Abstract
Mucin glycoproteins present a complex structural landscape arising from the multiplicity of glycosylation patterns afforded by their numerous serine and threonine glycosylation sites, often in clusters, and with variations in respective glycans. To explore the structural complexities in such glycoconjugates we used NMR to systematically analyze the conformational effects of glycosylation density within a cluster of sites. This allows correlation with molecular recognition through analysis of interactions between these and other glycopeptides, with antibodies, lectins, and sera, using a glycopeptide microarray. Selective antibody interactions with discrete conformational elements, reflecting aspects of the peptide and disposition of GalNAc residues are observed. Our results help bridge the gap between conformational properties and molecular recognition of these molecules, with implications for their physiological roles. Features of the native mucin motifs impact their relative immunogenicity and are accurately encoded in the antibody binding site, with the conformational integrity being preserved in isolated glycopeptides, as reflected in the antibody binding profile to array components.
The number of glycan structures of the human glycome is estimated to be many thousands.1 This diversity, arising from the variety of residues and multiple linkage options, is also amplified by glycan conjugation to other components including proteins and lipids. A large fraction of mammalian proteins are glycosylated,2 and the combinatorial possibilities of these glycoconjugates present a complexity unparalleled in genomics and proteomics, further compounded by the intrinsic heterogeneity in glycoproteins, thought to be a consequence of non-template driven glycosylation. Additionally, cellular regulation of glycan structures and patterns through differential enzyme expression in the normal or disease states allows cell surface glycoproteins to function as temporally regulated biomarkers. The endogenous presentation of aberrant glycosylation can also give rise to circulating antibodies that serve as secondary markers.3 Given their prominence in communication between cells and surroundings, understanding the contributions made by the protein and glycan components to molecular recognition are critical in how information is encoded for specific glycoprotein interactions and functions,
Mucin-type O-glycosylation, characterized by a prevalence of threonine and serine residues modified with N-acetylgalactosamine (GalNAc), constitutes a major and complex form of protein modification, encountered on the cell surface. Mucin O-glycan biosynthesis occurs in a stepwise fashion, initiated by members of a family of about two dozen polypeptide N-acetylgalactosaminyltransferases (ppGalNAcTs)4, followed by elaboration with other sugars to generate complex O-glycans. The glycosylation patterns generated by these enzymes are conferred by the catalytic domains,5 and if proximal O-GalNAcs are already in place, influenced by their lectin domains.6, 7 However, the lack of strictly defined consensus sequences for O-glycosylation,5 together with the known heterogeneity in O-glycan structures, creates challenges for defining discrete mucin recognition elements.
Characterization of O-glycosylation by mass spectral methods typically relies on chemically released glycans, with loss of crucial sequence-specific data on sites of modification. Non-destructive analysis with lectins of known carbohydrate epitope preferences is also commonly used,8 but lectins are largely insensitive to the glycoconjugate context. Their apparent affinities may reflect the degree to which pendant glycans are clustered, providing some basis for selectivity, but this can be rationalized in global thermodynamic terms without a detailed structural knowledge.9 These analytical limitations have given rise to an intrinsic ambiguity in defining the O-glycan epitopes. For instance, serine/threonine α-O-GalNAc is referred to as the Tn antigen, not discriminating the amino acid to which it is attached,10 providing insufficient definition of this epitope in the context of a glycoprotein. This structure, which is normally rare in humans is relevant because of the correlation of its aberrant appearance with poor prognosis in cancer, where altered densities and clustering are observed.11–13 It has been a target in diagnostic and therapeutic strategies, particularly in development of glycoconjugate anti–tumor vaccines.12, 14 Understanding the conformation of mucins even with the minimal Tn antigen, is broadly relevant to mucin structural biology since the α-O-GalNAc residue of their glycans is key in organizing the core glycoprotein scaffold15 underlying potentially more complex pendant glycans.
The relevance of more accurate characterization of epitopes with S/T-α-O-GalNAc is indicated by the differential recognition of glycosylation in isolated sites or in clusters by antibodies in normal immune responses, and those induced in therapeutic applications of glycoconjugate vaccines.3, 12, 16 Results from surface plasmon resonance17 and array binding studies 3, 18, 19 show that antibody recognition of mucin structures is influenced by presentation in the glycoconjugate environment, but these findings have not been accompanied by any structural studies. The importance of this is illustrated in a recent crystal structure of a T-α-O-GalNAc glycopeptide-antibody complex showing contacts between the antibody and both carbohydrate and peptide portions.20 Since material isolated from natural sources displays micro-heterogeneity even if isolated from a single cell type, we have employed chemical synthesis to provide homogeneous well-defined material needed for biophysical studies using NMR methods for a systematic analysis of mucin conformation as a function of glycosylation density. The constructs examined in this way and others have been assembled in a glycopeptide microarray to gain further insight into how conformational properties mediate binding of lectins and antibodies.
RESULTS AND DISCUSSION
Structural Analysis of Mucin Glycopeptides
The conformations of several glycopeptides based on the MUC2 related peptide sequence PTTTPLK, Ac-PTTTPLK-NH2 (PEP), Ac-PT*TTPLK-NH2 (A), Ac-PTT*TPLK-NH2 (B), Ac-PTTT*PLK-NH2 (C), Ac-PT*T*TPLK-NH2 (D), Ac-PT*TT*PLK-NH2 (E), Ac-PTT*T*PLK-NH2 (F), and Ac-PT*T*T*PLK-NH2 (G), where * indicates modification with α-O-GalNAc, were studied by NMR methods. These allow examination of incremental effects of glycosylation, and were previously biochemically characterized as ppGalNAcT substrates,21 showing differential reactivity. Their syntheses and preliminary NMR have been reported.22 More extensive NMR data, including NOEs and vicinal couplings have now been obtained and were used in the structure determination. Relationships to the features of other mucin motifs based on initial models were noted.23 The backbone 3JHN-Hα and threonine side-chain 3JHα-Hβ values measured (Table 1, Supporting Information Figure 1), are correlated with bond torsion angles.24, 25 Increasing values of the 3JHN-Hα coupling on the modified residues indicate a locally more extended arrangement, and being closer to the maximum, limit possible angular averaging for this bond. Upon glycosylation, the values of the 3JHα-Hβ coupling associated with the respective threonine side chains are near its minimum value, indicating both limited or no averaging, and an angle in the vicinity of 90° between the protons, with NOEs eliminating the other possible solution.
Table 1.
Peptide Backbone and Threonine Side Chain Vicinal Coupling Constants (Hz)a.
| Construct | PEP | A | B | C | D | E | F | G |
|---|---|---|---|---|---|---|---|---|
| 3JHN-Hα | ||||||||
| T2 | 7.8 | 8.8 | 7.9 | 8.0 | 8.9 | 8.6 | 7.8 | 8.9 |
| T3 | 8.0 | 8.3 | 8.9 | 8.1 | 9.1 | 8.3 | 8.9 | 9.5 |
| T4 | 7.4 | 6.8 | 7.6 | 8.3 | 7.2 | 8.0 | 8.5 | 8.5 |
| L6 | 6.6 | 6.3 | 6.3 | 5.9 | 6.1 | 5.7 | 5.5 | 5.5 |
| K7 | 7.2 | 7.3 | 7.3 | 6.7 | 7.2 | 6.7 | 7.3 | 7.0 |
| 3JHα-Hβ | ||||||||
| T2 | 4.5 | 2.2 | 5.0 | 4.4 | 2.6 | 2.6 | 5.1 | 2.4 |
| T3 | 4.4 | 3.7 | 2.0b | 5.1 | <2.0c | 4.2 | ND | ND |
| T4 | 6.2 | 5.9 | 5.7 | 2.2 | 6.1 | <2.0c | 2.2 | <2.0c |
glycosylated sites in bold, determined to better than +/− 0.2 Hz
coupling only partially resolved
coupling not resolved
ND not determined
Orientations of GalNAc residues on a given T residue relative to the peptide backbone largely appear unaffected by the presence of neighboring glycans. This is reflected in that the NOE contacts (Figure 1) between the GalNAc N-acetyl methyl groups and peptide backbone amides of each construct containing a single GalNAc, A, B, and C, are preserved for the respective sites in the fully glycosylated G, where all three sites are occupied by GalNAc. The N-acetyl group orientation appears largely fixed relative to the sugar ring, indicated by the large coupling constant of ~10 Hz between the N-acetyl amide proton and H2, so the NOEs provide information on the orientation of the GalNAc ring. With increasing glycosylation density there are a greater number of NOE contacts, arising from the additional protons introduced by the GalNAc residues, and among those in the peptide backbone, implying an increased organization at higher levels of glycosylation. Based on experimental constraints, full structures of the glycopeptides have been computed, revealing single well-defined families of structures for each construct with coordinates and NOE constraints deposited in the Protein Data Bank (Supporting Information Figure 2 and Table 1). The closest to the average solution of the TTT segment for the amino acid and pendant GalNAc of constructs A-F are shown in Figure 2 with the trace of the peptide in the plane of the paper, each superimposed on G. The backbone traces for the full length of the glycopeptides also show a remarkable degree of similarity with the possible exception of the N-terminal proline residue, which is not well constrained by inter-residue experimental parameters, leaving open the possibility that this residue is mobile. The twist in the peptide backbone (Figure 2) avoids clashes between neighboring GalNAc residues and permits the orientations in the singly glycosylated forms to persist in the more densely glycosylated constructs. Relative to the axis of the peptide backbone, the GalNAc residues on adjacent threonines are oriented roughly 120° counter clockwise relative to each other (Figure 3). Further independent validation of the structures for the constructs with two or three GalNAc substitutions (D–G) was obtained through determination of residual dipolar couplings (RDC) measured for CαHα, GalNAc C1H1, T methyl, Tβ, peptide NH and GalNAc N-acetyl NH in weakly aligning media.26 When these data, which are independent of the NOE and coupling constant information, were incorporated in a further round of structure refinement of these constructs, only slight changes in the molecular geometry were found, accompanied by additional reduction in the RMSDs for each family of structures. The structures of G before and after RDC refinement is shown in Figure 3B. Since the impact of motional averaging on the RDCs is different from that for NOEs, the consistent results reinforce the validity of a single conformational model, rather than possible interconversion between multiple conformers, implying the respective structures are quite energetically favorable and can be expected to maintain their conformational integrity when coupled to the array matrix. The conformations of the individual glycosylated amino acid residues, taken by themselves, are very similar. The results here show how the individual components assemble into clusters, an arrangement found in mucins. Interestingly, aligning the peptide Cα atoms for construct A in this work and the respective atoms of the glycopeptide extracted from the recent antibody-bound glycopeptide crystal structure,20 with the glycosylated T residues on each molecule in register, reveals very similar peptide backbone trace for each, with an RMSD of 0.655Å between the respective Cαs. Orientation of the GalNAc relative to the peptide chain is also similar in both cases, with a difference of ~30° relative to the backbones of the Cα aligned structures. While the O-GalNAc modification is near the N-terminus in both cases, the sequences, PT*TTPLK of A, and GT*KPPL in the antibody complex, are somewhat different. Qualitatively, results on a clustered Tn-glycophorin fragment are similar to our findings.27 The NMR structure of a MUC1 single repeat glycopeptide, modified on the T of the GVTSA segment, shows quite similar NMR and conformational features to those for the individual glycosylated amino acids reported here,28 however, it is difficult to make quantitative comparison with these or other reported NMR structures of MUC5AC29 and other mucin models since coordinates are not available in structure databases.
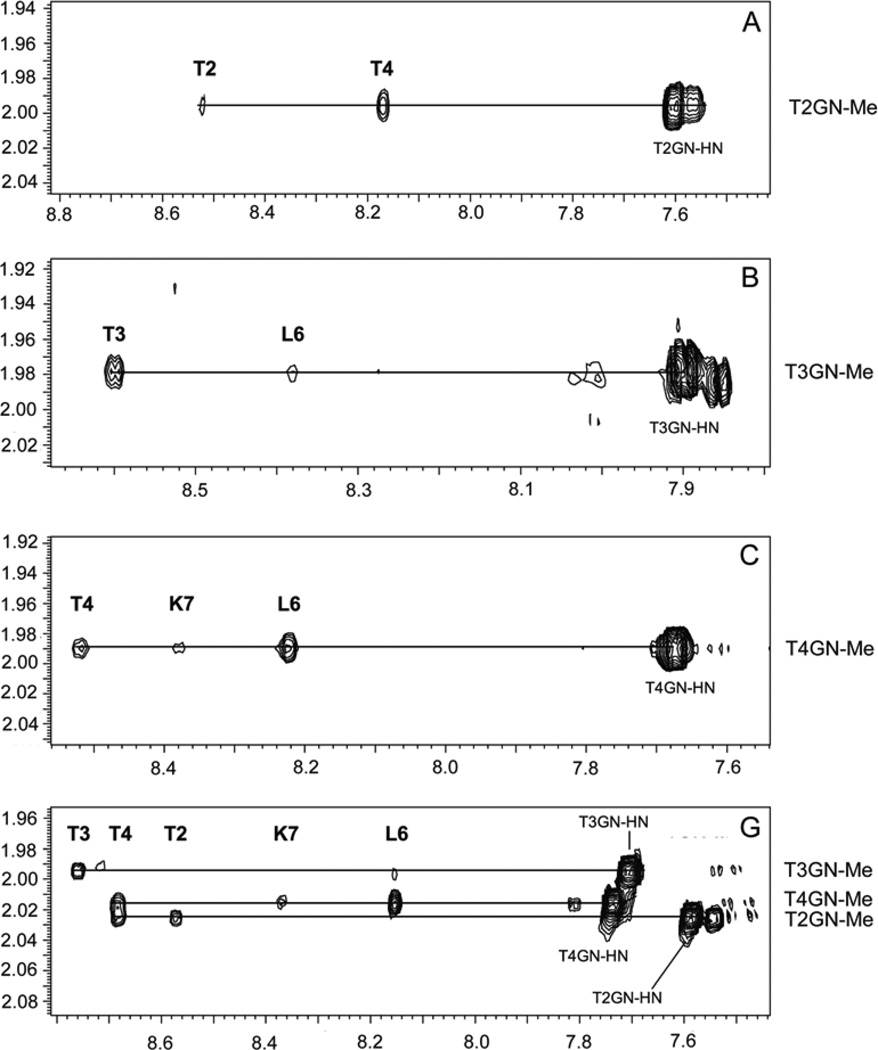
Figure 1.
NOE contacts between GalNAc methyl protons and amide backbone and N-acetyl amide protons for the individually glycosylated forms, A, B, C, and for the triglycosylated construct G of the PTTTPLK sequence.
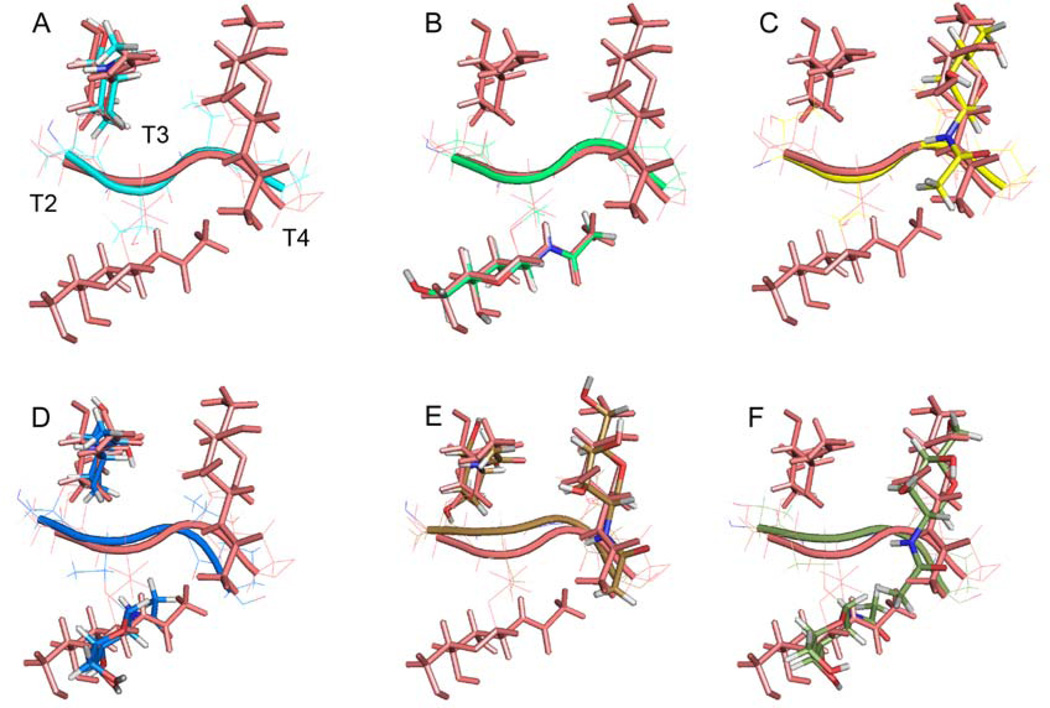
Figure 2.
The closest to the average structures of the TTTP segment for the mono- and diglycosylated PTTTPLK constructs A to F, in various colors, superimposed on the structure of the triglycosylated form G (salmon). RMSD to G for threonine and GalNAc residues heavy atoms are from A, 0.947Å; B, 0.740Å; C, 1.05Å; D, 1.323Å; E, 0.961Å; and F, 1.447Å. Direction of peptide backbone parallel to page. See Supporting Information Table 1 for structure statistics and PDB IDs.
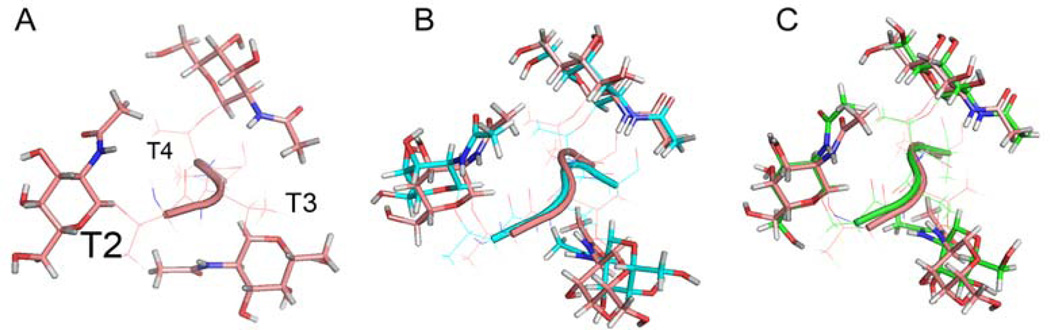
Figure 3.
View down the peptide backbone (perpendicular to page), N to C, (A) for the T*T*T* segment of the G construct. (B) superposition of the closest to the average of threonine and GalNAc heavy atoms for construct G with RDC refinement (cyan) and without RDC refinement (salmon). The RMSD between the structures is 1.194Å. (C) Superposition of threonine and GalNAc heavy atoms of G (salmon) with the serine, threonine and GalNAc heavy atoms of the S*T*T*AV structure (green) reported earlier.15 RMSD 1.665Å
Incremental glycosylation imparts enhanced rigidity to the motif as the number of both hydrogen bonding-like interactions between the GalNAc and the peptide backbone, and hydrophobic interactions between the methyl group on the GalNAc and amino acid side chains increases, similar to other systems.15 The arrangement of the GalNAc N-acetyl NH group and the carbonyl of its associated amino acid is consistent with intramolecular hydrogen bonding geometries15, 30 as found in the Cambridge Data Base.31 The same underlying organizational influences appear operative for construct G and the clustered triplet glycosylated motif solved earlier for the sequence S*T*T*AV,15 in spite of the difference of contexts in which these clusters appear (Figure 3C) and supports the contention of a consistent triplet cluster mucin motif. The profile of inhibition by several Tn bearing structures on the binding of sera from mice and primates, after challenge with a vaccine based on such a cluster, S*T*T*, supports this in a biological context.32 A similar target has also been identified for the monoclonal antibody MLS128 which can inhibit cancer cell growth.33 The determination of the structures here at atomic resolution and their stability are consistent with the extended organization of mucins.15 This precludes contributions from sequentially remote segments in the native environment, supporting the notion that the glycopeptides offer a faithful representation of their conformations in the larger mucin glycoprotein context. With the additional information developed here on mucin scaffolds, we turned to evaluating how these features are reflected in their molecular recognition using a microarray platform populated with those glycopeptides examined here by NMR, as well as others.
Microarray Analyses
Glycan microarrays, such as implemented by the Consortium for Functional Glycomics, have emerged as a key technology for efficient screening of carbohydrate-protein interactions.34–36 The slide-based format is attractive, requiring only minute amounts of ligand and binding proteins while providing rapid identification of carbohydrate-protein interactions. In extending this to glycoconjugate structures, arrays based on neoglycoproteins offer approximations of the local density and clustering of glycans found in mucins,35, 37, 38 and may be chemically more accessible, but are unlikely to be completely faithful in representing the organization of mucin motifs in vivo. Deviations from these native chemical structures perturb the organization15 and recognition by antibodies,32 particularly for clustered glycosylation. Arrays based on native mucin glycopeptide motifs, implemented here and also by others,3, 19 with direct ligation to the slide substrate, or through a carrier protein,39, 40 offer more natural targets for binding studies. For the most part, in previous studies with Tn glycopeptides immobilized in a slide or bead based array format, the Tn epitope has only been presented in isolated sites, or as in pairs in the MUC1 repeat sequence,3, 18, 19 with the aspect of clustering largely overlooked.
A microarray of glycopeptides with α-O-GalNAc S or T residues was assembled, Table 2, either clustered or in isolation. Included were those whose conformational properties and stability we have characterized in detail, described above (IDs 1–8). Additional biologically relevant glycopeptides with S/T-α-O-GalNAc in a variety of peptide contexts are present. These included a sequence from alpha-dystroglycan (IDs 9, 13), a MUC5AC sequence (IDs 10),7 a fragment of rat submandibular mucin (EA2) (IDs 11,12) that is a known substrate for ppGalNAcTs,41 and two segments from MUC1 (IDs 14–17). Clustered T-α-O-GalNAc without adjacent proline residues were also included (IDs 18–19), similar to a motif in the MUC2 construct G. To compare responses to those with the canonical Tn antigen, the structures Ac-T-(α-O-GalNAc)-NH2-(CH2)3-NH2)( ID 20) and S- and T-(α-O-GalNAc)-NH2 or -OH were present (IDs 23–26, 45,46). Additionally, there are glycosylated peptides from the hinge region of IgA1 in the glycopeptide array (IDs 27–44).42 Control glycans were also included (IDs 47–52).
Table 2.
List of Structures on Glycopeptide Arraya
| Chart ID |
Detail | Sequence |
|---|---|---|
| 1 | A-MUC2 | AcPT*TTPLK-NH2 |
| 2 | B-MUC2 | AcPTT*TPLK-NH2 |
| 3 | C-MUC2 | AcPTTT*PLK-NH2 |
| 4 | D-MUC2 | AcPT*T*TPLK-NH2 |
| 5 | E-MUC2 | AcPT*TT*PLK-NH2 |
| 6 | F-MUC2 | AcPTT*T*PLK-NH2 |
| 7 | G-MUC2 | AcPT*T*T*PLK-NH2 |
| 8 | R-MUC2 | AcPTTTPLK-NH2 |
| 9 | α-Dystroglycan | AcPPTTTTKKP-NH2 |
| 10 | MUC5AC | H2N-GTTPSPVPT*TSTTSAP-OH |
| 11 | EA2 | AcPTTDSTT*PAPTTKNH2 |
| 12 | EA2-R | AcPTTDSTTPAPTTKNH2 |
| 13 | α-Dystroglycan | AcPPT*T*T*T*KKP-NH2 |
| 14 | MUC1-1 | NH2-TSAPDT*RDAP-NH2 |
| 15 | MUC1-1R | NH2-TSAPDTRDAP-NH2 |
| 16 | MUC1-2 | H2N-APGS*T*APP-NH2 |
| 17 | MUC1-2R | H2N-APGSTAPP-NH2 |
| 18 | PADRE Tn3b | H2N-GaKcVAAWTLKAAaT*T*T*GCONH2 |
| 19 | Tn3 linker | Ac-T*T*T*-NH(CH2)3NH2 |
| 20 | Tn linker | Ac-T*-NH(CH2)3NH2 |
| 21 | Peptide-4 | H2N-KTTT-CONH2 |
| 22 | Peptide-5 | H2N-KTTTG-CONH2 |
| 23 | Ser-GalNAc1 | H2N-Ser(α-D-GalNAc)-NH2 |
| 24 | Ser-GalNAc2 | H2N-Ser(α-D-GalNAc)-OH |
| 25 | Thr-GalNAc1 | H2N-Thr(α-D-GalNAc)-NH2 |
| 26 | Thr-GalNAc2 | H2N-Thr(α-D-GalNAc)-OH |
| 27 | IgA-Pep01 | H2N-KPVPST*PPT*PS*C-OH |
| 28 | IgA-Pep02 | H2N-KPVPSTPPTPSC-OH |
| 29 | IgA-Pep03 | H2N-KPVPS*TPPTPSC-OH |
| 30 | IgA-Pep04 | H2N-KPST*PPT*PS*PS*C-OH |
| 31 | IgA-Pep05 | H2N-KPSTPPTPSPSC-OH |
| 32 | IgA-Pep06 | H2N-KT*PPT*PS*PS*TPC-OH |
| 33 | IgA-Pep07 | H2N-KTPPTPSPSTPC-OH |
| 34 | IgA-Pep08 | H2N-KTPPTPSPST*PC-OH |
| 35 | IgA-Pep09 | H2N-KPT*PS*PS*TPPT*C-OH |
| 36 | IgA-Pep10 | H2N-KPSPSTPPTPSC-OH |
| 37 | IgA-Pep11 | H2N-KPS*PS*TPPT*PSC-OH |
| 38 | IgA-Pep12 | H2N-KPSTPPTPSPSC-OH |
| 39 | IgA-Pep13 | H2N-KPS*TPPT*PSPSC-OH |
| 40 | IgA-Pep14 | H2N-KPSTPPTPSPSC-OH |
| 41 | IgA-Pep15 | H2N-KPST*PPTPS*PS*C-OH |
| 42 | IgA-Pep16 | H2N-KPSTPPTPS*PSC-OH |
| 43 | IgA-Pep17 | H2N-KPSTPPTPSPS*C-OH |
| 44 | IgA-Pep18 | H2N-KPST*PPTPSPSC-OH |
| 45 | Ser-GalNAc-2 | H2N-Ser(α-D-GalNAc)-OH |
| 46 | Thr-GalNAc-2 | H2N-Thr(α-D-GalNAc)-OH |
| 47 | Blood group A tetra | |
| 48 | Blood group A penta | |
| 49 | LNnT | |
| 50 | Man5 | |
| 51 | PBS | |
| 52 | Biotin |
* = GalNAc residue on Ser or Thr.
For PADRE sequence a=D-Ala, c=cyclohexylalanine.
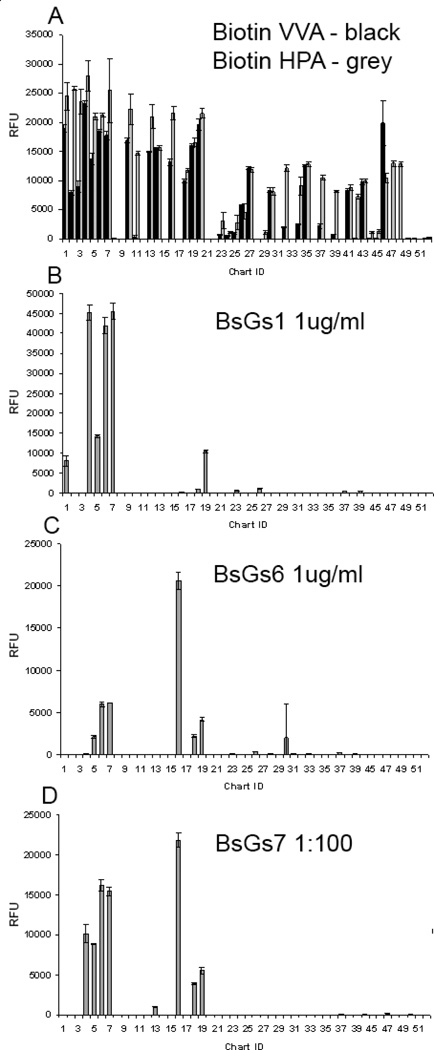
Presence and accessibility of the Tn-bearing structures on the array was established by binding of the lectins Helix pomatia agglutinin (HPA) and Vicia villosa agglutinin (VVA), which have broad specificity for the α-GalNAc structure.8 Representative responses for HPA and VVA at 1 µg/ml are plotted together in Figure 4A. HPA binding was consistent with the presence of α-GalNAc on the glycopeptides,8 although those to S/T-linked α-GalNAc (IDs 23–26) and IgA-Pep03 (ID 29) were weak. VVA binding was also consistent with the presence of α-GalNAc on the printed glycoconjugates, although apparently more selective than HPA. The crystal structures of HPA and VVA in complex with S-α-O-GalNAc43, 44 show shallow binding pockets that interact with the exposed GalNAc hydrophilic surface. This is consistent with their broad specificity and general use for detecting GalNAc.
Figure 4.
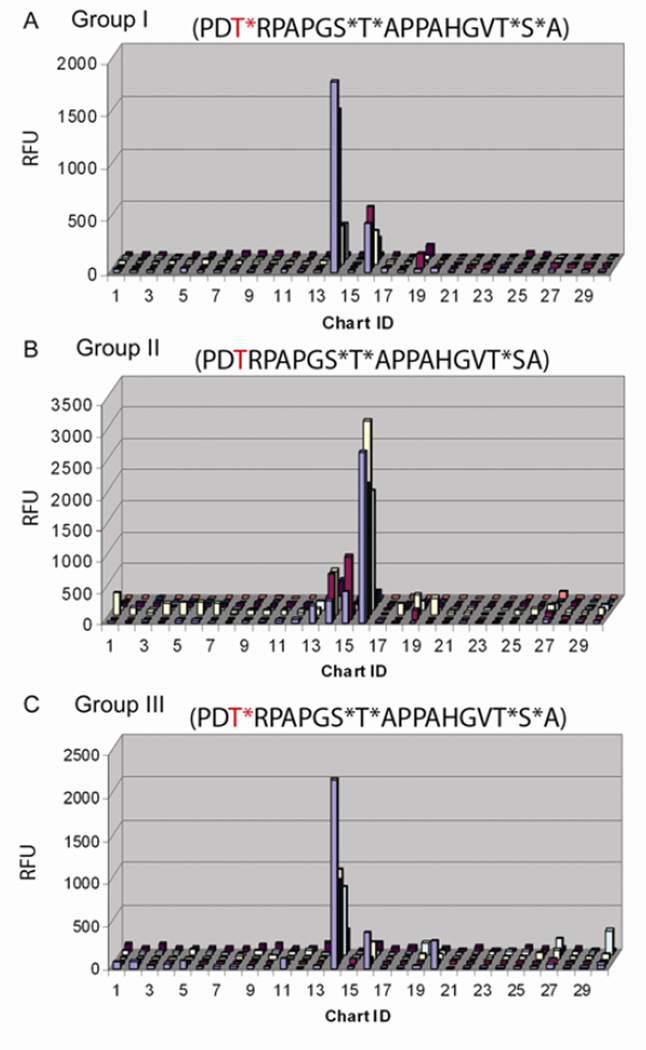
(A) Binding profile of biotinylated HPA lectin (grey bars) and biotinylated VVA lectin (solid black bars) both at 1 µg/ml detected with Cyanine5-labeled streptavidin. Binding profiles of (B) BaGs1 antibody representative of BaGs1, 2, 3, and 4, and (C) antibody BaGs6, representative of BaGs5 and 6 binding profiles. mAbs were assayed at10 µg/ml detected with AlexaFluor488-labeled anti-mouse IgM (5 µg/ml). (D) Binding profile of BaGs7 antibody (1:100 dilution of ascites fluid) detected with AlexaFluor488- anti-mouse IgM (5 µg/ml). Error bars represent +/− 1 SD. RFU = relative fluorescence units. ID corresponds to Table 2.
The array was further interrogated with a panel of seven anti-Tn monoclonal IgM antibodies (mAbs) from the laboratory of Georg Springer,45, 46 elicited from mice by Tn-bearing red blood cells and Tn components derived from O-type red blood cells.46 The seven mAbs were grouped into three subsets, BaGs 1–4, BaGs 5 and 6, and BaGs 7 based on the pattern of glycosylated structures recognized (Figure 4B–D). The mAbs show little preference for glycopeptides when the T-α-O-GalNAc (the canonical Tn structure) is presented at an isolated site of glycosylation, while strongly preferring adjacent pairs or triplets, but interestingly not a sequence of four T-α-O-GalNAc sites.
For the series of MUC2 glycopeptides (IDs 1–7) studied by NMR, antibodies BaGs 1–4 only recognize mono-glycosylated species when the GalNAc is on T2, but not on T3 or T4, in spite of the similarity of the conformation of the individual glycosylated amino acids. They recognize all three of the di-glycosylated species, and the fully glycosylated cluster, as well as more weakly, an isolated triplet of three T-α-O-GalNAc (Tn3) residues on a linker. The other singly or multiply glycosylated constructs in different contexts from MUC1, MUC5AC, EA2, and IgA are largely ignored. The preferences of BaGs5 and 6 are more restricted, favoring the glycopeptide where the C-terminal pair or all three sites are glycosylated in PTTTPLK, but more weakly interacting with the construct where both the first and last Thr are glycosylated. They also interact with an isolated Tn3, but favor an adjacent glycosylated S*T* pair in a MUC1 (ID 16) construct. BaGs7 has a similar profile to BaGs5 and 6, with the subtle difference that it also interacts with the PT*T*TPLK sequence. Interestingly, the antibodies did not recognize four GalNAc residues in a row. Detailed chemical structures of the immunogens that elicited the Springer monoclonal antibodies investigated here were not known, but were clearly able to induce antibodies targeted to a rather restricted range of mucin structures that allow us to infer aspects of their nature. The ability of antibodies to discriminate subtle differences in cluster glycosylation is found in surface plasmon resonance studies reported for two other anti-Tn mAbs, arising from immunization with tumor derived material, that bind to T-α-O-GalNAc glycopeptides with a strict requirement for adjacent glycosylation, either as a pair or in a triplet where the recognition can be abrogated when the central residue of a triplet is an unmodified T or a proline.17
With a better understanding of the conformational factors relating to the organization of mucin glycopeptides, we also addressed the level to which information on mucin epitopes persists through the immune response among members of a polyclonal distribution, where we have knowledge of the chemical structures of the antigen molecules. This provided an opportunity to establish responses to variations in the glycosylation motifs on the same peptide sequence. Sera from a trial evaluating the response to immunization with α-O-GalNAc containing MUC1 structures, anti-tumor therapeutic targets, were investigated on our array. Three MUC1 constructs were used in vaccination, distinguished by different glycosylation patterns on the several available S and T sites in the repeat units. Portions of the immunogens, compounds TSAPDT*RDAP (ID 14) and APGS*T*APP (ID 16) are present on the array. For Group I sera (immunogen: GVT*S*A(PDT*RPAPGS*T*APPAHGVT*S*A)5C) binding was dominated by both, with ID 14 binding greater than ID 16 even though both epitopes are contained in the vaccine (Figure 5A). Group II sera (immunogen: CHGVT*SA(PDTRPAPGS*T*APPAHGVT*SA)PDTRPA) with only the glycosylated epitopes corresponding to ID 16 present, generally demonstrated binding restricted to IDs 14 and 16, with the predominant response being to ID 16 (Figure 5B). Group III sera (immunogen: CHGVT*S*A(PDT*RPAPGS*T*APPAHGVT*S*A)PDT*RPA) generally demonstrated a restricted binding pattern, similar to Group I, with compound ID 14 much higher than ID 16 (Figure 5C). Only IgG antibodies, and not IgM from the sera were bound to the array. These IgGs show very little reactivity towards any other of the Tn-containing glycopeptides on the array. No binding of pre-immune sera was evident.
Figure 5.
Binding of IgG, in RFU, in each group of sera from individuals after immunization with the respective MUC1-constructs. (A) Binding of sera from 5 individuals from Group I. (B) Binding of sera from 7 individuals from Group II. (C) Binding of sera from 5 individuals from Group III. Only results for the first 30 array components (Table 2) are shown as none of the other components indicated significant binding. Sera were diluted 1:100 and detected with AlexaFluor555-labeled anti-human IgG (5 µg/ml).
All vaccine constructs had the glycosylation pattern of the second element APGS*T*APP. However, the central T residue of the first sequence TSAPDT*RDAP was glycosylated only for vaccine groups I and III. Interestingly, IgGs of sera from groups I and III favored the first sequence with the GalNAc present. In group II, the IgG response to the first sequence, with or without glycosylation, was significantly diminished, with a relatively robust response to the second fragment in glycosylated form. These results illustrate that specific glycoprotein features influence the editing, processing and presentation of the antigen. The monodisperse selectivity of these responses implies the stability of relevant structural information that is a composite of both peptide and carbohydrate components is retained in the cellular events, and is recapitulated in the short glycopeptide segments used on the array.
Sera samples from one of the groups in this trial, group I, have been previously analyzed3 on an array that included several 60aa MUC1 constructs (three 20 residue repeats), each with distinct glycosylation patterns. Serum antibodies preferred those including either GS*T*A, or this and PDT*R, in agreement with our observations for group I. When they applied group I sera to another glycopeptide array of single 20aa repeat MUC1 glycopeptides, antibody components also recognize both epitopes.19 These epitope preferences are further borne out by group I sera interactions with a randomized library of shorter glycopeptides.18 Importantly here, with group II sera, we have been able to extend this analysis to show the differential impact of glycosylation on the proximal PDTRP sequence for biasing the overall response, and the sensitivity of this array approach to evaluating outcomes of such vaccine therapies.
In this work, principles by which individual α-O-GalNAc-threonine units are assembled into larger clustered patterns of O-glycosylation, common in mucins, have been elucidated. Since geometries of the individual glycosylated threonines are shown to be quite similar, the selective antibody interactions observed in array screening imply the importance of relationships in the relative disposition of their glycans as presented on the multiple sites of the peptide scaffold, making up a significant portion of the molecular surface, along with components of the peptide itself. Variations in glycosylation density on the same peptide sequence in which the S/T-α-O-GalNAc is presented can be differentiated, a factor that has not been extensively investigated before. This enhances the value of the antibodies as reagents, but full explanation of the basis for the specificity awaits additional sequence and/or structural information on the antibodies. The affinity encoded in the mAbs, raised against natural material, for the synthetic constructs we have characterized and immobilized indicates the biological relevance of the conformations. Our microarray data, as well as that of others,3, 18, 19, 47 affirm that unlike lectins, antibodies broadly referred to as anti-Tn antibodies target not just the S/T-α-O-GalNAc structure, but surrounding features as well. They are unable to bind every potential Tn antigen site, even most presented on peptides, or the conventionally defined minimal Tn antigen. Further, the vaccination studies illustrate that in addition to ultimate antibody recognition, context and conformation play a role in mucin antigenicity, a consideration in optimal design of anti-mucin therapeutic agents.
METHODS
Glycopeptides
Synthesis of glycopeptides in the PTTTPLK series (ID 1–8), alpha-dystroglycan (ID 9 and 13), and MUC5AC (ID 10) have been reported previously.8, 22, 48 The IgA derived glycopeptides were synthesized following reported procedures.42 The MUC1 constructs (ID 14–17) and EA2 (ID 11 and 12) constructs were synthesized using standard procedures (See Supporting Information). The Tn-linker (ID 20), Tn3-linker (ID 19) and PADRE peptide Tn3 (ID 18), were provided by the laboratory of Geert-Jan Boons.
Glycopeptide Microarrays
The glycopeptide microarrays were prepared on N-hydroxysuccinimide (NHS) glass slides (Schott Nexterion) and immobilization of peptides and glycopeptides was achieved through amine functions. (Supporting Information) With concentrations adjusted to 100 µM in printing buffer (300 mM sodium phosphates, pH 8.5), 0.33nL of each solution was spotted using a piezoelectric printer. The microarray was printed in spot replicates of 6. Arrays were interrogated with monoclonal anti-Tn antibodies (a kind gift from the late Georg Springer), biotinylated lectins (Vector Labs), and serum samples from the laboratory of Dr. Phillip Livingston at Memorial Sloan-Kettering Cancer Center (MSKCC), described below, at the given concentrations or dilutions indicated in the figures, and detected with fluorescently labeled secondary antibodies and streptavidin as noted. Scanning and quantification were performed with ProScanArray scanner and ScanArray Express software (Perkin Elmer). The list of glycans/glycopeptides printed on the microarray is given in Table 2.
NMR Analysis
NMR data were collected on Varian INOVA 600, 800 and 900 MHz instruments using pulse sequence programs in the Varian software for double-quantum filtered COSY, TOCSY, NOESY, 13C and 15N HSQC, and 13C HMQC and HMBC experiments.49 Samples were run at various concentrations between 2 and 10 mM in D2O or 90% H2O/10% D2O. Most data were collected at 25°C. Because of overlap in amide signals, some experiments were repeated at other temperatures in the range of 15°C to 30°C. A 300ms mixing time was used for the NOESY experiments in 90% H2O and 350 ms for those in D2O. Couplings were measured from resolved peaks in 1-dimensional spectra. Structures were calculated with XPLOR-NIH 50 following the protocol described in the Supporting Information. Residual dipolar couplings were measured in didodecyl-phosphatidylcholine/dihexylphosphatidylcholine 3/1 molar ratio at 10% in 90% H2O/10% D2O 26 at in the range of 30–35°C using 1H-13C or 1H-15N HSQC sequences without 1H decoupling pulses in the heteronuclear evolution period, and couplings determined from splittings in the heteronuclear dimension.
Antibodies and Serum samples
The monoclonal anti-Tn antibodies used in this study were produced by the late Georg Springer.45, 46 These mouse monoclonal IgM antibodies are designated: BaGs1 (Ca3637), BaGs2 (Ca3239), BaGs3 (Ca3268), BaGs4 (Ca3342), BaGs5 (Ca3250), BaGs6 (Ca3638), and BaGs7 (Ca3749). BaGs1, 2, 3, 5, and 6 were purified by affinity chromatography, while BaGs4 and 7 were ascites fluid. Patients with breast cancer in remission were vaccinated (Supporting Information) in the adjuvant setting at MSKCC under IRB approved protocols and informed consent, with one of three Tn-MUC1-KLH (Keyhole Limpet Hemocyanin) conjugate vaccines plus immunological adjuvant QS-21.51 The enzymatically Tn glycosylated MUC1 constructs used, prepared in the Clausen laboratory from synthetic peptides,3, 52 contained five fully glycosylated MUC1 repeats (106 aa), GVT*S*A(PDT*RPAPGS*T*APPAHGVT*S*A)5C-KLH (Group I immunogen),1 ½ partially glycosylated MUC1 repeats, KLH-C HGVT*SA(PDTRPAPGS*T*APPAHGVT*SA)PDTRPA (Group II immunogen), or 1 ½ fully glycosylated MUC1 repeats KLH-CHGVT* S*A(PDT*RPAPGS*T*APPAHGVT*S*A)PDT*RPA (Group III immunogen).
Supplementary Material
ACKNOWLEDGMENTS
This research was funded by the Georgia Research Alliance and the NIH grants RO1GM066148, and P41RR005351. We are grateful to Dr. Geert-Jan Boons for providing several array molecules and valuable comments, and Dr. Phillip Livingston for valuable discussions.
Footnotes
ASSOCIATED CONTENT
Supporting Information
1-dimensional NMR spectra, overlay of families of structures for A to G, structure statisitics, RDC data, and additional methods details. This material is available free of charge via the Internet at http://pubs.acs.org.
Accession Codes
Protein Data Bank (PDB) coordinate file accession codes for MUC2 structures are A, 2LHV; B, 2LI2; C, 2LI1; D, 2LI0; E, 2LHZ; F, 2LHY; G, 2LHX, and are available free of charge via the Internet at http://www.rcsb.org.
Author Contributions
Notes
Dr. Ragupathi is a paid consultant and share holder in MabVax Therapeutics Inc. which has licensed the KLH-conjugate vaccines from MSKCC.
References
- 1.Cummings RD. The repertoire of glycan determinants in the human glycome. Mol. Biosyst. 2009;5:1087–1104. doi: 10.1039/b907931a. [DOI] [PubMed] [Google Scholar]
- 2.Agard NJ, Bertozzi CR. Chemical approaches to perturb, profile, and perceive glycans. Acc. Chem. Res. 2009;42:788–797. doi: 10.1021/ar800267j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wandall HH, Blixt O, Tarp MA, Pedersen JW, Bennett EP, Mandel U, Ragupathi G, Livingston PO, Hollingsworth MA, Taylor-Papadimitriou J, Burchell J, Clausen H. Cancer biomarkers defined by autoantibody signatures to aberrant O-glycopeptide epitopes. Cancer Res. 2010;70:1306–1313. doi: 10.1158/0008-5472.CAN-09-2893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ten Hagen KG, Fritz TA, Tabak LA. All in the family: The UDP-GalNAc: Polypeptide N-acetylgalactosaminyltransferases. Glycobiology. 2003;13:1R–16R. doi: 10.1093/glycob/cwg007. [DOI] [PubMed] [Google Scholar]
- 5.Gerken TA, Jamison O, Perrine CL, Collette JC, Moinova H, Ravi L, Markowitz SD, Shen W, Patel H, Tabak LA. Emerging paradigms for the initiation of mucin-type protein O-glycosylation by the polypeptide GalNAc transferase family of glycosyltransferases. J. Biol. Chem. 2011;286:14493–14507. doi: 10.1074/jbc.M111.218701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pedersen JW, Bennett EP, Schjoldager K, Meldal M, Holmer AP, Blixt O, Clo E, Levery SB, Clausen H, Wandall HH. Lectin domains of polypeptide GalNAc transferases exhibit glycopeptide binding specificity. J. Biol. Chem. 2011;286:32684–32696. doi: 10.1074/jbc.M111.273722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Raman J, Fritz TA, Gerken TA, Jamison O, Live D, Liu M, Tabak LA. The catalytic and lectin domains of UDP-GalNAc: Polypeptide alpha-N-acetylgalactosaminyltransferase function in concert to direct glycosylation site selection. J. Biol. Chem. 2008;283:22942–22951. doi: 10.1074/jbc.M803387200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wu AM, Lisowska E, Duk M, Yang ZG. Lectins as tools in glycoconjugate research. Glycoconjugate J. 2009;26:899–913. doi: 10.1007/s10719-008-9119-7. [DOI] [PubMed] [Google Scholar]
- 9.Dam TK, Brewer CF. Advances Carbohydr. Chem. Biochem. Vol 63. 2010. Multivalent lectin-carbohydrate interactions: Energetics and mechanisms of binding; pp. 139–164. [DOI] [PubMed] [Google Scholar]
- 10.Ju TZ, Otto VI, Cummings RD. The Tn antigen-structural simplicity and biological complexity. Angew. Chem. Int. Ed. 2011;50:1770–1791. doi: 10.1002/anie.201002313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Desai PR. Immunoreactive T and Tn antigens in malignancy: Role in carcinoma diagnosis, prognosis, and immunotherapy. Transfus. Med. Rev. 2000;14:312–325. doi: 10.1053/tmrv.2000.16229. [DOI] [PubMed] [Google Scholar]
- 12.Danishefsky SJ, Allen JR. From the laboratory to the clinic: A retrospective on fully synthetic carbohydrate-based anticancer vaccines. Angew. Chem. Int. Ed. 2000;39:836–863. doi: 10.1002/(sici)1521-3773(20000303)39:5<836::aid-anie836>3.0.co;2-i. [DOI] [PubMed] [Google Scholar]
- 13.Slovin SF, Ragupathi G, Musselli C, Olkiewicz K, Verbel D, Kuduk SD, Schwarz JB, Sames D, Danishefsky S, Livingston PO, Scher HI. Fully synthetic carbohydrate-based vaccines in biochemically relapsed prostate cancer: Clinical trial results with alpha-N-acetylgalactosamine-O-serine/threonine conjugate vaccine. J. Clin. Oncol. 2003;21:4292–4298. doi: 10.1200/JCO.2003.04.112. [DOI] [PubMed] [Google Scholar]
- 14.Springer GF. Immunoreactive T and Tn epitopes in cancer diagnosis, prognosis, and immunotherapy. J. Mol. Med. 1997;75:594–602. doi: 10.1007/s001090050144. [DOI] [PubMed] [Google Scholar]
- 15.Coltart DM, Royyuru AK, Williams LJ, Glunz PW, Sames D, Kuduk SD, Schwarz JB, Chen XT, Danishefsky SJ, Live DH. Principles of mucin architecture: Structural studies on synthetic glycopeptides bearing clustered mono-, di-, tri-, and hexasaccharide glycodomains. J. Am. Chem. Soc. 2002;124:9833–9844. doi: 10.1021/ja020208f. [DOI] [PubMed] [Google Scholar]
- 16.Kagan E, Ragupathi G, Yi SS, Reis CA, Gildersleeve J, Kahne D, Clausen H, Danishefsky SJ, Livingston PO. Comparison of antigen constructs and carrier molecules for augmenting the immunogenicity of the monosaccharide epithelial cancer antigen Tn. Cancer Immunol. Immunother. 2005;54:424–430. doi: 10.1007/s00262-004-0584-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Osinaga E, Bay S, Tello D, Babino A, Pritsch O, Assemat K, Cantacuzene D, Nakada H, Alzari P. Analysis of the fine specificity of Tn-binding proteins using synthetic glycopeptide epitopes and a biosensor based on surface plasmon resonance spectroscopy. FEBS Lett. 2000;469:24–28. doi: 10.1016/s0014-5793(00)01248-5. [DOI] [PubMed] [Google Scholar]
- 18.Kracun SK, Clo E, Clausen H, Levery SB, Jensen KJ, Blixt O. Random glycopeptide bead libraries for seromic biomarker discovery. J. Proteome Res. 2010;9:6705–6714. doi: 10.1021/pr1008477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Blixt O, Clo E, Nudelman AS, Sorensen KK, Clausen T, Wandall HH, Livingston PO, Clausen H, Jensen KJ. A high-throughput Oglycopeptide discovery platform for seromic profiling. J. Proteome Res. 2010;9:5250–5261. doi: 10.1021/pr1005229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Brooks CL, Schietinger A, Borisova SN, Kufer P, Okon M, Hirama T, MacKenzie CR, Wang LX, Schreiber H, Evans SV. Antibody recognition of a unique tumor-specific glycopeptide antigen. Proc. Nat. Acad. Sci. USA. 2010;107:10056–10061. doi: 10.1073/pnas.0915176107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Takeuchi H, Kato K, Hassan H, Clausen H, Irimura T. O-GalNAc incorporation into a cluster acceptor site of three consecutive threonines - distinct specificity of GalNAc-transferase isoforms. EurJBiochem. 2002;269:6173–6183. doi: 10.1046/j.1432-1033.2002.03334.x. [DOI] [PubMed] [Google Scholar]
- 22.Liu M, Barany G, Live D. Parallel solid-phase synthesis of mucin-like glycopeptides. Carbohydr. Res. 2005;340:2111–2122. doi: 10.1016/j.carres.2005.05.023. [DOI] [PubMed] [Google Scholar]
- 23.Barb AW, Borgert AJ, Liu MA, Barany G, Live D. Methods Enzymol., Vol 478: Glycomics. 2010. Intramolecular glycan-protein interactions in glycoproteins; pp. 365–388. [DOI] [PubMed] [Google Scholar]
- 24.Wang AC, Bax A. Determination of the backbone dihedral angles phi in human ubiquitin from reparametrized empirical Karplus equations. J. Am. Chem. Soc. 1996;118:2483–2494. [Google Scholar]
- 25.Schmidt JM. A versatile component-coupling model to account for substituent effects: Application to polypeptide phi and chi 1 torsion related 3J data. J. Magn. Reson. 2007;186:34–50. doi: 10.1016/j.jmr.2007.01.012. [DOI] [PubMed] [Google Scholar]
- 26.Ottiger M, Bax A. Bicelle-based liquid crystals for NMR-measurement of dipolar couplings at acidic and basic pH values. J. Biomol. NMR. 1999;13:187–191. doi: 10.1023/a:1008395916985. [DOI] [PubMed] [Google Scholar]
- 27.Schuster O, Klich G, Sinnwell V, Kranz H, Paulsen H, Meyer B. 'Wave-type' structure of a synthetic hexaglycosylated decapeptide: A part of the extracellular domain of human glycophorin A. J. Biomol. NMR. 1999;14:33–45. doi: 10.1023/a:1008397304851. [DOI] [PubMed] [Google Scholar]
- 28.Dziadek S, Griesinger C, Kunz H, Reinscheid UM. Synthesis and structural model of an alpha(2,6)-sialyl-T glycosylated MUC1 eicosapeptide under physiological conditions. Chem. Eur. J. 2006;12:4981–4993. doi: 10.1002/chem.200600144. [DOI] [PubMed] [Google Scholar]
- 29.Hashimoto R, Fujitani N, Takegawa Y, Kurogochi M, Matsushita T, Naruchi K, Ohyabu N, Hinou H, Gao XD, Manri N, Satake H, Kaneko A, Sakamoto T, Nishimura SI. An efficient approach for the characterization of mucin-type glycopeptides: The effect of O-glycosylation on the conformation of synthetic mucin peptides. Chem. Eur. J. 2011;17:2393–2404. doi: 10.1002/chem.201002754. [DOI] [PubMed] [Google Scholar]
- 30.Narimatsu Y, Kubota T, Furukawa S, Morii H, Narimatsu H, Yamasaki K. Effect of glycosylation on cis/trans isomerization of prolines in IgA1-hinge peptide. J. Am. Chem. Soc. 2010;132:5548–5549. doi: 10.1021/ja9106429. [DOI] [PubMed] [Google Scholar]
- 31.Kuhn B, Mohr P, Stahl M. Intramolecular hydrogen bonding in medicinal chemistry. J. Med. Chem. 2010;53:2601–2611. doi: 10.1021/jm100087s. [DOI] [PubMed] [Google Scholar]
- 32.Lo-Man R, Vichier-Guerre S, Perraut R, Deriaud E, Huteau V, BenMohamed L, Diop OM, Livingston PO, Bay S, Leclerc C. A fully synthetic therapeutic vaccine candidate targeting carcinoma-associated tn carbohydrate antigen induces tumor-specific antibodies in nonhuman primates. Cancer Res. 2004;64:4987–4994. doi: 10.1158/0008-5472.CAN-04-0252. [DOI] [PubMed] [Google Scholar]
- 33.Morita N, Yajima Y, Asanuma H, Nakada H, Fujita-Yamaguchi Y. Inhibition of cancer cell growth by anti-Tn monoclonal antibody MLS128. Biosci. Trends. 2009;3:32–37. [PubMed] [Google Scholar]
- 34.Blixt O, Head S, Mondala T, Scanlan C, Huflejt ME, Alvarez R, Bryan MC, Fazio F, Calarese D, Stevens J, Razi N, Stevens DJ, Skehel JJ, van Die I, Burton DR, Wilson IA, Cummings R, Bovin N, Wong CH, Paulson JC. Printed covalent glycan array for ligand profiling of diverse glycan binding proteins. Proc. Nat. Acad. Sci. USA. 2004;101:17033–17038. doi: 10.1073/pnas.0407902101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Oyelaran O, Gildersleeve JC. Glycan arrays: Recent advances and future challenges. Curr Opin Chem Biol. 2009;13:406–413. doi: 10.1016/j.cbpa.2009.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Smith DF, Song XZ, Cummings RD. Use of glycan microarrays to explore specificity of glycan-binding proteins. In: Fukuda M, editor. Methods Enzymol., Vol 480: Glycobiology. 2010. pp. 417–444. [DOI] [PubMed] [Google Scholar]
- 37.Godula K, Rabuka D, Nam KT, Bertozzi CR. Synthesis and microcontact printing of dual end-functionalized mucin-like glycopolymers for microarray applications. Angew. Chem. Int. Ed. 2009;48:4973–4976. doi: 10.1002/anie.200805756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gestwicki JE, Cairo CW, Mann DA, Owen RM, Kiessling LL. Selective immobilization of multivalent ligands for surface plasmon resonance and fluorescence microscopy. Anal. Biochem. 2002;305:149–155. doi: 10.1006/abio.2002.5652. [DOI] [PubMed] [Google Scholar]
- 39.Oyelaran O, Li Q, Farnsworth D, Gildersleeve JC. Microarrays with varying carbohydrate density reveal distinct subpopulations of serum antibodies. J. Proteome Res. 2009;8:3529–3538. doi: 10.1021/pr9002245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li QA, Rodriguez LG, Farnsworth DF, Gildersleeve JC. Effects of hapten density on the induced antibody repertoire. ChemBioChem. 2011;11:1686–1691. doi: 10.1002/cbic.201000235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fritz TA, Raman J, Tabak LA. Dynamic association between the catalytic and lectin domains of human UDP-GalNAc : Polypeptide alpha-N-acetylgalactosaminyltransferase-2. J. Biol. Chem. 2006;281:8613–8619. doi: 10.1074/jbc.M513590200. [DOI] [PubMed] [Google Scholar]
- 42.Bolscher JGM, Brevoord J, Nazmi K, Ju TZ, Veerman ECI, van Wijk JAE, Cummings RD, van Die I. Solid-phase synthesis of a pentavalent GalNAc-containing glycopeptide (Tn antigen) representing the nephropathy-associated IgA hinge region. Carbohydr. Res. 2010;345:1998–2003. doi: 10.1016/j.carres.2010.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Babino A, Tello D, Rojas A, Bay S, Osinaga E, Alzari PM. The crystal structure of a plant lectin in complex with the Tn antigen. FEBS Lett. 2003;536:106–110. doi: 10.1016/s0014-5793(03)00037-1. [DOI] [PubMed] [Google Scholar]
- 44.Sanchez JF, Lescar J, Chazalet V, Audfray A, Gagnon J, Alvarez R, Breton C, Imberty A, Mitchell EP. Biochemical and structural analysis of helix pomatia agglutinin - a hexameric lectin with a novel fold. J. Biol. Chem. 2006;281:20171–20180. doi: 10.1074/jbc.M603452200. [DOI] [PubMed] [Google Scholar]
- 45.Avichezer D, Springer GF, Schechter B, Arnon R. Immunoreactivities of polyclonal and monoclonal anti-T and anti-Tn antibodies with human carcinoma cells, grown in vitro and in a xenograft model. IntJCancer. 1997;72:119–127. doi: 10.1002/(sici)1097-0215(19970703)72:1<119::aid-ijc17>3.0.co;2-e. [DOI] [PubMed] [Google Scholar]
- 46.Springer GF, Chandrasekaran EV, Desai PR, Tegtmeyer H. Blood-group Tn-active macromolecules from human carcinomas and erythrocytes - characterization of and specific reactivity with monoclonal and polyclonal anti-Tn antibodies induced by various immunogens. Carbohydr. Res. 1988;178:271–292. doi: 10.1016/0008-6215(88)80118-6. [DOI] [PubMed] [Google Scholar]
- 47.Manimala JC, Roach TA, Li Z, Gildersleeve JC. High-throughput carbohydrate microarray profiling of 27 antibodies demonstrates widespread specificity problems. Glycobiology. 2007;17:17C–23C. doi: 10.1093/glycob/cwm047. [DOI] [PubMed] [Google Scholar]
- 48.Liu MA, Borgert A, Barany G, Live D. Conformational consequences of protein glycosylation: Preparation of O-mannosyl serine and threonine building blocks, and their incorporation into glycopeptide sequences derived from alphadystroglycan. Biopolymers. 2008;90:358–368. doi: 10.1002/bip.20847. [DOI] [PubMed] [Google Scholar]
- 49.van de Ven FJM. Multidimensional NMR in liquids. New York: Wiley-VCH; 1995. [Google Scholar]
- 50.Schwieters CD, Kuszewski JJ, Clore GM. Using Xplor-NIH for NMR molecular structure determination. Prog. Nucl. Mag. Reson. Spectrosc. 2006;48:47–62. [Google Scholar]
- 51.Gilewski T, Adluri S, Ragupathi G, Zhang SL, Yao TJ, Panageas K, Moynahan M, Houghton A, Norton L, Livingston PO. Vaccination of high-risk breast cancer patients with mucin-1 (MUC1) keyhole limpet hemocyanin conjugate plus QS-21. Clin. Cancer. Res. 2000;6:1693–1701. [PubMed] [Google Scholar]
- 52.Tarp MA, Sorensen AL, Mandel U, Paulsen H, Burchell J, Taylor-Papadimitriou J, Clausen H. Identification of a novel cancer-specific immunodominant glycopeptide epitope in the MUC1 tandem repeat. Glycobiology. 2007;17:197–209. doi: 10.1093/glycob/cwl061. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.