Abstract
MIP-2/CXCL2 is a murine chemokine related to human chemokines that possess the Glu-Leu-Arg (ELR) activation motif and activates CXCR2 for neutrophil chemotaxis. We determined the structure of MIP-2 to 1.9Å resolution and created a model with its receptor murine CXCR2 based on the coordinates of human CXCR4. Chemokine-induced migration of cells through specific G protein-coupled receptors is regulated by glycosaminoglycans (GAGs) that oligomerize chemokines. MIP-2 GAG-binding residues were identified that interact with heparin disaccharide I-S by NMR spectroscopy. A model a GAG:MIP-2:CXCR2 complex that supports a 2:2 complex between chemokine and receptor was created. Mutants of these disaccharide-binding residues were made and tested for heparin binding, in vitro neutrophil chemotaxis, and in vivo neutrophil recruitment to the mouse peritoneum and lung. The mutants have a 10-fold decrease in neutrophil chemotaxis in vitro. There is no difference in neutrophil recruitment between wild-type MIP-2 and mutants in the peritoneum but all activity of the mutants is lost in the lung supporting the concept that GAG regulation of chemokines is tissue-dependent.
Chemokines orchestrate leukocyte recruitment to sites of injury or infection. Some chemokines, particularly CXCL12, have roles during embryonic development (1). Chemotaxis is a complicated process that can be simplified into three events. First, chemokines are involved in transcytosis through the endothelium from the site of cellular secretion (2–5). Second, chemokines bind and activate specific G-protein coupled chemokine receptors (GPCRs) to initiate integrin-mediated adhesion and transmigration (6, 7). Finally, neutrophils respond to a chemokine gradient through the same GPCR to migrate through the matrix to the site of chemokine release (8). The interactions of glycosaminoglycans (GAGs) and chemokines regulate these individual processes (9, 10).
The chemokine superfamily is classified into four families based on the presence or absence of intervening amino acids between two conserved cysteines near the N-terminus that form disulfides with other cysteines in the protein sequence. Chemokines from three (CXC, CC, CX3C) of the four families essentially have the same monomeric structure, but tend to differ in their dimeric structure. The chemokine in the fourth family (XC) lacks one of the two cysteines, and forms two interconverting structures, a canonical chemokine monomer and a monomer with four β-strands lacking a C-terminal α-helix (11).
Chemokines activate their GPCRs in a family-dependent manner. While there may be a number of chemokines within a family that activate the same receptor, chemokines from different families never activate the same receptor. Structures of chemokines from all four families have been solved with most having a monomeric or dimeric structure, but higher oligomers up to a decamer have been determined (12, 13). The dimeric chemokines from each family are sufficiently different to explain family specificity, but most functional studies indicate that the activating chemokine is a monomer (14). There is also some evidence chemokine receptors can be activated by monomers and dimers, but monomers and dimers have different receptor affinities and activating properties, with the monomer having a higher binding affinity and eliciting most biological effects (15–17). More recently, structures of linear forms of oligomeric chemokines have been determined and proposed to have important roles in chemotaxis (13, 18–21). In contrast to the many structures available for chemokines, only one chemokine receptor (CXCR4) structure is available. CXCR4 was co-crystallized with two small molecule antagonists and is a dimer in five different space groups (22).
The interactions of chemokines with proteoglycans and glycosaminoglycans (GAGs) regulate biological activity (9, 10) but the mechanisms that lead to regulation are still unclear. GAGs enhance dimerization or oligomerization of chemokines (23–25) and may present chemokines to their receptors (4). It has also been suggested that GAG-mediated linear (as opposed to globular) oligomerization is important for presentation to receptors under flow conditions in the endothelium (19, 26). GAG-mediated oligomerization of chemokines (23–25) has also been proposed to mediate a chemokine gradient within the matrix that is necessary for chemotaxis (8). The functional effects of GAG-deficient mutant chemokines have been investigated in vitro and in vivo but the results are confounding (24, 27–29). The in vitro activities vary depending on chemokines. Some GAG-deficient mutants are as active as WT (30, 31), whereas GAG-deficient mutants of other chemokines are approximately 1000-fold less active (24). The in vivo activities also vary from increased cell recruitment (e.g., for CXCL8) to no activity at all (e.g., CCL2, CCL4, and CCL5) (30, 32) but, generally, there is no correspondence to the in vitro effects.
In the present study we determined the X-ray structure of MIP-2 to resolve whether the four monomers in the asymmetric (33) make linear oligomers recently reported for CCL4(34), CCL5 (21), CXCL10 (18, 26) and CXCL12 (13), speculated to be important for the function of chemokines (19). We were also interested in the interface between MIP-2 and murine CXCR2 (mCXCR2), and the structural and functional aspects of regulation by GAGs. A model of mCXCR2 was created based on the structure of human CXCR4 (22) and the MIP-2 X-ray structure to analyze the potential binding surface of MIP-2 and CXCR2. We also used NMR to identify MIP-2 residues that interact with a heparin disaccharide and developed a model of the GAG:MIP-2:CXCR2 complex. The GAG-binding residues identified by NMR were mutated to alanine and tested for in vitro chemotaxis and neutrophil recruitment to the mouse peritoneum and lung. We observe differences of GAG-deficient mutants between in vitro chemotaxis and the in vivo results, as well as differences of GAG-deficient mutants in in vivo neutrophil recruitment to the peritoneum and lung. This study was compared to neutrophil recruitment from CXCL8 GAG-deficient mutants to the murine peritoneum and lung (30, 31). This comparison of two chemokines in the ELR subfamily leads us to conclude GAG regulation is tissue- and chemokine-dependent and could differ dramatically in its effects, suggesting markedly different mechanisms.
EXPERIMENTAL PROCEDURES
Cloning, expression and purification of wild type and mutant MIP-2 in E. coli
For crystallization, recombinant MIP-2 was purified from Pichia pastoris with minor modifications of a protocol described previously (35). The P. pastoris clone expressing MIP-2 was grown at 30°C to an A600 of 2–6 in BMGY medium. For induction, the cells were transferred to BMMY media and induced at 30°C with 1% methanol every 24h for 72h. The protein was secreted into the medium and purified directly using a cation-exchange SP-sepharose column equilibrated with 25mM Tris-HCl, pH 7.4 and eluted with a linear gradient of 0–1M NaCl. The protein was further purified by size exclusion chromatography on a S75-sepharose column equilibrated with 0.1M ammonium acetate. The purified protein was concentrated to a final concentration of 10 mg/ml and used for crystallization.
For solution and functional studies, wild-type (WT) and mutants were made in E. coli. MIP-2 cDNA was cloned into pET-32a(+) between KpnI and NcoI sites, and expressed as an N-terminal His-tag and thioredoxin (Trx) fusion MIP-2 protein. The fusion protein carries an enterokinase cleavage site between the N-terminal tag and MIP-2. The cloning was designed to avoid any additional amino acids at the N-terminus of MIP-2 after enterokinase cleavage. MIP-2 carrying GAG-deficient mutations were generated using the multi-site mutagenesis kit (Stratagene). All clones were sequenced and transformed into BL21-Gold (DE3) E. coli cells for expression and purification. The protein expression was induced with 1mM IPTG at 37°C. N-terminal His-tag and thioredoxin (Trx) fusion MIP-2 protein was purified on a His Trap HP column (GE Healthcare) in a buffer containing 25mM Tris-HCl pH 7.4, 150mM NaCl, 5mM imidazole and EDTA-free protease inhibitor cocktail. The purified protein was dialyzed against enterokinase cleavage buffer (20mM Tris-HCl, pH 7.4, 50mM NaCl, 2mM CaCl2) and digested with enterokinase (Novagen) for 2–4hr at room temperature. The enterokinase-cleaved MIP-2 protein was collected in the flow through while the His-Trx product (and uncleaved protein) remained on the His Trap HP column. Fractions containing MIP-2 were pooled and were further purified on an YMC-Pack ODS-A C18 reverse phase-HPLC column equilibrated with 1% trifluoroacetic acid. The protein was eluted with a linear gradient of 0–55% acetonitrile. Purified MIP-2 was lyophilized and resuspended in sterile double distilled water. Due to the absence of aromatic amino acids, far UV absorbance using the formula 144 × (A215-A225) was used to determine protein concentration (36).
Crystallization, data collection, and processing
MIP-2 crystals were obtained by vapor diffusion in a hanging drop. An equal ratio of 10mg/ml of protein was mixed with reservoir solution containing 0.1M Tris, pH 8.5, and 30% PEG 3350. The crystals grew in 3 days and were briefly soaked in paratone oil (Hampton Research) as a cryo-protectant prior to X-ray diffraction data collection. Data from a single crystal were collected on an R-AXIS++ IV detector (Rigaku) to 1.9Å resolution at the Macromolecular X-ray Crystallography Facility at the Yale School of Medicine. The data set was processed and scaled using the HKL-2000 software package (37).
Structure determination and refinement
The structure of MIP-2 was solved by molecular replacement using the IL-8 structure (PDB code 3IL8) as a search model (38) and the program Molrep (39). The asymmetric unit contained two independent dimers. Model building and crystallographic refinement were carried out using the programs Coot (40) and Refmac (41), respectively. A bulk-solvent correction was applied with restrained individual B-factor refinement. The two dimers were refined and water molecules were added, resulting in a R and Rfree of 18.8% and 22.6%, respectively. The program PROCHECK (42) was used to check the model. All back bone torsion angles were within allowed regions of the Ramachandran plot (PDB code 3N52). Data and refinement statistics are shown in Table 1.
Table 1.
Summary of data-collection and refinement statistics for MIP-2.
| Data statistics | |
| Space group | P212121 |
| Unit-cell parameters (Å) | |
| a | 41.76 |
| b | 59.45 |
| c | 99.35 |
| Resolution (Å) | 1.9 |
| No. of reflections | 101689 |
| Unique reflections | 20213 |
| Completeness (%) | 99.9 (100) |
| Redundancy | 5.0 (5.0) |
| Rmerge (%)a | 3.9 (10.1) |
| I/σ(I) | 30.443(14.927) |
| Solvent content (%) | 37 |
| No of Protein molecules in ASU | 4 |
| Refinement statistics | |
| No. of reflections | 19059 |
| Rfactor (%)b | 18.7 |
| Rfree (%)b | 22.5 |
| No. of protein atoms | 2038 |
| No. of water molecules | 263 |
| Average B factors (Å2) | 45.6 |
| R.m.s.d. from ideal values | |
| Bond lengths (Å) | 0.030 |
| Bond angles (°) | 2.459 |
| Ramachandran statistics (%) | |
| Most favored | 96.5 |
| Favored | 3.5 |
| Additionally allowed | |
| Disallowed | |
Values in parentheses are for the high-resolution shell (1.97–1.90Å).
Rmerge = 100×ΣhΣi|(h)l−〈(h)〉|/ΣhΣi〈(hi)〉, where I is the observed intensity, and 〈I〉 is the average intensity of multiple observations of symmetry-related reflections.
R = Σ||Fo|−|Fc||/Σ|Fo|. Rfactor and Rfree were calculated using the working and test reflection sets, respectively. 5% of the entire reflection was randomly taken as a test set.
15N-labeled MIP-2 and NMR spectroscopy
For labeling with 15N, E. coli cells were grown in M9 minimal media containing 1.0 g/liters of 15NH4Cl (98% 15N) and purified as described above. The lyophilized protein was dissolved in 0.1M ammonium acetate, pH 6.0 and 5% D2O for NMR studies. Samples for NMR spectroscopy contained 300μM of 15N-MIP-2 with varying concentrations (0–2.4mM) of heparin disaccharide I-S (Sigma). Experiments were carried out at 25°C in a Varian INOVA 600 MHz spectrometer with a 5-mm triple resonance probe equipped with triple-axis (XYZ) pulsed magnetic field gradients. All pulse sequences were taken from the Varian BioPack user library. Spectra were processed and analyzed using the programs nmrPipe (43) and Sparky 3.114. Assignment of resonance peaks was done using 15N-NOESY-HSQC NMR spectra of MIP-2 and previously published resonance assignments (44). Absolute NMR chemical shift change of each residue for the disaccharide titration. were calculated with a normalizing factor of 6.5 adapted from Mulder et al (45).
Heparin sepharose and cation exchange chromatography
One hundred micrograms of WT and mutant MIP-2 proteins were loaded onto a 1ml heparin sepharose column (GE Healthcare) or 1ml SP-sepharose cation exchange column (GE Healthcare) in 20mM Tris-HCl, pH 7.4 buffer and eluted with a 25ml linear gradient (0–1M NaCl in 20mM Tris-HCl, pH 7.4 buffer) at a flow rate of 0.5ml/min. The protein was monitored by absorbance at 215nm, and the concentration of NaCl was determined using an in-line conductivity meter (46).
Structural alignments and models
The Cα comparison of dimers in the asymmetric unit (ASU) was performed using LSQKB from the CCP4 suite. For structural superposition the secondary structure matching (SSM) algorithm in CCP4 package was used. The model of the murine CXCR2 structure was created by homology modeling using the sequence and structure of CXCR4 (without the T4 lysoyme coordinates inserted between 229 and 230 of CXCR4) (22) in the SWISS-MODEL server (47). The sequence identity of the CXCR2 model generated with the CXCR4 template (without the N-terminal region not modeled in CXCR4) had an identity of 36% (Figure S4). The structure of CXCR2 was modeled from amino acid 40–329. The dimer of CXCR2 was obtained by aligning the monomer model of CXCR2 onto the CXCR4 template dimer. The orientation of the dimeric MIP-2 crystal structure relative to CXCR2 is derived from knowledge of each protein’s binding site, ELR for MIP-2 and the CXCR2 cavity as defined by the location of two antagonists for CXCR4. The MIP-2 GAG-binding site was determined by NMR and mutagenesis experiments. Positioning the GAGs was based on a similar model of CXCL8 (30, 48), which has basic residues at the α-helix. The MIP-2 molecule in the MIP-2:heparin oligosaccharide complex was aligned to the MIP-2 molecule in the CXCR2:MIP-2 complex to obtain the tertiary complex of GAG:MIP-2:CXCR2. The electrostatic potential maps were calculated from PyMol (49).
Measurement of endotoxin
The concentration of endotoxin in the recombinant proteins was measured using the Pyrogene Recombinant Factor C Endotoxin Detection system (Cambrex) following manufacturer’s instructions. All the proteins (20μg) had less than 1 EU of endotoxin.
In vitro chemotaxis of BM derived Neutrophils
8–10 weeks old female BALB/c were obtained from the Jackson Laboratory and housed in the Yale animal facility until the day of experiment. Murine bone marrow cells were harvested from femurs and tibias. Neutrophils were isolated from bone marrow by discontinuous percoll gradient as previously described (50). Chemotaxis was assayed in MultiScreen 96-well filtration plates (Millipore) of 3μm pore size. A total of 75 × 103 cells were placed in the upper chambers, chemokines were placed in the lower wells at different concentrations, and incubated for 2h at 37°C. The bottom wells containing the migrated cells (100μl) were transferred to a 96-well plate and mixed with an equal volume of cell titer-Glo reagent from a cell viability assay kit (Promega). The cell number was measured by luminescence and the experiments were performed in triplicates.
Peritoneal cell recruitment
Female BALB/c mice (8–10 weeks old) were injected intra-peritoneally with 100μl of 0.9% NaCl (lipopolysaccharide (LPS)-free) or LPS-free chemokine diluted in 0.9% NaCl. At 4h post injection the mice were sacrificed with CO2. Peritoneal lavage was performed 3 times with 5ml HBSS, and the lavages were pooled for individual mice. Cells were centrifuged at 1500 rpm for 5min, and washed twice in HBSS. The cells were stained with PE anti-mouse CD45 (leukocyte common antigen, Ly-5, e-biosciences, 12-0451) and APC anti-mouse Ly-6G (Gr-1) neutrophil specific antibodies (e-biosciences 17-5931) at concentrations recommended by the manufacturer for fluorescence-activated cell sorting analysis.
Broncho-alveoar lavage (BAL)
Female BALB/c mice (8–10 weeks old) were intranasally administered with 1 or 10μM of WT-MIP-2 or GAG-deficient MIP-2 mutants in 0.9% NaCl. After 4hrs, 3 lavages of 0.5ml were collected. The BAL fluids were pooled and immediately processed for differential cell counts. The differential cell counts were performed on cytocentrifuge preparations stained with HEMA 3(r) stain set (Fisher Scientific Company, MI). A minimum of 200 cells were counted for the differential cell count. For all mice experiments, the use and care of animals were approved by the Institutional Animal Care and Use Committee at Yale University.
Statistical analysis
All data were analyzed using Prism 4 software (GraphPad, San Diego, CA, USA). The Student two-tailed t test was used for statistical analysis.
RESULTS
The X-ray structure of MIP-2
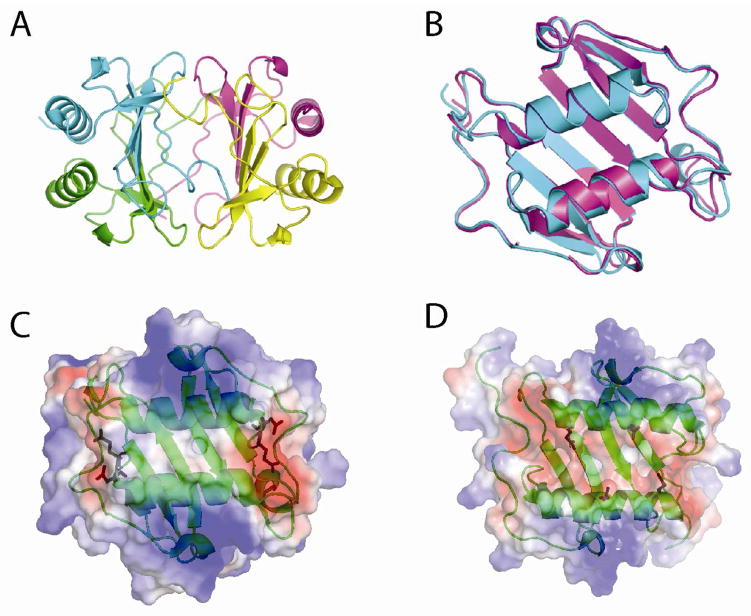
A preliminary study of MIP-2 crystals indicated there were four subunits in the asymmetric unit and a self-rotation function identified the presence of a non-crystallographic 2-fold axis (33). Possible solutions include a globular tetramer similar to structures of CXCL4, CXCL7 and CXCL10 with 2-fold or 222 symmetry (18, 51, 52). Another solution is two independent dimers similar to the NMR structure of the MIP-2 dimer (44). Another possibility included a linear structure similar to the CXCL10 and CCL5 tetramers (18, 21), CCL4 or the CXCL12 decamers (13, 34). Given the recent interest on the role of linear oligomerization (beyond a dimer) on chemotaxis, we were interested in determining the structure of MIP-2. The MIP-2 X-ray structure was solved to 1.9Å. The crystallographic statistics are shown in Table 1. The space group has two independent dimers in the asymmetric unit (Figure 1A). The structure of MIP-2 has the typical chemokine fold and dimerizes similar to other CXC chemokines. Using only Cα atoms and the Superpose module in the CCP4 (53), two MIP-2 dimers in the asymmetric unit were compared to each other and have a root mean square deviation (rmsd) of 0.54Å (Figure 1B). There were no contacts between the dimers. We calculated the surface electrostatic potential for both the MIP-2 monomer and dimer and found a highly electropositive potential with the exception of the dimeric interface, which is hydrophobic, and the loop prior to the C-terminal α-helix that is electronegative due to D53 and E55 (Figure 1C). CXCL8 has a similar electropositive potential on the outer part of the helices but an electronegative potential between the helices due to glutamic acid residues at positions 63 and 70 (Figure 1D).
Figure 1.
Tertiary structure of MIP-2. A, Four molecules present in the asymmetric unit are represented as cartoons and colored uniquely. Monomers in blue and cyan form one dimer, and the pink and yellow monomers form the other dimer. B, Ribbon structures of superposition of Cα atoms of two dimers in the asymmetric unit of the MIP-2 X-ray structure (RMSD 0.54Å). The superposition was based on matching the secondary structures and calculating an rmsd of the entire structure. C, The electrostatic potential surface of MIP-2. The electronegative region is shaded red and the blue color represents electropositive region. Side chains for D53 and E55 are shown with stick figures for each subunit. D, The electrostatic potential of CXCL8. Side chains for E63 and E70 are shown with stick figures in each subunit. MIP-2 in C is neutral between the helices with increased negative potential in the same location for CXCL8. The side chains for Figures 1C and 1D are the source of the electronegative potential for MIP-2 and CXCL8, respectively.
Comparison of the X-ray and NMR structure
For comparison of the X-ray and NMR structures (44), a representative model of the NMR ensemble determined by Cluster and domain composition (OLDERADO) was used. This OLDERADO program automatically clusters ensemble members into conformationally-related subfamilies and the centroid of the largest cluster is selected as the “most representative” model (54). Use of this method to select the model eliminates the loss of the most important conformations for molecular function represented in subsets of models within the final ensemble (54). Cα atoms derived from the MIP-2 X-ray and NMR structures (44) were superposed with a RMSD of 2.3Å. While the sheets superpose well, most deviations are found at the N–terminal sequences, 30s and 40s loops, and the C-terminal α-helix (Figure S3). In a recent comparative study of NMR and X-ray derived structures of the same protein, the RMSD of the compared structures is usually between 1.5 and 2.5Å and the β-strands, on average, match better than helices (55). Apart from the flexible regions, the structures are well conserved. The minimum and maximum distance of the receptor activation ELR motif in the two subunits of the NMR and X-ray MIP-2 dimer are between 42–49Å and 32–42Å respectively (see following section).
A three-dimensional model of the MIP-2 and mCXCR2 complex
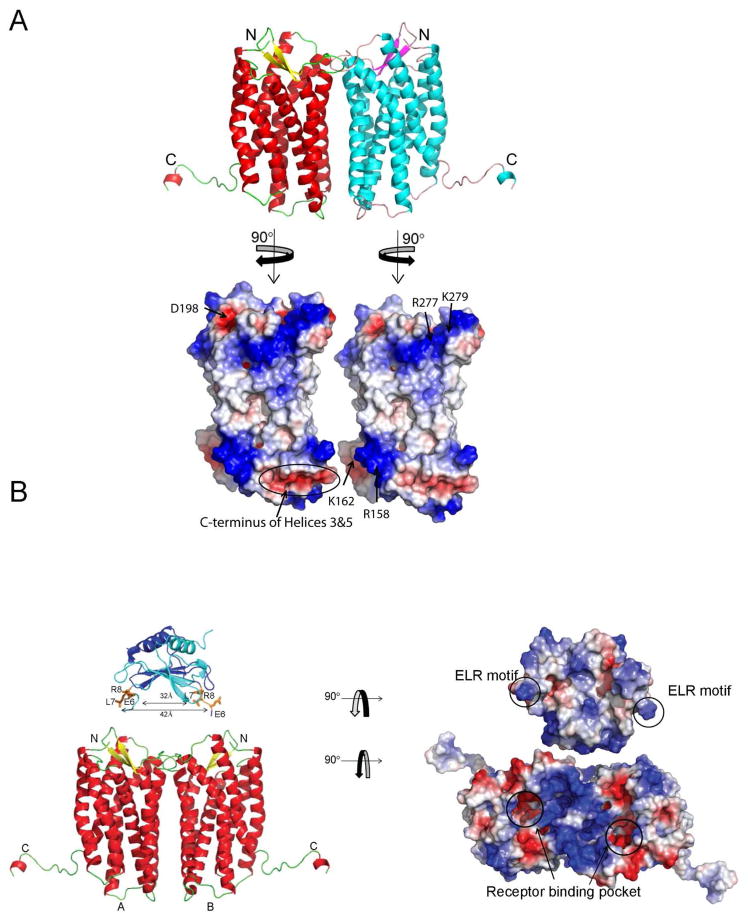
We generated a model of mCXCR2 (residues 40–329) based on the coordinates of human CXCR4, a dimer in five different space groups (22). The electrostatic potential of dimeric mCXCR2 is shown in Figure 2A. To analyze the feasibility of a dimeric structure for CXCR2, the electrostatic potential for the dimeric interface was explored for any unfavorable electrostatic interactions that would exclude the formation of a receptor dimer. The extracellular regions consisted of an electropositive potential from amino acids K279 and R277 interacting with D198. The intracellular regions had electrostatic interactions between R158 from one subunit with the electronegative dipoles at the C-terminal ends of helices 3 and 5 (Figure 2A, bottom panel). Due to the 2-fold symmetry these interactions occur twice, providing additional free energy for the formation of the dimer. Once the electrostatic potential for the dimeric form was shown to be feasible, the electrostatic potential of the chemokine dimer and receptor as well as distances between the dimer binding regions of each subunit for MIP-2 and CXCR2 were examined. The N-terminal region of MIP-2 is highly electropositive from residues R8, K12 and R17 and has one electronegative amino acid E7. The electrostatic potential from these residues complement two regions from mCXCR2 binding pocket, the electronegative potential contributed by E41, E197, D273, E286 D289, D290, D292 and electropositive residue K293 (Figure 2B right panel). The minimum and maximum distances of atoms in the ELR between monomers were 32Å and 42Å (42Å–49Å for the NMR structure), respectively. For CXCR2, the minimum and maximum distances from one binding site to the other ranged from 32Å and 49Å, respectively. These distances overlap and are consistent with the electrostatic complementarity of the MIP-2 ELR motif and the receptor binding sites. This analysis supports a 2:2 complex where each ELR could interact at the transmembrane binding pocket of each CXCR2 subunit. We note this model does not formally exclude MIP-2:CXCR2 models with a 1:1, 1:2, or 2:1 stoichiometry. In this regard, we note human CXCL8 studies functioning in vitro and in vivo as a monomer and dimer but models with human CXCR1 and CXCR2 are not described (16, 56, 57).
Figure 2.
Model of murine CXCR2 dimer bound to MIP-2 dimer. A, Murine CXCR2 was modeled using SWISS-MODEL server with CXCR4 as template (PDB code:3ODU) and is colored based on secondary structure. The electrostatic potential of the dimer interface is shown in the lower panel. Each monomer surface potential complements the other subunit. B, The orientation of the MIP-2 crystal structure above CXCR2 (A and B represent monomers) is derived from knowledge of the binding sites of each protein. The distance between the binding site cavity for each subunit of CXCR2 receptor overlaps with the distance between each ELR in the MIP-2 dimer. Monomers of MIP2 are represented as ribbon structures (dark blue and cyan), with the side chains of ELR motif as sticks (orange). The CXCR2 and MIP-2 were rotated 90° in opposite direction and the electrostatic potential calculated to reveal the potential interacting surfaces (right panel). ELR motifs in the MIP-2 dimers are circled and the arrows in the CXCR2 model represent the potential binding cavity.
Mapping the interaction of heparin disaccharide with MIP-2 by NMR spectroscopy
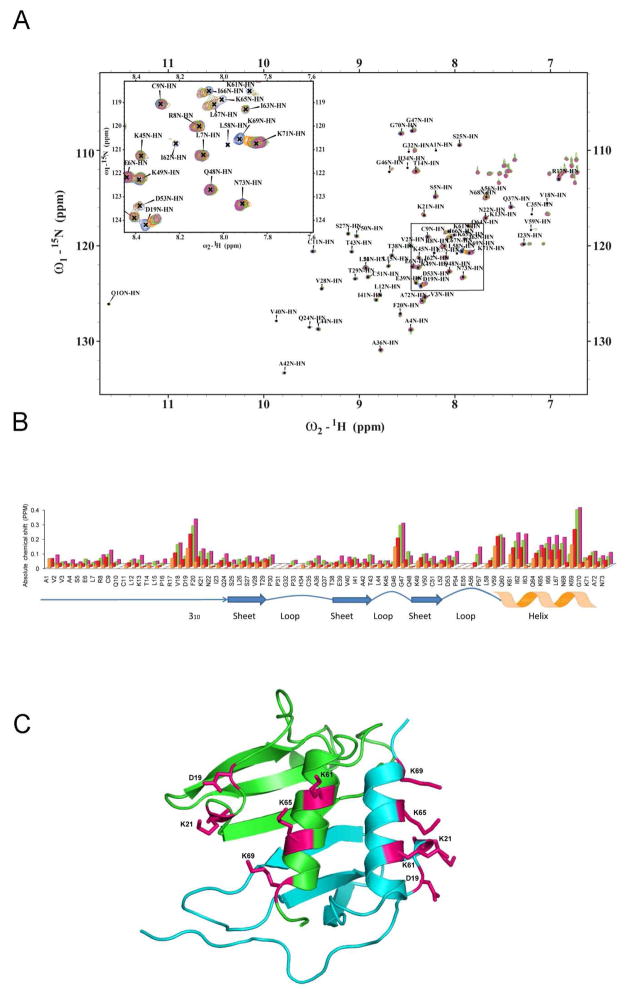
We were interested to determine the effects of glycosaminoglycan on the structure and function of the MIP-2/CXCR2 model. We first attempted to co-crystallize MIP-2 with various lengths of heparin (disaccharides, tetrasaccharides, and hexasaccharides) but all attempts were unsuccessful. The tetrasaccharides and hexasaccharides led to immediate precipitation. Attempts to soak MIP-2 crystals with an unsaturated heparin disaccharide I-S (α-4-deoxy-l-threo-hex-4-enopyranosyluronic acid-2-sulfate-(1→4)-d-glucosamine-N-sulfo-6-sulfate), a common model disaccharide to structurally model GAG interactions (Figure S1) (58, 59), were also unsuccessful. We therefore used the previously assigned chemical shifts of MIP-2 for heparin disaccharide binding experiments (44). 1H-15N-HSQC spectra were collected from solutions containing 300μM MIP-2 with various concentrations of heparin to probe chemical shift changes induced by addition of the disaccharide. Figure 3A shows the 1H-15N-HSQC spectrum of MIP-2 overlaid with spectra obtained from mixtures containing MIP-2:heparin disaccharide mixed in four different molar ratios (1:2, 1:4, 1.8 and 1:16). In this titration experiment the overall chemical shifts (and structure) of MIP-2 is maintained as seen in the overlay of the spectra. For each resonance peak observed in the apo-MIP-2 spectrum a corresponding peak occurs in each of the four spectra from the MIP-2:disaccharide titration. A number of peaks showed a concentration dependent change in resonance positions. Comparison of the absolute change in NMR chemical shift for each residue in the MIP-2:disaccharide complex (Figure 3B) reveals major changes in three regions: (1) the N-loop preceding the 310 helix, (2) two glycines of the 40s loop, and (3) a basic region on the C-terminal alpha helix (Figure 3C). These three groups of residues cluster to a region comparable with the binding site of heparin disaccharide in CXCL8 (46). Of these residues, the charged surface residues D19, K21, K61, K65 and K69 lie close to one another within a monomeric unit and are presumably being directly affected by disaccharide binding (Figure 3C). On the other hand changes in G46 and G47 may be due to indirect effects such as loop dynamics during binding as these are situated away from the major patch. Four alanine mutants were generated using combination of residues: (1) K61, K65 and K69 (3K-MIP-2); (2) K21, K61, K65, and K69 (4K-MIP-2); (3) D19, K61, K65, and K69 (3K1D-MIP-2); and (4) D19, K21, K61, K65 and K69 were (4K1D-MIP-2). Mutants of G46 and G47 were not made due the presumption that these chemical shifts were from indirect effects such as loop dynamics during binding.
Figure 3.
NMR spectroscopic studies of murine MIP-2 and heparin disaccharide interaction. A, NMR chemical shift changes induced by titration of heparin disaccharide I-S. 1H-15N-HSQC spectra of 300μM 15N-MIP-2 (blue) are overlaid with spectra from molar ratios of 15N-MIP-2 to disaccharide of 1:2, 1:4, 1:8, and 1:16 (orange, red, green, and pink), respectively. Peak labels indicate peaks for the uncomplexed (apo) protein. B, Absolute NMR chemical shift change of each residue for the disaccharide titration. Absolute NMR chemical shift change for each ratio are calculated as (((|Nppm-bound − Nppm-apo|) + (|Hppm-bound − Hppm-apo| × 6.5))/2). Uncomplexed 15N-MIP-2 compared to 15N-MIP-2:heparin disaccharide I-S ratios of 1:2 (orange), 1:4 (red), 1:8 (green), and 1:16 (pink). Changes in NMR chemical shifts for proline residues are reported as zero as they lack an amide proton. Regions of secondary structure are indicated below with block arrows representing β regions and zigzags representing helical regions. C, The chemical shifts are mapped and the residues are shown as sticks on a ribbon diagram of the crystallographic dimeric (green and blue monomers) structure of MIP-2.
Affinity of MIP-2 mutants to heparin
The specific heparin-binding affinity of the WT and each of the MIP-2 mutants was assessed by specific and non-specific sepharose chromatography as described by Kuschert and colleagues (46). The sodium chloride concentration required to elute the mutant proteins from a heparin-sepharose column was determined and compared to WT, yielding ΔNaClHep, where Δ[NaCl]Hep= [NaCl]Hep WT − [NaCl]Hep mutant. WT MIP-2 eluted from the heparin sepharose column at 0.51M NaCl. The four MIP-2 mutants eluted at much lower NaCl concentrations with the triple lysine mutant (3K-MIP-2) and quadruple lysine mutant (4K-MIP-2) eluting at 0.18 M and 0.17 M of NaCl, respectively. The addition of the aspartate to alanine mutant to each of the lysine mutants, 3K1D-MIP-2 and 4K1D-MIP-2, resulted in elution at 0.21M and 0.26M NaCl, respectively. To measure the non-specific, electrostatic interactions of the four mutants, the amount of NaCl required to elute mutants from a SP-sepharose column was compared to WT, yielding ΔNaClS, where Δ[NaCl]S = [NaCl]S WT − [NaCl]S mutant. The specificity of the interaction with heparin (ΔΔ[NaCl] = Δ[NaCl]H − Δ[NaCl]S) was determined as previously described (46) and all values for heparin binding, non-specific interactions, and the specificity values are show in Table 2. Substitution of lysines in the helices of the 3K–MIP-2 mutant decreased the affinity. The addition of a lysine (K21A) and aspartate (D19A) mutations to the 3K-MIP-2 mutant (3KD1-MIP-2, 4K-MIP-2, and 4KD1-MIP-2) does not have much effect on specificity compared to 3K-MIP-2 alone.
Table 2. Specificity of Heparin binding to MIP-2 mutants.
Specificity of each mutant is according to the method previously described (46). Δ[NaCl]Hep and Δ[NaCl]S is the difference in NaCl concentration required to elute the wild type MIP-2 compared to each mutant from heparin and S-Sepharose, respectively. ΔΔ[NaCl] is the specificity of heparin binding determined by heparin sepharose affinity after subtraction of nonspecific electrostatic binding as determined from S-Sepharose affinity (ΔΔ[NaCl] = Δ[NaCl]Hep − Δ[NaCl]S). All concentrations are molar.
| Heparin-Sepharose (M) | S-Sepharose (M) | ΔNaClHep (M) | ΔNaClS (M) | ΔΔNaCl (M) | |
|---|---|---|---|---|---|
| WT | 0.51 | 0.40 | |||
| 3K-MIP-2 | 0.18 | 0.26 | 0.33 | 0.14 | 0.19 |
| 3K1D-MIP-2 | 0.21 | 0.25 | 0.30 | 0.15 | 0.15 |
| 4K-MIP-2 | 0.17 | 0.25 | 0.34 | 0.15 | 0.19 |
| 4K1D-MIP-2 | 0.26 | 0.29 | 0.25 | 0.11 | 0.14 |
Model of GAG:MIP-2:CXCR2 complex
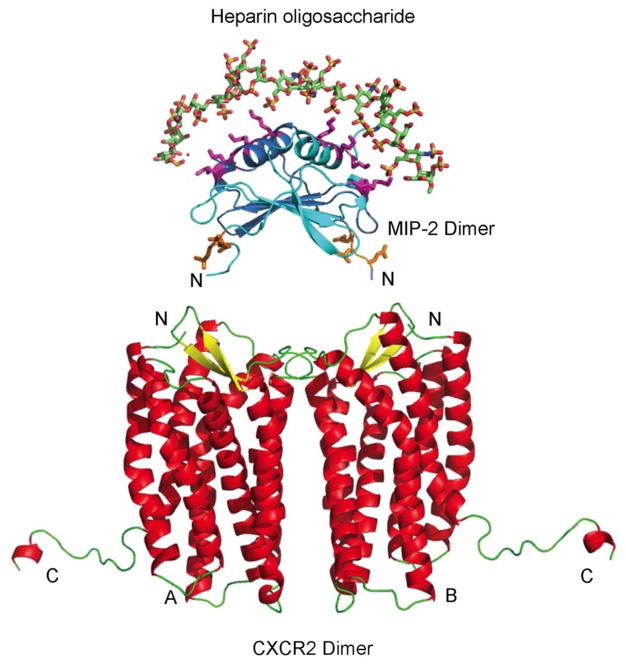
The disaccharide site provided a starting point to model a larger GAG. The placement of the lysine residue on the wild type MIP-2 allowed us to build GAG:MIP-2 interface across the two helices as shown in Figure 4, similar to what was previously predicted (48). To create the trimeric complex of a octadecameric glycosaminoglycan (alternating 2-N-sulfated, 6-O-sulfated α-D-Glc, and 2-O-sulfated β-L-IdoA repeating monosaccharides), MIP-2, CXCR2, the two complexes (GAG:MIP-2 and MIP-2:CXCL2) were superimposed using only the MIP-2 coordinates. The GAG was analyzed for potential effects on MIP-2 binding to CXCR2. First, the GAG interactions with MIP-2 do not interfere with the N-terminal region and the 30’s loop responsible for receptor binding, nor with ELR motif involved in receptor activation. Second, the GAG stabilizes the MIP-2 dimer and promotes the 2:2 stoichiometry between MIP-2 and CXCR2,
Figure 4.
GAG:MIP-2:CXCR2 interface model. The interface of heparin oligosaccharide, MIP-2 chemokine, and CXCR2 is shown in the model. The octadecameric heparin oligosaccharide encompasses the MIP-2 dimer over the helices. The monomers of MIP-2 are represented as ribbons (dark blue and light blue) with the side chains of heparin-interacting residues and the ELR motif as sticks (pink and orange, respectively). The heparin oligosaccharide MIP-2 model was aligned to MIP-2 of the MIP-2:CXCR2 complex (Figure 2) to obtain the tertiary model complex. The heparin oligosaccharide:MIP-2 interaction stabilizes MIP-2 dimer and does not interfere with binding to CXCR2.
In vitro and in vivo migration activities of MIP-2 mutants
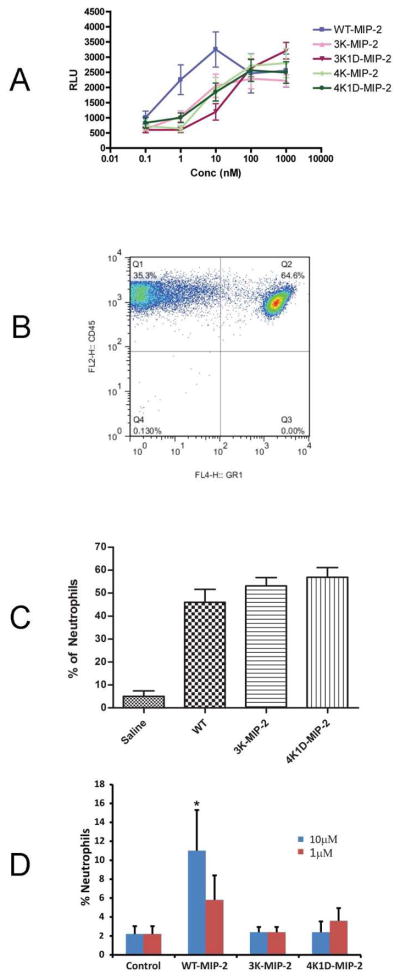
An in vitro migration assay was used to examine the chemotactic activity of WT and MIP-2 mutants in recruiting bone marrow derived neutrophils. A full dose-response experiment for bone marrow derived neutrophils was performed (Figure 5A). The study shows significant decreases in chemotaxis at 10nM from all four mutant (3K-MIP-2, 4K-MIP-2, 3K1D-MIP-2, and 4K1D-MIP-2) chemokines relative to WT MIP-2 (Figure 5A). At 100nM, WT and mutants reach an equivalent maximum of neutrophil chemotaxis. The EC50s of the MIP-2 mutants are approximately 50nM, 10-fold higher than the WT MIP-2 with an EC50 of 5nM. This effect is similar to in vitro chemotaxis studies of CC chemokine GAG-deficient binding mutants. RANTES GAG-deficient mutants have an EC50 1000-fold greater compared to WT, and MCP-1 and MIP-1β GAG-deficient mutants have a 10-fold increase in EC50 relative to their respective wild type proteins (24). Similarly, CXCL8 GAG-deficient mutants have a 10-fold increase in EC50 in vitro chemotaxis activity compared to WT (30).
Figure 5.
In vitro and in vivo chemotaxis effects of MIP-2 GAG-deficient mutants. A, In vitro chemotaxis of neutrophils in response to WT and MIP-2 mutants at various concentrations. The migrated neutrophils were measured by luminescence represented as relative luminescence unit (RLU). The graph represents the mean of three different experiments done in triplicate (***, p < 0.0001; *, p < 0.01). B, C and D, Effects of GAG-deficient mutations of MIP-2 in vivo. B, Representative FACS analysis of neutrophils (CD45+GR-1+) recruited to the peritoneum in response to 300ng IP injection of WT. C, Percentage of neutrophils recruited to the peritoneum in response to WT, 3K-MIP-2, and 4K1D-MIP-2. D, Percentage of neutrophils recruited in BAL in response to instillation of 1μM or 10 μM of WT MIP-2 or GAG-deficient mutants 3K-MIP-2, and 4K1D-MIP-2. The in vivo data represent the mean number of neutrophils from six female BALB/c mice of 8–10 weeks of age.
We next tested the heparin-binding mutants of MIP-2 for recruitment of neutrophils to the mouse peritoneum and lung. In previous dose-response study, 0.5μg of MIP-2 elicited the maximum recruitment of PMNs to the peritoneum 4h after injection (60). A dose of 0.1μg or 0.3μg of LPS-free WT or only two mutant proteins 3K-MIP-2 and 4K1D-MIP-2 (representing the areas of GAG binding identified by NMR titration) was injected into the peritoneum to determine the effect on neutrophil recruitment. (Figure 5C and Figure S2). Only neutrophils as determined by FACS analysis were measured (Figure 5B). Unlike the in vitro neutrophil migration assays that showed a 10-fold difference for all mutants, the in vivo experiments resulted in equal amounts of neutrophils after injection of WT MIP-2 or either of the two mutants (Figure 5C), suggesting that binding of GAGs is unnecessary for in vivo migration to the peritoneum.
We also tested the same two mutants and WT MIP-2 on neutrophil recruitment to the lung. We confirmed the results of Tanino et al that there is very little neutrophil recruitment to the lung at 1μM WT MIP-2 (30). At 10 μM WT MIP-2, however, there was significant increased neutrophil recruitment while the two mutants remained totally inactive (Figure 5D). This result was unlike the instillation of CXCL8 GAG-deficient mutants that resulted in an increase in PMN infiltration compared to WT CXCL8 (30). One of the CXCL8 mutants, K64A/K67A/R68A, has mutant residues similar to those of the mutant 3K-MIP-2 (K61A/K65A/K69A). These unexpected major differences between MIP2 and CXCL8 underscore the need to fully characterize GAG-chemokine structural and functional interactions.
DISCUSSION
Most but not all experiments support a chemokine monomer activating a receptor during in vitro chemotaxis but the role of monomers, dimers, or higher oligomers in vivo leads to confounding results when compared to the in vitro studies or to in vivo studies of other chemokines (24, 29–31, 61). Glycosaminoglycans are involved in chemokine function in vivo and would provide a reasonable explanation for the different observations if GAGs displayed a consistent effect. The roles of these heterogeneous molecules, composed of various small compounds linked to form larger molecules of various sizes, add to the complexities in understanding chemokine biology and its regulation. For example, GAGs oligomerize chemokines, which leads to either activation or inhibition of receptors (24, 30). The endothelial cells produce and coat the cell surface with GAGs promoting interactions with chemokines that bind to leukocytes and begin the process of rolling on the endothelium prior to extravasation (62). GAGs also protect N-terminal sequence from proteolysis that converts most chemokines from an agonist to an antagonist (63–66), supporting a role for maintaining the activated state of chemokines. In this study we focused on the role of GAGs on murine MIP-2, produced a model of the interface of MIP-2 with GAGs and the CXCR2 receptor, assayed the in vitro chemotactic activity and in vivo recruitment of neutrophils by wild-type MIP-2 and GAG-deficient mutants to the lung and peritoneum.
MIP-2 crystal structure and a model of the MIP-2:CXCR2 interface
Linear oligomerized forms of chemokines have been proposed to be important in the mechanism of in vivo cell recruitment (10–14). These oligomerized forms could be as short as a tetramer for CCL5 (24, 67) and CXCL10 (20) or as long as a decamer found in a crystal forms of CCL4 (34) and CXCL12 (13). Our interest in determining the structure of MIP-2 was partly based on a crystal form with four subunits in the asymmetric unit (33), suggesting it may form an additional oligomer chemokine. The X-ray crystal structure of MIP-2, however, consisted of two independent dimers with no structural similarity to the linear forms of chemokine oligomers (13, 20, 21). We compared the X-ray and NMR structure (44) and found very high structural similarity with the largest differences at the 30’s and 40’s loops, and the C-terminal helices (Figure. S3). The larger changes were at the helices rather than the β-sheet, consistent with an analysis of protein structures determined by both NMR and X-ray crystallography (55).
We were also interested in exploring the structural relationship between MIP-2 and its receptor mCXCR2. Given the difficulty in making accurate models of GPCRs based on the current number of available structures of GPCRs (68), we resorted to making a rudimentary model of CXCR2 and examining both the stoichiometry of the MIP-2:CXCR2 and the MIP-2 interface with the receptor and GAGs. A model of mCXCR2 was created based on the CXCR4 structure (68). The two sequences share 31% identity and have similar locations for cysteines that make disulfides in the extracellular region of CXCR4 (Figure S4). Based on the alignment with CXCR4, Cys 38 covalently bonds with Cys 285, and Cys 195 makes the second extracellular disulfide with Cys 118 in CXCR2. The model was created by replacing the CXCR4 residues with those of CXCR2 and by deleting the T4 lysozyme in the intracellular loop 2 (ICL2) that helps with crystallization. The electron density for the first 39 amino acids in CXCR4 is missing indicating this region is flexible. Therefore we could not model these residues for mCXCR2. The extracellular regions consisted of an electropositive potential from amino acids K279 and R277 interacting with D198. The intracellular regions had electrostatic interactions between R158 from one subunit with the electronegative dipoles at the C-terminal ends of helices 3 and 5. The transmembrane helices at the interface were, as expected, hydrophobic. Although this is a rudimentary model, we could not identify any regions that would be electrostatically repulsive and prevent dimer formation but these criteria do not eliminate a monomeric mCXCR2 as a functional subunit. The feasibility of a dimeric mCXCR2 was supported by experimental studies on human CXCR2 indicating this receptor is a ligand-independent dimer (69, 70). Having established a potential dimeric form for mCXCR2, knowledge of the binding sites for both chemokine and receptor was important in positioning the MIP-2 dimer and mCXCR2 in a proper orientation for interactions. The ELR signaling motif between the two subunits has a minimum and maximum distance of 32Å distance of 42Å, respectively. The more flexible N-terminal domain of the NMR structure provides distances of 42Å to 49Å. A similar analysis between the mCXCR2 binding pockets provides minimum and maximum distances of 32Å and 49Å, respectively, supporting a 2:2 stoichiometry. The maximal distance and volume of the binding site (3309Å3) for the receptor are consistent with accommodating the ELR motif within the MIP-2 N-terminal sequence AVVASELR. The length of this flexible peptide of MIP-2 is also consistent with the depth of the CXCR2 binding pocket (1177Å) and larger hexadecameric peptide antagonist (CVX15) complexed to CXCR4 in the crystal structure (22). The known orientation of MIP-2 binding to mCXCR2 dimer is also important in the analysis of electrostatic complementarity between chemokine and receptor. Figure 2B reveals a remarkable complementary that identifies residues potentially involved in these interactions. In addition, the unmodeled mCXCR2 N-terminal region important for chemokine binding contains seven electronegative residues and two lysine residues, resulting in a electronegative potential absent in the model that would complement MIP-2 even greater than shown in Figure 2.
The functional significance of the MIP-2:CXCR2 model remains unknown but a MIP-2 dimer is supported by studies of other CXC chemokines. A disulfide-locked dimer of CXCL12 induces some but not all of the signaling pathways of the WT protein in cellular studies (17). CXCL8 is more complicated because it activates both the human CXCR1 and CXCR2 receptors. A cellular study showed monomeric CXCL8 mutant is more active than a disulfide-locked dimer mutant in mediating a range of signaling pathways using CXCR1, but there is little difference between CXCL8 monomer and dimer using CXCR2. This was evident despite a ~5–6-fold higher affinity of the mutant monomer than the locked-dimer for both receptors, with WT CXCL8 having a Kd values between those of mutant monomer and locked-dimer for both hCXCR1 and hCXCR2 (71). In vivo neutrophil recruitment to the mouse lung by the monomer mutant or dimer mutant (or WT) were both necessary for activity, but the maximum number of recruited neutrophils is dependent on different doses and time points after instillation for WT and each mutant (31, 56). It is worthwhile noting another possibility for these different in vivo results between MIP-2 and CXCL8. MIP-2 and CXCL8 are potent and weak agonists for mCXCR2, respectively, with a Kd of 2–5 nM (35, 72, 73) for MIP-2 and a Kd of 65–400 nM for CXCL8 (73–75). Nonetheless, mCXCR2 is presumed to be the receptor that mediates human CXCL8 in published mice studies due to the inability of mCXCR2−/− mice to respond to CXCL8 (73, 74). However, a murine homologue of human CXCR1 has been cloned but role of CXCL8 as an agonist is controversial with one study showing no CXCL8 (75) and another study reporting a Kd of ~10 nM (and a Kd of 643 nM for MIP-2) (76). If murine CXCR1 and CXCR2 form heterodimers, as shown for human receptors (70), CXCL8 might target this complex in mice in these cross-species experiments using a human chemokine and a murine receptor. It may be necessary to re-evaluate the murine receptor for human CXCL8 activities in mice.
Structural and functional consequences of MIP-2 GAG-binding residues
The GAG-binding site of MIP-2 is also of interest in addressing the role of GAGs in MIP-2 mediated effects. Attempts to co-crystallize or soak various sizes of heparin with MIP-2 were unsuccessful. We therefore identified residues involved in interactions with heparin disaccharide I-S. Chemical shifts upon binding of disaccharide by MIP-2 occur at lysine residues in the C-terminal helix (residues 61, 65, and 69) as well at residues D19 and K21. Mutagenesis of residues 19 or 21 to alanine has no significant effect in heparin binding studies, in vitro neutrophil chemotaxis, and neutrophil recruitment in vivo relative to only the three lysine mutations at the of α-helix (3K-MIP-2). A model of the binding interface between GAGs and MIP-2 in the context of mCXCR2 is presented in Figure 4. The GAG-binding site across the two helices that would stabilize the dimer (48) and are distal to the N-terminal receptor binding region and the ELR activation motif. Having established the GAG:MIP-2:CXCR2 interface, it was important to understand the functional consequence of GAG-deficient mutants in vitro and in vivo. The in vitro EC50 of the GAG-deficient mutants are approximately 10-fold greater than wild-type, suggesting these mutants are slightly impaired in activating the receptor. This result is also supported by reduced Akt phosphorylation of the mutants (Figure S5).
The results from the in vivo studies were particularly interesting. There was no difference in neutrophil recruitment between the GAG-deficient mutants and wild-type MIP-2 in the peritoneum. Installation of WT and mutants to the lung, however, led to a dramatically different result. At the highest dose, the GAG-deficient mutants were unable to recruit any neutrophils, clearly establishing an important role for GAGs in MIP-2-mediated neutrophil recruitment to the lung. Based on these results the functional role of the GAG-binding site is tissue-dependent, presumably due to different forms of GAGs biosynthesized in each tissue (77) and their roles in the varying processes during migration to individual tissues. For example, GAGs could affect chemokines during transcytosis and binding to the vasculature (4), the steepness of the chemokine gradient (56), the kinetics of GAG interactions (30), the regulation of GAGs on the activating form (monomeric, dimeric, or oligomeric) of a chemokine (31), and the signaling intensity (78).
Comparison of the in vivo effects of the MIP-2:GAG studies with those in CXCL8 is also informative (30, 31). In the work of Tanino et al one of the GAG-deficient mutants, TM-CXCL8 (triple mutant of CXCL8) replacing two lysine residues and one arginine on the α-helix with alanine residues, is very similar to the 3K-MIP-2 mutant. The other mutant is a single mutant at the end of the helix (R68A). Both the single and triple GAG-deficient mutants of CXCL8 lead to dramatically enhanced neutrophil recruitment, the opposite of the MIP-2 mutants. In another in vivo study of CXCL8 single alanine mutants at the same locations were in delivered to the lung and peritoneum. Neutrophil recruitment to the lung was enhanced but to the peritoneum was slightly decreased relative to WT CXCL8 (31). Comparison of in vivo results in the peritoneum and lung between the current study on MIP-2 and previous studies with CXCL8 highlight different mechanisms involving GAGs on two similar chemokines in the ELR subfamily with GAG-binding sites on the α-helix. One likely explanation is the two “similar” GAG-binding sites are recognized by different tissue-dependent GAGs. The other mechanism, noted above, is that different cCXCR2 complexes may serve as the target receptors.
In summary, we have identified GAG-binding residues and verified the importance with functional studies of mutants. The in vivo MIP-2 data indicate GAG-deficient mutants have no difference in peritoneal neutrophil recruitment compared to WT but are unable to recruit neutrophils to the lungs. These results are in contrast to GAG-deficient CXCL8 mutants that show a decrease in the peritoneum and a dramatic increase in neutrophil recruitment in the lung (30, 31), and also highlight the subtleties of GAG interactions with ELR chemokines and the subsequent regulation of chemokine activity in different tissues. We developed a model of the GAG:MIP-2:CXCR2 and cited chemokine studies of dimeric chemokines involved in receptor interactions with different affinities and signaling activity (16, 17, 71). We suggest an additional function of the GAG-stabilized 2:2 complex. This complex most likely forms at the site of MIP-2 secretion where the chemokine concentration would be highest, favoring formation of dimers by mass action and the presence of GAGs. At this site recruited cells must change their function, ceasing migration (also due to lack of an increasing chemokine gradient) and initiating other activities associated with the innate response and wound healing. GAG stabilization of the 2:2 stoichiometry could be involved by preventing migration by the monomeric MIP-2, or limiting signaling pathways to only those that would be necessary for altered neutrophil functions.
Supplementary Material
Acknowledgments
This work was supported by NIH grants AI065029, AI082295 and UF06032.
We greatly appreciate the help of Tiffany Sun (Yale University) in using the flow cytometer and assistance in analyzing the FACS data. We thank Anne Imberty for coordinates of the heparin oliogosaccharide. We would also like to thank the Yale Keck Biotechnology Resource Laboratory for the use of mass spectrometry facility.
Footnotes
Figure S1, Chemical structure of heparin disaccharide I S. Figure S2 In vivo neutrophil recruitment by MIP-2 and GAG-binding deficient MIP-2 mutants at 100ng. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Raz E, Mahabaleshwar H. Chemokine signaling in embryonic cell migration: a fisheye view. Development. 2009;136:1223–1229. doi: 10.1242/dev.022418. [DOI] [PubMed] [Google Scholar]
- 2.Campanella GS, Lee EM, Sun J, Luster AD. CXCR3 and heparin binding sites of the chemokine IP-10 (CXCL10) J Biol Chem. 2003;278:17066–17074. doi: 10.1074/jbc.M212077200. [DOI] [PubMed] [Google Scholar]
- 3.Colditz IG, Schneider MA, Pruenster M, Rot A. Chemokines at large: in-vivo mechanisms of their transport, presentation and clearance. Thromb Haemost. 2007;97:688–693. [PubMed] [Google Scholar]
- 4.Middleton J, Neil S, Wintle J, Clark-Lewis I, Moore H, Lam C, Auer M, Hub E, Rot A. Transcytosis and Surface Presentation of IL-8 by Venular Endothelial Cells. Cell. 1997;91:385–395. doi: 10.1016/s0092-8674(00)80422-5. [DOI] [PubMed] [Google Scholar]
- 5.Pruenster M, Mudde L, Bombosi P, Dimitrova S, Zsak M, Middleton J, Richmond A, Graham GJ, Segerer S, Nibbs RJB, Rot A. The Duffy antigen receptor for chemokines transports chemokines and supports their promigratory activity. Nat Immunol. 2009;10:101–108. doi: 10.1038/ni.1675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gerszten RE, Garcia-Zepeda EA, Lim YC, Yoshida M, Ding HA, Gimbrone MA, Jr, Luster AD, Luscinskas FW, Rosenzweig A. MCP-1 and IL-8 trigger firm adhesion of monocytes to vascular endothelium under flow conditions. Nature. 1999;398:718–723. doi: 10.1038/19546. [DOI] [PubMed] [Google Scholar]
- 7.Rot A, von Andrian UH. Chemokines in innate and adaptive host defense: basic chemokinese grammar for immune cells. Annu Rev Immunol. 2004;22:891–928. doi: 10.1146/annurev.immunol.22.012703.104543. [DOI] [PubMed] [Google Scholar]
- 8.Rot A. Neutrophil attractant/activation protein-1 (interleukin-8) induces in vitro neutrophil migration by haptotactic mechanism. Eur J Immunol. 1993;23:303–306. doi: 10.1002/eji.1830230150. [DOI] [PubMed] [Google Scholar]
- 9.Celie JW, Beelen RH, van den Born J. Heparan sulfate proteoglycans in extravasation: assisting leukocyte guidance. Front Biosci. 2009;14:4932–4949. doi: 10.2741/3578. [DOI] [PubMed] [Google Scholar]
- 10.Proudfoot AE. The biological relevance of chemokine-proteoglycan interactions. Biochem Soc Trans. 2006;34:422–426. doi: 10.1042/BST0340422. [DOI] [PubMed] [Google Scholar]
- 11.Tuinstra RL, Peterson FC, Kutlesa S, Elgin ES, Kron MA, Volkman BF. Interconversion between two unrelated protein folds in the lymphotactin native state. Proc Natl Acad Sci U S A. 2008;105:5057–5062. doi: 10.1073/pnas.0709518105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Crump MP, Gong JH, Loetscher P, Rajarathnam K, Amara A, Arenzana-Seisdedos F, Virelizier JL, Baggiolini M, Sykes BD, Clark-Lewis I. Solution structure and basis for functional activity of stromal cell- derived factor-1; dissociation of CXCR4 activation from binding and inhibition of HIV-1. EMBO J. 1997;16:6996–7007. doi: 10.1093/emboj/16.23.6996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Murphy JW, Yuan H, Kong Y, Xiong Y, Lolis EJ. Heterologous quaternary structure of CXCL12 and its relationship to the CC chemokine family. Proteins. 2010;78:1331–1337. doi: 10.1002/prot.22666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rajarathnam K, Sykes BD, Kay CM, Dewald B, Geiser T, Baggiolini M, Clark-Lewis I. Neutrophil activation by monomeric interleukin-8. Science. 1994;264:90–92. doi: 10.1126/science.8140420. [DOI] [PubMed] [Google Scholar]
- 15.de Oliveira KB, Oda JM, Voltarelli JC, Nasser TF, Ono MA, Fujita TC, Matsuo T, Watanabe MA. CXCL12 rs1801157 polymorphism in patients with breast cancer, hodgkin’s lymphoma, and non-hodgkin’s lymphoma. J Clin Lab Anal. 2009;23:387–393. doi: 10.1002/jcla.20346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rajarathnam K, Prado GN, Fernando H, Clark-Lewis I, Navarro J. Probing receptor binding activity of interleukin-8 dimer using a disulfide trap. Biochemistry. 2006;45:7882–7888. doi: 10.1021/bi0605944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Veldkamp CT, Seibert C, Peterson FC, De la Cruz NB, Haugner JC, 3rd, Basnet H, Sakmar TP, Volkman BF. Structural basis of CXCR4 sulfotyrosine recognition by the chemokine SDF-1/CXCL12. Sci Signal. 2008;1:ra4. doi: 10.1126/scisignal.1160755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jabeen T, Leonard P, Jamaluddin H, Acharya KR. Structure of mouse IP-10, a chemokine. Acta Crystallogr D. 2008;64:611–619. doi: 10.1107/S0907444908007026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Salanga CL, Handel TM. Chemokine oligomerization and interactions with receptors and glycosaminoglycans: the role of structural dynamics in function. Exp Cell Res. 2011;317:590–601. doi: 10.1016/j.yexcr.2011.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Swaminathan GJ, Holloway DE, Colvin RA, Campanella GK, Papageorgiou AC, Luster AD, Acharya KR. Crystal structures of oligomeric forms of the IP-10/CXCL10 chemokine. Structure. 2003;11:521–532. doi: 10.1016/s0969-2126(03)00070-4. [DOI] [PubMed] [Google Scholar]
- 21.Wang X, Watson C, Sharp Joshua S, Handel Tracy M, Prestegard James H. Oligomeric Structure of the Chemokine CCL5/RANTES from NMR, MS, and SAXS Data. Structure. 2011;19:1138–1148. doi: 10.1016/j.str.2011.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wu B, Chien EYT, Mol CD, Fenalti G, Liu W, Katritch V, Abagyan R, Brooun A, Wells P, Bi FC, Hamel DJ, Kuhn P, Handel TM, Cherezov V, Stevens RC. Structures of the CXCR4 Chemokine GPCR with Small-Molecule and Cyclic Peptide Antagonists. Science. 2010;330:1066–1071. doi: 10.1126/science.1194396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hoogewerf AJ, Kuschert GS, Proudfoot AE, Borlat F, Clark-Lewis I, Power CA, Wells TN. Glycosaminoglycans mediate cell surface oligomerization of chemokines. Biochemistry. 1997;36:13570–13578. doi: 10.1021/bi971125s. [DOI] [PubMed] [Google Scholar]
- 24.Proudfoot AE, Handel TM, Johnson Z, Lau EK, LiWang P, Clark-Lewis I, Borlat F, Wells TN, Kosco-Vilbois MH. Glycosaminoglycan binding and oligomerization are essential for the in vivo activity of certain chemokines. Proc Natl Acad Sci U S A. 2003;100:1885–1890. doi: 10.1073/pnas.0334864100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Handel TM, Johnson Z, Crown SE, Lau EK, Proudfoot AE. Regulation of protein function by glycosaminoglycans--as exemplified by chemokines. Annu Rev Biochem. 2005;74:385–410. doi: 10.1146/annurev.biochem.72.121801.161747. [DOI] [PubMed] [Google Scholar]
- 26.Campanella GSV, Grimm J, Manice LA, Colvin RA, Medoff BD, Wojtkiewkz GR, Weissleder R, Luster AD. Oligomerization of CXCL10 is necessary for endothelial cell presentation and in vivo activity. J Immunol. 2006;177:6991–6998. doi: 10.4049/jimmunol.177.10.6991. [DOI] [PubMed] [Google Scholar]
- 27.Ali S, O’Boyle G, Hepplewhite P, Tyler JR, Robertson H, Kirby JA. Therapy with nonglycosaminoglycan-binding mutant CCL7: a novel strategy to limit allograft inflammation. Am J Transplant. 2010;10:47–58. doi: 10.1111/j.1600-6143.2009.02868.x. [DOI] [PubMed] [Google Scholar]
- 28.Peterson FC, Elgin ES, Nelson TJ, Zhang F, Hoeger TJ, Linhardt RJ, Volkman BF. Identification and characterization of a glycosaminoglycan recognition element of the C chemokine lymphotactin. J Biol Chem. 2004;279:12598–12604. doi: 10.1074/jbc.M311633200. [DOI] [PubMed] [Google Scholar]
- 29.Severin IC, Gaudry JP, Johnson Z, Kungl A, Jansma A, Gesslbauer B, Mulloy B, Power C, Proudfoot AE, Handel T. Characterization of the chemokine CXCL11-heparin interaction suggests two different affinities for glycosaminoglycans. J Biol Chem. 2010;285:17713–17724. doi: 10.1074/jbc.M109.082552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tanino Y, Coombe DR, Gill SE, Kett WC, Kajikawa O, Proudfoot AEI, Wells TNC, Parks WC, Wight TN, Martin TR, Frevert CW. Kinetics of Chemokine-Glycosaminoglycan Interactions Control Neutrophil Migration into the Airspaces of the Lungs. J Immunol. 2010;184:2677–2685. doi: 10.4049/jimmunol.0903274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gangavarapu P, Rajagopalan L, Kolli D, Guerrero-Plata A, Garofalo RP, Rajarathnam K. The monomer-dimer equilibrium and glycosaminoglycan interactions of chemokine CXCL8 regulate tissue-specific neutrophil recruitment. J Leukoc Biol. 2012;91:259–265. doi: 10.1189/jlb.0511239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Blanpain C, Doranz BJ, Bondue A, Govaerts C, De Leener A, Vassart G, Doms RW, Proudfoot A, Parmentier M. The core domain of chemokines binds CCR5 extracellular domains while their amino terminus interacts with the transmembrane helix bundle. J Biol Chem. 2003;278:5179–5187. doi: 10.1074/jbc.M205684200. [DOI] [PubMed] [Google Scholar]
- 33.Lolis E, Sweet RM, Cousens LS, Tekamp-Olson P, Sherry BA, Cerami A. Preliminary crystallographic analysis of murine macrophage inflammatory protein 2. J Mol Biol. 1992;225:913–915. doi: 10.1016/0022-2836(92)90411-c. [DOI] [PubMed] [Google Scholar]
- 34.Ren M, Guo Q, Guo L, Lenz M, Qian F, Koenen RR, Xu H, Schilling AB, Weber C, Ye RD, Dinner AR, Tang WJ. Polymerization of MIP-1 chemokine (CCL3 and CCL4) and clearance of MIP-1 by insulin-degrading enzyme. EMBO J. 2010;29:3952–3966. doi: 10.1038/emboj.2010.256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jerva LF, Sullivan G, Lolis E. Functional and receptor binding characterization of recombinant murine macrophage inflammatory protein 2: sequence analysis and mutagenesis identify receptor binding epitopes. Protein Sci. 1997;6:1643–1652. doi: 10.1002/pro.5560060805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Aitken A, Learmonth Ml. Protein Determination by UV Absorption. In: Walker JM, editor. The Protein Protocols Handbook. Humana Press Inc; Totowa, NJ: 1996. pp. 3–6. [Google Scholar]
- 37.Otwinowski Z, Minor W. Processing of X-Ray Diffraction Data Collected in Oscillation Mode. Methods Enzymol. 1997:307–325. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 38.Baldwin ET, Weber IT, St Charles R, Xuan JC, Appella E, Yamada M, Matsushima K, Edwards BF, Clore GM, Gronenborn AM, et al. Crystal structure of interleukin 8: symbiosis of NMR and crystallography. Proc Natl Acad Sci U S A. 1991;88:502–506. doi: 10.1073/pnas.88.2.502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vagin A, Teplyakov A. Molecular replacement with MOLREP. Acta Crystallogr D. 2010;66:22–25. doi: 10.1107/S0907444909042589. [DOI] [PubMed] [Google Scholar]
- 40.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 41.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 42.Laskowski RA, MacArthur MW, Moss DS, Thornton JM. PROCHECK: a program to check the stereochemical quality of protein structures. J Appl Cryst. 1993;26:283–291. [Google Scholar]
- 43.Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J Biomol NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 44.Shao W, Jerva LF, West J, Lolis E, Schweitzer BI. Solution structure of murine macrophage inflammatory protein-2. Biochemistry. 1998;37:8303–8313. doi: 10.1021/bi980112r. [DOI] [PubMed] [Google Scholar]
- 45.Mulder FA, Schipper D, Bott R, Boelens R. Altered flexibility in the substrate-binding site of related native and engineered high-alkaline Bacillus subtilisins. J Mol Biol. 1999;292:111–123. doi: 10.1006/jmbi.1999.3034. [DOI] [PubMed] [Google Scholar]
- 46.Kuschert GS, Hoogewerf AJ, Proudfoot AE, Chung CW, Cooke RM, Hubbard RE, Wells TN, Sanderson PN. Identification of a glycosaminoglycan binding surface on human interleukin-8. Biochemistry. 1998;37:11193–11201. doi: 10.1021/bi972867o. [DOI] [PubMed] [Google Scholar]
- 47.Arnold K, Bordoli L, Kopp J, Schwede T. The SWISS-MODEL workspace: a web-based environment for protein structure homology modelling. Bioinformatics. 2006;22:195–201. doi: 10.1093/bioinformatics/bti770. [DOI] [PubMed] [Google Scholar]
- 48.Lortat-Jacob H, Grosdidier A, Imberty A. Structural diversity of heparan sulfate binding domains in chemokines. Proc Natl Acad Sci U S A. 2002;99:1229–1234. doi: 10.1073/pnas.032497699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.DeLano WL. The PyMOL molecular graphics system. DeLano Scientific; Palo Alto, CA: 2002. [Google Scholar]
- 50.Dong X, Wu D. Methods for studying neutrophil chemotaxis. Methods Enzymol. 2006;406:605–613. doi: 10.1016/S0076-6879(06)06047-2. [DOI] [PubMed] [Google Scholar]
- 51.Malkowski MG, Wu JY, Lazar JB, Johnson PH, Edwards BF. The crystal structure of recombinant human neutrophil-activating peptide-2 (M6L) at 1.9-A resolution. J Biol Chem. 1995;270:7077–7087. doi: 10.1074/jbc.270.13.7077. [DOI] [PubMed] [Google Scholar]
- 52.St Charles R, Walz DA, Edwards BF. The three-dimensional structure of bovine platelet factor 4 at 3.0-A resolution. J Biol Chem. 1989;264:2092–2099. [PubMed] [Google Scholar]
- 53.Project CC. The CCP4 suite: programs for protein crystallography. Acta Crystallogr D. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 54.Kelley LA, Sutcliffe MJ. OLDERADO: on-line database of ensemble representatives and domains. On Line Database of Ensemble Representatives And DOmains. Protein Sci. 1997;6:2628–2630. doi: 10.1002/pro.5560061215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sikic K, Tomic S, Carugo O. Systematic comparison of crystal and NMR protein structures deposited in the protein data bank. Open Biochem J. 2010;4:83–95. doi: 10.2174/1874091X01004010083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Das ST, Rajagopalan L, Guerrero-Plata A, Sai J, Richmond A, Garofalo RP, Rajarathnam K. Monomeric and dimeric CXCL8 are both essential for in vivo neutrophil recruitment. PLoS One. 2010;5:e11754. doi: 10.1371/journal.pone.0011754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Schnitzel W, Monschein U, Besemer J. Monomer-dimer equilibria of interleukin-8 and neutrophil-activating peptide 2. Evidence for IL-8 binding as a dimer and oligomer to IL-8 receptor B. J Leukoc Biol. 1994;55:763–770. doi: 10.1002/jlb.55.6.763. [DOI] [PubMed] [Google Scholar]
- 58.Murphy JW, Cho Y, Sachpatzidis A, Fan C, Hodsdon ME, Lolis E. Structural and functional basis of CXCL12 (stromal cell-derived factor-1alpha) binding to heparin. J Biol Chem. 2007;282:10018–10027. doi: 10.1074/jbc.M608796200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Shaw JP, Johnson Z, Borlat F, Zwahlen C, Kungl A, Roulin K, Harrenga A, Wells TN, Proudfoot AE. The X-ray structure of RANTES: heparin-derived disaccharides allows the rational design of chemokine inhibitors. Structure. 2004;12:2081–2093. doi: 10.1016/j.str.2004.08.014. [DOI] [PubMed] [Google Scholar]
- 60.Mercer-Jones MA, Shrotri MS, Heinzelmann M, Peyton JC, Cheadle WG. Regulation of early peritoneal neutrophil migration by macrophage inflammatory protein-2 and mast cells in experimental peritonitis. J Leukoc Biol. 1999;65:249–255. doi: 10.1002/jlb.65.2.249. [DOI] [PubMed] [Google Scholar]
- 61.Ali S, Robertson H, Wain JH, Isaacs JD, Malik G, Kirby JA. A Non-Glycosaminoglycan-Binding Variant of CC Chemokine Ligand 7 (Monocyte Chemoattractant Protein-3) Antagonizes Chemokine-Mediated Inflammation. J Immunol. 2005;175:1257–1266. doi: 10.4049/jimmunol.175.2.1257. [DOI] [PubMed] [Google Scholar]
- 62.Wang L, Fuster M, Sriramarao P, Esko JD. Endothelial heparan sulfate deficiency impairs L-selectin- and chemokine-mediated neutrophil trafficking during inflammatory responses. Nat Immunol. 2005;6:902–910. doi: 10.1038/ni1233. [DOI] [PubMed] [Google Scholar]
- 63.Dean RA, Cox JH, Bellac CL, Doucet A, Starr AE, Overall CM. Macrophage-specific metalloelastase (MMP-12) truncates and inactivates ELR+ CXC chemokines and generates CCL2, -7, -8, and -13 antagonists: potential role of the macrophage in terminating polymorphonuclear leukocyte influx. Blood. 2008;112:3455–3464. doi: 10.1182/blood-2007-12-129080. [DOI] [PubMed] [Google Scholar]
- 64.Ellyard JI, Simson L, Bezos A, Johnston K, Freeman C, Parish CR. Eotaxin selectively binds heparin. An interaction that protects eotaxin from proteolysis and potentiates chemotactic activity in vivo. J Biol Chem. 2007;282:15238–15247. doi: 10.1074/jbc.M608046200. [DOI] [PubMed] [Google Scholar]
- 65.McQuibban GA, Butler GS, Gong JH, Bendall L, Power C, Clark-Lewis I, Overall CM. Matrix metalloproteinase activity inactivates the CXC chemokine stromal cell-derived factor-1. J Biol Chem. 2001;276:43503–43508. doi: 10.1074/jbc.M107736200. [DOI] [PubMed] [Google Scholar]
- 66.Sadir R, Imberty A, Baleux F, Lortat-Jacob H. Heparan sulfate/heparin oligosaccharides protect stromal cell-derived factor-1 (SDF-1)/CXCL12 against proteolysis induced by CD26/dipeptidyl peptidase IV. J Biol Chem. 2004;279:43854–43860. doi: 10.1074/jbc.M405392200. [DOI] [PubMed] [Google Scholar]
- 67.Murooka TT, Wong MM, Rahbar R, Majchrzak-Kita B, Proudfoot AE, Fish EN. CCL5-CCR5-mediated apoptosis in T cells: Requirement for glycosaminoglycan binding and CCL5 aggregation. J Biol Chem. 2006;281:25184–25194. doi: 10.1074/jbc.M603912200. [DOI] [PubMed] [Google Scholar]
- 68.Kufareva I, Rueda M, Katritch V, Stevens RC, Abagyan R. Status of GPCR modeling and docking as reflected by community-wide GPCR Dock 2010 assessment. Structure. 2011;19:1108–1126. doi: 10.1016/j.str.2011.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Trettel F, Di Bartolomeo S, Lauro C, Catalano M, Ciotti MT, Limatola C. Ligand-independent CXCR2 Dimerization. J Biol Chem. 2003;278:40980–40988. doi: 10.1074/jbc.M306815200. [DOI] [PubMed] [Google Scholar]
- 70.Wilson S, Wilkinson G, Milligan G. The CXCR1 and CXCR2 Receptors Form Constitutive Homo- and Heterodimers Selectively and with Equal Apparent Affinities. J Biol Chem. 2005;280:28663–28674. doi: 10.1074/jbc.M413475200. [DOI] [PubMed] [Google Scholar]
- 71.Nasser MW, Raghuwanshi SK, Grant DJ, Jala VR, Rajarathnam K, Richardson RM. Differential activation and regulation of CXCR1 and CXCR2 by CXCL8 monomer and dimer. J Immunol. 2009;183:3425–3432. doi: 10.4049/jimmunol.0900305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Bozic CR, Gerard NP, von Uexkull-Guldenband C, Kolakowski LF, Jr, Conklyn MJ, Breslow R, Showell HJ, Gerard C. The murine interleukin 8 type B receptor homologue and its ligands. Expression and biological characterization. J Biol Chem. 1994;269:29355–29358. [PubMed] [Google Scholar]
- 73.Lee J, Cacalano G, Camerato T, Toy K, Moore MW, Wood WI. Chemokine binding and activities mediated by the mouse IL-8 receptor. J Immunol. 1995;155:2158–2164. [PubMed] [Google Scholar]
- 74.Cacalano G, Lee J, Kikly K, Ryan AM, Pitts-Meek S, Hultgren B, Wood WI, Moore MW. Neutrophil and B cell expansion in mice that lack the murine IL-8 receptor homolog. Science. 1994;265:682–684. doi: 10.1126/science.8036519. [DOI] [PubMed] [Google Scholar]
- 75.Moepps B, Nuesseler E, Braun M, Gierschik P. A homolog of the human chemokine receptor CXCR1 is expressed in the mouse. Mol Immunol. 2006;43:897–914. doi: 10.1016/j.molimm.2005.06.043. [DOI] [PubMed] [Google Scholar]
- 76.Fan X, Patera AC, Pong-Kennedy A, Deno G, Gonsiorek W, Manfra DJ, Vassileva G, Zeng M, Jackson C, Sullivan L, Sharif-Rodriguez W, Opdenakker G, Van Damme J, Hedrick JA, Lundell D, Lira SA, Hipkin RW. Murine CXCR1 is a functional receptor for GCP-2/CXCL6 AND IL-8/CXCL8. J Biol Chem. 2006:M607705200. doi: 10.1074/jbc.M607705200. [DOI] [PubMed] [Google Scholar]
- 77.Shi X, Zaia J. Organ-specific Heparan Sulfate Structural Phenotypes. J Biol Chem. 2009;284:11806–11814. doi: 10.1074/jbc.M809637200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Sai J, Walker G, Wikswo J, Richmond A. The IL sequence in the LLKIL motif in CXCR2 is required for full ligand-induced activation of Erk, Akt, and chemotaxis in HL60 cells. J Biol Chem. 2006;281:35931–35941. doi: 10.1074/jbc.M605883200. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.