Abstract
We investigate protein–protein association using the associative-memory, water-mediated, structure, and energy model (AWSEM), a coarse-grained protein folding model that has been optimized using energy-landscape theory. The potential was originally parameterized by enforcing a funneled nature for a database of dimeric interfaces but was later further optimized to create funneled folding landscapes for individual monomeric proteins. The ability of the model to predict interfaces was not tested previously. The present results show that simulated annealing of the model indeed is able to predict successfully the native interfaces of eight homodimers and four heterodimers, thus amounting to a flexible docking algorithm. We go on to address the relative importance of monomer geometry, flexibility, and nonnative intermonomeric contacts in the association process for the homodimers. Monomer surface geometry is found to be important in determining the binding interface, but it is insufficient. Using a uniform binding potential rather than the water-mediated potential results in sampling of misbound structures that are geometrically preferred but are nonetheless energetically disfavored by AWSEM, as well as in nature. Depending on the stability of the unbound monomers, nonnative contacts play different roles in the association process. For unstable monomers, thermodynamic states stabilized by nonnative interactions correspond to productive, on-pathway intermediates and can, therefore, catalyze binding through a fly-casting mechanism. For stable monomers, in contrast, states stabilized by nonnative interactions generally correspond to traps that impede binding.
Keywords: binding interface prediction, swapped contacts
Protein–protein interfaces encode information that is key to a molecular understanding of biological functions. The folding of proteins is well understood in the framework of energy landscape theory and its principle of minimal frustration. Are binding landscapes also funneled? Mechanistic consequences of funneled binding landscapes have been investigated using structure-based models (1–5). The agreement of these mechanisms with observation suggests that binding landscapes are generally funneled, explaining why topology is indeed a major factor in determining binding mechanisms (1). A statistical analysis of a large database of protein complexes revealed that for many of the complexes, the binding energy gap is indeed larger than expected knowing the variance of the binding energy (6), the hallmark feature of a funneled landscape (7). When further testing this idea, Papoian et al. discovered that for other complexes, to have a funneled landscape for binding, unanticipated water-mediated interactions were required. They developed a water-mediated potential encoding these interactions (8). This transferable potential was later optimized to create funneled folding landscapes that successfully predict the structure of monomeric proteins (9, 10). Therefore, there is considerable support for the idea that, like folding landscapes, protein–protein recognition landscapes are funneled.
In this study, we test whether the associative-memory, water-mediated, structure, and energy model (AWSEM) potential can predict binding interfaces, the problem that motivated its original invention. Unlike rigid docking programs (11–13), our approach uses molecular dynamics with simulated annealing to search for structures energetically favored by the AWSEM potential. Whereas many docking protocols entail multiple stages (14) to accomplish interface prediction, including rigid body search to locate regions of interest (11, 13, 15) and refinement of docked structures and selecting the best models (12, 16, 17), simulated annealing of the AWSEM potential proves directly able to predict the binding interface of the dimers we have tested. The molecular dynamics implementation allows one also to compute free-energy profiles to predict mechanisms. Using this predictive transferable potential model, we now revisit the role of topology in determining binding mechanisms and explore the additional role played by nonnative contacts in coupled folding and binding reactions.
Binding-Interface Prediction
We used AWSEM to predict the binding interfaces of eight homodimers and four heterodimers. The homodimers were previously studied with pure structure–based models (1). The only structural information used by AWSEM was local backbone information of the monomers from the Protein Data Bank (PDB) structure of the dimeric complex; no information about dimeric contacts was included. The tertiary contacts within the monomers are also not used as input. The input of native monomeric information guides only local-in-sequence structure formation. Both the tertiary contacts within the monomers and between the two monomers are determined by the same transferrable tertiary contact potential, which is described briefly in Methods and previously by Davtyan et al. (10). The starting states of all simulations consisted of two completely unfolded and unbound monomers, and molecular dynamics with simulated annealing was performed to search for the bound state.
As shown in Figs. 1 and 2, the binding interfaces for the 12 dimers are generally very well predicted. One can argue that the interfaces of homodimers might be easier to predict because their binding interactions are usually stronger because of symmetry (18, 19). Homodimers are, in general, observed in a symmetric binding geometry, where strong contacts on the interface are doubled. We, therefore, also tested four heterodimers with relatively weak interfaces, and AWSEM was able to predict the interfaces to similar accuracy as the homodimers discussed herein. The heterodimers that we tested have mostly hydrophilic interfaces, which are relatively weak compared with hydrophobic interfaces and are, therefore, harder to predict. The water-mediated interactions (Methods) play a major role in predicting hydrophilic interfaces. When they are turned off, the prediction quality of dimers with hydrophilic interfaces get significantly worse, as shown in Figs. S2 and S3. Having successfully passed the prediction test for both homodimers and heterodimers, AWSEM was applied to study the mechanism of homodimer binding in greater detail, as described below.
Fig. 1.
Snapshots of best predicted structures (yellow) using AWSEM, compared with the PDB structure (blue). The name of the proteins, their PDB ID code, and the number of residues are shown in the figure. The first eight dimers are homodimers, and the last four are heterodimers.
Fig. 2.
The accuracy of the AWSEM predictions is measured by Q and rmsd of the Cα atoms of the complex. PDB ID codes for homodimers and heterodimes are in black and blue, respectively. Qcomplex is shown as a star symbol and Qinterface as a square symbol. Note that for 1LFB, there is a relatively large difference between the two Q values. This can be explained by its very small ratio of the number of interfacial contacts to the number of total contacts, as shown in Fig. S1. Dimer size refers to the number of residues in the dimer complex. Twenty or 40 independent annealing runs were performed for each dimer, starting from two monomers completely unfolded and separated. The final structure obtained at the end of the annealing runs with the best Q is selected for each dimer.
In Fig. 2, the seemingly worst prediction in our test set is for the homeodomain of liver transcription factor (LFB1) (PDB ID code 1LFB). Its intermonomeric contacts in the PDB structure are weaker than for all other proteins in the test set. Significant native or nonnative contacts only form at temperatures well below the binding temperatures of the other proteins. The PDB structure with which we compared our prediction turned out, in fact, only to be a model, not a directly determined crystal structure, as proposed by Ceska et al. (20), and involves a simple twofold rotation of the crystallographically determined monomer structure. It has been suggested that the homeodomain might be dimeric when bound to DNA (21, 22). However, we have been unable to find a crystal structure of the homeodomain of LFB1 in dimeric form in the presence or absence of DNA.
Inspired by the principle of minimal frustration, AWSEM was optimized by maximizing the ratio of the folding temperature to the glass transition temperature, similar to the Z-score optimization algorithm. However, the parameters found by optimization were developed using a training set containing only monomers. The success of the model in actually predicting binding structures buttresses the idea that the same energy landscape principles are applicable to binding processes as to monomeric folding. For Arc repressor (PDB ID code 1ARR) and Lambda repressor (PDB ID code 1LMB), Fig. 3 shows the total energy of the predicted complex at the end of each annealing simulation as a function of Qinterface, the fraction of native contacts formed on the interface. Low-energy structures are seen to correspond to near native states, and there appear to be few competing (low energy but low Qinterface) traps.
Fig. 3.
The total energies of the final complexes at the end of annealing simulations are plotted against Qinterface. Near-native bound structures have lower energy than nonspecific bound structures.
Experimental and Theoretical Descriptions of Protein Dimers
Homodimers are often categorized as being either obligatory or nonobligatory dimers, meaning that the monomers must associate to complete folding (obligatory) or are stably folded in isolation at physiological temperature (nonobligatory). This distinction can be made in the laboratory by performing equilibrium denaturation experiments. In these experiments, obligatory dimers show only two states [one with both monomers unfolded (or partially folded) and the other with the native dimer structure] and are, therefore, sometimes referred to as two-state dimers. Nonobligatory dimers have three populated states under physiological conditions: one with unfolded monomers, another with folded but unbound monomers, and yet another with the folded monomers bound together.
The binding-folding mechanism has been found to correlate with several global characterizations of the native dimer structure: interface hydrophobicity and the ratio of the number of interfacial contacts to the number of intramonomeric contacts being most important. A dimer with a highly hydrophobic interface and a large ratio of interfacial to monomeric contacts is typically two-state. Dimerization in these cases is, in some ways, reminiscent of monomeric protein folding insofar as the dimer as a whole can be thought of as a single domain folding cooperatively with the interface playing the part of the hydrophobic core. This type of folding mechanism is sometimes referred to as involving “induced fit” (23, 24), meaning that the presence of the binding partner is needed to induce the monomer to adopt its folded structure. Nonobligatory dimers typically have more hydrophilic interfaces and smaller ratios of interfacial contacts to monomeric contacts. These dimers associate via a lock-and-key–type mechanism (25) wherein complementary interfacial geometry and favorable contact energies drive association.
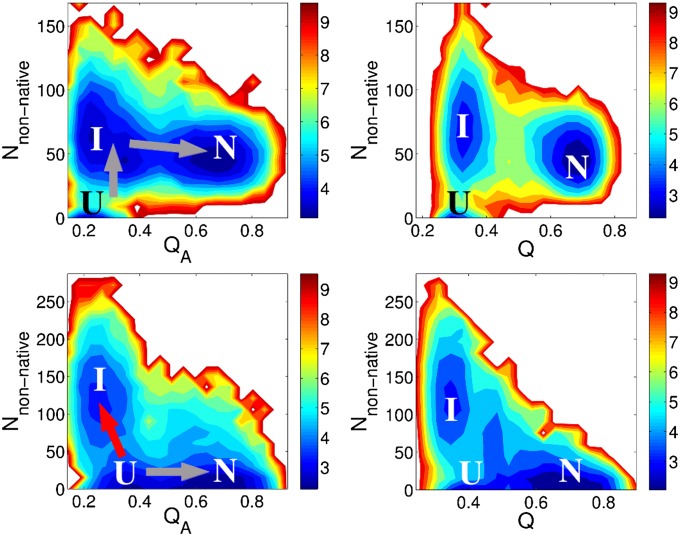
Knowing the size and shape of the interface has often proved sufficient to determine whether a homodimer will associate via a two-state or three-state mechanism (1). Using a structure-based model with uniform contact energies for only native interfacial contacts and native monomeric contacts, Levy et al. were able to accurately reconstruct the experimentally determined binding mechanisms for 11 homodimers. As shown in Fig. 4, the current model also correctly reproduces the observed pattern of two-state and three-state behaviors for these examples. Two stable states are observed for two-state dimer Arc repressor, the unfolded, unbound-state U and the native bound-state N. There is no stable intermediate state, indicating a folding-upon-binding mechanism. For the three-state dimer Lambda repressor, on the other hand, there is an additional intermediate state I, which consists of an ensemble of a variety of encounter complexes. These complexes have only one monomer folded and partially bound or are complexes in which both monomers have folded but remain unbound. The free-energy surfaces calculated using AWSEM are consistent with the experimental observations and previous theoretical modeling results. Unlike the previously used structure-based model, however, the current model that can predict dimer interfaces can also shed light on the role of nonnative interactions in the association process.
Fig. 4.
Free-energy surfaces of folding and binding of obligatory (two-state) and nonobligatory (three-state) dimers obtained using AWSEM. Free-energy surfaces are plotted as a function of the fraction of native contacts within the individual subunit QA (QB), Qinterface, Q of the complex, and the distance between the centers of mass of the two subunits (distanceCOM). State U refers to the unfolded and unbound state, and state N is the native bound state. The intermediate state I is observed only in the free-energy plot of the nonobligatory dimer 1LMB. For 1LMB, the macrobasins that contain a mixture of different states are left unlabeled. The simulations reproduce the binding mechanism inferred from experimental and previous theoretical modeling results (1). The free-energy surfaces are calculated at the folding temperature.
Role of Monomer Geometry in Interface Determination
One might argue that the successful predictions of dimer interfaces could be attributable to geometrical factors related to the limited number of ways that two dimers of prescribed geometry can associate. In this case, of course, the monomer is flexible and, therefore, does not have a fixed tertiary structure a priori. Nevertheless, to investigate the possible role of monomer geometry by itself, we changed the strength of any contacts between monomers to have a residue-independent, uniform value while retaining the transferable potential within the monomers. The strength of the intermonomeric interaction is rescaled so that the stability of the native bound state was the same as with the AWSEM potential. Prediction simulations using the uniform intermonomer contact strength with the same annealing schedule were performed. The simulations using the uniform intermonomer contact energy were significantly worse than the AWSEM predictions that used the optimized potential. Interestingly, the effects of changing the intermonomer contact strength to a uniform value are different for different dimers. For example, as shown in Fig. 5, for troponin C (PDB ID code 1CTA), with uniform contact energies the native bound state is no longer an energetically favored state. Instead, there are numerous misbound states with more favorable binding energies than the native. For troponin C, the native bound state is not the state with the maximal number of intermonomeric contacts. On the other hand, for arc repressor (PDB ID code 1ARR), the native bound state does still remain the lowest energy state when the contacts have uniform weight. Nevertheless, uniform intermonomeric contacts create an intermediate state I in this system, which drastically reduces the binding efficiency.
Fig. 5.
The AWSEM predictions (blue) vs. the predictions using a nonoptimized energy function with uniform intermonomer contact strength (red) for troponin C site III (PDB ID code 1CTA) and Arc repressor (PDB ID code 1ARR). In the plots on the left, the energies of the final configurations from each simulation are plotted as a function of Qinterface. In the plots on the right, the distribution of the number of samples collected from all simulations is plotted along Q. For 1CTA, the native bound state N is energetically less favored for uniform contact energy function than for AWSEM. For 1ARR, on the other hand, the native bound state is the lowest energy state for both energy functions. However, uniform intermonomeric contacts create an intermediate state I, which drastically reduces the binding efficiency.
The structure of the arc repressor monomer in the bound dimer consists of two helices and a β-strand, and the resulting dimer interface forces the two monomers to significantly intertwine. The large size of the interface allows the uniform interaction energy described above to still favor the correct bound structure, albeit with a rougher landscape as indicated by the presence of misbound structures encountered during annealing. Energetic heterogeneity is not the only contributor to high binding efficiency. In instances where the native binding-interface geometry forces the monomers to interweave, the flexibility of the local structure of the monomers also modulates the binding efficiency, as shown in Fig. S4. When the strength of the energetic term encoding the local in sequence structure bias is decreased, the percentage of successful binding simulations at first increases but finally decreases when the local bias becomes too weak. This is consistent with the suggestion that flexibility allows proteins to adjust to achieve optimal fit upon binding to perform specific biological functions (26). Binding is a dynamic process on a funneled landscape; geometry of the monomers alone does not completely explain the binding process.
Role of Nonnative Contacts in Dimer Formation and the Fly-Casting Mechanism
The water-mediated potential in AWSEM is a transferable potential that can be used to model the intermonomer tertiary interactions. This part of the model allows us to study the role of nonnative intermonomeric contacts in dimer formation. To discuss the role of nonnative interactions, it is informative to single out a special class of nonnative contacts called swapped contacts. The name comes from a type of intermonomer contact pair that is observed in domain-swapped dimers (27, 28). Swapped contacts are defined as nonnative intermonomeric contacts formed between the ith residue in monomer A and jth residue in monomer B that correspond to i and j being a native contact pair within the monomer. Note that sometimes there are pairs of residue indices (i, j) corresponding to a native monomer contact pair that are also native interfacial contact pairs. These contact pairs are excluded from the computation of the number of swapped contacts because they are considered to be native contacts. The swapped contacts are of special importance in dimer association because they are, on average, stronger than other random contacts that have no analog in the native monomer structure. According to the principle of minimal frustration, the native contacts within a stable monomeric protein are, on average, stronger than other random contacts; therefore, likewise, swapped contacts are more stable than random ones.
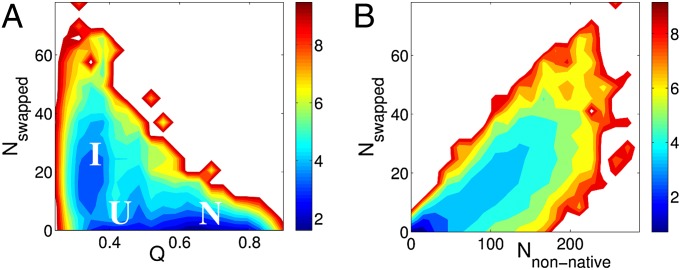
Nonnative interactions play different roles for obligatory and nonobligatory dimers as seen in Fig. 6. An example of an obligatory dimer, Arc repressor, is shown on the top of Fig. 6. States stabilized by nonnative interactions correspond to on-pathway intermediates that catalyze the association process through a fly-casting mechanism (29). The individual monomers, which are both in extended conformations before the association, have significantly larger capture radii than those of the folded monomers. The large capture radius increases the rate of binding. In the case of nonobligatory dimers, however, the states with nonnative contacts appear to be off-pathway and impede binding by acting as kinetic traps. We investigated further these off-pathway intermediates for the case of Lambda repressor (PDB ID code 1LMB). In Fig. 7, free-energy surfaces of Lambda repressor are plotted as functions of the number of swapped contacts Nswapped, Q, and Nnonnative. As in Fig. 6, we observe an off-pathway intermediate state stabilized by swapped contacts in the left plot of Fig. 7. The plot on the right shows a linear increase in the number of swapped contacts when the number of nonnative contacts increases. Fig. 7 suggests that the intermediate state I consists of a significant number of swapped contact pairs. These intermediate states stabilized by swapped contacts are kinetic traps in the binding of nonobligatory dimers. If both of the monomers of a nonobligatory dimer are significantly unfolded when they encounter each other, they may fall into the trap state I as shown in Figs. 6 and 7.
Fig. 6.
For Arc repressor (Upper) and Lambda repressor (Lower), free-energy surfaces at the folding temperature are plotted as a function of the number of nonnative intermonomeric contacts Nnonnative, QA, and Q of the complex. I, U, and N stand for intermediate, unbound, and native bound states, respectively. Nonnative interactions have different consequences for obligatory and nonobligatory dimers. In the case of obligatory dimers (Upper), states stabilized by nonnative interactions correspond to on-pathway (indicated as gray arrow) intermediates that can catalyze the association process through fly-casting mechanism. In the case of nonobligatory dimers, these states appear to be off-pathway (indicated as red arrow) and can, thereby, impede binding by acting as a trap.
Fig. 7.
Free-energy surfaces as a function of Nswapped for Lambda repressor (PDB ID code 1LMB). (A) Similar as in Fig. 6, we observe an off-pathway intermediate state, stabilized by the swapped contacts. (B) There is a linear increase of the number of swapped contacts when the number of nonnative contacts increases. The ratio of the number of swapped contacts to the number of nonnative contacts is about 10 to ∼20%. These suggest that the intermediate state stabilized by nonnative contact pairs contains a significant number of swapped contacts.
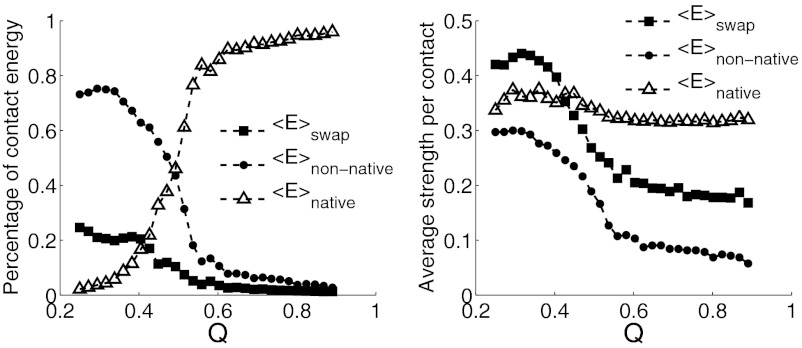
To summarize the roles of the different types of contacts during binding, we plot their average contact strength and their contributions to the total binding energy against Q in Fig. 8. As the complex approaches the native state, as shown in Fig. 8A, the major contributor to the binding energy switches from being nonnative contacts to native contacts, as expected. This change of contribution is steep around Q = 0.5, near the transition state region. At low Q region, total energy of swapped contacts is about 20% of the total binding energy. Consistent with the principle of minimal frustration, swapped contacts are on average stronger than other nonnative contacts throughout the whole binding process, as shown in Fig. 8B. At low Q, where the two monomers are first coming into contact, the average strength of the swapped contacts is even larger than the strength of the native contacts alone, suggesting their important role in stabilizing nonspecific bound structures at the start of the binding process. As binding progresses, the strengths of both swapped and nonnative contacts decrease, whereas the strength of the native contacts is, interestingly, more or less constant. These observations of the ubiquity of domain swapping are consistent with the experimental observation by Oliveberg of the universality of transient aggregation at high protein concentration (30).
Fig. 8.
For all three different types of contacts, their contributions to the total binding energy, and their average strength are plotted against Q of the Lambda repressor complex. Native contacts, swapped contacts, and nonnative contacts excluding swapped contacts are in triangle, square, and circle symbols, respectively. (Left) As the complex approaches the native state, the major contributor to the binding energy switches from nonnative contacts to native contacts. This change of contribution is steep around Q = 0.5, near the transition state region. Total energy of swapped contacts is about 20% of the total binding energy at the low Q region. (Right) Swapped contacts are on average stronger than other nonnative contacts throughout the whole Q region. As Q increases, the average strengths of both swapped and nonnative contacts decrease, whereas the strength of the native contacts is more or less constant. At the low Q region, where the two monomers are in initial encountering, the average strength of the swapped contacts is even larger than the native contacts.
Conclusions
The intent of this study was to investigate the extent to which protein–protein association is funneled by the same forces that determine the landscapes of monomeric proteins. We see that the association is well described by a funneled model but that there are residual effects of energetic frustration which allow nonnative interactions to play a role. The picture that emerges from the study is that folding and binding are dynamic processes that are often coupled and that both take place via diffusion on rugged but nevertheless largely funneled energy landscapes. Interactions that successfully predict the structure of monomeric proteins also prove sufficient to predict native dimeric interfaces. Monomer geometry alone does not lead to the successful prediction of binding modes: both energetic heterogeneity and flexibility of the monomers are important. Nonnative interactions can stabilize on-pathway or off-pathway conformations depending on the stability of the monomers, and swapped contacts in particular are stronger than other, nonspecific, nonnative contacts, in accordance with the principle of minimal frustration. Swapped contacts play an important role in stabilizing nonspecific bound structures at the start of the association process. Other nonnative interactions, on the other hand, also sometimes play a role, but, in general, dimeric proteins have evolved so as to eliminate traps on the combined folding and binding landscape.
Methods
AWSEM was described in detail recently (10). The tertiary contact energy function Vcontact consists of two terms, the direct contact Vdirect and the mediated contact Vwater. In simulations with multiple chains, the associative memory potential VAM, acts only locally in sequence within each monomer. On the other hand, Vcontact, the burial potential and the β hydrogen-bonding terms act both within and among the monomeric chains. When calculating the local density of residues, which is used by the helix and burial potentials as well as Vcontact, all chains are included. VAM is determined by a single memory, which is the structure of the monomer in the experimentally determined dimer structure. As mentioned above, this interaction includes only those pairs of residues that have a sequence separation of less than or equal to 9. No information about contacts within the monomers or on the dimer interface is included.
The predictions were performed using molecular dynamics with simulated annealing. The annealing simulations were initialized by completely unfolding the individual monomers and separating them. The temperature is then lowered to below the empirically determined binding temperature and a weak bias is applied between the centers of mass of the two monomers to ensure that contact is made during the course of the simulation. The free-energy surfaces were calculated using the weighted histogram analysis method (WHAM) (31) on the data collected from constant temperature simulations with umbrella sampling along Q.
Supplementary Material
Acknowledgments
We thank Patricio Craig and Mikael Oliveberg for stimulating discussions. We thank Yaakov Levy and Jin Wang for a critical reading of the manuscript. W.Z. and N.P.S. were supported by National Institute of General Medical Sciences Grants R01 GM44557 and P01 GM071862. Additional support was provided by the D. R. Bullard-Welch Chair at Rice University. A.D. and G.A.P. were supported by the Camille and Henry Dreyfus Foundation.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1216215109/-/DCSupplemental.
References
- 1.Levy Y, Wolynes PG, Onuchic JN. Protein topology determines binding mechanism. Proc Natl Acad Sci USA. 2004;101(2):511–516. doi: 10.1073/pnas.2534828100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Levy Y, Papoian GA, Onuchic JN, Wolynes PG. Energy landscape analysis of protein dimers. Isr J Chem. 2004;44(1-3):281–297. [Google Scholar]
- 3.Levy Y, Cho SS, Onuchic JN, Wolynes PG. A survey of flexible protein binding mechanisms and their transition states using native topology based energy landscapes. J Mol Biol. 2005;346(4):1121–1145. doi: 10.1016/j.jmb.2004.12.021. [DOI] [PubMed] [Google Scholar]
- 4.Lu Q, Lu HP, Wang J. Exploring the mechanism of flexible biomolecular recognition with single molecule dynamics. Phys Rev Lett. 2007;98(12):128105. doi: 10.1103/PhysRevLett.98.128105. [DOI] [PubMed] [Google Scholar]
- 5.Wang J, et al. Multi-scaled explorations of binding-induced folding of intrinsically disordered protein inhibitor IA3 to its target enzyme. PLOS Comput Biol. 2011;7(4):e1001118. doi: 10.1371/journal.pcbi.1001118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Papoian GA, Wolynes PG. The physics and bioinformatics of binding and folding-an energy landscape perspective. Biopolymers. 2003;68(3):333–349. doi: 10.1002/bip.10286. [DOI] [PubMed] [Google Scholar]
- 7.Bryngelson JD, Onuchic JN, Socci ND, Wolynes PG. Funnels, pathways, and the energy landscape of protein folding: A synthesis. Proteins. 1995;21(3):167–195. doi: 10.1002/prot.340210302. [DOI] [PubMed] [Google Scholar]
- 8.Papoian GA, Ulander J, Wolynes PG. Role of water mediated interactions in protein-protein recognition landscapes. J Am Chem Soc. 2003;125(30):9170–9178. doi: 10.1021/ja034729u. [DOI] [PubMed] [Google Scholar]
- 9.Papoian GA, Ulander J, Eastwood MP, Luthey-Schulten Z, Wolynes PG. Water in protein structure prediction. Proc Natl Acad Sci USA. 2004;101(10):3352–3357. doi: 10.1073/pnas.0307851100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Davtyan A, et al. AWSEM-MD: Protein structure prediction using coarse-grained physical potentials and bioinformatically based local structure biasing. J Phys Chem B. 2012;116(29):8494–8503. doi: 10.1021/jp212541y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.de Vries SJ, et al. HADDOCK versus HADDOCK: New features and performance of HADDOCK2.0 on the CAPRI targets. Proteins. 2007;69(4):726–733. doi: 10.1002/prot.21723. [DOI] [PubMed] [Google Scholar]
- 12.Pierce B, Weng Z. A combination of rescoring and refinement significantly improves protein docking performance. Proteins. 2008;72(1):270–279. doi: 10.1002/prot.21920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang C, Bradley P, Baker D. Protein-protein docking with backbone flexibility. J Mol Biol. 2007;373(2):503–519. doi: 10.1016/j.jmb.2007.07.050. [DOI] [PubMed] [Google Scholar]
- 14.Vajda S, Kozakov D. Convergence and combination of methods in protein-protein docking. Curr Opin Struct Biol. 2009;19(2):164–170. doi: 10.1016/j.sbi.2009.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fernández-Recio J, Totrov M, Abagyan R. ICM-DISCO docking by global energy optimization with fully flexible side-chains. Proteins. 2003;52(1):113–117. doi: 10.1002/prot.10383. [DOI] [PubMed] [Google Scholar]
- 16.Kozakov D, Schueler-Furman O, Vajda S. Discrimination of near-native structures in protein-protein docking by testing the stability of local minima. Proteins. 2008;72(3):993–1004. doi: 10.1002/prot.21997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mashiach E, Schneidman-Duhovny D, Andrusier N, Nussinov R, Wolfson HJ. FireDock: A web server for fast interaction refinement in molecular docking. Nucleic Acids Res. 2008;36(Web Server issue):W229–W232. doi: 10.1093/nar/gkn186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wolynes PG. Symmetry and the energy landscapes of biomolecules. Proc Natl Acad Sci USA. 1996;93(25):14249–14255. doi: 10.1073/pnas.93.25.14249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.André I, Strauss CEM, Kaplan DB, Bradley P, Baker D. Emergence of symmetry in homooligomeric biological assemblies. Proc Natl Acad Sci USA. 2008;105(42):16148–16152. doi: 10.1073/pnas.0807576105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ceska TA, et al. The X-ray structure of an atypical homeodomain present in the rat liver transcription factor LFB1/HNF1 and implications for DNA binding. EMBO J. 1993;12(5):1805–1810. doi: 10.2210/pdb1lfb/pdb. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Frain M, et al. The liver-specific transcription factor LF-B1 contains a highly diverged homeobox DNA binding domain. Cell. 1989;59(1):145–157. doi: 10.1016/0092-8674(89)90877-5. [DOI] [PubMed] [Google Scholar]
- 22.Tomei L, Cortese R, De Francesco R. A POU-A related region dictates DNA binding specificity of LFB1/HNF1 by orienting the two XL-homeodomains in the dimer. EMBO J. 1992;11(11):4119–4129. doi: 10.1002/j.1460-2075.1992.tb05505.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Koshland DE., Jr Application of a theory of enzyme specificity to protein synthesis. Proc Natl Acad Sci USA. 1958;44(2):98–104. doi: 10.1073/pnas.44.2.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Csermely P, Palotai R, Nussinov R. Induced fit, conformational selection and independent dynamic segments: An extended view of binding events. Trends Biochem Sci. 2010;35(10):539–546. doi: 10.1016/j.tibs.2010.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fischer E. Einfluss der configuration auf die wirkung der enzyme. Berichte der Deutschen Chemischen Gesellschaft. 1894;27(2):2985–2993. [Google Scholar]
- 26.Wang J, Xu L, Wang E. Optimal specificity and function for flexible biomolecular recognition. Biophys J. 2007;92(12):L109–L111. doi: 10.1529/biophysj.107.105551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yang S, et al. Domain swapping is a consequence of minimal frustration. Proc Natl Acad Sci USA. 2004;101(38):13786–13791. doi: 10.1073/pnas.0403724101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bennett MJ, Sawaya MR, Eisenberg D. Deposition diseases and 3D domain swapping. Structure. 2006;14(5):811–824. doi: 10.1016/j.str.2006.03.011. [DOI] [PubMed] [Google Scholar]
- 29.Shoemaker BA, Portman JJ, Wolynes PG. Speeding molecular recognition by using the folding funnel: The fly-casting mechanism. Proc Natl Acad Sci USA. 2000;97(16):8868–8873. doi: 10.1073/pnas.160259697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Oliveberg M. Alternative explanations for Multistate kinetics in protein folding: Transient aggregation and changing transition-state ensembles. Acc Chem Res. 1998;31(11):765–772. [Google Scholar]
- 31.Kumar S, Rosenberg JM, Bouzida D, Swendsen RH, Kollman PA. The weighted histogram analysis method for free-energy calculations on biomolecules. I. The method. J Comput Chem. 1992;13(8):1011–1021. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.