Abstract
Tissue morphogenesis is the process in which coordinated movements and shape changes of large numbers of cells form tissues, organs, and the internal body structure. Understanding morphogenetic movements requires precise measurements of whole-cell shape changes over time. Tissue folding and invagination are thought to be facilitated by apical constriction, but the mechanism by which changes near the apical cell surface affect changes along the entire apical–basal axis of the cell remains elusive. Here, we developed Embryo Development Geometry Explorer, an approach for quantifying rapid whole-cell shape changes over time, and we combined it with deep-tissue time-lapse imaging based on fast two-photon microscopy to study Drosophila ventral furrow formation. We found that both the cell lengthening along the apical–basal axis and the movement of the nucleus to the basal side proceeded stepwise and were correlated with apical constriction. Moreover, cell volume lost apically due to constriction largely balanced the volume gained basally by cell lengthening. The volume above the nucleus was conserved during its basal movement. Both apical volume loss and cell lengthening were absent in mutants showing deficits in the contractile cytoskeleton underlying apical constriction. We conclude that a single mechanical mechanism involving volume conservation and apical constriction-induced basal movement of cytoplasm accounts quantitatively for the cell shape changes and the nucleus movement in Drosophila ventral furrow formation. Our study provides a comprehensive quantitative analysis of the fast dynamics of whole-cell shape changes during tissue folding and points to a simplified model for Drosophila gastrulation.
Keywords: two-photon imaging, 4D reconstruction, segmentation
During development, sheets of cells undergo dramatic rearrangements to generate the complex 3D structures of the body and internal organs (1–3). This process, called tissue morphogenesis, results from coordinated cell shape changes and movements that collectively deform tissues (4–7). Although much is known about the role of cell patterning and signaling in tissue morphogenesis, our understanding of how tissues acquire their shapes is relatively immature. Shedding light on this central aspect of tissue morphogenesis requires detailed characterizations of the dynamics of shape changes on subcellular, cellular, and tissue levels. However, available methods for performing the necessary morphological analyses are still relatively rudimentary (8, 9).
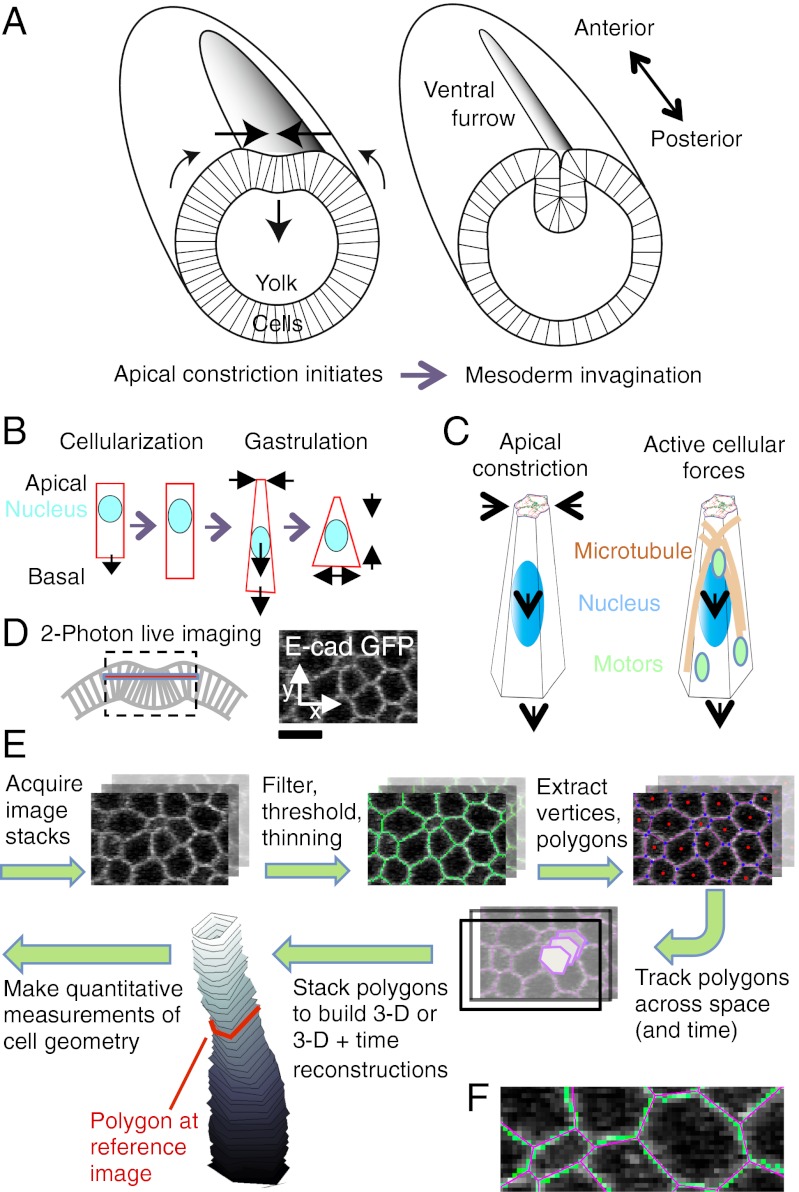
An important example of tissue morphogenesis is tissue folding and invagination. Gastrulation, the process in which the basic body plan is laid out, begins in Drosophila with the formation of the ventral furrow and subsequent invagination of the mesoderm primordium (Fig. 1A), a section of tissue that eventually gives rise to muscles and fat bodies (1). Before gastrulation, the Drosophila embryo undergoes cellularization, during which time the syncytial blastoderm is partitioned into individual cells. During cellularization, cell membranes are formed progressing from the apical to the basal end, resulting in cells of stereotypical columnar shape (Fig. 1B, Left). When gastrulation starts, individual cells constrict at their apical end and undergo elongation. Cells then shorten and widen their basal ends, transforming shapes from columnar to wedge-like (6, 10–12) (Fig. 1B, Right). Cell nuclei are initially close to the apical surface but move basally during the early phase of furrow formation (6, 13). These changes concerted across a large number of cells are thought to underlie ventral furrow formation in Drosophila (1). However, by what mechanism they are controlled and temporally coordinated with one another remained unclear from these studies, which were mostly based on fixed data.
Fig. 1.
Measuring whole-cell shape changes during ventral furrow formation in Drosophila. (A) Schematic cross-sections through the Drosophila embryo at the beginning of gastrulation (Left) and a few minutes later (Right), illustrating ventral furrow formation and invagination of mesodermal precursor cells. Modified from ref. 36. (B) Shape changes of ventral furrow cells (schematic). (C) Two alternative mechanisms that could underlie cell lengthening and basal movement of the nucleus. (D) Schematic of the imaging approach (Left) and visualized cell membranes in a representative z-slice at 15-μm depth (Right). (Scale bar: 10 μm.) (E) Pipeline for producing cell shape measurements based on EDGE. First, cell outlines are segmented in all image layers (green membranes), and each cell outline is reduced to a centroid with a set of vertices [i.e., a polygon (shown with pink edges, blue dots at the vertices, and red dots at the cell centroids)]. Second, polygons are tracked in time and space, starting from a predefined reference image (shown in red on the reconstructed cell), and a 3D reconstruction is built for each cell at each time (grayscale rendering; distance between adjacent polygons: Δz = 1 μm). (F) A representative image patch at high zoom illustrating the accuracy of the polygon approximation (Figs. S1 and S6).
Characterizing the detailed dynamics of cell shape changes helps to elucidate the mechanism governing these changes. In a recent study (14), we observed that apical constriction in ventral furrow cells in Drosophila is highly dynamic. By combining live imaging, image analysis, and genetic methods, we identified a pulsatile actin–myosin network, which reduces apical area in steps, suggestive of a ratchet-like mechanism.
It is conceivable that initial changes in cell length and nuclear position might be secondary consequences of apical constriction and that the associated contractile machinery might, therefore, constitute the major driving force of ventral furrow formation (3, 15) (Fig. 1C, Left). However, alternative scenarios seem plausible, in which separate, albeit sufficiently concerted, active cellular processes drive the sequence of apical and basal cell shape changes (16–19) and the relocation of the nucleus (20, 21) (Fig. 1C, Right). For instance, fibroblast elongation is believed to involve ends of growing microtubules promoting actin polymerization (19).
To shed light on the mechanisms underlying cell elongation and nuclear movement, a precise characterization of the fast spatiotemporal dynamics of whole-cell shape changes in a large number of cells is necessary. Deep-tissue live-imaging techniques provide the means for visualizing outlines of entire cells with a temporal resolution of a few seconds (8). However, most previously proposed methods for studying cell shape changes are restricted to 2D cross-sections of cells (22), whereas those methods enabling whole-cell reconstructions are limited to relatively simple and static shapes (23, 24) or rely heavily on manual editing (25). Here, we therefore developed a cell shape analysis tool, called Embryo Development Geometry Explorer (EDGE), which is optimized for analyzing the fast dynamics of whole-cell shape changes in three spatial dimensions plus time in planar sheets of cells. Using EDGE, we perform a detailed characterization of the dynamics of cell shape changes during ventral furrow formation, suggesting that apical constriction drives both cell lengthening and nucleus movement by volume conservation of the whole cell and above the nucleus.
Results
Measuring Cell Shapes with EDGE.
To visualize whole ventral furrow cells in live embryos, we used two-photon imaging, which is suitable for deep-tissue imaging (Materials and Methods and Fig. 1D). Cell membranes were visualized using a GFP-tagged transmembrane protein (E-Cadherin-GFP). Image stacks covered a section of tissue of about 90 μm in the ventral–dorsal directions and 45 μm in the anterior–posterior directions approximately centered on the ventral midline. The temporal resolution (12–18 s) was chosen to be sufficiently high to capture the dynamics of apical constriction observed in ventral furrow cells (14) (Materials and Methods).
We used EDGE to measure cell shapes from the two-photon image stacks (Materials and Methods and Fig. 1 E and F) (a detailed description of EDGE is provided in SI Text and Figs. S1–S4, and the software is open source and freely available at http://www.code.google.com/p/embryo-development-geometry-explorer/). EDGE identifies cell outlines in individual 2D images based on a sequence of image-processing steps, including band-pass filtering, thresholding, and morphological thinning. Vertices and centroids of individual 2D cell outlines are extracted, and cell outlines are then reduced to the polygons defined by the vertices. EDGE then tracks these polygons across z-stacks and time to form 4D (i.e., three space dimensions and one time dimension) representations of cells. The starting point of tracking is a reference image at some chosen depth and point in time (Fig. 1E). Tracking was checked by visual inspection, and sporadic tracking failures were hand-curated using EDGE’s built-in interface for automated and manual error correction (SI Text and Fig. S1). The accuracy of segmentation and tracking is assessed in Fig. S5.
To use the capabilities of EDGE in understanding whole-cell shape changes, we first automatically estimated the apical and basal limits of each cell at each point in time based on the labeling intensity difference between the cell’s membrane and interior region (Materials and Methods, SI Text, and Fig. S7). By visual inspection of six cells at 11 time points in one embryo, our automated method estimated the cell limits with an error of 1.2 μm (Fig. S7 D and E). Polygons tracked outside the identified cell limits were discarded, and parts within the limits that remained untracked were automatically repaired through extra- and interpolation (Materials and Methods and Fig. S8). We defined a curved coordinate in each cell, called the cell axis, that passes near the center of the cell’s cross-section at each depth from the center of the apical surface down to the center of the basal surface (SI Text, and Figs. S7 and S8). Cell length was defined as the length of this curve. Cell volume was calculated by multiplying the z-resolution of images, Δz (Fig. 1E), by the sum of all polygon areas, adding volumes of repaired cell ends where applicable. Apical area was measured as the cross-sectional area along a plane perpendicular to the cell axis 1 μm below the apical end. To improve tracking of cells moving in depth, segmentations were carried out independently for up to three reference images at different times and then matched based on overlap (SI Text). Finally, strong outliers in measurements were discarded (Fig. S9). Measurement errors were estimated from the fluctuations of measurements at subsequent time points and were typically in the range of 5–10% (e.g., 1.5 μm for cell length, 97 μm3 for volume) (Fig. S9). This estimate may be interpreted as an upper bound of the true (random) error given that quantities also change systematically over time.
Apical Constriction, Cell Length, and Cell Volume.
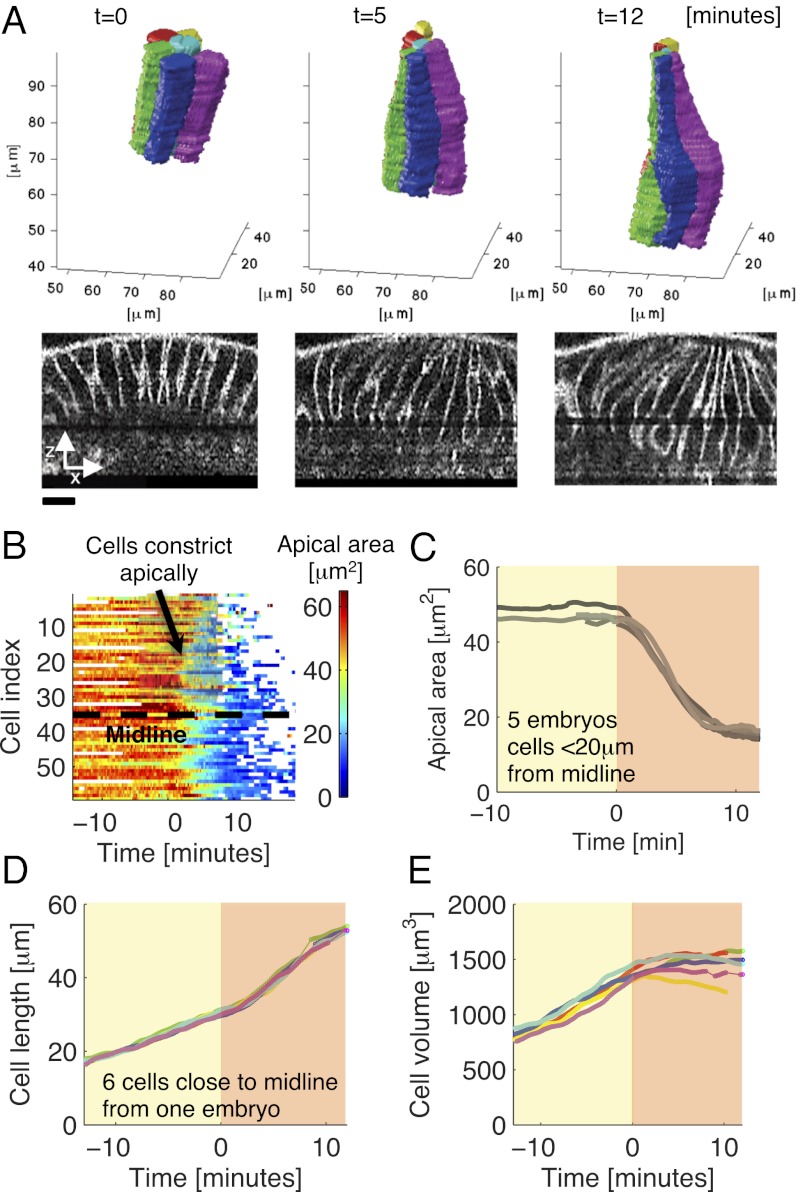
Fig. 2A shows six fully reconstructed cells tracked from the stage when gastrulation starts (Fig. 2A, Left) to the phase of near maximal elongation (Fig. 2A, Right). Most cells close to the midline started to constrict apically at about the same time (26) (Fig. 2B). For each embryo, we defined as t = 0 the time point at which the average apical area of cells within 20 μm from the midline started decreasing (Fig. 2C, SI Text, and Fig. S10). In this study, we call this time point the beginning of gastrulation. Shortly after t = 0, cells showed marked shape changes, initiating apically and proceeding more basally over time (Fig. 2A). Fig. 2D shows the lengths of the six cells from Fig. 2A. During cellularization, cells elongated with a near constant rate over time. At the beginning of gastrulation, rates increased considerably, manifested by an inflection point in cell length at t = 0 ± 1 min [mean ± SD; estimated by fitting a linear-constant-linear function to cell length (Fig. S11) in n = 147 cells; five embryos]. In contrast, cell volume, which also steadily increased during cellularization, slowed down in rate after t = 0 and stayed relatively constant after t = 5 min, despite the high elongation rate during this period (Fig. 2E). The maximal volume change in individual cells between t = 5 min and t = 12 min was 71 μm3 (5.3% of total volume; SD over same period = 1.7%; average over n = 147 cells; five embryos). This finding shows that the volume of individual cells is largely conserved during early ventral furrow formation. An important role of volume conservation in tissue morphogenesis has been emphasized in several model studies (27).
Fig. 2.
Whole-cell shape changes and measurements during ventral furrow formation. (A) Six reconstructed cells followed over time (Upper) with xz slices of the raw data (contrast enhanced; Lower). (Scale bar: 10 μm.) (B) Apical areas of all cells reconstructed in this embryo. (C) Mean apical cross-sectional area of cells close to the midline (distance < 20 μm) in five embryos. The onset of decrease in average apical area in each embryo was used to define the beginning of gastrulation, t = 0 (SI Text and Fig. S10), and to align different embryos in time. (D and E) Length (D) and volume (E) for the six cells from A. In C–E, shaded yellow and red backgrounds mark the periods before and after the beginning of gastrulation, respectively. Data in B–E are smoothed using a Gaussian kernel with σ = 30 s.
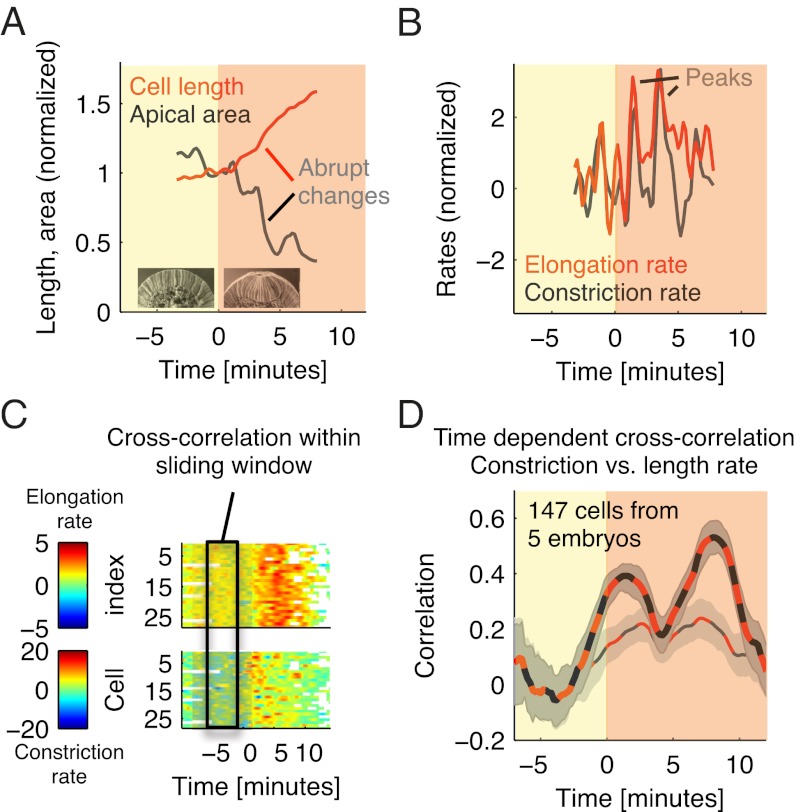
The near coincidence between the onset of apical constriction and a pronounced lengthening (Fig. 2 C and D) suggests a relation between these two processes. We observed that, often, considerable steps in lengthening (2–4 μm) occurred over relatively short periods of time (1–2 min) (Fig. 3A). Moreover, these steps tended to coincide with steps in apical area decrease, albeit showing slightly smoother dynamics (Fig. 3A). In the case of apical area, this step-like behavior was observed previously and interpreted as the result of the ratchet-like contractile dynamics of the actomyosin meshwork driving apical constriction in ventral furrow cells (14). As in ref. 14, we defined the constriction rate as the negative rate of change of apical surface area and compared it with the elongation rate (i.e., the rate of change of cell length) (Materials and Methods). With rates, steps translate into peaks, enabling a more precise comparison. We found that peaks in elongation rate, indeed, often coincided with peaks in constriction rate (Fig. 3B). To test for statistical significance, we analyzed the cross-correlation between the two rates at various points in time (Materials and Methods and Fig. 3C). The correlation was close to zero during cellularization but increased considerably at the beginning of gastrulation (thick red and black line in Fig. 3D). It exhibited two peaks: the first peak seems to reflect a steady increase in constriction and elongation rate at about t = 0, whereas the second peak shows a steady decrease at t = 8 min when both processes started saturating (compare Fig. 2 B–D). After high-pass filtering the constriction and elongation rates, the correlation was relatively small (thin red and black line in Fig. 3D), consistent with the observation that lengthening typically exhibits relatively smooth dynamics compared with apical constriction. However, the correlation and its confidence interval were still consistently larger than zero for 0 < t < 10 min, indicating that lengthening is statistically related even to the very rapid changes in apical area that are associated with the pulsed contractile actomyosin dynamics underlying apical constriction (14).
Fig. 3.
Phases of pronounced cell lengthening are correlated with pulses of apical constriction. (A and B) Cell length and apical surface area (A) and elongation and constriction rates (B) for a representative cell. To facilitate comparison, quantities were normalized by the length and apical area at t = 0, respectively. (C) To estimate the statistical dependency between the two rates in individual cells, we computed their (zero-lag) cross-correlation in a temporally moving window of length of 6 min. (D) The thick red/black dashed line marks the mean correlation, and the dark gray shade shows the (2, 98) percentile confidence interval obtained by bootstrap resampling based on 147 cells near the midline from five embryos (Materials and Methods). Also shown is the correlation after high-pass filtering the data using a Gaussian filter with σ = 2 min (thin red/black dashed line with bright gray shaded confidence intervals).
Constriction-Induced Basal Movement of Cytoplasm Seems to Drive Cell Lengthening by a Volume Conservation Mechanism.
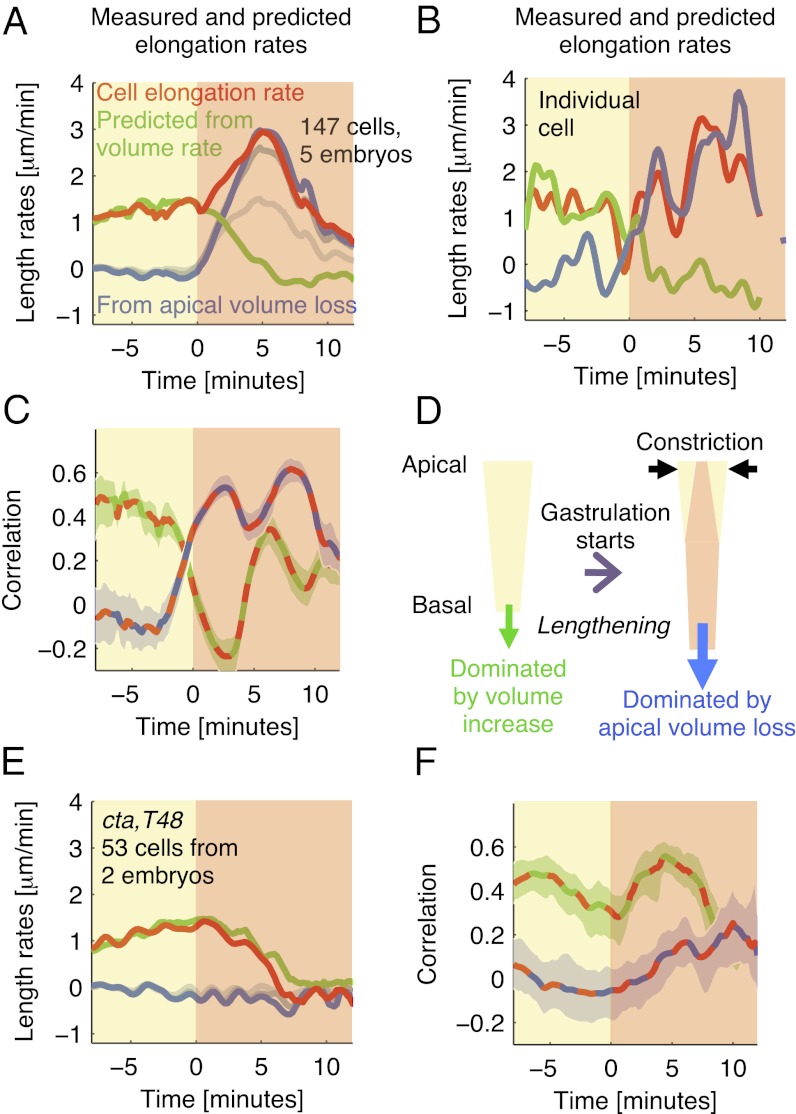
We next sought to shed light on the mechanism underlying cell lengthening. During cellularization, lengthening is caused by cell growth and associated with an increase in volume (Fig. 2 D and E). However, during gastrulation, cell volume changed much less (Fig. 2E), suggesting that lengthening was caused by a different mechanism. We estimated the rate of lengthening that was caused by growth by dividing the volume rate (i.e., the derivative of the volume with respect to time) by the basal surface area in each cell. During cellularization, this predicted elongation rate, on average, matched well with the real rate, but the two began to deviate considerably after gastrulation started (Fig. 4A). After t = 5 min, the predicted rate decayed close to zero, consistent with the small volume change during this period in individual cells (Fig. 2E). However, volume conservation suggests that the rate of lengthening could match the rate predicted from constriction-induced apical volume loss. Fig. 4A shows the average negative rate of change of apical volume divided by the basal surface area, where apical volume was defined as the apical 75% (blue curve), 50% (dark gray curve), or 25% (light gray curve) of the cell. During the first few minutes of gastrulation, constriction affected only the most apical cell part (Fig. S12), and thus, the three curves were nearly identical. However, at later times, more basal parts of the cell constricted (Fig. S12). We, therefore, computed the rate of apical volume loss based on the cell’s apical 75%. As expected, this rate, on average, was close to zero during cellularization, but it increased considerably after t = 0 and became well-matched to the real elongation rate at about the same time as when total cell volume became largely preserved (Fig. 4A). Consistent with Fig. 2E, elongation was still partly affected by cell growth over the first few minutes of gastrulation.
Fig. 4.
Constriction-induced apical volume loss predicts rate of cell lengthening. (A) Average measured cell elongation rate (red) compared to rates predicted from total volume change (green) and from constriction-induced apical volume loss [blue; gray curves, alternative definitions (see text)]. (B) The three rates for an individual representative cell. (C) Time dependent cross-correlations (described in Fig. 3C) between the cell elongation rate and the two predicted rates (red/green dashed line, prediction from total volume change; red/blue dashed line, prediction from apical volume loss). Analysis in A and C is based on same cells as in Fig. 3D. (D) Model: dominant factors in determining cell lengthening during cellularization and gastrulation. (E and F) Data for cta; T48 mutants presented as in A and C, respectively.
In individual cells, often even individual peaks in elongation rate coincided and matched in size with peaks predicted from apical volume loss, but not total volume change, during gastrulation (a representative example is shown in Fig. 4B). By computing the time-dependent cross-correlation described in Fig. 3C, we found that, whereas during cellularization, changes in cell elongation were correlated with those changes in total volume only, after gastrulation started, they were more strongly correlated with changes in apical volume loss (Fig. 4C). Our observations suggest a model in which apical constriction causes cytoplasm to move basally, which along with volume conservation, induces the pronounced increase in cell lengthening during the initial phase of gastrulation (Fig. 4D). A more thorough analysis shown in Fig. S13 suggests that the contribution of cell growth is slightly larger and that not all loss in apical volume is transformed into elongation during the early period of gastrulation (before t = 7 min). The latter seems to be a consequence of the yolk stalk closing only after this period (Fig. S13).
Because apical volume loss is associated with apical constriction, we next asked whether apical volume loss and signs of pronounced cell lengthening were absent in cta; T48 mutants (12, 28), in which the contractile actomyosin meshwork that normally drives apical constriction fails to stabilize (29). During cellularization, cells in cta; T48 mutants followed length (Fig. S10A) and rate (Fig. 4E) trajectories similar to those lengths and rates in WT cells, and we aligned both groups in time based on these measurements. Individual cells in cta; T48 mutants underwent fluctuations in apical surface area, but on average, the apical surface area changed very little (Fig. S10B) (14). Consistently, we found that cta; T48 mutant cells showed little apical volume loss and no phase of pronounced elongation (Fig. 4E). Therefore, volume rate continued dominating elongation into the period in which gastrulation would normally proceed (Fig. 4F) (the broad peak around t = 5 min again reflects the steady decrease in elongation rate over this period). These results show that stabilized actomyosin contractility leading to persistent apical constriction is necessary for the observed apical volume loss and cell lengthening.
Volume Conservation Principle Underlies Nucleus Movement.
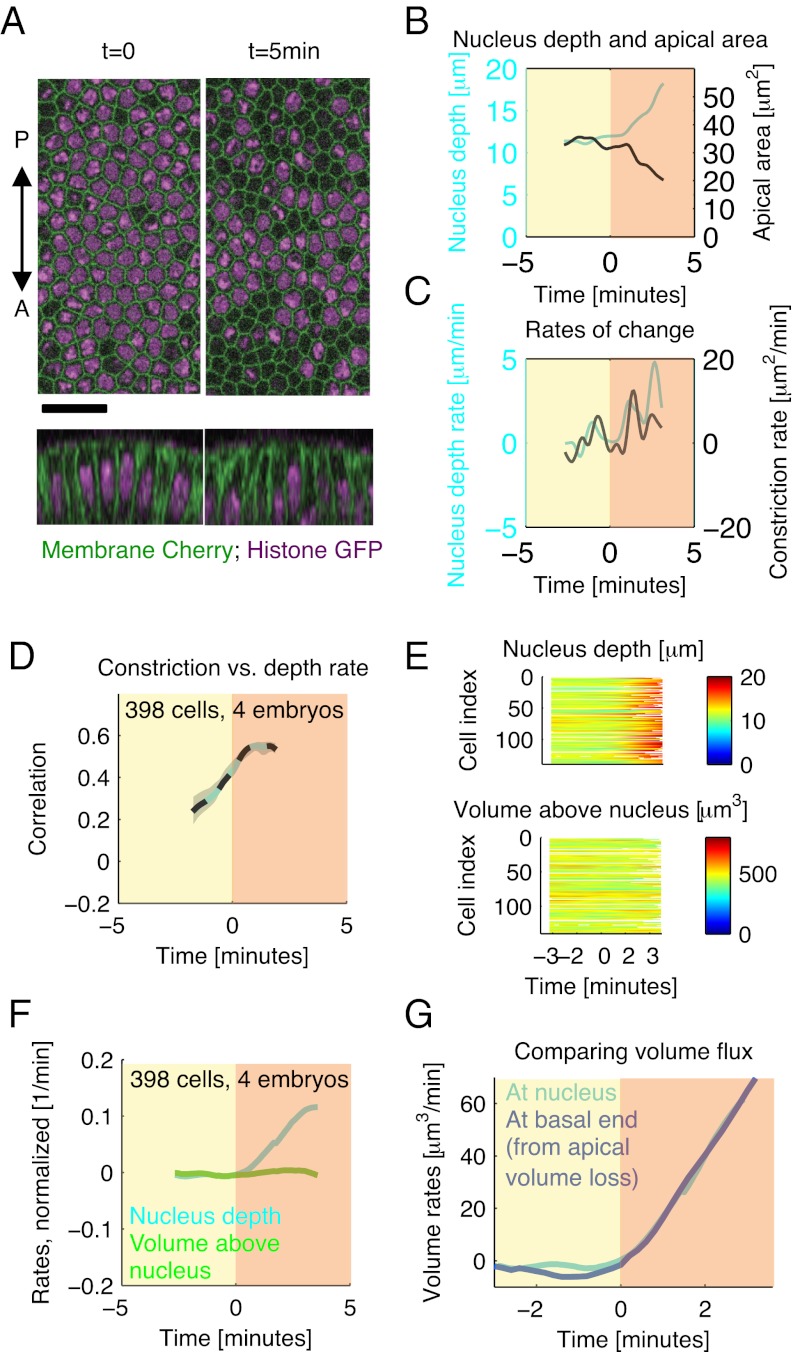
Next, we asked whether a mechanism involving volume conservation could also govern the basal movement of the nucleus during gastrulation. Such a mechanism could account for the fast and orchestrated movement of nuclei (6, 13) that seems necessary for the rapid folding of tissue during ventral furrow formation (Fig. 2A). To test this finding, we imaged embryos that were double-labeled with a membrane marker (membrane-Cherry) and a marker for nuclei (histone-GFP) using a confocal microscope (Materials and Methods and Fig. 5A). The onset of gastrulation, t = 0, was identified by the same method as in Fig. 2C.
Fig. 5.
Apical constriction predicts basal movement of nucleus while the volume above the nucleus is conserved. (A) Merged images of membrane-mCherry (green) and Histone-GFP labeling nuclei [purple; z-slices, 14 μm depth (Upper) and yz cross-sections (Lower)]. Nuclei started moving out of the image stacks at approximately t = 3.3 min. (Scale bar: 20 μm.) (B) Cross-sectional area below the apical surface and nucleus depth for a representative cell. (C) Constriction rate and nucleus drop rate (i.e., velocity) for this cell. (D) Time-dependent cross-correlation (described in Fig. 3C) between the two rates in sliding window of length of 2 min. (E) Nucleus depth and volume above the nucleus in cells from one representative embryo. Each row represents data (see colorbars) for an individual cell. (F) Their average rates (normalized in each cell by the nucleus depth and volume above nucleus at t = 0, respectively). (G) Test of volume conservation model: volume flux predicted from nucleus velocity (cyan) matches flux predicted from apical volume loss (blue; curves manually shifted in time to finely align t = 0 across datasets) (SI Text).
In individual cells, nucleus movement often proceeded in steps that coincided with periods of pronounced apical constriction (Fig. 5 B and C shows a representative example). Nucleus velocity and apical constriction became strongly correlated around the time that gastrulation started (Fig. 5D). In most cells, nuclei showed a significant basal movement of 6 μm within the first 3 min of gastrulation (corresponding to 50% of their original depth and 20% of the total cell length at this time) (Fig. 5E shows cells from a representative embryo). However, in most cells, the volume above the nucleus was largely conserved over this period (changes were typically less than 10%) (Fig. 5E, Lower). To better compare the two quantities, we calculated their rates of change and normalized them with their respective values at t = 0. The result shows that, after gastrulation started, the nucleus moved basally, whereas the volume above the nucleus remained largely unchanged (Fig. 5F). These observations suggest a simple model, in which the cytoplasm is an incompressible fluid and the nucleus acts like an impassable piston in a tube. When apical constriction occurs, the apical cytoplasm moves basally, and because it cannot pass the nucleus, it, instead, pushes the nucleus to the basal end of the cell.
Testing the Volume Conservation Principle.
This model and the model proposed for cell lengthening (Fig. 4D) make the simple prediction that the constriction-induced volume flux passing at the position of the nucleus should match the constriction-induced flux at the basal cell end. To test this prediction, we estimated the flux at the nucleus by multiplying the nucleus velocity by the cross-sectional area at the nucleus. The constriction-induced flux at the basal end of the cell is given by the rate of apical volume loss. Fig. 5G shows that the two fluxes, indeed, match very well over the period during which we can monitor nuclei positions. Note that the two curves are obtained for two different groups of embryos, in which cell membranes were labeled using different fluorescent markers and imaging was carried out with different types of microscopes. The good agreement between the two curves corroborates our hypothesis that volume conservation plays a key role in governing cell shape changes in ventral furrow formation.
Discussion
In this report, we introduce a method for quantitatively analyzing whole-cell shape changes in epithelia and combine it with fast two-photon microscopy and genetic approaches to study epithelial sheet bending and invagination in Drosophila gastrulation. We show that apical constriction is correlated with both cell lengthening and the basal movement of the nucleus. Moreover, we show that the cell volume lost apically because of constriction reappears basally in the form of lengthening and that the volume above the nucleus is conserved during its basal movement. We conclude that apical constriction-induced basal movement of cytoplasm and volume conservation are the key factors that control both cell lengthening and the nucleus movement. This mechanism differs from previously proposed mechanisms ascribing to microtubules a central role in controlling cell length in epithelial cells (18), fibroblasts (19), and myoblasts (17) as well as nucleus movement (20). Volume conservation seems to be an effective means for localized subcellular force-generating machineries to affect shape changes in much larger structures across entire cells and tissues. Assuming that the cytoplasm acts like a nearly incompressible fluid, this mechanism gives rise to a fast and nearly undiminished transmission of forces across large distances. It also provides cells with the possibility to expand and grow to certain directions, even if their active cytoskeletal components are purely contractive.
Materials and Methods
Fly Stocks and Genetics.
Fluorescent fusion protein stocks: Myosin–GFP (II or III; squ–GFP) (30), E-Cadherin–GFP (II; ubi-DE-cad–GFP) (31), membrane-mCherry (29), H2Av-GFP (32), and Spider-GFP (33) are described. Spider-GFP was recombined with cta/Cyo; T48 (28) to generate cta/Cyo; T48 Spider-GFP. Because Cta is maternally supplied, cta; T48 Spider-GFP flies were selected from the balanced stock to produce cta; T48 double mutant embryos.
Live Imaging.
Embryos used for live imaging were staged in Halocarbon 27 oil, dechorionated with 50% bleach, washed extensively with water, and mounted ventral side up on the glass surface of a glass-bottomed culture dish (P35G-1.5–10-C; MatTek Corporation). The glass surface was covered with embryo glue (Scotch tape resuspended in heptane) to immobilize the embryo during imaging. Water was added to the chamber of the culture dish that covered the embryos. All imaging was performed in water at room temperature (∼23 °C).
Live imaging is performed with a custom built two-photon scanning microscope (34) built around an upright Olympus BX51. Fluorescence photons are collected through both an N.A. 0.8 Olympus water immersion objective ×40 LUMPlanFl/IR and an N.A. 1.3 oil condenser lens and detected with high quantum efficiency hand-peaked GaAsP photomultipliers (Hamamatsu). The microscope is operated by the Matlab software ScanImage (35) modified to control a piezo objective (PI) and increase the laser power as we image deeper into the embryo. Images were taken with an excitation wavelength of 920 nm. Stacks of 60 images taken at 1-μm steps were recorded every 12 s (after cells started invaginating, 100 images were recorded every 18 s). The images are 256 × 128 pixels corresponding to 90 (ventral–dorsal) × 45 (anterior–posterior) -μm regions. The signal sampling time per pixel was 3.2 μs.
Preparation of embryos for two-channel live-cell imaging was performed as described previously in ref. 14. Membrane-mCherry (29) was crossed to Histone-GFP (32) to obtain flies with one copy of each marker. We imaged embryos from these females. Videos were obtained with a Leica SP5 confocal microscope, a 63×/1.3 N.A. glycerine immersion objective, an argon ion laser, and a 561-nm diode laser. Images were acquired using a pinhole setting from 1 to 2 Airy Units and the excitation band pass to 495–550 nm to detect GFP and 578–650 nm to detect mCherry. Stacks of 15 images taken at 2-μm steps were recorded every 5 s. The images are 190 × 512 pixels corresponding to 54 (ventral–dorsal) × 145 (anterior–posterior) -μm regions.
Data Processing.
Each image was clipped at ±2 SD, band pass-filtered [low pass, 2.5 (2.0) μm; high pass, 7.0 (10.0) μm; parameters are given for the two-photon data followed by the confocal data in parentheses if different], and thresholded at −0.4 (0.0) SD. Identified polygons with area <4.0 (2.0) μm2 were removed. Polygons in adjacent images were tracked if their centroids differed by <10 μm and their fractional overlap was >50% [50% (depth); 30% (time)], and each polygon contained the centroid of the other cell. If tracking failed, it was attempted sequentially for up to four subsequent images. After tracking was completed, missing polygons were added by linear interpolation in depth.
The apical (basal) cell limit was estimated from the positive (negative) peak of the derivative with respect to z of the difference in average signal intensity between the polygon’s border and central regions (Fig. S7). Polygons tracked beyond the cell limits were removed. Untracked regions at the apical or basal end within the cell limits were repaired by 3D interpolation between all pixels with labeling intensity above 0.5/0.7 (apical/basal) times the maximum intensity in that region (Fig. S8).
Measurements were interpolated to a 12-s (5-s) sample rate. All rates were estimated from the difference at subsequent time points after low pass-filtering measurements with a centered Gaussian of width σ = 30 s to improve robustness. Cross-correlations at time t were calculated on an individual cell basis in a sliding window of length of 6 min (2.5 min) centered at time t. One hundred bootstrap samples were drawn from the set of all considered cells. From this sample, the mean and (2, 98) percentile confidence interval were estimated at each time t.
Supplementary Material
Acknowledgments
This work was supported by National Institutes of Health Grant P50 GM 071508 (to M.A.G., S.Y.T., and M.K.; principle investigator: D. Botstein), and National Institute of Child Health and Human Development Grant 5R37HD15587 to E.F.W. B.H. was supported by the New Jersey Commission on Cancer Research Fellowship and Association for Computing Machinery Award R00GM089826 from the National Institute of General Medical Sciences. E.F.W. is an investigator of The Howard Hughes Medical Institute.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1205258109/-/DCSupplemental.
References
- 1.Leptin M. Gastrulation movements: The logic and the nuts and bolts. Dev Cell. 2005;8(3):305–320. doi: 10.1016/j.devcel.2005.02.007. [DOI] [PubMed] [Google Scholar]
- 2.Lecuit T, Lenne PF. Cell surface mechanics and the control of cell shape, tissue patterns and morphogenesis. Nat Rev Mol Cell Biol. 2007;8(8):633–644. doi: 10.1038/nrm2222. [DOI] [PubMed] [Google Scholar]
- 3.Quintin S, Gally C, Labouesse M. Epithelial morphogenesis in embryos: Asymmetries, motors and brakes. Trends Genet. 2008;24(5):221–230. doi: 10.1016/j.tig.2008.02.005. [DOI] [PubMed] [Google Scholar]
- 4.Holtfreter J. A study of the mechanics of gastrulation. Part 1. J Exp Zool. 1943;94(3):261–318. [Google Scholar]
- 5.Gustafson T, Wolpert L. Cellular mechanisms in the morphogenesis of the sea urchin larva. Change in shape of cell sheets. Exp Cell Res. 1962;27:260–279. doi: 10.1016/0014-4827(62)90229-x. [DOI] [PubMed] [Google Scholar]
- 6.Sweeton D, Parks S, Costa M, Wieschaus E. Gastrulation in Drosophila: The formation of the ventral furrow and posterior midgut invaginations. Development. 1991;112(3):775–789. doi: 10.1242/dev.112.3.775. [DOI] [PubMed] [Google Scholar]
- 7.Shih J, Keller R. Cell motility driving mediolateral intercalation in explants of Xenopus laevis. Development. 1992;116(4):901–914. doi: 10.1242/dev.116.4.901. [DOI] [PubMed] [Google Scholar]
- 8.Megason SG, Fraser SE. Imaging in systems biology. Cell. 2007;130(5):784–795. doi: 10.1016/j.cell.2007.08.031. [DOI] [PubMed] [Google Scholar]
- 9.Khairy K, Keller PJ. Reconstructing embryonic development. Genesis. 2011;49(7):488–513. doi: 10.1002/dvg.20698. [DOI] [PubMed] [Google Scholar]
- 10.Turner FR, Mahowald AP. Scanning electron microscopy of Drosophila melanogaster embryogenesis. II. Gastrulation and segmentation. Dev Biol. 1977;57(2):403–416. doi: 10.1016/0012-1606(77)90225-1. [DOI] [PubMed] [Google Scholar]
- 11.Leptin M, Grunewald B. Cell shape changes during gastrulation in Drosophila. Development. 1990;110(1):73–84. doi: 10.1242/dev.110.1.73. [DOI] [PubMed] [Google Scholar]
- 12.Parks S, Wieschaus E. The Drosophila gastrulation gene concertina encodes a G alpha-like protein. Cell. 1991;64(2):447–458. doi: 10.1016/0092-8674(91)90652-f. [DOI] [PubMed] [Google Scholar]
- 13.Kam Z, Minden JS, Agard DA, Sedat JW, Leptin M. Drosophila gastrulation: Analysis of cell shape changes in living embryos by three-dimensional fluorescence microscopy. Development. 1991;112(2):365–370. doi: 10.1242/dev.112.2.365. [DOI] [PubMed] [Google Scholar]
- 14.Martin AC, Kaschube M, Wieschaus EF. Pulsed contractions of an actin-myosin network drive apical constriction. Nature. 2009;457(7228):495–499. doi: 10.1038/nature07522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hardin J, Keller R. The behaviour and function of bottle cells during gastrulation of Xenopus laevis. Development. 1988;103(1):211–230. doi: 10.1242/dev.103.1.211. [DOI] [PubMed] [Google Scholar]
- 16.Jankovics F, Brunner D. Transiently reorganized microtubules are essential for zippering during dorsal closure in Drosophila melanogaster. Dev Cell. 2006;11(3):375–385. doi: 10.1016/j.devcel.2006.07.014. [DOI] [PubMed] [Google Scholar]
- 17.Straube A, Merdes A. EB3 regulates microtubule dynamics at the cell cortex and is required for myoblast elongation and fusion. Curr Biol. 2007;17(15):1318–1325. doi: 10.1016/j.cub.2007.06.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pope KL, Harris TJC. Control of cell flattening and junctional remodeling during squamous epithelial morphogenesis in Drosophila. Development. 2008;135(13):2227–2238. doi: 10.1242/dev.019802. [DOI] [PubMed] [Google Scholar]
- 19.Kharitonova MA, Vasiliev JM. Controlling cell length. Semin Cell Dev Biol. 2008;19(6):480–484. doi: 10.1016/j.semcdb.2008.07.008. [DOI] [PubMed] [Google Scholar]
- 20.Reinsch S, Gönczy P. Mechanisms of nuclear positioning. J Cell Sci. 1998;111(Pt 16):2283–2295. doi: 10.1242/jcs.111.16.2283. [DOI] [PubMed] [Google Scholar]
- 21.Gomes ER, Jani S, Gundersen GG. Nuclear movement regulated by Cdc42, MRCK, myosin, and actin flow establishes MTOC polarization in migrating cells. Cell. 2005;121(3):451–463. doi: 10.1016/j.cell.2005.02.022. [DOI] [PubMed] [Google Scholar]
- 22.Carpenter AE, et al. CellProfiler: Image analysis software for identifying and quantifying cell phenotypes. Genome Biol. 2006;7(10):R100. doi: 10.1186/gb-2006-7-10-r100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fernandez R, et al. Imaging plant growth in 4D: Robust tissue reconstruction and lineaging at cell resolution. Nat Methods. 2010;7(7):547–553. doi: 10.1038/nmeth.1472. [DOI] [PubMed] [Google Scholar]
- 24.Olivier N, et al. Cell lineage reconstruction of early zebrafish embryos using label-free nonlinear microscopy. Science. 2010;329(5994):967–971. doi: 10.1126/science.1189428. [DOI] [PubMed] [Google Scholar]
- 25.Tassy O, Daian F, Hudson C, Bertrand V, Lemaire P. A quantitative approach to the study of cell shapes and interactions during early chordate embryogenesis. Curr Biol. 2006;16(4):345–358. doi: 10.1016/j.cub.2005.12.044. [DOI] [PubMed] [Google Scholar]
- 26.Costa M, Wilson ET, Wieschaus E. A putative cell signal encoded by the folded gastrulation gene coordinates cell shape changes during Drosophila gastrulation. Cell. 1994;76(6):1075–1089. doi: 10.1016/0092-8674(94)90384-0. [DOI] [PubMed] [Google Scholar]
- 27.Odell GM, Oster G, Alberch P, Burnside B. The mechanical basis of morphogenesis. I. Epithelial folding and invagination. Dev Biol. 1981;85(2):446–462. doi: 10.1016/0012-1606(81)90276-1. [DOI] [PubMed] [Google Scholar]
- 28.Kölsch V, Seher T, Fernandez-Ballester GJ, Serrano L, Leptin M. Control of Drosophila gastrulation by apical localization of adherens junctions and RhoGEF2. Science. 2007;315(5810):384–386. doi: 10.1126/science.1134833. [DOI] [PubMed] [Google Scholar]
- 29.Martin AC, Gelbart M, Fernandez-Gonzalez R, Kaschube M, Wieschaus EF. Integration of contractile forces during tissue invagination. J Cell Biol. 2010;188(5):735–749. doi: 10.1083/jcb.200910099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Royou A, Sullivan W, Karess R. Cortical recruitment of nonmuscle myosin II in early syncytial Drosophila embryos: Its role in nuclear axial expansion and its regulation by Cdc2 activity. J Cell Biol. 2002;158(1):127–137. doi: 10.1083/jcb.200203148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Oda H, Tsukita S. Real-time imaging of cell-cell adherens junctions reveals that Drosophila mesoderm invagination begins with two phases of apical constriction of cells. J Cell Sci. 2001;114(Pt 3):493–501. doi: 10.1242/jcs.114.3.493. [DOI] [PubMed] [Google Scholar]
- 32.Clarkson M, Saint R. A His2AvDGFP fusion gene complements a lethal His2AvD mutant allele and provides an in vivo marker for Drosophila chromosome behavior. DNA Cell Biol. 1999;18(6):457–462. doi: 10.1089/104454999315178. [DOI] [PubMed] [Google Scholar]
- 33.Morin X, Daneman R, Zavortink M, Chia W. A protein trap strategy to detect GFP-tagged proteins expressed from their endogenous loci in Drosophila. Proc Natl Acad Sci USA. 2001;98(26):15050–15055. doi: 10.1073/pnas.261408198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Denk W, Strickler JH, Webb WW. Two-photon laser scanning fluorescence microscopy. Science. 1990;248(4951):73–76. doi: 10.1126/science.2321027. [DOI] [PubMed] [Google Scholar]
- 35.Pologruto TA, Sabatini BL, Svoboda K. ScanImage: Flexible software for operating laser scanning microscopes. Biomed Eng Online. 2003;2:13. doi: 10.1186/1475-925X-2-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mason FM, Martin AC. Tuning cell shape change with contractive ratchets. Curr Opin Genet Dev. 2011;21:671–679. doi: 10.1016/j.gde.2011.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.