Abstract
TAL1/SCL is a hematopoietic specific oncogene and its activity is regulated by associated transcriptional coactivators and corepressors. Dysregulation of TAL1 activity has been associated with T cell leukemogenesis. However, it remains unclear how the interactions between TAL1 and corepressors versus coactivators are properly regulated. Here we reported that PKA mediated phosphorylation regulates TAL1 interaction with the lysine specific demethylase (LSD1) that removes methyl group from methylated Lys 4 on histone H3 tails. Phosphorylation of serine 172 in TAL1 specifically destabilizes the TAL1-LSD1 interaction leading to promoter H3K4 hypermethylation and activation of target genes that have been suppressed in normal and malignant hematopoiesis. Knockdown of TAL1 or LSD1 led to a derepression of the TAL1 target genes in T cell acute lymphoblast leukemia (T-ALL) Jurkat cells, which is accompanied by elevating promoter H3K4 methylation. Similarly, treatment of PKA activator forskolin resulted in derepression of target genes by reducing its interaction with LSD1 while PKA inhibitor H89 represses them by suppressing H3K4 methylation levels. Consistent with the dual roles of TAL1 in transcription, TAL1 associated LSD1 is decreased while recruitment of hSET1 is increased at the TAL1 targets during erythroid differentiation. This process is accompanied by a dramatic increase in H3K4 methylation. Thus, our data revealed a novel interplay between PKA phosphorylation and TAL1 mediated epigenetic regulation that regulates hematopoietic transcription and differentiation programs during hematopoiesis and leukemogenesis.
Keywords: TAL1/SCL, LSD1, H3K4 demethylation, Serine phosphorylation, epigenetic regulation, leukemogenesis
INTRODUCTION
TAL1/SCL (hereafter referred as TAL1) is a hematopoietic specific member of the basic Helix-Loop-Helix (bHLH) family of transcription factors important for the development of all hematopoietic lineages and pathogenesis of T-cell acute lymphoblastic leukemia (T-ALL) (1–5). TAL1 heterodimerizes with ubiquitously expressed E2A/HEB proteins and binds a hexanucleotide sequence CANNTG termed E box to act as a regulator of transcription during hematopoiesis. TAL1 is required for specification and self-renewal of hematopoietic stem cells (HSCs), as well as for terminal differentiation of erythrocytes (6, 7). It is believed that TAL1 contributes mainly to transcriptional repression in early developmental stages and is then converted to an activator in complex with other erythroid specific transcription factors, such as GATA-1, LMO2, and LDB1, to activate erythroid specific transcription programs during differentiation (8, 9). Several studies have shown that TAL1 could function either as a transcription activator or as a repressor in a context dependent fashion. TAL1 represses transcription by recruiting ETO2, mSin3A, HDAC1/2 or LSD1. In contrast, it potentiates transcription by interacting with p300 and p300/CBP associated factor (PCAF) (10–16). Many of the TAL1 associated proteins possess intrinsic histone modification activities, thereby, altering enhancer/promoter chromatin structure and modulating the recruitment of RNA Pol II basal transcription machinery. In addition, there is another layer of regulation of TAL1 protein. Several threonine (Thr) and serine (Ser) residues of TAL1 can be phosphorylated by protein kinase B (Akt), mitogen activated protein kinase (MAPK), and protein kinase A (PKA) (17–20). In particular, serine 172 is phosphorylated in vitro and in vivo by PKA (18). Phosphorylation of this residue affects TAL1 target-dependent DNA recognition, therefore, potentially discriminating the DNA binding site selection during differentiation (18). The phosphorylation of TAL1 regulates its DNA binding and transcriptional activities (17, 19) suggesting that phosphorylation may dictate TAL1’s ability to associate with transcriptional corepressors and coactivators (21).
In T-cell leukemia, TAL1 activation has been frequently associated with this T-cell neoplastic disease (22, 23). Ectopic expression of TAL1 in T cells led to the development of leukemia and lymphoma in mice (24–26). Furthermore, the gene expression profiling analysis of TAL1 induced preleukemic thymocytes showed that aberrant activation of TAL1 in T-cells results in the repression of many more genes than genes being activated (27), suggesting that TAL1 mediated transcriptional repression plays a critical role in T cell leukemogenesis. Among them, pre-TCRα (pTα) and CD4, which are critical for T cell development (27, 28), as well as p16INK4A and p21, two important cell cycle regulators, are repressed by TAL1 in T-cell leukemia (15, 29–31). Consistent with these findings, it has been shown that TAL1 prevents erythroid progenitor differentiation by recruiting histone demethylase LSD1 and inhibits the TAL1 target genes required for differentiation (13). Interestingly, TAL1 decreases its interaction with LSD1 during differentiation of erythroid cells and is switched to a transcriptional activator to activate erythroid specific p4.2 gene expression (13). Apparently, the dynamic regulation of TAL1 and its associated coregulators may determine the onset of the hematopoietic differentiation program. However, it remains unknown how TAL1/LSD1 interaction is regulated during hematopoiesis.
During hematopoiesis, pluripotent HSCs are associated with both H3K4me3 and H3K27me3 to form so-called “bivalent domains” that facilitate differentiation into specific lineages by changing the relative level of specific modification and allow lineage-specific genes to be rapidly transcribed or vice-versa (32–34). LSD1 is the first identified histone lysine-specific demethylase involved in removing mono- or dimethylated H3K4 (35). As a putative transcriptional repressor, LSD1 associates with multiple corepressor complexes including COREST and NURD complexes (36–38). Histone deacetylase activities of these complexes facilitate the LSD1 demethylase enzymatic activity on the chromatin template and therefore, enhance its repressive function (39). LSD1 regulates a variety of target genes that are required for cell survival, growth, and differentiation through its ability to modulate promoter chromatin structure (38, 40). Dysregulation of LSD1 function has been associated with genome instability and tumor metastasis (37, 40, 41). Interestingly, knock down of LSD1 in hematopoietic progenitors impairs differentiation of multiple hematopoietic lineages indicating that LSD1 plays a critical role in hematopoiesis (13, 42).
To understand the detailed mechanism that regulates TAL1-associated coregulator complexes and how the dysregulation of this process contributes to T-cell leukemia, we investigated the underlying molecular mechanism that regulates the TAL1 and LSD1 interaction. We report here that the interaction between TAL1 and LSD1 is modulated by a PKA-mediated serine 172 phosphorylation located at the LSD1 interacting domain of TAL1. Phosphorylation mediated dynamic recruitment of LSD1 complex modulates TAL1 directed transcription of target genes that are essential for hematopoietic progenitor differentiation and T-cell development. Treatment of the PKA activator forskolin stimulates the expression of the TAL1 target genes by reducing its interaction with LSD1 while PKA inhibitor H89 further inhibits the TAL1 targets by suppressing promoter H3K4 methylation levels. Thus, our results suggest that the dynamic regulation of TAL1 and its associated histone demethylase LSD1 by PKA mediated phosphorylation controls the transcription of the TAL1 target genes via modulating the promoter H3K4 methylation.
RESULTS
PKA mediated Serine172 phosphorylation destabilizes the TAL1 and LSD1 interaction
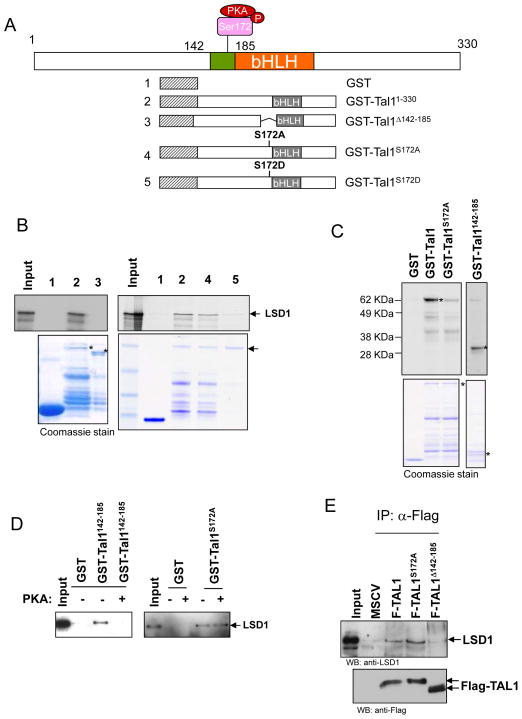
We reported recently that TAL1 dynamically interacts with histone demethylase LSD1 to negatively regulate TAL1 dependent transcription during erythroid differentiation (13). To further understand the molecular mechanism underlying the regulation of the TAL1-LSD1 interaction during hematopoiesis, we determined the domain of TAL1 required for LSD1 interaction. TAL1 directly interacts with LSD1, and the interacting domain encompasses amino acids (aa) 142-185 proximal to the bHLH domain (Figure 1A and B) (13), which contains a serine 172 residue that can be phosphorylated by protein kinase A (PKA) (Figure 1A) (18). To test whether serine 172 phosphorylation is important for TAL1 interaction with LSD1, the GST-TAL1 fusion mutants that delete the LSD1 interacting domain encompassing aa142-185, mutate serine 172 to aspartic acid (D), or change serine 172 to alanine (A) were conjugated with glutathione-sepharose beads and incubated with [35S]methionine-labeled LSD1 (Figure 1A). TAL1S172A, which mimicked unphosphorylated TAL1, interacts strongly with LSD1 (Figure 1B, right) while TAL1Δ142-185, which deleted the LSD1 interacting domain (Figure 1B, left), or TAL1S172D, which mimicked phosphorylated TAL1 (Figure 1B, right), completely loses its interaction with LSD1 (Figure 1B) suggesting that the Ser172 of TAL1 plays an important role in regulating TAL1 interaction with LSD1.
Figure 1. Serine 172 phosphorylation of TAL1 controls TAL1 interaction with LSD1.
(A) Schematic representation of TAL1 and GST-TAL1 fusions used in the GST-pull down assay. (B) GST control and GST-TAL1 fusion proteins preabsorbed to glutathione-sepharose beads were incubated with 35S-labeled LSD1. The bound LSD1 was resolved in SDS-PAGE and visualized by fluorograpgy (Top). Coomassie blue-stained gel shows the relative GST fusion protein loading marked by asterisks (Bottom). (C) Ser172 of TAL1 is phosphorylated by PKA in vitro. The purified GST-TAL1 and its mutants were incubated with the catalytic subunit of PKA in the presence of [γ-33P] ATP, resolved in SDS-PAGE, and visualized by autoradiography. (D) The effect of Ser 172 phosphorylation on the LSD1 recruitment. The purified GST-TAL1142-185 or GST-TAL1S172A was incubated with ATP in the presence or absence of the catalytic subunit of PKA. The reaction was further incubated with purified LSD1. The bound LSD1 was resolved in SDS-PAGE and visualized by Western blotting analysis. (E) Nuclear extracts from MEL cells transduced with vector control, TAL1, TAL1S172A, and TAL1Δ142-185 were precipitated with anti-Flag conjugated beads and analyzed by Western blot with LSD1 antibody (Top) and Flag antibody (Bottom).
It has been reported that Ser172 of TAL1 can be phosphorylated by PKA in vitro and in vivo (18). To test whether Ser172 of TAL1 is phosphorylated by PKA and whether PKA mediated phosphorylation alters the TAL1 and LSD1 interaction, we carried out in vitro phosphorylation assay using purified GST-TAL1 fusion protein or the corresponding TAL1 fusion with a ser172 to Ala substitution (GST-TAL1S172A) as substrate. Compared to GST alone control, GST-TAL1 fusion protein was strongly phosphorylated by PKA (Figuer 1C). Substitution of Ser172 to Ala dramatically reduced the PKA-mediated phosphorylation (Figure 1C) indicating that Ser172 in TAL1 is a major target of PKA. In addition, the LSD1 interacting domain of TAL1 (GST-TAL1142-185) was also strongly phosphorylated (Figure 1C). Having demonstrated that Ser172 of TAL1 is indeed phosphorylated by PKA, we then asked whether phosphorylation of Ser172 by PKA altered the interaction between TAL1 and LSD1. To test this hypothesis, the fusion protein containing wild-type TAL1 (GST-TAL1), the LSD1 interacting domain of TAL1 (GST-TAL1142-185), or the Ser172 to Ala substitution mutant (GST-TAL1S172A) was first phosphorylated in the presence or absence of PKA catalytic subunit, and then incubated with the purified LSD1. Phosphorylation of the LSD1 interacting domain of TAL1 or wt TAL1 completely lost its interaction with LSD1 (Figure 1D, left, and S1A) while incubation of GST-TAL1S172A with or without PKA catalytic subunit did not affect TAL1’s interaction with LSD1 (Figure 1D, right). Collectively, these results demonstrate that phosphorylation of serine 172 of TAL1 destabilizes its interaction with LSD1.
Furthermore, to test whether phosphorylation of Ser172 affects the TAL1 and LSD1 interaction in vivo, we stably expressed flag tagged TAL1 mutants in murine erythroleukemia (MEL) cells treated with DMSO for 24 hour. It has been reported that the DMSO treatment of MEL cells dramatically decreased the TAL1-LSD1 interaction (13). The expressed Flag tagged TAL1 mutants were then precipitated with Flag antibody and blotted with LSD1 antibody. While the TAL1S172A mutant enhances the interaction with LSD1 in comparison to the wild-type (wt) TAL1, TAL1Δ142-185 specifically loses its interaction with LSD1 (Figure 1E). Thus, both in vitro and in vivo data indicate that serine 172 phosphorylation of TAL1 plays an important role in regulating its interaction with LSD1.
Serine 172 is critical for regulating the recruitment of LSD1, H3K4methylation, and TAL1 mediated repression of p4.2 gene
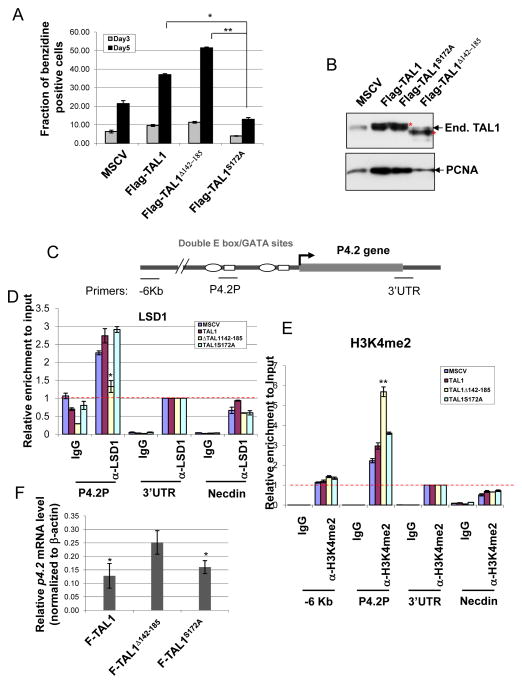
Next, we test whether phosphorylation of ser172 modulates erythroid progenitor differentiation by regulating TAL1 target gene transcription. To test this possibility, MEL cells expressing retroviral vectors encoded wt TAL1, TAL1S172A, or TAL1Δ142-185 cDNA were treated with 1.5% DMSO and then examined for erythroid differentiation by benzidine staining of the accumulation of hemoglobin. Compared to the wt TAL1 control, TAL1S172A which mimics unphosphorylated TAL1 and increases its interaction with LSD1 consistently inhibited DMSO-induced erythroid differentiation of MEL cells (Figure 2A). Meanwhile, the deletion of the LSD1 interacting domain in TAL1 significantly promoted erythroid differentiation (Figure 2A). The levels of Flag TAL1 expression were compatible between WT and mutants as well as that of endogenous TAL1 (Figure 2B). These data suggest that serine 172 of TAL1 regulates TAL1 directed erythroid differentiation by modulating its interaction with histone demethylase LSD1.
Figure 2. Serine 172 is critical for regulating the recruitment of LSD1, H3K4methylation, and TAL1 mediated repression of p4.2 gene.
(A) MEL cells were stably infected with retroviruses encoding the vector control, Flag-TAL1, Flag-TAL1Δ142-185, and Flag-TAL1S172A. The cells were induced to differentiation by addition of 1.5% DMSO for 3 and 5 days. The cells were collected for analysis of hemoglobin expression by using benzidine staining and counting consecutive 200 cells in the field. The data were collected from three independent experiments. (B) The levels of endogenous and exogenous TAL1 and their mutants expressed in MEL cells were determined by Western Blotting analysis (Top). The levels of PCNA (Bottom) were shown as a loading control. (C) Schematic representation of the mouse p4.2 gene locus. (D and E) Cross-linked chromatins from MEL cells infected with retrovirus encoding Flag-TAL1 and its mutants were precipitated with LSD1 (D) and H3K4me2 (E) antibodies. The precipitated chromatins were purified and analyzed by real-time PCR. The recruitment of LSD1 or enrichment of H3K4me2 is significantly difference in the Flag-TAL1Δ142-185 expressed cells in comparison to that of the MSCV, Flag-TAL1, and Flag-TAL1S172A expressed cells. (F) Total RNA from retrovirus infected MEL cells were isolated and analyzed for P4.2 expression by RT-qPCR. *P<0.05; **P<0.01 by student t test. Shown are the mean ± SEM of at least 3 independent experiments. The red dotted line represents background levels of signal.
Given that LSD1 is a H3K4 specific demethylase and modulates transcription by altering promoter chromatin modifications, the TAL1 mutants that change its ability to recruit LSD1 may affect its target gene transcription that is important for erythroid differentiation program. To test this possibility, a chromatin immunoprecipitation (ChIP) was carried out using antibodies against LSD1 and H3K4 dimethylation in MEL cells stably expressed Flag-tagged TAL1, TAL1S172A, and TAL1Δ142-185 to examine the effects of TAL1 mutants on LSD1 recruitment and promoter activity. As we expected, LSD1 interacted with the TAL1 binding sites, E box-GATA elements, at the proximal EPB4.2 (P4.2) promoter in the control and wt TAL1 transduced cells (Figure 2C and D). Deletion of the LSD1 interacting domain in TAL1 resulted in a significant loss of LSD1 recruitment while TAL1S172A didn’t affect its interaction with LSD1 at the P4.2 promoter in undifferentiated MEL cells (Figure 2D). As a result, H3K4 dimethylation is significantly elevated at the P4.2 promoter and the level of p4.2 mRNA is also increased in the TAL1Δ142-185 expressed MEL cells (Figure 2E and F). Taken together, the results reveal that the serine 172 of TAL1 which can be phosphorylated by PKA regulates TAL1 dependent transcriptional activity and erythroid differentiation program by modulating its dynamic association with H3K4 demethylase LSD1.
Serine 172 of TAL1 modulates TAL1 repressive activity in T-ALL cells
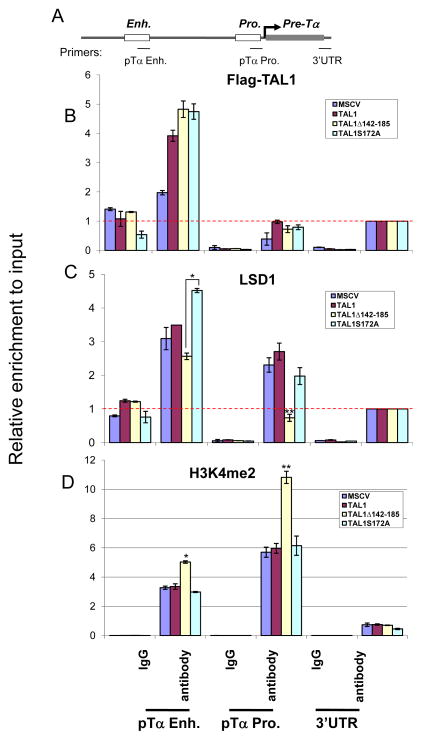
Activation of TAL1 is the most common gain-of-function mutation found in T-ALL patients (22, 43–45). In T-cell leukemia, TAL1 suppresses T cell differentiation and perturbs cell cycle progression during the double negative T cell stage (28). pre-TCRα (pTα) is exclusively expressed in developing thymocytes and is an essential selection and survival signal for the development of T thymocytes (30, 46). E-box motifs in the enhancer region of the pTα gene are required for its activation by E2A/HEB heterodimer in normal T-cell development and are occupied and repressed by TAL1 in Jurkat cells (Figure 3A) (46). Sequence comparison showed a 70.5% identity between mouse and human pTα enhancer sequences (Figure S1B). We further hypothesized that the TAL1 mutants that change TAL1’s ability to recruit LSD1 may affect T cell growth by altering the transcription of TAL1 target genes such as pTα and p21 in T-cell leukemia. To test this possibility, we stably expressed Flag-tagged TAL1 and its mutants, TAL1S172A and TAL1Δ142-185 that changed the TAL1’s affinity to LSD1, in T-ALL Jurkat cells (Figure S1C). ChIP assays were then performed in Jurkat cells that stably expressed Flag-TAL1 and its mutants using Flag, LSD1 and H3K4me2 antibodies to test the effects of mutations on LSD1 recruitment and H3K4 methylation at the pTα and p21 enhancers. Flag tagged TAL1 and its mutants bind equally to the enhancer region of the pTα gene (Figure 3B). Interestingly, correlated with TAL1 binding, LSD1 is also recruited to the same region of the pTα enhancer except for the TAL1Δ142-185 mutant, which lacks the LSD1 interacting domain and significantly reduces LSD1 recruitment at the enhancer (Figure 3C). The reduction of LSD1 recruitment was also seen at the promoter region (Figure 3C) (See discussion). Due to the decrease in the LSD1 recruitment, the expression of TAL1Δ142-185 led to an increase in H3K4me2 at the enhancers and the proximal promoters of the pTα gene (Figure 3D) as well as the p21 gene (Figure S2C), a cell cycle-dependent kinase inhibitor that blocks the cell cycle during G1 to S phase transition and is also targeted by TAL1 (15, 30). As a control, the TAL1 mutants didn’t affect the level of H3K4me2 at the promoter of TAL1 activated target, Runx1 (Figure S2E). These results suggest that LSD1 mediated epigenetic modification is important for TAL1 repressive action in leukemogenesis and the action of LSD1 may be also dependent on serine 172 phosphorylation.
Figure 3. Serine 172 of TAL1 modulates TAL1 repressive activity in T-ALL cells.
(A) Schematic representation of the pre-Tα gene locus. (B and C) ChIP analysis of TAL1 (B) and LSD1 (C) interactions with the Pre-Tα enhancer and promoter using cross-linked chromatin from Jurkat cells transduced with wide type Flag-TAL1, Flag-TAL1Δ142-185, and Flag-TAL1S172A. The recruitment of LSD1 is significantly reduced in the Flag-TAL1Δ142-185 expressed cells compared to that of the MSCV, Flag-TAL1, and Flag-TAL1S172A expressed cells. (D) ChIP analysis of H3K4me2 enrichment at the Pre-Tα enhancer and promoter from Jurkat cells transduced with wide type Flag-TAL1, Flag-TAL1Δ142-185, and Flag-TAL1S172A. The enrichment of H3K4me2 is significantly increased in the Flag-TAL1Δ142-185 expressed cells compared to that of the MSCV, Flag-TAL1, and Flag-TAL1S172A expressed cells. *P<0.05; **P<0.01 by student t test. Shown are the mean ± SEM of at least 3 independent experiments. The red dotted line represents background levels of signal.
PKA mediated phosphorylation modulates TAL1 repressive activity in T-ALL cells
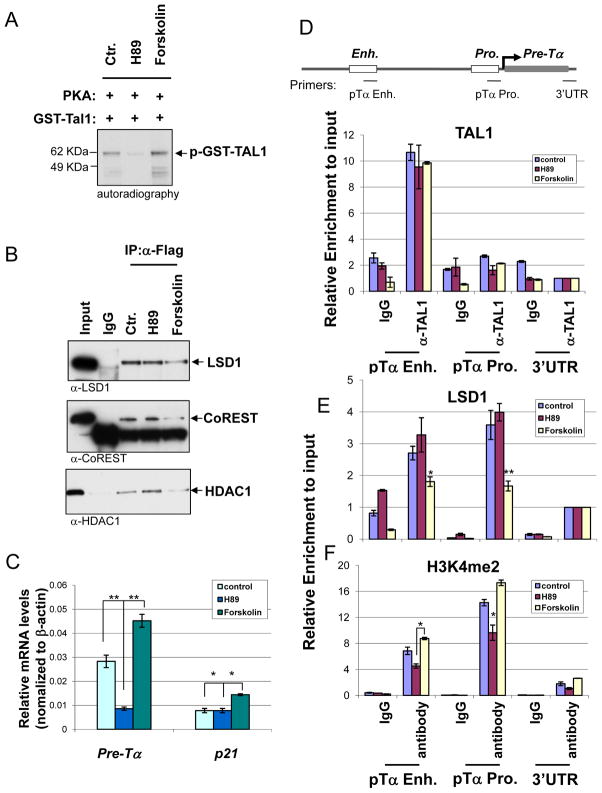
Next, we reasoned if PKA mediated phosphorylation of Ser172 in TAL1 modulates the recruitment of LSD1 and its repressive activity in T-ALL, PKA activator, forskolin, may relief the TAL1 repression while PKA inhibitor should enhance repression. To test this hypothesis, we first asked whether PKA inhibitor, H89, or activator, forskolin, inhibits or enhance PKA mediated TAL1 phosphorylation in vitro, respectively. Compared to the control, the treatment of PKA inhibitor, H89 (20 μM), abolished the phosphorylation of TAL1 while PKA activator, forskolin (40 μM), enhanced it (Figure 4A). To further test whether PKA mediated phosphorylation contributes to the regulation of the interaction between TAL1 and LSD1, the Flag tagged TAL1 expressed Jurkat cells were treated with PKA inhibitor H89 or PKA activator forskolin in the same concentration described above. Nuclear extracts were prepared and examined for the TAL1-LSD1 interaction by co-immunoprecipitation assays using antibody specific to Flag (Figure 4B) or TAL1 (Figure S2A). The PKA inhibitor H89 that prevents TAL1 phosphorylation, didn’t affect TAL1 interaction with the components of LSD1 complex, LSD1 (Top), CoREST (Middle), and HDAC1 (Bottom) (Figure 4B and S2A). In contrast, forskolin, an activator of PKA and stimulator of ser172 phosphorylation of TAL1 (Figure 1C, 4A, and S2A bottom), drastically decreased the TAL1-LSD1 complex interaction in Jurkat cells (Figure 4B and S2A). The treatment of H89 and forskolin didn’t affect the expression of LSD1 (Figure S2B). Thus, the data suggest that ser172 phosphorylation of TAL1 by PKA destabilizes TAL1 interaction with histone demethylase LSD1 complex in T-ALL Jurkat cells.
Figure 4. PKA mediated phosphorylation of TAL1 modulates TAL1 repressive activity in T-ALL cells.
(A) The purified GST-TAL1 was phosphorylated with the catalytic subunit of PKA and [γ-33P] ATP in the presence or absence of PKA inhibitor, H89 (20μM), or PKA activator, Foskolin (40μM). The phosphoprotein was resolved in SDS-PAGE and visualized by autoradiography. (B) Flag tagged TAL1 expressed Jurkat cells were treated with PKA inhibitor H89 or PKA activator forskolin for 2 hrs. The nuclear extracts were prepared and precipitated with Flag-specific antibody. The effect of PKA inhibitor or activator on the recruitment of LSD1 complex was analyzed by WB using antibodies specific to LSD1 (Top), CoREST (Middle), and HDAC1 (Bottom). (C) Total RNA from Jurkat cells treated with PKA inhibitor H89 or PKA activator forskolin for 2 hrs was isolated and analyzed for Pre-Tα and P21 expressions by RT-qPCR. (D and E) ChIP analysis of TAL1 (D) and LSD1 (E) interactions with the Pre-Tα enhancer and promoter using cross-linked chromatin from Jurkat cells treated with PKA inhibitor H89 and activator forskolin. The recruitment of LSD1 is significantly compromised in forskolin treatment compared to the control and H89 treatment. (F) ChIP analysis of TAL1 H3K4me2 enrichment at the Pre-Tα enhancer and promoter using cross-linked chromatin from Jurkat cells treated with PKA inhibitor H89 and activator forskolin. The enrichment of H3K4me2 is significantly increased in forskolin treatment compared to the H89 treatment. *P<0.05; **P<0.01 by student t test. Shown are the mean ± SEM of at least 3 independent experiments.
In addition, to test whether PKA mediated phosphorylation of serine 172 of TAL1 regulates TAL1 transcriptional activity in T-ALL, Jurkat cells were treated with 20 μM and 40 μM of PKA inhibitor H89 and PKA activator forskolin for 2 hrs, respectively. As expected, H89, which inhibits PKA mediated phosphorylation of TAL1 (Figure 4A) and enhances the TAL1-LSD1 interaction, suppresses pre-Tα transcription in comparison to the untreated control (Figure 4C). In contrast, forskolin that destabilizes TAL1-LSD1 interaction by activating PKA activity (Figure 4A and S2A bottom) stimulates the expression of the pTα and the p21 genes (Figure 4C). Although neither H89 nor forskolin affects TAL1 binding to the pre-Tα enhancer (Figure 4D), the recruitment of LSD1 to the pTα gene is significantly impaired by the PKA activator forskolin treatment (Figure 4E). As a consequence of the change of LSD1 recruitment, H89 inhibits H3K4 dimethylation while forskolin stimulates H3K4 methylation at the pTα gene (Figure 4F). The similar results have also been observed in the p21 locus (Figure S2D). It is interesting to note that although H89 slightly enhances the LSD1 recruitment but still inhibits H3K4 dimethylation. It is possible that the TAL1 phosphorylation level is low in Jurkat cells and H89 treatment would not increase the TAL1 associated LSD1, but strengthen their interaction. It is also possible that H89 has a negative effect on TAL1’s ability to recruitment coactivator such as hSET1. Nevertheless, the changes of TAL1 phosphorylation status dynamically regulate its interaction with LSD1 and the target promoter H3K4 methylation. Although TAL1 binds only to the pTα enhancer, the recruitment of LSD1 and reduction of H3K4me2 were observed in both enhancer and promoter (Figure 4D–F) that might be due to a possible enhancer and promoter interaction during transcriptional regulation (See Discussion). Taken together, our data indicate that modulation of PKA activity regulates the dynamic interactions between TAL1 and LSD1, which then controls transcription of the TAL1 target genes during leukemogenesis.
LSD1 is required for transcriptional repression of TAL1 target genes in T-ALL cells
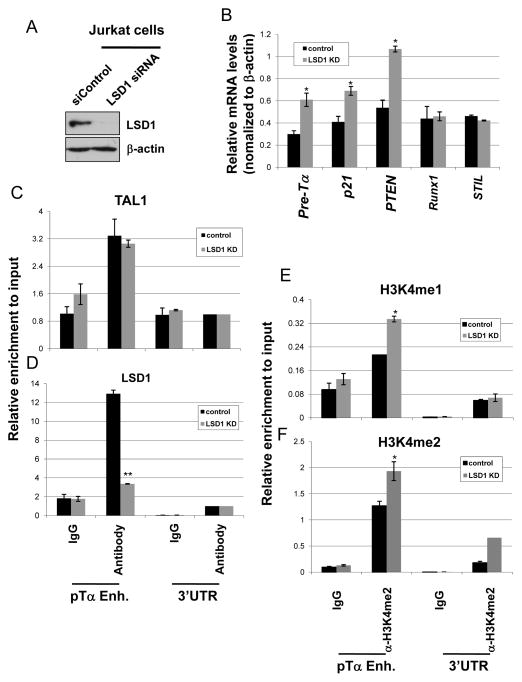
TAL1 is frequently activated in the majority of T-ALL patients (22, 43–45). The repressive activity of TAL1 has been linked to leukemogenesis (21, 47). In fact, the gene expression profiling analysis of the TAL1 induced preleukemic thymocytes shows that aberrant activation of TAL1 in T cells leads to the repression of many more genes than genes being activated (27). We then asked whether LSD1 is required for TAL1 dependent transcriptional repression in T-ALL leukemia cells. To test this hypothesis, LSD1 was knocked down in T-ALL Jurkat cells by SMART pool LSD1 siRNAs (Figure 5A). KD of LSD1 led to a strong derepression of the TAL1 targets, pTα and p21, as well as a known LSD1 target PTEN gene (Figure 5B) which was recently shown to be a direct TAL1 target in T-ALL cells (48). The effects of LSD1 appear to be specific for the TAL1 repressive targets because KD of LSD1 didn’t affect the TAL1 activated target, Runx1 (48), and unrelated STIL genes (Figure 5B). We further examined the TAL1 binding, LSD1 recruitment, and H3K4methylation at the pTα gene enhancer to test whether LSD1 is required for the inhibition of the TAL1 target genes, such as pTα and p21, in T-ALL cells. Figure 5 shows that KD of LSD1 didn’t affect the interaction of TAL1 with chromatin (Figure 5C). However, correlated with the derepression of the pTα and p21genes (Figure 5B), KD of LSD1 that eliminated LSD1 recruitment (Figure 5D) increases the levels of H3K4me1 and H3K4me2 at the TAL1 bound pTα enhancer (Figure 5E and F) as well as at the p21 enhancer (Figure S2F). Collectively, our data demonstrate that LSD1 recruitment is required for inhibiting TAL1 target genes that are required for the T-cell development in Jurkat T-ALL cells.
Figure 5. LSD1 is required for TAL1 mediated transcription repression in T-ALL cells.
(A) Western blot analysis of LSD1 and β-actin protein levels from the LSD1 KD Jurkat cells. (B) Total RNA from the vector control and LSD1 KD Jurkat cells was isolated and analyzed for the pre-Tα P21, PTEN, Runx1, and STIL expression by RT-qPCR. (C) ChIP analysis of TAL1 interaction with the Pre-Tα enhancer comparing the vector control and LSD1 KD Jurkat cells. (D) ChIP analysis of LSD1 interaction with the Pre-Tα enhancer comparing the vector control and LSD1 KD Jurkat cells. (E and F) ChIP analysis of H3K4me1 (E) and H3K4me2 (F) patterns at the Pre-Tα locus comparing the vector control and LSD1 KD Jurkat cells. *P<0.05; **P<0.01 by student t test. Shown are the mean ± SEM of at least 3 independent experiments.
LSD1 is recruited by TAL1 to repress genes required for T cell development
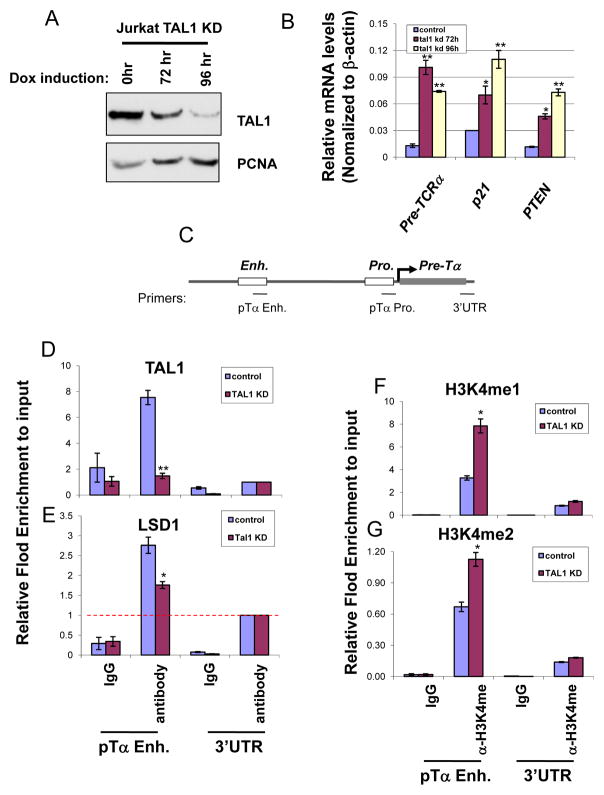
Next, we sought to determine whether LSD1 is recruited by TAL1 to regulate genes important for T cell development in Jurkat cells. The cells were stably expressed a doxycycline (Dox) inducible shRNA specifically targeting the human TAL1 sequence (48). TAL1 levels are dramatically diminished in the transduced Jurkat cells at 96 hrs upon Dox induction (Figure 6A). TAL1 knocked down (KD) Jurkat cells drastically decreased cell proliferation and colony formation abilities comparing to the uninduced Jurkat cells in the soft agar colony assay (Figure S3A), supporting that TAL1 is required for T-cell leukemic transformation (23). Furthermore, the loss of the colony formation may also be due to a significant increase in apoptotic cells in the Dox-induced TAL1 KD Jurkat cells (Figure S3B, bottom blue color). Importantly, KD of TAL1 resulted in a strong derepression of pTαp21, and PTEN genes (Figure 6B). As a control, KD of TAL1 in Jurkat cells reduced its activated target Runx1 (48) and did not affect unrelated STIL expression (Figure S3C). We, therefore, tested whether the loss of TAL1 leads to a decrease in LSD1 recruitment at the pTα enhancer (Figure 6C), which may contribute to the derepression of the pTα gene in the induced KD cells (Figure 6B). Although TAL1 and LSD1 are colocalized at the pTα enhancer in control cells (Figure 6D and E), the recruitment of LSD1 to the enhancer is dependent on the TAL1 DNA binding because the KD of TAL1 resulted in a significant decrease in LSD1 binding at the enhancer (Figure 6D and E). As a result of the loss of LSD1 recruitment, both H3K4me1 and H3K4me2 marks were increased at the pTα (Figure 6Fand G) and p21 (Figure S3D and E) enhancers, which correlate with the derepression of the pTα and p21 genes in the induced TAL1 KD Jurkat cells (Figure 6B). The results reveal that TAL1 represses target gene transcription by recruiting LSD1 and removing H3K4 methylation marks in T-cell leukemia.
Figure 6. KD of TAL1 leads to reduced recruitment of LSD1 and derepression of target genes.
(A) Inducible TAL1 knock down in Jurkat cells. TAL1 level is reduced in 96 hr. upon doxycycline treatment as determined by Western Blotting analysis. (B) Total RNA from Dox treated Jurkat cells for 0, 72, and 96 hrs was isolated and analyzed for the expression levels of pre-TαP21, and PTEN by RT-qPCR. (C) Schematic representation of the pre-Tα gene locus. (D and E) ChIP analysis showing TAL1 (D) and LSD1 (E) binding to the pre-Tα enhancer upon TAL1 knock down in Jurkat cells. (E) ChIP analysis showing H3K4me1 (F) and H3K4me2 (G) patterns at the pre-Tα enhancer upon TAL1 knock down in Jurkat cells. *P<0.05; **P<0.01 by student t test. Shown are the mean ± SEM of at least 3 independent experiments.
Activation of the TAL1 dependent p4.2 transcription and erythroid differentiation is associated with decreased LSD1 recruitment and increased H3K4 methylation
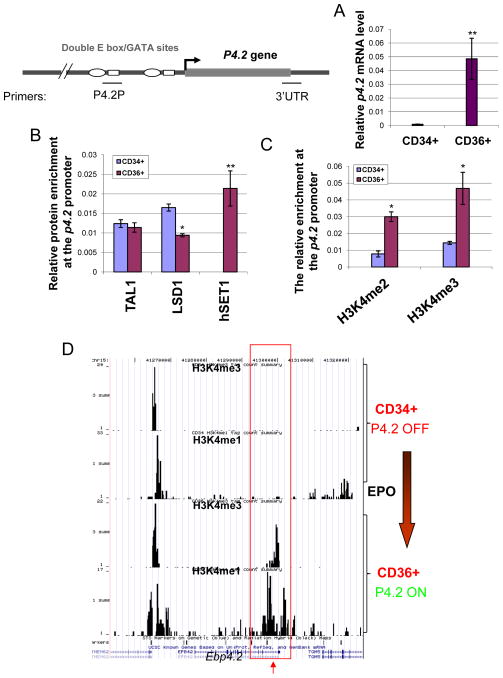
Finally, we tested whether the dynamic interaction between TAL1 and LSD1 is functionally important for TAL1 mediated erythroid differentiation of CD34+ HSCs. CD34+ HSCs were differentiated into CD36+ erythroid precursors with the addition of erythropoietin (EPO). P4.2 transcription is strongly induced in CD 36+ erythrocytes (Figure 7A). Consistent with the induction of P4.2 expression, ChIP assay showed that although TAL1 binds equally to the P4.2 proximal promoter in CD34 and CD36 cells, LSD1 recruitment decreased by 45% at the P4.2 promoter in CD36 cells (Figure 7B). Surprisingly, hSET1, a histone H3K4 methyltransferase, was recruited to the promoter only in CD36+ erythroid precursors, but not in CD34+ HSCs (Figure 7B). Furthermore, the inhibition of PKA activity by PKA inhibitor H89 in CD34+ HSCs blocks EPO induced differentiation and inhibits p4.2 transcription (Figure S4A and B). We observed a 10% decrease in CD36+ erythroid precursor in the H89 treated cells (Figure S4A). Together, these data are consistent with the notion that PKA mediated phosphorylation of TAL1 destabilizes the TAL1 and LSD1 interaction during hematopoiesis. The reduction of LSD1 recruitment and the increase in hSET1 binding strongly correlate with the elevation of H3K4me2 and H3K4me3 levels at the p4.2 promoter region upon differentiation (Figure 7C). We further performed an unbiased genome-wide ChIP-seq analysis of H3K4 mono- and tri methyl H3K4 modification patterns at the p4.2 locus comparing CD34+ HSCs to CD36+ erythroid precursors. Correlating with the transcriptional activity of the P4.2 gene and the hSET1 recruitment (Figure 7A and B), both H3K4 mono- and trimethylation are only enriched at the proximal P4.2 promoter in CD36+ erythroid precursors but not in the P4.2 silenced CD34+ HSCs (Figure 7D) consistent with the ChIP data (Figure 7C). Thus, our data suggest that the levels of H3K4 methylations play an important role in p4.2 activation during erythroid differentiation. Together, our results reveal that the dynamic regulation of H3K4 demethylase and methyltransferase, perhaps by PKA mediated phosphorylation of TAL1, plays a critical role in controlling erythroid-specific gene transcription during hematopoiesis.
Figure 7. The positive role of hSET1 and H3K4 methylation in TAL1 directed erythroid transcription program.
(A) Total RNA from CD34+ HSCs and CD36+ erythroid precursors were isolated and analyzed for the P4.2 expression by RT-qPCR. (B) ChIP analysis of TAL1, LSD1, and hSET1 binding patterns at the P4.2 promoter in CD34+ and CD36+ cells. (C) ChIP analysis of the levels of H3K4me2 and H3K4me3 at the p4.2 promoter in CD34+ HSC and CD 36+ erythrocytes. *P<0.05; **P<0.01 by student t test. Shown are the mean ± SEM of 3 independent experiments. (D) ChIP-seq genome browser data showing the patterns of H3K4me1 and H3K4me3 across the P4.2 locus in CD34+ HSCs versus CD36+erythroid precursors.
DISCUSSION
As a master regulator of hematopoiesis, TAL1 transcriptional activity has been attributed to its ability to recruit transcriptional corepressors and coactivators (10–15). Dynamic interactions between TAL1 and co-regulators result in different transcriptional consequences affecting hematopoietic cell differentiation and proliferation. TAL1 functions as an activator of erythroid differentiation programs while its repressive activity has been linked to oncogenesis in T- cell lineage (8, 9, 21, 27, 28, 49). It has been suggested that inappropriate associations of transcription factors with histone modifying enzymes often lead to aberrant activation or repression of normal gene expression patterns and perturb cellular differentiation and proliferation that may eventually contribute to malignancies (21, 49). How is the dynamic interaction of TAL1 with corepressors and coactivators properly modulated during hematopoiesis? What is the signal(s) that turns this molecular circuit on and off? We previously showed that TAL1 associates with histone demethylase LSD1 in erythroleukemia cells. LSD1 removes the methyl group from histone H3 lysine 4 of the TAL1 target genes and restricts TAL1 function in erythropoiesis (13). Upon induced erythroid differentiation, LSD1 may be released from the TAL1 complex and allow it to function as an activator, presumably recruiting coactivators for activating erythroid transcription programs (21) (Figure 7). Although the dramatically increase in hSET1 recruitment alone may be attributed to the elevation of H3K4me2 levels at the P4.2 promoter, The PKA inhibitor H89 blocks CD34+ HSC differentiation (Figure S4) (50) suggesting that CD34+ cell differentiation in part is mediated through the activation of the PKA pathway, perhaps by regulating TAL1 associated coregulators, such as LSD1. During T cell leukemogenesis, TAL1 and LSD1 also cooperatively inhibit the transcription of pTa, p21, and Pten genes, which play critical functions in T cell survival, proliferation, and differentiation (Figures 5 and 6). We further demonstrated that the dynamic interaction of TAL1 with H3K4 demethylase LSD1 is regulated by PKA mediated serine 172 phosphorylation of TAL1 protein. A key finding of this work is that PKA signal pathway regulates TAL1 association with LSD1 in the normal erythroid differentiation program and leukemogenesis of T-cell lineages. The phosphorylation of serine 172 of TAL1 specifically destabilizes TAL1 interaction with histone demethylase LSD1 and, therefore, leads to the activation of the certain TAL1 target genes in differentiated erythroid cells or T-cell leukemia. It was reported that TAL1 is phosphorylated in vitro and in vivo by PKA (18). Although the subcellular localization of TAL1 and its ability for the dimerization with E2A protein were not affected by this phosphorylation, The ser172 phosphorylation has a target-dependent effect on TAL1 DNA binding activity, thereby affecting transcription activities of TAL1 (18). The effect on the TAL1 and LSD1 interaction described in this report further illustrates the role of PKA mediated phosphorylation in TAL1’s function in transcriptional regulation. Taken together, these studies provide a potential explanation and underlying mechanism of how TAL1 dynamically interacts with corepressor LSD1 on certain targets in the same cells according to their differentiation stage.
TAL1 activity can be regulated by phosphorylation. Phosphorylations of Thr90 by Akt kinase and Ser122 by MAP kinase enhance transcriptional activity in TAL1 (17, 20). Interestingly, Ser172 which is located within the LSD1 interacting domain was also phosphorylated by PKA kinase in vitro and in vivo (18). In addition, we demonstrated in this report that in vitro phosphorylation of TAL1 by PKA abolished the interaction between TAL1 and LSD1 while substitution of Ser172 to Ala did not affect the interaction (Figure 1) indicating that phosphorylation of ser172 in TAL1 by PKA destabilizes the TAL1 ability to associate with corepressor LSD1. Consistent with the positive role of the phosphorylated TAL1, we found that Ser172 phosphorylation acts as an epigenetic switch that potentiates TAL1 transcription by altering TAL1-associated histone modifying enzymes, such as LSD1, and changes H3K4 methylation marks at TAL1 target genes. It is also interesting to note that the association between TAL1 and mSin3A is regulated through the acetylation of TAL1 by the histone acetyltransferase PCAF. mSin3A containing complexes repress transcription by removing the acetyl group, another active histone mark, from histone H3 and H4 tails (11). Thus, both Ser172 phosphorylation and acetylation of TAL1 destabilize different corepressors that restrict TAL1 function in hematopoietic differentiation. Cooperation between ser172 phosphorylation and lysine acetylation of TAL1 may play an important role in modulating TAL1 activity in hematopoietic proliferation and differentiation.
In addition to histone modification enzymes, corepressor ETO-2 was found in TAL1 containing multiple protein complexes (14–16). ETO-2 is a member of the ETO (eight-twenty-one) family of transcription repressors which are frequently involved in chromosome translocation in acute leukemia (49). ETO-2 is recruited by TAL1 to repress erythroid-specific target gene transcription in undifferentiated erythroid progenitors. Similar to the recruitment of LSD1, the presence of ETO-2 at the TAL1 targeted erythroid genes decreases during terminal erythroid differentiation (16). What is the signal(s) that turns off the interaction between TAL1 and ETO-2? Does PKA mediated phosphorylation of TAL1 also regulates this interaction? It has been reported that ETO-2 can mediate the interaction between TAL1 and Gfi-1b in erythroid cells (14). In fact, both TAL1 and Gfi-1b interact with histone demethylase LSD1 (13, 42). In addition, ETO-2 is present in the TAL1/LSD1 complexes from biochemical purification in K562 and Jurkat cells (13) suggesting that TAL1, ETO-2, and LSD1 might present in the same repressor complex to inhibit erythroid differentiation program. However, it remains to be further determined whether PKA signaling also regulates TAL1/ETO-2 interaction in erythropoiesis.
What are the roles of LSD1 in normal versus malignant hematopoiesis? Hematopoiesis is a complex process that involves transcription factors committing and differentiating HSCs into progenitor cells that progressively restrict their proliferation potential. The process of hematopoietic cell differentiation is frequently perturbed in leukemia. TAL1 is a master regulator of this process whose activity modulates hematopoietic cell proliferation and differentiation. In this regard, LSD1 controls hematopoietic differentiation by associating with hematopoietic specific transcription factor TAL1 and Gfi-1B (13, 42). TAL1 and LSD1 interaction limits the differentiation of erythroid progenitor cells (13, 42). Inhibiting the interaction between TAL1 and LSD1 is critical in allowing erythroid progenitors to differentiate. We demonstrated here that this process is regulated by PKA mediated phosphorylation. During erythroid differentiation, we observed that hSET1 is recruited to the p4.2 gene when LSD1 is released from the promoter (Figure 7B). hSET1 recruitment contributes to H3K4 methylation of the promoter and the activation of the p4.2 gene in CD36 erythroid precursors (Figure 7). It still remains to be determined whether hSET1 is recruited by TAL1 or other erythroid specific transcription factors. Regardless, our data suggest that the interplay between signal transduction pathways and histone modifying enzymes may play a critical role in controlling chromatin structure and the timing of hematopoietic programs.
In contrast to normal hematopoiesis, TAL1 was shown to inhibit pre-Tα and CD4 genes in T- cell leukemia (28, 30, 46), which play a central role in the transition of thymocytes from the DN to the DP stage during T-cell development. The repressive action of TAL1 requires the recruitment of LSD1 demethylase (Figures 5 and 6), as well as ETO2 and mSin3A (10, 13–15, 30). Although TAL1 only recognizes the pTα enhancer region, binding of TAL1 at enhancer apparently modulates the recruitment of LSD1 and demethylase activity at the pTα proximal promoter; therefore efficiently repress pTα expression in T-ALL cells (Figure 3 and 4). It is interesting to note that transcription factor binding at enhancer is able to control proximal promoter activity by modulating promoter chromatin modifications in the GATA-2 and β-globin loci (51, 52). Long-range chromatin looping has been shown to mediate efficient communication between enhancers and promoters (53). In this regard, enhancer occupied TAL1 may form a repressive chromatin loop to control the pTα promoter activity, perhaps, mediated by LSD1. The notion is supported by the evidences that LSD1 is required for nuclear receptor mediated epigenetic remodeling of a large chromosomal region through a LSD1-depedent inter- or intrachromatin interaction (54, 55). However, we could not rule out that possibility that the recruitment of LSD1 to the pTα promoter is mediated in part through other transcription factors or mechanisms.
On the other hand, TAL1 also activates a variety of genes affecting T-cell survival and growth, including CDK6, Runx1, and NKX3.1 (23, 48, 56). Given that PKA phosphorylation regulates the TAL1-LSD1 interaction in T-ALL Jurkat cells (Figures 3 and 4), it will be interesting to know if PKA phosphorylation of TAL1 could facilitate TAL1’s ability to recruit coactivators during leukemogenesis. Although it is unknown whether a coactivator(s) is involved in the TAL1 mediated activation of CDK6 and NKX3.1 genes in leukemogenesis, our study identified an underlying mechanism by which phosphorylation regulated demethylase LSD1 recruitment controls the timing of erythroid differentiation and oncogenesis of T-cell leukemia.
Materials and Methods
Cell lines, Virus mediated transduction, and siRNA knockdown
Murine erythroleukemia (MEL) and Jurkat cells were maintained as described (10). For MEL differentiation, 2×105 MEL cells/ml were incubated with 1.5% DMSO for specific time periods as indicated. The expression of hemoglobin was visualized by benzidine reagent (Sigma, St. Louis, MO) as described previously (57, 58). The fraction of hemoglobin-expressing cells was determined from counts of 200 cells in three independent experiments. Human CD34+ and CD36+ cells were enriched through positive immune-selection by flow cytometer and cultured according to the manufacturer’s instruction (Stem Cell Technologies, Vancouver, BC, CA). Plasmid constructs, retrovirus mediated transduction, and siRNA-mediated knockdown were described previously (13). The LSD1 knock-down in Jurkat cells was carried out by transfecting 40 pmol SMART pool LSD1 siRNAs (Cat#:M-009223-00-0050, Dharmacon RNAi technologies) using Nucleofector II. The cells were analyzed in 48 hrs post-transfection. The siRNA sequences are available in the Supplementary materials.
Site-directed mutagenesis and In vitro phosphorylation
Oligonucleotide mediated mutagenesis of Ser 172 to Ala or Ser 172 to Asp was performed on pGex-5X-Tal1 according to the manufacturer’s instructions (Stratagene, La Jolla, CA) and verified by DNA sequencing. For in vitro phosphorylation, 500 ng of the purified GST or GST-TAL1 fusion proteins were phosphorylated by 100 ng of the purified PKA catalytic subunit (65 KDa, Sigma, St. Louis, MO) in a 50 μl reaction volume containing 50 mM Tris-HCl, pH 7.5, 10 mM MgCl2, 200 μM ATP, and 20 μCi of [γ-33P] ATP at 30 °C for 40 min in the presence of phosphotase inhibitor cocktail (PhosStop, Roche, IN).
GST pull-down and in vitro transcription and translation
The GST pull-down assay was performed as described (12). Briefly, GST fusion proteins were synthesized in the BL-21 strain of E.coli and purified on glutathione-sepharose 4B following the manufacturer’s instruction (GE Healthcare). Equal amount of GST proteins were incubated with [35S]methionine labeled or baculoviral synthesized LSD1. The bound protein were resolved by SDS-PAGE and visualized by autoradiography or Western blotting analysis, respectively. [35S]methionine-labeled LSD1 was synthesized using a coupled in vitro transcription-translation system in reticulocyte lysates (Promega) in the presence of translation-grade [35S]methionine (Amersham).
RNA isolation and quantitative RT-PCR
Total RNA was prepared by using the SV total RNA isolation kit according to the manufacturer’s instruction (Promega, Madison, WI). A total of 1.5 μg RNA was treated with RNAse-free DNase I and then reverse transcribed by using the Superscript II reverse Transcriptase, following the manufacturer’s suggestion (Invitrogen). The quantity of cDNA was determined by qPCR analysis using a MyiQ Single-Color Real-Time PCR Detection System (Bio-Rad). Primer sequences and conditions for qPCR are available upon request.
Co-immunoprecipitation and Chromatin immunoprecipitationassay (ChIP)
The TAL1 complexes were immunoprecipitated from prepared nuclear extracts using Flag or TAL1 specific antibodies. The immunocomplexes were resolved in SDS-PAGE and analyzed by Western blotting analysis. TAL1 antibody was generated in rabbits by using human TAL1 fragments (aa 301–317; aa 276–289) as antigens.
Real-time PCR quantitative ChIP assays were carried out as described (57) using indicated antibodies with slight modifications. The fold difference was determined by the following equation: 2Ct(IP)-Ct(Ref). LSD1, H3K4me1 and H3K4me2 antibodies were purchased from Millipore (Lake Placid, NY). hSET1 antibody was purchased from Bethyl Laboratory (Montgomery, TX). TAL1 and HDAC1 antibodies were obtained from Santa Cruz Biotechnology (Santa Cruz, CA).
ChIP-Seq
Primary human CD34+ cells were isolated and differentiated to CD36+ cells as described (32). ChIP-Seq assays were performed as described. Sequence reads of 25-bp were obtained, mapped to human genome (hg18) and processed as described previously (59). The sequence reads have been deposited in the NCBI Short Read Archive (GSE12646). The BED files can be downloaded from the website (http://dir.nhlbi.nih.gov/papers/lmi/epigeneomes/hghscmethylation.html).
Supplementary Material
Acknowledgments
We are grateful to members of the Huang laboratory for their suggestions and comments. This work was supported by grants from the National Institute of Health (S.H., R01HL090589, R01HL091929, and R01HL091929-01A1S1-the ARRA Administrative supplement; Y.Q., R01HL095674). B.P. is supported by NIH T32 training grant (5T32-CA9126-34). K.Z. is supported by the Intramural Research programs, National Heart Lung Blood Institute, National Institute of Health.
Footnotes
Conflict of Interest
The authors declare no conflict of interest.
References
- 1.Porcher C, Swat W, Rockwell K, Fujiwara Y, Alt FW, Orkin SH. The T cell leukemia oncoprotein SCL/tal-1 is essential for development of all hematopoietic lineages. Cell. 1996 Jul 12;86(1):47–57. doi: 10.1016/s0092-8674(00)80076-8. [DOI] [PubMed] [Google Scholar]
- 2.Sanchez MJ, Bockamp EO, Miller J, Gambardella L, Green AR. Selective rescue of early haematopoietic progenitors in Scl(−/−) mice by expressing Scl under the control of a stem cell enhancer. Development. 2001 Dec;128(23):4815–27. doi: 10.1242/dev.128.23.4815. [DOI] [PubMed] [Google Scholar]
- 3.Robb L, Elwood NJ, Elefanty AG, Kontgen F, Li R, Barnett LD, et al. The scl gene product is required for the generation of all hematopoietic lineages in the adult mouse. Embo J. 1996 Aug 15;15(16):4123–9. [PMC free article] [PubMed] [Google Scholar]
- 4.Chen Q, Cheng JT, Tasi LH, Schneider N, Buchanan G, Carroll A, et al. The tal gene undergoes chromosome translocation in T cell leukemia and potentially encodes a helix-loop-helix protein. Embo J. 1990 Feb;9(2):415–24. doi: 10.1002/j.1460-2075.1990.tb08126.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Robb L, Begley CG. The SCL/TAL1 gene: roles in normal and malignant haematopoiesis. Bioessays. 1997 Jul;19(7):607–13. doi: 10.1002/bies.950190711. [DOI] [PubMed] [Google Scholar]
- 6.Orkin SH. Hematopoiesis: how does it happen? Curr Opin Cell Biol. 1995 Dec;7(6):870–7. doi: 10.1016/0955-0674(95)80072-7. [DOI] [PubMed] [Google Scholar]
- 7.Cantor AB, Orkin SH. Transcriptional regulation of erythropoiesis: an affair involving multiple partners. Oncogene. 2002 May 13;21(21):3368–76. doi: 10.1038/sj.onc.1205326. [DOI] [PubMed] [Google Scholar]
- 8.Kassouf MT, Hughes JR, Taylor S, McGowan SJ, Soneji S, Green AL, et al. Genome-wide identification of TAL1’s functional targets: insights into its mechanisms of action in primary erythroid cells. Genome Res. Aug;20(8):1064–83. doi: 10.1101/gr.104935.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tripic T, Deng W, Cheng Y, Zhang Y, Vakoc CR, Gregory GD, et al. SCL and associated proteins distinguish active from repressive GATA transcription factor complexes. Blood. 2009 Mar 5;113(10):2191–201. doi: 10.1182/blood-2008-07-169417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Huang S, Brandt SJ. mSin3A regulates murine erythroleukemia cell differentiation through association with the TAL1 (or SCL) transcription factor. Mol Cell Biol. 2000 Mar;20(6):2248–59. doi: 10.1128/mcb.20.6.2248-2259.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huang S, Qiu Y, Shi Y, Xu Z, Brandt SJ. P/CAF-mediated acetylation regulates the function of the basic helix-loop-helix transcription factor TAL1/SCL. Embo J. 2000 Dec 15;19(24):6792–803. doi: 10.1093/emboj/19.24.6792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Huang S, Qiu Y, Stein RW, Brandt SJ. p300 functions as a transcriptional coactivator for the TAL1/SCL oncoprotein. Oncogene. 1999 Sep 2;18(35):4958–67. doi: 10.1038/sj.onc.1202889. [DOI] [PubMed] [Google Scholar]
- 13.Hu X, Li X, Valverde K, Fu X, Noguchi C, Qiu Y, et al. LSD1-mediated epigenetic modification is required for TAL1 function and hematopoiesis. Proc Natl Acad Sci U S A. 2009 Jun 23;106(25):10141–6. doi: 10.1073/pnas.0900437106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schuh AH, Tipping AJ, Clark AJ, Hamlett I, Guyot B, Iborra FJ, et al. ETO-2 associates with SCL in erythroid cells and megakaryocytes and provides repressor functions in erythropoiesis. Mol Cell Biol. 2005 Dec;25(23):10235–50. doi: 10.1128/MCB.25.23.10235-10250.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Goardon N, Lambert JA, Rodriguez P, Nissaire P, Herblot S, Thibault P, et al. ETO2 coordinates cellular proliferation and differentiation during erythropoiesis. Embo J. 2006 Jan 25;25(2):357–66. doi: 10.1038/sj.emboj.7600934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cai Y, Xu Z, Xie J, Ham AJ, Koury MJ, Hiebert SW, et al. Eto2/MTG16 and MTGR1 are heteromeric corepressors of the TAL1/SCL transcription factor in murine erythroid progenitors. Biochem Biophys Res Commun. 2009 Dec 11;390(2):295–301. doi: 10.1016/j.bbrc.2009.09.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Palamarchuk A, Efanov A, Maximov V, Aqeilan RI, Croce CM, Pekarsky Y. Akt phosphorylates Tal1 oncoprotein and inhibits its repressor activity. Cancer Res. 2005 Jun 1;65(11):4515–9. doi: 10.1158/0008-5472.CAN-05-0751. [DOI] [PubMed] [Google Scholar]
- 18.Prasad KS, Brandt SJ. Target-dependent effect of phosphorylation on the DNA binding activity of the TAL1/SCL oncoprotein. J Biol Chem. 1997 Apr 25;272(17):11457–62. doi: 10.1074/jbc.272.17.11457. [DOI] [PubMed] [Google Scholar]
- 19.Prasad KS, Jordan JE, Koury MJ, Bondurant MC, Brandt SJ. Erythropoietin stimulates transcription of the TAL1/SCL gene and phosphorylation of its protein products. J Biol Chem. 1995 May 12;270(19):11603–11. doi: 10.1074/jbc.270.19.11603. [DOI] [PubMed] [Google Scholar]
- 20.Wadman IA, Hsu HL, Cobb MH, Baer R. The MAP kinase phosphorylation site of TAL1 occurs within a transcriptional activation domain. Oncogene. 1994 Dec;9(12):3713–6. [PubMed] [Google Scholar]
- 21.Hu X, Ybarra R, Qiu Y, Bungert J, Huang S. Transcriptional regulation by TAL1: a link between epigenetic modifications and erythropoiesis. Epigenetics. 2009 Aug 16;4(6):357–61. doi: 10.4161/epi.4.6.9711. [DOI] [PubMed] [Google Scholar]
- 22.Bash RO, Hall S, Timmons CF, Crist WM, Amylon M, Smith RG, et al. Does activation of the TAL1 gene occur in a majority of patients with T-cell acute lymphoblastic leukemia? A pediatric oncology group study. Blood. 1995 Jul 15;86(2):666–76. [PubMed] [Google Scholar]
- 23.Palomero T, Odom DT, O’Neil J, Ferrando AA, Margolin A, Neuberg DS, et al. Transcriptional regulatory networks downstream of TAL1/SCL in T-cell acute lymphoblastic leukemia. Blood. 2006 Aug 1;108(3):986–92. doi: 10.1182/blood-2005-08-3482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Larson RC, Lavenir I, Larson TA, Baer R, Warren AJ, Wadman I, et al. Protein dimerization between Lmo2 (Rbtn2) and Tal1 alters thymocyte development and potentiates T cell tumorigenesis in transgenic mice. Embo J. 1996 Mar 1;15(5):1021–7. [PMC free article] [PubMed] [Google Scholar]
- 25.Condorelli GL, Facchiano F, Valtieri M, Proietti E, Vitelli L, Lulli V, et al. T-cell-directed TAL-1 expression induces T-cell malignancies in transgenic mice. Cancer Res. 1996 Nov 15;56(22):5113–9. [PubMed] [Google Scholar]
- 26.Shank-Calvo JA, Draheim K, Bhasin M, Kelliher MA. p16Ink4a or p19Arf loss contributes to Tal1-induced leukemogenesis in mice. Oncogene. 2006 May 18;25(21):3023–31. doi: 10.1038/sj.onc.1209326. [DOI] [PubMed] [Google Scholar]
- 27.O’Neil J, Shank J, Cusson N, Murre C, Kelliher M. TAL1/SCL induces leukemia by inhibiting the transcriptional activity of E47/HEB. Cancer Cell. 2004 Jun;5(6):587–96. doi: 10.1016/j.ccr.2004.05.023. [DOI] [PubMed] [Google Scholar]
- 28.Herblot S, Steff AM, Hugo P, Aplan PD, Hoang T. SCL and LMO1 alter thymocyte differentiation: inhibition of E2A-HEB function and pre-T alpha chain expression. Nat Immunol. 2000 Aug;1(2):138–44. doi: 10.1038/77819. [DOI] [PubMed] [Google Scholar]
- 29.Dey S, Curtis DJ, Jane SM, Brandt SJ. The TAL1/SCL transcription factor regulates cell cycle progression and proliferation in differentiating murine bone marrow monocyte precursors. Mol Cell Biol. May;30(9):2181–92. doi: 10.1128/MCB.01441-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lacombe J, Herblot S, Rojas-Sutterlin S, Haman A, Barakat S, Iscove NN, et al. Scl regulates the quiescence and the long-term competence of hematopoietic stem cells. Blood. Jan 28;115(4):792–803. doi: 10.1182/blood-2009-01-201384. [DOI] [PubMed] [Google Scholar]
- 31.Hansson A, Manetopoulos C, Jonsson JI, Axelson H. The basic helix-loop-helix transcription factor TAL1/SCL inhibits the expression of the p16INK4A and pTalpha genes. Biochem Biophys Res Commun. 2003 Dec 26;312(4):1073–81. doi: 10.1016/j.bbrc.2003.11.030. [DOI] [PubMed] [Google Scholar]
- 32.Cui K, Zang C, Roh TY, Schones DE, Childs RW, Peng W, et al. Chromatin signatures in multipotent human hematopoietic stem cells indicate the fate of bivalent genes during differentiation. Cell Stem Cell. 2009 Jan 9;4(1):80–93. doi: 10.1016/j.stem.2008.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mikkelsen TS, Ku M, Jaffe DB, Issac B, Lieberman E, Giannoukos G, et al. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature. 2007 Aug 2;448(7153):553–60. doi: 10.1038/nature06008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bernstein BE, Mikkelsen TS, Xie X, Kamal M, Huebert DJ, Cuff J, et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006 Apr 21;125(2):315–26. doi: 10.1016/j.cell.2006.02.041. [DOI] [PubMed] [Google Scholar]
- 35.Shi Y, Lan F, Matson C, Mulligan P, Whetstine JR, Cole PA, et al. Histone demethylation mediated by the nuclear amine oxidase homolog LSD1. Cell. 2004 Dec 29;119(7):941–53. doi: 10.1016/j.cell.2004.12.012. [DOI] [PubMed] [Google Scholar]
- 36.Lee MG, Wynder C, Cooch N, Shiekhattar R. An essential role for CoREST in nucleosomal histone 3 lysine 4 demethylation. Nature. 2005 Sep 15;437(7057):432–5. doi: 10.1038/nature04021. [DOI] [PubMed] [Google Scholar]
- 37.Wang Y, Zhang H, Chen Y, Sun Y, Yang F, Yu W, et al. LSD1 is a subunit of the NuRD complex and targets the metastasis programs in breast cancer. Cell. 2009 Aug 21;138(4):660–72. doi: 10.1016/j.cell.2009.05.050. [DOI] [PubMed] [Google Scholar]
- 38.Lan F, Nottke AC, Shi Y. Mechanisms involved in the regulation of histone lysine demethylases. Curr Opin Cell Biol. 2008 Jun;20(3):316–25. doi: 10.1016/j.ceb.2008.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lee MG, Wynder C, Bochar DA, Hakimi MA, Cooch N, Shiekhattar R. Functional interplay between histone demethylase and deacetylase enzymes. Mol Cell Biol. 2006 Sep;26(17):6395–402. doi: 10.1128/MCB.00723-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Scoumanne A, Chen X. The lysine-specific demethylase 1 is required for cell proliferation in both p53-dependent and -independent manners. J Biol Chem. 2007 May 25;282(21):15471–5. doi: 10.1074/jbc.M701023200. [DOI] [PubMed] [Google Scholar]
- 41.Wang J, Hevi S, Kurash JK, Lei H, Gay F, Bajko J, et al. The lysine demethylase LSD1 (KDM1) is required for maintenance of global DNA methylation. Nat Genet. 2009 Jan;41(1):125–9. doi: 10.1038/ng.268. [DOI] [PubMed] [Google Scholar]
- 42.Saleque S, Kim J, Rooke HM, Orkin SH. Epigenetic regulation of hematopoietic differentiation by Gfi-1 and Gfi-1b is mediated by the cofactors CoREST and LSD1. Mol Cell. 2007 Aug 17;27(4):562–72. doi: 10.1016/j.molcel.2007.06.039. [DOI] [PubMed] [Google Scholar]
- 43.Aplan PD, Lombardi DP, Ginsberg AM, Cossman J, Bertness VL, Kirsch IR. Disruption of the human SCL locus by “illegitimate” V-(D)-J recombinase activity. Science. 1990 Dec 7;250(4986):1426–9. doi: 10.1126/science.2255914. [DOI] [PubMed] [Google Scholar]
- 44.Begley CG, Green AR. The SCL gene: from case report to critical hematopoietic regulator. Blood. 1999 May 1;93(9):2760–70. [PubMed] [Google Scholar]
- 45.Chen Q, Yang CY, Tsan JT, Xia Y, Ragab AH, Peiper SC, et al. Coding sequences of the tal-1 gene are disrupted by chromosome translocation in human T cell leukemia. J Exp Med. 1990 Nov 1;172(5):1403–8. doi: 10.1084/jem.172.5.1403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tremblay M, Herblot S, Lecuyer E, Hoang T. Regulation of pT alpha gene expression by a dosage of E2A, HEB, and SCL. J Biol Chem. 2003 Apr 11;278(15):12680–7. doi: 10.1074/jbc.M209870200. [DOI] [PubMed] [Google Scholar]
- 47.Izraeli S. Leukaemia -- a developmental perspective. Br J Haematol. 2004 Jul;126(1):3–10. doi: 10.1111/j.1365-2141.2004.04986.x. [DOI] [PubMed] [Google Scholar]
- 48.Palii CG, Perez-Iratxeta C, Yao Z, Cao Y, Dai F, Davison J, et al. Differential genomic targeting of the transcription factor TAL1 in alternate haematopoietic lineages. Embo J. Feb 2;30(3):494–509. doi: 10.1038/emboj.2010.342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Linggi BE, Brandt SJ, Sun ZW, Hiebert SW. Translating the histone code into leukemia. J Cell Biochem. 2005 Dec 1;96(5):938–50. doi: 10.1002/jcb.20604. [DOI] [PubMed] [Google Scholar]
- 50.Martinez MC, Larbret F, Zobairi F, Coulombe J, Debili N, Vainchenker W, et al. Transfer of differentiation signal by membrane microvesicles harboring hedgehog morphogens. Blood. 2006 Nov 1;108(9):3012–20. doi: 10.1182/blood-2006-04-019109. [DOI] [PubMed] [Google Scholar]
- 51.Demers C, Chaturvedi CP, Ranish JA, Juban G, Lai P, Morle F, et al. Activator-mediated recruitment of the MLL2 methyltransferase complex to the beta-globin locus. Mol Cell. 2007 Aug 17;27(4):573–84. doi: 10.1016/j.molcel.2007.06.022. [Research Support, N.I.H., Extramural Research Support, Non-U.S. Gov’t] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Grass JA, Boyer ME, Pal S, Wu J, Weiss MJ, Bresnick EH. GATA-1-dependent transcriptional repression of GATA-2 via disruption of positive autoregulation and domain-wide chromatin remodeling. Proc Natl Acad Sci U S A. 2003 Jul 22;100(15):8811–6. doi: 10.1073/pnas.1432147100. [Research Support, Non-U.S. Gov’t Research Support, U.S. Gov’t, P.H.S.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.de Laat W, Grosveld F. Spatial organization of gene expression: the active chromatin hub. Chromosome Res. 2003;11(5):447–59. doi: 10.1023/a:1024922626726. [Research Support, Non-U.S. Gov’t Review] [DOI] [PubMed] [Google Scholar]
- 54.Hsu PY, Hsu HK, Singer GA, Yan PS, Rodriguez BA, Liu JC, et al. Estrogen-mediated epigenetic repression of large chromosomal regions through DNA looping. Genome Res. 2010 Jun;20(6):733–44. doi: 10.1101/gr.101923.109. [Research Support, N.I.H., Extramural Research Support, Non-U.S. Gov’t] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hu Q, Kwon YS, Nunez E, Cardamone MD, Hutt KR, Ohgi KA, et al. Enhancing nuclear receptor-induced transcription requires nuclear motor and LSD1-dependent gene networking in interchromatin granules. Proc Natl Acad Sci U S A. 2008 Dec 9;105(49):19199–204. doi: 10.1073/pnas.0810634105. [Research Support, N.I.H., Extramural Research Support, Non-U.S. Gov’t Research Support, U.S. Gov’t, Non-P.H.S.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kusy S, Gerby B, Goardon N, Gault N, Ferri F, Gerard D, et al. NKX3.1 is a direct TAL1 target gene that mediates proliferation of TAL1-expressing human T cell acute lymphoblastic leukemia. J Exp Med. Sep 27;207(10):2141–56. doi: 10.1084/jem.20100745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Li X, Hu X, Patel B, Zhou Z, Liang S, Ybarra R, et al. H4R3 methylation facilitates beta-globin transcription by regulating histone acetyltransferase binding and H3 acetylation. Blood. Mar 11;11510:2028–37. doi: 10.1182/blood-2009-07-236059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Song SH, Hou C, Dean A. A positive role for NLI/Ldb1 in long-range beta-globin locus control region function. Mol Cell. 2007 Dec 14;28(5):810–22. doi: 10.1016/j.molcel.2007.09.025. [Research Support, N.I.H., Intramural] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z, et al. High-resolution profiling of histone methylations in the human genome. Cell. 2007 May 18;129(4):823–37. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.