Abstract
Post-translational modification of proteins by covalent attachment of ubiquitin or a polyubiquitin chain is involved in myriad of processes in eukaryotic cells. The particular outcome of ubiquitination is directed by the length of the ubiquitin conjugate and its linkage composition. Among seven possible isopeptide linkage sites in ubiquitin, K48 and K63 occur most commonly and act as distinct cellular signals. Strategies are reported here for analysis of linkage sites and complexity of K63-linked polyubiquitin chains, based on rapid chemical proteolysis at aspartate residues combined with immunoprecipitation and mass spectrometry. Rapid chemical proteolysis at aspartate residues results in K63-linked peptides with truncated branches, which enable identification and characterization of stretches of consecutive K63 linkages on generally available instruments. A characteristic cleavage pattern and a characteristic fragmentation pattern allow recognition of K63 oligomers in proteolytic mixtures. Engineered K63-linked polyubiquitin chains of defined lengths were used to evaluate and demonstrate the method. In-gel microwave-supported acid hydrolysis was used to observe peptides specific to K63-linked ubiquitin dimers and trimers. Acid hydrolysis in solution, used in conjunction with linkage-specific immunoprecipitation, allowed more complex K63-linked branches to be characterized. Finally a substrate protein, UbcH5b, was conjugated to mono-ubiquitin and to polyubiquitin chains containing only K63 linkages, and the sites of conjugation and chain lengths were characterized.
INTRODUCTION
The length, linkages, and consequent shapes of conjugated polyubiquitin chains direct the function and processing of many intracellular proteins in eukaryotes.1, 2 Consequently, there is great interest in the development of analytical methods that can define the primary structures of polyubiquitin conjugates. Bottom up,3, 4 middle out,5 and top down6 proteomic techniques have been applied to the problem. Trypsin-based bottom up strategies provide a GG tag, which allows sites of ubiquitin conjugation to be located on substrate proteins, however cannot distinguish between GG tags left from other proteins, e.g., Nedd8 and ISG15. Top down strategies (and also SDS-PAGE) can estimate the number of ubiquitin units attached to a protein although the distinction between polyubiquitination at a single site and mono-ubiquitination at multiple sites often cannot be made. Top down sequencing of large branched proteins is reported to be challenging 6 and will require special instrumentation.
Internal linkage sites within polyubiquitin chains are important biologically, however, determining the exact length, linkage composition, and sequence of linkages is an extreme analytical challenge. Although within the polyubiquitin chain isopeptide linkages can occur at any of the seven lysine residues (highlighted in the sequence shown), K48, and K63 linkages are the most abundant in vivo.7, 8 Chains with either homogeneous or mixed linkages are of great interest.9
MQIFVKTLTGKTITLEVEPSDTIENVKAKIQDKEGIPPDQQRLIFAGKQLEDGRTLSDYNIQKESTLHLVLRLRGG
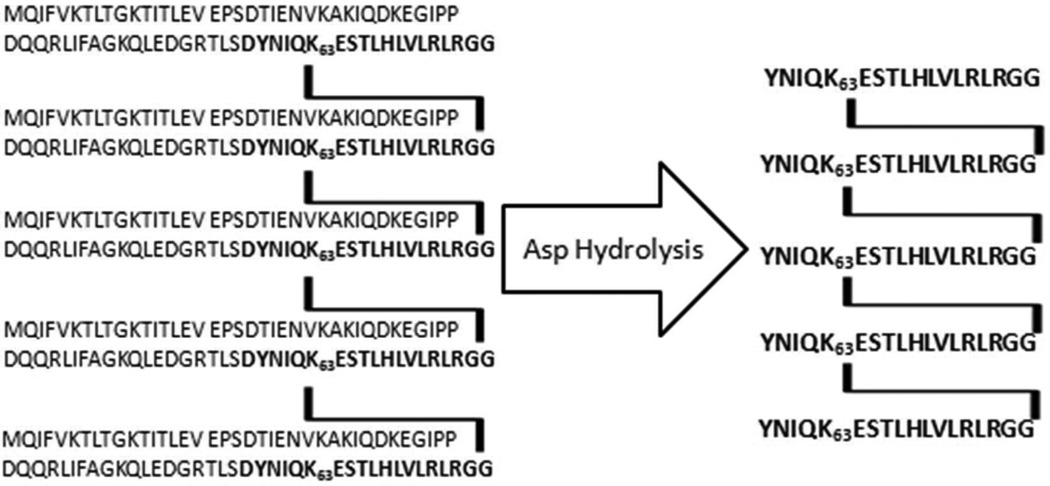
Collisionally induced dissociation (CID) of intact ubiquitin dimers has been reported to distinguish those linked at K48 from those linked through K63, however, the extension of this approach to longer ubiquitin polymers is reported to be challenging.6 We report here the implementation of a strategy designed to identify and characterize K63-linkages in unanchored as well as protein-attached polyubiquitin chains. This approach is based on the chemical or enzymatic cleavage at D52 or D58, which separates K63 from the other six lysine residues in each ubiquitin unit. Because there is no aspartate between D58 and the C-terminal G76 in ubiquitin, the product of interest from Asp-specific cleavage of a polyubiquitinated protein will contain a peptide from the protein, residues [58–76] or [53–76] of the proximal ubiquitin and any isopeptide branch attached to K63 of that ubiquitin. Figure 1 illustrates Asp-specific cleavage of K63-linked penta-ubiquitin in which multiple branch points are retained as long as each isopeptide linkage occurs at K63. Trimming the branches in this way reduces the molecular weight and brings the sample within the capabilities of widely used orbitrap mass spectrometers. In a mixed polymer with linkages at both K63 and other lysine residues, conjugated multi-[58–76] peptides will be recovered only from ubiquitin units connected by an uninterrupted stretch of two or more consecutive K63 isopeptide linkages. We will examine our strategy first by applying it to unanchored K63-linked ubiquitin chains of defined lengths, and then to a ubiquitinated model substrate protein.
Figure 1.
Sequence of a K63-linked pentamer of ubiquitin before and after Asp-specific proteolysis.
MATERIALS AND METHODS
Ubiquitin Expression
Competent E. coli BL-21(DE3) Rosetta™ cells were transformed to express wild type ubiquitin and several ubiquitin variants: K63R ubiquitin, K63 ubiquitin (with all lysines except K63 mutated to arginines), K0 ubiquitin (with all seven lysines mutated to arginines), and an additional mutant form of ubiquitin (D77) in which an extra aspartate residue is present on the C-terminus to prevent chain elongation.10 Following expression, cells were lysed by sonication, and the lysate was cleared via centrifugation at 22,000 rpm. The supernatant was precipitated by the addition of 1.5% (v/v) perchloric acid, and then again centrifuged. The ubiquitin-containing supernatant was then dialyzed using a 3 kDa molecular weight cutoff membrane against 2 L of 50 mM ammonium acetate (pH 4.5). The ubiquitin-containing solution was loaded onto a 5 mL cation exchange column (GE Healthcare Life Sciences, Pittsburgh PA) and eluted over a 22 column volume gradient from 0–40% solvent B (50 mM ammonium acetate, 1 M NaCl, pH 4.5). Ubiquitin-containing fractions were pooled and the purity was confirmed using SDS-PAGE.
Formation of Dimeric and Trimeric Ubiquitin
All polyubiquitin chains were synthesized enzymatically from the ubiquitin monomers described above using the method of Pickart et al10 with slight modifications. After expression in E. coli cells, His6-tagged UBE1 (E1 activating enzyme), UbcH5b (used as substrate), and Mms2 (necessary for Ubc13 E2 activity) were purified using a 5mL His Trap column (GE Healthcare Life sciences). The K63 specific E2, Ubc13, was expressed as a GST fusion protein in BL-21(DE3) cells. Yeast ubiquitin C-terminal hydrolase (YUH1) was expressed without an affinity tag and purified according to the method outlined by Johnston and co-workers.11 To generate dimeric K63-linked chains, 10 mg of each D77 and K63R monomers were allowed to react in the presence of 500 nm UBE1, 20 µM Mms2, 20 µM Ubc13, 5 mM TCEP and 15 mM ATP in a volume of 2 mL with a 50 mM Tris pH 8.0 buffer.12 Trimeric K63-linked chains were synthesized by removing the C-terminal residue D77 from dimeric K63 with YUH1, and then reacting the dimer with additional D77 monomer (2+1) under the same conditions. After twenty-four hours the ubiquitin chain reactions were stopped by the addition of 10 mL of cation buffer A (50 mM ammonium acetate, pH 4.5). The solution was spun at 13,000 rpm to remove precipitated E1 and E2 enzymes, and then slowly injected onto a 5 mL cation exchange column at 0.2 mL/min using an FPLC (GE Life Sciences). The polyubiquitin species were eluted with cation buffer B (50 mM ammonium acetate containing 1 M NaCl, pH 4.5) and exchanged into PBS pH 7.4. This solution was concentrated to 1 mL and the dimeric and trimeric species were separated by size exclusion chromatography (SEC). The SEC fractions were characterized using 15% SDS-PAGE and MALDI mass spectrometry.
Synthesis of Higher K63 Oligomer Mixtures
Longer K63-linked chains were created by reacting wild-type ubiquitin under the conditions described for dimeric and trimeric chains. The use of the Ubc13/Mms2 E2 complex heavily favors the formation of K63 isopeptide bonds.13 The mixture of ubiquitin oligomers was separated from the other reaction components using a cation exchange column and then was concentrated and exchanged into PBS pH 7.4 for SEC. Fractions were assessed using SDS-PAGE as well as mass spectrometry.
Mono and Polyubiquitination of UbcH5b
The C-terminally His6 tagged E2 enzyme, UbcH5b, was conjugated using the same reaction conditions as oligomerization, however with varying concentrations of monomeric ubiquitin and UbcH5b. Mono-ubiquitinated UbcH5b was recovered when the reaction outlined above was carried out with 75 µM UbcH5b and 1 mM of K0 ubiquitin in a 2 mL volume. K63-polyubiquitinated UbcH5b11 was synthesized under similar conditions with 75 µM UbcH5b and 1.5 mM K63 ubiquitin. After twenty-four hours the reaction was diluted with 10 mL of PBS containing 0.5 M NaCl, pH 7.4, and then loaded onto a 1 mL His-Trap column. The eluate, which contained a mixture of free UbcH5b and UbcH5b conjugated with varying numbers of ubiquitins, was concentrated and exchanged into PBS, 10 mM DTT, pH 7.4 to a final volume of 1 mL. This mixture was fractionated by SEC, and different oligomeric states of ubiquitinated UbcH5b were detected using 15% SDS-PAGE.
Microwave-supported acid digestion
For in solution digestion, the concentration of ubiquitin and ubiquitin containing samples was estimated based on ubiquitin’s molar extinction coefficient at 280 nm (1,490 cm−1 M−1). Total estimated protein was diluted to 0.1 mg/mL in 12.5% acetic acid and digested for between 90 seconds and 7 minutes. Digestion was carried out at 140°C using 300 W of microwave energy to maintain temperature in a CEM Discover microwave (Matthews, NC). After digestion the peptides were desalted using Millipore C-18 ZipTips (Billerica, MA) according to the manufacturer’s directions.
Enzymatic Digestions
Enzymatic digestion was performed with Asp-N endoprotease (Roche Diagnostics Indianapolis, IN) following the manufacturer’s directions at a 1:1000 enzyme to substrate ratio for between 18 and 24 hours. Sequencing grade trypsin was acquired from Promega (Madison, WI), and used following the manufacturer’s protocol.
Immunoprecipitation (IP)
K63-linkage specific antibody (Millipore, Billerica, MA) was conjugated to agarose beads using the ThermoPierce Direct IP kit (Rockford, IL) according to the manufacturer’s instructions. Immunoprecipitation and sample recovery were performed according the manufacturer’s protocol.
MALDI analysis
Mass spectra were acquired in linear mode on a Kratos Axima CFR MALDI-TOF MS (Shimadzu Biosciences, Columbia, MD). Laser power was varied to achieve optimal ion current of the higher mass branched peptides. One hundred scans (at ten laser pulses per scan) were integrated for each spectrum. The matrix was α-cyano-4-hydroxycinnamic acid at 10mg/mL in 70/30/0.1 acetonitrile/water/TFA.
LC-MS/MS using electrospray
. Mass spectrometry was carried out using an LTQ-Orbirap XL instrument (ThermoFisher, San Joe CA). Intact peptide masses were acquired at 100,000 resolution and product ions were measured at 15,000 resolution. Peptides were separated using reversed phase chromatography on a Profound system (Shimadzu BioSciences, Columbia MD) interfaced directly to the nanospray source of the mass spectrometer. Either a Zorbax C3 column (Agilent, Bethlehem, PA) or a Grace Vydac C18 column (Deerfield, IL) was used, at a flow rate of 500nL/min. Spectra acquired from products of tryptic digestion were searched with the MASCOT search engine (Matrix Science, London UK) against human proteins in the UniProtKB library. Spectra acquired from the products of acid cleavage of higher polymers and of Asn-N cleavage were interpreted manually.
NMR analysis
Heteronuclear transverse-relaxation optimized (TROSY) spectra were acquired on a Bruker Avance III 600 MHz NMR spectrometer equipped with a cryogenic TCI probe. Monomeric 15N-enriched ubiquitin was exchanged into PCR grade water and concentrated to 1 mM. This stock was used to make three different 450 μL NMR samples containing pure water, 12.5%(v/v) acetic acid, or 45%(v/v) acetic acid. All samples had 5%(v/v) D2O, 0.02%(w/v) sodium azide, and 0.4 mM ubiquitin. The [1H,15N]-TROSY spectra were recorded at 23°C using 16 scans (with 128 points in 15N and 4096 points in 1H). The spectra were processed using TopSpin v2.1 (Bruker Biospin Inc). Hydrogen bonding was analyzed using the PyMOL Molecular Graphics System, Version 1.5.0.4 (Schrodinger LLC, Portland, OR).
RESULTS AND DISCUSSION
Acid digestion and MALDI characterization
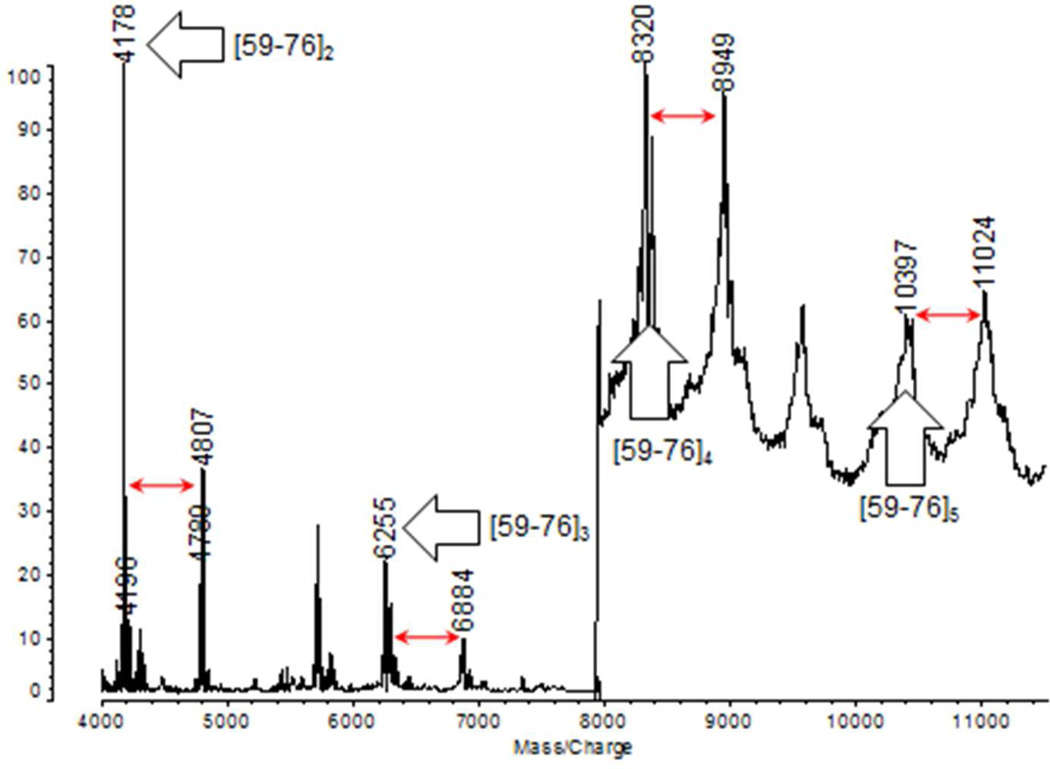
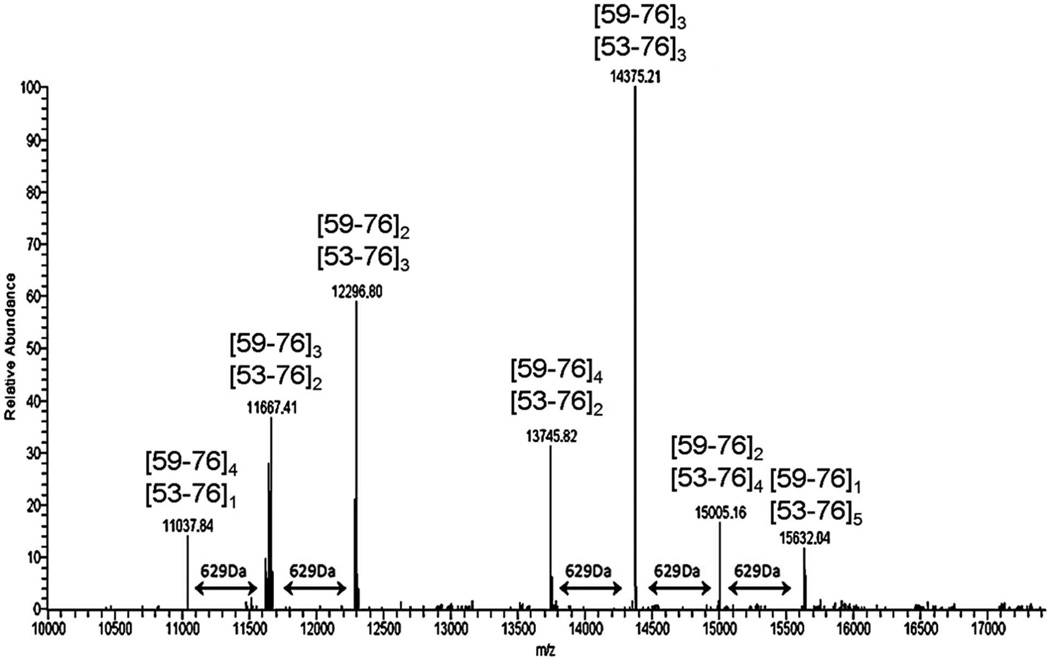
Partial MALDI spectra are shown in Figure 2 of peptide products produced by acid cleavage of the product mixture from polymerization of wild type ubiquitin with the Ubc13/Mms2 E2 complex. This complex favors formation of isopeptide bonds between the C-terminal carboxylate group of one ubiquitin molecule and K63 of another. The MALDI spectrum reveals the presence of dimers, trimers, tetramers, and pentamers analogous to the product shown in Figure 1. Each oligomer is accompanied by a second peak higher in mass by 629 Da. We propose that this corresponds to the presence of residues [52–57] in one of the chains where acid cleavage has occurred at D52/G53 instead of D58/Y59. This pattern of peaks separated by 629 Da has been observed consistently throughout our work and we propose that pairs of ions separated by 629 Da, the mass of GRTLSD, are indicators of K63-isopeptide bonds in ubiquitin oligomers cleaved by microwave-supported acid digestion.
Figure 2.
Partial MALDI spectra of acid cleavage products of K63 linked di-ubiquitin, tri-ubiquitin, tetra-ubiquitin and penta-ubiquitin. Diagnostic 629 Da mass shifts (see text) are indicated with red arrows. The structure of the pentamer [59–76]5 is shown in Figure 1.
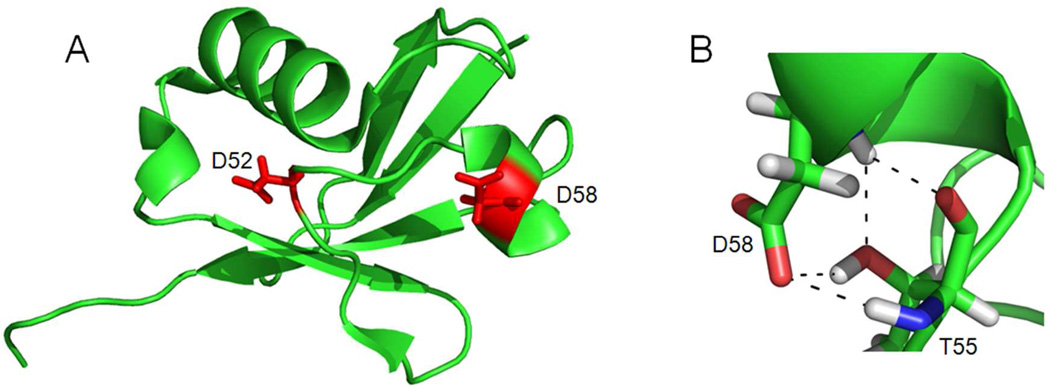
Peptides with missed cleavages at D58 are formed in high abundance from all four synthetic oligomers, and also from the wild type ubiquitin monomer (data not shown). Typically in microwave-supported acid proteolysis, peptides are formed without missed cleavages.14–16 These reactions are expected to proceed optimally with denatured proteins, in part because acid hydrolysis requires solvent accessibility to cleavage sites. In addition, the generally accepted mechanisms require cyclization of the carboxylic acid side chain of Asp to adjacent amide groups.17,18 Hydrogen bonding of these functional groups in a folded protein will inhibit cyclization and the formation of cleavage products. Nuclear magnetic resonance and X-ray crystal structures of ubiquitin19, 20 indicate that D58 is located in a 310-helix, while D52 is located in an extended and rather flexible loop21 (Figure 3A). Moreover, our PyMol analysis revealed several potential polar/hydrogen-bonding interactions between the backbone and side chain atoms of D58 and T55 (Figure 3B). No such interactions were found for D52. This helps explain the abundance of products in which cleavage has occurred at D52 and not at D58. To test the effect of acid on the tertiary structure of ubiquitin, we acquired two-dimensional [1H, 15N] NMR spectra of mono-ubiquitin in 12.5% acetic acid at room temperature (Supplementary Figure 1). It appears that in 12.5% acetic acid ubiquitin retains much of its native structure. The slow cleavage at D58 is interpreted as evidence that some hydrogen bonding is maintained during acid cleavage, even with elevated temperature. It should be mentioned here that ubiquitin is highly stable: it remains well folded up to ~100°C at neutral pH22 and is routinely purified using its solubility in perchloric acid.10 Therefore, it is no surprise that microwave-supported acid digestion is inhibited in some elements of secondary structure.
Figure 3.
Representation of the 3-D structure of ubiquitin (PDB ID: 1D3Z17). (A) D58 is located in a 310 helix while D52 is located in a loop. Both residues are colored red and shown as side chains. (B) PyMol revealed a network of polar or hydrogen-bonding interactions (dashed lines) between backbone and side chain atoms of D58 and T55. No such interactions are present for D52.
Characterization by LC-MS/MS and diagnostic ‘b’ ions
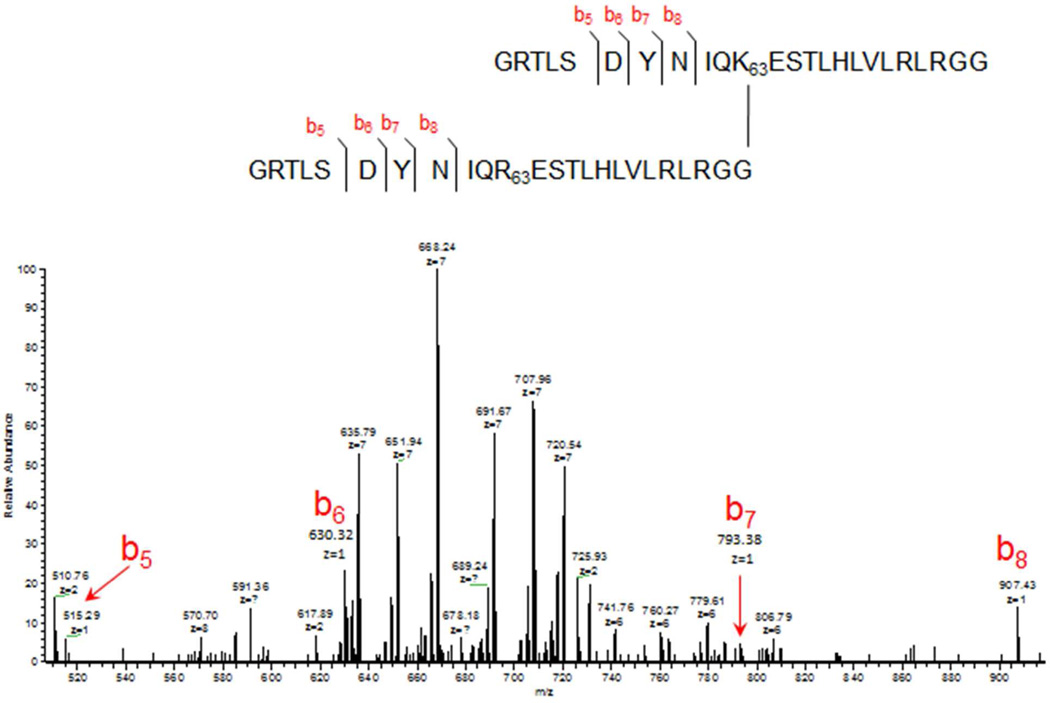
HPLC was used to fractionate branched peptides of interest for tandem mass spectrometry. Because of their size, both precursor and product ions from these mid-range peptides were analyzed with high resolution mass measurements and a “middle out” workflow.23 A collisionally induced dissociation product ion spectrum from a K63-linked dimer is shown in Figure 4, in which a series of ‘b’ ions is recorded that extends from the cleavage point D52/G53 in the truncated branch toward the isopeptide branching point at K63. This suite of low mass ions is characteristic of the truncated ubiquitin chains formed with acid cleavage and can be used to recognize such peptides. Isobaric coincidence is minimized by high-resolution accurate mass measurements. Similar use of diagnostic ‘b’ ions has been proposed by Warren et al using a 12-residue tag (in place of GG) to locate ubiquitination sites in substrate peptides.24 When multiple K63 branches are present these ions are abundant in the spectra, however they do not reveal the number of sequential K63 branches directly. This information can be deduced from the peptide molecular ion if the substrate peptide can be independently identified. One method for identification of the conjugated peptide would use a parallel analysis of GG-tagged peptides in a tryptic digest. Alternatively, an isolated conjugated protein could be identified by MS/MS analysis of unconjugated peptide products of acid cleavage.
Figure 4.
MS/MS spectrum of a branched peptide from a K63-linked ubiquitin dimer showing the series of ‘b’ ions from the GRTLSDYNIQ tails. Precursor charge state is +9. The annotated b ions have +1 charge states.
Immunoprecipitation for mass spectrometry
Analysis of polyubiquitin conjugates has benefited greatly from the production of antibodies that are specific for ubiquitin, and more recently, specific for certain ubiquitin-ubiquitin linkages. K63 linkage-specific peptides have been formulated as antigens in two different ways, using intact di-ubiquitin25 and using synthetic K63-linked peptides.26 We have used the antibody produced against K63-linked fragments of ubiquitin to purify peptides of interest from oligomers of wild-type ubiquitin (see Experimental Section). A mixture of ubiquitin oligomers was selected from SEC fractions for acid cleavage. Following digestion, K63-branched peptides were immunoprecipitated with the K63-linkage specific antibody. A mixture containing higher order ubiquitin oligomers was fractionated using reverse phase HPLC interfaced to a tandem mass spectrometer that provided high-resolution precursor and fragment ion detection. Examination of extracted ion chromatograms of diagnostic ‘b’ ions revealed a narrow range of elution times for the branched peptides. Precursor spectra were integrated over this range and decharged (deconvoluted) to reveal the heavy molecular ions shown in Figure 5. Charge state distributions included +12 to + 20 before deconvolution..
Figure 5.
Deconvoluted electrospray spectrum integrated over the course of isopeptide elution 28–35 min. of acid digestion products (see text).
Two sets of high molecular weight species in Figure 5 contain 629 Da mass shifts (GRTLSD) associated with missed cleavages in K63 branched peptides. A quartet of peaks is separated by increments of 629 Da, indicating cleavages at D52/G53 and/or D58/Y59 on at least three K63-linked branches. The triplet of peaks in the same Figure reveals another oligomer with at least two K63 branches.
Ubiquitination of UbcH5b
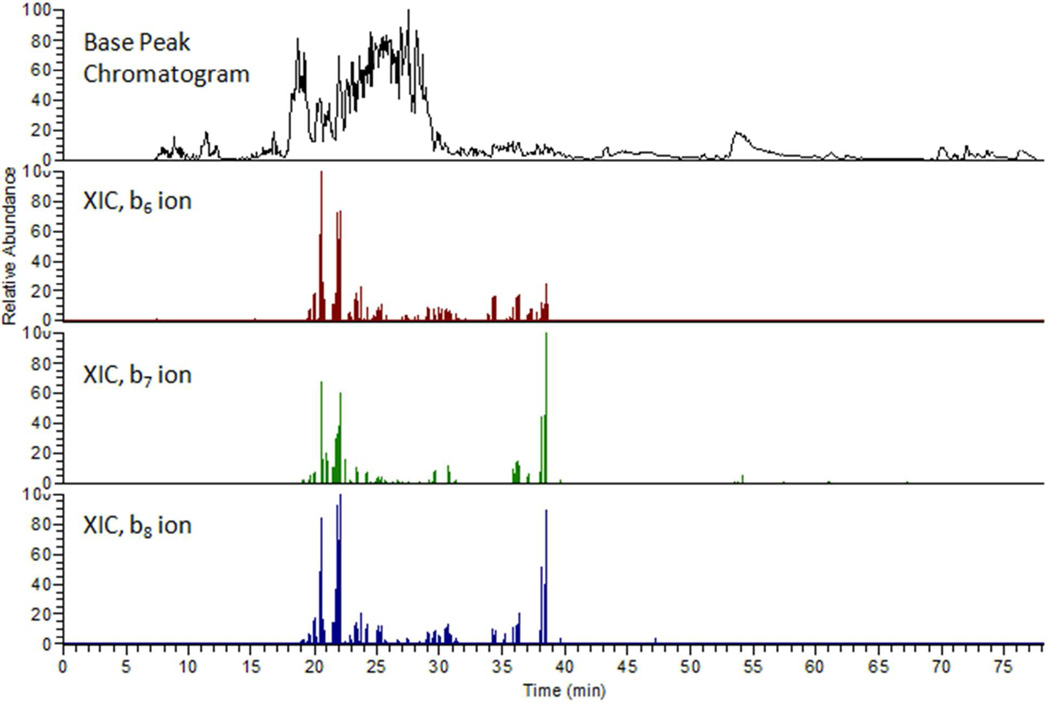
The E2 ubiquitin-conjugating enzyme UbcH5b, implicated in the ubiquitination of the p53 tumor suppressor, is itself a substrate for ubiquitination.27 UbcH5b was conjugated with a mono-ubiquitin variant (K0) having all lysines mutated to arginines, to test the hypothesis that sites of attachment by ubiquitin to a known substrate can be characterized by analyzing the branched peptide products of acid cleavage. Conjugated UbcH5b was cleaved by acid digestion for LC-MS/MS analysis. A suite of ‘b’ ions, shown in Figure 6 as extracted ion chromatograms, indicated the presence of several peptides eluting from the HPLC that contained ubiquitin truncated at G53. Masses of the precursor ions that produced the diagnostic ‘b’ ions matched within 5 ppm those predicted for peptides produced from the substrate protein by acid cleavage and attached to truncated mono-ubiquitin chains. These were the N terminal peptide [2–11] ALKIRHKELN containing K4 and K8, peptide [2–12] ALKIRHKELND, and peptide [117–129] DPLVPEIARIYKT with a conjugation site at K128. Peaks eluting at ~20 and ~22 minutes (Figure 6) correspond to the truncated conjugates of peptides [2–11] and [2–12].The peak eluting at ~38 min corresponds to UbcH5b peptide [117–129] conjugated at K128. Conjugation sites at K8 and K128 were independently confirmed using tryptic digestion and locating GG tags (Supplementary Figures 2 and 3).
Figure 6.
Base peak chromatogram (top) and extracted ion chromatograms (XICs) of three diagnostic ‘b’ ions from the product mixture from mono-ubiquitinated UbcH5b.
Next, a ubiquitin mutant (K63) containing only K63 with all other Lys residues mutated to Arg was allowed to react with UbcH5b under the conditions described above. The high masses of peptides that contain multiple K63 isopeptide bonds were exploited in the purification of the acid cleavage products. K63 linked di-ubiquitin, for example, produces a branched peptide trimmed by acid cleavage that weighs somewhat more than 4000 Da, and more mass is added by the conjugated substrate peptide. Commercially available molecular weight cutoff filters were used to remove peptides with masses below ~3 kDa from the acid cleavage products in order to reduce complexity and minimize nonspecific binding to the antibody.14 The sample was then immunoprecipitated, redissolved and injected into the LC-MS/MS, where sets of ions with intervals of 629 Da across multiple charge states identified a peptide with K63 isopeptide linkages (See supplementary Figure 4).
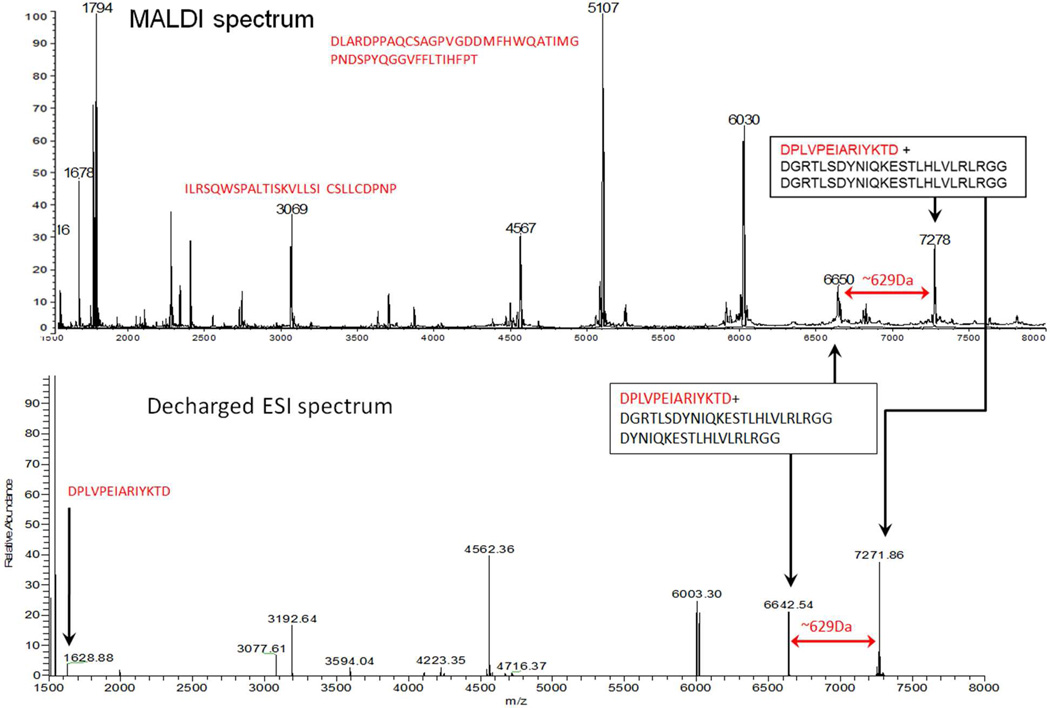
Figure 7 presents the decharged nanospray spectrum of this branched peptide obtained at high resolution (bottom) and a low resolution MALDI spectrum (top) of the same immunoprecipitate. The two spectra exhibit a similar pair of peaks separated by 629 Da. As expected, the average masses determined in the low resolution experiment are several Daltons higher than the corresponding monoisotopic masses recorded with high resolution. Since UbcH5b was known to be the substrate, an in silico digestion of the primary sequence provided candidate neutral masses to combine with the masses of possible combinations of ubiquitin chains cleaved at D52 or D58. Consideration of a decharged mono-isotopic neutral mass of 7271.86 Da led to the assignment of an acid-truncated di-ubiquitin chain attached to an acid digestion peptide product from UbcH5b [117–130] DPLVPEIARIYKTD (see Figure 7). The only lysine, K128, is assigned as the site of ubiquitination. A K63 isopeptide linkage is assigned in the ubiquitin chain, based on immunospecificity and the presence of acid cleavage products with mass differences of 629 Da (DGRTLS). Microwave-supported acid hydrolysis has been shown to cleave on either side of aspartate residues.14–16 and the masses of the peptides isolated in this experiment indicate that cleavage has occurred between E51/D52. Lastly the presence of unconjugated peptides from the substrate protein, shown in both spectra in red, would allow substrate candidates to be identified by MS/MS measurements if the substrate was not known.
Figure 7.
Partial mass spectra of UbcH5b peptide [117–130] conjugated by a K63 ubiquitin dimer truncated by acid cleavage. Top: partial MALDI spectrum recording near-average masses. Bottom: partial decharged nanospray spectrum showing monoisotopic masses. Peptide sequences in red originate from UbcH5b. Peptide sequences in black originate from ubiquitin.
Conclusions
Despite significant and on-going contributions to the ubiquitin field, proteomic methods based on trypsin digestion provide limited information28 regarding the linkage composition of polyubiquitin chains. Even when the lysine sites involved in polymerization can be identified with existing methods, determining the length and consecutive linkages remains a significant challenge. A method is presented here to recognize and identify consecutive K63 linkages in both unanchored and substrate-attached chains. K63 proximal to the conjugated substrate and any K63-linked branches are separated from other linkage sites by a very rapid Asp-selective chemical cleavage. This proteolysis is combined with immunoprecipitation and mass spectrometry to identify truncated K63-branched conjugates and to characterize a consecutive stretch of uninterrupted K63-linked ubiquitins, which is currently impossible with other methodologies. Truncation allows middle-out characterization of branched peptides on mass spectrometers that cannot provide top-down analysis of the very heavy polymers of ubiquitin. Enzymatic cleavage with Asp-N should also provide truncated branched peptides, however this is reported to be a slow and inefficient protease.29 In this report our strategy has been applied successfully to unanchored di- through penta-ubiquitin oligomers, and to mono-and di-ubiquitin moieties conjugated at specific positions on the protein UbcH5b. Characterization of poly-ubiquitination on conjugated proteins requires independent identification of the substrate protein, readily accomplished by MS/MS analysis of the unconjugated peptide products of acid cleavage (Fig.7), or by MS/MS identification of GG-tagged peptides in a parallel tryptic digestion (supporting Fig. 2 and 3).
Supplementary Material
ACKNOWLEDGMENTS
The authors thank Dr. Yan Wang, Director of the Proteomics Core Facility of the Maryland Pathogen Research Institute for discussion and advice, Dr. Rajesh Singh for kindly providing the UbcH5b plasmid, and Russell Knighton for assistance in an early stage of the project. The work was supported by NIH grants GM021248 (to CF), GM065334 (to DF), and a Bailey Fellowship (to JC) from the University of Maryland.
Footnotes
Supporting Information Available: This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Kerscher O, Felberbaum R, Hochstrasser M. Ann. Rev of Cell and Develop. Biol. 2006;22:159–180. doi: 10.1146/annurev.cellbio.22.010605.093503. [DOI] [PubMed] [Google Scholar]
- 2.Pickart CM, Fushman D. Curr.Opin.Chem.Biol. 2004;8:610–616. doi: 10.1016/j.cbpa.2004.09.009. [DOI] [PubMed] [Google Scholar]
- 3.Peng J, Schwartz D, Elias JE, Thoreen CC, Cheng D, Marsischky G, Roelofs J, Finley D, Gygi SP. Nat Biotech. 2003;21:921–926. doi: 10.1038/nbt849. [DOI] [PubMed] [Google Scholar]
- 4.Wagner SA, Beli P, Weinert BT, Nielsen ML, Cox JR, Mann M, Choudhary C. Mol. Cell. Prot. 2011;10:1074–1086. doi: 10.1074/mcp.M111.013284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Xu P, Peng J. Anal. Chem. 2008;80:3438–3444. doi: 10.1021/ac800016w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Strachan J, Roach L, Sokratous K, Tooth D, Long J, Garner TP, Searle MS, Oldham NJ, Layfield R. J. Proteome Res. 2012;11:1969–1980. doi: 10.1021/pr201167n. [DOI] [PubMed] [Google Scholar]
- 7.Xu P, Duong DM, Seyfried NT, Cheng D, Xie Y, Robert J, Rush J, Hochstrasser M, Finley D, Peng J. Cell. 2009;137:133–145. doi: 10.1016/j.cell.2009.01.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ziv I, Matiuhin Y, Kirkpatrick DS, Erpapazoglou Z, Leon S, Pantazopoulou M, Kin W, Gygi SP, Haguenauer-Tsapis R, Reis N, Glickman MN, Kleifeld O. Mol. Cell. Prot. 2011;10 doi: 10.1074/mcp.M111.009753. M111.009753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kratsova-Ivantsiv Y, Ciechanover A. J.Cell Sci. 2012;125:539–548. doi: 10.1242/jcs.093567. [DOI] [PubMed] [Google Scholar]
- 10.Pickart CM, Raasi S. Methods in Enzymology. 2005;399:21–36. doi: 10.1016/S0076-6879(05)99002-2. [DOI] [PubMed] [Google Scholar]
- 11.Johnston SC, Riddle SM, Cohen RE, Hill CP. EMBO J. 1999;18:3877–3887. doi: 10.1093/emboj/18.14.3877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Datta AB, Hura GL, Wolberger C. J. Molec.Biol. 2009;392:1117–1124. doi: 10.1016/j.jmb.2009.07.090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dong KC, Helgason E, Yu C, Phu L, Arnott DP, Bosanac I, Compana DM, Huang OW, Fedorova AV, Kirkpatrick DS, Hymowitz SG, Dueber EC. Structure. 2011;19:1053–1063. doi: 10.1016/j.str.2011.06.010. [DOI] [PubMed] [Google Scholar]
- 14.Cannon J. Asp-Selective Acid Hydrolysis for Proteomics. College Park, MD: University of Maryland, College Park; 20742. [Google Scholar]
- 15.Swatkoski S, Gutierrez P, Wynne C, Petrov A, Dinman JD, Edwards N, Fenselau C. J. Proteome Res. 2008;7:579–86. doi: 10.1021/pr070502c. [DOI] [PubMed] [Google Scholar]
- 16.Cannon J, Lohnes K, Wynne C, Wang Y, Edwards N, Fenselau C. J. Proteome Res. 2010;9:3886–90. doi: 10.1021/pr1000994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Inglis AS. Methods in Enzymology. 1983;91:324–332. doi: 10.1016/s0076-6879(83)91030-3. [DOI] [PubMed] [Google Scholar]
- 18.Li A, Sowder RC, Henderson LE, Moore SP, Garfinkel DJ, Fisher R. J. Anal.Chem. 2001;73:5395–5402. doi: 10.1021/ac010619z. [DOI] [PubMed] [Google Scholar]
- 19.Cornilescu G, Marquardt JL, Ottiger M, Bax A. J. Amer. Chem. Soc. 1998;120:6836–6837. [Google Scholar]
- 20.Vijay-Kumar S, Bugg CE, Cook WJ. J. Mol. Biol. 1987;194:531–544. doi: 10.1016/0022-2836(87)90679-6. [DOI] [PubMed] [Google Scholar]
- 21.Tjandra N, Feller SE, Pastor RW, Bax A. J. Amer. Chem. Soc. 1987;117:12562–12566. [Google Scholar]
- 22.Ibarra-Molero B, Loladze VV, Makhatadze GI, Sanchez-Ruiz JM. Biochem. 1999;38:8138–8149. doi: 10.1021/bi9905819. [DOI] [PubMed] [Google Scholar]
- 23.Cannon JR, Fenselau C. In: Proteomic and Metabolomic Approaches to Biomarker Discovery. Vennstra TD, Issaq HJ, editors. Chennai, India: Elsevier; 2013. [Google Scholar]
- 24.Warren MR, Parker CE, Mocanu V, Klapper D, Borchers C. H. Rapid Commun. Mass Spectrom. 2005;19:429–437. doi: 10.1002/rcm.1798. [DOI] [PubMed] [Google Scholar]
- 25.Newton K, Matsumoto ML, Wertz IE, Kirkpatrick DS, Lill JR, Tan J, Dugger D, Gordon N, Sidhu SS, Fellouse FA, Komives L, French DM, Ferrando RE, Lam C, Compaan D, Yu C, Bosanac I, Hymowitz SG, Kelley RF, Dixit VM. Cell. 2008;134:668–678. doi: 10.1016/j.cell.2008.07.039. [DOI] [PubMed] [Google Scholar]
- 26.EMD Millipore, Billerica MA. [Google Scholar]
- 27.Saville MK, Sparks A, Xirodimas DP, Wardrop J, Stevenson LF, Bourdon JC, Woods YL, Lane DP. J. Biol. Chem. 2004;279:42169–42181. doi: 10.1074/jbc.M403362200. [DOI] [PubMed] [Google Scholar]
- 28.Mirzaei H, Rogers RS, Grimes B, Eng J, Aderem A, Aebersold R. Molec. Systems. 2010;6:2004–2014. doi: 10.1039/c005242f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kim HT, Kim KP, Lledias F, Kisselev AF, Scaglione KM, Skowyra D, Gygi SP, Goldberg AL. J. Biolog. Chem. 2007;282:17375–17386. doi: 10.1074/jbc.M609659200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.