Abstract
The ragged topography created by orogenesis generates diversified habitats for plants in Taiwan. In addition to colonization from nearby mainland China, high species diversity and endemism of plants is also present in Taiwan. Five of the seven Scutellaria species (Lamiaceae) in Taiwan, for example, are endemic to the island. Hypotheses of multiple sources or in situ radiation have arisen to explain the high endemism of Taiwanese species. In this study, phylogenetic analyses using both nuclear and chloroplast markers revealed the multiple sources of Taiwanese Scutellaria species and confirmed the rapid and recent speciation of endemic species, especially those of the “indica group” composed of S. indica, S. austrotaiwanensis, S. tashiroi, and S. playfairii. The common ancestors of the indica group colonized first in northern Taiwan and dispersed regionally southward and eastward. Climate changes during glacial/interglacial cycles led to gradual colonization and variance events in the ancestors of these species, resulting in the present distribution and genetic differentiation of extant populations. Population decline was also detected in S. indica, which might reflect a bottleneck effect from the glacials. In contrast, the recently speciated endemic members of the indica group have not had enough time to accumulate much genetic variation and are thus genetically insensitive to demographic fluctuations, but the extant lineages were spatially expanded in the coalescent process. This study integrated phylogenetic and population genetic analyses to illustrate the evolutionary history of Taiwanese Scutellaria of high endemism and may be indicative of the diversification mechanism of plants on continental islands.
Introduction
Radiation and colonization are two major mechanisms for the development of high diversity in continental islands [1], [2]. The term radiation denotes a phenomenon of rapid speciation in a specific locality from a single origin, also defined as in situ diversification, which fits the propagule pool model in displaying single-source colonization [3]. By contrast, colonization is the phenomenon of species originating from multiple sources (or multiple origination), which can be illustrated by the migrant pool model [3]–e.g., the plant diversity of Taiwan and Ryukyu Archipelago [4]. However, whether through single or multiple originations, the appearance of endemic species on a continental island must involve reproductive isolation from the closely related species of continents.
Taiwan is a continental island situated off of Southeast Asia. The emergence of the shallow shelf of the Taiwan Strait during the Pleistocene glacial cycles connected Taiwan Island and mainland China, whereas the submergence of the shallow continental shelf during the interglacial periods separated them. The repeated topographic changes during the Pleistocene glacials resulted in several opportunities for colonization and isolation of organisms between China and Taiwan [4]. Such biogeographic events could cause that 52% of the native plant species of Taiwan have affinities to the flora of mainland China and high endemism of flora in Taiwan (c. 26.1% of natives) [5]. Therefore, high endemism in the flora of Taiwan can be explained by hypotheses of (1) continent-island colonization, (2) in situ radiation, or (3) synergy of colonization and radiation. The colonization hypothesis focuses on the phenomenon of multiple colonization events via the land bridge at the Taiwan Strait during the glacials, whereas the radiation hypothesis focuses on in situ speciation after colonization in Taiwan. The radiation hypothesis is usually related to local adaptation (i.e. adaptive radiation [6], [7]) or drift effect by geographic isolation and demographic change (i.e. non-adaptive radiation [8]). Although Taiwan is subtropical and was free of ice during the Pleistocene glacials, the unpleasant cold and dry conditions affected the demography of several Taiwanese organisms [9]–[15]. The rapid process of genetic drift of small-sized populations in glacial refugia can enlarge the gap between phylogenetically related “pre-species” [16], [17]. To explore the mechanism of formation of the endemics, we chose species of the genus Scutellaria as models in which to test the continent-island colonization hypothesis and the radiation (adaptive or non-adaptive) hypothesis.
Scutellaria, a genus commonly known as skullcaps, contains approximately 400 species around the world. Scutellaria is sister to Tinnea and together they form the well-supported group Scutellarioideae [18]. Several members of this genus–e.g., S. baicalensis, S. indica and S. lateriflora–are widely used in traditional medicine [19]–[23]. The specific floral types of both cleistogamous and chasmogamous flowers [24], [25] and restricted seed dispersal capabilities of bursting capsules [26]–[28] may have caused structured populations, especially in cases of widely distributed species, resulting in the high diversity of this genus. For example, predominant selfing, even in chasmogamous flowers, increases genetic differentiation between populations of S. indica [24]. Furthermore, the loss of pollinators for historical outcrossing populations has often been reported in Scutellaria montana, which do not produce cleistogamous flowers [28]. Similar patterns of restricted outcrossing are also observed in Taiwanese Scutellaria species (personal observation).
Five of seven Scutellaria species, S. taipenensis, S. playfairii, S. tashiroi, S. austrotaiwanensis, and S. taiwanensis, are endemic to Taiwan, while the other two species, S. barbata and S. indica, are widespread in Asia and are treated in the same section, Sect. Scutellaria [29]. Most of them are distributed between central and southern Taiwan [30]–[32] except the northerly distributed S. taipeiensis Huang, Hsiao, et Wu [33]. The Taiwanese species are morphologically similar and have identical chromosome numbers (2n = 26) [30]–[33], which fit the chromosome range of Sect. Scutellaria (2n = 24∼34) [29]. Mainland China, which contains 124 taxa (98 species and 26 varieties) [34], is probably the source of the Taiwanese species. Until recently, three endemic species, S. tashiroi, S. playfairii, and S. austrotaiwanensis, were considered different varieties under a species level [35],[31]. The same corolla type is also shared among S. indica, S. playfairii, and S. austrotaiwanensis [30], [31]. Some older specimens or records identified as S. indica in South Taiwan have been proposed to be misidentifications of S. taiwanensis [30]. These patterns imply a close phylogenetic relationship among S. tashiroi, S. playfairii, S. austrotaiwanensis, S. indica, and S. taiwanensis. These five species have more or less overlapping distributions but may differ in microhabitat preference. For example, S. playfairii is distributed in southern and eastern Taiwan, and S. tashiroi is distributed in eastern Taiwan and Lanyu Island. Both species can be found sympatrically in eastern Taiwan, but S. tashiroi prefers open rocky slopes, whereas S. playfairii prefers more or less shady slopes. Scutellaria austrotaiwanensis is mainly distributed in the Hengchun Peninsula, but some small patchy populations are also found in other region in southern Taiwan. Scutellaria taiwanensis grows in a habitat similar to that of S. austrotaiwanensis and S. playfairii in southern Taiwan but prefers more moist and shaded underforest habitats at slightly higher elevations. Scutellaria indica, which is not endemic to Taiwan, is distributed in northeastern and western Taiwan and is geographically differentiated from other species [30], [31].
To evaluate the hypotheses of diversification of Taiwanese Scutellaria, we designed both species-level and population-level studies to explore the evolutionary history of the genus. Phylogenetic analyses for Taiwanese Scutellaria were performed with several species collected from Japan, Asian mainland, Europe, North America, and South America to confirm the continent-island colonization hypothesis. Demographic change and biogeographic events in endemic Scutellaria species were further examined to test the radiation hypothesis. Based on these phylogenetic and population genetic analyses, we tried to determine the evolutionary history of Taiwanese Scutellaria species to elucidate mechanisms of diversification in continental island herbs.
Materials and Methods
Taxon Sampling and Study Populations
To resolve the hypotheses of multiple sources or single origin of Taiwanese Scutellaria species, we sampled all seven Taiwanese Scutellaria species and an additional 10 species in the field or from the Seed Bank, including S. amoena, S. amabilis, S. sessilifolia, S. galericulata, S. lateriflora, S. incana, S. alpine, S. baicalensis, S. salviifolia, S. diffusa, S. altissima, and S. zhongdianensis (Table S1). Tinnea rhodesiana was used as the outgroup. Population sampling was also performed for S. indica, S. tashiroi, S. austrotaiwanensis, and S. playfairii (namely, the indica group; see Results) and S. taiwanensis to reconstruct their biogeographic patterns (Table 1 and Figure 1A). In total, 269 individuals from 13 populations were collected. This study was conducted in accordance with the laws of Taiwan. The locations of field studies are not privately-owned or protected areas, and are not involved with endangered or protected species. No permits were required for this study. Leaves obtained from each individual were dried with silica gel immediately for preservation. The dried materials were ground to powder using liquid nitrogen and extracted with a cetyl trimethylammonium bromide method [36]. The extracted DNA was then dissolved in 1X TE buffer and stored at −20°C.
Table 1. Sampling areas and haplotype composition of Scutellaria species in the population study.
| Species/population | Population code | Sample size | Longitude | Latitude | Haplotype size | Haplotype compositionof each locus | ||||
| CHS | CAD | matK | ndh-rpl32 | rpl32-trnL | ||||||
| S. indica † | ||||||||||
| Awanda | ind1 | 9 | 121° 10′ 31′′E | 23° 57′ 12′′N | 18 | CHS-hap3 | CAD-hap5 | matK-hap2 | ndh/rpl-hap2 | rpl/trn-hap2 |
| Wulai | ind2 | 12 | 121° 34′ 27′′E | 24° 53 ′07′′N | 24 | CHS-hap4 | CAD-hap6 | matK-hap2 | ndh/rpl-hap3 | rpl/trn-hap3 |
| S. austrotaiwanensis * † | ||||||||||
| Hengchun peninsula | aus1 | 19 | 120° 52′ 24′′E | 22° 00′ 34′′N | 38 | CHS-hap2 | CAD-hap4 | matK-hap1 | ndh/rpl-hap1 | rpl/trn-hap1 |
| Lilungshan | aus2 | 33 | 120° 43′ 11′′E | 22° 10′ 35′′N | 42 | CHS-hap1 | CAD-hap1 | matK-hap1 | ndh/rpl-hap1 | rpl/trn-hap1 |
| 24 | CHS-hap1 | CAD-hap2 | matK-hap1 | ndh/rpl-hap1 | rpl/trn-hap1 | |||||
| Nanhua | aus3 | 14 | 120° 21′ 40′′E | 23° 01′ 07′′N | 28 | CHS-hap1 | CAD-hap3 | matK-hap1 | ndh/rpl-hap1 | rpl/trn-hap1 |
| S. playfairii * † | ||||||||||
| Dahan trail | pla1 | 5 | 120° 45′ 17′′E | 22° 24′ 46′′N | 10 | CHS-hap5 | CAD-hap7 | matK-hap1 | ndh/rpl-hap4 | rpl/trn-hap6 |
| Wutai | pla2 | 43 | 120° 43′ 05′′E | 22° 44′ 23′′N | 38 | CHS-hap5 | CAD-hap7 | matK-hap1 | ndh/rpl-hap4 | rpl/trn-hap4 |
| 14 | CHS-hap5 | CAD-hap7 | matK-hap1 | ndh/rpl-hap4 | rpl/trn-hap5 | |||||
| 22 | CHS-hap5 | CAD-hap7 | matK-hap1 | ndh/rpl-hap5 | rpl/trn-hap4 | |||||
| 4 | CHS-hap5 | CAD-hap7 | matK-hap1 | ndh/rpl-hap5 | rpl/trn-hap5 | |||||
| 4 | CHS-hap5 | CAD-hap8 | matK-hap1 | ndh/rpl-hap4 | rpl/trn-hap4 | |||||
| 2 | CHS-hap5 | CAD-hap8 | matK-hap1 | ndh/rpl-hap4 | rpl/trn-hap5 | |||||
| 2 | CHS-hap5 | CAD-hap8 | matK-hap1 | ndh/rpl-hap5 | rpl/trn-hap4 | |||||
| Wulu | pla3 | 35 | 121° 02′ 30′′E | 23° 10′ 26′′N | 46 | CHS-hap6 | CAD-hap9 | matK-hap1 | ndh/rpl-hap4 | rpl/trn-hap5 |
| 16 | CHS-hap6 | CAD-hap9 | matK-hap1 | ndh/rpl-hap4 | rpl/trn-hap6 | |||||
| 8 | CHS-hap6 | CAD-hap9 | matK-hap1 | ndh/rpl-hap6 | rpl/trn-hap5 | |||||
| S. tashiroi * † | ||||||||||
| Lanyu Island | tas1 | 15 | 121° 30′ 40′′E | 22° 04′ 32′′N | 30 | CHS-hap10 | CAD-hap12 | matK-hap1 | ndh/rpl-hap10 | rpl/trn-hap1 |
| Mugumuyu | tas2 | 16 | 121° 25′ 35′′E | 24° 00′ 10′′N | 32 | CHS-hap10 | CAD-hap13 | matK-hap1 | ndh/rpl-hap11 | rpl/trn-hap8 |
| Taroko | tas3 | 15 | 121° 30′ 51′′E | 24° 10′ 42′′N | 10 | CHS-hap10 | CAD-hap13 | matK-hap1 | ndh/rpl-hap11 | rpl/trn-hap8 |
| 20 | CHS-hap11 | CAD-hap13 | matK-hap1 | ndh/rpl-hap11 | rpl/trn-hap8 | |||||
| Wulu | tas4 | 22 | 121° 02′ 30′′E | 23° 10′ 26′′N | 12 | CHS-hap8 | CAD-hap11 | matK-hap1 | ndh/rpl-hap9 | rpl/trn-hap1 |
| 10 | CHS-hap9 | CAD-hap11 | matK-hap1 | ndh/rpl-hap1 | rpl/trn-hap1 | |||||
| 12 | CHS-hap9 | CAD-hap11 | matK-hap1 | ndh/rpl-hap4 | rpl/trn-hap1 | |||||
| 6 | CHS-hap9 | CAD-hap11 | matK-hap1 | ndh/rpl-hap9 | rpl/trn-hap1 | |||||
| 2 | CHS-hap12 | CAD-hap11 | matK-hap1 | ndh/rpl-hap4 | rpl/trn-hap1 | |||||
| 2 | CHS-hap13 | CAD-hap11 | matK-hap1 | ndh/rpl-hap4 | rpl/trn-hap1 | |||||
| S. taiwanensis * | ||||||||||
| Jin-Shui Camp | tai1 | 31 | 120° 43′ 42′′E | 22° 24′ 52′′N | 20 | CHS-hap7 | CAD-hap10 | matK-hap3 | ndh/rpl-hap7 | rpl/trn-hap7 |
| 42 | CHS-hap7 | CAD-hap10 | matK-hap3 | ndh/rpl-hap8 | rpl/trn-hap7 | |||||
endemic to Taiwan;
members of the indica group.
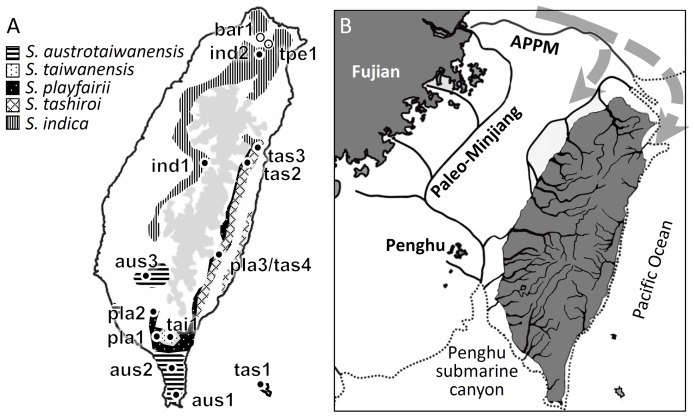
Figure 1. Species distribution and the paleo-drainage system in Taiwan.
(A) Distribution areas and sampling populations (solid dots) of five Taiwanese Scutellaria species used for demographic analyses. The sampling sites Yonghe (bar1) and Maokong (tpe1) for S. barbata and S. taipeiensis, respectively, are also indicated on this map (hollow dots). The gray areas are mountain ranges with altitudes of >2000 m. Aus, tai, pla, tas, ind, bar, and tpe indicate the sampling sites of S. austrotaiwanensis, S. taiwanensis, S. playfairii, S. tashiroi, S. indica, S. barbata, and S. taipeiensis, respectively. Detailed information of sampling sites is listed in Table 1. (B) The proposed paleo-drainage system around the Taiwan Strait during glacial periods of the Late Pleistocene [79]. The dotted arrows are possible colonization routes of ancestors of the indica group; the dotted line indicates the Late Pleistocene shoreline. APPM, the alternate path of the Paleo-Minjiang River.
Molecular Techniques
Polymerase chain reaction (PCR) was performed with 10–100 ng template DNA, 0.5–1 U Taq (Bernardo Scientific Corp., Taipei), 100 µM deoxyribonucleotide triphosphate, 0.2 µM each primer, and 0.1 µg/µL bovine serum albumin in a MultiGene thermal cycler (Labnet International, Inc.). The PCR program was set to 94°C for 3 min for enzyme activation, followed by 35 cycles of 94°C for 40 s, melting temperature for 40 s, and 72°C for 90 s, with a 5-min final extension at 72°C. PCR amplifications of five primer sets including three chloroplast regions (matK, ndhF-rpl32, and rpl32-trnL) and two low-copy nuclear regions (CHS and CAD) were performed. Optimal annealing temperatures were set at 47°C for chloroplast regions, 53°C for CHS, and 49°C for CAD regions. Within-population variation of all PCR products was screened with single-strand conformation polymorphism. Each PCR product was denatured for 10 min at 95°C and quickly moved into a −20°C cool box. Denatured products were separated by pre-cooling 10% polyacrylamide gel (acrylamide:bisacrylamide = 45∶1). PCR products with different fragment patterns were then sequenced directly in both directions using an ABI BigDye 3.1 Terminator Cycle Sequencing Kit (Applied Biosystems, Foster City, CA, USA). All sequence polymorphisms were visually rechecked from chromatograms with an ABI PRISM®3730XL DNA Sequencer (Perkin-Elmer, Foster City, CA, USA). PCR products were cloned with a yT&A cloning kit (Yeastern Biotech, Taipei, Taiwan) when they contained ambiguous nucleotides, and three to five clones were sequenced with the M13F and M13R primers to generate consensus sequences. The two sequences of a heterozygote were separated by comparing the sequences of the PCR product and the cloned sequence. Chromatograms were inspected by SeqMan implemented in DNASTAR ver. 7.0 (Lasergene, Germany). Gene confirmation and exon-intron junctions of each sequence were queried in the Nucleotide collection database at the National Center for Biotechnology Information website using the Nucleotide Basic Local Alignment Search Tool program and NetPlantGene server at the Center for Biological Sequence Analysis website (www.cbs.dtu.dk/services/NetPGene/). All sequences were deposited in the NCBI nucleotide sequence database under the following accession numbers: JX981343∼JX981446 and JX985445∼JX985457.
Phylogenetic Reconstruction
In addition to collecting sequences, we also downloaded the matK sequences of S. minor (HM850804.1), S. scordifolia (HQ839713.1), S. hirta (HQ911383.1), S. sieberi (HQ911384.1), S. viscidula (HQ676587.1), S. rehderiana (HQ676588.1), T. gracilis (HQ911386.1, the outgroup used for matK only), and the CHS sequence of S. viscidula (EU386767.1) from GenBank for phylogenetic analyses. Sequence alignments were performed with Clustal X [37] and manually edited using BioEdit ver. 7.0.9.0 [38]. Phylogenetic relationships were reconstructed using individual and combined sequences of five loci with the neighbor joining and Bayesian approaches implemented in Molecular Evolutionary Genetics Analysis v. 5.05 [39] and MrBayes ver. 3.1.2 [40], respectively. In the neighbor-joining analysis, the maximum composite likelihood model for substitutions and pairwise deletions for the treatment of gaps were set with 1000 bootstrap replications to measure the supporting values for grouping. In the Bayesian analysis, substitution models were set according to the evaluation of Akaike information criterion (AIC) and Bayesian information criterion (BIC) scores (Table S2), and two parallel runs in Markov chain Monte Carlo (MCMC) searches were performed for 10 million generations with sampling every 1000 generations for a total of 10,000 trees in each run. The first 10% of the generations were discarded (burn in). Bayesian posterior probabilities were estimated as the proportion of trees containing each node over the trees sampled during runs.
The species tree was also reconstructed using five loci with the assistance of BEAST ver. 1.6.1 [41]. Substitution models of each of loci were set according to the evaluation of AIC and BIC scores (see Table S2). Because we lacked fossil records for dating, we adopted the strict molecular clock in BEAST ver. 1.6.1 using a substitution rate of 2×10−9 per site per year [42] for chloroplast genome DNA (cpDNA) loci and 1.5×10−8 per site per year [43] for CHS and CAD with the Yule tree prior [41]. Three independent pre-runs of 10 million generations of the length of the MCMC searches were performed to obtain better parameter priors for the next five independent 10 million generations of the MCMC process. Genealogies were sampled every 1000 generations, with the first 10% discarded as burn in. All the statistics of the output values were summarized using TRACER ver. 1.5 [44] and both log and tree files of the last five runs were combined using LogCombiner ver. 1.6.1 [41]. TreeAnnotator ver. 1.6.1 [41] and FigTree ver. 1.3.1 [45] were used for summarizing and displaying the sampled trees, respectively.
Topological Tests for Origination Hypotheses
Given that the phylogenetic analyses indicated that the Taiwanese Scutellaria species are not monophyletic (see Results; Figure 2A), topological tests were performed to examine the hypothesis that the Taiwanese species are descendants of a single common ancestor colonizing Taiwan (Figure 3). The approximately unbiased (AU) test [46], the Kishino-Hasegawa (KH) test [47], and the Shimodaira-Hasegawa (SH) test [48], which are used to compare tree topologies with a null hypothesis, were performed using CONSEL [49], [50].
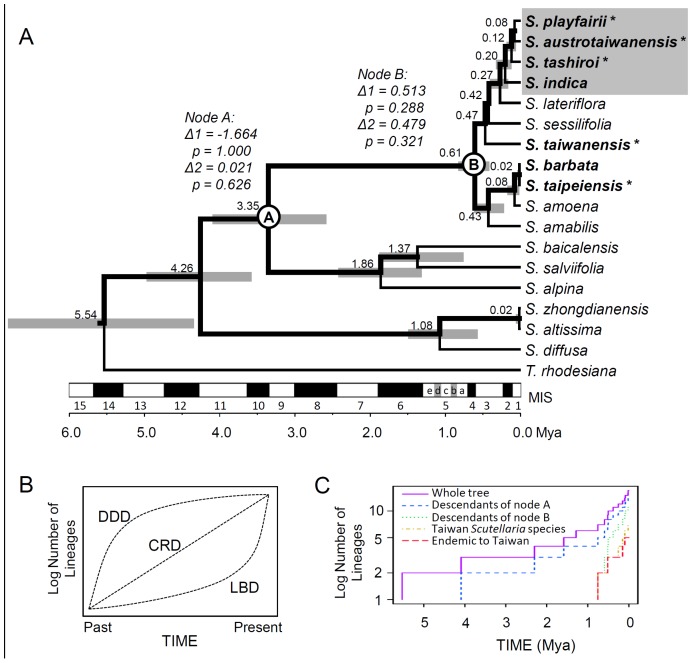
Figure 2. Phylogenetic relationships and the lineage through time (LTT) plots of Scutellaria samples in Taiwan.
(A) Phylogenetic tree reconstructed using CHS, CAD, matK, ndhF-rpl32, and rpl32-trnL under the Yule’s pure-birth speciation model. Bold lines indicate lineage grouping with a posterior probability of >90%; the node labels are the splitting time (unit: Mya); the node bar is the 95% highest posterior density interval of the splitting time; species displayed in bold are those distributed in Taiwan; and the stars indicate species endemic to Taiwan. Species inside the gray box are the indica group. The nodes have inferred diversification rate shifts labeled by nodes A and B. The results of the testing of diversification rate shifts of descendants of nodes A and B inferred by Δ-statistics are indicated with italics. Marine Isotope Stages (MIS) are indicated on the time scales. (B) Idealized log LTT plot showing the expected pattern under the hypotheses of density dependent diversification (DDD), constant rate of diversification (CRD), and late burst of diversification (LBD). (C) The lineage through time (LTT) plot of all Scutellaria samples used in the study. The graph reveals a delayed increase in the lineage accumulation rate in Taiwan species compared to that in descendants of node B, indicating a short history of species colonizing Taiwan.
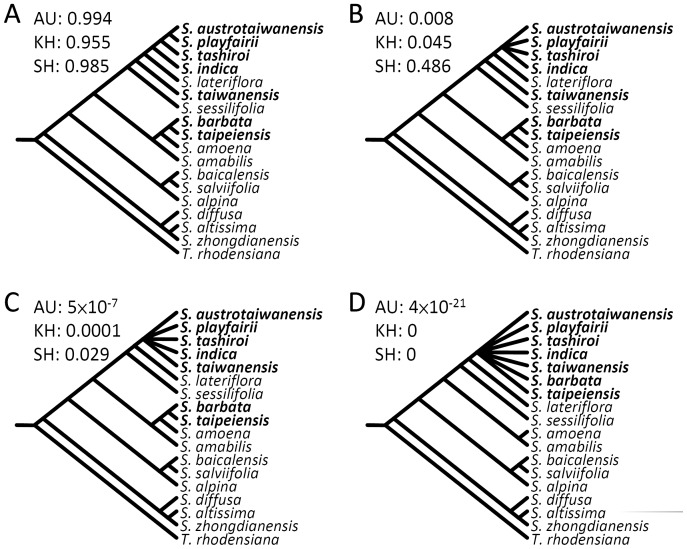
Figure 3. Topological tests of the origin hypotheses of the Taiwan Scutellaria species.
(A) the Yule speciation tree; (B) hypothesis of single origin of S. austrotaiwanensis, S. playfairii, S. tashiroi, and S. indica; (C) S. taiwanensis was hypothesized to be singly originated with S. austrotaiwanensis, S. playfairii, S. tashiroi, and S. indica; (D) hypothesis of single origin of all Taiwan Scutellaria species. Species native to Taiwan are marked in bold. P values of the approximately unbiased (AU), Kishino-Hasegawa (KH), and Shimodaira-Hasegawa (SH) tests are indicated.
Population Genetic Diversity of Five Scutellaria Species in Taiwan
To understand better the demographic history of the Scutellaria species in Taiwan after colonization, we used the gene markers for phylogenetic analysis and performed population sampling and single-strand conformation polymorphism experiments to obtain the gene frequencies of populations. After sequence alignment, indels were treated as the fifth character when calculating the indices of genetic diversity. The haplotype diversity (h), nucleotide diversity estimated by pairwise differences (π) and the θ W estimated by segregating sites were calculated using the DnaSP ver. 5.10.01 [51]. Both Tajima’s [52] D and Fu’s [53] Fs statistics, which evaluate the degrees of rare alleles and singletons, respectively, were estimated with 1000 coalescent simulations for both individual species and individual populations to assess demographic changes.
Biogeographic Inference from Statistical Dispersal-vicariance Analysis (S-DIVA)
To understand the biogeographic events of Taiwanese Scutellaria species and clarify their origin, we performed S-DIVA [54] to reconstruct the historical geographic ranges of species of the indica group using the RASP program [54]. S-DIVA reconstructs ancestral states with given phylogenetic tree based on Bayesian statistic dispersal-vicariance analysis. It optimized uncertainty of biogeographic events for each node. Because each population of the indica group has a haplotype that differs from the others, we used the “population” as the operational taxonomic units (OTUs) to reconstruct the prior tree of S-DIVA with BEAST using the same approaches as those for the Scutellaria species tree. Geographic distributions of the indica group in Taiwan were categorized into the north (population Wulai [ind2] of S. indica), the east (Mugumuyu [tas2] and Taroko [tas3] of S. tashiroi), the southeast (Wulu of S. tashiroi [tas4] and S. playfairii [pla3]), the west (Awanda [ind1] of S. indica), the south (Dahan Trail [pla1] and Wutai [pla2] of S. playfairii and Hengchun peninsula [aus1] and Lilungshan [aus2] of S. austrotaiwanensis), the southwest (Nanhua [aus3] of S. austrotaiwanensis), and Lanyu Island [tas1] of S. tashiroi. Population Jin-Shui Camp [tai1] of S. taiwanensis in the south was used as the outgroup to root the indica group. Only the neighboring ancestral areas were included to prevent the unreasonable inference of cross-Central-Mountain-Range dispersal. Information about geographic distributions was referred to the GBIF data portal (data.gbif.org) and specimen records. In total, 10,000 trees from the BEAST MCMC outputs were used, and the annotated tree devised using BEAST was set as the final condensed tree. The number of maximum areas was maintained at three.
Diversification Rate Shift
The prior tree used for S-DIVA was also used to infer the rate of species occurrence in Taiwan. Both topological and temporal strategies were used to infer diversification rate variation through time. First, the asymmetric rate shift of nodes of the species tree was estimated using single-tree analysis. The taxon-size-insensitive (TSI) and equal-rate Markov (ERM) random branching models were selected. The ERM branching process is a continuous-time method that uses discrete state and pure-birth processes to estimate diversification rate variation in supertrees. Colless’s tree imbalance index and the nodal probability product (M Π and its modified version, M Π*) and sum (M Σ and M Σ*) were used to display the diversification rate variation of the whole tree. Homogeneous evolutionary rates of descendant clades from the common ancestor (the node) were tested with delta-shift statistics (Δ1 and Δ2) at all nodes [55]. One million random resolutions with 1000 TSI-ERM resolved trees under one million ERM simulations were performed to estimate the probabilities of diversification rate shift for each node.
Second, a temporal analysis that accumulated speciation events (lineages) through time was performed to infer the time of species occurrence in Taiwan with lineage through time (LTT) analysis. The LTT analysis was performed in R using the APE package [56]. Chronograms of the species tree reconstructed using BEAST were used as input trees. Sets of total samples of Scutellaria, descendants of nodes A and B, and species distributed in Taiwan and endemic to Taiwan were plotted separately for comparison.
Population Structure
To examine the hierarchical genetic structure of Taiwanese Scutellaria, analysis of molecular variance (AMOVA) was performed using Arlequin v. 3.5.1.3 [57]. According to the grouping pattern determined in the phylogenetic analysis, AMOVA was performed using population data from five (the indica group and S. taiwanensis) and four (the indica group) species separately. One thousand permutations were executed to evaluate whether the variation distribution of populations/species departed from random variation.
Extended Bayesian Skyline Analysis
To infer changes in historical demography of five Taiwanese Scutellaria species, we drew extended Bayesian skyline plots (eBSPs) with BEAST ver. 1.6.1 [41] using the population samples. The monomorphic loci of individual species, which cannot supply demographic information, were excluded. The best substitution model of each locus in every species was reevaluated with AIC and BIC scores using population samples (see Table S2). Ten million MCMC simulations were run to obtain better setting parameters for the priors and operators. Then 50 million MCMC simulations were performed with sampling every 1000 generations and the first 10% discarded as burn in to obtain eBSP results.
Mismatch Analysis under a Spatial Expansion Model
Mismatch analysis was performed to evaluate the range expansions of five Scutellaria species under the spatial expansion model [58] using Arlequin var. 3.11 [59]. The infinite-island (demes) model with the assumption of constant population size (θ = θ 0 = θ 1) was assumed for the expectation, and each deme of a population exchanges a fraction m of migrants with other demes at time τ in this model [58]. The validity of the estimated expansion model was tested using the sum of squared deviations (SSD). Harpending’s [60] raggedness index r, which evaluates the smoothness of the observed mismatch curves, was also used to test demographic expansion. One thousand permutations were executed to evaluate the departures of observed data from expectations.
Results
Phylogenetic Relationships and Diversification Rate Assessment
Phylogenetic analysis of the set of 17 Scutellaria and other species illustrated close relationships among the seven Taiwanese species in the species tree, especially the group composed of S. indica, S. tashiroi, S. austrotaiwanensis, and S. playfairii (namely, the indica group; see Figure 2A) despite certain differences in the gene trees of a single locus (Figure S1). With the exception of the wide distribution of S. indica in Asia, the species of the indica group are endemic to Taiwan. The divergence times of these four species were short (79.8–204.1 Kya). Most interesting is that the American species S. lateriflora and inland Chinese species S. sessilifolia are epiphyletic to the indica group with divergence times of 274.6 Kya and 421.7 Kya, respectively, and another endemic Taiwanese species, S. taiwanensis, diverged with them at 472.2 Kya. The other two Taiwanese species, S. barbata and S. taipeiensis, split very recently (15.4 Kya) and are more deeply divergent with the other five Taiwanese species at 610.4 Kya. Despite the incomplete sampling of related species, this result provides evidence of various sources of Taiwanese species and recent and rapid speciation after colonization in Taiwan.
The late, rapid appearance of the LTT slopes in the Taiwanese species and endemic species also suggests a recent colonization or speciation of Scutellaria in Taiwan (Figure S2). Three LTT patterns, the density-dependent diversification (DDD), the constant rate of diversification (CRD), and the late burst of diversification (LBD) (Fig. 2B, see [61]), illustrated the basic hypotheses of diversification rates of species appearance by colonization and speciation. Based on the last 1000 postconvergence species trees devised using BEAST, the LTT analysis showed a delayed increase in the species accumulation rate in Taiwanese endemic Scutellaria species and suggested an LBD pattern of endemic species appearance in Taiwan, whereas the total Taiwanese Scutellaria species showed a constant rate of diversification pattern of gradual colonization/speciation in Taiwan (see Figure 2C). In addition to results of the LTT analysis, the test for heterogeneous diversification rates based on the topological analysis indicated significant among-lineage rate shift according to Colless’s imbalance index (P = 0.030), M Π and M Π* (P = 0.035 and 0.044, respectively) and M Σ and M Σ* (P = 0.026 and 0.033, respectively; Table S3). However, nodes A and B and other nodes did not shift diversification rate among descendant lineages according to Δ-statistics (see Figure 2A). Although the incomplete sampling of related species might interfere with this inference, the TSI-ERM model decreased the effect of sampling bias [55]. The finding of significant heterogeneous diversification rate in the whole tree without rate shift at any node reflects a gradual process of diversification rate change instead of punctuated rate shift [55].
Topological Tests of the Origin Hypotheses
To test whether the Taiwan Scutellaria species rapidly diverged after the colonization of their common ancestor in Taiwan, we used the species tree (see Figure 2A) to test the origin hypotheses (see Figure 3). The hypotheses of a single origin for all Taiwanese species and the indica group and S. taiwanensis were all rejected by the AU, KH, and SH tests, which means that the Taiwanese Scutellaria species have at least three sources: one that derived S. barbata and S. taipeiensis, one that derived S. taiwanensis, and one that derived the indica group. The topological hypothesis of collapsed lineages of the indica group (see Figure 3B) was also rejected by the AU and KH tests despite not being rejected by the SH test (P = 0.486). The tree topology inferred from the pure-birth branching process (the Yule model) using BEAST was not rejected by the three tests (see Figure 3A), implying a gradual instead of a punctuated speciation process for the indica group in Taiwan.
Biogeographic Inferences
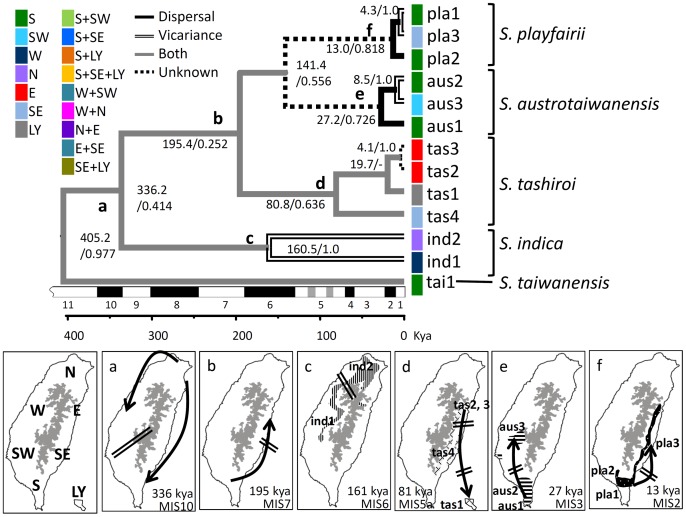
To understand the process of speciation and colonization of the rapidly divergent indica group, S-DIVA was performed. The ancestors of the indica group first colonized northern and eastern Taiwan around 336 Kya and then dispersed southward to western, southern, and southeast Taiwan. A vicariance event followed that has been inferred to have separated the southern and western ancestral populations (see Figure 4A). The northern and western lineages were ancestors of the extant populations of S. indica, which experienced another vicariance event at 161 Kya (see Figure 4C). The other lineage that colonized southern and southeastern Taiwan dispersed northward to eastern Taiwan at 195 Kya (see Figure 4B) and became the ancestor of S. tashiroi. The eastern and southeast populations of S. tashiroi then colonized southward to the Lanyu Islet, after which two vicariance events separated the eastern, southeast, and Lanyu populations at 81 Kya (see Figure 4D). Uncategorized events were identified for the divergence of the southern ancestral populations at 141 Kya, which originated S. austrotaiwanensis and S. playfairii. Two independent northward colonization events from the south to the southwest and southeast with later vicariances resulted in the present distribution of extant populations of S. austrotaiwanensis and S. playfairii at 27 Kya and 13 Kya, respectively (See Figure 4E and 4F). S-DIVA provides a clear biogeographic inference for the indica group in Taiwan.
Figure 4. Graphical depiction of the ancestral distribution of populations of the indica group inferred with the statistical dispersal-vicariance analysis (S-DIVA) of the BEAST tree.
Node labels are the divergence time (Kya) and the marginal probability of the node events; biogeographic events are indicated in different branch types; pie charts indicate the proportion of the ancestral ranges; and marine Isotope Stages (MIS) are indicated on the time scales. The pie chart with a black margin infers experienced extinction events. Biogeographic inferences of nodes a–f are presented in the lower panels. The arrow indicates the direction of dispersal, and the double lines indicate vicariance events. The distribution areas of extant populations of S. indica, S. tashiroi, S. austrotaiwanensis, and S. playfairii are marked in the panels c–f, respectively. The gray area in the middle of Taiwan indicates mountains with altitudes of >2000 m.
Genetic Diversity Estimation
Genetic variations within species and within populations were extremely low in species and population examination (Table 2 and S3). With the exception of two samples in population pla3 of S. tashiroi that were heterozygous in CHS (h = 9.09% and 2.94 for population pla3 and for S. tashiroi, respectively), the samples were homozygous. All samples of the five species were homozygous in locus CAD. No variation was discovered in 31 samples of S. taiwanensis at all loci. Among the indica group, S. austrotaiwanensis had the lowest diversity in nuclear loci and was monomorphic in chloroplast loci. The other two endemic species, S. playfairii and S. tashiroi, which sympatrically or parapatrically distributed in the eastern and southeastern Taiwan, had similar degrees of genetic diversity slightly higher than that of S. austrotaiwanensis. In contrast, the widely distributed S. indica had the highest genetic diversity (see Table 2). However, no variation was detected within populations of S. indica at all loci, whereas at least one locus was polymorphic at the population level in the other endemic species of the indica group (Table S4). These results indicate low genetic diversity of the indica group and suggest degrees of genetic differentiation between populations. In addition, positive but insignificant estimates of Tajima’s D and Fu’s Fs statistics were made at all loci of every species, implying a subdivision of populations of these species, whereas the statistical insignificance suggests a failure to reject population size change or obviously structured populations.
Table 2. Genetic diversity of populations of Scutellaria indica, S. austrotaiwanensis, S. playfairii, S. tashiroi, and S. taiwanensis.
| Locus | Species/Populations | N | H | S | Hd ± std | π ± std | θ ± std | Tajima’s D | p | Fu’s F | p |
| CAD | S. austrotaiwanensis * † | 66 | 3 | 2 | 0.671±0.015 | 0.001±4e−5 | 3.7e−4±2.7e−4 | 1.915 | 0.967 | 2.024 | 0.864 |
| CAD | S. indica † | 21 | 2 | 6 | 0.514±0.046 | 0.003±2e−4 | 0.001±0.001 | 2.671 | 0.999 | 7.641 | 0.995 |
| CAD | S. playfairii * † | 83 | 3 | 2 | 0.545±0.025 | 0.001±4e−5 | 4e−4±3e−4 | 0.739 | 0.791 | 1.032 | 0.739 |
| CAD | S. tashiroi * † | 68 | 3 | 2 | 0.648±0.025 | 0.001±3e−5 | 4e−4±3e−4 | 2.111 | 0.979 | 2.203 | 0.890 |
| CHS | S. austrotaiwanensis * † | 132 | 2 | 1 | 0.413±0.034 | 0.001±4e−5 | 2e−4±2e−4 | 1.363 | 0.900 | 2.011 | 0.864 |
| CHS | S. indica † | 42 | 2 | 4 | 0.502±0.027 | 0.003±1e−4 | 0.001±0.001 | 2.735 | 0.997 | 6.824 | 0.989 |
| CHS | S. playfairii * † | 166 | 2 | 1 | 0.491±0.013 | 0.001±2e−5 | 2e−4±2e−4 | 1.868 | 0.961 | 2.554 | 0.908 |
| CHS | S. tashiroi * † | 136 | 6 | 4 | 0.652±0.033 | 0.002±1e−4 | 0.001±0.001 | 2.078 | 0.977 | 1.362 | 0.764 |
| ndhF-rpl32 | S. indica † | 21 | 2 | 3 | 0.514±0.046 | 0.003±2e−4 | 0.001±0.001 | 2.234 | 0.991 | 4.389 | 0.975 |
| ndhF-rpl32 | S. playfairii * † | 83 | 3 | 3 | 0.360±0.058 | 0.001±2e−4 | 0.001±0.001 | 0.184 | 0.636 | 1.328 | 0.791 |
| ndhF-rpl32 | S. taiwanensis † | 31 | 2 | 1 | 0.452±0.063 | 0.001±1e−4 | 4e−4±4e−4 | 1.240 | 0.884 | 1.459 | 0.824 |
| ndhF-rpl32 | S. tashiroi * † | 68 | 5 | 5 | 0.717±0.038 | 0.003±3e−4 | 0.002±0.001 | 1.763 | 0.963 | 2.215 | 0.852 |
| rpl32-trnL | S. indica † | 21 | 2 | 6 | 0.514±0.046 | 0.005±5e−4 | 0.003±0.001 | 2.671 | 0.999 | 7.641 | 0.995 |
| rpl32-trnL | S. playfairii * † | 83 | 3 | 6 | 0.626±0.023 | 0.005±1e−4 | 0.002±0.001 | 3.018 | 0.999 | 8.057 | 0.986 |
| rpl32-trnL | S. tashiroi * † | 68 | 2 | 3 | 0.504±0.015 | 0.003±8e−5 | 0.001±0.001 | 2.728 | 0.996 | 6.085 | 0.982 |
Tajima’s D and Fu’s Fs were estimated based on 1000-repeat coalescent simulations in consideration of recombination.
N is the number of sequences; H is the number of haplotypes; Hd is the haplotype diversity; π and θ are the nucleotide diversity estimated by pairwise differences and number of segregating sites, respectively.
Chloroplast matK, which is monomorphic in every species, is excluded.
S. taiwanensis (N = 31), which is monomorphic at loci CAD, CHS, and rpl32-trmL, and S. austrotaiwanensis, which is monomorphic at loci ndhF-rpl32 and rpl32-trmL, are not shown.
endemic to Taiwan;
members of the indica group.
Detailed information about the genetic diversity of each sample population is given in Table S4.
Population Structure
Given the inference of population subdivision made through positive estimates of Tajima’s D and Fu’s Fs and the vicariance inferences of all species of the indica group made with S-DIVA, AMOVA was performed to examine whether the extant populations of Taiwanese Scutellaria species are structured (Table 3). Significant genetic differentiation was examined between species at all five loci (P<0.05) in both sets of comparisons (five-species including S. taiwanensis and the indica group only), but CAD (ΦCT = 0.060, P = 0.0899) was used for four-species examination. Significant population structures within species were also detected in every species at most loci except the intraspecies-monomorphic matK. The proportion of genetic variation was mostly contributed at this level (among populations within species), especially at the nuclear markers (except ndhF-rpl32). The high ΦSC and ΦST of most loci (except matK) and significant deviation from random variation (P<0.05) indicated that the extant populations of the indica group are differentiated between one other. In addition, the higher components of variation among populations within species compared with those within populations also suggest structured populations in every species that we examined.
Table 3. Summary of two comparisons for analysis of variance (AMOVA) using two nuclear loci and three chloroplast loci.
| Locus | Source of variation | df | S.S. | Var. Comp. | % Var. | Φ | P |
| Five species (the indica group + S. taiwanensis) | |||||||
| CAD | Among species | 4 | 47.590 | 0.037 | 7.32 | 0.073 | 0.0332 |
| Among populations within species | 8 | 62.309 | 0.420 | 83.89 | 0.905 | <0.0001 | |
| Within populations | 256 | 11.264 | 0.044 | 8.79 | 0.912 | <0.0001 | |
| CHS | Among species | 4 | 114.001 | 0.115 | 23.01 | 0.23 | 0.0049 |
| Among populations within species | 8 | 103.827 | 0.351 | 70.13 | 0.911 | <0.0001 | |
| Within populations | 525 | 18.030 | 0.034 | 6.86 | 0.931 | <0.0001 | |
| matK | Among species | 4 | 44.368 | 0.217 | 100 | 1 | 0.002 |
| Among populations within species | 8 | 0 | 0 | 0 | 0 | 1 | |
| Within populations | 256 | 0 | 0 | 0 | 1 | <0.0001 | |
| ndhF-rpl32 | Among species | 4 | 60.334 | 0.220 | 45.78 | 0.458 | <0.0001 |
| Among populations within species | 8 | 23.822 | 0.156 | 32.38 | 0.597 | <0.0001 | |
| Within populations | 256 | 26.895 | 0.105 | 21.84 | 0.782 | <0.0001 | |
| rpl32-trnL | Among species | 4 | 58.222 | 0.178 | 38.84 | 0.388 | 0.002 |
| Among populations within species | 8 | 33.839 | 0.226 | 49.37 | 0.807 | <0.0001 | |
| Within populations | 256 | 13.846 | 0.054 | 11.79 | 0.882 | <0.0001 | |
| Four species (the indica group only) | |||||||
| CAD | Among species | 3 | 32.372 | 0.030 | 6.03 | 0.060 | 0.0899 |
| Among populations within species | 8 | 62.309 | 0.420 | 84.00 | 0.894 | <0.0001 | |
| Within populations | 226 | 11.264 | 0.050 | 9.97 | 0.900 | <0.0001 | |
| CHS | Among species | 3 | 82.722 | 0.110 | 22.00 | 0.220 | 0.0059 |
| Among populations within species | 8 | 103.827 | 0.351 | 70.22 | 0.900 | <0.0001 | |
| Within populations | 464 | 18.030 | 0.039 | 7.78 | 0.922 | <0.0001 | |
| matK | Among species | 3 | 19.147 | 0.113 | 100 | 1 | 0.0108 |
| Among populations within species | 8 | 0 | 0 | 0 | 0 | 1 | |
| Within populations | 226 | 0 | 0 | 0 | 1 | <0.0001 | |
| ndhF-rpl32 | Among species | 3 | 49.691 | 0.232 | 48.56 | 0.4856 | 0.0010 |
| Among populations within species | 8 | 23.822 | 0.157 | 32.80 | 0.6376 | <0.0001 | |
| Within populations | 226 | 20.121 | 0.089 | 18.64 | 0.8136 | <0.0001 | |
| rpl32-trnL | Among species | 3 | 41.016 | 0.155 | 35.00 | 0.3500 | 0.0127 |
| Among populations within species | 8 | 33.839 | 0.226 | 51.14 | 0.7868 | <0.0001 | |
| Within populations | 226 | 13.846 | 0.061 | 13.86 | 0.8614 | <0.0001 | |
The indica group indicates S. indica, S. austrotaiwanensis, S. playfairii, and S. tashiroi.
Historical Demographics
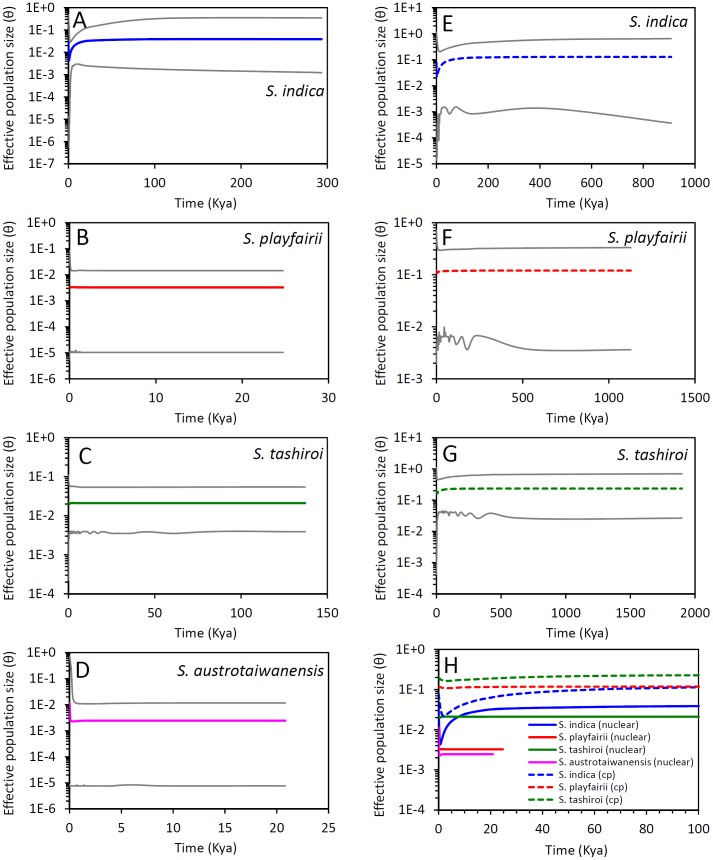
Demographic changes of the indica group were inferred based on eBSP analysis, which revealed long-term constant population sizes in these five species until thousands of years ago. Scutellaria indica revealed a serious bottleneck event beginning at approximately 20 Kya in the nuclear inference or earlier in the cpDNA inference (Figure 5). Both nuclear and cpDNA inferences of demographic history showed a very recent population-size recovery decades or hundreds of years ago. Differences in the inferences of the timing of the bottlenecks made using nuclear and cpDNA probably resulted from heterogeneous rates of lineage sorting of genomes, which are also reflected in differences in population size inferences. In contrast to the bottleneck of S. indica, that of S. austrotaiwanensis was inferred from evidence of a long-term constant population size followed by a very recent population size increase demonstrated by nuclear loci, although the wide range in the confidence interval caused by small variations might mislead the inference. The chloroplast eBSP was unable to construct S. austrotaiwanensis because of monomorphism in all chloroplast loci. In addition to demographic fluctuations in S. indica and S. austrotaiwanensis, permanent stable demography was found in the other two species of the indica group in both nuclear and cpDNA inferences (see Figure 5).
Figure 5. Extended Bayesian skyline plots (eBSPs) of members of the indica group in Taiwan.
A–D are eBSPs inferred from nuclear loci, and E–G are eBSPs of chloroplast (cp) loci. Figure H is a close look at the eBSPs of all species since 100 Kya. Solid and dotted lines indicate the median curves of the eBSPs inferred from nuclear and cpDNA, respectively. The gray curves are the 95% highest posterior density intervals of the eBSPs. Note that the very recent population size increase of S. austrotaiwanensis was inferred from nuclear loci, and a very recent recovery of the effective population sizes after bottlenecks was inferred in S. indica. Population sizes of both S. playfairii and S. tashiroi are constant in coalescent processes. The chloroplast eBSPs could not be inferred in S. austrotaiwanensis because of monomorphism.
For understanding better whether these Taiwanese Scutellaria species underwent historical range expansion, the SSD statistic and raggedness index were used to test the goodness of fit of the observed mismatch distributions to the expectations under the spatial expansion model using four polymorphic loci. Significant SSD values of four loci were taken as evidences of deviation from the estimated demographic model of spatial expansion in populations of S. indica. Significant SSD was also estimated for the nuclear loci of S. playfairii and CHS of S. austrotaiwanensis and the chloroplast locus rpl32-trnL of S. tashiroi and ndhF-rpl32 of S. taiwanensis. The significant values indicated that these loci cannot reflect the patterns of spatial expansion of these species. In contrast, the nuclear CAD of S. austrotaiwanensis and S. tashiroi, CHS of S. tashiroi as well as chloroplast ndhF-rpl32 of S. playfairii and S. tashiroi and rpl32-trnL of S. playfairii failed to reject the expectation of spatial expansion according to both SSD and raggedness indices. We also combined four members of the indica group for a mismatch analysis and obtained results of a failed rejection of range expansion in estimations of nuclear and chloroplast loci according to both SSD and raggedness indices (Table 4), which supports the inference of multiple dispersals suggested by S-DIVA.
Table 4. Summary of the test of spatial expansion model through the mismatch analyses.
| Parameter | indica group | S. indica † | S. tashiroi † ‡ | S. playfairii † ‡ | S. austrotaiwanensis † ‡ | S. taiwanensis ‡ | |
| CHS | τ (95% CI) | 4.9 (0.478–114.701) | 4.6 (0.000–66.882) | 2.2 (0.394–5.715) | 0.7 (0.405–0.922) | 0.6 (0.374–0.763) | – |
| t (95% CI) | 0.215(0.021–5.031) | 0.202(0.000–2.933) | 0.096(0.017–0.251) | 0.031(0.018–0.040) | 0.026(0.016–0.033) | – | |
| θ (95% CI) | 4.157 (0.001–10.094) | 0.001 (0.001–1.049) | 0.545 (0.001–1.541) | 0.001 (0.001–0.421) | 0.004 (0.001–0.351) | – | |
| M (95% CI) | 2.233 (0.451–99999) | 1.271 (0.000–5.866) | 1.899 (0.597–99999) | 99999 (6.037–99999) | 99999 (3.168–99999) | – | |
| SSD | 0.011 | 0.173* | 0.019 | 0.02* | 0.008* | – | |
| r | 0.033 | 0.752 | 0.088 | 0.241* | 0.201* | – | |
| CAD | τ (95% CI) | 5.0 (2.133–14.223) | 6.7 (0.000–50.500) | 1.3 (0.449–2.144) | 0.8 (0.423–1.106) | 0.6 (0.399–4.211) | – |
| t (95% CI) | 0.143(0.061–0.408) | 0.192(0.000–1.449) | 0.037(0.013–0.062) | 0.023(0.012–0.032) | 0.017(0.011–0.121) | – | |
| θ (95% CI) | 3.083 (0.001–7.201) | 0.001 (0.001–1.137) | 0.012 (0.001–0.921) | 0.006 (0.001–0.377) | 1.295 (0.001–3.151) | – | |
| M (95% CI) | 3.565 (0.761–14.919) | 1.28 (0.000–4.870) | 15.871 (1.675–99999) | 99999 (3.177–99999) | 99999 (0.812–99999) | – | |
| SSD | 0.019 | 0.196* | 0.008 | 0.031* | 0.004 | – | |
| r | 0.033 | 0.765 | 0.092 | 0.219* | 0.035 | – | |
| ndh-rpl32 | τ (95% CI) | 9.756 (0.789–20.893) | 3.488 (0.000–85.625) | 11.464 (0.430–25.197) | 11.858 (0.000–170.500) | – | 0.665 (0.307–1.342) |
| t (95% CI) | 4.302(0.348–9.212) | 1.538(0.000–37.754) | 5.055(0.190–11.110) | 5.228(0.000–75.176) | – | 0.293(0.135–0.592) | |
| θ (95% CI) | 2.555 (0.001–6.080) | 0.001 (0.001–1.647) | 2.172 (0.001–3.843) | 0.001 (0.001–0.570) | – | 0.004 (0.001–0.596) | |
| M (95% CI) | 1.448 (0.383–99999) | 1.442 (0.000–99999) | 0.377 (0.243–99999) | 0.624 (0.000–2.556) | – | 99999 (0.390–99999) | |
| SSD | 0.045 | 0.169* | 0.02 | 0.094 | – | 0.013* | |
| r | 0.114 | 0.765 | 0.061 | 0.646 | – | 0.213 | |
| rpl32-trnL | τ (95% CI) | 5.607 (2.595–9.123) | 14.922 (0.000–85.813) | 3.488 (0.306–51.131) | 5.536 (2.036–11.850) | – | – |
| t (95% CI) | 2.425(1.122–3.946) | 6.454(0.000–37.116) | 1.509(0.132–22.115) | 2.394(0.881–−5.125) | – | – | |
| θ (95% CI) | 0.106 (0.001–1.651) | 0.001 (0.001–0.865) | 0.001 (0.001–1.249) | 0.403 (0.001–1.856) | – | – | |
| M (95% CI) | 2.977 (0.780–10.551) | 1.21 (0.000–4.957) | 1.371 (0.093–99999) | 1.5 (0.297–6.441) | – | – | |
| SSD | 0.026 | 0.218* | 0.162* | 0.088 | – | – | |
| r | 0.139 | 0.765 | 0.754 | 0.273 | – | – | |
| matK | τ (95% CI) | 3.5 (0.000–95.739) | – | – | – | – | – |
| t (95% CI) | 1.195(0.000–32.698) | – | – | – | – | – | |
| θ (95% CI) | 0.001 (0.001–0.729) | – | – | – | – | – | |
| M (95% CI) | 0.217 (0.000–99999) | – | – | – | – | – | |
| SSD | 0.016 | – | – | – | – | – | |
| r | 0.761 | – | – | – | – | – |
Indices of monomorphic loci within species are shown in “–.” CI: confidence interval, α = 0.05; SSD: sum of squared deviations; r: raggedness index; τ: unscaled expansion time; t: scaled expansion time (unit: Mya); θ: unscaled population size.
P<0.05;
P<0.01;
P<0.00001 for the sum of squared deviations and raggedness index that reject the null hypothesis of expectation under a sudden expansion model.
members of the indica group;
endemic to Taiwan.
The time at which the range expansion events took place was dated using the expression t = τ/2µk, where the τ is the estimated number of generations after expansion, μ is the mutation rate per site per generation, and k is the sequence length. Mutation rates of 1.5% and 0.2% per site per million years were used for nuclear and chloroplast loci, respectively. A relatively short spatial expansion time at 0.017 Mya (95% confidence interval [95% CI]: 0.011–0.121 Mya) was estimated for S. austrotaiwanensis with CAD, and earlier expansion events occurred for S. tashiroi at 0.037 Mya (95% CI: 0.013–0.062 Mya) according to CAD or 0.096 Mya (95% CI: 0.017–0.251 Mya) according to CHS. Owing to its slower substitution rates, cpDNA may reflect events in the distant past. The spatial expansion time was estimated at 2.394 Mya (95% CI: 0.881∼5.125 Mya) with rpl32-trnF or 5.228 Mya (95% CI: 0–75.176 Mya) with ndhF-rpl32 for S. playfairii and 5.055 Kya (95% CI: 0.190–11.110 Mya) for S. tashiroi. These results suggest that the relatively recent spatial expansions of S. austrotaiwanensis and past expansions of S. playfairii and S. tashiroi were probably gradual events of long duration, a characteristic also reflected in the wide-ranging 95% CI associated with both nuclear and chloroplast markers. In addition, the spatial expansion time of whole members of the indica group was inferred as 0.215 Mya (95% CI: 0.021–5.031 Mya), 0.143 Mya (95% CI: 0.061–0.408 Mya), 4.302 Mya (95% CI: 0.348–9.212 Mya), 2.425 Mya (95% CI: 1.122–3.946 Mya), and 1.195 Mya (95% CI: 0–32.698 Mya) according to CHS, CAD, ndhF-rpl32, rpl32-trnF, and matK, respectively. Notably, the estimated expansion times were longer than the coalescent time estimated by the species trees (see Figures 2 and 4), especially for the chloroplast estimations, which could be explained by the slower evolutionary rates of the plastid genomes of plants and different coalescent rates of loci.
Discussion
Multiple Origins of Highly Endemic Scutellaria Species in Taiwan
Species on continental islands are mainly sourced from neighboring continents and local speciation. Taiwan is situated in southeastern Asia and is a component of the west Pacific Island arc. The enormous drop in sea level during the Pleistocene glacials formed land bridges that connected Taiwan, mainland Asia, and the Ryukyu Archipelago and accelerated organism colonization of Taiwan (e.g., weasels [62], freshwater fish [63], and damselfly [64]) and movement from Taiwan (e.g., Ophiorrhiza japonica [65]). Accordingly, all of the Taiwanese skullcaps are morphologically close and might be of single origin [29]. However, the results of phylogenetic and topological analyses (AU, KH, and SH tests) indicate at least three incidences of origination of Taiwanese endemic Scutellaria species (Figure 3) with resulting local speciation events between ∼0.2 Mya (the indica group) and 0.02 Mya (S. taipeiensis) (see Figure 2A). The rejection of collapsed phylogenetic topology at the common ancestor of the indica group by the AU and KH tests (see Figure 3B) suggests that the speciation of endemic S. austrotaiwanensis, S. playfairii, and S. tashiroi did not display temporal bursts of diversification [66], [67] but a gradual process of speciation. In other words, the hypothesis that a single radiation event could explain the high endemism of Taiwanese Scutellaria via various types of adaptation among species [67] is untenable. This rejection was also evidenced by the failure to detect diversification rate shifts at all nodes of the Taiwanese lineages according to Δ-statistics (see Figure 2A) and insignificant tail probabilities of asymmetric diversification rate indices of Taiwanese Scutellaria species (see Table S3). The steep and delayed increase in species accumulation rate in both clusters of the Taiwanese species and endemic species revealed in LTT curves (see Figure S2) indicate very recent species appearance (i.e., very recent colonization and speciation) of Scutellaria species in Taiwan, especially in endemic species that follow an LBD pattern (see Figure 2C). The rapid local speciation was probably involved in local adaptation and geographic isolation in the ragged topography and high Central Mountain Range of Taiwan Island, which provide diverse habitats and geographic barriers for distant populations.
With the exception of S. taiwanensis, the endemic Scutellaria species (i.e., S. taipeiensis and three endemic species of the indica group) were grouped with a corresponding widespread Asian species. Divergence times between endemic S. playfairii and S. austrotaiwanensis and between S. tashiroi and the most recent common ancestor (MRCA) of S. playfairii and S. austrotaiwanensis were ∼79.8 Kya and ∼121.6 Kya, respectively. These times roughly correspond to the interglacial Marine Isotope Stage (MIS) 5a and MIS5e, respectively. The divergence time between the widely distributed S. indica and endemic species of the indica group was ∼204.1 Kya, roughly the period of the interglacial MIS7. Notably, these species display geographically parapatric distribution, and S. indica is nearly allopatric with the endemic members of the indica group (see Figure 1A). Although the Taiwanese topography is ragged, the vicariance hypothesis of speciation is untenable because the divergent times of species occurred much later than the geographic events (e.g., orogenesis). In contrast, species divergence during the warm interglacial periods was probably caused by long-distance dispersal followed by geographic isolation [68], [69]. Such scenarios were also proved with S-DIVA (see Figure 4).
Scutellaria taipeiensis, which has the shortest coalescent history, diverged from the widespread Asian S. barbata at ∼15.4 Kya (see Figure 2A), or roughly during the last glacial maximum (LGM, the MIS2). Indeed, seed dispersal is presumably short if it relies on capsule burst. Long-distance dispersal may be difficult and occur only during flooding or via rivers [27], [70], but the hydrology of Taiwan was highly constrained during glacial periods. The extant population of S. taipeiensis is found only at low altitudes in northern Taiwan. The cold and dry climate during MIS2 might have played a role as the limiting factor for the distribution of common ancestors of S. taipeiensis and S. barbata. Populations that retreated into isolated refugia might have finally diverged from the others. Range expansion from refugia at the next warm interglacial (MIS1) explains the sympatric distribution of extant populations of S. barbata and S. taipeiensis in northern Taiwan.
Biogeographic Patterns of the Indica Group
The ragged topography of Taiwan provided multiple habitat choices for founders of species. According to the S-DIVA inference, founders of common ancestors of the indica group first colonized northern and northeastern Taiwan at approximately 336.2 Kya–the end of glacial stage MIS10 (see Figure 4). During the glacial periods, seabeds of the Taiwan Strait emerged owing to sea-level regression, connecting northern Taiwan and mainland Asia. Therefore, the seeds of founders likely colonized Taiwan through the paleo-river system on the emerged seabed of the northern Taiwan Strait. The ancestors of the indica group species, which grew in wet habitats, could have entered Taiwan via the water system of the alternate path of the Paleo-Minjiang River (APPM) to the northern part of Taiwan, one staying in the northwest part and another moving east (see Figure 1B), and consequently migrated to the southwest and southeast, respectively. However, the Penghu submarine canyon off of southwestern Taiwan separated southern Taiwan from mainland Asia [71] and might also have hindered southern colonizers into Taiwan. Similar effects of geographic barrier by the Paleo-Minjiang River can be seen in damselfly [64], freshwater crabs [72], and landlocked shrimp [73]. The warm interglacial period that followed (MIS9) accelerated southward dispersal throughout both the eastern and the western sides of the Central Mountain Range. The western lineage (descendants of node C, the extant populations of S. indica; see Figure 4) experienced a vicariance event during glacial stage MIS6 (∼160.5 Kya). The eastern lineage (descendants of node B; see Figure 4) further evolved gradually as three endemic species through serial processes of dispersal and vicariance. The southern ancestral population colonized northward through eastern Taiwan during interglacial MIS7 (∼195.4 Kya) and became isolated from its southern ancestors. The eastern population further speciated as S. tashiroi and dispersed southward again at interglacial MIS5a (∼80.8 Kya) with two vicariance events following. In contrast, the southern population somehow separated into two lineages (S. austrotaiwanensis and S. playfairii) and dispersed northward through the western and eastern Central Mountain Range during MIS3 (27.2 Kya) and the late MIS2 (13.0 Kya), respectively. Then, both dispersed northward and pre-species of S. austrotaiwanensis and S. playfairii separated from their southern ancestors and speciated further (see Figure 4).
S-DIVA illustrated a biogeographic pattern of dispersal/colonization during the warm interglacials with isolation following. Warm and wet weather during the interglacials could have led to greater water flow in rivers and more pollinators and may have prompted long-distance dispersal followed by colonization or vicariance. Furthermore, dispersal (colonization) promotes the establishment of novel niches and speciation [74]. Although an exceptional diversification rate, which is an essential condition of adaptive radiation [67], is absent in endemic Scutellaria, the temporally differential distribution consequence of niche allocation accelerated the speciation rate of Scutellaria in Taiwan. The flowering time of the Taiwanese Scutellaria is predominantly during summer [30], whereas S. indica has a longer and earlier flowering time (September to May) than that of other endemic members of the indica group (June to December) [30]. Scutellaria indica is distributed in northern and western Taiwan, in contrast to the southern and eastern distribution of the other endemic indica group members. Furthermore, S. tashiroi prefers more open rocky cliffs, whereas S. austrotaiwanensis and S. playfairii prefer slightly shaded environments (personal observation). Such niche allocation of the indica group could explain their high endemism and rapid speciation rate after colonization in Taiwan.
Demographic History of the Indica Group
In addition to elucidating the dispersal and fragmentation of populations of the indica group in Taiwan, we also found permanent constant population sizes in members of the indica group according to nonsignificant P values of Tajima’s D and Fu’s Fs test using coalescent simulations (see Table 2). This finding is also evidenced by the past long-term stable populations determined by eBSP analysis, with the exception of the recent bottleneck event of S. indica and the very recent population size increase of S. austrotaiwanensis (see Figure 5). The beginning of population size decrease in Taiwanese S. indica is roughly consistent with the period of the LGM that majorly affected Taiwanese vegetation between 21 and 15.8 Kya [75]. The average temperature of the LGM was roughly 5°C lower than that of the present [76], and the drier climate altered the vegetation distribution during the LGM such that drought-enduring herbs–e.g., the Artemisia, Apiaceae, and Poaceae–dominated the island vegetation [75]. The colder and drier climate of the LGM might have forced populations of S. indica to retreat to refugia as small populations or altered the reproductive strategy to cleistogamy [24], resulting in severe population size decrease (see Figure 5). In contrast, the population sizes of the other endemic members of the indica group did not decrease during the LGM, likely because the speciation times of these endemic species were too short to accumulate much genetic variation (see Table 2), i.e. insufficient duration for coalescence [77], and thus, were genetically insensitive to demographic fluctuation (e.g., [78]).
Although the population sizes of endemic indica-group members were permanently constant, the spatial expansion model was not rejected in S. austrotaiwanensis and S. tashiroi according to nuclear loci or in S. playfairii and S. tashiroi according to cpDNA markers under mismatch analysis (see Table 4). These results indicate that although no lineage expansion occurred in the coalescent process, but the extant lineages migrated and expanded geographically. Differing evolutionary rates of nuclear and plastid genomes result in different coalescent estimates (e.g., expansion time and demographic history). In other words, different rates of lineage sorting of various markers might result in different inferences of range expansion and demographic dynamics (see Table 4), which helps to infer the series time of range expansion. The higher evolutionary rate of nuclear DNA implies a shorter coalescent history of range expansion of S. austrotaiwanensis, whereas the lower evolutionary rate of cpDNA reflects a long-term spatial expansion of S. playfairii. Scutellaria tashiroi might have experienced a permanent spatial expansion after speciation for which the spatial expansion model is rejected by neither nuclear loci nor cpDNA ndhF-trnL32. Such inferences of range expansion are not only consistent with the inferences from S-DIVA but also reflect the various demographics in maternal and biparental inheritances–i.e., different dispersibility of seeds (reflected in both nuclear and cpDNA) and pollens (reflected in cpDNA only). Seed dispersal of Scutellaria is often mediated by gravity or water flow, which is affected by topography (e.g., river systems), whereas the pollination of Scutellaria is mediated by small Hymenoptera insects with short-distance pollen flow [35], [28]. The distance of pollen flow is presumably shorter than that of seed dispersal [27], [28], and therefore the migrants inferred by nuclear and plastid loci that effectively counteracted the drift effect could be different and lead to inconsistent demographic inferences.
Conclusions
Our study examined the genetic diversity and historical demographic change among Taiwanese skullcaps (Scutellaria). The high endemism and similarity in these species make them suitable examples of the effects of glacial-interglacial change on biodiversity in recently evolved endemic species on a continental island. The results imply that the original Taiwanese Scutellaria species evolved at least three times instead of a single time with radiation. Our finding also thoroughly demonstrates continual divergence in Taiwanese Scutellaria species and their close phylogenetic relationships. A delayed increase in LTT curve and recent divergence time indicate recent colonization and local adaptation for the endemic species. One of the Taiwanese clades, the indica group, diverged recently, and most members are distributed sympatrically or parapatrically. During the glacial period MIS10 (336 Kya), during which the seabed of the Taiwan Strait emerged, the common ancestor of the indica group may have colonized northwestern and northeastern Taiwan via the Paleo-Minjiang River and its alternate paths, respectively. Subsequently, warm and wet interglacials facilitated long-distance dispersal via pollen flow through increased numbers of pollinators or seed transport to colonize novel habitats via water flow; cold and dry glacials led populations to retreat into refugia and resulted in fragmentation. Repeated dispersal and isolation diversified populations or species of the indica group. The demography of S. indica, which has relatively high genetic diversity compared with that of other members of indica group, was more sensitive to climate change–i.e., the bottleneck at LGM. In contrast, the endemic species of the indica group revealed higher dispersibility for range expansion in coalescent processes but constant population size through time. The newly derived species have not accumulated enough genetic variation yet and present difficulty in reflecting genetic loss through population decline, but low variation could be irrelevant to the estimation of migration. Based on this case study, we suggest that the diversification of continental island herbs might be related to the sources of adjacent areas and past climatic and geographic changes. The results of this study also implied that island species can rapidly fill open niches caused by cyclic glacials/interglacials via rapid diversification through repeated dispersal and vicariance.
Supporting Information
Neighbor-joining (NJ) and Bayesian inference (BI) trees of Scutellaria species reconstructed by each of five loci. Species marked in red and red bold are distributed in Taiwan and endemic to Taiwan, respectively. Values indicated in the nodes are the bootstrap values and posterior probabilities for supporting the grouping of lineages in NJ trees and BI trees, respectively.
(DOCX)
Comparison of Lineage-Through-Time (LTT) plots, showing the CRD pattern of all Taiwan Scutellaria species (black curves) and the LBD pattern of the endemic Scutellaria species (gray curves) based on 1000 postconvergence Bayesian trees.
(DOCX)
List of Scutellaria species used in the phylogenetic analysis.
(DOCX)
Best substitution models for the five loci used in the Bayesian analyses.
(DOCX)
Tail probabilities of asymmetric values for the among-lineage diversification rate variation in the phylogenetic topologies reconstructed from total samples and Taiwanese samples, respectively, inferred from the species tree (BEAST).
(DOCX)
Genetic diversity of populations of Scutellaria species in Taiwan estimated using four polymorphic loci. The monomorphic matK in every population is not shown.
(DOCX)
Acknowledgments
The authors appreciate Chun-Ting Lin for his experimental assistance and thank Dr. Xun Gong and Li-Yuan Kuo for providing samples of S. amoena and S. sessilifolia (KW2135), respectively.
Funding Statement
Funding was provided by grants from the Forestry Bureau, Council of Agriculture (99AS-1.1.8-FB-e1) and the National Science Council, R.O.C (NSC 99-2621-B-020-001) to P-C Liao. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Pinto G, Mahler DL, Harmon LJ, Losos JB (2008) Testing the island effect in adaptive radiation: rates and patterns of morphological diversification in Caribbean and mainland Anolis lizards. P R Soc B 275(1652): 2749–2757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Givnish TJ (2010) Ecology of plant speciation. Taxon 59(5): 1326–1366. [Google Scholar]
- 3. Slatkin M (1977) Gene flow and genetic drift in a species subject to frequent local extinctions. Theor Popul Biol 12(3): 253–262. [DOI] [PubMed] [Google Scholar]
- 4. Chiang TY, Schaal BA (2006) Phylogeography of plants in Taiwan and the Ryukyu archipelago. Taxon 55(1): 31–41. [Google Scholar]
- 5. Hsieh TH (2002) Composition, endemism and phytogeographical affinities of the Taiwan flora. Taiwania 47(4): 298–310. [Google Scholar]
- 6. Schluter D (1996) Ecological causes of adaptive radiation. Am Nat 148: S40–S64. [Google Scholar]
- 7. Rainey PB, Travisano M (1998) Adaptive radiation in a heterogeneous environment. Nature 394(6688): 69–72. [DOI] [PubMed] [Google Scholar]
- 8. Gittenberger E (1991) What about non-adaptive radiation? Biol J Linn Soc 43(4): 263–272. [Google Scholar]
- 9. Lin TP (2001) Allozyme variations in Michelia formosana (Kanehira) Masamune (Magnoliaceae), and the inference of a glacial refugium in Taiwan. Theor Appl Genet 102(2–3): 450–457. [Google Scholar]
- 10. Huang SSF, Hwang SY, Lin TP (2002) Spatial pattern of chloroplast DNA variation of Cyclobalanopsis glauca in Taiwan and east Asia. Mol Ecol 11(11): 2349–2358. [DOI] [PubMed] [Google Scholar]
- 11. Hwang SY, Lin TP, Ma CS, Lin CL, Chung JD, et al. (2003) Postglacial population growth of Cunninghamia konishii (Cupressaceae) inferred from phylogeographical and mismatch analysis of chloroplast DNA variation. Mol Ecol 12(10): 2689–2695. [DOI] [PubMed] [Google Scholar]
- 12. Cheng YP, Hwang SY, Lin TP (2005) Potential refugia in Taiwan revealed by the phylogeographical study of Castanopsis carlesii Hayata (Fagaceae). Mol Ecol 14(7): 2075–2085. [DOI] [PubMed] [Google Scholar]
- 13. Cheng YP, Hwang SY, Chiou WL, Lin TP (2006) Allozyme variation of populations of Castanopsis carlesii (Fagaceae) revealing the diversity centres and areas of the greatest divergence in Taiwan. Ann Bot-London 98(3): 601–608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Lee YJ, Hwang SY, Ho KC, Lin TP (2006) Source populations of Quercus glauca in the last glacial age in taiwan revealed by nuclear microsatellite markers. Journal of Heredity 97(3): 261–269. [DOI] [PubMed] [Google Scholar]
- 15. Wu SH, Hwang CY, Lin TP, Chung JD, Cheng YP, et al. (2006) Contrasting phylogeographical patterns of two closely related species, Machilus thunbergii and Machilus kusanoi (Lauraceae), in Taiwan. J Biogeogr 33(5): 936–947. [Google Scholar]
- 16. Hewitt G (2000) The genetic legacy of the Quaternary ice ages. Nature 405(6789): 907–913. [DOI] [PubMed] [Google Scholar]
- 17. Carstens BC, Knowles LL (2007) Shifting distributions and speciation: species divergence during rapid climate change. Mol Ecol 16(3): 619–627. [DOI] [PubMed] [Google Scholar]
- 18. Wagstaff SJ, Hickerson L, Spangler R, Reeves PA, Olmstead RG (1998) Phylogeny in Labiatae s.l., inferred from cpDNA sequences. Plant Syst Evol 209(3–4): 265–274. [Google Scholar]
- 19. Janicsak G, Veres K, Kakasy AZ, Mathe I (2006) Study of the oleanolic and ursolic acid contents of some species of the Lamiaceae. Biochem Syst Ecol 34(5): 392–396. [Google Scholar]
- 20. Vidal F, Vidal JC, Gadelha AP, Lopes CS, Coelho MG, et al. (2007) Giardia lamblia: the effects of extracts and fractions from Mentha x piperita Lin. (Lamiaceae) on trophozoites. Exp Parasitol 115(1): 25–31. [DOI] [PubMed] [Google Scholar]
- 21. Charami MT, Lazari D, Karioti A, Skaltsa H, Hadjipavlou-Litina D, et al. (2008) Antioxidant and antiinflammatory activities of Sideritis perfoliata subsp. perfoliata (Lamiaceae). Phytother Res 22(4): 450–454. [DOI] [PubMed] [Google Scholar]
- 22. Schmiderer C, Grassi P, Novak J, Weber M, Franz C (2008) Diversity of essential oil glands of clary sage (Salvia sclarea L., Lamiaceae). Plant Biol 10(4): 433–440. [DOI] [PubMed] [Google Scholar]
- 23. Lukas B, Schmiderer C, Franz C, Novak J (2009) Composition of essential oil compounds from different Syrian populations of Origanum syriacum L. (Lamiaceae). J Agr Food Chem 57(4): 1362–1365. [DOI] [PubMed] [Google Scholar]
- 24. Sun M (1999) Cleistogamy in Scutellaria indica (Labiatae): effective mating system and population genetic structure. Mol Ecol 8(8): 1285–1295. [DOI] [PubMed] [Google Scholar]
- 25. Denda T (2002) Cleistogamy in Scutellaria rubropunctata Hayata (Labiatae) from the Ryukyu Archipelago. Biol Mag Okinawa 40: 7–14. [Google Scholar]
- 26. Olmstead RG (1990) Biological and historical factors influencing genetic diversity in the Scutellaria angustifolia complex (Labiatae). Evolution 44(1): 54–70. [DOI] [PubMed] [Google Scholar]
- 27. Williams PA (1992) Ecology of the endangered herb Scutellaria novae-zelandiae . New Zeal J Ecol 16(2): 127–135. [Google Scholar]
- 28. Cruzan MB (2001) Population size and fragmentation thresholds for the maintenance of genetic diversity in the herbaceous endemic Scutellaria montana (Lamiaceae). Evolution 55(8): 1569–1580. [DOI] [PubMed] [Google Scholar]
- 29. Paton AJ (1990) A global taxonomic investigation of Scutellaria (Labiatae). Kew Bull 45(3): 399–450. [Google Scholar]
- 30. Hsieh TH, Huang TC (1995) Notes on the Flora of Taiwan (20) – Scutellaria (Lamiaceae) in Taiwan. Taiwania 40(1): 35–56. [Google Scholar]
- 31. Hsieh TH, Huang TC (1997) Notes on the Flora of Taiwan (29) – Scutellaria austrotaiwanensis Hsieh & Huang sp. nov. (Lamiaceae) from Taiwan. Taiwania 42(2): 109–116. [Google Scholar]
- 32.Huang TC, Hsieh TH, Cheng WT (1998) Lamiaceae. In: Huang T-C, Boufford DE, Hsieh C-F, Lowry PPI, Ohashi H, et al.., editors. Flora of Taiwan, 2nd edition. Taipei, Taiwan, R.O.C.: Editorial Committee of the Flora of Taiwan, Department of Botany, National Taiwan University. 432–543.
- 33. Huang TC, Hsiao A, Wu MJ (2003) Note on the flora of Taiwan (35) –Scutellaria taipeiensis T.-C. Huang, A. Hsiao et M.-J. Wu sp. nov. (Laminaceae). Taiwania 48(2): 129–137. [Google Scholar]
- 34.Li XW, Hedge IC (1994) Scutellaria Linnaeus, Lamiaceae Lindley. St. Louis, MO and Cambridge, MA: Missouri Botanical Garden and Harvard University Herbaria. 75–103. Available: http://www.efloras.org. Accessed 2008 Feb 22.
- 35. Yamazaki T (1992) A revision of Scutellaria in Taiwan. J Jap Bot 67(6): 315–319. [Google Scholar]
- 36. Doyle JJ, Doyle JL (1987) A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull 19: 11–15. [Google Scholar]
- 37. Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG (1997) The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res 25(24): 4876–4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Hall TA (1999) BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucl Acids Symp Ser 41: 95–98. [Google Scholar]
- 39. Tamura K, Peterson D, Peterson N, Stecher G, Nei M, et al. (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28(10): 2731–2739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Ronquist F, Huelsenbeck JP (2003) MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19(12): 1572–1574. [DOI] [PubMed] [Google Scholar]
- 41. Drummond AJ, Rambaut A (2007) BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol Biol 7: 214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Wolfe KH, Li WH, Sharp PM (1987) Rates of Nucleotide Substitution Vary Greatly among Plant Mitochondrial, Chloroplast, and Nuclear DNAs. P Natl Acad Sci USA 84(24): 9054–9058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Koch MA, Haubold B, Mitchell-Olds T (2000) Comparative evolutionary analysis of chalcone synthase and alcohol dehydrogenase loci in Arabidopsis, Arabis, and related genera (Brassicaceae). Mol Biol Evol 17(10): 1483–1498. [DOI] [PubMed] [Google Scholar]
- 44.Rambaut A, Drummond AJ (2003) Tracer ver. 1.4.1. Edinburgh: Institute of Evolutionary Biology, University of Edinburgh. Available: http://tree.bio.ed.ac.uk/software/tracer/. Accessed 2012 Nov 5.
- 45.Rambaut A (2008) FigTree ver. 1.3.1. Edinburgh: Institute of Evolutionary Biology, University of Edinburgh. Available: http://tree.bio.ed.ac.uk/software/FigTree/. Accessed 2012 Nov 5.
- 46. Shimodaira H (2002) An approximately unbiased test of phylogenetic tree selection. Syst Biol 51(3): 492–508. [DOI] [PubMed] [Google Scholar]
- 47. Kishino H, Hasegawa M (1989) Evaluation of the maximum likelihood estimate of the evolutionary tree topologies from DNA sequence data, and the branching order in Hominoidea. J Mol Evol 29(2): 170–179. [DOI] [PubMed] [Google Scholar]
- 48. Shimodaira H, Hasegawa M (1999) Multiple comparisons of log-likelihoods with applications to phylogenetic inference. Mol Biol Evol 16(8): 1114–1116. [Google Scholar]
- 49. Shimodaira H, Hasegawa M (2001) CONSEL: for assessing the confidence of phylogenetic tree selection. Bioinformatics 17(12): 1246–1247. [DOI] [PubMed] [Google Scholar]
- 50. Shimodaira H (2004) Approximately unbiased tests of regions using multistep-multiscale bootstrap resampling. Ann Stat 32(6): 2616–2641. [Google Scholar]
- 51. Rozas J, Sanchez-DelBarrio JC, Messeguer X, Rozas R (2003) DnaSP, DNA polymorphism analyses by the coalescent and other methods. Bioinformatics 19(18): 2496–2497. [DOI] [PubMed] [Google Scholar]
- 52. Tajima F (1989) Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics 123(3): 585–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Fulford GR, Roberts MG, Heesterbeek JA (2002) The metapopulation dynamics of an infectious disease: tuberculosis in possums. Theor Popul Biol 61(1): 15–29. [DOI] [PubMed] [Google Scholar]
- 54.Yu Y, Harris AJ, He XJ (2011) RASP (Reconstruct Ancestral State in Phylogenies) 2.0 beta. Available: http://mnh.scu.edu.cn/soft/blog/RASP. Accessed 2012 Nov 5.
- 55.Moore BR, Chan KMA, Donoghue MJ (2004) Detecting diversification rate variation in supertrees. In: Bininda-Emonds ORP, editor. Phylogenetic Supertrees: Combining Information to Reveal the Tree of Life. Dordrecht, The Netherlands: Kluwer Academic. 487–533.
- 56. Paradis E, Claude J, Strimmer K (2004) APE: analyses of phylogenetics and evolution in R language. Bioinformatics 20(2): 289–290. [DOI] [PubMed] [Google Scholar]
- 57. Excoffier L, Lischer HEL (2010) Arlequin suite ver 3.5: a new series of programs to perform population genetics analyses under Linux and Windows. Mol Ecol Resour 10(3): 564–567. [DOI] [PubMed] [Google Scholar]
- 58. Excoffier L (2004) Patterns of DNA sequence diversity and genetic structure after a range expansion: lessons from the infinite-island model. Mol Ecol 13(4): 853–864. [DOI] [PubMed] [Google Scholar]
- 59. Excoffier L, Laval G, Schneider S (2005) Arlequin ver. 3.0: An integrated software package for population genetics data analysis. Evolutionary Bioinformatics Online 1: 47–50. [PMC free article] [PubMed] [Google Scholar]
- 60. Harpending HC (1994) Signature of ancient population-growth in a low-resolution mitochondrial DNA mismatch distribution. Hum Biol 66(4): 591–600. [PubMed] [Google Scholar]
- 61. Esselstyn JA, Timm RM, Brown RM (2009) Do geological or climatic processes drive speciation in dynamic archipelagos? The tempo and mode of diversification in Southeast Asian shrews. Evolution 63(10): 2595–2610. [DOI] [PubMed] [Google Scholar]
- 62. Hosoda T, Sato JJ, Lin LK, Chen YJ, Harada M, et al. (2011) Phylogenetic history of mustelid fauna in Taiwan inferred from mitochondrial genetic loci. Can J Zool 89(6): 559–569. [Google Scholar]
- 63. Wang CF, Hsieh CH, Lee SC, Wang HY (2011) Systematics and phylogeography of the Taiwanese endemic minnow Candidia barbatus (Pisces: Cyprinidae) based on DNA sequence, allozymic, and morphological analyses. Zool J Linn Soc-Lond 161(3): 613–632. [Google Scholar]
- 64. Huang JP, Lin CP (2011) Lineage-specific late pleistocene expansion of an endemic subtropical gossamer-wing damselfly, Euphaea formosa, in Taiwan. BMC Evol Biol 11: 94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Nakamura K, Denda T, Kokubugata G, Suwa R, Yang TYA, et al. (2010) Phylogeography of Ophiorrhiza japonica (Rubiaceae) in continental islands, the Ryukyu Archipelago, Japan. J Biogeogr 37(10): 1907–1918. [Google Scholar]
- 66.Simpson GG (1953) The Major Features of Evolution. New York: Columbia University Press.
- 67. Glor RE (2010) Phylogenetic insights on adaptive radiation. Annu Rev Ecol Evol Syst 41: 251–270. [Google Scholar]
- 68. McGlone MS, Duncan RP, Heenan PB (2001) Endemism, species selection and the origin and distribution of the vascular plant flora of New Zealand. J Biogeogr 28(2): 199–216. [Google Scholar]
- 69. Martin-Bravo S, Valcarcel V, Vargas P, Luceno M (2010) Geographical speciation related to Pleistocene range shifts in the western Mediterranean mountains (Reseda sect. Glaucoreseda, Resedaceae). Taxon 59(2): 466–482. [Google Scholar]
- 70. Middleton B (2000) Hydrochory, seed banks, and regeneration dynamics along the landscape boundaries of a forested wetland. Plant Ecol 146(2): 169–184. [Google Scholar]
- 71. Huang ZY, Yu HS (2003) Morphology and geologic implications of Penghu Channel off southwest Taiwan. Terr Atmos Ocean Sci 14(4): 469–485. [Google Scholar]
- 72. Shih HT, Fang SH, Ng PKL (2007) Phylogeny of the freshwater crab genus Somanniathelphusa Bott (Decapoda : Parathelphusidae) from Taiwan and the coastal regions of China, with notes on their biogeography. Invertebr Syst 21(1): 29–37. [Google Scholar]
- 73. Shih HT, Cai Y (2007) Two new species of the land-locked freshwater shrimps genus, Neocaridina Kubo, 1938 (Decapoda : Caridea : Atyidae), from Taiwan, with notes on speciation on the island. Zool Stud 46(6): 680–694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Levin DA (2005) Niche shifts: The primary driver of novelty within angiosperm genera. Syst Bot 30(1): 9–15. [Google Scholar]
- 75. Liew PM, Kuo CM, Huang SY, Tseng MH (1998) Vegetation change and terrestrial carbon storage in eastern Asia during the Last Glacial Maximum as indicated by a new pollen record from central Taiwan. Global Planet Change 17: 85–94. [Google Scholar]
- 76. Wei KY (2002) Environmental changes during the late Quaternary in Taiwan and adjacent seas: an overview of recent results of the past decade (1990–2000). West Pac Earth Sci 2: 149–160. [Google Scholar]
- 77.Hudson RR (1990) Gene genealogies and the coalescent process. In: Futuyma DJ, Antonovics J, editors. Oxford Survey of Evolutionary Biology. New York: Oxford University Press. 1–44.
- 78. Vilaca ST, Santos FR (2010) Biogeographic history of the species complex Basileuterus culicivorus (Ayes, Parulidae) in the Neotropics. Mol Phylogenet Evol 57(2): 585–597. [DOI] [PubMed] [Google Scholar]
- 79. Boggs SJ, Wang W-C, Lewis FS, Chen J-C (1979) Sediment properties and water characteristics of the Taiwan shelf and Slope. Acta Oceanographica Taiwanica 10: 10–49. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Neighbor-joining (NJ) and Bayesian inference (BI) trees of Scutellaria species reconstructed by each of five loci. Species marked in red and red bold are distributed in Taiwan and endemic to Taiwan, respectively. Values indicated in the nodes are the bootstrap values and posterior probabilities for supporting the grouping of lineages in NJ trees and BI trees, respectively.
(DOCX)
Comparison of Lineage-Through-Time (LTT) plots, showing the CRD pattern of all Taiwan Scutellaria species (black curves) and the LBD pattern of the endemic Scutellaria species (gray curves) based on 1000 postconvergence Bayesian trees.
(DOCX)
List of Scutellaria species used in the phylogenetic analysis.
(DOCX)
Best substitution models for the five loci used in the Bayesian analyses.
(DOCX)
Tail probabilities of asymmetric values for the among-lineage diversification rate variation in the phylogenetic topologies reconstructed from total samples and Taiwanese samples, respectively, inferred from the species tree (BEAST).
(DOCX)
Genetic diversity of populations of Scutellaria species in Taiwan estimated using four polymorphic loci. The monomorphic matK in every population is not shown.
(DOCX)