Summary
The ubiquitin system plays an important role in trafficking of signaling receptors from the plasma membrane to lysosomes. Triad1 is a ubiquitin ligase that catalyzes the formation of poly-ubiquitin chains linked via lysine-48 as well as lysine-63 residues. We show that depletion of Triad1 affects the sorting of both growth hormone and epidermal growth factor. Triad1-depleted cells accumulate both ligands in endosomes. While fluid phase transport to the lysosomes is reduced in the absence of Triad1, growth hormone receptor can recycle back to the plasma membrane together with transferrin. Using immune electron microscopy we show that Triad1 depletion results in enlarged endosomes with enlarged and irregular shaped intraluminal vesicles. The endosomes display prominent clathrin coats and show increased levels of growth hormone label. We conclude that Triad1 is required for the proper function of multivesicular bodies.
Keywords: EGF, GH, Triad1, Degradation, Endosome, Membrane receptors
Introduction
In mammalian cells, the involvement of ubiquitination in endosomal sorting has been well established (Clague and Urbé, 2010; Sorkin and Goh, 2009; Stuffers et al., 2009a; Varghese et al., 2008). Only in a few cases K63-linked ubiquitination was indicated specifically in endocytosis (Paiva et al., 2009; Kramer et al., 2010). Our model system, in which we study endocytosis and endosomal sorting of the growth hormone receptor (GHR), uses a K48 ubiquitin ligase for both endocytosis, and endosomal sorting (van Kerkhof et al., 2007; van Kerkhof et al., 2011). To learn more about the involvement of K63-linked ubiquitination, we used a limited library of silencing RNAs that specifically target ubiquitin ligases, known to be involved in K63 linkage assembly. From this screen TRIAD1 (two RING [really interesting new gene] fingers and DRIL [double RING finger linked] 1) emerged as being required for the degradation of both growth hormone (GH) and epidermal growth factor (EGF).
TRIAD domain-containing ubiquitin ligases contain a cysteine-rich DRIL/IBR domain that is flanked by two RING fingers. In several TRIAD proteins the N-terminal RING finger specifically binds K48 chain E2 conjugating enzymes, while the C-terminal RING finger binds the K63 E2 enzyme, Ubc13. The best studied TRIAD proteins are Parkin, Hoil-1, Hoip, Dorfin and Triad1 (Wenzel et al., 2011). TRIAD proteins regulate diverse biological processes (Huang et al., 2006; Ishigaki et al., 2007; Tokunaga et al., 2009). Parkin, the best studied TRIAD protein (reviewed by Chin et al., 2010), has been reported to regulate K48-dependent proteasomal degradation of several substrates (7) (Narendra et al., 2008). Triad1 got attention for its role in myeloid cell proliferation and by regulating the levels of the transcription factor Gfi-1 (Marteijn et al., 2007; Wang et al., 2011). For Triad1, direct interactions of the E2 enzymes UbcH7 and Ubc13 with RING1 and RING2, respectively, have been shown (Marteijn et al., 2009). Although the conserved combination of RING1 and RING2 together with the DRIL/IBR domain suggest a close collaboration between the K48 and K63 ubiquitin systems, its conservation remains elusive.
In the endosomal sorting system, both K63- and K48-linked ubiquitin play a role. The endosomal system collects membranes and proteins from the plasma membrane and the trans-Golgi network (TGN), and sorts them either back to the plasma membrane (default route) and to the TGN or, via multivesicular bodies (MVB) to the lysosome. Membrane coats of different compositions including clathrin collect the cargo and shape the membrane. Recently, Golgi localized γ-ear containing ARF binding adapter proteins (GGA) were attributed a role trapping the K63 ubiquitinated cargo from early endosome to the MVB (Lauwers et al., 2009). Rabs, a family of small GTPases, identify the formed vesicles and control their budding, uncoating, motility and fusion (reviewed by Markgraf et al., 2007; Stenmark, 2009). This is accomplished by recruiting tethering factors, sorting adapters, kinases, phosphatases and motor adapters. ESCRT (endosomal sorting complex required for transport) complexes receive the ubiquitinated cargo from coat proteins and GGAs and facilitate the formation of the intraluminal vesicles (ILV) containing the cargo. The Vps-C complexes, homotypic fusion and vacuole protein sorting (HOPS) and CORVET, convert Rab5 into Rab7-positive endosomes, thus creating mature MVBs (Rink et al., 2005; Woodman and Futter, 2008), tether membranes, guide SNARE assembly to drive membrane fusion (Richardson et al., 2004), and possibly act as ubiquitin ligases (Nickerson et al., 2009).
Since the dynamics of (de)ubiquitination plays a pivotal role in endosomal trafficking, ubiquitinating enzymes are expected to be part of the machinery. In particular, a role for c-Cbl, required both at the plasma membrane and in endosomes, has been extensively studied (Sorkin and Goh, 2009). Related to endosomal sorting, ESCRT0 contains K48 and K63-specific ubiquitin proteases, while components of HOPS, Vps-C and ESCRTs contain RING type E3 ligases. MVB sorting of most yeast membrane proteins depends on their ubiquitination by Rsp5p (Blondel et al., 2004; Dunn et al., 2004; Hettema et al., 2004; Katzmann et al., 2004; Morvan et al., 2004). In mammals, these roles are performed by its ortholog, the Nedd4 family, or SCF(β-transducing repeat-containing protein), and members of the MARCH family (Blot et al., 2004; Fukuda et al., 2006; Morokuma et al., 2007; Nakamura et al., 2005; Polo et al., 2002; van Kerkhof et al., 2007; van Kerkhof et al., 2011). In this study, we identified Triad1 as an E3 involved in endosomal trafficking. Depletion of Triad1 alters the fate of two different prototypic receptors, epidermal growth factor receptor (EGFR) and GHR. Both receptors accumulate at endosomes and remain in the recycling pathway. In addition, accumulation in lysosomes of the fluid phase marker, dextran, is diminished. In the absence of Triad1, the morphology of the sorting endosome and MVB changes, indicating a role in the sorting machinery of both membrane proteins and of fluid phase content.
Results
Triad1 affects post-endocytic trafficking of GHR
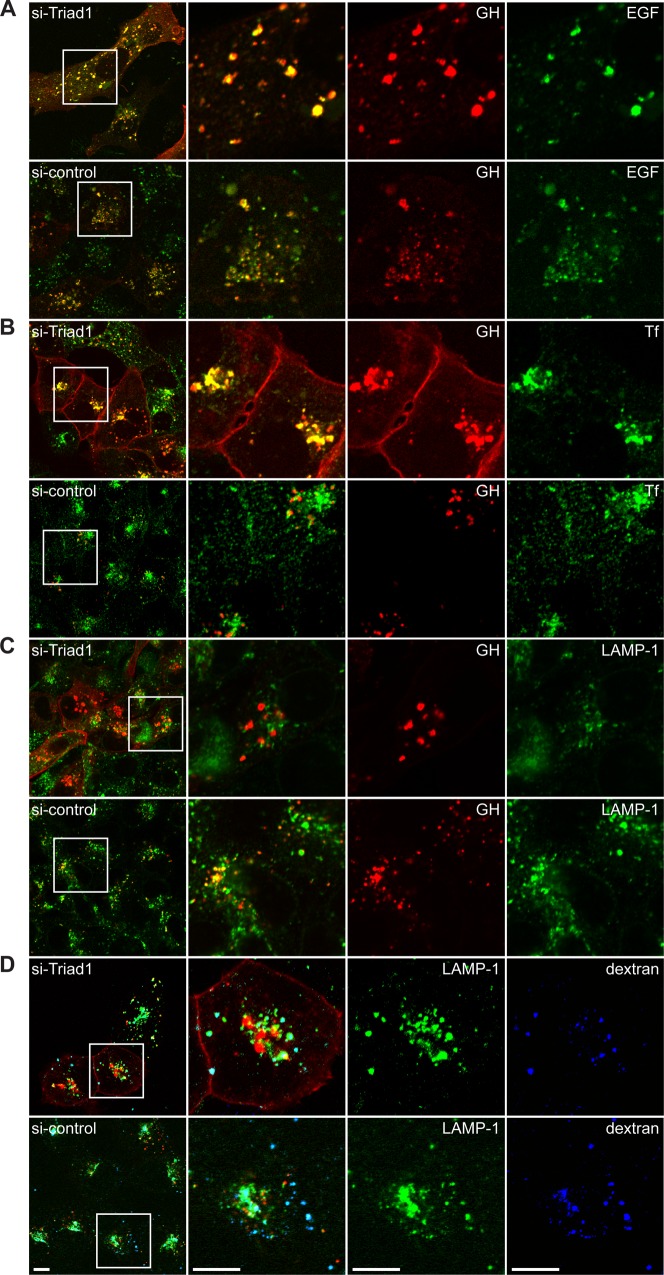
Triad1 is a ubiquitin ligase (E3) that binds both K48 and K63-specific ubiquitin E2 enzymes (Marteijn et al., 2009). Both types of chains are involved in receptor internalization. Therefore, we studied the role of Triad1 in protein sorting using different substrates to discriminate between endosomal sorting (EGF, GH), recycling between plasma membrane and endosomes (transferrin [Tf]) and fluid phase transport to lysosomes (dextran). U2OS cells, expressing both GHR and EGFR, were treated with a validated Triad1 siRNA for three days and analyzed for effects on uptake and distribution. A 30-min uptake of cy3-GH and Alexa 488-EGF in mock depleted (control) cells showed GH and EGF internalization mainly in endosomal vacuoles (Fig. 1A, lower panel). At this time point, lysosomal degradation of GH starts as was previously shown (van Kerkhof and Strous, 2001). In Triad1-depleted cells an increase in total label was observed and the fluorescence pattern changed from small dispersed punctae to larger and fewer spots (Fig. 1A, upper panel). Cy3-GH could clearly be identified at the plasma membrane (Fig. 1A–D, compare upper panels). Furthermore, large-sized vesicles, positive for both cy3-GH and Alexa 488-EGF, were observed. Closer examination of these vesicles revealed that they were clustered in groups, larger than in untreated cells (Fig. 1A, zoom). The overall subcellular localization of the vesicles did not seem to be altered compared to control cells. Two other validated siRNAs were equally effective in generating these phenotypes (data not shown). The data suggest that Triad1 depletion results in intracellular accumulation of both GH and EGF. The increase of cy3-GH at the plasma membrane indicates that Triad1 silencing might result in recycling.
Fig. 1. Effect of Triad1 depletion on EGF, GH, Tf and dextran.
(A) U2OS cells expressing GHR and EGFR were depleted for Triad1, incubated with cy3-GH (red) and Alexa-488 EGF (green) for 30 min, and the labels were visualized by confocal microscopy. (B) U2OS cells expressing GHR were depleted as in A and incubated with cy3-GH (red) for 30 min and chased for 30 min in presence of Alexa-488 Tf (green). (C) Cells as in B were incubated with cy3-GH (red) for 30 min, chased for 15 min and fixed. Permeabilized cells were incubated with anti-LAMP-1 antibodies and Alexa 488 (green), goat-anti-mouse antibodies. (D) Cells as in B were incubated with 647-dextran (blue) for 2 h, chased with cy3-GH (red) for 30 min, followed by a second chase without any fluorescent probe, stained with LAMP-1 antibodies (green) as in C. At the left side low resolution overviews are presented, at the right side phenotypic parts (boxed) of cells are enlarged to demonstrate the differences between the silencing effects of Triad1. Scale bars represent 10 µm. All data in this figure are representatives of three independent experiments.
Once established that Triad1-depletion had similar effects on both EGF and GH trafficking we used the GH system to investigate the nature of the vesicles in more detail. After a 30-min uptake of cy3-GH followed by 30 min of chase in the presence of Alexa 488-Tf, in control cells, Tf appeared in peri-nuclear, recycling endosomes, separated from cy3-GH (Fig. 1B). These endosomes were previously described as the recycling compartment that contains tubulovesicular structures and recycles EGF and Tf with slow kinetics (Sorkin et al., 1991; van Dam et al., 2002; van Kerkhof et al., 2011). In cells depleted for Triad1, the majority of cy3-GH co-localized with Tf in enlarged vesicular structures, possibly early endosomes. Normally, 80% of Tf receptors reside within early endosomes and recycle continuously between the plasma membrane and endosomes (Ciechanover et al., 1983). As we did neither observe a redistribution of Tf nor accumulation at the plasma membrane, we conclude that Triad1 does not affect the trafficking of Tf.
The accumulation of GHR in vesicles together with Tf in Triad1-depleted cells suggested that the receptors were not sorted to the lysosome anymore. To investigate this further, U2OS cells were incubated for 30 min with cy3-GH followed by a 15 min chase in absence of cy3-GH to allow GH to reach the late endosomes and lysosomes. After fixation, cells were permeabilized and stained with anti-LAMP-1 antibodies. In control cells, cy3-GH and LAMP-1 co-localized, indicating that GHR had reached the LAMP-1 positive late endosomes and lysosomes within 45 min (Fig. 1C, lower panel). In Triad1-depleted cells, cy3-GH and LAMP-1 did not co-localize, suggesting that GHR did not reach LAMP-1 positive compartments within this time frame. Therefore, we investigated whether a fluid phase endocytic marker was still able to reach LAMP-1-positive compartments. Cells were incubated with Alexa-647 dextran (10.000 Mr) for 2 h, followed by a 30-min uptake of cy3-GH, which was chased again for 15 min. As expected, in control cells, almost all dextran (∼90%) co-localized within LAMP-1-positive compartments, part of which could also be found in structures positive for cy3-GH (Fig. 1D, lower panel). In Triad1-depleted cells, dextran still co-localized with LAMP-1, but as observed in Fig. 1C, cy3-GH did not co-localize. Strikingly, the total amount of dextran present in Triad1-depleted cells was consistently lower. In supplementary material Fig. S3C, we repeated the experiment with a slightly different uptake protocol, directly comparing the uptake capacity via the GHR for 30 min with a 2 h accumulation of dextran. Together, the results show that fluid phase transport to lysosomes is affected by Triad1 depletion.
As both EGF and GH remain bound to their receptors until they arrive in the lysosomes, we conclude that both GH and EGF receptors take identical routes toward lysosomes (Fig. 1A). These data strongly suggest that in the absence of Triad1 both receptors accumulate in (recycling) endosomes together with the Tf receptor, and that fluid phase content, trafficking to LAMP-1-positive lysosomes, is impaired.
Triad1 acts at an endosomal compartment
Since uptake experiments with fluorescent ligands suggest that GHR transport to lysosomes is impaired, the effect of Triad1 depletion was evaluated biochemically. Quantitation of the western blot shown in Fig. 2A revealed a ∼4 times increase in the amount of GHR compared to cells treated with control siRNA. This suggests that the GHR degradation was strongly impeded. To investigate a possible effect on biosynthesis we performed pulse-chase labeling. Control and Triad1-depleted cells were pulse-labeled with [35S]methionine/cysteine and chased for 2 h (Fig. 2B). Quantification, corrected for differences in labeling efficiencies (see Materials and Methods), showed that in Triad1-depleted cells ∼20% less GHR was synthesized in 15 min compared to control cells. This indicates that GHR synthesis did not increase after depletion of Triad1. After a 2-h chase period ∼50% of GHR remained intact in control cells, whereas ∼65% remained in Triad1-depleted cells. Together, these data confirm that not synthesis of GHR is increased but its half-life is extended upon Triad1 depletion.
Fig. 2. Effect of Triad1 depletion on GHR synthesis, endocytosis, and degradation.
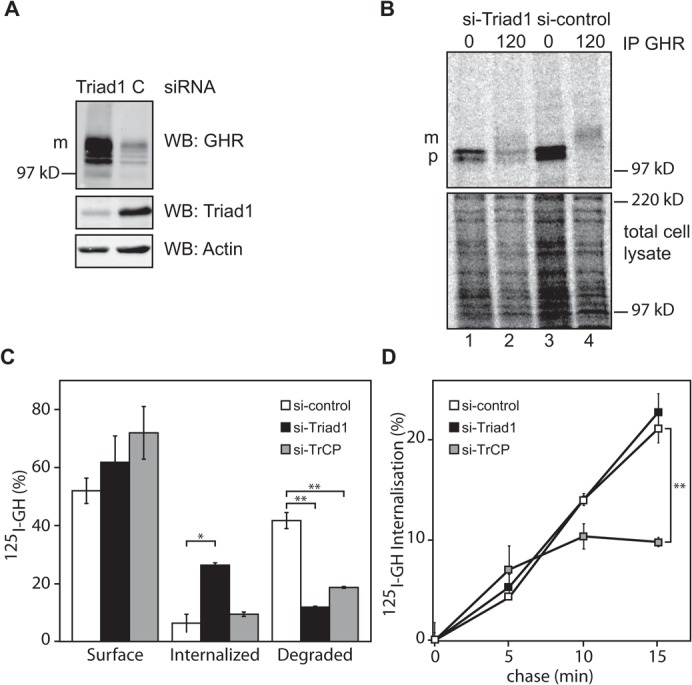
(A) Western blot of GHR, Triad1, and actin of U2OS lysates of cells treated with siRNA for Triad1 or control siRNA. In addition to the increased signal at the mature 130-kDa GHR band, the faster migrating band is due to intermediate degradation products. Both bands were taken to calculate the increase due to Triad1 depletion. m, mature GHR (130 kDa). Gene silencing of endogenous Triad1 is visualized in the middle panel, representative for the three siRNAs used in this study. (B) GHR-expressing U2OS cells depleted as in A were labeled with [35S]methionine for 15 min at 37°C and the radioactivity was chased for 2 h. GHR was immunoprecipitated with anti-GHR antibody (anti-T). p, precursor GHR (110 kDa); m, mature GHR (130 kDa); Triad1 (60 kDa). In the lower part total radioactivity was visualized. (C) GHR expressing U2OS cells, depleted as in A, were incubated with 180 ng/ml 125I-GH on ice for 2 h, and the cells were chased for 0 and 45 min. Surface-bound, internalized, and TCA-soluble label was expressed as percentage of total radioactivity. (D) Experiment as in C using a shorter time frame to measure only uptake kinetics. Intracellular and plasma membrane radioactivity were quantified. Error bars indicate ± 1 S.E.M. All data in this figure are representative of three independent experiments. The silencing efficiency as controlled by western blot was less than 30%. * (p<0.05) and ** (p<0.01) indicate significant differences.
To investigate this further, we performed uptake experiments using 125I-GH. Control, Triad1-, and TrCP-depleted U2OS cells were incubated for 2 h with 125I-GH on ice and subsequently chased at 37°C for 45 min. As expected, typically, the amount of initially bound GH was about 4.5 times higher in Triad1-depleted cells as compared to control cells (data not shown). After a 45-min chase, a slight increase of surface-bound GH was found in both Triad1 (p≤0.21) and TrCP (p≤0.093) depleted cells as compared to control depleted cells (Fig. 2C, left panel). Quantification of intracellular 125I-GH confirmed that Triad1-depleted cells accumulated significantly more (26%) GH compared to control (6%, p = 0.05), and TrCP-depleted cells (9%, p = 0.06), respectively (Fig. 2C, middle panel) (Fig. 1; supplementary material Fig. S3). In control cells, about 42% of the originally bound 125I-GH was degraded and secreted as free iodide within 2 h (Fig. 2C, right panel). In Triad1-depleted cells significantly less GH (12%, p = 0.004) was degraded, indicating that degradation was strongly inhibited. As shown before, depletion of TrCP, the ubiquitin ligase required for GHR endocytosis, reduced the degradation to 19% (p = 0.007) (van Kerkhof et al., 2011).
The relative large amount of intracellular GH in Triad1-depleted cells, compared to TrCP-depleted cells, combined with an unaffected synthesis rate of GHR, suggested that GHR was forced to recycle between plasma membrane and endosomes. This hypothesis requires that Triad1 is not involved in GHR endocytosis. To test this we performed a kinetic experiment in which 125I-GH was chased for short time periods (up to 15 min), a time frame in which only GH uptake occurs with little to no recycling (van Kerkhof et al., 2000). About 50% of the cells treated with siRNAs against TrCP or Triad1 showed the described phenotypes (supplementary material Fig. S3D). Under these conditions, GH uptake rates of Triad1-depleted and control cells were similar (p = 0.28). Silencing of TrCP resulted in a decreasing uptake rate within this short time frame, yielding significantly less intracellular GH (10%, p = 0.009) compared to control cells (Fig. 2D). Combined with the observed intracellular accumulation, these data strongly suggest that the increase in GHR at the plasma membrane is due to increased recycling rather than decreased endocytosis. We conclude that, following Triad1 silencing, endocytosed GHR is diverted from the route to lysosomes into the recycling pathway. This is supported by the co-localization of GHR with Tf, which implies that both GHR and EGFR accumulate in a recycling compartment.
Triad1 depletion accumulates GHR in endosomal vacuoles
Next, we analyzed cells showing the distinctive knockdown phenotype by correlative light-electron microscopy (van Rijnsoever et al., 2008). Control or Triad1-depleted U2OS cells were incubated for 30 min with biotinylated GH and processed for correlative microscopy. By light microscopy we selected cells showing a clustered localization of internalized GH-biotin in Triad1-depleted cells (Fig. 3A,D) and by correlative microscopy (supplementary material Fig. S2; Fig. 3B,C,E) we identified these spots as endosomes with many ILVs and prominent flat bi-layered clathrin coats, characteristic for early endosomes. The endosomes contained high levels of GH-biotin labeling that was associated with both the endosomal limiting membrane and the ILVs. In some cells, multiple endosomes were clustered (Fig. 3F). However, the endosomes appeared not to be tethered together like has been observed in USP8 depleted cells (Row et al., 2006). The endosomal vacuoles were surrounded by small vesicular and tubular membrane profiles also positive for GH-biotin (Fig. 3F). This localization differed from control cells, in which endosomes contained less GH-biotin label and were found, dispersed in the cytoplasm (supplementary material Fig. S2). Furthermore, in Triad1-depleted cells, the endosomes contained a more extended coat compared to endosomes from control cells. Together, these data show that depletion of Triad1 causes accumulation of endosomal vacuoles with surrounding vesicles and increases the number of ILVs.
Fig. 3. Ultrastructural characterization of the Triad1-depleted phenotype.
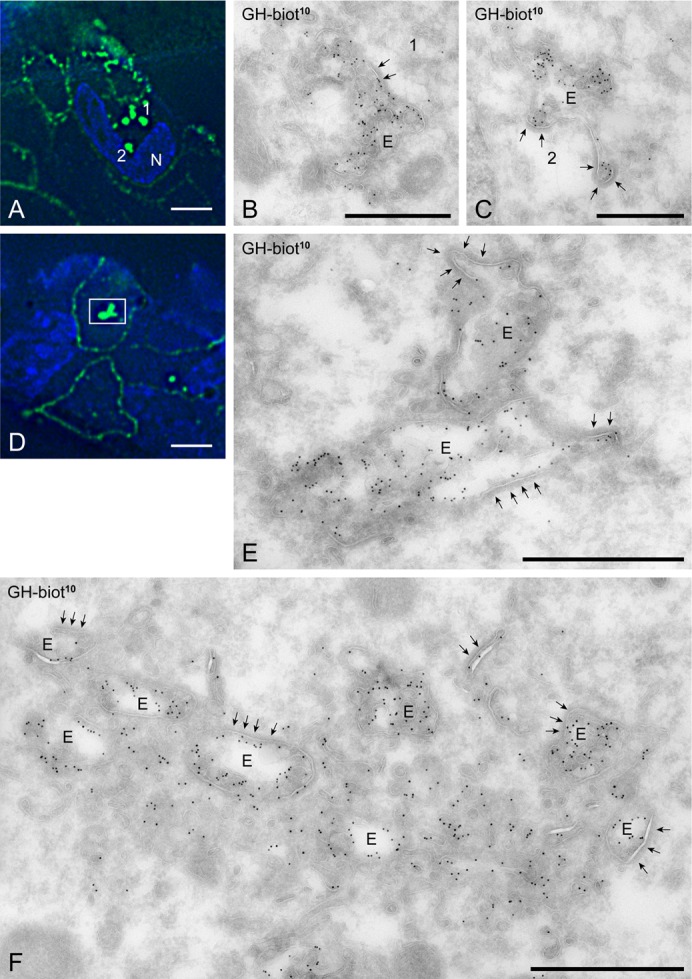
(A,D) U2OS cells were silenced for Triad1 for 3 days, incubated for 30 min with GH-biotin, fixed and prepared for ultrathin cryosectioning and labeling with anti-biotin. The anti-biotin antibody was subsequently marked with both Alexa488 and PAG10 to allow correlative microscopy. The large fluorescent spots identified in A (1 and 2) and D (square) are typically induced by Triad knockdown. Bars: 10 μm. (B,C) Applying correlative immune electron microscopy (for intermediate step in the procedure, see supplementary material Fig. S1), the fluorescent spots 1 and 2 shown in A were identified as endosomal vacuoles (E) with many ILVs, prominent bi-layered clathrin coats (arrows) and heavy labeling for GH-biotin (PAG10). (E) The bright fluorescent spot in D was found to consist of two closely apposed endosomes. (F) Example of a large cluster of endosomes induced by Triad knockdown. The endosomes are surrounded by many vesicular and tubular membrane profiles. Bars: B, 600; C, 500; E, 800; F, 800 nm.
Discussion
In this study we added yet another ubiquitin ligase to the growing list of ubiquitination enzymes involved in membrane traffic. Our confocal studies show that in Triad1-depleted cells GHR and EGFR accumulate in intracellular vesicles and at the plasma membrane. The intracellular vesicles are most probably early endosomes since Tf co-localized with GH in these vesicles. The confocal studies were confirmed by biochemical experiments that showed that lysosomal degradation of 125I-GH was indeed inhibited in Triad1 depleted cells. Electron microscopy studies of the endosomal sorting system showed that Triad1 depletion results in enlarged sorting endosomes with elongated protein coats. While the integral membrane proteins (the two receptors including their cargo) accumulated in MVBs as well as in the recycling pathway, limited amounts of dextran could still reach the LAMP-1 positive compartments. Previously, we have shown that proteasomal inhibitors block the transport of GH at the level of MVBs, while dissociating ligands as well as the fluid phase marker, colloidal-gold-labeled albumin, had free access to lysosomes (van Kerkhof et al., 2001). Formation of MVBs is still possible after depletion of ESCRT proteins (Stuffers et al., 2009b). Also under these conditions, EGFR could not progress to the lysosome, and the authors suggested a role for LBPA in the formation of the ESCRT-independent ILVs (Matsuo et al., 2004; Raiborg et al., 2008; Stuffers et al., 2009b). Assuming that growth factor and cytokine receptors use MVBs to travel to the lysosomes and take fluid phase content along, Triad1 is most likely a controlling factor in membrane trafficking between the recycling endosome and the lysosome. In its absence selection of membrane proteins at the MVB is inhibited as well as the throughput of endocytosed soluble content.
There are several reasons to assume that Triad1 works on the sorting machinery. First, Triad1 affects the sorting of both EGFR and GHR. The selection mechanisms of these receptors differ, both at the plasma membrane and in endosomes (TrCP for GHR via the ubiquitin-dependent endocytosis motif and c-Cbl for EGFR via phosphorylated Y1045 residue) (Govers et al., 1999; Levkowitz et al., 1999). At the level of MVBs Hrs seems to be involved in EGFR sorting, while Tsg101 is required for directing the GHR towards MVBs (van Kerkhof et al., 2011). Second, Triad1 depletion results in altered morphology of the endosomal system. Triad1 seems to affect either the selection of integral membrane proteins into MVBs (forcing them into a recycling mode back to the plasma membrane) or the disassembly of the sorting machinery, causing blockage of both membrane and content cargo to exit the MVB stage or fusion with the next category of lysosome-bound vesicles. The latter possibility is most relevant, as it would explain the electron microscopic observations: onset and fission of ILVs is unaffected and the blockage would induce accumulation of traffic to and from MVBs (recycling), ultimately resulting in GH stocked ILVs, leaving the majority of GH in the recycling pathway. The effect of Triad1 differs from the phenotypic effects of overexpressing the ATPase-deficient mutant hVPS4EQ (Sachse et al., 2004) under the latter conditions being less ILVs present. As VPS4 catalyzes ESCRT-III disassembly (Babst et al., 1998) and ILVs are still being formed including their cargo, it would imply that Triad1 acts in a later stage. An explanation for the increased size and irregular shape of both MVBs and ILVs would be the accumulation of unprocessed cargo that causes a shortage of both membrane and (ESCRT) factors.
The question how Triad1 is involved in endosomal transport remains open. Until now, endogenous substrates that are modified with ubiquitin by Triad1 are unknown (Marteijn et al., 2009). Given its localization in both cytosol and nucleus with no enrichment at the endosomal system, it is likely that this E3 ligase acts in multiple mechanisms (supplementary material Fig. S3A). Triad1 binds the transcription factors Gfi1 and 1B (Marteijn et al., 2007). Counter intuitively, forced Triad1 expression inhibits the ubiquitination and proteasomal degradation of Gfi1, whereas Triad1 knockdown results in increased ubiquitination. The inhibition of ubiquitination occurs also by an UbcH7 binding-defective Triad1 mutant as well as in the presence of proteasome inhibitors. This suggests that the inhibition of ubiquitination is not caused by the ubiquitination and proteasomal degradation of E3 ligases that mark Gfi1 with ubiquitin for proteasomal degradation. Thus, Triad1 may rather recruit a DUB to Gfi1 that may de-ubiquitinate the latter. The decrease in ubiquitination of binding partners is similar as reported for the TRIAD ubiquitin ligase Hoil-1 that inhibits ubiquitination of the transcription factor Dax-1 (Ehrlund et al., 2009) and the suppressor of cytokine signaling SOCS6 (Bayle et al., 2006). In light of these observations, Triad1 might control traffic through the endosomal compartment by ubiquitinating proteins implicated in MVB formation and fusion with lysosomes. Thus, a functional link between Triad1 and for instance the HOPS complex that is involved in lysosomal fusion events would be an obvious possibility, where Triad1 might cooperate with other RING proteins present in the HOPS complex such as Vps18 (Seals et al., 2000). In such scenario, less Triad1 activity would translate in increased signaling activity of growth factor receptors. This would be in line with a controlling function of the endocytic machinery in balancing the degradation of a variety of cytokine growth factor and tyrosine kinase receptors involved in migration, growth, survival, oncogenicity and immunity by endosome-ancillary factors like Cbl, Nedd4, Tsg101, and Triad1 (reviewed by Mosesson et al., 2008). Whether overexpression of Triad1, as shown previously to inhibit colony formation by myeloid progenitor cells, is directly regulated via Gfi1 ubiquitination or indirectly via cytokine receptor degradation remains an intriguing question.
Materials and Methods
Materials, antibodies and DNA constructs
The rabbit wild type and mutant GHR cDNAs in pcDNA3 have been described before (Govers et al., 1999). A plasmid expressing EGFR was obtained from Dr. Paul van Bergen en Henegouwen, Utrecht University. Protein A-beads were from Repligen. Antibody anti-GHR (T) was raised in rabbits against the cytoplasmic sequence between amino acids 271-381, as previously described (Strous et al., 1996). An antibody against Triad1 was generated by immunizing rabbits with a C-terminal peptide of the human Triad1 protein (NP_006312, amino acids 459-472, WKVERADSYDRGDL), coupled to the carrier protein LPH (C-WKV). After six immunization rounds with this peptide, antisera were collected and tested for specificity on cell lysates of Triad1 overexpressing and depleted cells. Antibody against the extracellular domain of the GHR (Mab5) was from Santa Cruz Biotechnology, Inc., goat anti-mouse IgG Alexa488 from Molecular Probes, and goat anti- rabbit IgG IRDye800 from Rockland Immunochemicals Inc, Gilbertsville (PA). Alexa488-Tf was from Molecular Probes, EGF-Alexa Fluor 488 streptavidin from Invitrogen, mouse monoclonal anti-FLAG (M2) from Sigma, and mouse monoclonal anti LAMP-1 antibody (CD107A) from BD Biopharming. Immobilized streptavidin was from Pierce, glutathione-sepharose from Amersham Biosciences, and Ni-NTA agarose from Qiagen. Culture media, fetal calf serum (FCS), L-glutamine, and antibiotics for tissue culture were purchased from Invitrogen.
Cell culture
Human osteosarcoma (U2OS) cells expressing the wild type and mutant GHR or EGFR were grown in DMEM (Invitrogen) with 10% FCS, 100 units/ml penicillin, 0.1 mg/ml streptomycin, supplemented with 150 µg/ml hygromycine (GHR) and/or 125 µg/ml zeocin (EGFR). Cells were cultured at 37°C with 5% CO2.
Transfections
To silence the expression of Triad1 cells were transfected with 50 nM of 3 validated small interfering RNAs directed against Triad1 (ARIH2 Ambion 135640, GCCTAATCCATCAAAACAT); UUGUGAGGAAGAGGAAGAA (Marteijn et al., 2007), and UUCUUCCUCUUCCUCACAA (Applied Biosystems Europe BV) using Dharmafect (Thermo Fisher Scientific). As all three RNAs showed identical phenotypes, the first was used in all studies. Three days after transfection, cells were used for experiments. Control cells were transfected with siRNA Control#1 (Ambion, AM4636). To control the efficiency of the gene silencing, antibodies against the Triad1 protein were used. Western blots were analyzed using an Odyssey infrared imaging system (Li-Cor Biosciences, Lincoln, Nebraska).
Pulse-chase analysis, immunoprecipitation, and SDS-PAGE
For metabolic labeling experiments, cells were grown in RPMI 1640 medium (BioWhittaker) without methionine and cysteine at 37°C for 2 h. The cells were labeled with 250 µCi of 35S-labeled Redivue Promix (a mixture of L-[35S]methionine and L-[35S]cysteine) (Amersham Biosciences) per 107 cells in RPMI 1640 medium. To chase, radioactive medium was replaced with medium supplemented with 1 mM methionine/cysteine (Sigma). The cells were lysed in 1% Triton X100, 1 mM EDTA in PBS in presence of protease inhibitors as described (van Kerkhof et al., 2000). After centrifugation immunoprecipitation were performed in lysates diluted 1:1 with Immunomix (1% Triton X100, 1% SDS, 0.5% DOC, 1% BSA, 1 mM EDTA in PBS). Immunoprecipitation was performed at 4°C for 2 h using specific antiserum and protein A-Sepharose beads (Amersham Biosciences). The beads were washed 3 times with immunomix and 3 times with 10-fold diluted PBS. The washed beads were boiled in Laemmli sample buffer for 5–10 min. A fraction of labeled total cell lysates was loaded on gel to correct for differences in cell number and labeling efficiency. Samples were loaded onto SDS-polyacrylamide gels and exposed to a storage phosphor imaging screen, which was scanned in a Personal Molecular Imager FX and analyzed with Quantity One software (Bio-Rad, Veenendaal, The Netherlands). Quantified data were corrected by dividing the specific signal by the signal obtained from total cell lysates.
Ligand binding, internalization, and degradation
125I-human GH was prepared with the use of chloramine T (Strous et al., 1996) and purified over a 500 µl Zeba spin column (Pierce, Thermoscientific). For internalization studies, cells were grown in 12-well plates, treated with siRNAs as described above, washed with MEM supplemented with 20 mM, HEPES pH 7.4 and 0.1% BSA, and incubated in a water bath. 125I-GH (180 ng/ml) was bound on ice for 2 h, the cells were washed free of unbound ligand, and incubated for 0–45 min at 37°C. Membrane-associated ligand was removed by acid wash (0.15 M NaCl, 50 mM glycine, 0.1% BSA, pH 2.5) on ice. Internalized ligand was determined by measuring the radioactivity after solubilization of the acid-treated cells in 1 N NaOH in a LKB gamma counter. For degradation studies, at the indicated times, the medium was collected and precipitated with 1 volume of ice-cold 20% TCA for 10 min on ice. Acid-soluble radioactivity was determined in the supernatant after centrifugation and was used as a measurement for degraded ligand. Membrane-associated and internalized ligands were determined as described above. Background was determined by subtracting initial intracellular and TCA precipitated medium from all other time points.
Immune fluorescence microscopy
Cy3-GH was prepared using a Fluorolink-Cy3 label kit according to the supplier's instructions (Amersham Biosciences). Cells, grown on coverslips, were incubated with Cy3-GH (1 µg/ml), Alexa488-labeled Tf (25 µg/ml), or EGF Alexafluor 488 streptavidin (100 ng/ml). Cells were washed with PBS to remove unbound label and fixed for 2 h in 3% paraformaldehyde in phosphate buffer, pH 7.4. After fixation, the cells were permeabilized in 0.2% Triton for 5 min and the coverslips were incubated with anti-Lamp1 and fluorescently labeled secondary antibodies. The coverslips were embedded in Mowiol, and visualization was performed using a Zeiss Ultraview system.
Correlative light electron microscopy (CLEM)
U2OS cells, expressing both GHR and EGFR were incubated with biotinylated GH for 30 min. Cells were fixed by adding freshly prepared 4% formaldehyde in 0.1 M phosphate buffer pH 7.4 to an equal volume of culture medium for 10 min, followed by post-fixation in 4% formaldehyde in 0.1 M phosphate buffer pH 7.4. Cells were stored until further processing in 1% formaldehyde at 4°C. Processing of cells for ultrathin cryosectioning was done as described (Slot and Geuze, 2007). In brief, fixed cells were washed with 0.05 M glycine in PBS, scraped gently from the dish in PBS containing 1% gelatin, resuspended and pelleted in 12% gelatin in PBS, 37°C. The cell pellet was solidified on ice and cut into small blocks. For cryoprotection, blocks were infiltrated overnight with 2.3 M sucrose at 4°C and afterwards mounted on aluminum pins and frozen in liquid nitrogen. To pick up ultrathin (65 nm) cryosections a 1:1 mixture of 2.3 M sucrose and 1.8% methylcellulose was used. Sections were placed on 75 mesh hexagonal cupper findergrids (Agar scientific), which contained a formvar film and were carbon coated.
Correlative microscopy of fluorescence- and immunogold-labeled ultrathin cryosections was done essentially as described (Vicidomini et al., 2008) with a few adaptations. Briefly, grids were incubated on 2% gelatin in PBS for 20 min at 37°C, then incubated with rabbit anti-Biotin (Rockland) in 1% BSA in PBS for 45 min, rinsed in 0.1% BSA in PBS, and incubated with goat anti-rabbit Alexa 488-conjugated antibodies (Molecular Probes) for 45 min. Grids were rinsed followed by incubation with protein A conjugated to 10 nanometer gold particles (PAG10; Department of Cell Biology, UMC, Utrecht) for 20 min, fixed for 5 min with 1% glutaraldehyde in PBS, rinsed in PBS and incubated in Hoechst 1 µg/ml in PBS. For fluorescence, grids were mounted between a microscope slide and coverslip with 50% glycerol and imaged in a Deltavision RT microscope (Applied Precision, Issaquah WA) using an EMCCD camera. For subsequent transmission electron microscopy, grids were unmounted, washed in water, incubated for 5 min in 2% uranyl acetate and 0.15 M oxalic acid and subsequently for 5 min in 0.4% uranyl acetate and 1.8% 25ctp methylcellulose, and viewed in a JEOL 1010 or in a Tecnai 12 (FEI) electron microscope.
Supplementary Material
Acknowledgments
This research was supported by the European Network of Excellence, Rubicon “Role of ubiquitin and ubiquitin-like modifiers in cellular regulation” (Grant LSHG-CT-2005-018683), the Marie Curie network, “UbiRegulators”, (Grant MRTN-CT-2006-034555), and The Netherlands Proteomics Centre, “Ubiquitin membrane trafficking”, proteomic analysis of ubiquitination in membrane trafficking, NPC3.1 to JS. We thank all other members of the GHR group for suggestions and helpful discussions.
Footnotes
Competing interests: The authors declare that there are no competing interests.
References
- Babst M., Wendland B., Estepa E. J., Emr S. D. (1998). The Vps4p AAA ATPase regulates membrane association of a Vps protein complex required for normal endosome function. EMBO J. 17, 2982–2993 10.1093/emboj/17.11.2982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayle J., Lopez S., Iwaï K., Dubreuil P., De Sepulveda P. (2006). The E3 ubiquitin ligase HOIL-1 induces the polyubiquitination and degradation of SOCS6 associated proteins. FEBS Lett. 580, 2609–2614 10.1016/j.febslet.2006.03.093 [DOI] [PubMed] [Google Scholar]
- Blondel M.-O., Morvan J., Dupré S., Urban-Grimal D., Haguenauer-Tsapis R., Volland C. (2004). Direct sorting of the yeast uracil permease to the endosomal system is controlled by uracil binding and Rsp5p-dependent ubiquitylation. Mol. Biol. Cell 15, 883–895 10.1091/mbc.E03-04-0202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blot V., Perugi F., Gay B., Prévost M. C., Briant L., Tangy F., Abriel H., Staub O., Dokhélar M. C., Pique C. (2004). Nedd4.1-mediated ubiquitination and subsequent recruitment of Tsg101 ensure HTLV-1 Gag trafficking towards the multivesicular body pathway prior to virus budding. J. Cell Sci. 117, 2357–2367 10.1242/jcs.01095 [DOI] [PubMed] [Google Scholar]
- Chin L. S., Olzmann J. A., Li L. (2010). Parkin-mediated ubiquitin signalling in aggresome formation and autophagy. Biochem. Soc. Trans. 38, 144–149 10.1042/BST0380144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciechanover A., Schwartz A. L., Dautry-Varsat A., Lodish H. F. (1983). Kinetics of internalization and recycling of transferrin and the transferrin receptor in a human hepatoma cell line. Effect of lysosomotropic agents. J. Biol. Chem. 258, 9681–9689. [PubMed] [Google Scholar]
- Clague M. J., Urbé S. (2010). Ubiquitin: same molecule, different degradation pathways. Cell 143, 682–685 10.1016/j.cell.2010.11.012 [DOI] [PubMed] [Google Scholar]
- Dunn R., Klos D. A., Adler A. S., Hicke L. (2004). The C2 domain of the Rsp5 ubiquitin ligase binds membrane phosphoinositides and directs ubiquitination of endosomal cargo. J. Cell Biol. 165, 135–144 10.1083/jcb.200309026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrlund A., Anthonisen E. H., Gustafsson N., Venteclef N., Robertson Remen K., Damdimopoulos A. E., Galeeva A., Pelto-Huikko M., Lalli E., Steffensen K. R. et al. (2009). E3 ubiquitin ligase RNF31 cooperates with DAX-1 in transcriptional repression of steroidogenesis. Mol. Cell. Biol. 29, 2230–2242 10.1128/MCB.00743-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuda H., Nakamura N., Hirose S. (2006). MARCH-III Is a novel component of endosomes with properties similar to those of MARCH-II. J. Biochem. 139, 137–145 10.1093/jb/mvj012 [DOI] [PubMed] [Google Scholar]
- Govers R., ten Broeke T., van Kerkhof P., Schwartz A. L., Strous G. J. (1999). Identification of a novel ubiquitin conjugation motif, required for ligand-induced internalization of the growth hormone receptor. EMBO J. 18, 28–36 10.1093/emboj/18.1.28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hettema E. H., Valdez-Taubas J., Pelham H. R. B. (2004). Bsd2 binds the ubiquitin ligase Rsp5 and mediates the ubiquitination of transmembrane proteins. EMBO J. 23, 1279–1288 10.1038/sj.emboj.7600137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y., Niwa J.-i., Sobue G., Breitwieser G. E. (2006). Calcium-sensing receptor ubiquitination and degradation mediated by the E3 ubiquitin ligase dorfin. J. Biol. Chem. 281, 11610–11617 10.1074/jbc.M513552200 [DOI] [PubMed] [Google Scholar]
- Ishigaki S., Niwa J., Yamada S., Takahashi M., Ito T., Sone J., Doyu M., Urano F., Sobue G. (2007). Dorfin-CHIP chimeric proteins potently ubiquitylate and degrade familial ALS-related mutant SOD1 proteins and reduce their cellular toxicity. Neurobiol. Dis. 25, 331–341 10.1016/j.nbd.2006.09.017 [DOI] [PubMed] [Google Scholar]
- Katzmann D. J., Sarkar S., Chu T., Audhya A., Emr S. D. (2004). Multivesicular body sorting: ubiquitin ligase Rsp5 is required for the modification and sorting of carboxypeptidase S. Mol. Biol. Cell 15, 468–480 10.1091/mbc.E03-07-0473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer L. B., Shim J., Previtera M. L., Isack N. R., Lee M. C., Firestein B. L., Rongo C. (2010). UEV-1 is an ubiquitin-conjugating enzyme variant that regulates glutamate receptor trafficking in C. elegans neurons. PLoS ONE 5, e14291 10.1371/journal.pone.0014291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauwers E., Jacob C., André B. (2009). K63-linked ubiquitin chains as a specific signal for protein sorting into the multivesicular body pathway. J. Cell Biol. 185, 493–502 10.1083/jcb.200810114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levkowitz G., Waterman H., Ettenberg S. A., Katz M., Tsygankov A. Y., Alroy I., Lavi S., Iwai K., Reiss Y., Ciechanover A. et al. (1999). Ubiquitin ligase activity and tyrosine phosphorylation underlie suppression of growth factor signaling by c-Cbl/Sli-1. Mol. Cell 4, 1029–1040 10.1016/S1097-2765(00)80231-2 [DOI] [PubMed] [Google Scholar]
- Markgraf D. F., Peplowska K., Ungermann C. (2007). Rab cascades and tethering factors in the endomembrane system. FEBS Lett. 581, 2125–2130 10.1016/j.febslet.2007.01.090 [DOI] [PubMed] [Google Scholar]
- Marteijn J. A., van der Meer L. T., van Emst L., van Reijmersdal S., Wissink W., de Witte T., Jansen J. H., Van der Reijden B. A. (2007). Gfi1 ubiquitination and proteasomal degradation is inhibited by the ubiquitin ligase Triad1. Blood 110, 3128–3135 10.1182/blood-2006-11-058602 [DOI] [PubMed] [Google Scholar]
- Marteijn J. A., van der Meer L. T., Smit J. J., Noordermeer S. M., Wissink W., Jansen P., Swarts H. G., Hibbert R. G., de Witte T., Sixma T. K. et al. (2009). The ubiquitin ligase Triad1 inhibits myelopoiesis through UbcH7 and Ubc13 interacting domains. Leukemia 23, 1480–1489 10.1038/leu.2009.57 [DOI] [PubMed] [Google Scholar]
- Matsuo H., Chevallier J., Mayran N., Le Blanc I., Ferguson C., Fauré J., Blanc N. S., Matile S., Dubochet J., Sadoul R. et al. (2004). Role of LBPA and Alix in multivesicular liposome formation and endosome organization. Science 303, 531–534 10.1126/science.1092425 [DOI] [PubMed] [Google Scholar]
- Morokuma Y., Nakamura N., Kato A., Notoya M., Yamamoto Y., Sakai Y., Fukuda H., Yamashina S., Hirata Y., Hirose S. (2007). MARCH-XI, a novel transmembrane ubiquitin ligase implicated in ubiquitin-dependent protein sorting in developing spermatids. J. Biol. Chem. 282, 24806–24815 10.1074/jbc.M700414200 [DOI] [PubMed] [Google Scholar]
- Morvan J., Froissard M., Haguenauer-Tsapis R., Urban-Grimal D. (2004). The ubiquitin ligase Rsp5p is required for modification and sorting of membrane proteins into multivesicular bodies. Traffic 5, 383–392 10.1111/j.1398-9219.2004.00183.x [DOI] [PubMed] [Google Scholar]
- Mosesson Y., Mills G. B., Yarden Y. (2008). Derailed endocytosis: an emerging feature of cancer. Nat. Rev. Cancer 8, 835–850 10.1038/nrc2521 [DOI] [PubMed] [Google Scholar]
- Nakamura N., Fukuda H., Kato A., Hirose S. (2005). MARCH-II is a syntaxin-6-binding protein involved in endosomal trafficking. Mol. Biol. Cell 16, 1696–1710 10.1091/mbc.E04-03-0216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narendra D., Tanaka A., Suen D. F., Youle R. J. (2008). Parkin is recruited selectively to impaired mitochondria and promotes their autophagy. J. Cell Biol. 183, 795–803 10.1083/jcb.200809125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nickerson D. P., Brett C. L., Merz A. J. (2009). Vps-C complexes: gatekeepers of endolysosomal traffic. Curr. Opin. Cell Biol. 21, 543–551 10.1016/j.ceb.2009.05.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paiva S., Vieira N., Nondier I., Haguenauer-Tsapis R., Casal M., Urban-Grimal D. (2009). Glucose-induced ubiquitylation and endocytosis of the yeast Jen1 transporter: role of lysine 63-linked ubiquitin chains. J. Biol. Chem. 284, 19228–19236 10.1074/jbc.M109.008318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polo S., Sigismund S., Faretta M., Guidi M., Capua M. R., Bossi G., Chen H., De Camilli P., Di Fiore P. P. (2002). A single motif responsible for ubiquitin recognition and monoubiquitination in endocytic proteins. Nature 416, 451–455 10.1038/416451a [DOI] [PubMed] [Google Scholar]
- Raiborg C., Malerød L., Pedersen N. M., Stenmark H. (2008). Differential functions of Hrs and ESCRT proteins in endocytic membrane trafficking. Exp. Cell Res. 314, 801–813 10.1016/j.yexcr.2007.10.014 [DOI] [PubMed] [Google Scholar]
- Richardson S. C., Winistorfer S. C., Poupon V., Luzio J. P., Piper R. C. (2004). Mammalian late vacuole protein sorting orthologues participate in early endosomal fusion and interact with the cytoskeleton. Mol. Biol. Cell 15, 1197–1210 10.1091/mbc.E03-06-0358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rink J., Ghigo E., Kalaidzidis Y., Zerial M. (2005). Rab conversion as a mechanism of progression from early to late endosomes. Cell 122, 735–749 10.1016/j.cell.2005.06.043 [DOI] [PubMed] [Google Scholar]
- Row P. E., Prior I. A., McCullough J., Clague M. J., Urbé S. (2006). The ubiquitin isopeptidase UBPY regulates endosomal ubiquitin dynamics and is essential for receptor down-regulation. J. Biol. Chem. 281, 12618–12624 10.1074/jbc.M512615200 [DOI] [PubMed] [Google Scholar]
- Sachse M., Strous G. J., Klumperman J. (2004). ATPase-deficient hVPS4 impairs formation of internal endosomal vesicles and stabilizes bilayered clathrin coats on endosomal vacuoles. J. Cell Sci. 117, 1699–1708 10.1242/jcs.00998 [DOI] [PubMed] [Google Scholar]
- Seals D. F., Eitzen G., Margolis N., Wickner W. T., Price A. (2000). A Ypt/Rab effector complex containing the Sec1 homolog Vps33p is required for homotypic vacuole fusion. Proc. Natl. Acad. Sci. USA 97, 9402–9407 10.1073/pnas.97.17.9402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slot J. W., Geuze H. J. (2007). Cryosectioning and immunolabeling. Nat. Protoc. 2, 2480–2491 10.1038/nprot.2007.365 [DOI] [PubMed] [Google Scholar]
- Sorkin A., Goh L. K. (2009). Endocytosis and intracellular trafficking of ErbBs. Exp. Cell Res. 315, 683–696 10.1016/j.yexcr.2008.07.029 [DOI] [PubMed] [Google Scholar]
- Sorkin A., Krolenko S., Kudrjavtceva N., Lazebnik J., Teslenko L., Soderquist A. M., Nikolsky N. (1991). Recycling of epidermal growth factor-receptor complexes in A431 cells: identification of dual pathways. J. Cell Biol. 112, 55–63 10.1083/jcb.112.1.55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stenmark H. (2009). Seeing is believing. Nat. Rev. Mol. Cell Biol. 10, 582 10.1038/nrm2747 [DOI] [PubMed] [Google Scholar]
- Strous G. J., van Kerkhof P., Govers R., Ciechanover A., Schwartz A. L. (1996). The ubiquitin conjugation system is required for ligand-induced endocytosis and degradation of the growth hormone receptor. EMBO J. 15, 3806–3812. [PMC free article] [PubMed] [Google Scholar]
- Stuffers S., Brech A., Stenmark H. (2009a). ESCRT proteins in physiology and disease. Exp. Cell Res. 315, 1619–1626 10.1016/j.yexcr.2008.10.013 [DOI] [PubMed] [Google Scholar]
- Stuffers S., Sem Wegner C., Stenmark H., Brech A. (2009b). Multivesicular endosome biogenesis in the absence of ESCRTs. Traffic 10, 925–937 10.1111/j.1600-0854.2009.00920.x [DOI] [PubMed] [Google Scholar]
- Tokunaga F., Sakata S., Saeki Y., Satomi Y., Kirisako T., Kamei K., Nakagawa T., Kato M., Murata S., Yamaoka S. et al. (2009). Involvement of linear polyubiquitylation of NEMO in NF-kappaB activation. Nat. Cell Biol. 11, 123–132 10.1038/ncb1821 [DOI] [PubMed] [Google Scholar]
- van Dam E. M., Ten Broeke T., Jansen K., Spijkers P., Stoorvogel W. (2002). Endocytosed transferrin receptors recycle via distinct dynamin and phosphatidylinositol 3-kinase-dependent pathways. J. Biol. Chem. 277, 48876–48883 10.1074/jbc.M206271200 [DOI] [PubMed] [Google Scholar]
- van Kerkhof P., Strous G. J. (2001). The ubiquitin-proteasome pathway regulates lysosomal degradation of the growth hormone receptor and its ligand. Biochem. Soc. Trans. 29, 488–493 10.1042/BST0290488 [DOI] [PubMed] [Google Scholar]
- van Kerkhof P., Govers R., Alves dos Santos C. M., Strous G. J. (2000). Endocytosis and degradation of the growth hormone receptor are proteasome-dependent. J. Biol. Chem. 275, 1575–1580 10.1074/jbc.275.3.1575 [DOI] [PubMed] [Google Scholar]
- van Kerkhof P., Alves dos Santos C. M., Sachse M., Klumperman J., Bu G., Strous G. J. (2001). Proteasome inhibitors block a late step in lysosomal transport of selected membrane but not soluble proteins. Mol. Biol. Cell 12, 2556–2566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Kerkhof P., Putters J., Strous G. J. (2007). The ubiquitin ligase SCF(betaTrCP) regulates the degradation of the growth hormone receptor. J. Biol. Chem. 282, 20475–20483 10.1074/jbc.M702610200 [DOI] [PubMed] [Google Scholar]
- van Kerkhof P., Westgeest M., Hassink G., Strous G. J. (2011). SCF(TrCP) acts in endosomal sorting of the GH receptor. Exp. Cell Res. 317, 1071–1082 10.1016/j.yexcr.2010.12.020 [DOI] [PubMed] [Google Scholar]
- van Rijnsoever C., Oorschot V., Klumperman J. (2008). Correlative light-electron microscopy (CLEM) combining live-cell imaging and immunolabeling of ultrathin cryosections. Nat. Methods 5, 973–980 10.1038/nmeth.1263 [DOI] [PubMed] [Google Scholar]
- Varghese B., Barriere H., Carbone C. J., Banerjee A., Swaminathan G., Plotnikov A., Xu P., Peng J., Goffin V., Lukacs G. L. et al. (2008). Polyubiquitination of prolactin receptor stimulates its internalization, postinternalization sorting, and degradation via the lysosomal pathway. Mol. Cell. Biol. 28, 5275–5287 10.1128/MCB.00350-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vicidomini G., Gagliani M. C., Canfora M., Cortese K., Frosi F., Santangelo C., Di Fiore P. P., Boccacci P., Diaspro A., Tacchetti C. (2008). High data output and automated 3D correlative light-electron microscopy method. Traffic 9, 1828–1838 10.1111/j.1600-0854.2008.00815.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H., Bei L., Shah C. A., Horvath E., Eklund E. A. (2011). HoxA10 influences protein ubiquitination by activating transcription of ARIH2, the gene encoding Triad1. J. Biol. Chem. 286, 16832–16845 10.1074/jbc.M110.213975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wenzel D. M., Lissounov A., Brzovic P. S., Klevit R. E. (2011). UBCH7 reactivity profile reveals parkin and HHARI to be RING/HECT hybrids. Nature 474, 105–108 10.1038/nature09966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodman P. G., Futter C. E. (2008). Multivesicular bodies: co-ordinated progression to maturity. Curr. Opin. Cell Biol. 20, 408–414 10.1016/j.ceb.2008.04.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.