Abstract
Despite the success of genome-wide association studies (GWAS) in identifying loci associated with common diseases, a significant proportion of the causality remains unexplained. Recent advances in genomic technologies have placed us in a position to initiate large-scale studies of human disease-associated epigenetic variation, specifically variation in DNA methylation (DNAm). Such Epigenome-Wide Association Studies (EWAS) present novel opportunities but also create new challenges that are not encountered in GWAS. We discuss EWAS study design, cohort and sample selections, statistical significance and power, confounding factors, and follow-up studies. We also discuss how integration of EWAS with GWAS can help to dissect complex GWAS haplotypes for functional analysis.
Keywords: Epigenomics, Disease Genetics, DNA Methylation, Epigenetics, Quantitative Trait
Introduction
Elucidating the genetic and non-genetic determinants of human complex diseases represents one of the principal challenges of biomedical research. In recent years, genome-wide association studies (GWAS) have uncovered >800 single nucleotide polymorphism (SNP) associations for more than 150 diseases and other traits1. Although the complete genetic basis is not yet known for any human complex disease, re-sequencing of exomes, and ultimately whole genomes, holds promise to identify most of the remaining causal genetic variations. However, there is now increasing interest in exploring how non-genetic variation, including epigenetic factors, could influence complex disease aetiology2-4.
The epigenome of a cell is highly dynamic, being governed by a complex interplay of genetic and environmental factors5. Normal cellular function relies on the maintenance of epigenomic homeostasis, which is further highlighted by numerous reported associations between epigenomic perturbations and human diseases, notably cancer4. However, most studies of such associations to date have been performed either with inadequate genome coverage (e.g. tens to hundreds of loci) but adequate sample size, or approaching genome-wide coverage (thousands of loci) but inadequate sample sizes. Consequently, for any given human complex disease, we remain unaware of the proportion of phenotypic variation that is attributable to inter-individual epigenomic variation. This problem can only be elucidated by large-scale, systematic epigenomic equivalents of GWAS – epigenome-wide association studies (EWAS) as first proposed in 20086. At least for DNAm, technology is now available that is directly comparable in resolution and throughput to the highly successful GWAS chips that allow genotyping of around 500K SNPs.
But how does one conduct an EWAS? In addition to considerations that are common to both GWAS and EWAS (e.g. adequate technology and sample size), the design of EWAS has specific considerations with respect to sample selection. DNAm patterns are specific to tissues and developmental stages, and also change over time. Furthermore, EWAS associations can be causal as well as consequential for the phenotype in question - a difference from GWAS that presents considerable challenges. Here, we discuss these considerations in the context of designing and analyzing an effective EWAS, keeping in mind that EWAS are likely to evolve, as did GWAS, as information and experience accumulate.
Epigenetic Variation and complex disease
Types of epigenetic information
Epigenetic information in mammals can be transmitted in multiple forms5, including mitotically stable DNAm, post-translational modifications of histone proteins, and ncRNAs. For DNAm, the predominant form is methylation of cytosines in the context of cytosine-guanine dinucleotides (CpG). However, recent results suggest that CpH methylation (where H = C/A/T) may be more common than previously appreciated7,8. Catalysed by Ten Eleven Translocation (TET) methylcytosine dioxygenases, 5-hydroxymethylation9,10 of cytosines (hmC) is yet another form of DNAm. Although details are still unclear, increasing evidence suggests a role of hmC in gene regulation and differentiation11 Histone modifications include, to name but a few, mono-, di- or tri-methylation, acetylation, and citrullination of one or more amino acids in the N-terminal tails of core histones5. More recently, it has been discovered that ncRNAs can self-propagate and be transmitted independently of the underlying DNA, in other words they can ‘epigenetically’ transmit regulatory information12,13. ncRNAs include short micro RNAs (miRNA), Piwi-interacting RNAs (piRNA), large intervening non-coding RNAs (lincRNA) and others12.
Epigenetic variation in health and disease
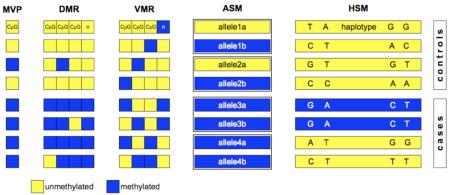
The full spectrum of epigenetic marks is currently unknown, but is potentially enormous, considering that the diploid human epigenome contains >108 Cs of which >107 are CpGs, and >108 histone tails, that can all potentially vary. The best-studied epigenetic mark is DNAm and Box 1 illustrates the most common features and contexts in which DNAm varies. DNAm variation at a single CpG site is known as a methylation variable position (MVP), which can be considered as the epigenetic equivalent of a SNP14. Very rarely, CpGs on only one of the two strands of DNA per allele are methylated. This is known as hemi-methylation, and probably reflects post-replication lag in DNAm maintenance in proliferating cells. If DNAm is altered at multiple adjacent CpG sites, this is referred to as a differentially methylated region (DMR); DMRs vary considerably in length, they are typically <1Kb but can exceed 1Mb15. Until recently, MVPs and DMRs were mostly studied in the context of core promoters, CpG islands (CGIs) and imprinted differentially methylated regions (iDMRs), however, it is becoming increasingly clear that DNAm is highly dynamic even outside of such regions. For example, a recent study found that tissue-, and cancer-specific DMRs preferentially occur in regions adjacent to CGIs, so-called CGI shores16. DNAm also plays a key role in silencing repeat elements, which may also impact on disease aetiology17,18.
Box 1. Definition of features known to vary in DNA methylation (DNAm).
This rapidly increasing list of features is not meant to be complete but intends to illustrate the key loci and contexts in which DNAm is known to vary.
Methylation variable position (MVP). A CpG site that shows differential methylation e.g. between different disease states as illustrated below. Given recent findings on non-CpG methylation, potentially all Cs could be MVPs.
Differentially methylated region (DMR):A region of the genome at which multiple adjacent CpG sites show differential methylation..DMRs can occur in many different contexts such as:
iDMR - imprinting-specific differentially methylated region
tDMR - tissue-specific differentially methylated region
rDMR - reprogramming-specific differentially methylated region
cDMR - cancer-specific differentially methylated region
aDMR -ageing-specific differentially methylated region
Variably methylated region (VMR). These are defined by increased variability rather than gain/loss of DNAm.
Allele-specific methylation (ASM). These are positions or regions that vary in DNAm depending on the parent-of-origin, the presence of a polymorphism or as a result of a stochastic event.
Haplotype-specific methylation (HSM). This is a differentially methylated region that is defined by a set of co-inherited SNPs (a haplotype).
CpG islands (CGIs). These are regions enriched for CpG sites. The majority of CGIs are unmethylated in all cell types.
CGI shores. These are regions immediately adjacent to CGIs and display higher variation in DNAm than CGIs despite their lower density of CpG sites.
The below figure shows different types of DNAm variation that can be identified with EWAS. For the purpose this simplified illustration, the cases and controls are assumed to have methylated or unmethylated CpG states only. Real samples will contain populations of different cells and hence display much more heterogeneous methylation levels across the full dynamic range between 0-100%.
The role of DNAm variation in complex disease has mainly been explored in the context of cancer, in what may be considered as early EWAS. Findings from these studies have been extensively discussed4,19, the key general conclusions being that tumour development is associated with gain of DNAm at CGIs, loss-of-imprinting, and epigenetic remodelling of repeat elements, particularly loss of DNAm at satellite DNA20,21. For non-malignant common complex diseases such as diabetes or autoimmunity, the epigenetic component is only just beginning to be investigated. Observations that support an epigenetic component in these diseases include the following. First, monozygotic twin (MZ) concordance for any complex disease is almost never 100% and recent small-scale EWAS of MZ twins discordant for systemic lupus erythematosus22 and autism spectrum disorders23 have found intra-MZ pair disease-associated epigenetic differences. Second, for several complex diseases, e.g. Type 1 Diabetes24, the incidence is rising in the general population and frequently altered in migrant populations, suggesting a role for non-genetic factors. Third, epidemiological evidence suggests that a sub-optimal in utero/early childhood environment can impact on disease outcomes (such as type 2 diabetes) in adulthood, a phenomenon termed developmental reprogramming25. Currently, the prime candidate for the molecular memory of the in utero environment is epigenetic modifications, including DNAm26-28.
Epigenetic variation as a consequence or cause of disease
As mentioned above, epigenetic variation can be causal for disease or can arise as a consequence of disease. Epigenetic variation could arise either directly or indirectly as a consequence of disease, and examples could include long-term alterations in immune-related cells in autoimmune disorders, altered metabolic regulation in type 2 diabetes, or somatic mutation-induced epigenetic alterations in cancer. However, distinguishing this from epigenetic variation that is causative of the disease process is not straightforward (as we discuss in greater detail below), but is critical since it will help elucidate the functional role of the disease-associated variation and potential utility in terms of diagnostics or therapeutics. A key step towards this goal is to determine whether the variation is present prior to any overt signs of disease. In this regard it is useful to consider how such epigenetic variation could arise prior to disease. Firstly, it could be inherited and hence be present in all tissues including the germline (i.e. transgenerational epigenetic inheritance), although the extent of this phenomenon is not fully known. Secondly, it could arise stochastically and be present soma-wide if it happens in early (e.g. in utero) development29,30 or be limited to one or a few tissues31,32 if it were to happen post-natally or during adult life. Thirdly, it could be environmentally-induced, either by adult life-style related factors such as diet or smoking33, or even in utero i.e. developmental reprogramming (described above).
It is also possible that the underlying genotype influences epigenetic variation, as recently demonstrated by several studies34-39. Loci harbouring genetic variants that influence methylation state have been termed methylation quantitative trait loci (methQTLs)34. In most methQTL, the correlations with cis-genotype are most pronounced. There is some evidence that genetic variation can also influence epigenetic states in trans, but this does not seem to be as prevalent as cis-effects38. Also, it is important to note that in most of these previous studies, the true causative genetic variant was not unequivocally identified, and the majority of methQTLs didn’t demonstrate a strict one-to-one relationship between cis-genotype and epigenotype. Rather, a given genotype generates an increased probability of methylation. Feinberg and Irizarry have recently argued for the existence of genetic variants in mouse and human genomes that do not change the mean phenotype but rather the variability of phenotype; this could be mediated epigenetically via variably methylated regions (VMR, see also Box 1)2. The existence of methQTLs provides a strong argument for integrated GWAS/EWAS to uncover genotypes that exert their function through epigenetic variation (discussed later).
methQTLs can also affect allele-specific methylation (ASM, see also Box 1). In this context, the steady-state methylation levels differ across the two alleles within the same cell. However, ASM can also occur in the absence of any specific genotype-epigenotype correlations. For example, parental imprinting, X-inactivation, random mono-allelic methylation of one allele, are all instances of ASM not due to differences in underlying genotype between methylated and unmethylated alleles.
Finally, it is also worth considering the possibility that in some cases disease-associated epigenetic variation could arise prior to disease-onset, but still not be causative for the disease per se. This type of epiphenomenon could be due to confounding, when an environmental factor such as smoking, or a genetic variant, induces both aberrant epigenetic states and disease.
These potential relationships between epigenetic variation and complex disease have important implications for the design and analysis of EWAS. First, they will determine the most relevant tissue and cell types to be sampled. Second, ‘reverse causation’ and confounding are particular issues for EWAS study design. Despite the considerable evidence of epigenetic perturbations in cancer4, and emerging evidence in other non-malignant diseases22,23,40-42, none of these studies has been able to conclusively distinguish causal from consequential epigenetic variants, a problem that has long been recognised43. Although any EWAS association with disease is potentially an advance, being able to identify the direction of causality will greatly aid in determining the utility of the epigenetic variation, e.g. as a marker of disease progression, as a target for reversal by treatment with an epi-drug, or as a measure of drug response by monitoring the kinetics of drug-induced epigenetic changes.
Profiling epigenetic variation
One of the major developments that enabled large-scale GWAS was of powerful but affordable genetic profiling technologies, in particular SNP arrays. Only recently have epigenomic profiling technologies reached the stage that large-scale EWAS have begun to be feasible. This requires that the mark/molecule is stable, amenable to high-throughput analysis, easily accessible in routine clinical samples, and that automatable whole-genome profiling methods are available. Currently, DNAm (and specifically CpG methylation) is the most suitable mark for EWAS. Other epigenetic marks may be as or more important, but are neither yet as easily accessible as DNAm in clinical specimens nor as amenable to high-throughput processing. In addition, there are numerous well-established correlations between different epigenetic marks, and hence profiling DNAm can, albeit indirectly, provide information about histone modification states and RNA dynamics5.
In principle, sequencing- and array-based profiling technologies can be used for EWAS. The most common of both these technologies have been extensively reviewed44 and independently benchmarked45,46, and are listed in Box 2. As is typical for this type of study, the choice comes down to balancing coverage, resolution, accuracy, specificity, throughput and cost47, Ultimately sequencing-based technologies are likely to prevail, but array-based methods like those used for GWAS are in our view the currently most suitable methods for EWAS. As described in Box 2, there are options for custom and off-the-self platforms covering the choices described above. Of these, the recently released Illumina 450K Infinium Methylation BeadChip looks in our view most promising for the first wave of EWAS, offering a good balance of genome-wide coverage (>450K CpG sites), resolution (single base pair) and throughput (12 samples per chip and up to 96 samples per run).
Box 2. Profiling technologies for EWAS.
Lack of suitable technology has been a major bottleneck for EWAS in the past. Fortunately, this is no longer the case and a variety of both array- and sequencing-based methods are now readily available. As these have been already been extensively reviewed44,47,80 and benchmarked45,46,81,82, they are only briefly described here along with some additional technologies that may also be suitable for EWAS as guidance for the variety of choices available.
Array-based technologies:
CHARM83: Comprehensive High-Throughput Relative Methylation; utilizes methylation-sensitive restriction enzymes.
Infinium84: The Infinium assay uses two different bead types (for methylated and unmethylated DNA) to detect CpG methylation of bisulfite treated DNA; utilizes chemical conversion of DNA.
Technologies that can be used in conjunction with arrays or sequencing:
HELP-chip/seq85: HpaII tiny fragment Enrichment by Ligation-mediated PCR; utilizes methylation-sensitive restriction enzymes.
MethylCap-chip/seq86: Methyl capture using the methyl binding domain of protein MeCP2; utilizes affinity enrichment.
MBD-chip/seq87,88: Methyl capture using complex of methyl binding proteins MBD2 and MBD3L1; utilizes affinity enrichment.
MeDIP-chip/seq89,90: Methylated DNA immunoprecipitation with antibody against 5-methylcytosine; utilizes affinity enrichment.
Sequencing-based technologies
BS-seq8: Whole-genome Bisulfite Sequencing; utilizes chemical conversion of DNA.
RRBS91: Reduced Representation Bisulfite Sequencing; utilizes chemical conversion of DNA.
Of these, the BS-seq approach - bisulfite conversion of randomly fragmented DNA followed by sequencing - provides the highest level of coverage and resolution, negligible bias towards CpG dense regions, and a direct read-out of non-CpG methylation92,93. Like all methods based on bisulfite conversion, BS-seq is not able to distinguish between methylated and hydroxymethylated cytosine bases94. Except for the reduced representation (RRBS) method which provides about 10% genome coverage, whole-genome BS-seq is currently too expensive for EWAS profiling, although costs keep falling rapidly. Affinity-based enrichment methods such as MeDIP-, MethylCap- and MBD-seq are more economical and highly automatable95 but are less quantitative and don’t provide single base resolution. In our view, the recently released Infiumium 450K BeadArrays seem well suited for EWAS profiling with respect to throughput, cost, resolution and accuracy. However, like other non sequencing-based methods this assay is susceptible to certain polymorphisms not known or considered at the time the array was designed.
Of course, the trade-off with all these methods is that many CpG sites are not profiled. As there is no epigenomic equivalent of the HapMap project which helped elucidate some of the genetic variation in the human genome77,78, we are not aware of the level of normal epigenetic variation that exists in human populations, or even which sites are the most relevant for disease aetiology. A true understanding of complex disease epigenomics will therefore only be realized when whole-genome methods become more affordable, possibly using techniques such as nanopore96 and single molecule real-time97 sequencing which are currently being developed. These will allow direct (i.e. no bisulfite, restriction or enrichment modifications required) and simultaneous determination of DNA methylation, DNA hydroxymethylation and DNA sequence in a single reaction.
Study designs for EWAS
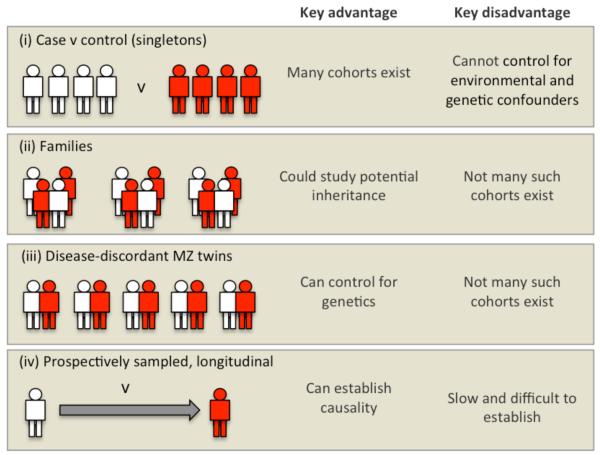
In this section, we discuss the most informative study designs for EWAS with respect to types of study subjects and addressing the issue of reverse causation. Figure 1 illustrates some of the advantages and disadvantages for the four examples discussed.
Figure 1. The different types of sample cohorts that could be used in an EWAS.
Refer to text for a full discussion.
Retrospective (case-control)
The most commonly used GWAS design involves unrelated individuals recruited on the basis of their phenotype (e.g. cases and controls). Many case-control samples are already available, in some cases with genotype and expression data that can be integrated with epigenomic data. However, a retrospective study cannot determine whether the identified epigenetic variants are due to disease-associated genetic differences, post-disease processes or disease-associated drug interventions. Early examples of using case-control studies to identify associations between epigenetic variation and clinically relevant phenotypes have included studies on metabolic dysfunction48 and treatment with tamoxifen49.
Parent-offspring pairs
These could be useful in EWAS that aim to identify transgenerational transmission of epigenetic marks (Box 3). It has recently been demonstrated that feeding F0 male mice either a high-fat or low-protein diet from weaning to the time of mating, results in F1 offspring with altered metabolic phenotypes28,50. Given that the sperm passes on very little, if any, cytoplasmic material to the offspring, these examples suggest the transgenerational transmission of epigenetic variants induced by the sub-optimal diet of the F0 males. A similar strategy using epigenomic profiling of parent-offspring trios could be used in humans. For example, if there is evidence to suggest that paternal environment influences phenotypic outcomes in the offspring, then one could perform integrated epigenomic and genomic profiling in the offspring to identify altered epigenetic variants, and the genetic information could be used to eliminate the possibility that genetic modifiers are causing the epigenetic variation. Such study designs will need to employ profiling methods able to detect allele-specific differences, be adequately powered, and have reliable measures of parental environmental exposures.
Box 3. Transgenerational Epigenetic Inheritance.
In mammals, epigenetic states are extensively reprogrammed between generations, and this is associated with the reinstatement of the pluripotent state that exists in very early development. However, a few studies have shown that occasionally epigenetic states are not completely reprogrammed, resulting in the transgenerational transmission of epigenetic states. The strongest evidence for this phenomenon in mammals comes from various mouse models such as Avy, and AxinFu (Refs29,30). In these models, the characteristic phenotype is associated with DNA methylation variation at the relevant locus. Interestingly, these states are not always completely reprogrammed between generations, thereby resulting in the range of phenotypes in the offspring being influenced by the phenotype of the parent, even in the absence of genetic heterogeneity. Establishing transgenerational epigenetic inheritance in humans is a far more challenging task since the outbred nature of human populations means that it is difficult to distinguish true epigenetic inheritance from the inheritance of genetic variants that determine variable epigenetic states. Nevertheless, several reports suggest that transgenerational epigenetic inheritance in humans may occur. If true, then we may need to reconsider whether some estimates of heritability are confounded by transgenerational epigenetic inheritance. For example, a given epigenetic state may be induced in the germline by environmental factors such as diet, and these states are passed on to the next generation, ultimately influencing phenotypic outcomes98. Indeed, in rats it has recently been demonstrated that a high-fat diet in fathers alters beta islet function in the daughters28. The true extent of this phenomenon is expected to become clearer in coming years.
Monozygotic (MZ) twins
MZ twins discordant for a disease of interest represent a useful resource for EWAS as any identified disease-associated epigenetic variant cannot be due to germline genetic variation32,51. However, unless the twins are recruited longitudinally, which is rarely possible, these studies cannot be used to distinguish between cause and consequence for the reasons discussed earlier. Recruiting large numbers of discordant MZ twins for a well-powered study is a potential problem, but some large twin resources are available (see under Links).
Longitudinal cohorts
Longitudinal cohort designs follow initially disease-free people (ideally from birth) over the course of many years, recording disease events and other phenotypic changes and taking biological samples. They are expensive to establish, but many such studies are already underway, some involving appropriate tissues for EWAS (see under Links). For example, the British 1946 birth cohort52 offers samples and data spanning 65 years so far for over 5000 individuals. Two major advantages of such studies, compared with many case-control designs, are the avoidance of confounding due to differences in the recruitment of cases and controls, and of bias due to case-control differences in the measurement of risk factors. Longitudinal studies can also be invaluable for establishing the temporal origins and stability of disease-associated epigenetic variation, and hence help to distinguish causal from consequential epigenetic variants. If environmental influences are also recorded, it may be possible to relate these to epigenetic changes.
Longitudinal cohorts of disease discordant MZ twins would convey the additional advantage of ruling out genetic influences on disease-associated epigenetic variation, but such cohorts are rarely available for EWAS of common diseases. A compromise two-phase study design, involving a disease-discordant MZ twin cohort for the discovery phase and a different longitudinal cohort for the replication phase is discussed below.
Choice of tissue for EWAS
In GWAS, most tissue types are suitable for identifying germline genetic variation and DNA extracted from patient blood or blood cell-derived cell lines is usually used. However, disease-associated epigenetic variation can be tissue-specific. Since the majority of EWAS use live individuals, DNA samples can only be easily accessed from certain sources such as blood, buccals, saliva, hair follicles, urine and faeces. Blood and blood subtypes for instance, are relevant for autoimmune diseases or blood-based cancers, and any tissue will suffice if the epigenetic variant is present soma-wide (as will be the case if induced during developmental reprogramming in early embryogenesis).
However, for many diseases alternative tissue sources need to be explored. These could include assaying cell-free serum DNA which comprises DNA from proliferating cells that is shed into the blood (as happens for most cancers), or post-mortem DNA, which is however less suitable if the aim is to establish causality. In fact, until epigenomic profiling can be routinely performed non-invasively (e.g. through imaging techniques53) and/or using very small tissue biopsies54, it will remain challenging to perform effective EWAS for brain-based and certain other diseases.
Another important issue is tissue heterogeneity. All tissues are composed of multiple cell types (e.g. blood contains >50 distinct cell types). If the disease-associated variation is restricted to a certain cell type that represents only a small proportion of the tissue sampled, then the variation may not be detected. The disease state itself can also alter the composition of cell types in a tissue (e.g. inflamed tissue will have a slightly different composition of cell types than non-inflamed tissue), and hence measured epigenetic differences between cases and controls may only reflect differences in cell type composition and not true epigenetic differences.
Finally, blood-spot (or Guthrie) cards are another valuable source of DNA. These are routinely created in many developed countries immediately after birth using either cord- or heel prick blood. Biobanks that include DNA and possibly other tissue, as well as phenotypic information, have been set up in several countries (see Links for examples).
Examples of EWAS study design
There isn’t a single EWAS design that will suit all purposes, but rather the most suitable design depends on the required outcome. This is best illustrated in the form of two hypothetical examples, from the many possible EWAS designs that could be conducted:
An EWAS for disease-risk epigenetic markers
Let’s assume that we are interested in identifying DNAm variants that arise prior to the onset of an autoimmune disease. We could start by performing genome-wide DNA methylation analysis of MZ twins discordant for the disease to identify disease-associated MVPs in immune-effector cells (i.e. a disease-relevant blood cell subset) that cannot be due to genetic variation. Then, we could take these MVPs and assay them in the same type of immune-effector cells from a prospective cohort, to look at DNAm at these sites in unrelated individuals sampled both before and after disease onset. Any MVPs that can be validated prior to disease onset are then candidate causal variations, and cannot be due to post-disease effects such as long-term medication or immune-related effects. Key follow-up studies could include correlation with gene expression and other epigenetic marks to investigate the affected pathways. Overall, this EWAS design combines analysis of a disease-relevant tissue from two independent cohorts that allow for discovery and validation of MVPs and elimination of various confounding factors.
An EWAS for drug-response epigenetic markers
Several cancer studies have identified epigenetic variants that can potentially be used to monitor disease progression and even response to treatment4. Some of these variants were detected by assaying DNA shed by the primary tumour into the patient’s serum, hence providing a relatively straightforward means of assessing progression55. An EWAS could also measure the DNAm state in serum from singleton patients that suffer from a given form of cancer, prior to, during, and following drug treatment. This could potentially identify epigenetic markers that predict the best response to treatment in real time. The root cause of the cancer-associated epigenetic variants (i.e. genetic or environmental) need not be known, nor would the primary tumour need to be directly analyzed, for the variant to be an effective measure of progression or response.
Statistical considerations for EWAS
Sample size and power
In 2005, just as the GWAS wave was about to break, Wang et al. published an influential review56 arguing for large sample sizes to detect small effects, and they highlighted the role of both minor allele frequency (MAF) and effect size in determining the power of a test of SNP association. They also discussed predictions from population genetics theory of the MAF spectrum over SNPs within a population, and the (limited) theory and data to predict effect size distributions. The corresponding arguments are no less compelling for EWAS, but the relevant parameters are even more difficult to predict, because of the paucity of data and relevant theory. DNA alleles do not typically vary across cells, and can now be typed with very low error rates. By contrast, methylation states may be tissue-specific, and can vary over cells within a tissue, over alleles within a cell (ASM) and in rare cases over DNA strands within an allele (hemi-methylation). Thus, for a tissue sample from one individual, the methylation state measured at a CpG site lies between zero and one, since it is an average over cells, alleles and strands and is further blurred by measurement error. Here, we use the limited available information about frequency spectra of DNA methylation variants, and their effect sizes for common disease, to tentatively propose power calculations under three scenarios. It remains unclear how realistic the proposed scenarios are but we hope at least to stimulate further discussion and investigation into this important aspect of EWAS study design.
A recent methylome analysis reported that on average 68% of CpG sites were methylated in human peripheral blood mononuclear cells57. There was great variation across genomic contexts: CpG sites in regions of high CpG density were almost always unmethylated, as were CGIs and 5′-UTRs; by contrast, 3′-UTRs, introns and repetitive elements were predominantly methylated. The rate of ASM was estimated to be between 0.3% and 0.6% (more than that attributable to imprinting alone). Hemi-methylation was found to be very rare (<0.2% which included non-CpG methylation and incomplete bisulfite conversion). The methylation spectrum was not symmetric: there were few sites close to being 100% methylated, but almost entirely unmethylated sites were not uncommon.
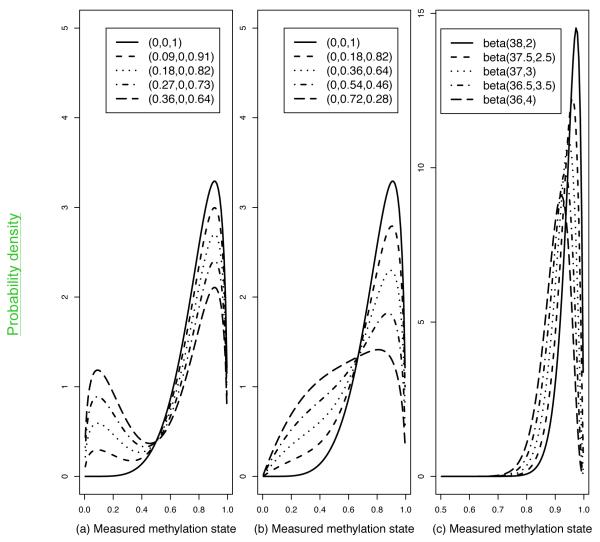
In Figure 2 (a,b) we have hypothesized methylation spectra for three different classes of individuals (“methylated”, “intermediate” and “unmethylated”) in order to generate overall frequency spectra in cases and controls. These form the basis of the power simulations, reported in Table 1. The difference in mean methylation rate between cases and controls provides a popular summary of effect size, but it does not reflect differences in variances or other features of the methylation spectrum. It also does not reflect the relative magnitude of methylation rates, whereas if a rare epigenotype in controls is almost absent in cases, this is likely to be more important than the same difference of mean rates for a more common epigenotype.
Figure 2. Hypothetical DNA methylation frequency spectra in cases and controls.
Methylation states in controls (solid curve) and cases for four effect sizes (other curves) are shown under three scenarios. For (a) and (b), the proportions of individuals who are, respectively, unmethylated, intermediate, or methylated in controls and the four sets of cases are shown in the keys. The distributions of measured methylation states are assumed to follow the following beta distributions (i) unmethylated individuals: beta(1.5,6) distribution, which has mean = 0.2, SD = 0.14; (ii) intermediate individuals: beta(2,2), mean = 0.50, SD = 0.22; (iii) methylated individuals: beta(6,1.5), mean = 0.80, SD = 0.14. For (c), the methylation spectrum is assumed to follow a single beta distribution for controls and each set of cases, and its parameters are shown in the key
Table 1. EWAS power simulation.
| (Number of cases, number of controls) |
Power (%) to detect an MVP | |||
|---|---|---|---|---|
| Scenario a |
meth OR = 1.24 md = 3.6% |
meth OR = 1.49 md = 7.2% |
meth OR = 1.78 md = 10.8% |
meth OR = 2.10 md = 14.4% |
| (100,100) | 0 0 | 0 0 | 2 0 | 18 1 |
| (200,200) | 0 0 | 4 0 | 55 11 | 99 78 |
| (400,400) | 1 0 | 66 21 | 100 98 | 100 100 |
| (800,800) | 20 3 | 100 99 | 100 100 | 100 100 |
| Scenario b |
meth OR = 1.24 md = 3.6% |
meth OR = 1.49 md = 7.2% |
meth OR = 1.78 md = 10.8% |
meth OR = 2.10 md = 14.4% |
| (100,100) | 0 0 | 1 0 | 13 1 | 60 19 |
| (200,200) | 0 0 | 16 2 | 84 46 | 100 97 |
| (400,400) | 2 0 | 85 51 | 100 100 | 100 100 |
| (800,800) | 33 8 | 100 100 | 100 100 | 100 100 |
| Scenario c |
meth OR = 1.27 md = 1.25% |
meth OR = 1.54 md = 2.5% |
meth OR = 1.82 md = 3.75% |
meth OR = 2.11 md = 5.0% |
| (100,100) | 1 0 | 37 10 | 95 77 | 100 99 |
| (200,200) | 7 1 | 95 78 | 100 100 | 100 100 |
| (400,400) | 50 19 | 100 100 | 100 100 | 100 100 |
| (800,800) | 98 88 | 100 100 | 100 100 | 100 100 |
Power (%) to detect an MVP at α = 10−6 (left entry in each cell) and α = 10−8 (right entry) for the sample sizes stated in column 1 under scenarios (a), (b) and (c) of Figure 2. methOR = methylation odds ratio, the odds for a random DNA strand in the tissue sample from a random case to be methylated, divided by the same odds for controls; md = difference in mean methylation rate between cases and controls. Analysis is via a Wald test in logistic regression implemented in the R software.
Odds ratios are well-established measures of genetic effect sizes for binary phenotypes. If we regard the mean methylation rate at a site in cases (or controls) to represent the methylation probability for a randomly-chosen DNA strand in the case (or control) tissue samples, then we can compute a methylation odds ratio. We call this methOR; it is the same as the ordinary OR except that the sampling unit is a DNA strand, rather than an individual. Thus, the methOR is the odds for a random DNA strand in the tissue sample from a random case to be methylated, divided by the same odds for controls. This provides a measure of effect size that incorporates relative magnitudes, but like the mean difference in rates it also does not allow for difference between cases and controls of features of the methylation spectrum such as its variance. As for other odds ratios, methOR is comparable across prospective and retrospective studies, and its value only measures association and does not imply causation.
Table 1 gives simulation-based power estimates for three sets of methylation spectra from Figure 2. They have similar methORs, while the case-control differences in mean methylation rates are the same for (a) and (b) but not (c). The fact that the power values differ between (a) and (b) emphasizes that there is no single-number measure of effect size since power depends on the entire methylation spectra in cases and controls. However, for the logistic regression analysis conducted in our simulations, methOR gives a better guide to power than the difference in rates. When methOR is around 1.25, a sample size of 800 cases + 800 controls is adequate to achieve 80% power at a significance level of α = 10−6 for scenario (c), but not (a) or (b) (see next section for a discussion of genome-wide significance for EWAS). When methOR is around 1.5, a sample size of 400 + 400 gives 80% power at α = 10−6 for (b) and (c), but not (a).
Very little is currently known about actual differences in methylation spectra at epigenetic variants implicated in disease, and recommendations about sample size will need to evolve with emerging data. A recent report58 on the effects of smoking on methylation identified one very strong association at a CpG site located in F2RL3, for which the median methylation rates were 95% for never-smokers and 83% for heavy smokers, giving a difference of 12% and methOR=2.7. Methylation status was much less variable in never smokers than in heavy smokers (inter-quartile ranges 0.94-0.96 and 0.78-0.88, respectively). For such a strong effect the sample size of 65 heavy smokers and 56 non-smokers was adequate to detect the association, but smoking is known to be among the most important environmental factors for health and other effect sizes of interest are likely to be much smaller. If we regard 1.5 to be a target methOR value, then it would seem to be not cost effective to pursue an EWAS with fewer than 400 cases and 400 controls, and 800 of each would be preferable to achieve good power. This is much less than the 2,000 cases and controls that became the de facto standard minimum sample size for GWAS following the Wellcome Trust Case Control Consortium (WTCCC) study59, reflecting the fact that effect sizes for EWAS and GWAS are not directly comparable. It seems likely that effect sizes and hence power will vary substantially according to genomic context, in which case genome-wide ranking by p-values is unsatisfactory60 and Bayesian measures of support that take power into account are more appropriate. Currently, however there remains little information to inform Bayesian prior distributions of effect sizes.
Genome-wide significance
In GWAS, the establishment of genome-wide thresholds for significance is complicated by correlations between the genotyped SNPs61. In EWAS, there are analogous correlations among DNAm sites in DMRs, but these correlations typically extend to at most a few kilobases, though to date they have only been reported in non-disease contexts. Based on what we discussed above on co-methylation, ASM and hemi-methylation, the vast majority of CpG methylation can be expected to be symmetric across strands and across alleles in somatic cells. Thus, the ~28 million CpG sites in the haploid human genome correspond, due to correlation within DMRs and methylation symmetry, to substantially fewer independent methylation states. If a set of 500K CpG sites were evenly spaced, the average spacing between sites may be large enough to allow an assumption of independence, in which case a significance level α = 10−6 per site gives probability 0.36 of no false positives (= type 1 error rate) under the null and this might be regarded as a liberal threshold for a possible EWAS association. If 5 million CpG sites were assayed, we would expect 5 false positives under the null at this α level. Correlation among neighbouring sites means that a specific calculation is required to identify a stringent standard for epigenome-wide significance (global type 1 error < 0.05), which will typically lie between 10−8 and 10−7.
Confounding in EWAS
GWAS can be affected by two sources of confounding. Firstly, with retrospective ascertainment there is a risk of systematic differences between cases and controls in the handling or processing of samples (known as technical confounding, which includes batch effects)62,63. Similar problems are possible for EWAS. Secondly, confounding can arise because the ancestry of cases differs systematically from that of controls (known as population structure and cryptic relatedness)64. This causes confounding in GWAS because any polygenic contribution to disease causation is correlated with ancestry, and environmental exposures may also be correlated with ancestry for example due to different geographic locations of ancestors. Whether or not “polyepigenetic” effects exist seems unclear, but environmental exposures correlated with ancestry seem likely to impact epigenetic studies.
Unlike GWAS, environmental factors can also directly confound an EWAS, by affecting both epigenotype and phenotype, which can inflate type 1 error and exaggerate effect size estimates. Potential confounders such as age65 and smoking behaviour should if possible be adjusted for in a regression analysis. Even if a measured covariate is not a confounder, but for example has an independent effect on phenotype, then adjusting for it can allow better delineation of the direct epigenetic effect.
Fortunately the large numbers of SNPs in a GWAS allow many possibilities to detect and correct confounding63, including genome-wide adjustment of association statistics, regression adjustment using principal coordinates and mixed regression models64. Similar methods are likely to be effective to detect and adjust for confounding in EWAS. For example, leading principle coordinates of genome-wide methylation states may encapsulate unmeasured confounders, so if these are also correlated with phenotype then it may be appropriate to include these as covariates in a regression analysis, as is common for GWAS analyses. Indeed if GWAS data is also available on the EWAS study individuals, it may be appropriate to adjust for leading principle coordinates of both genetic and epigenetic states.
Analysis of multi-stage studies
The values in Table 1 assume a single-stage study but as discussed above the possibilities of confounding, correlation with genotype and of reverse causation often argue for a two-stage study design, for example including a discordant MZ twin stage followed by a longitudinal cohort stage. In simple settings it is optimal if the sample size in each stage is inversely proportional to the square root of the cost per individual in that stage66. The question arises as to whether the second stage should assay all the sites from the first stage, or whether costs can be reduced by assaying in stage 2 only a limited set of “hits” from stage 1. The relatively low cost and additional information argue for the former strategy in general, unless stage 1 is large enough to eliminate all but a handful of potential hits. In either case it is broadly speaking optimal to conduct a single, joint analysis of results from both stages. If stage 1 involves MZ twin pairs, a paired analysis may be appropriate (such as a paired t-test) if there is substantially more variation among than within twin pairs. A combined two-sample case-control analysis is then not appropriate, but it is straightforward to combine test statistics from the two stages using standard meta-analysis techniques.
Replication for EWAS
Particularly in the early days of GWAS studies, replication of hits in an independent study was important in weeding out false positives that arose through technical or design flaws in the initial study. Arguably GWAS study design has improved to the extent that replication is less crucial now since there are many checks available on the quality of the primary study, but replication is still seen as highly desirable and is typically relatively easy to achieve. Ideally replication should be carried out by an independent group of researchers, preferably using a different study design and different laboratory techniques, yet studying the same polymorphism in the same population and with the same phenotype definition. In practice it is impossible to demand all this, and what constitutes a satisfactory compromise is a matter of debate, although there are some broad points of consensus67. For EWAS, the same issues arise and in addition the issues of correlation with genotype and reverse causation should both be addressed in replicate analyses. Thus a replication is potentially more demanding for EWAS than GWAS, yet limited availability of tissue samples and study subjects mean that replication will be harder to achieve. As EWAS begin to develop it would be inappropriate for reviewers and editors to impose overly strict replication requirements analogous to those used in the current mature phase of GWAS. In particular, we should avoid any encouragement for researchers to hold back samples or resources from the primary study in order to use them later to claim “replication”. Lessons should be learned from the GWAS experience: the primary study needs to be well powered, and rigorous quality checks imposed on the EWAS data. If replication is not immediately feasible this should not preclude publication, but the need for further confirmation of results should be acknowledged. The appropriate level of tolerance of false positives from the primary study depends on several factors, including the costs of follow-up analyses. If these costs are not too excessive it may be optimal to initially tolerate some false positives in order to minimise false negatives. The field of EWAS needs to develop similarly to GWAS, with standards tightening over time with progressive learning from accumulated experience.
Post-EWAS follow up studies
The ultimate aim of EWAS, like GWAS, is to provide a better understanding of disease aetiology, and to lead to the development of novel therapeutics and diagnostics. Typical follow-up experiments to determine the etiological role of disease-associated epigenetic variation could include correlation with other epigenetic modifications and collectively how they impact on gene expression. This could be achieved using ChIP-seq experiments, either for the many histone modifications known to correlate with DNAm68 or for transcription factors whose binding may be modulated – positively or negatively – by methylation at their target sites69. If a large effect size can be determined for a single site, then one could validate the link to the disease-associated phenotype by modulating the expression of the gene in question either in in vitro systems or model organism studies. However, a more likely scenario is of many disease-associated epigenetic variants each conferring only a small disease risk, as is suggested by the few small-scale EWAS to date22,23,40-42. In this case, it may be more fruitful to use approaches that integrate both computational and experimental methodologies to look at perturbations of entire transcriptional networks. The issue of reverse causation is also important in post-EWAS experiments, both in terms of which variants to follow-up, and the experimental approaches.
Even if the etiological role of any identified epigenetic variant proves elusive, it may still be possible to use them as predictive biomarkers. In this regard, the combination of chemical stability and ontogenetic plasticity make DNAm ideally suited as a biomarker. Translating any molecular marker including DNAm differences into clinically informative biomarkers has turned out to be more challenging70 than had been expected but progress has been made. Following earlier setbacks, a multi centre study identified, validated and replicated hypermethylation at SEPT9 as a blood-based DNAm biomarker for colorectal cancer in 200871, leading to a commercial test in early 201072. But enthusiasm is tempered with caution, as illustrated by the problems encountered by the cancer community in identifying biomarkers that predict which patients would benefit from a particular therapy70. The main problem has been the inability to select patients with a molecularly well-defined disease phenotypedue in large part to the heterogeneity of cancer tissues. Molecular heterogeneity is also an issue, though expected to be less important, for the common diseases that are being targeted by the first wave of EWAS.
Based on this experience, a systematic approach such as the recently launched OncoTrack project (see under Links) is needed to advance the field. Two bodies in particular - the Biomarkers Consortium and the AACR-FDA-NCI Cancer Biomarkers Collaborative - have recently issued a comprehensive report on the current state of affairs and future directions73. The response of the community has been positive with calls like ‘Bring on the biomarkers74” and pledging to replace the patched framework of fragmented research by a co-ordinated ‘big-science’ approach (such as OncoTrack) which has proved successful for efforts like the human and cancer genome projects. Based on this and other efforts, we can be cautiously optimistic that similar progress will also be made for epigenetic biomarkers.
Integration of EWAS and GWAS
The correlations that have been observed between genotype and epigenotype (methQTLs) are encouraging for the prospects of further integrated analysis. A recent study39 analysing SNPs, gene expression and DNAm in 77 HapMap cell lines identified SNPs that affect both gene expression and DNAm and provides evidence for shared genetic and epigenetic mechanisms affecting multiple QTLs. In this way, EWAS can be used to investigate genetic predispositions that exert their function through epigenetic mechanisms. A possible strategy involves designing of a custom array tiled across haplotypes identified by disease-associated GWAS SNPs, profiling it for differential DNAm and analysing the data stratified for risk SNPs rather than cases and controls. Using this strategy a recent study75 successfully integrated GWAS and EWAS data to identify haplotype-specific DNA methylation (HSM) in a Type 2 Diabetes and Obesity susceptibility locus. In the future, it may well be possible to do similar analyses for additional and combinatorial epigenetic marks to capture certain chromatin disease states e.g. based on altered bivalency status that are currently not easily captured by DNAm. Using multivariate Hidden Markov analysis of recurrent and spatially coherent combinations of epigenetic marks, a recent study76 reported 51 distinct chromatin states for human T cells that look highly promising for possible integration with GWAS data of blood-based diseases.
Conclusions and future directions
The success of GWAS in identifying disease-associated genetic variations clearly warrants the development of complementary approaches to identify additional variations that cannot be captured with GWAS. As outlined in this article, EWAS has the potential to do just that by capturing disease-associated epigenetic variations such as differential DNA methylation.
The single most useful resource empowering GWAS was the availability of a detailed SNP map of the human genome77,78 which allowed the selection of so-called tag SNPs for comprehensive variation coverage and cost-efficient profiling. DNAm is correlated over tissue-specific blocks of CpG sites spanning up to 1 Kb79. Knowledge of this block structure for different tissues and cell types has and will continue to improve the selection of CpG sites for EWAS as new methylome maps become available. Currently, such high-resolution maps are available for human embryonic stem cells, foetal fibroblasts and peripheral blood monocytes8,57, informing potential EWAS on early developmental disorders and blood-based diseases. As part of the recently launched International Human Epigenome Consortium (IHEC), 1000 reference epigenomes (including methylome maps) will be generated for human tissues and cell types over the coming years. In this context, these maps can be considered as the epigenetic equivalent to the human haplotype map and can be expected to significantly accelerate and improve our ability to conduct EWAS for many common diseases.
In addition to improving study design – for which we have discussed the key issues in this Review - the main challenge for EWAS will be access to appropriate samples. A useful starting point would be to establish the proposed Biobank Central (see under Links) which will allow researchers to electronically search for specific combinations of samples and associated data as required for EWAS. Initiation of new birth and other longitudinal cohorts should also be encouraged and existing collections should ensure that samples are suitable for EWAS and related studies that are likely to require chromatin (not just DNA) in the future. Finally, appropriately powered and designed EWAS need to be conducted to enable the development of tools for the analysis, interpretation and integration of EWAS data. To achieve this will require close cooperation between scientists, clinicians, resource providers and funding agencies as pioneered for GWAS. At the time of writing, the first wave of EWAS was still underway and an international conference (see under Links) has been arranged for later this year to discuss first results.
Acknowledgements
SB was supported by the Wellcome Trust (084071) and a Royal Society Wolfson Research Merit Award.
Glossary
- Alelle-specific Methylation (ASM)
The presence of DNA methylation on only one of the two alleles present in a cell. This could be due to parental imprinting, random methylation of one allele, or due to genetic effects.
- Bivalent chromatin
Chromatin that contains both activating and repressing epigenetic modifications at the same locus.
- Core promoters
Region upstream and downstream of Transcriptional Start Site (TSS), typically defined as the interval −60 to +40 bases from TSS.
- Core histones
The proteins that form the nucleosome which is composed of two copies each of histones 2A, 2B, 3 and 4. Together they form a histone octamer around which 147 bases of genomic DNA are wrapped.
- CpG islands (CGIs)
Regions of the genome (typically 500 bp - 2 kb) that contain a higher than expected frequency of CpG sites. CGIs are frequently unmethylated and found near promoter regions.
- Epigenome
The complete collection of epigenetic marks, such as DNA methylation and histone modifications, and other molecules that can transmit epigenetic information such as non-coding RNAs, that exist in a cell at any give point in time.
- Epimutation
A heritable aberrant epigenetic state.
- Exome
The part of a genome that encodes exons for translation into proteins.
- Genome-wide association studies (GWAS)
A genome-wide study of designed to identify genetic associations with observable trait/disease/condition e.g. diabetes.
- Imprinted genes
Genes that are expressed in a parent-of-origin specific manner.
- Loss-of-imprinting (LOI)
ParentalImprinting results in the epigenetic silencing of one allele of a gene due to its parental origin. Aberrant disruption of imprinting leads to both alleles being expressed i.e. loss-of-imprinting.
- Satellite DNA
Type of non-coding, repetitive DNA that is a component of functional centromeres and the main structural constituent of heterochromatin.
- Methylation quantitative trait loci (methQTL)
DNA variants that influence the DNA methylation state either in cis- or trans.
- Reverse causation
Refers to an association between A and B being due to B causing A rather than the presumed A causing B.
- Methylation-sensitive restriction enzyme digestion
Procedure to cleave double-stranded DNA depending on the methylation status of the enzyme’s recognition site. Some enzymes only cleave when recognition site is methylated and others only when site is unmethylated.
- Affinity enrichment
In this context, refers to a procedure to enrich methylated DNA fragments from a pool of methylated and unmethylated fragments using affinity reagents such as antibodies against 5-methylcytosine or other methyl-biding proteins.
- Reduced representation bisulfite sequencing (RRBS)
A procedure for single base resolution methylation analysis using bisulfite DNA sequencing of a representative part of a genome, typically 10%.
- Bayesian
The two main statistical schools are the Classical, or frequentist, school that dominated 20th century science and measures the strength of evidence against a hypothesis using p-values, and the Bayesian school, developed in the 19th century but currently undergoing a resurgence, which attempts to compute the posterior probability that the hypothesis is true.
- Population stratification
refers to any systematic pattern of mating in a population, which entails differences in allele frequencies between different parts of the population. These differences can be problematic if the different parts of the population are unequally represented in phenotypic groups, such as cases and controls, as this can lead to spurious associations between alleles and phenotypes.
- Principal co-ordinate analysis
a multivariate statistical technique that is related to Principal Components Analysis but investigates individuals rather than variables. It is often used to investigate population structure in a sample of individuals whose relatedness has been estimated from genome-wide genotype data.
Footnotes
Links
Biomarker Consortium: http://www.thebiomarkersconsortium.org/
Exemplar Project: http://www.oncotrack.org/
GWAS: http://www.genome.gov/26525384
IGGC: http://www.icgc.org/
IHEC: http://www.ihec-epigenomes.org/
Biobank Central: http://en.wikipedia.org/wiki/BioBank_Central
Biobank examples:
UK http://www.ukbiobank.ac.uk/
Canada http://biosample.ca/
Europe: http://www.eurobiobank.org/
International: http://www.p3gobservatory.org/
Longitudinal cohort examples:
UK: http://www.halcyon.ac.uk/?q=cohorts
http://www.bristol.ac.uk/alspac/
International: http://en.wikipedia.org/wiki/Longitudinal_study
Twin cohort examples:
UK: http://www.twinsuk.ac.uk/cohort.html
Europe: http://www.genomeutwin.org/
US: http://www.niehs.nih.gov/news/events/pastmtg/2005/twin/index.cfm
EWAS Conference: http://www.wellcome.ac.uk/conferences/epigenomics
Author’s web sites:
VKR: http://www.icms.qmul.ac.uk/Profiles/Diabetes/Rakyan%20Vardhman.htm
TAD: http://www.gurdon.cam.ac.uk/down.html
DJB: http://www.zebfontaine.eclipse.co.uk/djb.htm
SB: http://www.ucl.ac.uk/cancer/research-groups/medical-genomics
References
- 1.Hindorff LA, et al. Potential etiologic and functional implications of genome-wide association loci for human diseases and traits. Proc Natl Acad Sci U S A. 2009;106:9362–7. doi: 10.1073/pnas.0903103106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Feinberg AP, Irizarry RA. Evolution in health and medicine Sackler colloquium: Stochastic epigenetic variation as a driving force of development, evolutionary adaptation, and disease. Proc Natl Acad Sci U S A. 2010;107(Suppl 1):1757–64. doi: 10.1073/pnas.0906183107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Petronis A. Epigenetics as a unifying principle in the aetiology of complex traits and diseases. Nature. 2010;465:721–7. doi: 10.1038/nature09230. [DOI] [PubMed] [Google Scholar]
- 4.Kulis M, Esteller M. DNA methylation and cancer. Advances in genetics. 2010;70:27–56. doi: 10.1016/B978-0-12-380866-0.60002-2. [DOI] [PubMed] [Google Scholar]
- 5.Bernstein BE, Meissner A, Lander ES. The mammalian epigenome. Cell. 2007;128:66981. doi: 10.1016/j.cell.2007.01.033. [DOI] [PubMed] [Google Scholar]
- 6.MacArthur D. Why do genome-wide scans fail? Genetic Future. 2008 http://www.genetic-future.com/2008/03/why-do-genome-wide-scans-fail.html.
- 7.Ramsahoye BH, et al. Non-CpG methylation is prevalent in embryonic stem cells and may be mediated by DNA methyltransferase 3a. Proc Natl Acad Sci U S A. 2000;97:5237–42. doi: 10.1073/pnas.97.10.5237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lister R, et al. Human DNA methylomes at base resolution show widespread epigenomic differences. Nature. 2009;462:315–22. doi: 10.1038/nature08514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kriaucionis S, Heintz N. The nuclear DNA base 5-hydroxymethylcytosine is present in Purkinje neurons and the brain. Science. 2009;324:929–30. doi: 10.1126/science.1169786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tahiliani M, et al. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science. 2009;324:930–5. doi: 10.1126/science.1170116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Veron N, Peters AH. Epigenetics: Tet proteins in the limelight. Nature. 2011;473:293–4. doi: 10.1038/473293a. [DOI] [PubMed] [Google Scholar]
- 12.Zaratiegui M, Irvine DV, Martienssen RA. Noncoding RNAs and gene silencing. Cell. 2007;128:763–76. doi: 10.1016/j.cell.2007.02.016. [DOI] [PubMed] [Google Scholar]
- 13.Rassoulzadegan M, et al. RNA-mediated non-mendelian inheritance of an epigenetic change in the mouse. Nature. 2006;441:469–74. doi: 10.1038/nature04674. [DOI] [PubMed] [Google Scholar]
- 14.Rakyan VK, et al. DNA methylation profiling of the human major histocompatibility complex: a pilot study for the human epigenome project. PLoS Biol. 2004;2:e405. doi: 10.1371/journal.pbio.0020405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Frigola J, et al. Epigenetic remodeling in colorectal cancer results in coordinate gene suppression across an entire chromosome band. Nature genetics. 2006;38:540–9. doi: 10.1038/ng1781. [DOI] [PubMed] [Google Scholar]
- 16.Irizarry RA, et al. The human colon cancer methylome shows similar hypo- and hypermethylation at conserved tissue-specific CpG island shores. Nat Genet. 2009;41:178186. doi: 10.1038/ng.298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Edwards JR, et al. Chromatin and sequence features that define the fine and gross structure of genomic methylation patterns. Genome research. 2010;20:972–80. doi: 10.1101/gr.101535.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fabris S, et al. Biological and clinical relevance of quantitative global methylation of repetitive DNA sequences in chronic lymphocytic leukemia. Epigenetics: official journal of the DNA Methylation Society. 2011;6:188–94. doi: 10.4161/epi.6.2.13528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lechner M, Boshoff C, Beck S. Cancer epigenome. Advances in genetics. 2010;70:247–76. doi: 10.1016/B978-0-12-380866-0.60009-5. [DOI] [PubMed] [Google Scholar]
- 20.Ting DT, et al. Aberrant overexpression of satellite repeats in pancreatic and other epithelial cancers. Science. 2011;331:593–6. doi: 10.1126/science.1200801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Feber A, et al. Comparative methylome analysis of benign and malignant peripheral nerve sheath tumors. Genome research. 2011;21:515–24. doi: 10.1101/gr.109678.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Javierre BM, et al. Changes in the pattern of DNA methylation associate with twin discordance in systemic lupus erythematosus. Genome Res. 2010;20:170–9. doi: 10.1101/gr.100289.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nguyen A, Rauch TA, Pfeifer GP, Hu VW. Global methylation profiling of lymphoblastoid cell lines reveals epigenetic contributions to autism spectrum disorders and a novel autism candidate gene, RORA, whose protein product is reduced in autistic brain. The FASEB journal: official publication of the Federation of American Societies for Experimental Biology. 2010;24:3036–51. doi: 10.1096/fj.10-154484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bach JF. The effect of infections on susceptibility to autoimmune and allergic diseases. The New England journal of medicine. 2002;347:911–20. doi: 10.1056/NEJMra020100. [DOI] [PubMed] [Google Scholar]
- 25.Barker DJ. Maternal nutrition, fetal nutrition, and disease in later life. Nutrition. 1997;13:807–13. doi: 10.1016/s0899-9007(97)00193-7. [DOI] [PubMed] [Google Scholar]
- 26.Thompson RF, et al. Experimental intrauterine growth restriction induces alterations in DNA methylation and gene expression in pancreatic islets of rats. The Journal of biological chemistry. 2010;285:15111–8. doi: 10.1074/jbc.M109.095133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Heijmans BT, et al. Persistent epigenetic differences associated with prenatal exposure to famine in humans. Proc Natl Acad Sci US A. 2008;105:17046–9. doi: 10.1073/pnas.0806560105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ng SF, et al. Chronic high-fat diet in fathers programs beta-cell dysfunction in female rat offspring. Nature. 2010;467:963–6. doi: 10.1038/nature09491. [DOI] [PubMed] [Google Scholar]
- 29.Rakyan VK, et al. Transgenerational inheritance of epigenetic states at the murine Axin(Fu) allele occurs after maternal and paternal transmission. Proceedings of the National Academy of Sciences of the United States of America. 2003;100:2538–43. doi: 10.1073/pnas.0436776100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Morgan HD, Sutherland HG, Martin DI, Whitelaw E. Epigenetic inheritance at the agouti locus in the mouse. Nature genetics. 1999;23:314–8. doi: 10.1038/15490. [DOI] [PubMed] [Google Scholar]
- 31.Fraga MF, et al. Epigenetic differences arise during the lifetime of monozygotic twins. Proc Natl Acad Sci U S A. 2005;102:10604–9. doi: 10.1073/pnas.0500398102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kaminsky ZA, et al. DNA methylation profiles in monozygotic and dizygotic twins. Nat Genet. 2009;41:240–5. doi: 10.1038/ng.286. [DOI] [PubMed] [Google Scholar]
- 33.Christensen BC, et al. Aging and environmental exposures alter tissue-specific DNA methylation dependent upon CpG island context. PLoS Genet. 2009;5:e1000602. doi: 10.1371/journal.pgen.1000602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang D, et al. Genetic control of individual differences in gene-specific methylation in human brain. Am J Hum Genet. 2010;86:411–9. doi: 10.1016/j.ajhg.2010.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kerkel K, et al. Genomic surveys by methylation-sensitive SNP analysis identify sequence-dependent allele-specific DNA methylation. Nat Genet. 2008;40:904–8. doi: 10.1038/ng.174. [DOI] [PubMed] [Google Scholar]
- 36.Hellman A, Chess A. Extensive sequence-influenced DNA methylation polymorphism in the human genome. Epigenetics & chromatin. 2010;3:11. doi: 10.1186/1756-8935-3-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gibbs JR, et al. Abundant quantitative trait loci exist for DNA methylation and gene expression in human brain. PLoS Genet. 2010;6:e1000952. doi: 10.1371/journal.pgen.1000952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shoemaker R, Deng J, Wang W, Zhang K. Allele-specific methylation is prevalent and is contributed by CpG-SNPs in the human genome. Genome Res. 2010;20:883–9. doi: 10.1101/gr.104695.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bell JT, et al. DNA methylation patterns associate with genetic and gene expression variation in HapMap cell lines. Genome Biol. 2011;12:R10. doi: 10.1186/gb-2011-12-1-r10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Feinberg AP, et al. Personalized epigenomic signatures that are stable over time and covary with body mass index. Science translational medicine. 2010;2:49ra67. doi: 10.1126/scitranslmed.3001262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bell CG, et al. Genome-wide DNA methylation analysis for diabetic nephropathy in type 1 diabetes mellitus. BMC Med Genomics. 2010;3:33. doi: 10.1186/1755-8794-3-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mill J, et al. Epigenomic profiling reveals DNA-methylation changes associated with major psychosis. American journal of human genetics. 2008;82:696–711. doi: 10.1016/j.ajhg.2008.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Baylin S, Bestor TH. Altered methylation patterns in cancer cell genomes: cause or consequence? Cancer cell. 2002;1:299–305. doi: 10.1016/s1535-6108(02)00061-2. [DOI] [PubMed] [Google Scholar]
- 44.Laird PW. Principles and challenges of genome-wide DNA methylation analysis. Nat Rev Genet. 2010;11:191–203. doi: 10.1038/nrg2732. [DOI] [PubMed] [Google Scholar]
- 45.Harris RA, et al. Comparison of sequencing-based methods to profile DNA methylation and identification of monoallelic epigenetic modifications. Nature biotechnology. 2010;28:1097–105. doi: 10.1038/nbt.1682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bock C, et al. Quantitative comparison of genome-wide DNA methylation mapping technologies. Nature biotechnology. 2010;28:1106–14. doi: 10.1038/nbt.1681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Beck S. Taking the measure of the methylome. Nat Biotechnol. 2010;28:1026–8. doi: 10.1038/nbt1010-1026. [DOI] [PubMed] [Google Scholar]
- 48.Ulrey CL, Liu L, Andrews LG, Tollefsbol TO. The impact of metabolism on DNA methylation. Human molecular genetics. 2005;14(Spec No 1):R139–47. doi: 10.1093/hmg/ddi100. [DOI] [PubMed] [Google Scholar]
- 49.Widschwendter M, et al. Association of breast cancer DNA methylation profiles with hormone receptor status and response to tamoxifen. Cancer research. 2004;64:3807–13. doi: 10.1158/0008-5472.CAN-03-3852. [DOI] [PubMed] [Google Scholar]
- 50.Carone BR, et al. Paternally induced transgenerational environmental reprogramming of metabolic gene expression in mammals. Cell. 2010;143:1084–96. doi: 10.1016/j.cell.2010.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bell JT, Spector TD. A twin approach to unraveling epigenetics. Trends in genetics : TIG. 2011;27:116–25. doi: 10.1016/j.tig.2010.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pearson H. Epidemiology: Study of a lifetime. Nature. 2011;471:20–4. doi: 10.1038/471020a. [DOI] [PubMed] [Google Scholar]
- 53.Yamagata K. DNA methylation profiling using live-cell imaging. Methods. 2010;52:259–66. doi: 10.1016/j.ymeth.2010.04.008. [DOI] [PubMed] [Google Scholar]
- 54.Paliwal A, Vaissiere T, Herceg Z. Quantitative detection of DNA methylation states in minute amounts of DNA from body fluids. Methods. 2010;52:242–7. doi: 10.1016/j.ymeth.2010.03.008. [DOI] [PubMed] [Google Scholar]
- 55.Levenson VV. DNA methylation as a universal biomarker. Expert review of molecular diagnostics. 2010;10:481–8. doi: 10.1586/erm.10.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wang WY, Barratt BJ, Clayton DG, Todd JA. Genome-wide association studies: theoretical and practical concerns. Nature reviews. Genetics. 2005;6:109–18. doi: 10.1038/nrg1522. [DOI] [PubMed] [Google Scholar]
- 57.Li Y, et al. The DNA Methylome of Human Peripheral Blood Mononuclear Cells. PLoS Biol. 2010;8:e1000533. doi: 10.1371/journal.pbio.1000533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Breitling LP, Yang R, Korn B, Burwinkel B, Brenner H. Tobacco-Smoking-Related Differential DNA Methylation: 27K Discovery and Replication. American journal of human genetics. 2011;88:450–7. doi: 10.1016/j.ajhg.2011.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.The Wellcome Trust Case Control Consortium Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 2007;447:66178. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Stephens M, Balding DJ. Bayesian statistical methods for genetic association studies. Nature reviews. Genetics. 2009;10:681–90. doi: 10.1038/nrg2615. [DOI] [PubMed] [Google Scholar]
- 61.Hoggart CJ, Clark TG, De Iorio M, Whittaker JC, Balding DJ. Genome-wide significance for dense SNP and resequencing data. Genetic epidemiology. 2008;32:179–85. doi: 10.1002/gepi.20292. [DOI] [PubMed] [Google Scholar]
- 62.McCarthy MI, et al. Genome-wide association studies for complex traits: consensus, uncertainty and challenges. Nat Rev Genet. 2008;9:356–69. doi: 10.1038/nrg2344. [DOI] [PubMed] [Google Scholar]
- 63.Clayton DG, et al. Population structure, differential bias and genomic control in a large-scale, case-control association study. Nature genetics. 2005;37:1243–6. doi: 10.1038/ng1653. [DOI] [PubMed] [Google Scholar]
- 64.Astle W, Balding DJ. Population Structure and Cryptic Relatedness in Genetic Association Studies. Statistical Science. 2009;24:11. [Google Scholar]
- 65.Teschendorff AE, et al. Age-dependent DNA methylation of genes that are suppressed in stem cells is a hallmark of cancer. Genome Res. 2010;20:440–6. doi: 10.1101/gr.103606.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.van Belle G. Statistical Rules of Thumb. Wiley; 2008. [Google Scholar]
- 67.Chanock SJ, et al. Replicating genotype-phenotype associations. Nature. 2007;447:655–60. doi: 10.1038/447655a. [DOI] [PubMed] [Google Scholar]
- 68.Cedar H, Bergman Y. Linking DNA methylation and histone modification: patterns and paradigms. Nat Rev Genet. 2009;10:295–304. doi: 10.1038/nrg2540. [DOI] [PubMed] [Google Scholar]
- 69.Palacios D, Summerbell D, Rigby PW, Boyes J. Interplay between DNA methylation and transcription factor availability: implications for developmental activation of the mouse Myogenin gene. Molecular and cellular biology. 2010;30:3805–15. doi: 10.1128/MCB.00050-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Sawyers CL. The cancer biomarker problem. Nature. 2008;452:548–52. doi: 10.1038/nature06913. [DOI] [PubMed] [Google Scholar]
- 71.Grutzmann R, et al. Sensitive detection of colorectal cancer in peripheral blood by septin 9 DNA methylation assay. PloS one. 2008;3:e3759. doi: 10.1371/journal.pone.0003759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Payne SR. From discovery to the clinic: the novel DNA methylation biomarker mSETP9 for the detection of colrectoal cancer in blood. Epigenomics. 2010;2:11. doi: 10.2217/epi.10.35. [DOI] [PubMed] [Google Scholar]
- 73.AACR-FDA-NCI Cancer Biomarkers Collaborative advancing the use of biomarkers in cancer drug development. Clinical Cancer Research. 2010;16:19. doi: 10.1158/1078-0432.CCR-10-0880. [DOI] [PubMed] [Google Scholar]
- 74.Poste G. Bring on the biomarkers. Nature. 2011;469:2. doi: 10.1038/469156a. [DOI] [PubMed] [Google Scholar]
- 75.Bell CG, et al. Integrated Genetic and Epigenetic Analysis Identifies Haplotype-Specific Methylation in the FTO Type 2 Diabetes and Obesity Susceptibility Locus. PLoS One. 2010;5:e14040. doi: 10.1371/journal.pone.0014040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ernst J, Kellis M. Discovery and characterization of chromatin states for systematic annotation of the human genome. Nature biotechnology. 2010;28:817–25. doi: 10.1038/nbt.1662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Altshuler D, et al. A haplotype map of the human genome. Nature. 2005;437:1299–320. doi: 10.1038/nature04226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Frazer KA, et al. A second generation human haplotype map of over 3.1 million SNPs. Nature. 2007;449:851–61. doi: 10.1038/nature06258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Eckhardt F, et al. DNA methylation profiling of human chromosomes 6, 20 and 22. Nat Genet. 2006;38:1378–85. doi: 10.1038/ng1909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Beck S, Rakyan VK. The methylome: approaches for global DNA methylation profiling. Trends Genet. 2008;24:231–7. doi: 10.1016/j.tig.2008.01.006. [DOI] [PubMed] [Google Scholar]
- 81.Li N, et al. Whole genome DNA methylation analysis based on high throughput sequencing technology. Methods. 2010;52:203–12. doi: 10.1016/j.ymeth.2010.04.009. [DOI] [PubMed] [Google Scholar]
- 82.Robinson MD, Statham AL, Speed TP, Clark SJ. Protocol matters: which methylome are you actually studying? Epigenomics. 2010;2:587–598. doi: 10.2217/epi.10.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Irizarry RA, et al. Comprehensive high-throughput arrays for relative methylation (CHARM) Genome research. 2008;18:780–90. doi: 10.1101/gr.7301508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Bibikova M, et al. Genome-wide DNA methylation profiling using Infinium assay. Epigenomics. 2009;1:177–200. doi: 10.2217/epi.09.14. [DOI] [PubMed] [Google Scholar]
- 85.Suzuki M, Greally JM. DNA methylation profiling using HpaII tiny fragment enrichment by ligation-mediated PCR (HELP) Methods. 2010;52:218–22. doi: 10.1016/j.ymeth.2010.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Brinkman AB, et al. Whole-genome DNA methylation profiling using MethylCap-seq. Methods. 2010;52:232–6. doi: 10.1016/j.ymeth.2010.06.012. [DOI] [PubMed] [Google Scholar]
- 87.Rauch TA, Pfeifer GP. DNA methylation profiling using the methylated-CpG island recovery assay (MIRA) Methods. 2010;52:213–7. doi: 10.1016/j.ymeth.2010.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Serre D, Lee BH, Ting AH. MBD-isolated Genome Sequencing provides a high-throughput and comprehensive survey of DNA methylation in the human genome. Nucleic acids research. 2010;38:391–9. doi: 10.1093/nar/gkp992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Mohn F, Weber M, Schubeler D, Roloff TC. Methylated DNA immunoprecipitation (MeDIP) Methods in molecular biology. 2009;507:55–64. doi: 10.1007/978-1-59745-522-0_5. [DOI] [PubMed] [Google Scholar]
- 90.Down TA, et al. A Bayesian deconvolution strategy for immunoprecipitation-based DNA methylome analysis. Nat Biotechnol. 2008;26:779–85. doi: 10.1038/nbt1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Gu H, et al. Preparation of reduced representation bisulfite sequencing libraries for genome-scale DNA methylation profiling. Nature protocols. 2011;6:468–81. doi: 10.1038/nprot.2010.190. [DOI] [PubMed] [Google Scholar]
- 92.Cokus SJ, et al. Shotgun bisulphite sequencing of the Arabidopsis genome reveals DNA methylation patterning. Nature. 2008;452:215–9. doi: 10.1038/nature06745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Lister R, et al. Highly integrated single-base resolution maps of the epigenome in Arabidopsis. Cell. 2008;133:523–36. doi: 10.1016/j.cell.2008.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Huang Y, et al. The behaviour of 5-hydroxymethylcytosine in bisulfite sequencing. PLoS One. 2010;5:e8888. doi: 10.1371/journal.pone.0008888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Butcher LM, Beck S. AutoMeDIP-seq: a high-throughput, whole genome, DNA methylation assay. Methods. 2010;52:223–31. doi: 10.1016/j.ymeth.2010.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Clarke J, et al. Continuous base identification for single-molecule nanopore DNA sequencing. Nat Nanotechnol. 2009;4:265–70. doi: 10.1038/nnano.2009.12. [DOI] [PubMed] [Google Scholar]
- 97.Flusberg BA, et al. Direct detection of DNA methylation during single-molecule, realtime sequencing. Nat Methods. 2010;7:461–5. doi: 10.1038/nmeth.1459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Pembrey ME, et al. Sex-specific, male-line transgenerational responses in humans. European journal of human genetics: EJHG. 2006;14:159–66. doi: 10.1038/sj.ejhg.5201538. [DOI] [PubMed] [Google Scholar]