Abstract
Type 1 interferons (IFN1) elicit antiviral defenses by activating the cognate receptor composed of IFN-α/β receptor chain 1 (IFNAR1) and IFNAR2. Down-regulation of this receptor occurs through IFN1-stimulated IFNAR1 ubiquitination, which exposes a Y466-based linear endocytic motif within IFNAR1 to recruitment of the adaptin protein-2 complex (AP2) and ensuing receptor endocytosis. Paradoxically, IFN1-induced Janus kinase-mediated phosphorylation of Y466 is expected to decrease its affinity for AP2 and to inhibit the endocytic rate. To explain how IFN1 promotes Y466 phosphorylation yet stimulates IFNAR1 internalization, we proposed that the activity of a protein tyrosine phosphatase (PTP) is required to enable both events by dephosphorylating Y466. An RNAi-based screen identified PTP1B as a specific regulator of IFNAR1 endocytosis stimulated by IFN1, but not by ligand-independent inducers of IFNAR1 ubiquitination. PTP1B is a promising target for treatment of obesity and diabetes; numerous research programs are aimed at identification and characterization of clinically relevant inhibitors of PTP1B. PTP1B is capable of binding and dephosphorylating IFNAR1. Genetic or pharmacologic modulation of PTP1B activity regulated IFN1 signaling in a manner dependent on the integrity of Y466 within IFNAR1 in human cells. These effects were less evident in mouse cells whose IFNAR1 lacks an analogous motif. PTP1B inhibitors robustly augmented the antiviral effects of IFN1 against vesicular stomatitis and hepatitis C viruses in human cells and proved beneficial in feline stomatitis patients. The clinical significance of these findings in the context of using PTP1B inhibitors to increase the therapeutic efficacy of IFN against viral infections is discussed.
Type 1 interferons (IFN1, including IFN-α/β) are widely used to treat patients with viral infections (1–5). These cytokines elicit their antiviral effects by inducing IFN-stimulated genes (6, 7) whose transcription is activated as a result of a signal transduction pathway involving binding of IFN1 to its receptor [consisting of IFN-α/β receptor chain 1 (IFNAR1) and IFNAR2] followed by activation of Janus kinases (JAK; TYK2 and JAK1). These kinases induce tyrosine phosphorylation of signal transducers and activators of transcription (STAT1/2) and formation of transcriptionally active complexes that recognize IFN-stimulated regulatory elements (ISRE) within the IFN-stimulated genes, the products of which suppress viral replication and stimulate immune responses (reviewed in refs. 8–10).
The initial sensitivity of cells to IFN1 depends on cell surface receptor density that is regulated by endocytosis and subsequent lysosomal degradation (11). In human cells, endocytosis of this receptor is mediated by the interaction between the adaptin protein-2 complex (AP2) endocytic machinery complex and the tyrosine (Y466)-based linear endocytic motif within the IFNAR1 subunit (12). Such interaction is generally obscured by the IFNAR1-associated TYK2 kinase (13); accordingly, human cells lacking TYK2 exhibit a robust basal endocytosis and degradation of IFNAR1 (14, 15) as long as integrity of the Y466-based motif is preserved (13). Importance of this motif is further highlighted by reports that the human Y466F mutant is poorly endocytosed despite a robust ubiquitination (12) and that TYK2 knockout mice (whose IFNAR1 lacks an analogous motif) display normal levels of IFNAR1 (16, 17).
In human cells, unmasking of Y466 and its interaction with AP2 is stimulated by IFNAR1 ubiquitination (12) facilitated by the β-Trcp E3 ubiquitin ligase, which is recruited upon phosphorylation of Ser-535 within the IFNAR1 degron (18, 19). Such phosphorylation could be induced by IFN-α/β and mediated by activities of JAK (20, 21) and protein kinase D2 (22). Alternatively, a basal phosphorylation of Ser-535 by casein kinase 1α (23) can be stimulated by numerous inducers of ligand-independent IFNAR1 ubiquitination (20). These inducers—including activators of pathogen recognition receptors (24), the unfolded protein response (25), or proinflammatory cytokines such as interleukin-1 (IL-1) (26, 27)—act via p38 kinase-dependent priming phosphorylation that does not require JAK activity (28, 29). Both ligand/JAK-dependent and -independent pathways promote IFNAR1 ubiquitination, endocytosis, and degradation and restrict the extent of IFN1 signaling (reviewed in ref. 30; see Fig. 4H).
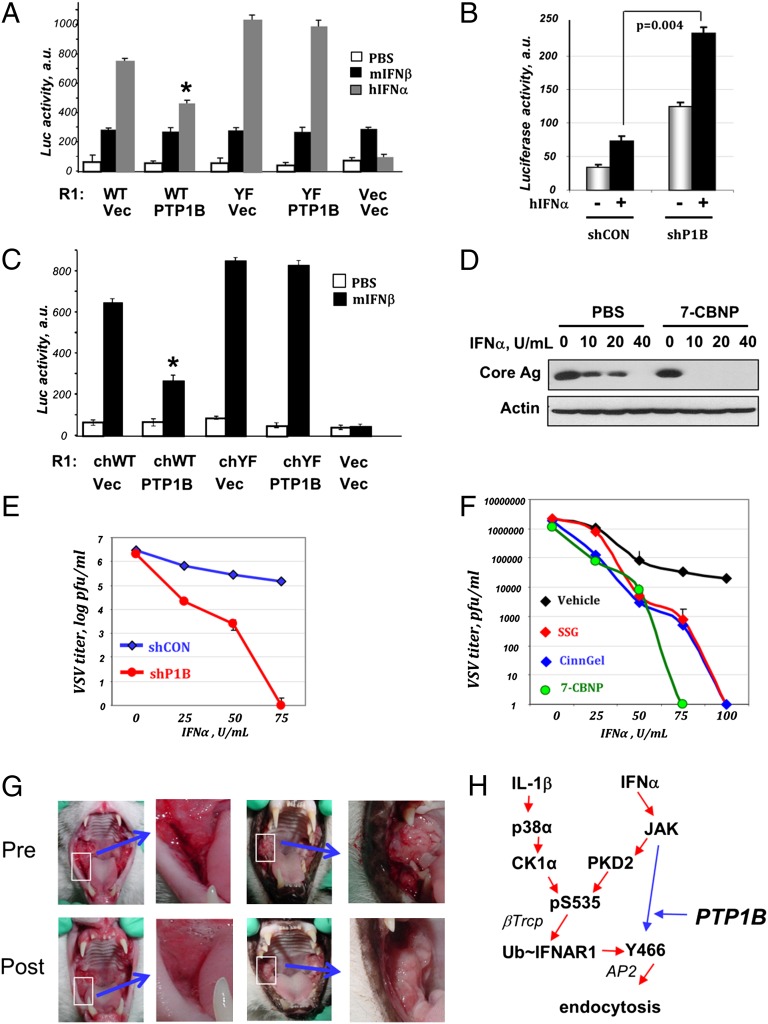
Fig. 4.
PTP1B regulates the extent of IFN1 signaling and antiviral defenses. (A) Relative activity of ISRE–luciferase reporter expressed in MEFs that received human IFNAR2 (R2) indicated types of human IFNAR1 and PTP1B where indicated. The cells were left untreated (white bars) or treated with mouse IFN-β (1,000 U/mL; black bars) or human IFN-α (1,000 U/mL; gray bars) for 5 h and harvested 40 h later. Normalized (per renilla lucifere activity) values from three independent experiments are presented. *P < 0.05 compared with all other groups. (B) ISRE–luciferase activity in human 2fTGH cells that received indicated shRNA and were treated or not with human IFN-α (1,000 U/mL for 30 min) and harvested 24 h thereafter was depicted as in A. (C) ISRE–luciferase activity in MEFs from IFNAR1 knockout mice reconstituted with chimeric IFNAR1 proteins (WT or YF, with or without PTP1B) and treated with mouse IFN-β where indicated. (D) Immunoblotting analysis of lysates from in Huh7.5 cells (pretreated with either PBS or 7-CBNP and subsequently treated with the indicated doses of IFN-α before being infected with HCV) was carried out using the anti-Core antigen antibody. Levels of β-actin are included as a loading control. (E) 2fTGH cells that received either shControl (blue line) or shRNA against PTP1B (red line) were pretreated with the indicated doses of IFN-α for 2 h before being exposed to VSV [multiplicity of infection (MOI) 0.1 for 8 h]. The VSV viral titers in cell supernatants collected 16 h later were analyzed and depicted as an average of five independent experiments (each in triplicate correct). (F) 2fTGH cells that received either vehicle (DMSO or PBS; black) or PTP1B inhibitors (CinnGel2Me, 10 µM, blue; SSG, 10 µg/mL, red; or 7-CBNP, 150 µM, green) and the indicated doses of IFN-α for 2 h before being exposed to VSV (MOI 0.1 for 8 h). VSV titer was assessed as in E. (G) Therapeutic effects of PTP1B inhibitor in feline patients with chronic viral stomatitis that failed standard anti-inflammatory treatment. Photos of selected lesions (white border area) pre- and postsubmucosal injection of 7-CBNP into the lesions are shown. (H) Scheme that summarizes signaling pathways that were established (30) and uncovered here. PTP1B counteracts JAK-induced phosphorylation of Y466-based endocytic motif whose exposure is required for efficient endocytosis upon IFNAR1 ubiquitination.
Our current work was triggered by an unexpected observation that, under conditions when IFN-α or IL-1β induced comparable increases in IFNAR1 ubiquitination, the rate of IFNAR1 endocytosis and degradation was lower in IFN-α–treated cells. Indeed, the Y466-dependent mechanism of IFN-α–stimulated IFNAR1 endocytosis appeared counterintuitive given an existing paradigm suggesting that this residue has to remain unphosphorylated to efficiently bind AP2 and enable endocytosis (31), and a report that Y466 undergoes JAK-mediated phosphorylation in response to IFN-α (32). To explain how IFN-α promotes Y466 phosphorylation yet stimulates IFNAR1 endocytosis, we proposed that the activity of a putative phosphatase enables both events by dephosphorylating Y466.
Here we describe the results of an RNAi-based screen that identified protein tyrosine phosphatase 1B (PTP1B) as a specific regulator of IFNAR1 endocytosis and thereby IFN1 signaling in human, but not in mouse, cells. PTP1B is known as an important regulator of intracellular signaling (33). PTP1B inhibitors are being sought to regulate leptin and insulin signaling and, accordingly, function as potential therapeutics for the treatment of type 2 diabetes mellitus and obesity (34–37). Novel, potent, selective, and orally available inhibitors of this enzyme were recently reported (34, 38, 39). We demonstrate that PTP1B regulates IFN1 signaling in human cells and that PTP1B inhibitors robustly synergize with IFN1 to protect human cells against hepatitis C virus (HCV) and vesicular stomatitis virus (VSV), and elicit a therapeutic effect in cats suffering from feline stomatitis. The clinical significance of these findings is discussed.
Results
The proteolytic turnover of IFNAR1 [assessed by a cycloheximide (CHX) chase assay] was more robustly stimulated by IL-1β than by IFN-α (Fig. 1A). As expected, IFN-α, but not IL-1β, stimulated phosphorylation of IFNAR1 on tyrosines (Fig. 1B). Given that treatment with either IL-1β or IFN-α led to a comparable increase in Ser-535 phosphorylation and ubiquitination of IFNAR1 (Fig. 1B), differences between efficacy of these two inducers in accelerating IFNAR1 degradation was likely conferred by events that occur downstream of IFNAR1 ubiquitination.
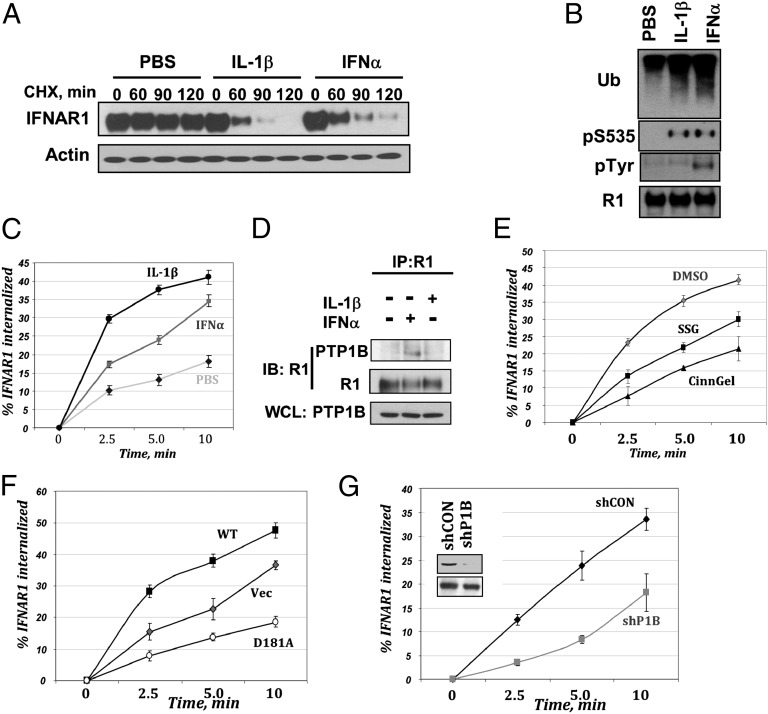
Fig. 1.
PTP1B regulates the ligand-inducible endocytosis of IFNAR1. (A) Degradation of IFNAR1 in human 293-IL-1RI cells treated with CHX (20 µg/mL) and either PBS, IFN-α (1,000 U/mL), or IL-1β (2 ng/mL) for indicated times. (B) Phosphorylation and ubiquitination of IFNAR1 immunopurified from 293-IL-1RI cells treated with PBS, IFN-α (1,000 U/mL), or IL-1β (2 ng/mL) for 10 min were analyzed by immunoblotting using the indicated antibodies. (C) Internalization rate of endogenous IFNAR1 in cells treated with PBS (diamonds), IFN-α (1,000 U/mL; squares), or IL-1β (2 ng/mL; circles) for indicated times was assessed from three independent experiments (each in six replicates) and depicted as average + SEM. (D) The interaction between endogenous IFNAR1 and PTP1B in 293-IL-1RI cells stimulated with either IFN-α (1,000 U/mL) or IL-1β (2 ng/mL) for 10 min was assessed by immunoprecipitation–immunoblotting using indicated antibodies. Levels of PTP1B in whole cell lysates (WCLs) were also analyzed. (E) Internalization of endogenous IFNAR1 in 293-IL-1RI cells treated with IFN-α (1,000 U/mL) in the presence of vehicle (DMSO; gray diamonds) or PTP1B inhibitors CinnGel2Me (10 µm; black triangles) or sodium stibogluconate (SSG; 10 µg/mL; black squares). (F) Internalization of endogenous IFNAR1 in IFN-α–treated 293-IL-1RI cells transfected with empty vector (Vec; gray diamonds) or constructs for expression of PTP1B (WT, black squares; D181A mutant, open circles). (G) Internalization of endogenous IFNAR1 in IFN-α–treated 293-IL-1RI cells that received control shRNA (black diamonds) or shRNA against PTP1B (shP1B; gray squares). Inset shows the effect of these shRNAs on the levels of PTP1B (Upper) or β-actin (Lower).
As ubiquitination of human IFNAR1 promotes its internalization by unmasking the Y466-based linear endocytic motif for its interaction with AP2 (12), we next analyzed the kinetics of IFNAR1 endocytosis. Consistent with published results (12, 19), treatment of cells with IFN-α increased the initial rate of IFNAR1 internalization as observed with either endogenous (Fig. 1C) or exogenously expressed (Fig. S1A) receptor. Intriguingly, treatment of cells with IFN-α was noticeably less efficient at stimulating endocytosis of endogenous IFNAR1 than treatment with IL-1β (Fig. 1C). Given that a Y466F mutation within the linear endocytic motif rendered IFNAR1 relatively resistant to an increase in its internalization rate in response to either IFN-α (Fig. S1A) or IL-1β (Fig. S1B), it is plausible that the putative postubiquitination event(s) that confers differential capability of these inducers to stimulate IFNAR1 internalization occurs either upstream or at the level of Y466 exposure. Because Y466 phosphorylation would be expected to decrease its affinity for AP2 (31), a lower efficacy of IFN-α as an endocytic stimulator might be attributed to a differential ability to activate JAK and cause IFNAR1 Tyr phosphorylation (as seen in Fig. 1B). Furthermore, the fact that IFN-α still stimulated IFNAR1 internalization suggested the existence of a putative PTP, the function of which is to dephosphorylate Y466 and enable endocytosis of the receptor.
To identify such a phosphatase, we carried out a limited RNAi screen targeted against human classical PTPs (as classified in ref. 40). Cells transfected with RNAi against PTP1B exhibited a decrease in IFN-α–stimulated endocytosis of IFNAR1, whereas the efficacy of IL-1β–induced endocytosis remained intact (Fig. S1C). Endogenous PTP1B and IFNAR1 could be coimmunoprecipitated from the lysates of cells treated with IFN1 (Fig. 1D and Fig. S1D), indicating that PTP1B is capable of interacting with IFNAR1.
Pretreatment of cells with pharmacological inhibitors of PTP1B such as CinnGel2Mel and sodium stibogluconate noticeably inhibited internalization of IFNAR1 in cells stimulated with IFN-α (Fig. 1E). Similar results were obtained using two other inhibitors, including a selective ZYZ inhibitor (referred to as Compound II in ref. 41) and a recently described potent and bioavailable derivative of cyano bromo naphthalene phosphonic acid (7-CBNP; referred to as compound 3g in ref. 42) (Fig. S1E). A lesser extent of inhibition was achieved in cells treated with a pharmacologic inhibitor of CD45 phosphatase (Fig. S1E). Overexpression of wild-type PTP1B noticeably augmented the IFNAR1 endocytic rate, whereas expression of the catalytically deficient substrate-trapping mutant PTP1BD181A (43) inhibited IFNAR1 endocytosis (Fig. 1F). Furthermore, shRNA-mediated knockdown of PTP1B slowed down the initial internalization of IFNAR1 (Fig. 1G). These results collectively suggest that PTP1B functions as a positive regulator of the ligand-induced IFNAR1 endocytosis.
PTP1B is known to catalyze the dephosphorylation of numerous substrates (44–48), and its effect on IFNAR1 endocytosis may be indirectly mediated by events upstream of unmasking Y466 or downstream of AP2 recruitment to IFNAR1. However, contrary to these possibilities, overexpression of PTP1B did not efficiently increase the initial rate of internalization of the IFNAR1Y466F mutant (Fig. 2A). Given that this mutant was able to bind to PTP1B with efficacy similar to that of wild-type IFNAR1 (Fig. 2B), it is likely that PTP1B, at least in part, regulates IFNAR1 endocytosis thorough an Y466-dependent mechanism. Consistent with this hypothesis, overexpression of PTP1B did not robustly increase IFNAR1 endocytosis in cells that received RNAi against AP2 (Fig. 2C). These results strongly suggest that regulation of IFNAR1 endocytosis by PTP1B is dependent on Y466 and AP2.
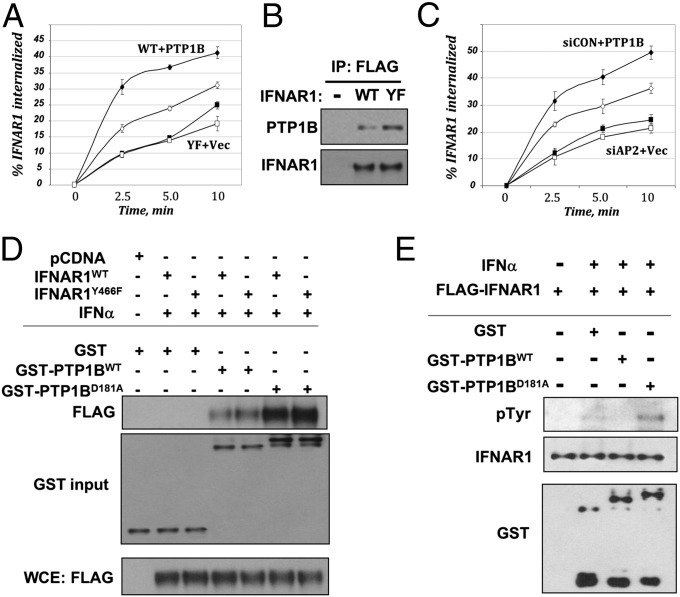
Fig. 2.
PTP1B regulates Tyr phosphorylation and protein stability of IFNAR1 in human cells. (A) Internalization of FLAG–IFNAR1 (WT, diamonds; or Y466F mutant, squares) in IFN-α–treated 293T cells that were cotransfected with empty vector (open symbols) or PTP1B (filled symbols). (B) Interaction between endogenous PTP1B and FLAG–IFNAR1 expressed in 293T cells treated with IFN-α was determined by immunoprecipitation–immunoblotting using the indicated antibodies. Loading was normalized to achieve comparable levels of FLAG–IFNAR1 proteins. (C) Internalization of endogenous IFNAR1 in IFN-α–treated 293T cells that received RNAi against AP2 (squares) or against luciferase (siCON; diamonds) and were cotransfected with either empty vector (open symbols) or PTP1B (filled symbols). (D) The interaction between bacterially produced GST-tagged PTP1B proteins (WT or D181A mutant) with FLAG-tagged IFNAR1 (WT or Y466F mutant) expressed in cells treated or not with IFN-α as indicated were analyzed by pulldown (using GSH beads) and immunoblotting. Normalized input levels of FLAG–IFNAR1 in the cell lysates used for binding experiments are also shown. (E) In vitro Tyr dephosphorylation of FLAG-tagged IFNAR1 purified from 293T cells treated or not with IFN-α upon the incubation with indicated bacterially produced GST-tagged protein (for 30 min at 37 °C) was analyzed by using the anti-phospho-Tyr antibody.
In line with coimmunoprecipitation analyses (Figs. 1D and 2B and Fig. S1D), the interaction between PTP1B (expressed in bacteria) and recombinant IFNAR1 purified from human cells treated with IFN1 was detected (Fig. 2D). Importantly, an inactive PTP1BD181A mutant was capable of efficiently trapping IFNAR1 as a putative substrate (Fig. 2D). In turn, incubation with wild-type, but not D181A mutant, PTP1B protein led to a decreased phospho-Tyr signal on IFNAR1 purified from IFN1-treated cells (Fig. 2E). These results suggest that PTP1B is capable of binding to IFNAR1 and dephosphorylating this receptor.
Knockdown of PTP1B caused a noticeable increase in IFN-α–induced Tyr phosphorylation of IFNAR1 yet did not affect Ser-535 phosphorylation or ubiquitination of this receptor (Fig. 3A), indicating that PTP1B regulates endocytosis of IFNAR1 downstream of its ubiquitination. Whereas turnover of IFNAR1 was stimulated by both IFN-α and IL-1β, PTP1B knockdown noticeably attenuated the extent of this stimulation only in IFN-α–treated cells (Fig. 3B), further suggesting that PTP1B specifically regulates the ligand-inducible pathway of IFNAR1 degradation in human cells. Given that, of all known species, murine IFNAR1 does not contain Y466 or its analog (Fig. 3C) and does not rely on this motif for IFNAR1 down-regulation (13), we further used mouse cells as a control. Importantly, the extent of IFN1-stimulated degradation (Fig. 3D) or internalization (Fig. S1F) of endogenous IFNAR1 was comparable between mouse embryonic fibroblasts (MEFs) from wild-type mice and those from animals lacking Ptpn1, the gene encoding PTP1B. This result indicates that PTP1B plays no apparent role in regulating endocytosis and stability of mouse IFNAR1 in mouse cells.
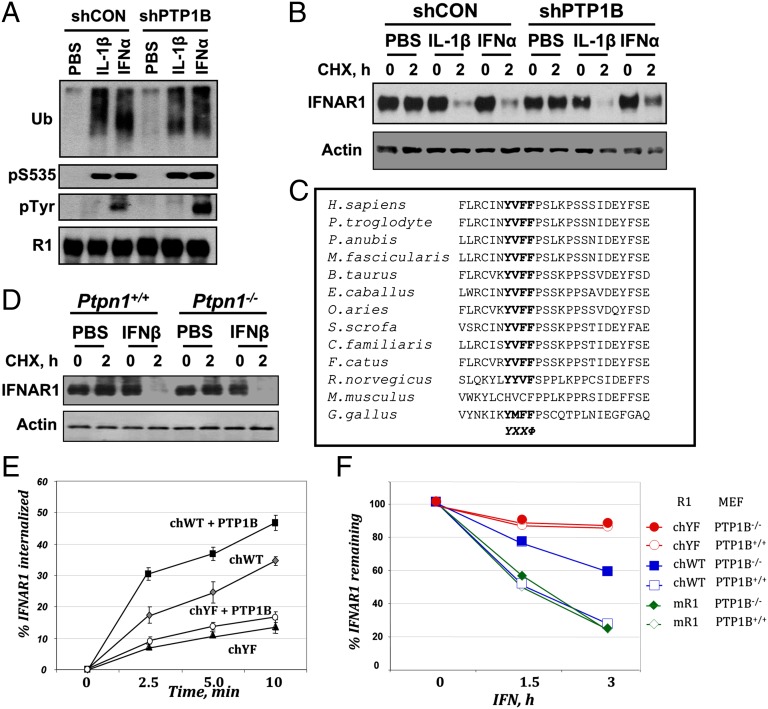
Fig. 3.
PTP1B regulates Tyr phosphorylation and the stability of IFNAR1 in human cells. (A) Endogenous IFNAR1 immunopurified from 293T cells that received indicated shRNA and were treated with IFN-α or IL-1β (as indicated) was analyzed by immunoblotting using the indicated antibodies. (B) Degradation of endogenous IFNAR1 in 293T cells transfected and treated as indicated was analyzed by immunoprecipitation–immunoblotting. Levels of actin in samples were also analyzed. (C) Primary sequence alignment of proximal fragments of the intracellular domains of IFNAR1 proteins from indicated species. Tyr-based endocytic motifs are depicted in bold. (D) MEFs from animals with indicated genotype were treated with PBS or murine IFN-β and CHX as indicated. Levels of murine IFNAR1 and actin were analyzed. (E) Internalization rate of chimeric mouse–human IFNAR1 proteins (chWT or chYF) coexpressed with PTP1B as indicated in MEFs from IFNAR1 knockout mice. (F) Turnover rate of FLAG-tagged mouse (mR1) or chimeric mouse–human IFNAR1 (chWT or chYF) expressed in WT or PTP1B-null MEFs treated with mIFN for indicated times.
To determine whether the latter reflects the properties of receptor or cells, we coexpressed human IFNAR2 with human IFNAR1 (wild-type or Y466F) in MEFs and treated them with species-specific IFN1. Internalization of the wild-type human IFNAR1 (but not of Y466F mutant) was increased upon coexpression of PTP1B. This effect was ligand-specific and not evident in the presence of mouse IFN1 (Fig. S1G), suggesting that presence of functional Y466 within IFNAR1 largely determines the extent of receptor endocytosis and its regulation by PTP1B even in mouse cells.
To further test this possibility, we generated mouse–human chimeric IFNAR1 proteins (Fig. S2A) whose expression enabled MEFs from IFNAR1 knockout mice to respond to murine IFN1 (Fig. S2B) and rendered receptor endocytosis sensitive to a PTP1B inhibitor (Fig. S2C). In this model, coexpression of PTP1B stimulated the endocytosis of chimeric IFNAR1 as long as the integrity of Y466 was preserved (Fig. 3E). Finally, we used CHX assays to compare the turnover rate of various IFNAR1 proteins expressed in MEFs from wild-type or PTP1B knockout mice. Whereas kinetics of degradation of either murine IFNAR1 or chimeric mouse–human IFNAR1Y466F mutant was comparable, the chimeric receptor harboring intact Y466 was noticeably more stable in PTP1B-null cells (Fig. 3F and Fig. S2D). Collectively, these data suggest that PTP1B regulates IFNAR1 stability in a manner that largely depends on the integrity of a Tyr-based endocytic motif.
If the latter conclusion is correct, it would be expected that the role of PTP1B in regulating the sensitivity to IFN1 should be much more important in human cells than in those of mouse origin. To test this possibility, we determined the transactivation of ISRE-driven luciferase in MEFs transfected with human IFNAR2 and various combinations of human IFNAR1 and PTP1B (similar to the setup of experiments shown in Fig. S1G). Without human IFNAR1, these cells could respond to mouse IFN-β, but not to human IFN-α (Fig. 4A and Fig. S2E). Expression of human IFNAR1 (wild type or Y466F mutant) conferred cell responsiveness to human IFN-α. Importantly, PTP1B overexpression significantly inhibited ISRE activation in cells harboring wild-type, but not the Y466F mutant of, human IFNAR1 (Fig. 4A and Fig. S2E). Given that the sensitivity to mouse IFN-β remained largely unaffected, these results suggest that PTP1B functions to attenuate IFN-α signaling in a manner dependent on the presence of human IFNAR1 and integrity of Y466. Importantly, the knockdown of PTP1B in human cells noticeably augmented ISRE activity conferred by endogenously produced IFN or stimulated by exogenously added ligand (Fig. 4B), further indicating that PTP1B negatively regulates the extent of responses to human IFN1. Coexpression of PTP1B suppressed ISRE activities in IFNAR1-null MEFs harboring chimeric mouse–human IFNAR1 but not a chimeric Y466F mutant (Fig. 4C), further emphasizing the importance of Y466 status in regulation of IFN1 signaling by PTP1B.
We next determined the role of PTP1B in modulating IFN-α–induced cellular defenses against HCV in human Huh7.5 cells adapted to HCV infection in vitro (49). Remarkably, pretreatment of these cells with PTP1B inhibitor 7-CBNP greatly decreased the doses of IFN-α required to suppress HCV replication assessed by levels of HCV Core antigen (Fig. 4D). We further tested the role of PTP1B in defenses against VSV in MEFs and human 2fTGH cells. VSV replication and its inhibition by IFN1 was comparable in MEFs from either wild-type or PTP1B-null mice; furthermore, the effect of IFN1 was not augmented by PTP1B inhibitor 7-CBNP in wild-type MEFs (Fig. S2F). Whereas VSV titer reached 1,925,000 × 103 in IFNAR1 knockout MEFs reconstituted with chimeric mouse–human IFNAR1, and IFN1 alone decreased this titer to 445 × 103, a combined pretreatment with 7-CBNP and IFN1 further decreased this titer to 45 × 103. These results indicate that PTP1B may limit the antiviral effects mediated by human IFNAR1. Indeed, knockdown of PTP1B in human 2fTGH robustly augmented the ability of human IFN-α to decrease in the replication of VSV in a dose-dependent manner (Fig. 4E). Furthermore, treatment of 2fTGH with three diverse pharmacologic inhibitors of PTP1B dramatically sensitized VSV to replication suppression even by low doses of IFN-α (Fig. 4F).
In the absence of relevant mouse in vivo models, we carried out a pilot clinical study on cats affected by chronic caudal stomatitis, a condition that is etiologically linked to feline calicivirus infection (50) and could be alleviated by injections of feline IFN-ω (51). Putative feline IFNAR1 sequence harbors a Tyr-based motif (Fig. 3C), and 7-CBNP robustly augments STAT1 phosphorylation in response to IFN1 in feline T cells (Fig. S3A). Accordingly, 2 wk after patients received a single submucosal injection of 7-CBNP, a notable decrease in inflammation score was observed in all five cats enrolled in this study (Fig. S3 B and C). That included a marked reduction in erythema, edema, and proliferative inflamed tissue volumes (Fig. 4G). Together, these data strongly suggest that inhibition of PTP1B augments IFN1-elicited signaling and antiviral defenses.
Discussion
Tyr-based linear endocytic motifs are known to serve as recognition sites for the AP2 complex (31). Whereas the role of the Y466-based motif in the internalization of IFNAR1 has been documented (12), the current paradigm predicted that the catalytic function of JAKs that determine the phosphorylation status of the Tyr residues within IFNAR1 [including Y466 (32)] might add another level of complexity to the regulation of IFNAR1 endocytosis and stability. Phosphorylation of Y466 is expected to reduce the affinity of AP2 components for the Tyr-based endocytic motif within IFNAR1 as it was described for CTLA4 (52–54). Indeed, current data reveal that IL-1β—an inducer of IFNAR1 ubiquitination that does not activate JAK and stimulate IFNAR1 Tyr phosphorylation—is a more efficient activator of IFNAR1 endocytosis and degradation than IFN-α (Fig. 1). Furthermore, given that IFN-α still stimulates IFNAR1 endocytosis, we postulated that phosphorylation of Y466 might be reversed by a putative PTP to enable IFNAR1 endocytosis and degradation (Fig. 4H). A genetic screen and subsequent genetic, pharmacologic, and biochemical analyses revealed PTP1B to be a key determinant of ligand-dependent IFNAR1 endocytosis (Figs. 1 and 2) and degradation (Fig. 3) and an important regulator of IFN1 signaling and antiviral defense (Fig. 4). Whereas IFN1 protects from viruses, it is known to mediate additional tissue damage in the context of many nonviral infections and autoimmune syndromes (55). It is plausible that PTP1B-dependent acceleration of IFNAR1 endocytosis may protect tissues from immunopathologic effects of IFN1.
PTP1B controls endocytosis of IFNAR1 downstream of its ubiquitination yet upstream of AP2 recruitment (Figs. 1 and 2). Given that PTP1B interacts with IFNAR1 and can dephosphorylate this receptor (Fig. 2), a direct removal of phosphate from Y466 and ensuing acceleration of IFNAR1 endocytosis would be the simplest explanation for PTP1B-dependent regulation of IFNAR1 stability and function (Fig. 4H). Although additional mechanisms such as stimulation of postinternalization sorting [proposed for the epidermal growth factor receptor (56)] or general interference with JAK–STAT phosphorylation (44, 57, 58) should not be ruled out, a direct involvement of Y466-driven endocytosis is evident from our studies. Indeed, mouse receptor lacks a Tyr-based motif (Fig. 3C), and PTP1B knockout mice display no abnormalities in the IFN1 pathway (59, 60), except for a modestly augmented STAT1/3 phosphorylation in MEFs attributed regulation of JAK2 and TYK2 (44). Accordingly, whereas interaction of IFNAR1 with TYK2 masks Y466 in unstimulated human cells to prevent basal down-regulation of this receptor (13–15, 61), mouse cells lacking TYK2 display normal levels of IFNAR1 (16, 17). Furthermore, replacement of Tyr residues within murine IFNAR1 does not affect its signaling (62). Collectively, these results and our current data clearly demonstrating the dependence of PTP1B-mediated effects on the integrity of Y466 in human and chimeric receptors strongly suggest that dephosphorylation of Y466 represents a major mode of regulation. Indeed, a robust increase of antiviral effects of IFN1 in vitro upon PTP1B inhibition seen in cells expressing human or chimeric IFNAR1 but not mutant lacking Y466 or mouse receptor (Fig. 4 and Fig. S2E).
The latter data are of medical significance because they suggest potential use of PTP1B inhibitors as antiviral drugs. Because we could not use mice for in vivo studies, the proof-of-principle clinical experiments were carried out in a small group of feline patients suffering from IFN-sensitive stomatitis (Fig. 4G and Fig. S3 B and C). Whereas the mechanism of beneficial effect of PTP1B inhibitor seen in these cats is likely to be complex, these data together with in vitro results provide a strong rationale for PTP1B inhibition for augmenting therapeutic effects of either endogenous or exogenous IFN1. This approach may optimize IFN1-based therapies in patients with viral infections (1–5), multiple sclerosis (63), and cancer (64). Future preclinical studies of PTP1B inhibitors using non-mouse-based in vivo models are warranted.
Methods
Methods are outlined in detail in SI Methods.
The siRNA oligonucleotides to knock down protein phosphatases or AP2 were from Qiagen. Human embryo kidney 293T cells, 293 cells that stably express the human IL-1 receptor (293 Il-1RI; provided by J. Ninomiya-Tsuji, North Carolina State University, Raleigh, NC), MEFs of diverse genotypes, 2fTGH cells (from G. Stark, Cleveland Clinic Foundation, Cleveland), and Huh7.5 (provided by C. Rice, Rockefeller University, New York) were grown in Dulbecco’s modified Eagle medium containing 10% (vol/vol) heat-inactivated FBS and penicillin/streptomycin. Constructs for the expression of IFNAR2 and ISRE-driven luciferase reporter (65) were gifts from J. Krolewski (University of California, Irvine, CA) and C. Horvath (Northwestern University, Evanstone, IL). The antiviral effects of IFN were determined by using VSV (Indiana serotype; a gift from R. Harty, University of Pennsylvania, Philadelphia) as described (66). Replication of HCV in Huh7.5 cells was monitored by immunoblot analysis of HCV Core Antigen. Antibodies that recognize endogenous IFNAR1 (67) and IFNAR1 phosphorylated on Ser-535 (18) as well as assays for immunoprecipitations, immunoblotting, and assessment of the kinetics of IFNAR1 degradation were described (13, 19, 68). IFNAR1 internalization assays were carried out by using a high-throughput fluorescence-based method as described (12).
The protein phosphatase inhibitors CinnGel2Me (Biomol International), sodium stibogluconate (Santa Cruz Biotechnology), and CD45 inhibitor (Santa Cruz) were purchased. The synthesis and properties of the ZYZ inhibitor (Compound II) were described (41). The 7-CBNP was synthesized and purified at Best West Laboratories as described (42) with the modification that used CuI/NaI/amine catalytic reaction condition, which enabled us to increase the yield of intermediate 6-bromo-7-iodo-naphthonitrile.
Supplementary Material
Acknowledgments
We thank J. Chernoff, R. Harty, C. Horvath, J. Krolewski, A. Mexas, B. Neel, J. Chernoff, J. Ninomiya-Tsuji, C. Rice, and G. Stark for reagents. This work was supported by National Institutes of Health Grants CA092900 and CA142425 (to S.Y.F.) and by a Mari Lowe Pilot grant (to S.Y.F. and J.R.L.).
Footnotes
Conflict of interest statement: D.P.B. is an employee and stockholder of Biogen Idec.
This article is a PNAS Direct Submission. D.E.L. is a guest editor invited by the Editorial Board.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1211491109/-/DCSupplemental.
References
- 1.Sato K, Takagi H, Ichikawa T, Kakizaki S, Mori M. Emerging therapeutic strategies for hepatitis C virus infection. Curr Mol Pharmacol. 2008;1(2):130–150. doi: 10.2174/1874467210801020130. [DOI] [PubMed] [Google Scholar]
- 2.Wohnsland A, Hofmann WP, Sarrazin C. Viral determinants of resistance to treatment in patients with hepatitis C. Clin Microbiol Rev. 2007;20(1):23–38. doi: 10.1128/CMR.00010-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Saracco G, Olivero A, Ciancio A, Carenzi S, Rizzetto M. Therapy of chronic hepatitis C: A critical review. Curr Drug Targets Infect Disord. 2003;3(1):25–32. doi: 10.2174/1568005033342127. [DOI] [PubMed] [Google Scholar]
- 4.De Francesco R, Rice CM. New therapies on the horizon for hepatitis C: Are we close? Clin Liver Dis. 2003;7(1):211–242. doi: 10.1016/s1089-3261(02)00069-7. [DOI] [PubMed] [Google Scholar]
- 5.Tellinghuisen TL, Rice CM. Interaction between hepatitis C virus proteins and host cell factors. Curr Opin Microbiol. 2002;5(4):419–427. doi: 10.1016/s1369-5274(02)00341-7. [DOI] [PubMed] [Google Scholar]
- 6.Brassard DL, Grace MJ, Bordens RW. Interferon-alpha as an immunotherapeutic protein. J Leukoc Biol. 2002;71(4):565–581. [PubMed] [Google Scholar]
- 7.Katze MG, He Y, Gale M., Jr Viruses and interferon: A fight for supremacy. Nat Rev Immunol. 2002;2(9):675–687. doi: 10.1038/nri888. [DOI] [PubMed] [Google Scholar]
- 8.Taniguchi T, Takaoka A. A weak signal for strong responses: Interferon-alpha/beta revisited. Nat Rev Mol Cell Biol. 2001;2(5):378–386. doi: 10.1038/35073080. [DOI] [PubMed] [Google Scholar]
- 9.Aaronson DS, Horvath CM. A road map for those who don’t know JAK-STAT. Science. 2002;296(5573):1653–1655. doi: 10.1126/science.1071545. [DOI] [PubMed] [Google Scholar]
- 10.Stark GR, Kerr IM, Williams BR, Silverman RH, Schreiber RD. How cells respond to interferons. Annu Rev Biochem. 1998;67:227–264. doi: 10.1146/annurev.biochem.67.1.227. [DOI] [PubMed] [Google Scholar]
- 11.Uzé G, Schreiber G, Piehler J, Pellegrini S. The receptor of the type I interferon family. Curr Top Microbiol Immunol. 2007;316:71–95. doi: 10.1007/978-3-540-71329-6_5. [DOI] [PubMed] [Google Scholar]
- 12.Kumar KG, et al. Site-specific ubiquitination exposes a linear motif to promote interferon-alpha receptor endocytosis. J Cell Biol. 2007;179(5):935–950. doi: 10.1083/jcb.200706034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kumar KG, et al. Basal ubiquitin-independent internalization of interferon alpha receptor is prevented by Tyk2-mediated masking of a linear endocytic motif. J Biol Chem. 2008;283(27):18566–18572. doi: 10.1074/jbc.M800991200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gauzzi MC, et al. The amino-terminal region of Tyk2 sustains the level of interferon alpha receptor 1, a component of the interferon alpha/beta receptor. Proc Natl Acad Sci USA. 1997;94(22):11839–11844. doi: 10.1073/pnas.94.22.11839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ragimbeau J, et al. The tyrosine kinase Tyk2 controls IFNAR1 cell surface expression. EMBO J. 2003;22(3):537–547. doi: 10.1093/emboj/cdg038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Karaghiosoff M, et al. Partial impairment of cytokine responses in Tyk2-deficient mice. Immunity. 2000;13(4):549–560. doi: 10.1016/s1074-7613(00)00054-6. [DOI] [PubMed] [Google Scholar]
- 17.Shimoda K, et al. Tyk2 plays a restricted role in IFN alpha signaling, although it is required for IL-12-mediated T cell function. Immunity. 2000;13(4):561–571. doi: 10.1016/s1074-7613(00)00055-8. [DOI] [PubMed] [Google Scholar]
- 18.Kumar KG, Krolewski JJ, Fuchs SY. Phosphorylation and specific ubiquitin acceptor sites are required for ubiquitination and degradation of the IFNAR1 subunit of type I interferon receptor. J Biol Chem. 2004;279(45):46614–46620. doi: 10.1074/jbc.M407082200. [DOI] [PubMed] [Google Scholar]
- 19.Kumar KG, et al. SCF(HOS) ubiquitin ligase mediates the ligand-induced down-regulation of the interferon-alpha receptor. EMBO J. 2003;22(20):5480–5490. doi: 10.1093/emboj/cdg524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu J, et al. Ligand-independent pathway that controls stability of interferon alpha receptor. Biochem Biophys Res Commun. 2008;367(2):388–393. doi: 10.1016/j.bbrc.2007.12.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Marijanovic Z, Ragimbeau J, Kumar KG, Fuchs SY, Pellegrini S. TYK2 activity promotes ligand-induced IFNAR1 proteolysis. Biochem J. 2006;397(1):31–38. doi: 10.1042/BJ20060272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zheng H, Qian J, Varghese B, Baker DP, Fuchs S. Ligand-stimulated downregulation of the alpha interferon receptor: Role of protein kinase D2. Mol Cell Biol. 2011;31(4):710–720. doi: 10.1128/MCB.01154-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu J, et al. Mammalian casein kinase 1alpha and its leishmanial ortholog regulate stability of IFNAR1 and type I interferon signaling. Mol Cell Biol. 2009;29(24):6401–6412. doi: 10.1128/MCB.00478-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Qian J, et al. Pathogen recognition receptor signaling accelerates phosphorylation-dependent degradation of IFNAR1. PLoS Pathog. 2011;7(6):e1002065. doi: 10.1371/journal.ppat.1002065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu J, et al. Virus-induced unfolded protein response attenuates antiviral defenses via phosphorylation-dependent degradation of the type I interferon receptor. Cell Host Microbe. 2009;5(1):72–83. doi: 10.1016/j.chom.2008.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Huangfu WC, et al. Inflammatory signaling compromises cell responses to interferon alpha. Oncogene. 2012;31(2):161–172. doi: 10.1038/onc.2011.221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.HuangFu WC, Qian J, Liu C, Rui H, Fuchs SY. Melanoma cell-secreted soluble factor that stimulates ubiquitination and degradation of the interferon alpha receptor and attenuates its signaling. Pigment Cell Melanoma Res. 2010;23(6):838–840. doi: 10.1111/j.1755-148x.2010.00770.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bhattacharya S, et al. Inducible priming phosphorylation promotes ligand-independent degradation of the IFNAR1 chain of type I interferon receptor. J Biol Chem. 2010;285(4):2318–2325. doi: 10.1074/jbc.M109.071498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bhattacharya S, et al. Role of p38 protein kinase in the ligand-independent ubiquitination and down-regulation of the IFNAR1 chain of type I interferon receptor. J Biol Chem. 2011;286(25):22069–22076. doi: 10.1074/jbc.M111.238766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Huangfu WC, Fuchs SY. Ubiquitination-dependent regulation of signaling receptors in cancer. Genes Cancer. 2010;1(7):725–734. doi: 10.1177/1947601910382901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bonifacino JS, Traub LM. Signals for sorting of transmembrane proteins to endosomes and lysosomes. Annu Rev Biochem. 2003;72:395–447. doi: 10.1146/annurev.biochem.72.121801.161800. [DOI] [PubMed] [Google Scholar]
- 32.Yan H, et al. Phosphorylated interferon-alpha receptor 1 subunit (IFNaR1) acts as a docking site for the latent form of the 113 kDa STAT2 protein. EMBO J. 1996;15(5):1064–1074. [PMC free article] [PubMed] [Google Scholar]
- 33.Bourdeau A, Dubé N, Tremblay ML. Cytoplasmic protein tyrosine phosphatases, regulation and function: The roles of PTP1B and TC-PTP. Curr Opin Cell Biol. 2005;17(2):203–209. doi: 10.1016/j.ceb.2005.02.001. [DOI] [PubMed] [Google Scholar]
- 34.Zhang S, Zhang ZY. PTP1B as a drug target: recent developments in PTP1B inhibitor discovery. Drug Discov Today. 2007;12(9-10):373–381. doi: 10.1016/j.drudis.2007.03.011. [DOI] [PubMed] [Google Scholar]
- 35.Lee S, Wang Q. Recent development of small molecular specific inhibitor of protein tyrosine phosphatase 1B. Med Res Rev. 2007;27(4):553–573. doi: 10.1002/med.20079. [DOI] [PubMed] [Google Scholar]
- 36.Zhang ZY, Lee SY. PTP1B inhibitors as potential therapeutics in the treatment of type 2 diabetes and obesity. Expert Opin Investig Drugs. 2003;12(2):223–233. doi: 10.1517/13543784.12.2.223. [DOI] [PubMed] [Google Scholar]
- 37.Ukkola O, Santaniemi M. Protein tyrosine phosphatase 1B: A new target for the treatment of obesity and associated co-morbidities. J Intern Med. 2002;251(6):467–475. doi: 10.1046/j.1365-2796.2002.00992.x. [DOI] [PubMed] [Google Scholar]
- 38.Jiang ZX, Zhang ZY. Targeting PTPs with small molecule inhibitors in cancer treatment. Cancer Metastasis Rev. 2008;27(2):263–272. doi: 10.1007/s10555-008-9113-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang ZY. Functional studies of protein tyrosine phosphatases with chemical approaches. Biochim Biophys Acta. 2005;1754(1-2):100–107. doi: 10.1016/j.bbapap.2005.09.005. [DOI] [PubMed] [Google Scholar]
- 40.Tonks NK. Protein tyrosine phosphatases: From genes, to function, to disease. Nat Rev Mol Cell Biol. 2006;7(11):833–846. doi: 10.1038/nrm2039. [DOI] [PubMed] [Google Scholar]
- 41.Xie L, et al. Cellular effects of small molecule PTP1B inhibitors on insulin signaling. Biochemistry. 2003;42(44):12792–12804. doi: 10.1021/bi035238p. [DOI] [PubMed] [Google Scholar]
- 42.Han Y, et al. Discovery of [(3-bromo-7-cyano-2-naphthyl)(difluoro)methyl]phosphonic acid, a potent and orally active small molecule PTP1B inhibitor. Bioorg Med Chem Lett. 2008;18(11):3200–3205. doi: 10.1016/j.bmcl.2008.04.064. [DOI] [PubMed] [Google Scholar]
- 43.Flint AJ, Tiganis T, Barford D, Tonks NK. Development of “substrate-trapping” mutants to identify physiological substrates of protein tyrosine phosphatases. Proc Natl Acad Sci USA. 1997;94(5):1680–1685. doi: 10.1073/pnas.94.5.1680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Myers MP, et al. TYK2 and JAK2 are substrates of protein-tyrosine phosphatase 1B. J Biol Chem. 2001;276(51):47771–47774. doi: 10.1074/jbc.C100583200. [DOI] [PubMed] [Google Scholar]
- 45.Salmeen A, Andersen JN, Myers MP, Tonks NK, Barford D. Molecular basis for the dephosphorylation of the activation segment of the insulin receptor by protein tyrosine phosphatase 1B. Mol Cell. 2000;6(6):1401–1412. doi: 10.1016/s1097-2765(00)00137-4. [DOI] [PubMed] [Google Scholar]
- 46.Sarmiento M, et al. Structural basis of plasticity in protein tyrosine phosphatase 1B substrate recognition. Biochemistry. 2000;39(28):8171–8179. doi: 10.1021/bi000319w. [DOI] [PubMed] [Google Scholar]
- 47.LaMontagne KR, Jr, Flint AJ, Franza BR, Jr, Pandergast AM, Tonks NK. Protein tyrosine phosphatase 1B antagonizes signalling by oncoprotein tyrosine kinase p210 bcr-abl in vivo. Mol Cell Biol. 1998;18(5):2965–2975. doi: 10.1128/mcb.18.5.2965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ferrari E, et al. Identification of new substrates of the protein-tyrosine phosphatase PTP1B by Bayesian integration of proteome evidence. J Biol Chem. 2011;286(6):4173–4185. doi: 10.1074/jbc.M110.157420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hardy RW, Marcotrigiano J, Blight KJ, Majors JE, Rice CM. Hepatitis C virus RNA synthesis in a cell-free system isolated from replicon-containing hepatoma cells. J Virol. 2003;77(3):2029–2037. doi: 10.1128/JVI.77.3.2029-2037.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dowers KL, et al. Association of Bartonella species, feline calicivirus, and feline herpesvirus 1 infection with gingivostomatitis in cats. J Feline Med Surg. 2010;12(4):314–321. doi: 10.1016/j.jfms.2009.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hennet PR, Camy GA, McGahie DM, Albouy MV. Comparative efficacy of a recombinant feline interferon omega in refractory cases of calicivirus-positive cats with caudal stomatitis: A randomised, multi-centre, controlled, double-blind study in 39 cats. J Feline Med Surg. 2011;13(8):577–587. doi: 10.1016/j.jfms.2011.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chuang E, et al. Interaction of CTLA-4 with the clathrin-associated protein AP50 results in ligand-independent endocytosis that limits cell surface expression. J Immunol. 1997;159(1):144–151. [PubMed] [Google Scholar]
- 53.Shiratori T, et al. Tyrosine phosphorylation controls internalization of CTLA-4 by regulating its interaction with clathrin-associated adaptor complex AP-2. Immunity. 1997;6(5):583–589. doi: 10.1016/s1074-7613(00)80346-5. [DOI] [PubMed] [Google Scholar]
- 54.Zhang Y, Allison JP. Interaction of CTLA-4 with AP50, a clathrin-coated pit adaptor protein. Proc Natl Acad Sci USA. 1997;94(17):9273–9278. doi: 10.1073/pnas.94.17.9273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Trinchieri G. Type I interferon: Friend or foe? J Exp Med. 2010;207(10):2053–2063. doi: 10.1084/jem.20101664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Eden ER, White IJ, Tsapara A, Futter CE. Membrane contacts between endosomes and ER provide sites for PTP1B-epidermal growth factor receptor interaction. Nat Cell Biol. 2010;12(3):267–272. doi: 10.1038/ncb2026. [DOI] [PubMed] [Google Scholar]
- 57.Lu X, et al. PTP1B is a negative regulator of interleukin 4-induced STAT6 signaling. Blood. 2008;112(10):4098–4108. doi: 10.1182/blood-2008-03-148726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Valentino L, Pierre J. JAK/STAT signal transduction: Regulators and implication in hematological malignancies. Biochem Pharmacol. 2006;71(6):713–721. doi: 10.1016/j.bcp.2005.12.017. [DOI] [PubMed] [Google Scholar]
- 59.Elchebly M, et al. Increased insulin sensitivity and obesity resistance in mice lacking the protein tyrosine phosphatase-1B gene. Science. 1999;283(5407):1544–1548. doi: 10.1126/science.283.5407.1544. [DOI] [PubMed] [Google Scholar]
- 60.Klaman LD, et al. Increased energy expenditure, decreased adiposity, and tissue-specific insulin sensitivity in protein-tyrosine phosphatase 1B-deficient mice. Mol Cell Biol. 2000;20(15):5479–5489. doi: 10.1128/mcb.20.15.5479-5489.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Richter MF, Duménil G, Uzé G, Fellous M, Pellegrini S. Specific contribution of Tyk2 JH regions to the binding and the expression of the interferon alpha/beta receptor component IFNAR1. J Biol Chem. 1998;273(38):24723–24729. doi: 10.1074/jbc.273.38.24723. [DOI] [PubMed] [Google Scholar]
- 62.Zhao W, et al. A conserved IFN-alpha receptor tyrosine motif directs the biological response to type I IFNs. J Immunol. 2008;180(8):5483–5489. doi: 10.4049/jimmunol.180.8.5483. [DOI] [PubMed] [Google Scholar]
- 63.Kieseier BC, Stüve O. A critical appraisal of treatment decisions in multiple sclerosis—old versus new. Nat Rev Neurol. 2011;7(5):255–262. doi: 10.1038/nrneurol.2011.41. [DOI] [PubMed] [Google Scholar]
- 64.Wang BX, Rahbar R, Fish EN. Interferon: Current status and future prospects in cancer therapy. J Interferon Cytokine Res. 2011;31(7):545–552. doi: 10.1089/jir.2010.0158. [DOI] [PubMed] [Google Scholar]
- 65.Parisien JP, Lau JF, Rodriguez JJ, Ulane CM, Horvath CM. Selective STAT protein degradation induced by paramyxoviruses requires both STAT1 and STAT2 but is independent of alpha/beta interferon signal transduction. J Virol. 2002;76(9):4190–4198. doi: 10.1128/JVI.76.9.4190-4198.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sharma S, et al. Triggering the interferon antiviral response through an IKK-related pathway. Science. 2003;300(5622):1148–1151. doi: 10.1126/science.1081315. [DOI] [PubMed] [Google Scholar]
- 67.Goldman LA, et al. Characterization of antihuman IFNAR-1 monoclonal antibodies: Epitope localization and functional analysis. J Interferon Cytokine Res. 1999;19(1):15–26. doi: 10.1089/107999099314379. [DOI] [PubMed] [Google Scholar]
- 68.Li Y, Gazdoiu S, Pan ZQ, Fuchs SY. Stability of homologue of Slimb F-box protein is regulated by availability of its substrate. J Biol Chem. 2004;279(12):11074–11080. doi: 10.1074/jbc.M312301200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.