoshima K, Fujimoto H, Kato N, Oishi I, Kondo T, Minami Y. Intrinsic kinase activity and SQ/TQ domain of Chk2 kinase as well as N-terminal domain of Wip1 phosphatase are required for regulation of Chk2 by Wip1. J Biol Chem. 2006;281(34):24847–62. doi: 10.1074/jbc.M600403200. M600403200 [pii] [DOI] [PubMed] [Google Scholar]
- 72.Xia Y, Ongusaha P, Lee SW, Liou YC. Loss of Wip1 sensitizes cells to stress- and DNA damage-induced apoptosis. J Biol Chem. 2009;284(26):17428–37. doi: 10.1074/jbc.M109.007823. M109.007823 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Oliva-Trastoy M, Berthonaud V, Chevalier A, Ducrot C, Marsolier-Kergoat MC, Mann C, Leteurtre F. The Wip1 phosphatase (PPM1D) antagonizes activation of the Chk2 tumour suppressor kinase. Oncogene. 2007;26(10):1449–58. doi: 10.1038/sj.onc.1209927. 1209927 [pii] [DOI] [PubMed] [Google Scholar]
- 74.Choi J, Nannenga B, Demidov ON, Bulavin DV, Cooney A, Brayton C, Zhang Y, Mbawuike IN, Bradley A, Appella E, Donehower LA. Mice deficient for the wild-type p53-induced phosphatase gene (Wip1) exhibit defects in reproductive organs, immune function, and cell cycle control. Mol Cell Biol. 2002;22(4):1094–105. doi: 10.1128/MCB.22.4.1094-1105.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Lu X, Nannenga B, Donehower LA. PPM1D dephosphorylates Chk1 and p53 and abrogates cell cycle checkpoints. Genes Dev. 2005;19(10):1162–74. doi: 10.1101/gad.1291305. gad.1291305 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Lindqvist A, de Bruijn M, Macurek L, Bras A, Mensinga A, Bruinsma W, Voets O, Kranenburg O, Medema RH. Wip1 confers G2 checkpoint recovery competence by counteracting p53-dependent transcriptional repression. EMBO J. 2009;28(20):3196–206. doi: 10.1038/emboj.2009.246. emboj2009246 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Li L, Neaves WB. Normal stem cells and cancer stem cells: the niche matters. Cancer Res. 2006;66(9):4553–7. doi: 10.1158/0008-5472.CAN-05-3986. 66/9/4553 [pii] [DOI] [PubMed] [Google Scholar]
- 78.Colmegna I, Diaz-Borjon A, Fujii H, Schaefer L, Goronzy JJ, Weyand CM. Defective proliferative capacity and accelerated telomeric loss of hematopoietic progenitor cells in rheumatoid arthritis. Arthritis Rheum. 2008;58(4):990–1000. doi: 10.1002/art.23287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Oliveras A, Soler MJ, Martinez-Estrada OM, Vazquez S, Marco-Feliu D, Vila JS, Vilaro S, Lloveras J. Endothelial progenitor cells are reduced in refractory hypertension. J Hum Hypertens. 2008;22(3):183–90. doi: 10.1038/sj.jhh.1002304. 1002304 [pii] [DOI] [PubMed] [Google Scholar]
- 80.Ruzankina Y, Pinzon-Guzman C, Asare A, Ong T, Pontano L, Cotsarelis G, Zediak VP, Velez M, Bhandoola A, Brown EJ. Deletion of the developmentally essential gene ATR in adult mice leads to age-related phenotypes and stem cell loss. Cell Stem Cell. 2007;1(1):113–26. doi: 10.1016/j.stem.2007.03.002. S1934-5909(07)00005-7 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Barker N, Ridgway RA, van Es JH, van de Wetering M, Begthel H, van den Born M, Danenberg E, Clarke AR, Sansom OJ, Clevers H. Crypt stem cells as the cells-of-origin of intestinal cancer. Nature. 2009;457(7229):608–11. doi: 10.1038/nature07602. nature07602 [pii] [DOI] [PubMed] [Google Scholar]
- 82.Alcantara Llaguno S, Chen J, Kwon CH, Jackson EL, Li Y, Burns DK, Alvarez-Buylla A, Parada LF. Malignant astrocytomas originate from neural stem/progenitor cells in a somatic tumor suppressor mouse model. Cancer Cell. 2009;15(1):45–56. doi: 10.1016/j.ccr.2008.12.006. S1535-6108(08)00409-1 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Molofsky AV, Slutsky SG, Joseph NM, He S, Pardal R, Krishnamurthy J, Sharpless NE, Morrison SJ. Increasing p16INK4a expression decreases forebrain progenitors and neurogenesis during ageing. Nature. 2006;443(7110):448–52. doi: 10.1038/nature05091. nature05091 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Itahana K, Zou Y, Itahana Y, Martinez JL, Beausejour C, Jacobs JJ, Van Lohuizen M, Band V, Campisi J, Dimri GP. Control of the replicative life span of human fibroblasts by p16 and the polycomb protein Bmi-1. Mol Cell Biol. 2003;23(1):389–401. doi: 10.1128/MCB.23.1.389-401.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Shibata KR, Aoyama T, Shima Y, Fukiage K, Otsuka S, Furu M, Kohno Y, Ito K, Fujibayashi S, Neo M, Nakayama T, Nakamura T, Toguchida J. Expression of the p16INK4A gene is associated closely with senescence of human mesenchymal stem cells and is potentially silenced by DNA methylation during in vitro expansion. Stem Cells. 2007;25(9):2371–82. doi: 10.1634/stemcells.2007-0225. 2007-0225 [pii] [DOI] [PubMed] [Google Scholar]
- 86.Molofsky AV, Pardal R, Iwashita T, Park IK, Clarke MF, Morrison SJ. Bmi-1 dependence distinguishes neural stem cell self-renewal from progenitor proliferation. Nature. 2003;425(6961):962–7. doi: 10.1038/nature02060. nature02060 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Wang Y, Yang J, Zheng H, Tomasek GJ, Zhang P, McKeever PE, Lee EY, Zhu Y. Expression of mutant p53 proteins implicates a lineage relationship between neural stem cells and malignant astrocytic glioma in a murine model. Cancer Cell. 2009;15(6):514–26. doi: 10.1016/j.ccr.2009.04.001. S1535-6108(09)00114-7 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Stecca B, Ruiz i Altaba A. A GLI1-p53 inhibitory loop controls neural stem cell and tumour cell numbers. EMBO J. 2009;28(6):663–76. doi: 10.1038/emboj.2009.16. emboj200916 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Meletis K, Wirta V, Hede SM, Nister M, Lundeberg J, Frisen J. p53 suppresses the self-renewal of adult neural stem cells. Development. 2006;133(2):363–9. doi: 10.1242/dev.02208. 133/2/363 [pii] [DOI] [PubMed] [Google Scholar]
- 90.Liu Y, Elf SE, Miyata Y, Sashida G, Huang G, Di Giandomenico S, Lee JM, Deblasio A, Menendez S, Antipin J, Reva B, Koff A, Nimer SD. p53 regulates hematopoietic stem cell quiescence. Cell Stem Cell. 2009;4(1):37–48. doi: 10.1016/j.stem.2008.11.006. S1934-5909(08)00577-8 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Zhu YH, Zhang CW, Lu L, Demidov ON, Sun L, Yang L, Bulavin DV, Xiao ZC. Wip1 regulates the generation of new neural cells in the adult olfactory bulb through p53-dependent cell cycle control. Stem Cells. 2009;27(6):1433–42. doi: 10.1002/stem.65. [DOI] [PubMed] [Google Scholar]
- 92.Fehrer C, Brunauer R, Laschober G, Unterluggauer H, Reitinger S, Kloss F, Gully C, Gassner R, Lepperdinger G. Reduced oxygen tension attenuates differentiation capacity of human mesenchymal stem cells and prolongs their lifespan. Aging Cell. 2007;6(6):745–57. doi: 10.1111/j.1474-9726.2007.00336.x. ACE336 [pii] [DOI] [PubMed] [Google Scholar]
- 93.Miranda-Vilela AL, Akimoto AK, Alves PC, Pereira LC, Goncalves CA, Klautau-Guimaraes MN, Grisolia CK. Dietary carotenoid-rich pequi oil reduces plasma lipid peroxidation and DNA damage in runners and evidence for an association with MnSOD genetic variant -Val9Ala. Genet Mol Res. 2009;8(4):1481–95. doi: 10.4238/vol8-4gmr684. gmr684 [pii] [DOI] [PubMed] [Google Scholar]
- 94.Lee JS, Lee MO, Moon BH, Shim SH, Fornace AJ, Jr, Cha HJ. Senescent growth arrest in mesenchymal stem cells is bypassed by Wip1-mediated downregulation of intrinsic stress signaling pathways. Stem Cells. 2009;27(8):1963–75. doi: 10.1002/stem.121. [DOI] [PubMed] [Google Scholar]
- 95.Wong ES, Le Guezennec X, Demidov ON, Marshall NT, Wang ST, Krishnamurthy J, Sharpless NE, Dunn NR, Bulavin DV. p38MAPK controls expression of multiple cell cycle inhibitors and islet proliferation with advancing age. Dev Cell. 2009;17(1):142–9. doi: 10.1016/j.devcel.2009.05.009. S1534-5807(09)00211-1 [pii] [DOI] [PubMed] [Google Scholar]
- 96.Le Guezennec X, Bulavin DV. WIP1 phosphatase at the crossroads of cancer and aging. Trends Biochem Sci. 2010;35(2):109–14. doi: 10.1016/j.tibs.2009.09.005. S0968-0004(09)00180-7 [pii] [DOI] [PubMed] [Google Scholar]
- 97.Sherr CJ. The INK4a/ARF network in tumour suppression. Nat Rev Mol Cell Biol. 2001;2(10):731–7. doi: 10.1038/35096061. 35096061 [pii] [DOI] [PubMed] [Google Scholar]
- 98.Rauta J, Alarmo EL, Kauraniemi P, Karhu R, Kuukasjarvi T, Kallioniemi A. The serine-threonine protein phosphatase PPM1D is frequently activated through amplification in aggressive primary breast tumours. Breast Cancer Res Treat. 2006;95(3):257–63. doi: 10.1007/s10549-005-9017-7. [DOI] [PubMed] [Google Scholar]
- 99.Natrajan R, Lambros MB, Rodriguez-Pinilla SM, Moreno-Bueno G, Tan DS, Marchio C, Vatcheva R, Rayter S, Mahler-Araujo B, Fulford LG, Hungermann D, Mackay A, Grigoriadis A, Fenwick K, Tamber N, Hardisson D, Tutt A, Palacios J, Lord CJ, Buerger H, Ashworth A, Reis-Filho JS. Tiling path genomic profiling of grade 3 invasive ductal breast cancers. Clin Cancer Res. 2009;15(8):2711–22. doi: 10.1158/1078-0432.CCR-08-1878. 1078-0432.CCR-08-1878 [pii] [DOI] [PubMed] [Google Scholar]
- 100.Hu X, Stern HM, Ge L, O’Brien C, Haydu L, Honchell CD, Haverty PM, Peters BA, Wu TD, Amler LC, Chant J, Stokoe D, Lackner MR, Cavet G. Genetic alterations and oncogenic pathways associated with breast cancer subtypes. Mol Cancer Res. 2009;7(4):511–22. doi: 10.1158/1541-7786.MCR-08-0107. 7/4/511 [pii] [DOI] [PubMed] [Google Scholar]
- 101.Hirasawa A, Saito-Ohara F, Inoue J, Aoki D, Susumu N, Yokoyama T, Nozawa S, Inazawa J, Imoto I. Association of 17q21-q24 gain in ovarian clear cell adenocarcinomas with poor prognosis and identification of PPM1D and APPBP2 as likely amplification targets. Clin Cancer Res. 2003;9(6):1995–2004. [PubMed] [Google Scholar]
- 102.Saito-Ohara F, Imoto I, Inoue J, Hosoi H, Nakagawara A, Sugimoto T, Inazawa J. PPM1D is a potential target for 17q gain in neuroblastoma. Cancer Res. 2003;63(8):1876–83. [PubMed] [Google Scholar]
- 103.Ehrbrecht A, Muller U, Wolter M, Hoischen A, Koch A, Radlwimmer B, Actor B, Mincheva A, Pietsch T, Lichter P, Reifenberger G, Weber RG. Comprehensive genomic analysis of desmoplastic medulloblastomas: identification of novel amplified genes and separate evaluation of the different histological components. J Pathol. 2006;208(4):554–63. doi: 10.1002/path.1925. [DOI] [PubMed] [Google Scholar]
- 104.Mendrzyk F, Radlwimmer B, Joos S, Kokocinski F, Benner A, Stange DE, Neben K, Fiegler H, Carter NP, Reifenberger G, Korshunov A, Lichter P. Genomic and protein expression profiling identifies CDK6 as novel independent prognostic marker in medulloblastoma. J Clin Oncol. 2005;23(34):8853–62. doi: 10.1200/JCO.2005.02.8589. 23/34/8853 [pii] [DOI] [PubMed] [Google Scholar]
- 105.Castellino RC, De Bortoli M, Lu X, Moon SH, Nguyen TA, Shepard MA, Rao PH, Donehower LA, Kim JY. Medulloblastomas overexpress the p53-inactivating oncogene WIP1/PPM1D. J Neurooncol. 2008;86(3):245–56. doi: 10.1007/s11060-007-9470-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Loukopoulos P, Shibata T, Katoh H, Kokubu A, Sakamoto M, Yamazaki K, Kosuge T, Kanai Y, Hosoda F, Imoto I, Ohki M, Inazawa J, Hirohashi S. Genome-wide array-based comparative genomic hybridization analysis of pancreatic adenocarcinoma: identification of genetic indicators that predict patient outcome. Cancer Sci. 2007;98(3):392–400. doi: 10.1111/j.1349-7006.2007.00395.x. CAS395 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Lambros MB, Natrajan R, Geyer FC, Lopez-Garcia MA, Dedes KJ, Savage K, Lacroix-Triki M, Jones RL, Lord CJ, Linardopoulos S, Ashworth A, Reis-Filho JS. PPM1D gene amplification and overexpression in breast cancer: a qRT-PCR and chromogenic in situ hybridization study. Mod Pathol. 2010;23(10):1334–45. doi: 10.1038/modpathol.2010.121. modpathol2010121 [pii] [DOI] [PubMed] [Google Scholar]
- 108.Hu W, Feng Z, Modica I, Klimstra DS, Song L, Allen PJ, Brennan MF, Levine AJ, Tang LH. Gene Amplifications in Well-Differentiated Pancreatic Neuroendocrine Tumors Inactivate the p53 Pathway. Genes Cancer. 2010;1(4):360–368. doi: 10.1177/1947601910371979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Lu X, Bocangel D, Nannenga B, Yamaguchi H, Appella E, Donehower LA. The p53-induced oncogenic phosphatase PPM1D interacts with uracil DNA glycosylase and suppresses base excision repair. Mol Cell. 2004;15(4):621–34. doi: 10.1016/j.molcel.2004.08.007. S1097276504004721 [pii] [DOI] [PubMed] [Google Scholar]