Abstract
Background
Elevated microsatellite alterations at selected tetranucleotide repeats (EMAST) is a genetic signature observed in 60% of sporadic colorectal cancers (CRCs). Unlike microsatellite unstable CRCs where hypermethylation of the DNA mismatch repair (MMR) gene hMLH1’s promoter is causal, the precise cause of EMAST is not clearly defined but points towards hMSH3 deficiency.
Aim
To examine if hMSH3 deficiency causes EMAST, and to explore mechanisms for its deficiency.
Methods
We measured −4 bp framshifts at D8S321 and D20S82 loci within EGFP-containing constructs to determine EMAST formation in MMR-proficient, hMLH1−/−, hMSH6−/−, and hMSH3−/− CRC cells. We observed the subcellular location of hMSH3 with oxidative stress.
Results
D8S321 mutations occurred 31-and 40-fold higher and D20S82 mutations occurred 82-and 49-fold higher in hMLH1−/− and hMSH3−/− cells, respectively, than in hMSH6−/− or MMR-proficient cells. hMSH3 knockdown in MMR-proficient cells caused higher D8S321 mutation rates (18.14 and 11.14×10−4 mutations/cell/generation in two independent clones) than scrambled controls (0 and 0.26×10−4 mutations/cell/generation; p<0.01). DNA sequencing confirmed the expected frameshift mutations with evidence for ongoing mutations of the constructs. Because EMAST-positive tumors are associated with inflammation, we subjected MMR-proficient cells to oxidative stress via H2O2 to examine its effect on hMSH3. A reversible nuclear-to-cytosol shift of hMSH3 was observed upon H2O2 treatment.
Conclusion
EMAST is dependent upon the MMR background, with hMSH3−/− more prone to frameshift mutations than hMSH6−/−, opposite to frameshift mutations observed for mononucleotide repeats. hMSH3−/− mimics complete MMR failure (hMLH1−/−) in inducing EMAST. Given the observed heterogeneous expression of hMSH3 in CRCs with EMAST, hMSH3-deficiency appears to be the event that commences EMAST. Oxidative stress, which causes a shift of hMSH3’s subcellular location, may contribute to an hMSH3 loss-of-function phenotype by sequestering it to the cytosol.
Introduction
DNA mismatch repair (MMR) dysfunction drives the process of microsatellite instability (MSI), observed in nearly all Lynch Syndrome colorectal cancers (CRCs) and in 15–20% of sporadic CRCs [1]. MSI is observed when both alleles of a major DNA MMR gene is inactivated by mutation in Lynch tumors or by hypermethylation in sporadic tumors, causing MMR protein production or function to fail, and allowing frameshifts to occur at DNA microsatellite sequences throughout the genome [1]–[3]. Classic MSI is determined by the occurrence of frameshifts in mono/dinucleotide markers, and has a defined panel for which to compare studies [4]. This panel, or variations of these mono/dinucleotide markers, readily detects hMLH1- and hMSH2-deficiency (as dysfunction of these two proteins completely inactivates DNA MMR), and occasionally hMSH6-deficiency [1], [4]. The MSI panel is not as specific to detect hMSH3-deficiency due to the nature of repair that this protein contributes to MMR, based on yeast studies [5], [6]. DNA MMR is an enzyme system whose main cellular function is to repair nucleotide mispairs and insertion/deletion loops (IDLs) during DNA replication. The specificity of mismatch recognition is directed by hMutSα and hMutSβ; hMutSα (hMSH2-hMSH6 heterodimer) binds to single mispairs and small IDLs (≤2 nucleotides), whereas hMutSβ (hMSH2-hMSH3 heterodimer) binds larger IDLs [1], [5]–[7]. We and others have observed DNA lesion binding specificities with purified hMutSα and hMutSβ as well as made specific observations on binding using human CRC cells of differing DNA MMR-deficient backgrounds harboring constructs containing specified-length IDLs or nucleotide mispairs [8]–[12].
Recently, a new form of MSI has been described in up to 60% of sporadic colon [13]–[15] and over 30% of rectal cancers [16]. Unlike classic MSI, elevated microsatellite alterations at selected tetranucleotide repeats (EMAST) is determined by a panel of tetranuleotide markers [13]–[17]. EMAST appears to be more common than classic MSI and seems to be an acquired feature associated with inflammation in CRCs [14], [16]. CRC patients who have EMAST-positive tumors have a poorer prognosis than those with non-EMAST or MSI tumors [16], [18]. EMAST has been observed in various tumors, including non-small cell lung [17], bladder [19], ovarian [20], prostate [21], skin [22], and colorectal [13]–[16], but these studies do not provide full evidence as to the cause of EMAST. In the initial reports of EMAST in colorectal cancers, the authors observed nuclear heterogeneity of hMSH3 within EMAST-positive tumors [13], [14]. Our data, combined with yeast [5], [6], and other human studies [13]–[16], suggests that hMSH3-deficiency might be the driver of EMAST in CRCs.
In order to test if hMSH3-deficiency is a principal cause of EMAST, we generated tetranucleotide repeat-containing constructs that were fused out-of-frame with EGFP, such that when frameshifted by −4 bp (i.e. deletion of one tetranucleotide unit), EGFP would be expressed. This allowed observation of the generation of EMAST in real-time, and afforded us the ability to calculate mutation frequencies and mutation rates in multiple MMR-deficient backgrounds. In this report, our data clearly shows that hMSH3-deficiency is a cause of EMAST in human CRC cells and we further provide a possible mechanism that may contribute to the acquired hMSH3-deficiency in human CRCs.
Results
Establishment of a Measuring System for EMAST Tetranucleotide Frameshifts
Because some studies have indicated that EMAST is associated with heterogeneous expression of hMSH3 in colorectal tumors [13]–[16], our approach was to generate a system to measure EMAST in a quantitative fashion as a means to directly examine if hMSH3-deficiency is the cause of EMAST. Human tetranucleotide D8S321 and D20S82 loci sequences (containing 12 and/or 16 repeats of AAAG, respectively) have shown the highest frequency for a −4 bp frameshift in colorectal tumors [14], [16]. These two loci were therefore selected for study using CRC cells with different MMR genetic backgrounds to ascertain subsequent mutation. We have previously established a similar experimental system where deletion of 1 bp of (A)n would lead to the expression of the reporter gene EGFP when a construct was placed +1 bp out-of-frame [10]–[12].
To generate the experimental constructs, we inserted the D8S321 (D8) and D20S82 (D20) loci as 60-mers containing the tetranucleotide repeats and surrounding nucleotides immediately after the EGFP start codon [Table S1]. Experimental constructs [D8-OF (D8-out-of-frame) and D20-OF] were made +1 bp out-of-frame such that deletion of one AAAG repeat will shift the EGFP reading frame (+1 bp to −3 bp) into the correct frame to cause EGFP expression. Mutation resistant (MR) plasmids (MR-D8 and MR-D20), serving as negative controls, were constructed by replacing the middle AA nucleotides within the AAAG repeats with C/G nucleotides in every third AAAG, interrupting the series of tetranucleotide repeats to prevent frameshift mutations. The MR-D8 and MR-D20 plasmids were placed out-of frame (OF; D8/D20-MR-OF) and/or in-frame (IF; D8/D20-MR-IF) to be used as negative and positive controls for EGFP expression (also see Materials and Methods). These constructs were transfected into human CRC cell lines with different MMR genetic backgrounds and stable cell lines were established via hygromycin B selection. Single cell clones were subsequently established. DNA sequencing was employed to correctly confirm the transfected EMAST constructs (Table S1). As expected, cells carrying MR-IF (positive control) were EGFP positive, whereas cells containing MR-OF (negative control) were EGFP negative.
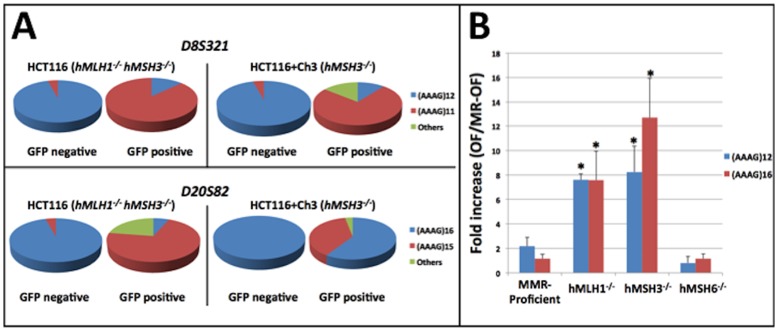
To test if hMSH3-deficiency could cause EMAST, non-fluorescent cells carrying the different stably transfected EMAST constructs were sorted by flow cytometry and exponentially grown for four to six weeks. Each week, cells were analyzed in duplicate by flow cytometry for EGFP expression, presumably resulting from deletion of one AAAG repeat from the transfected D8S321 and/or D20S82 loci. EGFP positive cells began emerging as soon as one week after EGFP negative cells were sorted and plated (data not shown). To ensure that our experimental system allowed us to correctly assess the occurrence of EMAST, the EGFP negative and positive cells from hMLH1−/− and/or hMSH3 −/− cells harboring the experimental constructs (D8-OF and D20-OF) were sorted to isolate genomic DNA for sequencing. As expected, essentially all clones (greater than 96%) from EGFP negative cells retained wild type (WT) sequences for each of D8S321 and D20S82 loci in both hMLH1−/− and hMSH3−/− cell lines, whereas clones from EGFP positive cells acquired deletion of one AAAG repeat ( Figure 1A and Figure S1A and S1B). We also observed expansion frameshift mutations at each locus by DNA sequencing (included in “others” in Figure 1A ). The most frequent one (ranging from 12.5% to as high as 30% of total EGFP-positive cells) was insertion of two AAAG repeats [(AAAG)12 to (AAAG)14] in D8S321 that changed the reading frame from +1 bp to +9 bp (listed as “others” in Figure 1A and Figure S1A). In addition, more than one AAAG repeat deletion (e.g. 4 or 10 AAAG repeat deletion) was rarely observed (<8% of total, listed in “others” in Figure 1A ). Importantly, deletion/insertion mutations were always detected within the microsatellite sequences while no mutation occurred in the flanking sequences surrounding AAAG repeat (Figure S1). These observations indicate that we have established a reliable experimental system to monitor the occurrence of EMAST in CRC cells.
Figure 1. DNA sequencing results and EMAST mutation frequencies in CRC cells with different MMR backgrounds.
(A) Genomic DNA isolated from EGFP-negative and -positive cells carrying experimental constructs (D8-OF and/or D20-OF) was used to amplify DNA fragments containing EMAST loci to examine if the frameshift mutations had occurred. (B) Mutation frequencies were calculated by dividing the percentage of EGFP positive population in the experimental group (OF) by the percentage of EGFP positive in its negative control (MR-OF) to compute the increase of the EGFP-positive population in folds. There were significantly more EGFP-positive cells in hMLH1- and hMSH3-deficient cells compared to MMR-proficient and/or hMSH6-deficient cells. *: p<0.05.
Frameshift Mutations at Tetranucleotide Repeats of D8S321 and D20S82 Loci are Dependent on the MMR Background
To examine the effect of MMR background on frameshift mutations at the tetranucleotide D8S321 and D20S82 loci, we calculated mutation frequencies of the markers by comparing each experimental group (OF) to its negative control (MR-OF) using the following formula: “(EGFP positive cells/total live cells from OF)/(EGFP positive cells/total live cells from MR-OF)”. hMLH1−/− cells (completely devoid of DNA MMR activity) and hMSH3 −/− cells showed 7–12 fold higher mutation frequencies compared to the controls for both loci ( Figure 1B ). In particular, the mutation frequencies observed in hMSH3 −/− were comparable to that in hMLH1 −/− for both loci ( Figure 1B ). The −4 bp mutation rates for both loci in each MMR background were also calculated ( Table 1 ). With complete MMR-deficiency (hMLH1−/−), mutation rates for both loci were 27 and 56×10−4 mutation/cell/generation. Similarly, hMSH3−/− cells generated 33 and 34×10−4 mutations/cell/generation. Thus, the mutation rates with hMSH3-deficiency are very comparable to those observed for complete MMR-deficiency. This is in sharp contrast to hMSH6-deficiency (<1×10−4 mutation/cell/generation), essentially identical to MMR-proficient cells. These findings strongly suggest the hMSH3-deficiency is the driver for EMAST formation.
Table 1. Calculated EMAST mutation rates for D8S321 and D20S82 in various MMR backgrounds.
| MMR Background | D8S321 (AAAG)12 | D20S82 (AAAG)16 |
| MMR-Proficient | 2.45×10−4±1.04×10−4 | 0.31×10−4±0.31×10−4 |
| hMLH1−/− | 26.96×10−4±1.20×10−4a, b | 56.4×10−4±5.89×10−4a, b |
| hMSH3−/− | 34.82×10−4±1.26×10−4c, d | 33.86×10−4±4.25×10−4c, d |
| hMSH6−/− | 0.86×10−4±0.86×10−4 | 0.69×10−4±0.69×10−4 |
Data from the EGFP-positive population at weeks 4, 5, and 6 time points were used for -4 bp frameshift mutation rate analysis. Single mutation rates were calculated by combining and averaging time-specific mutation rates. Rates are expressed as mutations at microsatellite sequence per cell per generation. Data shown are mean±SEM.
a, b, c, and d represent significant difference in the mutation rate (P<0.05).
hMLH1−/− vs. MMR-proficient (D8S321 P = 0.00012, D20S82 P = 0.01069);
hMLH1−/− vs. hMSH6−/− (D8S321 P = 0.00012, D20S82 P = 0.01021);
hMSH3−/− vs. MMR-proficient (D8S321 P = 0.00008, D20S82 P = 0.01525);
hMSH3−/− vs. hMSH6−/− (D8S321 P = 0.0001, D20S82 P = 0.01415). Mutations rates were calculated based on the “method of the mean” developed by Luria and Delbruck [25].
Direct Knockdown of hMSH3 Expression in MMR-proficient Cells Causes EMAST
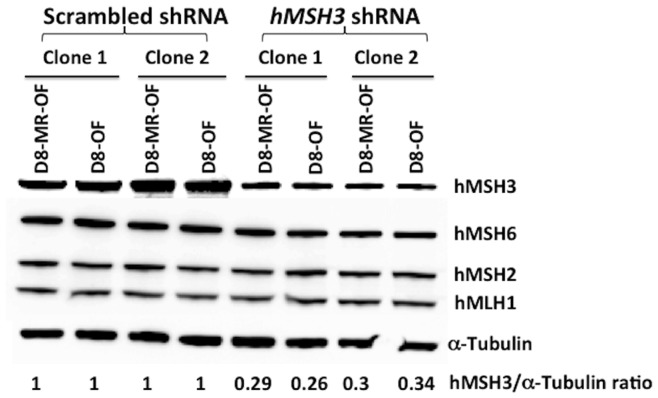
To confirm a role of hMSH3 in EMAST, we knocked down hMSH3 expression in MMR-proficient cells harboring the D8S321-EGFP construct by transfecting plasmids carrying hMSH3-specific shRNA and/or scrambled control, then measured D8S321 tetranucleotide frameshift mutations as above. Prior to shRNA transfection, hMSH3 expression levels in chosen MMR-proficient clones were comparable to each other (Figure S2). After generation of shRNA stable cell lines (single cell clones), hMSH3 expression was significantly reduced in hMSH3 shRNA transfected cells ( Figure 2 ; varying between 26–34% of the protein levels of the scramble controls).
Figure 2. Expression levels of MMR proteins in hMSH3 knockdown clones.
hMSH3 shRNA transfection successfully and specifically reduced the hMSH3 expression levels in the CRC cell clones. Note that the expression levels of other DNA mismatch repair genes remain unchanged.
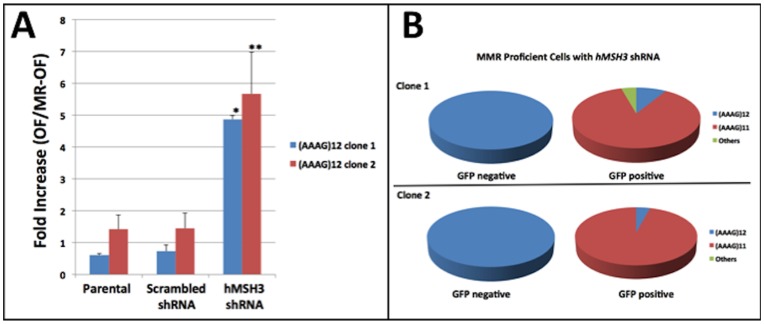
Using the shRNA stably-transfected cells containing the D8S321 locus, we examined the cell lines for occurrence of EMAST by flow cytometry. An EGFP-positive population appeared in cells where hMSH3 expression was knocked down. hMSH3 KD cells showed 5-fold (clone 1) and 6-fold (clone 2) increase in mutation frequencies ( Figure 3A ) compared to scrambled shRNA control cells. Reduction in hMSH3 expression greatly increased mutation rates of the D8S321 construct to 12.7 and 18.4×10−4 mutation/cell/generation, compared to 1×10−4 mutation/cell/generation in scramble controls ( Table 2 ). EGFP-negative and -positive cells were separated out for DNA sequencing to examine frameshift mutations. In both clones, greater than 90% of the EGFP negative cells retained WT sequence [(AAAG)12], whereas approximately 90% of the EGFP-positive cells revealed a deletion of one repeat of AAAG ( Figure 3B ). Expansion of the microsatellite repeats was also detected (included as “others” in Figure 3B ). Overall, our observations indicate that direct KD of hMSH3 causing hMSH3-deficiency alone is able to generate EMAST in previous MMR-proficient CRC cells.
Figure 3. EMAST mutation frequencies in hMSH3 knockdown CRC single-clone cells.
(A) SW480 cells (MMR proficient) carrying the EMAST construct (D8-OF or D8-MR-OF) were transfected with hMSH3 shRNA construct (hMSH3 KD) or scramble control (scramble) separately. Two independent hMSH3 and scramble KD clones as well as the parental cells were included for this study. EMAST mutation frequencies were calculated as described in Figure 1B for parental, scramble, and/or hMSH3 KD cells. There were significantly more EGFP-positive cells in hMSH3 KD cells compared to parent and/or scramble control cells. *: p<0.001; **: p<0.05. (B) EGFP-negative and -positive cells from hMSH3 KD cells carrying D8-OF were sorted for genomic DNA isolation to examine frameshift mutations by sequencing.
Table 2. Calculated EMAST mutations rates for D8S321 clones with knockdown of hMSH3.
| D8S321 (AAAG)12clone 1 | D8S321 (AAAG)12clone 2 | |
| Parental | 0×10−4±0×10−4 | 1.2×10−4±0.62×10−4 |
| Scrambled shRNA | 0×10−4±0×10−4 | 1.2×10−4±0.57×10−4 |
| hMSH3 shRNA | 18.4×10−4±0.18×10−4 a | 12.7×10−4±1.14×10−4a |
Data from the EGFP-positive population at weeks 4, 5 and 6 time points were used for -4 bp frameshift mutation rate analysis. Single mutation rates were calculated by combining and averaging time-specific mutation rates. Rates are expressed as mutations at microsatellite sequence per cell per generation. Data shown are mean±SEM.
represents significant difference in the mutation rate (P<0.05). hMSH3 shRNA vs. scramble shRNA (clone 1 P = 0.000082, clone 2 P = 0.003086).
Oxidative Stress Causes Subcellular Compartmental Shift of hMSH3 from the Nucleus to the Cytosol
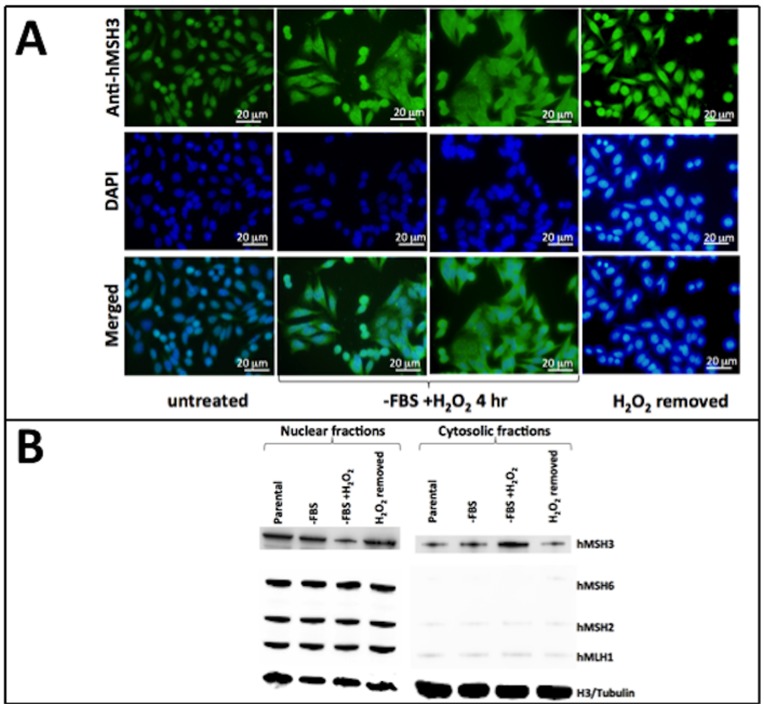
EMAST has been observed to be tightly associated with heterogeneous loss of hMSH3 expression as well as nuclear heterogeneous expression (Haugen et al., 2008; Lee et al., 210, Devaraj et al., 2010). Our studies above further provide evidence that reduction of hMSH3 expression alone can induce EMAST in human CRC cells. We began to explore how hMSH3 might become deficient in EMAST-positive tumors with the observed reduction from full to heterogeneous expression. In CRC, EMAST is associated with inflammatory cells [13], [14], [16] that may facilitate oxidative stress, which has been shown to impair MMR function [23]. Using H2O2 as a surrogate, we investigated the effect(s) of oxidative stress on hMSH3 expression. MMR-proficient cells were treated with increasing concentrations of H2O2 (50 µM –500 µM) for up to 24 hours. We did not detect any reductions in total hMSH3 protein levels in any conditions we tried (data not shown). However, upon treating cells with 50 µM of H2O2 for 4 hours, hMSH3 was found homogenously in both the nucleus and the cytosol in a significant percentage of cells (40–60%; second panel column in Figure 4A ). Strikingly, hMSH3 was observed to completely vacate nuclei in some cells [10–20% of total cells (third panel column in Figure 4A ], in sharp contrast to the untreated cells where hMSH3 predominately remained localized in the nucleus (left panels in Figure 4A ). The shift of hMSH3 localization could be detected as soon as 2 hours into treatment, and the effect peaked between 4 and 8 hours after H2O2 treatment started (data not shown). Unlike hMSH3, hMLH1, hMSH2, and hMSH6 did not alter their cellular localization and remained in nuclei ( Figure 4B and Figure S3A, S3B, and S3C). Cell fractionation to separate nuclear and cytosolic fractions matched our immunocytochemistry observations (Lanes 1–3 and 5–7 in Figure 4B ).
Figure 4. Oxidative stress causes subcellular compartmental shift from the nucleus to the cytosol for hMSH3.
(A) MMR-proficient SW480 cells were serum-starved for 24 hours and then treated with 50 µM of H2O2 for 4 hours. After fixation, cells were stained with anti-hMSH3 antibody and then Alexa 488 conjugated anti-mouse antibody to observe the subcellular location of hMSH3. To test if the effect was reversible, cells were cultured for additional 24 hours after the removal of H2O2. (B) SW480 cells were serum-starved for 24 hours and then treated with H2O2 for 4 hours (−FBS+H2O2). Parental cells (Parental) and cells that were serum-starved without the subsequent H2O2 treatment (−FBS) were used as controls. Cellular proteins were separated into nuclear and cytosolic fractions using a nuclear fractionation kit. Equal volume of proteins from each group was used for SDS-PAGE and subsequent Western Blotting to check levels of hMSH3. Anti-histone H3 was used for nuclear fraction loading equality, and tubulin was used for cytosolic fraction loading equality.
To examine if the effect of H2O2 on hMSH3 was reversible, we replaced H2O2 containing media with fresh media after 4 hours of treatment. hMSH3 was found to be nuclear by 24 hours after removal of H2O2 (right panels in Figure 4A and lanes 4 and 8 in Figure 4B ). This result may indicate that the effect of H2O2 on hMSH3 was reversible, where hMSH3 is allowed to re-enter into the nucleus once oxidative stress abated. Alternatively, the hMSH3 that was in the cytosol might have been damaged and degraded and thus was incapable of re-entering into nuclei, and might indicate that the observed nuclear hMSH3 was newly-synthesized. To determine which scenario could be correct, we treated cells with 20 µg/ml of cyclohexamide upon removal of H2O2 to stop protein synthesis for 24 hours. hMSH3 was found to be in the nucleus (data not shown), demonstrating that the hMSH3 that was shifted to the cytosol was indeed able to re-enter into the nucleus.
Discussion
DNA MMR studies from bacteria and yeast indicate that hMutSβ, a heterodimer of hMSH2 and hMSH3, binds and directs repair of insertion-deletion loops ±2 nucleotides [1], [5], [6]. Thus, hMSH3-deficiency would be expected to allow large insertion-deletion loop frameshift mutations, which has been strongly suggested in previous studies [13]–[16]. Determining the consequence of hMSH3-deficiency was not previously of prime importance, as there has never been described a germline mutation for hMSH3 to cause Lynch Syndrome, and there was no correlation with sporadic CRC until the description of EMAST in CRCs [13]–[16]. The linkage of EMAST with heterogeneous loss of hMSH3 expression in sporadic CRCs suggested that this DNA MMR protein could drive EMAST. In our study, we set out to test this by creating a human cell model to measure EMAST formation in a quantitative fashion. Our study shows that (a) EMAST formation is dependent on the MMR background, with hMSH3-deficiency matching complete MMR-deficiency in causing EMAST, (b) hMSH3-deficiency mutation rates for EMAST formation are markedly higher compared to that of hMSH6-deficiency (essentially null for EMAST formation), (c) knockdown of hMSH3 alone initiates formation of EMAST, and (d) oxidative stress may be one of the factors causing deficiency of hMSH3, as this MMR protein shifts its subcellular location with H2O2. Altogether, our study offers strong support to the concept that hMSH3-deficiency drives EMAST formation and provides a possible mechanism that may contribute to hMSH3-deficiency in human CRCs.
Tetranucleotide frameshifts that define EMAST in CRC tumors can be by insertion or deletion of tetranucleotide repeats. In our experiments, we designed constructs to detect a −4 bp deletion; however upon DNA sequencing some insertions (expansion) of the microsatellites were also detected. Indeed, the majority of frameshift mutations detected in our experiments were due to deletion of one AAAG unit ( Figure 1A and 3B ). We selected the D8S321 and D20S82 loci for our experiments because deletion of one AAAG unit had been shown to be the most prevailing mutation found among loci utilized to detect EMAST [14], [16]. Because our system was designed to only detect one type of mutation, our rates calculated are likely an underestimation of the mutability of these loci with hMSH3-deficiency. Individual sequence context and flanking sequences may also contribute to the different outcome of the mutation (e.g. insertion vs. deletion) [11], [12], [24]. At the D20S82 locus, only about one-third of EGFP-positive hMSH3−/− cells demonstrated one AAAG unit deletion and this could be due to this particular clone containing multiple copies of the construct. This is contrasted by more than 70% of the EGFP positive cells carrying the same construct in hMLH1−/− showing deletion of one AAAG unit, indicating the construct behaved as designed. For hMSH3−/− cells, mutation on one of the copies would lead to EGFP expression, while the rest remained WT, confounding sequencing analysis. This, however, would not affect the mutation frequency and mutation rate as the calculation was based on the number of the EGFP-positive cells.
It is not fully clear for EMAST-positive tumors how hMSH3 is downregulated to initiate EMAST. There is clear evidence that EMAST-positive tumors are strongly associated with inflammatory cells, particularly within the epithelial components of the tumor and in the stroma surrounding the tumor (so called “tumor nests”) [16]. Thus, one hypothesis is that inflammation triggers an acquired reduction of hMSH3, which subsequently drives EMAST. Interestingly, H2O2 treatment (simulating oxidative stress) leads to a shift of hMSH3 into the cytosol away from its functional site in the nucleus without diminishing its overall expression levels. With short-term H2O2 treatment, we could not demonstrate detectable levels of EMAST formation in our model (data not shown). A longer and consistent presence of oxidative stress may be required to commence EMAST, considering EMAST appears to be associated with chronic inflammation and with advanced histology of tumors [14]. Alternatively, other components of inflammation and/or the tumor milieu may work synergistically with oxidative stress to cause EMAST. EMAST appears to be a poor prognostic factor for patients with CRC [16], [18]. This is in contrast to classic MSI for which patients with CRC have better survival at the same staged tumor of patients with MSS tumors [1]. It remains to be determined if this is intrinsic to the type of MMR defect (hMLH1 vs. hMSH3) or some other factors, such as the degree or type of inflammation in the tumor.
In summary, we show through the creation of a novel cell model that hMSH3-deficiency drives EMAST. Our data strongly complements the observation of heterogeneous loss of hMSH3 expression in EMAST-positive tumors, and suggests that acquired hMSH3-deficiency, driven by oxidative stress, may be the most common DNA MMR defect among human CRCs.
Materials and Methods
Cell Lines and Reagents
The human colon cancer cell lines, HCT116 (hMLH1−/− hMSH3−/−), DLD-1 (hMSH6−/−), HCT116+ch3 (hMLH1-restored but hMSH3−/−), and SW480 (MMR-proficient) cells were cultured as described previously [10]. HCT116, DLD-1, and SW480 were purchased from American Type Culture Collection (ATCC; Manassas; Virginia). HCT116+ch3 cells were kindly provided by Dr. Minoru Koi, Baylor University Medical Center, Dallas, TX [10]. Anti-hMLH1, hMSH2, hMSH3, and hMSH6 antibodies were purchased from BD Pharmigen (San Diego, CA). Anti-tubulin antibody was from Sigma (St. Louis, MO). HRP-conjugated anti-mouse antibody was purchased from Cell Signaling (Danvers, MA). Alexa 488 conjugated anti-mouse antibody was from Invitrogen (Grand Island, NY).
Cloning of pIREShyg2-D8S321-EGFP and pIREShyg2-D20S82-EGFP Plasmids
The two human loci, D8S321 and D20S82, have been reported to have the highest mutation frequency in EMAST colonic tumors [13]–[16], and were selected to construct plasmids. D8S321 and D20S82 sequences contain 12 and/or 16 repeats of AAAG, respectively. Sense and anti-sense oligos containing D8S321 or D20S82, surrounding sequences, as well as PmeI and AscI cloning sites (Table S1) were synthesized (Integrated DNA Technologies, Inc., San Diego, CA) and subjected to T4-polynucleotide kinase reactions to phosphorylate the 5′ ends (Invitrogen). The sense and anti-sense oligos were allowed to anneal and cloned into pIREShyg2-EGFP plasmids via PmeI-AscI sites immediately after the start codon of the EGFP gene as described previously [10]–[12], [24]. Experimental plasmids were constructed +1 bp out of frame, such that one deletion of a tetranucleotide repeat within D8S321 or D20S82 (−4 bp frameshift mutation) would shift the reading frame into the correct frame to allow EGFP expression. Mutation resistant plasmids [MR-out-of-frame (OF)] were constructed by using sense and anti-sense oligos in which two nucleotides within every third repeat of AAAG were changed to prevent frameshift mutations. The mutation resistant in frame (MR-D8-IF) plasmid, a positive control for EGFP expression, was constructed by deleting 1 bp from D8S321 sequence in the MR-D8-OF plasmids using Quikchange II site-directed mutagenesis kit (Stratagene, La Jolla, CA).
hMSH3 shRNA Construction
hMSH3 shRNA construct and its paired empty vector were kindly provided by Dr. Ajay Goel, Baylor University Medical Center, Dallas, TX. The GFP sequence of this construct was removed by digesting the construct with ApaI and PacI, filling-in using Klenow fragment to form blunt ends, and then performing blunt-end ligation. To make a scrambled control, the GFP sequence from the empty vector was first removed as described above and the scrambled sequence, removed from a commercially available construct (GeneCopoeia, Rockville, MD), was inserted onto the vector via EcoRI and XhoI sites. The existence and the accuracy of the scramble and/or hMSH3 shRNA sequences on the plasmids were confirmed by DNA sequencing.
Transfection and Generation of Stable Cell Lines
Cells were transfected with the different pIREShyg2-D8S321-EGFP and pIREShyg2-D20S82-EGFP plasmids (described in Table S1) by using Nucleofector kits (Amaxa, Cologne, Germany). Stable cell lines were established by hygromycin B (Invitrogen) selection. Each cell line was subjected to single cell sorting using FACS ARIA (Becton Dickinson, San Jose, CA) to establish individual clones. All single cell clones were examined by DNA sequencing to ensure the existence and accuracy of the EMAST construct they carried.
For hMSH3 knockdown (KD) cells, MMR-proficient cells (SW480) carrying D8-OF and/or D8-MR-OF constructs were transfected with plasmids harboring scramble or hMSH3-specific shRNA using Fugene 6 (Roche, Mannheim, Germany), followed by puromycin (Invitrogen) selection to establish stable lines. Two independent clones from each group were used for the studies.
Mutation Analysis by Flow Cytometry
Four hundred thousand non-fluorescent stable-transfected cells were sorted onto each well on 6-well plates by FACS ARIA (Becton Dickinson) and cultured for 4 to 6 weeks. Cells were maintained to keep them in exponential growth. To quantitate EGFP-positive populations, cells from each 100-mm dish were trypsinized, collected, and resuspended in a total volume of 2 ml of cold PBS containing 1 µg/ml of propidium iodide (PI; Sigma) right before analysis. Each group was analyzed in duplicate on a FACSCalibur (Becton Dickinson) for each time point.
For the hMSH3 KD experiments, non-fluorescent MMR-proficient cells containing D8-MR-OF or D8-OF were sorted one week before scramble or hMSH3-specific shRNA transfection. Puromycin-selected scramble or hMSH3 KD cells were analyzed by flow cytometry to quantitate EGFP-positive cells as described above.
Polymerase Chain Reactions and DNA Sequencing
Genomic DNA from sorted EGFP-negative and/or -positive cells was PCR-amplified using a cloned Pfu DNA polymerase (Stratagene). The PCR products were subjected to TA cloning (Promega, Madison, WI) to obtain individual clones. Multiple clones from EGFP negative and/or positive cells were sequenced to examine the frameshift mutations. The primers used for sequencing are as following: Forward primer: 5′-GAGCTCGGATCTGTACAGGC-3′; Reverse primer: 5′TGCCGTCGTCCTTGAAGAAGA-3′.
Determination of Frameshift Mutation Rates within D8S321 and D20S82 Loci
Mutation rates were calculated by the “method of the mean” developed by Luria and Delbruck [25] as described previously [10]–[12], [24].
Western Blotting
Cells were seeded onto 6-well plates a day before protein preparation. Cells were rinsed with cold PBS and directly homogenized on the plates in cell lysis buffer (150 mM NaCl, 10 mM Tris-HCl, pH. 7.2, 0.1% IGEPAL, 0.5% Na-deoxycholate, and 5 mM EDTA) using 1 ml pipette tips. After a 15 min-incubation on ice, lysed cells were centrifuged at 10,000 rpm for 5 min at 4°C, from which clear protein lysates were collected. Protein concentration was determined using the BCA Protein Assay kit (Piece, Rockford, IL). Protein lysates were separated by 4–20% Tris-Acetate SDS-PAGE (Invitrogen) and transferred onto nitrocellulose membrane. Immunodetection was done using primary antibodies against hMLH1 (1∶1000), hMSH2 (1∶1000), hMSH3 (1∶500), hMSH6 (1∶2000), Tubulin (1∶8000), and horseradish peroxidase-conjugated secondary antibody (1∶5000). Signals were detected using Immobilon Western (Millipore) and captured by ImageQuant LAS4000 (General Electric, New York City, NY). To strip off the antibody for re-probing with a different antibody, membranes were incubated with strip-off buffer (1.5% Glycine/1% SDS/1% Tween-20, pH 2.2) at room temperature for 30 min, followed by washing/neutralizing with 1X TBST.
Immunocytochemistry
Twenty-five thousand cells were seeded onto each well on 8-well chamber slides (Invitrogen) and incubated overnight. After serum starvation for 24 hr, cells were treated with 50 µM H2O2 for 4 hours. Cells were fixed with cold acetone on ice for 5 min, air-dried, and stored at 4°C until staining. During the staining process, slides were kept in a humidified chamber at room temperature. Slides were re-hydrated with 5% FBS/PBS (blocking/washing buffer) for 20 min before incubating with antibodies against hMLH1 (1∶100), hMSH2 (1∶100), hMSH3 (1∶100), or hMSH6 (1∶500) for 90 min and then Alexa 488 conjugated anti-mouse antibody (1∶2000) for 1 hr in dark. Slides were mounted with Prolong Gold mounting media with DAPI (Invitrogen). Pictures were taking using an Olympus DP72 Fluorescent microscope.
Supporting Information
Chromatographs for DNA sequencing of tetranucleotide microsatellites. (A) The EMAST construct carrying D8S321 marker contained 12 copies of AAAG, which underwent deletion of one copy of the AAAG unit or insertion of two copies of AAAG repeat as shown. (B) The D20S82 construct harboring 16 copies of AAAG underwent deletion of one repeat of AAAG. Please note that the flanking sequences remained identical to the wild-type when EMAST markers underwent deletion/insertion.
(TIF)
Expression levels of MMR proteins in clones prior to shRNA transfection. Note that clones contained equivalent amounts of hMSH3 protein before attempted knockdown of hMSH3.
(TIFF)
Subcellular localization of hMLH1, hMSH2, and hMSH6 upon H2O2 treatment. After 24 hours serum-starvation and then 4 hours treatment with H2O2, cells were fixed and stained with antibodies against (A) hMLH1, (B) hMSH2, and/or (C) hMSH6, and then Alexa 488 conjugated anti-mouse antibody. No subcellular shift was observed for these MMR proteins.
(TIF)
Primer sequences (underlined) and restriction sites ( italicized ) used to insert D8S321 and D20S82 loci sequences onto pIREShyg2-EGFP plasmids to make the EMAST constructs.
(TIFF)
Acknowledgments
The authors thank the DNA Sequencing and the Flow Cytometry Cores at the University of Michigan Medical School for their assistance.
Funding Statement
This work was supported by the United States Public Health Service (DK067287 and CA162147) and the SDSU/UCSD Comprehensive Cancer Center Partnership (CA132379 and CA132384). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Grady WM, Carethers JM (2008) Genomic and epigenetic instability in colorectal cancer pathogenesis. Gastroenterology 135: 1079–1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Kane MF, Loda M, Gaida GM, Lipman J, Mishra R, et al. (1997) Methylation of the hMLH1 promoter correlates with lack of expression of hMLH1 in sporadic colon tumors and mismatch repair-defective human tumor cell lines. Cancer Res 57: 808–811. [PubMed] [Google Scholar]
- 3. Veigl ML, Kasturi L, Olechnowicz J, Ma AH, Lutterbaugh JD, et al. (1998) Biallelic inactivation of hMLH1 by epigenetic gene silencing, a novel mechanism causing human MSI cancers. Proc Natl Acad U S A 95: 8698–8702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Boland CR, Thibodeau SN, Hamilton SR, Sidransky D, Eshleman JR, et al. (1998) A National Cancer Institute Workshop on Microsatellite Instability for cancer detection and familial predisposition: development of international criteria for the determination of microsatellite instability in colorectal cancer. Cancer Res 58: 5248–57. [PubMed] [Google Scholar]
- 5. Clark AB, Cook ME, Tran HT, Gordenin DA, Resnick MA, et al. (1999) Functional analysis of human MutSalpha and MutSbeta complexes in yeast. Nucleic Acids Res 27: 736–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Harrington JM, Kolodner RD (2007) Saccharomyces cerevisiae Msh2-Msh3 acts in repair of base -base mispair. Mol Cell Biol 27: 6546–6554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Acharya S, Wilson T, Gradia S, Kane MF, Guerrette S, et al. (1996) hMSH2 forms specific mispair-binding compleses with hMSH3 amd hMSH6. Proc Natl Acad U S A 93: 13629–13634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Gendvhel J, Littman SJ, Drummond JT, Modrich P (1998) Isolation of MutSbeta from human cells and comparison of the mismatch repair specificities of MutSbeta and MutSalpha. J Biol Chem 273: 19895–19901. [DOI] [PubMed] [Google Scholar]
- 9. Tajima A, Iwaizumi M, Tseng-Rogenski S, Cabrera BL, Carethers JM (2011) Both hMutSα and hMutSß DNA mismatch repair complexes participate in 5-fluorouracil cytotoxicity. PLoS One 6: e28117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Chung H, Young DJ, Lopez CG, Le TA, Lee JK, et al. (2008) Mutation rates of TGFBR2 and ACVR2 coding microsatellite in human cells with defective DNA mismatch repair. PLoS One 3: e3463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Chung H, Lopez CG, Holmstrom J, Young DJ, Lai JF, et al. (2010) Both microsatellite length and sequence context determine frameshift mutation rates in defective DNA mismatch repair. Hum Mol Genet 19: 2638–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Chung H, Lopez CG, Young DJ, Lai JF, Holmstrom J, et al. (2010) Flanking sequence specificity determines coding microsatellite heteroduplex and mutation rates with defective DNA mismatch repair (MMR). Oncogene 29: 2172–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Haugen AC, Goel A, Yamada K, Marra G, Nguyen TP, et al. (2008) Genetic instability caused by loss of MutS homologue 3 in human colorectal cancer. Cancer Res 68: 8465–8472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Lee SY, Chung H, Devaraj B, Iwaizumi M, Han HS, et al. (2010) Microsatellite alterations at selected tetranucleotide repeats are associated with morphologies of colorectal neoplasias. Gastroenterology 139: 1519–1525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Yamada K, Kanazawa S, Koike J, Sugiyama H, Xu C, et al. (2010) Microsatellite instability at tetranucleotide repeats in sporadic colorectal cancer in Japan. Oncol Rep 23: 551–561. [PMC free article] [PubMed] [Google Scholar]
- 16. Devaraj B, Lee A, Cabrera BL, Miyai K, Luo L, et al. (2010) Relationship of EMAST and microsatellite instability among patients with rectal cancer. J Gastrointest Surg 14: 1521–1528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Ahrendt SA, Decker PA, Doffek K, Wang B, Xu L, et al. (2000) Microsatellite instability at selected tetranucleotide repeats is associated with p53 mutations in non-small cell lung cancer. Cancer Res 60: 2488–2491. [PubMed] [Google Scholar]
- 18. Garcia M, Choi C, Kim HR, Daoud Y, Toiyama Y, et al. (2012) Association between recurrent metastasis from stage II and III primary colorectal and moderate microsatellite instability. Gastroenterology 143: 48–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Catto JW, Azzouzi AR, Amira N, Rehman I, Feeley KM, et al. (2003) Distinct patterns of microsatellite instability are seen in tumors of the urinary tract. Oncogene 22: 8699–8706. [DOI] [PubMed] [Google Scholar]
- 20. Singer G, Kallinowski T, Hartmann A, Dietmaier W, Wild PJ, et al. (2004) Different types of microsatellite instability in ovarian carcinoma. Int J Cancer 112: 643–646. [DOI] [PubMed] [Google Scholar]
- 21. Burger M, Denzinger S, Hammerschmied CG, Tannapfel A, Obermann EC, et al. (2006) Elevated microsatellite alternations at selected tetranucleotides (EMAST) and mismatch repair gene expression in prostate cancer. J Mol Med 84: 833–841. [DOI] [PubMed] [Google Scholar]
- 22. Danaee H, Nelson HH, Karagas MR, Schned AR, Ashok TD, et al. (2002) Microsatellite instability at tetranucleotide repeats in skin and bladder cancer. Oncogene 21: 4894–4899. [DOI] [PubMed] [Google Scholar]
- 23. Chang CL, Marra G, Chauhan DP, Ha HT, Chang DK, et al. (2002) Oxidative stress inactivates the human DNA mismatch repair system. Am J Physiol Cell Physiol 283: C148–154. [DOI] [PubMed] [Google Scholar]
- 24. Chung H, Chaudhry J, Lai JF, Young DJ, Carethers JM (2012) Flanking nucleotide specificity for DNA mismatch repair-deficient frameshifts within activin receptor 2 (ACVR2). Mutat Res 729: 73–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Luria SE, Delbruck M (1943) Mutations of Bacteria from Virus Sensitive to Virus Resistance. Genetics 28: 491–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Chromatographs for DNA sequencing of tetranucleotide microsatellites. (A) The EMAST construct carrying D8S321 marker contained 12 copies of AAAG, which underwent deletion of one copy of the AAAG unit or insertion of two copies of AAAG repeat as shown. (B) The D20S82 construct harboring 16 copies of AAAG underwent deletion of one repeat of AAAG. Please note that the flanking sequences remained identical to the wild-type when EMAST markers underwent deletion/insertion.
(TIF)
Expression levels of MMR proteins in clones prior to shRNA transfection. Note that clones contained equivalent amounts of hMSH3 protein before attempted knockdown of hMSH3.
(TIFF)
Subcellular localization of hMLH1, hMSH2, and hMSH6 upon H2O2 treatment. After 24 hours serum-starvation and then 4 hours treatment with H2O2, cells were fixed and stained with antibodies against (A) hMLH1, (B) hMSH2, and/or (C) hMSH6, and then Alexa 488 conjugated anti-mouse antibody. No subcellular shift was observed for these MMR proteins.
(TIF)
Primer sequences (underlined) and restriction sites ( italicized ) used to insert D8S321 and D20S82 loci sequences onto pIREShyg2-EGFP plasmids to make the EMAST constructs.
(TIFF)