Abstract
Classical bovine spongiform encephalopathy is a transmissible prion disease that is fatal to cattle and is a human health risk due to its association with a strain of Creutzfeldt-Jakob disease (vCJD). Mutations to the coding region of the prion gene (PRNP) have been associated with susceptibility to transmissible spongiform encephalopathies in mammals including bovines and humans. Additional loci such as the retinoic acid receptor beta (RARB) and stathmin like 2 (STMN2) have also been associated with disease risk. The objective of this study was to refine previously identified regions associated with BSE susceptibility and to identify positional candidate genes and genetic variation that may be involved with the progression of classical BSE. The samples included 739 samples of either BSE infected animals (522 animals) or non-infected controls (207 animals). These were tested using a custom SNP array designed to narrow previously identified regions of importance in bovine genome. Thirty one single nucleotide polymorphisms were identified at p < 0.05 and a minor allele frequency greater than 5%. The chromosomal regions identified and the positional and functional candidate genes and regulatory elements identified within these regions warrant further research.
Keywords: BSE, Susceptibility, SNP, Candidate Genes
Introduction
Classical Bovine Spongiform Encephalopathy (BSE) is the variant of transmissible spongiform encephalopathies (TSEs) found in Bos taurus or bovines. The TSEs are progressive degenerative neurological disorders caused by the conversion of endogenous prion protein (PRPc) to an abnormally folded form (PrPRes)1,2 and are always fatal.3 TSEs are unique in that they may be genetic, sporadic or transmitted but, in all cases the disease state is associated with accumulation of the abnormal PrPRes in the brain and central nervous system (CNS) causing degeneration.4 Classical BSE is a form of TSE acquired by the consumption of meat and/or bone meal contaminated with the (PrPRes) or infectious prion agent5 and was first observed in the United Kingdom (UK) in 1986.6 A large number of infected animals entered the food chain in the preclinical state and were consumed by the human population in the UK and elsewhere.7 This consumption has been linked to a variant form of Creutzfeldt-Jakob disease (vCJD)8,9 with approximately 200 reported cases in Europe , Asia, and North America (data from National Creutzfeldt-Jakob Disease Surveillance Unit (NCJDSU), Western General Hospital Edinburgh, Scotland: Worldwide vCJD statistics 2010).
Prion disease pathogenesis occurs when endogenous PrPRes auto catalyzes the conversion of PrPc into the PrPRes form. Exposure to the PrPRes promotes or facilitates this conversion through a mechanism that is not yet understood.10 PrPRes is highly resistant to breakdown by the proteases and other proteolytic machinery of the cell.11,12 This resistance results in the accumulation of the non-degraded form of the protein which results in the neural degeneration and spongiform appearance of infected CNS tissues.4 In classical BSE, the pathogenesis of the disease is the result of the transference of the infectious prion from the digestive tract into the peripheral nervous system and then to the brain.5 Studies of the pathogenesis have concluded that the route of transmission is from the ileal Peyer’s patches and the tonsils through the parasympathetic and sympathetic nerve fibers of the autonomous nervous system.5 It has been hypothesized that a number of cellular proteins and the genes may be involved in this progression.13,14 Structural variation in prion protein locus, the gene for the PrP protein, is strongly associated with the risk of all categories of prion disease in humans including vCJD.15 In addition, specific PRNP alleles have been associated with TSE susceptibility in sheep.16,17 In classical BSE, variation in the promoter region of the PRNP locus has also been shown to be associated with risk of disease incidence.18,19 In addition to the PRNP locus, single nucleotide polymorphisms (SNPs) associated with other genes such as the retinoic acid receptor beta (RARB) and stathmin-like 2 proteins (STMN2) have been shown to be related to human susceptibility to prion disease.15,20 Genome wide association studies have also identified several other genomic regions associated with BSE incidence13,14,21,22 but little is known about how these regions and the genes within them that may affect the pathogenesis of classical BSE. Additionally, genome wide association studies in humans have identified other loci of modest overall effect as research targets.23 The objective of this study was to refine previously identified regions associated with BSE susceptibility and to identify positional candidate genes and genetic variation that may be involved with the progression of classical BSE.
Results and Discussion
Candidate gene identification
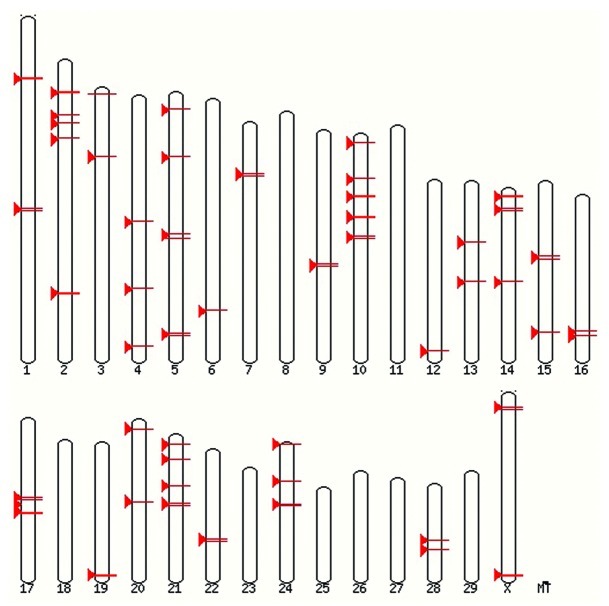
Fifty seven regions on BTAU 4.024 associated with BSE incidence and susceptibility were identified utilizing data generated previously within our lab and throughout published literature.13,14,21,22 These regions are shown in Figure 1. A window of 1MB was created around each previously identified marker (500,000 bases to either side) in order to capture all of the genes closely linked to identified associations. Known genes within each 1MB pair window in BTAU 4.0 were then obtained using BIOMART.25 All previously reported candidate genes for prion disease susceptibility,21,22 were included as well as newly identified candidate genes based on potential functional role in BSE. Candidate genes were identified using NCBI Genbank,26 Gene Ontology27 and KEGG Pathway28 databases and were included if the gene was known to be expressed in nervous tissue, was involved in proteolysis, protein folding or unfolding or protein transport.
Figure 1.
Regions of BTAU 4.0 previously identified as associated to incidence of classical BSE in cattle.
Marker identification for candidate genes and genome regions
Transcript sequences from liver, hypothalamus, muscle, adipose, duodenum, kidney, lung blood, cortex and Peyer’s patch were evaluated to identify SNPs in each of the 89 candidate genes mapped to the 57 regions of the bovine genome thought to be related to BSE susceptibility. Whole-genome sequences from a Holstein bull and a Black Angus bull29were also used to identify structural variation within candidate genes. Non-synonymous mutations were selected when available as were SNPs predicted to introduce a stop codon or splice variation.
Overall structural variation was identified in 87 of 89 candidate genes. Additional SNPs were added to increase the resolution within the 57 one MB pair windows of BTAU 4.0.
The origin of all SNPs in the constructed panel is shown in Additional File 1. Whenever possible, markers were selected that were identified in more than one dataset.
Genotyping
A custom panel of 384 SNP markers was designed to evaluate 87 candidate genes identified in underlying QTLs for BSE incidence. The total number of SNP markers included in the analysis after quality control measures was 240. This panel was used to genotype 739 samples of either BSE infected animals (522 animals) or non-infected controls (207 animals). The animal samples came from a population of half-sibs and a set of unrelated case and control samples. The samples were combined for this study in a modified sib-TDT test using the DFAM procedure in the PLINK software30 that allows for addition of unrelated individuals. The combined population was examined for stratification and none was observed.
A total of 31 markers were associated with incidence of BSE in this population at a significance level of p > 0.05 and a minor allele frequency greater than 5% (Table 1). Some of these SNPs are located close to one another and are potentially within LD in the population.
Table 1. Significance of associated marker loci using the DFAM Procedure in PLINK for a sib-TDT with unrelated individuals.
| Chromosome | Position | Reference Allele | Alternate Allele | Chi Square | P-value | Minor Allele Frequency |
|---|---|---|---|---|---|---|
| 1 |
29642496 |
A |
C |
6.386 |
0.0115 |
0.1555 |
| 1 |
89510456 |
G |
A |
10.56 |
0.001154 |
0.4095 |
|
2 |
14990009 |
A |
G |
5.877 |
0.01534 |
0.1843 |
|
2 |
15112384 |
G |
A |
4.316 |
0.03776 |
0.1385 |
| 2 |
108616181 |
A |
G |
3.948 |
0.04692 |
0.1953 |
| 3 |
32165127 |
A |
G |
5.328 |
0.02099 |
0.1926 |
|
6 |
98267764 |
G |
A |
4.033 |
0.04463 |
0.2006 |
|
6 |
98467430 |
G |
A |
4.634 |
0.03134 |
0.09362 |
| 7 |
24913972 |
G |
A |
3.922 |
0.04766 |
0.1796 |
| 9 |
63199430 |
A |
G |
5.111 |
0.02378 |
0.4367 |
| 10 |
4459856 |
G |
A |
4.17 |
0.04115 |
0.1166 |
|
10 |
21273459 |
A |
G |
7.068 |
0.007846 |
0.1389 |
|
10 |
21653055 |
T |
A |
9.434 |
0.00213 |
0.3812 |
|
10 |
38243823 |
G |
A |
4.889 |
0.02703 |
0.2597 |
|
10 |
38355911 |
A |
G |
5.93 |
0.01488 |
0.1598 |
|
10 |
38725566 |
A |
G |
5.49 |
0.01913 |
0.4004 |
|
10 |
47743503 |
C |
G |
6.311 |
0.012 |
0.3567 |
|
10 |
48274239 |
A |
G |
5.917 |
0.015 |
0.4836 |
| 12 |
80176597 |
G |
A |
5.801 |
0.01602 |
0.308 |
| 13 |
46715840 |
C |
A |
5.136 |
0.02343 |
0.4889 |
|
14 |
43784117 |
A |
G |
5.381 |
0.02036 |
0.3536 |
|
14 |
44086806 |
G |
A |
4.206 |
0.04028 |
0.3295 |
| 15 |
35224440 |
A |
G |
8.253 |
0.004069 |
0.4907 |
| 17 |
40657770 |
A |
G |
6.493 |
0.01083 |
0.1669 |
| 17 |
41455808 |
A |
G |
8.599 |
0.003363 |
0.1543 |
|
20 |
39198878 |
A |
G |
8.721 |
0.003146 |
0.465 |
|
20 |
39329256 |
G |
A |
5.716 |
0.01682 |
0.3464 |
| 21 |
13354965 |
G |
A |
8.603 |
0.003357 |
0.2177 |
| 21 |
24945877 |
G |
C |
5.042 |
0.02475 |
0.3529 |
| 22 |
42219360 |
G |
A |
7.385 |
0.006578 |
0.1735 |
| 28 | 26571763 | A | C | 27.32 | 1.73E-07 | 0.07955 |
Bold indicates clusters of significantly associated markers
Classical BSE as a phenotype
Previous studies21,22 have suggested that it is difficult to test for the genetic association for the clinical presentation of BSE. The explanation provided is that this could be attributed to the fact that while animals that developed BSE are clearly susceptible to disease and the control samples may represent animals that were either not exposed to enough infectious material to develop disease or are in fact resistant to infection. This phenotypic variation may reduce the power to detect disease association. Also as noted,21,22 the use of stringent multiple testing corrections such as Bonferroni to this type of a phenotype may be overly prone to type II errors and thus discard real, though subtle associations in favor of reducing or eliminating type I or false positive results. In contrast, this study was designed to refine the previously identified results13,14,21,22by evaluating a reduced number of markers in a larger pooled population of animals. However, with the total of 240 markers included in the analysis and the criteria of p < 0.05 means that approximately 12 of the identified 31 markers may be significant by chance.
Regions previously identified for BSE susceptibility
This work utilized genomic regions previously identified or suggested to be important to classical BSE susceptibility. However, this work refined previous associations and identified novel candidate genes (Table 2). As shown in Table 2, the position of the associated markers in this study are close but not identical to those shown previously. The resolution of this work was also higher as the aim was to have markers every 100,000 base pairs in the target areas. The increased resolution allowed further refinement of candidate genes and the identification of regulatory regions that may impact disease susceptibility.
Table 2. Previously identified and currently refined genomic regions related to BSE susceptibility.
| Chromosome | Position (Mb) | P-value | Observed in outside population | Reference | Location Mb |
|---|---|---|---|---|---|
| 1 |
29.6 |
0.012 |
Yes |
14, 21 |
29.1 |
| 1 |
89.5 |
0.001 |
Yes |
14, 21, 22 |
89.7 |
| 2 |
14.9 |
0.015 |
No |
|
|
| 2 |
15.1 |
0.038 |
21 |
15.3 |
|
| 2 |
108.6 |
0.047 |
No |
21, 22 |
108.6 |
| 3 |
32.1 |
0.021 |
No |
22 |
32.4 |
| 6 |
98.2 |
0.045 |
No |
21 |
98.8 |
| 6 |
98.4 |
0.031 |
|||
| 7 |
24.9 |
0.048 |
No |
21 |
24.6 |
| 9 |
63.1 |
0.024 |
No |
21 |
62.9 |
| 10 |
4.4 |
0.041 |
No |
21 |
4.4 |
| 10 |
21.2 |
0.008 |
Yes |
13, 21, 22 |
29.2 |
| 10 |
21.6 |
0.002 |
|||
| 10 |
38.2 |
0.027 |
No |
21 |
38.7 |
| 10 |
38.3 |
0.015 |
|||
| 10 |
38.7 |
0.019 |
|||
| 10 |
47.7 |
0.012 |
|
21, 13 |
40.0 and 48.0 |
| 10 |
48.2 |
0.015 |
Yes |
||
| 12 |
80.1 |
0.016 |
No |
21 |
80.1 |
| 13 |
46.7 |
0.023 |
No |
21 |
47.2 |
| 14 |
43.7 |
0.02 |
Yes |
15, 21, 22 |
44 |
| 14 |
44.1 |
0.04 |
|||
| 15 |
35.2 |
0.004 |
No |
21, 22 |
35.7 |
| 17 |
40.6 |
0.011 |
Yes |
14, 21, 22 |
41.0 and 44.2 |
| 17 |
41.4 |
0.003 |
|||
| 20 |
39.2 |
0.003 |
Yes |
21, 13 |
38.8 |
| 20 |
39.3 |
0.017 |
|||
| 21 |
13.3 |
0.003 |
No |
21, 22 |
13.2 and 33.2 |
| 21 |
24.9 |
0.025 |
No |
21 |
24.8 |
| 22 |
42.2 |
0.007 |
No |
21 |
42.4 |
| 28 | 26.6 | 0.0000002 | No | 21 | 26.9 |
New candidate genes
Several pathogenesis studies have concluded that the route of transmission of classical BSE is through consumption of the misfolded PRNP which is then transported into ileal Peyer’s patches and the tonsils and then through the parasympathetic and sympathetic nerve fibers of the autonomous nervous system.5 The misfolded PRNP then leads to the misfolding of native PRNP, which resists degradation and accumulates in the CNS resulting in neurodegeneration.5 It has been hypothesized that a number of cellular proteins and genes play an important role in the progression of the disease.13,14 Several proteins (Table 3) are known to participate in transport of the prion protein across the gut epithelium, transport to dendritic cells, accumulation in lymph tissues and progression from lymph tissues to CNS tissues and specifically to the brain.
Table 3. Positional and functional candidate genes for BSE susceptibility.
| Significant Marker |
|
|
|
|---|---|---|---|
| Chromosome | Position | Candidate Gene (position) | Relationship to BSE |
| 6 |
98267764 |
ANTXR2 (6:98000385-98236768) |
Mutation results in hyalinosis, deposition of extracellular clear tissue protein lesions50 |
| 6 |
98467430 |
||
| 7 |
24913972 |
SLC12A2 (7:24685293-24776754) |
Functions to maintain ion balance, related to schizophrenia51 |
| 9 |
63199430 |
MDN1-1 (9:63073546-63213136) |
Unfolded protein binding31 |
| 10 |
21273459 |
NEDD8 (10:2121578-21224714) |
Conjugates scaffold proteins to allow proteosome mediated protein degradation pathway,31,32 amyloid precursor protein binding,52 accumulates in inclusions during neurodegenerative disease,52 increased in Alzheimer's disease.35 |
| PSME2 (10:21283913-21287173) |
Proteosome activation,53 downregulation associated with neurodegenerative disease.54 |
||
| PSME1(10:21292117-21294778) |
Proteosome activation,53 downregulation associated with neurodegenerative disease.54 |
||
| CHMP4A (10:21227527-21230632) |
Chaperone protein associated with transmembrane transport in neurons.55 |
||
| 10 |
38243823 |
UBR1 (10:38136589-38243823) |
Recognition component of ubiquitin protein degradation pathway, mutations associated with Johanson-blizzard syndrome56 and bipolar disorder.57 |
| 10 |
38355911 |
||
| 10 |
38725566 |
||
| 20 |
39198878 |
GDNF (20:38861046-38881932) |
Suppresses neurodegeneration36 in Parkinson's,37 mutation related to Down's Syndrome and Schizophrenia,38 variant active in Alzheimer's Disease.58 |
| 20 |
39329256 |
||
| 21 |
24945877 |
MORF4L1 (21: 24920985-24938434) |
NEDD8 pathway, histone acetylation, regulation of splice variants.59 |
| 22 |
42219360 |
FHIT (22:40968128-42518027) |
Mutations and splice variants related to incidence of Alzheimer disease,60 Multiple sclerosis,61 and Parkinson's disease.62 |
| 28 | 26571763 | PCBD1 (28:26334706) | Strong Association to Alzheimer's disease.63 |
An example of a functional candidate gene that may be linked to the pathogenesis of classical BSE is NEDD8 (Neural precursor cell expressed, developmentally down-regulated 8), which is located at approximately 21.2CM on BTAU 10. It is close to a marker (10: 21273459) that was significant at p <0.005 and would likely be linked to that marker. NEDD8 encodes an 81 amino acid polypeptide which is 60% identical and 80% homologous to ubiquitin.31 Amyloid precursor protein binding protein-1 (APP-BP1) binds to the carboxyl terminus of the amyloid precursor protein (APP) and serves as the activation enzyme for the ubiquitin-like protein, NEDD8.32 NEDD8 conjugation pathway has shown to be essential for proteolytic targeting by ubiquitination.33 Impairment of this process may contribute to the amyloid plaques evident in classical BSE. NEDD8 has been shown to accumulate in both glial and neuronal inclusions in several neurodegenerative conditions such as Parkinson disease, Alcoholic liver disease and Astrocytoma.34 Co-location of APP-BP1 and NEDD8 is shown in lipid rafts in the hippocampus of Alzheimer’s patients but not in other less affected regions of the brain.35 Further investigation is needed to validate a relationship between NEDD8 and to determine the nature of the potential relationship with classical BSE susceptibility.
Another important positional and functional candidate gene identified in this study is GDNF (glial cell derived neurotrophic factor). GDNF has been shown to mitigate neuronal degeneration in a range of neurodegenerative conditions and was one of the first investigated targets for gene therapy in Parkinson disease.36,37 GDNF is thought to promote survival and differentiation of developing neurons and to protect mature neurons.36 Mutations in or impairments of GDNF action have been shown contribute to the symptoms of Down syndrome and Schizophrenia.38 The action of GDNF could play a role in innate resistance to neurodegeneration in classical BSE. As with NEDD8, future research is needed to establish a relationship and to determine the potential role of GDNF as a target for BSE resistance.
Candidate markers near regulatory regions or CpG Islands
Several markers that were significantly associated with classical BSE susceptibility are positioned close to annotated CpG islands in the bovine genome. Vertebrate CpG islands are short DNA sequences that are significantly richer in GC bases as compared to the rest of the genome and are predominantly non-methylated.39 Most, if not all, CpG islands are sites of transcription initiation though many are not currently annotated promoters39. Shared DNA sequence features adapt CpG islands for promoter function by destabilizing nucleosomes and attracting proteins that create a transcriptionally preferable chromatin state.39 The significantly associated SNP markers in close proximity to CpG islands identified in this study are shown in Table 4.
Table 4. Associated loci in a regulatory region and nearby candidate genes.
| Significant Marker |
|
|
|
|
|---|---|---|---|---|
| Chromosome | Position | CPG Island Position | Nearby Genes | Function |
| 14 |
44086806 |
14:44086435-44087067 |
MED30 (14:44067815-44086895) |
Thyroid hormone binding,64 RNA polymerase cofactor activity, regulation of transcription.65 |
| EXT1 (14:43445834-43759710) |
Heparan sulfate proteoglycan biosynthetic process and disease process,66,67 recognition and binding of prion protein.68 |
|||
| 10 |
21273459 |
10:21283889-21284116 |
NEDD8 (10:2121578-21224714) |
Conjugates scaffold proteins to allow proteosome mediated protein degradation pathway,31,32 amyloid precursor protein binding,52 accumulates in inclusions during neurodegenerative disease,52 increased in Alzheimer's disease.35 |
| 10:21282710-21283083 |
PSME2 (10:21283913-21287173) |
Proteosome activation,53 downregulation associated with neurodegenerative disease.54 |
||
| 10:21289609-21290219 |
PSME1(10:21292117-21294778) |
Proteosome activation,53 downregulation associated with neurodegenerative disease.54 |
||
| 10:21294367-21294916 |
CHMP4A (10:21227527-21230632) |
Chaperone protein associated with transmembrane transport in neurons.55 |
||
| 10 |
38243823 |
10:38122791-38123802 |
UBR1 (10:38136589-38243823) |
Recognition component of ubiquitin protein degradation pathway, mutations associated with Johanson-blizzard syndrome56 and bipolar disorder.57 |
| 10 |
38355911 |
10:38287535-38287874 |
||
| 20 |
39329256 |
20:39285739-39286473 |
GDNF (20:38861046-38881932) |
Suppresses neurodegeneration36 in Parkinson's,37 mutation related to Down's Syndrome and Schizophrenia,38 variant active in Alzheimer's Disease.58 |
| 21 | 24945877 | 21:24920950-24922062 | MORF4L1 (21: 24920985-24938434) | NEDD8 pathway, histone acetylation, regulation of splice variants.59 |
One example of a candidate gene that could be relevant to classical BSE pathogenesis and is in close proximity to both a significant associated marker and CpG island is exostosin 1 (EXT1). As noted previously,22 EXT1 is an endoplasmic reticulum transmembrane glycoprotein responsible for the synthesis and display of cell surface heparin sulphate glycosaminoglycans (GAGs).40 The N terminus of PRNP contains a GAG-binding motif and that may play a functional role in the uptake and transport of aberrant PRNP in gastrointestinal and neural cells in classical BSE transmission and infection.21,41 This relationship needs to be evaluated in the bovine and the link to classical BSE disease needs to be shown in further research.
In conclusion, this research has identified the most exhaustive list of candidate genes for classical BSE susceptibility to date and has supported an association of 31 markers shown to be related to classical BSE susceptibility in a large dataset of 729 case and control animals. It has improved the resolution and power of previous analyses and provided strong functional and positional candidate genes which may lead to better understanding of this disease in its natural host and provide candidate gene and regulatory targets for further research.
Materials and Methods
Animal information
This study used two sets of animals including both case (affected) and control (unaffected) samples from two populations with different population structures. The first data set is family based and includes 225 BSE affected and 193 BSE unaffected half-sibs Holsteins from 30 sire families. These were paternal half-sibs with different dams from the UK which were collected during the BSE outbreak in the mid-1990s. The second population included 297 BSE affected and 14 BSE unaffected animals. The control animals are contemporaries of the cases and were collected from the same farms. In these two populations, BSE status was determined both by examination by qualified veterinarians and then post-mortem by histology (Veterinary Laboratories Agency, New Haw, Surrey, UK). The control animals are assumed to have been exposed to the same environment, did not exhibit any signs of disease and are assumed to have been disease free.
Genomic DNA was isolated from the family based population using a phenol chloroform extraction as described by Hernandez-Sanchez et al.13 Genomic DNA from the case-control population was isolated using a high salt phenol/chloroform extraction as described by Sherman et al.42
mRNA-seq library construction and sequencing using Illumina platform
RNA was prepared for liver (7 animals), hypothalamus (11 animals), muscle and duodenum (12 animals), kidney and lung (14 animals) and adipose (10 animals) tissues. Total RNA was extracted using TRIzol reagent (Invitrogen) from the frozen tissues. The quality and quantity of RNA was determined using a Nanodrop (Nanodrop technologies) and Agilent Bioanalyzer (Agilent).
The RNAs from each tissue were pooled and cDNA libraries were constructed from each tissue pool according to the protocol (Illumina). The first step in the process of Illumina library construction involved purifying the poly-A containing mRNA molecules from total RNA using poly-T oligo-attached magnetic beads. Poly (A) RNA was fragmented followed by random hexamer reverse transcription and second strand synthesis. These cDNA fragments were end repaired prior to ligation of the Illumina GEX PE adapters according to the standard library generation protocol (Illumina). A ~85 bp size range was excised from the library on 6% agarose gel and the resultant library was subjected to 15 cycles of PCR followed by quantification using Nanodrop (Nanodrop Technologies).
Sequencing was performed on Genome Analyzer II (Illumina) which uses a massively parallel sequencing-by-synthesis four-dye approach, to generate billions of bases of high-quality DNA sequence per run. Cluster generation and sequencing was performed on a PE 75nt X 75nt flow cell generating ~1.7-6.0 GB of sequence.
Maq (version 0.7.1) was used to map reads and to generate SNP lists.43 First, the reads were converted from the Illumina export format to the Sanger FASTQ format using the “fq_all2std.pl export2std” command. The “match” command was used to align the paired-end reads to bovine transcripts from Ensembl (release 57).44 The resulting alignments were then merged using the “mapmerge” command and consensus sequences were generated using the “assemble” command. The “cns2snp” command was used to build a list of potential SNP sites from the consensus sequences. A second shorter list of SNPs was also generated using the “maq.pl SNPfilter” command. Default settings were used in all cases, except that the -N option was used with the “assemble” command to specify the number of haplotypes present in each sample. The shorter, filtered SNP list was annotated using NGS-SNP45 and release 57 of Ensembl,44 which includes SNPs from dbSNP build 130.46
Total RNA-Seq- library construction and sequencing using SOLiD platform
For each Cortex (8), Blood (10), Peyer’s Patch (6) libraries samples were pooled into 10ul before depleting the ribosomal RNA using the RiboMinus Eukaryote kit (Invitrogen) and concentrated with a RiboMinus Concentration Module. Five hundred nanograms (as determined by Agilent RNA Nano Chip on the Agilent Bioanalyzer) were carried through library prep using the whole transcriptome RNA-Seq Kit (Applied Biosystems). Libraries were quantified by QRTPCR using the Taqman kit (Applied Biosystems). Fragment sizes were checked using the DNA 1000 chip (Agilent) with the Agilent Bioanalyzer. Beads for Solid 4 were prepared using an EZBead system (Applied Biosystems). Sequencing was conducted on the SOLiD 4 system (ABI). Reads (50bp) were aligned to bovine transcripts from Ensembl (release 57)44 and SNPs were identified by GEOSPIZA.
Whole genome sequencing of Angus and Holstein bull
Genomic DNA from a Black Angus bull and a Holstein bull was sequenced using the Applied Biosystems SOLiD 3 sequencer (Life Technologies Corporation), using a combination of fragment and mate-paired libraries. The libraries were prepared using the reagents and protocols provided by Applied Biosystems, and as reported in previous studies.29
Sequence reads were mapped to the Btau4.0 bovine genome assembly using the Bioscope 1.0 software suite (Life Technologies Corporation). A list of putative SNPs was generated for each animal from the mapped reads, using the diBayes SNP Detection module (with the “med-coverage” stringency setting) included with Bioscope. The lists were subjected to additional filtering, to remove SNPs with particularly high read depth (higher than 95% of the other SNPs from the same animal), and to remove SNPs that could not be unambiguously placed on bovine genome assembly UMD3.147 using the megablast algorithm48 in BLAST+,49 100 bp of flanking sequence, and an e-value threshold of 1e-35. SNPs were annotated using NGS-SNP45and release 57 of Ensembl,44 which includes SNPs from dbSNP build 130.46
Genotyping and data quality control
The total of 729 samples were genotyped with a custom Illumina Golden Gate Vera Code Assay containing 384 SNPs following the manufacturers recommended protocol (Illumina Inc.). Genotypes were called using the BeadStudio software (Illumina Inc.) and processed through the automated genotype calling. Genotypes were then subjected to data quality control parameters. Seventeen SNPs were removed for being monomorphic. Overall, a genotyping success rate of 96% was achieved. Forty seven SNPs from the original set of 384 SNPs were excluded on the basis of assay failure or poor genotype clustering. The remaining 320 SNPs were analyzed through the summary statistics of the PLINK program.30 Additionally, 80 SNPs were removed due to a minor allele frequency (MAF) less than 0.01. In total 240 SNPs were included in the analysis.
Statistical analysis
PLINK software v1.07 was used to perform the statistical analysis.30 The genotypic data from both populations was combined and was analyzed using the DFAM procedure in PLINK. The DFAM procedure in PLINK implements the sib-TDT and also allows for unrelated individuals to be included (via a clustered-analysis using the Cochran-Mantel-Haesnzel). This data was combined instead of being analyzed separately in the two populations due to the unbalanced nature of the second case-control population. This was the most extensive candidate gene list tested for BSE including more number of animals than previous work.
Supplementary Material
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgements
Jennifer Thomson constructed the custom marker panel, performed analysis, and wrote the manuscript; Victoria Bowles and Yan Meng participated in sequencing library construction; Urmila Basu contributed to the library construction, sequencing and manuscript writing; Victoria Bowles, Jung-Woo Choi contributed to SNP identification and marker panel construction, Paul Stothard and Stephen Moore were involved in experimental design, and manuscript writing.
Glossary
Abbreviations:
- BSE
bovine spongiform encephalopathy
- PrP
prion protein
- TSE
transmissible spongiform encephalopathy
- vCJD
variant Creutzfeldt-Jakob disease
- SNP
single nucleotide polymorphism
- CNS
central nervous system
Supplemental Material
Supplemental materials may be found here:
http://www.landesbioscience.com/journals/prion/article/21866/
Footnotes
Previously published online: www.landesbioscience.com/journals/prion/article/21866
References
- 1.Prusiner SB. Novel proteinaceous infectious particles cause scrapie. Science. 1982;216:136–44. doi: 10.1126/science.6801762. [DOI] [PubMed] [Google Scholar]
- 2.Legname G, Baskakov IV, Nguyen HO, Riesner D, Cohen FE, DeArmond SJ, et al. Synthetic mammalian prions. Science. 2004;305:673–6. doi: 10.1126/science.1100195. [DOI] [PubMed] [Google Scholar]
- 3.Prusiner SB. Prions. Proc Natl Acad Sci U S A. 1998;95:13363–83. doi: 10.1073/pnas.95.23.13363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Novakofski J, Brewer MS, Mateus-Pinilla N, Killefer J, McCusker RH. Prion biology relevant to bovine spongiform encephalopathy. J Anim Sci. 2005;83:1455–76. doi: 10.2527/2005.8361455x. [DOI] [PubMed] [Google Scholar]
- 5.Balkema-Buschmann A, Fast C, Kaatz M, Eiden M, Ziegler U, McIntyre L, et al. Pathogenesis of classical and atypical BSE in cattle. Prev Vet Med. 2011;102:112–7. doi: 10.1016/j.prevetmed.2011.04.006. [DOI] [PubMed] [Google Scholar]
- 6.Wells GA, Scott AC, Johnson CT, Gunning RF, Hancock RD, Jeffrey M, et al. A novel progressive spongiform encephalopathy in cattle. Vet Rec. 1987;121:419–20. doi: 10.1136/vr.121.18.419. [DOI] [PubMed] [Google Scholar]
- 7.Donnelly CA, Ferguson NM, Ghani AC, Anderson RM. Implications of BSE infection screening data for the scale of the British BSE epidemic and current European infection levels. Proc Biol Sci. 2002;269:2179–90. doi: 10.1098/rspb.2002.2156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bruce ME, Will RG, Ironside JW, McConnell I, Drummond D, Suttie A, et al. Transmissions to mice indicate that ‘new variant’ CJD is caused by the BSE agent. Nature. 1997;389:498–501. doi: 10.1038/39057. [DOI] [PubMed] [Google Scholar]
- 9.Hill AF, Desbruslais M, Joiner S, Sidle KC, Gowland I, Collinge J, et al. The same prion strain causes vCJD and BSE. Nature. 1997;389:448–50, 526. doi: 10.1038/38925. [DOI] [PubMed] [Google Scholar]
- 10.Atarashi R, Moore RA, Sim VL, Hughson AG, Dorward DW, Onwubiko HA, et al. Ultrasensitive detection of scrapie prion protein using seeded conversion of recombinant prion protein. Nat Methods. 2007;4:645–50. doi: 10.1038/nmeth1066. [DOI] [PubMed] [Google Scholar]
- 11.Telling GC, Parchi P, DeArmond SJ, Cortelli P, Montagna P, Gabizon R, et al. Evidence for the conformation of the pathologic isoform of the prion protein enciphering and propagating prion diversity. Science. 1996;274:2079–82. doi: 10.1126/science.274.5295.2079. [DOI] [PubMed] [Google Scholar]
- 12.Riesner D. Biochemistry and structure of PrP(C) and PrP(Sc) Br Med Bull. 2003;66:21–33. doi: 10.1093/bmb/66.1.21. [DOI] [PubMed] [Google Scholar]
- 13.Hernández-Sánchez J, Waddington D, Wiener P, Haley CS, Williams JL. Genome-wide search for markers associated with bovine spongiform encephalopathy. Mamm Genome. 2002;13:164–8. doi: 10.1007/BF02684022. [DOI] [PubMed] [Google Scholar]
- 14.Zhang C, De Koning DJ, Hernández-Sánchez J, Haley CS, Williams JL, Wiener P. Mapping of multiple quantitative trait loci affecting bovine spongiform encephalopathy. Genetics. 2004;167:1863–72. doi: 10.1534/genetics.104.026401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mead S, Poulter M, Uphill J, Beck J, Whitfield J, Webb TE, et al. Genetic risk factors for variant Creutzfeldt-Jakob disease: a genome-wide association study. Lancet Neurol. 2009;8:57–66. doi: 10.1016/S1474-4422(08)70265-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Laegreid WW, Clawson ML, Heaton MP, Green BT, O’Rourke KI, Knowles DP. Scrapie resistance in ARQ sheep. J Virol. 2008;82:10318–20. doi: 10.1128/JVI.00710-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Alvarez L, Arranz JJ, San Primitivo F. Identification of a new leucine haplotype (ALQ) at codon 154 in the ovine prion protein gene in Spanish sheep. J Anim Sci. 2006;84:259–65. doi: 10.2527/2006.842259x. [DOI] [PubMed] [Google Scholar]
- 18.Murdoch BM, Clawson ML, Yue S, Basu U, McKay S, Settles M, et al. PRNP haplotype associated with classical BSE incidence in European Holstein cattle. PLoS One. 2010;5:10.1371.. doi: 10.1371/journal.pone.0012786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sander P, Hamann H, Drögemüller C, Kashkevich K, Schiebel K, Leeb T. Bovine prion protein gene (PRNP) promoter polymorphisms modulate PRNP expression and may be responsible for differences in bovine spongiform encephalopathy susceptibility. J Biol Chem. 2005;280:37408–14. doi: 10.1074/jbc.M506361200. [DOI] [PubMed] [Google Scholar]
- 20.Lloyd S, Mead S, Collinge J. Genetics of prion disease. Top Curr Chem. 2011;305:1–22. doi: 10.1007/128_2011_157. [DOI] [PubMed] [Google Scholar]
- 21.Murdoch BM, Clawson ML, Laegreid WW, Stothard P, Settles M, McKay S, et al. A 2cM genome-wide scan of European Holstein cattle affected by classical BSE. BMC Genet. 2010;11:20. doi: 10.1186/1471-2156-11-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Murdoch BM, Murdoch GK, Settles M, McKay S, Williams JL, Moore SS. Genome-wide scan identifies loci associated with classical BSE occurrence. PLoS One. 2011;6:e26819. doi: 10.1371/journal.pone.0026819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mead S, Uphill J, Beck J, Poulter M, Campbell T, Lowe J, et al. Genome-wide association study in multiple human prion diseases suggests genetic risk factors additional to PRNP. Hum Mol Genet. 2012;21:1897–906. doi: 10.1093/hmg/ddr607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu Y, Qin X, Song XZ, Jiang H, Shen Y, Durbin KJ, et al. Bos taurus genome assembly. BMC Genomics. 2009;10:180.1471-2164-10-180. doi: 10.1186/1471-2164-10-180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Guberman JM, Ai J, Arnaiz O, Baran J, Blake A, Baldock R, et al. BioMart Central Portal: an open database network for the biological community. Database (Oxford) 2011 [DOI] [PMC free article] [PubMed]
- 26.Benson DA, Karsch-Mizrachi I, Lipman DJ, Ostell J, Sayers EW. GenBank. Nucleic Acids Res. 2011;39(Database issue):D32–7. doi: 10.1093/nar/gkq1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Barrell D, Dimmer E, Huntley RP, Binns D, O’Donovan C, Apweiler R. The GOA database in 2009--an integrated Gene Ontology Annotation resource. Nucleic Acids Res. 2009;37(Database issue):D396–403. doi: 10.1093/nar/gkn803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ogata H, Goto S, Sato K, Fujibuchi W, Bono H, Kanehisa M. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res. 1999;27:29–34. doi: 10.1093/nar/27.1.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stothard P, Choi JW, Basu U, Sumner-Thomson JM, Meng Y, Liao X, et al. Whole genome resequencing of black Angus and Holstein cattle for SNP and CNV discovery. BMC Genomics. 2011;12:559. doi: 10.1186/1471-2164-12-559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–75. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kim W, Bennett EJ, Huttlin EL, Guo A, Li J, Possemato A, et al. Systematic and quantitative assessment of the ubiquitin-modified proteome. Mol Cell. 2011;44:325–40. doi: 10.1016/j.molcel.2011.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kamitani T, Kito K, Nguyen HP, Yeh ET. Characterization of NEDD8, a developmentally down-regulated ubiquitin-like protein. J Biol Chem. 1997;272:28557–62. doi: 10.1074/jbc.272.45.28557. [DOI] [PubMed] [Google Scholar]
- 33.Podust VN, Brownell JE, Gladysheva TB, Luo RS, Wang C, Coggins MB, et al. A Nedd8 conjugation pathway is essential for proteolytic targeting of p27Kip1 by ubiquitination. Proc Natl Acad Sci U S A. 2000;97:4579–84. doi: 10.1073/pnas.090465597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mori F, Nishie M, Piao YS, Kito K, Kamitani T, Takahashi H, et al. Accumulation of NEDD8 in neuronal and glial inclusions of neurodegenerative disorders. Neuropathol Appl Neurobiol. 2005;31:53–61. doi: 10.1111/j.1365-2990.2004.00603.x. [DOI] [PubMed] [Google Scholar]
- 35.Chen Y, Liu W, McPhie DL, Hassinger L, Neve RL. APP-BP1 mediates APP-induced apoptosis and DNA synthesis and is increased in Alzheimer’s disease brain. J Cell Biol. 2003;163:27–33. doi: 10.1083/jcb.200304003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Connor B, Dragunow M. The role of neuronal growth factors in neurodegenerative disorders of the human brain. Brain Res Brain Res Rev. 1998;27:1–39. doi: 10.1016/S0165-0173(98)00004-6. [DOI] [PubMed] [Google Scholar]
- 37.Grondin R, Gash DM. Glial cell line-derived neurotrophic factor (GDNF): a drug candidate for the treatment of Parkinson’s disease. J Neurol. 1998;245(Suppl 3):35–42. doi: 10.1007/PL00007744. [DOI] [PubMed] [Google Scholar]
- 38.Carter CJ. eIF2B and oligodendrocyte survival: where nature and nurture meet in bipolar disorder and schizophrenia? Schizophr Bull. 2007;33:1343–53. doi: 10.1093/schbul/sbm007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Deaton AM, Bird A. CpG islands and the regulation of transcription. Genes Dev. 2011;25:1010–22. doi: 10.1101/gad.2037511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.McCormick C, Leduc Y, Martindale D, Mattison K, Esford LE, Dyer AP, et al. The putative tumour suppressor EXT1 alters the expression of cell-surface heparan sulfate. Nat Genet. 1998;19:158–61. doi: 10.1038/514. [DOI] [PubMed] [Google Scholar]
- 41.Yin S, Pham N, Yu S, Li C, Wong P, Chang B, et al. Human prion proteins with pathogenic mutations share common conformational changes resulting in enhanced binding to glycosaminoglycans. Proc Natl Acad Sci U S A. 2007;104:7546–51. doi: 10.1073/pnas.0610827104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chomczynski P, Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem. 1987;162:156–9. doi: 10.1016/0003-2697(87)90021-2. [DOI] [PubMed] [Google Scholar]
- 43.Li H, Ruan J, Durbin R. Mapping short DNA sequencing reads and calling variants using mapping quality scores. Genome Res. 2008;18:1851–8. doi: 10.1101/gr.078212.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hubbard TJ, Aken BL, Ayling S, Ballester B, Beal K, Bragin E, et al. Ensembl 2009. Nucleic Acids Res. 2009;37(Database issue):D690–7. doi: 10.1093/nar/gkn828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Grant JR, Arantes AS, Liao X, Stothard P. In-depth annotation of SNPs arising from resequencing projects using NGS-SNP. Bioinformatics. 2011;27:2300–1. doi: 10.1093/bioinformatics/btr372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sherry ST, Ward M, Sirotkin K. dbSNP-database for single nucleotide polymorphisms and other classes of minor genetic variation. Genome Res. 1999;9:677–9. [PubMed] [Google Scholar]
- 47.Zimin AV, Delcher AL, Florea L, Kelley DR, Schatz MC, Puiu D, et al. A whole-genome assembly of the domestic cow, Bos taurus. Genome Biol. 2009;10:R42. doi: 10.1186/gb-2009-10-4-r42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhang Z, Schwartz S, Wagner L, Miller W. A greedy algorithm for aligning DNA sequences. J Comput Biol. 2000;7:203–14. doi: 10.1089/10665270050081478. [DOI] [PubMed] [Google Scholar]
- 49.Camacho C, Coulouris G, Avagyan V, Ma N, Papadopoulos J, Bealer K, et al. BLAST+: architecture and applications. BMC Bioinformatics. 2009;10:421. doi: 10.1186/1471-2105-10-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shieh JT, Swidler P, Martignetti JA, Ramirez MC, Balboni I, Kaplan J, et al. Systemic hyalinosis: a distinctive early childhood-onset disorder characterized by mutations in the anthrax toxin receptor 2 gene (ANTRX2) Pediatrics. 2006;118:e1485–92. doi: 10.1542/peds.2006-0824. [DOI] [PubMed] [Google Scholar]
- 51.Potkin SG, Turner JA, Guffanti G, Lakatos A, Fallon JH, Nguyen DD, et al. FBIRN A genome-wide association study of schizophrenia using brain activation as a quantitative phenotype. Schizophr Bull. 2009;35:96–108. doi: 10.1093/schbul/sbn155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Joo Y, Ha S, Hong BH, Kim J, Chang KA, Liew H, et al. Amyloid precursor protein binding protein-1 modulates cell cycle progression in fetal neural stem cells. PLoS One. 2010;5:e14203. doi: 10.1371/journal.pone.0014203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ravikumar B, Rubinsztein DC. Role of autophagy in the clearance of mutant huntingtin: a step towards therapy? Mol Aspects Med. 2006;27:520–7. doi: 10.1016/j.mam.2006.08.008. [DOI] [PubMed] [Google Scholar]
- 54.Rubinsztein DC. The roles of intracellular protein-degradation pathways in neurodegeneration. Nature. 2006;443:780–6. doi: 10.1038/nature05291. [DOI] [PubMed] [Google Scholar]
- 55.Lee JA, Liu L, Javier R, Kreitzer AC, Delaloy C, Gao FB. ESCRT-III subunits Snf7-1 and Snf7-2 differentially regulate transmembrane cargos in hESC-derived human neurons. Mol Brain. 2011;4:37. doi: 10.1186/1756-6606-4-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zenker M, Mayerle J, Lerch MM, Tagariello A, Zerres K, Durie PR, et al. Deficiency of UBR1, a ubiquitin ligase of the N-end rule pathway, causes pancreatic dysfunction, malformations and mental retardation (Johanson-Blizzard syndrome) Nat Genet. 2005;37:1345–50. doi: 10.1038/ng1681. [DOI] [PubMed] [Google Scholar]
- 57.Wellcome Trust Case Control Consortium Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 2007;447:661–78. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Airavaara M, Pletnikova O, Doyle ME, Zhang YE, Troncoso JC, Liu QR. Identification of novel GDNF isoforms and cis-antisense GDNFOS gene and their regulation in human middle temporal gyrus of Alzheimer disease. J Biol Chem. 2011;286:45093–102. doi: 10.1074/jbc.M111.310250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Larance M, Kirkwood KJ, Xirodimas DP, Lundberg E, Uhlen M, Lamond AI. Characterization of MRFAP1 turnover and interactions downstream of the NEDD8 pathway. Mol Cell Proteomics. 2012;11:M111–, 014407. doi: 10.1074/mcp.M111.014407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Li H, Wetten S, Li L, St Jean PL, Upmanyu R, Surh L, et al. Candidate single-nucleotide polymorphisms from a genomewide association study of Alzheimer disease. Arch Neurol. 2008;65:45–53. doi: 10.1001/archneurol.2007.3. [DOI] [PubMed] [Google Scholar]
- 61.Hafler DA, Compston A, Sawcer S, Lander ES, Daly MJ, De Jager PL, et al. International Multiple Sclerosis Genetics Consortium Risk alleles for multiple sclerosis identified by a genomewide study. N Engl J Med. 2007;357:851–62. doi: 10.1056/NEJMoa073493. [DOI] [PubMed] [Google Scholar]
- 62.Fung HC, Scholz S, Matarin M, Simón-Sánchez J, Hernandez D, Britton A, et al. Genome-wide genotyping in Parkinson’s disease and neurologically normal controls: first stage analysis and public release of data. Lancet Neurol. 2006;5:911–6. doi: 10.1016/S1474-4422(06)70578-6. [DOI] [PubMed] [Google Scholar]
- 63.Grupe A, Li Y, Rowland C, Nowotny P, Hinrichs AL, Smemo S, et al. A scan of chromosome 10 identifies a novel locus showing strong association with late-onset Alzheimer disease. Am J Hum Genet. 2006;78:78–88. doi: 10.1086/498851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wang Q, Sharma D, Ren Y, Fondell JD. A coregulatory role for the TRAP-mediator complex in androgen receptor-mediated gene expression. J Biol Chem. 2002;277:42852–8. doi: 10.1074/jbc.M206061200. [DOI] [PubMed] [Google Scholar]
- 65.Rienzo M, Nagel J, Casamassimi A, Giovane A, Dietzel S, Napoli C. Mediator subunits: gene expression pattern, a novel transcript identification and nuclear localization in human endothelial progenitor cells. Biochim Biophys Acta. 2010;1799:487–95. doi: 10.1016/j.bbagrm.2010.05.001. [DOI] [PubMed] [Google Scholar]
- 66.Kim BT, Kitagawa H, Tamura J, Saito T, Kusche-Gullberg M, Lindahl U, et al. Human tumor suppressor EXT gene family members EXTL1 and EXTL3 encode alpha 1,4- N-acetylglucosaminyltransferases that likely are involved in heparan sulfate/ heparin biosynthesis. Proc Natl Acad Sci U S A. 2001;98:7176–81. doi: 10.1073/pnas.131188498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Nadanaka S, Kitagawa H. Heparan sulphate biosynthesis and disease. J Biochem. 2008;144:7–14. doi: 10.1093/jb/mvn040. [DOI] [PubMed] [Google Scholar]
- 68.Warner RG, Hundt C, Weiss S, Turnbull JE. Identification of the heparan sulfate binding sites in the cellular prion protein. J Biol Chem. 2002;277:18421–30. doi: 10.1074/jbc.M110406200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.