Abstract
The catabolite control protein CcpA is a pleiotropic regulator that mediates the global transcriptional response to rapidly catabolizable carbohydrates, like glucose in Gram-positive bacteria. By whole transcriptome analyses, we characterized glucose-dependent and CcpA-dependent gene regulation in Clostridium difficile. About 18% of all C. difficile genes are regulated by glucose, for which 50% depend on CcpA for regulation. The CcpA regulon comprises genes involved in sugar uptake, fermentation and amino acids metabolism, confirming the role of CcpA as a link between carbon and nitrogen pathways. Using combination of chromatin immunoprecipitation and genome sequence analysis, we detected 55 CcpA binding sites corresponding to ∼140 genes directly controlled by CcpA. We defined the C. difficile CcpA consensus binding site (creCD motif), that is, ‘RRGAAAANGTTTTCWW’. Binding of purified CcpA protein to 19 target creCD sites was demonstrated by electrophoretic mobility shift assay. CcpA also directly represses key factors in early steps of sporulation (Spo0A and SigF). Furthermore, the C. difficile toxin genes (tcdA and tcdB) and their regulators (tcdR and tcdC) are direct CcpA targets. Finally, CcpA controls a complex and extended regulatory network through the modulation of a large set of regulators.
INTRODUCTION
Clostridium difficile, a Gram-positive, anaerobic, spore forming bacterium, is an emerging pathogen. It is the leading cause of antibiotic-associated diarrhoea and is implicated in almost all cases of pseudomembranous colitis in humans. A higher morbidity and mortality rate because of C. difficile has been described in recent years (1,2). The development of C. difficile infection (CDI) is usually associated with antibiotic treatment, which disrupts the gut flora, allowing this bacterium to colonize the intestine (3). The spectrum of diseases caused by C. difficile is highly variable and depends not only on host factors but also, for the severe forms, on the level of toxin production (4).
The pathology process of C. difficile is divided in three main steps as follows: spore germination, host colonization and toxin production. After host infection by C. difficile spores, germination occurs, and the vegetative forms multiply in the gut. Clostridium difficile can adhere to the mucus layer and colonize the human gut (5). Pathogenic strains then produce two toxins (TcdA and TcdB), which are considered as the major virulence factors, that lead to the disruption of the actin cytoskeleton of intestinal epithelium cells (6,7), therefore, conferring the CDI symptoms (diarrhoea, epithelial apoptosis and ulceration). The toxin genes lie within a 19.6-kb pathogenicity locus (PaLoc) that also includes three accessory genes encoding the specific transcriptional regulators of toxin genes (TcdR and TcdC), and a holin-like pore forming protein (TcdE) involved in toxin secretion (8–10).
Among the other potential virulence factors, Clostridium difficile binary toxin (CDT), which is produced by 35% of C. difficile strains, has been recently shown to play a role in adherence to the host intestinal epithelial cells (11). Other factors, such as the adhesin Cwp66, the flagellum, the fibronectin-binding protein FbpA and the surface layer protein SlpA also play a role in cell adherence [for review see reference (5)]. Clostridium difficile also displays some surface proteolytic activities that seem to contribute to nutrients release (12), such as the cysteine protease Cwp84, which is also involved in SlpA maturation (13). Nevertheless, in vivo virulence processes of CDI are still poorly understood.
Many bacterial pathogens produce virulence factors to overcome the drastic changes in environment that they encounter during infection. Consequently, the regulation of the virulence genes is an important step of the pathogenicity process. In C. difficile, toxin production increases as cells enter into stationary phase and is modulated in response to different environmental signals, such as temperature, biotin limitation, the presence of antibiotics, of butyric acid or butanol and of certain amino acids like cysteine or proline (14). This indicates a correlation between environmental signals and virulence factors regulation. The transcription of tcdA and tcdB is also repressed by the presence of rapidly metabolizable carbohydrates, such as glucose or other sugars taken up by the phosphoenolpyruvate-dependent phosphotransferase system (PTS) (15). The PTS is composed of enzyme I and histidine-containing phosphocarrier protein (HPr), present in the cytoplasm, and the carbohydrate specific transporter enzyme II, consisting usually of three domains (EIIA, EIIB and EIIC). The carbon catabolite repression (CCR) in bacteria is generally regarded as a regulatory mechanism to ensure sequential utilization of carbohydrates (16). CCR has been extensively studied in Bacillus subtilis [for review see references (16,17)]. The global mechanism of CCR is mediated by the pleiotropic regulator CcpA, a member of the LacI/GalR family of transcription factors (16). CcpA represses the transcription of several hundred catabolic genes and activates the expression of some genes of overflow metabolism (17). Positive and negative transcriptional regulation of CcpA-controlled genes involves the binding of CcpA to the cis-acting catabolite responsive element named cre site. In Bacilli, the DNA-binding activity of CcpA is enhanced by its interaction with HPr-Ser-P, a phosphorylated form of HPr (16). Although CcpA controls most genes subjected to catabolite control in B. subtilis, transcriptomic data have also revealed the existence of CcpA-independent CCR mechanisms (18,19).
CcpA plays a central role in the control of metabolism in several Firmicutes, such as Lactococcus lactis, Bacillus cereus, Clostridium acetobutylicum and several streptococci and staphylococci (20–24). CcpA also regulates the expression of virulence-associated genes. In Staphylococcus aureus, Streptococcus pneumoniae, Clostridium perfringens and Bacillus anthracis, CcpA participates in the control of toxin production, colonization, capsule synthesis, biofilm formation and/or antibiotic resistance (25–29). Moreover, ccpA mutants of S. aureus and B. anthracis are attenuated on virulence in animal models of infection (24,28). The coordinate CcpA-dependent regulation of virulence and carbon utilization genes might be critical for fitness when pathogens compete with other microbes for niche colonization (23).
Recently, we have demonstrated that CcpA is involved in the glucose-dependent repression of C. difficile toxin production. This repression is because of a direct binding of CcpA to the regulatory region of the tcdA and tcdB genes (30). This is the first evidence that CcpA acts as a link between carbon source utilization and virulence gene expression in C. difficile. This bacterium has a large repertoire of genes dedicated to carbohydrate catabolism. Clostridium difficile ability to use an extended range of carbohydrates might be important during the pathogenesis process by promoting survival and growth in the intestine (31). Moreover, other factors important for infection and pathogenesis (besides the two major toxins) may be under CCR. The aim of this study was to analyse the global impact of glucose on gene expression and the regulatory role of CcpA in C. difficile. To define the processes coordinately controlled by glucose mediated or not by CcpA, we performed a transcriptome analysis of JIR8094 and its isogenic ccpA mutant strains grown in the presence or absence of glucose. We further combined a genome scale in vivo determination of CcpA direct targets to an in silico analysis and an in vitro protein-DNA interaction study to define the CcpA DNA-binding motif, therefore, characterizing the CcpA regulon in C. difficile. The transcriptomic data were also correlated with phenotypic tests concerning the impact of glucose addition and CcpA inactivation on the fermentation process and sporulation efficiency. The data obtained in this study provide new insights into understanding the links between nutrient acquisition and virulence in C. difficile.
MATERIALS AND METHODS
Bacterial strains and growth conditions
The C. difficile strain JIR8094 (32) and its isogenic ΔccpA mutant (30) were grown anaerobically (5% H2, 5% CO2 and 90% N2) in tryptone–yeast extract (TY) or TY medium containing 0.5% glucose (TYG) in Freter chamber (15). Sporulation medium (SM) (33) was used for sporulation assays. SM medium contained per liter: 90 g Bacto–tryptone, 5 g Bacto–peptone, 1 g (NH4)2SO4, 1.5 g Tris base and pH at 7.4. When necessary, cefoxitin (25 µg/ml) or erythromycin (2.5 µg/ml) was added to C. difficile cultures.
Sporulation assay
Mid-exponential growth cultures of C. difficile JIR8094 and its isogenic ccpA mutant in SM medium were used to inoculate 5 ml of SM broth or SM broth supplemented with 0.5% glucose. After 24, 48 and 96 h of growth, 350 µl of culture was divided into two samples. To determine the number of colony forming unit (CFU), one sample was serially diluted and plated on Brain–Heart Infusion (BHI) with 0.1% taurocholate (Sigma). To determine the number of spores in the culture, the other sample was heated at 60°C for 30 min before plating in BHI with 0.1% taurocholate. The percentage of sporulation was determined as the ratio of the number of spores/ml and the number of bacteria per millilitre.
Volatile fatty acid analysis
The end products of bacteria fermentation were detected by gas–liquid chromatography. Strain JIR8094 and the ccpA mutant were grown in TY or TYG for 48 h at 37°C. After centrifugation, supernatants were recovered and mixed with sulphuric acid and ether to extract the volatile fatty acids. For each sample, 5 µl of the supernatants was injected in a gas chromatograph (CP-3380; Varian) equipped with a flame ionization detector and connected to an integrator (model C-55A; Shimadzu). A glass column (2 m × 4 mm) packed with 10% SP 1000 plus 1% H3PO4 on Chromosorb W AW (100/120 mesh) was used. The instrument was operated at 170°C for 18 min. The operating conditions were as follows: injector temperature, 200°C; detector temperature, 200°C; carrier gas (nitrogen) flow rate, 30 ml min−1. Each peak of the gas–liquid chromatography patterns was identified by the retention time compared with that obtained with standard (2-methylpentanoic acid at 10 mM). The amounts of fatty acids were calculated by comparison with an internal standard (34).
Microarray design, DNA-array hybridization
The microarray of C. difficile 630 genome was designed as previously described (35) [Gene Expression Omnibus (GEO) database accession number GPL10556]. Transcriptome was performed using four different RNA preparations for each condition (JIR8094 in TY and TYG, ccpA mutant in TY and TYG at 10 h of growth). Labelled DNA hybridization to microarrays and array scanning were done according to Saujet et al. (35). The complete experimental data set was deposited in the GEO database with a superseries record accession number GSE35152.
Chromatin immunoprecipitation followed by microarray hybridization (ChIP-on-Chip) experiments
The C. difficile strain 630 genome was obtained from the European Molecular Biology Laboratory (EMBL) database. Probe design for the microarray was performed by using OligoArray 2.0 software (36). One to three oligonucleotides were designed for each 3774 intergenic region, including 100 bp of the 3′ and 5′ ends of genes edge. We could not design oligonucleotides for 123 intergenic regions. Probes were replicated twice on the Agilent produced microarrays to reach a final density of 15 200 probes per array. A total of 459 positive controls and 77 negative controls were also included. The description of the microarray design was submitted to the GEO database (accession number GPL15104).
The chromatin immunoprecipitations (ChIP) were performed using three independent cultures for each strain. The C. difficile JIR8094 and its isogenic ccpA mutant (negative control) strains were grown in TYG medium until an optic density (OD)600 of 0.6 (37). The cells were treated with formaldehyde (final concentration: 1%) and incubated for 10 min at 37°C. Cross-linking was quenched by addition of glycine (final concentration: 400 mM). Cells were then collected by centrifugation, washed twice with Tris-buffered saline (20 mM Tris–HCl pH 7.5, 150 mM NaCl) and stored at −80°C. Pellets were resuspended in 10 ml of buffer A [50 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES)–KOH pH 7.5, 150 mM NaCl, 1 mM ethylenediaminetetraacetic acid (EDTA), 1% Triton X-100, 0.1% sodium dodecyl sulphate (SDS), 0.1% sodium deoxycholate and Complete antiprotease] and sonicated in a water bath sonicator (Bioruptor, Diagenode) to shear DNA to an average size of 400–600 bp. After centrifugation, the supernatant was incubated for 3 h at 4°C in the presence of 40 µl of Dynabeads® Pan mouse IgG (Life Technologies™) on a rotating wheel. Dynabeads were then removed (non-specific immunoprecipitation), and supernatant was incubated at 4°C overnight with polyclonal anti-CcpA antibodies (1/500 dilution). The mixture was then incubated with 50 µl of Dynabeads® Pan mouse IgG (Life Technologies™) for 90 min at 4°C and washed twice with buffer A, once with buffer A plus 500 mM NaCl, once with buffer B (10 mM Tris–HCl pH 8.0, 1 mM EDTA, 0.1% Nonidet-P40,0.5% sodium deoxycholate) and once with 10 mM Tris–HCl pH 7.5, 1 mM EDTA. Antibodies bound to the magnetic beads were then eluted for 20 min at 65°C with 100 µl of elution buffer (50 mM Tris–HCl pH 7.5, 10 mM EDTA, 1% SDS). Samples were then treated with proteinase K (50 µg) for 2 h at 56°C and 6 h at 65°C in a 0.5× elution buffer. Immunoprecipitated DNA samples were finally purified using the QIAquick polymerase chain reaction (PCR) purification kit (QIAGEN). DNA obtained from immunoprecipitation was then labelled with Cy5 or Cy3 using the Bioprime plus array CGH indirect genomic labelling system (Invitrogen) as recommended by the manufacturer. Labelled DNA hybridization to microarrays and array scanning were done according to Saujet et al. (35). The complete experimental data set were deposited in the GEO database with the accession number GSE34971.
Microarrays or ChIP-on-Chip data analysis
All the slides, from microarray or ChIP-on-Chip experiment, were analysed using R and linear model for microarray data software from Bioconductor project (www.bioconductor.org). For each slide, we corrected background with the ‘normexp’ method (38), resulting in strictly positive values and reducing variability in the log ratios for genes with low levels of hybridization signal. Then, we normalized each slide with the ‘loess’ method (39). To test for differential expression, we used the Bayesian adjusted t-statistics, and we performed a multiple testing correction of Benjamini and Hochberg (40) based on the false discovery rate. For the transcriptome data, a gene is considered as differentially expressed when the P-value is <0.05. For the ChIP-on-Chip experiment, a DNA region is considered as significantly enriched with a P-value <0.05. The enrichment factor for a given intergenic region was calculated as the ratio of hybridization of immunoprecipitated DNA in the JIR8094 compared with the ccpA mutant strains.
Bioinformatic analysis of CcpA binding regions
For the prediction of CcpA binding sites in C. difficile genome, we searched for a common motif in the promoter region of 20 direct target genes identified in ChIP-on-Chip experiments with a high ratio of enrichment (CD0201, CD0279A, CD0395, CD0576, CD0833 CD1477, CD1536, CD1768, CD1893, CD2241, CD2280, CD2327, CD2347, CD2600, CD2625, CD2702, CD3218, CD3244, CD3377, CD3664). The de novo motif extraction program MEME (41,42) was used for the creCD motif identification. A positional weight matrices (PWM)-based model of the CcpA binding motif was generated and further used for genome-wide detection of additional CcpA targets in genomic upstream regions. The screening of additional CcpA binding sites upstream of the genes identified in the ChIP-on-Chip analysis and/or the CcpA transcriptomic analysis was carried out using the RegPredict web server (43). The motif was searched in the leader regions from position −250 to +50 relative to the translational start site for the MEME program and −300 and +300 for RegPredict. Scores of candidate sites were defined as the sum of positional nucleotide weights. The score threshold for the search of candidate binding sites was selected as a minimal score in the training set of known CcpA target sites (score value 4.0). The details of the identified CcpA taget binding sites and corresponding genes are captured and displayed in the RegPrecise database (regprecise.lbl.gov).
DNA techniques and electrophoretic mobility shift assay
DNA fragments containing the putative CcpA binding site (creCD site) of genes CD0395, CD0833, CD1477, CD1536, CD2327 and CD2600 were amplified by PCR from genomic DNA of C. difficile 630 strain with primers OBD713–716, OBD646–649, OBD678–681, OBD658–661, OBD706–709 and OBD664–667, respectively (Supplementary Table S1). Deletion of the creCD site was performed on these amplified DNA fragments as described in Supplementary Figure S1. All PCR products were then used as templates to amplify and to radioactively label a smaller DNA fragment of 200–300 bp containing or not containing the putative creCD site with the corresponding primers listed in Supplementary Table S1. The radioactively labelled DNA fragments correspond to positions −97 to +82 with respect to the translational start site of CD0395, +213 to +434 of CD0833, −146 to +75 of CD1477, +132 to +357 of CD1536, −215 to +73 of CD2327 and −183 to +22 of CD2600.
The other DNA fragments containing a creCD site of genes CD0576 (position +44 to +300 with respect to the translational start site), CD0659 (−338 to −168 and −220 to −52), CD0664 (−18 to +205), CD0770 (−3 to +179 and +149 to +300), CD1214 (−107 to +106), CD1768 (−216 to −6), CD1893 (−193 to +30), CD1935 (−267 to −58), CD2241 (−197 to +46), CD2357 (+118 to +368), CD3218 (+197 to +405), CD3244 (−385 to −197) and CD3664 (−162 to +52) were amplified and radioactively labelled by PCR from chromosomal DNA with corresponding primers listed in Supplementary Table S1. Promoter regions of CD1893 and CD3218 and internal fragments of genes CD2327 and CD1477 without putative creCD sites (negative controls) were also amplified and radioactively labelled by PCR from chromosomal DNA with primers OBD684–685 (position −193 to −43 with respect to the translational start site), OBD612–613 (−117 to +84), OBD710–711 (+159 to +328) and OBD704–705 (+57 to +208), respectively.
For the radioactive labelling of PCR fragments, all forward primers were end-labelled with T4 polynucleotide kinase (Fermentas and γ-32P- adenosine triphosphate (3000 Ci.mM−1; Perkin Elmer) as recommended by the manufacturer. After PCR, amplified labelled fragments were then purified by QIAquick® Nucleotide Removal kit (Qiagen™). The labelled fragments were incubated with increasing amounts of the purified CcpA–His6 at room temperature for 15 min in 20 µl of reaction volume containing binding buffer (10 mM Tris pH 7.5, 1 mM EDTA; 50 mM KCl; 50 μg/ml bovine serum albumin, 0.05% Nonidet P-40, 10% glycerol, 1 mM Dithiothreitol (DTT), 10 μg/ml polydI-dC) as described in Antunes et al. (30). Samples were loaded during electrophoresis onto a 5% non-denaturing polyacrylamide gel in Tris–glycine buffer (25 mM Tris–HCl pH 8, 190 mM glycine, 1 mM EDTA) (30). After electrophoresis, the gel was dried and analysed by autoradiography.
RESULTS AND DISCUSSION
Overview of the transcriptome data
The ability of bacteria to adapt to environmental changes involves a number of physiological responses and complex molecular regulatory circuits. In many pathogenic bacteria, carbohydrate substrates constitute an important signal used for compartment-specific modulation of virulence genes during infection (44). Previously, we have shown that glucose regulates C. difficile toxin genes expression through a direct binding of CcpA to the tcdA and tcdB regulatory regions (30), suggesting that CcpA seems to link virulence gene expression and carbohydrate availability in this bacterium. However, a comprehensive study of the role of CcpA in C. difficile physiology has not been undertaken so far.
To determine the role of CcpA in CCR and the global impact of glucose on gene expression, we performed a transcriptional analysis of the wild-type strain JIR8094 and its isogenic ccpA mutant strain grown in TY or TYG at the onset of stationary phase. Based on analysis performed (TY versus TYG in JIR8094 or in ccpA and ccpA versus JIR8094 in TYG or in TY), ∼21% of the genes (782 genes) were differentially expressed with a fold change ≥2 (Supplementary Table S2). In the presence of glucose, ∼18% of the global genome (667 genes) was differently expressed in strain JIR8094. Among them, 349 and 318 genes were up- and downregulated, respectively (Table 1). As described in B. subtilis and other Firmicutes, such as B. cereus or Streptococcus pyogenes (19,21,45), we observed that C. difficile modulates a large number of genes in response to glucose availability.
Table 1.
Number of genes regulated by glucose and/or CcpA
| Genes groupa | Number of genesb |
|---|---|
| Glucose-dependent | 667 (18%) |
| Upregulated genes | 349 |
| Downregulated genes | 318 |
| CcpA-dependent | 407 (11%) |
| Upregulated by CcpA | 197 |
| Downregulated by CcpA | 210 |
| Regulated by glucose in a CcpA-dependent manner | 361 (10%) |
| Upregulated genes | 170 |
| Downregulated genes | 191 |
| Regulated by glucose in a CcpA-independent manner | 303 (8%) |
| Upregulated genes | 177 |
| Downregulated genes | 126 |
| Regulated by CcpA in absence of glucose | 43 (1%) |
| Upregulated by CcpA | 8 |
| Downregulated by CcpA | 35 |
| Regulated by CcpA independent of glucose | 3 (0.08%) |
| Upregulated by CcpA | 2 |
| Downregulated by CcpA | 1 |
| Atypical regulation | 112 (0.3%) |
| Upregulated genes | 39 |
| Downregulated genes | 73 |
Number of genes subject to regulation by glucose and/or CcpA in in vitro growth conditions. The percentage of genes regulated was calculated considering 3759 genes analysed in the transcriptome analysis.
aGroups partly overlap.
bA gene was considered to be regulated if transcription was induced or repressed at least 2-fold (P < 0.05).
The CcpA regulon consists of ∼11% of all genes (407 genes, Table 1), and these results are in accordance to what has been reported in other Firmicutes (17,20). However, only 50% of the glucose-regulated genes are controlled by CcpA in C. difficile in the conditions tested, whereas in B. subtilis >80% of the genes are controlled by CcpA in response to glucose (19). This indicates the existence of still uncharacterized signalling pathways that react to glucose in C. difficile. A total of 182 genes were regulated by glucose through a mechanism that is fully or partially CcpA dependent. It is worth noting that CcpA positively controls half of them in the presence of glucose. Comparable observations have been described for S. aureus (46). However, for most Firmicutes, the number of genes repressed by glucose in a CcpA-dependent manner largely exceeds the genes activated by CcpA (17,20,45). Finally, we noted that the expression of some genes was regulated by CcpA independently of glucose. All genes differentially expressed in response to glucose and/or controlled by CcpA are listed in Supplementary Data (Supplementary Table S2). To confirm the results obtained in the microarrays expression data, we selected a subset of 15 representative genes of different cell functions regulated by CCR and performed quantitative reverse-transcriptase (qRT)-PCR with RNA preparations from identical experimental conditions of the transcriptome. qRT-PCR results confirmed array data for majority of the tested genes (Supplementary Table S4).
In C. difficile, glucose and/or CcpA mainly regulate the carbohydrate and energy metabolisms. Moreover, the glucose adaptation response also implicates the regulation of genes involved in carbon sources transport, amino acids and peptide catabolism, stress response, sporulation, proteins of the cell envelope, toxin synthesis and transcriptional regulation. CcpA can control genes directly by binding in the regulatory region of the controlled genes or indirectly through other mechanisms (see later in the text). To discriminate between these two possibilities, we needed to identify the CcpA binding motif in C. difficile.
Identification of direct CcpA targets by ChIP-on-Chip
In B. subtilis, CcpA directly controls transcription of the target genes by interacting with cre sites located within or downstream from the promoter regions (47). Thus, we first tried to identify the direct CcpA targets by screening the genome for the B. subtilis cre consensus motif (creBS), and we then compared the list obtained with that of genes controlled by CcpA in transcriptome. However, we did not find correlation between the presence of a creBS motif and the CcpA-dependent control. Moreover, we have previously demonstrated that the deletion of creBS-like sites in the regulatory region of tcdA and tcdB does not abolish CcpA binding (30). This strongly suggests that the cre motif recognized by CcpA in C. difficile (creCD) is significantly different of the creBS motif. To identify the CcpA direct targets in vivo and define the creCD consensus sequence, we performed ChIP-on-Chip experiments. For this purpose, the C. difficile JIR8094 strain was grown in TYG and then cross-linked with formaldehyde and further immunoprecipitated with a specific anti-CcpA antibody (30).
A total of 244 oligonucleotides were differentially enriched in the presence of CcpA with a P-value of <0.05 and an enrichment factor ≥1.5. We identified 55 direct targets of CcpA in vivo corresponding to ∼140 genes by ChIP-on-Chip experiments (Table 2). Most of these genes encode proteins involved in (i) fermentation, such as acetoacetate production (scoB), hydrogen production (hymB) and Stickland reactions (hadA and prdA); (ii) energy metabolism (CD2429A, CD1170 and CD1807); (iii) amino acids metabolism (CD1536, CD3664 and gcvPB), including peptidases (CD0779 and CD2347) and amino acid or peptide transporters (oppC, CD2173 and brnQ); (iv) PTS carbohydrate transporters and associated enzymes involved in sugar utilization (CD0468, CD0494, CD2280, gatA, celA and CD1336); and (v) non-PTS sugar utilization (rbsR and CD1586), sialic acid degradation (nanE) or ethanolamine transport (CD0742). Genes implicated in sporulation, such as the major response regulator Spo0A, were also directly controlled by CcpA. Moreover, genes encoding six regulatory proteins, and 12 proteins of unknown function have been identified as direct CcpA targets. Among the targets depicted by the ChIP-on-Chip experiments, 29 genes (53%) were regulated by glucose and/or CcpA in transcriptome. The overlap of genes identified in transcriptome and by ChIP-on-Chip analysis is similar to that of other studies (48,49).
Table 2.
Direct CcpA targets identified by ChIP-on-Chip experiments
| Gene | Name | FCa | Oligonucleotide 5′ position | Operon | creCD motif | Positionb | Glucose regulationc | CcpA regulationc |
|---|---|---|---|---|---|---|---|---|
| CD0201 | 11 | 260578 | CD0201 | GAGAAAATGTTTACAG | 77 | NR | NR | |
| CD0279A or CD0279 | 7.7 | 340693 | AAGAAAACGTTATTAC | −370 | − | − | ||
| CD0298d | rbsR | 1.9 | 359108 | CD0298–CD0302 | AGTAAAACGGTTTCCT | −28 | − | − |
| CD0395e | hadAe | 7.1 | 467014 | CD0395–CD0401 | AGGAAAACGTTATGCT | −12 | − | − |
| CD0468 | 3 | 556433 | CD0467–CD0469 | TCAAAAAGATTTTCAT | −255 | NR | NR | |
| CD0494d | 4 | 583410 | CD0490–CD0494 | AAGAAAACGTTAAGCA | 26 | NR | NR | |
| CD0530d | 4.6 | 635577 | CD0530–CD0528 | AGGAAAACGTATTATA | −78 | NR | NR | |
| CD0576e | virSe | 33 | 687951 | GAGAAAAGGATTTCTA | 92 | NR | NR | |
| CD0580d | gapN | 2.1 | 692989 | AGAAAAACGTTAACTT | −124 | NR | NR | |
| CD0679d | 4 | 821606 | ATGATAACATTTTCTT | −169 | NR | NR | ||
| CD0742 | 6.5 | 905449 | CD0741–CD0742 | GGGAAAACGATACCAA | −76 | NR | NR | |
| CD0779d | 1.7 | 949162 | CD0778–CD0780 | GAGAAAAAGACATCAT | 50 | − | NR | |
| CD0833e | acnBe | 9.7 | 1009212 | CD0832–CD0834 | AGGAAATGGTATTTGT | 284 | − | − |
| CD0854d | oppC | 5.7 | 1029471 | CD0853–CD0856 | AAGAAAACCTTATCTA | 77 | − | − |
| CD1116 | 36 | 1312695 | GAGAAAAGGTTTCCAA | 167 | NR | + | ||
| CD1137 | 5.2 | 1336232 | CD1136–CD1137 | GAAAAAATGTTTTTTT | −37 | NR | NR | |
| CD1170 | 5.9 | 1368750 | CD1170–CD1173 | ND | ND | + | NR | |
| CD1214e | spo0Ae | 1.5 | 1412498 | TGTAAAAAGTTTAGTT | −42 | − | − | |
| CD1321d | 1.6 | 1531839 | AGGAAATAGTTAACTT | 24 | − | − | ||
| CD1336 | 1.5 | 1550212 | TAGAAAACGTTTTAAA | −41 | NR | NR | ||
| CD1385 | 3.6 | 1603687 | CD1384–CD1387 | AAGAAAGCGTTTTGAA | −111 | − | − | |
| CD1477e | feoAe | 13 | 1714290 | CD1477–CD1480 | AAGATAACGATTGCTT | −52 | NR | + |
| CD1536e | 15.5 | 1781719 | CD1536–aspB | AAGAAAACGATTTTGT | 231 | NR | − | |
| CD1586 | 6.6 | 1838850 | CD1584–CD1589 | TAGAAAAGGTTATCAT | 111 | + | NR | |
| CD1599 | thiD | 3.8 | 1854754 | CD1599–CD1601 | TTGAAAACGTTTAGTG | 76 | NR | – |
| CD1658 | gcvPB | 1.8 | 1925192 | CD1657–CD1658 | AGGAAATAGATAAGTT | −40 | − | − |
| CD1768d,e | 10 | 2045870 | GAGAAAAGGTTTTGTT | −93 | + | + | ||
| CD1807 | 3.2 | 2091015 | ND | ND | − | − | ||
| CD1893d,e | 6.8 | 2198560 | TATAAAACGTTTTCTT | −75 | NR | NR | ||
| CD1935d,e | spoVSe | 1.8 | 2233439 | AAATAAAGGTTTTCTT | −142 | NR | − | |
| CD2111d | 6 | 2440328 | TAGAAAATGTTTGCAG | −7 | NR | NR | ||
| CD2173 | 1.8 | 2515137 | CD2177–CD2172 | GAGAAAAAATTATAAA | 56 | − | − | |
| CD2241d,e | nanEe | 7.8 | 2592979 | CD2241–CD2239 | AAGAAAAAGTTTTCAT | −72 | NR | NR |
| CD2280d | 4.5 | 2646506 | CD2282–CD2279 | AAGAAATCGTTTCTTT | 100 | NR | NR | |
| CD2327d,e | gatAe | 30 | 2689528 | CD2327–CD2325 | GGGAAAACATTTTCTT | −113 | − | − |
| CD2344 | 1.7 | 2714126 | CD2344–CD2341 | AAGATAAAATTTTTTT | −152 | − | − | |
| CD2347 | 8 | 2716984 | CAGAAAAGGTTTGCAA | 176 | + | + | ||
| CD2429A or CD2429d | 11 | 2804869 | CD2429A–CD2427 | GAGAAAAGGTATGCAA | −9 or 217 | NR | NR | |
| CD2481d | ung | 3.6 | 2863759 | CD2482–CD2481 | TTGAAAAAGATTACTA | 47 | NR | NR |
| CD2600e | cstAe | 5.5 | 3009085 | TGGAAATAGTTTTCTT | −67 | NR | NR | |
| CD2625d | 4.4 | 3031999 | AGGATAACGTTATCAT | −61 | NR | NR | ||
| CD2626d | 6 | 3034769 | CD2627–CD2626 | GAGAAAAAATATACAA | 131 | NR | NR | |
| CD2678 | scoB | 15 | 3093914 | CD2676–CD2679 | ATGTAAACGTTATCTT | 120 | NR | NR |
| CD2693 | 1.5 | 3113024 | AAGAAAAAAATAGGTT | 4 | − | − | ||
| CD2702 | brnQ | 4.5 | 3124254 | GGGAAAACGTTAACAA | −104 | − | NR | |
| CD2879 | 3.8 | 3368263 | TAGATAAGGTTTTCTT | −128 | NR | NR | ||
| CD2884d | celA | 1.6 | 3372956 | CD2884–CD2880 | ATGAAAAAGTTTTCTT | −186 | NR | NR |
| CD3175d | cggR | 1.5 | 3711205 | AGGAGAACATTATCAA | 109 | + | − | |
| CD3184 | dpaL | 8.5 | 3726740 | TAGATTTGGTTTAAAT | 206 | NR | NR | |
| CD3218e | 5 | 3766712 | GAGTTAACGTTTTCAA | 254 | + | − | ||
| CD3244d,e | prdAe | 6.5 | 3796160 | CD3244–CD3236 | GAGAAAATGTTTACAA | −293 | + | + |
| CD3377 | mgtA | 7.4 | 3950193 | CD3377–CD3376 | ND | ND | NR | + |
| CD3406d | hymB | 3 | 3984409 | CD3405–CD3407 | CTGATAACGTTTTCTA | 270 | NR | NR |
| CD3649 | rlmH | 13 | 4258421 | AAGAAAAGGCTTTCTA | 70 | NR | NR | |
| CD3664e | 30 | 4275636 | AAGAAAACATTTTCAT | −30 | NR | NR |
aFold Change (FC) corresponding to enrichment factor in ChIP-on-Chip experiments.
bcreCD motif position from ATG start codon. ND = not detected.
cGene regulation resulting from transcriptome and qRT-PCR analysis. ‘–’ for downregulation, ‘+’ for upregulation and ‘NR’ for absence of regulation. For more details, see Supplementary Table S2.
dIndicates the existence of a second putative creCD motif.
ecreCD sites confirmed by EMSA experiments.
In silico definition of a creCD consensus
The region of C. difficile genes determined as direct CcpA targets by ChIP-on-Chip experiments with an enrichment factor >5 was searched for common CcpA binding sites to define the creCD motif using the MEME software. A conserved 16-bp pseudopalindromic motif (RRGAAAASGTTTTCWT, where R stands for adenine or guanine, S stands for guanine or cytosine and W stands for adenine or thymine) was identified in the upstream regions of 12 CcpA target genes. Then, we used this motif and the RegPredict web server (43) to search for similar CcpA binding sites for all genes identified by the ChIP-on-Chip experiment. Among the 55 CcpA direct target genes, we failed to identify a creCD motif for only three genes (CD1170, CD1807, CD3377), despite the fact that CD1807 and CD3377 were controlled by CcpA in transcriptome (Table 2). This could be explained by a degeneration of their binding sites as proposed for some targets of the Escherichia coli NsrR repressor identified in vivo using ChIP-on-Chip technology (48).
Interestingly, for some target sites that reside inside of operons, we observed a clear correlation between the position of the oligonucleotides bound by CcpA in ChIP-on-Chip and the location of a putative creCD site. This is the case for acnB, oppC, CD1385, gcvPB, scoB, CD2626 and hymB (Table 2). The alignment of all probable CcpA binding motifs listed in Table 2 allows the establishment of a creCD consensus (Figure 1). The creCD motif identified in this work for C. difficile has limited similarity with the cre consensus previously identified in various bacteria from the bacilli class of Firmicutes including Bacillus, Lactococcus, Enterococcus and Streptococcus species (20,21,47,50). Besides the guanine and the cytosine at 3rd and 14th positions, respectively, which are identical in both motifs, the creCD consensus is not similar to the creBS motif (Figure 1). Importantly, the creCD motif lacks the essential central CG bases (16) present in the creBS consensus (WTGNNARCGNWWWCAW). These observations correlate with the weak similarities of CcpA helix-turn-helix (HTH)-DNA binding domain from these bacteria (47% identity), whereas, for instance, the HTH motif of CcpA of L. lactis has 70% identity with that of B. subtilis, and both have similar cre sites.
Figure 1.
C. difficile creCD consensus of the CcpA binding motif. The motif sequence logo was created based on the alignment of all the proposed direct targets of CcpA listed in Table 2, using the weblogo website (http://weblogo.berkeley.edu). The height of the letters is proportional to their frequency.
CcpA directly binds to the creCD motif of genes identified by ChIP-on-Chip
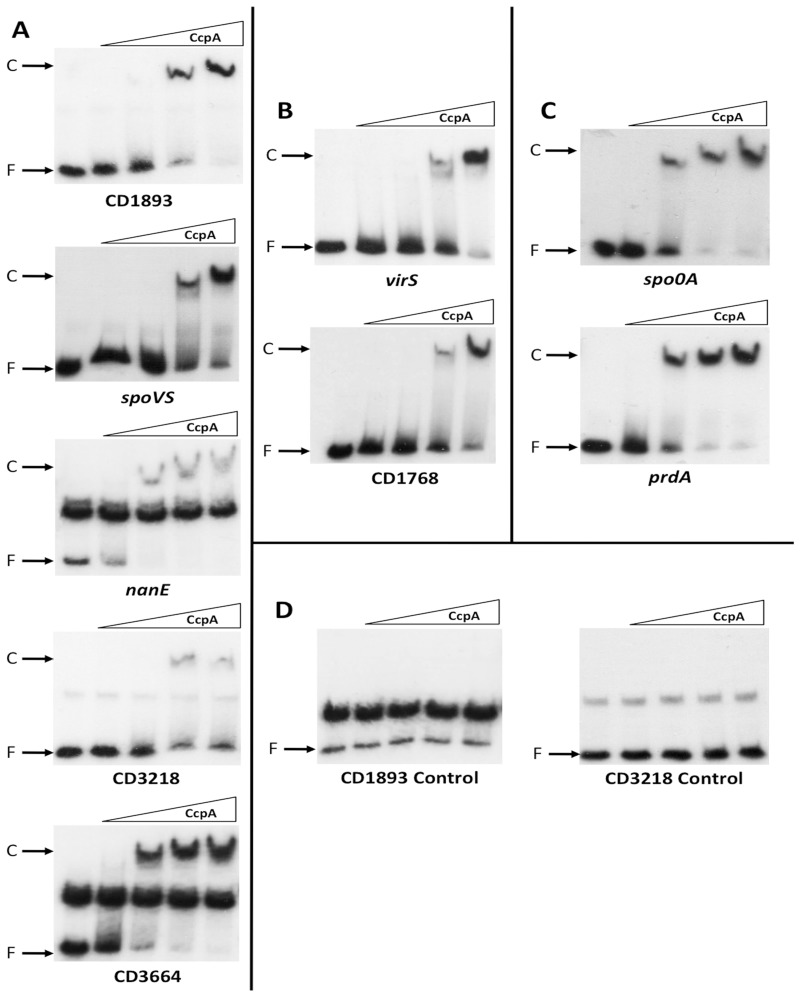
To validate the ChIP-on-Chip experiment and confirm the role of the creCD site in the CcpA binding activity, electrophoretic mobility shift assays (EMSAs) were performed using purified C. difficile CcpAHis6 (30) and DNA fragments containing a putative creCD site of 15 CcpA targets (Table 2). The CcpA transcriptional regulator induced a shift in mobility of DNA fragments containing a creCD site of all genes tested (CD1893, spoVS, nanE, CD3218, CD3664, virS, CD1768, spo0A, prdA, hadA, feoA, gatA, acnB, CD1536 and cstA) (Figures 2 and 3). However, the concentration of CcpA required to obtain a full shift differed depending on the target genes. To demonstrate the specificity of CcpA binding to the creCD site-containing DNA fragments, we first showed that no shift was induced by CcpA with promoter regions of CD1893 and CD3218 or with internal DNA fragments of feoA and gatA that do not contain a creCD site (Figures 2D and 3C). Moreover, the formation of the CcpA–DNA complex was prevented by the addition in excess of unlabelled DNA fragments containing the creCD site of acnB, CD1893 and CD3218 used as a competitor (data not shown). These results validated the binding of CcpA to the targets identified by ChIP-on-Chip, including those with low-enrichment factors like spo0A or spoVS, and showed the specific interaction of CcpA with creCD sites.
Figure 2.
Binding of CcpA to different targets with a putative creCD sequence identified by ChIP-on-Chip experiments. EMSAs of DNA fragments containing the creCD binding sites of the CD1893, spoVS (CD1935), nanE (CD2241), CD3218, CD3664, virS (CD0576), CD1768, spo0A (CD1214) and prdA (CD3244) genes. The 32P-labelled DNA fragments were incubated with increasing concentrations of purified CcpA–His6 as described in the ‘Materials and Methods’ section: (A) 0, 32, 65, 97 and 129 nM of CcpA–His6; (B) 0, 97, 129, 194 and 259 nM of CcpA–His6; (C) 0, 129, 259, 388 and 518 nM of CcpA–His6. (D) Negative controls corresponding to the DNA fragments containing promoter regions of CD1893 and CD3218 without putative creCD sites were incubated with 0, 65, 97 and 129 nM of CcpA–His6, and 0, 97, 129, 194 and 259 nM of CcpA–His6, respectively. The autoradiograms of the gels are shown. F = free DNA; C = protein–DNA complex.
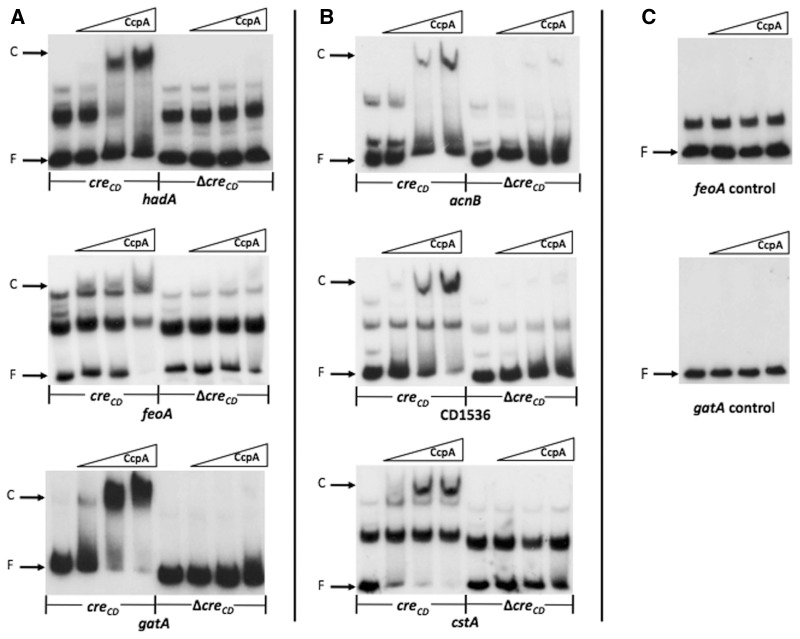
Figure 3.
Impact of creCD deletion on the binding of CcpA to the hadA, acnB, feoA, CD1536, gatA and cstA target genes. EMSAs of DNA fragments containing the creCD binding site (creCD) or the same DNA fragment where the creCD site has been deleted (ΔcreCD) of the hadA (CD0395), feoA (CD1477), gatA (CD2327), acnB (CD0833), CD1536 and cstA (CD2600) genes. The 32P-labelled DNA fragments were incubated with increasing concentrations of purified CcpA–His6 as described in the ‘Materials and Methods’ sections: (A) 0, 13, 32 and 65 nM of CcpA–His6; (B) 0, 65, 97 and 129 nM of CcpA–His6. (C) Internal 32P-labelled DNA fragments of feoA and gatA genes without putative creCD sites were incubated with 0, 65, 97 and 129 nM of CcpA–His6 (negative controls). The autoradiograms of the gels are shown. F: free DNA; C: protein–DNA complex.
To further demonstrate that the creCD site alone is required for the binding of CcpA, we performed EMSAs using DNA fragments deleted or not for the creCD site of the hadA, acnB, feoA, CD1536, gatA and cstA genes. As shown in Figure 3, the deletion of the creCD site abolished the shift induced by CcpA for the regulatory regions tested. We confirm that CcpA can interact in vitro with the creCD site-containing DNA fragments of 15 genes, and we show that this binding disappears when the creCD site is deleted for six genes. Interestingly, CcpA interacts in vivo with creCD sites that are located inside of operons, and we clearly demonstrate that in the case of acnB, this creCD sequence is recognized and bound by CcpA in vivo (ChIP-on-Chip) and in vitro (EMSA) leading to the strong repression of the aksA-acnB-icd operon by CcpA. This site internal to an operon is functional as previously shown for the iol operon of B. subtilis (51).
Identification of additional candidates for a direct CcpA control
Using the RegPredict web server (43), we extended the search of the creCD motif to the complete C. difficile genome of strain 630. We detected 901 putative creCD sites corresponding to 783 genes with a score >4, located between position −300 to +300 from the translational start sites (Supplementary Table S3). This in silico analysis allowed us to identify 143 putative creCD sites corresponding to about half of the CcpA-controlled genes in transcriptome (Supplementary Table S2). Most of these sites are adjacent or inside genes involved in carbon and energy metabolisms (35 genes), in amino acids biosynthesis or degradation (31 genes), in sporulation (7 genes), in regulation (10 genes), in cell envelope (10 genes) or encoding proteins of unknown function (29 genes).
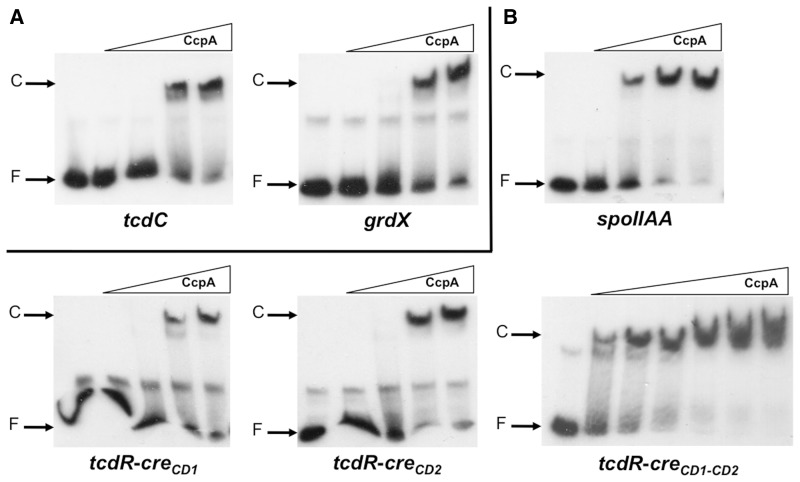
To show whether at least some of these predicted creCD sites are functional, we performed EMSAs with creCD site-containing DNA fragments of two genes (spoIIAA and grdX) negatively regulated by CcpA in transcriptome, but not retained by CcpA in our ChIP-on-Chip experiment conditions (Supplementary Table S2). A shift of a DNA fragment containing the creCD motif at position +198 relative to the translational start site of grdX was observed in the presence of 97 nM of CcpA (Figure 4A). This interaction disappeared in the presence of an excess of the same unlabelled DNA fragment (data not shown). For the spoIIAA gene, two different creCD sites located at position +87 and +194 from the translational start site were found with RegPredict. We showed that 388 nM of CcpA induced a shift with the DNA fragment containing the site at position +87 (Figure 4B) and not with a DNA fragment containing the site at position +194 (data not shown). However, when we performed EMSA with a DNA fragment containing both creCD sites, 97 nM of CcpA was sufficient to induce a shift (data not shown). These results indicated that the binding affinity of CcpA to the spoIIAA regulatory region increased when both creCD sequences are present. Thus, despite the fact that we did not see the binding of CcpA to the spoIIAA and grdX genes in the ChIP-on-Chip conditions used, these genes could also be direct physiological CcpA targets. This suggests that CcpA might directly control additional genes with creCD motifs deduced from the in silico search.
Figure 4.
Binding of CcpA to the creCD sites of tcdC, grdX, tcdR and spoIIAA target genes. EMSAs of DNA fragments containing the creCD sites of the tcdC (CD0664), grdX (CD2357), tcdR (CD0659) and spoIIAA (CD0770) genes. The 32P-labelled DNA fragments were incubated with increasing concentrations of purified CcpA–His6 as described in the ‘Materials and Methods’ section: (A) 0, 32, 65, 97 and 129 nM of CcpA–His6 for tcdC and grdX; (B) 0, 129, 259, 388 and 518 nM of CcpA–His6 for tcdR-creCD1,tcdR-creCD2 and spoIIAA and 0, 65, 97, 129, 259, 388 and 518 nM of CcpA–His6 for tcdR-creCD1-CD2. The autoradiograms of the gels are shown.
Interestingly, five putative creCD sequences are detected in the PaLoc as follows: upstream of the translational start site of tcdR (positions −235 and −169) or tcdB (position −211) and inside the tcdA gene (position +277) or tcdC gene (position +58). The binding of CcpA to the promoter region of tcdB and to the 5′ end of the tcdA gene, which has been already demonstrated (30), is consistent with the position of the creCD motif. Thus, we tested the possible interaction of CcpA with the creCD sites of the promoter region of tcdR and of the 5′ end of the tcdC gene. We detected a shift with the creCD site-containing fragment of tcdC in the presence of 97 nM of CcpA (Figure 4A). We also observed the formation of a protein–DNA complex with a DNA fragment containing the two putative creCD sites of tcdR when we used 65 nM of CcpA, whereas 388 nM was needed for a shift of the DNA fragment containing only one of each site (Figure 4B) suggesting a cooperative effect of the creCD sites. Therefore, with the exception of tcdE, CcpA interacts with the promoter region or with the 5′ end of the PaLoc genes. These results show the complex regulation of toxin synthesis in response to glucose availability mediated by CcpA.
Role of glucose and CcpA in the control of C. difficile carbohydrate metabolism
In this work, we used genome-wide transcriptome, ChIP-on-Chip experiments and in silico analysis to characterize the CcpA regulon. We discussed later in the text in more details the involvement of glucose and CcpA in the global control of key metabolic pathways and cellular processes of C. difficile.
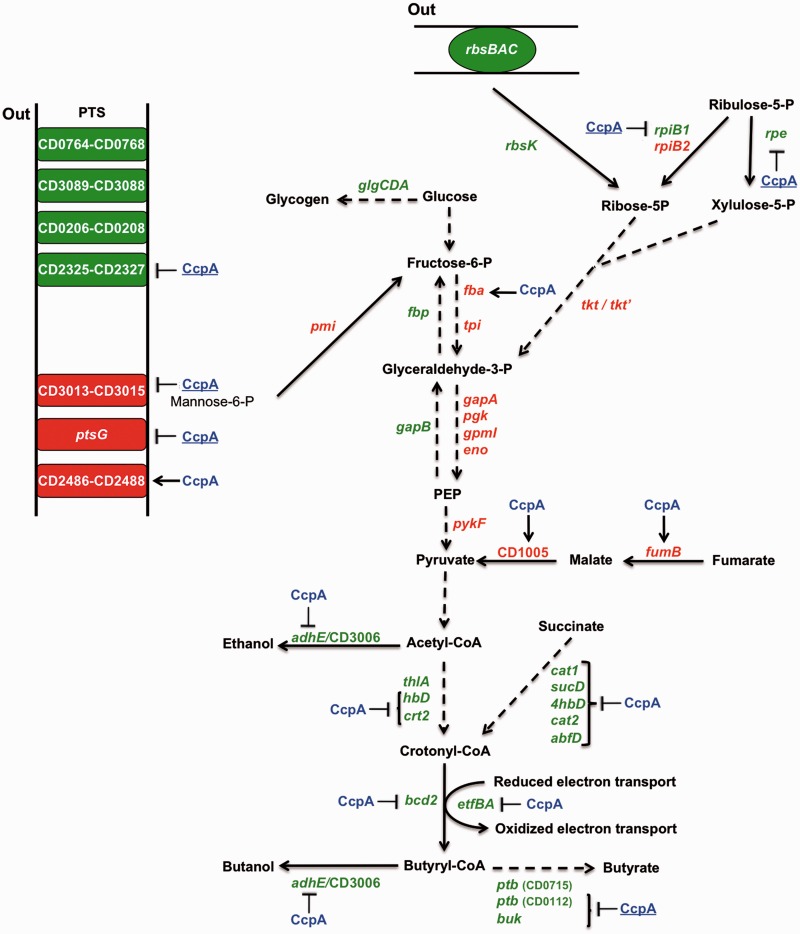
Compared with other pathogenic clostridia, C. difficile has a larger repertoire of EII PTS (51 PTS in strain 630), and this bacterium is able to ferment several sugars taken up by PTS, such as fructose, glucose, mannitol and mannose (52). In the presence of glucose, we found that C. difficile preferentially expresses glucose (CD2667–CD2666), fructose (CD2486–CD2488) and mannose (CD3015–CD3013) PTS, whereas the synthesis of some PTS of secondary carbon sources, such as sorbitol (CD0764–CD0768), galactitol (CD2327–CD2325) or cellobiose (CD3089–CD3088), was repressed by glucose (Figure 5). All PTS transporters induced by glucose were regulated by CcpA. Among the PTS systems repressed by glucose, only the galactitol PTS was controlled by CcpA. We further demonstrated by qRT-PCR that during exponential growth, the expression of the gatABC operon was strongly repressed by glucose through CcpA (Supplementary Figure S2). The expression of the rbsRKBAC (CD0298–CD0302) operon encoding the ribose repressor, a ribose ABC transporter and the ribokinase was repressed by glucose in C. difficile. Moreover, when we looked at the rbsR expression at mid-exponential growth phase by qRT-PCR analysis, we observed that rbsR expression was partially repressed by CcpA in response to glucose (Supplementary Figure S2). We demonstrated in ChIP-on-Chip experiment that the rbs operon and genes encoding cellobiose or galactitol PTS (CD2884-CD2880 and CD2327-CD2325) and two other PTS systems (CD0467–CD0469 and CD0490–CD0494 operons) were direct CcpA targets (Table 2 and Figure 3A). Only a few PTS and non-PTS sugar utilization systems were regulated by glucose and/or CcpA in transcriptome in the conditions we used. However, other genes involved in sugar assimilation systems were bound by CcpA in ChIP-on-Chip. We cannot exclude that genes involved in carbohydrate utilization are not detected in transcriptome, as they are expressed at basal level in the absence of sugar inducers.
Figure 5.
Role of glucose and CcpA in the regulation of carbohydrate utilization, central carbon metabolism and fermentation. Genes encoding enzymes or transporters, which are regulated by CcpA and/or glucose in transcriptome. Green: gene downregulated by glucose; red: gene upregulated by glucose; black: gene not regulated by glucose; blue CcpA: regulation by glucose through CcpA; blue and underlined CcpA: other type of control by CcpA; perpendicular: gene downregulated by CcpA; arrow: gene upregulated by CcpA. glgCDA (CD0882–CD0884), glycogen biosynthesis; fbp (CD1191), fructose-1,6-bisphosphatase; fba (CD0403), fructose-1,6-bisphosphate aldolase; tpi (CD3172), triosephosphate isomerase; gapA (CD3174) and gapB (CD1767), glyceraldehyde-3-phosphate dehydrogenase; pgk (CD3173), phosphoglycerate kinase; gpmI (CD3171), 2,3-bisphosphoglycerate-mutase; eno (CD3170), enolase; pykF (CD3394), pyruvate kinase; fumB (CD1004), fumarate hydratase subunit B; CD1005, putative nicotinamide adenine dinucleotide-dependent malic enzyme; rpe (CD2319), putative ribulose-phosphate 3-epimerase; rpiB1 (CD2320), ribose-5-phosphate isomerase B1; rpiB2 (CD3480), ribose-5-phosphate isomerase B2; rbsK (CD0299), ribokinase; tkt’ (CD2321), transketolase, central and C-terminal; tkt (CD2322), transketolase, N-terminal; CD0764–CD0768, PTS sorbitol IIC, IIB, IIA; CD3089–CD3088, PTS cellobiose IIBC; CD2327–CD2325, PTS galactitol IIA, IIB, IIC; CD0208–CD0206, PTS fructose-like IIA, IIB, IIC; CD2486–CD2488, PTS fructose-like IIC, IIB, IIA; CD3015–CD3013, PTS mannose-specific IIA, IIB, IIC; ptsG (CD2667, CD2666), PTS glucose-specific IIA, IICB; pmi (CD2491), mannose-6-phosphate isomerase; rbsBAC (CD0300–CD0302), ribose ABC transporter; adhE (CD2966) and CD3006, aldehyde-alcohol dehydrogenase; thlA (CD1059), acetyl-CoA acetyltransferase; hbd (CD1058), 3-hydroxybutyryl-CoA dehydrogenase; crt2 (CD1057), 3-hydroxybutyryl-CoA dehydratase; cat1 (CD2343), succinyl-CoA: coenzyme A transferase; sucD (CD2342), succinate-semialdehyde dehydrogenase; 4hbd (CD2338), 4-hydroxybutyrate dehydrogenase; cat2 (CD2339), 4-hydroxybutyrate CoA transferase; abfD (CD2341), vinylacetyl-coa-δ-isomerase; bcd2 (CD1054), butyryl-CoA dehydrogenase; etfBA (CD1055–CD1056), electron transfer flavoproteins; ptb (CD0715, CD0112), phosphate butyryltransferase; buk (CD0113), butyrate kinase.
After sugar uptake, the carbohydrates are metabolized through the central carbon metabolism to produce energy and precursors for anabolism. Glucose addition regulates several pathways of the central carbon metabolism, such as glycolysis, gluconeogenesis and pentose-phosphate pathway (PPP) in C. difficile. The expression of fba encoding the fructose-1,6-bisphosphate aldolase, a key enzyme of glycolysis, was strongly induced by glucose and positively regulated by CcpA (Figure 5). However, most genes encoding for the central glycolysis/gluconeogenesis pathway were regulated by glucose independently of CcpA. In fact, the gapA-pgk-tpi-gpmI-eno operon (CD3174–CD3170) was strongly induced by glucose, whereas the expression of gapB (CD1767) and fbp (CD1191) was repressed. Likewise, in B. subtilis, the expression of the gapA operon is induced by glucose, whereas the gapB and pckA genes involved in gluconeogenesis are repressed by glucose through a CcpA-independent mechanism (18). The PPP is an alternative route of glucose degradation that generates nicotinamide adenine dinucleotide phosphate (NADPH) and pentoses (Figure 5). Among the genes involved in this pathway, synthesis of the transketolase from the tkt’ and tkt genes (CD2321, CD2322) and the ribose-5-P isomerase from rpiB2 (CD3480) was induced by glucose in strain JIR8094 (Figure 5). In B. subtilis and S. aureus, only the gluconate conversion enzymes of the PPP to provide ribulose 5-P are weakly induced by glucose (18,46).
Glucose and/or CcpA regulate the synthesis of several other enzymes involved in carbon metabolism. The aksA-acnB-icd operon (CD0832–CD0834), which encodes a trans-homoaconitate synthase, an aconitate hydratase and an isocitrate/3-isopropylmalate dehydrogenase, respectively, was strongly repressed by glucose through CcpA. As mentionned previously, we found a creCD site inside acnB, and we showed that it is a direct CcpA target in vivo (Table 2) and in vitro (Figure 3B). Moreover, expression of fumB (CD1004) involved in malate formation from fumarate and of a gene encoding a protein sharing 57% identity with YtsJ of B. subtilis involved in malate to pyruvate interconversion (CD1005) was induced by glucose in a CcpA-dependent manner (Figure 5, Supplementary Table S2). However, these genes lack creCD sites, therefore, suggesting an indirect effect of CcpA. Finally, the pyruvate carboxylase pycA gene (CD0021) leading to oxaloacetate production from pyruvate was induced in the presence of glucose by a CcpA-independent mechanism.
The role of glucose and CcpA in the regulation of energy metabolism
The principal source of energy in C. difficile comes from the catabolism of carbohydrates and amino acids. Clostridium difficile metabolizes carbohydrates through fermentation, leading to the production of short-chain fatty acids (lactate, acetate, butyrate, propionate … ) and minor quantity of alcohols (12,53). The pathways allowing the synthesis of butyryl-CoA from acetyl-CoA (CD1054–CD1059) or succinate (CD2344–CD2338) and the further production of butyrate (CD0112–CD0113) or butanol (CD2966) from butyryl-CoA were repressed by glucose through a CcpA-dependent mechanism (Figure 5, Supplementary Tables S2 and S4). Among these genes, creCD sites are present inside the bcd2 operon (CD1054–CD1059) and upstream of CD2344 and adhE (CD2966) (Figure 5 and Supplementary Table S2).
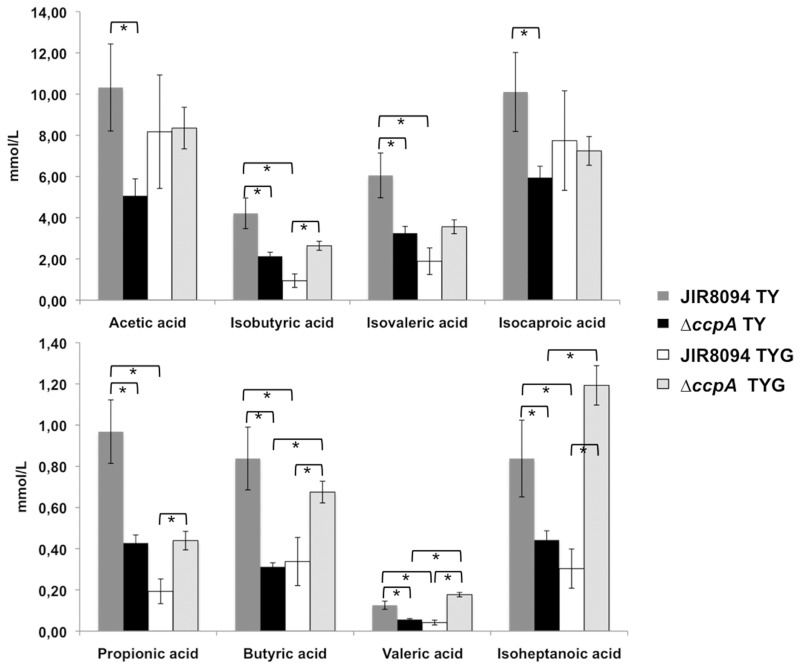
To confirm the impact of glucose addition and CcpA regulation on the fermentation processes, we analysed by gas–liquid chromatography the end products of fermentation of strain JIR8094 and the ccpA mutant grown in TY or in TYG. We could not detect butanol nor ethanol by these assays. The formation of butyric, isobutyric, valeric, propionic and isoheptanoic acids was repressed by glucose, and this downregulation was fully or partially abolished in a ccpA mutant in TYG (Figure 6). These results are in agreement with our transcriptome data, indicating that key steps of fermentation are repressed by glucose through CcpA. It is intriguing to note that the ccpA gene disruption led to a decreased production of the end products of fermentation in the absence of glucose. This effect could result from a change in the metabolism balance because of the lack of CcpA, to an indirect influence of CcpA in the fermentation pathways through the modulation of intracellular concentration of metabolites or a cascade of regulation by uncharacterized mechanisms.
Figure 6.
Fermentation end products of the strain JIR8094 and the ccpA mutant grown in the presence or absence of glucose. Gas–liquid chromatography analysis of the metabolic end products from JIR8094 and ΔccpA mutant strains after 48 h of growth in TY or TYG medium was performed. The mean and standard error of three experiments are shown. Asterisk corresponds to significative statistical difference analysed by the student t-test (P < 0.05).
Clostridium difficile can also use amino acids as energy sources through Stickland reactions. These reactions consist in the coupled fermentation of two amino acids, in which one is oxidatively deaminated or decarboxylated (Stickland donor) and another is reductively deaminated or reduced (Stickland acceptor). The most efficient Stickland donors are valine, leucine, isoleucine and alanine, whereas the most efficient acceptors are proline, glycine and leucine (54,55).
The expression of prdE, prdF and prdA genes (CD3237, CD3239 and CD3244), required for proline reduction, was induced by glucose and positively controlled by CcpA (Figure 7, Supplementary Table S2 and Figure S2). Actually, prdA harbours a creCD site in the promoter region to which CcpA was able to bind in vivo and in vitro (Table 2 and Figure 2C). The expression of ldhA, hadA, hadI, acdB and etfB1 genes (CD0394, CD0395, CD0396, CD0399 and CD0400) required for the leucine reductive branch leading to the formation of isocaproate was 4-fold repressed by glucose by a CcpA-dependent mechanism (Figure 7). We also identified hadA as a CcpA direct target by ChIP-on-Chip analysis (Table 2) and demonstrated that the creCD site located upstream of hadA was essential for CcpA binding, as its deletion no longer showed a shift by EMSA analysis (Figure 3A). Finally, most of the genes of the trxA2-trxB3-grdXEABCD operon involved in glycine reduction (CD2358–CD2348) and gcv operon involved in glycine oxidation (CD1657–CD1658) were repressed by CcpA in reponse to glucose (Figure 7) (56). Moreover, CcpA binds to the creCD motif internal to grdX (CD2357) (Figure 4A) and a creCD site is present upstream of gcvPB (CD1658) (Table 2). Despite the fact that the utilization of serine to produce energy has not been described in C. difficile, this amino acid is preferentially degraded with threonine, arginine, cysteine and proline by Clostridium sticklandii that is closely related to C. difficile. Interestingly, the expression of sdaB (CD3222) involved in serine degradation to form pyruvate that can be further metabolized by fermentation was induced by glucose and positively controlled by CcpA (Figure 7).
Figure 7.
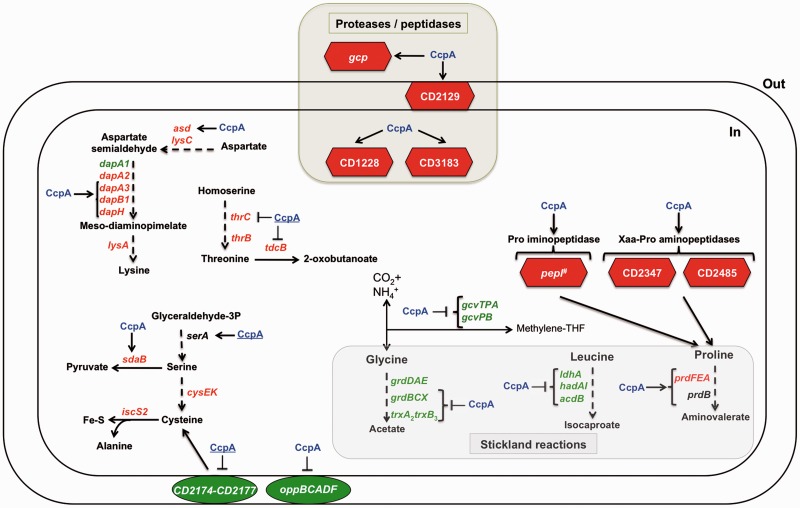
Role of glucose and CcpA in the control of peptide and amino acid metabolisms. Genes encoding enzymes or transporters, which are regulated by CcpA and/or glucose in transcriptome. Green: gene downregulated by glucose; red: gene upregulated by glucose; black: gene not regulated by glucose; blue CcpA: regulation by glucose through CcpA; blue and underlined CcpA: other type of control by CcpA; hash: localization unknown; perpendicular: gene downregulated by CcpA; arrow: gene upregulated by CcpA. lysC (CD2054), aspartokinase; asd (CD3224), aspartate-semialdehyde dehydrogenase; dapA1 (CD3000), dihydrodipicolinate synthase 1; dapA2 (CD3223), dihydrodipicolinate synthase 2; dapA3 (CD3225), dihydrodipicolinate synthase 3; dapB1 (CD3226), dihydrodipicolinate reductase; dapH (CD3227), 2,3,4,5-tetrahydropyridine-2,6-dicarboxylate; lysA (CD2053), diaminopimelate decarboxylase; thrC (CD2118), threonine synthase, thrB (CD2119), homoserine kinase; tdcB (CD2514), threonine dehydratase II; serA (CD0995), putative D-3-phosphoglycerate dehydrogenase; sdaB (CD3222), L-serine dehydratase; cysE (CD1595), serine acetyltransferase; cysK (CD1594), O-acetylserine sulphydrylase; iscS2 (CD1279), cysteine desulphurase; oppBCADF (CD0853–CD0857), oligopeptide ABC transporter; CD2177–CD2174, cystine ABC transporter; prdF (CD3237), proline racemase; prdE (CD3239), proline reductase; prdA (CD3244), D-proline reductase proprotein; prdB (CD3241), proline reductase; trxB3 (CD2356), thioredoxin reductase 3; trxA2 (CD2355), thioredoxin 2; grdX (CD2357), glycine reductase complex component; grdE (CD2354), glycine reductase complex component B subunits α and β; grdA (CD2352), glycine reductase complex selenoprotein A; grdB (CD2351), glycine reductase complex component B γ subunit; grdC (CD2349), glycine reductase complex component C subunit β; grdD (CD2348), glycine reductase complex component C subunit α; ldhA (CD0394), 2-hydroxyisocaproate dehydrogenase; hadA (CD0395), 2-hydroxyisocaproate CoA transferase; hadI (CD0396), activator of dehydratase; acdB (CD0399), acyl-CoA dehydrogenase; gcvTPA (CD1657), bi-functional glycine dehydrogenase/aminomethyl transferase protein; gcvPB (CD1658), Glycine decarboxylase; CD1228, putative protease; CD3183, putative peptidase; CD2485, putative Xaa-Pro aminopeptidase; CD2347, putative Xaa-Pro dipeptidase; gcp (CD0152), putative O-sialoglycoprotein endopeptidase; CD2129, putative membrane-associated peptidase; pepI (CD3041), proline iminopeptidase.
Thus, CcpA seems to play a key role in the hierarchy of amino acid utilization in C. difficile. It directly regulates the genes involved in Stickland reactions by using preferentially proline as an energy source in detriment of leucine or glycine. It is worth adding that addition of proline represses toxin synthesis, whereas genes encoding enzymes for the reductive pathway of glycine and leucine are upregulated during growth conditions that favour toxin production (57). This observation suggests a connection between proline utilization and the control of toxin synthesis that probably involves a regulation by CcpA.
Amino acids metabolism
In C. difficile, the synthesis of several proteases and peptidases was regulated by CcpA in response to glucose (Figure 7). Among them, the expression of gcp (CD0152) encoding an O-sialoglycoprotein endopeptidase and three other peptidases encoding genes (CD1228, CD2129 and CD3183) was induced by glucose in a CcpA-dependent manner. Similarly, the same type of regulation was found for the proline iminopeptidase gene pepI (CD3041) and peptidases encoding genes (CD2347 and CD2485) that share similarities with Xaa-Pro dipeptidases. So, we observed a coordinated regulation of the peptidases (release of free proline) and the proline reductase encoding genes (54). By contrast, the expression of the oppBCADF operon (CD0853–CD0857) encoding an oligopeptides ABC transporter was repressed by glucose through a CcpA-dependent mechanism. Furthermore, CD2347 and oppBCADF operons are CcpA direct targets bound by CcpA in vivo and harbouring a creCD site (Table 2).
In C. difficile, the glucose induces the biosynthetic pathways of lysine, threonine, serine and cysteine, and CcpA plays a role in the control of some of them (Figure 7, Supplementary Table S2). The expression of asd-dapA3-dapB1 and dapH (CD3224–CD3227) involved in lysine biosynthesis from aspartate was positively controlled by CcpA in response to glucose availability (Figure 7). Expression of the cysK-cysE (CD1594–CD1595) and the iscR-iscS2-iscU (CD1278–CD1280) operons involved in cysteine synthesis and iron–sulphur biogenesis from cysteine (sulphur donor), respectively, was increased in the presence of glucose independently of CcpA (Figure 7, Supplementary Table S2). Thus, in the presence of glucose, the increased expression of pathways required for iron-sulphur production might be related to the crucial role of iron–sulphur proteins in electron transfer and fermentation pathways (58). In conclusion, we show that CcpA in C. difficile is not only involved in the control of carbon metabolism but is also implicated in the regulation of nitrogen metabolism as described for B. subtilis (17).
Control of stress response and cell wall metabolism by glucose and CcpA
During host infection, C. difficile is exposed to several stresses and responds by inducing different mechanisms of defense, such as the synthesis of the heat shock proteins (Hsps), which are also induced by acid and oxidative stresses in C. difficile (59). We observed that CcpA regulated the expression of genes involved in heat stress response in the presence of glucose likely through an indirect control, as we did not detect creCD sites upstream of these genes (Supplementary Table S2). The expression of certain genes implicated in oxidative stress, such as norV, encoding a protein involved in nitric oxide (NO) detoxification, and trxB (CD1691), encoding a thioredoxin reductase was induced by glucose through CcpA. Furthermore, the presence of a creCD site in norV (CD1157) indicated that this gene is probably directly regulated by CcpA (Supplementary Table S2). The involvement of CcpA in the regulation of stress proteins has also been reported in Lactobacillus plantarum (60) and in Enterococcus faecalis (61).
Previous studies showed the involvement of cell surface-associated proteins in C. difficile colonization (5). We observed that CcpA in response to glucose or CcpA alone was implicated in the control of a large number of genes encoding cell wall proteins and peptidoglycan hydrolases potentially implicated in C. difficile adhesion to the host cells (Supplementary Table S2). Moreover, a creCD site is present in the dltDABC operon that was repressed by CcpA (Supplementary Table S2). This operon encodes for proteins responsible for the teichoic acids D-alanylation, playing an important role in the bacterial autolytic activity (62). In S. pneumoniae, CcpA has also been reported to be implicated in the regulation of genes encoding for cell surface proteins important for colonization (23).
Involvement of glucose and CcpA in sporulation
It has been recently described that CcpA is one of the few regulators present in the proteome of C. difficile spores (63). Sporulation and germination are key events of the infectious cycle of C. difficile (31), and spores mainly contribute to nosocomial transmission. In C. difficile, the global response regulator Spo0A (CD1214) and the associated sensor histidine kinase CD1579 (64) are involved in the sporulation initiation. We observed from the transcriptome that CcpA negatively regulated spo0A and CD1579 expression in the presence of glucose (Supplementary Table S2). By qRT-PCR, we further demonstrated that spo0A was also 2–3-fold repressed by glucose (Supplementary Table S4). Moreover, the expression of several genes involved in different stages of sporulation, such as sigE (CD2643) and sigG (CD2642) and spoIIAA-spoIIAB-sigF (CD0770–CD0772), spoIIE (CD3490), spoIIIAG (CD1198), spoIVA (CD2629) and spoVS (CD1935), was repressed by glucose in the absence of CcpA. CcpA also negatively regulated most of these spo genes in the absence of glucose (Supplementary Table S2). Furthermore, we found that the spo0A gene encoding the key regulator of the initiation of sporulation and the spoIIAA-spoIIAB-sigF operon, which is responsible for the synthesis and the activation/inactivation of SigF (involved in the endospore formation), are direct targets that contain creCD sites (Table 2) to which CcpA is able to bind (Figures 2 and 4B).
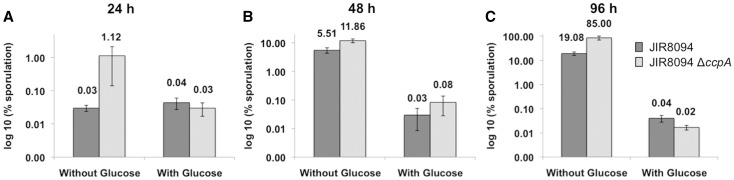
We further tested the involvement of glucose and CcpA inactivation on the sporulation efficiency during 96 h of growth (Figure 8). After 48 h of growth, we observed that the sporulation efficiency of JIR8094 and ccpA mutant strains was similar and strongly reduced when glucose was added. This indicates that sporulation is strongly repressed by glucose and that this repression does not involve CcpA. This has already been reported for C. perfringens where glucose-mediated catabolite repression of sporulation is not because of CcpA activity (26). Therefore, like in C. perfringens, glucose-repression of C. difficile sporulation implicates a still uncharacterized CcpA-independent pathway. In addition, CcpA is required for sporulation in C. perfringens in the absence of glucose, with a 60-fold drop in sporulation efficiency in a ccpA mutant at 24 h of growth in DSSM medium (26). On the contrary, in C. difficile, the rate of sporulation efficiency in the absence of glucose was 8–37-fold higher in the ccpA mutant compared with the JIR8094 strain at 24 h of growth in SM medium (Figure 8A). This is in agreement with the transcriptome results showing that CcpA is implicated in the repression of genes involved in the control of the first stages of sporulation like the SpoIIAA operon and the SpoIIE phosphatase (Supplementary Table S2).
Figure 8.
Effect of glucose addition or CcpA inactivation on the sporulation efficiency. Comparison of sporulation efficiency of C. difficile JIR8094 and ccpA mutant strains grown in SM medium supplemented or not supplemented with 0.5% of glucose at 24 h (A), 48 h (B) and 96 h (C). Total cell counts were obtained by enumeration of CFUs derived from serial dilutions of the initial culture plated on BHI agar supplemented with 0.1% of taurocholate. The same procedure was applied for spore counts with an additional step of heat treatment at 60°C for 30 min, to select for heat resistant spores. Percentage of sporulation is the ratio between the spore titer and total cell titer at each time point. The mean and standard error of three experiments are shown.
Complex regulatory network mediated by CcpA
There are few studies concerning the combination of regulators that control gene expression during host infection. The data generated herein demonstrated that glucose and/or CcpA controlled the expression of 67 regulators (Supplementary Table S2). Twenty of them were regulated by glucose through a CcpA-dependent mechanism. We deduced from the transcriptome analysis that CcpA controlled 30 regulators, and we revealed by ChIP-on-Chip experiments that five transcriptional regulators (rbsR, cggR, CD1584, CD2111 and spo0A) and a sensor histidine–kinase (virS) were direct CcpA targets (Table 2). However, regulation of expression of genes encoding the DeoR type regulator CD2111 and VirS was not observed in the transcriptome conditions used, and this may be because of the low level of their expression. In addition, the CcpA-controlled regulatory genes CD1428 and CD3031 contain a creCD site (Supplementary Table S2). We suggest that these seven CcpA direct targets may mediate through a cascade of regulation part of the indirect regulation by CcpA observed in our analysis. Finally, CcpA indirectly controlled several additional regulators (Supplementary Table S2). These regulators might be under the control of direct CcpA regulators or their activity might be modulated by other mechanisms, including changes in the availability of metabolic effectors as proposed for CggR and CcpC in B. subtilis (65). Interestingly, in B. subtilis, a cre sequence is located upstream of cggR, but cggR transcription is affected by a ccpA deletion independently of this cre (65,66).
The ability of CcpA to promote different regulatory outcomes, leading to activation or repression, suggests that additional modulating signals and/or regulatory pathways might interact with CcpA in the regulation of specific target genes. CodY is a pleiotropic transcriptional regulator that monitors the nutrient sufficiency of the environment. In C. difficile, CodY regulates the toxin genes, and genes implicated in amino acids biosynthesis, nutrient transport, fermentation and sporulation (67). Several genes are found directly regulated by CcpA and CodY. It is the case of toxin specific regulator gene tcdR, the oppBCADF operon for oligopeptide transport, the butyrate metabolism operon (CD1054–CD1059) or genes implicated in the early stages of sporulation, such as the spoIIAA operon and spoIIE (67). This means that CcpA and CodY play a major role in several circuits of regulation in C. difficile and that these circuits could overlap between them rendering even more complex the regulatory network.
CONCLUSION
In conclusion, our results show the importance of the global regulator CcpA in C. difficile as a key factor controlling several processes, such as selective utilization of carbon sources, differential fermentation pathways, amino acid metabolism and toxin production in response to glucose availability. The identification of CcpA targets in vivo and in vitro, and also the definition of the C. difficile cre consensus allow us to dissociate direct and indirect effects of CcpA. The CcpA direct targets are mainly involved in the utilization of alternative sugar sources (PTS or non-PTS sugar transporters), in fermentation pathways leading to the production of butyrate, in Stickland reactions, in toxin synthesis and in regulatory functions including two key steps of the initiation of sporulation.
Phylogeny analysis shows that CcpA of C. difficile clusters with that of clostridia, but not with CcpA of bacilli (30). Interestingly, we found putative creCD-like sites in the regulatory regions of genes known to be controlled by CcpA in other clostridia, including the xylose utilization gene xylB in C. acetobutylicum (22) and the perfringolysin O gene pfoA in C. perfringens (29). A comparative genomic analysis of distribution of CcpA binding sites in bacteria from the bacilli and clostridia groups reinforces these observations on the divergence of the cre motifs in these two phylogenetically distant groups of Firmicutes and the relatively good conservation of the cre motifs inside of the clostridia and bacilli groups (D.A.R and N.V.S, unpublished observation). The characterization of a cre consensus motif for clostridia will probably help for the determination of the CcpA regulons, which remain poorly characterized in clostridia compared with other Firmicutes.
The high number of regulators controlled by CcpA either directly or indirectly suggests that CcpA plays a central role in the hierarchical organization of the genetic regulatory network of C. difficile as shown for B. subtilis (68). We are also aware that C. difficile virulence depends on coordinate interactions between multiple regulatory circuits. As an example, CodY has been shown to have a direct role in the regulation of sporulation and of toxin genes (67). The next step will be to understand how CcpA and CodY, two major global regulators of metabolism, interact to coordinate virulence in response to environmental signals. In S. aureus, the trichloroacetic acid cycle seems to be intimately integrated into regulating virulence factor synthesis and biofilm formation through specific or global regulators like CodY and CcpA (69). In C. difficile, we might propose that the fermentation processes are similarly correlated with the signal transduction pathways controlling virulence. Indeed, addition of butyric acid induces toxin synthesis, and during high-toxin synthesis, genes coding for the enzymes involved in the formation of butyrate from succinate or acetyl-CoA are upregulated (53,57). CodY and CcpA are implicated in the regulation of genes involved in these fermentation pathways and in toxin production linking these two processes. Finally, it is possible that CcpA might also contribute to the ability of this pathogen to obtain key nutrients in the host environment and to colonize the gut. Experiments are now in progress to further characterize the role of CcpA and its regulon during growth and survival in the animal models of CDI.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Tables 1–4 and Supplementary Figures 1 and 2.
FUNDING
Research grant US Public Health Service [AI057637]; predoctoral fellowship Fundação para a Ciência e a Tecnologia [SFRH//BD/16399/2004 to A.A.] and ERA-NET PathoGenoMics/CDIFFGEN (to A.A. and E.C.). U.S. Department of Energy [DE-SC0004999] and Russian Foundation for Basic Research [10-04-01768] (to D.A.R.). Funding for open access charge: Institut Pasteur.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENT
The authors would like to thank Philippe Bouvet for his help on the gas-liquid chromatography analysis of C. difficile products of fermentation. They also acknowledge J. Rood for the gift of the JIR8094 strain.
REFERENCES
- 1.Ricciardi R, Rothenberger DA, Madoff RD, Baxter NN. Increasing prevalence and severity of Clostridium difficile colitis in hospitalized patients in the United States. Arch. Surg. 2007;142:624–631; discussion 631. doi: 10.1001/archsurg.142.7.624. [DOI] [PubMed] [Google Scholar]
- 2.Loo VG, Poirier L, Miller MA, Oughton M, Libman MD, Michaud S, Bourgault AM, Nguyen T, Frenette C, Kelly M, et al. A predominantly clonal multi-institutional outbreak of Clostridium difficile-associated diarrhea with high morbidity and mortality. N. Engl. J. Med. 2005;353:2442–2449. doi: 10.1056/NEJMoa051639. [DOI] [PubMed] [Google Scholar]
- 3.Bartlett JG. Narrative review: the new epidemic of Clostridium difficile-associated enteric disease. Ann. Intern. Med. 2006;145:758–764. doi: 10.7326/0003-4819-145-10-200611210-00008. [DOI] [PubMed] [Google Scholar]
- 4.Akerlund T, Svenungsson B, Lagergren A, Burman LG. Correlation of disease severity with fecal toxin levels in patients with Clostridium difficile-associated diarrhea and distribution of PCR ribotypes and toxin yields in vitro of corresponding isolates. J. Clin. Microbiol. 2006;44:353–358. doi: 10.1128/JCM.44.2.353-358.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Deneve C, Janoir C, Poilane I, Fantinato C, Collignon A. New trends in Clostridium difficile virulence and pathogenesis. Int. J. Antimicrob. Agents. 2009;33(Suppl. 1):S24–S28. doi: 10.1016/S0924-8579(09)70012-3. [DOI] [PubMed] [Google Scholar]
- 6.Just I, Selzer J, Wilm M, von Eichel-Streiber C, Mann M, Aktories K. Glucosylation of Rho proteins by Clostridium difficile toxin B. Nature. 1995;375:500–503. doi: 10.1038/375500a0. [DOI] [PubMed] [Google Scholar]
- 7.Just I, Wilm M, Selzer J, Rex G, von Eichel-Streiber C, Mann M, Aktories K. The enterotoxin from Clostridium difficile (ToxA) monoglucosylates the Rho proteins. J. Biol. Chem. 1995;270:13932–13936. doi: 10.1074/jbc.270.23.13932. [DOI] [PubMed] [Google Scholar]
- 8.Mani N, Lyras D, Barroso L, Howarth P, Wilkins T, Rood JI, Sonenshein AL, Dupuy B. Environmental response and autoregulation of Clostridium difficile TxeR, a sigma factor for toxin gene expression. J. Bacteriol. 2002;184:5971–5978. doi: 10.1128/JB.184.21.5971-5978.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Matamouros S, England P, Dupuy B. Clostridium difficile toxin expression is inhibited by the novel regulator TcdC. Mol. Microbiol. 2007;64:1274–1288. doi: 10.1111/j.1365-2958.2007.05739.x. [DOI] [PubMed] [Google Scholar]
- 10.Govind R, Dupuy B. Secretion of Clostridium difficile toxins A and B requires the holin-like protein TcdE. PLoS Pathog. 2012;8:e1002727. doi: 10.1371/journal.ppat.1002727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schwan C, Stecher B, Tzivelekidis T, van Ham M, Rohde M, Hardt WD, Wehland J, Aktories K. Clostridium difficile toxin CDT induces formation of microtubule-based protrusions and increases adherence of bacteria. PLoS Pathog. 2009;5:e1000626. doi: 10.1371/journal.ppat.1000626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Seddon SV, Hemingway I, Borriello SP. Hydrolytic enzyme production by Clostridium difficile and its relationship to toxin production and virulence in the hamster model. J. Med. Microbiol. 1990;31:169–174. doi: 10.1099/00222615-31-3-169. [DOI] [PubMed] [Google Scholar]
- 13.Kirby JM, Ahern H, Roberts AK, Kumar V, Freeman Z, Acharya KR, Shone CC. Cwp84, a surface-associated, cysteine protease, plays a role in the maturation of the surface layer of Clostridium difficile. J. Biol. Chem. 2009;284:34666–34673. doi: 10.1074/jbc.M109.051177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.O’Connor JR, Johnson S, Gerding DN. Clostridium difficile infection caused by the epidemic BI/NAP1/027 strain. Gastroenterology. 2009;136:1913–1924. doi: 10.1053/j.gastro.2009.02.073. [DOI] [PubMed] [Google Scholar]
- 15.Dupuy B, Sonenshein AL. Regulated transcription of Clostridium difficile toxin genes. Mol. Microbiol. 1998;27:107–120. doi: 10.1046/j.1365-2958.1998.00663.x. [DOI] [PubMed] [Google Scholar]
- 16.Deutscher J, Francke C, Postma PW. How phosphotransferase system-related protein phosphorylation regulates carbohydrate metabolism in bacteria. Microbiol. Mol. Biol. Rev. 2006;70:939–1031. doi: 10.1128/MMBR.00024-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fujita Y. Carbon catabolite control of the metabolic network in Bacillus subtilis. Biosci. Biotechnol. Biochem. 2009;73:245–259. doi: 10.1271/bbb.80479. [DOI] [PubMed] [Google Scholar]
- 18.Blencke HM, Homuth G, Ludwig H, Mader U, Hecker M, Stulke J. Transcriptional profiling of gene expression in response to glucose in Bacillus subtilis: regulation of the central metabolic pathways. Metab. Eng. 2003;5:133–149. doi: 10.1016/s1096-7176(03)00009-0. [DOI] [PubMed] [Google Scholar]
- 19.Moreno MS, Schneider BL, Maile RR, Weyler W, Saier MH., Jr Catabolite repression mediated by the CcpA protein in Bacillus subtilis: novel modes of regulation revealed by whole-genome analyses. Mol. Microbiol. 2001;39:1366–1381. doi: 10.1111/j.1365-2958.2001.02328.x. [DOI] [PubMed] [Google Scholar]
- 20.Zomer AL, Buist G, Larsen R, Kok J, Kuipers OP. Time-resolved determination of the CcpA regulon of Lactococcus lactis subsp. cremoris MG1363. J. Bacteriol. 2007;189:1366–1381. doi: 10.1128/JB.01013-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.van der Voort M, Kuipers OP, Buist G, de Vos WM, Abee T. Assessment of CcpA-mediated catabolite control of gene expression in Bacillus cereus ATCC 14579. BMC Microbiol. 2008;8:62. doi: 10.1186/1471-2180-8-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ren C, Gu Y, Hu S, Wu Y, Wang P, Yang C, Yang S, Jiang W. Identification and inactivation of pleiotropic regulator CcpA to eliminate glucose repression of xylose utilization in Clostridium acetobutylicum. Metab. Eng. 2010;12:446–454. doi: 10.1016/j.ymben.2010.05.002. [DOI] [PubMed] [Google Scholar]
- 23.Carvalho SM, Kloosterman TG, Kuipers OP, Neves AR. CcpA ensures optimal metabolic fitness of Streptococcus pneumoniae. PLoS One. 2011;6:e26707. doi: 10.1371/journal.pone.0026707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li C, Sun F, Cho H, Yelavarthi V, Sohn C, He C, Schneewind O, Bae T. CcpA mediates proline auxotrophy and is required for Staphylococcus aureus pathogenesis. J. Bacteriol. 2010;192:3883–3892. doi: 10.1128/JB.00237-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Seidl K, Stucki M, Ruegg M, Goerke C, Wolz C, Harris L, Berger-Bachi B, Bischoff M. Staphylococcus aureus CcpA affects virulence determinant production and antibiotic resistance. Antimicrob. Agents Chemother. 2006;50:1183–1194. doi: 10.1128/AAC.50.4.1183-1194.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Varga J, Stirewalt VL, Melville SB. The CcpA protein is necessary for efficient sporulation and enterotoxin gene (cpe) regulation in Clostridium perfringens. J. Bacteriol. 2004;186:5221–5229. doi: 10.1128/JB.186.16.5221-5229.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Varga JJ, Therit B, Melville SB. Type IV pili and the CcpA protein are needed for maximal biofilm formation by the gram-positive anaerobic pathogen Clostridium perfringens. Infect. Immun. 2008;76:4944–4951. doi: 10.1128/IAI.00692-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chiang C, Bongiorni C, Perego M. Glucose-dependent activation of Bacillus anthracis toxin gene expression and virulence requires the carbon catabolite protein CcpA. J. Bacteriol. 2011;193:52–62. doi: 10.1128/JB.01656-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mendez MB, Goni A, Ramirez W, Grau RR. Sugar inhibits the production of the toxins that trigger clostridial gas gangrene. Microb. Pathog. 2012;52:85–91. doi: 10.1016/j.micpath.2011.10.008. [DOI] [PubMed] [Google Scholar]
- 30.Antunes A, Martin-Verstraete I, Dupuy B. CcpA-mediated repression of Clostridium difficile toxin gene expression. Mol. Microbiol. 2011;79:882–899. doi: 10.1111/j.1365-2958.2010.07495.x. [DOI] [PubMed] [Google Scholar]
- 31.Sebaihia M, Wren BW, Mullany P, Fairweather NF, Minton N, Stabler R, Thomson NR, Roberts AP, Cerdeno-Tarraga AM, Wang H, et al. The multidrug-resistant human pathogen Clostridium difficile has a highly mobile, mosaic genome. Nat. Genet. 2006;38:779–786. doi: 10.1038/ng1830. [DOI] [PubMed] [Google Scholar]
- 32.O’Connor JR, Lyras D, Farrow KA, Adams V, Powell DR, Hinds J, Cheung JK, Rood JI. Construction and analysis of chromosomal Clostridium difficile mutants. Mol. Microbiol. 2006;61:1335–1351. doi: 10.1111/j.1365-2958.2006.05315.x. [DOI] [PubMed] [Google Scholar]
- 33.Wilson KH, Kennedy MJ, Fekety FR. Use of sodium taurocholate to enhance spore recovery on a medium selective for Clostridium difficile. J. Clin. Microbiol. 1982;15:443–446. doi: 10.1128/jcm.15.3.443-446.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Carlier JP, Sellier N. Gas chromatographic-mass spectral studies after methylation of metabolites produced by some anaerobic bacteria in spent media. J. Chromatogr. 1989;493:257–273. doi: 10.1016/s0378-4347(00)82733-4. [DOI] [PubMed] [Google Scholar]
- 35.Saujet L, Monot M, Dupuy B, Soutourina O, Martin-Verstraete I. The key sigma factor of transition phase, SigH, controls sporulation, metabolism, and virulence factor expression in Clostridium difficile. J. Bacteriol. 2011;193:3186–3196. doi: 10.1128/JB.00272-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rouillard JM, Zuker M, Gulari E. OligoArray 2.0: design of oligonucleotide probes for DNA microarrays using a thermodynamic approach. Nucleic Acids Res. 2003;31:3057–3062. doi: 10.1093/nar/gkg426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Grainger DC, Overton TW, Reppas N, Wade JT, Tamai E, Hobman JL, Constantinidou C, Struhl K, Church G, Busby SJ. Genomic studies with Escherichia coli MelR protein: applications of chromatin immunoprecipitation and microarrays. J. Bacteriol. 2004;186:6938–6943. doi: 10.1128/JB.186.20.6938-6943.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ritchie ME, Silver J, Oshlack A, Holmes M, Diyagama D, Holloway A, Smyth GK. A comparison of background correction methods for two-colour microarrays. Bioinformatics. 2007;23:2700–2707. doi: 10.1093/bioinformatics/btm412. [DOI] [PubMed] [Google Scholar]
- 39.Smyth GK, Speed T. Normalization of cDNA microarray data. Methods. 2003;31:265–273. doi: 10.1016/s1046-2023(03)00155-5. [DOI] [PubMed] [Google Scholar]
- 40.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and poweful approach to multiple testing. J. R. Stat. Soc. Series B Stat. Methodol. 1995;57:289–300. [Google Scholar]
- 41.Pons N, Batto JM, Ehrlich SD, Renault P. Development of software facilities to characterize regulatory binding motifs and application to streptococcaceae. J. Mol. Microbiol. Biotechnol. 2008;14:67–73. doi: 10.1159/000106084. [DOI] [PubMed] [Google Scholar]
- 42.Bailey TL, Boden M, Buske FA, Frith M, Grant CE, Clementi L, Ren J, Li WW, Noble WS. MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res. 2009;37:W202–W208. doi: 10.1093/nar/gkp335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Novichkov PS, Rodionov DA, Stavrovskaya ED, Novichkova ES, Kazakov AE, Gelfand MS, Arkin AP, Mironov AA, Dubchak I. RegPredict: an integrated system for regulon inference in prokaryotes by comparative genomics approach. Nucleic Acids Res. 2010;38:W299–W307. doi: 10.1093/nar/gkq531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gorke B, Stulke J. Carbon catabolite repression in bacteria: many ways to make the most out of nutrients. Nat. Rev. Microbiol. 2008;6:613–624. doi: 10.1038/nrmicro1932. [DOI] [PubMed] [Google Scholar]
- 45.Kietzman CC, Caparon MG. Distinct time-resolved roles for two catabolite-sensing pathways during Streptococcus pyogenes infection. Infect. Immun. 2011;79:812–821. doi: 10.1128/IAI.01026-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Seidl K, Muller S, Francois P, Kriebitzsch C, Schrenzel J, Engelmann S, Bischoff M, Berger-Bachi B. Effect of a glucose impulse on the CcpA regulon in Staphylococcus aureus. BMC Microbiol. 2009;9:95. doi: 10.1186/1471-2180-9-95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schumacher MA, Sprehe M, Bartholomae M, Hillen W, Brennan RG. Structures of carbon catabolite protein A-(HPr-Ser46-P) bound to diverse catabolite response element sites reveal the basis for high-affinity binding to degenerate DNA operators. Nucleic Acids Res. 2011;39:2931–2942. doi: 10.1093/nar/gkq1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Partridge JD, Bodenmiller DM, Humphrys MS, Spiro S. NsrR targets in the Escherichia coli genome: new insights into DNA sequence requirements for binding and a role for NsrR in the regulation of motility. Mol. Microbiol. 2009;73:680–694. doi: 10.1111/j.1365-2958.2009.06799.x. [DOI] [PubMed] [Google Scholar]
- 49.Anbalagan S, McShan WM, Dunman PM, Chaussee MS. Identification of Rgg binding sites in the Streptococcus pyogenes chromosome. J. Bacteriol. 2011;193:4933–4942. doi: 10.1128/JB.00429-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Suarez CA, Blancato VS, Poncet S, Deutscher J, Magni C. CcpA represses the expression of the divergent cit operons of Enterococcus faecalis through multiple cre sites. BMC Microbiol. 2011;11:227. doi: 10.1186/1471-2180-11-227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Miwa Y, Fujita Y. Involvement of two distinct catabolite-responsive elements in catabolite repression of the Bacillus subtilis myo-inositol (iol) operon. J. Bacteriol. 2001;183:5877–5884. doi: 10.1128/JB.183.20.5877-5884.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Nakamura S, Nakashio S, Yamakawa K, Tanabe N, Nishida S. Carbohydrate fermentation by Clostridium difficile. Microbiol. Immunol. 1982;26:107–111. doi: 10.1111/j.1348-0421.1982.tb00159.x. [DOI] [PubMed] [Google Scholar]
- 53.Karlsson S, Lindberg A, Norin E, Burman LG, Akerlund T. Toxins, butyric acid, and other short-chain fatty acids are coordinately expressed and down-regulated by cysteine in Clostridium difficile. Infect. Immun. 2000;68:5881–5888. doi: 10.1128/iai.68.10.5881-5888.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Jackson S, Calos M, Myers A, Self WT. Analysis of proline reduction in the nosocomial pathogen Clostridium difficile. J. Bacteriol. 2006;188:8487–8495. doi: 10.1128/JB.01370-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kim J, Darley D, Selmer T, Buckel W. Characterization of (R)-2-hydroxyisocaproate dehydrogenase and a family III coenzyme A transferase involved in reduction of L-leucine to isocaproate by Clostridium difficile. Appl. Environ. Microbiol. 2006;72:6062–6069. doi: 10.1128/AEM.00772-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Fonknechten N, Chaussonnerie S, Tricot S, Lajus A, Andreesen JR, Perchat N, Pelletier E, Gouyvenoux M, Barbe V, Salanoubat M, et al. Clostridium sticklandii, a specialist in amino acid degradation: revisiting its metabolism through its genome sequence. BMC Genomics. 2010;11:555. doi: 10.1186/1471-2164-11-555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Karlsson S, Burman LG, Akerlund T. Induction of toxins in Clostridium difficile is associated with dramatic changes of its metabolism. Microbiology. 2008;154:3430–3436. doi: 10.1099/mic.0.2008/019778-0. [DOI] [PubMed] [Google Scholar]
- 58.Meyer J. Clostridial iron-sulphur proteins. J. Mol. Microbiol. Biotechnol. 2000;2:9–14. [PubMed] [Google Scholar]
- 59.Emerson JE, Stabler RA, Wren BW, Fairweather NF. Microarray analysis of the transcriptional responses of Clostridium difficile to environmental and antibiotic stress. J. Med. Microbiol. 2008;57:757–764. doi: 10.1099/jmm.0.47657-0. [DOI] [PubMed] [Google Scholar]
- 60.Zotta T, Ricciardi A, Guidone A, Sacco M, Muscariello L, Mazzeo MF, Cacace G, Parente E. Inactivation of ccpA and aeration affect growth, metabolite production and stress tolerance in Lactobacillus plantarum WCFS1. Int. J. Food Microbiol. 2012;155:51–59. doi: 10.1016/j.ijfoodmicro.2012.01.017. [DOI] [PubMed] [Google Scholar]
- 61.Leboeuf C, Leblanc L, Auffray Y, Hartke A. Characterization of the ccpA gene of Enterococcus faecalis: identification of starvation-inducible proteins regulated by ccpA. J. Bacteriol. 2000;182:5799–5806. doi: 10.1128/jb.182.20.5799-5806.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Camiade E, Peltier J, Bourgeois I, Couture-Tosi E, Courtin P, Antunes A, Chapot-Chartier MP, Dupuy B, Pons JL. Characterization of Acp, a peptidoglycan hydrolase of Clostridium perfringens with N-acetylglucosaminidase activity that is implicated in cell separation and stress-induced autolysis. J. Bacteriol. 2010;192:2373–2384. doi: 10.1128/JB.01546-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lawley TD, Croucher NJ, Yu L, Clare S, Sebaihia M, Goulding D, Pickard DJ, Parkhill J, Choudhary J, Dougan G. Proteomic and genomic characterization of highly infectious Clostridium difficile 630 spores. J. Bacteriol. 2009;191:5377–5386. doi: 10.1128/JB.00597-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Underwood S, Guan S, Vijayasubhash V, Baines SD, Graham L, Lewis RJ, Wilcox MH, Stephenson K. Characterization of the sporulation initiation pathway of Clostridium difficile and its role in toxin production. J. Bacteriol. 2009;191:7296–7305. doi: 10.1128/JB.00882-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Doan T, Aymerich S. Regulation of the central glycolytic genes in Bacillus subtilis: binding of the repressor CggR to its single DNA target sequence is modulated by fructose-1,6-bisphosphate. Mol. Microbiol. 2003;47:1709–1721. doi: 10.1046/j.1365-2958.2003.03404.x. [DOI] [PubMed] [Google Scholar]
- 66.Ludwig H, Rebhan N, Blencke HM, Merzbacher M, Stulke J. Control of the glycolytic gapA operon by the catabolite control protein A in Bacillus subtilis: a novel mechanism of CcpA-mediated regulation. Mol. Microbiol. 2002;45:543–553. doi: 10.1046/j.1365-2958.2002.03034.x. [DOI] [PubMed] [Google Scholar]
- 67.Dineen SS, McBride SM, Sonenshein AL. Integration of metabolism and virulence by Clostridium difficile CodY. J. Bacteriol. 2010;192:5350–5362. doi: 10.1128/JB.00341-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Goelzer A, Bekkal Brikci F, Martin-Verstraete I, Noirot P, Bessieres P, Aymerich S, Fromion V. Reconstruction and analysis of the genetic and metabolic regulatory networks of the central metabolism of Bacillus subtilis. BMC Syst. Biol. 2008;2:20. doi: 10.1186/1752-0509-2-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Somerville GA, Proctor RA. At the crossroads of bacterial metabolism and virulence factor synthesis in Staphylococci. Microbiol. Mol. Biol. Rev. 2009;73:233–248. doi: 10.1128/MMBR.00005-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.