Abstract
Dormancy is a state of metabolic arrest that facilitates the survival of organisms during environmental conditions incompatible with their regular course of life. Many organisms have deep dormant stages to promote an extended life span (increased longevity). In contrast, plants have seed dormancy and seed longevity described as two traits. Seed dormancy is defined as a temporary failure of a viable seed to germinate in conditions that favor germination, whereas seed longevity is defined as seed viability after dry storage (storability). In plants, the association of seed longevity with seed dormancy has not been studied in detail. This is surprising given the ecological, agronomical, and economic importance of seed longevity. We studied seed longevity to reveal its genetic regulators and its association with seed dormancy in Arabidopsis (Arabidopsis thaliana). Integrated quantitative trait locus analyses for seed longevity, in six recombinant inbred line populations, revealed five loci: Germination Ability After Storage1 (GAAS1) to GAAS5. GAAS loci colocated with seed dormancy loci, Delay Of Germination (DOG), earlier identified in the same six recombinant inbred line populations. Both GAAS loci and their colocation with DOG loci were validated by near isogenic lines. A negative correlation was observed, deep seed dormancy correlating with low seed longevity and vice versa. Detailed analysis on the collocating GAAS5 and DOG1 quantitative trait loci revealed that the DOG1-Cape Verde Islands allele both reduces seed longevity and increases seed dormancy. To our knowledge, this study is the first to report a negative correlation between seed longevity and seed dormancy.
Dormancy describes a state of apparent metabolic arrest during which the normal progression of life activities and development is dramatically reduced or brought to a halt. Dormancy facilitates the survival of organisms during environmental conditions that cannot support the regular course of life. Many organisms have dormant stages, which in different species have different names, such as dauer stage in Drosophila spp., diapause in water flea and fish embryos, akinetes in cyanobacteria, spores in yeast (Saccharomyces cerevisiae), and dormancy in plant seeds and flower buds. Organisms can enter the dormant state due to environmental cues such as a lack of water by undergoing desiccation, low temperature, or through developmentally programmed arrest, as occurs, for example, in yeast spores and plant seeds. In most organisms, dormancy has been related to an extension of their life span (increasing longevity; Lubzens et al., 2010).
In contrast to most other organisms described above, in plant seeds dormancy and longevity have been described as two separate traits. Seed dormancy is defined as a temporal failure of a seed to germinate in conditions that favor germination (Bewley, 1997). Seed dormancy can be overcome by environmental cues (i.e. seed dry storage [after ripening] and cold stratification). Seed dormancy has been studied extensively (Finch-Savage and Leubner-Metzger, 2006; Holdsworth et al., 2008), and recently, a quantitative trait locus (QTL) analysis in combination with transcriptome analyses in Arabidopsis (Arabidopsis thaliana) has revealed that natural variation for seed dormancy is controlled by independent genetic and molecular pathways (Bentsink et al., 2010).
Seed longevity is defined as seed viability after seed dry storage (storability) and, therefore, describes the total seed life span (Rajjou and Debeaujon, 2008). This storability period includes both the dormant and nondormant states. During seed storage, seeds deteriorate, lose vigor, and, as a result, become more sensitive to stresses during germination, and ultimately die. The rate of this aging depends on the seed moisture content, temperature, and initial seed quality (Walters, 1998; Walters et al., 2005). Seed longevity is a quantitative trait for which variation is present among naturally occurring accessions. QTLs for seed longevity have been identified after natural aging in Arabidopsis (Bentsink et al., 2000; Clerkx et al., 2004b), lettuce (Lactuca sativa; Schwember and Bradford, 2010), and rice (Oryza sativa; Sasaki et al., 2005) and after artificial aging imposed by a controlled deterioration test (CDT) in Arabidopsis (Bentsink et al., 2000; Clerkx et al., 2004b), rice (Miura et al., 2002), and wheat (Triticum aestivum; Landjeva et al., 2009). The ability of the CDT to predict seed longevity was shown by the colocalization of major QTLs after artificial and natural aging in two different studies using the Arabidopsis recombinant inbred line (RIL) populations Landsberg erecta (Ler)/Cape Verde Islands (Cvi; Bentsink et al., 2000) and Ler/Shakdara (Sha; Clerkx et al., 2004b). Besides these genetic analyses, also proteome studies in Arabidopsis have shown that similar molecular events occur during natural and artificial (CDT) aging (Rajjou et al., 2008). In contrast with this, Schwember and Bradford (2010) did not find overlap between seed longevity QTLs under conventional and controlled deterioration storage conditions in lettuce.
The genetic basis of seed longevity is unclear. However, there are several groups of mutants that have altered seed longevity. The majority of mutants with known effects on seed longevity are the seed developmental mutants. Mutations in the key regulators of seed maturation lead to rapid loss of viability upon storage, as has been shown for leafy cotyledon1 (lec1) and abscisic acid intensitive3 (abi3) mutants (Ooms et al., 1993; Clerkx et al., 2004a; Sugliani et al., 2009). Another group of mutants with a seed longevity phenotype consists of the testa mutants. The seed coat or testa acts as a structural barrier to protect the embryo and seed reserves from biotic and abiotic stresses. The testa-defective mutants, including transparent testa (tt) and aberrant testa shape (ats; Debeaujon et al., 2000), display considerably reduced seed longevity. Moreover, mutations in protection and repair systems that prevent seed vigor loss lead to decreased seed longevity. Arabidopsis mutants affected in vitamin E (lipophilic antioxidant) biosynthesis, vte1 and vte2, exhibited significantly reduced seed longevity (Sattler et al., 2004). Waterworth et al. (2010) showed that DNA LIGASEVI and DNA LIGASEIV, which are essential to maintain genome integrity in plants, are major determinants of Arabidopsis seed quality and longevity. The atlig6 mutant and atlig6 atlig4 double mutant are more sensitive to controlled seed aging than wild type.
Proteins and enzymes are also described as factors that may determine seed longevity. Heat stress transcription factor-overaccumulating seeds of transgenic Arabidopsis display enhanced accumulation of Heat Stress Protein and improved tolerance to aging (Prieto-Dapena et al., 2006). Protein repair appears to play a key role in the long-term survival of seeds in the dry state. PIMT (for protein l-isoaspartyl methyltransferase), which limits and repairs age-damaged aspartyl and asparaginyl residues in proteins, has been associated with greater seed longevity because it is highly accumulated in sacred lotus seed (Nelumbo nucifera), one of the world’s longest living seeds (1,300 years; Shen-Miller, 2002). Overexpression of PIMT1 in Arabidopsis enhanced both seed longevity and germination vigor, whereas reduced PIMT1 expression led to increased sensitivity to aging treatments and loss of seed vigor under stressful germination conditions (Ogé et al., 2008). However, PIMT exhibited a decreased activity in naturally aged barley (Hordeum vulgare) seeds (Mudgett et al., 1997). In addition, enzymes playing roles in the detoxification of reactive oxygen species, such as glutathione peroxidase and glutathione reductase (Bailly et al., 1996), and toxic cyanide compounds, such as β-mercaptopyruvate sulfurtransferase (Rajjou et al., 2008), are important to prolong seed longevity.
Given the important precondition of many organisms to become dormant before exposure to and survival of long-term (desiccation) stress, as well as the ecological, agronomical, and economic importance of seed longevity, it is surprising that the association of seed longevity and seed dormancy has not been studied in much detail. The current idea is that seed dormancy and seed longevity are positively correlated. This hypothesis is based mainly on the performance of the earlier mentioned lec1, abi3 (Ooms et al., 1993; Clerkx et al., 2004a; Sugliani et al., 2009), tt, and ats mutants (Debeaujon et al., 2000) but also on the loss-of-function mutant in the DOG1 gene (Bentsink et al., 2006) and the green seed mutant (enhancer of abi3-1; Clerkx et al., 2003). All these mutants have a reduced dormancy level that correlates with reduced seed longevity.
Here, we study natural variation for seed longevity in order to reveal its genetic regulators and their possible association with seed dormancy. We have performed integrated QTL analyses for seed longevity, measured as germination ability after storage at ambient conditions, in six RIL populations. These populations were derived from crosses between the Arabidopsis standard laboratory accession Ler and the accessions Cvi, Antwerpen (An-1), St. Maria do Feira (Fei-0), Kashmir (Kas-2), Kondara (Kond), and Sha, which were previously used for seed dormancy analyses (Bentsink et al., 2010). The major seed longevity QTLs colocated with the earlier identified seed dormancy QTLs. QTLs and colocation have been validated by near-isogenic lines (NILs). The results are discussed in the context of the current knowledge of seed dormancy and seed longevity.
RESULTS
Seed Longevity of the Parental Accessions and Their RIL Populations
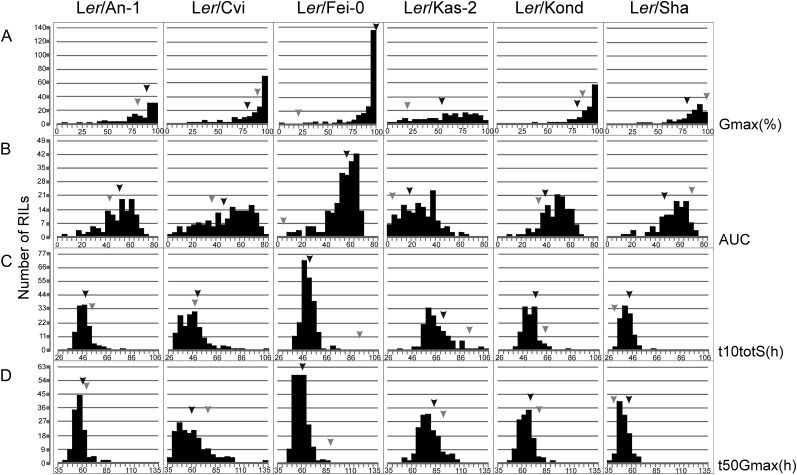
Seed longevity was measured as germination ability after seed dry storage at ambient conditions, which is referred to as “natural aging” in this work. We have used seeds of seven accessions (Ler, An-1, Cvi, Fei-0, Kas-2, Kond, and Sha) and six RIL populations that were constructed from crosses between Ler and the other accessions for this study. The RILs were grown between 2002 and 2005 (Supplemental Table S1). After harvest, seed dormancy behavior was measured until germination reached 100% (Bentsink et al., 2010). In early 2010 (after 4–7 years of dry storage), the same RILs were assessed for seed longevity (germination ability after aging) using the Germinator tool developed by Joosen et al. (2010). As a result of aging, the maximal germination percentage (Gmax) decreased. Overall, the six populations exhibited variation for Gmax (ranging from 0% to 100%; Fig. 1A). Transgression beyond values of both parents was observed in all populations, indicating that both parental lines carried allele-increasing and -decreasing seed longevity (Fig. 1A). The Ler/Kas-2 population harvested in 2002 was the most aged population; approximately one-third of the RILs germinated to less than 50%, whereas the majority of the RILs in Ler/Fei-0 (harvested in 2004) and Ler/Kond (harvested in 2003) populations had a Gmax higher than 75%.
Figure 1.
Frequency distributions of seed longevity presented by four germination parameters in six RIL populations. The different RIL populations are indicated at the top. The x axis contains the trait values for Gmax (%; A), AUC (B), t10totS (h; C), and t50Gmax (h; D). Arrowheads depict the values of parental lines (black arrowheads for Ler and gray arrowheads for other accessions).
The Ler parent, which was grown together with each population, had a Gmax ranging from 54% to 100% (Supplemental Table S1). The Ler parent grown with the Ler/Cvi population harvested in 2005 had a much lower germination ability (Gmax of 80%) than the longer stored Ler parents grown in earlier years (2003 and 2004). This might be the consequence of different growing environments, since it is known that environmental conditions during seed maturation strongly affect seed quality (Contreras et al., 2008).
Seed germination after storage is not only analyzed by Gmax but also by other germination parameters, such as germination rates (time to reach 10% germination of the total number of seeds [t10totS] and time to reach 50% germination of the total number of germinated seeds [t50Gmax]) and area under the curve (AUC; a parameter that describes the germination curve based on the germination rate and the Gmax), which were also measured by the Germinator tool. There was a lot of variation for these three additional parameters in the six populations: t10totS ranged from 30 to 120 h, t50Gmax from 30 to 150 h, and AUC from 0 to 85 (Fig. 1, B–D). For these germination parameters, we do not have the initial values, since the Germinator tool that allows scoring of large populations was not developed at the time the seeds were harvested. Moreover, as a result of aging, a reduction of germination behavior becomes first apparent in a lower germination rate (t10totS and t50Gmax), followed by a decrease of Gmax, which are both reflected by a reduction in AUC. For this reason, these parameters might still contain valuable information for the seed longevity analyses. Therefore, we will focus our study mainly on QTLs found for Gmax but will compare these also with QTLs identified for AUC and use these together to identify colocation with seed dormancy.
Integrated QTL Analyses for Seed Longevity in Six RIL Populations
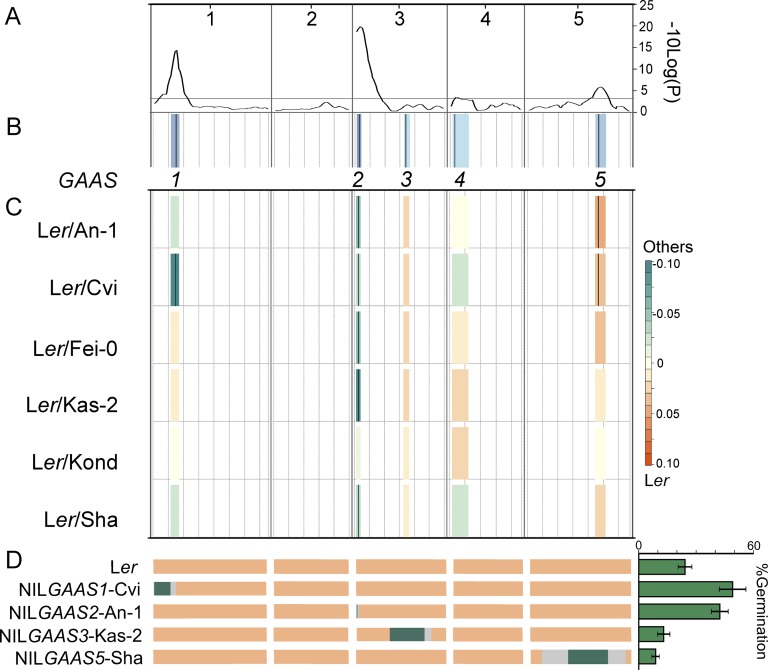
In order to identify loci controlling seed longevity, mixed-model QTL analyses were performed as described by Bentsink et al. (2010). The six RIL populations were explored simultaneously, and the different allele effects were examined for each QTL and every population. In total, five QTLs for seed longevity measured as Gmax were identified. We named these QTLs Germination Ability After Storage1 (GAAS1) to GAAS5 (Fig. 2). These loci showed strong additive effects accounting for an average of 23.3% of the total explained variance (Table I); no epistatic interaction among loci was detected. The mapping results revealed three major loci, GAAS1, GAAS2, and GAAS5, which were also detected for the other parameters (Table I; Supplemental Table S3). The genome-wide significance values of the major QTLs, calculated on a −10Log(P) scale using a final QTL model after backward selection from the Composite Interval Mapping model, were 15.6, 20.5, and 7.2, respectively, while those of the minor QTLs were below 7.2.
Figure 2.
Integrated QTL analyses for seed longevity expressed as Gmax in six RIL populations. A, Genome-wide profiling of simple interval mapping with a genome-wide threshold of 2.74 on the −10Log(P) scale. B, Composite interval mapping with a fixed cofactor revealed five QTL loci (GAAS1–GAAS5). Cofactor positions are depicted by black vertical bars. The confidence interval of each locus is presented by the blue columns: the darker the color, the more significant the QTL. C, QTL effects for every single population. Orange indicates that the Ler allele increases seed longevity (Gmax), and cyan indicates that the Ler allele decreases seed longevity. The intensity of the color corresponds to the size of the QTL effect: the higher the intensity, the stronger the effect. Black vertical bars indicate the significance of the QTL effect in that population. D, Confirmation of the major seed longevity loci (GAAS1, GAAS2, GAAS3, and GAAS5). The genotypes of NILGAAS1-Cvi, NILGAAS2-An-1, NILGAAS3-Kas-2, and NILGAAS5-Sha as well as Ler are schematically presented. Orange indicates the Ler alleles, and cyan indicates the alleles of the other accessions; missing marker data are indicated in light gray. Seed longevity (Gmax) values of the four NILs and Ler after natural aging for 5 years are indicated at the right. The Gmax values of the NILs are significantly different from that of Ler (P < 0.05).
Table I. QTLs for Gmax in six populations, as obtained by integrated analyses comprising composite interval mapping and backward selection.
The genome-wide threshold in composite interval mapping was 2.74 on the −10Log(P) scale, with P representing the P value. QTL name, chromosome, position, significance expressed on a −10Log(P) scale, and a ±1.5 dropoff interval on the −10Log(P) scale (support interval) are presented. P values are taken from the final multi-QTL model after backward selection. Dropoff intervals are assessed on the composite interval mapping profile. QTL allele substitution effects (Gmax) are given in the right part of the table. A negative value indicates that Ler is decreasing the Gmax, whereas a positive value indicates that Ler increases the Gmax. Significant effects are indicated in boldface; these are the effects of the QTLs indicated in Figure 1. In the bottom part of the table, for each population's mean Gmax, the explained variance and the average of explained variance by main-effect QTLs are presented in percentages. The last column shows the range of explained effects of every locus in percentages.
|
Gmax
QTL Name |
Chromosome |
Position |
−10Log(P) |
Support Interval |
QTL Effects | Range of Explained Variance by Locus |
||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
Ler/An-1 |
Ler/Cvi | Ler/Fei-0 | Ler/Kas-2 | Ler/Kond | Ler/Sha | |||||||
| % | ||||||||||||
| GAAS1 | 1 | 29.8 | 15.6 | 27.6–29.8 | −0.027 | −0.101 | 0.011 | 0.007 | −0.005 | −0.026 | 0.6–12.5 | |
| GAAS2 | 3 | 5.4 | 20.5 | 2.6–7.8 | −0.061 | −0.044 | −0.059 | −0.095 | −0.017 | −0.043 | 1.9–15.5 | |
| GAAS3 | 3 | 67.9 | 4.1 | 65.6–72.9 | 0.019 | 0.028 | 0.027 | 0.031 | 0.009 | 0.011 | 1.0–3.5 | |
| GAAS4 | 4 | 6.2 | 4.0 | 3.3–75.4 | −0.002 | −0.025 | 0.009 | 0.024 | 0.031 | −0.028 | 0.2–3.9 | |
| GAAS5 | 5 | 93.3 | 7.2 | 89.5–102.7 | 0.045 | 0.041 | 0.032 | 0.016 | −0.004 | 0.028 | 0.4–5.4 | |
| Mean Gmax | 0.83 | 0.81 | 0.91 | 0.61 | 0.90 | 0.84 | ||||||
| Explained variance by main-effect QTLs (%) | 22.3 | 49.2 | 22.4 | 18.4 | 7.9 | 19.8 | ||||||
| Average of explained variance by main-effect QTLs (%) | 23.3 | |||||||||||
The major locus GAAS2 explained 1.9% to 15.5% of the phenotypic variation in the individual populations and had a significant effect in almost all populations (Table I; Fig. 2). The Ler allele of this locus decreased seed longevity (lower Gmax) in all cases. GAAS1, the second strongest QTL, accounting for 0.6% to 12.5% of the effect in the individual populations, only showed a significant effect in the Ler/Cvi population; also for this QTL, the Ler allele decreased seed longevity. GAAS5, explaining 0.4% to 5.4% of phenotypic variation in the single populations, had a significant effect in two of the six populations (Ler/An-1 and Ler/Cvi); for this QTL, the Ler allele increased seed longevity.
Seven additional minor QTLs identified for the other parameters (AUC, t10totS, and t50Gmax) are named GAAS6 to GAAS12 (Supplemental Table S2).
Confirmation of the GAAS Loci
To characterize the GAAS loci, NILs carrying single genomic fragments of different accessions into the Ler genetic background were used (Table II). The germination behavior of these lines was analyzed after 5 years of seed dry storage. We could confirm four of the five GAAS (Gmax) loci. For the two strongest QTLs, GAAS1 and GAAS2, we had only one NIL each available (NILGAAS1-Cvi and NILGAAS2-An-1, respectively). The seeds of these lines performed significantly better after storage when compared with their genetic background Ler (Table II; Fig. 2D). GAAS3 was validated by NILGAAS3-Kas-2, which showed a significant reduction in Gmax when compared with Ler. For the GAAS5 locus, we had five NILs available (NILGAAS5-Cvi, NILGAAS5-Fei-0, NILGAAS5-Kas-2, NILGAAS5-Kond, and NILGAAS5-Sha), of which only NILGAAS5-Fei-0 and NILGAAS5-Sha showed a significant reduction for Gmax in comparison with Ler. Nongerminating seeds were stained with 2,3,5-triphenyl tetrazolium chloride to determine whether they were dead (Moore, 1985). We concluded that the seeds were deteriorated, since the majority of the seeds did not stain (Supplemental Fig. S1).
Table II. Confirmation of GAAS loci by NILs.
The germination behavior of a set of NILs grown in 2007 (QTL locus, NIL name, and original name [Bentsink et al., 2010]) after 5 years of natural aging is presented by Gmax, AUC, t10totS, t50Gmax, and percentage of normal seedlings. Average values and se (as indicated for each NIL) are indicated. Germination behaviors that are significantly different from that of Ler are indicated by asterisks (*P < 0.05, **P < 0.01).
| QTL Locus | NIL Name | Original Name | Gmax | AUC | t10totS | t50Gmax | Normal Seedling |
|---|---|---|---|---|---|---|---|
| % | h | h | % | ||||
| GAAS1 | NILGAAS1-Cvi | NILDOG2-Cvi | 49.1 ± 6.9** | 22.7 ± 4.9* | 65.4 ± 6.4 | 74.5 ± 5.4 | 21.4 ± 4.9* |
| GAAS2 | NILGAAS2-An-1 | NILDOG22-An-1 | 42.4 ± 4.4** | 18.4 ± 2.1** | 62.7 ± 2.7 | 73.9 ± 1.3 | 16.1 ± 2.6* |
| GAAS3 | NILGAAS3-Kas-2 | NILDOG6-Kas-2 | 13.0 ± 3.2* | 4.1 ± 1.0* | 107.9 ± 22.7 | 93.4 ± 7.7 | 4.3 ± 1.1 |
| GAAS5 | NILGAAS5-Cvi | NILDOG1-Cvi | 14.6 ± 3.3 | 4.3 ± 1.2* | 84.2 ± 3.0 | 95.3 ± 4.2 | 2.2 ± 0.5** |
| GAAS5 | NILGAAS5-Fei-0 | NILDOG1-Fei-0 | 4.4 ± 2.3** | 1.4 ± 0.9** | 80.5 ± 0.0 | 100.9 ± 10.3 | 0.6 ± 0.4** |
| GAAS5 | NILGAAS5-Kas-2 | NILDOG1-Kas-2 | 22.5 ± 3.4 | 8.1 ± 1.6 | 88.4 ± 8.6 | 84.8 ± 3.6 | 6.1 ± 1.6 |
| GAAS5 | NILGAAS5-Kond | NILDOG1-Kond | 24.3 ± 4.1 | 11.0 ± 2.2 | 68.9 ± 4.3 | 74.4 ± 3.1 | 8.3 ± 2.3 |
| GAAS5 | NILGAAS5-Sha | NILDOG1-Sha | 8.6 ± 1.6** | 2.3 ± 0.5** | 125.1 ± 10.1** | 97.6 ± 9.4 | 2.3 ± 0.6** |
| GAAS7 | NILGAAS7-Fei-0 | NILDOG20-Fei-0 | 34.3 ± 5.5 | 15.7 ± 3.0 | 68.6 ± 6.5 | 74.0 ± 4.5 | 14.9 ± 3.1 |
| Ler | 24.2 ± 3.6 | 9.6 ± 1.9 | 81.8 ± 6.8 | 80.7 ± 3.8 | 8.9 ± 1.7 |
Comparison between Seed Longevity and Seed Dormancy
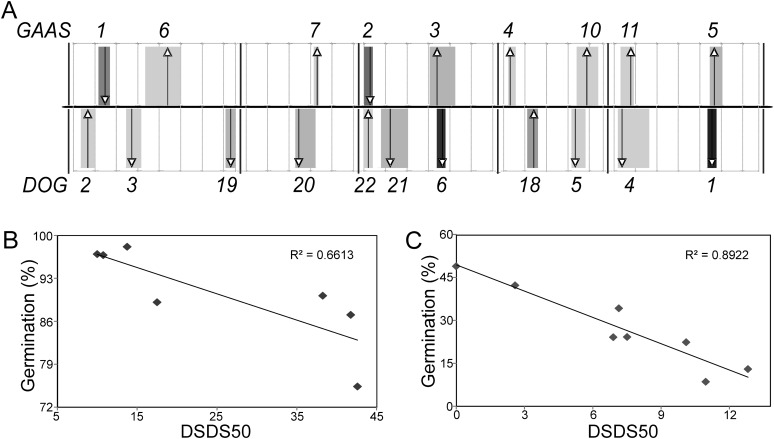
In order to explore the relationship between seed longevity and seed dormancy, we investigated the overlap between QTLs that have been mapped for these two traits. Integrated QTL analyses for both seed longevity and seed dormancy (Delay of Germination [DOG]) have been performed in the same populations (Bentsink et al., 2010), which allows a neat comparison between those two traits. Four of the five GAAS loci identified for Gmax overlapped with DOG loci (GAAS2/DOG22, GAAS3/DOG6, and GAAS5/DOG1) or mapped in very close proximity (GAAS1/DOG2; Fig. 3A). All five GAAS Gmax QTLs were also identified using AUC as a longevity parameter, which can be explained by the high correlation between these two parameters (r2 = 0.75–0.90; Supplemental Table S3). AUC showed the highest variation (Fig. 1B), which provided more statistical power in the QTL analyses and led to the identification of four additional QTLs (Supplemental Table S3). Seven of the nine GAAS AUC QTLs colocated with seed dormancy QTLs (Fig. 3A). The three additional genomic regions that showed colocation are GAAS7/DOG20, GAAS10/DOG5, and GAAS11/DOG4 (Fig. 3A). Unexpectedly, a negative relationship between seed dormancy and seed longevity was observed. Deep dormancy correlated with low storability and shallow dormancy with high storability, shown by the direction of the arrows in Figure 3A. Since we had NILs available for most of the QTLs, we were able to analyze this correlation in more detail (Fig. 3, B and C). The NILs were grown in two independent experiments in the greenhouse (in 2006 and 2007) and analyzed for their seed dormancy (directly after harvest) and seed longevity behavior (after 5 years of storage; Table II; Supplemental Table S4). The negative correlation between seed longevity (Gmax) and seed dormancy (days of seed dry storage required to reach 50% germination [DSDS50]) that was identified by the QTL analyses was proven to be very significant, given the very high correlation coefficients (r2) of 0.66 (P = 0.03) and 0.89 (P < 0.01), respectively, for the two experiments (Fig. 3, B and C).
Figure 3.
Colocation and correlation between seed longevity (GAAS) and seed dormancy (DOG) QTLs. A, Integrated composite interval mapping profiles of seed longevity (AUC; top) and seed dormancy (DSDS50; bottom) performed in the six RIL populations. The confidence interval of each locus is presented by the gray columns: the darker the color, the more significant the QTL. Cofactor positions are depicted by arrows: arrows pointing up indicate that the Ler allele is increasing the trait value, and when the arrows point down, Ler decreases the trait value. B and C, Correlation of seed longevity (germination percentage) and seed dormancy (DSDS50) in NILs. B, Experiment harvested in 2006 including NILDOG1/GAAS5-Sha, NILDOG22/GAAS2-An-1, NILDOG2/GAAS1-Cvi, NILDOG6/GAAS3-Fei-0, NILDOG6/GAAS3-Kas-2, NILDOG20/GAAS7-Fei-0, and Ler. C, Experiment harvested in 2007 including NILDOG20/GAAS7-Fei-0, NILDOG1/GAAS5-Sha, NILDOG1/GAAS5-Kond, NILDOG1/GAAS5-Kas-2, NILDOG22/GAAS2-An-1, NILDOG2/GAAS1-Cvi, NILDOG6/GAAS3-Kas-2, and Ler.
Next we investigated the correlation between seed longevity and seed dormancy in the RILs; however, no correlation was found (r2 between 0.01 and 0.12 for the six populations). This lack of correlation might be explained by the fact that total genetic variation was not fully explained by the identified QTLs (Table I) and by the low variation for seed longevity in RIL populations (Fig. 1). Furthermore, we have ended up with a random combination of seed longevity and dormancy loci in the RILs due to the broken linkage (which is the nature of this type of population). This results in a random combination of phenotypes, since some of these loci have a stronger longevity effect and others a stronger dormancy effect. To remove the above-mentioned noise, we have performed correlation analyses on RILs that have been selected for either seed longevity increasing or decreasing alleles at the position of the strongest four QTLs (GAAS1, GAAS2, GAAS3, and GAAS5) in two of the populations that show the largest variation for seed longevity (Ler/Kas-2) or the maximum possibility of QTL colocation within a population (Ler/Cvi; Supplemental Table S5 and S6). These analyses showed again the negative relationship between seed longevity and dormancy (Supplemental Fig. S4), with r2 of 0.58 for Ler/Kas-2 (P < 0.01) and 0.78 for Ler/Cvi (P < 0.01).
Seed Longevity and Dormancy Are Regulated by One Gene at the Position of GAAS5/DOG1
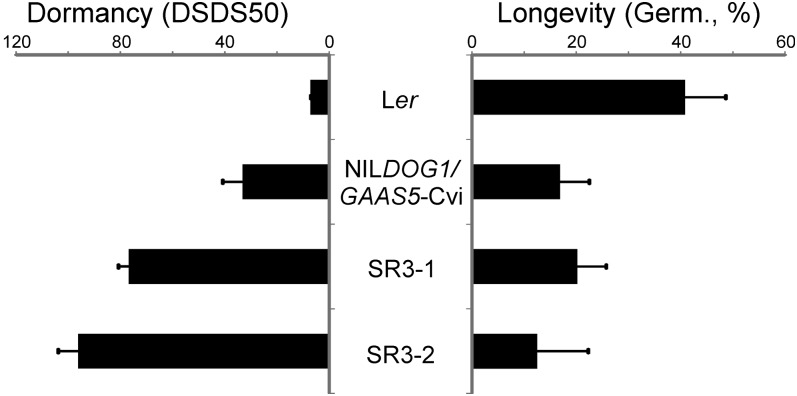
The colocation between seed longevity and seed dormancy QTLs can be caused by a single gene or by separate linked genes. The genetic nature of the colocation can only be studied when the genes underlying the QTLs are identified. So far, the only identified QTL is the dormancy locus DOG1 (collocating with GAAS5). We used transgenic lines that carry the DOG1 Cvi allele in the Ler genetic background to investigate whether these lines, in addition to their increased dormancy level, also showed a seed longevity phenotype. The Gmax of seeds (Ler, NILGAAS5/DOG1-Cvi, and two independent transformants) that had been naturally aged for 7 years was still above 70% and did not reveal clear differences between any of the genotypes (Supplemental Fig. S3). In order to accelerate the aging, we stored these seeds at 75% relative humidity for 59 d and analyzed the Gmax at several time points during this storage. After 59 d of storage in 75% relative humidity, all seeds of all genotypes were completely deteriorated. However, the two transformants were significantly less storable than the Ler control, which shows that DOG1 does not only control seed dormancy but also seed longevity (Fig. 4; Supplemental Fig. S3). This result supports the negative correlation between seed longevity and seed dormancy.
Figure 4.
Seed dormancy and longevity phenotypes of Ler, NILDOG1/GAAS5-Cvi, and two independent transformants. Seed dormancy measured as DSDS50 and longevity after 7 years of storage and 45 d in 75% relative humidity (germination percentage) is shown for Ler, NILDOG1/GAAS5-Cvi, and two independent transformants (SR3-1 and SR3-2), which contain the DOG1 Cvi allele in the Ler genetic background.
DISCUSSION
In order to study natural variation for seed longevity, we used seed batches that had been stored for 4 to 7 years at ambient conditions. Seed longevity had been analyzed in Arabidopsis populations previously, using artificial (CDT; Bentsink et al., 2000; Clerkx et al., 2004b; Joosen et al., 2012) and natural (Bentsink et al., 2000) aging. Bentsink et al. (2000) showed that naturally aged seeds of the Ler/Cvi RIL population that had been stored for 4 years led to the detection of one QTL on chromosome 1 (GAAS1 region). This locus was also the major QTL in our experiment here for the Ler/Cvi population. However, with this long-term-aged seed, we were able to identify two additional QTLs (GAAS2 and GAAS5) in this Ler/Cvi genetic background. GAAS1 and GAAS2 or colocating QTLs were also detected after artificial aging in the Ler/Cvi, Ler/Sha, and Bayreuth/Sha (not for GAAS2) RIL populations (Bentsink et al., 2000; Clerkx et al., 2004b; Joosen et al., 2012). GAAS5 appears to be specific for natural aging and does not show a QTL after controlled deterioration in the Ler/Cvi and Ler/Sha populations, which indicates that the CDT does not completely mimic natural aging. Overall, we were able to identify many more loci than in earlier work (Bentsink et al., 2000; Clerkx et al., 2004b; Joosen et al., 2012), probably due to the much longer time that the seeds had been stored and because of the integrated approach on the multiple populations that was taken.
The parents of every population were grown together with the RIL populations. Ler differed the most from Fei-0 and Kas-2 accessions for seed longevity (Supplemental Table S2). In both cases, Ler seeds were better storable, which might be explained by the fact that the Ler allele contributes to better storability for almost all GAAS loci except for GAAS2 (Fig. 2). The seed longevity of Ler was not very different from those of the accessions An-1, Cvi, Kond, and Sha, but transgressions beyond these parents were identified for those populations (Fig. 1), which resulted in the identification of seed longevity QTLs for which alleles of both parents contributed to better storability (Fig. 2).
GAAS2 and GAAS5 had significant allelic effects in more than one population (Table I; Supplemental Table S2), which may indicate the importance of these seed longevity loci under natural selection. For GAAS1 and GAAS2, the Ler allele decreased seed longevity, while the GAAS5 Ler allele increased seed longevity. Epistatic interactions between the seed longevity loci were not identified, which indicated that natural variation for seed longevity in these populations is determined by additive loci. However, part of the differences might result from genotype-environment interactions due to differences in the growing environments of each population.
Candidate Genes
Several GAAS genomic regions identified here contain genes previously associated with seed longevity. Most obvious is the colocation of GAAS2 with the vitamin E locus. Sattler et al. (2004) have shown that vitamin E (tocopherol) is essential for seed longevity in an artificial aging assay, as it prevents lipid oxidation during seed germination. Genetic analysis of seed vitamin E levels in the Cvi/Ler and Columbia/Ler populations exhibited a common QTL on the top of chromosome 3 (GAAS2 region), namely QVE7 and QVE8, respectively (Sattler et al., 2004).
The DNA ligase AtLIG4 coincides within the confidence interval of GAAS6 in the middle of chromosome 1. Repair of DNA damage in seeds to maintain genome integrity is one of the mechanisms to prevent seed deterioration (Waterworth et al., 2010). DNA damage is associated with single and double strand breaks, which can be rejoined by DNA ligase. Two DNA ligase genes, AtLIG4 and AtLIG6, were shown to be involved in DNA repair upon seed imbibition in Arabidopsis. Mutations in these genes lead to decreased seed viability and seed germination vigor after a CDT.
PIMT genes might be the underlying loci for GAAS3 and GAAS5. PIMT1 and PIMT2, which are located on the lower arms of chromosome 3 and 5, respectively, play a role in the repair of age-related protein damage in Arabidopsis (Ogé et al., 2008). The accumulation of the PIMT1 enzyme enhances resistance to seed vigor loss induced by CDT.
Colocation between Seed Longevity QTLs and Seed Dormancy QTLs
Several QTL studies have been performed for seed longevity (Bentsink et al., 2000; Miura et al., 2002; Clerkx et al., 2004b; Sasaki et al., 2005; Landjeva et al., 2009; Schwember and Bradford, 2010) and seed dormancy (Bentsink et al., 2007); however, none of these discuss the relationship (colocation and/or correlation) of both traits. Therefore, we report, to our knowledge for the first time, a negative correlation between seed longevity and seed dormancy QTLs. Lower storability levels correlated with higher seed dormancy levels, and conversely, better storability correlated with lower seed dormancy. This finding is unexpected, since current seed literature only describes correlations of low longevity with low dormancy and high longevity with high dormancy. However, these correlations were mainly based on mutants such as lec1, abi3, tt, ats, dog1, and the green seed mutant. We assume that these contrasting observations are based on the nature of the mutations. The induced mutants all have defective seed maturation and consequently did not become dormant and desiccation tolerant, as these features are acquired during seed maturation. Very likely, these mutants represent artifacts that do not survive in nature. Moreover, none of the earlier mentioned seed longevity mutants (i.e. atlig4, atlig6, pimt1, and pimt2) have been investigated for their seed dormancy behavior. Such an analysis would reveal whether the same processes could affect seed longevity and seed dormancy and thereby also provide insight in the underlying mechanisms. We expect that the natural variants that we used in our study display the ecologically relevant germination behavior. The GAAS5/DOG1-Fei-0 allele (Supplemental Fig. S2) is special in that it has lower longevity and lower seed dormancy when compared with Ler. The Fei-0 DOG1 allele has an opposite allelic effect as compared with the other alleles for this QTL (Cvi, Kas-2, Kond, Sha), indicating that this allele is even weaker than the Ler allele, which is not a null allele (Bentsink et al., 2006, 2010). The Fei-0 allele of DOG1, therefore, might result in a nonfunctional DOG1 gene, since it has a similar phenotype (lack of dormancy and low storability) to the dog1 mutant (Bentsink et al., 2006).
The colocation between seed longevity and seed dormancy QTLs can be caused by a single gene or by separate linked genes, and this remains to be investigated. Detailed analyses on GAAS5/DOG1 showed that DOG1 is controlling both seed longevity and seed dormancy. DOG1 has been cloned, and the Ler transformant containing the DOG1 allele of Cvi shows complementation of the seed dormancy phenotype of NILDOG1 (Bentsink et al., 2006) and also reduction in seed longevity (Fig. 4). This result shows that GAAS5 is actually the same locus as DOG1 and that the DOG1-Cvi allele leads to both higher seed dormancy and lower seed longevity. The molecular mechanism by which DOG1 controls seed dormancy is still unclear; however, recently it has been proposed that DOG1 protein abundance in freshly harvested seeds acts as a timer for seed dormancy release (Nakabayashi et al., 2012). How the DOG1 protein affects seed storability remains to be investigated.
The novel observation of a negative correlation between dormancy and longevity strongly suggests that seeds are able to extend their life span either by dormancy (and dormancy cycling) or by an active longevity mechanism. Selection for the different mechanisms could be based on the natural environments in which the seeds are dispersed, dry environments resulting in active longevity mechanisms and humid environments resulting in dormancy cycling during which aging damage may be prevented or repaired. The presence of loci that either improve longevity or increase seed dormancy within one accession will allow adaptive plasticity, resulting in the expression of the optimal phenotype over a range of environments (i.e. dry to humid; Simons, 2011).
CONCLUSION
We performed integrated QTL analyses on natural variation that exists for seed longevity after natural aging. We used six RIL populations that were stored between 4 and 7 years at ambient conditions. The major QTLs could be confirmed by NILs that also had been stored for 5 years. Seed longevity and dormancy data revealed a negative correlation, which contrasts with the common notion in seed biology research. High storability correlated with shallow seed dormancy, and low storability correlated with high levels of seed dormancy.
MATERIALS AND METHODS
Plant and Seed Materials
RILs
The six RIL populations derived from crosses between the Arabidopsis (Arabidopsis thaliana) standard laboratory accession Ler and the accessions An-1, Cvi, Fei-0, Kas-2, Kond, and Sha were used. These populations have been analyzed previously for seed dormancy as described by Bentsink et al. (2010). The RILs were grown and harvested between 2003 and 2005 as described by Bentsink et al. (2010). Seeds of every RIL were stored in 6- × 13-cm cellophane flat bags at room temperature without humidity control until seed longevity was analyzed (Table I).
NILs
The NILs we used in this work were originally developed by the introgression of the identified dormancy QTL regions into the Ler genetic background (Table II; Supplemental Table S3; Alonso-Blanco et al., 2003; Bentsink et al., 2010). In 2006, seven NILs, NILGAAS5-Sha, NILGAAS2-An-1, NILGAAS1-Cvi, NILGAAS3-Cvi, NILGAAS3-Fei-0, NILGAAS3-Kas-2, and NILGAAS5-Fei-0, were grown and harvested together with Ler. In 2007, nine NILs, NILGAAS1-Cvi, NILGAAS2-An-1, NILGAAS3-Kas-2, NILGAAS5-Cvi, NILGAAS5-Fei-0, NILGAAS5-Kas, NILGAAS5-Kond, NILGAAS5-Sha, and Ler were grown and harvested. Growing and harvesting methods were as described by Bentsink et al. (2010), and the storage method was as described for the RILs.
The DOG1 Transformants
The two transformants, SR3-1 and SR3-2, containing the DOG1 Cvi allele into the Ler genetic background were obtained as described by Bentsink et al. (2006). These lines and their control lines (Ler and NILDOG1-Cvi) were grown as in the study of Bentsink et al. (2006) and stored as described for the RILs.
Seed Longevity Measurement
Natural Aging
Seed longevity was evaluated as germination ability after several years of storage in natural conditions. We have used the same seed batches described by Bentsink et al. (2010). The germination percentage after dormancy release was 100% for all lines, as was described by these authors. Germination assays after aging were performed according to Joosen at al. (2010) over a period of 7 d. Briefly, six samples, 50 to 200 seeds each, were sown on two layers of blue germination papers equilibrated with 43 mL of demineralized water in plastic trays (15 × 21 cm). Trays were piled and wrapped in a closed and transparent plastic bag. Germination was incubated in a 22°C incubator under continuous light (30 W m−2).
Seed longevity was confirmed by viability test with 2,3,5-triphenyl tetrazolium chloride according to the International Seed Testing Association (Moore, 1985). After 7 d of germination assay, nongerminated seeds were taken out for staining. Seeds were punched gently by sharp forceps to make staining solution easily penetrate the embryo, placed on filter paper (Sartorius filter discs 3hw) soaked with 1% 2,3,5-triphenyl tetrazolium chloride, sealed, and incubated at 28°C for 2 d. Seeds that are viable stain red, and seeds that are dead do not stain.
Artificial Aging
In order to accelerate aging, we stored Ler, NILDOG1-Cvi, and the two independent transformants (SR3-1 and SR3-2) above a saturated NaCl solution in a closed desiccator (relative humidity of 75%) for 59 d. At 0, 13, 31, 38, 45, 52, and 59 d, we performed germination assays as described above. Two-way ANOVA (P < 0.05) was performed in order to identify the differences in Gmax between the lines.
Seed Dormancy Measurement
Germination assays during after-ripening were performed on the same seed lots as described for the seed longevity measurements. The data and methods used are presented by Bentsink et al. (2010). Germination data were fitted to logistic curves by nonlinear regression analysis to determine DSDS50 as described by Bentsink et al. (2010).
Germination Parameters
Images from germination assays were taken twice per day over a 7-d period. Automatic scoring and curve fitting were analyzed by the Germinator package (Joosen et al., 2010). Four parameters, Gmax, AUC, t10totS, and t50Gmax, representing the germination ability were extracted. Gmax is the final germination percentage at the end of the germination assay. The AUC parameter was measured at 120 h after sowing. AUC describes the germination curve by combining germination rate and Gmax. The Germinator curve-fitting script, a part of the Germinator package, enables one to calculate averages and to perform statistical Student’s t test. At the end of the germination assay, the percentage of normal seedlings was scored.
Integrated QTL Analyses
Integrated QTL analyses of the six RIL populations were carried out for every germination parameter defined above. The analysis method was performed as described by Bentsink et al. (2010).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Viability test for seeds after natural aging with 2,3,5-triphenyl tetrazolium chloride.
Supplemental Figure S2. Seed germination behavior of Ler, dog1 mutant, and NILDOG1/GAAS5-Fei-0 during seed dry storage.
Supplemental Figure S3. Seed life span of Ler, NILDOG1/GAAS5-Cvi, and two independent transformants.
Supplemental Figure S4. Correlation analyses between seed longevity and seed dormancy of the RILs.
Supplemental Table S1. Details of the six RIL populations.
Supplemental Table S2. QTLs for AUC, t10totS, and t50Gmax in six RIL populations.
Supplemental Table S3. Correlation coefficient of germination parameters: Gmax, AUC, t10totS, and t50Gmax in six RIL populations.
Supplemental Table S4. Seed longevity and dormancy phenotypes of NILs of two independent experiments.
Supplemental Table S5. Genotypes of the RILs selected for seed longevity increasing and decreasing alleles for the Ler/Kas-2 population.
Supplemental Table S6. Genotypes of the RILs selected for seed longevity increasing and decreasing alleles for the Ler/Cvi populations.
Acknowledgments
We thank Corrie Hanhart and Maarten Koornneef (Laboratory of Genetics, Wageningen University) for storing the seeds of the RIL populations over all these years. We also thank Henk Hilhorst (Laboratory of Plant Physiology, Wageningen University), Bas Dekkers (Department of Molecular Plant Physiology, Utrecht University), and two anonymous reviewers for useful comments and critical reading of the manuscript.
Glossary
- QTL
quantitative trait locus
- CDT
controlled deterioration test
- RIL
recombinant inbred line
- Ler
Landsberg erecta
- Cvi
Cape Verde Islands
- Sha
Shakdara
- NIL
near-isogenic line
- An-1
Antwerpen
- Fei-0
St. Maria do Feira
- Kas-2
Kashmir
- Kond
Kondara
- Gmax
maximal germination percentage
- AUC
area under the curve
- t10totS
time to reach 10% germination of the total number of seeds
- t50Gmax
time to reach 50% germination of the total number of germinated seeds
- DSDS50
days of seed dry storage required to reach 50% germination
References
- Alonso-Blanco C, Bentsink L, Hanhart CJ, Blankestijn-de Vries H, Koornneef M. (2003) Analysis of natural allelic variation at seed dormancy loci of Arabidopsis thaliana. Genetics 164: 711–729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailly C, Benamar A, Corbineau F, Come D. (1996) Changes in malondialdehyde content and in superoxide dismutase, catalase and glutathione reductase activities in sunflower seeds as related to deterioration during accelerated aging. Physiol Plant 97: 104–110 [Google Scholar]
- Bentsink L, Alonso-Blanco C, Vreugdenhil D, Tesnier K, Groot SPC, Koornneef M. (2000) Genetic analysis of seed-soluble oligosaccharides in relation to seed storability of Arabidopsis. Plant Physiol 124: 1595–1604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentsink L, Hanson J, Hanhart CJ, Blankestijn-de Vries H, Coltrane C, Keizer P, El-Lithy M, Alonso-Blanco C, de Andrés MT, Reymond M, et al. (2010) Natural variation for seed dormancy in Arabidopsis is regulated by additive genetic and molecular pathways. Proc Natl Acad Sci USA 107: 4264–4269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentsink L, Jowett J, Hanhart CJ, Koornneef M. (2006) Cloning of DOG1, a quantitative trait locus controlling seed dormancy in Arabidopsis. Proc Natl Acad Sci USA 103: 17042–17047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentsink L, Soppe W, Koornneef M. (2007) Genetics aspects of seed dormancy. In KJ Bradford, H Nonogaki, eds, Seed Development, Dormancy and Germination. Blackwell Publishing, Oxford, pp 113–127 [DOI] [PubMed]
- Bewley JD. (1997) Seed germination and dormancy. Plant Cell 9: 1055–1066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clerkx EJ, Vries HB, Ruys GJ, Groot SP, Koornneef M. (2003) Characterization of green seed, an enhancer of abi3-1 in Arabidopsis that affects seed longevity. Plant Physiol 132: 1077–1084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clerkx EJM, Blankestijn-De Vries H, Ruys GJ, Groot SPC, Koornneef M. (2004a) Genetic differences in seed longevity of various Arabidopsis mutants. Physiol Plant 121: 448–461 [Google Scholar]
- Clerkx EJM, El-Lithy ME, Vierling E, Ruys GJ, Blankestijn-De Vries H, Groot SPC, Vreugdenhil D, Koornneef M. (2004b) Analysis of natural allelic variation of Arabidopsis seed germination and seed longevity traits between the accessions Landsberg erecta and Shakdara, using a new recombinant inbred line population. Plant Physiol 135: 432–443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Contreras S, Bennett MA, Metzger JD, Tay D. (2008) Maternal light environment during seed development affects lettuce seed weight, germinability, and storability. HortScience 43: 845–852 [Google Scholar]
- Debeaujon I, Léon-Kloosterziel KM, Koornneef M. (2000) Influence of the testa on seed dormancy, germination, and longevity in Arabidopsis. Plant Physiol 122: 403–414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finch-Savage WE, Leubner-Metzger G. (2006) Seed dormancy and the control of germination. New Phytol 171: 501–523 [DOI] [PubMed] [Google Scholar]
- Holdsworth MJ, Bentsink L, Soppe WJJ. (2008) Molecular networks regulating Arabidopsis seed maturation, after-ripening, dormancy and germination. New Phytol 179: 33–54 [DOI] [PubMed] [Google Scholar]
- Joosen RVL, Arends D, Willems LAJ, Ligterink W, Jansen RC, Hilhorst HWM. (2012) Visualizing the genetic landscape of Arabidopsis seed performance. Plant Physiol 158: 570–589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joosen RVL, Kodde J, Willems LAJ, Ligterink W, van der Plas LHW, Hilhorst HWM. (2010) GERMINATOR: a software package for high-throughput scoring and curve fitting of Arabidopsis seed germination. Plant J 62: 148–159 [DOI] [PubMed] [Google Scholar]
- Landjeva S, Lohwasser U, Börner A. (2009) Genetic mapping within the wheat D genome reveals QTL for germination, seed vigour and longevity, and early seedling growth. Euphytica 171: 129–143 [Google Scholar]
- Lubzens E, Cerdà J, Clark MS. (2010) Introduction. In E Lubzens, J Cerdà, MS Clark, eds, Topics in Current Genetics: Dormancy and Resistance in Harsh Environments, Vol 21. Springer, Heidelberg, pp 1–4
- Miura K, Lin Y, Yano M, Nagamine T. (2002) Mapping quantitative trait loci controlling seed longevity in rice (Oryza sativa L.). Theor Appl Genet 104: 981–986 [DOI] [PubMed] [Google Scholar]
- Moore RP. (1985) Handbook on Tetrazolium Testing. International Seed Testing Association, Zurich, p 99
- Mudgett MB, Lowenson JD, Clarke S. (1997) Protein repair l-isoaspartyl methyltransferase in plants: phylogenetic distribution and the accumulation of substrate proteins in aged barley seeds. Plant Physiol 115: 1481–1489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakabayashi K, Bartsch M, Xiang Y, Miatton E, Pellengahr S, Yano R, Seo M, Soppe WJ. (2012) The time required for dormancy release in Arabidopsis is determined by DELAY OF GERMINATION1 protein levels in freshly harvested seeds. Plant Cell 24: 2826–2838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogé L, Bourdais G, Bove J, Collet B, Godin B, Granier F, Boutin JP, Job D, Jullien M, Grappin P. (2008) Protein repair l-isoaspartyl methyltransferase 1 is involved in both seed longevity and germination vigor in Arabidopsis. Plant Cell 20: 3022–3037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ooms JJJ, Léon-Kloosterziel KM, Bartels D, Koornneef M, Karssen CM. (1993) Acquisition of desiccation tolerance and longevity in seeds of Arabidopsis thaliana: a comparative study using abscisic acid-insensitive abi3 mutants. Plant Physiol 102: 1185–1191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prieto-Dapena P, Castaño R, Almoguera C, Jordano J. (2006) Improved resistance to controlled deterioration in transgenic seeds. Plant Physiol 142: 1102–1112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajjou L, Debeaujon I. (2008) Seed longevity: survival and maintenance of high germination ability of dry seeds. C R Biol 331: 796–805 [DOI] [PubMed] [Google Scholar]
- Rajjou L, Lovigny Y, Groot SPC, Belghazi M, Job C, Job D. (2008) Proteome-wide characterization of seed aging in Arabidopsis: a comparison between artificial and natural aging protocols. Plant Physiol 148: 620–641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki K, Fukuta Y, Sato T. (2005) Mapping of quantitative trait loci controlling seed longevity of rice (Oryza sativa L.) after various periods of seed storage. Plant Breed 124: 361–366 [Google Scholar]
- Sattler SE, Gilliland LU, Magallanes-Lundback M, Pollard M, DellaPenna D. (2004) Vitamin E is essential for seed longevity and for preventing lipid peroxidation during germination. Plant Cell 16: 1419–1432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwember AR, Bradford KJ. (2010) Quantitative trait loci associated with longevity of lettuce seeds under conventional and controlled deterioration storage conditions. J Exp Bot 61: 4423–4436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen-Miller J. (2002) Sacred lotus, the long-living fruits of China Antique. Seed Sci Res 12: 131–143 [Google Scholar]
- Simons AM. (2011) Modes of response to environmental change and the elusive empirical evidence for bet hedging. Proc Biol Sci 278: 1601–1609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugliani M, Rajjou L, Clerkx EJM, Koornneef M, Soppe WJJ. (2009) Natural modifiers of seed longevity in the Arabidopsis mutants abscisic acid insensitive3-5 (abi3-5) and leafy cotyledon1-3 (lec1-3). New Phytol 184: 898–908 [DOI] [PubMed] [Google Scholar]
- Walters C. (1998) Understanding the mechanisms and kinetics of seed aging. Seed Sci Res 8: 223–244 [Google Scholar]
- Walters C, Hill LM, Wheeler LJ. (2005) Dying while dry: kinetics and mechanisms of deterioration in desiccated organisms. Integr Comp Biol 45: 751–758 [DOI] [PubMed] [Google Scholar]
- Waterworth WM, Masnavi G, Bhardwaj RM, Jiang Q, Bray CM, West CE. (2010) A plant DNA ligase is an important determinant of seed longevity. Plant J 63: 848–860 [DOI] [PubMed] [Google Scholar]