Abstract
Delivery of small interfering RNA (siRNA) targeted to specific cell types is a significant challenge for the development of RNA interference-based therapeutics. Recently, PTD-DRBD, a double-stranded RNA binding domain (DRBD) fused to the TAT protein transduction domain (PTD), was shown to be effective at delivering siRNA in a non-cell type-specific manner. Here, we evaluated the potential of DRBD as a general protein platform for targeted small interfering RNA (siRNA) delivery. We found that a single DRBD was insufficient to stably complex siRNA when fused to targeting peptides other than PTD, which facilitated nonspecific nucleic acid binding. In contrast to PTD-DRBD, fusion proteins containing two DRBDs (2× DRBD) yielded specific and stable siRNA binding. These proteins could mediate the cellular uptake of siRNA in vitro, though compared with PTD-DRBD gene silencing was attenuated by endosomal entrapment. Our findings suggest that unlike a single DRBD, 2× DRBD inhibits siRNA escape into the cytoplasm and/or induces an internalization pathway distinct from that of PTD-DRBD. Collectively, these data indicate that while 2× DRBD retains siRNA-binding activity when fused to different cell surface-interacting peptides, the utility of 2× DRBD for cell-specific RNA interference is limited without further protein engineering to enhance the bioavailability of the delivered siRNAs.
Keywords: DRBD, protein transduction domain, RNA interference, siRNA delivery
Introduction
Therapeutics based on RNA interference hold great promise for the treatment of numerous diseases that are not currently druggable. In recent years, both viral and nonviral methods for delivery and expression of interfering RNA molecules have been explored.1 Examples of nonviral delivery vectors include peptides, proteins, aptamers, cationic polymers, lipoplexes, and liposomes.2,3,4,5,6,7 Some of these agents are not suitable for systemic in vivo delivery due to factors such as instability in serum, undesirable pharmacokinetics, or the expense or feasibility of scaling-up to obtain therapeutic quantities. In addition to meeting these criteria, an effective delivery agent requires targeting to disease-relevant tissue to avoid potentially toxic side effects of either the agent or the associated interfering RNA.
One approach for delivering RNA to specific cells is to use small proteins or polypeptides that contain a receptor-binding or cell-homing domain to direct the complex to the surface of the desired cell population. These vectors also require RNA-binding capability; a property sometimes conferred using cationic peptide sequences that electrostatically interact with negatively charged RNA backbones.2,8 As an alternative to cationic peptides, there are many examples of both prokaryotic and eukaryotic proteins that contain canonical double-stranded RNA (dsRNA)-binding motifs.9 These proteins can bind to RNA duplexes both specifically (i.e., they do not bind duplexed DNA) and with high affinity.10,11 A well-studied example of these is protein kinase R (PKR; also known as dsRNA-activated protein kinase DAI), which contains two dsRNA-binding motifs.12,13 In isolation, both motifs can bind dsRNA, with affinities of Kd = 3.8 × 10−7 mol/l and Kd = 2 × 10−7 mol/l for the N-terminal motif 1 and C-teminal motif 2, respectively.11,14 Together, the motifs act cooperatively to enhance the affinity of dsRNA binding by 100-fold (Kd = 4 × 10−9 mol/l).11 In a recent study, PKR DRBD motif 1 was fused to a protein transduction domain (PTD) composed of three repeats of the cell-penetrating peptide (CPP) TAT.15 This fusion protein, termed PTD-DRBD, and now commercially available as Transductin, facilitated robust gene silencing in cultured cells and effectively reduced luciferase reporter gene expression in the nasal and tracheal passages of a transgenic mouse.15 While PTD-DRBD is effective at small interfering RNA (siRNA) delivery in vitro or when directly administered to target tissues in vivo, it is not suitable for intravenous systemic delivery; TAT motifs interact with serum molecules such as glycosaminoglycans, which can block its internalizing activity.16 Further, because TAT is a general cell-binding peptide and CPP, it does not target specific cells or tissue types. We reasoned that targeting DRBD with cell-homing peptides or receptor ligands in place of TAT would provide a strategy for cell type-specific delivery of siRNAs.
In the present study, we evaluated DRBDs derived from PKR as a general siRNA delivery vector when fused with either cell-penetrating domains or cell-homing peptides. We found that PTD-DRBD, which contains the cationic cell-binding domain TAT and a single DRBD, can non-specifically interact with siRNA, but that stable and specific binding of siRNA by fusion proteins lacking TAT requires two DRBDs. Unexpectedly, while fusion proteins containing two DRBDs can mediate internalization of siRNA directed by both CPP and cell-homing peptides, gene silencing is attenuated by the inclusion of the larger RNA-binding domain. In addition, regardless of the number of DRBD motifs, we found that the concentration of protein required for maximal gene silencing exceeds the amount required for siRNA binding. These studies demonstrate that the structural properties of DRBD, and possibly other protein delivery vectors, can significantly affect the accessibility of siRNA to the RNA-induced silencing complex and, ultimately, the efficacy of gene silencing.
Results
Complex formation with siRNA and DRBD fusion proteins
We first tested whether DRBD could complex siRNA when fused to a peptide with affinity for a specific cell type. For this, we generated a protein, PPS-DRBD, in which the three tandem copies of the TAT peptide in PTD-DRBD were replaced with the brain vascular targeting peptide DSPAHPS (PPS), a motif that our lab previously identified by in vivo biopanning of mouse brain endothelium17 (Figure 1). For comparison we also generated DRBD alone, which lacks a cell-binding domain (Figure 1). The ability of each protein to bind siRNA was measured by electrophoretic mobility shift assay (Figure 2). Both PPS-DRBD and DRBD failed to induce a marked siRNA mobility shift, even at a 40-fold molar excess of protein relative to siRNA (Figure 2a,b). To address the possibility that the purification method caused a loss of RNA-binding activity, we also tested DRBD purified under denaturing conditions followed by refolding; this protein preparation yielded a comparable result (Supplementary Figure S1). In contrast, incubating PTD-DRBD with siRNA resulted in a mobility shift in a dose-dependent manner (Figure 2c). We hypothesized that the PTD domain might be mediating nonspecific interaction with siRNA. To test this, we performed an electrophoretic mobility shift assay using dsDNA of the same sequence as the siRNA (Figure 2c). PTD-DRBD induced a dsDNA shift at a 4:1 molar ratio, though, in contrast to siRNA, the dsDNA complex failed to migrate into the gel, indicating possible aggregation (Figure 2c). To test if the polybasic TAT motifs within the PTD domain facilitate binding, we incubated synthetic peptides with siRNA and measured electrophoretic mobility. siRNA migration was shifted when incubated with TAT, but not control peptides, including the polybasic TLH and B2 peptides (Figure 2d). These data indicate that DRBD alone or fused to the PPS-targeting domain is not sufficient to stably bind siRNA. However, the TAT motifs in PTD-DRBD enable nonspecific nucleic acid binding.
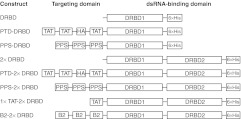
Figure 1.
Structure of DRBD fusion proteins. N-terminal targeting domains are fused to either PKR RNA-binding motif 1 (DRBD1) or an RNA-binding domain containing both PKR motifs 1 and 2 (DRBD2). Short linker regions are indicated by solid, black lines. A 6x histidine tag (6xHis) at the C-terminus is used for purification. DRBD, double-stranded RNA-binding domain; PKR, protein kinase R.
Figure 2.
Electrophoretic mobility gel shift assays of siRNA and dsDNA binding to DRBD fusion proteins or synthetic peptides. 1 μmol/l siRNA or dsDNA was incubated with varying concentrations of protein and their interaction measured by migration on a 6% acrylamide gel followed by staining with ethidium bromide. (a,b) Gel shift of siRNA binding to DRBD or PPS-DRBD with increasing molar ratios of protein to siRNA. (c) Gel shift of either siRNA or dsDNA binding to PTD-DRBD at different molar ratios. (d) Gel shift of 1 μmol/l siRNA interacting with synthetic peptides at either 20 or 40 μmol/l. DRBD, double-stranded RNA-binding domain; dsDNA, double-stranded DNA; PTD, protein transduction domain; siRNA, small interfering RNA.
It was previously reported that proteins containing the complete PKR RNA-binding motif (2× DRBD) can complex with small dsRNA (~20–25 base pairs) and siRNA.10,18 Given this, we next tested whether the PPS domain (PPS-2× DRBD) fused to 2× DRBD could interact with siRNA. Both 2× DRBD alone and PPS-2× DRBD yielded near complete siRNA binding at a 2:1 molar ratio (Figure 3a,b). An additional mobility shift was observed at 4:1 molar ratios, which is consistent with previous studies indicating that the length of a small dsRNA is sufficient to allow binding of two molecules of 2× DRBD in a concentration-dependent manner.10,11 Increasing the concentration of protein did not result in a further mobility shift or aggregation as evidenced by the same band migration pattern at ratios as high at 16:1 (Figure 3c). Unlike PTD-DRBD, neither of these constructs bound to dsDNA (Figure 3a,b). PTD fused to 2× DRBD also did not bind to dsDNA, and it displayed a siRNA-binding profile nearly identical to 2× DRBD (Figure 3d). In a previous report, PTD-DRBD was complexed with siRNA at relatively high molar ratios (20-60:1) for in vitro gene silencing.15 At these concentrations, complexes formed with PTD-2× DRBD migrate into the gel but those with PTD-DRBD are retained in the gel well, indicative of aggregation (Figure 3e). Interestingly, at ratios of 40 and 60:1, the complex undergoes a third mobility shift compared to that at 4:1 (Figure 3d versus Figure 3e). This suggests that at high concentrations, a third protein molecule may be able to associate with the siRNA. Collectively, these results indicate that 2× DRBD fused to either PTD or cell-targeting peptides can specifically and stably bind siRNA, but not dsDNA.
Figure 3.
Electrophoretic mobility gel shift assays of siRNA and dsDNA binding to 2× DRBD fusion proteins. Nucleic acid binding was performed as in Figure 2. (a,b,d) Gel shift of 1 μmol/l siRNA or dsDNA incubated with 0, 1, 2, or 4 μmol/l protein before electrophoresis. (c) Gel shift of 1 μmol/l siRNA incubated with PPS-2× DRBD at concentrations up to 16 μmol/l. (e) Gel shift of siRNA incubated with PTD-2× DRBD and PTD-DRBD at a 20-, 40-, and 60-fold relative molar excess. DRBD, double-stranded RNA-binding domain; dsDNA, double-stranded DNA; siRNA, small interfering RNA.
siRNA delivery and gene silencing with DRBD fusion proteins
Having established that our 2× DRBD fusion proteins could bind to siRNA, we next evaluated their ability to deliver siRNA and mediate gene silencing in vitro. We hypothesized that the greater specificity and affinity of the siRNA interaction with PTD-2× DRBD would confer more potent knockdown relative to PTD-DRBD. To test this, we measured knockdown of the housekeeping gene hypoxanthine-guanine phosphoribosyltransferase (HPRT) in HeLa cells treated with siRNA complexes formed with each protein (Figure 4). PPS-2× DRBD served as a negative control as PPS peptide does not bind to HeLa cells (Supplementary Figure S2). As previously mentioned, at least a 20–60-fold molar excess of PTD-DRBD is typically complexed with siRNA for in vitro gene silencing, despite our observation that much less is required for siRNA binding.15 In order to directly compare silencing efficacy of PTD-DRBD to PTD-2× DRBD, we chose to form complexes with a 60-fold excess of protein over siRNA. After 24 hours, HPRT message was reduced ~82% in cells treated with PTD-DRBD, and no knockdown was observed from treatment with PPS-2× DRBD (Figure 4). Unexpectedly, treatment with PTD-2× DRBD only resulted in ~19% knockdown (Figure 4). To test if the size of the transduction domain might somehow be inhibitory when fused to 2× DRBD, we also tested a truncated form of PTD that only contained a single TAT motif (1× TAT-2× DRBD). However, this protein did not mediate any knockdown of HPRT (Figure 4). To determine whether a CPP other than TAT might yield knockdown, we tested 2× DRBD fused to the CPP, penetratin;19 however this construct also failed to mediate knockdown (Supplementary Figure S3). Together, these findings suggest that 2× DRBD, but not DRBD, inhibits robust gene silencing in vitro.
Figure 4.
Gene silencing of HPRT mediated by different DRBD fusion proteins. HeLa cells were incubated with complexes at a final concentration of 50 nmol/l siRNA and 3 μmol/l protein for 24 hours and HPRT mRNA measured by reverse transcription and quantitative PCR (RT-qPCR). Expression is normalized to cells treated with NC1 siRNA, which is set equal to 1. Data are derived from three independent experiments and presented as mean ± SD. *P < 0.01 and **P < 0.0001, Student's t-test. DRBD, double-stranded RNA-binding domain; HPRT, hypoxanthine-guanine phosphoribosyltransferase; PTD, protein transduction domain; siRNA, small interfering RNA.
A possible explanation for lack of gene silencing is that the interaction between the PTD and the larger RNA-binding domain prevents efficient binding and/or internalization into cells. To address this, we incubated HeLa cells with complexes containing protein and fluorescently labeled siRNA (Figure 5). After incubation, cells were acid washed to remove extracellular complexes and siRNA localization was monitored by fluorescence microscopy. Cells treated with PTD-DRBD showed punctate, vesicular signal, localized near the nucleus (Figure 5a). Treatment with PTD-2× DRBD resulted in a similar pattern but the puncta were larger and there was a higher overall level of internal cell fluorescence (Figure 5a,b). This enhanced signal may be due to the higher affinity binding of PTD-2× DRBD. Treatment using PPS-2× DRBD or siRNA alone did not result in appreciable intracellular siRNA localization (Figure 5a). Since siRNA needs to escape into the cytoplasm to interact with the silencing machinery, we also evaluated whether the vesicular localization of siRNA changed over time when treated with the different proteins. After 24 hours of incubation, siRNA delivered with both PTD-DRBD and PTD-2× DRBD remained primarily punctate with perinuclear staining (Figure 5c). There was no clear diffusion of siRNA throughout the cell with PTD-DRBD. These results show that siRNA complexed to both PTD-DRBD and PTD-2× DRBD can bind to and be internalized into cells.
Figure 5.
Binding and internalization of siRNA/DRBD complexes. (a) Complexes containing fluorescent Cy3-labeled siRNA and PTD-DRBD, PTD-2× DRBD, PPS-2× DRBD, or Cy3 siRNA alone, were incubated with HeLa cells and then acid washed to remove extracellular complexes. Internalized siRNA was visualized by fluorescence microscopy. (b) Mean fluorescence intensity of internalized Cy3-labeled siRNA measured under the conditions used in a. Error bars are SD. (c) Internalized siRNA visualized 24 h after treatment with complexes in a. Bar of low magnification images in a is 200 μm and in the high magnification inset images is 25 μm. Bars in c are 25 μm. a.u., arbitrary units; DRBD, double-stranded RNA-binding domain; PTD, protein transduction domain; siRNA, small interfering RNA.
We hypothesized that minimal gene silencing resulted from limited dissociation of the siRNA from the high affinity RNA-binding domain and/or that the siRNA that did dissociate remained trapped in endocytic compartments. To test these possibilities, we treated cells with siRNA complexes in the presence or absence of chloroquine, an endosomolytic agent used to enhance gene delivery20,21,22 (Figure 6a). In contrast to treatment without chloroquine, HPRT message was reduced 92% in cells treated with either PTD-DRBD or PTD-2× DRBD (Figure 6a). HPRT levels also decreased after treatment with 1× TAT-2× DRBD, though the silencing was more modest (59%) (Figure 6a). A comparable level of knockdown (~60%) was achieved with penetratin-2× DRBD, indicating that siRNA delivery can be achieved with a distinct CPP when fused to 2× DRBD (Supplementary Figure S3). Minimal knockdown (12%) was observed using PPS-2× DRBD (Figure 6a). As our initial hypothesis was that PTD-2× DRBD's higher affinity for siRNA would enhance silencing activity (i.e., less protein is required to deliver the same amount of siRNA), we tested whether lower concentrations of protein (0.4 versus 3.0 μmol/l in Figure 6a) could mediate knockdown (Figure 6b). In the absence of chloroquine, no protein constructs, including PTD-DRBD, reduced HPRT mRNA levels (Figure 6b). However, in the presence of chloroquine, cells treated with PTD-2× DRBD but not PTD-DRBD had significantly lower levels of HPRT message (63 versus 4%, respectively) (Figure 6b). Together, these results suggest that lack of silencing with PTD-2× DRBD in the absence of chloroquine is due to restricted endosomal escape of siRNA rather than a lack of dissociation from the RNA-binding domain. Further, on a molar basis, PTD-2× DRBD has a greater ability to deliver siRNA into cells as compared with PTD-DRBD.
Figure 6.
Silencing of HPRT mediated by DRBD fusion proteins in the presence or absence of chloroquine. (a) HeLa cells were treated with complexes at a concentration of 50 nmol/l siRNA and 3 μmol/l protein in the presence of 50 μmol/l chloroquine. HPRT levels were measured by reverse transcription-quantitative PCR 24 hours after treatment. (b) HeLa cells were treated with complexes at a concentration of 50 nmol/l siRNA and 0.4 μmol/l protein in the presence or absence of chloroquine. Data are derived from three independent experiments and presented as mean ± SD. *P < 0.01, Student's t-test. DRBD, double-stranded RNA-binding domain; HPRT, hypoxanthine-guanine phosphoribosyltransferase; PTD, protein transduction domain; siRNA, small interfering RNA.
Transferrin receptor-targeted delivery of siRNA with a peptide 2× DRBD fusion protein
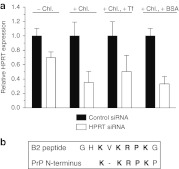
It is possible that targeting 2× DRBD fusion proteins to specific receptors, rather than using nonspecific CPPs, may result in an altered trafficking pathway that yields greater localization of cargo to the cytoplasm versus the endosome. To test this and to evaluate siRNA delivery with a receptor-targeted fusion protein, we generated a 2× DRBD fusion protein containing three repeats of the B2 peptide sequence (GHKVKRPKG) in place of the TAT PTD domains (Figure 1a). We previously identified the B2 peptide by phage display panning against recombinant transferrin receptor (TfR) and showed that adenovirus capsids engineered to present B2 display enhanced transduction of cells in a TfR-mediated manner.23 Coincidentally, the B2 peptide also shares a high degree of similarity with the mature prion protein (PrP) N-terminal polybasic domain (KKRPKP), which has been shown to have membrane interacting and cell-internalizing properties (Figure 7b).24,25 We first tested delivery of siRNA and HPRT knockdown in HeLa cells, which express TfR and have been used to study transferrin trafficking.26 Interestingly, in the absence of chloroquine, treatment with B2-2× DRBD siRNA complexes resulted in a 30% reduction in HPRT message, which is 12% greater knockdown than that obtained with PTD-2× DRBD (Figure 7a versus Figure 4). In the presence of chloroquine, HPRT knockdown was increased to 65% (Figure 7a). To test whether siRNA delivery and knockdown occurs through the TfR, we competed binding by pre-incubating cells with transferrin or bovine serum albumin control protein before adding the siRNA complex. While there was a decrease in HPRT knockdown with transferrin pre-treatment relative to the control (67 versus 49%), this difference was variable and not statistically significant (Figure 7a). These results indicate that B2-2× DRBD can mediate gene silencing but that internalization might be occurring through multiple pathways which include binding TfR and a cell-penetrating mechanism involving the PrP-like internalization motif present within the B2 sequence.
Figure 7.
Silencing of HPRT mediated by a DRBD fusion protein displaying the transferrin receptor-binding peptide B2. (a) HeLa cells were treated with complexes at a concentration of 50 nmol/l siRNA and 3 μmol/l protein in the presence or absence of 50 μmol/l chloroquine (Chl.), transferrin (Tf), and/or bovine serum albumin (BSA). Cells were preincubated with 5 μmol/l Tf or BSA 30 minutes before treatment with complexes. HPRT levels were measured by reverse transcription-quantitative PCR 24 hours after treatment. (b) Alignment of the B2 peptide sequence with the N-terminal polybasic domain of the mature prion protein (PrP). Data are derived from ≥3 independent experiments and presented as mean ± SD. DRBD, double-stranded RNA-binding domain; HPRT, hypoxanthine-guanine phosphoribosyltransferase; siRNA, small interfering RNA.
Discussion
In this study, we sought to determine whether PKR-derived DRBD could serve as a protein platform for cell type-specific targeted delivery of siRNA. For this, we compared knockdown mediated by cell-binding peptides fused to either a single DRBD or the complete PKR RNA-binding motif that contains two DRBDs. Our evaluation revealed that PTD-DRBD, which can mediate robust gene silencing as shown in earlier work, does not stably bind siRNA in the absence of PTD. This result is consistent with previous studies of the RNA-binding activity of PKR demonstrating that although the N-terminal DRBD motif 1 can bind short RNAs (20–25 bp), the binding is of low affinity and unstable.10,11 This suggests that the TAT peptides in PTD-DRBD are acting in tandem with the DRBD to enable stable siRNA binding through electrostatic interaction, which is further supported by the observation that dsDNA also binds to PTD-DRBD when in molar excess. The requirement for PTD raises the question as to what degree the DRBD contributes to the siRNA-binding activity of PTD-DRBD. It could be that the DRBD is not absolutely required and that a polypeptide composed of only the PTD or several tandem repeats of TAT may also have siRNA complexation properties that could confer silencing activity.
In the contrast to PTD-DRBD, we showed that the inclusion of both PKR-derived DRBDs results in stable and dsRNA-specific binding, even when the DRBDs are fused to the TAT-containing PTD. This means that 2× DRBD can be fused to various cell-binding peptides, including highly positively charged sequences, and still maintain binding of the siRNA restricted to the DRBD. This property is important as it prevents the cell-binding peptide from interacting with the siRNA, which could interfere with its ability to associate with cell surface moieties. Both PTD-DRBD and PTD-2× DRBD can complex with siRNA and be internalized, however, PTD-2× DRBD complexes are more readily internalized at lower concentrations compared with PTD-DRBD, as inferred from a greater degree of gene silencing. This may occur because at lower molar ratios of protein to siRNA, the siRNA binds to the PTD domain of PTD-DRBD and this impedes uptake of the complex into the cell. In contrast, at the same molar ratios, siRNA binding to PTD-2× DRBD is restricted to the DRBDs, allowing the unencumbered PTD to mediate more effective cell surface binding and internalization.
Despite the improved siRNA binding and internalization properties of 2× DRBD fusion proteins, we found that these complexes are not nearly as robust at mediating gene silencing as PTD-DRBD. This was true for both TAT and penetratin fusions, which supports the hypothesis that it is not the specific CPP that is restricting silencing but rather a property of the 2× DRBD domain. Penetratin has been used to mediate rapid internalization of siRNA when directly conjugated to modified siRNA via a disulfide bond.27 The resulting molecule is smaller than the complexes tested in our studies, which may explain its greater silencing activity.
Interestingly, despite the observation that siRNA can bind to 2× DRBD proteins at a 2–4 molar excess, maximal silencing requires a greater than eightfold molar excess. It may be that excess, unbound protein facilitates knockdown in a manner that doesn't necessarily require direct interaction with the siRNA. One possibility is that at high concentrations, free CPP stimulates cargo uptake as reported for TAT-DNA complexes.28 Alternatively, the high protein concentration may prevent siRNA dissociation from DRBD proteins after dilution into the transfection media.
Treatment of cells with chloroquine resulted in silencing comparable with PTD-DRBD, which indicates that the 2× DRBD complexes are sequestered to endosomal/lysosomal compartments and do not efficiently escape into the cytoplasm. This is consistent with earlier studies showing that siRNA complexed with a protein containing the PKR DRBDs could facilitate gene silencing, but only when admixed with the fusogenic peptide KALA.18 One explanation for this observation is that both PTD-DRBD and 2× DRBD fusion proteins traffic through the same compartments but that the larger relative size and/or structural features of the 2× DRBD fusion protein complex hinder endosomal escape. Alternatively, the 2× DRBD constructs might enter the cell through a pathway distinct from PTD-DRBD. It has been reported that PTD-DRBD and other TAT fusion proteins utilize a macropinocytic uptake mechanism that promotes cytoplasmic cargo delivery.16 However, there are differing reports on the exact mechanism of TAT uptake and recently it was shown that TAT peptide fused to relatively small cargo could directly translocate across the plasma membrane whereas fusion to larger cargo results in endocytic internalization.29 When considering the relationship between complex size and bioactivity, it is of note that at the molar concentrations of protein used for in vitro gene silencing (20-60:1), PTD-DRBD complexes are aggregated. Counter-intuitively, aggregates are more efficient at mediating gene silencing than soluble or lower-order complexes. Thus, 2× DRBD complexes might preferentially traffic through endocytic compartments that lead to entrapment while PTD-DRBD trafficking allows for enhanced cytoplasmic localization of bound siRNA. In support of the concept that utilizing different internalization mechanisms may affect gene-silencing efficacy, our transferrin receptor-targeted protein provided enhanced knockdown relative to PTD-2× DRBD in the absence of chloroquine. However, because transferrin did not compete with B2-2× DRBD-mediated gene knockdown, the B2 peptide may be utilizing both the transferrin receptor as well as other cell surface moieties, via interaction with the B2 polybasic motif, for internalization. One possibility is that the net positive charge of the B2 domain facilitates nonspecific electrostatic interaction with negative charges on the cell surface (e.g., from glycosaminoglycans). In support of this, internalization of the PrP cell-penetrating motif is inhibited by exogenous heparin and chondroitin sulfate.24 This property may, in part, explain the incomplete inhibition of silencing by transferrin receptor competition. It would be interesting to evaluate whether the B2 peptide, in addition to binding TfR, is a novel CPP.
Overall, our study demonstrates that protein-based siRNA delivery vectors utilizing canonical DRBDs display several properties that could be used for cell type-specific targeting. However, the effect of the DRBD structure on the bioavailability of siRNA delivered with the 2× DRBD proteins evaluated in this study poses a significant limitation to their utility. It would be interesting to test whether further vector engineering could overcome these shortcomings. For example, a membrane-destabilizing fusogenic/endosomolytic peptide could be introduced into the 2× DRBD fusion protein. Alternatively, chemical modification of the proteins with endosome-disrupting groups might enhance cytoplasmic cargo delivery.30 Future studies will show whether these, or other strategies, will yield more potent 2× DRBD-based vectors for delivery of siRNA targeted to specific cell types.
Materials and Methods
Cell culture. HeLa cells were cultured in complete medium (Dulbecco's modified Eagle's medium containing 10% fetal bovine serum) at 37 °C, 5% CO2.
Cloning of fusion protein expression vectors. DNA-encoding DRBD fusion proteins was generated by PCR-based gene synthesis. The primary amino acid sequence of all constructs is shown in Supplementary Figure S4. Unlike the other constructs, penetratin-2× DRBD contains a flexible linker previously used to generate a penetratin-mCherry protein for cellular delivery.31 Overlapping oligonucleotides encoding the proteins were designed using NIH Helix Systems DNAWorks version 3.1.32 Codons were optimized for expression in Escherichia coli. Oligonucleotides (IDT, Coralville, IA) were annealed, extended, and PCR amplified. Full-length synthesized genes contained 5′ NcoI and 3′ BamHI restriction sites. Digested PCR products were ligated into pET11d expression vector (Stratagene, Santa Clara, CA) backbone generated by digesting with NcoI and BamHI restriction enzymes (New England Biolabs, Ipswich, MA). The sequence of each expression plasmid was confirmed by sequencing performed at the University of Iowa DNA Core Facility.
Protein expression and purification. All proteins were expressed and purified except for PTD-DRBD, which was obtained from IDT (commercially sold as Transductin). pET11d expression plasmids were transformed into BL21 Star E. coli (Life Technologies, Carlsbad, CA). A 25 ml starter culture grown using a freshly transformed colony was inoculated into 500 ml of LB containing 100 μg/ml ampicillin and grown at 37 °C, 220 rpm shaking, until the OD600 reached between 0.6 and 0.8. Protein expression was induced with 1 mmol/l IPTG and cultures were grown an additional 4 hours at 37 °C, 220 rpm shaking. Cultures were centrifuged at 5,000 rpm at 4 °C for 15 minutes to pellet bacteria. Protein constructs containing 2× DRBD required denaturing purification and on-column refolding in order to remove bacterial RNA bound to the protein. For purification under denaturing conditions, the pellet was resuspended in 5 ml of Buffer A (50 mmol/l sodium phosphate, pH 7.4) containing Complete Mini protease inhibitors (Roche, Basel, Switzerland); 30K units of rLysozyme (Novagen, Madison, WI), 150 units of benzonase (Sigma-Aldrich, St. Louis, MO), and MgCl2 to a final concentration of 2 mmol/l and the suspension was incubated on ice for 20 minutes. Ten milliliter of 1.1× Buffer B (50 mmol/l sodium phosphate, 555.5 mmol/l NaCl, 22.2 mmol/l imidazole, 6.66 mol/l guanidine HCL, 2.2 mmol/l β-mercaptoethanol; pH 7.4) was added to the suspension that was then sonicated on ice seven times for 30 seconds with the amplitude set at 50 using a Model GE50 Ultrasonic Processor (Sonics & Materials, Newton, CT); 35 ml of Buffer B was added for a final volume of 50 ml and the lysate was allowed to incubate on ice for 25 minutes to solubilize proteins. The lysate was clarified by centrifugation for 40 minutes at 16,000g, 4 °C. The supernatant was then filtered through a 0.4 μm PVDF filter (Millipore, Billerica, MA). Using a BioRad BioLogic LP System (Bio-Rad, Hercules, CA), denatured proteins were bound to a HiTrap IMAC column (GE Healthcare, Waukesha, WI) charged with cobalt chloride. The column was washed with 10 ml of Buffer B diluted to 1× with Buffer A. Bound protein was refolded on-column by applying a 0–100% linear gradient over 80 minutes at 0.5 ml/minute flow rate starting with 100% Buffer B/0% Buffer C (50 mmol/l sodium phosphate, 500 mmol/l NaCl, 20 mmol/l imidazole; pH 7.4) and ending with 0% BufferB/100% Buffer C. Purified, refolded proteins were eluted with Buffer D (50 mmol/l sodium phosphate, 300 mmol/l NaCl, 150 mmol/l imidazole; pH 7.4) and concentrated using Amicon Ultra 10K MWCO centrifugal filters (Millipore). Concentrated eluates were buffer exchanged into phosphate-buffered saline + 10% glycerol using Zeba desalting columns (Thermo Fisher Scientific, Waltham, MA). For non-denaturing purification of the DRBD and PPS-DRBD proteins used in Figure 2a,b, benzonase- and lysozyme-treated lysates were clarified by centrifugation and then combined with 1× Buffer B containing 750 mmol/l NaCl. Extracts were applied to the IMAC cobalt column and washed with Buffer B containing 1 mol/l NaCl. After elution, extracts were treated again with benzonase to remove co-purifying bacterial nucleic acid. Benzonase was removed by re-purifying the protein on an IMAC cobalt column. All protein preparations were aliquoted and stored at −80 °C. Coomassie staining of all proteins was performed and is shown in Supplementary Figure S5.
Electrophoretic mobility shift assays. The siRNA used was Ambion Silencer REST siRNA (catalog no. AM16708) obtained from Life Technologies. dsDNA was prepared using DNA oligonucleotides (IDT) of the same sequence as the siRNA. DNA oligonucleotides were annealed by heating to 95 °C for 4 minutes in annealing buffer (100 mmol/l potassium acetate, 2 mmol/l magnesium acetate, 30 mmol/l HEPES, pH 7.4) and then allowed to cool to room temperature. In all gel shifts except for in Figure 3e, 8 pmol of siRNA or dsDNA were combined with varying amounts of each protein in a final volume of 8 μl, yielding concentrations of 1 μmol/l siRNA or dsDNA. In Figure 3e, 1 pmol of siRNA was complexed with proteins in order to achieve the higher molar protein ratios in a small volume. Mixtures were incubated at room temperature for 15 minutes and then combined with 2 μl of 5× TBE Loading Buffer (Life Technologies). The entire reaction volumes were electrophoresed on a 0.5× TBE, 6% acrylamide gel at 90 V for 45 minutes in 0.5× TBE running buffer. The gel was stained with 0.5 μg/ml ethidium bromide (Bio-Rad) and visualized using a VersaDoc 5000 MP (Bio-Rad). For gel shifts with synthetic peptides, N-terminally biotinylated TAT (RKKRRQRRR), PPS (DSPAHPS), TLH (GWTLHNK), or B2 (GHKVKRPKG) peptides (American Peptide Company, Sunnyvale, CA) were incubated at a final concentration of either 20 or 40 μmol/l with 1 μmol/l siRNA and analyzed as described above.
siRNA delivery and HPRT gene silencing in vitro. Knockdown experiments were performed as previously described.15 HeLa cells were plated in 48-well dishes at 2–3 × 104 cells/well in regular media (Dulbecco's modified Eagle's medium with 10% fetal bovine serum) 14–16 hours before siRNA treatment. Complexes were formed by incubating 6 pmol of HPRT S1 Dicer-substrate siRNA (IDT) or Universal Negative Control (NC1) Dicer-substrate siRNA (IDT) with either 48 or 360 pmol of protein on ice for 30 minutes. Dulbecco's modified Eagle's medium containing 10% Q-serum (Q-Media) prepared as previously described15 was added to complexes to yield a final concentrations of 50 nmol/l siRNA and 0.4 or 3 μmol/l protein. Media and siRNA/protein mixtures were added to cells that were previously washed one time with Q-Media and then preincubated in Q-Media for 30 minutes at 37 °C. Q-Media was removed from cells before adding the siRNA/protein mixture. Complexes were incubated with cells for 4 hours and then removed and replaced with fresh complete medium. Cells were incubated for 24 hours and then harvested for RNA. For chloroquine experiments, chloroquine was added during both the siRNA incubation step and during the 24-hour incubation period. For transferrin receptor blocking experiments, HeLa cells were pretreated for 30 minutes with 5 μmol/l holo-transferrin (Sigma-Aldrich) or bovine serum albumin (New England Biolabs) in Q-Media before knockdown. Trizol Reagent (Life Technologies) was used to extract and purify total RNA from treated cells. cDNA was synthesized from RNA using the MultiScribe High Capacity cDNA Reverse Transcription Kit (Applied Biosystems, Foster City, CA) according to the manufacturer's protocol. Quantitative PCR was performed using TaqMan Assays (Applied Biosystems). GAPDH was used as the endogenous control gene. Cycling was performed on a 7900HT Fast Real Time PCR System (Applied Biosystems). Relative HPRT gene expression was calculated using the ΔΔCT method.33 GAPDH was used to calculate the ΔCT for HPRT in HeLa cells, knockdown with HPRT siRNA was calculated as the average relative quantity from three independent experiments and variation is represented as the SD of the three relative quantities. The variation of HPRT in cells treatment with NC1 siRNA was calculated as the population SD of the ΔCT s for each treatment condition in the three independent experiments.
Internalization of fluorescent siRNA/protein complexes in vitro. HeLa cells were incubated with complexes containing 50 nmol/l scrambled sequence Cy3-siRNA (IDT) and 2 μmol/l protein prepared as described above for HPRT gene-silencing experiments. After 3 hours of incubation, the medium containing complexes was removed and the cell nuclei were stained for 10 minutes at 37 °C with Hoechst 3342 dye (Life Technologies) diluted into complete media. Cells were then washed three times with complete media followed by washing with low pH acid buffer (0.2 mol/l glycine, pH 2.8). After the acid wash, cells were washed one additional time with Dulbecco's PBS (Life Technologies) before examination by fluorescence microscopy. The brightness and contrast of the primary images were adjusted equivalently using Image J software (NIH, Bethesda, MD). Measurement of internal Cy3 fluorescence was performed using Image J software. The fluorescence intensity (integrated density) was measured for 16 cells per analyzed image. The integrated density was calculated and corrected by subtracting background signal from regions proximal to the measured cells. Mean fluorescence intensity and SD was calculated from two independent experiments.
Peptide cell binding and recombinant protein characterization. See Supplementary Materials and Methods.
SUPPLEMENTARY MATERIAL Figure S1. Electrophoretic mobility shift assay of siRNA binding to DRBD prepared under denaturing purification conditions. Figure S2. Binding of synthetic TAT and PPS peptides to HeLa cells. Figure S3. Silencing of HPRT mediated by penetratin-2× DRBD fusion protein in the presence or absence of chloroquine. Figure S4. Primary amino acid sequence of each fusion protein. Figure S5. SDS-PAGE and Coomassie Blue staining of purified fusion proteins. Materials and Methods.
Acknowledgments
The authors would like to acknowledge the members of the Davidson Lab for discussion and technical advice. We would also like to thank Anna Okulist for assistance with cell culture experiments. This work was supported by National Institutes of Health grants NS063246, DK54759, NS50210, HL007638, the University of Iowa Institute for Clinical and Translational Science, and the Roy J Carver Trust. The authors declared no conflict of interest.
Supplementary Material
Electrophoretic mobility shift assay of siRNA binding to DRBD prepared under denaturing purification conditions.
Binding of synthetic TAT and PPS peptides to HeLa cells.
Silencing of HPRT mediated by penetratin-2× DRBD fusion protein in the presence or absence of chloroquine.
Primary amino acid sequence of each fusion protein.
SDS-PAGE and Coomassie Blue staining of purified fusion proteins.
References
- Davidson BL., and, McCray PB., Jr Current prospects for RNA interference-based therapies. Nat Rev Genet. 2011;12:329–340. doi: 10.1038/nrg2968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar P, Wu H, McBride JL, Jung KE, Kim MH, Davidson BL.et al. (2007Transvascular delivery of small interfering RNA to the central nervous system Nature 44839–43. [DOI] [PubMed] [Google Scholar]
- Kuwahara H, Nishina K, Yoshida K, Nishina T, Yamamoto M, Saito Y.et al. (2011Efficient in vivo delivery of siRNA into brain capillary endothelial cells along with endogenous lipoprotein Mol Ther 192213–2221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis ME, Zuckerman JE, Choi CH, Seligson D, Tolcher A, Alabi CA.et al. (2010Evidence of RNAi in humans from systemically administered siRNA via targeted nanoparticles Nature 4641067–1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNamara JO., 2nd, , Andrechek ER, Wang Y, Viles KD, Rempel RE, Gilboa E.et al. (2006Cell type-specific delivery of siRNAs with aptamer-siRNA chimeras Nat Biotechnol 241005–1015. [DOI] [PubMed] [Google Scholar]
- Santel A, Aleku M, Keil O, Endruschat J, Esche V, Durieux B.et al. (2006RNA interference in the mouse vascular endothelium by systemic administration of siRNA-lipoplexes for cancer therapy Gene Ther 131360–1370. [DOI] [PubMed] [Google Scholar]
- Yonenaga N, Kenjo E, Asai T, Tsuruta A, Shimizu K, Dewa T.et al. (2012RGD-based active targeting of novel polycation liposomes bearing siRNA for cancer treatment J Control Release 160177–181. [DOI] [PubMed] [Google Scholar]
- Lundberg P, El-Andaloussi S, Sütlü T, Johansson H., and, Langel U. Delivery of short interfering RNA using endosomolytic cell-penetrating peptides. FASEB J. 2007;21:2664–2671. doi: 10.1096/fj.06-6502com. [DOI] [PubMed] [Google Scholar]
- Tian B, Bevilacqua PC, Diegelman-Parente A., and, Mathews MB. The double-stranded-RNA-binding motif: interference and much more. Nat Rev Mol Cell Biol. 2004;5:1013–1023. doi: 10.1038/nrm1528. [DOI] [PubMed] [Google Scholar]
- Bevilacqua PC., and, Cech TR. Minor-groove recognition of double-stranded RNA by the double-stranded RNA-binding domain from the RNA-activated protein kinase PKR. Biochemistry. 1996;35:9983–9994. doi: 10.1021/bi9607259. [DOI] [PubMed] [Google Scholar]
- Schmedt C, Green SR, Manche L, Taylor DR, Ma Y., and, Mathews MB. Functional characterization of the RNA-binding domain and motif of the double-stranded RNA-dependent protein kinase DAI (PKR) J Mol Biol. 1995;249:29–44. doi: 10.1006/jmbi.1995.0278. [DOI] [PubMed] [Google Scholar]
- Patel RC., and, Sen GC. Identification of the double-stranded RNA-binding domain of the human interferon-inducible protein kinase. J Biol Chem. 1992;267:7671–7676. [PubMed] [Google Scholar]
- Green SR., and, Mathews MB. Two RNA-binding motifs in the double-stranded RNA-activated protein kinase, DAI. Genes Dev. 1992;6 12B:2478–2490. doi: 10.1101/gad.6.12b.2478. [DOI] [PubMed] [Google Scholar]
- Tian B., and, Mathews MB. Functional characterization of and cooperation between the double-stranded RNA-binding motifs of the protein kinase PKR. J Biol Chem. 2001;276:9936–9944. doi: 10.1074/jbc.M007328200. [DOI] [PubMed] [Google Scholar]
- Eguchi A, Meade BR, Chang YC, Fredrickson CT, Willert K, Puri N.et al. (2009Efficient siRNA delivery into primary cells by a peptide transduction domain-dsRNA binding domain fusion protein Nat Biotechnol 27567–571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wadia JS, Stan RV., and, Dowdy SF. Transducible TAT-HA fusogenic peptide enhances escape of TAT-fusion proteins after lipid raft macropinocytosis. Nat Med. 2004;10:310–315. doi: 10.1038/nm996. [DOI] [PubMed] [Google Scholar]
- Chen YH, Chang M., and, Davidson BL. Molecular signatures of disease brain endothelia provide new sites for CNS-directed enzyme therapy. Nat Med. 2009;15:1215–1218. doi: 10.1038/nm.2025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Lee SH, Choe J., and, Park TG. Intracellular small interfering RNA delivery using genetically engineered double-stranded RNA binding protein domain. J Gene Med. 2009;11:804–812. doi: 10.1002/jgm.1365. [DOI] [PubMed] [Google Scholar]
- Derossi D, Joliot AH, Chassaing G., and, Prochiantz A. The third helix of the Antennapedia homeodomain translocates through biological membranes. J Biol Chem. 1994;269:10444–10450. [PubMed] [Google Scholar]
- Dean RT, Jessup W., and, Roberts CR. Effects of exogenous amines on mammalian cells, with particular reference to membrane flow. Biochem J. 1984;217:27–40. doi: 10.1042/bj2170027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luthman H., and, Magnusson G. High efficiency polyoma DNA transfection of chloroquine treated cells. Nucleic Acids Res. 1983;11:1295–1308. doi: 10.1093/nar/11.5.1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veldhoen S, Laufer SD, Trampe A., and, Restle T. Cellular delivery of small interfering RNA by a non-covalently attached cell-penetrating peptide: quantitative analysis of uptake and biological effect. Nucleic Acids Res. 2006;34:6561–6573. doi: 10.1093/nar/gkl941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia H, Anderson B, Mao Q., and, Davidson BL. Recombinant human adenovirus: targeting to the human transferrin receptor improves gene transfer to brain microcapillary endothelium. J Virol. 2000;74:11359–11366. doi: 10.1128/jvi.74.23.11359-11366.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wadia JS, Schaller M, Williamson RA., and, Dowdy SF. Pathologic prion protein infects cells by lipid-raft dependent macropinocytosis. PLoS ONE. 2008;3:e3314. doi: 10.1371/journal.pone.0003314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundberg P, Magzoub M, Lindberg M, Hällbrink M, Jarvet J, Eriksson LE.et al. (2002Cell membrane translocation of the N-terminal (1-28) part of the prion protein Biochem Biophys Res Commun 29985–90. [DOI] [PubMed] [Google Scholar]
- Macia E, Ehrlich M, Massol R, Boucrot E, Brunner C., and, Kirchhausen T. Dynasore, a cell-permeable inhibitor of dynamin. Dev Cell. 2006;10:839–850. doi: 10.1016/j.devcel.2006.04.002. [DOI] [PubMed] [Google Scholar]
- Davidson TJ, Harel S, Arboleda VA, Prunell GF, Shelanski ML, Greene LA.et al. (2004Highly efficient small interfering RNA delivery to primary mammalian neurons induces MicroRNA-like effects before mRNA degradation J Neurosci 2410040–10046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ignatovich IA, Dizhe EB, Pavlotskaya AV, Akifiev BN, Burov SV, Orlov SV.et al. (2003Complexes of plasmid DNA with basic domain 47-57 of the HIV-1 Tat protein are transferred to mammalian cells by endocytosis-mediated pathways J Biol Chem 27842625–42636. [DOI] [PubMed] [Google Scholar]
- Mishra A, Lai GH, Schmidt NW, Sun VZ, Rodriguez AR, Tong R.et al. (2011Translocation of HIV TAT peptide and analogues induced by multiplexed membrane and cytoskeletal interactions Proc Natl Acad Sci USA 10816883–16888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varkouhi AK, Scholte M, Storm G., and, Haisma HJ. Endosomal escape pathways for delivery of biologicals. J Control Release. 2011;151:220–228. doi: 10.1016/j.jconrel.2010.11.004. [DOI] [PubMed] [Google Scholar]
- Cronican JJ, Thompson DB, Beier KT, McNaughton BR, Cepko CL., and, Liu DR. Potent delivery of functional proteins into Mammalian cells in vitro and in vivo using a supercharged protein. ACS Chem Biol. 2010;5:747–752. doi: 10.1021/cb1001153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoover DM., and, Lubkowski J. DNAWorks: an automated method for designing oligonucleotides for PCR-based gene synthesis. Nucleic Acids Res. 2002;30:e43. doi: 10.1093/nar/30.10.e43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ., and, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Electrophoretic mobility shift assay of siRNA binding to DRBD prepared under denaturing purification conditions.
Binding of synthetic TAT and PPS peptides to HeLa cells.
Silencing of HPRT mediated by penetratin-2× DRBD fusion protein in the presence or absence of chloroquine.
Primary amino acid sequence of each fusion protein.
SDS-PAGE and Coomassie Blue staining of purified fusion proteins.