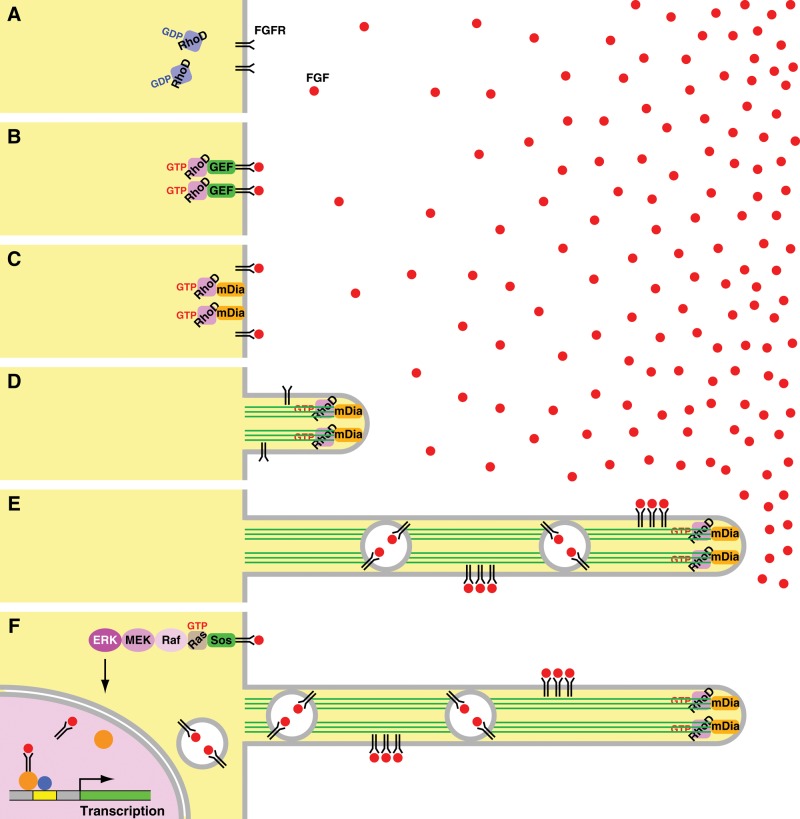
The small GTPase RhoD, activated by fibroblast growth factor (FGF) signaling, forms actin-based, cytoneme-like, thin and long cellular protrusions through activating mDia3C. These protrusions transmit FGF receptors toward the cell body. They are likely to be responsible for intercellular communication between FGF-producing cells and target cells.
Abstract
The small GTPase RhoD regulates actin cytoskeleton to collapse actin stress fibers and focal adhesions, resulting in suppression of cell migration and cytokinesis. It also induces alignment of early endosomes along actin filaments and reduces their motility. We show here that a constitutively activated RhoD generated two types of actin-containing thin peripheral cellular protrusions distinct from Cdc42-induced filopodia. One was longer, almost straight, immotile, and sensitive to fixation, whereas the other was shorter, undulating, motile, and resistant to fixation. Moreover, cells expressing wild-type RhoD extended protrusions toward fibroblast growth factor (FGF) 2/4/8–coated beads. Stimulation of wild-type RhoD-expressing cells with these FGFs also caused formation of cellular protrusions. Nodules moved through the RhoD-induced longer protrusions, mainly toward the cell body. Exogenously expressed FGF receptor was associated with these moving nodules containing endosome-like vesicles. These results suggest that the protrusions are responsible for intercellular communication mediated by FGF and its receptor. Accordingly, the protrusions are morphologically and functionally equivalent to cytonemes. RhoD was activated by FGF2/4/8. Knockdown of RhoD interfered with FGF-induced protrusion formation. Activated RhoD specifically bound to mDia3C and facilitated actin polymerization together with mDia3C. mDia3C was localized to the tips or stems of the protrusions. In addition, constitutively activated mDia3C formed protrusions without RhoD or FGF stimulation. Knockdown of mDia3 obstructed RhoD-induced protrusion formation. These results imply that RhoD activated by FGF signaling forms cytoneme-like protrusions through activation of mDia3C, which induces actin filament formation.
INTRODUCTION
Diverse types of cell protrusions play roles in a variety of cellular functions, including cell migration, cell differentiation, and intercellular communication. Among these protrusions, filopodia and lamellipodia have been extensively and intensively studied. In consequence, molecular mechanisms of their generation and their structures and functions are relatively well clarified (Small et al., 2002; Mattila and Lappalainen, 2008; Faix et al., 2009; Mellor, 2010). Actin filament formation is responsible for generation of these protrusions. Filopodia contain tight parallel bundles of actin filaments, whereas lamellipodia are filled with branched or cross-linked actin filaments. Filopodia serve as sensory antennae or tentacles to probe microenvironment of cells, although their roles are diverse and not fully understood. On the other hand, lamellipodia are essential for cell migration by crawling. Both of these protrusions adhere to the substratum through integrin-based focal complexes or nascent focal adhesions (Nobes and Hall, 1995; Geiger et al., 2009).
Cytonemes are actin-based, thin, long cell protrusions first reported in Drosophila wing imaginal disk cells and mouse embryo limb bud cells (Ramírez-Weber and Kornberg, 1999), although cytonemes of limb bud cells have been inadequately characterized. Wing or eye imaginal disk cells and tracheal cells extend cytonemes toward signaling center cells in the organizers, producing specific morphogen or growth factor signaling proteins, such as Decapentaplegic (Dpp)/bone morphogenetic protein (BMP), Spitz/epidermal growth factor (EGF), and Branchless (Bnl)/fibroblast growth factor (FGF; Hsiung et al., 2005; Roy et al., 2011). Nodules or punctae containing the receptors of each morphogen/growth factor are specifically segregated in cytonemes and move through cytonemes toward the cell body. Accordingly, cytonemes are proposed to be involved in long-distance intercellular communication by directly transferring specific morphogens/growth factors with their receptors. Similar protrusions have been reported in a variety of cell types, including human neutrophils, mouse B lymphocytes, and mast cells treated in various ways (Galkina et al., 2001; Gupta and DeFranco, 2003; Fifadara et al., 2010). In addition, viral cytonemes, which extend from uninfected target cells to an infected cell by the interaction between viral envelope glycoprotein with the receptor, transmit retroviruses from infected to uninfected cells (Sherer et al., 2007).
The Rho family of small GTPases participate in the formation of filopodia and lamellipodia by regulating actin filament formation. Cdc42 binds to activate neural Wiskott-Aldrich syndrome protein (N-WASP) together with phosphatidylinositol 4,5-bisphosphate (PIP2) or Src homology 3 (SH3) domain–containing proteins. Activated N-WASP induces Arp2/3 complex–mediated, branched actin filament formation (Takenawa and Suetsugu, 2007; Campellone and Welch, 2010). Cdc42 also binds and activates mDia2, which nucleates and elongates unbranched actin filaments. A current working model hypothesizes that a subset of uncapped actin filaments of the Arp2/3 complex–nucleated dendritic networks is targeted for continued actin filament elongation by mDia2 (Yang et al., 2007; Mattila and Lappalainen, 2008; Faix et al., 2009; Mellor, 2010). mDia2 also nucleates to form new unbranched actin filaments. The barbed ends of these elongating actin filaments are converged through the motor activity of myosin-X, leading to the formation of filopodia. The actin filaments are cross-linked to become stiff bundles by fascin in the shaft of filopodia. RhoQ (TC10) and RhoJ (RhoT or TCL), both of which are phylogenetically close to Cdc42, also activate N-WASP and form longer filopodial protrusions than does Cdc42 (Abe et al., 2002), although involvement of mDia proteins in actin filament formation for the protrusion formation is unclear.
In contrast, the mechanisms of cytoneme formation remain poorly understood. It is unclear whether any Rho family proteins, N-WASP and the Arp2/3 complex, mDia proteins, other actin nucleation factors, or elongation factors (Dominguez, 2009; Campellone and Welch, 2010; Chesarone et al., 2010; Takano et al., 2010) are involved in actin filament formation giving rise to cytonemes. RhoD induces alignment of early endosomes along actin filaments and reduces their movement through activating hDia2C (corresponding to mDia3C) and Src (Murphy et al., 1996; Gasman et al., 2003). It also regulates the actin cytoskeleton to disassemble actin stress fibers and focal adhesions by antagonizing RhoA, resulting in suppression of cell migration and cytokinesis (Tsubakimoto et al., 1999). Furthermore, overexpressed, constitutively activated RhoD forms actin-containing thin cell protrusions reaching 20–30 μm in length, with some being attached to the substratum but others being motile due to a lack focal adhesion–associated proteins (Murphy et al., 1996). Fluorescently labeled transferrin is internalized through these protrusions and colocalized with RhoD.
Thus we addressed the question of whether the protrusions induced by RhoD correspond to cytonemes and investigated the mechanisms of actin filament formation by RhoD. The morphological and functional properties of RhoD-induced protrusions in cultured mesenchymal cells were equivalent to those of cytonemes. RhoD was activated by FGF signaling, and RhoD-activated mDia3C was responsible for the actin filament formation in the protrusion.
RESULTS
RhoD forms two types of actin-based thin protrusions
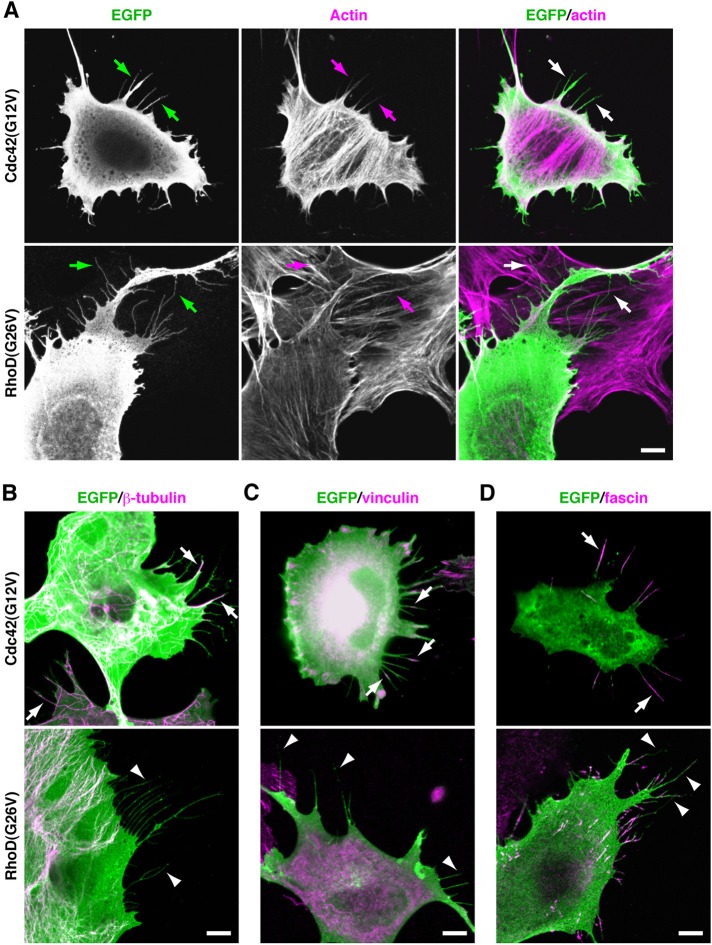
To assess properties of cell protrusions induced by the constitutively active RhoD(G26V) (Gly26 is converted to Val) in comparison with those by Cdc42(G12V), we transfected mouse mesenchymal C3H/10T1/2 (10T1/2) cells with enhanced green fluorescent protein (EGFP)-fused cDNAs encoding each protein. Cdc42(G12V)-formed filopodia were usually <10 μm in length, thicker, and straight and contained actin filament bundles (Figure 1A and Supplemental Figure S1A). Some, if not all, also contained microtubules detected by staining with an anti–β-tubulin monoclonal antibody (mAb; Figure 1B). They attached to the substratum as indicated by staining with an anti-vinculin mAb detecting focal adhesions and focal complexes (Figure 1C). RhoD(G26V)-formed protrusions also contained actin filaments and were often >10 μm long, thinner, and undulating in paraformaldehyde (PFA)-fixed cells (Figures 1A and S1A). They neither contained microtubules nor had vinculin-containing focal complexes (Figure 1, B and C), indicating that they were unattached to the substratum. This is consistent with findings by Murphy et al. (1996) and findings that activated RhoD collapses focal adhesions as well as stress fibers (Tsubakimoto et al., 1999).
FIGURE 1:
RhoD gives rise to actin filament–based thin protrusions distinct from Cdc42-induced filopodia detected in PFA-fixed cells. (A) Distinct morphology between Cdc42-induced filopodia and RhoD-induced protrusions. The 10T1/2 cells transfected with EGFP–Cdc42(G12V) or EGFP–RhoD(G26V) were fixed with PFA. EGFP fluorescence (green) and actin filaments detected with Alexa Fluor 546–phalloidin (magenta) are shown. Arrows point to Cdc42-induced filopodia and RhoD-induced thin protrusions. (B) Presence and absence of microtubules in Cdc42-induced filopodia and in RhoD-induced protrusions, respectively. Microtubules were detected by anti–β-tubulin mAb staining (magenta). Arrows and arrowheads point to microtubule-containing filopodia and the protrusions lacking microtubules, respectively. (C) Presence and absence of vinculin-containing focal complexes in Cdc42-induced filopodia and in RhoD-induced protrusions, respectively. Vinculin was detected by anti-vinculin mAb staining (magenta). Arrows and arrowheads point to vinculin-containing focal complexes in filopodia and the protrusions lacking vinculin, respectively. (D) Distribution of fascin in Cdc42-induced filopodia and RhoD-induced protrusions. Fascin was detected by anti-fascin mAb staining (magenta). Arrows and arrowheads point to fascin-containing filopodia and the protrusions lacking fascin, respectively. Scale bars: 10 μm.
Fascin tightly cross-links actin filaments in filopodia and is postulated to give stiffness to filopodia (Hashimoto et al., 2005; Vignjevic et al., 2006). Fascin was localized along the entire length of almost all Cdc42-induced filopodia (Figure 1D). In contrast, although some RhoD-induced protrusions intermittently contained fascin, other protrusions were devoid of fascin (Figure 1D). This incomplete association with fascin, as well as the lack of focal complexes, seems to render the protrusions flexible.
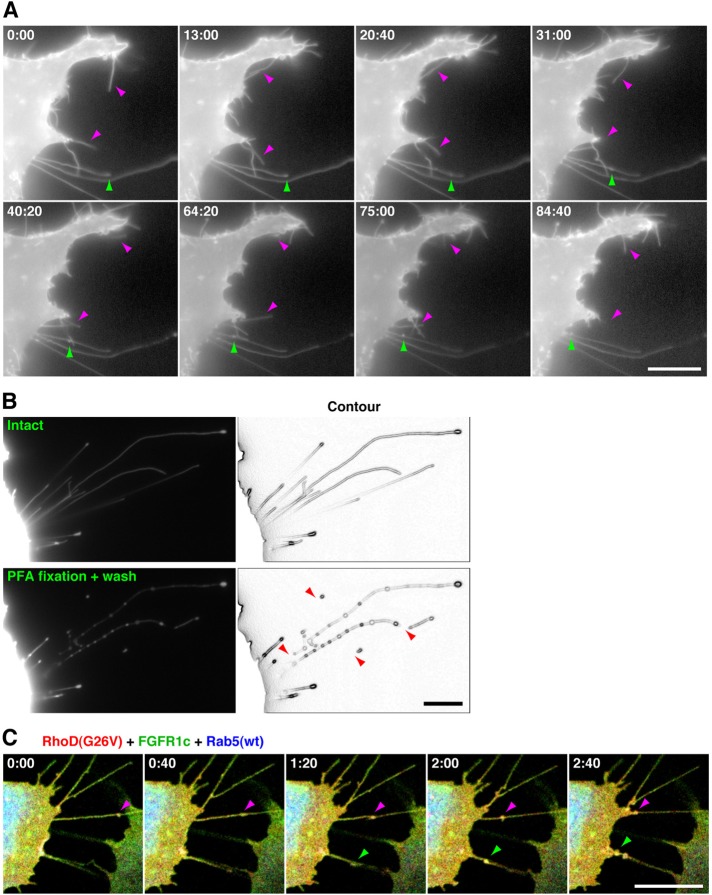
EGFP–RhoD(G26V)-transfected cells without PFA fixation showed two types of protrusions (Figure 2A). Some were relatively short (usually <20 μm) and undulating, which appeared to correspond to the protrusions in the fixed cells, and others were longer (some were >60 μm; Figure S1B) and almost straight. Time-lapse microscopy showed that the former protrusions moved flexibly and dynamically but that the latter were immotile (Figure 2A and Supplemental Movie S1). The latter contained fluorescent nodules or punctae. Although the nodules moved in both anterograde and retrograde directions, most of them eventually moved toward the cell body (Movies S1 and S2). The velocity of movement of the nodules varied, and each nodule occasionally changed velocity during movement (Movie S2). Some never or hardly moved during observation. We assessed whether the former shorter protrusions elongated to form the latter longer ones. However, the shorter protrusions barely elongated during the observation of EGFP fluorescence (see Discussion).
FIGURE 2:
RhoD generates two types of thin protrusions detected in unfixed cells. (A) Time-lapse images of two types of RhoD-induced protrusions. The 10T1/2 cells transfected with EGFP–RhoD(G26V) were analyzed by time-lapse imaging at the time indicated (min:s). Magenta arrowheads indicate shorter protrusions moving flexibly and dynamically. Green arrowheads point to a nodule moving in a retrograde direction through a longer immotile protrusion. (B) Breakdown of RhoD-induced protrusions by PFA fixation and washing. The 10T1/2 cells transfected with EGFP–RhoD(G26V) were observed before (top) and after (bottom) 15-min PFA fixation and washing with PBS. Arrowheads indicate collapsed or fragmented longer protrusions. (C) Time-lapse images of FGFR1c- and Rab5-containing nodules moving through the RhoD(G26V)-induced protrusions. The 10T1/2 cells cotransfected with mOrange2–RhoD(G26V) (red), FGFR1c–EGFP (green), and Cerulean–Rab5(wt) (blue) were analyzed by time-lapse imaging at the time indicated. Arrowheads point to FGFR1c- and Rab5-containing nodules moving through protrusions. Scale bars: 10 μm.
When the EGFP–RhoD(G26V)-transfected cells were fixed with PFA and washed to be processed for immunofluorescence microscopy, the longer protrusions were particularly collapsed or fragmented into shorter protrusions (Figure 2B). Thus the protrusions are sensitive to physical and chemical stresses. At least some of the shorter protrusions detected in the fixed cells (Figures 1 and S1B) were therefore artifactually generated as a result of fixation and washing. The properties of longer protrusions, that is, their dimension, inclusion of nodules or punctae moving in both directions, and sensitivity to fixation, are reminiscent of cytonemes (Ramírez-Weber and Kornberg, 1999; Hsiung et al., 2005; Roy et al., 2011).
RhoD-induced protrusions are extended toward FGFs and transmit FGF receptors
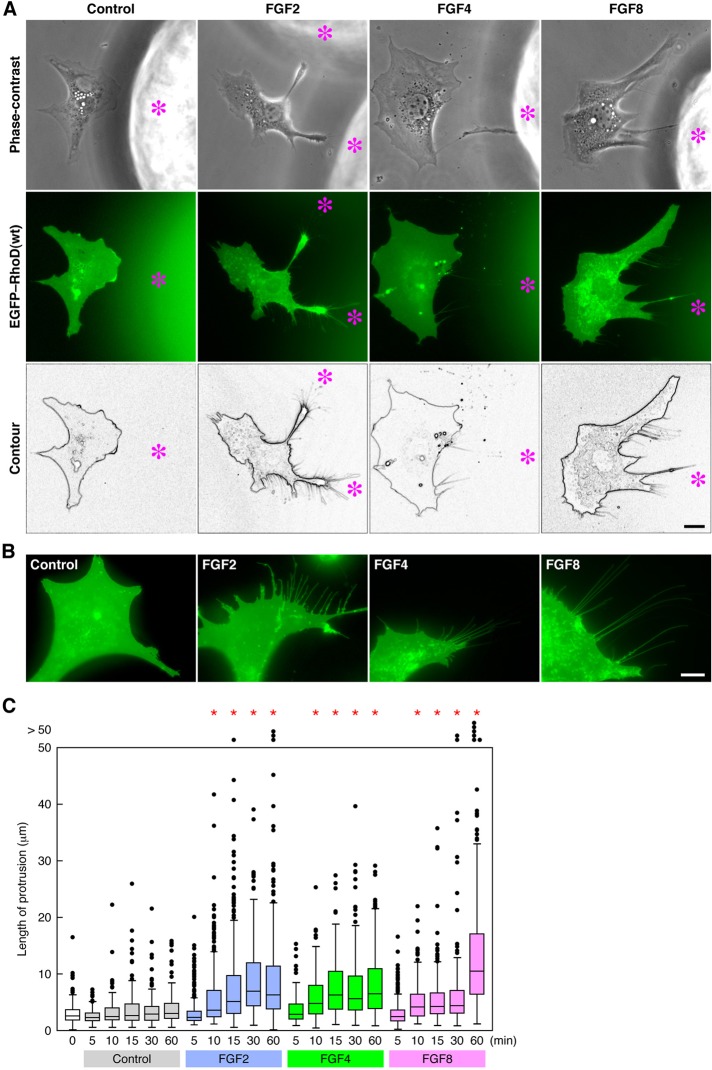
The RhoD(G26V)-induced protrusions were often extended toward neighboring cells (Figures 1A and S1C). These observations and those that the protrusions contained moving nodules suggest that the protrusions participate in intercellular communication, as do cytonemes. Because cytonemes are formed in response to morphogens/growth factors such as Dpp/BMP, Spitz/EGF, and Bnl/FGF in Drosophila (Hsiung et al., 2005; Roy et al., 2011), we examined whether RhoD-induced protrusions are extended toward FGFs. Heparin-conjugated acrylic beads soaked in FGF2, FGF4, or FGF8 were placed on dishes culturing 10T1/2 cells, which had been previously transfected with EGFP-tagged wild-type (wt) RhoD and subjected to serum starvation for 24 h. Although the cells close to the beads without FGF coating inefficiently extended protrusions, the cells near the beads coated with each FGF extended thin protrusions toward the beads by 3 h after the beads were added (Figure 3A). In addition, protrusions extending in a forward direction to the beads were much longer than those extending in a backward direction (Figure S2A).
FIGURE 3:
RhoD(wt)-transfected cells extend thin protrusions by FGF stimulation. (A) Protrusion extension of RhoD(wt)-transfected cells toward FGF-coated beads. FGF2/4/8-coated heparin–acrylic beads were placed for ∼3 h in the culture of 10T1/2 cells transfected with EGFP–RhoD(wt) and then serum-starved. Asterisks indicate the position of FGF-coated beads. (B) Protrusion formation of RhoD(wt)-transfected cells stimulated with FGFs. EGFP–RhoD(wt)-transfected and serum-starved 10T1/2 cells were stimulated for > 60 min with FGF2/4/8. Scale bars: 10 μm. (C) The length of protrusions induced by FGF stimulation in RhoD(wt)-transfected cells. EGFP–RhoD(wt)-transfected and serum-starved 10T1/2 cells were stimulated with FGF2/4/8 for the time indicated. The length is shown as box-and-whisker plots with boxes and whiskers encompassing 75th/25th and 95th/5th percentiles, respectively. Dots out of the frame represent the number (but not the length) of >50 μm of protrusions. n > 110. *, p < 0.0001 by t test compared with the control.
To determine whether FGFs diffused from the beads participate in the protrusion extension, we then added FGF2, FGF4, or FGF8 to the culture medium of EGFP–RhoD(wt)-transfected and serum-starved 10T1/2 cells. Although we tried to record a time-lapse image of protrusion elongation, the protrusions scarcely elongated during the observation of EGFP fluorescence (see Discussion). Thus we measured the length of thin protrusions at fixed time points after the FGF stimulation. The cells stimulated with each FGF started to extend thin and long protrusions 10 min after the stimulation, and the protrusions elongated or were maintained for at least 60 min (Figure 3, B and C). These results imply that FGFs diffused from the beads or added to the medium are responsible for the protrusion formation.
Mammalian FGFs bind and activate alternatively spliced forms of four FGF receptors (FGFR1–4). Among them, FGF2 and FGF4 exert mitogenic activity through FGFR1c (Ornitz et al., 1996). When we coexpressed mOrange2-tagged RhoD(G26V), C-terminally EGFP-tagged FGFR1c, and Cerulean-tagged wild-type Rab5 in 10T1/2 cells, FGFR1c and Rab5, which is localized to the early endosome membranes (Zerial and McBride, 2001), were associated with the nodules in the RhoD-formed protrusions (Figures 2C and S1D and Movie S3). The nodules moved through the protrusions toward the cell body. Collectively these results suggest that cells extend protrusions toward FGF-sourcing cells through activation of RhoD and that the protrusions transmit FGFs via FGFRs to the cell body. At least some of these FGFs–FGFRs might be transported in association with early endosome-like vesicles. Therefore these protrusions are likely to be involved in intercellular communication and morphologically and functionally equivalent to cytonemes.
RhoD is activated by FGF signaling and required for FGF signaling–mediated protrusion formation
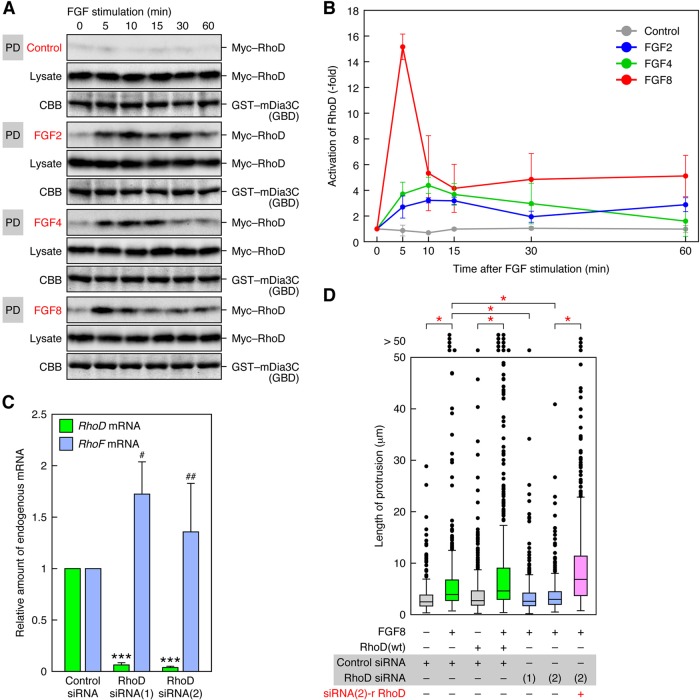
Because the RhoD(wt)-expressing cells extended protrusions toward FGFs, and FGF stimulation facilitated protrusion formation in the cells, we addressed whether RhoD is activated by FGFs. The 10T1/2 cells were transfected with Myc-tagged wild-type RhoD, serum-starved, and stimulated with FGFs. Activation of RhoD was analyzed by a pulldown assay with glutathione S-transferase (GST)-tagged GTPase-binding domain (GBD) of mDia3C (see Figure 6B, discussed later in this article), which is a target protein of RhoD (Gasman et al., 2003). Any of FGF2, FGF4, and FGF8 activated RhoD within 5 min after stimulation, and the activation was sustained for >30 min (Figure 4, A and B). The peak time of RhoD activation (5–15 min) may reflect the initiation time of protrusion formation (∼10 min; see Figure 3C). Because FGF8 activated RhoD more strongly and formed longer protrusions at 60 min than did FGF2 and FGF8 (see Figure 3C), there might be a correlation between the strength of RhoD activation and the length of the protrusions.
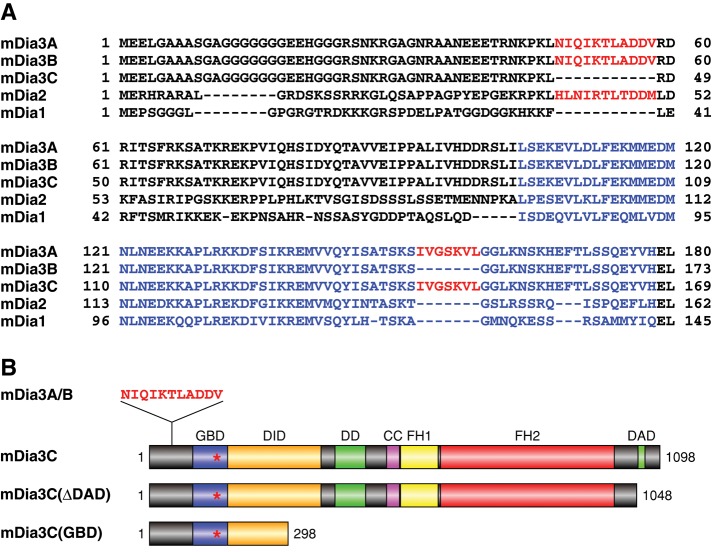
FIGURE 6:
Mouse mDia3 isoforms generated by alternative splicing. (A) Alignment of N-terminal amino acid sequences of mDia3A/B/C in comparison with those of mDia1 and mDia2. The sequences encoded by two alternative cassette exons are shown in red. The sequences of GBD are shown in blue. (B) Functional domains of mDia3 isoforms and deletion mutants of mDia3C. Red asterisks indicate the position of amino acid sequence encoded by the second cassette exon. DID, diaphanous inhibitory domain; DD, dimerization domain; CC, coiled-coil domain; FH1 and FH2, formin homology domains 1 and 2, respectively.
FIGURE 4:
RhoD is activated by FGF stimulation and required for FGF-induced protrusion formation. (A) Activation of RhoD by FGF stimulation. Activation of RhoD was analyzed by a pulldown assay with GST–mDia3C(GBD) and lysates of Myc–RhoD(wt)-transfected 10T1/2 cells stimulated with FGF2/4/8. Myc–RhoD was detected by immunoblotting with the anti-Myc mAb. (B) The degree of RhoD activation by FGF stimulation in the analysis of (A). The values are mean ± SD of three independent experiments. (C) Specific suppression of RhoD expression by RhoD siRNAs. The 10T1/2 cells were transfected with RhoD Stealth siRNA(1) or (2). The levels of endogenous RhoD and RhoF mRNAs were analyzed 48 h after the transfection by real-time PCR. The values are normalized with β-actin mRNA levels. ***, p < 0.001; #, p = 0.095; ##, p = 0.36 (not significant) by t test compared with control siRNA transfection. (D) Requirement of RhoD for FGF8-induced protrusion formation. The 10T1/2 cells were first transfected with Stealth siRNAs and 48 h later with EGFP–RhoD(wt) or siRNA(2)-resistant EGFP–RhoD. The cells were stimulated for ∼60 min with FGF8 48 h after the RhoD transfection, and the protrusion length was analyzed. n > 390. *, p < 0.0001 by t test.
To determine whether RhoD is essential for FGF-induced protrusion formation, we knocked down RhoD expression by RNA interference (RNAi). Either expression of two distinct Stealth small interfering RNAs (siRNAs) of RhoD in 10T1/2 cells prominently reduced the endogenous RhoD mRNA level, whereas neither significantly affected the endogenous RhoF mRNA level (Figure 4C). Thus both these siRNAs efficiently knock down RhoD, but neither has an off-target effect on RhoF, which is most closely related to RhoD among small GTPases. As shown above, FGF8 stimulation highly promoted protrusion formation if RhoD(wt) was exogenously expressed (Figure 4D). The stimulation still facilitated protrusion formation, however, even without exogenous RhoD(wt) expression (Figure 4D). When the RhoD siRNAs were expressed in the presence of FGF8 stimulation, the protrusion formation was reduced almost to levels comparable with those seen without FGF8 stimulation and exogenous RhoD(wt) expression (Figure 4D). If an siRNA-resistant RhoD (Figure S2B) was expressed, the cells were released from the siRNA-induced suppression of the protrusion formation and formed longer protrusions (Figure 4D). In addition, expression of the dominant-negative RhoD(T31K) also interfered with the FGF8-induced protrusion formation (Figure S2C). These results indicate that endogenous RhoD is indispensable for FGF8-induced protrusion formation.
N-WASP is not required for RhoD-induced protrusion formation
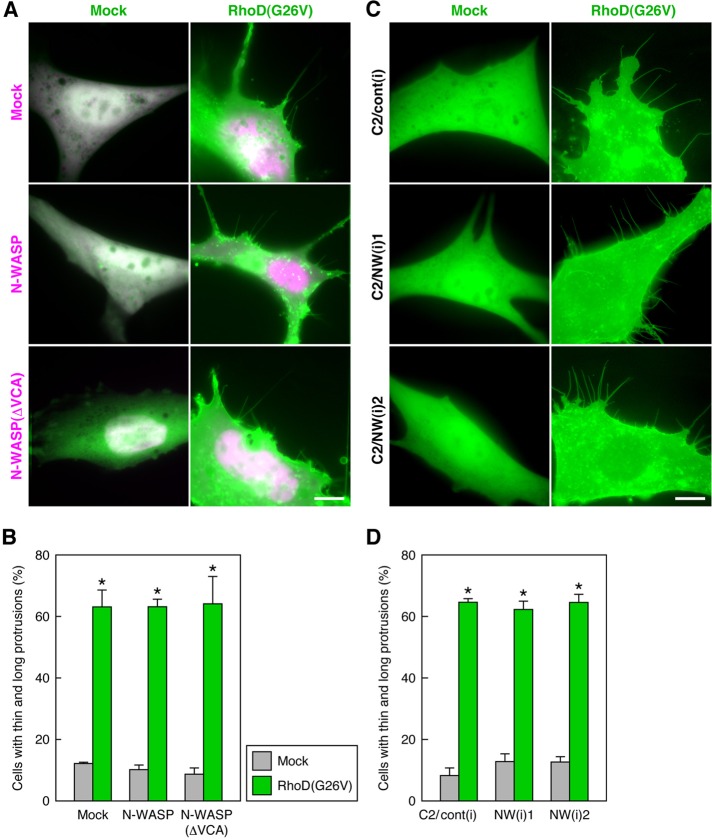
Because the RhoD-induced protrusions contain actin filaments, actin filament formation seems to be a prerequisite for protrusion formation. Intracellular actin filament formation necessitates factors that nucleate and elongate actin filaments (Dominguez, 2009; Campellone and Welch, 2010; Chesarone et al., 2010). N-WASP activated by Cdc42 and PIP2 or SH3 domain–containing proteins activates the Arp2/3 complex to form filopodia (Takenawa and Suetsugu, 2007; Campellone and Welch, 2010). N-WASP, in collaboration with nebulin, also nucleates unbranched actin filaments in skeletal muscle myofibrils (Takano et al., 2010). Thus we examined whether N-WASP was involved in the formation of RhoD-induced protrusions. Neither expression of wild-type N-WASP nor that of the dominant-negative N-WASP(ΔVCA) (Oikawa et al., 2008) affected the RhoD(G26V)-induced protrusion formation in 10T1/2 cells (Figure 5, A and B). Furthermore, the protrusion formation by the expression of RhoD(G26V) was not hindered in mouse C2C12 myoblast clones stably expressing siRNAs of N-WASP (Takano et al., 2010; Figure 5, C and D). Accordingly, N-WASP does not participate in the RhoD-induced protrusion formation, probably because it is not involved in actin filament formation in the protrusions.
FIGURE 5:
N-WASP does not participate in RhoD-induced protrusion formation. (A) Neither N-WASP nor a dominant-negative N-WASP affects RhoD-induced protrusion formation. The 10T1/2 cells were cotransfected with mCherry–RhoD(G26V) (green) and EGFP–N-WASP or EGFP–N-WASP(ΔVCA) (magenta) and analyzed 48 h after the transfection. (B) Ratio of the cells extending thin and long protrusions in the analysis of (A). The values are mean ± SD of three independent experiments. *, p < 0.001 by t test compared with respective mock transfection (control). (C) Knockdown of N-WASP does not interfere with RhoD-induced protrusion formation. C2C12 myoblast clones stably expressing control and N-WASP siRNAs were transfected with EGFP–RhoD(G26V) and analyzed 48 h after the transfection. Scale bars: 10 μm. (D) Ratio of the cells extending thin and long protrusions in the analysis of (C). The values are mean ± SD of three independent experiments. *, p < 0.001 by t test compared with respective mock transfection (control).
RhoD binds to mDia3C, which is associated with the RhoD-induced protrusions
Formins exert their functions in both actin nucleation and elongation (Chesarone et al., 2010). Members of the Dia subfamily of mammalian formins bind to and are activated by several Rho family proteins. mDia2, which is activated by Cdc42 or RhoF, and mDia1, which is activated by RhoF, induce filopodial protrusions (Peng et al., 2003; Pellegrin and Mellor, 2005; Goh et al., 2011). On the other hand, RhoD has been shown to specifically bind to mDia3C to regulate endosome movement by aligning early endosomes along actin filaments (Gasman et al., 2003). Thus we tried to clone mouse mDia3 cDNAs to assess whether any mDia proteins play essential roles in the formation of RhoD-induced protrusions. We obtained three alternatively spliced forms of mDia3 cDNAs by reverse transcriptase PCR (RT-PCR). The longest cDNA contained two alternative cassette exons, whereas the other two cDNAs contained one or the other of the two cassette exons (Figure 6A). The cDNAs containing only the first cassette exon and only the second one correspond to mDia3B and mDia3C, respectively (Gasman et al., 2003; Mellor, 2010). We designated the isoform coded by the cDNA containing both the cassette exons as mDia3A, which had not been registered in the DNA databases. The sequence of the second cassette exon codes for a part of the GBD.
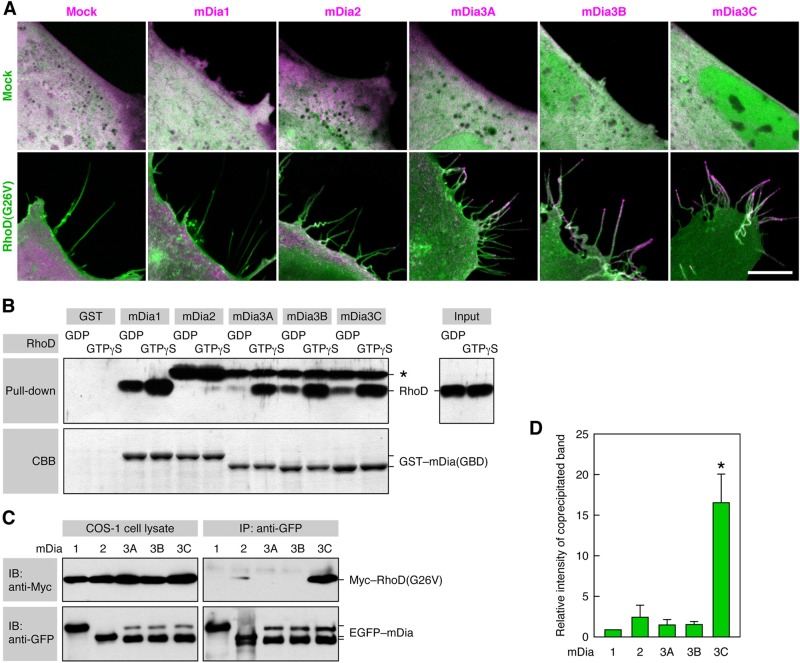
We analyzed the localization of EGFP-tagged mDia proteins in Cerulean–RhoD(G26V)-transfected 10T1/2 cells. All of the mDia1, mDia2, and mDia3A/B/C were diffusely distributed throughout the cytoplasm without RhoD(G26V) expression (Figure 7A). When mDia1 was coexpressed with RhoD(G26V), it was present in the cell body but not in the RhoD(G26V)-induced protrusions (Figure 7A). On the other hand, although mDia2 was predominantly distributed in the cell body, it was also located to the tips of some, but not all, of the protrusions. By contrast, mDia3A/B/C were highly concentrated at the tips or stems of the protrusions and less prominently present in the cell body (Figure 7A). Endogenous mDia3 was also localized in the RhoD(G26V)-induced protrusions, as detected by the staining of PFA-fixed cells with an anti-mDia3 polyclonal antibody (pAb; Figure S2D).
FIGURE 7:
mDia3C is localized to the RhoD-induced protrusions and binds to RhoD. (A) Distribution of mDia proteins in RhoD-transfected cells. The 10T1/2 cells were cotransfected with Cerulean–RhoD(G26V) (green) and EGFP–mDia proteins (magenta). Scale bar: 10 μm. (B) Binding of active RhoD to mDia1 and mDia3A/B/C in a pulldown assay. The binding of GDP- or GTPγS-loaded RhoD to GST–mDia(GBD) was analyzed by immunoblotting with the anti-RhoD pAb. *, an unidentified protein present in Sf21 cell lysates that was detected by the anti-RhoD pAb. (C) Specific binding of RhoD(G26V) to mDia3C in a coimmunoprecipitation assay. COS-1 cells were cotransfected with Myc–RhoD(G26V) and EGFP–mDia proteins, cell lysates were immunoprecipitated with anti-GFP pAb, and coprecipitated Myc–RhoD(G26V) was detected with the anti-Myc mAb. (D) Relative intensity of coprecipitated Myc–RhoD(G26V) band in the analysis of (C). The values are mean ± SD of three independent experiments. *, p < 0.05 by t test compared with mDia1.
A pulldown assay with GST-tagged GBD of mDia proteins showed that GTPγS-loaded active RhoD strongly bound to mDia1 and mDia3A/B/C but not to mDia2 (Figure 7B). GDP-loaded inactive RhoD only marginally or much less strongly bound to these mDia proteins. On the other hand, when EGFP-tagged mDia proteins were immunoprecipitated with an anti-GFP pAb from lysates of COS-1 cells cotransfected with Myc–RhoD(G26V) and EGFP–mDia proteins, RhoD(G26V) was markedly coprecipitated with mDia3C but only marginally with mDia1, mDia2, mDia3A, or mDia3B (Figure 7, C and D). These results imply that activated RhoD can bind to mDia1 and mDia3 isoforms in vitro but that it is specifically associated with mDia3C in vivo. Indeed, mDia3C was exclusively concentrated in the RhoD-induced protrusions but not in the cell body. In contrast, mDia1 existed solely in certain areas of the cell body in which RhoD may not be present.
RhoD-activated mDia3C is essential for protrusion formation
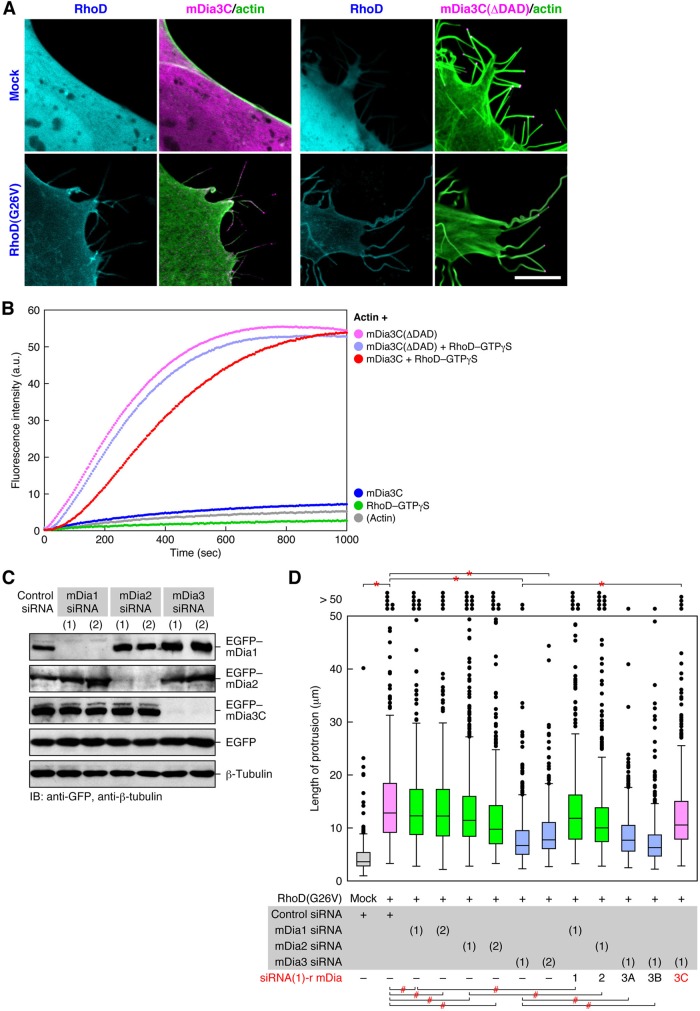
We also addressed whether RhoD-bound mDia3C is involved in the thin and long protrusion formation. As shown above, 10T1/2 cells transfected with wild-type mDia3C in the absence of activated RhoD barely formed such protrusions, whereas the cells coexpressing mDia3C and RhoD(G26V) efficiently formed the protrusions containing actin filaments (Figures 7A and 8A). When mDia3C(ΔDAD), a constitutively active form lacking Dia autoregulatory domain (DAD; Figure 6B; Block et al., 2008), was transfected to 10T1/2 cells, the cells formed protrusions regardless of whether RhoD(G26V) was cotransfected (Figure 8A). These protrusions contained actin filament bundles thicker than those in the RhoD(G26V)-induced protrusions. mDia3C(ΔDAD) was restricted to the tips of the protrusions, although mDia3C was associated with both tips and stems of the protrusions. These results suggest that regulated activation of mDia3C mediated by RhoD leads to thin actin filament bundle formation generating the thin and long protrusions but that constitutively activated mDia3C gives rise to thicker actin filament bundles.
FIGURE 8:
Activated mDia3C is essential for actin filament formation–mediated protrusion formation. (A) A constitutively active mDia3C(ΔDAD) forms protrusions independently of RhoD. The 10T1/2 cells were cotransfected with EGFP–mDia3C or EGFP–mDia3C(ΔDAD) (magenta) and mOrange2-Lifeact (green) together with or without Cerulean–RhoD(G26V) (cyan). Scale bar: 10 μm. (B) Facilitation of actin polymerization by mDia3C together with RhoD or solely by mDia3C(ΔDAD). Actin polymerization was analyzed by a fluorometric actin polymerization assay. a.u., arbitrary units. (C) Specific knockdown of mDia isoforms by respective mDia siRNAs. The 10T1/2 cells were first transfected with mDia Stealth siRNAs and 48 h later with EGFP–mDia1, mDia2, or mDia3C. The levels of proteins at 48 h after the second transfection were analyzed by immunoblotting with anti-GFP pAb. (D) Requirement of mDia3C but not mDia1, mDia2, or mDia3A/B for the RhoD-induced protrusion formation. The 10T1/2 cells were first transfected with mDia Stealth siRNAs and 48 h later with mCherry–RhoD(wt) and siRNA(1)-resistant EGFP–mDia1, mDia2, or mDia3A/B/C. The protrusion length was analyzed 48 h after the second transfection. n > 420. *, p < 0.0001, #, p > 0.01 (not significant) by t test.
We therefore analyzed whether mDia3C activated by RhoD promotes actin polymerization by a fluorometric actin polymerization assay. Neither mDia3C nor RhoD–GTPγS solely facilitated actin polymerization (Figure 8B). By contrast, mDia3C, together with RhoD–GTPγS, prominently accelerated actin polymerization (Figure 8B). Accordingly, mDia3C activated by RhoD is likely to form actin filaments responsible for protrusion formation. On the other hand, mDia3C(ΔDAD) alone promoted actin polymerization remarkably, and addition of RhoD–GTPγS no longer increased the enhancement by mDia3C(ΔDAD) (Figure 8B). These results are consistent with the above results showing the expression of mDia3C(ΔDAD) generated protrusions containing thick actin filament bundles irrespective of the presence of RhoD(G26V).
Next we asked whether mDia3C among mDia proteins plays a primary role in the RhoD-induced protrusion formation by applying RNAi. Expression of Stealth siRNAs of mDia1, mDia2, and mDia3 in 10T1/2 cells specifically reduced mDia1, mDia2, and mDia3 protein levels, respectively, but did not have off-target effects on the other mDia isoforms (Figure 8C). Expression of the siRNAs of mDia1 or mDia2 together with RhoD(G26V) in 10T1/2 cells did not significantly affect the RhoD-induced protrusion formation (Figure 8D). In contrast, expression of mDia3 siRNAs reduced the length of thin protrusions (Figure 8D). When an siRNA-resistant mDia1, mDia2, mDia3A, or mDia3B (Figure S2E) was expressed together with each corresponding siRNA, the protrusion length was not affected (Figure 8D). By contrast, expression of an siRNA-resistant mDia3C (Figure S2E) rescued the cells from the mDia3 siRNA-induced reduction of protrusion length (Figure 8D). Consequently, mDia3C is responsible for the RhoD-induced protrusion formation, but mDia1, mDia2, mDia3A, or mDia3B is not. We conclude that these data, taken together, indicate that activated RhoD forms the protrusions by directly binding to activate mDia3C, which induces actin polymerization in the protrusions.
DISCUSSION
Several Rho family proteins, including Cdc42, RhoQ, RhoJ, and RhoF, bring about filopodial cell protrusions (Abe et al., 2002; Mattila and Lappalainen, 2008; Faix et al., 2009; Mellor, 2010). These protrusions are relatively thick and stable and resistant to fixation. In contrast, cytonemes are thinner and sensitive to fixation (Ramírez-Weber and Kornberg, 1999). Although molecular mechanisms of cytoneme formation have been obscure, we have shown here that RhoD-mediated signaling is responsible for the formation of cytoneme-like protrusions. Live imaging of unfixed EGFP–RhoD(G26V)-transfected cells detected two types of protrusions, shorter motile and longer immotile protrusions. The shorter protrusions barely elongated during our observation of EGFP fluorescence. However, we cannot exclude the possibility that the shorter protrusions elongate to become the longer ones. The longer protrusions were sensitive to physical and chemical stresses, such as washing and PFA fixation, and collapsed or fragmented. Thus both types of the protrusions might be unable to elongate under UV illumination applied to detect EGFP fluorescence. The protrusions induced by FGF stimulation in RhoD(wt)-transfected cells also hardly elongated during the observation of EGFP fluorescence. This result favors the above notion that the protrusions are sensitive to physical and chemical stresses. Accordingly, live imaging of unfixed cells is required to detect and analyze such structures.
Some of the RhoD-induced longer protrusions were >60 µm in length. They were much longer and thinner than filopodia induced by Cdc42 subfamily members or RhoF. The dimensional difference between the RhoD-induced protrusions and filopodia suggest that they are generated by distinct mechanisms and exert distinct functions. Although the length of Drosophila cytonemes is variable, some cytonemes are as long as 80 μm (Hsiung et al., 2005; Roy et al., 2011). Thus the length of RhoD-induced protrusions is comparable with that of Drosophila cytonemes, despite the difference between mammalian cell culture system and in developing Drosophila tissues. Wing imaginal disk cells, eye disk cells, and tracheal cells extend cytonemes toward Dpp/BMP-, Spitz/EGF-, and Bnl/FGF-producing cells, respectively. These cytonemes segregate receptors of each morphogen/growth factor (Roy et al., 2011). The receptors move in both anterograde and retrograde directions in cytonemes (Hsiung et al., 2005). RhoD(wt)-expressing 10T1/2 cells extended thin and long protrusions toward FGF2/4/8-coated beads or in response to stimulation with each of the FGFs separately. The RhoD(G26V)-induced protrusions also contained nodules moving in both directions, and most of the nodules eventually moved toward the cell body. FGFR1c coexpressed with RhoD(G26V) was associated with the nodules and moved through the protrusions. Therefore RhoD-induced protrusions are likely to be morphologically and functionally equivalent to cytonemes. Although there is no RhoD orthologue in Drosophila, some Rho family protein, such as Rho-1 or Rac-1, which is most closely related to mammalian RhoD, might be involved in cytoneme formation in Drosophila.
Why is FGFR transported to the cell body through cytonemes or the RhoD-induced protrusions? Canonically, plasma membrane–associated receptor tyrosine kinases activated by their growth factor ligands transduce signals into the Ras–ERK, phosphatidylinositol 3-kinase (PI3K)–Akt, or phospholipase C-γ (PLCγ) signaling, which is associated with the plasma membrane or the cytosol (Lemmon and Schlessinger, 2010). On the other hand, several growth factors and their receptor complexes, such as EGFs–EGFRs and FGFs–FGFRs, are also translocated to the nucleus (Johnson et al., 2004; Bryant and Stow, 2005; Carpenter and Liao, 2009; Dunham-Ems et al., 2009). Following activation by FGF, plasma membrane–associated FGFR is internalized into early endosomal compartments, which is an essential early step in nuclear translocation of FGFR. FGF and FGFR are then released from the early endosomes to the cytosol, although it is unknown whether they are linked or separated at this time. They interact with nuclear import machinery, including importin β, in the cytosol and are translocated to the nucleus. In the nucleus, FGFs–FGFRs interact with transcription regulators, such as CREB-binding protein and ribosomal S6 kinase 1, and participate in transcription of genes involved in cell growth, differentiation, or development. However, FGFs–FGFRs should be unable to induce these nuclear events when they are on cytonemes or the RhoD-induced protrusions. This seems to be the reason why FGFR (as well as EGFR) is transported to the cell body through the protrusions (Figure 9).
FIGURE 9:
Postulated mechanisms of the FGF–RhoD–mDia3C-induced protrusion formation and the transport of FGFRs through the protrusions. (A) RhoD is inactive without FGF stimulation. (B) If FGF, even at a low concentration, binds to its receptors, a GEF for RhoD is activated by FGFR signaling. The GEF then activates RhoD. (C) Activated RhoD binds to activate mDia3C. (D) Activated mDia3C nucleates and elongates actin filaments to form protrusions. (E) Cells extend the protrusions toward high concentrations of FGF or the source of FGF. Higher concentrations of FGF can efficiently bind to FGFRs. FGF–FGFRs are transported through the protrusions either on the plasma membrane or on the early endosome-like vesicle membrane toward the cell body. (F) FGF–FGFRs transported to the cell body activates the canonical Ras–ERK, PI3K–Akt, or PLCγ signaling. FGF–FGFRs are also translocated to the nucleus through early endosomal compartments and participate in transcription of genes involved in cell growth, differentiation, or development.
FGF2/4/8 activated RhoD, and activated RhoD was required for the FGF-induced protrusion formation. Drosophila has three FGF genes, including Bnl, whereas mouse or human has 22 FGF genes (Itoh and Ornitz, 2011). Mammalian FGF2/4/8 function in a paracrine manner and act as morphogens or growth/differentiation factors in developing embryos. Analyses of gene-knockout mice have shown that the FGFs are involved in the development of particular tissues, such as the limb (Sun et al., 2002; Beenken and Mohammadi, 2009; Itoh and Ornitz, 2011). Therefore cells in these tissues of developing embryos might also generate the cytoneme-like protrusions induced by these FGFs through the activation of RhoD.
Activated RhoD bound to mDia1 and mDia3A/B/C but not to mDia2 in vitro, whereas activated RhoD bound only to mDia3C in vivo. Although mDia1 and mDia2 were predominantly present in the cell body even when RhoD(G26V) was expressed, exogenously expressed mDia3A/B/C were concentrated in the RhoD-induced protrusions. These results may imply that mDia1 is not colocalized with RhoD in the cell body, even if it can bind to RhoD in vitro. On the other hand, although mDia3A/B have the potential to bind to RhoD, endogenous mDia3A/B might neither be associated with the protrusions nor be involved in the protrusion formation. Nevertheless, exogenously overexpressed mDia3A/B might be ectopically localized to the protrusions and displace endogenous mDia3C. The siRNA-resistant mDia3C, but not the siRNA-resistant mDia3A/B, rescued the cells from the interference with the RhoD(G26V)-induced protrusion formation by the mDia3 siRNA. These results imply that activated RhoD forms the protrusions through activating mDia3C but not mDia3A/B. Indeed, mDia3C (hDia2C) has been reported to be a specific effector of RhoD (Gasman et al., 2003). Activated RhoD binds to mDia3C and associates with early endosomes in the cell body. They induce an alignment of early endosomes along actin filaments and markedly reduce the endosome motility by depending on Src tyrosine kinase activity. Thus RhoD is likely to utilize mDia3C for both the protrusion formation and the regulation of endosome movement through actin filaments. mDia3 is also localized to the kinetochores and involved in kinetochore–microtubule attachment required for correct chromosome alignment and segregation, which is regulated by Cdc42 or Aurora B (Yasuda et al., 2004; Cheng et al., 2011). Although it remains to be determined which mDia3 isoform(s) is responsible for this function, each of the mDia3 isoforms is likely to be regulated by specific signaling molecules and to exert specific functions.
Both mDia3C, together with active RhoD, and the constitutively active mDia3C(ΔDAD) prominently enhanced actin polymerization. In addition, mDia3C(ΔDAD) formed protrusions regardless of the expression of RhoD(G26V). mDia3C(ΔDAD) was confined to the tips of the protrusions. These results may indicate that actin nucleation/elongation function of mDia3C, which is activated by RhoD, is essential for the protrusion formation. However, the protrusions formed by mDia3C(ΔDAD) were thicker and contained actin filament bundles thicker than those in the RhoD(G26V)-induced protrusions. Accordingly, although mDia3C is responsible for the thin actin filament bundle formation in the RhoD-induced protrusions, regulated activation of mDia3C mediated by RhoD may be required for the thin and long protrusion formation. Alternatively, cooperation between mDia3C and other effector proteins of RhoD might be necessary for the thin and long protrusion formation. The thicker protrusions are reminiscent of those formed by mDia2(ΔDAD) expression (Block et al., 2008). Although the mDia2(ΔDAD)-induced protrusions are club-shaped, however, mDia3C(ΔDAD)-induced protrusions were rod-shaped or tapered and appeared to be longer than the mDia2(ΔDAD)-induced protrusions. The difference might be ascribed to different nucleation/elongation abilities exhibited by these mDia proteins.
Figure 9 summarizes postulated mechanisms of the protrusion formation by FGF-activated RhoD, elongation of the protrusions toward the source of FGF, and the transport of FGFRs through the protrusions. This study has substantiated the molecular mechanisms of cytoneme or cytoneme-like protrusion formation. Investigation of whether RhoD and mDia3C play central roles in the formation of similar protrusions distinct from filopodia in other cell types (Galkina et al., 2001; Gupta and DeFranco, 2003; Sherer et al., 2007; Fifadara et al., 2010) may generalize the findings in this study. If inactivating mutations occur in the RhoD, RhoD GEF (guanine nucleotide exchange factor for RhoD), or mDia3 gene, cytonemes or cytoneme-like protrusions may not be formed. Therefore it is intriguing to determine whether these mutations lead to developmental abnormalities due to a lack of cytoneme-mediated intercellular communication.
MATERIALS AND METHODS
Cell culture
Mouse C3H/10T1/2 (10T1/2) mesenchymal progenitor cells (Reznikoff et al., 1973) were cultured as described previously (Watanabe-Takano et al., 2010). Mouse skeletal muscle C2C12 myoblast clones stably transfected with N-WASP siRNA1, siRNA2, and control siRNA expression vectors (C2/NW(i)1, C2/NW(i)2, and C2/cont(i), respectively) were established and cultured as described previously (Takano et al., 2010).
cDNA cloning and plasmid construction
Mouse mDia2 cDNA (mKIAA4117) was obtained from Kazusa DNA Research Institute. Mouse mDia3 cDNA fragments were cloned from mouse brain mRNA by RT-PCR using Omniscript Reverse Transcriptase and ProofStart DNA Polymerase (Qiagen, Valencia, CA). Three primer sets used were:
5´-AGAGAAAGATGGAGGAGCTCG-3´ and 5´-AGTTCAGCTGCAGCAGTGATG-3´;
5´-CTGCTATTTGCATTGTTGGGG-3´ and 5´-GAAGGAAATGTCAAGGCCAGT-3´;
5´-GGTCCAAGATTGAGCCTAAAG-3´ and 5´-CTTAAATGTCTCCATTCTCCC-3´.
Three distinct cDNA fragments encoding the N-terminal portion of mDia3A, mDia3B, and mDia3C were obtained with primer set 1. Primer sets 2 and 3 generated cDNA fragments encoding the common central and C-terminal portions, respectively. Three cDNA fragments obtained with the respective primer sets were digested with XbaI and BglII and ligated into single cDNAs encoding full-length mDia3A, mDia3B, and mDia3C. The sequence of mouse mDia3A cDNA has been deposited to the DDBJ/GenBank/EMBL database with the accession number AB713187.
siRNA-resistant (r) RhoD, mDia1, mDia2, mDia3A, mDia3B, and mDia3C cDNAs were constructed by introducing point mutations into wild-type cDNAs by PCR. The primer sets used were:
siRNA(2)-r RhoD: 5´-GTTACAACGCCACTCTGCAGATGAAGG-3´ and 5´-GCTCGAACACTGTGGGACTGTAGCTCTC-3´;
siRNA(1)-r mDia1: 5´-CTTCCTGTGCAAGCTTCGAGACACCAAGTC-3´ and 5´-GAGATATTGAAGCCGAAAGCACCAGCATTC-3´;
siRNA(1)-r mDia2: 5´-CAGAACCTTATAAAGCACCTTCCTGATGAG-3´ and 5´-GATCATTGACTCTGACAACTGTGTTTCATC-3´;
siRNA(1)-r mDia3A/B/C: 5´-CCGCGAACAGTATGACAAACTGTCCACC-3´ and 5´-GCGTTCTGTGTAAAGCTCATCATCTTTTCC-3´.
A cDNA encoding mDia3C(ΔDAD) was constructed by substituting the codon GAG for Glu-1049 with the termination codon TAG by PCR with Phusion High-Fidelity DNA Polymerase (New England BioLabs, Ipswich, MA). The cDNA of mouse RhoD(wt), RhoD(G26V) (Tsubakimoto et al., 1999), Rab5b(wt) (Sun et al., 2003), mDia2, mDia3A, mDia3B, mDia3C, mDia3C(ΔDAD), or human Cdc42(G12V) (Abe et al., 2002) was inserted into pEGFP-C1, pmCherry-C1, pmOrange2-C1, or pCerulean-C1 vector (Clontech, Mountain View, CA) in frame with each fluorescent protein tag.
Mouse FGFR1c cDNA fragments were cloned from mRNA of mouse embryonic stem cell line R1 by RT-PCR. Two primers sets used were:
5´-ACGGATCCTTGTTACCAACCTCTAACCG-3´ and 5´-TGCCTAAGACCAGTCTGTCTCGTGGC-3´;
5´-TGGAGTCTCCGAATATGAGCTCCCTG-3´ and 5´-GGGGATCCGGCGCCGTTTGAGTCCACTG-3´.
Two cDNA fragments encoding the N- and C-terminal portions of FGFR1c were digested with BamHI and ligated into a single cDNA encoding full-length FGFR1c. The FGFR1c cDNA was inserted into the BglII–BamHI site of pEGFP-N2 vector in frame with the EGFP tag.
Oligo DNA of Lifeact, a 17–amino acid peptide of Abp140 (Riedl et al., 2008), was prepared by annealing complementary 56-base synthetic deoxyribonucleotides. It was inserted into the BamHI–HindIII site of pmOrange2-C1 vector in frame with the fluorescent protein tag.
Fluorescence microscopy and live imaging
The recombinant plasmids were transfected to 10T1/2 cells with GeneJammer Transfection Reagent (Stratagene, Agilent, Santa Clara, CA). The transfected cells were fixed with 4% PFA in phosphate-buffered saline (PBS) for 15 min and permeabilized with 0.2% Triton X-100 in PBS for 5 min. They were incubated with anti-vinculin mAb hVIN-1 (Sigma-Aldrich, St. Louis, MO), anti-fascin mAb (Merck Millipore, Billerica, MA), anti–β-tubulin mAb E7 (Developmental Studies Hybridoma Bank, University of Iowa, Iowa City, IA), or anti-mDia3 pAb (Santa Cruz Biotechnology, Santa Cruz, CA) and then incubated with Alexa Fluor 555–goat anti-mouse or rabbit immunoglobulin G or Alexa Fluor 546–phalloidin (Life Technologies, Carlsbad, CA). The specimens were observed with a Carl Zeiss (Oberkochen, Germany) confocal laser-scanning microscope LSM 410 or with an Olympus (Tokyo, Japan) confocal laser-scanning microscope FV-1000D.
For live imaging, cells were cultured on 35-mm glass base dishes and maintained at 37°C in 5% CO2 during observation. For stimulation of RhoD(wt)-transfected 10T1/2 cells with FGFs, the cells were cultured for 24 h in serum-free DMEM before addition of FGFs. The cells were stimulated with 100 ng/ml FGF2, FGF4, or FGF8 (Wako Pure Chemical Industries, Osaka, Japan) premixed with 15 μg/ml heparin sodium (Wako Pure Chemical Industries). For analysis of the protrusions extending toward FGF-coated beads, >100-μm-diameter heparin–acrylic beads (Sigma-Aldrich) were sorted out with Cell Strainer (BD Biosciences, Franklin Lakes, NJ). The beads were soaked in 100 ng/ml FGF and washed once with PBS. They were placed in the culture of RhoD(wt)-transfected and serum-starved 10T1/2 cells. These cells were observed with either the Olympus confocal laser-scanning microscope FV-1000D or a Carl Zeiss inverted microscope Axiovert 200 equipped with a CoolSNAP cf charge-coupled device camera (Photometrics, Tucson, AZ) operated with Openlab software (PerkinElmer, Waltham, MA). Time-lapse images were processed with ImageJ software.
RNAi
Stealth RNAi siRNAs (Life Technologies) were used to knock down mouse RhoD and mDia1/2/3. The target sequences of the siRNAs were:
RhoD siRNA(1): 5´-CCAAACAGCTTTGACAACGTCTCCA-3´ (nt 374–398 from the initiation codon);
RhoD siRNA(2): 5´-CCCACAGTGTTTGAGCGCTATAATG-3´ (nt 142–166);
mDia1 siRNA(1): 5´-GCTTCAATATCAGCTTCCTTTGTAA-3´ (nt 2942–2966);
mDia1 siRNA(2): 5´-GGTTGATCAAATGATTGATAAAACA-3´ (nt 1365–1389);
mDia2 siRNA(1): 5´-CAGAGTCCATGATTCAGAACTTAAT-3´ (nt 2186–2210);
mDia2 siRNA(2): 5´-AGAAGATTGAATTGGTTAAAGATCG-3´ (nt 1876–1900);
mDia3 siRNA(1): 5´-GAGCTTTACCCAGAATGCCAGAGAA-3´ (nt 2868–2892);
mDia3 siRNA(2): 5´-GAAAGCATTTATGAATAATAAGTTT-3´ (nt 738–762);
negative control siRNA: 5´-GAGTTCACCGAAAGTCCGAATCGAA-3´.
The siRNAs were mixed with Lipofectamine RNAiMAX Reagent (Life Technologies) in OPTI-MEM (Life Technologies) and transfected to 10T1/2 cells.
Real-time PCR
The amounts RhoD and RhoF mRNAs were quantitated by real-time PCR. The cDNAs were synthesized from cytoplasmic RNA with SuperScript VILO cDNA Synthesis Kit (Life Technologies). The primer sets used were:
RhoD: 5´-GAAGACGTCACTGATGATGGTCTTCG-3´ and 5´-AAGCAGAGGAGCAAGACATTGGCATC-3´;
RhoF: 5´-AGTGTGACAGTGGGTAACAAGGAG-3´ and 5´-TGATGAGGACGTTGTCGTAACTGG-3´.
The quantitation was conducted with Applied Biosystems 7300 Real-Time PCR System by using SYBR Green I Dye (Life Technologies).
Pulldown assay
Activation of RhoD by FGF stimulation was analyzed by a pulldown assay with the GST-tagged GBD of mDia3C. A cDNA fragment encoding the N-terminal region (aa 1–298) containing the GBD domain of mDia3C was fused in frame with the GST tag in pGEX-6P vector (GE Healthcare, Waukesha, WI). The GST–mDia3C(GBD) cDNA was amplified and inserted into pFastBac1 vector (Life Technologies). The recombinant GST–mDia3C(GBD) was expressed in Sf21 cells with Bac-to-Bac Baculovirus Expression System (Life Technologies). GST–mDia3C(GBD) was immobilized to glutathione Sepharose 4B (GE Healthcare) and affinity-purified. RhoD(wt) cDNA was fused in frame with the N-terminal Myc-tag in pEF-BOS/Myc vector and transfected to 10T1/2 cells. The transfected cells stimulated with FGFs after serum starvation were lysed, and the lysates were applied to a GST pulldown assay, as described previously (Takano et al., 2010). Bound Myc–RhoD was detected by immunoblotting with anti-Myc mAb Myc1-9E10 (American Type Culture Collection, Manassas, VA).
The recombinant GST–mDia2(GBD) (aa 1–307), GST–mDia3A(GBD) (aa 1–309), and GST–mDia3B(GBD) (aa 1–302) were expressed in Sf21 cells with Bac-to-Bac Baculovirus Expression System as GST–mDia3C(GBD). GST–mDia1(GBD) (aa 1–373) and RhoD(wt) cDNAs were fused in frame with the GST tag in pGEX-6P vector and expressed in the Escherichia coli strain XL1-Blue. The GST tag of GST–RhoD was excised with PreScission protease (GE Healthcare) in 50 mM Tris-HCl (pH 7.5), 150 mM NaCl, 5 mM EDTA, 1 mM dithiothreitol (DTT), 1 μg/ml leupeptin, and 1 μg/ml pepstatin A at 4°C overnight. RhoD (10 μM) was mixed with 2 μM GTPγS or GDP at 37°C for 10 min, and the loading was stopped by adding 15 mM MgCl2. GTPγS or GDP-loaded RhoD was applied to a GST–mDia(GBD) pulldown assay. Bound RhoD was detected by immunoblotting with anti-RhoD rabbit pAb, which was prepared as anti–M-Ras pAb (Sun et al., 2006) and affinity-purified with RhoD coupled to Formyl-Cellulofine (Seikagaku, Tokyo, Japan).
Coimmunoprecipitation assay
COS-1 cells were cotransfected with pEF-BOS/Myc-RhoD and pEGFP-C1/mDia1, mDia2, mDia3A, mDia3B, or mDia3C. Anti-GFP pAb (MBL, Nagoya, Japan) was coupled to Protein G Sepharose 4 Fast Flow (GE Healthcare). The transfected cells were lysed as described previously (Takano et al., 2010), and the lysates were incubated with the antibody-coupled Sepharose for 60 min at 4°C. After a thorough washing with the lysis buffer, bound proteins were dissociated with the SDS sample buffer and detected by immunoblotting with the anti-GFP pAb and anti-Myc mAb (Yokoyama et al., 2007).
Actin polymerization assay
A fluorometric actin polymerization assay with pyrene-labeled actin was conducted as described previously (Takano et al., 2010), using 1 μM actin (10% pyrene-labeled), 100 nM mDia3C, 100 nM mDia3C(ΔDAD), and 100 nM RhoD–GTPγS. Pyrene fluorescence was monitored at 25°C with an FP-8300DS spectrofluorometer (JASCO).
Supplementary Material
Acknowledgments
We thank Y. Takai for providing recombinant plasmids of Cdc42. This work was supported by Grants-in-Aid for Scientific Research on Priority Areas (18057005 and 20054004 to T.E.) and for Scientific Research on Innovative Areas (23117506 to T.E.) from the Ministry of Education, Culture, Sports, Science, and Technology of Japan; Grants-in-Aid for Scientific Research (B) (23300144 to T.E.) and for Young Scientists (B) (24770118 to K.T.) from the Japan Society for the Promotion of Science; and an Intramural Research Grant (23-5) for Neurological and Psychiatric Disorders of National Center of Neurology and Psychiatry.
Abbreviations used:
- 10T1/2
C3H/10T1/2
- BMP
bone morphogenetic protein
- Bnl
Branchless
- DAD
Dia autoregulatory domain
- Dpp
Decapentaplegic
- DTT
dithiothreitol
- EGF
epidermal growth factor
- EGFP
enhanced green fluorescent protein
- FGF
fibroblast growth factor
- FGFR
FGF receptor
- GBD
GTPase-binding domain
- GEF
guanine nucleotide exchange factor
- GST
glutathione S-transferase
- mAb
monoclonal antibody
- N-WASP
neural Wiskott-Aldrich syndrome protein
- pAb
polyclonal antibody
- PBS
phosphate-buffered saline
- PFA
paraformaldehyde
- PI3K
phosphatidylinositol 3-kinase
- PIP2
phosphatidylinositol 4,5-bisphosphate
- PLCγ
phospholipase C-γ
- RNAi
RNA interference
- RT-PCR
reverse transcriptase PCR
- SH3
Src homology 3
- siRNA
small interfering RNA
- wt
wild type
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E12-04-0315) on October 3, 2012.
*These authors contributed equally to this work.
†Present address: Department of Biochemistry and Molecular Biology, Kobe University Graduate School of Medicine, Kobe 650-0017, Japan.
REFERENCES
- Abe T, Kato M, Miki H, Takenawa T, Endo T. Small GTPase Tc10 and its homologue RhoT induce N-WASP-mediated long process formation and neurite outgrowth. J Cell Sci. 2002;116:155–168. doi: 10.1242/jcs.00208. [DOI] [PubMed] [Google Scholar]
- Beenken A, Mohammadi M. The FGF family: biology, pathophysiology and therapy. Nat Rev Drug Discov. 2009;8:235–253. doi: 10.1038/nrd2792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Block J, Stradal TE, Hänisch J, Geffers R, Köstler SA, Urban E, Small JV, Rottner K, Faix J. Filopodia formation induced by active mDia2/Drf3. J Microsc. 2008;231:506–517. doi: 10.1111/j.1365-2818.2008.02063.x. [DOI] [PubMed] [Google Scholar]
- Bryant DM, Stow JL. Nuclear translocation of cell-surface receptors: lessons from fibroblast growth factor. Traffic. 2005;6:947–954. doi: 10.1111/j.1600-0854.2005.00332.x. [DOI] [PubMed] [Google Scholar]
- Campellone KG, Welch MD. A nucleator arms race: cellular control of actin assembly. Nat Rev Mol Cell Biol. 2010;11:237–251. doi: 10.1038/nrm2867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carpenter G, Liao H-J. Trafficking of receptor tyrosine kinases to the nucleus. Exp Cell Res. 2009;315:1556–1566. doi: 10.1016/j.yexcr.2008.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng L, Zhang J, Ahmad S, Rozier L, Yu H, Deng H, Mao Y. Aurora B regulates formin mDia3 in achieving metaphase chromosome alignment. Dev Cell. 2011;20:342–352. doi: 10.1016/j.devcel.2011.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chesarone MA, DuPage AG, Goode BL. Unleashing formins to remodel the actin and microtubule cytoskeletons. Nat Rev Mol Cell Biol. 2010;11:62–74. doi: 10.1038/nrm2816. [DOI] [PubMed] [Google Scholar]
- Dominguez R. Actin filament nucleation and elongation factors—structure-function relationships. Crit Rev Biochem Mol Biol. 2009;44:351–366. doi: 10.3109/10409230903277340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunham-Ems SM, Lee YW, Stachowiak EK, Pudavar H, Claus P, Prasad PN, Stachowiak MK. Fibroblast growth factor receptor-1 (FGFR1) nuclear dynamics reveal a novel mechanism in transcription control. Mol Biol Cell. 2009;20:2401–2412. doi: 10.1091/mbc.E08-06-0600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faix J, Breitsprecher D, Stradal TE, Rottner K. Filopodia: complex models for simple rods. Int J Biochem Cell Biol. 2009;41:1656–1664. doi: 10.1016/j.biocel.2009.02.012. [DOI] [PubMed] [Google Scholar]
- Fifadara NH, Beer F, Ono S, Ono SJ. Interaction between activated chemokine receptor 1 and FcɛRI at membrane rafts promotes communication and F-actin-rich cytoneme extensions between mast cells. Int Immunol. 2010;22:113–128. doi: 10.1093/intimm/dxp118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galkina SI, Sud'ina GF, Ullrich V. Inhibition of neutrophil spreading during adhesion to fibronectin reveals formation of long tubulovesicular cell extensions (cytonemes) Exp Cell Res. 2001;266:222–228. doi: 10.1006/excr.2001.5227. [DOI] [PubMed] [Google Scholar]
- Gasman S, Kalaidzidis Y, Zerial M. RhoD regulates endosome dynamics through Diaphanous-related formin and Src tyrosine kinase. Nat Cell Biol. 2003;5:195–204. doi: 10.1038/ncb935. [DOI] [PubMed] [Google Scholar]
- Geiger B, Spatz JP, Bershadsky AD. Environmental sensing through focal adhesions. Nat Rev Mol Cell Biol. 2009;10:21–33. doi: 10.1038/nrm2593. [DOI] [PubMed] [Google Scholar]
- Goh WI, Sudhaharan T, Lim KB, Sem KP, Lau CL, Ahmed S. Rif-mDia1 interaction is involved in filopodium formation independent of Cdc42 and Rac effectors. J Biol Chem. 2011;286:13681–13694. doi: 10.1074/jbc.M110.182683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta N, DeFranco AL. Visualizing lipid raft dynamics and early signaling events during antigen receptor-mediated B-lymphocyte activation. Mol Biol Cell. 2003;14:432–444. doi: 10.1091/mbc.02-05-0078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto Y, Skacel M, Adams JC. Roles of fascin in human carcinoma motility and signaling: prospects for a novel biomarker. Int J Biochem Cell Biol. 2005;37:1787–1804. doi: 10.1016/j.biocel.2005.05.004. [DOI] [PubMed] [Google Scholar]
- Hsiung F, Ramirez-Weber FA, Iwaki DD, Kornberg TB. Dependence of Drosophila wing imaginal disc cytonemes on Decapentaplegic. Nature. 2005;437:560–563. doi: 10.1038/nature03951. [DOI] [PubMed] [Google Scholar]
- Itoh N, Ornitz DM. Fibroblast growth factors: from molecular evolution to roles in development, metabolism and disease. J Biochem. 2011;149:121–130. doi: 10.1093/jb/mvq121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson HM, Subramaniam PS, Olsnes S, Jans DA. Trafficking and signaling pathways of nuclear localizing protein ligands and their receptors. Bioessays. 2004;26:993–1004. doi: 10.1002/bies.20086. [DOI] [PubMed] [Google Scholar]
- Lemmon MA, Schlessinger J. Cell signaling by receptor tyrosine kinases. Cell. 2010;141:1117–1134. doi: 10.1016/j.cell.2010.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattila PK, Lappalainen P. Filopodia: molecular architecture and cellular functions. Nat Rev Mol Cell Biol. 2008;9:446–454. doi: 10.1038/nrm2406. [DOI] [PubMed] [Google Scholar]
- Mellor H. The role of formins in filopodia formation. Biochim Biophys Acta. 2010;1803:191–200. doi: 10.1016/j.bbamcr.2008.12.018. [DOI] [PubMed] [Google Scholar]
- Murphy C, Saffrich R, Grummt M, Gournier H, Rybin V, Rubino M, Auvinen P, Lütcke A, Parton RG, Zerial M. Endosome dynamics regulated by a Rho protein. Nature. 1996;384:427–432. doi: 10.1038/384427a0. [DOI] [PubMed] [Google Scholar]
- Nobes CD, Hall A. Rho, rac, and cdc42 GTPases regulate the assembly of multimolecular focal complexes associated with actin stress fibers, lamellipodia, and filopodia. Cell. 1995;81:53–62. doi: 10.1016/0092-8674(95)90370-4. [DOI] [PubMed] [Google Scholar]
- Oikawa T, Itoh T, Takenawa T. Sequential signals toward podosome formation in NIH-src cells. J Cell Biol. 2008;182:157–169. doi: 10.1083/jcb.200801042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ornitz DM, Xu J, Colvin JS, McEwen DG, MacArthur CA, Coulier F, Gao G, Goldfarb M. Receptor specificity of the fibroblast growth factor family. J Biol Chem. 1996;271:15292–15297. doi: 10.1074/jbc.271.25.15292. [DOI] [PubMed] [Google Scholar]
- Pellegrin S, Mellor H. The Rho family GTPase Rif induces filopodia through mDia2. Curr Biol. 2005;15:129–133. doi: 10.1016/j.cub.2005.01.011. [DOI] [PubMed] [Google Scholar]
- Peng J, Wallar BJ, Flanders A, Swiatek PJ, Alberts AS. Disruption of the Diaphanous-related formin Drf1 gene encoding mDia1 reveals a role for Drf3 as an effector for Cdc42. Curr Biol. 2003;13:534–545. doi: 10.1016/s0960-9822(03)00170-2. [DOI] [PubMed] [Google Scholar]
- Ramírez-Weber F-A, Kornberg TB. Cytonemes: cellular processes that project to the principal signaling center in Drosophila imaginal discs. Cell. 1999;97:599–607. doi: 10.1016/s0092-8674(00)80771-0. [DOI] [PubMed] [Google Scholar]
- Reznikoff CA, Brankow DW, Heidelberger C. Establishment and characterization of a cloned line of C3H mouse embryo cells sensitive to postconfluence inhibition of division. Cancer Res. 1973;33:3231–3238. [PubMed] [Google Scholar]
- Riedl J, et al. Lifeact: a versatile marker to visualize F-actin. Nat Methods. 2008;5:605–607. doi: 10.1038/nmeth.1220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy S, Hsiung F, Kornberg TB. Specificity of Drosophila cytonemes for distinct signaling pathways. Science. 2011;332:354–358. doi: 10.1126/science.1198949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherer NM, Lehmann MJ, Jimenez-Soto LF, Horensavitz C, Pypaert M, Mothes W. Retroviruses can establish filopodial bridges for efficient cell-to-cell transmission. Nat Cell Biol. 2007;9:310–315. doi: 10.1038/ncb1544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Small JV, Stradal T, Vignal E, Rottner K. The lamellipodium: where motility begins. Trends Cell Biol. 2002;12:112–120. doi: 10.1016/s0962-8924(01)02237-1. [DOI] [PubMed] [Google Scholar]
- Sun P, Watanabe H, Takano K, Yokoyama T, Fujisawa J, Endo T. Sustained activation of M-Ras induced by nerve growth factor is essential for neuronal differentiation of PC12 cells. Genes Cells. 2006;11:1097–1113. doi: 10.1111/j.1365-2443.2006.01002.x. [DOI] [PubMed] [Google Scholar]
- Sun P, Yamamoto H, Suetsugu S, Miki H, Takenawa T, Endo T. Small GTPase Rah/Rab34 is associated with membrane ruffles and macropinosomes and promotes macropinosome formation. J Biol Chem. 2003;278:4063–4071. doi: 10.1074/jbc.M208699200. [DOI] [PubMed] [Google Scholar]
- Sun X, Mariani FV, Martin GR. Functions of FGF signalling from the apical ectodermal ridge in limb development. Nature. 2002;418:501–508. doi: 10.1038/nature00902. [DOI] [PubMed] [Google Scholar]
- Takano K, Watanabe-Takano H, Suetsugu S, Kurita S, Tsujita K, Kimura S, Takenawa T, Endo T. Nebulin and N-WASP cooperate to cause IGF-1-induced sarcomeric actin filament formation. Science. 2010;330:1536–1540. doi: 10.1126/science.1197767. [DOI] [PubMed] [Google Scholar]
- Takenawa T, Suetsugu S. The WASP-WAVE protein network: connecting the membrane to the cytoskeleton. Nat Rev Mol Cell Biol. 2007;8:37–48. doi: 10.1038/nrm2069. [DOI] [PubMed] [Google Scholar]
- Tsubakimoto K, Matsumoto K, Abe H, Ishii J, Amano M, Kaibuchi K, Endo T. Small GTPase RhoD suppresses cell migration and cytokinesis. Oncogene. 1999;18:2431–2440. doi: 10.1038/sj.onc.1202604. [DOI] [PubMed] [Google Scholar]
- Vignjevic D, Kojima S, Aratyn Y, Danciu O, Svitkina T, Borisy GG. Role of fascin in filopodial protrusion. J Cell Biol. 2006;174:863–875. doi: 10.1083/jcb.200603013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe-Takano H, Takano K, Keduka E, Endo T. M-Ras is activated by bone morphogenetic protein-2 and participates in osteoblastic determination, differentiation, and transdifferentiation. Exp Cell Res. 2010;316:477–490. doi: 10.1016/j.yexcr.2009.09.028. [DOI] [PubMed] [Google Scholar]
- Yang C, Czech L, Gerboth S, Kojima S, Scita G, Svitkina T. Novel roles of formin mDia2 in lamellipodia and filopodia formation in motile cells. PLoS Biol. 2007;5:2624–2645. doi: 10.1371/journal.pbio.0050317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasuda S, Oceguera-Yanez F, Kato T, Okamoto M, Yonemura S, Terada Y, Ishizaki T, Narumiya S. Cdc42 and mDia3 regulate microtubule attachment to kinetochores. Nature. 2004;428:767–771. doi: 10.1038/nature02452. [DOI] [PubMed] [Google Scholar]
- Yokoyama T, Takano K, Yoshida A, Katada F, Sun P, Takenawa T, Andoh T, Endo T. DA-Raf1, a competent intrinsic dominant-negative antagonist of the Ras–ERK pathway, is required for myogenic differentiation. J Cell Biol. 2007;177:781–793. doi: 10.1083/jcb.200703195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zerial M, McBride H. Rab proteins as membrane organizers. Nat Rev Mol Cell Biol. 2001;2:107–117. doi: 10.1038/35052055. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.