Abstract
The bacterial ribosome consists of three rRNA molecules and 57 proteins and plays a crucial role in translating mRNA-encoded information into proteins. Because of the ribosome’s structural and mechanistic complexity, it is believed that each ribosomal component coevolves to maintain its function. Unlike 5S rRNA, 16S and 23S rRNAs appear to lack mutational robustness, because they form the structural core of the ribosome. However, using Escherichia coli Δ7 (null mutant of operons) as a host, we have recently shown that an active hybrid ribosome whose 16S rRNA has been specifically substituted with that from non–E. coli bacteria can be reconstituted in vivo. To investigate the mutational robustness of 16S rRNA and the structural basis for its functionality, we used a metagenomic approach to screen for 16S rRNA genes that complement the growth of E. coli Δ7. Various functional genes were obtained from the Gammaproteobacteria and Betaproteobacteria lineages. Despite the large sequence diversity (80.9–99.0% identity with E. coli 16S rRNA) of the functional 16S rRNA molecules, the doubling times (DTs) of each mutant increased only modestly with decreasing sequence identity (average increase in DT, 4.6 s per mutation). The three-dimensional structure of the 30S ribosome showed that at least 40.7% (628/1,542) of the nucleotides were variable, even at ribosomal protein-binding sites, provided that the secondary structures were properly conserved. Our results clearly demonstrate that 16S rRNA functionality largely depends on the secondary structure but not on the sequence itself.
Keywords: functional plasticity
The ribosome plays a crucial role in translating mRNA-encoded information into proteins, which consists of 2 (large and small) subunits. In prokaryotes, the small 30S subunit consists of 16S rRNA and 21 proteins, whereas the large 50S subunit consists of 23S rRNA, 5S rRNA, and 36 proteins. Both in bacteria and archaea, 16S or 23S rRNA shapes the structural core of the subunit particle, whereas many ribosomal proteins are often found on the surface of the subunit, with extensions that protrude into the RNA core (1–3). In contrast, 5S rRNA (small RNA with 120 bases) simply attaches to the 50S subunit and constitutes a structure called a central protuberance (1, 3). Given this structural and mechanistic complexity, as well as their biological significance, it is generally believed that the rRNA genes coevolved with their cognate sets of ribosomal protein genes. In other words, horizontal gene transfer should not occur in rRNA genes because such radical changes would compromise the structural integrity of the ribosome, leading to immediate cell death (4, 5). Taking advantage of this species-specific characteristic (as well as their omnipresence and adequate mutation rates and the adequate amount of information available), researchers have used 16S rRNA genes for the last 30 y to infer the relationships among prokaryotes (4, 6).
Despite the apparent conservative nature of rRNA, some genetic studies using Escherichia coli have reported the mutational robustness of rRNA even at protein-binding sites (7, 8). In addition, there is growing evidence to suggest that horizontal gene transfer of 16S rRNA genes takes place in nature (9–13). Recently, using experimental horizontal gene transfer (i.e., the interspecies exchange of E. coli 16S rRNA genes with a foreign counterpart), we have also shown that the active hybrid ribosome could be reconstituted in vivo by using E. coli Δ7, a null mutant of the ribosomal RNA (rrn) operon, as a host (14). Surprisingly, the 16S rRNA from not only the Gammaproteobacteria species Serratia ficaria (94.6% identity) but also from the evolutionarily distant Betaproteobacteria species Ralstonia pickettii (81.6% identity) have been found to form ribosomes active enough to support the growth of E. coli Δ7 (14). All these studies strongly suggest that the ribosome may be much more plastic than had previously been believed, perhaps enough to accommodate foreign 16S rRNA from diverse organisms.
To investigate the mutational robustness of 16S rRNA in this study, we screened for functional genes in E. coli. This study uses a functional metagenomic approach (15) to extensively study the possible sequences of RNA-encoding genes. Various 16S rRNA genes were obtained from a diverse lineage of bacteria whose molecular basis for functionality has been discussed based on the primary and secondary structures of 16S rRNA as well as the three-dimensional structure of the ribosome.
Results and Discussion
Sequence Diversity of Functional 16S rRNA Genes in E. coli.
To rigorously determine the extent to which 16S rRNA genes are interchangeable with those in E. coli, we used environmental DNA (metagenome) as a source of the 16S rRNA genes. This approach takes advantage of the large sequence (and the secondary structure’s) diversity of the 16S rRNA genes in nature (16) and selects those that are functional in E. coli. Genetic selection was carried out as described previously (14) (Fig. S1). Briefly, E. coli strain KT101 lacking all seven rRNA operons (rrnA, B, C, D, E, G, H) in its chromosome (14, 17) was used as the host; its growth was complemented by the rrnB operon encoded in the rescue plasmid pRB101 (pSC101 ori, ampicillin resistant). The plasmid contains the counter selectable marker sacB and thus can be conditionally eliminated on addition of sucrose into the medium (14, 17). Using general universal primers (18), we amplified approximately the entire length of the 16S rRNA gene (8–1,541, E. coli numbering) from environmental DNA. The amplicons were cloned into another rrnB-encoding plasmid pRB103 (pSC101 ori, Zeocin resistant) (Fig. S1), and an expression library of diverse 16S rRNA genes was created. We then screened this library (∼15,000 clones) for functional sequences that complemented the growth of KT101 on sucrose-containing plates.
After counterselection, ∼200 clones of KT103-derivatives (carrying pRB103 whose 16S rRNA gene was substituted with foreign genes) were obtained, from which 33 nonredundant 16S rRNA genes (A01–H03) were identified. Through multiple alignment of E. coli 16S rRNA and our metagenomically retrieved 16S rRNA sequences, it was found that at least 628 (40.7%) of the 1,542 nucleotides were variable, indicating marked mutational robustness of the 16S rRNA. Strikingly, the functional 16S rRNA sequences (except A10 and F02, which were 99.0% identical to E. coli 16S rRNA) obtained in this study showed only 80.9–89.3% identity to E. coli 16S rRNA, which was well below the value reported thus far (Proteus vulgaris 16S rRNA, 94% identity to E. coli 16S rRNA) (19).
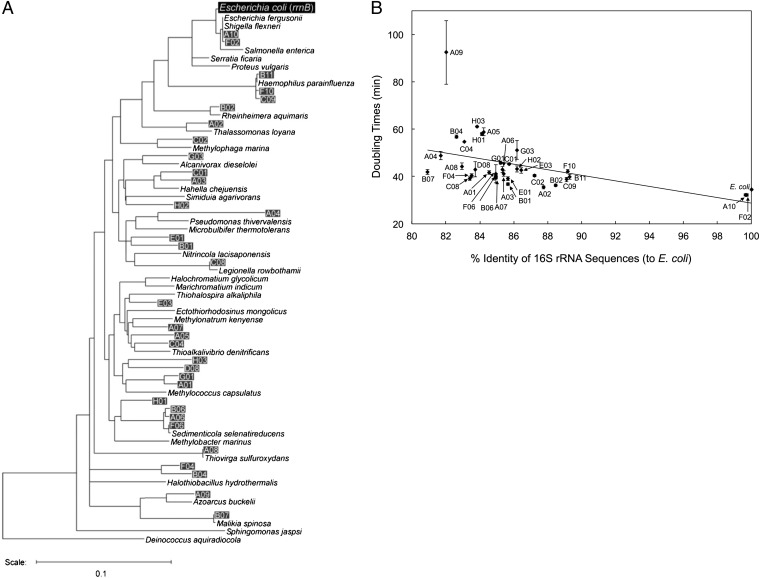
The phylogenetic relationships of these functional 16S rRNA genes, as well as those of several relevant bacteria, are shown in Fig. 1A. Among the 33 clones, 31 were derived from the diverse lineages of Gammaproteobacteria and two clones (B07 and A09) were from Betaproteobacteria. No clones were obtained from other Proteobacteria classes or other non-Proteobacteria phyla, suggesting the presence of a distinct threshold for 16S rRNA functionality.
Fig. 1.
(A) Neighbor-joining phylogenetic tree of the 16S rRNA genes. Environmental 16S rRNA genes that were functional in E. coli KT103 are shown as clone ID (A01–H03) with their closest relatives (as nomenclature). Several other relevant strains are also shown. (B) DTs (in minutes) versus sequence identity (%, compared with the E. coli sequence) of KT103 derivatives harboring foreign 16S rRNA genes. The black slope was fitted to the data. The average increase in DT was calculated to be 4.6 s per point mutation.
The growth phenotypes, defined by the doubling times (DTs) of the mutants, showed a weak negative correlation with sequence identity to the E. coli 16S rRNA gene (Fig. 1B). The DTs of each mutant increased only modestly with decreasing sequence identity; the average increase in DT was 4.6 s per point mutation. The Betaproteobacteria clone B07, whose 16S rRNA sequence showed the most distant relation to E. coli (differing by 274 nucleotides, 80.9% identity), had a DT of 42.0 min, which was still comparable to that of E. coli (34.5 min).
Structural Basis for the Mutational Robustness in 16S rRNA.
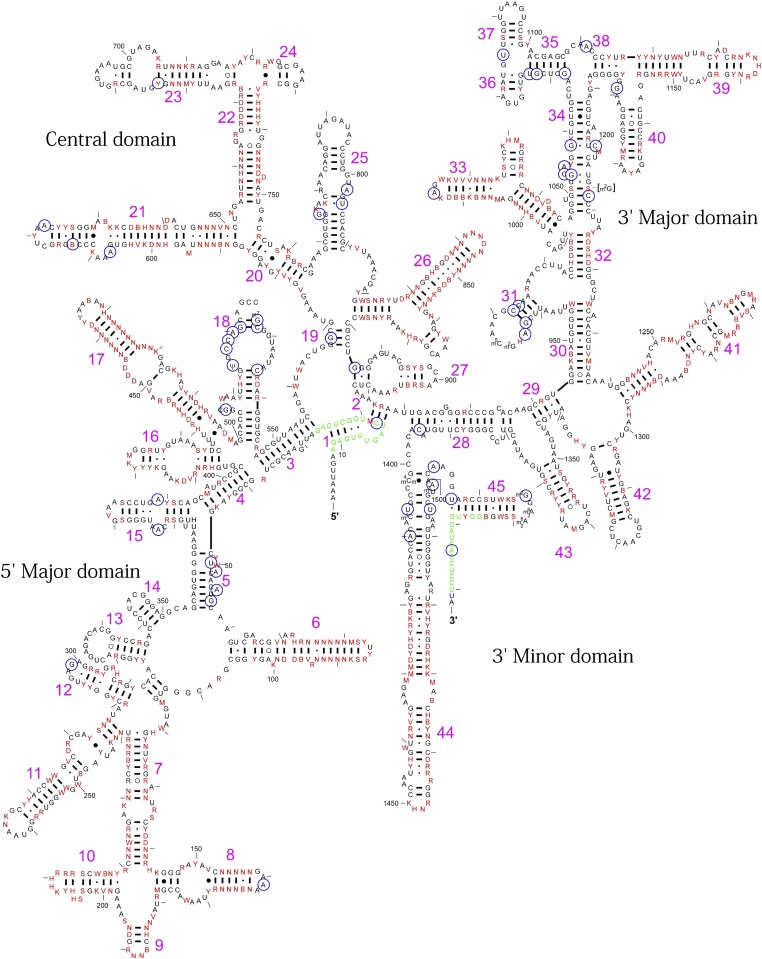
We then questioned why the ribosome was so tolerant to the interspecies exchange of 16S rRNA. From in vitro studies, it has already been observed that 30S and 50S subunits can be exchanged between distantly related species (e.g., E. coli and Bacillus subtilis), suggesting the sufficient compatibility of bridges between subunits (20, 21). Recently, it has also been reported that the B. subtilis 70S ribosome is functional in an E. coli in vitro translation system, suggesting the flexibility of interactions between the ribosome and its interacting partners (e.g., initiation factors, elongation factors, release factors, and tRNAs) (22). In this article, we have therefore mainly focused on analyzing the interactions between 16S rRNA and ribosomal proteins. The 16S rRNA is typically recognized by ribosomal proteins via salt bridges between phosphate oxygen atoms of the RNA backbone and positively charged protein residues, where nucleotide bases are not strictly discriminated (2, 8). Coincidently, all functional 16S rRNAs, irrespective of their identities (80.9–99.7%), retained their secondary structures; their consensus sequence could easily be superimposed onto the secondary structure map of E. coli 16S rRNA (16) (Fig. 2). For example, although the individual sequence of helix (h) 21, which provides a binding site for ribosomal protein S8 (7), varied among functional sequences (0–25.0%) (Fig. S2), compensatory interactions between nucleotide pairs (including both Watson–Crick and noncanonical base pairs) were conserved to form an identical secondary structure (Fig. S3).
Fig. 2.
Variable 16S rRNA nucleotides in E. coli. Thirty-three nonredundant functional 16S rRNA sequences that were obtained using a functional metagenomic approach were aligned, and their consensus sequences were superimposed onto the E. coli secondary structure map. The original E. coli 16S rRNA map was obtained from the Comparative RNA Web site (16). The 628 variable nucleotides are shown in red. Base nomenclature: R can refer to A or G; Y can refer to U or C; W can refer to A or U; K can refer to U or G; B can refer to all bases except A; D can refer to all bases except C; H can refer to all bases except G; V can refer to all bases except U; and N can refer to all bases (A, U, G, or C). The nucleotides in green are the binding sites of the PCR primers—that is, Bac8f at the 5′-end or UN1541r at the 3′-end (14, 18). The nucleotides circled in blue are the invariable nucleotides reported by Mankin’s group (27). The number of RNA helices (h1–h45) is indicated in pink.
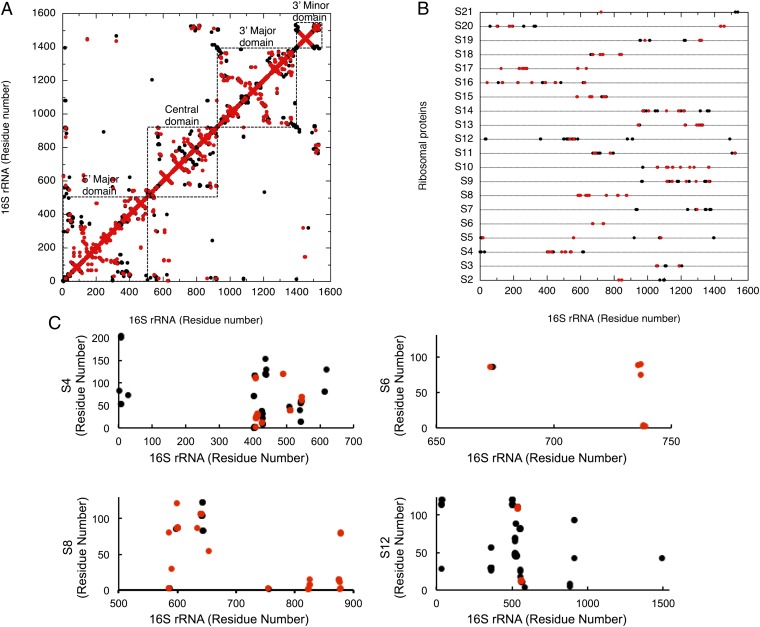
To analyze the contribution of the 628 variable nucleotides to ribosomal structure formation in more detail, we constructed contact maps based on the crystal structure of the E. coli ribosome (PDB code: 3R8O) (23) (Fig. 3). In these maps, all atomic interactions between oxygen, phosphorus, or nitrogen atoms within 3.4 Å were extracted and dotted in black. These interactions included both specific electrostatic interactions and hydrogen bonds. We then overlaid the points for the 628 variable nucleotides (red dots) onto the RNA–RNA (intra-16S rRNA) (Fig. 3A) and RNA–protein (16S rRNA-ribosomal proteins) (Fig. 3 B and C) contact maps. In the RNA–RNA contact map, the interactions of the compensatory nucleotides within the RNA helices were seen as patterns that protruded perpendicular to the diagonal line (Fig. 3A). In this map, 376 nucleotides, representing 79.7% of the total of 472 variable nucleotides, occurred in this protruding pattern, confirming the compensatory and conservative nature of the RNA secondary structure.
Fig. 3.
Variable nucleotides involved in (A) intra-16S rRNA and (B) 16S rRNA–ribosomal protein interactions. Contact maps were constructed based on the crystal structure of the E. coli ribosome (PDB ID code 3R8O) (23). Atomic interactions between oxygen, phosphorus, or nitrogen atoms within 3.4 Å distances were extracted and dotted in black. Contacts that involve variable nucleotides are dotted in red. (C) Detailed two-dimensional contact maps of 16S rRNA and some ribosomal proteins. The dots have the same color as those in A and B. The lengths of the ribosomal proteins S4, S6, S8, and S12 are 206 aa, 131 aa, 130 aa, and 124 aa, respectively.
In contrast, 133 of the total 410 RNA–protein interactions involved variable nucleotides (Fig. 3B). Thus, at least 32.4% of all RNA nucleotides at protein-binding sites are not functionally recognized by ribosomal proteins in a base-specific manner. Specifically, more than half of all interactions involving ribosomal proteins S6 (83.3%), S8 (75.0%), S10 (75.0%), S17 (63.2%), and S2 (50%) did not require base specificity (Fig. 3 B and C). Moreover, ribosomal protein S20, which connects the 5′ and 3′ minor domains of 16S rRNA, was also base-nonspecific at the 3′ minor domain (Fig. 3B). Many residues involved in the binding sites of the ribosomal proteins such as S4 (33.3%), S5 (15.8%), and S12 (8.1%), which are located in the functional center of the 30S subunit (1, 2, 24), were also found to be more or less flexible (Fig. 3 B and C). These data strongly suggest that the correct formation of the secondary structure of 16S rRNA is critical for the establishment of the 30S subunit’s tertiary structure and function; the primary structure of the 16S rRNA itself is less important. This finding is consistent with previous genetic studies (7, 8, 19) but conflicts with the complexity hypothesis (5), which stresses the importance of conserving the primary structure of rRNA.
Overall, we found that the conservation of secondary structures is critical for the functionality of 16S rRNA but with some exceptions; radical changes were found, including some deletions and insertions. Compared with the RNA helices of E. coli, h6, h10, and h17 of H01 were shorter by 7, 15, and 25 bases, respectively, whereas h6 of B01 was longer by 13 bases (Fig. S4A). However, these changes were found at the tips of the RNA helices that were not involved in protein binding but instead were exposed to the surface, providing basis for the exceptional flexibility (Fig. S4B).
Conclusion and Perspectives.
The architecture of the ribosome is one of the most complex and sophisticated among that of biological machineries. The assembly process as well as the assembled form is highly complex. This extreme ribosomal complexity is believed to be the basis for the species specificity in 16S rRNA, although solid experimental evidence is not available in this regard (4, 6). It should be noted that 5S rRNA genes, which have less complex structures, are known to be freely exchangeable in bacteria, at least in vitro (25). Using E. coli as a host, Fox’s group has also shown that 5S rRNA from any Vibrio species, except for Vibrio gazogenes, could be horizontally transferred (26). In this study, through an evolutionary experiment in E. coli, we have tried to clarify the constraints on horizontal gene transfer of 16S rRNA and observed the unexpected plasticity of 16S rRNA. Our finding suggests that ribosomal proteins recognize their binding sites primarily based on the secondary structure of the 16S rRNA. Thus, the establishment of the secondary structure seems to precede the assembly of the ribosome. This base-nonspecific recognition mechanism likely reflects the origin of molecular recognition between RNAs and proteins, which may also apply to the evolution of 23S rRNA. In fact, our preliminary experiment suggested that at least 645 of a total of 2,904 E. coli 23S rRNA bases can undergo mutations. This high mutational robustness should provide the basis for the horizontal transfer of 16S rRNA genes observed in some microorganisms in nature (9–13).
By screening mutations that are deleterious to the functionality of the 16S rRNA, Mankin et al. created a list of conservative nucleotides (27) (Fig. 2). Our approach is complementary to their approach but additionally screens directly for mutable nucleotides, taking advantage of the vast sequence diversity of the metagenome (15). The list of variable nucleotides identified in our functional 16S rRNA sequences is highly valuable for mutational studies of the ribosome as the number, positions, and patterns far exceed those reported thus far. For example, the Ribosomal Mutation Database (http://www.oxfordjournals.org/nar/database/summary/229) contains only 321 variable nucleotides. One direct practical application is the use of our mutant strains to evaluate the probability of the emergence of resistance mutations during the development of novel antimicrobial agents that target the ribosomal 30S subunit.
Materials and Methods
Functional Metagenomic Approach.
E. coli strains.
SQ171 (ΔrrnG ΔrrnA ΔrrnD ΔrrnE ΔrrnH ΔrrnB ΔrrnC/pTRNA67, pKK3535), a Δ7 prrn strain, was kindly provided by the Suzuki laboratory (University of Tokyo), with the permission of Drs. Selwyn Quan (Stanford University, Stanford, CA) and Catherine L. Squires (Tufts University, Medford, MA), who originally constructed the strain (28) and plasmids (19). The plasmid pKK3535 (E. coli rrnB, ampicillin resistant, pBR322 ori) in the SQ171 strain was replaced by pRB101 (E. coli rrnB, sacB, ampicillin resistant, pSC101 ori) to generate strain KT101 (14, 17). KT101 was maintained at 37 °C in 2× YT [1.6% (wt/vol) peptone, 1% (wt/vol) yeast extract, and 0.5% (wt/vol) NaCl] medium supplemented with 50 µg/mL ampicillin (14, 17).
Metagenome samples.
Genomic DNA, extracted from various environments (soils, fermented products, and seawater), was used as the source of the 16S rRNA genes (15). These samples were mixed and served as polymerase chain reaction (PCR) templates. The mixed metagenomes were found to comprise genomes from diverse lineages of bacteria on the basis of the 16S rRNA sequence analysis: 5.4% from Firmicutes, 5.4% from Spirochetes, 2.7% from Alphaproteobacteria, 32.4% from Betaproteobacteria, 24.3% from Gammaproteobacteria, 8.1% from Deltaproteobacteria, and 21.6% from Epsilonproteobacteria.
Cloning and selection.
To amplify the 16S rRNA genes from the environmental metagenome by PCR, we used the KOD-Plus-Ver.2 DNA polymerase (Toyobo) and the primer pairs Bac8f(19A)–UN1541r(1527U) or Bac8f(19C)–UN1541r(1527C) (14, 18). The primer sequences used are as follows: Bac8f(19A), 5′-AGAGTTTGATCATGGCTCAG-3′; UN1541r(1527U), 5′-AAGGAGGTGATCCAACC-3′; Bac8f(19C), 5′-AGAGTTTGATCCTGGCTCAG-3′; and UN1541r(1527C), 5′-AAGGAGGTGATCCAGCC-3′. The resulting amplicons were cloned into pRB103 (containing the entire rrnB operon, pSC101 ori, and Zeocin resistance marker) (14, 17) by using the In-fusion procedure (14), and the products were used directly to transform KT101 (Fig. S1). Transformants were then selected on LB [1% (wt/vol) peptone, 0.5% (wt/vol) yeast extract, and 0.5% (wt/vol) NaCl] agar plates containing 100 µg/mL Zeocin (Invitrogen). At this stage, we identified healthy (2 mm diameter after 16 h), midsized, small, and tiny colonies (the great majority indicating a dominant-negative phenotype) at a ratio of ∼2:1:1:150, suggesting the functional diversity of the cloned environmental 16S rRNAs. We then randomly picked ∼15,000 colonies, suspended them in LB liquid medium, and spotted them onto LB agar plates containing 100 µg/mL Zeocin and 5% (wt/vol) sucrose (14, 17). After this counterselection, we obtained 89 viable KT103 derivatives, which were colony purified. All of the KT103 derivatives obtained could no longer grow on LB agar plates containing 50 µg/mL ampicillin, indicating complete removal of pRB101.
Phylogenetic Analysis.
Plasmid DNA was extracted from the 89 KT103 derivatives, and the entire lengths of the 16S rRNA genes were sequenced. Multiple sequence alignment was carried out using the CLUSTAL W program (http://clustalw.ddbj.nig.ac.jp/top-j.html). A neighbor-joining tree was constructed and drawn using the TreeView software. Because of several near-identical sequences, we excluded all redundant sequences, except one from each group, to yield 33 representative clones. To further characterize the 16S rRNA sequences, we searched for their closest relatives in the database by using the SEQMATCH service supplied by the Ribosomal Database Project at http://rdp.cme.msu.edu/ (29) with the filter setting “isolated type strains, with a length of 1200 bp∼”. Then, we used the TREE BUILDER service to construct the neighbor-joining tree. The 16S rRNA genes of E. coli, S. ficaria, and R. pickettii, together with some other relevant 16S rRNA genes, such as those from Salmonella enterica or P. vulgaris (19), were also included in the analysis. The sequence of Deinococcus aquiradiocola was used as an outgroup.
Growth Phenotype Analysis.
Phenotypes of the KT103 derivatives were quantitatively analyzed from the DT as described previously (8, 14, 17). Briefly, 0.4 µL (0.2%, vol/vol) of overnight preculture was inoculated into 200 µL of 2× YT supplemented with 100 µg/mL Zeocin and 5% (wt/vol) sucrose in a flat-bottom 96-well microplate. The microplate was incubated at 37 °C with vigorous agitation in a VersaMax plate reader (Molecular Devices), and the absorbance at 600 nm (A600) was monitored every 15 min. The average DT was calculated from at least five independent cultures.
Calculation of Atomic Contacts in the 30S Ribosome.
Interatomic contacts within 16S rRNA and between 16S rRNA and ribosomal proteins were determined with the CONTACT software (30) by using the atomic coordinates of the E. coli 30S ribosome (PDB ID code, 3R8O) (23). The contacts were defined with an interatomic distance of <3.4 Å. To sort out specific interactions (i.e., electrostatic and hydrogen-bonding interactions), carbon atoms were excluded from the calculation.
Supplementary Material
Acknowledgments
We thank Drs. Selwyn Quan, Catherine Squires, and Tsutomu Suzuki for providing strains and plasmids. This work was partly supported by the Japan Society for the Promotion of Science (JSPS) Grant-in-Aid for Scientific Research (B) 23380197 (to K.M.), Grant-in-Aid for Scientific Research on Innovative Areas 24119515 to (K.M.), and Grant-in-Aid for Challenging Exploratory Research 24651231(to K.M.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The sequences reported in this paper have been deposited in the DNA Data Bank of Japan (DDBJ) database, http://www.ddbj.nig.ac.jp (accession nos. AB597523–AB597555).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1213609109/-/DCSupplemental.
References
- 1.Schuwirth BS, et al. Structures of the bacterial ribosome at 3.5 A resolution. Science. 2005;310(5749):827–834. doi: 10.1126/science.1117230. [DOI] [PubMed] [Google Scholar]
- 2.Brodersen DE, Clemons WM, Jr, Carter AP, Wimberly BT, Ramakrishnan V. Crystal structure of the 30 S ribosomal subunit from Thermus thermophilus: structure of the proteins and their interactions with 16 S RNA. J Mol Biol. 2002;316(3):725–768. doi: 10.1006/jmbi.2001.5359. [DOI] [PubMed] [Google Scholar]
- 3.Klein DJ, Moore PB, Steitz TA. The roles of ribosomal proteins in the structure assembly, and evolution of the large ribosomal subunit. J Mol Biol. 2004;340(1):141–177. doi: 10.1016/j.jmb.2004.03.076. [DOI] [PubMed] [Google Scholar]
- 4.Woese CR. Bacterial evolution. Microbiol Rev. 1987;51(2):221–271. doi: 10.1128/mr.51.2.221-271.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jain R, Rivera MC, Lake JA. Horizontal gene transfer among genomes: The complexity hypothesis. Proc Natl Acad Sci USA. 1999;96(7):3801–3806. doi: 10.1073/pnas.96.7.3801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Woese CR, Kandler O, Wheelis ML. Towards a natural system of organisms: Proposal for the domains Archaea, Bacteria, and Eucarya. Proc Natl Acad Sci USA. 1990;87(12):4576–4579. doi: 10.1073/pnas.87.12.4576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Moine H, Squires CL, Ehresmann B, Ehresmann C. In vivo selection of functional ribosomes with variations in the rRNA-binding site of Escherichia coli ribosomal protein S8: Evolutionary implications. Proc Natl Acad Sci USA. 2000;97(2):605–610. doi: 10.1073/pnas.97.2.605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kitahara K, Kajiura A, Sato NS, Suzuki T. Functional genetic selection of Helix 66 in Escherichia coli 23S rRNA identified the eukaryotic-binding sequence for ribosomal protein L2. Nucleic Acids Res. 2007;35(12):4018–4029. doi: 10.1093/nar/gkm356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang Y, Zhang Z. Comparative sequence analyses reveal frequent occurrence of short segments containing an abnormally high number of non-random base variations in bacterial rRNA genes. Microbiology. 2000;146(Pt 11):2845–2854. doi: 10.1099/00221287-146-11-2845. [DOI] [PubMed] [Google Scholar]
- 10.Schouls LM, Schot CS, Jacobs JA. Horizontal transfer of segments of the 16S rRNA genes between species of the Streptococcus anginosus group. J Bacteriol. 2003;185(24):7241–7246. doi: 10.1128/JB.185.24.7241-7246.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Acinas SG, Marcelino LA, Klepac-Ceraj V, Polz MF. Divergence and redundancy of 16S rRNA sequences in genomes with multiple rrn operons. J Bacteriol. 2004;186(9):2629–2635. doi: 10.1128/JB.186.9.2629-2635.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Eardly BD, Nour SM, van Berkum P, Selander RK. Rhizobial 16S rRNA and dnaK genes: Mosaicism and the uncertain phylogenetic placement of Rhizobium galegae. Appl Environ Microbiol. 2005;71(3):1328–1335. doi: 10.1128/AEM.71.3.1328-1335.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Miller SR, et al. Discovery of a free-living chlorophyll d-producing cyanobacterium with a hybrid proteobacterial/cyanobacterial small-subunit rRNA gene. Proc Natl Acad Sci USA. 2005;102(3):850–855. doi: 10.1073/pnas.0405667102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kitahara K, Miyazaki K. Specific inhibition of bacterial RNase T2 by helix 41 of 16S ribosomal RNA. Nat Commun. 2011;2:549. doi: 10.1038/ncomms1553. [DOI] [PubMed] [Google Scholar]
- 15.Uchiyama T, Miyazaki K. Functional metagenomics for enzyme discovery: Challenges to efficient screening. Curr Opin Biotechnol. 2009;20(6):616–622. doi: 10.1016/j.copbio.2009.09.010. [DOI] [PubMed] [Google Scholar]
- 16.Cannone JJ, et al. The comparative RNA web (CRW) site: An online database of comparative sequence and structure information for ribosomal, intron, and other RNAs. BMC Bioinformatics. 2002;3:2. doi: 10.1186/1471-2105-3-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kitahara K, Suzuki T. The ordered transcription of RNA domains is not essential for ribosome biogenesis in Escherichia coli. Mol Cell. 2009;34(6):760–766. doi: 10.1016/j.molcel.2009.05.014. [DOI] [PubMed] [Google Scholar]
- 18.Weisburg WG, Barns SM, Pelletier DA, Lane DJ. 16S ribosomal DNA amplification for phylogenetic study. J Bacteriol. 1991;173(2):697–703. doi: 10.1128/jb.173.2.697-703.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Asai T, Zaporojets D, Squires C, Squires CL. An Escherichia coli strain with all chromosomal rRNA operons inactivated: Complete exchange of rRNA genes between bacteria. Proc Natl Acad Sci USA. 1999;96(5):1971–1976. doi: 10.1073/pnas.96.5.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Takeda M, Lipmann F. Comparison of amino acid polymerization in B. Subtilis and E. coli cell-free systems; hybridization of their ribosomes. Proc Natl Acad Sci USA. 1966;56(6):1875–1882. doi: 10.1073/pnas.56.6.1875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chang FN, Sih CJ, Weisblum B. Lincomycin, an inhibitor of aminoacyl sRNA binding to ribosomes. Proc Natl Acad Sci USA. 1966;55(2):431–438. doi: 10.1073/pnas.55.2.431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chiba S, et al. Recruitment of a species-specific translational arrest module to monitor different cellular processes. Proc Natl Acad Sci USA. 2011;108(15):6073–6078. doi: 10.1073/pnas.1018343108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dunkle JA, et al. Structures of the bacterial ribosome in classical and hybrid states of tRNA binding. Science. 2011;332(6032):981–984. doi: 10.1126/science.1202692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ogle JM, Ramakrishnan V. Structural insights into translational fidelity. Annu Rev Biochem. 2005;74:129–177. doi: 10.1146/annurev.biochem.74.061903.155440. [DOI] [PubMed] [Google Scholar]
- 25.Wrede P, Erdmann VA. Activities of B. stearothermophilus 50 S ribosomes reconstituted with prokaryotic and eukaryotic 5 S RNA. FEBS Lett. 1973;33(3):315–319. doi: 10.1016/0014-5793(73)80219-4. [DOI] [PubMed] [Google Scholar]
- 26.Lee YH, Dsouza L, Fox GE. Experimental investigation of an RNA sequence space. Orig Life Evol Biosph. 1993;23(5–6):365–372. doi: 10.1007/BF01582086. [DOI] [PubMed] [Google Scholar]
- 27.Yassin A, Fredrick K, Mankin AS. Deleterious mutations in small subunit ribosomal RNA identify functional sites and potential targets for antibiotics. Proc Natl Acad Sci USA. 2005;102(46):16620–16625. doi: 10.1073/pnas.0508444102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cruz-Vera LR, Rajagopal S, Squires C, Yanofsky C. Features of ribosome-peptidyl-tRNA interactions essential for tryptophan induction of tna operon expression. Mol Cell. 2005;19(3):333–343. doi: 10.1016/j.molcel.2005.06.013. [DOI] [PubMed] [Google Scholar]
- 29.Cole JR, et al. The Ribosomal Database Project: Improved alignments and new tools for rRNA analysis. Nucleic Acids Res. 2009;37(Database Issue):D141–D145. doi: 10.1093/nar/gkn879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Collaborative Computational Project, Number 4 The CCP4 suite: Programs for protein crystallography. Acta Crystallogr D Biol Crystallogr. 1994;50(Pt 5):760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.