Abstract
The inactive X chromosome’s (Xi) physical territory is microscopically devoid of transcriptional hallmarks and enriched in silencing-associated modifications. How these microscopic signatures relate to specific Xi sequence is unknown. Therefore, we profiled Xi gene expression and chromatin states at high resolution via allele-specific sequencing in mouse trophoblast stem cells. Most notably, X-inactivated transcription start sites harbored distinct epigenetic signatures relative to surrounding Xi DNA. These sites displayed H3-lysine27-trimethylation enrichment and DNaseI hypersensitivity, similar to autosomal Polycomb targets, yet excluded Pol II and other transcriptional hallmarks, similar to non-transcribed genes. CTCF bound X-inactivated and escaping genes, irrespective of measured chromatin boundaries. Escape from X-inactivation occurred within, and X-inactivation was maintained exterior to, the area encompassed by Xist in cells subject to imprinted and random X-inactivation. The data support a model whereby inactivation of specific regulatory elements, rather than a simple chromosome-wide separation from transcription machinery, governs gene silencing over the Xi.
INTRODUCTION
X-chromosome inactivation (XCI) equalizes X-linked gene dosage between mammalian sexes, resulting in transcriptional silencing of one of two female X chromosomes during early development. XCI is critical for mammalian development, and epigenetic processes required for XCI, most notably gene silencing mediated by Polycomb group proteins and non-coding RNA, play important roles in many biological phenomena (Surface et al., 2010). As such, XCI is a paradigm for epigenetic silencing mediated by non-coding RNA.
Two waves of XCI occur in the mouse. The first, imprinted XCI, initiates at the 8-cell stage of development and results in inactivation of the paternally inherited X-chromosome (Kalantry et al., 2009; Patrat et al., 2009). Imprinted XCI is maintained in extra-embryonic cells, while cells from the inner mass reactivate their paternal X during blastocyst maturation (Williams et al., 2011). XCI then reoccurs within the inner cell mass, randomly selecting the paternal or maternal X for silencing (Escamilla-Del-Arenal et al., 2011).
The inactive X chromosome (Xi) is distinguished from autosomes by several salient features. A ~17 kb non-coding RNA, Xist, is expressed from and coats the Xi in cis, and is required for the early maintenance of XCI (Kalantry et al., 2009; Namekawa et al., 2010). Xist coating results in widespread Xi deposition of H3-lysine27-trimethylation (H3K27me3), catalyzed by the Polycomb Repressive Complex 2 (PRC2). After coating, the Xi can be visualized microscopically with antibodies recognizing H3K27me3 or PRC2 components (Mak et al., 2002; Plath et al., 2003; Silva et al., 2003). PRC2 is required for maintenance of XCI during differentiation of extra-embryonic lineages (Kalantry et al., 2006; Wang et al., 2001), and acts redundantly with PRC1 to maintain XCI in the embryo (Schoeftner et al., 2006).
The Xi’s physical territory is microscopically devoid of transcription-associated hallmarks, including RNA polymerase II (Pol II), histone H3-acetylation, and histone H3-lysine4-methylation (H3K4me) (Escamilla-Del-Arenal et al., 2011). Exclusion of these marks from the Xi’s territory is Xist-dependent, and occurs during initiation of XCI. Movement of genes into the territory is coincident with silencing (Chaumeil et al., 2006). Whether relocation itself causes gene silencing, or is simply correlated with XCI, is unclear (Escamilla-Del-Arenal et al., 2011).
The content of X-linked DNA is additionally noteworthy. ~35% of human and mouse X-linked DNA is derived from LINE repeats, compared to ~20% of autosomal DNA (Fujita et al., 2010). Due to this enrichment, LINEs were thought to be conduits for Xist and associated silencing factors as they coat the Xi, although recent work argues a more indirect role for LINEs in this process (Tattermusch and Brockdorff, 2011). In this regard, LINEs have been proposed to nucleate formation of a transcriptionally silent spatial core within the Xi, into which X-linked genes are recruited as they are silenced (Chaumeil et al., 2006; Chow et al., 2010; Namekawa et al., 2010).
Finally, while the majority of X-linked genes are silenced by XCI, a minority escapes X-inactivation. The proportion and identity of escaping genes differs between cell types, and ranges from 3 to 15% of X-linked genes (Carrel and Willard, 2005; Patrat et al., 2009; Yang et al., 2010). Mechanistic models suggest escaping genes are positioned exterior to the Xi’s silent domain, in contrast to X-inactivated genes, allowing escapers to efficiently access transcription machinery (Chaumeil et al., 2006). The CTCF insulator protein may also play a critical role in licensing escape (Filippova et al., 2005).
Though well studied on a microscopic level, there is little quantitative information regarding Xi chromatin at sub-microscopic resolution. Understanding the epigenetic states of individual regulatory elements over the Xi is critical to a complete mechanistic understanding of XCI. Therefore, via a combination of allele-specific RNA-, ChIP-, FAIRE-, and DNase-Seq, we profiled X-linked chromatin patterns at the sub-microscopic scale in mouse trophoblast stem cells (TSCs), which are subject to imprinted XCI. The resulting gene expression and chromatin maps solidify TSCs as a platform for understanding the maintenance of XCI in a stem cell population. Our analysis revealed unexpected properties of X-inactivated and escaping genes, both in terms of their epigenetic signatures, as well as their sub-nuclear localization patterns in TSCs and also in cells subject to random XCI. Together, our results suggest a model whereby the major mechanism of transcriptional silencing associated with maintenance of XCI is not a simple chromosome-wide separation from transcription machinery but rather localized occlusion of Pol II from specific sites along the Xi.
RESULTS
Quantitative allele-specific expression map of the TSC Xi
In order to study XCI in a natural context, and still differentiate between the active X (Xa) and Xi when analyzing gene expression and chromatin patterns, we chose to study XCI in female TSCs, where inactivation patterns are non-random. Because TSCs maintain imprinted XCI, their Xa and Xi are maternally and paternally inherited, respectively, and SNP-overlapping sequence reads in these cells reliably trace chromosome of origin (Quinn et al., 2006).
As a prerequisite for understanding the relationship between Xi chromatin patterns and gene silencing, we began our study by measuring allelic gene expression via strand-specific RNA-Seq in female TSC lines derived from crosses between CAST/EiJ (Cast) and C57BL/6J (B6) mice. Considering our goal of relating expression to Xi chromatin patterns, and that the most accurate build of the genome is B6-derived, downstream chromatin analyses focused on a TSC line with a Cast Xa and B6 Xi (C/B). RNA-Seq was performed on an additional TSC line carrying a B6 Xa and Cast Xi (B/C), in order to help differentiate between strain- and parent-of-origin-specific expression biases.
The two female TSC lines were selected for RNA-Seq based on normal karyotypes and expression of TSC-specific markers (not shown). TSC poly-adenylated RNA was collected, and replicate cDNA libraries were prepared and sequenced ((Ingolia et al., 2009); Table S1). SNP-overlapping reads were identified, and allelic expression was calculated by dividing the number of B6- or Cast-overlapping reads by the total number of allelic reads per gene. The resulting expression maps served as a necessary baseline for the subsequent interpretation of X-linked chromatin patterns.
Internal consistency and experimental validation indicated RNA-Seq data to be of high quality. R2 values comparing gene expression between C/B and B/C replicates were 0.970 and 0.985, respectively; this same comparison between the two TSC lines gave an R2 of 0.917. Moreover, 9 genes were selected for validation via a quantitative allele-specific RT-PCR assay and in all cases measured allelic ratios and sequencing data were concordant (Figure S1).
We next determined what genes were subject to and escaped TSC XCI, examining genes with ≥20 SNP-overlapping reads in both TSC lines. Previous analyses used a cutoff of 10% Xi expression to differentiate between X-inactivated and escaping genes (Carrel and Willard, 2005; Yang et al., 2010). We sought to empirically define TSC escape, reasoning that a 10% cutoff might exclude true escapers. Allelic expression data were fit to a mixture of beta binomial distributions, allowing us to determine the probability of escape per gene (Supplemental Methods). For each gene, the proportion of Xi expression, the total number of escaping reads, and the sequence quality scores of SNP-overlapping bases within these reads, were accounted for in escape determination.
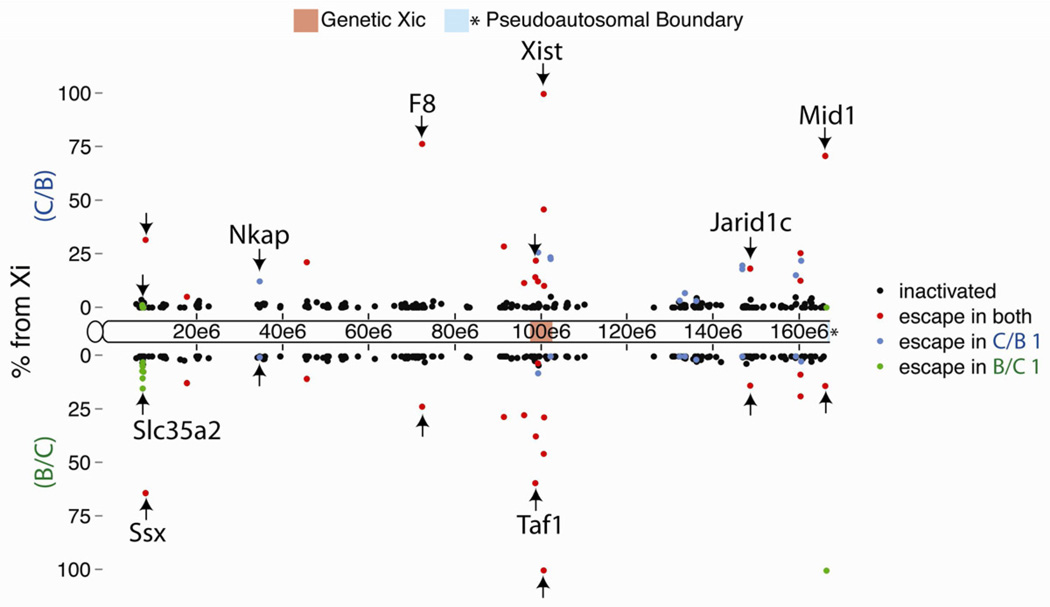
13% of genes (35 from 262 eligible) escaped XCI to varying degrees (Figure 1; Table S2). Escape did not occur in predictable patterns along the Xi, with genes escaping in isolation or in groups. With Xist as the major exception, escapers remained predominantly Xa expressed, with a median of ~15% expression from the Xi. 27 escapers exhibited at least 10% Xi expression. In contrast, X-inactivated genes had a median of ~0.5% Xi expression, with no correlation between inactivation strength and gene distance from Xist (R2 0.07, 0.04 for C/B, B/C; Figure 1).
Figure 1. Allele-specific expression map of the TSC Xi.
Dots denote X location and XCI status of evaluated TSC genes. Y-axis is % Xi expression in TSC lines C/B (positive scale) and B/C (negative scale). Arrows mark representative escapers.
See also Figure S1.
Genes that escaped XCI in TSCs also escaped in other tissues. We analyzed allelic expression via PCR in day 6.5 embryonic tissue and found the 4 genes tested escaped in the ectoplacental cone and extraembryonic ectoderm (not shown). Additionally, about ½ of escapers identified in mouse tissues subject to random XCI also escaped TSC XCI; 7 of 13 escapers from embryonic kidney cells (Yang et al., 2010), and 10 of 20 escapers from neural precursor cells (Splinter et al., 2011), escaped in TSCs. Finally, we noted that Xi genetic background significantly affected TSC escape profiles. About ½ of escapers did so in both TSC lines; however, 11 genes escaped only in C/B TSCs, and 8 only in B/C TSCs (blue and green dots, Figure 1). Allelic PCR in independently derived TSC lines verified that the majority of differential escape was heritable and not TSC-line specific (Figure S1).
High-resolution analysis of TSC Xi chromatin environments
Having established an allelic expression map of the TSC X-chromosome, we initiated our analysis of X-linked epigenetic patterns at high resolution, via ChIP-Seq in C/B TSCs. The overarching goal of these experiments was to characterize the sub-microscopic chromatin signatures over the TSC Xi, and quantitatively examine how these signatures related to gene expression, DNA features, and known microscopic properties of the chromosome (Escamilla-Del-Arenal et al., 2011). The survey initially centered on H3K27me3, which is required for XCI maintenance in extra-embryonic cells (Kalantry et al., 2006; Wang et al., 2001). Additional ChIPSeq datasets were generated for repression-associated histone H4-lysine20-monomethylation (H4K20me1), which also microscopically coats the TSC Xi, as well as marks associated with active transcription, such as Pol II, H3K4me2, and histone H3-lysine36-trimethylation (H3K36me3), and total histone H3 (H3).
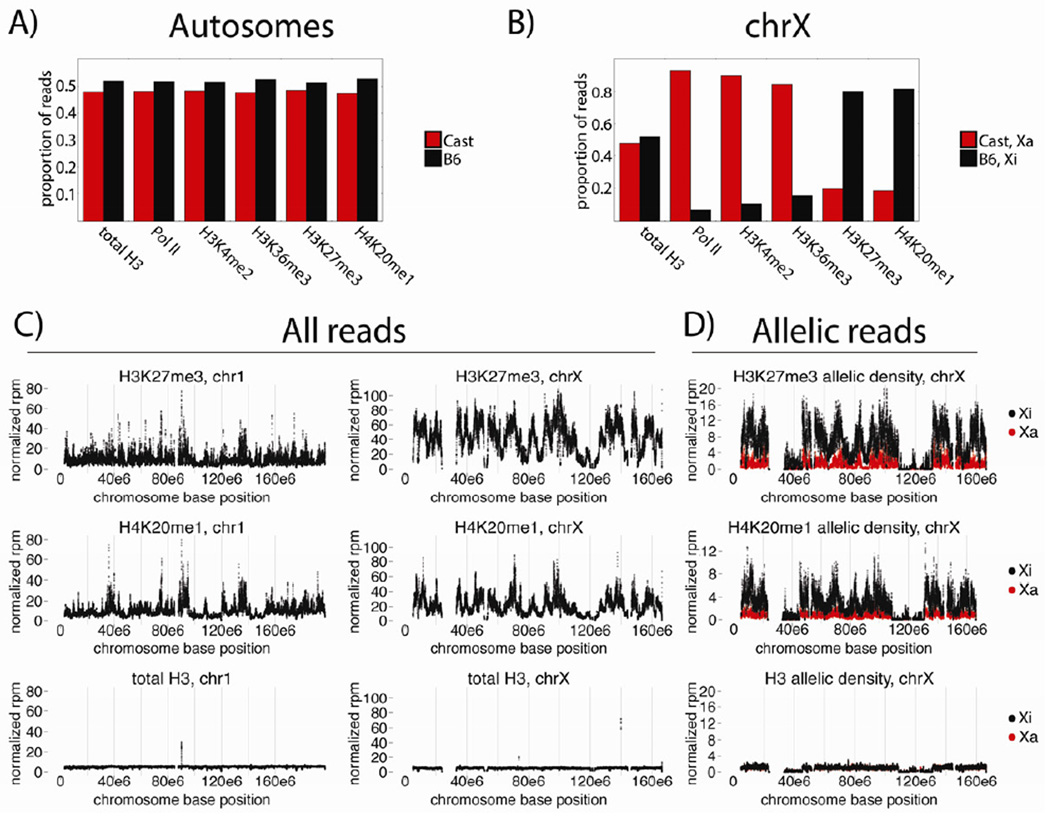
Both autosomal and X-linked ChIP-Seq data showed expected allelic distributions. Autosomal Cast-to-B6 allelic ratios were approximately 1:1 in all datasets, with a slight bias towards the reference genome (Figure 2A). In contrast, allelic biases were skewed over the X-chromosomes in a manner consistent with data from classical immunofluorescence (IF) microscopy experiments (Escamilla-Del-Arenal et al., 2011), with active marks biased towards the Cast genome, or Xa, and repressive marks biased towards the B6 genome, or Xi (Figure 2B).
Figure 2. ChIP-Seq shows X-linked epigenetic biases.
(A), (B) Proportional allelic distributions over autosomes (A) and chrX (B).
(C) chr1 and chrX non-allelic tiling density plots, in normalized reads per million per 40kb bin (rpm). Only bins with ≥50% alignability are shown.
(D) chrX allelic tiling density plots, scales as in (C).
The tiling densities of H3K27me3 and H4K20me1 were next examined, in order to quantitatively address how their microscopic Xi enrichments related to specific chromosomal regions (Figure 2C). Both marks exhibited large-scale density fluctuation over the X, with megabase-sized regions of enrichment and depletion (Figure 2C, right panels). These X-linked profiles had a Spearman coefficient of 0.88, higher than their autosomal correlation of 0.46. X-linked H3K27me3 and H4K20me1 patterns also contrasted with chromosome 1 patterns, for example, where enriched regions were more punctate (Figure 2C, left panels). Importantly, allelic data showed the major patterns of X-linked H3K27me3 and H4K20me1 enrichment reflected those of the Xi (Figure 2C, right panels vs. 2D).
LINE-dense regions of the Xi are exterior to H3K27me3 and Xist domains
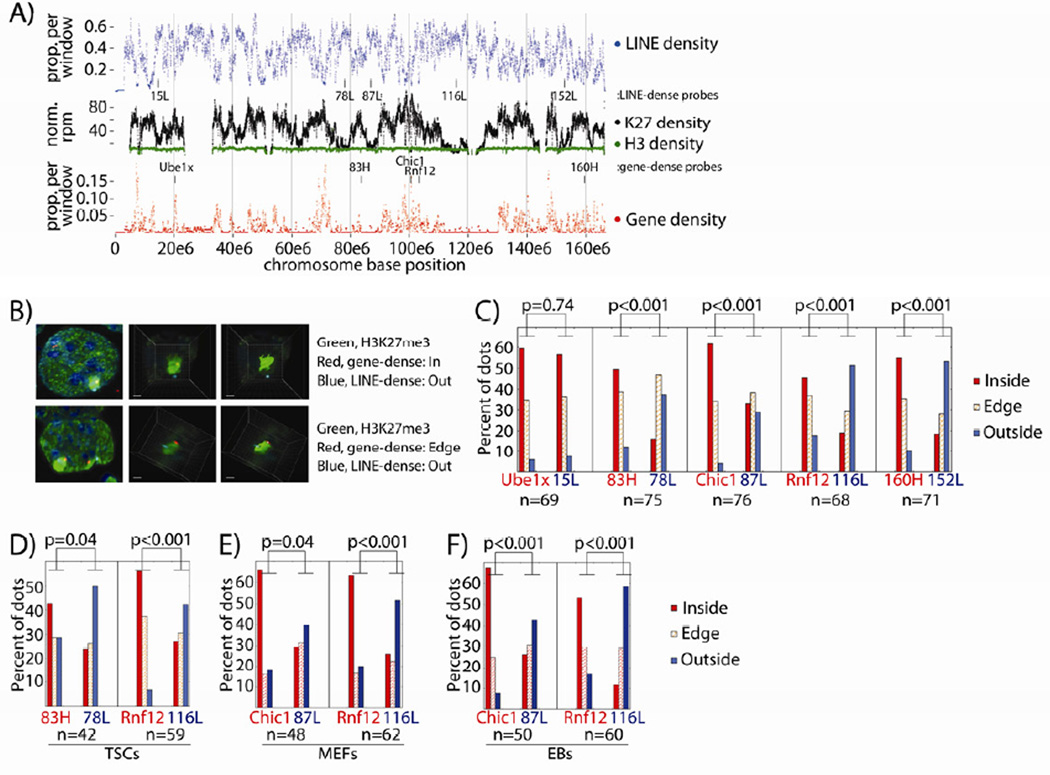
Previous studies have shown both H3K27me3 and Xist co-localize with gene- and not LINE-dense regions of the Xi (Chadwick, 2007; Duthie et al., 1999; Mak et al., 2002; Marks et al., 2009). We addressed the relationship between X-linked H3K27me3 density, genes, and LINEs, continuing to fit our data in context with previous observations before examining more detailed epigenetic aspects of the TSC Xi. Tiling density plots over the TSC X showed reciprocal relationships between H3K27me3 levels and genes, and LINEs (Figure 3A). H3K27me3 and gene density positively correlated over the X (Spearman coefficient 0.29; Figure 3A, middle vs. bottom). In contrast, X-linked LINE and H3K27me3 density were inversely correlated (Spearman coefficient −0.57; Figure 3A, middle vs. top). Almost invariably, peaks of LINE density co-localized with valleys of H3K27me3 density, and vice versa (Figures 3A, S2A,B). The continuity of H3 density within LINE dense regions validated our ability to detect and normalize for H3K27me3 within these regions.
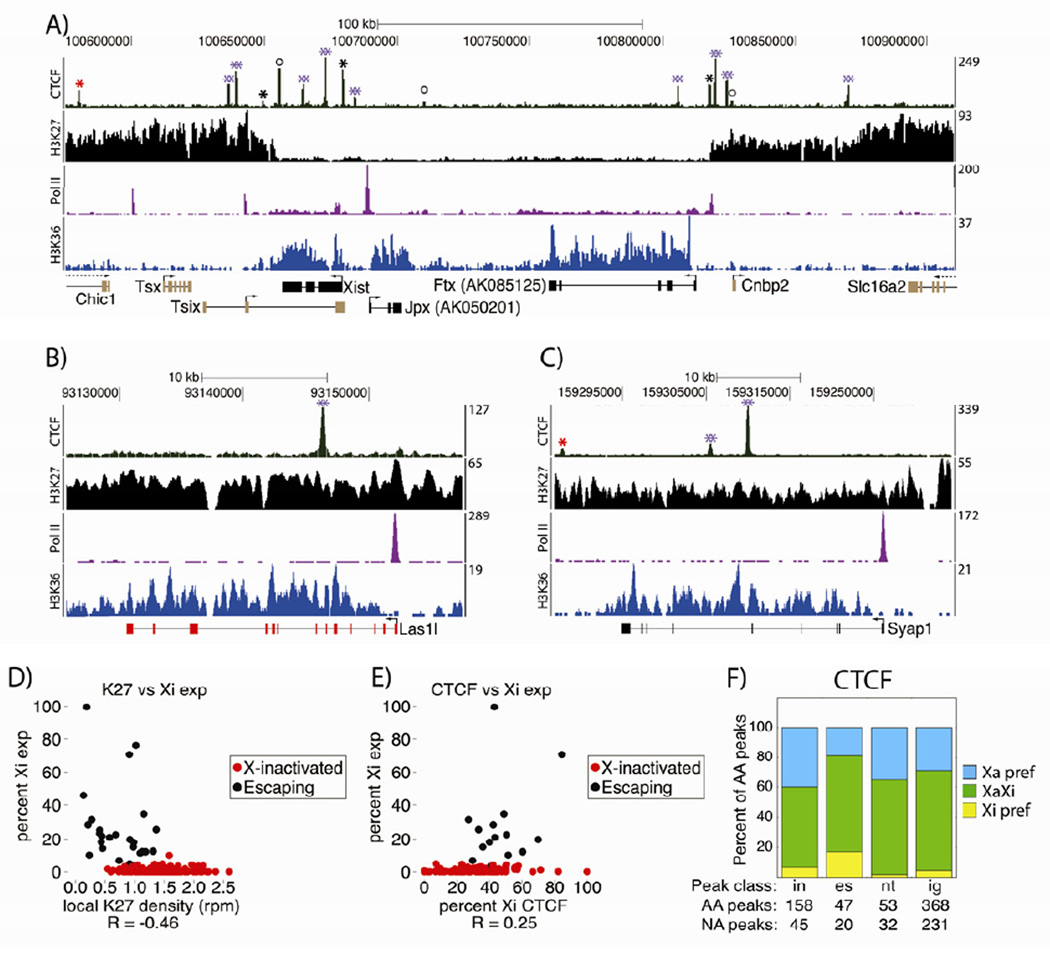
Figure 3. H3K27me3 and LINE density inversely correlate over the Xi.
(A) Tiling LINE and gene density over the X. H3K27me3/H3 density are as in Figure 2C. Only bins with ≥50% alignability are shown. FISH probe locations from (B–F) are marked.
(B) DNA FISH quantification process. H3K27me3 IF is green, with gene-dense / LINE-dense probes in red and blue. Left to right shows the progression: a whole nucleus, an extracted Xi, a 3D-rendered Xi image.
(C–F) FISH probe location relative to Xi H3K27me3 (C) or Xist (D–F) domains, by percentage. n, Xi’s counted per probe pair. P-values are from χ2 tests comparing dots inside/on the edge of vs. outside the Xi domain.
See also Figure S2.
LINEs and other repeats have been proposed to make up the spatial core of the Xi from the initiation of XCI onward, with X-inactivated genes moving into the proposed core as they are silenced (Chaumeil et al., 2006; Chow et al., 2010; Clemson et al., 2006; Namekawa et al., 2010). Considering the different H3K27me3 levels between LINE- and gene-dense regions of the TSC Xi, it was difficult to reconcile how these sequence classes would occupy the same nuclear space. We therefore directly examined the spatial relationship between the gene- and LINE-dense regions of the TSC Xi. Using H3K27me3 IF combined with DNA FISH, we examined the locations of five X-linked FISH probe pairs relative to the H3K27me3 coat. Pairs consisted of probes in neighboring gene- and LINE-dense regions (Figure 3A, labeled tick marks; Figure S2C). H3K27me3 IF and DNA FISH were performed, Z-stack images were collected and deconvolved, and the areas surrounding individual Xi’s were subjected to 3D reconstruction. After imaging, probes were scored as either inside, on the edge of, or exterior to the Xi’s H3K27me3 coat (Figure 3B).
Surprisingly, LINE-dense regions were more frequently exterior to the H3K27me3 domain than gene-dense regions, for four of five probe pairs (Figure 3C). Similar results were obtained with two probe pairs relative to the Xist coat (Figure 3D). One of five LINE-dense probes, 15L, was most frequently interior to the H3K27me3 domain, potentially due to higher H3K27me3 levels or proximity to the centromere (Figure S2C). We also examined the spatial relationship between LINE- and gene-dense regions in cell types subject to random XCI: mouse embryonic fibroblasts (MEFs) and embryoid bodies (EBs). The LINE-dense regions examined in these cells were external to Xist at a frequency similar to that found in TSCs (Figures 3E,F). We conclude that in TSCs, MEFs, and EBs, a majority of the most LINE-dense Xi regions are spatially separated from gene-dense regions of the chromosome, which are encompassed by Xist and H3K27me3. Given this spatial separation, our results suggest maintenance of gene silencing during XCI is not associated with translocation into a LINE-dense spatial core.
X-inactivated promoters exclude active chromatin marks and are enriched in H3K27me3 and H4K20me1
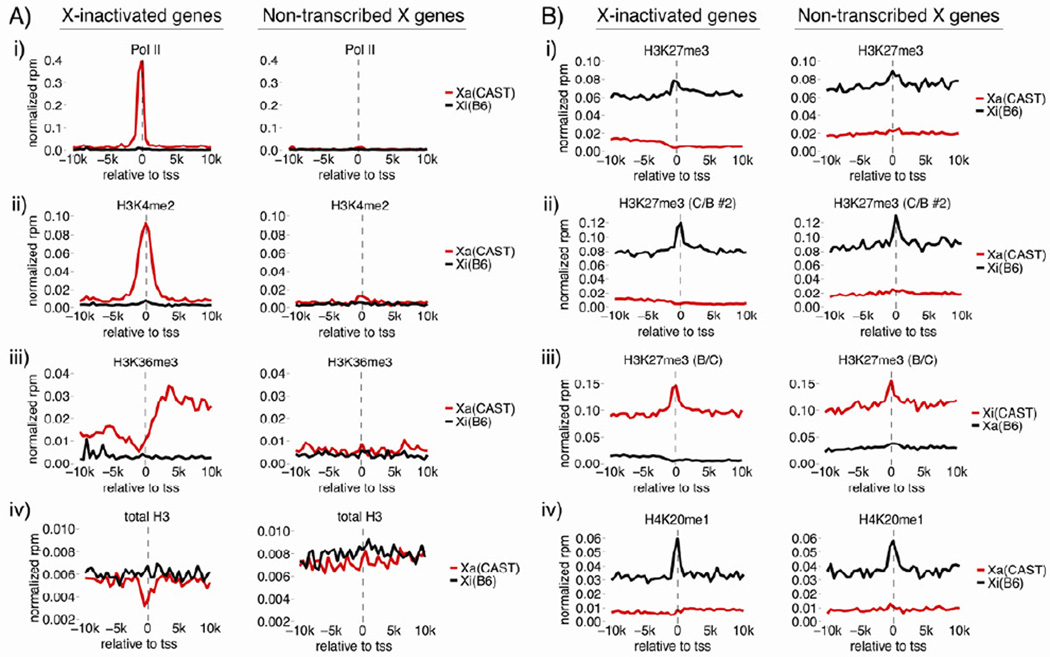
Having established the large-scale relationship between histone marks and DNA features of the TSC Xi, we began a more high-resolution study of X-linked chromatin. We initially compared two X-linked gene categories that differ in their transcriptional status: (1) X-inactivated genes, on average expressed at 99.5% from the Xa, and (2) non-transcribed genes, defined by their complete lack of expression as assessed by RNA-Seq. Comparison of metagene data between these two gene categories required normalization for gene-set size, as well as SNP density (Figure S3).
Figure 4 shows allelic metagene plots in the 10kb surrounding X-inactivated and non-transcribed transcription start sites (TSS). As expected from RNA-Seq data, nearly all Pol II, H3K4me2, and H3K36me3 signal over X-inactivated genes derived from the Xa, indicating Pol II binding to X-inactivated TSS is blocked (Figure 4A, panels i-iii). H3K27me3 density at X-inactivated genes was uniformly higher on the Xi relative to the Xa; however, there was a modest peak of H3K27me3 density in the 4kb surrounding both X-inactivated and non-transcribed TSS (Figure 4B, panel i). A similar H3K27me3 peak was present in two independently derived TSC lines, confirming TSS enrichment as a general feature of H3K27me3 accumulation over the TSC Xi (Figure 4B, panels ii, iii). H4K20me1 enrichment over Xi TSS mirrored H3K27me3 patterns (Figure 4B, panel iv), consistent with the high positive correlation between these two modifications in tiling density plots (Figure 2C).
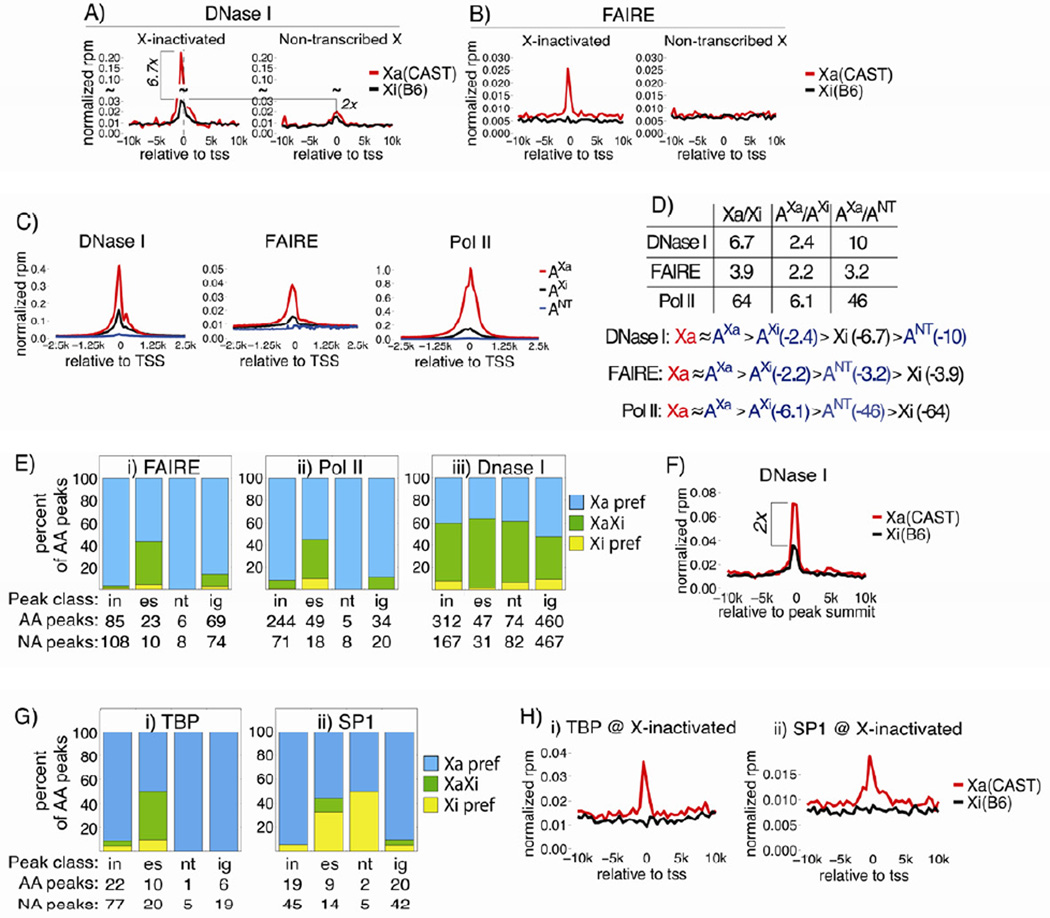
Figure 4. Xi promoters exclude active chromatin marks and are enriched in H3K27me3 and H4K20me1.
Allelic metagene profiles of X-inactivated and non-transcribed X-linked genes. Cast (red) and B6 (black) data are shown as rpm per 500bp bin. (C/B #2), a C/B TSC line not used for RNA-Seq. (B/C), the TSC line from Figure 1 RNA-Seq. See also Figure S3.
Xi H3K27me3 and H4K20me1 TSS enrichment were similar between X-inactivated and non-transcribed genes (Figure 4B, panels i-iv). This contrasts with previous work in EBs, which found H3K27me3 density to be higher on X-inactivated compared to non-transcribed genes, perhaps highlighting a difference between cell types or stages of XCI analyzed (Marks et al., 2009). Our results indicate that TSS-proximal nucleosomes, regardless of Xa transcriptional status, are more likely to be modified with H3K27me3 and H4K20me1 than surrounding Xi sequence, suggesting these sites either have increased capacity to recruit the modifications, or they more stably retain them post-deposition.
Regulatory elements display DNaseI hypersensitivity across the TSC Xi
The absence of transcription-associated chromatin signatures at X-inactivated TSS suggested these regions exist in a locally closed state, lacking the nucleosome depletion typically found at utilized TSS. To address this hypothesis, we profiled allelic nucleosome density with two antibody-independent methods, DNase-and FAIRE-Seq (Giresi and Lieb, 2009; Song et al., 2011). DNase-Seq detects genomic regions that are hypersensitive to DNaseI digestion, whereas FAIRE-Seq uses formaldehyde treatment to enrich for genomic regions not crosslinked to proteins. Both techniques identify nucleosome-depleted sites typically found at active promoters and regulatory elements, but are also known to detect non-overlapping sites (Song et al., 2011).
DNase- and FAIRE-Seq patterns were first examined over autosomes, to verify both methods performed as expected in TSCs. Indeed, for both techniques, signal was present at highly expressed genes and absent from non-transcribed genes (Figure S4A,B). Autosomal Polycomb targets, though lowly expressed in TSCs, accumulated signal from both methods, expected given the H3 depletion at their TSS (Figure S4C).
We next examined the X-linked DNase-Seq metagene profile. A significant enrichment of DNase-Seq signal was observed at X-inactivated TSS on the Xi (Figure 5A). This result was surprising considering the near-total exclusion of Pol II and other active chromatin marks from these same sites. While the DNaseI signal surrounding X-inactivated TSS was 6.7 fold lower on the Xi compared to the Xa, Xi signal was well above background, and 2 fold greater than Xi signal at non-transcribed genes (Figure 5A). Importantly, allelic profiles were due to small signal contributions from many genes, and likely represent the average DNaseI hypersensitivity (DHS) found at X-inactivated promoters (Figure S4D). In total, 198 genes contributed TSS-associated signal to the Xi profile, with median and maximum signal contributions at 0.3% and 1.9%, respectively. These numbers were similar to those on the Xa, where 201 genes contributed signal, and the median and maximum contributions were 0.2% and 3.4%, respectively. Therefore, X-inactivated TSS exhibited a level of Xi DHS above surrounding sequence, and what would have been expected for a non-transcribed gene.
Figure 5. DNaseI hypersensitivity without FAIRE enrichment at X-inactivated TSS.
(A), (B) DNase-Seq and FAIRE allelic metagene profiles, as in Figure 4.
(C) Metagene profiles of autosomal genes, expression-matched to X-inactivated Xa genes (AXa), X-inactivated Xi genes (AXi), and non-transcribed genes (ANT).
(D) DNaseI, FAIRE, and Pol II ratios between Xa and Xi genes, AXa and AXi genes, and AXa and ANT genes. Classes were compared assuming that signal from Xa and AXa genes would be equivalent, given their similar expression levels.
(E) Allelic, X-linked distribution of FAIRE (i), Pol II (ii), and DNaseI (iii) peaks. Categorization is relative to genes falling within 5 kb of a peak start or end; “in”, “es”, “nt”, and “ig” peaks associate with X-inactivated, escaping, non-transcribed, or intergenic regions, respectively. ‘Xa/Xi pref’, peaks detected on one X but not the other; ‘XaXi’, peaks detected on both X’s; ‘AA peaks’, peaks that were Allelically Assigned to at least one X via SNP-overlapping reads. ‘NA peaks’, peaks that were Not Assignable to either X due to lack of allelic data.
(F) Allelic metagene profile of X-linked, intergenic sites of DHS.
(G) Distribution of TBP (i) and SP1 (ii) peaks over the Xa and Xi, as in (E).
(H) Allelic metagene profiles of TBP (i) and SP1 (ii) surrounding X-inactivated genes, as in (A).
See also Figures S4,5.
In contrast, FAIRE-Seq did not detect Xi-associated signal at TSS, although robust signal was seen over X-inactivated TSS on the Xa (Figure 5B), and over autosomal Polycomb targets (Figure S4C). The lack of TSS-associated FAIRE signal was consistent with our total H3 profiling, which lacked Xi, TSS-localized H3 depletion (Figure 4A, panel iv). TSS signal-to-noise estimates from metagene profiles suggested that DNase-Seq had a higher dynamic range than either FAIRE- or total H3-Seq, with a signal-to-noise ratio of 20, as compared to 4 and 2, respectively. Therefore, the lack of FAIRE enrichment and H3 depletion at Xi TSS, despite the presence of DHS, suggests the existence of a nucleosome-depleted site on the Xi that is smaller or less persistent than the equivalent Xa site.
RNA-Seq analysis indicated that X-inactivated genes were expressed from the TSC Xi at low levels, with a median expression value of 0.037 reads-per-kilobase-million (rpkm), similar to the median of 0.089 rpkm for autosomal Polycomb targets. We therefore examined whether the amounts of DNaseI, FAIRE, and Pol II observed at X-inactivated TSS were the same as, or different than, what would be expected for an equivalently expressed autosomal gene. If different than expected, these features might suggest distinct epigenetic properties of X-inactivated TSS that may yield insight into XCI’s mechanism. To make the comparison, we selected 3 autosomal genes classes: those expressed at levels similar to X-inactivated genes on the Xa (AXa) and Xi (AXi), and non-transcribed genes (ANT). Metagene profiles of DNaseI, FAIRE, and Pol II were created for each (Figure 5C), and the ratio of TSS-associated signal between X-linked and autosomal classes was compared (Figure 5D). Allelic and total data were not directly compared because of their different enrichment scales; instead, ratios between X linked and expression-matched autosomal genes were calculated separately, and data were compared assuming that signal from Xa and AXa genes would be equivalent, given their similarly robust expression. The relative Xi DHS of X-inactivated TSS was higher than that observed at non-transcribed autosomal genes, and lower than that observed at expression-matched autosomal genes (Figure 5D; AXi > Xi > ANT). In contrast, Xi FAIRE and Pol II signal at X-inactivated TSS was comparable to autosomal non-transcribed genes, showing levels approaching background (Figure 5D; AXi > ANT > Xi). Therefore, X-inactivated TSS harbor an Xi epigenetic state distinct from lowly expressed and non-transcribed autosomal genes, showing DHS but lacking FAIRE or Pol II enrichment. The transcriptional difference between X-inactivated and non-transcribed genes could be explained if a low level of Xi Pol II binding resulted in efficient target gene elongation.
We next examined whether regulatory elements across the Xi had epigenetic properties similar to those of X-inactivated TSS. MACS-defined peaks of DNaseI, FAIRE, and Pol II were separated into 4 classes based on relation to their most proximal gene. Individual peaks associated with X-inactivated, escaping, non-transcribed, or intergenic regions, defined as genomic space greater than 5kb away from a known gene (Figure 5E; “in”, “es”, “nt”, and “ig” peaks, respectively). Allelic binding events were then determined via an empirical background model (Table S3). Consistent with the exclusion observed at X-inactivated TSS (Figures 5B and 4A, panel i), FAIRE and Pol II peaks were rarely detected surrounding X-inactivated, non-transcribed, and intergenic Xi regions, while Xi detection increased around escaping genes (Figure 5E, panels i,ii). In contrast, but in line with our TSS analysis (Figure 5A), DNaseI peaks were detected across the Xi, mostly at sites shared with the Xa, regardless of associating region (Figure 5E, panel iii).
Intergenic DHS frequently marks regulatory elements that participate in transcriptional control of nearby and distal genes (Song et al., 2011). To date, such sites have not been investigated in relation to XCI. Of the 460 X-linked, allelically assignable intergenic DNaseI peaks, ~50% maintained Xi DHS despite the near transcriptional silence of most X-linked genes (Figure 5E, panel iii).
To understand how Xi regulatory elements are processed by XCI, the average chromatin state at intergenic DHS sites was examined via metagene analysis, using MACS-defined peak summits as reference points. The analysis focused on the 176 intergenic DHS sites detected on both X’s, to avoid confounding contributions from peaks specific to the Xa or Xi. These sites showed significant Xi DHS, with an average Xa:Xi ratio of 2 (Figure 5F). Excluding DHS sites that bound CTCF from this analysis did not change the Xa:Xi ratio (not shown). In contrast, other active marks were excluded from Xi intergenic DHS sites, similar to that observed at X-inactivated TSS (Figure S5A). Consistent with intergenic DHS marking active regulatory elements on the Xa but not Xi, we observed Xa-only enrichment of H3K4me1 and H3K27-acetylation, two marks associated with utilized transcription factor binding (Figure S5B). Intergenic DHS sites also showed small but detectable levels of Xi H3K27me3 and H4K20me1 enrichment (Figure S5C).
Considering that DHS is often indicative of transcription factor binding, we examined whether such binding occurred on the Xi. ChIP-Seq was performed for the general transcription factor TBP, and the gene-specific transcription factor SP1, whose motif was significantly enriched over DHS peaks on the X (not shown). In neither case was binding detected over the Xi, although robust signal was present on the Xa, as expected (Figures 5G,H).
In summary, Xi regulatory elements retained DHS, similar to Xa counterparts and autosomal Polycomb targets, yet excluded Pol II and transcription factors, and were not detected as open chromatin via FAIRE, similar to non-transcribed genes (Figures 5, S4). These data indicate X-inactivated regulatory elements maintain chromatin states distinct from both autosomal Polycomb targets and non-transcribed genes. Moreover, the observed TBP and SP1 exclusion suggests that XCI operates at least in part by preventing efficient transcription factor binding to the Xi.
Variable H3K27me3 micro-environments surrounding genes escaping XCI
We next examined chromatin modifications surrounding genes that escaped XCI, in an effort to understand how localized transcription occurs within the repressive Xi environment. As expected, escapers associated with marks of active transcription (Figure S6A). Additionally, many escapers existed in H3K27me3-depleted microenvironments, such as those in Figure 6A. Generally, escape level inversely correlated with local H3K27me3 density (Figure 6D; r, −0.46, Pearson’s coefficient), although not all escapers were locally insulated from the mark. Several escapers, such as Syap1, had H3K27me3 levels that closely resembled those found at X-inactivated genes (Figure 6B,C). Previous works suggest that the DNA binding protein CTCF, and nuclear position relative to the Xist domain, may play important roles in escape (Chaumeil et al., 2006; Filippova et al., 2005). We therefore examined whether these two features might better correlate with Xi expression than local H3K27me3 levels.
Figure 6. Variable H3K27me3 levels associated with escape, and widespread CTCF binding to the Xi.
(A), (B), (C) Genomic windows surrounding the X-inactivation center (A), Las1l (B), and Syap1 (C). non-allelic ChIP-Seq data is shown above genome annotations. Black genes escaped, red genes were X-inactivated, gold genes were not allelically assigned. Red/black *’s mark CTCF peaks on the Xa/Xi; purple **’s, those on both X’s; circles, non-assignable peaks.
(D) Xi expression vs. genic H3K27me3 density, for X-linked genes with ≥20 allelic RNA-Seq reads.
(E) Xi expression vs. Xi CTCF binding levels, for genes in (D) that contain a CTCF peak within 5kb of their start or end.
(F) Allelic distribution of CTCF peaks, as in Figure 5E.
See also Figure S6.
CTCF binds sites present on both X’s
We addressed CTCF’s role in facilitating escape by localizing Xi binding sites via ChIP-Seq. CTCF binding correlates with insulation between chromatin states genome-wide (Song et al., 2011). Furthermore, CTCF binds the Jarid1c gene only in species where it escapes XCI, suggesting a role for CTCF in escape licensing (Filippova et al., 2005). We therefore hypothesized that local CTCF binding would be associated with increased levels of escape. Indeed, there was a moderate positive correlation between CTCF binding and Xi expression, supporting a role for CTCF in escape (Figure 6E; r = 0.25; Pearson’s correlation). However, we observed CTCF binding across the Xi, regardless of X-inactivation status or gene presence (Figure 6A–C, green ‘CTCF’ track). 59, 81, 68, and 71 percent of allelically assignable CTCF peaks within X-inactivated, escaping, non-transcribed, and intergenic regions, respectively, were present on the Xi or both X’s (Figure 6F). These results indicate CTCF binding alone is not predictive of escape or local insulation from H3K27me3.
Also unexpectedly, the majority of Xi CTCF peaks were located at sites shared with the Xa and not Xi specific (394 of 428 Xi peaks; Figure 6A–C, purple stars; Figure 6F, green bars). Xi-specific CTCF binding did exist, for example at the H3K27me3 boundaries flanking the X-inactivation center (Figure 6A, black *’s), but made up a minority of peaks (34 of 428 Xi peaks; Figure 6F, yellow bars). The presence of shared CTCF peaks across varied Xi environments, regardless of gene or chromatin boundary location, suggests complex utilization of the protein in XCI.
Transcriptional competence and position relative to the Xist domain are uncoupled
Lastly, we examined the nuclear position of escaping and X-inactivated genes relative to the Xi’s H3K27me3 and Xist domains. Previous work has shown the area encompassed by Xist to be microscopically devoid of transcription-associated features, such as Pol II, and that escaping genes are preferentially located outside of the Xist domain (Escamilla-Del-Arenal et al., 2011). Considering these data, it has been hypothesized that externalization relative to the Xist domain may place genes in an environment permissive to transcription, potentially playing a causal role in escape.
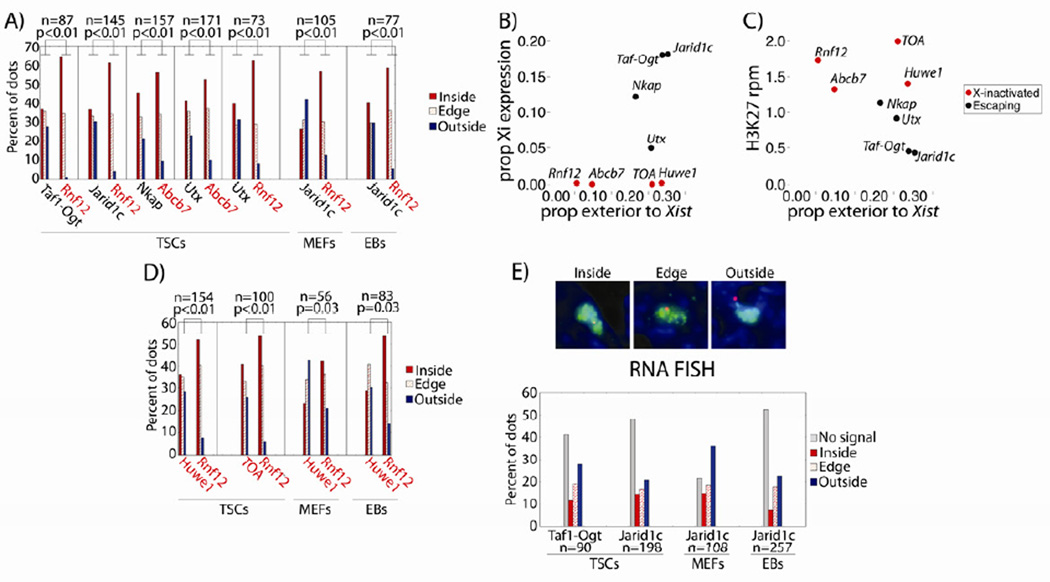
To examine the relationship between escape and Xist externalization in TSCs, we selected 4 escaping loci expressed at varying levels from the Xi: Taf1-Ogt (18% average Xi exp.), Jarid1c (18% Xi exp.), Nkap (12% Xi exp.), and Utx (5% Xi exp.). The nuclear position of these genes relative to the Xi’s Xist and H3K27me3 coat was examined together with an X-inactivated gene (Rnf12 or Abcb7), using the DNA FISH assay described in Figure 3. This assay allowed escaping and X-inactivated gene location to be quantified in tandem, providing an internal control per experiment.
As expected, escaping genes were more frequently exterior to the Xist domain than X-inactivated genes, in TSCs, MEFs, and EBs (Figure 7A). Localization differences were not found when examining position relative to the Xi’s H3K27me3 coat (Figure S6B). For these 6 loci, externalization frequency correlated well with TSC Xi expression and H3K27me3 levels (r = 0.87 and −0.95, respectively, Pearson’s correlation; Figure 7B), with some exceptions: Utx and Nkap had similar externalization frequencies but different Xi expression levels, and Rnf12 and Abcb7 had different externalization frequencies but similar Xi expression levels (p = 0.03 for externalization, χ2 test; Figure 7B).
Figure 7. Externalization relative to the microscopic Xist domain does not induce escape.
(A) Gene location relative to Xist domain, by percentage. Vertical lines separate probe pairs of escaping (black font) and X-inactivated (red font) genes. n, Xi’s counted per probe pair. P-values are from χ2 tests comparing dots inside/on the edge of vs. outside the Xi domain.
(B,C) Xist domain externalization frequency vs. (B) Xi expression, or (C) local H3K27me3 density.
(D) Gene location relative to Xist domain, as in (A).
(E) RNA FISH signal relative to Xist domain. ‘No Signal’, mono-allelic expression from the Xa. Representative images are above the bar graph.
See also Figure S6.
Considering these exceptions, we further explored the relationship between Xist domain externalization and Xi expression, attempting to parse the difference between causality and correlation. The positions of two X-inactivated loci adjacent to escaping genes were examined relative to the Xist domain: Huwe1, adjacent to the escaper Jarid1c and displaying 0.1% Xi expression, and a ~200kb locus, referred to as TOA, for Taf1 and Ogt Associated, which contained 4 X-inactivated genes adjacent to the escapers Taf1 and Ogt and displayed 0% aggregate Xi expression. In TSCs, these two loci had externalization frequencies similar to neighboring escaping loci, despite lacking Xi expression and maintaining high levels of H3K27me3 (Figure 7B–D). The same localization pattern was observed in MEFs and EBs (Figure 7D). RNA FISH confirmed the monoallelic expression of these loci (Figure S6C). Together, these results indicate that gene externalization relative to the Xist domain is positively correlated with, but insufficient to induce, escape from XCI.
In light of this result, we examined where escaper transcription occurred on the single-cell level, via RNA FISH and 3D reconstruction of the Xist domain. If the microscopic region encompassed by Xist is truly impermeable to transcription, as previous analyses would predict (Escamilla-Del-Arenal et al., 2011), then escaping loci would only express when located on the boundary of, or exterior to, the Xist domain, where Pol II and associated factors could be accessed. We chose two of the most robustly escaping TSC loci for this analysis, Jarid1c and Taf1-Ogt, given their high levels of bi-allelism and clarity of RNA FISH signal. In contrast to what would be predicted from existing models, 28% and 20% of the total observed RNA FISH dots for Jarid1c and Taf1-Ogt localized within the TSC Xist domain (representing 14% and 12% of the total Xi’s counted; Figure 7E). Similar co-localization frequencies were observed for Jarid1c and Xist in MEFs (21%) and EBs (15%) (Figure 7E). The detection of RNA FISH signal within the region encompassed by Xist suggests that the microscopic domain is not an obligate silent compartment in TSCs, MEFs, or EBs.
Together, two lines of evidence indicated that transcriptional competence is uncoupled from gene position relative to the microscopic Xist domain, in TSCs, MEFs, and EBs. First, X-inactivated loci adjacent to escapers remained inactive despite their frequent localization outside of the microscopic Xist domain (Figure 7D). Second, escape frequently co-localized with the Xist domain, as assessed via RNA FISH (Figure 7E). We conclude that XCI is unlikely to be maintained solely by a chromosome-wide separation from transcription machinery. Rather, our results suggest a role for the locus-specific silencing of regulatory elements in the inactivation process.
DISCUSSION
Our allelic gene expression and chromatin analyses revealed a complex set of epigenetic environments over the TSC Xi. Most notably, X-inactivated regulatory elements harbored an epigenetic signature distinct from autosomal Polycomb targets and non-transcribed genes, displaying DHS without enrichment of other transcriptional hallmarks. Together with microscopic analyses examining gene location relative to the Xist domain, our data suggest a central role for the site-specific silencing of regulatory elements in maintenance of both random and imprinted XCI.
We present the first global analysis of allelic gene expression over an imprinted Xi, finding that 13% of TSC genes escaped XCI. This percentage was similar to that estimated for human cells (Carrel and Willard, 2005), but different from the 3% observed in mouse kidney (Yang et al., 2010), indicating different cell types have varying escape frequency. Degree of TSC escape could be partly, though not perfectly, predicted by local H3K27me3 levels. Considering this imperfect correlation, we hypothesize that many escaping genes fluctuate between H3K27me3 insulated and un-insulated states, with expression occurring most robustly during times of insulation. Alternatively, some genes may escape even in the presence of high local H3K27me3 levels.
Considering its classification as an insulator, we addressed CTCF’s role in establishing escape from XCI. Perhaps not surprisingly given its widespread binding patterns (Song et al., 2011), the X-linked distribution of CTCF was complex. Xi CTCF binding positively correlated with escape, supporting a role in the process, as has been proposed (Filippova et al., 2005). However, 92% of the 428 CTCF peaks detected over the Xi were at sites shared with the Xa. Accordingly, Xi CTCF binding did not predict the location of escapers or H3K27me3 boundaries, consistent with results from a previous study examining transgene insulation during XCI (Ciavatta et al., 2006). The function of mirrored CTCF binding over diverse X-linked environments is unknown. We favor the possibility that CTCF binds certain sites without discrimination between X’s, but these sites are differentially utilized in the creation of allele-specific chromatin structures.
Regarding the epigenetic properties of Xi regulatory elements, we describe evidence indicating these sites are recognized from surrounding sequence, yet rendered non-functional by the XCI machinery. On average, Xi regulatory elements showed enrichment of H3K27me3 and H4K20me1 relative to neighboring Xi DNA, supporting the notion that Xi regulatory elements are recognized by cellular machinery. Remarkably, regulatory elements across the TSC Xi showed DHS, despite excluding Pol II and other transcriptional hallmarks. These data suggest that nucleosomes surrounding Xi regulatory elements are recognized and displaced, but an unidentified property of these loci – a certain conformation, non-canonical nucleosome, or associated protein or modification – precludes efficient binding of Pol II to the Xi.
Binding of two transcription factors, TBP and SP1, was not detected over Xi regulatory elements despite these elements harboring the DHS typical of transcription factor binding. This lack of detectable binding indicates that XCI operates at least in part by preventing the stable association of Pol II recruitment factors to targets. In the complete absence of transcription factor binding, the DHS observed at Xi regulatory elements could be the byproduct of a mechanism excluding such factors from the Xi. This mechanism would be PRC2-independent, considering Xi DHS peaks do not invariably coincide with H3K27me3 enriched regions (not shown), and TSC autosomal PRC2 targets bind TBP, SP1, and Pol II (Figure S4C). It is also possible that transcription factors bind the TSC Xi at low levels, resulting in DHS, but our allelic ChIP-Seq lacked the sensitivity needed to detect such binding. In this scenario, an estimated upper bound of Xi transcription factor binding might be on par with DHS levels at Xi TSS, about 7-fold less on the Xi as compared to the Xa. Were this the case, XCI could operate combinatorially through inhibition of transcription factor binding and function, given that X-inactivated genes are repressed ~200-fold on the TSC Xi and essentially lack Pol II binding. Continued study of Xi chromatin states will likely yield additional insight into XCI’s mechanism.
Informed by our genomic analyses, spatial properties of the Xi were examined relative to pre-existing mechanistic models of XCI. These models suggest that the Xi’s spatial core, marked by Xist, is a transcriptionally silent, repeat-dense region, into which genes are recruited as they are inactivated (Chaumeil et al., 2006; Chow et al., 2010; Clemson et al., 2006; Namekawa et al., 2010). Entry into this domain may induce, or at a minimum, help maintain silencing by preventing access to transcription machinery. Accordingly, escapers are thought to be actively maintained at the domain’s exterior, allowing them access to Pol II (Escamilla-Del-Arenal et al., 2011).
Our work suggests significant revisions to the above model. Site-specific DNA FISH demonstrated that LINE-dense regions did not invariably make up the spatial core of the Xi, as has been proposed (Chaumeil et al., 2006; Chow et al., 2010; Clemson et al., 2006; Namekawa et al., 2010). Rather, gene- and LINE-dense regions occupied separate nuclear territories in TSCs, MEFs, and EBs, with LINE-dense DNA most frequently adjacent to the Xist domain. This spatial separation suggests that maintenance of gene silencing during XCI does not require colocalization with a LINE-dense core, and supports an indirect role for the Xi’s most LINE-dense regions in Xist-mediated silencing (Tattermusch and Brockdorff, 2011). Previous works defining the Xi’s core as repeat-dense have relied on site-non-specific FISH probes such as Cot-1, which cannot differentiate between repetitive sequence in genic and intergenic regions, perhaps explaining the observed discrepancies.
Our data also suggest that gene externalization relative to the measured Xist domain is a consequence rather than cause of escape. At two separate loci, escapers and adjacent X-inactivated genes were found outside of the Xist domain at similar frequencies. If externalization were a primary factor in inducing escape, then these X-inactivated loci would have exhibited increased Xi expression upon externalization. Instead, they maintained silencing regardless of location relative to the measured Xist domain, indicating that licensing of escape occurs on a gene-specific level and is not strictly determined by chromosome topology. In support of this, escape was detected within the microscopic Xist domain, as assessed via RNA FISH. This notion is further supported by the observation that differential gene regulation can occur within topologically associated genomic regions (Dixon et al., 2012; Nora et al., 2012). We hypothesize escapers and topologically associated X-inactivated genes are externalized due to escaper interactions with transcription factories, which are abundant outside of the measured Xist domain.
Considered together, our results support a model for XCI whereby individual regulatory elements are maintained in a silent state by a mechanism that persists regardless of their location relative to a larger X-linked domain. While a chromosome-wide exclusion of transcription machinery from the Xi’s physical territory may play a role in XCI, it ultimately appears secondary to site-specific silencing during XCI maintenance; genes escaping XCI were expressed within the Xi’s interior, and X-inactivated genes remained silent when separated from microscopic Xist domain. Separation from the microscopic domain may occur dynamically, resulting in temporary loss of local Xist coating at externalized X-inactivated genes. Alternatively, Xist binding may persist over externalized regions, but at levels that are not detectable by conventional RNA FISH. In either case, silencing is stable throughout externalization, as gene expression levels of external and internal X-inactivated genes were indistinguishable.
The distinct sub-microscopic epigenetic signatures of the TSC Xi lend additional credence to a model of XCI where gene silencing is governed by inactivation of individual regulatory elements rather than a chromosome-scale, spatial segregation away from transcription machinery. Our surprising observation that X-inactivated regulatory elements display DHS and proximal H3K27me3 and H4K20me1 enrichment indicates that these sites on the Xi are recognized as such from surrounding DNA. The absence of transcription-associated signals from genes along the Xi, despite their apparent exposure to a nuclear environment permissive to transcription, indicates that XCI-induced epigenetic signatures can be stably maintained independent of a chromosome-scale nuclear compartment dedicated to transcriptional silencing.
METHODS
TSC derivation and culture
TSC lines were derived and cultured as previously described (Quinn et al., 2006).
RNA-Seq and validation
Strand-specific cDNA libraries were prepared from polyA-purified TSC RNA as described in (Ingolia et al., 2009). Analysis details are included in the Supplemental Information, Figure S7, and Tables S4 and 5. Quantitative allele-specific RT-PCR was performed as described in (Kalantry et al., 2009).
ChIP-, DNase-, and FAIRE-Seq
ChIP was performed using 10 – 40 million feeder-free TSCs and 10 µg of antibody per IP. FAIRE-Seq and DNase-Seq were performed as described (Giresi and Lieb, 2009; Song et al., 2011).
DNA and RNA FISH
DNA and RNA FISH / IF experiments were performed essentially as described in (Chaumeil et al., 2006; Kalantry et al., 2006).
Allelic analysis
SNP data was obtained from (http://www.sanger.ac.uk/resources/mouse/genomes/) on 1/15/10, and used to create an in silico Cast genome build. Only reads uniquely aligning to B6/Cast genomes were used for downstream analyses.
Supplementary Material
RNA-Seq analysis finds 13% of genes escape X-inactivation in trophoblast stem cells
Inactive X regulatory elements lack Pol II binding yet retain DNaseI hypersensitivity
CTCF binds across the inactive X, mostly to locations shared with the active X
X-inactivated genes near escapers are often separated from the measured Xist cloud
ACKNOWLEDGEMENTS
Thanks to Magnuson lab members for critical comments, to Terry Furey and Darin London for DNase-Seq help, and to Victor Lobanenkov for kindly providing the CTCF antibody mixture. This work was supported by the NIH grant R01-GM10974 and UNC Cancer Research Fund. JMC was supported by the American Cancer Society post-doctoral fellowship 117571-PF-09-124-01-DDC.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
SUPPLEMENTAL INFORMATION
Supplemental Information includes detailed procedures, 7 figures, and 5 tables. Raw data are available under the NCBI accession number GSE39406.
References
- Carrel L, Willard HF. X-inactivation profile reveals extensive variability in X-linked gene expression in females. Nature. 2005;434:400–404. doi: 10.1038/nature03479. [DOI] [PubMed] [Google Scholar]
- Chadwick BP. Variation in Xi chromatin organization and correlation of the H3K27me3 chromatin territories to transcribed sequences by microarray analysis. Chromosoma. 2007;116:147–157. doi: 10.1007/s00412-006-0085-1. [DOI] [PubMed] [Google Scholar]
- Chaumeil J, Le Baccon P, Wutz A, Heard E. A novel role for Xist RNA in the formation of a repressive nuclear compartment into which genes are recruited when silenced. Genes Dev. 2006;20:2223–2237. doi: 10.1101/gad.380906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chow JC, Ciaudo C, Fazzari MJ, Mise N, Servant N, Glass JL, Attreed M, Avner P, Wutz A, Barillot E, et al. LINE-1 activity in facultative heterochromatin formation during X chromosome inactivation. Cell. 2010;141:956–969. doi: 10.1016/j.cell.2010.04.042. [DOI] [PubMed] [Google Scholar]
- Ciavatta D, Kalantry S, Magnuson T, Smithies O. A DNA insulator prevents repression of a targeted X-linked transgene but not its random or imprinted X inactivation. Proc Natl Acad Sci U S A. 2006;103:9958–9963. doi: 10.1073/pnas.0603754103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clemson CM, Hall LL, Byron M, McNeil J, Lawrence JB. The X chromosome is organized into a gene-rich outer rim and an internal core containing silenced nongenic sequences. Proc Natl Acad Sci U S A. 2006;103:7688–7693. doi: 10.1073/pnas.0601069103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon JR, Selvaraj S, Yue F, Kim A, Li Y, Shen Y, Hu M, Liu JS, Ren B. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature. 2012;485:376–380. doi: 10.1038/nature11082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duthie SM, Nesterova TB, Formstone EJ, Keohane AM, Turner BM, Zakian SM, Brockdorff N. Xist RNA exhibits a banded localization on the inactive X chromosome and is excluded from autosomal material in cis. Hum Mol Genet. 1999;8:195–204. doi: 10.1093/hmg/8.2.195. [DOI] [PubMed] [Google Scholar]
- Escamilla-Del-Arenal M, da Rocha ST, Heard E. Evolutionary diversity and developmental regulation of X-chromosome inactivation. Hum Genet. 2011 doi: 10.1007/s00439-011-1029-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filippova GN, Cheng MK, Moore JM, Truong JP, Hu YJ, Nguyen DK, Tsuchiya KD, Disteche CM. Boundaries between chromosomal domains of X inactivation and escape bind CTCF and lack CpG methylation during early development. Dev Cell. 2005;8:31–42. doi: 10.1016/j.devcel.2004.10.018. [DOI] [PubMed] [Google Scholar]
- Fujita PA, Rhead B, Zweig AS, Hinrichs AS, Karolchik D, Cline MS, Goldman M, Barber GP, Clawson H, Coelho A, et al. The UCSC Genome Browser database: update 2011. Nucleic Acids Res. 2010 doi: 10.1093/nar/gkq963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giresi PG, Lieb JD. Isolation of active regulatory elements from eukaryotic chromatin using FAIRE (Formaldehyde Assisted Isolation of Regulatory Elements) Methods. 2009;48:233–239. doi: 10.1016/j.ymeth.2009.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingolia NT, Ghaemmaghami S, Newman JR, Weissman JS. Genome-wide analysis in vivo of translation with nucleotide resolution using ribosome profiling. Science. 2009;324:218–223. doi: 10.1126/science.1168978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalantry S, Mills KC, Yee D, Otte AP, Panning B, Magnuson T. The Polycomb group protein Eed protects the inactive X-chromosome from differentiation-induced reactivation. Nat Cell Biol. 2006;8:195–202. doi: 10.1038/ncb1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalantry S, Purushothaman S, Bowen RB, Starmer J, Magnuson T. Evidence of Xist RNA-independent initiation of mouse imprinted X-chromosome inactivation. Nature. 2009;460:647–651. doi: 10.1038/nature08161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mak W, Baxter J, Silva J, Newall AE, Otte AP, Brockdorff N. Mitotically stable association of polycomb group proteins eed and enx1 with the inactive x chromosome in trophoblast stem cells. Curr Biol. 2002;12:1016–1020. doi: 10.1016/s0960-9822(02)00892-8. [DOI] [PubMed] [Google Scholar]
- Marks H, Chow JC, Denissov S, Francoijs KJ, Brockdorff N, Heard E, Stunnenberg HG. High-resolution analysis of epigenetic changes associated with X inactivation. Genome Res. 2009;19:1361–1373. doi: 10.1101/gr.092643.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Namekawa SH, Payer B, Huynh KD, Jaenisch R, Lee JT. Two-step imprinted X inactivation: repeat versus genic silencing in the mouse. Mol Cell Biol. 2010;30:3187–3205. doi: 10.1128/MCB.00227-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nora EP, Lajoie BR, Schulz EG, Giorgetti L, Okamoto I, Servant N, Piolot T, van Berkum NL, Meisig J, Sedat J, et al. Spatial partitioning of the regulatory landscape of the X-inactivation centre. Nature. 2012;485:381–385. doi: 10.1038/nature11049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patrat C, Okamoto I, Diabangouaya P, Vialon V, Le Baccon P, Chow J, Heard E. Dynamic changes in paternal X-chromosome activity during imprinted X-chromosome inactivation in mice. Proc Natl Acad Sci U S A. 2009;106:5198–5203. doi: 10.1073/pnas.0810683106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plath K, Fang J, Mlynarczyk-Evans SK, Cao R, Worringer KA, Wang H, de la Cruz CC, Otte AP, Panning B, Zhang Y. Role of histone H3 lysine 27 methylation in X inactivation. Science. 2003;300:131–135. doi: 10.1126/science.1084274. [DOI] [PubMed] [Google Scholar]
- Quinn J, Kunath T, Rossant J. Mouse trophoblast stem cells. Methods Mol Med. 2006;121:125–148. doi: 10.1385/1-59259-983-4:123. [DOI] [PubMed] [Google Scholar]
- Schoeftner S, Sengupta AK, Kubicek S, Mechtler K, Spahn L, Koseki H, Jenuwein T, Wutz A. Recruitment of PRC1 function at the initiation of X inactivation independent of PRC2 and silencing. EMBO J. 2006;25:3110–3122. doi: 10.1038/sj.emboj.7601187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva J, Mak W, Zvetkova I, Appanah R, Nesterova TB, Webster Z, Peters AH, Jenuwein T, Otte AP, Brockdorff N. Establishment of histone h3 methylation on the inactive X chromosome requires transient recruitment of Eed-Enx1 polycomb group complexes. Dev Cell. 2003;4:481–495. doi: 10.1016/s1534-5807(03)00068-6. [DOI] [PubMed] [Google Scholar]
- Song L, Zhang Z, Grasfeder LL, Boyle AP, Giresi PG, Lee BK, Sheffield NC, Graf S, Huss M, Keefe D, et al. Open chromatin defined by DNaseI and FAIRE identifies regulatory elements that shape cell-type identity. Genome Res. 2011;21:1757–1767. doi: 10.1101/gr.121541.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Splinter E, de Wit E, Nora EP, Klous P, van de Werken HJ, Zhu Y, Kaaij LJ, van Ijcken W, Gribnau J, Heard E, et al. The inactive X chromosome adopts a unique three-dimensional conformation that is dependent on Xist RNA. Genes Dev. 2011;25:1371–1383. doi: 10.1101/gad.633311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surface LE, Thornton SR, Boyer LA. Polycomb group proteins set the stage for early lineage commitment. Cell Stem Cell. 2010;7:288–298. doi: 10.1016/j.stem.2010.08.004. [DOI] [PubMed] [Google Scholar]
- Tattermusch A, Brockdorff N. A scaffold for X chromosome inactivation. Hum Genet. 2011 doi: 10.1007/s00439-011-1027-4. [DOI] [PubMed] [Google Scholar]
- Wang J, Mager J, Chen Y, Schneider E, Cross JC, Nagy A, Magnuson T. Imprinted X inactivation maintained by a mouse Polycomb group gene. Nat Genet. 2001;28:371–375. doi: 10.1038/ng574. [DOI] [PubMed] [Google Scholar]
- Williams LH, Kalantry S, Starmer J, Magnuson T. Transcription precedes loss of Xist coating and depletion of H3K27me3 during X-chromosome reprogramming in the mouse inner cell mass. Development. 2011;138:2049–2057. doi: 10.1242/dev.061176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang F, Babak T, Shendure J, Disteche CM. Global survey of escape from X inactivation by RNA-sequencing in mouse. Genome Res. 2010;20:614–622. doi: 10.1101/gr.103200.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.