Abstract
PPM1D (PP2Cδ or Wip1) was identified as a wild type p53-induced Ser/Thr phosphatase that accumulates after DNA damage and classified into the PP2C family. It dephosphorylates and inactivates several proteins critical for cellular stress responses, including p38 MAPK, p53, and ATM. Furthermore, PPM1D is amplified and/or overexpressed in a number of human cancers. Thus, inhibition of its activity could constitute an important new strategy for therapeutic intervention to halt the progression of several different cancers. Previously, we reported the development of a cyclic thioether peptide with low micromolar inhibitory activity towards PPM1D. Here, we describe important improvements in the inhibitory activity of this class of cyclic peptides and also present a binding model based upon the results. We found that specific interaction of an aromatic ring at the X1 position and negative charge at the X5 and X6 positions significantly increased the inhibitory activity of the cyclic peptide, with the optimized molecule having Ki = 110 nM. To the best of our knowledge, this represents the highest inhibitory activity reported for an inhibitor of PPM1D. We further developed an inhibitor selective for PPM1D over PPM1A with Ki = 2.9 μM. Optimization of the cyclic peptide and mutagenesis experiments suggest that a highly basic loop unique to PPM1D is related to substrate specificity. We propose a new model for the catalytic site of PPM1D and inhibition by the cyclic peptides that will be useful both for the subsequent design of PPM1D inhibitors and for identification of new substrates.
Kinases and phosphatases are important regulators of protein function in biological systems and thus constitute good targets for the development of new drugs. While the human genome encodes 518 kinases (1), there are estimated to be only 147 phosphatases; of those, only 40 are serine/threonine phosphatases (1–3). The PP2C family in humans consists of seven monomeric serine/threonine phosphatases (4, 5). This includes PPM1D (also called PP2Cδ or Wip1), which was first identified as induced by wild type p53 after DNA damage (6). Consistent with other members of the PP2C family, PPM1D is a monomeric enzyme that requires divalent cations, either Mn2+ or Mg2+, for catalytic activity and is insensitive to oakadaic acid (7). This phosphatase is composed of two major domains: a highly conserved N-terminal phosphatase domain and a less-conserved, non-catalytic domain at the C terminus (7). The known substrates of PPM1D include several proteins critical for cellular stress responses, namely: p38 MAPK (8), Chk1 (9), Chk2 (10–12), ATM (13), and p53 (9). Dephosphorylation of each of these proteins by PPM1D results in its inactivation. PPM1D is amplified and/or over-expressed in a number of human cancers, such as breast cancer (14–16), neuroblastoma (17), medulloblastoma (18), ovarian clear cell adenocarcinoma (19), and pancreatic adenocarcinoma (20). In addition, PPM1D-null mice show a dramatic tumor-resistant phenotype (21). Thus, inhibition of PPM1D activity could constitute an important new strategy for therapeutic intervention to halt the progression of several different cancers.
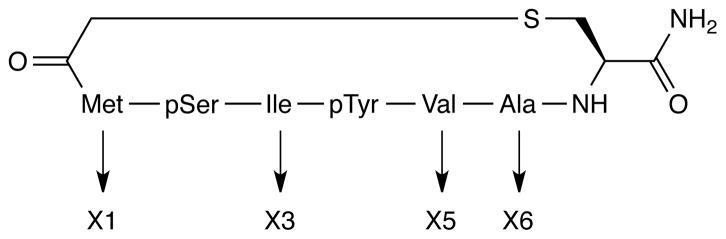
PPM1D dephosphorylates phosphoserine (pS) or phosphothreonine (pT) as a part of two different peptide motifs: pT-X-pY (22) and pS/pT-Q (23). In a study of the pT-X-pY motif, we observed that PPM1D preferentially dephosphorylates pT from a diphosphorylated sequence compared to a monophosphorylated one and that amino acids adjacent to the motif do not significantly affect the substrate specificity (24). Additionally, it was found that pS substitution of the pT in the pT-X-pY sequence from p38 MAPK resulted in PPM1D inhibition. This result raised the possibility that a pS-substituted peptide could be developed as an effective inhibitor of PPM1D phosphatase activity. After extensive optimization, a cyclic thioether peptide of sequence M-pS-I-pY-VAC was identified with a Ki of approximately 5 μM (Figure 1).
Figure 1.
Schematic representation of the cyclic thioether peptide. The thioether bond as formed between the acylated N-terminal residue and the cysteine sidechain at the C terminus.
Combining this result with mutagenesis studies of the protein and a NMR solution structure of the cyclic peptide, we were able to propose a structural model of the complex at the active site (24). For this, we developed a homology model of PPM1D from the crystal structure of the related PPM1A (PP2Cα) protein in humans (25). Although this model incorporated the pS and pY residues of the cyclic peptide in key charge-charge interactions with the protein, it did not provide obvious roles for the Met, Ile and Ala residues. We suggested that at least some of them may interact with a relatively long loop adjacent to the active site (residues 237–268) that is unique to PPM1D. Unfortunately, due to its absence in the PPM1A template, the loop could not be included in our homology model. Although the precise start of the insertion is uncertain, a sub-segment of it (residues 245–268) has been dubbed the “B-loop” by Chuman et al. (26) because of the preponderance of positively charged amino acids. Interestingly, the B-loop sprouts from a conserved sub-domain (residues 165–194 of PPM1A) that has been designated a flap, presumably due to its different conformations observed in the crystal structures of both eukaryotic and prokaryotic homologues (27–33) (Supporting Information Figure S1). It has been postulated that movement of the flap regulates enzymatic activity through modulation of the binding of a third metal ion, substrate recognition and/or steric availability to the catalytic site. Thus, that the B-loop is part of the flap sub-domain in PPM1D supported the idea that it plays a role in regulation.
In the current work, we describe the development of a new cyclic peptide with significantly enhanced inhibitory activity. While keeping the pS-I-pY core residues, the sidechains at the other positions were extensively modified to identify an optimized molecule with a Ki for PPM1D of approximately 100 nM, reflecting a 50-fold improvement over the starting peptide. Combining the results of the optimization with the results of strategic mutations in the B-loop enabled further refinement of the model of the PPM1D catalytic domain. We also describe improvements obtained in selectivity for PPM1D over PPM1A by modification of the pS residue of the cyclic peptide. These results further demonstrate the possibility of developing selective inhibitors for this neglected class of phosphatase targets.
Experimental Procedures
Chemical Reagents
Resins and N-α-Fmoc-protected amino acids, including phosphorylated amino acids and Fmoc succinimide, were purchased from Novabiochem (San Diego, CA). N-α-Fmoc-protected unusual amino acids, including L-2-amino-4-phosphono-4,4-difluorobutyric acid (F2Pab), were obtained from Anaspec (San Jose, CA). Chloroacetic anhydride, solvents, and the amino acid standard solution for amino acid analysis were obtained from Sigma-Aldrich (Milwaukee, WI).
Peptide synthesis
Peptides were synthesized by the solid phase method utilizing 9-.fluorenylmethoxycarbonyl (Fmoc)/tert-butyl or tert-butoxycarbonyl (Boc)/benzyl chemistry. Trityl (Trt) and p-methoxybenzyl groups were utilized as Cys sidechain protecting groups for Fmoc- and Boc-chemistry, respectively. Peptides were assembled on NovaPEG Rink amide resin or Wang resin for Fmoc chemistry or MBHA resin for Boc chemistry. The amino group of O-sulfo-L-serine was protected by an Fmoc group (34). Phosphorylated Ser and Tyr were incorporated as Fmoc-Ser[PO(OBzl)OH]-OH and Fmoc-Tyr(PO3H2)-OH, respectively. In the synthesis of peptides 46 and 48, O-Sulfo-L-serine and homoserine (Hse) were assembled as Boc-Ser(SO3H)-OH and Fmoc-Hse(Trt)-OH on MBHA resin, respectively. The coupling reactions were carried out by means of the HBTU-HOBt method. To introduce a phosphate group onto the hydroxyl group of the Hse residue in peptide 48, the trityl group was removed by treatment with trifluoroacetic acid/dichloromethane/triisopropylsilane (1/94/5 v/v) after the amino group of the Phe residue at position X1 was protected with a Boc group. Phosphorylation was achieved using dibenzyl-N,N′-diisopropylphosphoramidate and anhydrous tert-butylhydroperoxid. The N terminus of each peptide was chloroacetylated using chloroacetic anhydride for downstream cyclization. Cleavage of the peptide from the resin was achieved with trifluoroacetic acid/water/triisopropylsilane (92.5/5/2.5 v/v) or trifluoromethanesulfonic acid/trifluoroacetic acid/water/triisopropylsilane (10/82.5/5/2.5 v/v) for 2 h at room temperature. After removing the resin by filtration, the filtrate was concentrated by flushing with nitrogen gas and crude peptides were precipitated by diethyl ether. The crude peptides were then dissolved in 1% triethylamine-containing water (pH 8–9, approximately 3 mM) and stored at room temperature overnight to allow cyclization to occur. The cyclization reaction was quenched by acidifying using acetic acid. Crude peptides were purified using reversed-phase high-performance liquid chromatography (RP-HPLC) on a preparative C4 column (BioAdvantage Pro 300, Thomson Liquid Chromatography) with a water-acetonitrile solvent system containing trifluoroacetic acid. Purified peptides were characterized by matrix-associated laser desorption ionization time-of-flight mass spectrometry (MALDI micro MX, Waters) and RP-HPLC on an analytical C18 column (Eclipse XDB-C18, Agilent). The purity of all peptides was found to be > 95%.
Amino acid composition analysis and determination of concentration
Amino acid composition analysis was carried out using the phenylthiocarbamyl method (35, 36). Peptide stock solutions in water (ca. 5 mM) were transferred to a glass tube for hydrolysis, resulting in ca. 10 nmol per glass tube. The peptide was hydrolyzed with 6 N hydrogen chloride solution at 105 °C in a sealed tube for 24 h. After cooling, the solution was evaporated with nitrogen gas and a water bath. A solution (10 μl) of ethanol:water:triethylamine (2:1:1 v/v) was added to each tube. After removing the solution in vacuo, a phenylisothiocyanate-containing solution (ethanol:water:triethylamine:phenylisothiocyanate, 7:1:1:1 v/v) was added to the lyophilized sample. The tube was left for 30 min at room temperature. Subsequently, the sample was redried in vacuo to remove excess reagent. The dried sample was dissolved in 1 ml 60 mM sodium acetate buffer (pH 6.0) and 10% of the total was analyzed by RP-HPLC as follows: BioAdvantage Pro 300 C18 5 μm (4.6 × 250 mm) column; solution A, 60 mM sodium acetate buffer (pH 6.0); solution B, acetonitrile; gradient, 5–50%B/0–30 min. Identification and quantitation of each amino acid in the solution was performed on the basis of retention times and peak area integration as compared with that of an amino acid standard solution. Peptide concentration was calculated on the basis of a standard curve made by the peptide stock solution described above.
Protein expression and purification
The cloning and purification of the human PPM1D catalytic domain (residues 1–420) containing an N-terminal His-tag was performed as previously described (22). The plasmid containing the catalytic domain was used for construction of mutants using the QuickChange protocol as described by the manufacturer (Stratagene, La Jolla, CA). The sequences of the oligonucleotides used for PPM1D R243A were as follows: 5′-GAGTGTAATGAACAAGTCTGGGGTTAACGCTGTAGTTTGGAAACGACCTCGACTC -3′ and 5′-GAGTCGAGGTCGTTTCCAAACTACAGCGTTAACCCCAGACTTGTTCATTACACTC-3′. An HpaI restriction enzyme site was created in the oligonucleotides (underlined) to facilitate screening of mutant clones. For PPM1D K247A, the following oligonucleotides were used: 5′-CTGGGGTGAATCGTGTAGTTTGGGCCCGACCTCGACTCACTCACAATGGACCTG-3′ and 5′-CAGGTCCATTGTGAGTGAGTCGAGGTCGGGCCCAAACTACACGATTCACCCCAG-3′. An ApaI site (underlined) was created in the oligonucleotides for mutant screening.
Individual mutant plasmids were co-transformed with pACYC- Duet (Novagen, Madison, WI) expressing Skp and DsbC proteins to improve solubility of the proteins into BL21-star competent cells (Invitrogen, Carlsbad, CA). Transformed colonies were selected on LB plates containing ampicillin (100 μg/mL) and chloramphenicol (15 μg/mL). Cells were grown (800 mL) to mid-log phase in Circle Grow (Q-Biogen, Irvine, CA) at 37 °C before transfer to 16 °C. After incubation for approximately 15 min, cells were induced with 0.5 mM isopropyl 1-thio-α-D-galactopyranoside for 16 h. Cells were harvested and the His-tagged mutant PPM1D proteins purified by metal affinity chromatography, as described for the wild type protein (22). Activity of the purified protein was confirmed by measuring the kinetic properties for dephosphorylation of a substrate peptide from ATM ([S1981pS] human ATM(1976–1986)-GY).
Phosphatase activity
Phosphatase activities of wild type and mutant PPM1D were measured by a malachite green/molybdate based assay (Millipore), as described previously (22). A substrate peptide ([S1981pS] human ATM(1976–1986), AFEEG-pS-QSTTIGY or [T180pT, Y182pY] human p38α MAPK(175–185), TDDEM-pT-G-pY-VAT) was incubated with 60 ng of PPM1D in 50 mM Tris-HCl pH 7.5, 0.1 mM EGTA, 0.02% 2-mercaptoethanol, 40 mM NaCl, and 30 mM MgCl2 for 7 min at 30 °C, and the amount of free phosphoric acid released from the substrate was detected by measuring the absorbance at 650 nm from the molybdate:malachite green:phosphate reaction complex Determination of the kinetic parameters Km and Vmax were performed with a range of peptide concentrations.
Inhibitory activity of the cyclic peptides was measured as previously described (22). PPM1D was incubated with 30 μM substrate peptide ([S1981pS] human ATM(1976–1986) or [T180pT, Y182pY] human p38α MAPK) and a cyclic peptide under the same conditions as the phosphatase activity assay described above. For most assays, peptides were included at 10 μM, with some peptides tested also at 1 μM. Data was expressed as mean ± S.D. Statistical analysis of the inhibitory activity was performed by Student’s t-test with p<0.05 considered statistically significant. The determination of the apparent inhibitory constant Ki was performed with a range of cyclic peptide concentrations. For most peptides, the concentration was determined from the powder weight of the cyclic peptide; the peptide concentration of peptides 1 and 37 was calculated using amino acid composition analysis.
PPM1A phosphatase activity was measured using the malachite green/molybdate assay described above. Cyclic peptides were incubated with 12 ng or 24 ng of PPM1A under the same conditions described above. For assays of PPM1A inhibition, cyclic peptide 40 (50 μM) was used as the substrate.
Inhibition mechanism
The mechanism of inhibition of cyclic thioether peptide 37 was estimated from the initial velocities of dephopshorylation measured at different concentrations of cyclic peptide and [S1981pS] human ATM(1976–1986) peptide, as described above. The observed initial velocities were graphed on double-reciprocal plots and fitted by the Lineweaver-Burk equation (eq. 1) using GraphPad Prism 4 (GraphPad Software, La Jolla, CA).
| (1) |
In equation 1, V is the reaction velocity, Km is the Michaelis–Menten constant, Vmax is the maximum reaction velocity, and [S] is the substrate concentration.
Molecular Modeling
The molecular model of PPM1D was updated from that used previously (24). A new sequence alignment with the PPM1A structural template is presented in Supporting Information Figure S2, which also depicts the 7 regions of conservation defined by Chuman et al. (26). The only changes to the previous model were to regions I and VII, which now make all regions consistent with the model of Chuman et al. (26). Although changes to the model in region I involve residues that comprise the active site, the sequence changes are conservative and the impact on the predictive properties of the model are minimal. Specifically, in the previous model PPM1D residue Arg76 was at the position of the functionally important, phosphate-binding Arg33 residue of PPM1A (25, 37). In the current model, this is replaced by PPM1D residue Arg18. The change also added metal-binding PPM1D residue Glu22, which substitutes for PPM1A residue Glu37. However, this residue is on the opposite side of the active site from which the substrates approach and bind. Addition of PPM1D residues Lys19 and Tyr20 to positions proximal to (above) the metal center is significant, as they feature in the optimization of the cyclic peptide inhibitor (see Results).
Models of the cyclic peptide inhibitor bound to PPM1D were energy minimized with the CHARMM software package (38). The backbone of the cyclic peptide was the same as in the structure determined by NMR in our previous study (24). Residue topology files for the different sidechains tested on the inhibitor were developed manually using the “all22_prot” force field of MacKerell et al. (39). Since our focus was on the interactions with substrate or inhibitors, we did not attempt to model the inserted loops and deletions of the PPM1D homology model distant from the active site.
Structural similarity searches of the Protein Data Bank (40) were conducted with the VAST conformational search engine (41). Structural alignments, conformational adjustments and figures were generated with the UCSF Chimera molecular modeling software package (42). Sequence alignments and figures were made with BioEdit Sequence Alignment Editor (43).
Circular Dichroism (CD) Spectroscopy
The CD spectra of wild type and mutant PPM1D were recorded on a JASCO J-715 spectropolarimeter (Easton, MD) with a cylindrical cell of 1 mm path length at room temperature. The CD cell was washed with an aqueous NaOH solution before each new measurement to remove any material that might have adhered to the inner surface. Sample solutions (1.8–2.1 μM PPM1D in 50 mM Tris-HCl, 30 mM MgCl2, 5% glycerol, pH 7.5) were prepared 5 min before measurement. All spectra shown are the average of eight repeated measurements obtained by collecting data from 260 to 190 nm at 0.2 nm intervals with a response time of 2 sec for each point. The results are expressed as the mean residue ellipticity.
Results
Phe substitution at position X1 increases the inhibitory activity of the cyclic peptide against PPM1D
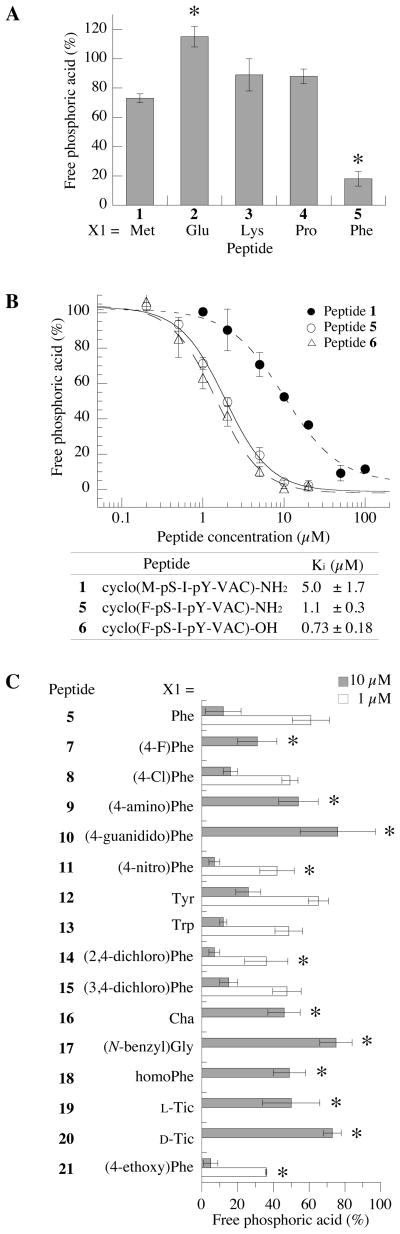
Previously, we identified a cyclic thioether peptide (M-pS-I-pY-VAC) (peptide 1) (Figure 1) that functions as an inhibitor of the Ser/Thr phosphatase PPM1D (24). In that peptide, only the X3 position between the pS and pY residue was optimized. Here, we explored the structure-activity relationship for each of the other positions of the peptide with the goals of improving the inhibitory activity and learning more about PPM1D substrate specificity. Initially, position X1 (Figure 1) was varied by replacing the Met of peptide 1 with different amino acids, and then the inhibitory activity of the analogs were determined. Inhibitory activity was calculated from the ability of the cyclic peptide to suppress dephosphorylation of pS in a peptide derived from ATM (residues 1976–1986 with phosphorylation of Ser1981) using a malachite green/molybdate assay to detect the amount of free phosphoric acid in solution. Performing this assay in the presence of 10 μM peptide 1, we observed a 30% decrease in PPM1D dephosphorylation activity (Figure 2A), resulting in a Ki value for peptide 1 of 5.0 ± 1.7 μM (Figure 2B). This value is 5-fold reduced as compared with the previous report of this inhibitor (24). However, in that study the peptide concentration was calculated from pY absorbance at 280 nm with 640 M−1 cm−1 as the molar extinction coefficient (24). As reported, the molar extinction coefficients for pY range from 600–700 M−1 cm−1 at pH 7.0 (44) and they are strongly pH dependent (45). In this study, we determined the cyclic peptide concentration from the dry weight, which by amino acid composition analysis we found to be more accurate. The concentration of peptide 1 determined in this way was approximately 6-fold higher than the concentration calculated from the molar extinction coefficient used previously, resulting in overestimation of the Ki value. Due to the difficulty associated with purifying large quantities of PPM1D, each new inhibitor were tested at 10 μM, with some also used at 1 μM for greater discrimination of the inhibitors.
Figure 2.
An aromatic residue at position X1 of the cyclic peptide is important for inhibitory activity. (A) Inhibitory activity of cyclic peptides (10 μM) substituted at position X1 in the presence of [S1981pS] human ATM(1976–1986) peptide (30 μM) as a substrate (*p<0.05 as compared with peptide 1). (B) Concentration dependence of inhibition by cyclic peptides: peptide 1, cyclo(M-pS-I-pY-VAC)-amide (closed circle); peptide 5, cyclo(F-pS-I-pY-VAC)-amide (open circle); peptide 6, cyclo(F-pS-pY-I-VAC)-OH (open triangle). (C) Inhibitory activity of cyclic peptides used at 10 μM and 1 μM, as indicated (*p<0.05 as compared with peptide 5).
In our original model of peptide 1 bound to PPM1D, the Met sidechain of peptide 1 was proximal to Arg76 (24). Thus, we initially hypothesized that mutation of the Met residue to an acidic residue would allow formation of a charge-charge interaction that could improve the inhibitory activity of the cyclic peptide. In contrast to the hypothesis, a peptide in which Glu was substituted at position X1 (peptide 2) abrogated inhibitory activity of the peptide at 10 μM and also functioned as a weak substrate for PPM1D (Figure 2A). Further substitution at the X1 position with a Lys residue (peptide 3) or a Pro residue (peptide 4) also did not significantly alter the inhibitory activity compared to peptide 1. In contrast, a Phe-substituted analog (peptide 5) significantly decreased PPM1D phosphatase activity in the presence of 10 μM peptide (Figure 2A). From measurements at several concentrations of peptide 5, we determined that the Ki value of peptide 5 was 1.1 ± 0.3 μM (Figure 2B), about 5-fold more potent than the original peptide 1. A second cyclic peptide terminating in a carboxylic acid group (peptide 6) rather than an amide as in peptide 5 showed a similar inhibitory activity (Ki = 0.73 ± 0.18 μM, Figure 2B) as peptide 5, indicating that a negative charge at the extreme C terminus does not affect inhibitory activity.
To further explore the role of the Phe residue in the X1 position for inhibitory activity, a series of peptides was synthesized containing Phe derivatives and their inhibitory activities were determined (Figure 2C). Substitution of position X1 with halogenated phenylalanine derivatives (peptides 7, 8, 14, and 15) did not significantly change the inhibitory activity as compared with peptide 5. Likewise, substitution of the Phe at position X1 with 4-nitrophenylalanine (peptide 11), Tyr (peptide 12), Trp (peptide 13), and 4-ethoxyphenyalanine (peptide 21) did not greatly improve the inhibitory activity relative to peptide 5. These results indicate that addition of a functional group to the 3- and/or 4-position of the aromatic ring does not affect the interaction between the cyclic peptide and PPM1D. In contrast, peptides 9 and 10, containing basic functional groups on the Phe (4-aminophenylalanine and 4-guanididophenylanalnine, respectively) showed decreased inhibitory activities at 10 μM (Figure 2C). These results indicate that a basic functional group at position X1 disrupts the interaction of the cyclic peptide with PPM1D.
To determine if the position of the aromatic ring of the Phe had an effect on inhibitory activity, position X1 was substituted with N-benzylglycine (peptide 17), homophenylalanine (peptide 18), and L- and D-Tic [(S)- and (R)-1,2,3,4-tetrahydroisoquinoline-3-carboxylic acid] (peptides 19 and 20, respectively) (structures in Supporting Information Figure S3). The inhibitory activities of these peptides were reduced relative to peptide 5 at 10 μM (Figure 2C). Peptide 16, containing beta-cyclohexylalanine (Cha), which has a cyclohexyl ring instead of the aromatic ring of Phe, also showed decreased activity as compared with the Phe-substituted peptide, with approximately 50% inhibition of dephosphorylation observed in the presence of 10 μM peptide 16. Thus, these results suggest that a simple phenyl ring at position X1 of the peptide is important for inhibition, but additional groups attached to the aromatic ring do not further enhance activity.
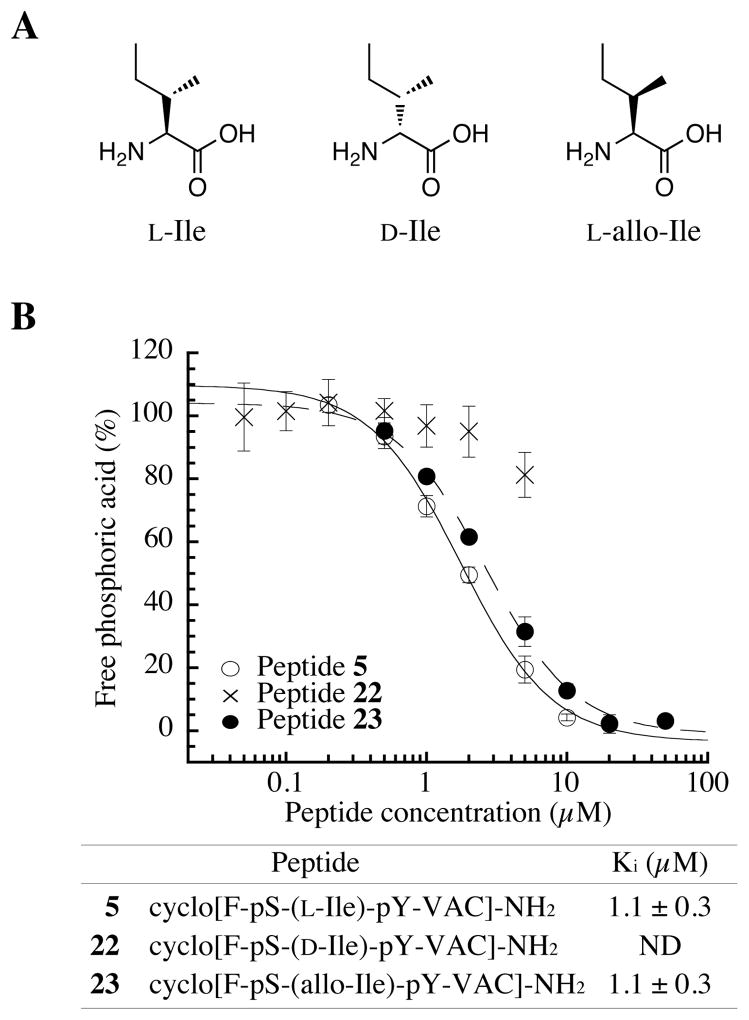
The chirality of the Ile residue at position X3 affects PPM1D inhibition
Our previous work showed that the Ile residue at position X3 is critical for inhibition (24). However, the importance of the stereochemistry of the Ile was not investigated. Therefore, D-Ile and L-allo-Ile (Figure 3A) substituted peptides (22 and 23, respectively) were synthesized, and their inhibitory activity was measured. The D-Ile peptide 22 showed greatly reduced inhibitory activity (Figure 3B). In contrast, the Ki value of the L-allo-Ile analog peptide 23 was 1.1 ± 0.3 μM, similar to that of peptide 5, which has L-Ile (Figure 3B). These results indicate that the chirality at the beta position of the Ile is not important for inhibition, while that of the alpha position is important.
Figure 3.
The enantiomer of the Ile at position X3 is important for inhibition of PPM1D phosphatase activity. (A) Chemical structure of isoleucine analogs. (B) Concentration dependence of the inhibition of PPM1D by cyclic peptide [F-pS-(L-Ile)-pY-V-A] (peptide 5, open circle), [F-pS-(D-Ile)-pY-VAC] (peptide 22, cross), and [F-pS-(L-allo-Ile)-pY-VAC] (peptide 23, closed circle) in the presence of [S1981pS] human ATM(1976–1986) peptide (30 μM) as a substrate.
Negative charges at positions X5 and X6 are preferred for inhibition
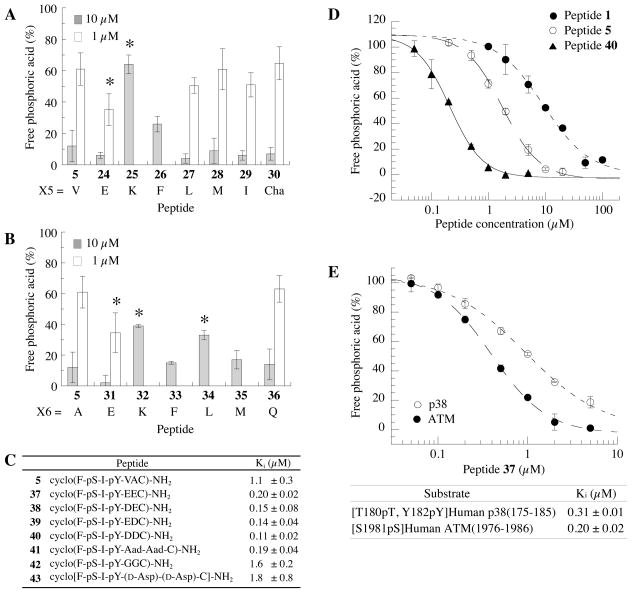
Figure 4 shows the inhibitory activities of X5- and X6-substituted cyclic peptides in which the Val and Ala residues were modified to aromatic, acidic, basic, neutral, and aliphatic amino acids. In position X5, the substitution of Val in the original peptide with Lys (peptide 25) or Phe (peptide 26) reduced the inhibitory activity of the cyclic peptide (Figure 4A). Substitution with Leu (peptide 27), Met (peptide 28), Ile (peptide 29), or Cha (peptide 30) did not have any significant effects (Figure 4A). In contrast, a peptide containing Glu (peptide 24) at position X5 showed improved inhibitory activity, decreasing PPM1D phosphatase activity by >90% and > 60% at 10 and 1 μM, respectively.
Figure 4.
Acidic residues at positions X5 and X6 increase the inhibitory activity of the cyclic peptides. Inhibitory activity of cyclic peptide (F-pS-I-pY-X5-X6-C) in the presence of [S1981pS] human ATM(1976–1986) peptide (30 μM) as a substrate. Inhibitory activities of (A) cyclo(F-pS-I-pY-X5-AC)-amide and (B) cyclo(F-pS-I-pY-V-X6-C)-amide peptides were measured at 10 μM (closed bar) and 1 μM (open bar) (*p<0.05 as compared with peptide 5). (C) Ki values of cyclic thioether peptides substituted by acidic residues at both position X5 and X6. (D) Concentration dependence of cyclic thioether peptide (M-pS-I-pY-VAC)-amide (peptide 1, closed circle), (F-pS-I-pY-VAC)-amide (peptide 5, open circle), (F-pS-I-pY-DDC)-amide (peptide 40, closed triangle). (E) Inhibitory activity of cyclic peptide 37 in the presence of different Wip1 substrates (open circle, [T180pT, Y182pY]Human p38(175–185); closed circle, [S1981pS]Human ATM(1976–1986)).
Substitutions at position X6 with hydrophobic amino acids, as in peptides 33 (Phe), 34 (Leu), and 35 (Met), either did not have any significant effect or significantly reduced inhibitory activity as peptide 34 (Figure 3B), while incorporation of Lys at position X6 (peptide 32) also significantly reduced inhibitory activity (Figure 4B). In contrast, increased inhibitory activity was observed when a Glu residue was put in position X6 (peptide 31), with PPM1D dephosphorylation activity decreasing by 95% and 80% when the cyclic peptide was used at 10 μM and 1 μM, respectively. Combined with the results of the screen at position X5, these data suggest that an acidic residue is preferred at positions X5 and X6 for PPM1D inhibition. Indeed, a cyclic peptide containing Gln at position X6 (peptide 36) did not have improved inhibitory activity relative to peptide 5 (Figure 4B). Moreover, the improvement associated with introduction of an acidic residue was moderately larger in the X6 position as compared with the X5 position.
Finally, we determined the effect of having acidic residues at both X5 and X6 on inhibitory activity. As expected, inclusion of Glu residues at both positions X5 and X6 further increased the inhibitory activity of the peptide (peptide 37). The Ki value of peptide 37 was 0.20 ± 0.02 μM, an improvement of 4-fold relative to peptide 5 (Figure 4C). A similar Ki value for this peptide was observed in the presence of a different substrate derived from human p38 MAPK (residues 175–185, [T180pT, Y182pY]) (Figure 4E), demonstrating that PPM1D inhibition was not dependent on the substrate motif. To evaluate whether any acidic residue would improve the inhibitory activity or if a Glu residue was specifically required, peptides were tested in which X5 or X6 were individually substituted with Asp instead of Glu (peptides 38 and 39, respectively), X5 and X6 were both Asp (peptide 40), and X5 and X6 were both homoglutamic acid (Aad, peptide 41). All peptides containing an acidic group at one or both of positions X5 and X6 showed similar inhibitory activities (Figure 4C), although peptide 40, containing two Asp residues, did show slightly improved activity relative to peptide 37, which has two Glu residues. The Ki value for peptide 40 was found to be 110 nM, with 10- and 50-fold increased relative to peptides 5 and 1, respectively (Figure 4D). In contrast, when positions X5 and X6 were substituted with Gly residues (peptide 42), the inhibitory activity was not affected as compared with peptide 5 (Figure 4C). Thus, a negative charge at positions X5 and X6 is critical, although the carbon chain length of the sidechain with the acidic residues does not seem to affect inhibitory activity.
To investigate if the stereochemistry affects the inhibitory activity, a cyclic peptide containing D-Asp at positions X5 and X6 (peptide 43) was synthesized and its inhibitory activity was estimated (Figure 4C). The Ki value of peptide 43 was found to be 1.8 ± 0.8 μM, approximately 20-fold lower than peptide 40, containing L-Asp at positions X5 and X6 (Figure 4C). Thus, the chirality at positions X5 and X6 affects the inhibitory activity, however the effect is not as large as that observed at position X3.
Binding of the cyclic peptides to PPM1D is competitive with substrate
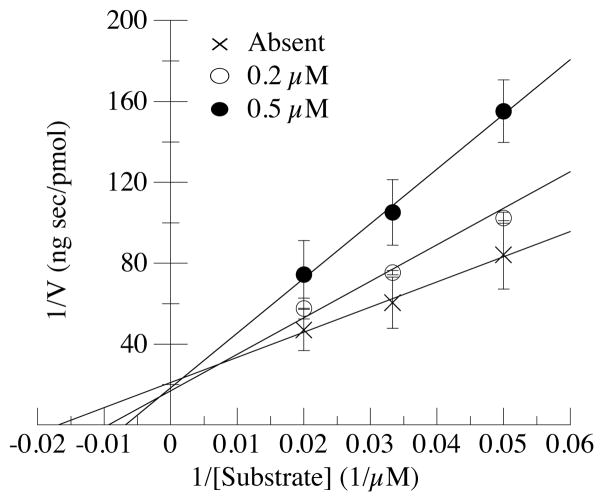
Having identified an optimized inhibitor, we next investigated its mechanism of inhibition to better understand its binding site on PPM1D. Figure 5 shows the double-reciprocal plot of PPM1D dephosphorylation of human ATM(1976–1986) [S1981pS] in the absence or presence of cyclic peptide inhibitor 37. The inverse of V, the rate of the PPM1D-mediated reaction, is shown on the vertical axis, while the inverse of the substrate concentration is plotted on the abscissa. Fitting of the data using the Lineweaver-Burk equation provided the maximum reaction velocity Vmax from the inverse of the intercept of the vertical axis and the Michaelis constant Km, which indicates the binding affinity of the substrate to PPM1D, from the inverse of the intercept on the abscissa. The slope of the best fit line is equal to the value of Vmax/Km. The data show that the value of Vmax/Km changed at different cyclic peptide concentrations, whereas the Vmax value was independent of the cyclic peptide concentration. These results indicate that the presence of the cyclic peptide decreased the substrate binding affinity in PPM1D but not the rate of reaction. Thus, these results demonstrate that the cyclic peptide is a competitive inhibitor of PPM1D and, as such, binds to the catalytic site.
Figure 5.
The cyclic peptide is a competitive inhibitor of PPM1D. Double-reciprocal plot of PPM1D phosphatase activity against [S1981pS]human ATM(1976–1986) peptide concentration during the inhibition of PPM1D with different concentrations of cyclic thioether peptide (F-pS-I-pY-EEC)-amide (peptide 37) (cross, absent; open circle, 0.2 μM; closed circle, 0.5 μM).
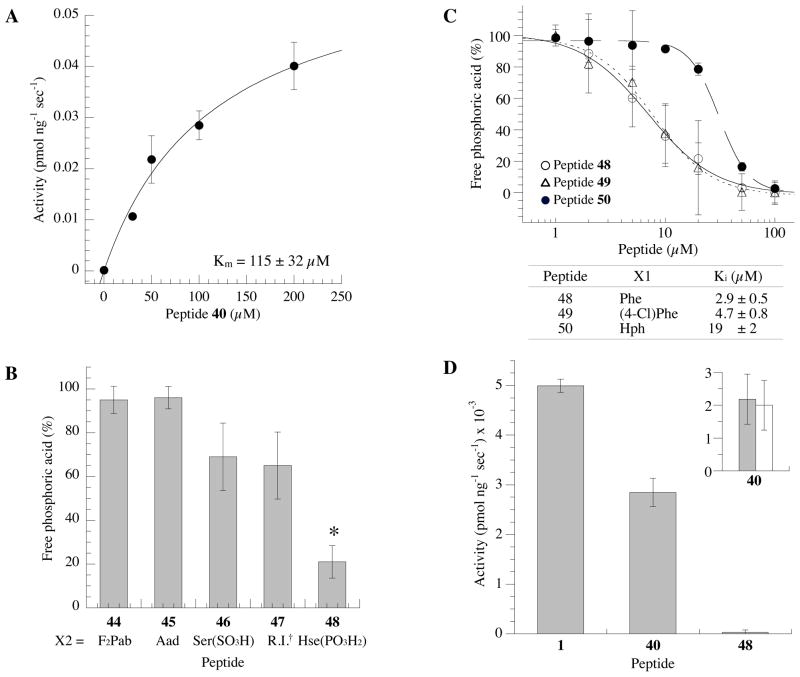
Substitution of pS with phospho-homoserine confers specificity towards PPM1D over PPM1A
As specific inhibitory activity is critical for drug development, we assessed whether peptide 40 was specific for PPM1D. Previously, we found that peptide 1 was recognized as a substrate by PPM1A (PP2Cα) (Km = 11 μM) (24). Similarly, PPM1A dephosphorylated 40 with Km = 115 ± 32 μM, 10-fold less than peptide 1 (Figure 6A). We synthesized a series of peptides containing non-hydrolyzable mimetics of phosphoserine to try to identify a peptide that would not be recognized by PPM1A as either a substrate or an inhibitor. We first substituted the pS with L-2-amino-4-phosphono-4,4-difluorobutyric acid (F2Pab), in which the bridging oxygen of the phosphate group in pS is replaced with a CF2-moiety. In contrast to previous results in which a F2Pab-substituted peptide was partially active against PPM1D (5-fold weaker than the parent peptide) (24), peptide 44, cyclo(F-F2Pab-I-pY-DDC)-amide, did not show inhibitory activity against PPM1D at 25 μM (Figure 6B). Peptide 45, containing Aad, also did not have any inhibitory activity towards PPM1D (Figure 6B). Similarly, peptides containing Ser(SO3H) in place of pS (peptide 46) and the retro-inverse peptide 47, cyclo[(D-Asp)-(D-Asp)-(D-pY)-(D-I)-(D-pS)-(D-F)-C]-amide, showed no significant inhibitory activity toward PPM1D at 25 μM (Figure 6B). In contrast, when the pS was substituted with phosphorylated homoserine (pHse, peptide 48), the resultant peptide showed good inhibitory activity toward PPM1D, with Ki = 2.9 ± 0.5 μM (Figure 6B and 6C). Importantly, peptide 48 did not show any activity, either as a substrate or as an inhibitor, towards PPM1A (Km > 100 μM) (Figure 6D). Thus, although the Ki value of peptide 48 was approximately 30-fold reduced in comparison to peptide 40, it was highly selective. One possible reason for the decreased inhibitory activity of peptide 48 is that the longer sidechain of pHse may disrupt or weaken critical stabilizing interactions between PPM1D and other resides on the cyclic peptide. To determine if this was occurring, we examined cyclic peptides containing (4-Cl)Phe (peptide 49, cyclo[(4-Cl)Phe-Hse(PO3H2)-I-pY-DDC]-amide) or homophenylalanine (Hph, peptide 50, cyclo[Hph-Hse(PO3H2)-I-pY-DDC]-amide) with the hypothesis that the increased length of the Phe sidechain would improve inhibitory activity. While the Ki value of peptide 49 was to be found 4.7 ± 0.8 μM, similar to that of peptide 48, peptide 50 showed decreased inhibitory activity (Ki = 19 ± 2 μM) (Figure 6C). These results suggest that the orientation of the aromatic ring at position X1 of peptide 48 is still critical for inhibition, as described earlier.
Figure 6.
Peptide 48 is a selective inhibitor of PPM1D. (A) Dephosphorylation of peptide 40 by PPM1A. (B) Inhibitory activity of cyclic peptides (25 μM) substituted at position X2 in the presence of [S1981pS] human ATM(1976–1986) peptide (30 μM) as a substrate (*p<0.05 as compared with a no-inhibitor control). †Retro-inverse peptide. (C) Concentration dependence of cyclic thioether peptides 48 (open circle), 49 (open triangle), and 50 (closed circle). (D) Phosphatase activity of PPM1A against cyclic peptides 1, 40 and 48 at 100 μM. Inset: PPM1A phosphatase activity against peptide 40 (50 μM) in the presence (open bar) or in the absence of peptide 48 (closed bar).
Molecular Modeling
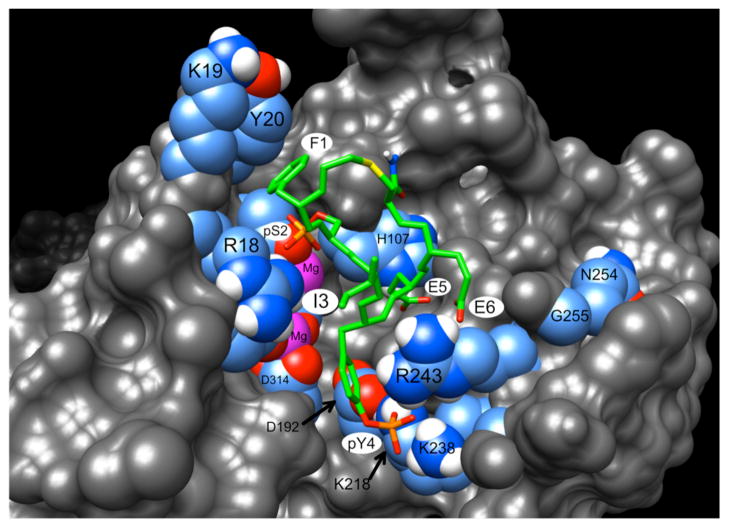
To understand the physical basis for the enhanced binding affinity, we undertook to model one of the best cyclic peptides (37) bound to the active site of PPM1D. We knew from our previous experimental and computer docking studies with substrates and peptide inhibitor 1 that the interaction is dominated by electrostatic interactions involving the two phosphate groups (22, 24). Specifically, the phosphate group of the pS residue interacts with the metal center and Arg18 and the phosphate of pY interacts with the sidechains of Lys218 and Lys238 (this latter confirmed by mutagenesis studies (24)). Thus, we maintained these interactions as the core of the updated model. As seen in Figure 7 (and the alternate view in Supporting Information Figure S4), this locates the phenylalanine at position X1 of peptide 37 (F1), next to the non-polar regions of the Lys19 and Tyr20 of PPM1D, thus forming a stabilizing, hydrophobic interaction. This explains why, as demonstrated in Figure 2, highly charged sidechains at X1 lead to reduced inhibition. The fact that Tyr, Trp and some derivatized Phe residues were also favored, and not proline and cyclohexane (peptide 16), suggests that the stabilization is enhanced by aromatic, pi-pi interactions. This is also consistent with the inhibitor being selective for PPM1D over PPM1A, since the corresponding residues in the latter are the non-aromatic Val34 and Glu35. This conformation also buries the sidechain of the X5 peptide residue (E5) on top of PPM1D His107. This would explain why, as shown in Figure 4A, the best inhibition was obtained with negatively-charged residues at this position. Given the electrostatic influence of the carboxylate group, the histidine can be expected to be protonated, thus forming a stabilizing salt-bridge.
Figure 7.
Model of cyclic peptide inhibitor 37 bound to the active site of PPM1D. Residues discussed in the text are labeled. Highlighted residues of the protein are colored cyan, blue, red and white, for carbon, nitrogen, oxygen and hydrogen. The metal ions are magenta. The unique colors of the cyclic peptide are green, yellow and orange, for carbon, sulfur and phosphorus.
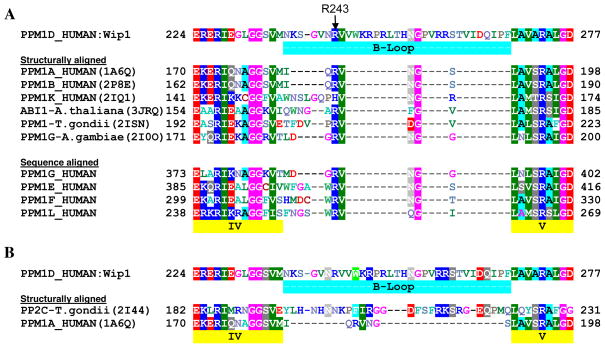
Unfortunately, the effects of substitutions at the X3 and X6 positions (i.e., preference for an isoleucine and a negatively charged residue, respectively) are more complicated to understand. Given the conformation of the bound cyclic peptide, these two sidechains project away from the conserved, catalytic core of PPM1D and are either solvent exposed or interact with the unique B-loop insert of the flap subdomain (24, 43). In an attempt to find a structural template for the B-loop, the Protein Data Bank (40) was searched for all structures similar to PPM1A, the prototypic PP2C family member (41). The results were clustered into 14 unique structures by visual inspection (Supporting Information Table S1). These were then superimposed to identify the range of conformational changes and sequence insertions/deletions in the flap subdomain. The results are displayed in Figure 8A, which aligns the sequence of PPM1D with that of PPM1A and the subset of structures close enough to be structurally aligned, and with homologous human sequences that lack known structure. The most prominent feature in the B-loop region is the conservation of a RV motif (residues Arg243 and Val244 in PPM1D) among all the sequences. It should be noted that human PPM1H (SwissProt: Q9ULR3) and PPM1J (SwissProt: Q5JR12) proteins also have an insertion in the flap region with RV motifs, but the sequences are too divergent to align unambiguously. While this procedure failed to provide a clear template for the B-loop, it did highlight the importance of the RV motif, so as an initial hypothesis, we chose the simplest option of keeping the RV residues of PPM1D structurally aligned with that of PPM1A. As seen in Figure 7, this places Arg243 in an ideal position to interact with the X6 residue (E6) of the cyclic peptide. This explains the preference for a negatively-charged sidechain at this position of the cyclic peptide and the decreased inhibitory effect of positively-charged residues (Figure 4B). Although of a greater distance, this position of Arg243 also allows for interaction with the pY sidechain of the cyclic peptide. In contrast, structurally aligning the similarly conserved Asn254 and Gly255 residues of PPM1D positions them too distant from the active site to appear relevant to regulation. We also attempted automated computer modeling of the B-loop to investigate how it might fold over the active site and interact with the I3 peptide sidechain. Unfortunately, this was unsuccessful, and we are still unable to credibly position these additional B-loop residues in the model.
Figure 8.
Putative alignment of the B-loop region of PPM1D. (A) Comparison with PPM1A and structurally aligned PP2C homologues with similar flap sub-domain conformations, and with human PP2C proteins with unknown 3D structure. References for the structures are given in Supporting Table 1. The Swiss-Prot database (http://expasy.org/sprot/) accession numbers for PPM1D and the other human sequences are: PPM1D_HUMAN (O15297), PPM1A_HUMAN (P35813), PPM1B_HUMAN (O75688), PPM1E_HUMAN (Q8WY54), PPM1F_HUMAN (P49593), PPM1G_HUMAN (O15355), PPM1K_HUMAN (Q8N3J5), PPM1L_HUMAN (Q5SGD2). (B) Comparison with the structurally aligned sequences of PPM1A and PP2Ctg.
Finally, of all the homologous structures identified, only PP2Ctg from Taxoplasma gondii (2I44.pdb) (31) had a B-loop insertion similar to PPM1D. This is shown in Figure 8B, which aligns the flap subdomain of PPM1D with that of the structurally aligned PP2Ctg and PPM1A. In addition to being of similar lengths, they have the highlighted similarity of the WXR(246–248), RRS(258–260) and DQ(264–265) subsequences of PPM1D. However, because the overall degree of conservation is relatively low and it would reposition the highly conserved RV residues away from the location in the PPM1A structure, we disfavored the use of PP2Ctg as a template for the B-loop. Nonetheless, the structural overlay of PP2Ctg and PPM1A shown in Supporting Information Figure S5 provides a good, general hypothesis for how the B-loop emerges from the conserved flap subdomain.
Arg243 and Lys247 in PPM1D are crucial for phosphatase activity
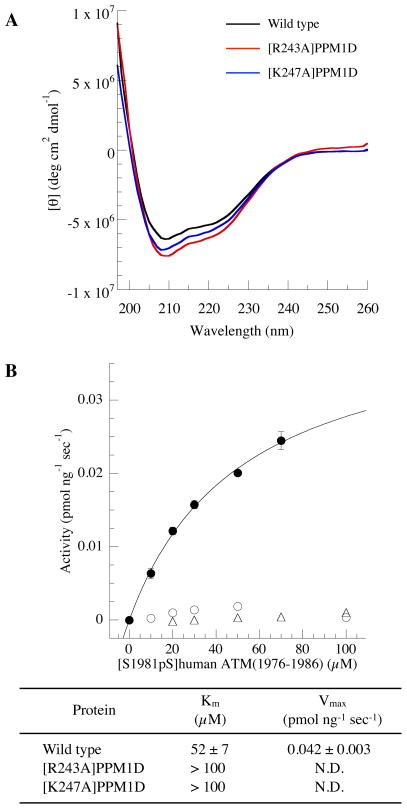
The model of PPM1D in complex with the cyclic peptides, combined with the increased inhibitory activity of cyclic peptides containing acidic groups at positions X5 and X6, suggests the importance of the basic residues in the B-loop for PPM1D activity. Among these amino acids, Arg243 is proposed by the model to interact with the acidic amino acid at position X6; Lys247, located proximal to Arg243, may also interact with acidic amino acids in the cylic peptide. To investigate if such interactions were occurring, we individually mutated these residues to alanine and looked at the effect of the mutations on PPM1D activity. We first used CD spectroscopy to ensure that the mutations did not alter the PPM1D structure. The CD spectrum of wild type PPM1D was characterized by double minima of negative ellipticity at approximately 200 and 225 nm and positive ellipticity at shorter wavelengths in the presence of Mg2+. The two mutants, [R243A]PPM1D and [K247A]PPM1D, had very similar CD spectra (Figure 9A), indicative of similar structures for the wild type and mutant proteins. Next, we estimated the phosphatase activities of the two mutants using the human ATM substrate peptide. Suprisingly, both mutants showed no phosphatase activity. These results demonstrate that Arg243 and Lys247 are crucial for PPM1D catalytic activity, likely through recognition of the phosphorylated substrate.
Figure 9.
Characterization of the structure and activity of PPM1D mutants. (A) CD spectra of wild type and mutant PPM1D; wild type (black), [R243A]PPM1D (red), [K247A]PPM1D (blue). Protein concentration was 1.8–2.1 μM in 50 mM Tris-HCl, 30 mM MgCl2, and 5% glycerol (pH 7.5). (B) Km values and Vmax values for wild type and mutant PPM1D. Phosphatase activities were measured using [S1981pS]human ATM(1976–1986) substrate peptide (closed circle, wild type; open circle, [R243A]PPM1D; open triangle, [K247A]PPM1D).
Discussion
In this study, the activity of a cyclic peptide inhibitor of PPM1D was improved 50-fold relative to previously-reported inhibitors of this type (24). Alteration of specific sidechains in the cyclic peptide resulted in marked improvements in activity and also revealed new structural information about the catalytic site and substrate specificity of PPM1D. At position X1, an aromatic residue (especially Phe) generally improves the inhibitory activity. At position X3, only the L-enantiomer of the Ile alpha carbon leads to good inhibitory activity, while either enantiomer at the beta position is tolerated. At positions X5 and X6, a negatively-charged residue increases the inhibitory activity dramatically. Combined, the optimal cyclic peptide inhibited PPM1D with Ki = 110 nM.
Previously, we found that a Leu at position X3 converts the cyclic peptide into a substrate for PPM1D, whereas Val- and Cha-substituted analogs had similar inhibitory activities as the Ile-substituted analog (24). Branching at the beta position is required for inhibitory activity, possibly by providing steric hindrance that prevents the pS sidechain from appropriately positioning in the binding pocket for hydrolysis. Furthermore, hydrophobic interactions in this region appear to be required, as a Gly-substituted linear peptide is a much weaker inhibitor as compared with the Ile-substituted linear peptide (24). The observation that the peptides had similar activity indicates that the methyl group at the beta position does not bind in one specific pocket of PPM1D.
As described above, substitution of acidic residues at positions X5 and X6 greatly improved inhibitory activity, showing a 10-fold increase relative to peptide 5 (Figure 4C, D). This observation is consistent with previous research on PPM1D substrate specificity, which reported that mutation of two Glu residues to alanines in the substrate sequence of ATM led to a 3-fold decrease in Km relative to the wild type sequence (23). A separate study investigating p53 substrate peptides reported that the introduction of acidic residues around p53 Ser15 resulted in high substrate affinity to PPM1D (26). Combined, these results demonstrate the importance for negative charge in the substrate or inhibitor for binding to PPM1D, likely through formation of electrostatic interactions with the enzyme. PPM1D is the only known PP2C family members to have a highly-basic loop inserted into the flap sub-domain, which may confer a preference for acidic substrates. Our molecular model suggests that Arg243 could interact with the Glu at position X6 on the cyclic peptide if it has the same position as in the PPM1A template. The interaction of the Glu at X6 is further suggested by the similar inhibitory activities of other peptides containing an L-acidic residue at this position and the poor inhibitory activities of those with basic residues. Thus, the uniqueness of the B-loop makes it an interesting target for development of a highly specific inhibitor.
As PPM1D is a member of a family of phosphatases, it is important to understand the selectivity of our inhibitors. We previously demonstrated that peptide 1 had no activity towards PP2A, but was a substrate for PPM1A (24). Likewise, although peptide 40 showed significantly improved PPM1D inhibition, it was dephosphorylated by PPM1A, albeit 10–fold less than peptide 1. Therefore, to obtain an inhibitor selective for PPM1D, five different peptides were synthesized, four containing a pS mimetic and the fifth a retro-inverse peptide. This process proved difficult, as four of the five showed weak to no PPM1D inhibitory activity. Only peptide 48, in which pS was substituted with pHse, was found to be active against PPM1D, although its inhibitory activity was approximately 30-fold decreased relative to peptide 40 (Figure 6C). This peptide was not dephosphorylated by PPM1A, nor was it an inhibitor of it. These results indicate that the inhibitory activity of PPM1D is strongly dependent on the carbon chain length of the pS sidechain. The dramatic changes in PPM1A dephosphorylation that resulted from relatively minor chemical variations likely reflect changes to the conformation of the complex that determine whether the phosphate bond is positioned correctly for hydrolysis by the enzyme’s metal center. Thus, we show it is possible to develop a selective inhibitor of PPM1D, but detailed structural information will likely be required to further improve the inhibitory activity.
For quite a while, the structure of PPM1A from Das et al. (25) was the only one available for this class of enzymes. However, recently published related structures have revealed that the so-called “flap sub-domain” adjacent to the active site is likely not a static unit, but rather changes conformation to accommodate the binding of different substrates (27–30, 32, 33) (Supporting Information Figure S1). This makes accurate modeling of substrate and inhibitor binding much more difficult. This is especially true for PPM1D, since it has the added complication of the relatively long B-loop. Despite this difficulty, we can surmise a number of structural features important for activity from the available data. The first is the conservation of the Arg243-Val244 sequence of PPM1D in all eukaryotic PP2C sequences and some prokaryotic ones (Figure 8A). The second is the correspondence of His107 in PPM1D with His62 in PPM1A (Supporting Information Figure S2), which has been implicated as the acid which protonates the leaving group oxygen and is thus crucial for the phosphatase activity. A corresponding histidine residue is conserved in most of the other eukaryote homologues (except PP2CH/J) and in some prokaryotic ones. In the model of the PPM1D-inhibitor complex, a negatively-charged residue at the cyclic peptide X5 position forms a stabilizing salt bridge and sterically blocks His107 of PPM1D, thus enhancing inhibition.
As the B-loop is unique to PPM1D in the PP2C family, it is important to know how it confers the unique substrate specificity of this enzyme and, thus, its physiological functions. The elimination of activity observed here for the R243A and K247A point mutations is consistent with the results of Chuman et al. (26), who substituted B-loop residues 245–268 with the corresponding residues from the PPM1A sequence (NGS). In this case, both the Km and kcat of the mutant were approximately 3-fold reduced relative to wild type PPM1D. The difference between complete abrogation and 3-fold reduction may reflect the different substrates used (ATM vs. p53 phosphopeptides) and their differing patterns of negatively charged residues. This raises the question of the purpose of these and the other positively charged residues in the B-loop. Presumably they function to attract and correctly position the highly negatively charged mono- and di-phosphorylated substrates for catalysis. Fundamental to this understanding is to know exactly where the B-loop starts in the flap subdomain sequence. As seen in Figure 8, PPM1D and the other proteins display a range of insertions before the RV motif compared to PPM1A. Thus, it is uncertain whether the highly conserved RV residues are located at the same position in the structure as in PPM1A. If the locations are conserved, as in the model presented here and as defined by Chuman et al. (26), then the arginines corresponding to Arg243 likely play similarly important roles in all the other PP2C proteins that share this motif. If not, then Arg243 plays a unique role in PPM1D along with Lys247 and potentially some or all of the other positively charged residues in the B-loop. Further research is needed to discover how these residues coordinate in a possibly mobile flap-subdomain mechanism.
Here, we report a cyclic thioether peptide (F-pS-I-pY-DDC-amide) characterized by good inhibitory activity (Ki=110 nM). To date, only a few compounds have been reported as PPM1D inhibitors (26, 43, 46, 47), and the cyclic peptides presented here show the highest inhibitory activity, to the best of our knowledge. We further identified a second cyclic peptide (F-pHse-I-pY-DDC-amide) that showed selectivity for inhibition of PPM1D over PPM1A. Although the two phosphoric acid groups on the cyclic peptide severely limit the cellular bioavailability of the compounds, their substitution with non-phosphorus-based mimetics, such as a tetrazole scaffold, a sulfhydantoin scaffold, or a pyridine-based scaffold (48), could be an effective way to overcome this issue. In addition, although the Lipinski rules suggest that a cyclic peptide would have poor absorption or permeation into a cell because of its large molecular weight, use of a drug delivery system, such as nanoparticles, including liposomes (49–52), and cell-penetrating peptide conjugation (53–55), could overcome this issue. In the absence of detailed structural information, the findings in this study will be valuable in designing the next generation of small molecule inhibitors of PPM1D and in understanding the substrate specificity of PPM1D as a serine/threonine phosphatase.
Supplementary Material
Abbreviations
- Aad
homoglutamic acid
- Boc
tert-butoxycarbonyl
- CD
circular dichroism
- Cha
beta-cyclohexylalanine
- Fmoc
9-fluorenylmethoxycarbonyl
- F2Pab
L-2-amino-4-phosphono-4,4-difluorobutyric acid
- Hph
L-homophenylalanine
- Hse
L-homoserine
- pS
phosphoserine
- pT
phosphothreonine
- RP-HPLC
reversed-phase high-performance liquid chromatography
- Tic
1,2,3,4-tetrahydroisoquinoline-3-carboxylic acid
- Trt
trityl
Footnotes
This research was supported by the Intramural Research Program of the National Cancer Institute and National Institute of Diabetes and Digestive and Kidney Diseases, National Institutes of Health.
Supporting Information Available: Supporting Figures 1–6 show the full alignment of PPM1A and PPM1D along with additional views of the PPM1D model and Supporting Table 1 contains PDB accession numbers and references for proteins used in modeling. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Alonso A, Sassing J, Bottini N, Friedberg I, Osterman A, Godzik A, Hunter T, Dixon J, Mustelin T. Protein tyrosine phosphatases in the human genome. Cell. 2004;117:699–711. doi: 10.1016/j.cell.2004.05.018. [DOI] [PubMed] [Google Scholar]
- 2.Cohen PT. Protein phosphatase 1--targeted in many directions. J Cell Sci. 2002;115:241–256. doi: 10.1242/jcs.115.2.241. [DOI] [PubMed] [Google Scholar]
- 3.Andersen JN, Jansen PG, Echwald SM, Mortensen OH, Fukada T, del Vecchio R, Tonks NK, Moller NP. A genomic perspective on protein tyrosine phosphatases: gene structure, pseudogenes, and genetic disease linkage. FASEB J. 2004;18:8–30. doi: 10.1096/fj.02-1212rev. [DOI] [PubMed] [Google Scholar]
- 4.Moorhead GB, Trinkle-Mulcahy L, Ulke-Lemee A. Emerging roles of nuclear protein phosphatases. Nat Rev Mol Cell Biol. 2007;8:234–244. doi: 10.1038/nrm2126. [DOI] [PubMed] [Google Scholar]
- 5.Gallego M, Virshup DM. Protein serine/threonine phosphatases: life, death, and sleeping. Curr Opin Cell Biol. 2005;17:197–202. doi: 10.1016/j.ceb.2005.01.002. [DOI] [PubMed] [Google Scholar]
- 6.Fiscella M, Zhang H, Fan S, Sakaguchi K, Shen S, Mercer WE, Vande Woude GF, O’Connor PM, Appella E. Wip1, a novel human protein phosphatase that is induced in response to ionizing radiation in a p53-dependent manner. Proc Natl Acad Sci U S A. 1997;94:6048–6053. doi: 10.1073/pnas.94.12.6048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lu X, Nguyen TA, Moon SH, Darlington Y, Sommer M, Donehower LA. The type 2C phosphatase Wip1: an oncogenic regulator of tumor suppressor and DNA damage response pathways. Cancer Metastasis Rev. 2008;27:123–135. doi: 10.1007/s10555-008-9127-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Takekawa M, Adachi M, Nakahata A, Nakayama I, Itoh F, Tsukuda H, Taya Y, Imai K. p53-inducible wip1 phosphatase mediates a negative feedback regulation of p38 MAPK-p53 signaling in response to UV radiation. EMBO J. 2000;19:6517–6526. doi: 10.1093/emboj/19.23.6517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lu X, Nannenga B, Donehower LA. PPM1D dephosphorylates Chk1 and p53 and abrogates cell cycle checkpoints. Genes Dev. 2005;19:1162–1174. doi: 10.1101/gad.1291305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fujimoto H, Onishi N, Kato N, Takekawa M, Xu XZ, Kosugi A, Kondo T, Imamura M, Oishi I, Yoda A, Minami Y. Regulation of the antioncogenic Chk2 kinase by the oncogenic Wip1 phosphatase. Cell Death Differ. 2006;13:1170–1180. doi: 10.1038/sj.cdd.4401801. [DOI] [PubMed] [Google Scholar]
- 11.Yoda A, Xu XZ, Onishi N, Toyoshima K, Fujimoto H, Kato N, Oishi I, Kondo T, Minami Y. Intrinsic kinase activity and SQ/TQ domain of Chk2 kinase as well as N-terminal domain of Wip1 phosphatase are required for regulation of Chk2 by Wip1. J Biol Chem. 2006;281:24847–24862. doi: 10.1074/jbc.M600403200. [DOI] [PubMed] [Google Scholar]
- 12.Oliva-Trastoy M, Berthonaud V, Chevalier A, Ducrot C, Marsolier-Kergoat MC, Mann C, Leteurtre F. The Wip1 phosphatase (PPM1D) antagonizes activation of the Chk2 tumour suppressor kinase. Oncogene. 2007;26:1449–1458. doi: 10.1038/sj.onc.1209927. [DOI] [PubMed] [Google Scholar]
- 13.Shreeram S, Demidov ON, Hee WK, Yamaguchi H, Onishi N, Kek C, Timofeev ON, Dudgeon C, Fornace AJ, Anderson CW, Minami Y, Appella E, Bulavin DV. Wip1 phosphatase modulates ATM-dependent signaling pathways. Mol Cell. 2006;23:757–764. doi: 10.1016/j.molcel.2006.07.010. [DOI] [PubMed] [Google Scholar]
- 14.Li J, Yang Y, Peng Y, Austin RJ, van Eyndhoven WG, Nguyen KC, Gabriele T, McCurrach ME, Marks JR, Hoey T, Lowe SW, Powers S. Oncogenic properties of PPM1D located within a breast cancer amplification epicenter at 17q23. Nat Genet. 2002;31:133–134. doi: 10.1038/ng888. [DOI] [PubMed] [Google Scholar]
- 15.Sinclair CS, Rowley M, Naderi A, Couch FJ. The 17q23 amplicon and breast cancer. Breast Cancer Res Treat. 2003;78:313–322. doi: 10.1023/a:1023081624133. [DOI] [PubMed] [Google Scholar]
- 16.Harrison M, Li J, Degenhardt Y, Hoey T, Powers S. Wip1-deficient mice are resistant to common cancer genes. Trends Mol Med. 2004;10:359–361. doi: 10.1016/j.molmed.2004.06.010. [DOI] [PubMed] [Google Scholar]
- 17.Saito-Ohara F, Imoto I, Inoue J, Hosoi H, Nakagawara A, Sugimoto T, Inazawa J. PPM1D is a potential target for 17q gain in neuroblastoma. Cancer Res. 2003;63:1876–1883. [PubMed] [Google Scholar]
- 18.Ehrbrecht A, Muller U, Wolter M, Hoischen A, Koch A, Radlwimmer B, Actor B, Mincheva A, Pietsch T, Lichter P, Reifenberger G, Weber RG. Comprehensive genomic analysis of desmoplastic medulloblastomas: identification of novel amplified genes and separate evaluation of the different histological components. J Pathol. 2006;208:554–563. doi: 10.1002/path.1925. [DOI] [PubMed] [Google Scholar]
- 19.Hirasawa A, Saito-Ohara F, Inoue J, Aoki D, Susumu N, Yokoyama T, Nozawa S, Inazawa J, Imoto I. Association of 17q21-q24 gain in ovarian clear cell adenocarcinomas with poor prognosis and identification of PPM1D and APPBP2 as likely amplification targets. Clin Cancer Res. 2003;9:1995–2004. [PubMed] [Google Scholar]
- 20.Loukopoulos P, Shibata T, Katoh H, Kokubu A, Sakamoto M, Yamazaki K, Kosuge T, Kanai Y, Hosoda F, Imoto I, Ohki M, Inazawa J, Hirohashi S. Genome-wide array-based comparative genomic hybridization analysis of pancreatic adenocarcinoma: identification of genetic indicators that predict patient outcome. Cancer Sci. 2007;98:392–400. doi: 10.1111/j.1349-7006.2007.00395.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bulavin DV, Phillips C, Nannenga B, Timofeev O, Donehower LA, Anderson CW, Appella E, Fornace AJJ. Inactivation of the Wip1 phosphatase inhibits mammary tumorigenesis through p38 MAPK-mediated activation of the p16(Ink4a)-p19(Arf) pathway. Nat Genet. 2004;36:343–350. doi: 10.1038/ng1317. [DOI] [PubMed] [Google Scholar]
- 22.Yamaguchi H, Minopoli G, Demidov ON, Chatterjee DK, Anderson CW, Durell SR, Appella E. Substrate specificity of the human protein phosphatase 2Cdelta, Wip1. Biochemistry. 2005;44:5285–5294. doi: 10.1021/bi0476634. [DOI] [PubMed] [Google Scholar]
- 23.Yamaguchi H, Durell SR, Chatterjee DK, Anderson CW, Appella E. The Wip1 phosphatase PPM1D dephosphorylates SQ/TQ motifs in checkpoint substrates phosphorylated by PI3K-like kinases. Biochemistry. 2007;46:12594–12603. doi: 10.1021/bi701096s. [DOI] [PubMed] [Google Scholar]
- 24.Yamaguchi H, Durell SR, Feng H, Bai Y, Anderson CW, Appella E. Development of a substrate-based cyclic phosphopeptide inhibitor of protein phosphatase 2Cdelta, Wip1. Biochemistry. 2006;45:13193–13202. doi: 10.1021/bi061356b. [DOI] [PubMed] [Google Scholar]
- 25.Das AK, Helps NR, Cohen PT, Barford D. Crystal structure of the protein serine/threonine phosphatase 2C at 2.0 A resolution. EMBO J. 1996;15:6798–6809. [PMC free article] [PubMed] [Google Scholar]
- 26.Chuman Y, Yagi H, Fukuda T, Nomura T, Matsukizono M, Shimohigashi Y, Sakaguchi K. Characterization of the active site and a unique uncompetitive inhibitor of the PPM1-type protein phosphatase PPM1D. Protein Pept Lett. 2008;15:938–948. doi: 10.2174/092986608785849236. [DOI] [PubMed] [Google Scholar]
- 27.Pullen KE, Ng HL, Sung PY, Good MC, Smith SM, Alber T. An alternate conformation and a third metal in PstP/Ppp, the M. tuberculosis PP2C-Family Ser/Thr protein phosphatase. Structure. 2004;12:1947–1954. doi: 10.1016/j.str.2004.09.008. [DOI] [PubMed] [Google Scholar]
- 28.Bellinzoni M, Wehenkel A, Shepard W, Alzari PM. Insights into the catalytic mechanism of PPM Ser/Thr phosphatases from the atomic resolution structures of a mycobacterial enzyme. Structure. 2007;15:863–872. doi: 10.1016/j.str.2007.06.002. [DOI] [PubMed] [Google Scholar]
- 29.Wehenkel A, Bellinzoni M, Schaeffer F, Villarino A, Alzari PM. Structural and binding studies of the three-metal center in two mycobacterial PPM Ser/Thr protein phosphatases. J Mol Biol. 2007;374:890–898. doi: 10.1016/j.jmb.2007.09.076. [DOI] [PubMed] [Google Scholar]
- 30.Rantanen MK, Lehtio L, Rajagopal L, Rubens CE, Goldman A. Structure of Streptococcus agalactiae serine/threonine phosphatase. The subdomain conformation is coupled to the binding of a third metal ion. FEBS J. 2007;274:3128–3137. doi: 10.1111/j.1742-4658.2007.05845.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Almo SC, Bonanno JB, Sauder JM, Emtage S, Dilorenzo TP, Malashkevich V, Wasserman SR, Swaminathan S, Eswaramoorthy S, Agarwal R, Kumaran D, Madegowda M, Ragumani S, Patskovsky Y, Alvarado J, Ramagopal UA, Faber-Barata J, Chance MR, Sali A, Fiser A, Zhang ZY, Lawrence DS, Burley SK. Structural genomics of protein phosphatases. J Struct Funct Genomics. 2007;8:121–140. doi: 10.1007/s10969-007-9036-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schlicker C, Fokina O, Kloft N, Grune T, Becker S, Sheldrick GM, Forchhammer K. Structural analysis of the PP2C phosphatase tPphA from Thermosynechococcus elongatus: a flexible flap subdomain controls access to the catalytic site. J Mol Biol. 2008;376:570–581. doi: 10.1016/j.jmb.2007.11.097. [DOI] [PubMed] [Google Scholar]
- 33.Shi Y. Serine/threonine phosphatases: mechanism through structure. Cell. 2009;139:468–484. doi: 10.1016/j.cell.2009.10.006. [DOI] [PubMed] [Google Scholar]
- 34.LAPATSANIS L, MILIAS G, FROUSSIOS K, KOLOVOS M. Synthesis of N-2,2,2-(Trichloroethoxycarbonyl)-L-Amino Acids and N-(9-Fluorenylmethoxycarbonyl)-L-Amino Acids Involving Succinimidoxy Anion as a Leaving Group in Amino-Acid Protection. Synthesis(Stuttgart) 1983:671–673. [Google Scholar]
- 35.Janssen PSL, Vannispen JW, Melgers PATA, Vandenbogaart HWM, Hamelinck RLAE, Goverde BC. Hplc Analysis of Phenylthiocarbamyl (Ptc) Amino-Acids .1. Evaluation and Optimization of the Procedure. Chromatographia. 1986;22:345–350. [Google Scholar]
- 36.Janssen PSL, Vannispen JW, Melgers PATA, Vandenbogaart HWM, Vanaalst GWM, Goverde BC. Hplc Analysis of Phenylthiocarbamyl (Ptc) Amino-Acids .2. Application in the Analysis of (Poly)Peptides. Chromatographia. 1986;22:351–357. [Google Scholar]
- 37.Jackson JL, Young MR. Protein phosphatase-2A regulates protein tyrosine phosphatase activity in Lewis lung carcinoma tumor variants. Clin Exp Metastasis. 2003;20:357–364. doi: 10.1023/a:1024012000009. [DOI] [PubMed] [Google Scholar]
- 38.Brooks BR, Brooks CL3, Mackerell ADJ, Nilsson L, Petrella RJ, Roux B, Won Y, Archontis G, Bartels C, Boresch S, Caflisch A, Caves L, Cui Q, Dinner AR, Feig M, Fischer S, Gao J, Hodoscek M, Im W, Kuczera K, Lazaridis T, Ma J, Ovchinnikov V, Paci E, Pastor RW, Post CB, Pu JZ, Schaefer M, Tidor B, Venable RM, Woodcock HL, Wu X, Yang W, York DM, Karplus M. CHARMM: the biomolecular simulation program. J Comput Chem. 2009;30:1545–1614. doi: 10.1002/jcc.21287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.MacKerell AD, Bashford D, Bellott M, Dunbrack RL, Evanseck JD, Field MJ, Fischer S, Gao J, Guo H, Ha S, Joseph-McCarthy D, Kuchnir L, Kuczera K, Lau FTK, Mattos C, Michnick S, Ngo T, Nguyen DT, Prodhom B, Reiher WE, Roux B, Schlenkrich M, Smith JC, Stote R, Straub J, Watanabe M, Wiorkiewicz-Kuczera J, Yin D, Karplus M. All-atom empirical potential for molecular modeling and dynamics studies of proteins. J Phys Chem B. 1998;102:3586–3616. doi: 10.1021/jp973084f. [DOI] [PubMed] [Google Scholar]
- 40.Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE. The Protein Data Bank. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gibrat JF, Madej T, Bryant SH. Surprising similarities in structure comparison. Curr Opin Struct Biol. 1996;6:377–385. doi: 10.1016/s0959-440x(96)80058-3. [DOI] [PubMed] [Google Scholar]
- 42.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. UCSF Chimera--a visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 43.Bang J, Yamaguchi H, Durell SR, Appella E, Appella DH. A small molecular scaffold for selective inhibition of Wip1 phosphatase. ChemMedChem. 2008;3:230–232. doi: 10.1002/cmdc.200700281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.McNemar C, Snow ME, Windsor WT, Prongay A, Mui P, Zhang R, Durkin J, Le HV, Weber PC. Thermodynamic and structural analysis of phosphotyrosine polypeptide binding to Grb2-SH2. Biochemistry. 1997;36:10006–10014. doi: 10.1021/bi9704360. [DOI] [PubMed] [Google Scholar]
- 45.Aposto I, Kuciel R, Wasylewska E, Ostrowski WS. Phosphotyrosine as a substrate of acid and alkaline phosphatases. Acta Biochim Pol. 1985;32:187–197. [PubMed] [Google Scholar]
- 46.Belova GI, Demidov ON, Fornace AJJ, Bulavin DV. Chemical inhibition of Wip1 phosphatase contributes to suppression of tumorigenesis. Cancer Biol Ther. 2005;4:1154–1158. doi: 10.4161/cbt.4.10.2204. [DOI] [PubMed] [Google Scholar]
- 47.Rayter S, Elliott R, Travers J, Rowlands MG, Richardson TB, Boxall K, Jones K, Linardopoulos S, Workman P, Aherne W, Lord CJ, Ashworth A. A chemical inhibitor of PPM1D that selectively kills cells overexpressing PPM1D. Oncogene. 2008;27:1036–1044. doi: 10.1038/sj.onc.1210729. [DOI] [PubMed] [Google Scholar]
- 48.Rye CS, Baell JB. Phosphate isosteres in medicinal chemistry. Curr Med Chem. 2005;12:3127–3141. doi: 10.2174/092986705774933452. [DOI] [PubMed] [Google Scholar]
- 49.Ruoslahti E, Bhatia SN, Sailor MJ. Targeting of drugs and nanoparticles to tumors. J Cell Biol. 2010;188:759–768. doi: 10.1083/jcb.200910104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Heidel JD, Davis ME. Clinical developments in nanotechnology for cancer therapy. Pharm Res. 2011;28:187–199. doi: 10.1007/s11095-010-0178-7. [DOI] [PubMed] [Google Scholar]
- 51.Franzen S. A comparison of peptide and folate receptor targeting of cancer cells: from single agent to nanoparticle. Expert Opin Drug Deliv. 2011;8:281–298. doi: 10.1517/17425247.2011.554816. [DOI] [PubMed] [Google Scholar]
- 52.Falciani C, Accardo A, Brunetti J, Tesauro D, Lelli B, Pini A, Bracci L, Morelli G. Target-Selective Drug Delivery through Liposomes Labeled with Oligobranched Neurotensin Peptides. ChemMedChem. 2011;6:678–685. doi: 10.1002/cmdc.201000463. [DOI] [PubMed] [Google Scholar]
- 53.Patel LN, Zaro JL, Shen W. Cell penetrating peptides: intracellular pathways and pharmaceutical perspectives. Pharm Res. 2007;24:1977–1992. doi: 10.1007/s11095-007-9303-7. [DOI] [PubMed] [Google Scholar]
- 54.Hansen M, Kilk K, Langel U. Predicting cell-penetrating peptides. Adv Drug Deliv Rev. 2008;60:572–579. doi: 10.1016/j.addr.2007.09.003. [DOI] [PubMed] [Google Scholar]
- 55.Stewart KM, Horton KL, Kelley SO. Cell-penetrating peptides as delivery vehicles for biology and medicine. Org Biomol Chem. 2008;6:2242–2255. doi: 10.1039/b719950c. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.