Abstract
Acute myeloid leukemia (AML) is a heterogeneous group of hematopoietic malignancies with variable response to treatment. AMLs bearing MLL (mixed lineage leukemia) rearrangements are associated with intermediate or poor survival. MicroRNAs (miRNAs), a class of small noncoding RNAs, have been postulated to be important gene expression regulators virtually in all biological processes, including leukemogenesis. Through a large-scale, genome-wide miRNA expression profiling assay of 85 human AML and 15 normal control samples, we show that among 48 miRNAs that are significantly differentially expressed between MLL- and non–MLL-rearranged AML samples, only one (miR-495) is expressed at a lower level in MLL-rearranged AML than in non–MLL-rearranged AML; meanwhile, miR-495 is also significantly down-regulated in MLL-rearranged AML samples compared with normal control samples. Through in vitro colony-forming/replating assays and in vivo bone marrow transplantation studies, we show that forced expression of miR-495 significantly inhibits MLL-fusion-mediated cell transformation in vitro and leukemogenesis in vivo. In human leukemic cells carrying MLL rearrangements, ectopic expression of miR-495 greatly inhibits cell viability and increases cell apoptosis. Furthermore, our studies demonstrate that PBX3 and MEIS1 are two direct target genes of miR-495, and forced expression of either of them can reverse the effects of miR-495 overexpression on inhibiting cell viability and promoting apoptosis of human MLL-rearranged leukemic cells. Thus, our data indicate that miR-495 likely functions as a tumor suppressor in AML with MLL rearrangements by targeting essential leukemia-related genes.
Leukemia arises as a result of genetic lesions that cause uncontrolled proliferation in cells of the hematopoietic lineage (1, 2). Chromosome translocations are frequently observed in both acute myeloid leukemia (AML) (2–4) and other hematologic malignancies. The MLL (mixed lineage leukemia; HRX, ALL-1, Htrx) gene, located at 11q23, is frequently involved in chromosome translocations with more than 60 different partner genes (5–8). The critical feature of these chromosomal rearrangements is the generation of an in-frame fusion transcript consisting of 5′ MLL and 3′ sequences of the gene on the partner chromosome (7, 8). MLL-rearranged leukemia occurs in approximately 10% of patients with de novo or treatment-related acute leukemia (9–11). MLL-rearranged leukemia is classified as a disease with an intermediate or poor risk of prognosis (1, 12).
The most well-studied downstream target genes of MLL fusion proteins are HOXA9 and MEIS1 (13–15), and their aberrant overexpression has been shown to be required for the induction and maintenance of MLL-rearranged leukemia (16–21). HOX proteins can form heterodimers or heterotrimers with members of the three-amino-acid loop extension family of cofactors, including PBX and MEIS proteins, to regulate the transcription of multiple downstream targets directly (22–24). Similar to HOXA9 and MEIS1, overexpression of PBX3 has also been frequently observed in various subtypes of AML with unfavorable prognosis, particularly in MLL-rearranged leukemia (13, 18, 25–28). Our recent study showed that PBX3 also functions as an oncogene in MLL-rearranged leukemia (29). Thus, both MEIS1 and PBX3 likely function as essential cofactors of HOXA9 and play critical oncogenic roles in the pathogenesis of MLL-rearranged leukemia.
MicroRNAs (miRNAs) are a class of small, noncoding RNAs that are important for posttranscriptional gene regulation in both health and disease (30, 31). Although aberrant expression of many miRNAs has been observed in various subtypes of AML (8, 29, 32–40), the biological functions of most of them in leukemogenesis have not been characterized.
In the present study, we show first that a miRNA, namely miR-495, is the only miRNA that is significantly down-regulated in MLL-rearranged AML compared with both non–MLL-rearranged AML and normal controls. We then used both in vitro and in vivo models to study the biological function of miR-495 and to identify its critical target genes in MLL-rearranged leukemia.
Results
Expression of miR-495 Is Significantly Down-Regulated in MLL-Rearranged AML.
We and others previously showed that a number of miRNAs, such as miR-196b and the miR-17-92 cluster, were aberrantly overexpressed in MLL-rearranged AML compared with normal controls, and these miRNAs likely play important oncogenic roles in MLL-fusion–mediated cell transformation and leukemogenesis (32, 39, 41, 42). To identify potential tumor-suppressor miRNAs in MLL-rearranged AML, we performed a large-scale, genome-wide miRNA expression profiling assay of 85 AML (including 10 MLL- and 75 non–MLL-rearranged AML) and 15 normal control bone marrow (BM) [including 6 CD34+ hematopoietic stem/progenitor cell, 5 CD33+ myeloid progenitor cell, and 4 mononuclear cell (MNC)] samples by use of Exiqon miRNA arrays (Methods).
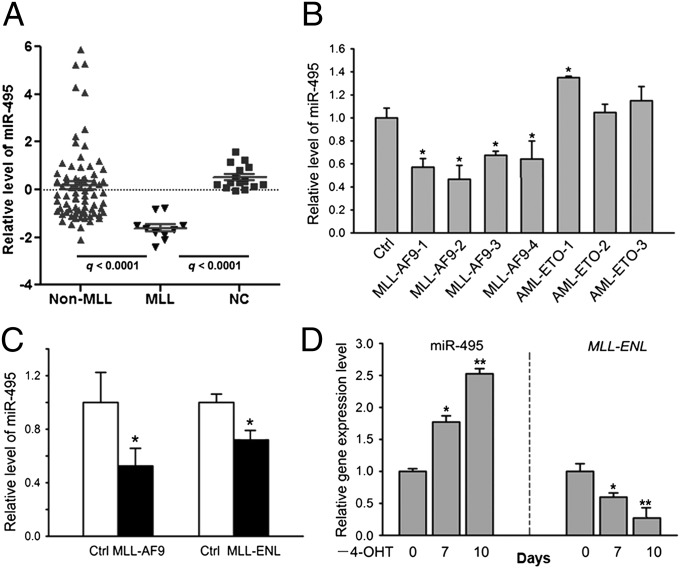
Using significance analysis of microarrays (SAM) (43), we identified 48 miRNAs that were significantly differentially expressed between the MLL-rearranged (n = 10) and non–MLL-rearranged (n = 75) AML samples; very strikingly, 47 of them (including miR-196b and the individual miRNAs of the miR-17-92 cluster) had a significantly higher expression level, whereas only one (miR-495) had a significantly lower expression level in MLL-rearranged AML than in non–MLL-rearranged AML [q < 0.05, false discover rate (FDR) < 0.05; Fig. 1A and Fig. S1]. We next compared miRNA expression between MLL-rearranged AML and normal controls and found that 44 miRNAs (including miR-196b and the individual miRNAs of the miR-17-92 cluster) had a significantly higher expression level, whereas 32 miRNAs (including miR-495) had a significantly lower level of expression in MLL-rearranged AML than in the normal controls (q < 0.05, FDR < 0.05; Fig. 1A and Fig. S2). Thus, miR-495 is the only miRNA that is expressed at a significantly lower level in MLL-rearranged AML compared with both non–MLL-rearranged AML and normal controls.
Fig. 1.
miR-495 is significantly down-regulated in human MLL-rearranged AML compared with non–MLL-rearranged AML or normal controls. (A) Relative expression levels of miR-495 in 10 human MLL-rearranged AML (MLL), 75 non–MLL-rearranged AML (non-MLL), and 15 normal control (NC, including 6 CD34+ hematopoietic stem/progenitor cell, 5 CD33+ myeloid progenitor cell, and 4 MNC) samples, as detected by Exiqon miRNA microarray assays. (B) Quantitative PCR (qPCR) analysis of miR-495 expression level in human cord blood CD34+ cells that were retrovirally transduced with MSCV-MLL-AF9, MSCV-AML-ETO, or empty vector (Ctrl). (C) Down-regulation of miR-495 in MLL-AF9 or MLL-ENL–transduced mouse BM progenitor cells. (D) Withdrawal of 4-OHT (days 0, 7, and 10 are shown) results in a significant increase of miR-495 expression but a significant decrease of MLL-ENL expression in MLL-ENL-ERtm cells. *P < 0.05; **P < 0.01, two-tailed t test.
miR-495 Is Down-Regulated by MLL-Fusion Proteins.
To determine whether MLL fusions directly regulate the level of miR-495, we retrovirally transduced MLL-AF9, a fusion gene resulting from a common translocation between chromosome 9 and 11 (44), into human normal CD34+ cord blood stem cells (45) and observed that the expression level of miR-495 was significantly (P < 0.05) down-regulated by forced expression of MLL-AF9 (Fig. 1B). Similarly, a significant down-regulation (P < 0.05) of miR-495 was also observed in mouse normal BM progenitor cells after retroviral transduction of MSCVneo-MLL-AF9 or MSCVneo-MLL-ENL (Fig. 1C). We also used the MLL-ENL-ERtm cell line, a cell line stably expressing the conditional MLL-ENL derivative (13, 46), to investigate the dependence of miR-495 expressional repression on the presence of MLL fusion proteins further. As shown in Fig. 1D, after withdrawal of 4-hydroxytamoxifen (4-OHT) for 7–10 d, miR-495 expression was significantly increased (P < 0.05), along with a diminishing of MLL-ENL expression.
miR-495 Functions as a Tumor-Suppressor miRNA in Vitro.
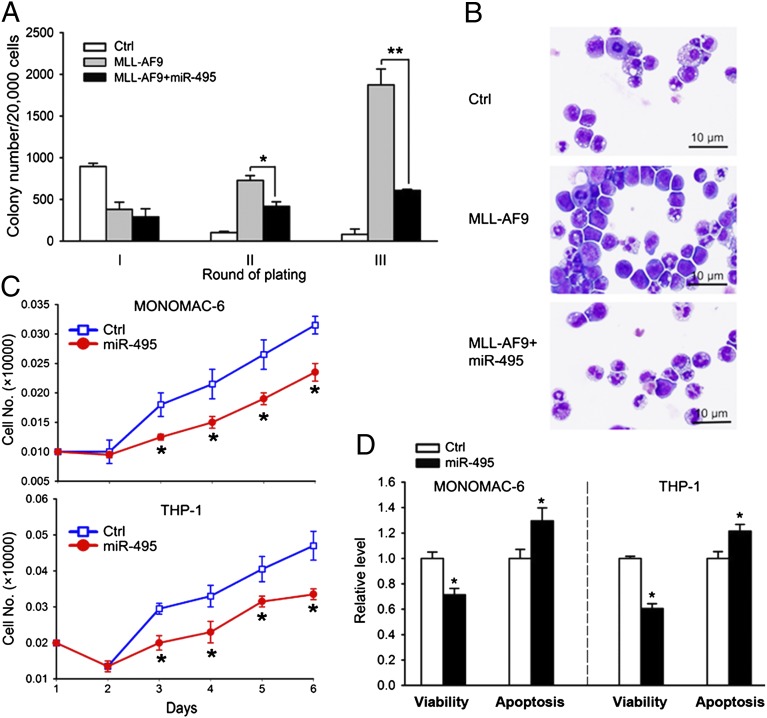
To investigate the biological function of miR-495, we performed a colony-forming/replating assay. Normal mouse BM progenitor cells were transduced with MSCV-PIG [empty vector bearing a phosphoglycerate kinase (PGK)-puromycin-internal ribosome entry site (IRES)-GFP cassette (47) as a negative control] or MSCV-PIG-miR-495, together with MSCVneo or MSCVneo-MLL-AF9, and were then plated onto methylcellulose medium. The colonies were replated every 7 d under the same conditions. As shown in Fig. 2A, forced expression of miR-495 significantly inhibited (P < 0.05) the colony-forming capacity induced by MLL-AF9 after replating (i.e., in the second and third rounds of plating). Notably, forced expression of miR-495 substantially promoted cell differentiation as shown by cytospin analysis (Fig. 2B). We next investigated the biological function of miR-495 in human MLL-rearranged leukemia cells. As shown in Fig. 2C, overexpression of miR-495 consistently inhibited the growth/proliferation of MONOMAC-6/t (9, 11) and THP-1/t (9, 11) cells from day 3 after transfection. Forced expression of miR-495 also significantly inhibited cell viability and promoted apoptosis in MONOMAC-6 and THP-1 cells (Fig. 2D). These results indicate that miR-495 has an inhibitory effect on leukemia cell growth in vitro.
Fig. 2.
miR-495 functions as a tumor-suppressor miRNA in vitro. (A) miR-495 inhibits MLL-AF9–induced cell transformation of normal mouse BM progenitor cells. Normal mouse BM cells were retrovirally transduced with MSCVneo+MSCV-PIG (as control), MSCVneo-MLL-AF9+MSCV-PIG (MLL-AF9), or MSCVneo-MLL-AF9+MSCV-PIG-miR-495 (MLL-AF9+miR-495), and colony forming/replating assays were done thereafter. (B) miR-495 promotes the differentiation of MLL-AF9–transduced mouse BM progenitor cells. Colony-forming cells were collected for cytospin analysis from the secondary round of replating. (C) Ectopic expression of miR-495 inhibits cell growth/proliferation of MONOMAC-6 and THP-1 cells. Cells were transfected with MSCV-PIG (Ctrl) or MSCV-PIG-miR-495 (miR-495). Cell numbers were counted every day after transfection for 6 d. (D) Forced expression of miR-495 decreases cell viability and promotes apoptosis of MONOMAC-6 and THP-1 cells. Cells were transfected with MSCV-PIG (Ctrl) or MSCV-PIG-miR-495 (miR-495), and cell viability and apoptosis were assessed 48 h after transfection. *P < 0.05; **P < 0.01, two-tailed t test.
miR-495 Inhibits MLL-Fusion–Mediated Leukemogenesis in Vivo.
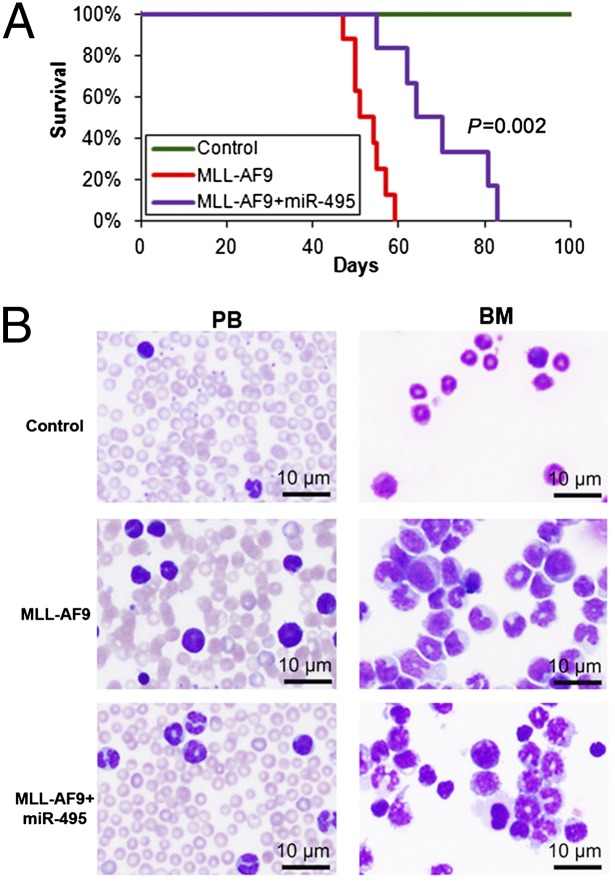
To assess the in vivo function of miR-495 in leukemogenesis, we retrovirally cotransduced MSCV-PIG or MSCV-PIG-miR-495 together with MSCVneo or MSCVneo-MLL-AF9 into normal BM progenitor cells isolated from B6.SJL donor mice (CD45.1; the mice were killed 5 d after 5-fluorouracil treatment) and then plated onto methylcellulose medium. After 7 d of selection of double-transduction-positive cells with puromycin and G418, we collected and washed the colony cells and then transplanted the cells into lethally irradiated C57BL/6 recipient mice. We found that forced expression of miR-495 significantly delayed leukemogenesis mediated by MLL-AF9 (median overall survival, 67 d vs. 52 d; P = 0.002) (Fig. 3A). Forced expression of miR-495 remarkably reduced the proportion of immature blast cells in both peripheral blood and BM (Fig. 3B). These findings suggest that miR-495 does play a tumor-suppressor role in MLL-rearranged leukemia.
Fig. 3.
miR-495 inhibits MLL-fusion–mediated leukemogenesis in vivo. (A) Mouse BM transplantation assay of the control group (Control; MSCVneo+MSCV-PIG; n = 5), the MLL-AF9 group (MSCVneo-MLL-AF9+MSCV-PIG; n = 8), and the MLL-AF9+miR-495 group (MSCVneo-MLL-AF9+MSCV-PIG-miR495; n = 6). The MLL-AF9+miR-495 group developed leukemia significantly (P = 0.002, log–rank test) slower than the MLL-AF9 alone group. (B) miR-495 increased cell differentiation in both peripheral blood (PB) and BM of the recipient mice.
Identification of Potential Target Genes of miR-495.
To identify potential target genes of miR-495, we have also performed (protein-coding) gene expression profiling of 79 of the above 100 human samples, including 70 AML (composed of 9 MLL- and 61 non-MLL-rearranged AML cases) and 9 normal controls (composed of 3 CD34+, 2 CD33+, and 4 MNC samples) by use of an Agilent custom-design microarray platform (Methods). Through correlation of expression of miR-495 with that of genes across the 79 samples, we found that 471 genes exhibited a significantly inverse correlation of expression with miR-495 (r < −0.2, P < 0.05, Pearson correlation). Of the 471 genes, 128 are predicted target genes of miR-495 in both human and mouse genomes (Table S1). Through Gene Ontology analysis, we found that these candidate target genes are significantly enriched in biological process categories such as “response to stimulus,” “signal transduction,” “cell activation,” “chromatin modification,” and “hemopoietic or lymphoid organ development” (Fig. S3). Through gene set enrichment analysis (48), we show that these genes are significantly enriched in gene sets that are up-regulated in leukemic stem cells or normal hematopoietic stem cells, in those that are up-regulated in MLL/t(11q23)-associated AML, and in those that are potential direct targets of MAZ, LEF1, and SP1, three stem-cell-self-renewal-related genes (Table S2).
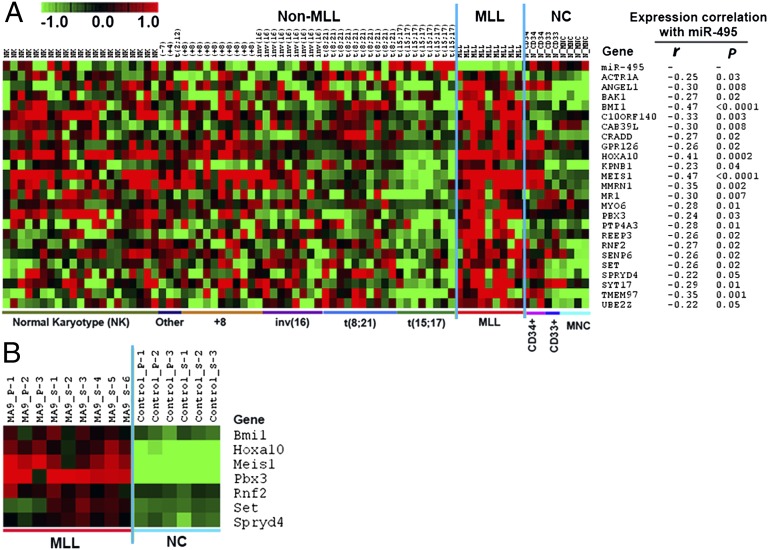
Because we are interested in MLL-rearranged AML, we next narrowed down the number of candidate target genes of miR-495 from 128 to 24 (i.e., ACTR1A, ANGEL1, BAK1, BMI1, C10ORF140, CAB39L, CRADD, GPR126, HOXA10, KPNB1, MEIS1, MMRN1, MR1, MYO6, PBX3, PTP4A3, REEP3, RNF2, SENP6, SET, SPRYD4, SYT17, TMEM97, and UBE2Z), because these genes are expressed at a significantly higher level in the 9 human MLL-rearranged leukemia samples compared with both the 9 normal controls and the 61 non–MLL-rearranged AML samples, in an inverse relationship to miR-495 (Fig. 4A). Furthermore, in the analysis of Affymetrix gene arrays of 9 MLL-AF9–mediated mouse leukemia samples and 6 control samples (39), we observed that 7 (i.e., Bmi1, Hoxa10, Meis1 , Pbx3 , Rnf2 , Set , and Spryd4) of the above 24 candidate target genes were significantly overexpressed (q < 0.05; FDR < 0.01; SAM) in MLL-AF9 mouse leukemia samples relative to normal controls (Fig. 4B). Thus, these seven genes are the highly possible target genes of miR-495 in MLL-rearranged leukemia.
Fig. 4.
Expression profiles of candidate target genes of miR-495. (A) Expression profiles of the 24 candidate target genes of miR-495 and their expression correlation with miR-495 in the set of 79 human samples (including 9 MLL-rearranged AML, 61 nonMLL-rearranged AML, and 9 normal control). r, correlation coefficient; P, P value. Pearson correlation was applied to analyze the correlation. (B) Expression profiles of the seven potential target genes of miR-495 in the set of 15 mouse BM cell samples collected from mouse BM transplantation assays, which include 9 MLL-AF9 mouse leukemic BM cell samples (3 from primary BM transplantation and 6 from secondary BM transplantation recipient mice) and 6 normal control BM cell samples (3 each from primary and secondary BM transplantation recipient mice). MA9, MLL-AF9; _P, primary transplantation recipient mouse; _S, secondary transplantation recipient mouse. Expression data were mean centered, and the relative value for each sample is represented by a color, with red representing a high expression and green representing a low expression (scale shown at upper left).
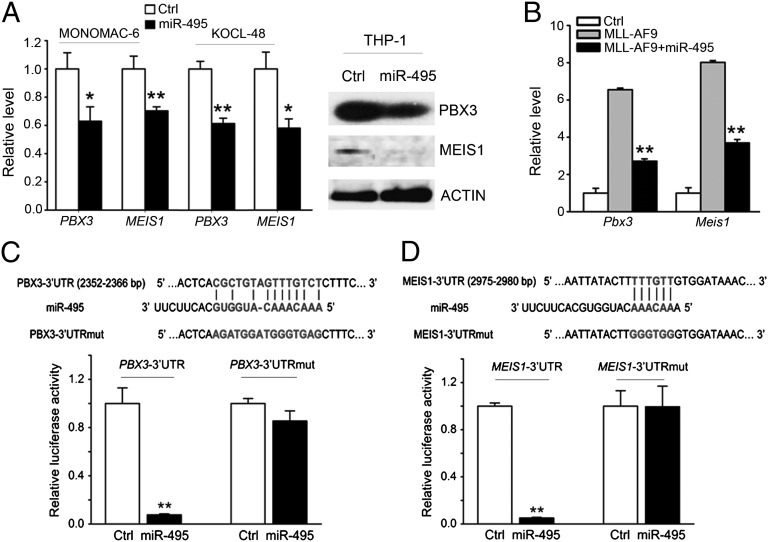
Both PBX3 and MEIS1 Are Direct Targets of miR-495.
As shown in Fig. 4B, PBX3 and MEIS1 are the two potential target genes of miR-495 that exhibited the most significant up-regulation in MLL-AF9 leukemic BM cells relative to the normal control BM cells. Moreover, these two genes have proven to be broadly involved in multiple processes of hematopoiesis and leukemogenesis, as cofactors of HOX genes (13, 16, 18, 22, 23, 49, 50). Therefore, we sought to determine whether PBX3 and MEIS1 are genuine direct target genes of miR-495. As expected, we observed that forced expression of miR-495 significantly reduced (P < 0.05) the endogenous expression of PBX3 and MEIS1 in human MLL-rearranged leukemic cell lines, including MONOMAC-6, KOCL-48/t (4, 11), and THP-1 (Fig. 5A). Similarly, MLL-AF9 transduction resulted in a six- to eightfold increase in endogenous expression of PBX3 and MEIS1 in mouse BM progenitor cells, whereas coexpression of miR-495 reduced their levels to approximately 50% (Fig. 5B). Finally, our luciferase reporter and mutagenesis assays showed that miR-495 targeted the 3′ UTR of both PBX3 and MEIS1 directly (Fig. 5 C and D). Thus, several lines of evidence indicate that both PBX3 and MEIS1 are direct target genes of miR-495.
Fig. 5.
PBX3 and MEIS1 are direct target genes of miR-495. (A) Ectopic expression of miR-495 significantly (P < 0.05) represses endogenous expression of PBX3 and MEIS1 in MLL-rearranged AML cells. The cells were transfected with MSCV-PIG (control) or MSCV-PIG-miR-495, and then the effect of miR-495 overexpression was analyzed 48 h after transfection at both mRNA (Left; detected by qPCR, and the GAPDH expression level was used for normalization) and the protein (Right; detected by Western blot) levels. (B) Inhibitory effect of miR-495 on the endogenous expression of Pbx3 and Meis1 in BM cells of the BM transplantation recipient mice shown in Fig. 3A. Gene levels were normalized to the level of endogenous Gapdh. (C and D) miR-495 directly targets PBX3 (C) and MEIS1 (D) as detected by luciferase reporter and mutagenesis assays. In HEK293T cells, plasmids encoding the wild-type 3′ UTR of PBX3 or MEIS1 (namely, PBX3/MEIS1-3′UTR) or the mutant 3′ UTR in which the predicted miR-495 binding site was mutated (namely, PBX3/MEIS1-3′UTRmut), together with MSCV-PIG or MSCV-PIG-miR-495, were cotransfected with β-gal reporter control vector. Luciferase reporter assays were done 48 h after transfection. Forced expression of miR-495 could significantly repress luciferase activity of the reporter gene bearing the 3′ UTR of PBX3 or MEIS1 in human 293T cells, whereas mutation at the predicted target site in the 3′ UTR abrogated the repression. The normalized luciferase activities represent the firefly: β-gal ratios normalized to the control sample. Error bars present SD obtained from three independent experiments. *P < 0.05; **P < 0.01, two-tailed t test.
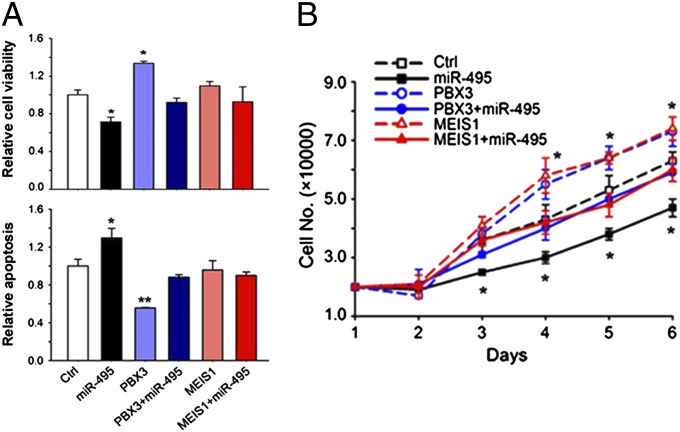
Forced Expression of Either PBX3 or MEIS1 Can Reverse the Effects of miR-495 in Human MLL-Rearranged Leukemic Cells.
We then investigated whether PBX3 and MEIS1 are truly functionally important target genes of miR-495 in MLL-rearranged leukemic cells. We found that forced expression of PBX3 alone could significantly (P < 0.05) increase cell viability and promote cell growth/proliferation, whereas decreasing apoptosis of MONOMAC-6 cells, in a manner opposite to miR-495; forced expression of MEIS1 alone could also significantly promote cell growth/proliferation, although its forced expression showed no significant effect on cell viability and apoptosis (Fig. 6). More importantly, cotransfection of PBX3 or MEIS1 with miR-495 could completely reverse the effects of miR-495 on cell viability, apoptosis, and cell growth in MONOMAC-6 cells (Fig. 6). A similar phenomenon was also observed in THP-1, a different human leukemia cell line with MLL rearrangements (Fig. S4). Therefore, our data suggest that both PBX3 and MEIS1 are functionally important target genes of miR-495 in MLL-rearranged leukemic cells.
Fig. 6.
Both PBX3 and MEIS1 are functionally important target genes of miR-495 in MLL-rearranged leukemic cells. (A) Analysis of the effects of forced expression of miR-495 (MSCV-PIG-miR-495+MSCVneo), PBX3 (MSCV-PIG+MSCVneo-PBX3), PBX3+miR-495 (MSCV-PIG-miR-495+MSCVneo-PBX3), MEIS1 (MSCV-PIG+MSCVneo-MEIS1), and MEIS1+miR-495 (MSCV-PIG-miR-495+MSCVneo-MEIS1), respectively, on cell viability (Upper) and apoptosis (Lower) of MONOMAC-6 cells. Cell viability and apoptosis were detected 48 h after transfection. (B) Analysis of their effects on cell growth/proliferation of MONOMAC-6 cells. Cell numbers were counted every day after transfection for 6 d. The coding regions (CDS) of PBX3 and MEIS1 were cloned into MSCVneo, and thus their ectopic expression would not be repressed by endogenous or cotransfected miR-495. The cells transfected with MSCV-PIG+MSCVneo (Ctrl) were used as controls. *P < 0.05; **P < 0.01, two-tailed t test.
Discussion
In the present study, we identified only one miRNA (miR-495) that was expressed at a significantly lower level in human MLL-rearranged AML (n = 10) than in non–MLL-rearranged AML (n = 75); in contrast, 47 miRNAs were expressed at a significantly higher level in the former than in the latter (Fig. S1). This observation is in accordance with the notion that MLL fusion proteins usually promote expression of downstream target genes at the transcriptional level (13–15, 32, 39, 41, 42). Compared with human normal hematopoietic cell controls, miR-495 (along with 31 other miRNAs) is significantly down-regulated in MLL-rearranged AML (Fig. S2). We then showed that ectopic expression of MLL fusion genes in both human and mouse normal hematopoietic stem/progenitor cells could significantly down-regulate endogenous expression of miR-495 and that the depletion of MLL fusions resulted in the up-regulation of miR-495 (Fig. 1). Thus, our data suggest that there is an MLL-fusion–mediated negative regulation of the production of miR-495 in hematopoietic cells.
Thus far, although the research on the role of miR-495 in cancer is still in its infancy, miR-495 has been implicated in both oncogenic (in breast cancer) and tumor suppressor (in gastric cancer) roles in solid tumors (51, 52). Here we show that miR-495 functions as a critical tumor suppressor in MLL-rearranged leukemia. First of all, we demonstrated that forced expression of miR-495 could significantly inhibit colony-forming capacity of mouse normal BM progenitor cells mediated by MLL fusions (Fig. 2A) and induce cell differentiation (Fig. 2B). In human leukemic cells with MLL rearrangements (e.g., MONOMAC-6 and THP-1 cells), we found that ectopic expression of miR-495 could significantly inhibit cell growth/proliferation (Fig. 2C) and increase apoptosis while decreasing cell viability (Fig. 2D). Furthermore, we also performed mouse BM transplantation assays and showed that forced expression of miR-495 could significantly inhibit MLL-AF9–mediated leukemogenesis in transplanted mice (Fig. 3).
Through a series of correlation analyses, we identified 24 candidate target genes of miR-495 that exhibited a significantly inverse correlation of expression with miR-495 in 79 human samples (including 9 MLL-rearranged and 61 non–MLL-rearranged AML samples, along with 9 normal control samples) and were expressed at a significantly higher level in the MLL-rearranged AML samples compared with both the non–MLL-rearranged AML and normal control samples, in a manner completely opposite to miR-495 (Fig. 4A). Of the 24 candidate target genes, 7 (including Bmi1, Hoxa10, Meis1, Pbx3, Rnf2, Set, and Spryd4) were also significantly overexpressed in MLL-AF9 mouse leukemia samples relative to normal controls (Fig. 4B). Previous studies from us and others indicated that both MEIS1 and PBX3 are important downstream target genes of MLL fusion proteins and play a critical oncogenic role (likely through cooperating with HOXA genes, e.g., HOXA9) in the development and maintenance of MLL-rearranged leukemia (13, 14, 16, 17, 21, 29). Thus, we focused on MEIS1 and PBX3 for further studies. We showed that forced expression of miR-495 could significantly repress endogenous expression of MEIS1 and PBX3 (Fig. 5 A and B), and our luciferase reporter/mutagenesis assays confirmed that both genes are genuine direct targets of miR-495 (Fig. 5 C and D). Furthermore, we showed that coexpressing MEIS1 or PBX3 (the expressional construct contains only coding region, with no 3′ UTR of the gene) could reverse the effects of ectopically expressed miR-495 in human leukemic cells with MLL rearrangements (Fig. 6 and Fig. S4). Therefore, PBX3 and MEIS1 are two functionally important direct target genes of mir-495 in MLL-rearranged leukemia. In the future, it would also be important to determine whether some other potential targets we identified in Fig. 4 are essential direct target genes of miR-495 in MLL-associated leukemia, and indeed some of those genes such as BMI1 and HOXA10 have been shown to play critical oncogenic roles in MLL-rearranged leukemia (15, 21, 53).
We and others have shown previously that MLL fusion proteins could up-regulate expression of the miR-17-92 cluster and miR-196b (28, 39, 41, 42, 54), and here we show that MLL fusions could also repress expression of miR-495. Tumor suppressor genes such as RASSF2 and CDKN1A (i.e., p21) have been identified as direct target genes of the miR-17-92 cluster in MLL-rearranged leukemia, and it is possible that some other well-known tumor-suppressor target genes (e.g., PTEN and BIM) (55–57) may also be genuine targets of this miRNA cluster in MLL-rearranged leukemia. Interestingly, we have shown that miR-196b targets both oncogenes (e.g., HOXA9 and MEIS1) and tumor-suppressor genes (e.g., FAS) in MLL-rearranged leukemia, although its repression of expression of the tumor-suppressor target genes likely plays a predominant role in leukemogenesis, and thereby overall miR-196b functions as an oncogenic miRNA (39). In the present study, we show that as a tumor suppressor miRNA, miR-495 directly targets critical oncogenic target genes such as MEIS1 and PBX3, both of which are transcriptionally up-regulated by MLL fusions and play essential roles in the development and maintenance of MLL-rearranged leukemia.
Collectively, results from our present study together with those from previous studies (28, 39, 41, 42, 54) delineate a complex signaling network mediated by MLL fusions in MLL-rearranged leukemia, in which miRNAs contributed as essential gene expression modulators (a model is shown in Fig. S5). Thus, these studies not only substantially broaden our understanding of the complex mechanisms underlying the pathogenesis of MLL-rearranged leukemia but also implicate potential new therapeutic strategies to treat MLL-rearranged leukemia, a type of disease with resistance to present therapy. For example, in the future, we can treat MLL-rearranged leukemia with clinically applicable nanoparticles that packaged with both mimic oligos for miR-495 and the antagomiR oligos for the miR-17-92 cluster, so that we cannot only deplete expression of critical oncogenes (e.g., MEIS1 and PBX3) but also restore expression of essential tumor suppressor genes (e.g., RASSF2 and p21, and probably also PTEN and BIM) (55–57) to reach effective antitumor effects.
Materials and Methods
miRNA and mRNA Expression Profiling Assays.
The miRNA expression profiling assays of the 100 human samples (including 10 MLL-rearranged, 75 nonMLL rearranged AML, and 15 normal controls) were conducted by use of Exiqon miRCURY LNA arrays (v10.0; covering 757 human miRNAs). The mRNA microarrays of the 79 human samples (including 9 MLL-rearranged AML, 61 nonMLL-rearranged AML, and 9 normal control) and the 15 mouse BM samples (including 9 MLL-AF9-induced AML samples and 6 negative control samples) were conducted by use of Agilent’s custom-design microarrays and Affymetrix GeneChip Mouse Gene 1.0 ST arrays, respectively. Part of the array data have been reported elsewhere recently (29, 39, 58). The microarray data sets have been deposited in the Gene Expression Omnibus (GEO) database (accession nos. GSE30258 and GSE34185).
Cell Culture and Transfection.
MONOMAC-6, THP-1, and KOCL-48 cells were grown in RPMI medium 1640 and transfected using the Amaxa Nucleofector Technology (Amaxa Biosystems). More details are provided in SI Materials and Methods.
Cell Apoptosis and Viability Assay.
Cell apoptosis and viability were assessed 48 h after transfection using the ApoLive-Glo Multiplex Assay Kit (Promega) according to the manufacturer’s manuals.
Luciferase Reporter and Mutagenesis Assays.
Luciferase reporter and mutagenesis assays were conducted as described previously (39), with some modifications (SI Materials and Methods).
Colony-Forming/Replating Assay.
These experiments were conducted as described previously (32) with some modifications (SI Materials and Methods).
BM Transplantation.
Normal BM cells of B6.SJL (CD45.1) mice were retrovirally transduced with corresponding constructs, through two rounds of spinoculation (39), and then injected by tail vein into lethally irradiated (960 rads) 8- to 10-wk-old C57BL/6 (CD45.2) recipient mice with 3 × 105 donor cells per mouse plus a radioprotective dose of 1 × 106 whole BM cells. All experiments on mice were approved by the Institutional Animal Care and Use Committee of the University of Chicago.
Histopathology and Immunohistochemistry.
Tissue samples were fixed in formalin, embedded in paraffin, sectioned, and stained with H&E. Cytospins of peripheral blood and BM were stained with Wright-Giemsa.
Supplementary Material
Acknowledgments
We thank Gregory Hannon, Scott Hammond, Lin He, and Scott Armstrong for providing retroviral constructs. This work was supported in part by National Institutes of Health (NIH) Gant R01 CA127277 (to J.C.), a Leukemia and Lymphoma Society Translational Research Grant (to J.D.R. and J.C.), an American Cancer Society Research Scholar grant (to J.C.), the G. Harold and Leila Y. Mathers Charitable Foundation (J.C.), the Fidelity Foundation (J.D.R. and J.C.), the University of Chicago Committee on Cancer Biology Fellowship Program (X.J.), a Leukemia and Lymphoma Society Special Fellowship (to Z.L.), Gabrielle’s Angel Foundation for Cancer Research (X.J., H.H., Z.L., and J.C.,), NIH R01 CA118319 Sub-Award (to J.C.M. and J.C.), and NIH Grants P01 CA40046 (to M.M.L.B.) and P30 CA014599 Cancer Center Support Grant (CCSG) (to M.M.L.B.). J.C.M. is a Leukemia and Lymphoma Society Scholar.
Footnotes
The authors declare no conflict of interest.
Data deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession nos. GSE30258 and GSE34185).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1217519109/-/DCSupplemental.
References
- 1.Löwenberg B, Downing JR, Burnett A. Acute myeloid leukemia. N Engl J Med. 1999;341(14):1051–1062. doi: 10.1056/NEJM199909303411407. [DOI] [PubMed] [Google Scholar]
- 2.Zhang Y, Rowley JD. Chromatin structural elements and chromosomal translocations in leukemia. DNA Repair (Amst) 2006;5(9-10):1282–1297. doi: 10.1016/j.dnarep.2006.05.020. [DOI] [PubMed] [Google Scholar]
- 3.Rowley JD. Chromosome translocations: Dangerous liaisons revisited. Nat Rev Cancer. 2001;1(3):245–250. doi: 10.1038/35106108. [DOI] [PubMed] [Google Scholar]
- 4.Peterson LF, et al. Acute myeloid leukemia with the 8q22;21q22 translocation: Secondary mutational events and alternative t(8;21) transcripts. Blood. 2007;110(3):799–805. doi: 10.1182/blood-2006-11-019265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ziemin-van der Poel S, et al. Identification of a gene, MLL, that spans the breakpoint in 11q23 translocations associated with human leukemias. Proc Natl Acad Sci USA. 1991;88(23):10735–10739. doi: 10.1073/pnas.88.23.10735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tkachuk DC, Kohler S, Cleary ML. Involvement of a homolog of Drosophila trithorax by 11q23 chromosomal translocations in acute leukemias. Cell. 1992;71(4):691–700. doi: 10.1016/0092-8674(92)90602-9. [DOI] [PubMed] [Google Scholar]
- 7.Rowley JD. Chromosomal translocations: Revisited yet again. Blood. 2008;112(6):2183–2189. doi: 10.1182/blood-2008-04-097931. [DOI] [PubMed] [Google Scholar]
- 8.Chen J, Odenike O, Rowley JD. Leukaemogenesis: more than mutant genes. Nat Rev Cancer. 2010;10(1):23–36. doi: 10.1038/nrc2765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rowley JD, Olney HJ. International workshop on the relationship of prior therapy to balanced chromosome aberrations in therapy-related myelodysplastic syndromes and acute leukemia: Overview report. Genes Chromosomes Cancer. 2002;33(4):331–345. doi: 10.1002/gcc.10040. [DOI] [PubMed] [Google Scholar]
- 10.Pui CH, et al. Clinical heterogeneity in childhood acute lymphoblastic leukemia with 11q23 rearrangements. Leukemia. 2003;17(4):700–706. doi: 10.1038/sj.leu.2402883. [DOI] [PubMed] [Google Scholar]
- 11.Krivtsov AV, Armstrong SA. MLL translocations, histone modifications and leukaemia stem-cell development. Nat Rev Cancer. 2007;7(11):823–833. doi: 10.1038/nrc2253. [DOI] [PubMed] [Google Scholar]
- 12.Rowley JD. Seminars from the University of Minnesota. Chromosome translocations: Dangerous liaisons. J Lab Clin Med. 1998;132(4):244–250. doi: 10.1016/s0022-2143(98)90036-1. [DOI] [PubMed] [Google Scholar]
- 13.Zeisig BB, et al. Hoxa9 and Meis1 are key targets for MLL-ENL-mediated cellular immortalization. Mol Cell Biol. 2004;24(2):617–628. doi: 10.1128/MCB.24.2.617-628.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kroon E, et al. Hoxa9 transforms primary bone marrow cells through specific collaboration with Meis1a but not Pbx1b. EMBO J. 1998;17(13):3714–3725. doi: 10.1093/emboj/17.13.3714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Smith LL, et al. Functional crosstalk between Bmi1 and MLL/Hoxa9 axis in establishment of normal hematopoietic and leukemic stem cells. Cell Stem Cell. 2011;8(6):649–662. doi: 10.1016/j.stem.2011.05.004. [DOI] [PubMed] [Google Scholar]
- 16.Wong P, Iwasaki M, Somervaille TC, So CW, Cleary ML. Meis1 is an essential and rate-limiting regulator of MLL leukemia stem cell potential. Genes Dev. 2007;21(21):2762–2774. doi: 10.1101/gad.1602107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kumar AR, et al. A role for MEIS1 in MLL-fusion gene leukemia. Blood. 2009;113(8):1756–1758. doi: 10.1182/blood-2008-06-163287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Faber J, et al. HOXA9 is required for survival in human MLL-rearranged acute leukemias. Blood. 2009;113(11):2375–2385. doi: 10.1182/blood-2007-09-113597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ayton PM, Cleary ML. Transformation of myeloid progenitors by MLL oncoproteins is dependent on Hoxa7 and Hoxa9. Genes Dev. 2003;17(18):2298–2307. doi: 10.1101/gad.1111603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Roth JJ, Crist RC, Buchberg AM. Might as well face it: MLL’s addicted to HOX. Blood. 2009;113(11):2372–2373. doi: 10.1182/blood-2009-01-197616. [DOI] [PubMed] [Google Scholar]
- 21.Orlovsky K, et al. Down-regulation of homeobox genes MEIS1 and HOXA in MLL-rearranged acute leukemia impairs engraftment and reduces proliferation. Proc Natl Acad Sci USA. 2011;108(19):7956–7961. doi: 10.1073/pnas.1103154108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shah N, Sukumar S. The Hox genes and their roles in oncogenesis. Nat Rev Cancer. 2010;10(5):361–371. doi: 10.1038/nrc2826. [DOI] [PubMed] [Google Scholar]
- 23.Milech N, Kees UR, Watt PM. Novel alternative PBX3 isoforms in leukemia cells with distinct interaction specificities. Genes Chromosomes Cancer. 2001;32(3):275–280. doi: 10.1002/gcc.1190. [DOI] [PubMed] [Google Scholar]
- 24.Thorsteinsdottir U, Kroon E, Jerome L, Blasi F, Sauvageau G. Defining roles for HOX and MEIS1 genes in induction of acute myeloid leukemia. Mol Cell Biol. 2001;21(1):224–234. doi: 10.1128/MCB.21.1.224-234.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Armstrong SA, et al. MLL translocations specify a distinct gene expression profile that distinguishes a unique leukemia. Nat Genet. 2002;30(1):41–47. doi: 10.1038/ng765. [DOI] [PubMed] [Google Scholar]
- 26.Bullinger L, et al. Use of gene-expression profiling to identify prognostic subclasses in adult acute myeloid leukemia. N Engl J Med. 2004;350(16):1605–1616. doi: 10.1056/NEJMoa031046. [DOI] [PubMed] [Google Scholar]
- 27.Krivtsov AV, et al. Transformation from committed progenitor to leukaemia stem cell initiated by MLL-AF9. Nature. 2006;442(7104):818–822. doi: 10.1038/nature04980. [DOI] [PubMed] [Google Scholar]
- 28.Li Z, et al. Consistent deregulation of gene expression between human and murine MLL rearrangement leukemias. Cancer Res. 2009;69(3):1109–1116. doi: 10.1158/0008-5472.CAN-08-3381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li Z, et al. Up-regulation of a HOXA-PBX3 homeobox-gene signature following down-regulation of miR-181 is associated with adverse prognosis in patients with cytogenetically abnormal AML. Blood. 2012;119(10):2314–2324. doi: 10.1182/blood-2011-10-386235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.He L, Hannon GJ. MicroRNAs: small RNAs with a big role in gene regulation. Nat Rev Genet. 2004;5(7):522–531. doi: 10.1038/nrg1379. [DOI] [PubMed] [Google Scholar]
- 31.Xiao C, Rajewsky K. MicroRNA control in the immune system: Basic principles. Cell. 2009;136(1):26–36. doi: 10.1016/j.cell.2008.12.027. [DOI] [PubMed] [Google Scholar]
- 32.Li Z, et al. Distinct microRNA expression profiles in acute myeloid leukemia with common translocations. Proc Natl Acad Sci USA. 2008;105(40):15535–15540. doi: 10.1073/pnas.0808266105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mi S, et al. MicroRNA expression signatures accurately discriminate acute lymphoblastic leukemia from acute myeloid leukemia. Proc Natl Acad Sci USA. 2007;104(50):19971–19976. doi: 10.1073/pnas.0709313104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Marcucci G, et al. MicroRNA expression in cytogenetically normal acute myeloid leukemia. N Engl J Med. 2008;358(18):1919–1928. doi: 10.1056/NEJMoa074256. [DOI] [PubMed] [Google Scholar]
- 35.Jongen-Lavrencic M, Sun SM, Dijkstra MK, Valk PJ, Löwenberg B. MicroRNA expression profiling in relation to the genetic heterogeneity of acute myeloid leukemia. Blood. 2008;111(10):5078–5085. doi: 10.1182/blood-2008-01-133355. [DOI] [PubMed] [Google Scholar]
- 36.Garzon R, et al. Distinctive microRNA signature of acute myeloid leukemia bearing cytoplasmic mutated nucleophosmin. Proc Natl Acad Sci USA. 2008;105(10):3945–3950. doi: 10.1073/pnas.0800135105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Garzon R, et al. MicroRNA signatures associated with cytogenetics and prognosis in acute myeloid leukemia. Blood. 2008;111(6):3183–3189. doi: 10.1182/blood-2007-07-098749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Marcucci G, Mrózek K, Radmacher MD, Garzon R, Bloomfield CD. The prognostic and functional role of microRNAs in acute myeloid leukemia. Blood. 2011;117(4):1121–1129. doi: 10.1182/blood-2010-09-191312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li Z, et al. miR-196b directly targets both HOXA9/MEIS1 oncogenes and FAS tumour suppressor in MLL-rearranged leukaemia. Nat Commun. 2012;3:688. doi: 10.1038/ncomms1681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Arnold CP, et al. MicroRNA programs in normal and aberrant stem and progenitor cells. Genome Res. 2011;21(5):798–810. doi: 10.1101/gr.111385.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mi S, et al. Aberrant overexpression and function of the miR-17-92 cluster in MLL-rearranged acute leukemia. Proc Natl Acad Sci USA. 2010;107(8):3710–3715. doi: 10.1073/pnas.0914900107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wong P, et al. The miR-17-92 microRNA polycistron regulates MLL leukemia stem cell potential by modulating p21 expression. Cancer Res. 2010;70(9):3833–3842. doi: 10.1158/0008-5472.CAN-09-3268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci USA. 2001;98(9):5116–5121. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Super HJ, Martinez-Climent J, Rowley JD. Molecular analysis of the Mono Mac 6 cell line: Detection of an MLL-AF9 fusion transcript. Blood. 1995;85(3):855–856. [PubMed] [Google Scholar]
- 45.Wei J, et al. Microenvironment determines lineage fate in a human model of MLL-AF9 leukemia. Cancer Cell. 2008;13(6):483–495. doi: 10.1016/j.ccr.2008.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mueller D, et al. Misguided transcriptional elongation causes mixed lineage leukemia. PLoS Biol. 2009;7(11):e1000249. doi: 10.1371/journal.pbio.1000249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.He L, et al. A microRNA polycistron as a potential human oncogene. Nature. 2005;435(7043):828–833. doi: 10.1038/nature03552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Subramanian A, et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102(43):15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chang CP, et al. Pbx proteins display hexapeptide-dependent cooperative DNA binding with a subset of Hox proteins. Genes Dev. 1995;9(6):663–674. doi: 10.1101/gad.9.6.663. [DOI] [PubMed] [Google Scholar]
- 50.Chang CP, Brocchieri L, Shen WF, Largman C, Cleary ML. Pbx modulation of Hox homeodomain amino-terminal arms establishes different DNA-binding specificities across the Hox locus. Mol Cell Biol. 1996;16(4):1734–1745. doi: 10.1128/mcb.16.4.1734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hwang-Verslues WW, et al. miR-495 is upregulated by E12/E47 in breast cancer stem cells, and promotes oncogenesis and hypoxia resistance via downregulation of E-cadherin and REDD1. Oncogene. 2011;30(21):2463–2474. doi: 10.1038/onc.2010.618. [DOI] [PubMed] [Google Scholar]
- 52.Li Z, et al. miR-495 and miR-551a inhibit the migration and invasion of human gastric cancer cells by directly interacting with PRL-3. Cancer Lett. 2012;323(1):41–47. doi: 10.1016/j.canlet.2012.03.029. [DOI] [PubMed] [Google Scholar]
- 53.Xia ZB, Anderson M, Diaz MO, Zeleznik-Le NJ. MLL repression domain interacts with histone deacetylases, the polycomb group proteins HPC2 and BMI-1, and the corepressor C-terminal-binding protein. Proc Natl Acad Sci USA. 2003;100(14):8342–8347. doi: 10.1073/pnas.1436338100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Popovic R, et al. Regulation of mir-196b by MLL and its overexpression by MLL fusions contributes to immortalization. Blood. 2009;113(14):3314–3322. doi: 10.1182/blood-2008-04-154310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Xiao C, et al. Lymphoproliferative disease and autoimmunity in mice with increased miR-17-92 expression in lymphocytes. Nat Immunol. 2008;9(4):405–414. doi: 10.1038/ni1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Olive V, et al. miR-19 is a key oncogenic component of mir-17-92. Genes Dev. 2009;23(24):2839–2849. doi: 10.1101/gad.1861409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Koralov SB, et al. Dicer ablation affects antibody diversity and cell survival in the B lymphocyte lineage. Cell. 2008;132(5):860–874. doi: 10.1016/j.cell.2008.02.020. [DOI] [PubMed] [Google Scholar]
- 58.He C, et al. Young intragenic miRNAs are less coexpressed with host genes than old ones: Implications of miRNA-host gene coevolution. Nucleic Acids Res. 2012;40(9):4002–4012. doi: 10.1093/nar/gkr1312. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.