Abstract
Adeno-associated virus (AAV)-derived vectors are promising gene delivery systems, and a number of design strategies have been pursued to improve their performance. For example, genetic insertion of proteins into the capsid may be used to achieve vector retargeting, reduced immunogenicity, or to track vector transport. Unfortunately, rational approaches to genetic insertion have experienced limited success due to the unpredictable context-dependent nature of protein folding and the complexity of the capsid's macroassembly. We report the construction and use of a frame-enriched DNase-based random insertion library based on AAV2 cap, called pAAV2_RaPID (Random Peptide Insertion by DNase). The fluorescent mCherry protein was inserted randomly throughout the AAV2 capsid and the library was selected for fluorescent and infectious variants. A capsid site was identified in VP3 that can tolerate the large protein insertion. In contrast to previous efforts to incorporate fluorescent proteins into the AAV2 capsid, the isolated mCherry mutant maintains native infectivity while displaying robust fluorescence. Collectively, these results demonstrate that the pAAV2_RaPID platform library can be used to create fully infectious AAV vectors carrying large functional protein domains on the capsid.
Keywords: AAV, directed evolution, random domain insertion
Introduction
Adeno-associated virus (AAV) is a small (20–25 nm), nonenveloped, single-stranded DNA virus with an icosahedral (T = 1) 60-mer capsid formed from the assembly of three overlapping subunits VP1:VP2:VP3, typically in a 1:1:10 ratio. Its 4.7 kb nucleotide (nt) genome contains two open-reading frames (ORFs)—rep and cap—which encode overlapping protein sequences. Rep encodes four proteins responsible for viral genome replication, packaging, and site-specific integration into the S1 locus of human chromosome 19.1,2 Cap encodes the three structural proteins, VP1, VP2, and VP3. Assembly-activating protein, recently found in an alternate reading frame of the cap gene, mediates assembly of the VP proteins into the capsid before genome packaging occurs.3 The primary cell surface receptors for AAV serotype 2 (AAV2) are heparan sulfate (HS) proteoglycans, while various coreceptors have been identified.4,5
AAV is being studied intensively for its utility as a safe and effective gene delivery vector. It transduces both dividing and nondividing cells6 and elicits a comparatively mild adaptive immune response.7 Alterations to the virus capsid have been made to retarget native tropism,8,9,10 map the infection pathway,11 or evade host antibody detection.12,13,14 Capsid modifications can be achieved via genetic mutation,8 chemical conjugation,11,15,16 or noncovalent attachment.17 Of these approaches, genetic alterations have the advantages of relatively uniform end products, ease of production, reduced cost, and avoidance of potentially toxic synthetic chemicals.
Genetic modification of AAV has been attempted, including insertion of cell-targeting peptides8 and fluorescent proteins.18,19 However, these rationally designed vectors have seen mixed success. For example, insertion of relatively large fluorescent proteins into the AAV2 capsid has been limited to the N-terminus of VP2, resulting in viruses that are fluorescent but display decreased infectivity.8,18,19 Labor intensive and costly mutagenesis studies have been employed to create maps of insertion-tolerant regions of the AAV capsid,9,20,21 but functional insertion varies widely even for similarly sized small peptides of different primary sequence at the same capsid location.21 Difficulties in genetic insertion of peptides/proteins into the AAV capsid likely arise from the unpredictable, context-dependent nature of protein subunit folding. Furthermore, in contrast to monomeric protein design, genetic insertion into AAV capsids must maintain efficient supramolecular assembly of the capsid, genome packaging, and dynamic conformational changes.22 Therefore, instead of purely rational design strategies, a high-throughput combinatorial method may be valuable to identify optimal insertion sites for the incorporation of desired peptides and proteins. In particular, a useful approach would be used to create highly diverse libraries of capsid genes with randomly inserted peptides followed by selection/screening to identify virus variants with desired phenotypic properties.
A number of directed evolution approaches have been utilized to generate improved AAV vectors.10,14,23,24 For the random insertion of peptides/proteins, transposon-based mutagenesis has been used to build an AAV2 gene library with a hexahistidine motif inserted randomly throughout the cap ORF.25,26 However, no library mutants were isolated that conformed to the selective pressure—having both affinity for nickel while retaining infectivity. Alternatively, DNase-based random domain insertion is a useful complement to transposon-based methods as it introduces additional sequence diversity through a range of deletions and tandem duplications (indels) at each site of insertion.27 Since accompanying deletions have been shown in some cases to drastically improve the assembly of AAV insertional mutants,28 the additional indel sequence diversity may make DNase-based approaches more powerful. In addition, a DNase-based method was previously used to discover sites in the AAV5 capsid permissive to deletions and duplications.29 To date, however, there have been no reports describing random insertion of large functional proteins into the AAV capsid.
In this study, we utilize a DNase-based random domain insertion strategy to build a modular, functionally enriched AAV2 capsid gene library named pAAV2_RaPID (Random Peptide Insertion by DNase). The platform library allows the insertion of a desired peptide/protein randomly throughout the VP3 region of AAV2. We demonstrate the utility and power of pAAV2_RaPID by isolating a virus variant with the mCherry gene genetically inserted into a capsid location never before tested for the insertion of relatively large protein domains. This mCherry-AAV2 hybrid vector maintains high genome packaging titer, wild type (wt)-like binding to heparin, displays robust fluorescence, and in contrast to previous attempts of inserting fluorescent proteins into AAV2, exhibits no loss in infectivity. Our pAAV2_RaPID library may be a useful tool to not only generate enhanced AAV vectors with modified functionalities but to also uncover fundamental structure–function relationships of the AAV capsid.
Results
Construction of random insertion library
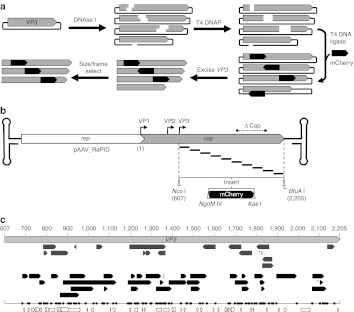
A library of AAV2 cap genes was constructed containing a modular DNA sequence randomly inserted into the VP3-shared capsid structural domain (Figure 1a). The mCherry ORF30–which was used for selection and further study in this work–was chosen as a modular placeholder, since its size was large enough to permit size selection during library cloning. The mCherry placeholder was flanked by directional restriction endonuclease sites (NgoM IV and Kas I) to allow efficient construction of other random insertion libraries by simple subcloning procedures. The resulting DNA library was transformed into Escherichia coli and plated on selective media, yielding ~5 × 106 individual colonies. After purification from E. coli, the plasmid library was resolved by agarose gel electrophoresis to isolate plasmid-mCherry hybrids containing a single insert (~30% of total band intensity) and to remove self-religated and non-cut plasmids. DNA sequencing analysis of individual clones from the size-selected gene library reveals insertions are dispersed throughout VP3, with deletions and duplications occurring at sites of mCherry insertion (Figure 1c). In addition, sequences were inserted in-frame at both cap-mCherry junctions (upstream and downstream) in 12% of the sampled variants, similar to previous reports.27 Thus, a large parental library of cap genes was created with a modular DNA sequence randomly inserted throughout the VP3 domain, which can be easily replaced with sequences encoding other peptides of interest.
Figure 1.
Construction of pAAV2_RaPID, a modular, enriched random domain insertion library. (a) The random insertion library was constructed by brief DNase I treatment of a plasmid containing the cap gene, resulting in a library of randomly linearized vectors. After blunting by T4 DNA polymerase, a modular linker (mCherry ORF) flanked by unique RE sites, NgoM IV and Kas I, encoding small amino acids alanine-glycine and glycine-alanine respectively, was ligated blunt-wise into the library. The resulting cap-insert hybrids were size selected by excision and purification using agarose gel electrophoresis, resulting in a library of capsid genes with a randomly inserted linker. These genes were then enriched for in-frame variants using a pInSALect derivative.31 The enriched gene pool was cloned into the VP3 portion (between Nco I and BfuA I sites) of an AAV2 expression vector, yielding the pAAV2_RaPID library. (b) A scaled diagram of the pAAV2_RaPID library illustrates an array of random deletions/duplications at insertion sites of mCherry–shown below with flanking RE sites NgoM IV and Kas I–throughout the VP3 ORF. The hairpin loops represent the ITRs (not drawn to scale) and ΔCap ind icates the region of cap deleted from the vector into which the library genes were cloned. Start codons of capsid proteins VP1, VP2, and VP3 are indicated by bent arrows. Nucleotide numbering used in this study is shown, starting with 1 at the adenosine of the VP1 start codon, continuing with 607 at the adenosine of the VP3 start codon (at a Nco I site), and ending with 2205 at the final nucleotide of the TAA stop codon (next to a BfuA I cut site). (c) Scaled representation of insertions found in sequenced clones from the pAAV2_RaPID library, aligned with the VP3 ORF (light gray arrow, nucleotide positions in VP1 numbering). Arrows indicate both the direction of insert and size of deletion for each clone in the library before (upper, dark gray) and after (lower, black arrows) frame selection. *Indicates a duplication. The one-dimensional scatter plot shows the distribution of all crossover sites (upstream and downstream, naive and enriched). Below, structural features of VP3 are illustrated with arrows (sheets) and cylinders (coils). AAV, adeno-associated virus; ITR, inverted terminal repeat; ORF, open-reading frame; RE, restriction endonuclease.
Frame selection to enrich virus plasmid library
Since a limited number of AAV2 mutants can be selected for traits such as packaging and infectivity, we investigated whether the variants having mCherry out-of-frame from VP3 could be removed from the parental gene library before performing selection. To accomplish this, the ensemble of VP3-mCherry gene fusions were subcloned into a derivative of pInSALect plasmid to create vectors that express our VP3-mCherry hybrids as N-terminal fusions to β-lactamase.31 Selection in E. coli was then performed to isolate in-frame VP3-mCherry variants. DNA sequencing analysis of clones from the “frame-selected” library revealed 100% in-frame VP3-mCherry gene fusions (n = 34) (Figure 1c). Furthermore, insertion sites within VP3 were dispersed throughout the VP3 ORF and did not correlate with secondary structural elements, suggesting frame enrichment was unbiased. The frame-selected pool of VP3-mCherry gene fusions was subcloned into an AAV2 expression vector, yielding the final enriched, modular library pAAV2_RaPID (Figure 1b).
Collectively, our results show that we have generated a highly diverse combinatorial library with a modular linker sequence randomly inserted throughout the VP3 region of the cap gene. In contrast to transposon-based libraries, the pAAV2_RaPID library contains additional diversity through deletions and duplications of varying sizes, which may be useful in identifying additional phenotypic functionality.27 Simple and efficient subcloning of alternate inserts can be achieved through the use of the directional NgoM IV and Kas I sites. Furthermore, the frame-enriched pAAV2_RaPID library increases the number of in-frame variants that can be included in phenotypic selection schemes that are limited in population size. Ultimately, this may lead to more effective directed evolution experiments when using random domain insertion in AAV.
Selection and high-throughput screening for fluorescent AAV2-mCherry hybrid
As a demonstration of the utility of the pAAV2_RaPID library, we sought to identify sites in the VP3 domain of the AAV2 capsid protein that would tolerate insertion of a large protein domain but retain the ability to form infectious capsids. mCherry was chosen as a model insertion because of its comparatively large size (26.7 kDa, ~45% the size of VP3), proximity of its N- and C-termini,32 potential utility in virus-trafficking studies,18,19,33 and it can be selected via fluorescence-activated cell sorting (FACS).
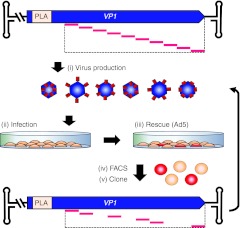
We reasoned that the probability of finding successfully assembled, infectious virions carrying the mCherry protein would be inversely proportional to the copy number of mCherry per capsid. Therefore, we used a capsid mosaic approach34 in which the pAAV2_RaPID gene library was used to produce a pool of AAV2 hybrids with mCherry randomly inserted in the VP3 region of VP1 only. To achieve this, VP2/VP3 expression was ablated from the pAAV2_RaPID library by knocking out the major splice acceptor site of the cap gene.35 Thus, during virus production, only VP1-mCherry hybrids are expressed from the pAAV2_RaPID library. Wt VP2 and VP3 proteins were expressed in trans to allow complete assembly of the mosaic virions with all three VP proteins (VP1-mCherry, VP2, VP3). To facilitate Darwinian evolution during selection, the AAV2 inverted terminal repeat (ITR) sequences were placed in cis with RaPID genomes containing the VP1-mCherry gene library (Figure 2), but left absent from the wt VP2/VP3 genes. Therefore, VP1-mCherry containing capsids will package their respective hybrid genomes, resulting in a phenotype (infectivity and fluorescence) and genotype (mCherry insertion site within VP1) link.
Figure 2.
Selection for fluorescent VP1-mCherry hybrids. (i) The pAAV2_RaPID library was used to produce a pool of AAV2 hybrids with mCherry inserted in the VP3 region of VP1 only, which contains the PLA2 domain required for infectivity; wt VP2/3 genes were supplemented in trans. (ii) A monolayer of HEK-293T cells were infected with the virus pool. (iii) Successfully infectious variants were then rescued by superinfection with Ad5. (iv) Cells were harvested and sorted by FACS to isolate those expressing fluorescent VP1-mCherry proteins. (v) DNA was extracted from the sorted cells, amplified by PCR, and subcloned back into the VP1 expression vector, generating an enriched library gene pool. The selection procedure was iterated twice more using the enriched gene pools for a total of three rounds of selection. FACS, fluorescence-activated cell sorting.
A selection scheme was devised to isolate fluorescent AAV2-mCherry hybrids that retain infectivity in HEK-293T cells (referred to hereinafter as 293T cells) (Figure 2, see Methods); 293T cells were infected with the mosaic AAV2-mCherry virus library and coinfected with adenovirus to amplify successfully infectious AAV2-mCherry hybrids. Cells positive for mCherry fluorescence were then isolated via FACS.
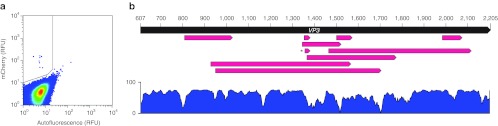
Infection by the naive virus pool yields a low percentage of mCherry-positive cells (<0.005%) as determined by FACS and epifluorescence microscopy (Figure 3a). Overall, 1,460 mCherry-positive cells were sorted in the first round. Interestingly, sampling of 10 clones after selection revealed a dominant wt population (data not shown). The minority mutant fraction, containing the randomly inserted mCherry ORF, was recovered by cloning an ampicillin resistance cassette into the upstream NgoM IV linker site, an restriction endonuclease site not present in wt genes (Supplementary Figure S1) followed by selection in E. coli. After enrichment, sampling of the gene pool returned 100% mutant sequences. Sequencing of 10 variants from the selected gene pool after round 1 identified two clones (r1c3, r1c10) with mCherry inserted into hypervariable region 5 (HVR5) of VP1.36 r1c3 contains mCherry in place of the deleted amino acid sequence 453-GTTTQSR (VP1 numbering), and r1c10 contains a similar deletion: 452-SGTTTQSR.
Figure 3.
Screening of fluorescent VP1-mCherry hybrids isolated after one round of selection. (a) FACS data from round 1 selection. Y-axis indicates the mCherry signal in relative fluorescence units (RFU). X-axis represents cell autofluorescence in the green spectrum. Gate in upper left quadrant defines the region where cells were collected during selection. (b) Clones recovered after the first round of selection are illustrated as in Figure 2. Each clone is represented by a red arrow that indicates the region of VP3 deleted at the site of mCherry insertion. Clone r1c3 is marked with an asterisk. A similarity plot is shown below, indicating the regions of VP3 that are conserved across AAV serotypes 1-12, generated from an alignment in Vector NTI. Bar at left indicates percent similarity. Note r1c3 lies in a wide valley of the similarity plot located at the GH2/3 loop. AAV, adeno-associated virus; FACS, fluorescence-activated cell sorting.
Subsequent rounds of selection resulted in an increased mCherry-positive frequency in FACS measurements (~0.04%). However, sequencing of selected RaPID variants indicated convergence on large deletions at sites of insertion in VP1, averaging 488 amino acids by the third round (data not shown). Thus, under the given selection pressures, the virus gene pool appears to have reached its optimum after the first round of selection.
Detection of mCherry in capsid of selected clone r1c3
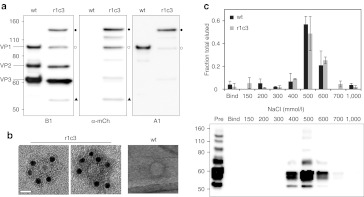
The two selected clones harboring mCherry within HVR5 of VP1 differ by only one deleted residue, so we focused on characterizing the properties of only one of these mutant clones, r1c3. Western blot analysis of the purified mutant indicates wt-like stoichiometric incorporation of the VP1-mCherry protein into the virus capsid (Figure 4a). Several additional protein bands are observed that stain positive for mCherry and VP-specific antibodies, suggesting either mild proteolysis or alternate translation, but the full-length VP1-mCherry protein appears to be the dominant VP1 species. To detect the presence of mCherry directly on assembled virus capsids, we used immunoelectron microscopy to obtain high resolution images of the AAV2-mCherry virions (Figure 4b). Immunostaining of r1c3 with a polyclonal mCherry antibody resulted in successful labeling of the capsid. Wt AAV2 capsid control samples were not labeled under identical conditions. These results provide two lines of direct evidence that mCherry is incorporated into the VP1 subunit and is displayed on the capsid surface of the r1c3 clone.
Figure 4.
Physical characterization of r1c3. (a) Western blot analysis of clone r1c3. Staining with B1 (left panel), which binds the C-terminus of VP1/VP2/VP3, shows a typical pattern for wt AAV2. In comparison, B1 staining shows normal VP2/VP3 expression for r1c3 with a higher molecular weight VP1-mCherry hybrid protein (closed circles), confirmed by mCherry (mCh) and A1 staining (middle and right panels, respectively). A fragment sized similarly to wt VP1 was also seen in r1c3 by B1 staining (open circles; left panel), but appears positive for mCherry staining (middle panel). In addition, r1c3 showed a lower molecular weight band (closed triangles) with B1 and mCherry staining that was negative for A1, likely a C-terminal proteolytic fragment. These and additional bands in the blots may indicate mild proteolysis of r1c3, but the full-length VP1-mCherry chimera appears to be the dominant VP1 species. (b) Immunoelectron microscopy of r1c3 demonstrates incorporation of the VP1-mCherry protein into the capsid. Transmission electron micrographs were taken after staining with a polyclonal mCherry antibody followed by a secondary antibody conjugated to gold nanoparticles (12 nm, black dots). Wt AAV2 is shown at right under identical staining conditions. Mutant r1c3 is shown as an empty (left panel) and full (middle panel) capsid, both containing VP1-mCherry chimeric proteins. Bar = 20 nm. (c) Affinity chromatography shows r1c3 retains affinity for heparin, the primary cell surface receptor for AAV2. qPCR analysis (top panel) of eluates with increasing NaCl show wt AAV2 (black bars) and r1c3 (gray bars) both elute in peak fractions near 500 mmol/l NaCl. Error bars indicate SD from three (wt) or two (r1c3) independent experiments. Western blot analysis (bottom panel) of samples run under non-reducing conditions, stained with B1, verifies presence of capsid subunits in elutions. AAV, adeno-associated virus; qPCR, quantitative PCR.
mCherry-capsid hybrids display wt-like infectivity and robust fluorescence
To establish how mCherry insertion influences the functional properties of AAV2, we characterized the binding and infectivity of the mosaic capsids built using the mCherry-VP1 hybrid r1c3. First, we evaluated the binding affinity of r1c3 for its primary cell surface receptor, HS proteoglycans, using heparin-sepharose column chromatography (Figure 4c). Affinity of AAV2 for heparin is correlated with the salt concentration required to disrupt ionic interactions, thus eluting virions from the heparin column. Our results indicate the mutant retains wt-like affinity for heparin, since both samples display similar elution profiles with peak fractions near 500 mmol/l NaCl.
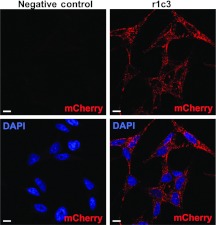
To determine whether mutant r1c3 particles are able to bind cells and become internalized, the mosaic virus was added to a monolayer of HeLa cells (Figure 5). Confocal microscopic examination reveals a punctate mCherry signal throughout the cytoplasm, indicating successful internalization and fluorescent properties of the VP1-mCherry hybrid capsid.
Figure 5.
Mosaic AAV2-mCherry mutant r1c3 is fluorescent and internalizes into HeLa cells. HeLa cells were infected at a MOI of 7 × 105 vg/cell for 6 hours at 37 °C, fixed and stained with DAPI. The negative control (left column) is cells only. Confocal imaging reveals a punctate signal in the mCherry emission spectra (red) throughout the cytoplasm. DAPI (blue) marks cell nuclei. Bar = 10 µmol/l. AAV, adeno-associated virus, DAPI, 4′,6-diamidino-2-phenylindole; MOI, multiplicity of infection; vg, vector genome.
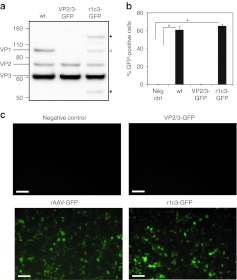
To compare the infectivity of mosaic capsids containing the AAV2-mCherry hybrid with native AAV2 capsids, the r1c3 capsid was packaged with a cytomegalovirus-green fluorescent protein (GFP) transgene reporter and applied to HeLa cells, followed by flow cytometric analysis. As a control, virions assembled from VP2 and VP3 only were packaged with the identical GFP reporter (VP2/3-GFP) and applied at the same multiplicity of infection (MOI). As expected, negligible fluorescence was detected by flow cytometry for the VP2/3-GFP virions (Figure 6; Supplementary Figure S2). Remarkably, the r1c3 VP1-mCherry hybrid fully restored infectivity of the VP2/3 particles to levels comparable to wt. Collectively, our results indicate the r1c3 mutant maintains wt-like affinity for heparin, is internalized into cells in vitro, and displays robust mCherry fluorescence with no loss of infectivity.
Figure 6.
The r1c3 mutant transduces HeLa cells with wt efficiency. (a) Western blot analysis shows the expression of VPs in mosaic r1c3 produced with a 1:1 molar ratio of VP1-mCherry:VP2/VP3 helper plasmids. VP2/3 capsids packaged with a GFP transgene show no expression of VP1. (b,c) HeLa cells were transduced at MOI = 2,000 and analyzed by flow cytometry after 36 hours. Transduction is visualized by epifluorescence microscopy (in c) and quantified by flow cytometry (in b). The incorporation of VP1-mCherry in r1c3-GFP completely recovers infectivity compared with wt, while VP2/3 only virions display negligible transduction. Each transduction was performed in triplicate (n = 3), with error bars indicating SD. *P value <0.001. Bar = 10 µmol/l. GFP, green fluorescent protein; MOI, multiplicity of infection.
Structural modeling of mCherry-capsid hybrids
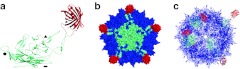
To visualize the theoretical structure of the mCherry-capsid hybrid, molecular modeling was performed (Figure 7). Based on modeling analysis and prior studies of the GH2/3 loop,9,21,37 the structural effects of insertion into HVR5 (surface loop IV) on the capsid should be tolerated.38 This loop is on the surface of the capsid at the tips of the threefold protrusions that extend from the surface of the AAV capsid. Interestingly, the positioning of mCherry on the AAV2 capsid in the r1c3 model containing randomly placed mCherry-cap hybrid subunits (Figure 7c) bears a remarkable resemblance to the immunoelectron micrographs of this mutant (Figure 4b).
Figure 7.
Molecular model of AAV2-mCherry hybrid r1c3. (a) VP3 (green) with inserted mCherry (red) as a monomer; symmetry axes are labeled as triangle (threefold), pentagon (fivefold), and oval (twofold). (b) A special pentamer in the context of the capsid (blue) showing the VP3-mCherry subunit in green-red. (c) A ribbon diagram showing five VP1-mCherry monomers randomly placed in the context of the capsid (blue). AAV, adeno-associated virus.
Discussion
We have built a modular, diverse AAV2 capsid gene platform library, named pAAV2_RaPID. Preselection of pAAV2_RaPID for in-frame insertions was performed to allow for more efficient screening of functional variants (see Supplementary Discussion). This may be critical in future applications where additional sequence complexity is desired in the random insertion library. For example, randomized linkers connecting cap and heterologous sequences may improve tolerance of difficult insertions or facilitate allosteric regulation of viral behavior by inserted domains.27 Using this library, a DNA sequence encoding a desired peptide can be quickly and efficiently inserted randomly throughout the AAV2 cap gene. Selection can then be performed to identify insertion location and deletion/duplication combinations that are optimal for the proper functioning of the desired peptide.
Using infective selection on 293T cells and FACS, we successfully isolated a novel mutant, r1c3, containing a functional mCherry protein genetically inserted into the VP3 domain of the capsid protein VP1. All previous reports indicate that genetic incorporation of large proteins, including GFP, into the AAV2 capsid interferes with infectivity significantly.8,18,19 Thus, the preservation of native infectivity in r1c3 is a substantial finding. Since AAV-trafficking patterns have been shown to be vector dose-dependent,39 the use of improved fluorophore-tagged AAV vectors such as r1c3 may help identify infection pathways at lower MOI, which may be more clinically relevant. Furthermore, encoding of large protein domains in general in the GH2/3 loop may offer improved vector performance such as in luciferase-displaying AAV vectors useful for in vivo tracking applications.40 Although other approaches to generate labeled AAV capsids have been demonstrated, genetic incorporation offers several advantages, such as ease of production, reduced cost, and higher purity.19
The library selection scheme used in this work takes advantage of a mosaic capsid approach in order to accommodate the large mCherry protein on the capsid while maintaining genome packaging and infectivity. VP1 was selected to harbor the randomly inserted mCherry throughout the VP3-shared region, since it is required for infectivity (Figure 6b).41 With the emergence of large deletions in the first round of selection (Figure 3b) and convergence in later rounds, the pressure to assemble genome-containing VP1-mCherry hybrids may have been counteracted by the requirement to maintain the genome packaging limit of AAV2 (~5.2 kb).42 Recovery of oversized clones such as r1c3 may have been due to the packaging of partial genomes containing the full-length cap gene.42 Isolation of viral clones with large genome deletions suggests the inclusion of wt VP2/VP3 in trans may have been sufficient to rescue defunct library genes during production, as VP2/VP3 alone are capable of assembly and packaging and have marginal infectivity (Figure 6a).41 Although this directed evolution study was successful at identifying a functional mutant, r1c3, the selection scheme used here may be improved in future studies by reduction of the total AAV2 genome size. For example, this may be achieved by deleting portions of rep in library genomes and delivering full-length rep copies in trans during replication, potentially helping to balance the selective pressures in favor of functional VP1 hybrids. Furthermore, domination of the library population by wt cap genes after the first round of selection was likely due to the advantage in reproductive fitness of wt over the average library member. Although recombination with VP2/3 plasmids cannot be ruled out, it is unlikely this plays a dominant role since later rounds of selection did not return any wt cap genes.
Despite the occurrence of off-target selective pressures, two successful clones, r1c3 and r1c10, were isolated after just one round of selection. Mutant r1c3 was chosen for further analysis because the insertion was located in a region of low-sequence homology with serotypes 1-12, known as HVR5,36 located in the GH2/3 loop.43 Though this region is known to tolerate mutation,21,44 insertion of a large protein domain has not been demonstrated previously. To our knowledge, the largest protein successfully inserted into any VP3-shared crystallized region of the AAV2 capsid protein has been limited to the 34 amino acid Z34C immunoglobulin-binding domain inserted into the GH12/13 loop.28 Interestingly, the packaging efficiency of this mutant was improved an order of magnitude by deleting 9 amino acids (581-589) at the site of insertion. This previous result corroborates the utility of our DNase-based random insertion library method as demonstrated here, where deletions of various sizes normally accompany insertions and can improve tolerance to heterologous domains. Later work from Gigout et al.45 demonstrated a mosaic approach to incorporate the Z34C peptide with improved packaging efficiency. In our work, we confirm the utility of mosaic capsid approaches in combination with random insertion by inserting a significantly larger protein by almost an order of magnitude: mCherry (240 aa) versus Z34C (34 aa). Taken together, our pAAV2_RaPID library approach identified the GH2/3 loop of the AAV2 capsid as a region tolerant to protein domains of size in magnitude not previously demonstrated.
Binding of HS proteoglycans is an important event in the life cycle of AAV2, as it has been shown to induce conformational changes in the capsid structure46 and is the primary cell-binding step during infection.4 Although the main HS-binding region on the capsid sits in the valley between the threefold spikes,5 directly below the inserted mCherry protein, r1c3 retains high affinity for heparin (Figure 4c). This result is consistent with the length and flexibility of HS chains, facilitating access to capsid-binding sites in the presence of potential sterically obstructive mCherry appendages. Alternatively, HS interactions with HS-binding domains on the virus capsid that are free of mCherry may be sufficient to achieve wt affinity, in agreement with the reported non-linearity of HS binding by mosaic capsids composed of varying ratios of capsid subunits from HS-positive and HS-negative serotypes.47 Furthermore, the folding of mCherry and its linking sequences may provide enough flexibility or steric access to allow unimpeded interactions with HS-binding sites, as supported by the model (Figure 7). Remarkably, r1c3 maintains full infectivity similar to wt AAV2 capsids. This result suggests that the AAV2-mCherry hybrid may also maintain wt interactions with its coreceptors and follow a similar intracellular pathway, although we cannot rule out at this time that r1c3 may follow an alternative intracellular pathway with similar efficiency. Overall, the retention of wt-like heparin binding and infectivity by the VP1-mCherry hybrid virus is a clear demonstration of the potential utility of our pAAV2_RaPID library for constructing virus nanoparticles with large protein VP hybrids.
In summary, we have constructed a modular, diverse, and enriched random insertion library, pAAV2_RaPID. The library can be used for the facile insertion of desired peptide domains randomly throughout the AAV2 capsid gene, followed by selection/screening for optimal sites. We demonstrated the utility of the pAAV2_RaPID library by identifying a novel mCherry-AAV2 hybrid containing the largest reported protein insertion in the VP3 domain. Strikingly, in addition to producing high titers, the hybrid virus vector retains full infectivity and displays robust fluorescent properties. This fluorescently labeled AAV2 variant may prove useful in further elucidating mechanisms of viral infection and pave the way for the combination of imaging and therapy in AAV-mediated gene delivery. Furthermore, the successful insertion of mCherry into the surface of the AAV2 capsid without affecting virus infectivity has important implications for future vector design with other protein domains once thought too large to incorporate into AAV.
Materials and Methods
Plasmids. pRepCap was created by subcloning the AAV genome (without ITRs) from pSub201 into a pBlueScript SK II (+) backbone using Xba I. The plasmid vector was then subjected to site-directed mutagenesis to remove existing Kas I, NgoM IV, Nco I, and BfuA I (RaPID restriction sites) restriction sites with silent mutations. The VP3 start codon was mutagenized to produce a Nco I site and a BfuA I site was inserted just downstream of the stop codon, yielding pRepCap_RR (RR: RaPIDready). The modified genome was then subcloned into pSub201ΔNgoMIV (pSub201 with the NgoM IV site removed from the vector backbone) using Xba I sites to regain the ITRs, yielding pSub201_RR. To create the pAAV2_RR expression vector, the entire modified genome (with ITRs) was then cut from pSub201_RR using Pvu II, and ligated to the pET28a(+) backbone between the Pvu II and Xmn I sites. To generate pAAV2_RRΔcap, pSub201_RR was digested with BsiW I and Xcm I, blunted with T4 DNA polymerase and religated. The partially deleted cap gene was then subcloned into pAAV2_RR using Nco I and BfuA I sites. pMIN_VP3 was created by excising VP3 from pRepCap_RR using Nco I and BfuA I sites, and ligating to a minimized plasmid backbone generated by sequence overlap extension48 using pBAD24 as a template. pInSALect_RR was created by removal of RaPID restriction sites from pInSALect, followed by insertion of Nco I and BfuA I sites between the intein sequences. Mosaic expression vectors were constructed as previously described. Briefly, pAAV2_RR was mutagenized to knock out the major splice acceptor site downstream of the VP1 start codon, giving VP1-only expression vector pAAV2_VP1_RR. For wt VP2/VP3 expression, pRC_RR was mutagenized to knock out the VP1 start codon, yielding pRC_RR_VP2/3.
RaPID library construction. pMIN_VP3 was digested with dilute DNase I (New England Biolabs, Ipswich, MA) in 1 mmol/l MnCl2, resulting in a pool of randomly linearized plasmids. This pool was resolved by agarose gel electrophoresis with a restriction endonuclease-linearized control to identify the near full-length linearized fraction, which was excised and purified using a Zymoclean Gel DNA recovery kit (Zymo Research, Irvine, CA). The randomly linearized pool was then blunted with T4 DNA polymerase and dephosphorylated with T4 Polynucleotide Kinase (NEB, Ipswitch, MA). The mCherry ORF was amplified by sequence overlap extension48 to remove Nco I and Kas I sites and to add an NgoM IV site upstream and a Kas I site downstream of the coding sequence. Pfu DNA polymerase was used for mCherry amplification to generate blunt ends, allowing ligation to the linearized pool of cap genes. Transformation into DH10β E. coli (Life Technologies, Grand island, NY) was followed by amplification on agar plates, and purification using a Zyppy Plasmid Miniprep Kit (Zymo Research, Irvine, CA). The resulting plasmid library was then size-selected by agarose gel electrophoresis to enrich the single insert-containing fraction (Supplementary Figure S1). The size-selected naive pool was then subcloned into pInSALect_RR using Nco I and BfuA I restriction sites, followed by three rounds of selection on agar containing carbenicillin (100 µg/ml) and chloramphenicol (50 µg/ml). The frame-selected plasmid pool was purified and the mutant cap genes were subcloned into pAAV2_RRΔcap using Nco I and BfuA I sites to create pAAV2_RaPID. The VP1 only library, pAAV2_VP1_RaPID was created by cloning the VP3 library from pAAV2_RaPID into VP1-only expression vector pAAV2_VP1_RRΔcap using Nco I and BfuA I.
Virus production. The AAV2_RaPID_mCherry virus libraries were generated by linear polyethyleneimine-mediated (molecular weight 25 kD) triple transfection of pAAV2_VP1_RaPID_mCherry (10 ng), pRC_RR_VP2/3 (5 µg), and pXX6 (25 µg) into each of 10 × 15 cm poly-L-lysine–coated plates of 70% confluent 293T. The cells were harvested 46–48 hours post-transfection, lysed by three cycles of freeze/thaw in 1% protease inhibitor cocktail (Sigma-Aldrich, St Louis, MO) and treated with 4 U/ml Benzonase (Sigma-Aldrich) for 30 minutes. The lysate was then clarified by centrifugation and loaded onto an iodixanol step gradient (15/25/40/54%) in Beckman Ultra-Clear QuickSeal Tubes (Beckman Coulter, Brea, CA). The lysate was resolved by centrifugation at 48,000 rpm for 1.75 hours in a 70Ti rotor, and the purified virus library extracted from the 40% layer. The genomic titer was determined by qPCR using SYBR green (Life Technologies) and primers against rep (forward: AGGACCAGGCCTCATACATCTC, reverse: TGTCC AAGGCAGCCTTGATT) on a BIO-RAD CFX96. Clonal production of r1c3 packaged with its genome for transmission electron microscopy studies was performed similarly using pAAV2_VP1_RaPID_r1c3 (2 pmol), pRC_RR_VP2/3 (2 pmol), and XX6 (2 pmol). For confocal and transduction studies, r1c3 was packaged with a GFP transgene. Its genome was cloned into pBlueScript SK II (+) using Xba I to remove ITRs, generating pRC_RR_VP1_r1c3; 293T cells were then transfected as above with pRC_RR_VP1_r1c3 (1 pmol), pRC_RR_VP2/3 (1 pmol), pAAV_GFP (containing a cytomegalovirus-GFP reporter flanked by AAV2 ITRs) (1 pmol), and XX6 (1.65 pmol) and purified as above. Virus titers were quantified via qPCR using primers against the cytomegalovirus promoter in the GFP transgene cassette (forward: TCACGGGGATTTCCAA GTCTC, reverse: AATGGGGCGGAGTTGTTACGAC).
Virus selection. The virus library was applied to 70% confluent 293T cells at a MOI of ~10 viral genomes per cell. Four hours after infection, the library was rescued with adenovirus serotype 5 at an MOI of ~10 IU/cell; 48 hours post-infection, the cells were harvested by trypsinization, pelleted and resuspended in phosphate-buffered saline (PBS) (5 mmol/l EDTA). The cells were then filtered through a 40 µmol/l cell strainer (BD, Franklin Lakes, NJ) and sorted using a DakoMoFlo cell sorter. mCherry-positive cells were lysed by three cycles of freeze/thaw and purified using a Zyppy Plasmid Miniprep Kit after adding 1 µg of salmon sperm DNA as a carrier. Cap genes were amplified by PCR using a Roche Expand Hi-Fidelity PCR kit (Roche, Indianapolis, IN) and subcloned into pAAV2_VP1_RRΔcap using Nco I and BfuA I.
Western blot. Virus samples were resolved in 4–12% or 12% Bis-TrisNuPAGE gels (Life Technologies) and transferred to nitrocellulose (GE Healthcare, Pittsburgh, PA) at 40 V for 90 minutes, otherwise following manufacturer guidelines. Gels were run under reducing conditions, except for the heparin fractions (Figure 4). Blocking was performed in 5% skim milk in PBS with 0.1% Tween-20 (PBS-T) for 1 hour while rocking. To wash, blots were rinsed three times and rocked for 20 minutes in PBS-T. Primary antibodies were applied to blots overnight at 4 °C in PBS with 3% BSA (3% BSA-PBS) at the following dilutions. A1 (monoclonal mouse anti-VP1 from American Research Products, Belmont, MA) was diluted 1:200; B1 (monoclonal mouse anti-VP1, 2, 3 from American Research Products) diluted 1:50; and mCherry (polyclonal rabbit from BioVision, Milpitas, CA) diluted 1:100. After washing, goat anti-mouse (Jackson ImmunoResearch, West Grove, PA) or rat anti-rabbit (American Research Products) peroxidase-conjugated secondary antibodies were applied at a 1:2,000 dilution in 5% skim milk in PBS-T for 1 hour. Blots were then washed four times for 15 minutes with PBS-T while rocking. Imaging was performed on a Fujifilm LAS 4000 after applying Lumi-Light western blotting substrate (Roche).
Immunoelectron microscopy. Purified virus samples were applied to charged carbon grids (Ted Pella, Redding, CA) for 5 minutes; wt was adsorbed to a 300 mesh grid with lacey carbon type-A (product no. 01890-F), while r1c3 was applied to a 300 mesh grid with continuous carbon (product no. 01753-F). Grids were then blocked for 45 minutes with 3% BSA-PBS, washed in 3 drops of PBS and stained with the polyclonal anti-mCherry antibody (see above) for 1 hour. After washing, gold nanoparticle (12 nm) conjugated goat anti-rabbit secondary antibodies (Jackson ImmunoResearch) were applied at a 1:30 dilution in 3% BSA-PBS for 1 hour to visualize mCherry proteins attached to the virions. Samples were then washed and negative stained with 0.75% uranylformate to highlight the viral capsids before imaging on a JEOL 2010 transmission electron microscope operating at 120 kV (JEOL, Tokyo, Japan).
Heparin chromatography. Virus samples were applied to a HiTrap heparin-sepharosecolumn (GE Healthcare) equilibrated with 50 mmol/l Tris-Cl, pH 7.6, 10 mmol/l MgCl2, and 150 mmol/l NaCl. Fractionation was performed by passing increasing concentrations of NaCl in 1 ml increments and collecting the flow-through. Fractions were quantified by qPCR or analyzed by western blot as above.
Confocal microscopy. HeLa cells were seeded onto poly-L-lysine–coated glass coverslips in a 24-well tissue culture plate at a density of 6 × 104 cells per well in phenol-red free Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum and 1% penicillin-streptomycin. The following day, r1c3-GFP was applied in serum-free media for 6 hours at 37 °C, 5% CO2. Cells were then washed twice with PBS, fixed in 4% paraformaldehyde for 15 minutes, washed twice and stained with DAPI (1 µg/ml) for 15 minutes. After washing twice more in PBS, cells were mounted onto glass slides in 5 µl of Fluoromount-G (SouthernBiotech, Birmingham, AL). Samples were allowed to dry at 4 °C overnight before examination on a Zeiss LSM 710. Images were processed using Zen 2010 software (Carl Zeiss MicroImaging, Jena, Germany).
Transduction assay. HeLa cells were seeded at 1 × 105 cells per well in a PLL-coated 24-well tissue culture plate and incubated for 18 hours at 37 °, 5% CO2. Virus samples were diluted in serum-free DMEM and added to cells after removing serum-containing media. Cells were incubated with virus for 4 hours at 37 °C, followed by replacement of media to serum-containing DMEM. Cells were incubated for another 36 hours and harvested by trypsin digestion, followed by neutralization with serum-containing DMEM. Cells were washed once with PBS (5 mmol/l EDTA), passed through a 40 µmol/l cell strainer and analyzed on a FACSCanto II flow cytometer (BD Biosciences, San Jose, CA). Data was processed using FlowJo software (TreeStar, Ashland, OR). Epifluorescent images were taken using a Canon EOS Rebel XS camera adapted to a Zeiss Axiovert 40 CFL (Carl Zeiss MicroImaging).
Molecular modeling. The r1c3 capsid model was generated in SWISS MODEL using the crystal structure AAV2 (PDB# 1LP3) supplied as a template (Model I) and using the AAV1 (PDB# 3NG9), mCherry (PDB# 2H5Q), and red fluorescent protein (PDB# 3NEZ) when a template was not supplied (Model II). Following evaluation of the models in COOT, Model II was selected for further analysis because the two components of the chimera–the AAV2 VP3 and mCherry structures–were consistent with this model (Figure 7a). To depict the mCherry insertion, which is in VP1 only, in the context of the capsid, two options were used. One capsid model was generated with a unique AAV2-mCherry pentamer (Figure 7b), and the other had five AAV2-mCherry monomers randomly assembled on an AAV2 capsid (Figure 7c).
SUPPLEMENTARY MATERIAL Figure S1. Removal of wt genes from pAAV2_RaPID library. Figure S2. Raw flow cytometry data from Figure 6. Discussion.
Acknowledgments
We would like to acknowledge the University of North Carolina at Chapel Hill Gene Therapy Center Vector Core for providing us with pSub201, Stefan Lutz (Emory University) for providing us with pInSALect and Roger Tsien (University of California, San Diego) for providing the mCherry plasmid. This material is based upon work supported by the National Science Foundation under grant no. 0955536 and UTMDACC SPORE in Ovarian Cancer P50-CA083639 to J.S., Institute of Biosciences and Bioengineering Hamill Innovations Award to J.S. and J.J.S., Robert A. Welch Foundation (C-1614) to J.J.S., National Institutes of Health under grants P01-HL59412 and R01-GM082946 to M.A.-M., and National Institutes of Health Biotechnology Training Program T32 GM008362 to J.J. The authors declared no conflict of interest.
Supplementary Material
Removal of wt genes from pAAV2_RaPID library.
Raw flow cytometry data from Figure 6.
References
- Kotin RM, Siniscalco M, Samulski RJ, Zhu XD, Hunter L, Laughlin CA.et al. (1990Site-specific integration by adeno-associated virus Proc Natl Acad Sci USA 872211–2215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samulski RJ, Zhu X, Xiao X, Brook JD, Housman DE, Epstein N.et al. (1991Targeted integration of adeno-associated virus (AAV) into human chromosome 19 EMBO J 103941–3950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonntag F, Schmidt K., and, Kleinschmidt JA. A viral assembly factor promotes AAV2 capsid formation in the nucleolus. Proc Natl Acad Sci USA. 2010;107:10220–10225. doi: 10.1073/pnas.1001673107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Summerford C., and, Samulski RJ. Membrane-associated heparan sulfate proteoglycan is a receptor for adeno-associated virus type 2 virions. J Virol. 1998;72:1438–1445. doi: 10.1128/jvi.72.2.1438-1445.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qing K, Mah C, Hansen J, Zhou S, Dwarki V., and, Srivastava A. Human fibroblast growth factor receptor 1 is a co-receptor for infection by adeno-associated virus 2. Nat Med. 1999;5:71–77. doi: 10.1038/4758. [DOI] [PubMed] [Google Scholar]
- Kaplitt MG, Leone P, Samulski RJ, Xiao X, Pfaff DW, O'Malley KL.et al. (1994Long-term gene expression and phenotypic correction using adeno-associated virus vectors in the mammalian brain Nat Genet 8148–154. [DOI] [PubMed] [Google Scholar]
- Zaiss AK, Liu Q, Bowen GP, Wong NC, Bartlett JS., and, Muruve DA. Differential activation of innate immune responses by adenovirus and adeno-associated virus vectors. J Virol. 2002;76:4580–4590. doi: 10.1128/JVI.76.9.4580-4590.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Q, Mamounas M, Yu G, Kennedy S, Leaker B, Merson J.et al. (1998Development of novel cell surface CD34-targeted recombinant adeno-associated virus vectors for gene therapy Hum Gene Ther 91929–1937. [DOI] [PubMed] [Google Scholar]
- Girod A, Ried M, Wobus C, Lahm H, Leike K, Kleinschmidt J.et al. (1999Genetic capsid modifications allow efficient re-targeting of adeno-associated virus type 2 Nat Med 51438. [DOI] [PubMed] [Google Scholar]
- Müller OJ, Kaul F, Weitzman MD, Pasqualini R, Arap W, Kleinschmidt JA.et al. (2003Random peptide libraries displayed on adeno-associated virus to select for targeted gene therapy vectors Nat Biotechnol 211040–1046. [DOI] [PubMed] [Google Scholar]
- Seisenberger G, Ried MU, Endress T, Büning H, Hallek M., and, Bräuchle C. Real-time single-molecule imaging of the infection pathway of an adeno-associated virus. Science. 2001;294:1929–1932. doi: 10.1126/science.1064103. [DOI] [PubMed] [Google Scholar]
- Le HT, Yu QC, Wilson JM., and, Croyle MA. Utility of PEGylated recombinant adeno-associated viruses for gene transfer. J Control Release. 2005;108:161–177. doi: 10.1016/j.jconrel.2005.07.019. [DOI] [PubMed] [Google Scholar]
- Lee GK, Maheshri N, Kaspar B., and, Schaffer DV. PEG conjugation moderately protects adeno-associated viral vectors against antibody neutralization. Biotechnol Bioeng. 2005;92:24–34. doi: 10.1002/bit.20562. [DOI] [PubMed] [Google Scholar]
- Maheshri N, Koerber JT, Kaspar BK., and, Schaffer DV. Directed evolution of adeno-associated virus yields enhanced gene delivery vectors. Nat Biotechnol. 2006;24:198–204. doi: 10.1038/nbt1182. [DOI] [PubMed] [Google Scholar]
- Joo KI, Fang Y, Liu Y, Xiao L, Gu Z, Tai A.et al. (2011Enhanced real-time monitoring of adeno-associated virus trafficking by virus-quantum dot conjugates ACS Nano 53523–3535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei F, McConnell KI, Yu TK., and, Suh J. Conjugation of paclitaxel on adeno-associated virus (AAV) nanoparticles for co-delivery of genes and drugs. Eur J Pharm Sci. 2012;46:167–172. doi: 10.1016/j.ejps.2012.02.022. [DOI] [PubMed] [Google Scholar]
- Bartlett JS, Kleinschmidt J, Boucher RC., and, Samulski RJ. Targeted adeno-associated virus vector transduction of nonpermissive cells mediated by a bispecific F(ab'gamma)2 antibody. Nat Biotechnol. 1999;17:181–186. doi: 10.1038/6185. [DOI] [PubMed] [Google Scholar]
- Warrington KH, Jr, Gorbatyuk OS, Harrison JK, Opie SR, Zolotukhin S., and, Muzyczka N. Adeno-associated virus type 2 VP2 capsid protein is nonessential and can tolerate large peptide insertions at its N terminus. J Virol. 2004;78:6595–6609. doi: 10.1128/JVI.78.12.6595-6609.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lux K, Goerlitz N, Schlemminger S, Perabo L, Goldnau D, Endell J.et al. (2005Green fluorescent protein-tagged adeno-associated virus particles allow the study of cytosolic and nuclear trafficking J Virol 7911776–11787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabinowitz JE, Xiao W., and, Samulski RJ. Insertional mutagenesis of AAV2 capsid and the production of recombinant virus. Virology. 1999;265:274–285. doi: 10.1006/viro.1999.0045. [DOI] [PubMed] [Google Scholar]
- Wu P, Xiao W, Conlon T, Hughes J, Agbandje-McKenna M, Ferkol T.et al. (2000Mutational analysis of the adeno-associated virus type 2 (AAV2) capsid gene and construction of AAV2 vectors with altered tropism J Virol 748635–8647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kronenberg S, Böttcher B, von der Lieth CW, Bleker S., and, Kleinschmidt JA. A conformational change in the adeno-associated virus type 2 capsid leads to the exposure of hidden VP1 N termini. J Virol. 2005;79:5296–5303. doi: 10.1128/JVI.79.9.5296-5303.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perabo L, Büning H, Kofler DM, Ried MU, Girod A, Wendtner CM.et al. (2003In vitro selection of viral vectors with modified tropism: the adeno-associated virus display Mol Ther 8151–157. [DOI] [PubMed] [Google Scholar]
- Li W, Asokan A, Wu Z, Van Dyke T, DiPrimio N, Johnson JS.et al. (2008Engineering and selection of shuffled AAV genomes: a new strategy for producing targeted biological nanoparticles Mol Ther 161252–1260. [DOI] [PubMed] [Google Scholar]
- Koerber JT., and, Schaffer DV. Transposon-based mutagenesis generates diverse adeno-associated viral libraries with novel gene delivery properties. Methods Mol Biol. 2008;434:161–170. doi: 10.1007/978-1-60327-248-3_10. [DOI] [PubMed] [Google Scholar]
- Koerber JT, Jang JH, Yu JH, Kane RS., and, Schaffer DV. Engineering adeno-associated virus for one-step purification via immobilized metal affinity chromatography. Hum Gene Ther. 2007;18:367–378. doi: 10.1089/hum.2006.139. [DOI] [PubMed] [Google Scholar]
- Guntas G., and, Ostermeier M. Creation of an allosteric enzyme by domain insertion. J Mol Biol. 2004;336:263–273. doi: 10.1016/j.jmb.2003.12.016. [DOI] [PubMed] [Google Scholar]
- Ried MU, Girod A, Leike K, Büning H., and, Hallek M. Adeno-associated virus capsids displaying immunoglobulin-binding domains permit antibody-mediated vector retargeting to specific cell surface receptors. J Virol. 2002;76:4559–4566. doi: 10.1128/JVI.76.9.4559-4566.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hida K, Won SY, Di Pasquale G, Hanes J, Chiorini JA., and, Ostermeier M. Sites in the AAV5 capsid tolerant to deletions and tandem duplications. Arch Biochem Biophys. 2010;496:1–8. doi: 10.1016/j.abb.2010.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaner NC, Campbell RE, Steinbach PA, Giepmans BN, Palmer AE., and, Tsien RY. Improved monomeric red, orange and yellow fluorescent proteins derived from Discosoma sp. red fluorescent protein. Nat Biotechnol. 2004;22:1567–1572. doi: 10.1038/nbt1037. [DOI] [PubMed] [Google Scholar]
- Gerth ML, Patrick WM., and, Lutz S. A second-generation system for unbiased reading frame selection. Protein Eng Des Sel. 2004;17:595–602. doi: 10.1093/protein/gzh068. [DOI] [PubMed] [Google Scholar]
- Shu X, Shaner NC, Yarbrough CA, Tsien RY., and, Remington SJ. Novel chromophores and buried charges control color in mFruits. Biochemistry. 2006;45:9639–9647. doi: 10.1021/bi060773l. [DOI] [PubMed] [Google Scholar]
- Borovjagin AV, McNally LR, Wang M, Curiel DT, MacDougall MJ., and, Zinn KR. Noninvasive monitoring of mRFP1- and mCherry-labeled oncolytic adenoviruses in an orthotopic breast cancer model by spectral imaging. Mol Imaging. 2010;9:59–75. [PMC free article] [PubMed] [Google Scholar]
- Gigout L, Rebollo P, Clement N, Warrington KH, Jr, Muzyczka N, Linden RM.et al. (2005Altering AAV tropism with mosaic viral capsids Mol Ther 11856–865. [DOI] [PubMed] [Google Scholar]
- Muralidhar S, Becerra SP., and, Rose JA. Site-directed mutagenesis of adeno-associated virus type 2 structural protein initiation codons: effects on regulation of synthesis and biological activity. J Virol. 1994;68:170–176. doi: 10.1128/jvi.68.1.170-176.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao G, Alvira MR, Somanathan S, Lu Y, Vandenberghe LH, Rux JJ.et al. (2003Adeno-associated viruses undergo substantial evolution in primates during natural infections Proc Natl Acad Sci USA 1006081–6086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi W, Arnold GS., and, Bartlett JS. Insertional mutagenesis of the adeno-associated virus type 2 (AAV2) capsid gene and generation of AAV2 vectors targeted to alternative cell-surface receptors. Hum Gene Ther. 2001;12:1697–1711. doi: 10.1089/104303401750476212. [DOI] [PubMed] [Google Scholar]
- Govindasamy L, Padron E, McKenna R, Muzyczka N, Kaludov N, Chiorini JA.et al. (2006Structurally mapping the diverse phenotype of adeno-associated virus serotype 4 J Virol 8011556–11570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding W, Zhang LN, Yeaman C., and, Engelhardt JF. rAAV2 traffics through both the late and the recycling endosomes in a dose-dependent fashion. Mol Ther. 2006;13:671–682. doi: 10.1016/j.ymthe.2005.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asokan A, Johnson JS, Li C., and, Samulski RJ. Bioluminescent virion shells: new tools for quantitation of AAV vector dynamics in cells and live animals. Gene Ther. 2008;15:1618–1622. doi: 10.1038/gt.2008.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grieger JC, Johnson JS, Gurda-Whitaker B, Agbandje-McKenna M., and, Samulski RJ. Surface-exposed adeno-associated virus Vp1-NLS capsid fusion protein rescues infectivity of noninfectious wild-type Vp2/Vp3 and Vp3-only capsids but not that of fivefold pore mutant virions. J Virol. 2007;81:7833–7843. doi: 10.1128/JVI.00580-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Z, Yang H., and, Colosi P. Effect of genome size on AAV vector packaging. Mol Ther. 2010;18:80–86. doi: 10.1038/mt.2009.255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Q, Bu W, Bhatia S, Hare J, Somasundaram T, Azzi A.et al. (2002The atomic structure of adeno-associated virus (AAV-2), a vector for human gene therapy Proc Natl Acad Sci USA 9910405–10410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boucas J, Lux K, Huber A, Schievenbusch S, von Freyend MJ, Perabo L.et al. (2009Engineering adeno-associated virus serotype 2-based targeting vectors using a new insertion site-position 453-and single point mutations J Gene Med 111103–1113. [DOI] [PubMed] [Google Scholar]
- Gigout L, Rebollo P, Clement N, Warrington KH, Jr, Muzyczka N, Linden RM.et al. (2005Altering AAV tropism with mosaic viral capsids Mol Ther 11856–865. [DOI] [PubMed] [Google Scholar]
- Levy HC, Bowman VD, Govindasamy L, McKenna R, Nash K, Warrington K.et al. (2009Heparin binding induces conformational changes in Adeno-associated virus serotype 2 J Struct Biol 165146–156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabinowitz JE, Bowles DE, Faust SM, Ledford JG, Cunningham SE., and, Samulski RJ. Cross-dressing the virion: the transcapsidation of adeno-associated virus serotypes functionally defines subgroups. J Virol. 2004;78:4421–4432. doi: 10.1128/JVI.78.9.4421-4432.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho SN, Hunt HD, Horton RM, Pullen JK., and, Pease LR. Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene. 1989;77:51–59. doi: 10.1016/0378-1119(89)90358-2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Removal of wt genes from pAAV2_RaPID library.
Raw flow cytometry data from Figure 6.