Abstract
Calsequestrins (CSQ) are high capacity, medium affinity, calcium-binding proteins present in the sarcoplasmic reticulum (SR) of cardiac and skeletal muscles. CSQ sequesters Ca2+ during muscle relaxation and increases the Ca2+-storage capacity of the SR. Mammalian CSQ has been well studied as a model of human disease, but little is known about the environmental adaptation of CSQ isoforms from poikilothermic organisms. The mummichog, Fundulus heteroclitus, is an intertidal fish that experiences significant daily and seasonal environmental fluctuations and is an interesting study system for investigations of adaptation at the protein level. We determined the full-length coding sequence of a CSQ isoform from skeletal muscle of F. heteroclitus (FCSQ) and characterized the function and structure of this CSQ. The dissociation constant (Kd) of FCSQ is relatively insensitive to changes in temperature and pH, thus indicating that FCSQ is a eurytolerant protein. We identified and characterized a highly conserved salt bridge network in FCSQ that stabilizes the formation of front-to-front dimers, a process critical to CSQ function. The functional profile of FCSQ correlates with the natural history of F. heteroclitus suggesting that the eurytolerant function of FCSQ may be adaptive.
Introduction
Fluctuating thermal environments pose a significant challenge to poikilothermic organisms. Animals that can exploit these variable thermal habitats are termed eurythermal. The eurythermal mummichog, Fundulus heteroclitus (Teleostei; Cyprinodontiformes), is a common killifish found in intertidal zones along the eastern coast of North America where it encounters fluctuating environmental, and thus physiological, temperatures during both seasonal and daily tidal cycles. Additionally, along its distribution, populations of F. heteroclitus experience an extreme thermal gradient: for every one degree change in latitude the mean annual water temperature changes by 1°C [1], [2]. Changes in temperature affect all biological and chemical processes, including the functions of and interactions between proteins [3]. The non-catalytic Ca2+-binding protein calsequestrin (CSQ), the focus of this study, undergoes large scale structural changes including folding and polymerization during Ca2+-binding [4] making it an interesting study system for investigations of environmental effects on protein structure and function.
Contraction and relaxation in skeletal and cardiac muscles are mediated in large part by the release and uptake of Ca2+ by the sarcoplasmic reticulum (SR). In vertebrates, CSQ is found in the SR where it is a high capacity Ca2+-binding protein localized to the terminal cisternae [5]–[8]. In mammals, CSQ sequesters a relatively large amount of Ca2+ (40–50 moles Ca2+ per mole CSQ) [5], [6], keeping levels of free Ca2+ low (0.2 mM–0.5 mM) within the SR during relaxation [9], [10]. Royer and Ríos [11] calculated that the total concentration of Ca2+ in the terminal cisternae of SR is between 35 mM and 175 mM. During a single twitch contraction there is a relatively small (∼10%) decrease in free Ca2+ in the SR, but more Ca2+ is released from the SR during repetitive contraction or tetanic events [5], [6], [9], [10], [12], [13]. In vertebrates, Ca2+ sequestration by CSQ enables fast twitch glycolytic and cardiac muscles to undergo repetitive high frequency cycles of contraction and relaxation without a cycle-by-cycle depression in developed tension, a key characteristic of both of these muscle types [14]–[18]. Additionally, CSQ may play a regulatory role in Ca2+ release through interactions with the ryanodine receptor (RyR) mediated through junctional SR proteins triadin and junctin [19], [20].
CSQ isoforms consist of three, contiguous thioredoxin-like domains each composed of a hydrophobic core surrounded by acidic residues that form a negatively charged surface [4], [21]. A model that is based on X-ray crystallographic evidence from mammalian cardiac and skeletal isoforms of CSQ illustrates the Ca2+-dependent conformational changes at the secondary, tertiary, and quaternary levels [4], [21], [22]. In the absence of Ca2+, CSQ has a random coil structure. As Ca2+ concentration increases to 10 µM, CSQ gains α-helical content as the three thioredoxin-like domains fold. Above 10 µM Ca2+, the full monomeric structure is reached. Front-to-front followed by back-to-back dimers form at 10 µM–1 mM Ca2+. Higher order linear polymers form above 1 mM Ca2+ [22], [23]. Back-to-back and front-to-front dimers form electronegative pockets where Ca2+ ions are transiently coordinated by pairs of negatively charged residues [12]. This transient binding may also allow faster diffusion of Ca2+ along the CSQ polymer and more effective delivery of Ca2+ to the RyR [11].
A number of non-covalent interactions among amino acid residues within mammalian CSQ have been identified that may be required for proper monomer folding, polymerization, and interaction with other proteins [4], [21]. Beard et al. [12] summarized the current understanding of CSQ polymerization and the role of the previously identified non-covalent contacts in polymerization. Interactions between the highly acidic C-terminal tails of two CSQ molecules lead to the formation of a back-to-back dimer, and a cluster of salt bridge interactions (Glu215-Lys86, Glu25-Lys24 and Glu169-Lys85) tethers this back-to-back formation. Front-to-front dimerization is stabilized by the salt bridge Glu55-Lys49, and similar to the back-to-back dimer, an acidic pocket forms between the monomers upon dimerization [12].
Paralogous isoforms of CSQ are expressed in vertebrate fast twitch skeletal muscle (CSQ1) and slow twitch skeletal and cardiac muscles (CSQ2). While a significant amount of research has focused on CSQ structure and function in mammals, much less is known about CSQ in fishes. CSQ has been identified in skeletal muscle and heater cells of fishes including carp (Cyprinus carpio) and blue marlin (Makaira nigricans), respectively [24], [25], but the CSQ isoform was not determined. Additionally, the sole (Solea senegalensis) expresses two of each CSQ isoform (e.g. CSQ2A and CSQ2B) [26]. In the present study, we determined the full-length coding sequence of a CSQ isoform from F. heteroclitus skeletal muscle (FCSQ), and subsequently expressed, purified, and characterized the function and structure of this CSQ. Interestingly, FCSQ is relatively insensitive to changes in temperature and pH, which indicates FCSQ is a eurytolerant protein [27]. The function of FCSQ correlates with the natural history of F. heteroclitus; proteins that are relatively insensitive to changes in environmental and physiological conditions may be advantageous to organisms that live in dynamic environments. Additionally, computational modeling of FCSQ reveals a highly conserved, charged interaction network that appears to be critical for proper CSQ function. This interaction network is equivalent to one identified in mammalian CSQ2 that is required for proper polymerization and Ca2+-binding [28]. The high level of conservation at the primary, secondary, and tertiary structural levels across CSQ isoforms and species indicates that conclusions based on studies of FCSQ will be broadly applicable to isoforms from a variety of vertebrates.
Materials and Methods
Ethical Statement
Animal Use Protocols (00014-2008 and 00038-2011) were approved by Valdosta State University’s Institutional Animal Care and Use Committee (Animal Welfare Assurance Number A4578-01). Care was taken to minimize pain and discomfort of animals.
Collection of Specimens
Fundulus heteroclitus were caught off the coast of Jekyll Island, Georgia, USA (31°2′27.37′′N and 81°25′21.66′′W) with minnow traps. Fish were transported to Valdosta State University and held in re-circulating seawater (28 ppt) tanks until sacrificed. They were fed daily with flake food. Fish were euthanized with Finquel (MS-222) followed by spinal transection and pithing. Following removal of the skin, glycolytic skeletal muscle samples were taken from the axial musculature on the dorsal side of each individual immediately below or anterior to the dorsal fin. Muscle was either immediately used for RNA isolation or frozen at −80°C until use.
RNA Isolation, cDNA Preparation, and Sequencing
Total RNA from F. heteroclitus was isolated from skeletal muscle using TRIzol reagent (Invitrogen) following manufacturer’s instructions. Single-strand cDNA was made from total RNA (5 µg) using oligo (dT) primer (Table 1) and Superscript III (Invitrogen) as per manufacturer’s protocol. cDNA was stored at −20°C until used to amplify F. heteroclitus CSQ cDNA (FCSQ).
Table 1. Primers used for amplification of Fundulus heteroclitus CSQ cDNA.
| aPrimer Name | bPrimer Sequence | cDNA positions |
| RT-PCR Primer | ||
| Oligo (dT) | 5′-GCTTTTTTTTTTTTTTTTTTTT-3′ | Poly(A) tail |
| Degenerate primers | ||
| CSQ8F | 5′-GCWGCYCARGTCTTGGARGA-3′ | 241 → 260 |
| CSQ11R | 5′-CCWGAMAGRACATCYTCKATCCA-3′ | 1084 ← 1106 |
| Fundulus specific primers | ||
| CSQAF | 5′-GAGGAGCCAGTGGAGGTCATTG-3′ | 430 → 451 |
| CSQBR | 5′-AGATGATGCTGAGGTCAGGAAGG-3′ | 895 ← 917 |
| CSQF1 | 5′-ACATTTCTCCATCCCTTCCAAG-3′ | −112 → −91 |
| CSQR1 | 5′-ATTTCATTTTGACCCCACCA-3′ | 1505 ← 1524 |
| CSQF2 | 5′-ATGGAAAAGGGCCTGGAG-3′ | 58 → 76 |
| CSQR2 | 3′-TTACTCATCATCATCATCATCATCA-3′ | 1280 ←1305 |
F indicates forward primers; R indicates reverse primers.
M = A or C; R = A or G; Y = C or T; W = A or T; K = G or T.
Locations of primers are based on cDNA consensus sequence of Fundulus heteroclitus CSQ. Position 1 indicates the putative start site (ATG). Negative positions indicate positions upstream of the cDNA start site.
Initial amplifications of partial FCSQ single-strand cDNA were performed in 50 µl reaction volumes containing 100 ng/µl cDNA, 1X Advantage 2 polymerase buffer, 1X Advantage 2 polymerase mix (Clontech), 200 µM dNTPs, and 500 nM degenerate primers (Table 1). Degenerate primers, CSQ8F and CSQ11R, were designed from an alignment of available teleost CSQ1 and CSQ2 sequences (NCBI Accession Numbers: Danio rerio (zebrafish) CSQ2- NM_001002682; F. heteroclitus (mummichog) CSQ2- CV824367; Takifugu rubripes (pufferfish) CSQ1- CK829454 and CK829229. Reactions were as follows: initial denaturation of 2 min at 94°C, followed by 35 cycles of 45 sec at 94°C with an annealing step of 1 min at 62°C and an elongation step of 45 sec at 72°C; the reaction concluded with a final 5 min elongation at 72°C. Purified PCR products were ligated in pCR 2.1 TOPO vector, and TOP10 One Shot cells (Invitrogen) were transformed as per manufacturer’s instructions. Plasmids containing inserts were sequenced at the Florida State University Sequencing Facility.
5′ and 3′ RACE-ready cDNAs from FCSQ were prepared using the SMARTer RACE Amplification Kit (Clontech) following the manufacturer’s instructions. Based on partial FCSQ nucleotide sequences, gene specific primers CSQAF and CSQBR (Table 1) were designed for use in 3′ and 5′ RACE-PCR, respectively. Amplification was performed with Advantage 2 DNA polymerase with the gene specific primer and Universal primer. The 5′ and 3′ cDNA samples were prepared under the following conditions in 50 µl reactions: 2.5 µl first-strand cDNA, 1X Universal Primer Mix (UPM), 200 µM gene specific primer, 1X Advantage 2 polymerase buffer, 200 µM dNTPs, 1X Advantage 2 DNA polymerase mix. Touchdown PCR was performed as follows: 5 cycles of denaturation at 94°C for 30 sec and annealing and extension at 72°C for 3 min; 5 cycles of denaturation at 94°C for 30 sec, annealing at 70°C for 30 sec, and extension at 72°C for 3 min; and 25 cycles of denaturation at 94°C for 30 sec, annealing at 68°C for 30 sec, and extension at 72°C for 3 min. Samples were cloned, sequenced, and aligned as described above. The full-length coding sequence of FCSQ cDNA was then amplified with gene specific primers, CSQF1 and CSQR1 (Table 1). Reaction mixtures were identical to those used to obtain partial FCSQ sequence, and PCR reactions were similar except the extension time for each cycle was 75 sec. PCR products were cloned and sequenced as described above. A consensus sequence of FCSQ was generated from an alignment of full-length FCSQ from three individuals.
Sequence Comparisons
To visualize the evolutionary relationship of FCSQ to other teleost CSQ isoforms, a phylogenetic tree was constructed. A multiple sequence alignment of teleost CSQ amino acid sequences including the SR targeting sequence (NCBI Accession Numbers: Solea senegalensis (sole) CSQ1A- BAG49512.1, CSQ1B- BAH85793.1, CSQ2A- BAG49513.1, CSQ2B- BAH85794.1; Oreochromis niloticus (tilapia) CSQ1A- XP_003457488.1, CSQ1B- XP_003454795.1, CSQ2A- XP_003455497.1, CSQ2B- XP_003440974.1; Danio rerio (zebrafish) CSQ1A- NP_001003620.2, CSQ1B- NP_001070192.1, CSQ2A- AAH75775.1; Oncorhynchus mykiss (trout) CSQ2A- NP_001153971.1; Salmo salar (salmon) CSQ1A- ACH70680.1) was performed using ClustalW on the EBI server with default parameters [29]. The phylogenetic tree was estimated from the sequence alignment using the neighbor-joining method in ClustalW with default parameters.
Expression and Purification of Recombinant Teleost FCSQ
Full-length coding sequence of FCSQ minus the predicted signal sequence was amplified via PCR as described above using the gene specific primers CSQF2 and CSQR2 (Table 1). PCR products of expected length were ligated into PetBlue-1 AccepTor Vector (Novagen). Colonies containing plasmid with correct insert orientation were identified by digestion with XmnI, and the sequence was confirmed by sequencing. Tuner(DE3)pLacI cells (Novagen) were transformed with plasmid containing FCSQ as per manufacturer’s instructions. LB broth containing 100 µg/ml ampicillin and 1% glucose was inoculated and grown at 37°C with agitation until OD600 of 1. Temperature of the cultures was decreased to 15°C prior to induction of expression with 500 µM IPTG for 24 hours. Cells were harvested by centrifugation and stored at −80°C until FCSQ purification.
FCSQ was purified following the protocol described in Cala and Jones [30] with minor modifications. Briefly, cells were resuspended in Buffer A (10 mM MOPS, 0.1 mM EGTA, 500 mM NaCl, 1 mM DTT, pH 7.0) and lysed by intermittent sonication for 15–20 sec with cooling on ice after each sonication. The homogenate was centrifuged for 10 min at 14,000×g, and the supernatant was applied to a phenyl sepharose column (2×13 cm) equilibrated with Buffer A. The column was washed with 100 mL Buffer A. Protein was eluted with 100 mL Buffer A containing 10 mM CaCl2 followed by 100 mL 10 mM MOPS, pH 7.0. Fractions containing recombinant FCSQ were identified using 10% Tris/Glycine SDS-PAGE; gels were stained with Coomassie Blue. Fractions containing partially purified FCSQ were combined, exhaustively dialyzed against 0.1 M potassium phosphate, 1 mM EGTA, pH 7.1, and applied to a DEAE sepharose column (2.5×8 cm) equilibrated with 0.1 M potassium phosphate, 1 mM EGTA, pH 7.1. The column was washed with the same buffer, and FCSQ was eluted using a linear gradient of 0–1.0 M NaCl in 0.1 M potassium phosphate, 1 mM EGTA, pH 7.1.
Fractions containing FCSQ were identified using 10% Tris/Glycine SDS-PAGE as described above and combined; purified FCSQ was concentrated using Amicon Ultra-15 centrifugal filter devices with a 10 kDa cutoff (Millipore). FCSQ identity and purity were verified by 10% Tris/Glycine SDS-PAGE and subsequent staining with either Stains-all, which stains Ca2+-binding proteins blue [31], [32], or Coomassie Blue. In addition, N-terminal Edman sequencing of purified recombinant FCSQ was conducted at Florida State University’s Analytical Lab and Research Facility.
Decalcification of FCSQ and Buffer Solutions
Divalent cations were removed from recombinant FCSQ by dialysis against 10 mM Tris-HCl, pH 7.5, containing 5% Chelex (w/v) at 4°C for 4 days with continuous stirring, changing buffer every 24 hours. Divalent cations were removed from buffer used in the fluorometric assay by the addition of 5% Chelex with continuous stirring at 4°C for 4 days, changing Chelex every 24 hours. Because decalcification with Chelex increases pH of buffers, the pH was adjusted immediately prior to assay using 1.0 N and 0.5 N HCl in 10 mM Tris-HCl that had been decalcified with 5% Chelex. Decalcification of FCSQ and buffers was confirmed by atomic absorption spectroscopy at Valdosta State University’s Advanced Spectroscopy and Biotechnology Facility.
Intrinsic Fluorescence Assay and Calculation of Kd
Concentration of decalcified FCSQ was determined using the bicinchoninic acid (BCA) protein assay kit (Sigma) prior to fluorescence assays. An intrinsic fluorescence assay that relies on tryptophan fluorescence was modified from Park et al. [23] and Hidalgo et al. [33]. For the temperature dependence experiments, CaCl2 standard (0.1 M; Orion) was added stepwise to FCSQ (15 µg/ml) in 10 mM Tris-HCl, pH 7.5, at 25°C. The pH was allowed to vary with temperature of the assay (10, 15, 25, or 35°C). For the pH dependence experiments, the pH of 10 mM Tris-HCl was adjusted to pH 6.6, 6.8, 7.2, or 7.5 at 25°C, and the assays were run at 15°C. The sample was allowed to equilibrate five minutes after each addition of Ca2+. Emission fluorescence was measured with an LS 55 Fluorescence Spectrometer (PerkinElmer). Excitation wavelength was 282 nm, and emission wavelength was 331 nm; 5 nm slit width was used for both excitation and emission.
A minimum of three FCSQ preparations was used for each experimental condition, and assays using each of the protein preparations were run in triplicate. Estimates for dissociation constant (Kd) were calculated using the following equation [34]:
| (1) |
where Y represents fluorescence, KH represents the Hill constant, n represents the index of cooperativity or Hill coefficient, and [Ca] represents the calcium concentration. The Hill constant indicates the concentration of ligand that accounts for ½ the maximal response, which in the case of fluorescence assays is the concentration of calcium that results in ½ the maximal fluorescence. The Kd values were calculated as Kd = [KH]n. Standard error was calculated from the averages obtained for each FCSQ protein preparation. One-way ANOVA, followed by Tukey HSD multiple pairwise comparisons with significance set at 5% was used to determine significant differences in dissociation constants (Kd) and Hill coefficients (n).
Molecular Modeling
Sequences were aligned using ClustalW version 2.0 housed on the EBI server [29]. A homology model of FCSQ was built using the crystal structure of CSQ2 from dog (Canis lupus familiaris), DCSQ2 (PDB ID: 1SJI) as a template [4]. The first 20 residues of the coding sequence were identified as the SR targeting signal sequence and were removed prior to modeling. The C-terminal tail was trimmed to match the primary structure of DCSQ2.
The model was built using SwissModel [35]. The first two N-terminal residues and the C-terminal tail of FCSQ were truncated to match the template structure, producing a monomer with 350 residues and dimer with 700 residues (chain A residues 1–350 and chain B residues 351–700, equivalently 1′-350’). Model quality was validated using SwissModel built-in evaluation procedures. Visual Molecular Dynamics (VMD) [36] and Swiss-PDB Viewer [37] were used to visualize the molecules. A molecular model of FCSQ front-to-front dimer was made using PyMol [38].
The VMD Autopsf plugin was used to add missing hydrogen atoms and to create atom connectivity files (psf files) with the Charmm force field topology file top_all22_prot.inp [39]. The Salt Bridge plugin of VMD was used to calculate the distance between oxygen and nitrogen atoms in the protein and to determine which residues are involved in salt bridging based on a 3.2 Å distance constraint. Angles between participating oxygen and nitrogen atoms were calculated manually in VMD. An explicit TIP3P water box with 10 Å padding was created for each system, which was ionized with K+ ions to neutralize charge [40].
Electrostatic interaction energies, U(r), in solvated systems are estimated with Coulomb’s law screened by an effective dielectric constant:
| (2) |
where ke is the Coulomb constant 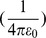 , q is the charge, r is the distance between the basic nitrogen (donor) and the acidic oxygen closest to the donor (acceptor), θ is the angle between N-H••O, and Deff is the effective dielectric constant. Indeed, the net contribution of a salt bridge interaction to protein folding free energy can only be obtained through computation [41].
, q is the charge, r is the distance between the basic nitrogen (donor) and the acidic oxygen closest to the donor (acceptor), θ is the angle between N-H••O, and Deff is the effective dielectric constant. Indeed, the net contribution of a salt bridge interaction to protein folding free energy can only be obtained through computation [41].
A linear distance-dependent dielectric (DDD) [42] was chosen to estimate Deff:
| (3) |
where ε(r) is the functional form of Deff in Equation 2, A is 2, and r is the distance between donor and acceptor. The constant, A, describes the level of solvent screening effect and is based on the level of exposure of the interaction to solvent. Since the interactions are buried in the dimer interface a value of 2 was chosen for A, indicating minor solvent exposure.
Results and Discussion
F. heteroclitus CSQ Primary Structure
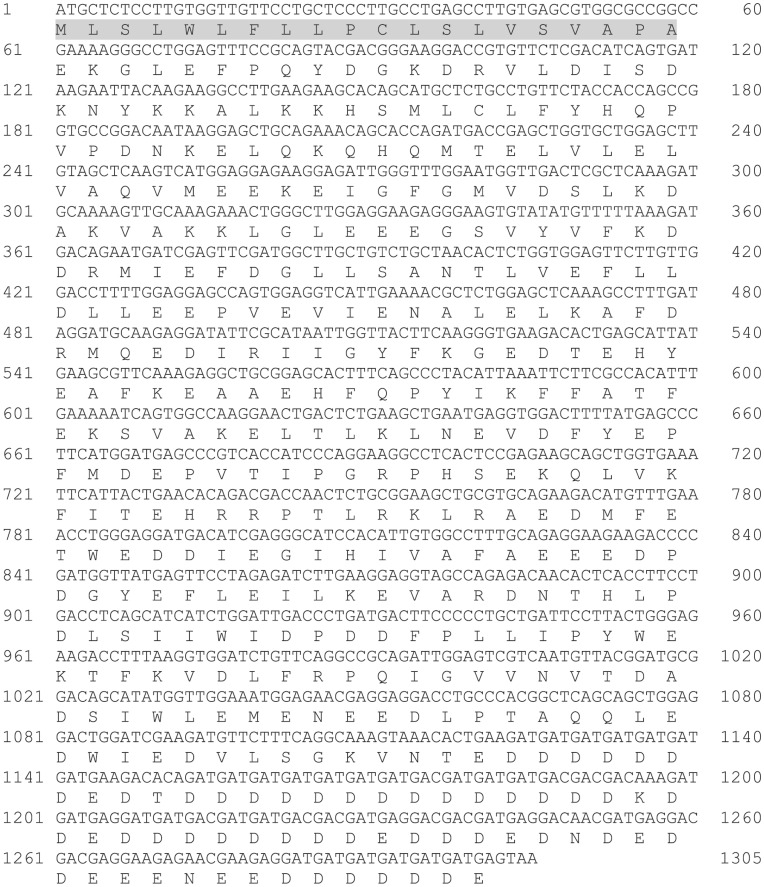
We isolated and determined the full-length coding sequence of a CSQ isoform from the southern population of F. heteroclitus (FCSQ; NCBI Accession Number: HQ615687) with an open-reading frame of 1,305 bases (Figure 1). The first 20 amino acids, the SR targeting sequence, are cleaved after import into the SR [43]. The mature FCSQ deduced amino acid sequence is predicted to be 414 amino acids with a calculated molecular weight of 48,474 Da. Similar to other vertebrate CSQs, FCSQ has a high proportion of acidic residues with 137 of the 414 amino acid residues (33%) being aspartic acid and glutamic acid. These acidic amino acid residues are responsible for forming clusters of 2–3 amino acids that coordinate Ca2+, thus playing a critical role in the Ca2+-binding capacity of CSQ [44].
Figure 1. Nucleotide and deduced amino acid sequence of the coding region of FCSQ cDNA.
Sequences were reconstructed from overlapping sequences of PCR amplified fragments. Primer sequences were removed prior to alignment. The putative SR targeting sequence is shaded.
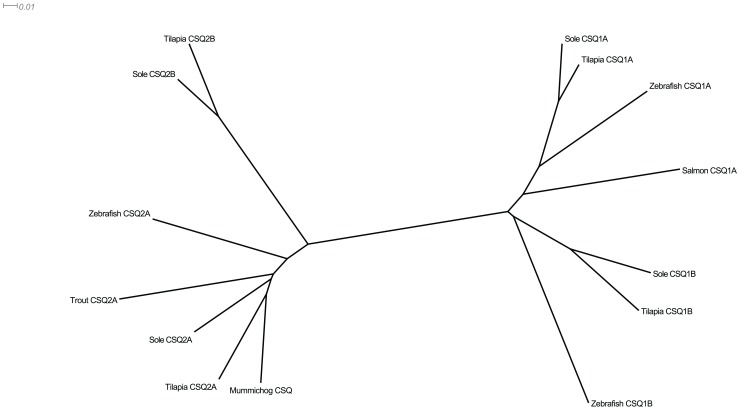
A phylogenetic analysis of FCSQ with other fish CSQ sequences shows FCSQ to be most similar to the oxidative skeletal muscle and cardiac isoform, CSQ2A (Figure 2). Primary sequence comparisons between FCSQ and other teleost CSQs show 56–60% identity (78–80% similarity) and 71–88% identity (88–97% similarity) for CSQ1 and CSQ2 isoforms, respectively. FCSQ cDNA was amplified from F. heteroclitus skeletal muscle cDNA using degenerate primers that were based on an alignment of CSQ1 and CSQ2 isoforms from D. rerio, F. heteroclitus, and T. rubripes, as described above. Due to the arrangement of oxidative and glycolytic fibers in teleosts, we cannot discount the presence of oxidative fibers in muscle samples that were used to isolate total RNA, which was subsequently used in RT-PCR and CSQ amplification. Therefore, based on sequence analysis, it is likely that the PCR reactions using degenerate primers amplified a CSQ isoform expressed in oxidative skeletal muscles, specifically, CSQ2A.
Figure 2. Neighbor-joining tree displaying relationships between CSQ orthologs and paralogs.
Teleost fishes express multiple isoforms of CSQ in a variety of tissues. FCSQ (mummichog CSQ) aligns most closely with CSQ2A isoforms.
Recent research on CSQ expression in poikilothermic fishes has found that there are more than two isoforms of CSQ. The naming of CSQ isoforms in fishes seems to be based more on similarity to either mammalian CSQ1 or CSQ2 rather than tissue specific expression. Korajoki and Vornanen [45] identified multiple transcripts of CSQ2 in cardiac muscle of rainbow trout (Oncorhynchus mykiss) that are products of a single gene and not paralogous genes. In addition, Infante et al. [26] isolated two isoforms each of CSQ1 and CSQ2 from ESTs and cDNA libraries from the Senegalese sole (Solea senegalensis) that were not tissue specific. They also found that all four isoforms were expressed in ten different tissues [26]. An area of future research is to identify isoforms and expression patterns of CSQ in F. heteroclitus skeletal and cardiac muscles and to characterize the functions of these isoforms in this eurythermal teleost.
Functional Characterization
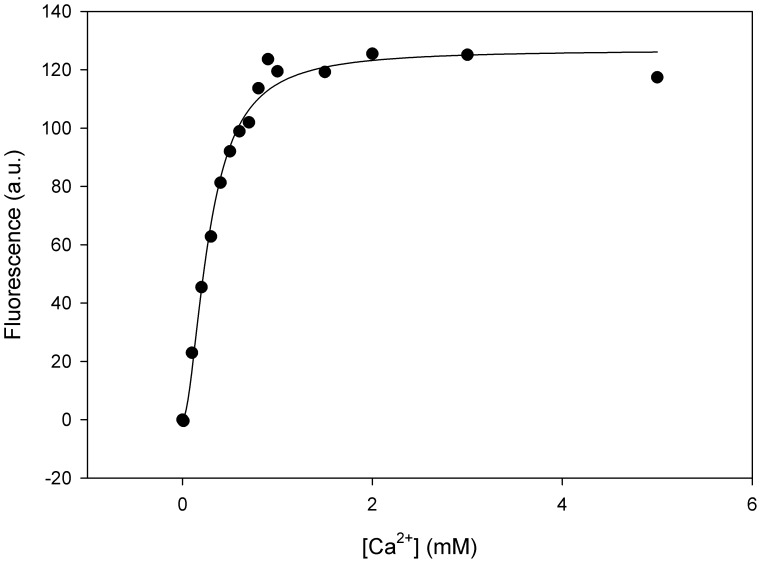
This study is the first to investigate the sensitivity of FCSQ function to changes in temperature and pH. Dissociation constants of Ca2+ were determined using an assay that monitors intrinsic tryptophan fluorescence. Change in intrinsic tryptophan fluorescence in response to Ca2+ is interpreted as a conformational change of CSQ during Ca2+-binding and has been demonstrated in human cardiac CSQ [46]–[48] and dog cardiac CSQ [23]. Intrinsic fluorescence has also been used to determine the equilibrium binding constants [33] and dissociation kinetics of rabbit skeletal CSQ [20], [49]. This study is the first to apply this technique to CSQ from a teleost and to show that the intrinsic fluorescence of FCSQ is sensitive to Ca2+ (Figure 3).
Figure 3. Representative fluorescence data of Ca2+-binding by FCSQ.
Fitting of this data yields a Kd value of 102 µM and a Hill coefficient of 1.8.
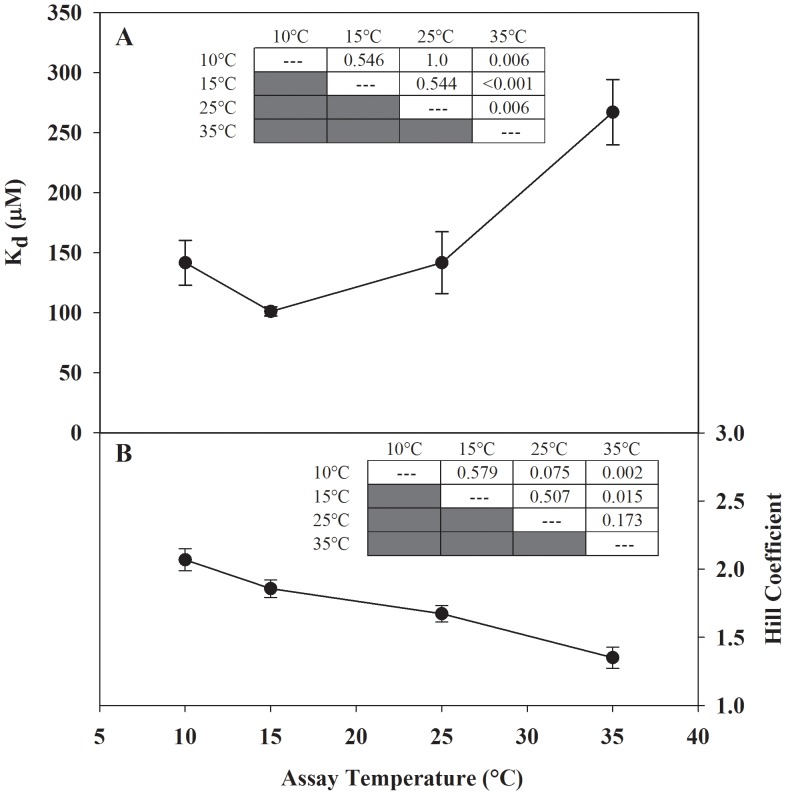
The FCSQ isoform examined in the current study is relatively insensitive to changes in temperature and pH. In the physiological temperature range for F. heteroclitus, 10°C to 25°C, there are no statistical differences in Kd, but at 35°C the Kd is significantly different (higher) than the Kd values at the lower temperatures (Figure 4A). In laboratory acclimated animals a heat shock response is elicited at 35°C [50], which is at the high end of the thermal range of F. heteroclitus. The increase in Kd at 35°C, which correlates with the heat shock response, may be a result of partial unfolding or increased flexibility within regions of the protein that leads to a reduced affinity for Ca2+, and thus decreased Ca2+-binding ability at higher temperatures.
Figure 4. Effect of temperature on the apparent dissociation constant (Panel A) and Hill coefficient (Panel B) of Ca2+-binding by FCSQ.
Each data point represents a minimum of three protein preparations run in triplicate. Significant differences were determined using one way ANOVA followed by Tukey HSD multiple pairwise comparisons with significance set at 5%. Inset tables show P values from all pairwise comparisons. Error bars represent S.E.M.
Interestingly, the longjaw mudsucker, Gillichthys mirabilis, which is distantly related to F. heteroclitus, has a similar eurythermal lifestyle and latitudinal distribution, but on the Pacific Coast. The apparent Km vs. temperature curves for the eurytolerant pyruvate kinase (PK) isoform from G. mirabilis binding to its substrate, phosphoenolpyruvate, show a trend that is qualitatively similar to the Kd vs. temperature curve for FCSQ Ca2+-binding. In the physiological range of temperatures of G. mirabilis (10°C–30°C) the Km of PK shows very little thermal sensitivity, but outside the range Km increases rapidly [27], [51]. These convergent phenotypes from G. mirabilis PK and F. heteroclitus CSQ suggest that thermal insensitivity may be an adaptive response utilized in proteins from organisms that experience significant and rapid environmental variability.
The change in Hill coefficient (n) in response to temperature shows a similar trend to the Kd measurements (Figure 4B). The Hill coefficients determined at 10°C and 35°C are significantly different. There are no significant differences among Hill coefficients determined from 10°C to 25°C. There is, however, a qualitative decreasing trend in Hill coefficient values with increasing temperature. A similar trend has been reported for an oligomeric alcohol dehydrogenase from the archaeon Sulfolobus solfataricus [52]. The Hill coefficient is a measure of the sigmoidicity of the binding curve, which is a reflection of the level of cooperativity of the binding mechanism [34]. An n value of 1 indicates a non-cooperative binding mechanism, while n <1 indicates negative cooperativity, and n >1 indicates positive cooperativity. The cooperative mechanism of CSQ Ca2+-binding can be easily described using the scheme outlined in the Introduction. As [Ca2+] increases, CSQ folds and forms front-to-front and back-to-back dimers where the majority of Ca2+ is coordinated [22]. As the number of dimer interfaces increases, Ca2+-binding capacity of CSQ increases. Making the assumptions that the Hill coefficient reports on cooperativity, and cooperativity is mediated by polymerization, the decrease in Hill coefficient in response to increased temperature suggests that higher temperature inhibits FCSQ polymerization.
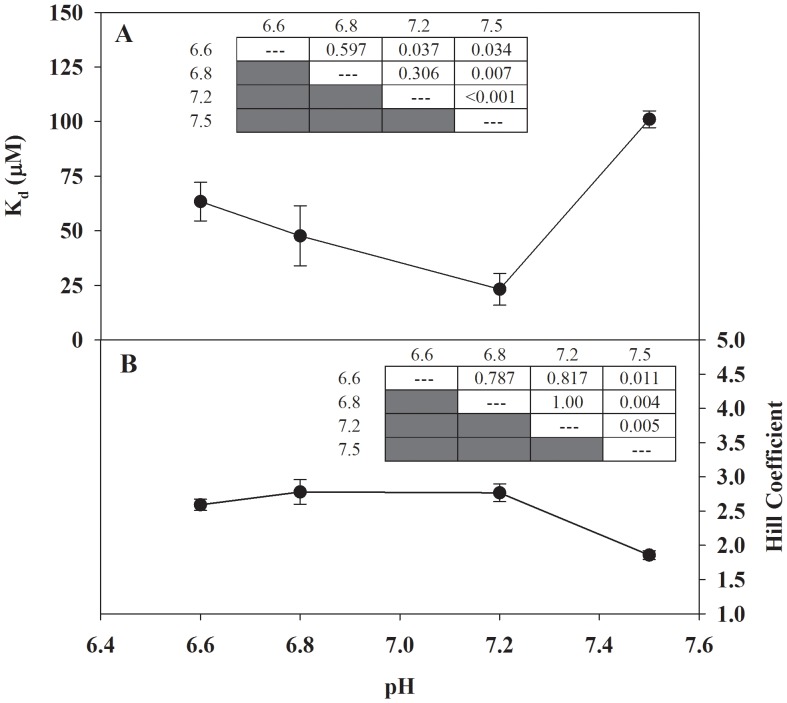
As intracellular temperature changes in a poikilothermic vertebrate, the intracellular pH also changes [53]–[55]; generally, for every 1°C increase in temperature, pH decreases by approximately 0.017 units [53]. Both temperature and pH have been shown to affect F. heteroclitus protein function [56], and the pH values used in our Kd vs. pH experiments are physiologically relevant to changes in pH that occur in this teleost. There is no significant difference in Kd values between pH 7.2 and 6.8, or between 6.8 and 6.6, but the Kd value calculated at pH 7.5 is statistically different (higher) then the Kd values calculated at lower pH (i.e. 6.6, 6.8, and 7.2; Figure 5A). The trend in Hill coefficients mirrors that for Kd values (Figure 5B). Overall, FCSQ function appears to be relatively insensitive to pH changes. This relative insensitivity of FCSQ function to pH variation contrasts with the function of rabbit skeletal CSQ [33]. While the Hill constant (KH) of rabbit CSQ remained relatively constant (∼300 µM), the Hill coefficient (n) decreased from 4.4 at pH 7.5 to ∼1 at pH 6.0 [33], which results in an inverse relationship between pH and Kd. The changes in Ca2+-binding ability and cooperativity of rabbit CSQ as a function of pH suggest that the contrasting functional profile of FCSQ may be an adaptive response of F. heteroclitus to the variable intertidal environment.
Figure 5. Effect of pH on the apparent dissociation constant (Panel A) and Hill coefficient (Panel B) of Ca2+-binding by FCSQ.
Each data point represents a minimum of three protein preparations run in triplicate. Significant differences were determined using one way ANOVA followed by Tukey HSD multiple pairwise comparisons with significance set at 5%. Inset tables show P values from all pairwise comparisons. Error bars represent S.E.M.
Structural Characterization
To investigate the structural basis of polymerization in FCSQ we characterized the electrostatic interactions at the front-to-front dimeric interface. The crystal structure of dog cardiac CSQ (DCSQ2) was used as the template for a homology model of FCSQ. The sequence identity between the target (FCSQ) and template (DCSQ2) sequences is 68.3%, well above the “twilight zone” threshold for homology modeling [57]. At this level of sequence identity SwissModel produces reliable models with resolutions similar to experimentally determined structures [58]. The root mean square deviation (RMSD), a measure of difference between two macromolecular structures, is 1.10 Å between the template and target structures, which is within the resolution of the template structure, 2.40 Å.
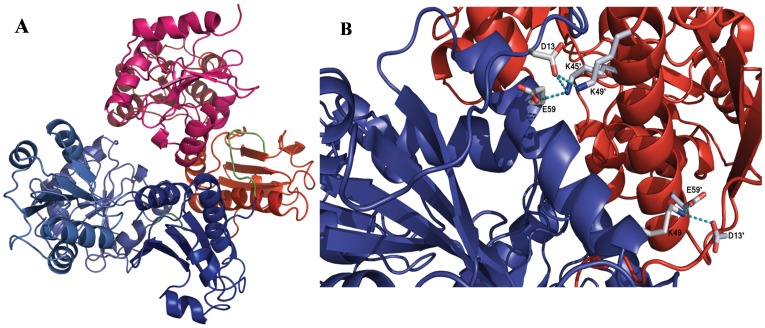
Based on the homology model of FCSQ, monomers of FCSQ will form front-to-front dimers (Figure 6A). Table 2 shows the residues involved in intermonomeric salt bridges and distances found in this analysis in both the template and target structures. Interestingly, in this model and in the crystal structure of DCSQ2 the residues are symmetrical, but the interactions are not. While the salt bridge between D13–K45’ (45′ indicates the opposing monomer) is present on one side of the dimer (Figure 6B), the reverse interaction, D13’-K45, is not fully formed. Similarly, the salt bridge between E300–K68’ is fully formed, while E300’-K68 is not present. Although not a symmetric interaction, this pair of salt bridges seems to be cooperative. When D13–K45’ and E300–K68’ are fully formed, D13’-K45 and E300’-K68 are not, causing one fully formed salt bridge interaction point between Domain I of monomer A and Domain I of monomer B. Most likely during a dynamic breathing mode of the protein, the intact salt bridge pair will swap back and forth while the more stable interactions between residues 13 and 49, and 49 and 59 of both monomers remain intact.
Figure 6. Model of FCSQ front-to-front dimer.
Panel A shows Chain A (blue) and Chain B (red) interacting to form an electronegative pocket where Ca2+ is coordinated by pairs of negatively charged residues. During front-to-front dimerization Domain I from each monomer interacts at the N-terminal end of the molecules. The N-terminal tails that participate in dimerization are shown in green. Panel B shows the salt bridge network that stabilizes the interactions between Domain I of Chains A and B. Participating side chains are shown in CPK coloring. Chain coloring as in panel A. Electrostatic interactions (salt bridges) are shown as dashed, cyan lines.
Table 2. Intermonomeric salt bridges of FCSQ and DCSQ2.
| FCSQ | Dog CSQ2 | |||||||
| Salt Bridge | Distance (Å) | U(r) (kcal/mol) | ε(r) = Deff | U(ε(r)) (kcal/mol) | Distance (Å) | U(r) (kcal/mol) | ε(r) = Def | U(ε(r)) (kcal/mol) |
| D13–K45’ | 3.01 | 18.79 | 6.0 | 3.12 | 2.95 | 21.47 | 5.9 | 3.64 |
| D13–K49’ | 2.86 | 23.61 | 5.7 | 4.13 | 2.91 | 23.85 | 5.8 | 4.10 |
| D13’–K49 | 2.90 | 15.43 | 5.8 | 2.66 | 2.89 | 15.84 | 5.8 | 2.74 |
| E300–K68’ | 3.11 | 22.94 | 6.2 | 3.69 | 2.95 | 22.54 | 5.9 | 3.82 |
| E59–K49’ | 3.08 | 22.29 | 6.2 | 3.62 | 3.02 | 22.67 | 6.0 | 3.75 |
| E59’-K49 | 2.74 | 26.76 | 5.5 | 4.88 | 2.83 | 26.47 | 5.7 | 4.68 |
Distance is measured from the center of mass of the nitrogen basic residue (donor) and the center of mass of the oxygen in the acidic residue (acceptor). U(r) is the interaction energy with no solvent screening effect. Deff is as described in the methods using a value of A = 2. U(ε(r)) is the interaction energy calculated using Deff that incorporates solvent screening.
Confirmation of the importance of the residues involved in these salt bridges is provided by the work of Bal et al. [28]. Using rat cardiac CSQ they found two N-terminal charge clusters that are critical for CSQ polymerization and function. They found that residues D32, K64, and K68 are involved in an intermonomeric salt bridge network that stabilizes front-to-front dimers [28]. These residues correspond to residues E13, K45, and K49 in our model of FCSQ. That these residues are conserved from fish to mammal is strong evidence that they are important for CSQ function across isoforms and species. This network acts as an important tether holding front-to-front dimers together (Figure 6A, B) [28]. In conjunction with the N-terminal arm, this tether helps position opposing CSQ monomers to allow pairs of negatively charged residues to bind Ca2+ molecules. Removal of any of these salt bridges should alter Ca2+-binding affinity and capacity, most likely negatively. Indeed, Bal et al. [28] found that alteration of the corresponding residues in rat cardiac CSQ diminished Ca2+-binding ability.
The strength of salt bridge interactions depends on pH, angle of the interaction, and distance between interacting residues, as well as the indirect effects of the solvent [59]. The interaction energies (U(r)) are given for both FCSQ and DCSQ2 using a Deff of 1 (Table 2), which is essentially using Coulomb’s law to measure the interaction of the residues in vacuum and thus yields high values for the salt bridge energy. U(ε(r)), in contrast, incorporates solvent screening effects by using a constant, A, as described by Mehler and Eichele [42]. This functional form of Deff is based on empirical data and provides a method of accounting for the shielding effects of the solvent when calculating salt bridge interaction energies. An A of 1 indicates an interaction buried in the protein interior with very little screening effects of the solvent; an A of 2 indicates a moderately solvent exposed interaction, and an A of 4 indicates a surface interaction that is highly exposed to solvent. While the residues involved in the tethering site are separated from solvent by only about 3 Å or more, the specific salt bridging atoms are protected from solvent by non-salt bridging atoms, giving at least one van der Waals layer of protection from solvent effects, and therefore are considered buried with moderate solvent exposure. The most solvent exposed residues are K68 (K68’) and E300 (E300’), but again, the specific salt bridging atoms are protected, in this case from other protein residues. Since all of the salt bridge interactions identified here are buried in the dimer interface, an A of 2 was chosen. While this model is an approximation method, the interaction energies calculated in this study correspond to values derived by experimental methods and other computational methods, and as such support our methodologies employed in this research [59], [60].
FCSQ Function as an Adaptation
Temperature fluctuations can affect all aspects of protein structure and function including ligand binding and oligomerization [3], two processes that are critical to CSQ function. The typical pattern of amino acid substitutions for thermal adaptation found in enzymatic systems leads to small changes in structure away from the active sites. These changes appear to modulate function through changes in conformational flexibility [3], [61], [62], [63]. Recently, this paradigm of thermal adaptation elucidated for enzymes was extended to the non-catalytic protein parvalbumin (PV), which binds Ca2+ in the nanomolar range [64]–[66]. Parvalbumin is a member of the canonical, EF-hand family of Ca2+- binding proteins. EF-hand proteins have a highly conserved helix-loop-helix motif that coordinates Ca2+ with high specificity and high affinity [67]. In addition, a similar mode of thermal adaptation has been demonstrated in troponin C (TnC), which is also an EF-hand Ca2+-binding protein [68]–[71]. However, it has yet to be established if the same underlying principles governing thermal adaptation of PV and enzymes apply to the non-canonical CSQ. In contrast to PV and TnC, CSQ binds Ca2+ in a non-specific manner with clusters of acidic residues transiently coordinating Ca2+ ions in the electronegative pockets formed between dimers [22].
Previously, thermal compensation has been demonstrated in proteins for which polymerization is critical to function (e.g. actin and tubulin). Actins from a variety of vertebrates adapted to different temperatures show thermal stability and polymerization thermodynamics that correlate with physiological temperature [72]. While a specific primary structure-based mechanism was not determined, it is likely that the differences found among different actins are due to amino acid substitutions that lead to differences in structure, which conserve polymerization ability at physiological temperature. A specific structural mechanism has been found for polymerization of microtubules from cold-adapted Antarctic fish. Longitudinal interfaces of microtubule monomers are highly conserved, while adaptive substitutions occur in the monomer core and on the lateral interfaces. These substitutions affect a small number of non-covalent interactions that conserve the polymerization ability of the microtubules at physiological temperature [73].
The functional results reported here suggest that the in vitro function of FCSQ correlates with the natural history of F. heteroclitus. The residues involved in polymerization and Ca2+- coordination in CSQ are highly conserved suggesting substitutions that modulate function in response to environmental, and thus physiological, temperature occur elsewhere in the molecule. Further comparative studies of FCSQ and CSQ isoforms from fishes native to other thermal habitats will be needed to determine if the paradigm of thermal adaptation in enzymes and EF-hand proteins applies to CSQ.
Acknowledgments
We thank Drs. H. Lee Woodcock, III, W. Ross Ellington, and Jonghoon Kang for comments on the manuscript, Dr. Steve Miller at the Florida State University Sequencing Facility for sample sequencing, and Dr. Gretchen K. Bielmyer at Valdosta State University for confirming decalcification using atomic absorption spectroscopy.
Funding Statement
This work was supported by a Valdosta State University Faculty Seed Research grant to TJG and a National Science Foundation grant (IOS-0817805) to TJG and TJF. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Powers DA (1989) Fish as model systems. Science 246: 352–358. [DOI] [PubMed] [Google Scholar]
- 2. Powers DA, Schulte PM (1998) Evolutionary adaptations of gene structure and expression in natural populations in relation to a changing environment: a multidisciplinary approach to address the million-year saga of a small fish. J Exp Zool 282: 71–94. [PubMed] [Google Scholar]
- 3.Hochachka PW, Somero GN (2002) Biochemical adaptation: mechanism and process in physiological evolution. New York, NY: Oxford University Press. 480 p.
- 4. Park H, Park IY, Kim E, Youn B, Fields K, et al. (2004) Comparing skeletal and cardiac calsequestrin structures and their calcium binding: a proposed mechanism for coupled calcium binding and protein polymerization. J Biol Chem 279: 18026–18033. [DOI] [PubMed] [Google Scholar]
- 5. MacLennan DH, Wong PTS (1971) Isolation of a calcium-sequestering protein from sarcoplasmic reticulum. Proc Natl Acad Sci U S A 68: 1231–1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Ikemoto N, Bhatnagar GM, Nagy B, Gergely J (1972) Interaction of divalent cations with the 55,000-dalton protein component of the sarcoplasmic reticulum. J Biol Chem 247: 7835–7837. [PubMed] [Google Scholar]
- 7. Cozens B, Reithmeier RAF (1984) Size and shape of rabbit skeletal muscle calsequestrin. J Biol Chem 259: 6248–6252. [PubMed] [Google Scholar]
- 8. Franzini-Armstrong C, Kenney LJ, Varriano-Marston E (1987) The structure of calsequestrin in triads of vertebrate skeletal muscle: a deep etch study. J Cell Biol 105: 49–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Rudolf R, Magalhães PJ, Pozzan T (2006) Direct in vivo monitoring of sarcoplasmic reticulum Ca2+ and cytoplasmic cAMP dynamics in mouse skeletal muscle. J Cell Biol 173: 187–193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Canato M, Scorzeto M, Giacomello M, Protasi F, Reggiani C, et al. (2010) Massive alterations of sarcoplasmic reticulum free calcium in skeletal muscle fibers lacking calsequestrin revealed by a genetically encoded probe. Proc Natl Acad Sci U S A 107: 22326–22331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Royer L, Ríos E (2009) Deconstructing calsequestrin. Complex buffering in the calcium store of skeletal muscle. J Physiol 587: 3101–3111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Beard NA, Laver DR, Dulhunty AF (2004) Calsequestrin and the calcium release channel of skeletal and cardiac muscle. Prog Biophys Mol Biol 85: 33–69. [DOI] [PubMed] [Google Scholar]
- 13. Ríos E, Launikonis BS, Royer L, Brum G, Zho J (2006) The elusive role of store depletion in the control of intracellular calcium release. J Muscle Res Cell Motil 27: 337–350. [DOI] [PubMed] [Google Scholar]
- 14. Hasenfuss G, Reinecke H, Studer R, Pieske B, Meyer M, et al. (1996) Calcium cycling proteins and force-frequency relationship in heart failure. Basic Res Cardiol 91 Suppl 217–22. [DOI] [PubMed] [Google Scholar]
- 15. Paolini C, Quarta M, Nori A, Boncompagni S, Canato M, et al. (2007) Reorganized stores and impaired calcium handling in skeletal muscle of mice lacking calsequestrin-1. J Physiol 583: 767–784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Knollmann BC (2009) New roles of calsequestrin and triadin in cardiac muscle. J Physiol 587: 3081–3087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Protasi F, Paolini C, Canato M, Reggiani C, Quarta M (2011) Lessons from calsequestrin-1 ablation in vivo: much more than a Ca(2+) buffer after all. J Muscle Res Cell Motil 32: 257–270. [DOI] [PubMed] [Google Scholar]
- 18. Kornyeyev D, Petrosky AD, Zepeda B, Ferreiro M, Knollmann B, et al. (2012) Calsequestrin 2 deletion shortens the refractoriness of Ca2+ release and reduces rate-dependent Ca2+-alternans in intact mouse hearts. J Mol Cell Cardiol 52: 21–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Beard NA, Sakowska MM, Dulhunty AF, Laver DR (2002) Calsequestrin is an inhibitor of skeletal muscle ryanodine receptor calcium release channels. Biophys J 82: 310–320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Wei L, Varsányi M, Dulhunty AF, Beard NA (2006) The conformation of calsequestrin determines its ability to regulate skeletal ryanodine receptors. Biophys J 91: 1288–1301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Wang S, Trumble WR, Liao H, Wesson CR, Dunker AK, et al. (1998) Crystal structure of calsequestrin from rabbit skeletal muscle sarcoplasmic reticulum. Nat Struct Biol 5: 476–483. [DOI] [PubMed] [Google Scholar]
- 22. Sanchez EJ, Lewis KM, Danna BR, Kang C (2012) High-capacity Ca2+ binding of human skeletal calsequestrin. J Biol Chem 287: 11592–11601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Park H, Wu S, Dunker AK, Kang C (2003) Polymerization of calsequestrin. Implications for Ca2+ regulation. J Biol Chem 278: 16176–16182. [DOI] [PubMed] [Google Scholar]
- 24. Watabe S, Ushio H, Hashimoto K (1991) Purification and characterization of a calsequestrin-like calcium-binding protein from carp (Cyprinus carpio) sarcoplasmic reticulum. Comp Biochem Physiol B 99: 545–552. [DOI] [PubMed] [Google Scholar]
- 25. Block BA, O’Brien J, Meissner G (1994) Characterization of the sarcoplasmic reticulum proteins in the thermogenic muscles of fish. J Cell Biol 127: 1275–1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Infante C, Ponce M, Manchado M (2011) Duplication of calsequestrin genes in teleosts: molecular characterization in the Senegalese sole (Solea senegalensis). Comp Biochem Physiol B Biochem Mol Biol 158: 304–314. [DOI] [PubMed] [Google Scholar]
- 27. Somero GN, Low PS (1977) Eurytolerant proteins: mechanisms for extending the environmental tolerance tange of enzyme-ligand interactions. Am Nat 111: 527–538. [Google Scholar]
- 28. Bal NC, Sharon A, Gupta SC, Jena N, Shaikh S, et al. (2010) The catecholaminergic polymorphic ventricular tachycardia mutation R33Q disrupts the N-terminal structural motif that regulates reversible calsequestrin polymerization. J Biol Chem 285: 17188–17196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, et al. (2007) Clustal W and Clustal X version 2.0. Bioinformatics 23: 2947–2948. [DOI] [PubMed] [Google Scholar]
- 30. Cala SE, Jones LR (1983) Rapid purification of calsequestrin from cardiac and skeletal muscle sarcoplasmic reticulum vesicles by Ca2+-dependent elution from phenyl-sepharose. J Biol Chem 258: 11932–11936. [PubMed] [Google Scholar]
- 31. King LE, Morrison M (1976) The visualization of human erythrocyte membrane proteins and glycoproteins in SDS polyacrylamide gels employing a single staining procedure. Anal Biochem 71: 223–230. [DOI] [PubMed] [Google Scholar]
- 32. Campbell KP, MacLennan DH, Jorgensen AO (1983) Staining of the Ca2+-binding proteins, calsequestrin, calmodulin, troponin C, and S-100, with the cationic carbocyanine dye “Stains-all”. J Biol Chem 258: 11267–11273. [PubMed] [Google Scholar]
- 33. Hidalgo C, Donoso P, Rodriguez PH (1996) Protons induce calsequestrin conformational changes. Biophys J 71: 2130–2137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Goutelle S, Maurin M, Rougier F, Barbaut X, Bourguignon L, et al. (2008) The Hill equation: a review of its capabilities in pharmacological modelling. Fundam Clin Pharmacol 22: 633–648. [DOI] [PubMed] [Google Scholar]
- 35. Guex N, Peitsch MC (1997) SWISS-MODEL and the Swiss-PdbViewer: an environment for comparative protein modeling. Electrophoresis 18: 2714–2723. [DOI] [PubMed] [Google Scholar]
- 36.Humphrey W, Dalke A, Schulten K (1996) VMD: visual molecular dynamics. J Mol Graph 14: 33–38, 27–28. [DOI] [PubMed]
- 37. Arnold K, Bordoli L, Kopp J, Schwede T (2006) The SWISS-MODEL workspace: a web-based environment for protein structure homology modelling. Bioinformatics 22: 195–201. [DOI] [PubMed] [Google Scholar]
- 38.The PyMOL Molecular Graphics System, Version 1.5.0.4 Schrödinger, LLC.
- 39. MacKerell AD Jr, Bashford D, Dunbrack RL, Evanseck JD, Field MJ, et al. (1998) All-atom empirical potential for molecular modeling and dynamics studies of proteins. J Phys Chem B 102: 3586–3616. [DOI] [PubMed] [Google Scholar]
- 40. Jorgensen WL, Chandrasekhar J, Madura JD, Impey RW, Klein ML (1983) Comparison of simple potential functions for simulating liquid water. J Chem Phys 79: 926–935. [Google Scholar]
- 41. Bosshard HR, Marti DN, Jelesarov I (2004) Protein stabilization by salt bridges: concepts, experimental approaches and clarification of some misunderstandings. J Mol Recognit 17: 1–16. [DOI] [PubMed] [Google Scholar]
- 42. Mehler EL, Eichele G (1984) Electrostatic effects in water-accessible regions of proteins. Biochemistry 23: 3887–3891. [Google Scholar]
- 43. Reithmeier RA, de Leon S, MacLennan DH (1980) Assembly of the sarcoplasmic reticulum. Cell-free synthesis of the Ca2++Mg2+-adenosine triphosphatase and calsequestrin. J Biol Chem 255: 11839–11846. [PubMed] [Google Scholar]
- 44. Yano K, Zarain-Herzberg A (1994) Sarcoplasmic reticulum calsequestrins: structural and functional properties. Mol Cell Biochem 135: 61–70. [DOI] [PubMed] [Google Scholar]
- 45. Korajoki H, Vornanen M (2009) Expression of calsequestrin in atrial and ventricular muscle of thermally acclimated rainbow trout. J Exp Biol 212: 3403–3414. [DOI] [PubMed] [Google Scholar]
- 46. Houle TD, Ram ML, Cala SE (2004) Calsequestrin mutant D307H exhibits depressed binding to its protein targets and a depressed response to calcium. Cardiovasc Res 64: 227–233. [DOI] [PubMed] [Google Scholar]
- 47. Kim E, Youn B, Kemper L, Campbell C, Milting H, et al. (2007) Characterization of human cardiac calsequestrin and its deleterious mutants. J Mol Biol 373: 1047–1057. [DOI] [PubMed] [Google Scholar]
- 48. Valle G, Galla D, Nori A, Priori SG, Gyorke S, et al. (2008) Catecholaminergic polymorphic ventricular tachycardia-related mutations R33Q and L167H alter calcium sensitivity of human cardiac calsequestrin. Biochem J 413: 291–303. [DOI] [PubMed] [Google Scholar]
- 49. Beltrán M, Barrientos G, Hidalgo C (2006) Fast kinetics of calcium dissociation from calsequestrin. Biol Res 39: 493–503. [DOI] [PubMed] [Google Scholar]
- 50. Healy TM, Tymchuk WE, Osborne EJ, Schulte PM (2010) Heat shock response of killifish (Fundulus heteroclitus): candidate gene and heterologous microarray approaches. Physiol Genomics 41: 171–184. [DOI] [PubMed] [Google Scholar]
- 51. Somero GN (1975) Temperature as a selective factor in protein evolution: the adaptational strategy of “compromise”. J Exp Zool 194: 175–188. [DOI] [PubMed] [Google Scholar]
- 52. Giordano A, Febbraio F, Russo C, Rossi M, Raia CA (2005) Evidence for co-operativity in coenzyme binding to tetrameric Sulfolobus solfataricus alcohol dehydrogenase and its structural basis: fluorescence, kinetic and structural studies of the wild-type enzyme and non-co-operative N249Y mutant. Biochem J 388: 657–667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Reeves RB (1972) An imidazole alphastat hypothesis for vertebrate acid-base regulation: tissue carbon dioxide content and body temperature in bullfrogs. Respir Physiol 14: 219–236. [DOI] [PubMed] [Google Scholar]
- 54. Moerland T, Egginton S (1998) Intracellular pH of muscle and temperature: Insight from in vivo 31P NMR measurements in a stenothermal Antarctic teleost (Harpagifer antarcticus). J Therm Biol 23: 275–282. [Google Scholar]
- 55. Sartoris FJ, Bock C, Pörtner HO (2003) Temperature-dependent pH regulation in eurythermal and stenothermal marine fish: an interspecies comparison using 31P-NMR. J Therm Biol 28: 363–371. [Google Scholar]
- 56. Grove TJ, McFadden LA, Chase PB, Moerland TS (2005) Effects of temperature, ionic strength and pH on the function of skeletal muscle myosin from a eurythermal fish, Fundulus heteroclitus . J Muscle Res Cell Motil 26: 191–197. [DOI] [PubMed] [Google Scholar]
- 57. Nayeem A, Sitkoff D, Krystek S (2006) A comparative study of available software for high-accuracy homology modeling: from sequence alignments to structural models. Protein Sci 15: 808–824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Sánchez R, Sali A (1997) Advances in comparative protein-structure modelling. Curr Opin Struct Biol 7: 206–214. [DOI] [PubMed] [Google Scholar]
- 59. Warshel A, Sharma PK, Kato M, Parson WW (2006) Modeling electrostatic effects in proteins. Biochim Biophys Acta 1764: 1647–1676. [DOI] [PubMed] [Google Scholar]
- 60. Anderson DE, Becktel WJ, Dahlquist FW (1990) pH-induced denaturation of proteins: a single salt bridge contributes 3–5 kcal/mol to the free energy of folding of T4 lysozyme. Biochemistry 29: 2403–2408. [DOI] [PubMed] [Google Scholar]
- 61. Závodszky P, Kardos J, Svingor, Petsko GA (1998) Adjustment of conformational flexibility is a key event in the thermal adaptation of proteins. Proc Natl Acad Sci U S A 95: 7406–7411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Jaenicke R (2000) Stability and stabilization of globular proteins in solution. J Biotechnol 79: 193–203. [DOI] [PubMed] [Google Scholar]
- 63. Somero GN (2004) Adaptation of enzymes to temperature: searching for basic “strategies”. Comp Biochem Physiol B Biochem Mol Biol 139: 321–333. [DOI] [PubMed] [Google Scholar]
- 64. Erickson JR, Sidell BD, Moerland TS (2005) Temperature sensitivity of calcium binding for parvalbumins from Antarctic and temperate zone teleost fishes. Comp Biochem Physiol A Mol Integr Physiol 140: 179–185. [DOI] [PubMed] [Google Scholar]
- 65. Erickson JR, Moerland TS (2006) Functional characterization of parvalbumin from the Arctic cod (Boreogadus saida): similarity in calcium affinity among parvalbumins from polar teleosts. Comp Biochem Physiol A Mol Integr Physiol 143: 228–233. [DOI] [PubMed] [Google Scholar]
- 66. Whittington AC, Moerland TS (2012) Resurrecting prehistoric parvalbumins to explore the evolution of thermal compensation in extant Antarctic fish parvalbumins. J Exp Biol 215: 3281–3292. [DOI] [PubMed] [Google Scholar]
- 67. Lewit-Bentley A, Réty S (2000) EF-hand calcium-binding proteins. Curr Opin Struct Biol 10: 637–643. [DOI] [PubMed] [Google Scholar]
- 68. Gillis TE, Marshall CR, Xue XH, Borgford TJ, Tibbits GF (2000) Ca(2+) binding to cardiac troponin C: effects of temperature and pH on mammalian and salmonid isoforms. Am J Physiol Regul Integr Comp Physiol 279: R1707–15. [DOI] [PubMed] [Google Scholar]
- 69. Gillis TE, Tibbits GF (2002) Beating the cold: the functional evolution of troponin C in teleost fish. Comp Biochem Physiol A Mol Integr Physiol 132: 763–772. [DOI] [PubMed] [Google Scholar]
- 70. Blumenschein TMA, Gillis TE, Tibbits GF, Sykes BD (2004) Effect of temperature on the structure of trout troponin C. Biochemistry. 43: 4955–4963. [DOI] [PubMed] [Google Scholar]
- 71. Gillis TE, Marshall CR, Tibbits GF (2007) Functional and evolutionary relationships of troponin C. Physiol Genomics. 32: 16–27. [DOI] [PubMed] [Google Scholar]
- 72. Swezey RR, Somero GN (1982) Polymerization thermodynamics and structural stabilities of skeletal muscle actins from vertebrates adapted to different temperatures and hydrostatic pressures. Biochemistry 21: 4496–4503. [DOI] [PubMed] [Google Scholar]
- 73. Detrich HW, Parker SK, Williams RC, Nogales E, Downing KH (2000) Cold adaptation of microtubule assembly and dynamics. Structural interpretation of primary sequence changes present in the alpha- and beta-tubulins of Antarctic fishes. J Biol Chem 275: 37038–37047. [DOI] [PubMed] [Google Scholar]