Abstract
Pathogenicity of many Gram-negative bacteria depends on a type III secretion (T3S) system which translocates bacterial effector proteins into eukaryotic cells. The membrane-spanning secretion apparatus is associated with a cytoplasmic ATPase complex and a predicted cytoplasmic (C) ring structure which is proposed to provide a substrate docking platform for secreted proteins. In this study, we show that the putative C ring component HrcQ from the plant pathogenic bacterium Xanthomonas campestris pv. vesicatoria is essential for bacterial pathogenicity and T3S. Fractionation studies revealed that HrcQ localizes to the cytoplasm and associates with the bacterial membranes under T3S-permissive conditions. HrcQ binds to the cytoplasmic T3S-ATPase HrcN, its predicted regulator HrcL and the cytoplasmic domains of the inner membrane proteins HrcV and HrcU. Furthermore, we observed an interaction between HrcQ and secreted proteins including early and late T3S substrates. HrcQ might therefore act as a general substrate acceptor site of the T3S system and is presumably part of a larger protein complex. Interestingly, the N-terminal export signal of the T3S substrate AvrBs3 is dispensable for the interaction with HrcQ, suggesting that binding of AvrBs3 to HrcQ occurs after its initial targeting to the T3S system.
Introduction
Many Gram-negative pathogenic bacteria employ a type III secretion (T3S) system to translocate effector proteins into eukaryotic cells. T3S systems are conserved among plant and animal pathogenic bacteria and are evolutionarily related to the bacterial flagellum, which is the key bacterial motility organelle and hereafter is referred to as flagellar T3S system [1], [2], [3]. Electron microscopy studies of isolated flagellar and translocation-associated T3S systems from Salmonella spp. and Shigella flexneri, respectively, revealed that the membrane-spanning portions of both T3S systems share a similar architecture consisting of ring structures in the inner membrane (IM) and outer membrane (OM) [4]-[9]. These architectural similarities are reflected by homologies in the amino acid sequences of at least eight components of the secretion machinery that presumably constitute the core structural elements of the T3S system. In translocation-associated T3S systems the nomenclature of these proteins refers to the Ysc proteins from the animal pathogenic bacterium Yersinia [10]. They include three cytoplasmic proteins (YscL, N and Q), five IM proteins (YscR, S, T, U and V) and the OM secretin (YscC), which is absent from flagellar T3S systems.
The membrane-spanning portions of flagellar and translocation-associated T3S systems are associated with extracellular appendages including the flagellar hook and filament as well as the pilus (in plant pathogenic bacteria; up to 2 µm long) or needle (in animal pathogenic bacteria; 40-80 nm long) of translocation-associated T3S systems [11]. The pilus and needle are essential for T3S and presumably provide protein transport channels to the host-pathogen interface. They are directly or indirectly connected to the T3S translocon, which is a predicted oligomeric protein channel in the host plasma membrane and mediates effector protein translocation into the host cell [12]-[14].
One of the model systems to study T3S is the Gram-negative bacterium Xanthomonas campestris pv. vesicatoria, which causes bacterial spot disease of pepper and tomato plants. The T3S system from X. campestris pv. vesicatoria translocates approximately 30 to 40 effector proteins into the plant cell where they interfere with host cellular processes such as gene expression, signal transduction cascades and the suppression of host defense responses to the benefit of the pathogen [15]. Effector protein translocation is activated in planta by a yet unknown signal and depends on the chromosomal hrp (hypersensitive response and pathogenicity) gene cluster, which encodes the components of the T3S system [15],[16]. Mutant studies with individual hrp genes revealed that efficient T3S does not only depend on predicted components of the T3S system but also on control proteins – designated Hpa (Hrp associated) - that presumably regulate T3S substrate specificity and recognition. Among the control proteins is the general T3S chaperone HpaB which binds to and promotes the efficient secretion and translocation of multiple effector proteins [17]-[19]. HpaB presumably targets effector proteins to the ATPase HrcN of the T3S system which can dissociate HpaB-effector protein complexes and thus might facilitate the entry of effector proteins into the inner channel of the T3S system [20].
In addition to HpaB, the efficient translocation of effector proteins depends on HpaC, which is a T3S substrate specificity switch (T3S4) protein. HpaC promotes the secretion of translocon and effector proteins but suppresses the efficient secretion of HrpB2, which is required for T3S pilus formation [21]-[23]. Given the architecture of the T3S system, pilus assembly likely occurs prior to the secretion of translocon and effector proteins, suggesting that the substrate specificity of the T3S system switches from “early“ to “late“ substrates [14], [24], [25]. The switch is mediated by T3S4 proteins that interact with the cytoplasmic domains of members of the YscU family of IM proteins. It was proposed that T3S4 proteins induce a conformational change in the cytoplasmic domains of YscU family members that leads to an alteration in substrate recognition [3], [14], [24]. In agreement with this model, HpaC interacts with the C-terminal domain of the YscU homolog HrcU (HrcUC). Furthermore, the hpaC mutant phenotype can be suppressed by a point mutation in HrcUC that likely mimicks the predicted conformational change [21], [26]. HrcUC interacts with HrpB2, suggesting that it provides a docking site for early T3S substrates. However, an interaction between HrcUC and late T3S substrates has not yet been observed [21]. It is therefore still unclear how late substrates are recognized by the T3S system.
In the present study, we analyzed a possible contribution of the YscQ homolog HrcQ to T3S and substrate docking. HrcQ belongs to the family of putative cytoplasmic (C) ring components of the T3S system that are proposed to form a cup-like structure with a diameter of approximately 40 nm. The predicted C ring of translocation-associated T3S systems has not yet been visualized because it presumably easily disconnects from the membrane-spanning secretion apparatus during the purification procedure. However, the C ring was visualized by electron microscopy of isolated flagellar T3S systems [27], [28]. Flagellar C rings consist of three proteins (FliG, M and N) that connect the C ring to the IM components of the T3S system such as the ATPase complex or the ring components in the IM [4], [27]-[31]. FliM and FliN share amino acid sequence similarities with predicted C ring components of translocation-associated T3S systems. Given the finding that YscQ and homologous proteins from animal pathogenic bacteria interact with effector proteins or effector-chaperone complexes, the predicted C ring of translocation-associated T3S systems was proposed to act as a recruitment platform for secreted proteins [32], [33]. We therefore investigated the contribution of HrcQ from X. campestris pv. vesicatoria to T3S and substrate binding. Mutant and protein studies revealed that HrcQ is an essential component of the T3S system and associates with the bacterial membranes under T3S-permissive conditions. In vitro pull-down assays showed that HrcQ interacts with conserved components of the T3S system at the IM including the IM proteins HrcV and HrcU, the ATPase HrcN and its predicted regulator HrcL. Furthermore, HrcQ binds to early and late T3S substrates and therefore is likely involved in T3S substrate recognition. Interestingly, the analysis of derivatives of the effector protein AvrBs3 suggests that the N-terminal T3S and translocation signal is dispensable for the interaction of AvrBs3 with HrcQ.
Materials and Methods
Bacterial Strains and Growth Conditions
Bacterial strains and plasmids used in this study are listed in Table 1. Escherichia coli cells were grown at 37°C in lysogeny broth (LB) or Super medium (Qiagen, Hilden, Germany). X. campestris pv. vesicatoria strains were cultivated at 30°C in nutrient-yeast-glycerol (NYG) medium [47] or in minimal medium A [48] supplemented with sucrose (10 mM) and casamino acids (0.3%). Plasmids were introduced into E. coli by transformation and into X. campestris pv. vesicatoria by conjugation, using pRK2013 as a helper plasmid in triparental matings [49]. Antibiotics were added to the media at the following final concentrations: ampicillin, 100 µg/ml; kanamycin, 25 µg/ml; rifampicin, 100 µg/ml; spectinomycin, 100 µg/ml; gentamycin, 7.5 µg/ml.
Table 1. Bacterial strains and plasmids used in this study.
| Relevant characteristicsa | Reference or source | ||||
| X. campestris pv. vesicatoria | |||||
| 85-10 | Pepper-race 2; wild type; Rifr | [51], [73] | |||
| 85-10ΔhrcQ | hrcQ deletion mutant of strain 85-10 | This study | |||
| 85-10ΔhrcQ::hrcQ | Derivative of strain 85-10ΔhrcQ carrying hrcQ-c-myc under control of the native promoter inserted into the hpaFG region | This study | |||
| 85* | 85-10 derivative containing the hrpG* mutation | [34] | |||
| 85*ΔhrcQ | hrcQ deletion mutant of strain 85* | This study | |||
| 85*ΔhrcQ::hrcQ | Derivative of strain 85*ΔhrcQ carrying hrcQ-c-myc under control of the native promoter inserted into the hpaFG region | This study | |||
| E. coli | |||||
| BL21 (DE3) | F- ompT hsdSB (rB - mB -) gal dcm (DE3) | Stratagene, Heidelberg, Germany | |||
| Top10 | F- mcrA Δ(mrr-hsdRMS-mcrBC) Φ80lacZΔM15 ΔlacX74 recA1 araD139 Δ(araleu) 7697 galU galK rpsL (StrR) endA1 nupG | Invitrogen, Karlsruhe, Germany | |||
| DH5α | F- recA hsdR17(rk-,mk+) Φ80dlacZ ΔM15 | Bethesda Research Laboratories, Bethesda, Md. | |||
| DH5α λpir | F- recA hsdR17(rk-,mk+) Φ80dlacZ ΔM15 [λpir]α | [74] | |||
| Plasmids | |||||
| pBlueskript(II) KS | Phagemid, pUC derivative; Apr | Stratagene | |||
| pBRM | Golden Gate-compatible derivative of pBBR1MCS-5 for lacpromoter-driven gene expression; contains a 3× c-Mycepitope-encoding sequence | [75] | |||
| pBRM-P | Derivative of pBRM without lac promoter | This study | |||
| pBRMhrcN | pBRM derivative encoding HrcN-c-Myc | [75] | |||
| pBRMhrcQ | pBRM derivative encoding HrcQ-c-Myc | This study | |||
| pBRMhrcQStop | pBRM derivative encoding HrcQ | This study | |||
| pBRM-PhrcQ | pBRM-P derivative containing hrcQ-c-myc and 299 bp upstream region | This study | |||
| pBRM-PhrcQStop | pBRM-P derivative containing hrcQ and 299 bp upstream region | This study | |||
| pBRMhrcV | pBRM derivative encoding HrcV-c-Myc | N. Hartmann and D. Büttner, unpublished | |||
| pBRMhrcV324-645 | pBRM derivative encoding HrcV324-645-c-Myc | N. Hartmann and D. Büttner, unpublished | |||
| pBRMxopJ | pBRM derivative encoding XopJ-c-Myc | [26] | |||
| pDGW4MhpaA | Derivative of pDGW4M encoding HpaA-c-Myc | [80] | |||
| pDMhrcL | Derivative of pDSK602 encoding HrcL-c-Myc | [20] | |||
| pGEX-2TKM | GST expression vector; ptac GST lacI q pBR322 ori; Apr, derivative ofpGEX-2TK with polylinker of pDSK604 | Stratagene[38] | |||
| pGhpaA | pGEX-2TKM derivative encoding GST-HpaA | [76] | |||
| pGhpaB | pGEX-2TKM derivative encoding GST-HpaB | [18] | |||
| pGhpaC | pGEX-2TKM derivative encoding GST-HpaC | [18] | |||
| pGhrpB2 | pGEX-2TKM derivative encoding GST-HrpB2 | [62] | |||
| pGhrpE | pGEX-2TKM derivative encoding GST-HrpE | [55] | |||
| pGhrcL | pGEX-2TKM derivative encoding GST-HrcL | [20] | |||
| pGhrcN | pGEX-2TKM derivative encoding GST-HrcN | [76] | |||
| pGhrcQ | pGEX-2TKM derivative encoding GST-HrcQ | This study | |||
| pGhrcU | pGEX-2TKM derivative encoding GST-HrcU | [21] | |||
| pGhrcU255-357 | pGEX-2TKM derivative encoding GST-HrcU255-357 | [21] | |||
| pGxopA | pGEX-2TKM derivative encoding GST-XopA | [17] | |||
| pGxopJ | pGEX-2TKM derivative encoding GST-XopJ | This study | |||
| pGxopF1 | pGEX-2TKM derivative encoding GST-XopF1 | [18] | |||
| pG300 | pGEX-2TKM derivative encoding GST-AvrBs3 | [77] | |||
| pG356F | pGEX-2TKM derivative encoding GST-AvrBs3Δ2 which lacks theN-terminal 152 amino acids | [17] | |||
| pGavrBs350 | pGEX-2TKM derivative encoding GST-AvrBs31-50 | [17] | |||
| pLAND-P | Derivative of pOK1 carrying fragments of the hpaFG region flanking lacZ, the lac promoter and a triple c-Myc epitope-encoding sequence | This study | |||
| pLAND-PhrcQ | Derivative of pLAND-P carrying hrcQ and 299 bp upstream region | This study | |||
| pOK1 | Suicide vector; sacB sacQ mobRK2 oriR6K; Smr | [78] | |||
| pOKΔhrcQ | Derivative of pOK1 carrying the flanking regions of hrcQ | This study | |||
| pRK2013 | ColE1 replicon, TraRK+ Mob+; Kmr | [49] | |||
| pUC119 | ColE1 replicon; Apr | [79] | |||
Ap, ampicillin; Km, kanamycin; Rif, rifampicin; Sm, spectinomycin; Gm, gentamycin; r, resistant.
Plant Material and Plant Inoculations
The near-isogenic pepper cultivars Early Cal Wonder (ECW) and ECW-10R [50], [51] were grown as described previously [52]. X. campestris pv. vesicatoria strains were inoculated with a needle-less syringe into the intercellular spaces of leaves at concentrations of 2×108 colony-forming units (CFU) ml-1 in 1 mM MgCl2 if not stated otherwise. The appearance of disease symptoms and the HR were scored over a period of one to nine days post inoculation (dpi). For the better visualization of the HR, leaves were bleached in 70% ethanol. Experiments were repeated at least two times. For in planta growth curves, bacteria were inoculated at a density of 104 CFU/ml into leaves of susceptible ECW plants. Bacterial counts were determined over a period of 11 dpi as described [52].
Generation of Expression Plasmids
For the generation of hrcQ expression constructs, hrcQ with or without stop codon was amplified by PCR from X. campestris pv. vesicatoria strain 85-10 and cloned into the Golden Gate-compatible expression vector pBRM using the type IIs restriction enzyme BsaI in a single restriction/ligation reaction [53]. The broad host-range vector pBRM is a derivative of plasmid pBBR-MCS5 and allows the expression of genes under control of the lac promoter in frame with a 3× c-Myc epitope-encoding sequence. Alternatively, we cloned hrcQ including its native promoter (299 bp upstream of the translation initiation codon GTG) into plasmid pBRM-P, which is a derivative of pBRM that lacks the lac promoter (Table 1). To obtain GST fusion constructs, hrcQ and xopJ were amplified by PCR from X. campestris pv. vesicatoria strain 85-10 and cloned into the EcoRI/XhoI sites of vector pGEX, in frame with a GST-encoding sequence. Primer sequences are available upon request.
Construction of X. campestris pv. vesicatoria Deletion Mutants
For the generation of a genomic hrcQ deletion mutant, 700-bp and 750-bp regions flanking hrcQ and spanning part of the 5′- and 3′- region of hrcQ were amplified by PCR and cloned into the BamHI and ApaI sites of plasmid pOK1. The resulting construct pOKΔhrcQ was conjugated into strains 85-10 and 85*. Double cross-overs resulted in strains 85-10ΔhrcQ and 85*ΔhrcQ that were selected as described previously [44] and contain a deletion of codons 11 to 243 of hrcQ followed by a nonsense mutation.
Construction of the Suicide Vector pLAND-P
To generate plasmid pLAND-P, we amplified 850- and 770-bp fragments spanning hpaF and the 5′-region of hpaG as well as the 3′-region of hpaG, respectively, by primers hpaF-for (5′-TTTGGTCTCTCATGCATGCGGCGATGGCAGTC-3′, hpaF-rev (5′-TTTGGTCTCTAGACCCCATGGCAGCGAGAGGTTGCGAAG), hpaG-for (5′-TTTGGTCTCTGTCTCTAATTATCGTTGAGCTGAGCAG-3′) and hpaG-rev (5′-TTTGGTCTCTGATC CTCCTGCGTGTGCATG-3′). PCR amplicons were digested with BsaI (restriction sites are underligned in primer sequences, overhangs are written in italics) and ligated into the BamHI and NcoI sites of the suicide vector pOK1, thus generating pOKhpaFG. The ligation reaction led to a mutation in the internal NcoI site of pOK1 and to the generation of a BsaI and NcoI site between both ligated amplicons. Next, we amplified the lacZ gene including the lac promoter and the 3× c-Myc epitope-encoding sequence from plasmid pBRM-P using primers lacZ-P-for (5′-TTTCGTCTCTAATTCAGAGACCGCAGCTGGCACGACAG-3′) and Myc-rev (5′-TTTCGTCTCTCATGGTCAGTTCAAGTCTTCTTC-3′). The amplicon was digested with Esp3I and ligated into the BsaI/NcoI-digested pOKhpaFG, resulting in pLAND-P. The introduction of a BsaI site by primer lacZ-P-for and the presence of a BsaI site upstream of the 3× c-Myc epitope-encoding sequence allows the targeted ligation of genes of interest in the opposite orientation of the hpaFG transcription. To generate pLAND-PhrcQ, hrcQ including its native promoter was amplified and cloned into pLAND-P in a single restriction/ligation reaction using BsaI and ligase.
Glutathione S-transferase (GST) Pull-down Assays
For GST pull-down assays, GST and GST fusion proteins were synthesized in E. coli BL21(DE3). Bacterial cells from 50 ml cultures were resuspended in phosphate-buffered saline (PBS) and broken with a French press. Insoluble cell debris were removed by centrifugation, and soluble GST and GST fusion proteins were immobilized on a glutathione sepharose matrix according to the manufacturer’s instructions (GE Healthcare, Munich, Germany). Unbound proteins were removed by washing twice with PBS, and the glutathione sepharose matrix was incubated with 600 µl E. coli cell lysates containing the putative interaction partners for 1 to 2 h at 4°C. Unbound proteins were removed by washing four times with PBS and bound proteins were eluted with 10 mM reduced glutathione at room temperature for 2 h. 10 µl total protein lysates and 20 µl eluted proteins were analyzed by SDS-PAGE and immunoblotting using antibodies specific for the c-Myc epitope and GST, respectively (Roche Applied Science, Mannheim, Germany; GE Healthcare) [22]. Horseradish peroxidase-labeled anti-mouse and anti-goat antibodies (GE Healthcare) were used as secondary antibodies. Antibody reactions were visualized by enhanced chemiluminescence (GE Healthcare).
Generation of Polyclonal HrcQ Antibodies
For the generation of HrcQ antibodies, rabbits were immunized with a HrcQ-specific peptide (AEVIAFERDAEPDD, amino acids 82 to 95) (Biogenes, Berlin, Germany). The serum after the second booster injection was used for immunoblot analysis.
Protein Secretion Studies and Immunoblot Analysis
T3S assays were performed as described previously [54]. Briefly, bacteria were incubated in secretion medium and equal amounts of bacterial total cell extracts and culture supernatants were analyzed by SDS-PAGE and immunoblotting. We used polyclonal antibodies specific for HrpF [55], HrpB2 [22] and HrcQ (this study), and monoclonal anti-c-Myc (Roche Applied Science, Mannheim, Germany) and anti-GST antibodies (Amersham Pharmacia Biotech, Freiburg, Germany). Horseradish peroxidase-labeled anti-rabbit antibodies (GE Healthcare) were used as secondary antibodies. Experiments were repeated at least two times. Blots were routinely reacted with an antibody specific for the intracellular proteins HrcN or HrcJ to ensure that no bacterial lysis had occurred [22] (data not shown).
Subcellular Localisation Studies
For subcellular localization of proteins, bacteria were grown overnight in minimal medium A supplemented with sucrose and casamino acids. Bacterial cells from 50 ml cultures were resuspended in 3 ml 100 mM HEPES (4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid) and lysed with a French press. Cell debris were removed by centrifugation and 1 ml of each lysate was centrifuged at 200.000 g for 90 minutes at 4°C. Similar protein levels were adjusted according to OD600. The pellet, which corresponds to the membrane fraction, was resuspended in 1 ml 100 mM HEPES. Membrane and soluble fractions were mixed with Laemmli buffer and 20 µl total bacterial lysate, membrane and soluble fractions were analyzed by SDS-PAGE, Coomassie staining (data not shown) and immunoblotting.
Results
HrcQ from X. campestris pv. vesicatoria is Essential for Pathogenicity and T3S
hrcQ from X. campestris pv. vesicatoria strain 85-10 is the first gene of the hrpD operon of the hrp gene cluster and encodes a predicted protein of 304 amino acids that is conserved in Xanthomonas spp. (91 to 93% amino acid identity with HrcQ proteins from Xanthomonas oryzae pv. oryzae, X. oryzae pv. oryzicola, Xanthomonas axonopodis pv. citri and X. axonopodis pv. glycines; 76% amino acid identity with HrcQ from X. campestris pv. campestris). The C-terminal portion of HrcQ shares weak sequence similarity (27 to 33% amino acid sequence identity) with members of the YscQ protein family from animal pathogenic bacteria. To study the contribution of HrcQ to pathogenicity of X. campestris pv. vesicatoria, we generated a hrcQ deletion mutant carrying an in-frame deletion of codons 11 to 243 followed by a nonsense mutation. The hrcQ deletion was introduced into the genome of the wild-type strain 85-10 and its derivative 85-10hrpG* (85*), which contains HrpG*, a constitutively active version of the key regulatory protein HrpG. HrpG* activates – in most cases via the transcriptional regulator HrpX – the expression of a genome-wide regulon including hrp, putative virulence and effector genes [19], [34]-[36].
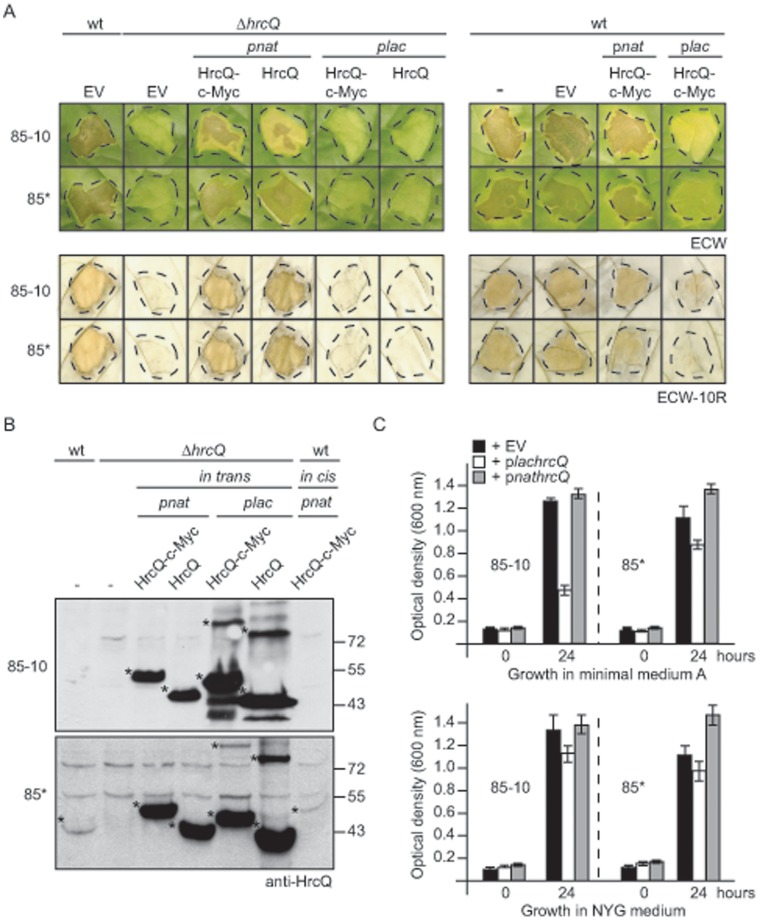
For infection studies, hrcQ wild-type and deletion mutant strains were inoculated into leaves of susceptible ECW and resistant ECW-10R pepper plants. ECW-10R plants carry the Bs1 resistance gene and induce the hypersensitive response (HR) upon recognition of the type III effector AvrBs1, which is delivered by strain 85-10 [37], [38]. The HR is a rapid local cell death at the infection site that restricts bacterial ingress and is activated upon detection of individual bacterial effector proteins (also designated Avr [avirulence] proteins) by the plant surveillance system [39]. As expected, strains 85-10 and 85* induced disease symptoms in ECW and the HR in ECW-10R pepper plants whereas no reactions were observed for the respective hrcQ deletion mutants (Fig. 1A). Ectopic expression of HrcQ-c-Myc or untagged HrcQ derivatives under control of the native promoter (see Materials and Methods) complemented the mutant phenotype of strain 85*ΔhrcQ with respect to HR induction and disease symptoms when the bacteria were inoculated at densities of 108 CFU/ml. At lower inoculation densities, however, expression of hrcQ from the native promoter only partially restored symptom formation (Fig. S1). This was also observed in strain 85-10ΔhrcQ, even with higher inoculation densities (Fig. 1A; data not shown). To confirm that HrcQ derivatives were stably synthesized, protein extracts of bacterial cells grown in minimal medium were analyzed by immunoblotting, using HrcQ-specific antibodies. As expected, lac promoter-driven expression of hrcQ and hrcQ-c-myc led to increased protein amounts when compared with hrcQ derivatives expressed under control of the native promoter in strain 85-10ΔhrcQ. The amounts of the native HrcQ protein were significantly lower and were only detectable in protein extracts of strain 85* which constitutively expresses the hrp genes (Fig. 1B).
Figure 1. HrcQ contributes to bacterial pathogenicity and in planta growth.
(A) Ectopic expression of hrcQ under control of the native promoter partially restores pathogenicity of hrcQ deletion mutant strains. X. campestris pv. vesicatoria strains 85-10 (wt), 85-10ΔhrcQ (ΔhrcQ), 85* (wt) and 85*ΔhrcQ (ΔhrcQ) without expression construct (-) or carrying plasmid pBRM (EV) or derivatives thereof expressing hrcQ or hrcQ-c-myc under control of the native (pnat) or the lac (plac) promoter as indicated were inoculated into leaves of susceptible ECW and resistant ECW-10R pepper plants. Bacterial strains in the left panel (complementation studies) were inoculated at a density of 108 CFU ml-1, strains in the right panel (analysis of dominant-negative effects) at a density of 2×108 CFU ml-8. Disease symptoms were photographed 8 dpi. For the better visualization of the HR, leaves were bleached in ethanol 2 dpi. Dashed lines mark the infiltrated areas. (B) Immunodetection of HrcQ and HrcQ-c-Myc. X. campestris pv. vesicatoria strains 85-10 (wt), 85-10ΔhrcQ (ΔhrcQ), 85* (wt) and 85*ΔhrcQ (ΔhrcQ) without expression construct (-) or encoding HrcQ or HrcQ-c-Myc in trans or in cis under control of the native (pnat) or the lac (plac) promoter as indicated were grown in minimal medium A. Equal amounts of total cell extracts were analyzed by immunoblotting, using HrcQ-specific antibodies. HrcQ-specific signals are marked with asterisks, additional signals result from unspecific binding of the antibodies. The HrcQ-specific signals above 72 kDa presumably correspond to HrcQ protein complexes that were not dissociated by SDS-PAGE. (C) Overexpression of hrcQ leads to reduced bacterial in vitro growth. X. campestris pv. vesicatoria strains 85-10 and 85* carrying plasmid pBRM (EV) or expressing hrcQ-c-myc under control of the lac (plachrcQ) or the native (pnathrcQ) promoter were grown over night in complex NYG medium and resuspended in minimal medium A or NYG medium at an optical density (OD600 nm) of 0.2. The cultures were incubated at 30°C and the optical density was measured over a period of 24 h. Error bars represent standard deviations.
We also introduced an expression construct encoding a C-terminally c-Myc epitope-tagged derivative of the ATPase HrcN under control of the lac promoter into strains 85-10ΔhrcQ and 85*ΔhrcQ. It was previously shown that the phenotype of flagellar C ring mutants can be partially suppressed upon ectopic expression of the ATPase-encoding gene fliI or upon upregulation of the regulatory genes flhDC [40], [41]. Notably, however, expression of hrcN-c-myc did not alter the hrcQ deletion mutant phenotype (Fig. S2). This was probably not caused by a potential negative effect of elevated HrcN levels because we have previously shown that overexpression of hrcN in strain 85-10 does not alter the wild-type in planta phenotype [20]. We therefore conclude that the loss of hrcQ cannot be counteracted by elevated levels of the T3S-ATPase.
Overexpression of hrcQ Exerts a Dominant-negative Effect on Pathogenicity and in vitro Bacterial Growth
Given the lack of complementation of the hrcQ mutant phenotype by lac promoter-driven expression of hrcQ, we analyzed whether the overexpression of hrcQ is detrimental for pathogenicity. For this, the wild-type strain 85-10 carrying plasmid pBRM or hrcQ expression constructs was inoculated into susceptible and resistant pepper plants. Fig. 1A shows that the expression of hrcQ-c-myc under control of the native promoter did not significantly interfere with pathogenicity of strain 85-10. By contrast, the lac promoter-driven expression of hrcQ-c-myc led to severely reduced plant reactions (Fig. 1A). Similar phenotypes were observed with derivatives of strain 85*, suggesting that the dominant-negative effect of hrcQ overexpression could not be counteracted by the constitutive expression of T3S genes (Fig. 1A).
To investigate whether the ectopic expression of hrcQ exerts a general negative effect on bacterial fitness, we also analyzed bacterial growth in vitro. When strains 85-10 and 85* carrying plasmid pBRM or hrcQ expression constructs were grown in minimal medium, the overexpression of hrcQ from the lac promoter led to a significant reduction of bacterial growth (Fig. 1C). This was also observed when bacteria were grown in complex medium NYG. However, the negative effect of hrcQ overexpression was less pronounced in NYG than in minimal medium, suggesting that the interference of increased HrcQ levels with bacterial fitness also depends on the culture medium (Fig. 1C).
HrcQ is Essential for T3S in vitro
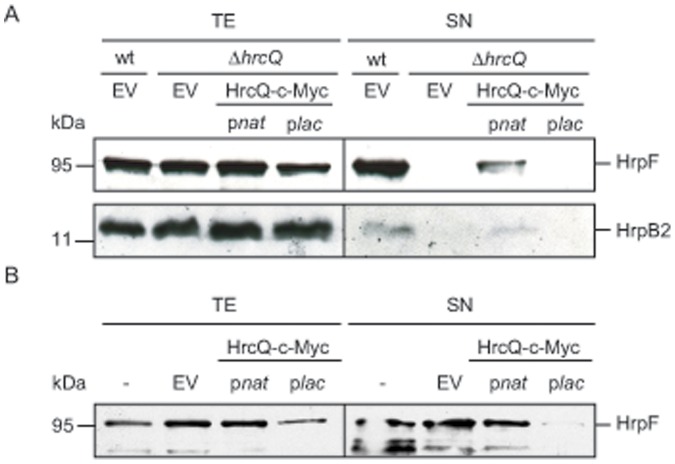
To investigate a potential influence of HrcQ on T3S in vitro, strains 85* and 85*ΔhrcQ were incubated in secretion medium. Total cell extracts and culture supernatants were analyzed by immunoblotting, using specific polyclonal antisera against the translocon protein HrpF and the early T3S substrate HrpB2, respectively. As expected, HrpF and HrpB2 were secreted by strain 85* but were not detectable in the supernatant of strain 85*ΔhrcQ, suggesting that HrcQ is essential for T3S (Fig. 2A). The secretion deficiency was partially complemented by ectopic expression of hrcQ-c-myc under control of the native but not of the lac promoter (Fig. 2A).
Figure 2. HrcQ is essential for T3S of the translocon protein HrpF and the early T3S substrate HrpB2.
(A) Strains 85* (wt) and 85*ΔhrcQ (ΔhrcQ) carrying plasmid pBRM (EV) or expression constructs encoding HrcQ-c-Myc under control of the native (pnat) or the lac (plac) promoter were incubated in secretion medium. Total cell extracts (TE) and culture supernatants (SN) were analyzed by immunoblotting using antibodies specific for HrpF and HrpB2, respectively. (B) lac promoter-driven expression of hrcQ-c-myc exerts a negative effect on T3S of HrpF. Strain 85* without expression construct (-), with plasmid pBRM (EV) or derivatives thereof encoding HrcQ-c-Myc as described in panel A were incubated in secretion medium. TE and SN were analyzed by immunoblotting using HrpF-specific antibodies.
To test a possible negative effect of hrcQ overexpression on T3S, we performed additional secretion assays with strain 85* carrying pBRM or hrcQ expression constructs. The analysis of culture supernatants by immunoblotting revealed that the lac promoter-driven expression of hrcQ-c-myc in strain 85* interfered with the efficient secretion of the translocon protein HrpF (Fig. 2B). By contrast, expression of hrcQ-c-myc under control of the native promoter did not significantly affect HrpF secretion (Fig. 2B). This is in agreement with the results of the infection experiments (see above) and suggests that the lac promoter-driven expression of hrcQ derivatives exerts a negative effect on T3S and pathogenicity.
Insertion of hrcQ into a Genomic "Landing Platform" Fully Restores Pathogenicity of the hrcQ Deletion Mutant
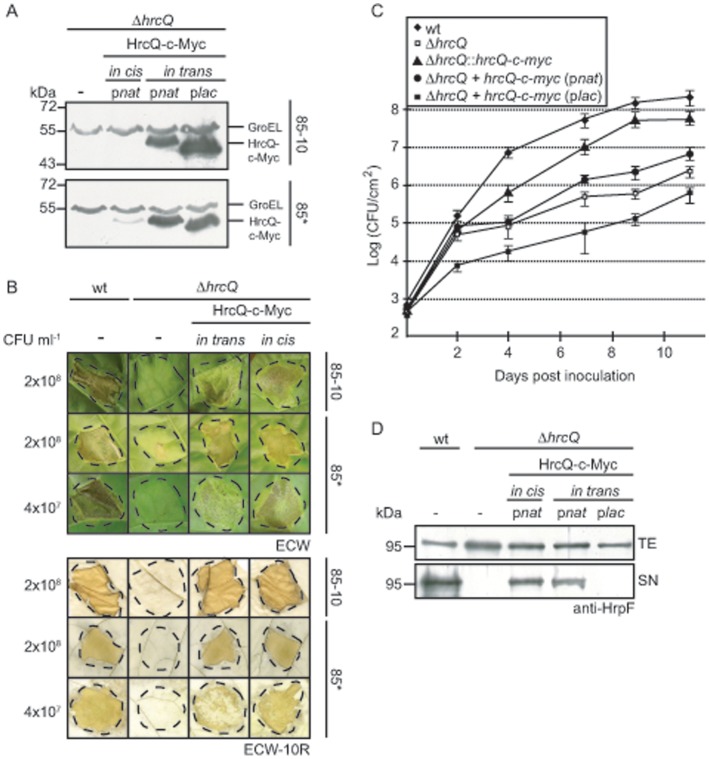
The negative effect of increased HrcQ levels prompted us to perform additional complementation studies with hrcQ deletion strains in which hrcQ was expressed in cis under control of the native promoter. For this, hrcQ was inserted into the genomic hpaF-hpaG locus in the flanking region of the hrp gene cluster by homologous recombination. hpaF and hpaG encode proteins with homology to the N- and C-terminal regions, respectively, of the leucine-rich repeat-containing effector protein XopAE from Xanthomonas spp. It was previously shown that mutations in the hpaFG region do not significantly interfere with bacterial pathogenicity [42], [43]. For the insertion of hrcQ into the hpaFG region of X. campestris pv. vesicatoria, we generated a novel Golden Gate-compatible suicide vector (pLAND-P; see Materials and Methods) that contains fragments of hpaF and hpaG flanking the lacZ gene and a triple c-Myc epitope-encoding sequence. Two recognition sites for the type IIs restriction enzyme BsaI upstream and downstream of lacZ allowed the targeted insertion of genes by a Golden Gate reaction (see Materials and Methods) in the opposite orientation to the direction of the hpaFG transcription which excludes that their expression is controlled by the hpaFG promoter (Fig. S3). hrcQ including its native promoter was cloned into pLAND-P and inserted into the hpaFG region of strains 85-10ΔhrcQ and 85*ΔhrcQ, respectively, by homologous recombination as was described previously [44]. Immunoblot analysis of bacterial protein extracts showed that chromosomal hrcQ-c-myc was expressed in strain 85*ΔhrcQ::hrcQ-c-myc and that the amounts of HrcQ-c-Myc were significantly reduced when compared with strain 85*ΔhrcQ carrying hrcQ-c-myc expression plasmids (Fig. 1B and 3A). As expected, chromosomally encoded HrcQ-c-Myc was not detectable in protein extracts of strain 85-10ΔhrcQ::hrcQ-c-myc which contains the wild-type hrpG gene and therefore does not efficiently express the hrp genes when bacteria are cultivated in non-inducing minimal medium (Fig. 1B and 3A).
Figure 3. Complementation studies with chromosomally encoded hrcQ-c-myc.
(A) Immunodetection of in cis-encoded HrcQ-c-Myc. X. campestris pv. vesicatoria strains 85-10ΔhrcQ (ΔhrcQ) and 85*ΔhrcQ (ΔhrcQ) carrying plasmid pBRM (-) or encoding HrcQ-c-Myc in the chromosome (in cis) or on an expression plasmid (in trans) under control of the native (pnat) or the lac (plac) promoter as indicated were grown in minimal medium A. Equal amounts of total cell extracts were analyzed by immunoblotting, using c-Myc epitope- and GroEL-specific antibodies, respectively. GroEL was analyzed as a loading control. (B) Insertion of hrcQ-c-myc into the hpaFG region restores wild-type symptom formation in hrcQ deletion mutants. Strains 85-10 (wt), 85-10ΔhrcQ (ΔhrcQ), 85* (wt) and 85*ΔhrcQ (ΔhrcQ) carrying plasmid pBRM (-) or encoding HrcQ-c-Myc under control of the native promoter from plasmid pBRM-P (in trans) or in the chromosome (in cis) were inoculated at bacterial cell densities of 2×108 or 4×107 CFU/ml as indicated into leaves of susceptible ECW and resistant ECW-10R pepper plants. Disease symptoms were photographed 9 dpi. For the better visualization of the HR, leaves were bleached in ethanol 2 dpi. Dashed lines mark the infiltrated areas. (C) In planta growth of a hrcQ deletion mutant strain can be partially restored upon expression of hrcQ in cis or in trans. X. campestris pv. vesicatoria strains 85-10 (wt), 85-10ΔhrcQ (ΔhrcQ), 85-10ΔhrcQ::hrcQ-c-myc (ΔhrcQ::hrcQ-c-myc) and 85-10ΔhrcQ expressing hrcQ-c-myc under control of the native (pnat) or the lac (plac) promoter as indicated were inoculated into leaves of susceptible ECW pepper plants. Bacterial growth was analyzed over a period of 11 days. Values are the mean of three samples from three plants. Error bars represent standard deviations. The experiment was repeated two times. One representative experiment is shown. (D) In vitro T3S assays with hrcQ deletion mutants encoding HrcQ-c-Myc on the chromosome or on expression plasmids. Strains 85* (wt) and 85*ΔhrcQ (ΔhrcQ) carrying plasmid pBRM (-) or encoding HrcQ-c-Myc in the chromosome (in cis) or on an expression plasmid (in trans) under control of the native (pnat) or the lac (plac) promoter as indicated were incubated in secretion medium. Total cell extracts (TE) and culture supernatants (SN) were analyzed by immunoblotting using HrpF-specific antibodies.
Plant infection experiments revealed that the in cis expression of hrcQ-c-myc fully complemented the hrcQ mutant phenotype of both strains 85-10ΔhrcQ and 85*ΔhrcQ, even at low inoculation densities (Fig. 3B). We also studied in planta growth of strain 85-10ΔhrcQ carrying different hrcQ expression constructs over a period of eleven days. Fig. 3C shows that the wild-type strain 85-10 grew to a density of more than 108 CFU/cm2 of the infected plant tissue, whereas growth of the hrcQ deletion mutant was significantly reduced. Expression of hrcQ under control of the lac promoter led to a further reduction of in planta growth, suggesting that hrcQ overexpression has a dominant-negative effect on in planta growth (Fig. 3C). Bacterial growth of strain 85-10ΔhrcQ was slightly increased upon ectopic expression of hrcQ under control of the native promoter. By contrast, a more significant increase of bacterial in planta multiplication was observed when hrcQ-c-myc was introduced into the genome of strain 85-10ΔhrcQ, however, wild-type bacterial growth levels were not restored (Fig. 3C). We conclude that the in cis expression of hrcQ under control of the native promoter complements the phenotype of the hrcQ deletion mutant with respect to plant reactions and partially complements the defect in in planta multiplication. A partial complementation was also observed for the in vitro T3S of the translocon protein HrpF (Fig. 3D).
HrcQ Localizes to the Bacterial Membranes upon Activation of the T3S System
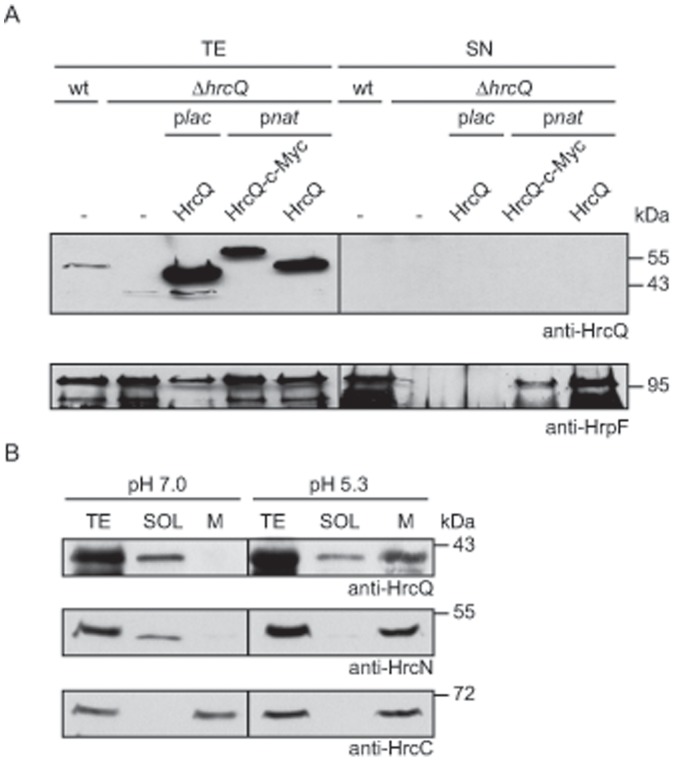
Given the homology of HrcQ to predicted C ring components, we investigated the localization of HrcQ in X. campestris pv. vesicatoria. For this, we first performed secretion studies with strains 85* and 85*ΔhrcQ carrying hrcQ or hrcQ-c-myc expression constructs. Fig. 4A shows that HrcQ and derivatives were present in the total cell extracts but not detectable in the culture supernatants when bacteria were incubated in secretion medium, suggesting that HrcQ is not secreted by the T3S system. Next, we separated membrane fractions and soluble proteins of strain 85* by ultracentrifugation after cultivation of the bacteria in miminal medium A at pH 7.0 (nonpermissive conditions for T3S) or pH 5.3 (T3S-permissive conditions). At pH 7.0, HrcQ was predominantly present in the soluble fraction. By contrast, at pH 5.3 the amounts of HrcQ in the membrane fraction were significantly increased, suggesting a membrane association of HrcQ upon activation of the T3S system (Fig. 4B). When the blots were reprobed with antibodies specific for the ATPase HrcN and the OM secretin HrcC, HrcC was mainly present in the membrane fraction under both secretion-permissive and nonpermissive conditions. By contrast, the ATPase HrcN was detected in the soluble fraction at pH 7.0 whereas the amounts of HrcN in the membrane fraction were significantly increased under T3S-permissive conditions as was reported previously [20] (Fig. 4B).
Figure 4. Secretion and subcellular fractionation studies with HrcQ.
(A) HrcQ is not detectable in the culture supernatant under T3S-inducing conditions. Strains 85* (wt) and 85*ΔhrcQ (ΔhrcQ) carrying plasmid pBRM (-) or expressing hrcQ or hrcQ-c-myc under control of the native (pnat) or the lac (plac) promoter as indicated were incubated in secretion medium. Total cell extracts (TE) and culture supernatants (SN) were analyzed by immunoblotting, using HrcQ- and HrpF-specific antibodies. HrpF was analyzed as a positive control for T3S. (B) HrcQ preferentially associates with the bacterial membranes under T3S-permissive conditions. Strain 85* was grown in minimal medium A supplemented with sucrose and casamino acids under secretion-permissive (pH 5.3) and non-permissive (pH 7.0) conditions. Membrane (M) and soluble (SOL) fractions were separated by ultracentrifugation and analyzed by immunoblotting, using antibodies directed against HrcQ, HrcN and HrcC, respectively.
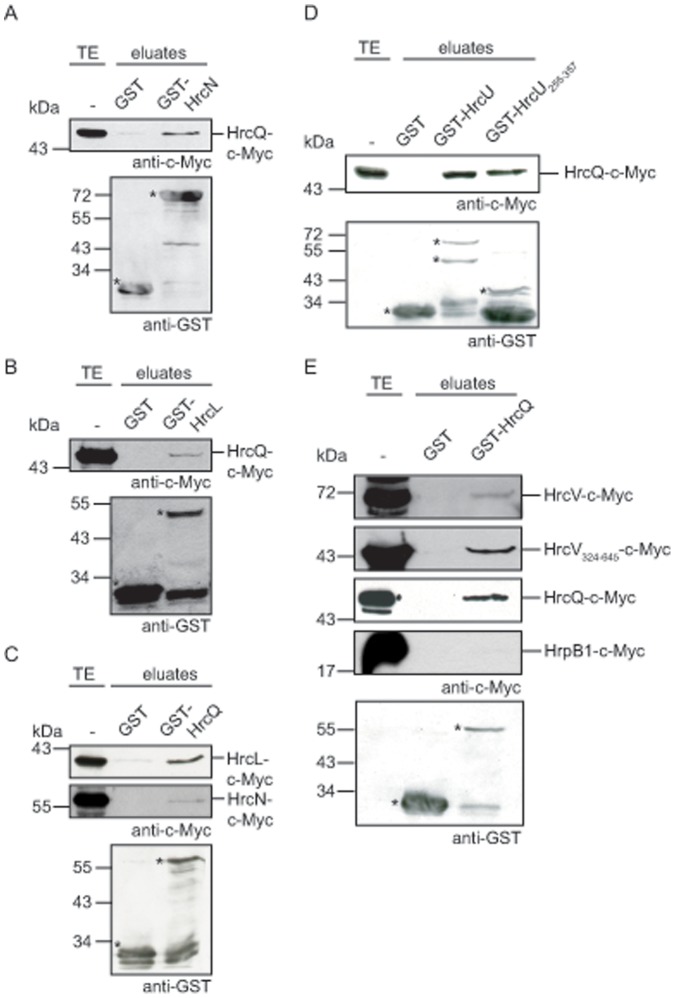
HrcQ Interacts with itself and with IM-associated Components of the T3S System
Given the predicted role of HrcQ as a C ring component, HrcQ is likely to interact with other cytoplasmic or IM-associated proteins of the T3S system. To test this possibility, GST fusions of the cytoplasmic T3S ATPase HrcN and its predicted regulator HrcL were immobilized on glutathione sepharose and incubated with a bacterial lysate containing HrcQ-c-Myc. Bound proteins were eluted from the matrix and analyzed by immunoblotting using c-Myc epitope- and GST-specific antibodies. HrcQ-c-Myc specifically coeluted with GST-HrcN and GST-HrcL but not with GST alone, suggesting that it interacts with the ATPase HrcN and its predicted regulator HrcL (Fig. 5A and B). Both interaction were also shown in the opposite direction with GST-HrcQ and C-terminally c-Myc epitope-tagged derivatives of HrcL and HrcN (Fig. 5C).
Figure 5. HrcQ interacts with itself, HrcN, HrcL and the cytoplasmic domains of HrcU and HrcV.
(A) GST pull-down assays with HrcN. GST and GST-HrcN were immobilized on glutathione sepharose and incubated with a bacterial lysate containing HrcQ-c-Myc. Total cell extracts (TE) and eluted proteins (eluates) were analyzed by immunoblotting using c-Myc epitope- and GST-specific antibodies. GST and GST fusion proteins are marked with asterisks, additional bands correspond to degradation products. (B) GST pull-down assays with HrcL. GST and GST-HrcL were immobilized on glutathione sepharose and incubated with a bacterial lysate containing HrcQ-c-Myc. TE and eluates were analyzed as is described in panel A. (C) GST-HrcQ interacts with HrcL and HrcN. GST and GST-HrcQ were immobilized on glutathione sepharose and incubated with bacterial lysates containing HrcL-c-Myc and HrcN-c-Myc, respectively. TE and eluates were analyzed as is described in panel A. One representative blot incubated with GST-specific antibodies is shown. (D) HrcQ interacts with the cytoplasmic domain of HrcU. GST, GST-HrcU and GST-HrcU255-357 were immobilized on glutathione sepharose and incubated with a bacterial lysate containing HrcQ-c-Myc. TE and eluates were analyzed as is described in panel A. GST-HrcU is cleaved at the conserved NPTH motif. The signals detected by the GST-specific antibody therefore correspond to GST-HrcU and the N-terminal cleavage product [26]. (E) HrcQ interacts with the cytoplasmic domain of HrcV and with itself. GST and GST-HrcQ were immobilized on glutathione sepharose and incubated with bacterial lysates containing HrcV-c-Myc, HrcV324-645-c-Myc, HrcQ-c-Myc or HrpB1-c-Myc. TE and eluates were analyzed as is described in panel A. One representative blot incubated with GST-specific antibodies is shown.
We also analyzed a possible interaction of HrcQ with the IM proteins HrcV and HrcU. These proteins consist of eight and four transmembrane domains, respectively, and cytoplasmic domains that might be involved in T3S substrate docking [45]. GST pull-down assays revealed that HrcQ-c-Myc coelutes with GST fusions of HrcU and the C-terminal cytoplasmic domain of HrcU, respectively, spanning amino acids 255 to 357 (Fig. 5D). This region of HrcU was previously shown to interact with the early T3S substrate HrpB2, the T3S4 protein HpaC and the ATPase HrcN [20], [21]. We also observed interactions between GST-HrcQ and C-terminally c-Myc epitope-tagged derivatives of HrcV and the HrcV deletion derivative HrcV324-645-c-Myc, which corresponds to the cytoplasmic domain of HrcV [45], suggesting that HrcQ interacts with the cytoplasmic domains of both HrcU and HrcV (Fig. 5E).
As HrcQ is homologous to predicted C ring components that likely form a ring complex, we also investigated a possible self-interaction of HrcQ. When analyzed by GST pull-down assays, HrcQ-c-Myc was detected in the eluate of GST-HrcQ but not of GST, suggesting that HrcQ can interact with itself (Fig. 5E). Notably, no interaction was detected between GST-HrcQ and a C-terminally c-Myc epitope-tagged derivative of HrpB1, which is a yet uncharacterized predicted component of the T3S system [22] (Fig. 5E). This suggests that the observed protein-protein interactions were not caused by unspecific protein binding to GST-HrcQ.
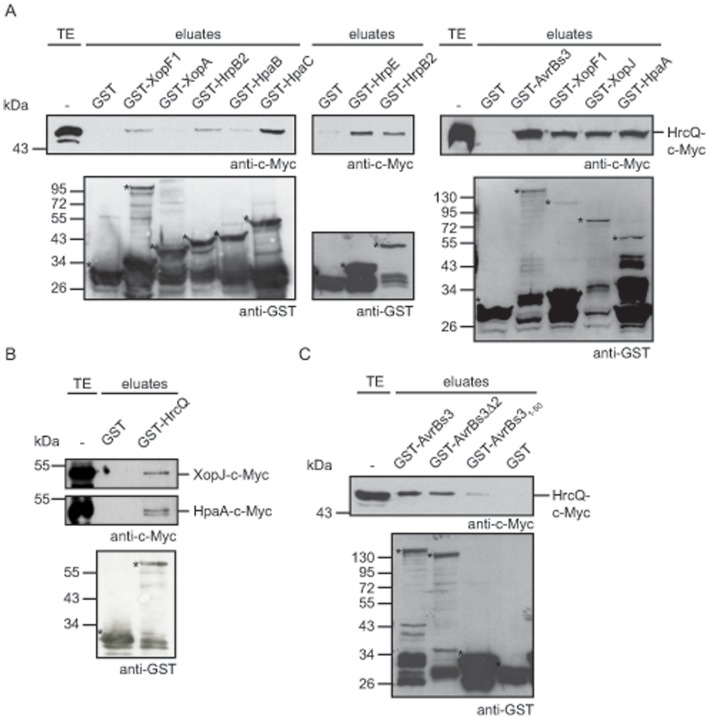
HrcQ Interacts with the T3S4 Protein HpaC and T3S Substrates
It was previously reported that predicted C ring components from animal pathogenic bacteria interact with effector proteins or effector-chaperone complexes [32], [33], [46]. We therefore tested a possible interaction of HrcQ with secretion substrates as well as with the general T3S chaperone HpaB and the T3S4 protein HpaC, which are probably both involved in the targeting of secreted proteins to the T3S system [17], [18], [21]. All candidate interaction partners were immobilized as GST fusion proteins and incubated with bacterial lysates containing HrcQ-c-Myc. Eluted proteins were analyzed by immunoblotting as described above. Fig. 6A shows that HrcQ-c-Myc coeluted with GST-HpaC and GST fusions of the early T3S substrate HrpB2, the pilus protein HrpE and the effector protein XopF1. By contrast, only small amounts of HrcQ-c-Myc were detected in the eluates of GST fusions of the predicted translocon protein XopA and the T3S chaperone HpaB (Fig. 6A).
Figure 6. HrcQ provides a docking site for early and late T3S substrates.
(A) HrcQ interacts with T3S substrates and the T3S4 protein HpaC. GST, GST-XopF1, GST-XopA, GST-HrpB2, GST-HpaB, GST-HpaC, GST-HrpE, GST-AvrBs3, GST-XopJ and GST-HpaA were immobilized on glutathione sepharose and incubated with bacterial lysates containing HrcQ-c-Myc. Total cell extracts (TE) and eluted proteins (eluates) were analyzed by immunoblotting using c-Myc epitope- and GST-specific antibodies. GST and GST fusion proteins are marked with asterisks, additional bands correspond to degradation products. (B) HrcQ interacts with XopJ and HpaA. GST and GST-HrcQ were immobilized on glutathione sepharose and incubated with bacterial lysates containing XopJ-c-Myc and HpaA-c-Myc, respectively. TE and eluates were analyzed as is described in panel A. One representative blot incubated with GST-specific antibodies is shown. (C) The N-terminal region of AvrBs3 is dispensable for the interaction with HrcQ. GST, GST-AvrBs3, GST-AvrBs3Δ2 and GST-AvrBs31-50 were immobilized on glutathione sepharose and incubated with a bacterial lysate containing HrcQ-c-Myc. TE and eluates were analyzed as is described in panel A.
The interaction between HrcQ and the effector protein XopF1 prompted us to investigate whether HrcQ can also bind to additional effector proteins. We therefore performed pull-down assays with GST fusions of the effector proteins AvrBs3, XopJ and HpaA. Notably, HrcQ-c-Myc was detected in the eluates of all GST-effector fusions but not of GST alone, suggesting that it provides a binding site for several effector proteins (Fig. 6A). We also performed the interaction studies in the opposite direction with GST-HrcQ and C-terminally c-Myc epitope-tagged derivatives of putative HrcQ interaction partners. Fig. 6B shows that the effector proteins XopJ-c-Myc and HpaA-c-Myc were detected in the eluate of GST-HrcQ but not of GST alone. By contrast, XopF1-c-Myc and HrpB2-c-Myc were not detected in the eluate of GST-HrcQ, suggesting that they did not interact with the GST-HrcQ fusion protein (data not shown). However, it cannot be excluded that the C-terminal c-Myc epitope of XopF1 and HrpB2 masks a protein region and/or affects the folding of both proteins and thus the interaction with HrcQ. Notably, we have previously observed that a C-terminally c-Myc epitope-tagged derivative of HrpB2 does not complement the hrpB2 mutant phenotype, suggesting that the c-Myc epitope interferes with protein function [21].
The N-terminal Region of AvrBs3 is Dispensable for the Interaction with HrcQ
As it is assumed that the targeting of T3S substrates and/or their recognition by components of the secretion apparatus often depends on a secretion signal in the N-terminal regions of secreted proteins, we wondered whether this signal is required for the binding of effector proteins to HrcQ. T3S signals of effector proteins from X. campestris pv. vesicatoria have not yet been studied in detail, but a T3S signal and HpaB-binding site were previously localized in the N-terminal 50 amino acids of the TAL effector AvrBs3 [17]. To investigate the contribution of this protein region to the interaction of AvrBs3 with HrcQ, we performed additional pull-down assays with HrcQ-c-Myc and GST fusion proteins containing the full-length AvrBs3 protein, the N-terminal 50 amino acids or an N-terminally truncated derivative of AvrBs3, designated AvrBs3Δ2, which lacks the first 152 amino acids. HrcQ-c-Myc coeluted with GST-AvrBs3 and GST-AvrBs3Δ2, suggesting that the N-terminal region of AvrBs3 is dispensable for the interaction with HrcQ. By contrast, the amounts of HrcQ-c-Myc that were detected in the eluate of GST-AvrBs31-50 were significantly reduced (Fig. 6C). Thus, the N-terminal 50 amino acids of AvrBs3 are probably not sufficient for the efficient binding of HrcQ. Taken together, we conclude from our findings that HrcQ interacts with components and substrates of the T3S system and that the T3S signal of AvrBs3 is not required for the interaction with HrcQ.
Discussion
In this study we identified HrcQ as an essential component and a potential substrate docking site of the T3S system from X. campestris pv. vesicatoria. HrcQ is homologous to predicted C ring components of T3S systems (YscQ protein family) that are assumed to form a cytoplasmic cup-like structure and might provide a binding site for chaperone-effector complexes [14]. Given that putative C ring components have mainly been studied in animal pathogenic bacteria, our data present the first detailed functional characterization of an YscQ family member from a plant pathogen. Genetic approaches revealed that HrcQ from X. campestris pv. vesicatoria is essential for T3S of early and late substrates as well as for pathogenicity and bacterial in planta growth. Loss of pathogenicity was also observed for hrcQ mutants that express hrpG* and thus encode a constitutively active version of the key regulator HrpG (Fig. 1). Furthermore, the hrcQ mutant phenotype could not be complemented by overexpression of the ATPase-encoding hrcN gene, suggesting that enhanced expression levels of T3S genes and/or overexpression of hrcN cannot compensate for the loss of HrcQ. This is in contrast to the finding that the phenotype of flagellar C ring mutants can be partially complemented by overexpression of the ATPase-encoding gene fliI or by upregulation of the regulatory genes flhDC [40], [41]. The C ring of flagellar T3S systems is therefore probably not essential for secretion, which is different from the crucial role of HrcQ from X. campestris pv. vesicatoria.
Loss of pathogenicity of the hrcQ deletion mutant was specifically caused by the absence of HrcQ because the mutant phenotype was partially complemented by ectopic expression of hrcQ in trans. Overexpression of hrcQ under control of the lac promoter did not complement the hrcQ mutant phenotype and significantly reduced T3S and pathogenicity of hrcQ wild-type strains. Surprisingly, lac promoter-driven expression of hrcQ also affected bacterial growth in vitro, suggesting that elevated HrcQ levels are detrimental for bacterial fitness (Fig. 1). The latter observation was unexpected because the in vitro growth of X. campestris pv. vesicatoria is assumed to be independent of the T3S system. The negative effect of hrcQ overexpression on bacterial growth was less pronounced in complex medium, suggesting that it can be partially compensated by favourable environmental conditions. To circumvent the dominant-negative effects caused by increased HrcQ amounts, we inserted hrcQ under control of the native promoter into the hpaFG region next to the hrp gene cluster of X. campestris pv. vesicatoria hrcQ deletion mutants. Chromosomally encoded HrcQ-c-Myc fully complemented the phenotype of hrcQ deletion mutants with respect to plant reactions, which confirms the above hypothesis that increased copy numbers of hrcQ are not favourable for pathogenicity (Fig. 3).
Given the homology of HrcQ with predicted C ring components from animal pathogenic bacteria, we assumed that HrcQ is a cytoplasmic component of the T3S system. In line with this hypothesis, fractionation studies revealed that HrcQ is predominantly present in cytoplasmic fractions and specifically associates with the bacterial membranes under T3S-permissive conditions (Fig. 4). This is reminiscent of the membrane association of the HrcQ homologs HrcQA and HrcQB from Pseudomonas syringae that are translated as two separate proteins [56]. Notably, we have previously shown that also the cytoplasmic ATPase HrcN and its predicted regulator HrcL from X. campestris pv. vesicatoria specifically associate with the bacterial membranes under T3S-activating conditions [20]. Our in vitro interaction studies showed that both HrcN and HrcL interact with HrcQ (Fig. 5), suggesting that all three proteins are present in a complex which associates with the membrane-spanning components of the T3S system under secretion-permissive conditions. This is in agreement with previously reported interactions between predicted C ring components and the ATPase complex of T3S systems from animal pathogenic bacteria [32], [57]-[60]. Our interaction studies also revealed a self-interaction of HrcQ, suggesting that HrcQ can form homo-oligomers as is expected for a predicted C ring component. Complex formation was also described for HrcQA and HrcQB from P. syringae and the two translation products of yscQ from Yersinia [56], [61].
In addition to HrcN and HrcL, HrcQ interacts with the IM proteins HrcU and HrcV that are essential components of the membrane-spanning export apparatus (Fig. 5) [14]. HrcU contains four transmembrane helices and a cytoplasmic domain (HrcUC) that is proteolytically cleaved and interacts with the T3S4 protein HpaC [26], [45], [62]. HrcUC is presumably involved in the T3S substrate specificity switch and in substrate binding because it interacts with the early T3S substrate HrpB2 [21], [26]. As HrcU also interacts with HrcQ (Fig. 5) and the ATPase HrcN (via the cytoplasmic domain) as well as with the predicted regulator of the ATPase, HrcL (via the transmembrane region) [20], it is presumably not only required for the control of substrate specificity but additionally contributes to the docking of the predicted HrcQ-HrcL-HrcN complex to the secretion apparatus at the IM.
In contrast to HrcU, the IM protein HrcV has not yet been intensively characterized in X. campestris pv. vesicatoria. While previous studies revealed an interaction of HrcV with the general T3S chaperone HpaB and the T3S4 protein HpaC, it is yet unknown whether HrcV can directly bind to secreted proteins as was shown for HrcV homologs from flagellar T3S systems (FlhA protein family) [18], [63]-[66]. The new finding of an interaction between the cytoplasmic domain of HrcV and HrcQ suggests that HrcV is involved in the recruitment of cytoplasmic components of the T3S system as is postulated for HrcU. An interaction between HrcV and the ATPase complex still needs to be investigated.
Notably, HrcQ also interacts with the T3S4 protein HpaC, which might be involved in the targeting of T3S substrates to the secretion system (Fig 6). As HpaC additionally binds to the ATPase HrcN as well as to the IM proteins HrcV and HrcU [18], [20], it is possible that the T3S apparatus contains multiple binding sites for HpaC. In agreement with this hypothesis, T3S substrates or chaperone-substrate complexes from animal pathogenic bacteria interact with the predicted C ring as well as with the ATPase and/or the cytoplasmic domains of members of the YscV family of IM proteins [27], [32], [33], [46], [63], [65], [67]-[70]. In future studies, it has to be investigated whether the interaction of HpaC with HrcQ is essential for the T3S substrate specificity switch. Notably, additional interaction studies showed that HrcQ also interacts with different T3S substrates including the early substrate HrpB2 and effector proteins (Fig. 6). This was not caused by unspecific binding because HrcQ did not significantly interact with GST, GST-HpaB and GST-XopA. Given the interaction of HrcQ with secretion substrates, HrcQ is (besides HrcUC) the second known potential substrate binding site of the T3S system from X. campestris pv. vesicatoria. As the early T3S substrate HrpB2 interacts with both HrcQ and HrcUC, it is possible that the T3S apparatus provides several binding sites for a given secretion substrate. In future studies we will investigate whether the binding of HrpB2 and other T3S substrates to HrcQ is required for their efficient secretion.
The recruitment and/or secretion of T3S substrates by the T3S system is assumed to depend on an N-terminal secretion signal that is not conserved on the amino acid level [71], [72]. T3S signals have been localized in several T3S substrates from plant and animal pathogenic bacteria and presumably are required for the targeting and docking of these protein to the secretion apparatus. Notably, however, in most cases the contribution of the T3S signal to substrate docking has not yet been analyzed. We have previously shown that the T3S signal of the TAL effector AvrBs3 is located within the N-terminal 50 amino acids that also provide a binding site for HpaB [17]. Surprisingly, however, pull-down assays with HrcQ and AvrBs3 derivatives revealed that the N-terminal region of AvrBs3 is dispensable for the efficient interaction with HrcQ (Fig. 6), suggesting that the T3S signal from AvrBs3 is not required for the binding to HrcQ. This indicates that the interaction of AvrBs3 with HrcQ occurs after the T3S signal-mediated targeting of AvrBs3 to the T3S system. It remains to be investigated how the T3S signal is recognized and whether it is required for substrate binding.
In summary, we conclude from our findings that HrcQ is likely an important substrate docking site of the T3S system but is presumably not the primary binding site for secreted proteins.
Supporting Information
Complementation studies with strain 85*ΔhrcQ. X. campestris pv. vesicatoria strains 85* (wt) and 85*ΔhrcQ (ΔhrcQ) carrying plasmid pBRM (-) or derivatives thereof expressing hrcQ or hrcQ-c-myc under control of the native (pnat) or the lac (plac) promoter as indicated were inoculated at a density of 4×107 CFU ml-1 into leaves of susceptible ECW and resistant ECW-10R pepper plants. Disease symptoms were photographed 9 dpi. For the better visualization of the HR, leaves were bleached in ethanol 2 dpi. Dashed lines mark the infiltrated areas.
(TIF)
Ectopic expression of hrcN-c-myc does not complement the hrcQ mutant phenotype. (A) Infection studies with derivatives of strain 85*ΔhrcQ. Strains 85-10 (wt), 85* (wt), 85-10ΔhrcQ (ΔhrcQ) and 85*ΔhrcQ (ΔhrcQ) carrying plasmid pBRM (-) or encoding HrcN-c-Myc as indicated were inoculated at a density of 4×108 CFU ml-1 into leaves of susceptible ECW and resistant ECW-10R pepper plants. Disease symptoms were photographed 9 dpi. For the better visualization of the HR, leaves were bleached in ethanol 2 dpi. Dashed lines mark the infiltrated areas. Expression of hrcN-c-myc under control of the lac promoter was previously shown to complement the hrcN mutant phenotype [20]. (B) HrcN-c-Myc is stably synthesized in strain 85*ΔhrcQ. X. campestris pv. vesicatoria strains 85* (wt) and 85*ΔhrcQ (ΔhrcQ) carrying plasmid pBRM (-) or encoding HrcN-c-Myc as indicated were grown in NYG medium and total cell extracts were analyzed by immunoblotting, using a c-Myc epitope-specific antibody.
(TIF)
Generation of the suicide vector pLAND-P. DNA fragments of the hpaFG region flanking the lacZ gene, the lac promoter (lacP) and the 3× c-Myc epitope-encoding sequence were cloned into the suicide vector pOK1 (see Materials and Methods). BsaI sites upstream of lacP and downstream of lacZ allow the directional cloning of genes of interest in frame with the 3× c-Myc epitope-encoding sequence. Genes are represented by arrows. The DNA sequences given in the boxes refer to the overhangs that are generated after restriction of the DNA with BsaI.
(TIF)
Acknowledgments
We are grateful to M. Jordan for technical assistance, to N. Hartmann for generating constructs pBRMhrcV and pBRMhrcV324-645 and to U. Bonas for helpful comments on the manuscript.
Funding Statement
This work was supported by grants from the Deutsche Forschungsgemeinschaft (BU 2145/5-1; Sonderforschungsbereich SFB 648) to D.B. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Pallen MJ, Penn CW, Chaudhuri RR (2005) Bacterial flagellar diversity in the post-genomic era. Trends Microbiol 13: 143–149. [DOI] [PubMed] [Google Scholar]
- 2. Desvaux M, Hebraud M, Henderson IR, Pallen MJ (2006) Type III secretion: what's in a name? Trends Microbiol 14: 157–160. [DOI] [PubMed] [Google Scholar]
- 3. Chevance FFV, Hughes KT (2008) Coordinating assembly of a bacterial macromolecular machine. Nature 6: 455–465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Francis MR, Sosinsky GE, Thomas D, DeRosier DJ (1994) Isolation, characterization and structure of bacterial flagellar motors containing the switch complex. J Mol Biol 235: 1261–1270. [DOI] [PubMed] [Google Scholar]
- 5. DePamphilis ML, Adler J (1971) Fine structure and isolation of the hook-basal body complex of flagella from Escherichia coli and Bacillus subtilis . J Bacteriol 105: 384–395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Blocker A, Jouihri N, Larquet E, Gounon P, Ebel F, et al. (2001) Structure and composition of the Shigella flexneri "needle complex", a part of its type III secreton. Mol Microbiol 39: 652–663. [DOI] [PubMed] [Google Scholar]
- 7. Marlovits TC, Kubori T, Sukhan A, Thomas DR, Galan JE, et al. (2004) Structural insights into the assembly of the type III secretion needle complex. Science 306: 1040–1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Kubori T, Matsushima Y, Nakamura D, Uralil J, Lara-Tejero M, et al. (1998) Supramolecular structure of the Salmonella typhimurium type III protein secretion system. Science 280: 602–605. [DOI] [PubMed] [Google Scholar]
- 9. Sani M, Allaoui A, Fusetti F, Oostergetel GT, Keegstra W, et al. (2007) Structural organization of the needle complex of the type III secretion apparatus of Shigella flexneri. . Micron 38: 291–301. [DOI] [PubMed] [Google Scholar]
- 10. He SY, Nomura K, Whittam TS (2004) Type III protein secretion mechanism in mammalian and plant pathogens. Biochim Biophys Acta 1694: 181–206. [DOI] [PubMed] [Google Scholar]
- 11. Erhardt M, Namba K, Hughes KT (2010) Bacterial nanomachines: the flagellum and type III injectisome. Cold Spring Harb Perspect Biol 2: a000299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Mueller CA, Broz P, Cornelis GR (2008) The type III secretion system tip complex and translocon. Mol Microbiol 68: 1085–1095. [DOI] [PubMed] [Google Scholar]
- 13. Mattei PJ, Faudry E, Job V, Izore T, Attree I, et al. (2010) Membrane targeting and pore formation by the type III secretion system translocon. Febs J 278: 414–426. [DOI] [PubMed] [Google Scholar]
- 14. Büttner D (2012) Protein export according to schedule – architecture, assembly and regulation of type III secretion systems from plant and animal pathogenic bacteria. Microbiol Mol Biol Rev 76: 262–310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Büttner D, Bonas U (2010) Regulation and secretion of Xanthomonas virulence factors. FEMS Microbiol Rev 34: 107–133. [DOI] [PubMed] [Google Scholar]
- 16. Büttner D, Bonas U (2002) Getting across-bacterial type III effector proteins on their way to the plant cell. EMBO J 21: 5313–5322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Büttner D, Gürlebeck D, Noel LD, Bonas U (2004) HpaB from Xanthomonas campestris pv. vesicatoria acts as an exit control protein in type III-dependent protein secretion. Mol Microbiol 54: 755–768. [DOI] [PubMed] [Google Scholar]
- 18. Büttner D, Lorenz C, Weber E, Bonas U (2006) Targeting of two effector protein classes to the type III secretion system by a HpaC- and HpaB-dependent protein complex from Xanthomonas campestris pv. vesicatoria . Mol Microbiol 59: 513–527. [DOI] [PubMed] [Google Scholar]
- 19. Schulze S, Kay S, Büttner D, Egler M, Eschen-Lippold L, et al. (2012) Analyses of new type III effectors from Xanthomonas uncover XopB and XopS as suppressors of plant immunity. New Phytol 195: 894–911. [DOI] [PubMed] [Google Scholar]
- 20. Lorenz C, Büttner D (2009) Functional characterization of the type III secretion ATPase HrcN from the plant pathogen Xanthomonas campestris pv. vesicatoria. J Bacteriol 191: 1414–1428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Lorenz C, Schulz S, Wolsch T, Rossier O, Bonas U, et al. (2008) HpaC controls substrate specificity of the Xanthomonas type III secretion system. PLoS Pathog 4: e1000094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Rossier O, Van den Ackerveken G, Bonas U (2000) HrpB2 and HrpF from Xanthomonas are type III-secreted proteins and essential for pathogenicity and recognition by the host plant. Mol Microbiol 38: 828–838. [DOI] [PubMed] [Google Scholar]
- 23. Hartmann N, Schulz S, Lorenz C, Fraas S, Hause G, et al. (2012) Characterization of HrpB2 from Xanthomonas campestris pv. vesicatoria identifies protein regions that are essential for type III secretion pilus formation. Microbiol 158: 1334–1349. [DOI] [PubMed] [Google Scholar]
- 24. Deane JE, Abrusci P, Johnson S, Lea SM (2010) Timing is everything: the regulation of type III secretion. Cell Mol Life Sci 67: 1065–1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Cornelis GR, Agrain C, Sorg I (2006) Length control of extended protein structures in bacteria and bacteriophages. Curr Opin Microbiol 9: 201–206. [DOI] [PubMed] [Google Scholar]
- 26. Lorenz C, Büttner D (2011) Secretion of early and late substrates of the type III secretion system from Xanthomonas is controlled by HpaC and the C-terminal domain of HrcU. Mol Microbiol 79: 447–467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Gonzalez-Pedrajo B, Minamino T, Kihara M, Namba K (2006) Interactions between C ring proteins and export apparatus components: a possible mechanism for facilitating type III protein export. Mol Microbiol 60: 984–998. [DOI] [PubMed] [Google Scholar]
- 28. McMurry JL, Murphy JW, Gonzalez-Pedrajo B (2006) The FliN-FliH interaction mediates localization of flagellar export ATPase FliI to the C ring complex. Biochemistry 45: 11790–11798. [DOI] [PubMed] [Google Scholar]
- 29. Oosawa K, Ueno T, Aizawa S (1994) Overproduction of the bacterial flagellar switch proteins and their interactions with the MS ring complex in vitro. J Bacteriol 176: 3683–3691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Brown PN, Mathews MA, Joss LA, Hill CP, Blair DF (2005) Crystal structure of the flagellar rotor protein FliN from Thermotoga maritima . J Bacteriol 187: 2890–2902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Zhao R, Pathak N, Jaffe H, Reese TS, Khan S (1996) FliN is a major structural protein of the C-ring in the Salmonella typhimurium flagellar basal body. J Mol Biol 261: 195–208. [DOI] [PubMed] [Google Scholar]
- 32. Morita-Ishihara T, Ogawa M, Sagara H, Yoshida M, Katayama E, et al. (2006) Shigella Spa33 is an essential C-ring component of type III secretion machinery. J Biol Chem 281: 599–607. [DOI] [PubMed] [Google Scholar]
- 33. Spaeth KE, Chen YS, Valdivia RH (2009) The Chlamydia type III secretion system C-ring engages a chaperone-effector protein complex. PLoS Pathog 5: e1000579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Wengelnik K, Rossier O, Bonas U (1999) Mutations in the regulatory gene hrpG of Xanthomonas campestris pv. vesicatoria result in constitutive expression of all hrp genes. J Bacteriol 181: 6828–6831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Wengelnik K, Van den Ackerveken G, Bonas U (1996) HrpG, a key hrp regulatory protein of Xanthomonas campestris pv. vesicatoria is homologous to two-component response regulators. Mol Plant-Microbe Interact 9: 704–712. [DOI] [PubMed] [Google Scholar]
- 36. Noël L, Thieme F, Nennstiel D, Bonas U (2001) cDNA-AFLP analysis unravels a genome-wide hrpG-regulon in the plant pathogen Xanthomonas campestris pv. vesicatoria . Mol Microbiol 41: 1271–1281. [DOI] [PubMed] [Google Scholar]
- 37. Ronald PC, Staskawicz BJ (1988) The avirulence gene avrBs1 from Xanthomonas campestris pv. vesicatoria encodes a 50-kDa protein. Mol Plant-Microbe Interact 1: 191–198. [PubMed] [Google Scholar]
- 38. Escolar L, Van den Ackerveken G, Pieplow S, Rossier O, Bonas U (2001) Type III secretion and in planta recognition of the Xanthomonas avirulence proteins AvrBs1 and AvrBsT. Mol Plant Pathol 2: 287–296. [DOI] [PubMed] [Google Scholar]
- 39. Jones JD, Dangl JL (2006) The plant immune system. Nature 444: 323–329. [DOI] [PubMed] [Google Scholar]
- 40. Erhardt M, Hughes KT (2010) C-ring requirement in flagellar type III secretion is bypassed by FlhDC upregulation. Mol Microbiol 75: 376–393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Konishi M, Kanbe M, McMurry JL, Aizawa S (2009) Flagellar formation in C-ring-defective mutants by overproduction of FliI, the ATPase specific for flagellar type III secretion. J Bacteriol 191: 6186–6191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Noël L, Thieme F, Nennstiel D, Bonas U (2002) Two novel type III system-secreted proteins of Xanthomonas campestris pv. vesicatoria are encoded within the hrp pathogenicity island. J Bacteriol 184: 1340–1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Tamir-Ariel D, Navon N, Burdman S (2007) Identification of genes in Xanthomonas campestris pv. vesicatoria induced during its interaction with tomato. J Bacteriol 189: 6359–6371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Kaniga K, Delor I, Cornelis GR (1991) A wide-host-range suicide vector for improving reverse genetics in gram-negative bacteria: inactivation of the blaA gene of Yersinia enterocolitica . Gene 109: 137–141. [DOI] [PubMed] [Google Scholar]
- 45. Berger C, Robin GP, Bonas U, Koebnik R (2010) Membrane topology of conserved components of the type III secretion system from the plant pathogen Xanthomonas campestris pv. vesicatoria . Microbiol 156: 1963–1974. [DOI] [PubMed] [Google Scholar]
- 46. Lara-Tejero M, Kato J, Wagner S, Liu X, Galan JE (2011) A sorting platform determines the order of protein secretion in bacterial type III systems. Science 331: 1188–1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Daniels MJ, Barber CE, Turner PC, Sawczyc MK, Byrde RJW, et al. (1984) Cloning of genes involved in pathogenicity of Xanthomonas campestris pv. campestris using the broad host range cosmid pLAFR1. EMBO J 3: 3323–3328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, et al. (1996) Current protocols in molecular biology. New York: John Wiley & Sons, Inc.
- 49. Figurski D, Helinski DR (1979) Replication of an origin-containing derivative of plasmid RK2 dependent on a plasmid function provided in trans . Proc Natl Acad Sci USA 76: 1648–1652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Minsavage GV, Dahlbeck D, Whalen MC, Kearny B, Bonas U, et al. (1990) Gene-for-gene relationships specifying disease resistance in Xanthomonas campestris pv. vesicatoria - pepper interactions. Mol Plant-Microbe Interact 3: 41–47. [Google Scholar]
- 51. Kousik CS, Ritchie DF (1998) Response of bell pepper cultivars to bacterial spot pathogen races that individually overcome major resistance genes. Plant Disease 82: 181–186. [DOI] [PubMed] [Google Scholar]
- 52. Bonas U, Schulte R, Fenselau S, Minsavage GV, Staskawicz BJ, et al. (1991) Isolation of a gene-cluster from Xanthomonas campestris pv. vesicatoria that determines pathogenicity and the hypersensitive response on pepper and tomato. Mol Plant-Microbe Interact 4: 81–88. [Google Scholar]
- 53. Engler C, Kandzia R, Marillonnet S (2008) A one pot, one step, precision cloning method with high throughput capability. PLoS ONE 3: e3647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Rossier O, Wengelnik K, Hahn K, Bonas U (1999) The Xanthomonas Hrp type III system secretes proteins from plant and mammalian pathogens. Proc Natl Acad Sci USA 96: 9368–9373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Büttner D, Nennstiel D, Klüsener B, Bonas U (2002) Functional analysis of HrpF, a putative type III translocon protein from Xanthomonas campestris pv. vesicatoria. J Bacteriol 184: 2389–2398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Fadouloglou VE, Tampakaki AP, Glykos NM, Bastaki MN, Hadden JM, et al. (2004) Structure of HrcQB-C, a conserved component of the bacterial type III secretion systems. Proc Natl Acad Sci U S A 101: 70–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Biemans-Oldehinkel E, Sal-Man N, Deng W, Foster LJ, Finlay BB (2011) Quantitative proteomic analysis reveals formation of an EscL-EscQ-EscN type III complex in enteropathogenic Escherichia coli . J Bacteriol 193: 5514–5519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Jackson MW, Plano GV (2000) Interactions between type III secretion apparatus components from Yersinia pestis detected using the yeast two-hybrid system. FEMS Microbiol Lett 186: 85–90. [DOI] [PubMed] [Google Scholar]
- 59. Johnson DL, Stone CB, Mahony JB (2008) Interactions between CdsD, CdsQ, and CdsL, three putative Chlamydophila pneumoniae type III secretion proteins. J Bacteriol 190: 2972–2980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Johnson S, Blocker A (2008) Characterization of soluble complexes of the Shigella flexneri type III secretion system ATPase. FEMS Microbiol Lett 286: 274–278. [DOI] [PubMed] [Google Scholar]
- 61. Bzymek KP, Hamaoka BY, Ghosh P (2012) Two translation products of Yersinia yscQ assemble to form a complex essential to type III secretion. Biochemistry 51: 1669–1677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Schulz S, Büttner D (2011) Functional characterization of the type III secretion substrate specificity switch protein HpaC from Xanthomonas . Infect Immun 79: 2998–3011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Bange G, Kummerer N, Engel C, Bozkurt G, Wild K, et al. (2010) FlhA provides the adaptor for coordinated delivery of late flagella building blocks to the type III secretion system. Proc Natl Acad Sci U S A 107: 11295–11300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Minamino T, Kinoshita M, Hara N, Takeuchi S, Hida A, et al. (2012) Interaction of a bacterial flagellar chaperone FlgN with FlhA is required for efficient export of its cognate substrates. Molecular Microbiology 83: 775–788. [DOI] [PubMed] [Google Scholar]
- 65. Minamino T, MacNab RM (2000) Interactions among components of the Salmonella flagellar export apparatus and its substrates. Mol Microbiol 35: 1052–1064. [DOI] [PubMed] [Google Scholar]
- 66. Creasey EA, Delahay RM, Daniell SJ, Frankel G (2003) Yeast two-hybrid system survey of interactions between LEE-encoded proteins of enteropathogenic Escherichia coli . Microbiol 149: 2093–2106. [DOI] [PubMed] [Google Scholar]
- 67. Akeda Y, Galan JE (2005) Chaperone release and unfolding of substrates in type III secretion. Nature 437: 911–915. [DOI] [PubMed] [Google Scholar]
- 68. Boonyom R, Karavolos MH, Bulmer DM, Khan CM (2010) Salmonella pathogenicity island 1 (SPI-1) type III secretion of SopD involves N- and C-terminal signals and direct binding to the InvC ATPase. Microbiol 156: 1805–1814. [DOI] [PubMed] [Google Scholar]
- 69. Gauthier A, Finlay BB (2003) Translocated intimin receptor and its chaperone interact with ATPase of the type III secretion apparatus of enteropathogenic Escherichia coli . J Bacteriol 185: 6747–6755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Thomas J, Stafford GP, Hughes C (2004) Docking of cytosolic chaperone-substrate complexes at the membrane ATPase during flagellar type III protein export. Proc Natl Acad Sci U S A 101: 3945–3950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Arnold R, Jehl A, Rattei T (2010) Targeting effectors: the molecular recognition of type III secreted proteins. Microbes Infect 12: 346–358. [DOI] [PubMed] [Google Scholar]
- 72. McDermott JE, Corrigan A, Peterson E, Oehmen C, Niemann G, et al. (2011) Computational prediction of type III and IV secreted effectors in gram-negative bacteria. Infect Immun 79: 23–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Canteros BI (1990) Diversity of plasmids and plasmid-encoded phenotypic traits in Xanthomonas campestris pv. vesicatoria. PhD thesis, University of Florida.
- 74. Ménard R, Sansonetti PJ, Parsot C (1993) Nonpolar mutagenesis of the ipa genes defines IpaB, IpaC, and IpaD as effectors of Shigella flexneri entry into epithelial cells. J Bacteriol 175: 5899–5906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Szczesny R, Jordan M, Schramm C, Schulz S, Cogez V, et al. (2010) Functional characterization of the Xps and Xcs type II secretion systems from the plant pathogenic bacterium Xanthomonas campestris pv. vesicatoria . New Phytol 187: 983–1002. [DOI] [PubMed] [Google Scholar]
- 76.Lorenz C (2009) Functional characterization of the conserved components HrcN und HrcU of the type III secretion system from Xanthomonas campestris pv. vesicatoria. PhD thesis, Martin Luther University Halle-Wittenberg.
- 77. Szurek B, Marois E, Bonas U, Van den Ackerveken G (2001) Eukaryotic features of the Xanthomonas type III effector AvrBs3: protein domains involved in transcriptional activation and the interaction with nuclear import receptors from pepper. Plant J 26: 523–534. [DOI] [PubMed] [Google Scholar]
- 78. Huguet E, Hahn K, Wengelnik K, Bonas U (1998) hpaA mutants of Xanthomonas campestris pv. vesicatoria are affected in pathogenicity but retain the ability to induce host-specific hypersensitive reaction. Mol Microbiol 29: 1379–1390. [DOI] [PubMed] [Google Scholar]
- 79. Vieira J, Messing J (1987) Production of single-stranded plasmid DNA. Methods Enzymol 153: 3–11. [DOI] [PubMed] [Google Scholar]
- 80. Lorenz C, Kirchner O, Egler M, Stuttmann J, Bonas U, et al. (2008) HpaA from Xanthomonas is a regulator of type III secretion. Mol Microbiol 69: 344–360. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Complementation studies with strain 85*ΔhrcQ. X. campestris pv. vesicatoria strains 85* (wt) and 85*ΔhrcQ (ΔhrcQ) carrying plasmid pBRM (-) or derivatives thereof expressing hrcQ or hrcQ-c-myc under control of the native (pnat) or the lac (plac) promoter as indicated were inoculated at a density of 4×107 CFU ml-1 into leaves of susceptible ECW and resistant ECW-10R pepper plants. Disease symptoms were photographed 9 dpi. For the better visualization of the HR, leaves were bleached in ethanol 2 dpi. Dashed lines mark the infiltrated areas.
(TIF)
Ectopic expression of hrcN-c-myc does not complement the hrcQ mutant phenotype. (A) Infection studies with derivatives of strain 85*ΔhrcQ. Strains 85-10 (wt), 85* (wt), 85-10ΔhrcQ (ΔhrcQ) and 85*ΔhrcQ (ΔhrcQ) carrying plasmid pBRM (-) or encoding HrcN-c-Myc as indicated were inoculated at a density of 4×108 CFU ml-1 into leaves of susceptible ECW and resistant ECW-10R pepper plants. Disease symptoms were photographed 9 dpi. For the better visualization of the HR, leaves were bleached in ethanol 2 dpi. Dashed lines mark the infiltrated areas. Expression of hrcN-c-myc under control of the lac promoter was previously shown to complement the hrcN mutant phenotype [20]. (B) HrcN-c-Myc is stably synthesized in strain 85*ΔhrcQ. X. campestris pv. vesicatoria strains 85* (wt) and 85*ΔhrcQ (ΔhrcQ) carrying plasmid pBRM (-) or encoding HrcN-c-Myc as indicated were grown in NYG medium and total cell extracts were analyzed by immunoblotting, using a c-Myc epitope-specific antibody.
(TIF)
Generation of the suicide vector pLAND-P. DNA fragments of the hpaFG region flanking the lacZ gene, the lac promoter (lacP) and the 3× c-Myc epitope-encoding sequence were cloned into the suicide vector pOK1 (see Materials and Methods). BsaI sites upstream of lacP and downstream of lacZ allow the directional cloning of genes of interest in frame with the 3× c-Myc epitope-encoding sequence. Genes are represented by arrows. The DNA sequences given in the boxes refer to the overhangs that are generated after restriction of the DNA with BsaI.
(TIF)