Abstract
Mammalian long intergenic noncoding (linc)RNAs are best known for modulating transcription. Here we report a post-transcriptional function for lincRNA-p21 as a modulator of translation. Association of the RNA-binding protein HuR with lincRNA-p21 favored the recruitment of let-7/Ago2 to lincRNA-p21, leading to lower lincRNA-p21 stability. Under reduced HuR levels, lincRNA-p21 accumulated in human cervical carcinoma HeLa cells, increasing its association with JUNB and CTNNB1 mRNAs and selectively lowering their translation. With elevated HuR, lincRNA-p21 levels declined, which in turn derepressed JunB and β-catenin translation and increased the levels of these proteins. We propose that HuR controls translation of a subset of target mRNAs by influencing lincRNA-p21 levels. Our findings uncover a role for lincRNA as a post-transcriptional inhibitor of translation.
INTRODUCTION
Gene expression is robustly regulated at the post-transcriptional level by RNA-binding proteins (RBPs) and by noncoding (nc)RNAs. Small ncRNAs, particularly microRNAs (miRNAs), partially base-pair with specific target mRNAs and repress their expression by lowering mRNA stability and/or translation (Chekulaeva and Filipowicz, 2009; Guo et al., 2010). Gene repression by microRNAs is accomplished through the recruitment of RNA-induced silencing complex (RISC) components such as argonaute (Ago)2, which cleaves target mRNA, and Rck/p54, which facilitates the formation of cytoplasmic processing bodies (PBs), remodels mRNA-associated ribonucleoprotein complexes (mRNPs), and influence mRNA translation, storage, and degradation (Weston and Sommerville, 2006; Chu and Rana, 2006; Bartel 2009). Long ncRNAs (lncRNAs) have been implicated in numerous gene transcription processes, as indicators of transcription factor activity, decoys that titrate away RBPs, functional guides for RNP complexes, and scaffolds for the assembly of functionally related proteins like transcriptional regulators (Wang and Chang, 2011). LncRNAs have also been reported to participate in a limited number of post-transcriptional processes: the lncRNA metastasis-associated lung adenocarcinoma transcript 1 (MALAT1) was implicated in splicing, the cytoplasmic half-Staufen 1-binding site lncRNAs (1/2-sbsRNAs) in Staufen 1-mediated mRNA decay, and an antisense lncRNA (BACE1-AS) interacts with and stabilizes the mRNA encoding the enzyme BACE1 (Faghihi et al., 2008; Tripathi et al., 2010; Gong and Maquat, 2011).
Recently, PAR-CLIP (photoactivatable-ribonucleoside-enhanced crosslinking and immunoprecipitation) analysis (Mukherjee et al., 2011) revealed that the RBP HuR associates with many mRNAs in human cervical carcinoma HeLa cells (~75% of PAR-CLIP RNA tags), and with numerous ncRNAs (~25% of tags, identified as described by Cabili et al., 2011). Among these, the vast majority were lncRNAs, including long intergenic (li)ncRNA-p21, MALAT1, NEAT1, and lncRNAs involved in X chromosome inactivation (Cabili et al., 2011; Mukherjee et al., 2011). HuR is a ubiquitous RBP that influences cell proliferation, survival, carcinogenesis, and the stress and immune responses. HuR performs these functions mainly by associating with subsets of mRNAs and increasing their stability and/or modulating their translation (Hinman and Lou, 2008; Abdelmohsen and Gorospe, 2010). For a few HuR target mRNAs, HuR affects mRNA stability and translation by competing or cooperating with mRNA decay-promoting RBPs [e.g., AUF1, TTP (Lal et al., 2004; Young et al., 2009)] and with microRNAs [e.g., miR-122, let-7 (Bhattacharyya et al., 2006; Kim et al., 2009)]. However, for most target mRNAs, the molecular effectors of HuR’s post-transcriptional fate are unknown.
RESULTS and DISCUSSION
HuR associates with lincRNA-p21, recruits let-7/RISC, accelerates lincRNA-p21 degradation
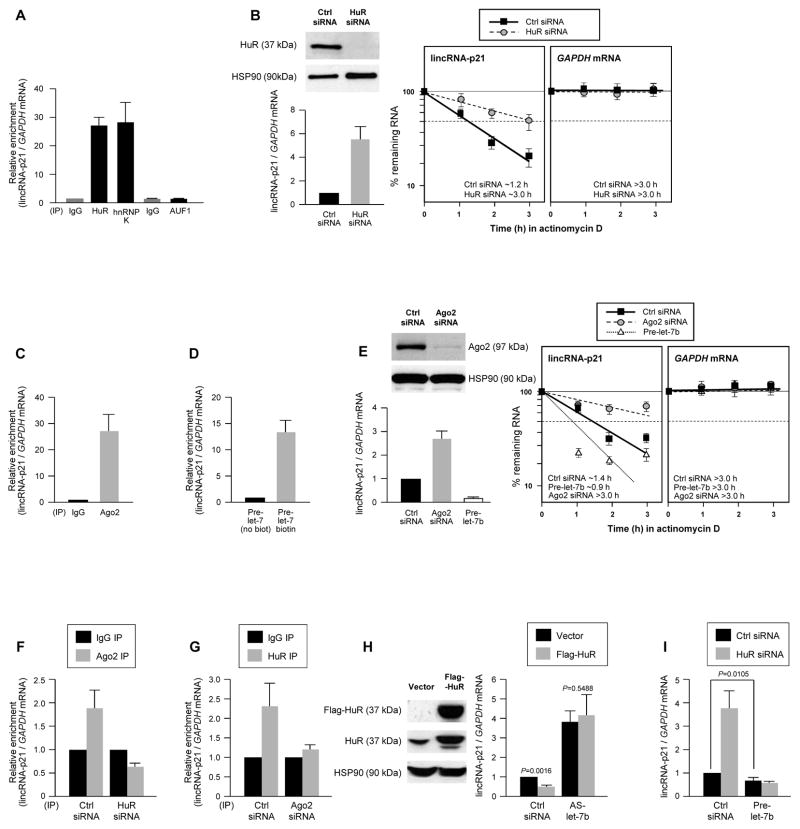
An association between HuR with lincRNA-p21 was detected using the RNP immunoprecipitation (RIP) assay (Experimental Procedures). IP reactions were carried out using HeLa cell lysates and anti-HuR antibody, RNA was extracted from the IP material and analyzed by RT-qPCR using primers specific to human lincRNA-p21, a transcript expressed from a locus between CDKN1A and SFSR3 (Fig. S1A). The human lincRNA-p21 was readily detectable in HeLa cells and was ~3.0 kb in length, like the mouse counterpart (Figs. S1A–C). We sought to investigate this interaction further, given the role of HuR and lincRNA-p21 in the stress response (Abdelmohsen and Gorospe, 2010; Huarte et al., 2010). As shown, lincRNA-p21 was strongly enriched in anti-HuR IP reactions and in control anti-hnRNP K IP reactions (Huarte et al., 2010), but not in anti-AUF1 IP reactions (Fig. 1A). HuR-lincRNA-p21 interactions were also detected in mouse cells (Figs. S1D, S1E).
Figure 1. HuR and let-7/Ago cooperatively promote lincRNA-p21 decay.
(A) RIP analysis of HeLa cell lysates using IgG and antibodies recognizing HuR, AUF1 or hnRNP K. LincRNA-p21 and housekeeping GAPDH mRNA abundance was quantified using RT-qPCR and represented as enrichment in RBP RIP compared with IgG RIP. (B) 48 h after transfecting HeLa cells with Ctrl or HuR siRNAs, HuR and loading control HSP90 levels were assessed by WB (top left), the steady-state lincRNA-p21 and GAPDH mRNA levels quantified by RT-qPCR (bottom left), and the half-life of lincRNA-p21 by measuring the decline in transcript levels after actinomycin D treatment. (C) RIP analysis of the interaction of Ago2 with lincRNA-p21, performed as in (A). (D) 48 h after transfection of HeLa cells with pre-let-7 or biotin-pre-let-7, the relative enrichment of endogenous lincRNA-p21 was assessed by biotin pulldown. (E) 48 h after silencing Ago2 or overexpressing pre-let-7b in HeLa cells, the steady-state levels and half-life of lincRNA-p21 were assessed as in (B). (F, G) 48 h after transfecting HeLa cells with HuR siRNA (F) or Ago2 siRNA (G), the association of lincRNA-p21 with Ago2 (F) and HuR (G) was assessed by RIP analysis. (H, I) 48 h after overexpressing Flag-HuR (H) in HeLa cells, silencing HuR (I), expressing let-7b antagomir (AS-let-7b) (H), or overexpressing let-7b (I), lincRNA-p21 abundance was assessed by RT-qPCR analysis. In all panels, the data represent the means and S.D. (error bars) from 3 independent experiments. Western blots in (B, E, H) are representative of 3 independent experiments.
We hypothesized that HuR might stabilize lincRNA-p21, as HuR stabilizes many mRNAs (Hinman and Lou, 2008). Forty-eight h after silencing HuR using small interfering (si)RNA in HeLa cells, we measured the steady-state lincRNA-p21 levels, as well as the lincRNA-p21 half-life after inhibiting transcription by incubating cells with actinomycin D and measuring the rate of lincRNA-p21 clearance using RT-qPCR. Contrary to prediction, lincRNA-p21 levels were higher and its half-life longer in HuR-silenced cells (t1/2~3 h) than in control cells (t1/2~1.2 h) (Fig. 1B), indicating that HuR destabilized the lincRNA-p21. Accordingly, lincRNA-p21 expression levels were significantly higher in embryonic fibroblasts (MEFs) derived from a mouse lacking both HuR alleles (HuR−/−; Fig. S2A) (Katsanou et al., 2009). These results indicate that HuR enhances lincRNA-p21 decay.
Given earlier evidence that HuR suppressed target c-Myc mRNA expression by facilitating its interaction with let-7/RISC (Kim et al., 2009), we examined whether a similar repression mechanism controlled lincRNA-p21 levels. Mouse lincRNA-p21 was predicted to associate with several miRNAs, with let-7 showing a prominent effect among them (Fig. S2B, S2C). These interactions appeared to be functional, as MEFs deficient in Ago2, a necessary component of let-7/RISC (Cheloufi et al., 2010), displayed higher lincRNA-p21 levels (Fig. S2D). In HeLa cells, Ago2 RIP analysis showed robust enrichment in lincRNA-p21 (Fig. 1C), while transfection of biotinylated precursor (pre)-let-7b followed by pulldown analysis of bound endogenous target mRNAs using streptavidin beads and RT-qPCR analysis (Lal et al., 2011) revealed a marked enrichment in lincRNA-p21 compared with a control transcript (GAPDH mRNA), but not in pulldowns using non-biotinylated control pre-let-7b (Fig. 1D). These interactions affected lincRNA-p21 stability, as its half-life was higher after Ago2 silencing (t1/2~3 h) and was lower after overexpressing pre-let-7b (t1/2~0.9 h) (Fig. 1E). Collectively, these data indicate that HuR and let-7/Ago2 lower lincRNA-p21 stability.
As assessed by RIP analysis, silencing Ago2 in HeLa cells reduced the interaction of HuR with lincRNA-p21, while silencing HuR lowered the interaction of Ago2 with lincRNA-p21 (Fig. 1F, G). Overexpression of Flag-tagged HuR significantly reduced lincRNA-p21 levels, but did not reverse the elevated lincRNA-p21 levels observed after inhibition of endogenous let-7 using an antagomir (AS-let-7b) (Fig. 1H). Together with evidence that the heightened lincRNA-p21 after HuR silencing was prevented by overexpressing pre-let-7 (Fig. 1I), and that mutating let-7 sites can block the HuR-elicited repression (Fig. S1C) our findings suggest that HuR and let-7/Ago2 repress lincRNA-p21 expression cooperatively, and that HuR and let-7/Ago2 binding to lincRNA-p21 is crucial for lincRNA-p21 decay.
LincRNA-p21 selectively interacts with target CTNNB1 and JUNB mRNAs
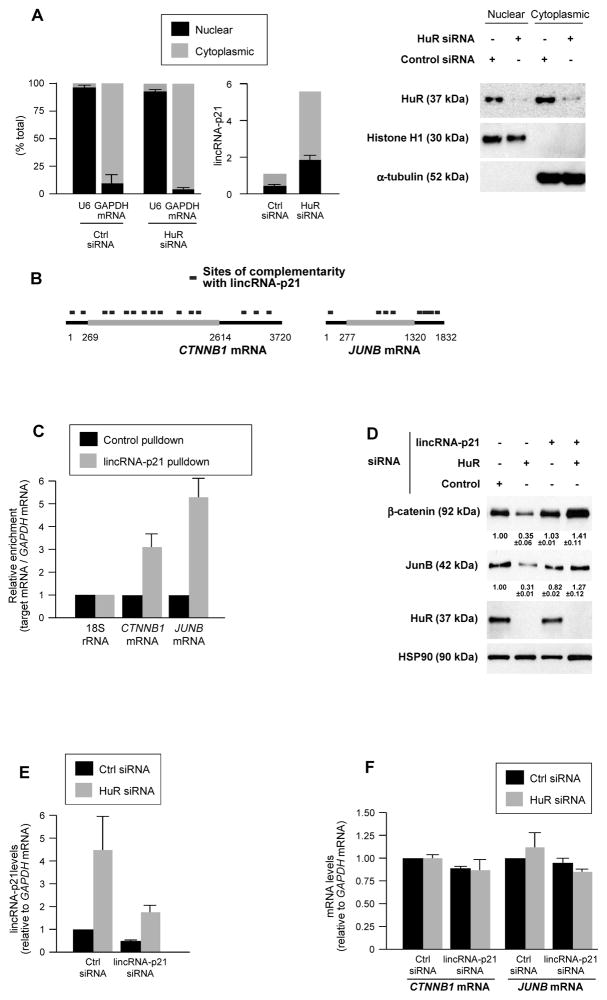
LincRNA-p21 was moderately more abundant in the cytoplasm than in the nucleus of fractionated HeLa cells and its levels increased proportionately after silencing HuR (Fig. 2A). LincRNA-p21 subcellular localization was further analyzed by tagging lincRNA-p21 with MS2 RNA hairpins, tracked intracellularly by fluorescent fusion protein MS2-YFP [(Bertrand et al., 1998; Lee et al., 2010) Fig. S3A]. We postulated that this distribution could impact upon cytoplasmic gene regulatory events and further hypothesized that lincRNA-p21 might elicit some of HuR’s effects on target mRNAs.
Figure 2. lincRNA-p21 associates with β-catenin and JunB mRNAs, lowers their expression.
(A) 48 h after transfecting siRNAs in HeLa cells, the levels of nuclear control transcript (U6), cytoplasmic control transcript (GAPDH mRNA) and lincRNA-p21 were assessed by RT-qPCR in nuclear and cytoplasmic fractions (left), and WB analysis was performed (right). (B) Regions of predicted interaction between lincRNA-p21 and human CTNNB1 and JUNB mRNAs; details in Table S1. (C) HeLa cell lysates were incubated with 5′-end biotin-labeled antisense lincRNA-p21 oligo (lincRNA-p21 pulldown) and sense oligo (control pulldown); after pulldown, RNA was extracted and CTNNB1 and JUNB mRNAs, as well as normalization control 18S rRNA were assessed by RT-qPCR. (D, E) 48 h after transfecting HeLa cells with siRNAs, the levels of β-catenin, JunB, HuR, and HSP90 were assessed by WB analysis and densitometry (D), and lincRNA-p21 (E), CTNNB1, and JUNB mRNAs (F) were quantified by RT-qPCR. In (A, C–F), data represent the means and S.D. (error bars) from at least 3 independent experiments. Western blots in (A, D) are representative of 3 independent experiments.
To test these possibilities, we focused on mRNAs encoding β-catenin (CTNNB1) and JunB (JUNB), identified as being translationally repressed after HuR was silenced (López de Silanes et al., 2003; Lebedeva et al., 2011). Several regions of high complementarity with lincRNA-p21 were identified for CTNNB1 mRNA (15 sites) and for JUNB mRNA (8 sites), but only 2 for GAPDH mRNA (Fig. 2B; Table S1). In HeLa cells, the interaction of endogenous lincRNA-p21 with CTNNB1 and JUNB mRNAs was quantified by affinity pulldown of endogenous lincRNA-p21 using a biotinylated RNA antisense to lincRNA-p21 (Experimental Procedures). As shown in Fig. 2C, CTNNB1 and JUNB mRNAs showed significantly greater interaction with lincRNA-p21 than GAPDH mRNA (used for normalization of sample input) and 18S rRNA (used as reference for enrichment). Similarly, biotinylated mouse lincRNA-p21 incubated with MEF lysates, followed by RNA extraction and detection by RT-qPCR, revealed its selective interaction with mouse ctnnb1 and junb mRNAs (Figs. S3B, C); conversely, in vitro-transcribed unlabeled lincRNA-p21, was selectively pulled down using biotinylated mouse ctnnb1and junb RNAs (Fig. S3D).
Lowering HuR in HeLa cells decreased β-catenin and JunB levels, as assessed by Western blotting (Fig. 2D). Strikingly, however, simultaneous silencing lincRNA-p21 by using a specific siRNA that lowered lincRNA-p21 levels to ~40–45% of the levels seen in Ctrl siRNA cells (Fig. 2E) and preferentially silenced cytoplasmic lincRNA-p21 (Fig. S3F), prevented the decline in β-catenin and JunB levels (Fig. 2D). Simply silencing lincRNA-p21 or Ago2 in HeLa cells did not affect β-catenin or JunB levels (Fig. S3G), supporting the notion that repression required HuR silencing. These effects were not due to changes in CTNNB1 or JUNB mRNA levels (Fig. 2F), nor were they due to changes in β-catenin or JunB protein stability (not shown), suggesting that lincRNA-p21 likely reduced translation of CTNNB1 and JUNB mRNAs.
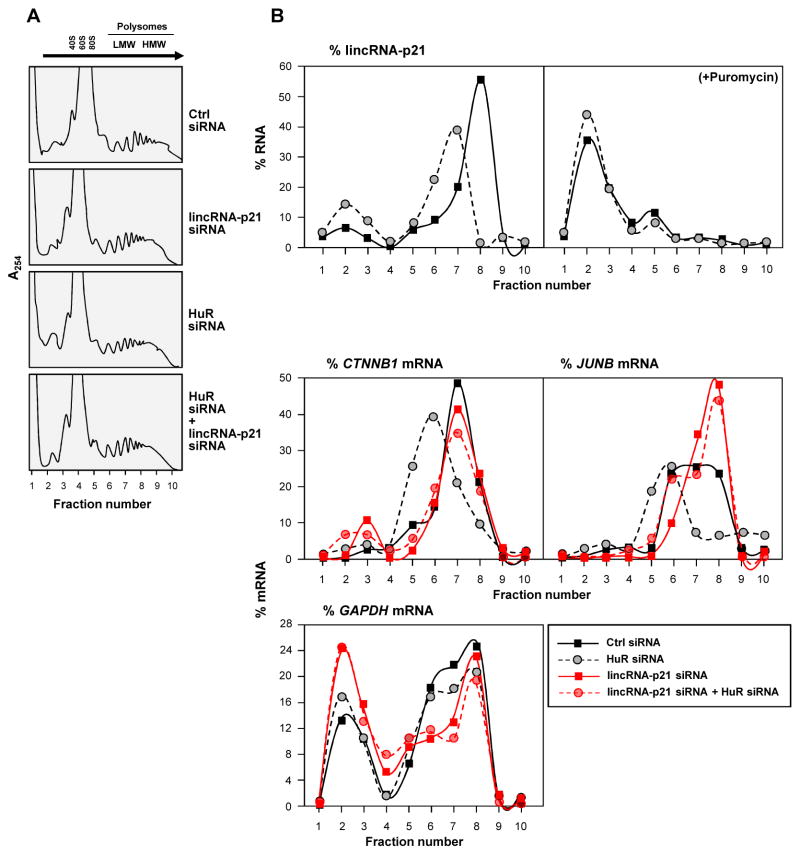
LincRNA-p21 associates with translational apparatus, diminishes CTNNB1 and JUNB polysomes
To directly test the possibility that lincRNA-p21 may influence translation, HeLa cell lysates expressing different lincRNA-p21 and HuR levels were fractionated through sucrose gradients. The lightest components sedimented at the top (fractions 1,2), small (40S) and large (60S) ribosomal subunits and monosomes (80S) in fractions 3–5, and progressively larger polysomes, ranging from low- to high-molecular-weight (LMW, HMW) in fractions 6–10 (Fig. 3A). Silencing HuR and/or lincRNA-p21 did not change the polysome distribution profiles or eIF2α phosphorylation (Fig. 3A, Fig. S4A), indicating that these interventions did not affect global translation. After isolating RNA from each fraction, RT-qPCR analysis indicated that lincRNA-p21 was abundant in fractions 6–9; although silencing HuR elevated lincRNA-p21 levels overall, its distribution shifted towards smaller polysomes (Fig. 3B, top left). LincRNA-p21 associated with polysomes and did not simply cosediment with polysomes, as puromycin treatment, which disrupts polysomes, markedly shifted leftward the distribution of lincRNA-p21 (Fig. 3B, top right). The polysomal sizes of CTNNB1 and JUNB mRNAs also shifted leftward after HuR silencing, in keeping with reduced translation (Fig. 3B, bottom). Interestingly, silencing lincRNA-p21 totally prevented the reduction in polysomes seen after silencing HuR (Fig. 3B, bottom), in agreement with the increased β-catenin and JunB abundance (Fig. 2E). The distribution of the housekeeping GAPDH mRNA did not show this pattern (Fig. 3B, bottom), indicating that silencing HuR siRNA and/or lincRNA-p21 specifically affected CTNNB1 and JUNB mRNAs. It remains to be determined whether other mRNAs are translationally repressed by lincRNA-p21 in this manner, as well as the fractions of JUNB and CTNNB1 mRNA pools that associate with lincRNA-p21.
Figure 3. lincRNA-p21 associates with polysomes, suppresses β-catenin and JunB translation.
Forty-eight h after siRNA transfection of HeLa cells, (A) polysomes in cytoplasmic extracts were fractionated through sucrose gradients (arrow: direction of sedimentation; –, no ribosomal components), and (B) the relative distribution of lincRNA-p21 on polysome gradients +/− puromycin (top), and relative levels of CTNNB1, JUNB, and GAPDH mRNAs (bottom), were studied by RT-qPCR analysis of RNA in gradient fractions, and represented as % of total RNA in the gradient. Data are representative of 3 independent experiments.
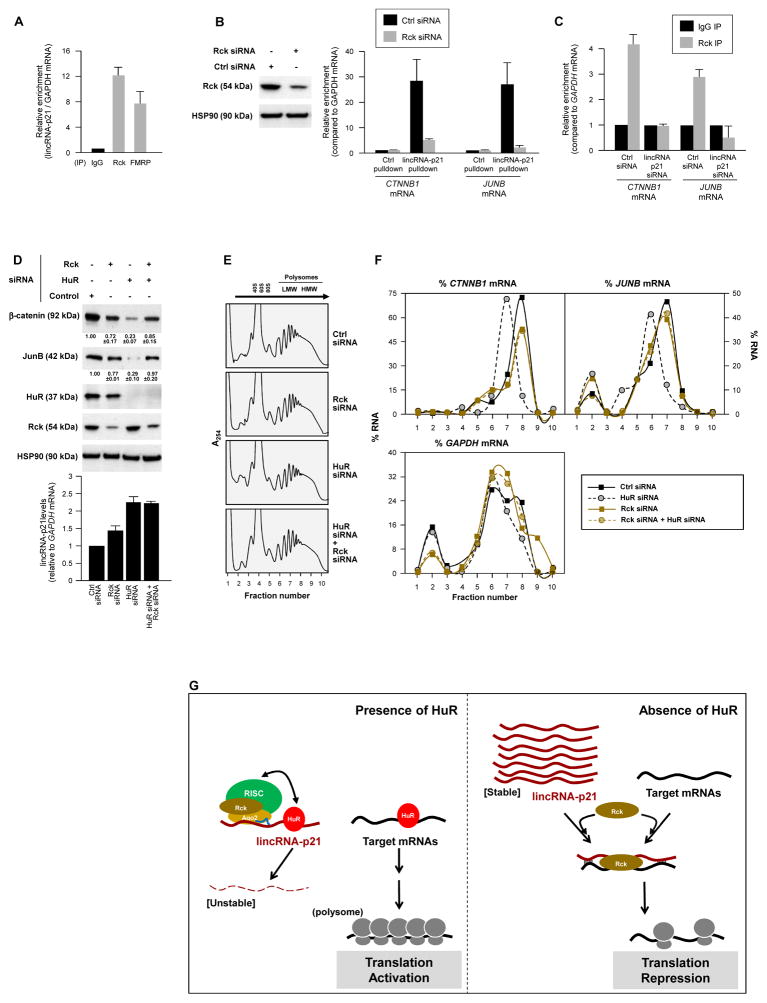
Finally, we investigated if lincRNA-p21 inhibited the translation of CTNNB1 and JUNB mRNAs by enhancing their interaction with translational repressors. By RIP analysis, the translational repressors Rck and FMRP, were found to interact with lincRNA-p21 in HeLa cells (Fig. 4A) and MEFs (Fig. S4B). In HeLa lysates, Rck and FMRP (but not TIAR) also associated with MS2-tagged lincRNA-p21 (immobilized on beads via MS2-GST; Fig. S4C). Interestingly, the interaction of endogenous lincRNA-p21 with endogenous CTNNB1 and JUNB mRNAs in HeLa cells (measured as in Fig. 2C) was potently reduced if Rck was silenced (Fig. 4B), indicating that Rck facilitated these interactions. A similar effect of Rck was seen with a tagged mouse lincRNA-p21 (Fig. S4D). Conversely, when lincRNA-p21 was silenced, Rck did not associate with CTNNB1 or JUNB mRNAs (Fig. 4C). In turn, silencing Rck in HeLa cells reversed the inhibition of β-catenin and JunB expression seen after HuR silencing (Fig. 4D top); lincRNA-p21 levels were not markedly changed by Rck silencing (Fig. 4D graph). These findings indicated that the repression of β-catenin and JunB translation by lincRNA-p21 required Rck function (Chu and Rana, 2006). Supporting this possibility, the decline in the sizes of polysomes associated with CTNNB1 and JUNB mRNAs after silencing HuR [previously attributed to the higher lincRNA-p21 levels (Fig. 3B)], was only seen when Rck was expressed (Fig. 4E, F); lincRNA-p21 followed a similar distribution pattern (Fig. S4E). Whether Rck reduces the translation of other mRNAs in a similar fashion remains to be studied.
Figure 4. Translation inhibition by lincRNA-p21 involves recruitment of translation repressor Rck.
All experiments were done in HeLa cells. (A) RIP analysis of the interaction of endogenous lincRNA-p21 with Rck and FMRP. (B) 48 h after transfecting the siRNAs shown, the relative interaction of lincRNA-p21 and CTNNB1 or JUNB mRNAs was studied by RIP analysis. (C) RIP analysis of Rck interaction with CTNNB1 or JUNB mRNAs in cells expressing normal levels or silenced lincRNA-p21. (D) WB analysis and densitometric quantification (top) and lincRNA-p21 RT-qPCR analysis (bottom) 48 h after silencing Rck and/or HuR. (E, F) 48 h after transfecting the siRNAs shown, polysomes were prepared (E) and the relative distribution of CTNNB1, JUNB, and GAPDH mRNAs (F) was studied as explained in Fig. 3. (F) Schematic of the proposed mechanism whereby lincRNA-p21, under negative control by HuR, represses the translation of CTNNB1 and JUNB mRNAs; see text for details. Data in (A–D) represent the means and S.D. (error bars) from 3 independent experiments. Data in (B, D, E, F) are representative of 3 independent experiments.
Perspective: lincRNA-p21 inhibits translation of target mRNAs
Based on these results, we propose that in the presence of HuR, lincRNA-p21 is unstable through the recruitment of let-7/Ago2. HuR then promotes the translation of targets CTNNB1 and JUNB mRNAs by favoring their association with polysomes (López de Silanes et al., 2003; Lebedeva et al., 2011) (Fig. 4G). In the absence of HuR, lincRNA-p21 is stable and accumulates, and Rck promotes the association of lincRNA-p21 with CTNNB1 and JUNB mRNAs, repressing their translation through a mechanism that includes reduced polysome sizes (Fig. 4G); in addition, base-pair interactions of lincRNA-p21 with target mRNAs may result in ribosome ‘drop-off’. In sum, HuR-dependent translation activation requires rapid degradation of lincRNA-p21 in order to prevent the recruitment of translation repressors onto target mRNAs. Similar regulation may affect other mRNAs whose translation increases by HuR (Abdelmohsen and Gorospe, 2010). Through these regulatory processes, HuR can help implement a well-established pro-oncogenic, cell-protective program (Fig. S4F, G) which includes pro-survival proteins β-catenin and JunB (Shaulian, 2010; Fu et al., 2011). With rising recognition that lncRNAs play pivotal roles in disease processes (Wapinski and Chang, 2011), other proteins regulated by the orchestrated influence of RBPs, lncRNAs and microRNAs are likely to emerge.
EXPERIMENTAL PROCEDURES
Cell culture, transfection, small interfering RNAs, microRNAs and plasmids
Human HeLa cells and mouse embryonic fibroblast (MEF) were cultured in DMEM (Invitrogen) supplemented with 10% (v/v) FBS and antibiotics. All siRNAs, including control (Ctrl) siRNA (UUCUCCGAACGUGUCACGUdTdT), and siRNAs to lower lincRNA-p21 (CTGCAAGGCCGCATGATGAdTdT), HuR (CGUAAGUUAUUUCCUUUAAdTdT), Ago2, and Rck (sc-44409 and sc-72246, respectively, Santa Cruz Biotechnology), were transfected at 20 nM final concentration using lipofectamine 2000 (Invitrogen) and analyzed 48 h later. Pre- and anti-let-7b (Ambion) were transfected at 10 nM final concentration. The oligomers for affinity pulldown of endogenous human lincRNA-p21 [GGGTGGCTCACTCTTCTGGC (antisense) and GCCAGAAGAGTGAGCCACCC (sense)] were biotinylated at the 5′ end. Actinomycin D (Sigma) was used at 2.5 μg/μl. A plasmid expressing lincRNA-p21 (Huarte et al., 2010), was used to construct plasmid plincRNA-p21-MS2. Plasmid pMS2-YFP was previously reported (Lee et al., 2010); pMS2-GST was a kind gift from J.A. Steitz.
Western blot analysis
Whole-cell lysates, prepared in RIPA buffer, were separated by SDS-polyacrylamide gel electrophoresis (SDS-PAGE), and transferred onto PVDF membranes (Invitrogen iBlot Stack). Primary antibodies recognizing β-catenin, JunB, α-tubulin, histone H1, HSP90, HuR, Rck, FMRP, GFP, eIF2α and phospho-eIF2α were from Santa Cruz Biotechnology. Antibodies recognizing Ago2, MBP, AUF1, TIAR and Flag were from Abcam, Cell Signaling Technology, Millipore, BD Biosciences and Sigma respectively. HRP-conjugated secondary antibodies were from GE Healthcare.
Immunoprecipitation assays
For immunoprecipitation (IP) of endogenous RNP complexes from whole-cell extracts (Abdelmohsen et al., 2010), cells were lysed in 20 mM Tris-HCl at pH 7.5, 100 mM KCl, 5 mM MgCl2 and 0.5% NP-40 for 10 min on ice and centrifuged at 10,000 × g for 15 min at 4°C. The supernatants were incubated with protein A-Sepharose beads coated with antibodies that recognized HuR, Rck or FMRP (Santa Cruz Biotechnology), Ago2 (Abcam) or AUF1 (Millipore), or with control IgG (Santa Cruz Biotechnology) for 1 h at 4°C. After the beads were washed with NT2 buffer (50 mM Tris-HCl at pH 7.5, 150 mM NaCl, 1 mM MgCl2 and 0.05% NP-40), the complexes were incubated with 20 units of RNase-free DNase I (15 min at 37°C) and further incubated with 0.1% SDS/0.5 mg/ml Proteinase K (15 min at 55 °C) to remove DNA and proteins, respectively. The RNPs isolated from the IP materials was further assessed by RT-qPCR analysis.
RNA analysis
Trizol (Invitrogen) was used to extract total RNA and acidic phenol (Ambion) was used to extract RNA for RIP analysis (Abdelmohsen et al., 2010). Reverse transcription (RT) was performed using random hexamers and reverse transcriptase (SSII, Invitrogen) and real-time, quantitative (q)PCR using gene-specific primers (supplemental Table S2) and SYBR green master mix (Kapa Biosystems), using the Applied Biosystems 7300 instrument.
Biotin pulldown assay
To synthesize biotinylated transcripts, PCR fragments were prepared using forward primers that contained the T7 RNA polymerase promoter sequence (Abdelmohsen et al., 2010). Primers used to prepare biotinylated transcripts are listed below (supplemental Table S2). After purification of the PCR products, biotinylated transcripts were synthesized using MaxiScript T7 kit (Ambion) and whole-cell lysates (50 μg per sample) were incubated with 1 μg of purified biotinylated transcripts for 1 h at 25°C; complexes were isolated with Streptavidin-coupled Dynabeads (Invitrogen). The proteins present in the pulldown material were detected by Western blot analysis and the RNA present in the pulldown material by RT-qPCR analysis.
Biotinylated lincRNA-p21 was synthesized using T7 RNA polymerase and plasmid pcDNA3 lincRNA-p21 (Huarte et al., 2010). Forward PCR primers contained the T7 RNA polymerase promoter sequence (CCAAGCTTCTAATACGACTCACTATAGGGAGA [T7]). Primers used are listed in the supplemental Table S2.
For antisense oligomer pulldown, biotin-labeled DNA against human lincRNA-p21 (0.5 μg) was incubated with HeLa cell lysates for 2 h and the complexes were isolated with Streptavidin-coupled Dynabeads (Invitrogen).
Polysome analysis
Forty-eight h after transfection with siRNAs, HeLa cells were pre-incubated with cycloheximide (Sigma; 100 μg/ml for 15 min) and cytoplasmic lysates were prepared and fractionated by ultracentrifugation through 15–60% linear sucrose gradients; 10 fractions were collected and RNA extracted from each fraction was used for RT-qPCR analysis, as described (Lee et al., 2010).
Subcellular Fractionation
Cytosolic and nuclear fractions were collected as described previously (Lal et al., 2004). Briefly, cells were lysed with a buffer containing 10 mM Tris-HCl, pH 7.4, 100 mM, NaCl, 2.5 mM MgCl2 and 40 μg/ml digitonin for 10 min and the resulting lysates were centrifuged with 2,060 g for 10 min at 4°C. The supernatant was used for the cytosolic fraction. The pellets were washed, incubated with RIPA buffer at 4°C for 10 min and the nuclear fraction collected after centrifugation at 4°C for 10 min at 21000 g.
Bioinformatic analysis of lincRNA-p21 interaction sites with mRNAs
We used BLAST (http://blast.ncbi.nlm.nih.gov/) to identify local regions of sequence similarity between lincRNA-p21 (supplemental text) and CTNNB1 mRNA (NM_001904.3), JUNB mRNA (NM_002229.2), and GAPDH mRNA (NM_002046.3). The similarity regions with a length ≥ 20 bp, E value ≤ 210 and matching to the reverse complementary sequence of lincRNA-p21 were selected as and considered as possible interaction regions through base-paring between lincRNA-p21 and each mRNA. The supplemental Table S1 lists the putative interaction regions identified.
Supplementary Material
Acknowledgments
We thank D.L. Kontoyiannis, G.J. Hannon, N. Mukherjee, and J.D. Keene for providing reagents and information. JHY, KA, SS, XY, JLM, SD, KGB, and MG were supported by the NIA-IRP, NIH. MZ was supported by NIH U54CA149169.
References
- Abdelmohsen K, Pullmann R, Jr, Lal A, Kim HH, Galban S, Yang X, Blethrow JD, Walker M, Shubert J, Gillespie DA, et al. Phosphorylation of HuR by Chk2 regulates SIRT1 expression. Mol Cell. 2007;25:543–557. doi: 10.1016/j.molcel.2007.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abdelmohsen K, Gorospe M. Posttranscriptional regulation of cancer traits by HuR. Wiley Interdiscip Rev RNA. 2010;1:214–229. doi: 10.1002/wrna.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertrand E, Chartrand P, Schaefer M, Shenoy SM, Singer RH, Long RM. Localization of ASH1 mRNA particles in living yeast. Mol Cell. 1998;2:437–445. doi: 10.1016/s1097-2765(00)80143-4. [DOI] [PubMed] [Google Scholar]
- Bhattacharyya SN, Habermacher R, Martine U, Closs EI, Filipowicz W. Relief of microRNA-mediated translational repression in human cells subjected to stress. Cell. 2006;125:1111–1124. doi: 10.1016/j.cell.2006.04.031. [DOI] [PubMed] [Google Scholar]
- Cabili MN, Trapnell C, Goff L, Koziol M, Tazon-Vega B, Regev A, Rinn JL. Integrative annotation of human large intergenic noncoding RNAs reveals global properties and specific subclasses. Genes Dev. 2011;25:1915–1927. doi: 10.1101/gad.17446611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chekulaeva M, Filipowicz W. Mechanisms of miRNA-mediated post-transcriptional regulation in animal cells. Curr Opin Cell Biol. 2009;21:452–460. doi: 10.1016/j.ceb.2009.04.009. [DOI] [PubMed] [Google Scholar]
- Cheloufi S, Dos Santos CO, Chong MM, Hannon GJ. A dicer-independent miRNA biogenesis pathway that requires Ago catalysis. Nature. 2010;465:584–589. doi: 10.1038/nature09092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu CY, Rana TM. Translation repression in human cells by microRNA-induced gene silencing requires RCK/p54. PLoS Biol. 2006;4:e210. doi: 10.1371/journal.pbio.0040210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faghihi MA, Modarresi F, Khalil AM, Wood DE, Sahagan BG, Morgan TE, Finch CE, St Laurent G, 3rd, Kenny PJ, Wahlestedt C. Expression of a noncoding RNA is elevated in Alzheimer’s disease and drives rapid feed-forward regulation of beta-secretase. Nat Med. 2008;14:723–730. doi: 10.1038/nm1784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Y, Zheng S, An N, Athanasopoulos T, Popplewell L, Liang A, Li K, Hu C, Zhu Y. β-catenin as a potential key target for tumor suppression. Int J Cancer. 2011;129:1541–1551. doi: 10.1002/ijc.26102. [DOI] [PubMed] [Google Scholar]
- Gong C, Maquat LE. lncRNAs transactivate STAU1-mediated mRNA decay by duplexing with 3′ UTRs via Alu elements. Nature. 2011;470:284–288. doi: 10.1038/nature09701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo H, Ingolia NT, Weissman JS, Bartel DP. Mammalian microRNAs predominantly act to decrease target mRNA levels. Nature. 2010;466:835–840. doi: 10.1038/nature09267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinman MN, Lou H. Diverse molecular functions of Hu proteins. Cell Mol Life Sci. 2008;65:3168–3181. doi: 10.1007/s00018-008-8252-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huarte M, Guttman M, Feldser D, Garber M, Koziol MJ, Kenzelmann-Broz D, Khalil AM, Zuk O, Amit I, Rabani M, et al. A large intergenic noncoding RNA induced by p53 mediates global gene repression in the p53 response. Cell. 2010;142:409–419. doi: 10.1016/j.cell.2010.06.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katsanou V, Milatos S, Yiakouvaki A, Sgantzis N, Kotsoni A, Alexiou M, Harokopos V, Aidinis V, Hemberger M, Kontoyiannis DL. The RNA-binding protein Elavl1/HuR is essential for placental branching morphogenesis and embryonic development. Mol Cell Biol. 2009;29:2762–2776. doi: 10.1128/MCB.01393-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HH, Kuwano Y, Srikantan S, Lee EK, Martindale JL, Gorospe M. HuR recruits let-7/RISC to repress c-Myc expression. Genes Dev. 2009;23:1743–1748. doi: 10.1101/gad.1812509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lal A, Mazan-Mamczarz K, Kawai T, Yang X, Martindale JL, Gorospe M. Concurrent versus individual binding of HuR and AUF1 to common labile target mRNAs. EMBO J. 2004;23:3092–3102. doi: 10.1038/sj.emboj.7600305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lal A, Thomas MP, Altschuler G, Navarro F, O’Day E, Li XL, Concepcion C, Han YC, Thiery J, Rajani DK, et al. Capture of microRNA-bound mRNAs identifies the tumor suppressor miR-34a as a regulator of growth factor signaling. PLoS Genet. 2011;7:e1002363. doi: 10.1371/journal.pgen.1002363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lebedeva S, Jens M, Theil K, Schwänhausser B, Selbach M, Landthaler M, Rajewsky N. Transcriptome-wide analysis of regulatory interactions of the RNA-binding protein HuR. Mol Cell. 2011;43:340–352. doi: 10.1016/j.molcel.2011.06.008. [DOI] [PubMed] [Google Scholar]
- Lee EK, Kim HH, Kuwano Y, Abdelmohsen K, Srikantan S, Subaran SS, Gleichmann M, Mughal MR, Martindale JL, Yang X, et al. hnRNP C promotes APP translation by competing with FMRP for APP mRNA recruitment to P bodies. Nat Struct Mol Biol. 2010;17:732–739. doi: 10.1038/nsmb.1815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- López de Silanes I, Fan J, Yang X, Zonderman AB, Potapova O, Pizer ES, Gorospe M. Role of the RNA-binding protein HuR in colon carcinogenesis. Oncogene. 2003;22:7146–7154. doi: 10.1038/sj.onc.1206862. [DOI] [PubMed] [Google Scholar]
- Mukherjee N, Corcoran DL, Nusbaum JD, Reid DW, Georgiev S, Hafner M, Ascano M, Jr, Tuschl T, Ohler U, Keene JD. Integrative regulatory mapping indicates that the RNA-binding protein HuR couples pre-mRNA processing and mRNA stability. Mol Cell. 2011;43:327–339. doi: 10.1016/j.molcel.2011.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaulian E. AP-1--The Jun proteins: Oncogenes or tumor suppressors in disguise? Cell Signal. 2010;22:894–899. doi: 10.1016/j.cellsig.2009.12.008. [DOI] [PubMed] [Google Scholar]
- Tripathi V, Ellis JD, Shen Z, Song DY, Pan Q, Watt AT, Freier SM, Bennett CF, Sharma A, Bubulya PA, et al. The nuclear-retained noncoding RNA MALAT1 regulates alternative splicing by modulating SR splicing factor phosphorylation. Mol Cell. 2010;39:925–938. doi: 10.1016/j.molcel.2010.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang KC, Chang HY. Molecular mechanisms of long noncoding RNAs. Mol Cell. 2011;43:904–914. doi: 10.1016/j.molcel.2011.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wapinski O, Chang HY. Long noncoding RNAs and human disease. Trends Cell Biol. 2011;21:354–361. doi: 10.1016/j.tcb.2011.04.001. [DOI] [PubMed] [Google Scholar]
- Weston A, Sommerville J. Xp54 and related (DDX6-like) RNA helicases: roles in messenger RNP assembly, translation regulation and RNA degradation. Nucleic Acids Res. 2006;34:3082–3094. doi: 10.1093/nar/gkl409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young LE, Sanduja S, Bemis-Standoli K, Pena EA, Price RL, Dixon DA. The mRNA binding proteins HuR and tristetraprolin regulate cyclooxygenase 2 expression during colon carcinogenesis. Gastroenterology. 2009;136:1669–1679. doi: 10.1053/j.gastro.2009.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.